Abstract
The homeostatic chemokine CCL17, also known as thymus and activation regulated chemokine (TARC), has been associated with various diseases such as asthma, idiopathic pulmonary fibrosis, atopic dermatitis and ulcerative colitis. Neutralization of CCL17 by antibody treatment ameliorates the impact of disease by blocking influx of T cells. Monoclonal antibody M116 derived from a combinatorial library shows potency in neutralizing CCL17-induced signaling. To gain insight into the structural determinants of antigen recognition, the crystal structure of M116 Fab was determined in complex with CCL17 and in the unbound form. Comparison of the structures revealed an unusual induced-fit mechanism of antigen recognition that involves cis-trans isomerization in two CDRs. The structure of the CCL17-M116 complex revealed the antibody binding epitope, which does not overlap with the putative receptor epitope, suggesting that the current model of chemokine-receptor interactions, as observed in the CXCR4-vMIP-II system, may not be universal.
Abbreviations: mAb, monoclonal antibody; DTT, dithiothreitol; PEG, polyethylene glycol; EDTA, ethylenediaminetetraacetic acid; HEPES, 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid; CDR, complementarity determining region; VH, variable domain of the heavy chain; VL, variable domain of the light chain; PDB, Protein Data Bank; RMSD, root-mean-square deviation
Keywords: CCL17, Antibody, Crystal structure, Epitope, Neutralization, Cis-trans isomerization
Graphical abstract
Highlights
-
•
Crystal structure of CCL17 in complex with neutralizing M116 Fab was determined.
-
•
An induced-fit binding mechanism involves cis-trans isomerization in two CDRs.
-
•
The antibody binding epitope does not overlap with the putative receptor epitope.
1. Introduction
Chemokines are small, secreted molecules that regulate leukocyte trafficking. The human chemokine system currently includes about 50 chemokines and 20 chemokine receptors [1]. Based on the position of the first two of the four conserved cysteine residues, chemokines are divided into four subfamilies: CXC, CC, C and CX3C [2]. Chemokines bind to leukocytes via their corresponding seven transmembrane-spanning G-protein coupled receptors that have been grouped according to the structure of their chemokine ligands (CXCR, CCR, XCR and CX3CR).
Despite sequence variability among chemokines, their tertiary structures are remarkably similar. A common ‘chemokine’ fold consists of the N-terminal loop followed by a three-stranded β-meander and a C-terminal α-helix, which covers one face of the meander. The structure is stabilized by two disulfide bonds connecting the N-loop to the core. A large number of the CC and CXC chemokines has been structurally characterized [3]. Approximately half of them form dimers, the functional significance of which is unclear.
Thymus and activation regulated chemokine (TARC), known as CCL17, is constitutively expressed in the thymus and is produced by dendritic cells, endothelial cells, keratinocytes, bronchial epithelial cells and fibroblasts [4]. It is a ligand for CCR4, which is predominantly expressed on Th2 lymphocytes, basophils and natural killer cells [5]. The CC chemokines that are associated with a Th2 profile (CCL17 and CCL22) have an important role in the development of pulmonary diseases [6], [7]. A specific monoclonal antibody (mAb) against CCL17 was shown to attenuate ovalbumin-induced airway eosinophilia in mice and diminish the degree of airway hyperresponsiveness with a concomitant decrease in Th2 cytokine levels [8]. Anti-CCL17 mAbs may provide an improved safety profile in comparison to anti-CCR4 mAbs by selectively blocking CCL17 without interaction with the CCR4 expressing platelets. In addition, such mAbs will not block the beneficial innate immune effects of CCL22 on CCR4 [9].
Anti-CCL17 mAb M116 was isolated from a phage display combinatorial library [10]. M116 is derived from human germlines IGHV5-51 for VH and IGKV4-1 for VL. The antibody binds human CCL17 with high affinity and blocks signaling through CCR4 [11]. To gain insight into molecular interactions, we have determined the crystal structure of the M116 Fab in complex with CCL17 and in the unbound form. Comparison of the structures revealed an unusual induced-fit mechanism of antigen recognition that involves cis-trans isomerization in two CDRs.
2. Materials and methods
2.1. Expression and purification
The G7T mutant of human chemokine CCL17 (residues 1–71 corresponding to 24–94 of UniProtKB entry CCL17_HUMAN) was expressed in Escherichia coli strain BL21 (DE3), isolated from inclusion bodies, and refolded as previously described [12]. Briefly, inclusion bodies were collected in the solubilization buffer consisting of 8 M urea, 5 mM EDTA, 10 mM DTT, and 20 mM Tris HCl, pH 7. Solubilized inclusion bodies were clarified by centrifugation at 18,000×g for 10 min at 4 °C and loaded onto an SP Sepharose Fast Flow column (GE Healthcare). Protein was eluted using a 0–100% gradient of the buffer composed of 8 M urea, 1 M NaCl, and 10 mM potassium phosphate, pH 6.8. Pooled fractions were diluted by the refolding buffer (0.1 M NaHCO3, 1.5 M guanidinium chloride, 3 mM cysteine, and 0.3 mM cystine) and incubated at room temperature for 48 h followed by incubation at 4 °C for 66 h.
The M116 Fab is composed of 220 residues of the light chain and 230 residues of the heavy chain including a hexahistidine tag at the C-terminus of the heavy chain. The constant domains are human κ (light chain) and IgG1 (heavy chain). The Fab was transiently expressed in HEK 293 F cells using Lonza-based vectors and was purified by affinity and ion exchange chromatography using, respectively, HisTrap and Source 15 S columns (GE Healthcare).
2.2. Crystallization, X-ray data collection, and structure determination
Crystallization was carried out by the vapor-diffusion method at 20 °C using an Oryx4 robot (Douglas Instruments). The experiments were composed of equal volumes of protein and reservoir solution in a sitting drop format in 96-well Corning 3550 plates. The initial screening was performed with the PEGs kit (Qiagen) and in-house screens. The crystal hits were optimized by microseed matrix screening [13]. The Fab crystals suitable for X-ray analysis were obtained from 18% PEG 3350 in 0.1 M HEPES, pH 7.5. The Fab-CCL17 complex was crystallized from 20% PEG 3350, 0.2 M K/Na tartrate at pH 7.4.
For X-ray data collection, one crystal of each the Fab and the complex was soaked for a few seconds in the respective mother liquor supplemented with 20% glycerol, and flash-cooled in liquid nitrogen. Diffraction data were collected with a Pilatus 6 M detector at the Advanced Photon Source (Argonne, IL) with the exposures of 1 s per half-degree image and were processed with XDS [14]. X-ray data statistics are given in Table 1.
Table 1.
X-ray data and refinement statistics.
| Data set | M116 Fab | CCL17+M116 Fab |
|---|---|---|
| X-Ray Data | ||
| Space group | P21 | P1 |
| Unit cell (Å) | 53.74, 64.92, 73.79 | 51.51, 81.93, 130.50 |
| Unit cell angles (°) | 90, 107.29, 90 | 93.96, 99.21, 104.19 |
| Asymmetric unit content | one Fab | Four 1:1 complexes |
| Vm (Å3/Da)/solvent (%) | 2.54/52 | 2.34/47 |
| Resolution (Å) | 30–1.50 (1.54–1.50)a | 30–1.9 (1.95–1.90)a |
| Number of measured reflections | 234,637 (8696) | 409,919 (24,336) |
| Number of unique reflections | 73,827 (3956) | 153,743 (10,386) |
| Completeness (%) | 95.1 (68.8) | 96.2 (87.9) |
| Multiplicity | 3.2 (2.2) | 2.7 (2.3) |
| Rsym (I) | 0.036 (0.285) | 0.075 (0.480) |
| Mean I/σ(I) | 18.6 (3.3) | 9.3 (2.3) |
| B factor from Wilson plot (Å2) | 26.5 | 30.9 |
| Refinement | ||
| Resolution (Å) | 15–1.5 | 15–1.9 |
| Total number of non-hydrogen atoms | 3770 | 16,264 |
| Number of water molecules | 362 | 1020 |
| Rcryst (%) | 18.0 | 18.0 |
| Rfree (%) | 20.8 | 23.1 |
| RMSD bond lengths (Å) | 0.009 | 0.008 |
| RMSD bond angles (°) | 1.3 | 1.2 |
| Mean B-factor from model (Å2) | 27.1 | 34.6 |
| Ramachandran plot, most favored (%) | 90.0 | 90.5 |
| Ramachandran plot, disallowed (%) | 0.3b | 0.2b |
Values for highest resolution shell are in parentheses.
One residue, Ala57(L), is in a left-handed helical conformation in all copies of the Fab.
The structure of the Fab-CCL17 complex was determined by molecular replacement with Phaser [15] using the structures of CCL17 (PDB entry 1nr4) [16] and of human germline antibody 5-51/B3 (PDB entry 5i1l) [17] as search models. The refined Fab structure was then used in structure determination of the unbound Fab. Both structures were refined with Refmac5 [18]. Manual adjustments were performed using Coot [19]. All crystallographic calculations were performed with the CCP4 suite of programs [20]. Ramachandran statistics were calculated with PROCHECK [21]. Figures were prepared with PyMol (Schrödinger).
2.3. Accession numbers
The coordinates and structure factors for M116 Fab alone and in complex with CCL17 have been deposited in the Protein Data Bank under accession codes 5wk2 and 5wk3, respectively.
2.4. Nomenclature
The Chothia numbering scheme of antibody residues [22] is used throughout the manuscript.
3. Results and discussion
3.1. CCL17 structure
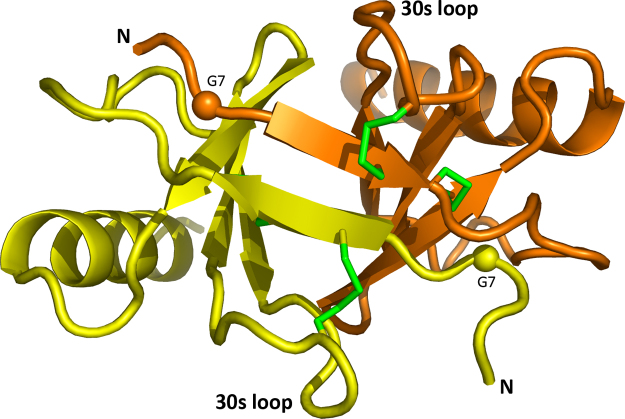
Previous crystallographic and dynamic light scattering studies [16] have shown that CCL17 forms dimers similar to those observed for some other CC chemokines including CCL2 (MCP1) and CCL5 (RANTES). To facilitate crystallization of the CCL17-Fab complex, a monomeric variant of CCL17 was designed based on the crystal structure of CCL17 in the unbound form (PDB entry 1nr4) [16]. A significant part of the dimer interface involves a β-bridge between the N-terminal sections (Fig. 1). A G7T mutation was introduced in order to disrupt the packing of the β-strands. An amino acid with a branched side chain (threonine) was selected to ensure steric clashes in a hydrophobic pocket at Gly7 formed by Phe15 and Ile49 of the other chain. As expected, the CCL17 mutant exists in a monomeric form in the present structure.
Fig. 1.
CCL17 dimer from the structure 1nr4 [16]. Positions of G7T mutations are indicated by spheres, disulfides by green sticks.
The crystal structure of the CCL17-M116 Fab complex was determined at 1.9 Å resolution. There are four copies of the complex in the asymmetric unit in the crystal. Six residues at the N-terminus of CCL17 are disordered in all four molecules. Pairwise superposition of CCL17 molecules gives an RMSD of 0.7 Å for Cα atoms, indicating no major differences between them. However, small deviations occur all over, particularly in the 30 s loop connecting the first two β-strands and in a loop preceding the α-helix. These observations suggest that the CCL17 G7T mutant exhibits a substantial degree of conformational flexibility.
Wild-type unliganded CCL17 (PDB entry 1nr4) [16] was crystallized in space group P1 with four dimers in the asymmetric unit that are virtually identical (RMSD is about 0.2 Å for the entire dimer). A peculiar feature of the CCL17 dimer is that it is asymmetric. Superposition of the subunits gives an RMSD of 1.3 Å. Major deviations occur in the N-terminal β-strand and the 30 s loop, which has a completely different conformation in the two subunits. This is accompanied by a conformational switch from a gauche(+) to gauche(-) configuration at Cys34 of the Cys10-Cys34 disulfide.
Interestingly, all four independent CCL17 molecules in the CCL17-M116 complex correspond to only one subunit of the CCL17 dimer, even though the regions of conformational flexibility are not involved in contacts with the Fab. The RMSD between CCL17 in the complex and the structurally related subunit of the dimer is about 0.6 Å, whereas it is 1.3 Å when compared with the other subunit. The CCL17 conformation observed in the M116 complex also corresponds to that of the unbound CCL17 in the tetragonal crystal form, where the chemokine was found to be a monomer (PDB entry 1nr2) [16].
We hypothesize that the two conformations of CCL17 observed in the dimer reflect the ‘relaxed’ and the ‘tense’ states of the molecule. The tension develops following the formation of an intermolecular β-bridge between N-terminal residues 8–12. The β-bridge itself does not impose any restrictions on the symmetry as is evident in many dimeric proteins including another CC chemokine, CCL2, where a symmetric dimer obeys exact crystallographic symmetry (PDB entry 1dol) [23]. The asymmetry of CCL17 is probably owing to Glu9 in the middle of the β-strand. Dimerization brings two acidic residues, buried at the dimer interface, next to each other. A possible role of Glu9 as a destabilizing factor, which may be abated by the acidic pH of the medium to promote the protonation of the carboxyl group, was noted earlier [16]. But even if the dimer is formed at pH 4.6 in the triclinic crystal, the two Glu9 residues need more space than would be allowed by the symmetric dimer. Yet, under certain conditions, the dimer forms at the expense of the second subunit adopting a different, presumably tense, conformation. The breach of symmetry involves residues around Glu9 and in the 30 s loop, which are fastened to the β-strand through the disulfide bond Cys10-Cys34. The driving force for the asymmetric dimerization is likely the formation of the extensive intersubunit interface covering almost 1000 Å2 on each subunit.
Following this hypothesis, monomeric CCL17 represents the relaxed state whereas the tense conformation may exist only in the context of a complex, either with the second subunit in the CCL17 dimer or with another molecule.
3.2. CCL17-M116 complex
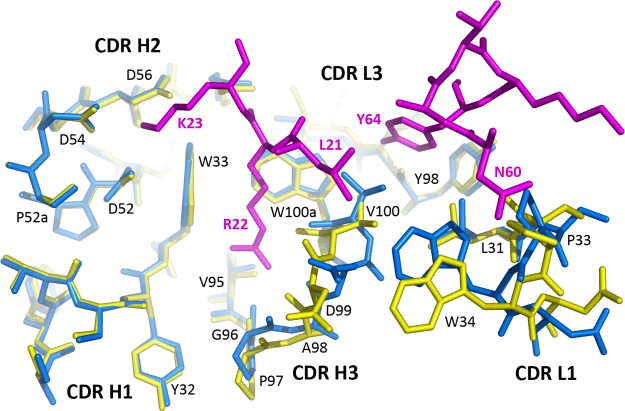
The crystal structure of the CCL17-M116 complex reveals the interacting residues, i.e. the epitope (on the antigen) and the paratope (on the antibody). M116 recognizes a conformational epitope, which spans 3 segments of the CCL17 polypeptide chain, namely two loops (residues 21–23 and 44–45) and the C-terminal helix (residues 60–68) (Fig. 2A). Based on the number of intermolecular contacts, the key epitope residues are Arg22, Tyr64, and Lys23 (Fig. 3 and Table 2). Arg22 fits in between Trp33(H) and Trp100a(H) and makes hydrogen bonds to three main-chain carbonyl groups of CDR H3. Tyr64 fits in a hydrophobic pocket formed by Val100(H), Trp100a(H), Tyr98(L) and Leu31(L). Lys23 is engaged in electrostatic interactions with a cluster of acidic residues at CDR H2 involving Asp52, Asp54 and Asp56. CCL17-M116 interactions involve 9 residues of CCL17 and 18 residues of M116 representing 5 CDRs out of 6 (all except CDR L2). The surface area buried in the complex is relatively small; it covers 660 Å2 on CCL17 and only 500 Å2 on M116.
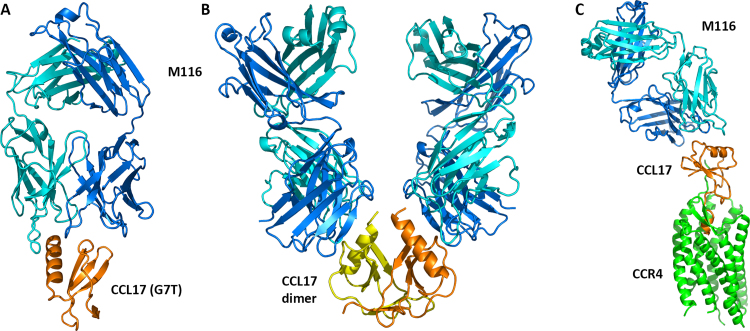
Fig. 2.
Interactions between M116, CCL17 and its receptor CCR4. (A) Crystal structure of the CCL17-M116 complex. (B) A model of the CCL17-M116 dimer based on the structures of monomeric CCL17-M116 complex and free CCL17 dimer (PDB entry 1nr4) [16]. (C) A model of CCL17-CCR4-M116 ternary complex. The model was obtained by superposition of the CCL17-M116 structure on the CXCR4-vMIP-II structure (PDB entry 4rws) [24]. CCL17 is in orange/yellow, Fab light chain in cyan, Fab heavy chain in blue.
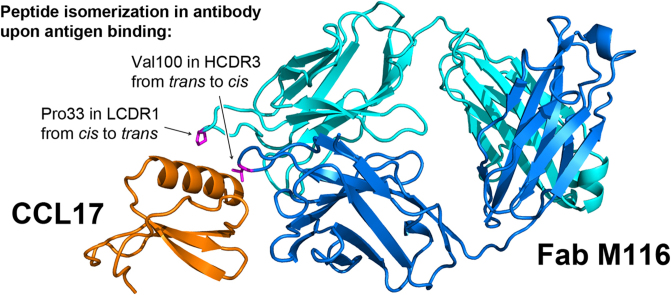
Fig. 3.
Conformational changes in M116 upon binding CCL17. The unbound form of the Fab is shown in yellow, the bound form is shown in blue, CCL17 in magenta. Upon CCL17 binding Pro33 of CDR L1 switches from cis to trans, Val100 of CRD H3 switches from trans to cis.
Table 2.
Hydrogen bonds and salt bridges between CCL17 and M116.
| CCL17 | M116 | Distance (Å) | |||
|---|---|---|---|---|---|
| Arg22 | NE | – | Val100(H) | O | 2.83 |
| Arg22 | NH2 | – | Gly96(H) | O | 2.77 |
| Arg22 | NH2 | – | Ala98(H) | O | 2.92 |
| Lys23 | NZ | – | Asp52(H) | OD2 | 2.79 |
| Lys23 | NZ | – | Asp54(H) | OD2 | 2.78 |
| Lys23 | NZ | – | Asp56(H) | OD2 | 2.81 |
| Tyr64 | OH | – | Tyr98(L) | O | 2.68 |
| Ser67 | OG | – | Tyr98(L) | OH | 2.64 |
M116 binds CCL17 on the side opposite to the dimerization surface, which involves the N-terminal section (residues 5–15) and the free surface of the β-meander (not covered by the α-helix). Therefore, M116 would not block dimerization of CCL17 and should bind the chemokine dimer at a 2:2 ratio. However, the geometry of the CCL17-M116 complex indicates that one M116 mAb cannot bind both subunits of the same dimer simultaneously because the C-terminal ends of the Fabs, which are proximal to the hinge region, are too far apart (Fig. 2B). The only option for M116 mAb to engage both Fab arms is to bind two CCL17 dimers.
M116 inhibits CCL17-mediated chemotaxis, calcium mobilization and β-arrestin recruitment [11]. An apparent explanation of the neutralization effect of M116 is that the binding site of M116 overlaps, at least partially, with the CCR4 epitope, thus preventing CCL17 from coupling with its receptor. While no structural information is available for the CCL17-CCR4 complex, a recent structure of CXCR4 in complex with vMIP-II [24] may serve as a paradigm for chemokine-receptor interactions. On the one hand, viral chemokine antagonist vMIP-II exhibits a broad-spectrum interaction with both CC and CXC chemokine receptors [25] and therefore mimics both types of chemokines. On the other hand, CXCR4 and CCR4 belong to the same family of G-protein coupled receptors and share substantial sequence similarity.
CXCR4 binds vMIP-II with 1:1 stoichiometry. The N-terminus of vMIP-II fits in the receptor transmembrane funnel while the N-terminus of CXCR4 protrudes to interact with the core of vMIP-II. The dimeric form of the chemokine is inconsistent with this mode of binding. In fact, the N-terminus of the receptor replaces the second subunit of the chemokine dimer in forming the intersubunit β-bridge. Although the contacts are numerous, they are not particularly specific, implying that this model may be applicable to other chemokine-receptor pairs. If so, CCL17 would bind CCR4 as a monomer, and the binding epitope would involve the N-terminus and the 30 s loop. Then M116 would bind on the opposite side of the chemokine molecule (Fig. 2C), which raises the question of how this antibody fulfills its neutralizing potential.
Of course, the CCR4-CCL17 system may differ from CXCR4-vMIP-II in how the receptor recognizes its ligand. Another possible explanation of the puzzle is that CCR4 may preferentially bind the tense conformation of CCL17. Mab M116 stabilizes the relaxed conformation of CCL17, thus hampering the binding of CCL17 by its receptor. This hypothesis may also allege the dimerization of CCL17 as a means to facilitate binding to the receptor since the tense form of the chemokine exists only in the dimer. Further studies are needed to clarify the interplay between CCR4, CCL17 and mAb M116.
3.3. Induced fit in M116
The crystal structure of M116 Fab in the unbound form was determined at 1.5 Å resolution. Comparison of this structure to the Fab in the CCL17 complex shows no changes in the quaternary structure monitored by the VL/VH packing angle. Superposition of the two structures gives an RMSD of 0.36 Å for all 234 Cα atoms of the variable domains. The largest deviations, up to 1.5–2 Å, are observed in CDR L1 and CDR H3. When residues 32–35 of VL and 97–100 of VH are excluded from superposition, the RMSD goes down to 0.22 Å, indicating no relative rotation of the VL and VH domains.
The differences observed in CDR L1 and CDR H3 are indicative of an induced-fit mechanism of antigen recognition and, surprisingly, both are related to cis-trans isomerization. Binding of CCL17 causes the transition of Ser32-Pro33(L) peptide bond from cis to trans and Asp99-V100(H) peptide bond from trans to cis (Fig. 3), respectively. In the presence of CCL17, Val100(H) is pulled into a hydrophobic pocket between Leu21 and Tyr64 of the antigen, which is achieved by trans-cis isomerization. The unusual for valine cis-configuration is stabilized by hydrogen bonds between Arg22 of CCL17 and carbonyl oxygens of Val100 and Ala98. The cis-trans isomerization of Pro33(L) facilitates a better fit of CDR L1, particularly of Pro33 and Trp34, to the α-helix of CCL17.
The energy barrier for the cis-trans transition in peptide bonds is about 13 kcal/mol for proline residues and close to 20 kcal/mol for non-proline residues [26]. For CCL17 and M116, the association may be driven mostly by enthalpy owing to a high density of hydrogen bonds and salt bridges at the molecular interface (Table 2). Crystal structures reveal a relatively small number of water molecules expelled upon complex formation, suggesting that entropy probably plays a minor role in this case. While the antibody-antigen interaction is apparently strong enough to overcome the cis-trans transition barrier, the association may proceed via a two-step mechanism, with one isomerization step at a time.
At least one antibody with a non-proline cis-peptide in CDR H3 has been reported based on the structure of SM3 Fab with a peptide from tumor antigen MUC1 [27]. However, it is unknown if the Gly96-Gln97 bond adopts a cis-conformation in the unbound antibody.
4. Conclusions
The crystal structure of M116 Fab in complex with its target CCL17 identified the binding epitope of the antibody consisting of three segments of the polypeptide chain. Comparison to the Fab structure in the unbound form revealed substantial conformational rearrangements in CDR H3 and CDR L1 to accommodate the antigen. Unexpectedly, upon binding the antigen, both CDRs undergo cis-trans isomerizations, which involve a valine in CDR H3 and a proline in CDR L1. This type of an induced fit is rarely observed in antibodies and probably never involved two CDRs simultaneously. The findings demonstrate yet another mechanism, by which antibodies adjust to their targets.
Acknowledgements
We are grateful to Bingyuan Wu and Yonghong Zhao for producing proteins for this study. The X-ray experiment used resources of the Advanced Photon Source, a U.S. Department of Energy Office of Science User Facility operated by Argonne National Laboratory under Contract no. DE-AC02-06CH11357.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2017.11.005.
Appendix A. Transparency document
Supplementary material
References
- 1.Griffith J.W., Sokol C.L., Luster A.D. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu. Rev. Immunol. 2014;32:659–702. doi: 10.1146/annurev-immunol-032713-120145. [DOI] [PubMed] [Google Scholar]
- 2.Zlotnik A., Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12:121–127. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 3.Wang X., Sharp J.S., Handel T.M., Prestegard J.H. Chemokine oligomerization in cell signaling and migration. Prog. Mol. Biol. Transl. Sci. 2013;117:531–578. doi: 10.1016/B978-0-12-386931-9.00020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saeki H., Tamaki K. Thymus and activation regulated chemokine (TARC)/CCL17 and skin diseases. J. Dermatol. Sci. 2006;43:75–84. doi: 10.1016/j.jdermsci.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Imai T., Baba M., Nishimura M., Kakizaki M., Takagi S., Yoshie O. The T cell-directed CC chemokine TARC is a highly specific biological ligand for CC chemokine receptor 4. J. Biol. Chem. 1997;272:15036–15042. doi: 10.1074/jbc.272.23.15036. [DOI] [PubMed] [Google Scholar]
- 6.Belperio J.A., Dy M., Murray L., Burdick M.D., Xue Y.Y., Strieter R.M., Keane M.P. The role of the Th2 CC chemokine ligand CCL17 in pulmonary fibrosis. J. Immunol. 2004;173:4692–4698. doi: 10.4049/jimmunol.173.7.4692. [DOI] [PubMed] [Google Scholar]
- 7.Yogo Y., Fujishima S., Inoue T., Saito F., Shiomi T., Yamaguchi K., Ishizaka A. Macrophage derived chemokine (CCL22), thymus and activation-regulated chemokine (CCL17), and CCR4 in idiopathic pulmonary fibrosis. Respir. Res. 2009;10:80. doi: 10.1186/1465-9921-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawasaki S., Takizawa H., Yoneyama H., Nakayama T., Fujisawa R., Izumizaki M., Imai T., Yoshie O., Homma I., Yamamoto K., Matsushima K. Intervention of thymus and activation-regulated chemokine attenuates the development of allergic airway inflammation and hyperresponsiveness in mice. J. Immunol. 2001;166:2055–2062. doi: 10.4049/jimmunol.166.3.2055. [DOI] [PubMed] [Google Scholar]
- 9.Matsukawa A., Hogaboam C.M., Lukacs N.W., Lincoln P.M., Evanoff H.L., Kunkel S.L. Pivotal role of the CC chemokine, macrophage-derived chemokine, in the innate immune response. J. Immunol. 2000;164:5362–5368. doi: 10.4049/jimmunol.164.10.5362. [DOI] [PubMed] [Google Scholar]
- 10.Shi L., Wheeler J.C., Sweet R.W., Lu J., Luo J., Tornetta M., Whitaker B., Reddy R., Brittingham R., Borozdina L., Chen Q., Amegadzie B., Knight D.M., Almagro J.C., Tsui P. De novo selection of high-affinity antibodies from synthetic fab libraries displayed on phage as pIX fusion proteins. J. Mol. Biol. 2010;397:385–396. doi: 10.1016/j.jmb.2010.01.034. [DOI] [PubMed] [Google Scholar]
- 11.K. Boakye, A. Del Vecchio, J. Kehoe, E. Lacy, L. Murray, M. Ryan, S. Santulli-Marotto, J. Wheeler, B. Whitaker, A. Teplyakov, Anti-CCL17 Antibodies, US patent US2015/0125458, 2015.
- 12.Santulli-Marotto S., Fisher J., Petley T., Boakye K., Panavas T., Luongo J., Kavalkovich K., Rycyzyn M., Wu B., Gutshall L., Coelho A., Hogaboam C.M., Ryan M. Surrogate antibodies that specifically bind and neutralize CCL17 but not CCL22. Monoclon. Antib. Immunodiagn. Immunother. 2013;32:162–171. doi: 10.1089/mab.2012.0112. [DOI] [PubMed] [Google Scholar]
- 13.Obmolova G., Malia T.J., Teplyakov A., Sweet R., Gilliland G.L. Promoting crystallization of antibody-antigen complexes via microseed matrix screening. Acta Crystallogr. 2010;D66:927–933. doi: 10.1107/S0907444910026041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kabsch W. XDS. Acta Crystallogr. 2010;D66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asojo O.A., Boulègue C., Hoover D.M., Lu W., Lubkowski J. Structures of thymus and activation-regulated chemokine (TARC) Acta Crystallogr. 2003;D59:1165–1173. doi: 10.1107/s0907444903009454. [DOI] [PubMed] [Google Scholar]
- 17.Teplyakov A., Obmolova G., Malia T.J., Luo J., Muzammil S., Sweet R., Almagro J.C., Gilliland G.L. Structural diversity in a human antibody germline library. MAbs. 2016;8:1045–1063. doi: 10.1080/19420862.2016.1190060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 19.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., McNicholas S.J., Murshudov G.N., Pannu N.S., Potterton E.A., Powell H.R., Read R.J., Vagin A., Wilson K.S. Overview of the CCP4 suite and current developments. Acta Crystallogr. 2011;D67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 22.Chothia C., Lesk A.M. Canonical structures for the hypervariable regions of immunoglobulins. J. Mol. Biol. 1987;196:901–917. doi: 10.1016/0022-2836(87)90412-8. [DOI] [PubMed] [Google Scholar]
- 23.Lubkowski J., Bujacz G., Boqué L., Domaille P.J., Handel T.M., Wlodawer A. The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions. Nat. Struct. Biol. 1997;4:64–69. doi: 10.1038/nsb0197-64. [DOI] [PubMed] [Google Scholar]
- 24.Qin L., Kufareva I., Holden L.G., Wang C., Zheng Y., Zhao C., Fenalti G., Wu H., Han G.W., Cherezov V., Abagyan R., Stevens R.C., Handel T.M. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine. Science. 2015;347:1117–1122. doi: 10.1126/science.1261064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kledal T.N., Rosenkilde M.M., Coulin F., Simmons G., Johnsen A.H., Alouani S., Power C.A., Lüttichau H.R., Gerstoft J., Clapham P.R., Clark-Lewis I., Wells T.N., Schwartz T.W. A broad-spectrum chemokine antagonist encoded by Kaposi's sarcoma-associated herpesvirus. Science. 1997;277:1656–1659. doi: 10.1126/science.277.5332.1656. [DOI] [PubMed] [Google Scholar]
- 26.Schulz G.D., Schirmer R.H. Springer-Verlag; New York: 1984. Principles of Protein Structure. [Google Scholar]
- 27.Bates P.A., Dokurno P., Freemont P.S., Sternberg M.J. Conformational analysis of the first observed non-proline cis-peptide bond occurring within the complementarity determining region (CDR) of an antibody. J. Mol. Biol. 1998;284:549–555. doi: 10.1006/jmbi.1998.2210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material