Abstract
Riemerella anatipestifer is an important pathogenic bacterium that infects ducks. It exhibits resistance to multiple classes of antibiotics. Multidrug efflux pumps play a major role as a mechanism of antimicrobial resistance in Gram-negative pathogens and they are poorly understood in R. anatipestifer. In this study, a gene encoding the B739_0873 protein in R. anatipestifer CH-1, which belongs to the resistance-nodulation-cell division (RND) efflux pump family, was identified. With respect to the substrate specificity of B739_0873, the antibiotic susceptibility testing showed that the B739_0873 knockout strain was more sensitive to aminoglycosides and detergents than the wild-type strain. The transcription of B739_0873 was up-regulated when R. anatipestifer CH-1 was exposed to sub-inhibitory levels of these substrates. From the gentamicin accumulation assay, we concluded that B739_0873 was coupled to the proton motive force to pump out gentamicin. Furthermore, site-directed mutagenesis demonstrated that Asp 400, Asp 401, Lys 929, Arg 959, and Thr 966 were the crucial function sites of B739_0873 in terms of its ability to extrude aminoglycosides and detergents. Finally, we provided evidence that B739_0873 is co-transcribed with B739_0872, and that both B739_0872 and B739_0873 are required for aminoglycoside and detergent resistance. In view of these results, we designate B739_0873 as RaeB (Riemerella anatipestifer efflux).
Keywords: Riemerella anatipestifer, B739_0873 gene, raeB gene, RND efflux pump, aminoglycoside, detergent, resistance
Introduction
Riemerella anatipestifer is a Gram-negative bacterium that belongs to the Flavobacteriaceae family (Segers et al., 1993). It infects ducks, geese, turkeys, chickens, and other birds, leads to contagious septicemia, and causes large economic losses in the duck industry (Ryll et al., 2001; Hess et al., 2013). Currently, at least 21 serotypes have been described worldwide, but there is no cross-protection between the different serotypes (Pathanasophon et al., 1996, 2002). Thus, antibiotics are still the major preventative and therapeutic approach against R. anatipestifer infection in poultry. Previous reports showed that the use of ceftiofur, novobiocin, penicillin, oxytetracycline, and streptomycin could reduce the mortality in ducks infected with R. anatipestifer (Sandhu and Dean, 1980; Chang et al., 2003). Unfortunately, the widespread use of antibiotics to treat R. anatipestifer infection has resulted in multidrug resistance in R. anatipestifer. Based on clinical investigation, R. anatipestifer is known to exhibit a very wide spectrum of drug resistance, including resistance to aminoglycosides, cephalosporins, lincosamides, macrolides, nalidixic acid, penicillins, rifampicin, and sulfonamides (Zhong et al., 2009; Sun et al., 2012). Using antibiotic therapy to achieve a good therapeutic effect has become more challenging. Thus, it is necessary to understand the multidrug resistance mechanisms of R. anatipestifer to find a way to prevent and treat R. anatipestifer infection.
In bacteria, multidrug efflux pumps are generally encoded by genetic elements capable of mediating intrinsic and acquired resistance to antibiotics (Li et al., 2015). Among these multidrug efflux pumps, the resistance-nodulation-cell division (RND) family members appear to be the most effective efflux systems in Gram-negative bacteria (Nikaido, 1994, 1996). RND transporters form a tripartite complex, consisting of an RND efflux pump, a periplasmic membrane fusion protein (MFP) and an outer membrane channel protein (OMP) to export the toxic compounds (Nikaido, 2011). To drive the export of the toxic substances, members of the RND family utilize the proton motive force (PMF) as the energy source (Paulsen et al., 1996). In recent years, numerous functions of RND efflux pumps have been identified; in addition to antibiotic extrusion, they are involved in bacterial pathogenicity and the bacterial stress response (Piddock, 2006). However, up to now, no reports have been published on the RND efflux pumps of R. anatipestifer.
In this study, we elucidated the biological functions of an RND efflux pump, the B739_0873 protein, namely RaeB for the first time, and obtained information to help to increase understanding of the multidrug-resistance mechanism of R. anatipestifer.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
All the plasmids and strains used in this study are listed in Table 1. R. anatipestifer CH-1 (GenBank accession number: NC_018609) was isolated from the brains of diseased ducks from Chengdu, Sichuan, China, identified by our laboratory (Wang et al., 2014) and cultured in tryptic soybean broth (TSB, Oxoid) or tryptic soy agar (TSA, Oxoid) medium at 37°C in 5% CO2. Escherichia coli strains were grown in Luria-Bertani (LB, Oxoid) broth or LB agar (1.5% agar powder, Solarbio) at 37°C. The antimicrobial agents that were added to the media used for strain construction and selection were purchased from Sigma, and they were added to reach the following final concentrations: ampicillin (Amp), 100 μg/ml; cefoxitin (Cfx), 1 μg/ml; chloramphenicol (Cm), 25 μg/ml; kanamycin (Kan), 50 μg/ml; and spectinomycin (Spc), 80 μg/ml.
Table 1.
Bacterial strains and plasmids used in this study.
| Strains and plasmids | Description | Source or reference |
|---|---|---|
| Strains | ||
| R. anatipestifer CH-1 | Serotype 1, CfxS, KanR, SpcS | Wang et al., 2014 |
| RA-CH-1 ΔraeB | raeB deletion mutant of R. anatipestifer CH-1 strain, SpcR | This study |
| RA-CH-1 ΔraeB pLMF03::raeB | RA-CH-1 ΔraeB carrying pLMF03::raeB, KanR, CfxR | This study |
| RA-CH-1 ΔraeA ΔraeB | raeA-raeB deletion mutant of R. anatipestifer CH-1 strain, SpcR | This study |
| RA-CH-1 ΔraeA ΔraeB pLMF03::raeB | RA-CH-1 ΔraeA ΔraeB carrying pLMF03::raeB, KanR, CfxR | This study |
| RA-CH-1 ΔraeB pD400A | RA-CH-1 ΔraeB carrying pLMF03::raeBD400A, KanR, CfxR | This study |
| RA-CH-1 ΔraeB pD401A | RA-CH-1 ΔraeB carrying pLMF03::raeBD401A, KanR, CfxR | This study |
| RA-CH-1 ΔraeB pK929E | RA-CH-1 ΔraeB carrying pLMF03::raeBK929E, KanR, CfxR | This study |
| RA-CH-1 ΔraeB pR959A | RA-CH-1 ΔraeB carrying pLMF03::raeBR959A, KanR, CfxR | This study |
| RA-CH-1 ΔraeB pT966E | RA-CH-1 ΔraeB carrying pLMF03::raeBR966E, KanR, CfxR | This study |
| E. coli K-12 X7232 | endA1 hsdR17(rK- mK+) glnV44 thi-1 recA1 gyrA relA1Δ(lacZYA-argF)U169λpir deoR (Φ80dlac Δ(lacZ)M15) | Roland et al., 1999 |
| E. coli K-12 X7232 pRE112::raeBUSD | E. coli K-12 X7232 carrying pRE112::raeBUSD, SpcR, CmR | This study |
| E. coli K-12 X7232 pRE112::raeB-raeAUSD | E. coli K-12 X7232 carrying pRE112::raeA-raeBUSD, SpcR, CmR | This study |
| E. coli K-12 X7213 | thi-1 thr-1 leuB6 glnV44 fhuA21 lacY1 recA1 RP4-2-Tc::Mu λpirΔasdA4Δzhf-2::Tn10 | Roland et al., 1999 |
| E. coli K-12 X7213 pRE112::raeBUSD | E. coli K-12 X7213 carrying pRE112::raeBUSD, SpcR, CmR | This study |
| E. coli K-12 X7213 pRE112::raeA-raeBUSD | E. coli K-12 X7213 carrying pRE112::raeA-raeBUSD, SpcR, CmR | This study |
| E. coli S17-1 | thi-1 thr leu tonA lac Y supE recA::RP4-2-Tc::Mu KanR | Miller and Mekalanos, 1988 |
| E. coli S17-1 pLMF03::raeB | E. coli S17-1 carrying pLMF03::raeB, AmpR, CfxR | This study |
| E. coli S17-1 pLMF03::raeBD400A | E. coli S17-1 carrying pLMF03::raeBD400A, AmpR, CfxR | This study |
| E. coli S17-1 pLMF03::raeBD401A | E. coli S17-1 carrying pLMF03::raeBD401A, AmpR, CfxR | This study |
| E. coli S17-1 pLMF03::raeBK929E | E. coli S17-1 carrying pLMF03::raeBK929E, AmpR, CfxR | This study |
| E. coli S17-1 pLMF03::raeBR959A | E. coli S17-1 carrying pLMF03::raeBR959A, AmpR, CfxR | This study |
| E. coli S17-1 pLMF03::raeBT966E | E. coli S17-1 carrying pLMF03::raeBT966E, AmpR, CfxR | This study |
| E. coli BL21(DE3) | Expressing host cell | Invitrogen |
| E. coli BL21(DE3) pET32a (+)::raeB | E. coli BL21(DE3) carrying pET32a (+)::raeB, AmpR | This study |
| Plasmids | ||
| pYES1 | YAC-BAC shuttle plasmid, SpcR | Invitrogen |
| pRE112 | sacB mobRP4 R6K ori, CmR, pRE112-T-vector | Kong et al., 2011 |
| pRE112::raeBUSD | pRE112 carrying raeBUSD from R. anatipestifer CH-1 and pYES1, SpcR, CmR | This study |
| pRE112::raeA-raeBUSD | pRE112 carrying raeA-raeBUSD from R. anatipestifer CH-1 and pYES1, SpcR, CmR | This study |
| pET32a (+) | pBR322 lacZ, IPTG-inducible promoter, AmpR | Invitrogen |
| pET32a (+)::raeB | pET32a (+) carrying the truncated raeB from R. anatipestifer CH-1, AmpR | This study |
| pLMF03 | B739_0921 promoter, ori ColE1, ori pRA0726, AmpR, CfxR | Liu et al., 2016 |
| pLMF03::raeB | pLMF03 carrying raeB from R. anatipestifer CH-1, CfxR | This study |
| pLMF03::raeBD400A | pLMF03 carrying raeB with mutation D400A, CfxR | This study |
| pLMF03::raeBD401A | pLMF03 carrying raeB with mutation D401A, CfxR | This study |
| pLMF03::raeBK929E | pLMF03 carrying raeB with mutation K929E, CfxR | This study |
| pLMF03::raeBR959A | pLMF03 carrying raeB with mutation R959A, CfxR | This study |
| pLMF03::raeBT966E | pLMF03 carrying raeB with mutation T966E, CfxR | This study |
R, resistance; S, sensitive; Amp, ampicillin; Cfx, cefoxitin; Cm, chloramphenicol; Kan, kanamycin; Spc, spectinomycin.
Polymerase Chain Reaction Method
The DNA fragments were obtained by polymerase chain reaction (PCR). PCR was carried out in a Hybaid PCR thermocycler (Bio-Rad) using the DNA Polymerase (Takara) according to manufacturer’s instructions. All primers used in this study were synthesized by Invitrogen.
Quantitative Real-Time-PCR Analysis
To monitor the mRNA expression levels, quantitative real-time (qRT)-PCR was used. Total RNA was extracted from R. anatipestifer CH-1 strains in the logarithmic phase using RNAiso Plus (Takara) according to the manufacturer’s instructions. The RNA was then reverse transcribed into cDNA using the PrimeScriptTM RT Reagent Kit (Takara) with gDNA Eraser (Takara). PCR with SYBR® Premix Ex TaqTM II (Takara) was performed using the gene-specific primers P1-16S rRNA-F/P2-16S rRNA-R P3-raeBRT-F/P4-raeBRT-R and P5-B739_0874-F/P6-B739_0874-R. In the qRT-PCR analysis, 16S rRNA gene was used as an internal control. The relative gene expression was calculated using the 2-Δ ΔCt method with Bio-Rad CFX Manager software (Schmittgen and Livak, 2008). Experiments were performed in triplicate.
Construction of Knockout Strains
Recombinant suicide plasmids pRE112::B739_0873USD and pRE112::B739_0872-B739_0873USD were used to delete the B739_0873 and both the B739_0872 and B739_0873 genes, respectively, by allelic exchange, according to previously described methods (Luo et al., 2015). Briefly, the primers P7-raeBup-F/P8-raeBup-R, P13-raeA-raeBup-F/P14-raeA-raeB up-R, and P11-raeBdown-F/P12-raeBdown-R were used to amplify the upstream and downstream homologous arm regions of the B739_0873 and B739_0872-B739_0873 genes in the R. anatipestifer CH-1 genome, respectively. The primers P9-raeBspc-F/P10-raeBspc-R and P15-raeA-raeBspc-F/P10-raeBspc-R were used to amplify Spc resistant (SpcR) cassettes from the plasmid pYES1. Each set of three PCR fragments was integrated using overlap PCR (Xiong et al., 2006) with primer pairs P7-raeBup-F/P12-raeBdown-R and P13-raeA-raeBup-F/P12-raeBdown-R. The fusion segments were then cloned into pRE112 to construct pRE112::B739_0873USD and pRE112::B739_0872-B739_0873USD. Subsequently, the recombinant plasmids were introduced into R. anatipestifer CH-1 by conjugation as described previously (Liao et al., 2015). The transconjugants were selected on TSA plates containing Spc (80 μg/ml) and Cfx (1 μg/ml), and they were then confirmed using PCR with the conserved 16S rRNA gene primers P1-16S rRNA-F/P2-16S rRNA-R and the corresponding identifying primers P16-raeBIdent-F/P17-raeBIdent-R and P18-raeA-raeBIdent-F/P19-raeA-raeBIdent-R. The resultant gene-deletion mutant strains were designated RA-CH-1 ΔB739_0873 and RA-CH-1 ΔB739_0872 ΔB739_0873.
Construction of Complemented Strains
To confirm that the RA-CH-1 ΔB739_0873 and RA-CH-1 ΔB739_0872 ΔB739_0873 phenotypes were due to the B739_0873 gene deletion, the E. coli–R. anatipestifer shuttle plasmid pLMF03 was used to construct the recombinant plasmid pLMF03::B739_0873 that contained an intact B739_0873 gene. Briefly, the primers P20-raeB-F/P21-raeB-R were used to amplify the B739_0873 open reading frame (ORF), which was ligated into pLMF03 at NcoI and XhoI sites to generate pLMF03::B739_0873. For the complementation analysis, the pLMF03::B739_0873 plasmid was introduced into the two R. anatipestifer CH-1 mutant strains by conjugation, as described previously (Wang et al., 2017). The transconjugants were selected on TSA plates containing Kan (50 μg/ml) and Cfx (1 μg/ml), and they were then confirmed using PCR with the conserved 16S rRNA gene primers P1-16S rRNA-F/P2-16S rRNA-R and the Cfx-identifying primers P24-cfx-F/P25-cfx-R. The resultant complemented mutant strains were designated RA-CH-1 ΔB739_0873 pLMF03::B739_0873 and RA-CH-1 ΔB739_0872 ΔB739_0873 pLMF03::B739_0873.
Bacterial Growth Curves and Competition Experiments in Vitro
To evaluate the growth rates under non-competitive conditions, we monitored the growth curves for the wild-type strain, the RA-CH-1 ΔB739_0873 mutant, and the RA-CH-1 ΔB739_0873 complemented strain, according to the previously described method (Luo et al., 2015). In vitro competition experiments were performed with the wild-type strain and the RA-CH-1 ΔB739_0873 mutant (Perez et al., 2012). Briefly, both strains were mixed in a 1:1 ratio when they were in the exponential phase. Subsequently, approximately 10-5 cells from the mixtures were added to 10 ml TSB and grown at 37°C. After 16 h, 10-fold serial dilutions of the cells were spread onto both TSA and TSA containing 80 μg/ml Spc in duplicate and incubated overnight at 37°C. The competition index (CI) was defined as the ratio between the number of mutant and wild-type CFUs. All experiments were performed in triplicate.
Minimal Inhibitory Concentration Determination
A twofold serial dilution assay was used to measure the minimal inhibitory concentration (MIC) of antimicrobial agents for R. anatipestifer CH-1 strains according to the Clinical and Laboratory Standards Institute guidelines (CLSI, 2015). The general method from the CLSI document was used and there was no specific guidance document for R. anatipestifer. The E. coli American Type Culture Collection (ATCC) 25922 strain was used for quality control. Antimicrobial agents were serially diluted twofold in TSB broth with concentrations ranging between 1 and 512 μg/ml. The turbidity of the inoculum was adjusted to 107 CFU/ml and 100 μl was added into every well. The 96-well microtiter plates were incubated at 37°C for 24 h. The lowest concentration that inhibited bacterial growth was considered to be the MIC. All tests were performed in triplicate.
Gentamicin Accumulation Assay
To determine whether the B739_0873 protein was driven by PMF, the PMF inhibitor carbonyl cyanide m-chlorophenylhydrazone (CCCP, Sigma) was used to attempt to inhibit B739_0873 activity. First, a series of CCCP concentrations (1.25, 2.5, 5, and 10 μM) were prepared to determine the optimal concentration for the full survival of the wild-type strain, the RA-CH-1 ΔB739_0873 mutant and the complemented strain RA-CH-1 ΔB739_0873 pLMF03::B739_0873. Subsequently, the MICs of aminoglycosides and detergents for these R. anatipestifer CH-1 strains were determined with or without CCCP.
To further confirm that the B739_0873 protein was the inhibited efflux pump, a gentamicin accumulation assay was performed. Cells of R. anatipestifer CH-1, RA-CH-1 ΔB739_0873 and RA-CH-1 ΔB739_0873 pLMF03::B739_0873 were grown in TSB broth and harvested at the exponential phase of growth, and then washed twice at room temperature with 50 mM phosphate buffer (pH 7.0), containing 1 mM MgSO4 and 0.4% (wt/vol) glucose. The washed cells were resuspended in the same buffer at a density of 1 mg (dry weight) per ml, including 32 μg/ml gentamicin. To determine the dry weight, cells harvested at the exponential phase of growth by centrifugation for 10 min at 8,000 rpm, removed the supernatant and dried to a constant weight at 60°C. At the 24-min time point, 5 μM CCCP was added to the cells suspensions to assess energy-dependent efflux. The samples were collected every 8 min, centrifuging at 12,000 g and 4°C for 1 min and the samples were immediately diluted into ice-cold phosphate buffer, followed by two washes and sonication on ice. Gentamicin uptake was measured using a Gentamicin ELISA Test Kit following the manufacturer’s instructions (Reagen).
Site-Directed Mutagenesis
Subsequently, we investigated whether five conserved amino acid residues (Asp 400, Asp 401, Lys 929, Arg 959, and Thr 966) (Guan and Nakae, 2001; Su et al., 2006) in the R. anatipestifer CH-1 B739_0873 protein are vital for it to function. The point mutations were introduced into plasmid pLMF03::B739_0873 using the overlap PCR method (Xiong et al., 2006). The mutant sites were designed in primers. As illustrated in Table 2, primer pairs P26-raeBD400A-R/P27-raeBD400A-F, P28-raeBD401A-R/P29- raeBD401A-F, P30-raeBK929E-R/P31-raeBK929E-F, P32-raeBR959A-R/P33-raeBR959A-F, and P34-raeBT966E-R/P35-raeBT966E-F, were reverse complementary. The upstream fragments were amplified from the R. anatipestifer CH-1 genome using primer pairs P20-raeB-F/P26-raeBD400A-R, P20-raeB-F/P28-raeBD401A, P20-raeB-F/P30-raeBK929E-R, P20-raeB-F/P32-raeBR959A-R, and P20-raeB-F/P34-raeBT966E-R, and the downstream fragments were amplified using primer pairs P27-raeBD400A-F/P21-raeB-R, P29-raeBD401A-F/P21-raeB-R, P31-raeBK929E-F/P21-raeB-R, P33-raeBR959A-F/P21-raeB-R, and P35-raeBT966E-F/P21-raeB-R. The two PCR fragments were integrated using overlap PCR with primer pair P20/P21 to generate versions of the B739_0873 gene, each with a single mutation site. The B739_0873 mutant fragments were purified by MiniBEST DNA fragment Purification Kit (Takara) following the manufacturer’s instructions and then cloned into the pLMF03 plasmid to generate mutant recombinant plasmids, pLMF03::B739_0873D400A, pLMF03::B739_0873D401A, pLMF03::B739_0873K929E, pLMF03::B739_0873R959A, and pLMF03::B739_0873T966E. They were introduced into RA-CH-1 ΔB739_0873 by conjugation, as described previously (Wang et al., 2017). The transconjugants were selected on TSA plates containing Kan (50 μg/ml) and Cfx (1 μg/ml), and they were then confirmed using PCR with the conserved 16S rRNA gene primers P1-16S rRNA-F/P2-16S rRNA-R and the Cfx-identifying primers P24-cfx-F/P25-cfx-R. The resultant complemented mutant strains were designated RA-CH-1 ΔB739_0873 pD400A, RA-CH-1 ΔB739_0873 pD401A, RA-CH-1 ΔB739_0873 pK929E, RA-CH-1 ΔB739_0873 pR959A, and RA-CH-1 ΔB739_0873 pT966A.
Table 2.
Primers used in this study.
| Primer | Sequence (5′–3′) | Source |
|---|---|---|
| P1-16S rRNA-F | CGAAAGTGATAAGTTAGCCACCT | This study |
| P2-16S rRNA-R | GCAGCACCTTGAAAATTGTCC | This study |
| P3-raeBRT-F | AAGAGCCTTCGTTATCACAGT | This study |
| P4-raeBRT-R | AATTTCTCGCTCTTGCCCTC | This study |
| P5-B739_0874-F | TCACACGAATACAATGGTT | This study |
| P6-B739_0874-R | AGGCTGTACTTTGATAACTCT | This study |
| P7-raeBup-F | CGCGGATCCCACTACAACTAGATCAAGCG | This study |
| P8-raeBup-R | AATAAGGGCGACACGGAAATGTTAATGGCGTTGATTTTCCTT | This study |
| P9-raeBspc-F | AAGGAAAATCAACGCCATTAACATTTCCGTGTCGCCCTTATT | This study |
| P10-raeBspc-R | ATCTTCCTTAGCCAGTTTTCTGAGGCCATCAAACCACGTCA | This study |
| P11-raeBdown-F | TGACGTGGTTTGATGGCCTCAGAAAACTGGCTAAGGAAGAT | This study |
| P12-raeBdown-R | CGGGGTACCACCTACGATATGACGGTTC | This study |
| P13-raeA-raeBup-F | CGCGGATCCGCCGTCTGTACTTATTTCG | This study |
| P14-raeA-raeBup-R | AATAAGGGCGACACGGAAATGTTTATTTTTCTGTTAAAGTTCT | This study |
| P15-raeA-raeBspc-F | AGAACTTTAACAGAAAAATAAACATTTCCGTGTCGCCCTTATT | This study |
| P16-raeBIdent-F | GGATCCATGAATAAAAAAACATTATTATCTATTATAG | This study |
| P17-raeBIdent-R | TAACTTTGTTTTAGGGCGACT | This study |
| P18-raeA-raeBIdent-F | TAGACAGGCTTATCTTGGACA | This study |
| P19-raeA-raeBIdent-R | TAACAAAATACCATCAAGGCTA | This study |
| P20-raeB-F | CATGCCATGGATGAAATTAGCAGAAGTATCC | This study |
| P21-raeB-R | CCGCTCGAGCTCCATTTTTTGAAGCCTCT | This study |
| P22-Truncated raeB-F | CGCGGATCCATGACCATATATCCAGGGGCATC | This study |
| P23-Truncated raeB-R | CCGCTCGAGGTTCTGTTGATTGATACGAGCA | This study |
| P24-cfx-F | CTCGCCAGAATCATAGACAAG | This study |
| P25-cfx-R | ATAGCGCATAAGACAGGTTC | This study |
| P26-raeBD400A-R | CACAATGGCATCAGCTACCAATATCCCT | This study |
| P27-raeBD400A-F | AGGGATATTGGTAGCTGATGCCATTGTG | This study |
| P28-raeBD401A-R | CACAATGGCGGCGTCTACCAATATCCCT | This study |
| P29-raeBD401A-F | AGGGATATTGGTAGACGCCGCCATTGTG | This study |
| P30-raeBK929E-R | GCATTTTCCGCCACCAAACCAATCAAC | This study |
| P31-raeBK929E-F | GTTGATTGGTTTGGTGGCGGAAAATGC | This study |
| P32-raeBR959A-R | CATCAAAATAGGACGAAGAGCAGC | This study |
| P33-raeBR959A-F | GCTGCTCTTCGTCCTATTTTGATG | This study |
| P34-raeBT966E-R | CCATCGCTATCGTTTCCATCAAAA | This study |
| P35-raeBT966E-F | TTTTGATGGAAACGATAGCGATGG | This study |
-, mutation sites underlined.
Antibody Preparation
The recombinant plasmid pET32a (+)::B739_0873 was used to express B739_0873 protein. Briefly, the primers P22-Truncated raeB-F/P23-Truncated raeB-R were used to amplify the truncated B739_0873 ORF, which was ligated into pET32a (+) at BamHI and XhoI sites to generate pET32a (+)::B739_0873.
Strain E. coli BL21 (DE3) pET32a (+)::B739_0873 was grown overnight at 37°C in LB broth containing 100 μg/ml Amp. Subsequently, the LB broth containing 100 μg/ml Amp was inoculated with the overnight culture to an optical density at 600 nm (OD600) of 0.05 and grown at 37°C. Expression was induced by adding 0.6 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at an OD600 of 0.6 for 6 h at 37°C. Cells harvested by centrifugation for 10 min at 8,000 rpm and 4°C were suspended in lysis buffer (20 mM Tris–HCl, pH 8.0) and then sonicated on ice. After centrifugation at 10,000 rpm and 4°C for 10 min, the supernatant of the lysate was loaded onto an equilibrated (with 20 mM Tris–HCl, pH 8.0) nickel-nitrilotriacetic acid (NTA) column (7sea Biotech, China). By washing with lysis buffer (20 mM Tris–HCl, pH 8.0) containing 20 and 50 mM imidazole, non-specific contaminants were removed. Subsequently, the B739_0873 protein was eluted with lysis buffer (20 mM Tris–HCl, pH 8.0) containing 200 mM imidazole. Then the purified protein was placed in the dialysis membrane (Solarbio), soaked in phosphate-buffered saline (PBS) buffer (pH 7.4) and changed the buffer every other 1, 2, 4 h, the final dialyzed overnight, to eliminate any residual imidazole.
The purified B739_0873 His-tagged protein was used to generate a polyclonal antibody. Approximately 2 mg of B739_0873 protein emulsified in complete Freund’s adjuvant (Sigma) was used to immunize four rabbits (the local rabbit industry in Ya’an, Sichuan) via intradermal injections. Subsequently, booster doses of 3 and 4 mg of B739_0873 protein were prepared in incomplete Freund’s adjuvant (Sigma), and the immunization was given after 2 and 3 weeks, respectively, using subcutaneous injections. A week after the last immunization, the antibody was collected from the ear vein of the rabbits and frozen at -80°C.
Immunoblot Analysis
Strains RA-CH-1 ΔB739_0873 pLMF03, RA-CH-1 ΔB739_0873 pLMF03::B739_0873, RA-CH-1 ΔB739_0873 pD400A, RA-CH-1 ΔB739_0873 pD401A, RA-CH-1 ΔB739_0873 pK929E, RA-CH-1 ΔB739_0873 pR959A, and RA-CH-1 ΔB739_0873 pT966A were grown at 37°C in TSB containing 1 μg/ml Cfx. Bacteria were harvested by centrifugation until they grew to exponential phase, suspended in the PBS buffer and then sonicated on ice. The sonicated cells were suspended in loading buffer and heated for 5 min at 100°C. Proteins were separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) using TGX Stain-FreeTM FastCastTM Acrylamide Kit (Bio-Rad) and subsequently transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad). Non-specific binding sites were blocked with 5% skim milk (Solarbio) in TBS-Tween 20 (0.05%). The PVDF membranes were probed with B739_0873 rabbit polyclonal antibody (1:200), followed by a 1:3,000 dilution of a goat anti-rabbit IgG alkaline phosphatase-conjugated secondary antibody (Bio-Rad). The binding of antibodies to protein was revealed using a substrate for horseradish peroxidase (HRP)-based chemiluminescence Western blot detection following the manufacturer’s instructions (Takara).
Ethics Statement
All animals were handled in strict accordance with good animal practices, as defined by the local animal welfare bodies. The protocol for the animal work to be performed at Sichuan Agriculture University was reviewed and approved by the Sichuan Agriculture University ethics committee in September 2014.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism version 7 software for Windows. The significance of the between-group differences was ascertained using Student’s t-test. A value of P < 0.05 was considered significant.
Results
Sequence Analysis of R. anatipestifer CH-1 raeB
The B739_0873 gene of R. anatipestifer CH-1 consists of an ORF with 3,156 bp, which encodes a 1,051-amino acid protein with a putative molecular mass of 115 kDa. The B739_0873 protein is annotated as a cation/multidrug efflux pump in the National Center for Biotechnology Information (NCBI) database. A protein–protein Basic Local Alignment Search Tool (BLASTP) analysis of the B739_0873 amino acid sequence of different R. anatipestifer strains indicated 99–100% identity. Homologs of B739_0873 are also present in Riemerella columbina, Riemerella columbipharyngis, Flavobacteriaceae bacterium 3519-10, Salinimicrobium terrae, Chryseobacterium treverense, Chryseobacterium solincola, Salegentibacter salegens, and Salegentibacter agarivorans, and identities range from 77 to 83%. However, the function of all these homologs has not previously been characterized.
Further analysis of the B739_0873 protein revealed that it was located in the cytoplasmic membrane and there were 12 trans-membrane domains (TMD) and two extremely large periplasmic loops between helix 1 and 2 and between helix 7 and 8. These results are consistent with the general features of RND efflux pumps (Tseng et al., 1999). The Clustal comparison showed that B739_0873 shares 28% identity with E. coli AcrB and 27% identity with Pseudomonas aeruginosa MexB. Furthermore, the essential residues in E. coli AcrB (Asp 407, Asp 408, Lys 940, Arg 971, and Thr 978) (Su et al., 2006; Takatsuka and Nikaido, 2006) and P. aeruginosa MexB (Asp 407, Asp 408, Lys 939, Arg 969, and Thr 976) (Guan and Nakae, 2001) that are responsible for the proton relay pathway are well conserved in B739_0873 (corresponding to Asp 400, Asp 401, Lys 929, Arg 959, and Thr 966).
Analysis of the upstream regions of the B739_0873 gene showed that the B739_0872 gene encodes a protein belonging to the MFP family (Dinh et al., 1994), the B739_0871 gene encodes a protein belonging to the outer membrane factor (OMF) family (Paulsen et al., 1997) and the B739_0870 gene encodes a tetracycline resistance repressor protein (TetR) family transcriptional regulator (Ramos et al., 2005). These three genes occur together with the B739_0873 gene on the R. anatipestifer CH-1genome (Figure 1). Sequence alignments showed that B739_0870 shares 31% identity with E. coli AcrR, B739_0872 shares 24% identity with E. coli AcrA and 25% identity with P. aeruginosa MexA, and B739_0871 shares 20% identity with E. coli TolC and 23% identity with P. aeruginosa OprM.
FIGURE 1.
Schematic representation of the position and size of the resistance-nodulation-cell division (RND) efflux pump genes in the genomes of R. anatipestifer CH-1, E. coli, and P. aeruginosa. The RND efflux pump genes are described in yellow, membrane fusion protein (MFP) in blue, the outer membrane protein (OMP) in red, and the regulator in green.
Overall, the bioinformatic analysis suggests that B739_0873 is a putative RND efflux pump. Thus, we designated the B739_0870-0871-0872-0873 genes as raeR-raeC-raeA-raeB and the B739_0870-0871-0872-0873 proteins as RaeR-RaeC-RaeA-RaeB described below.
Increased Aminoglycoside and Detergent Susceptibility of raeB Mutant
To investigate the role of raeB, a mutated raeB was constructed in R. anatipestifer CH-1. To determine whether the inactivation of raeB affected the growth state of R. anatipestifer CH-1, both the growth curves and the CI values were evaluated. The results showed that RA-CH-1 ΔraeB had the same growth rate and CI value as the wild-type strain (Supplementary Figures S1, S2). To determine whether the inactivation of raeB affected the virulence of R. anatipestifer CH-1, the median lethal dose (LD50) was measured as described previously by Wang et al. (2017). The results showed that there were no significant LD50 changes between the wild-type strain and RA-CH-1 ΔraeB (data not shown).
In a second set of experiments, the susceptibility of the RA-CH-1 ΔraeB mutant and the wild-type strain to a variety of antimicrobial agents with dissimilar structures (including Amp, acriflavine, azithromycin, aztreonam, carbenicillin, cefradine, ceftiofur, cefuroxime, cephalothin, chloromycetin, ciprofloxacin, enrofloxacin, erythromycin, ethidium bromide, florfenicol, gentamicin, Kan, lincomycin, nalidixic acid, rifampicin, sodium dodecyl sulfate (SDS), streptomycin, sulfamethoxazole, tetracycline, trimethoprim, Triton X-100, and vancomycin) was compared. It was shown that deletion of raeB increased susceptibility to all the aminoglycosides tested [streptomycin (eightfold), Kan (eightfold), and gentamicin (16-fold)] and all the detergents tested [Triton X-100 (fourfold) and SDS (16-fold)] (Table 3). There were no MIC changes between the wild-type strain and the mutant strain for any of the tested cephalosporins, chloramphenicols, cationic dyes, quinolones, glycopeptides, lincosamides, macrolides, penicillins, rifampicin, sulfonamides, and tetracyclines (Supplementary Table S1). To exclude the possibility that gene inactivation had a polar effect on the transcription of adjacent genes, RT-PCR was performed to measure the mRNA levels of the downstream gene, B739_0874, and it was shown that there was no significant difference in B739_0874 gene transcription between the wild-type strain and the RA-CH-1 ΔraeB mutant (data not shown). This result revealed that the changes were caused solely by raeB. Complementation of the RA-CH-1 ΔraeB mutant with plasmid pLMF03::raeB restored resistance to aminoglycosides and detergents (Table 3). This result indicated that raeB is involved in aminoglycoside and detergent resistance.
Table 3.
MICs of aminoglycosides and detergents for R. anatipestifer CH-1 strains.
| Strains | MIC (μg/ml) | ||||
|---|---|---|---|---|---|
| Gentamicin | Streptomycin | Kanamycin | Sodium dodecyl sulfate | Triton X-100 | |
| R. anatipestifer CH-1 | 32 | 128 | 256 | 640 | 160 |
| RA-CH-1 ΔraeB | 2 | 16 | 32 | 40 | 40 |
| RA-CH-1 ΔraeB pLMF03::raeB | 32 | 128 | 256 | 640 | 160 |
| RA-CH-1 ΔraeA ΔraeB | 2 | 16 | 32 | 40 | 40 |
| RA-CH-1 ΔraeA ΔraeB pLMF03::raeB | 2 | 16 | 32 | 40 | 40 |
| RA-CH-1 ΔraeB pD400A | 2 | 16 | 32 | 40 | 40 |
| RA-CH-1 ΔraeB pD401A | 2 | 16 | 32 | 40 | 40 |
| RA-CH-1 ΔraeB pK929E | 2 | 16 | 32 | 40 | 40 |
| RA-CH-1 ΔraeB pR959A | 2 | 16 | 32 | 40 | 40 |
| RA-CH-1 ΔraeB pT966E | 2 | 16 | 32 | 40 | 40 |
Induction of raeB Transcription by Aminoglycoside and Detergent Exposure
To address whether raeB is regulated by aminoglycosides and detergents, sub-inhibitory concentrations of aminoglycosides and detergents were added to an R. anatipestifer CH-1 growth culture and the transcription of raeB was measured using qRT-PCR. As shown in Figure 2, the level of raeB expression was up-regulated by two- to sevenfold after treatment with Triton X-100, SDS, streptomycin, Kan, or gentamicin.
FIGURE 2.
Relative fold changes of raeB mRNA expression levels in R. anatipestifer CH-1 after treatment with aminoglycosides and detergents. The sub-inhibitory concentrations of aminoglycosides and detergents were added to the TSB broth (gentamicin, 16 μg/ml; Kan, 128 μg/ml; streptomycin, 64 μg/ml; Triton X-100, 80 μg/ml; SDS, 320 μg/ml). SDS, sodium dodecyl sulfate. Error bars indicate the standard deviation (n = 3).
Inhibition of RaeB Activity by CCCP
The final concentration of CCCP in TSB broth was 5 μM, which did not affect the growth of the wild-type strain, the RA-CH-1 ΔB739_0873 mutant and the complemented strain RA-CH-1 ΔB739_0873 pLMF03::B739_0873. The results showed that the addition of CCCP (5 μM) decreased the MICs of streptomycin (eightfold), gentamicin (16-fold), and Kan (eightfold) for the wild-type strain and RA-CH-1 ΔraeB pLMF03::raeB. Similarly, the MICs of SDS and Triton X-100 for the wild-type strain and RA-CH-1 ΔraeB pLMF03::raeB decreased by 4- and 16-fold, respectively, in the presence of CCCP. In contrast, CCCP addition did not modify the MICs of streptomycin, gentamicin, Kan, SDS, and Triton X-100 for RA-CH-1 ΔraeB. The MICs of streptomycin, gentamicin, Kan, SDS, and Triton X-100 for the wild-type strain and RA-CH-1 ΔraeB pLMF03::raeB in the presence of CCCP were consistent with those evidenced for RA-CH-1 ΔraeB in the absence or presence of CCCP. These data indicated that an aminoglycoside and detergent efflux pump exists in R. anatipestifer CH-1.
As shown in Figure 3, at the 24-min incubation time point, the accumulation level of gentamicin achieved a steady-state for all strains. The gentamicin accumulation level in the mutant strain RA-CH-1 ΔraeB was about six times higher than that in the wild-type strain and RA-CH-1 ΔraeB pLMF03::raeB. After CCCP was added to the cells containing gentamicin at 24-min, the accumulation of gentamicin increased in the wild-type strain and RA-CH-1 ΔraeB pLMF03::raeB; these accumulation levels were lower than that in RA-CH-1 raeB. In contrast, under our conditions, CCCP had no significant effect on the level of gentamicin accumulation in the mutant strain RA-CH-1 ΔraeB. In addition, the wild-type strain, RA-CH-1 ΔraeB, and RA-CH-1 ΔraeB pLMF03::raeB with no CCCP added at 24-min served as controls. These data indicated that RaeB pumped out gentamicin in an energy-dependent process, presumably coupled to the PMF.
FIGURE 3.
Accumulation of gentamicin by R. anatipestifer CH-1 cells. CCCP, carbonyl cyanide m-chlorophenylhydrazone; WT, R. anatipestifer CH-1; ΔraeB, RA-CH-1 ΔraeB; ΔraeB pLMF03::raeB, RA-CH-1 ΔraeB pLMF03::raeB complemented strain. Error bars indicate the standard deviation (n = 3). The arrow indicate CCCP was added at 24-min.
Identification of the Functional Sites in RaeB
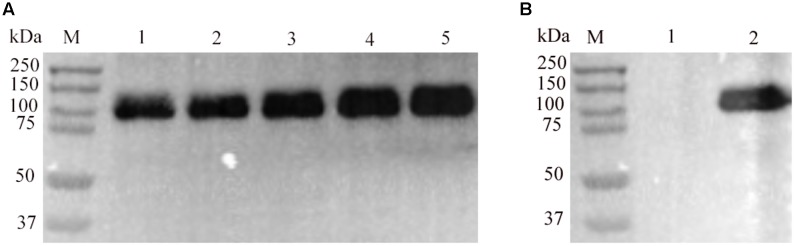
We investigated whether amino acid residues Asp 400, Asp 401, Lys 929, Arg 959, and Thr 966 are vital for the function of the RaeB protein of R. anatipestifer CH-1, five mutant recombinant plasmids, pLMF03::raeBD400A, pLMF03::raeBD401A, pLMF03::raeBK929E, pLMF03::raeBR959A, and pLMF03::raeBT966E, were constructed. These plasmids were introduced into RA-CH-1 ΔB739_0873, and the MICs for RA-CH-1 ΔraeB pD400A, RA-CH-1 ΔraeB pD401A, RA-CH-1 ΔraeB pK929E, RA-CH-1 ΔraeB pR959A, and RA-CH-1 ΔraeB pT966A were examined. It was shown that none of these mutant strains exhibited restored resistance to aminoglycosides and detergents (Table 3). To exclude the possibility that these results occurred because these mutant raeB genes were not expressed, we detected the expression of these mutant proteins by immunoblotting with antibody against the RaeB protein. The immunoblot analysis showed that all these RaeB mutant proteins expressed in RA-CH-1 ΔraeB (Figure 4A) were detectable and indistinguishable from the RaeB protein expressed in RA-CH-1 ΔraeB (Figure 4B).
FIGURE 4.
Immunoblotting analysis of RaeB protein and mutant RaeB proteins produced from pLMF03 in RA-CH-1 ΔraeB. (A) Lanes consist of molecular size marker (Bio-Rad) (M), RA-CH-1 ΔraeB pD400A (1), RA-CH-1 ΔraeB pD401A (2), RA-CH-1 ΔraeB pK929E (3), RA-CH-1 ΔraeB pR959A (4), and RA-CH-1 ΔraeB pT966E (5). (B) Lanes consist of molecular size marker (M), RA-CH-1 ΔraeB pLMF03 (1), and RA-CH-1 ΔraeB pLMF03::raeB (2).
Requirement of raeA and raeB for Aminoglycoside and Detergent Resistance
We confirmed that raeB is co-transcribed with raeA (Supplementary Figure S3) as previously reported by Liu et al. (2017). To understand more about raeB, the RA-CH-1 ΔraeA ΔraeB mutant was constructed. Antibiotic susceptibility testing showed that this mutant displayed MICs that were decreased by eightfold for streptomycin, 16-fold for gentamicin, eightfold for Kan, 16-fold for SDS, and fourfold for Triton X-100 (Table 3). However, complementation of the RA-CH-1 ΔraeA ΔraeB mutant with plasmid pLMF03::raeB did not restore resistance to any of these compounds (Table 3). These results indicate that both raeA and raeB are required for aminoglycoside and detergent resistance.
Discussion
RND efflux pumps are widespread among Gram-negative bacteria and play an important role in producing multidrug resistance (Nikaido, 2011). They are located in the cytoplasmic membrane, capturing structurally and functionally dissimilar substrates either from the cytoplasm or from the periplasmic space and then transporting them to the extracellular medium (Nikaido, 2011). As reported, there are total six RND efflux pumps in E. coli (Anes et al., 2015) and 10–12 in P. aeruginosa (Poole, 2008; Fernando and Kumar, 2013). Among these RND efflux pumps, AcrB of E. coli and MexB of P. aeruginosa are the best studied members (Puzari and Chetia, 2017). Here, genome sequence analysis suggested that the raeB gene in R. anatipestifer CH-1 encodes a putative RND efflux pump, and we investigated the biological function of RaeB in R. anatipestifer CH-1, as well as its energy source and functional site.
As is well known, RND efflux pumps are responsible for the extrusion of a very wide range of antimicrobial agents (Nikaido, 2011). The substrate specificity study revealed that inactivation of the raeB gene in R. anatipestifer CH-1 decreased resistance to the aminoglycosides and detergents. According to previous studies, aminoglycoside efflux pumps have been identified in many bacteria; well-characterized examples include AcrD in E. coli (Rosenberg et al., 2000) and MexY in P. aeruginosa (Morita et al., 2012). Comparing the substrate specificity of these three efflux pumps, it is clear that they all export aminoglycosides and detergents. Although there were no MIC changes between the wild-type strain and the RA-CH-1 ΔraeB mutant for quinolones and β-lactams, it was still not sure that whether RaeB exported quinolones and β-lactams or not because other resistance factors could compromise RaeB-mediated resistance to quinolones and β-lactams (Morita et al., 2001). The substrate specificity of E. coli AcrD is determined predominantly by the two large periplasmic loops (Elkins and Nikaido, 2002). In P. aeruginosa MexY, a region corresponding to a proximal binding pocket connected to a periplasm-linked cleft, part of a drug export pathway of E. coli AcrB, was identified and proposed to play a role in aminoglycoside recognition (Lau et al., 2014). Thus, it was speculated that the periplasmic loops between helix 1 and 2 and between helix 7 and 8 and the corresponding aminoglycoside recognition region of MexY in RaeB are related to its substrate specificity.
According to the reports, the expression of efflux pump genes is induced by the addition of the export substrate (Guglierame et al., 2006; Morita et al., 2006; Pletzer and Weingart, 2014). Similarly, it was found that raeB in R. anatipestifer CH-1 was up-regulated in the presence of aminoglycosides and detergents. In Gram-negative bacteria, regulatory proteins can interact with inducers and therefore increase the transcription of efflux pump genes (Ramos et al., 2005). Usually, the regulatory protein genes are located next to the efflux pump genes in the chromosome (Cuthbertson and Nodwell, 2013). In E. coli, acrR is located in a region downstream of acrB and AcrR functions as a repressor of the AcrB efflux pump (Ma et al., 1996). Not surprisingly, the raeR gene, which is located in a region upstream of raeB in R. anatipestifer CH-1, encodes a TetR family transcriptional regulator. This finding is meaningful for future research on the regulatory mechanism underlying R. anatipestifer resistance.
It has been demonstrated that that AcrB of E. coli utilizes PMF as energy for its transport function (Paulsen et al., 1996). In this study, RaeB of R. anatipestifer CH-1 was proved to transport gentamicin by PMF. Additionally, in AcrB of E. coli, five charged residues Asp 407, Asp 408, Lys 940, Arg 971, and Thr 978 produce a proton-relay network, and the protonation and deprotonation of the residues disturbs the network and initiates a series of conformational changes that result in substrate transport (Su et al., 2006). We also determined that these corresponding amino acid residues in RaeB of R. anatipestifer CH-1 (Asp 400, Asp 401, Lys 929, Arg 959, and Thr 966) played an essential role in aminoglycoside and detergent transport. However, no structural information about the RaeB protein is currently available, so further studies determining whether these five amino acid residues are relevant to the proton transport process in R. anatipestifer CH-1 are needed.
In some Gram-negative bacteria, RND efflux pumps have a role not only in antibiotic resistance, but also in bacterial fitness and virulence (Alvarez-Ortega et al., 2013). As reported for Enterobacter cloacae, growth curves and competition experiments in vitro demonstrate that lack of the AcrB efflux pump poses a fitness cost to E. cloacae (Perez et al., 2012). However, deleting the raeB gene did not alter the fitness of R. anatipestifer CH-1. This may reflect a possible adaptation, in that other efflux pump systems may exist in R. anatipestifer CH-1 and were overproduced to compensate for the deletion. Additionally, efflux pumps AcrB in E. cloacae (Perez et al., 2012), AcrB in Klebsiella pneumoniae (Padilla et al., 2010), VexM and VexF in Vibrio cholerae (Bina et al., 2008), MmpL11 in Mycobacterium tuberculosis (Tullius et al., 2011), and AcrB in Salmonella typhimurium (Buckley et al., 2006) contribute to bacterial virulence. Interestingly, in this study, LD50 was measured between the wild-type strain and the mutant strain RA-CH-1 ΔraeB, the result showed that deletion of the raeB gene had no significant impact on the LD50 of R. anatipestifer CH-1. The possibility that raeB is not the main virulence factor in R. anatipestifer CH-1 may account for this result. All these data suggest that RND efflux pumps have multiple different physiological functions in different species of bacteria.
Author Contributions
A-CC and XZ conceived of and designed the project. XZ and M-FL constructed the raeB and raeA-raeB R. anatipestifer CH-1 deletion mutants and assessed their antimicrobial resistance. D-KZ and XZ assessed the mRNA levels of the raeB and B739_0874 genes using RT-PCR. XZ, M-SW, and M-FL constructed the mutant recombinant plasmids, pLMF03:: raeBD400A, pLMF03::raeBD401A, pLMF03::raeBK929E, pLMF03::raeBR959A, and pLMF03::raeBT966E. XZ and M-FL constructed the RA-CH-1 ΔraeB complemented strain and the RA-CH-1 ΔraeA ΔraeB complemented strain. XZ, QY, and YW performed the CCCP inhibition assay and gentamicin accumulation assay. M-SW, K-FS, and X-YC constructed the bacterial growth curves and performed the in vitro competition experiments. X-XZ, R-YJ, and SC performed the LD50 determination for R. anatipestifer CH-1 and RA-CH-1 ΔraeB. XZ, A-CC, and FB drafted and revised the manuscript. All the authors read and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the National Key Research and Development Program of China (2017YFD0500800), the China’s Agricultural Research System (CARS-42-17), and the Special Fund for Key Laboratory of Animal Disease and Human Health of Sichuan Province(2016JPT0004).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02435/full#supplementary-material
FIGURE S1 | Growth curves for R. anatipestifer CH-1 strains. WT, R. anatipestifer CH-1; ΔraeB, RA-CH-1 ΔraeB; ΔraeB pLMF03::raeB, RA-CH-1 ΔraeB pLMF03::raeB complemented strain. Error bars indicate the standard deviation (n = 3).
FIGURE S2 | Competition experiments in vitro for the wild-type strain R. anatipestifer CH-1 and the RA-CH-1 ΔB739_0873 mutant. The mixed cells in the exponential phase were diluted to 10-6, 10-7, 10-8, and 10-9 and then spread onto both TSA and TSA containing 80 μg/ml spectinomycin and incubated overnight at 37°C. The competition index (CI) was defined as the ratio between the number of mutant and wild-type CFUs. Error bars indicate the standard deviation (n = 3).
FIGURE S3 | PCR of the raeA-raeB truncated fragment using different templates. The lanes consist of a molecular size marker (Takara) (M), a positive-control with the R. anatipestifer CH-1 genome DNA as the template (1), a negative-control with H2O as the template (2), a positive-control with total RNA of R. anatipestifer CH-1 before removing the DNA as the template (3), a negative-control with total RNA of R. anatipestifer CH-1 after removing the DNA as the template (4), and cDNA of R. anatipestifer CH-1 as the template (5).
References
- Alvarez-Ortega C., Olivares J., Martinez J. L. (2013). RND multidrug efflux pumps: what are they good for? Front. Microbiol. 4:7 10.3389/fmicb.2013.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anes J., McCusker M. P., Fanning S., Martins M. (2015). The ins and outs of RND efflux pumps in Escherichia coli. Front. Microbiol. 6:587 10.3389/fmicb.2015.00587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina X. R., Provenzano D., Nguyen N., Bina J. E. (2008). Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. Infect. Immun. 76 3595–3605. 10.1128/IAI.01620-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley A. M., Webber M. A., Cooles S., Randall L. P., La Ragione R. M., Woodward M. J., et al. (2006). The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell. Microbiol. 8 847–856. 10.1111/j.1462-5822.2005.00671.x [DOI] [PubMed] [Google Scholar]
- Chang C. F., Lin W. H., Yeh T. M., Chiang T. S., Chang Y. F. (2003). Antimicrobial susceptibility of Riemerella anatipestifer isolated from ducks and the efficacy of ceftiofur treatment. J. Vet. Diagn. Invest. 15 26–29. 10.1177/104063870301500106 [DOI] [PubMed] [Google Scholar]
- CLSI (2015). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-fifth Informational Supplement. CLSI Document M100-S25. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Cuthbertson L., Nodwell J. R. (2013). The TetR family of regulators. Microbiol. Mol. Biol. Rev. 77 440–475. 10.1128/MMBR.00018-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh T., Paulsen I. T., Saier M. H. (1994). A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of gram-negative bacteria. J. Bacteriol. 176 3825–3831. 10.1128/jb.176.13.3825-3831.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins C. A., Nikaido H. (2002). Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominantly by two large periplasmic loops. J. Bacteriol. 184 6490–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando D. M., Kumar A. (2013). Resistance-nodulation-division multidrug efflux pumps in Gram-negative bacteria: role in virulence. Antibiotics 2 163–181. 10.3390/antibiotics2010163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L., Nakae T. (2001). Identification of essential charged residues in transmembrane segments of the multidrug transporter MexB of Pseudomonas aeruginosa. J. Bacteriol. 183 1734–1739. 10.1128/JB.183.5.1734-1739.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglierame P., Pasca M. R., De Rossi E., Buroni S., Arrigo P., Manina G., et al. (2006). Efflux pump genes of the resistance-nodulation-division family in Burkholderia cenocepacia genome. BMC Microbiol. 6:66. 10.1186/1471-2180-6-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess C., Enichlmayr H., Jandreski-Cvetkovic D., Liebhart D., Bilic I., Hess M. (2013). Riemerella anatipestifer outbreaks in commercial goose flocks and identification of isolates by MALDI-TOF mass spectrometry. Avian Pathol. 42 151–156. 10.1080/03079457.2013.775401 [DOI] [PubMed] [Google Scholar]
- Kong Q. K., Yang J., Liu Q., Alamuri P., Roland K. L., Curtiss R. (2011). Effect of deletion of genes involved in lipopolysaccharide core and O-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovar typhimurium. Infect. Immun. 79 4227–4239. 10.1128/IAI.05398-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau C. H., Hughes D., Poole K. (2014). MexY-promoted aminoglycoside resistance in Pseudomonas aeruginosa: involvement of a putative proximal binding pocket in aminoglycoside recognition. mBio 5:e01068–14. 10.1128/mBio.01068-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Z., Plesiat P., Nikaido H. (2015). The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 28 337–418. 10.1128/CMR.00117-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H. B., Cheng X. J., Zhu D. K., Wang M. S., Jia R. Y., Chen S., et al. (2015). TonB energy transduction systems of Riemerella anatipestifer are required for iron and hemin utilization. PLOS ONE 10:e0127506. 10.1371/journal.pone.0127506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M. F., Huang M., Zhu D. K., Wang M. Y., Jia R. Y., Chen S., et al. (2017). Identifying the genes responsible for iron-limited condition in Riemerella anatipestifer CH-1 through RNA-Seq-based analysis. Biomed Res. Int. 2017:8682057. 10.1155/2017/8682057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M. F., Wang M. Y., Zhu D. K., Wang M. S., Jia R. Y., Chen S., et al. (2016). Investigation of TbfA in Riemerella anatipestifer using plasmid-based methods for gene over-expression and knockdown. Sci. Rep. 6:37159. 10.1038/srep37159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H. Y., Liu M. F., Wang L. Y., Zhou W. S., Wang M. S., Cheng A. C., et al. (2015). Identification of ribosomal RNA methyltransferase gene ermF in Riemerella anatipestifer. Avian Pathol. 44 162–188. 10.1080/03079457.2015.1019828 [DOI] [PubMed] [Google Scholar]
- Ma D., Alberti M., Lynch C., Nikaido H., Hearst J. E. (1996). The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19 101–112. [DOI] [PubMed] [Google Scholar]
- Miller V. L., Mekalanos J. J. (1988). A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170 2575–2583. 10.1128/jb.170.6.2575-2583.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y., Kimura N., Mima K., Mizushima T., Tsuchiya T. (2001). Roles of MexXY- and MexAB-multidrug efflux pumps in intrinsic multidrug resistance of Pseudomonas aeruginosa PAO1. J. Gen. Appl. Microbiol. 47 27–32. 10.2323/jgam.47.27 [DOI] [PubMed] [Google Scholar]
- Morita Y., Sobel M. L., Poole K. (2006). Antibiotic inducibility of the MexXY multidrug efflux system of Pseudomonas aeruginosa: involvement of the antibiotic-inducible PA5471 gene product. J. Bacteriol. 188 1847–1855. 10.1128/JB.188.5.1847-1855.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y., Tomida J., Kawamura Y. (2012). MexXY multidrug efflux system of Pseudomonas aeruginosa. Front. Microbiol. 3:408 10.3389/fmicb.2012.00408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. (1994). Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 264 382–388. 10.1126/science.8153625 [DOI] [PubMed] [Google Scholar]
- Nikaido H. (1996). Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178 5853–5859. 10.1128/jb.178.20.5853-5859.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. (2011). Structure and mechanism of RND-type multidrug efflux pumps. Adv. Enzymol. Relat. Areas Mol. Biol. 77 1–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla E., Llobet E., Domenech-Sanchez A., Martinez-Martinez L., Bengoechea J. A., Alberti S. (2010). Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 54 177–183. 10.1128/AAC.00715-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathanasophon P., Phuektes P., Tanticharoenyos T., Narongsak W., Sawada T. (2002). A potential new serotype of Riemerella anatipestifer isolated from ducks in Thailand. Avian Pathol. 31 267–270. 10.1080/03079450220136576 [DOI] [PubMed] [Google Scholar]
- Pathanasophon P., Sawada T., Pramoolsinsap T., Tanticharoenyos T. (1996). Immunogenicity of Riemerella anatipestifer broth culture bacterin and cell-free culture filtrate in ducks. Avian Pathol. 25 705–719. 10.1080/03079459608419176 [DOI] [PubMed] [Google Scholar]
- Paulsen I. T., Brown M. H., Skurray R. A. (1996). Proton-dependent multidrug efflux systems. Microbiol. Rev. 60 575–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen I. T., Park J. H., Choi P. S., Saier M. H. (1997). A family of gram-negative bacterial outer membrane factors that function in the export of proteins, carbohydrates, drugs and heavy metals from gram-negative bacteria. FEMS Microbiol. Lett. 156 1–8. 10.1016/S0378-1097(97)00379-0 [DOI] [PubMed] [Google Scholar]
- Perez A., Poza M., Fernandez A., Fernandez Mdel C., Mallo S., Merino M., et al. (2012). Involvement of the AcrAB-TolC efflux pump in the resistance, fitness, and virulence of Enterobacter cloacae. Antimicrob. Agents Chemother. 56 2084–2090. 10.1128/AAC.05509-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock L. J. (2006). Multidrug-resistance efflux pumps - not just for resistance. Nat. Rev. Microbiol. 4 629–636. 10.1038/nrmicro1464 [DOI] [PubMed] [Google Scholar]
- Pletzer D., Weingart H. (2014). Characterization of AcrD, a resistance-nodulation-cell division-type multidrug efflux pump from the fire blight pathogen Erwinia amylovora. BMC Microbiol. 14:13. 10.1186/1471-2180-14-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. (2008). Bacterial multidrug efflux pumps serve other functions. Microbe 3 179–185. 10.1128/microbe.3.179.1 [DOI] [Google Scholar]
- Puzari M., Chetia P. (2017). RND efflux pump mediated antibiotic resistance in Gram-negative bacteria Escherichia coli and Pseudomonas aeruginosa: a major issue worldwide. World J. Microbiol. Biotechnol. 33 1–8. 10.1007/s11274-016-2190-5 [DOI] [PubMed] [Google Scholar]
- Ramos J. L., Martinez-Bueno M., Molina-Henares A. J., Teran W., Watanabe K., Zhang X., et al. (2005). The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69 326–356. 10.1128/MMBR.69.2.326-356.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland K., Curtiss R., III, Sizemore D. (1999). Construction and evaluation of a delta cya delta crp Salmonella typhimurium strain expressing avian pathogenic Escherichia coli O78 LPS as a vaccine to prevent airsacculitis in chickens. Avian Dis. 43 429–441. [PubMed] [Google Scholar]
- Rosenberg E. Y., Ma D., Nikaido H. (2000). AcrD of Escherichia coli is an aminoglycoside efflux pump. J. Bacteriol. 182 1754–1756. 10.1128/JB.182.6.1754-1756.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryll M., Christensen H., Bisgaard M., Christensen J. P., Hinz K. H., Kohler B. (2001). Studies on the prevalence of Riemerella anatipestifer in the upper respiratory tract of clinically healthy ducklings and characterization of untypable strains. J. Vet. Med. B Infect. Dis. Vet. Public Health 48 537–546. 10.1111/j.1439-0450.2001.00471.x [DOI] [PubMed] [Google Scholar]
- Sandhu T. S., Dean W. F. (1980). Effect of chemotherapeutic agents on Pasteurella anatipestifer infection in White Pekin ducklings. Poult. Sci. 59 1027–1030. [DOI] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3 1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Segers P., Mannheim W., Vancanneyt M., De Brandt K., Hinz K. H., Kersters K., et al. (1993). Riemerella anatipestifer gen. nov., comb. nov., the causative agent of septicemia anserum exsudativa, and its phylogenetic affiliation within the Flavobacterium-Cytophaga rRNA homology group. Int. J. Syst. Bacteriol. 43 768–776. 10.1099/00207713-43-4-768 [DOI] [PubMed] [Google Scholar]
- Su C. C., Li M., Gu R., Takatsuka Y., McDermott G., Nikaido H., et al. (2006). Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway. J. Bacteriol. 188 7290–7296. 10.1128/jb.00684-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N., Liu J. H., Yang F., Lin D. C., Li G. H., Chen Z. L., et al. (2012). Molecular characterization of the antimicrobial resistance of Riemerella anatipestifer isolated from ducks. Vet. Microbiol. 158 376–383. 10.1016/j.vetmic.2012.03.005 [DOI] [PubMed] [Google Scholar]
- Takatsuka Y., Nikaido H. (2006). Threonine-978 in the transmembrane segment of the multidrug efflux pump AcrB of Escherichia coli is crucial for drug transport as a probable component of the proton relay network. J. Bacteriol. 188 7284–7289. 10.1128/JB.00683-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng T. T., Gratwick K. S., Kollman J., Park D., Nies D. H., Goffeau A., et al. (1999). The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1 107–125. [PubMed] [Google Scholar]
- Tullius M. V., Harmston C. A., Owens C. P., Chim N., Morse R. P., McMath L. M., et al. (2011). Discovery and characterization of a unique mycobacterial heme acquisition system. Proc. Natl. Acad. Sci. U.S.A. 108 5051–5056. 10.1073/pnas.1009516108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. Y., Zhang P. Y., Zhu D. K., Wang M. S., Jia R. Y., Chen S., et al. (2017). Identification of the ferric iron utilization gene B739_1208 and its role in the virulence of R. anatipestifer CH-1. Vet. Microbiol. 201 162–169. 10.1016/j.vetmic.2017.01.027 [DOI] [PubMed] [Google Scholar]
- Wang X. J., Liu W. P., Zhu D. K., Yang L. F., Liu M. F., Yin S. J., et al. (2014). Comparative genomics of Riemerella anatipestifer reveals genetic diversity. BMC Genomics 15:479. 10.1186/1471-2164-15-479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong A. S., Yao Q. H., Peng R. H., Duan H., Li X., Fan H. Q., et al. (2006). PCR-based accurate synthesis of long DNA sequences. Nat. Protoc. 1 791–797. 10.1038/nprot.2006.103 [DOI] [PubMed] [Google Scholar]
- Zhong C. Y., Cheng A. C., Wang M. S., Zhu D. K., Luo Q. H., Zhong C. D., et al. (2009). Antibiotic susceptibility of Riemerella anatipestifer field isolates. Avian Dis. 53 601–607. 10.1637/8552-120408-ResNote.1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 | Growth curves for R. anatipestifer CH-1 strains. WT, R. anatipestifer CH-1; ΔraeB, RA-CH-1 ΔraeB; ΔraeB pLMF03::raeB, RA-CH-1 ΔraeB pLMF03::raeB complemented strain. Error bars indicate the standard deviation (n = 3).
FIGURE S2 | Competition experiments in vitro for the wild-type strain R. anatipestifer CH-1 and the RA-CH-1 ΔB739_0873 mutant. The mixed cells in the exponential phase were diluted to 10-6, 10-7, 10-8, and 10-9 and then spread onto both TSA and TSA containing 80 μg/ml spectinomycin and incubated overnight at 37°C. The competition index (CI) was defined as the ratio between the number of mutant and wild-type CFUs. Error bars indicate the standard deviation (n = 3).
FIGURE S3 | PCR of the raeA-raeB truncated fragment using different templates. The lanes consist of a molecular size marker (Takara) (M), a positive-control with the R. anatipestifer CH-1 genome DNA as the template (1), a negative-control with H2O as the template (2), a positive-control with total RNA of R. anatipestifer CH-1 before removing the DNA as the template (3), a negative-control with total RNA of R. anatipestifer CH-1 after removing the DNA as the template (4), and cDNA of R. anatipestifer CH-1 as the template (5).