Abstract
The transcription factor NRF2 is a master regulator of the cellular antioxidant response and is often genetically activated in Non-Small Cell Lung Cancers (NSCLCs) by, for instance, mutations in the interacting protein KEAP1. While direct pharmacological inhibition of NRF2 has proven challenging, its aberrant activation rewires biochemical networks in cancer cells that may create special vulnerabilities. Here, we use chemical proteomics to map druggable proteins that are selectively expressed in KEAP1-mutant NSCLC cells. Principal among these was NR0B1, an atypical orphan nuclear receptor that we show engages in a multimeric protein complex to regulate the transcriptional output of KEAP1-mutant NSCLC cells. We further identify small molecules that covalently target a conserved cysteine within the NR0B1 protein interaction domain and demonstrate that these compounds disrupt NR0B1 complexes and impair the anchorage-independent growth of KEAP1-mutant cancer cells. Our findings designate NR0B1 as a druggable, transcriptional regulator that supports NRF2-dependent lung cancers.
Cancer cells rewire central metabolic networks to provide a steady source of energy and building blocks needed for cell division and rapid growth (Possemato et al., 2011; Vander Heiden et al., 2009). This demand for energy produces toxic metabolic byproducts, including reactive oxygen species (ROS), that, if left unchecked, can promote oxidative stress and impair cancer cell viability (Gorrini et al., 2013; Schafer et al., 2009). Many cancers counter a rise in oxidative stress by activating the NRF2 pathway, a master regulator of the cellular antioxidant response (Gorrini et al., 2013; Leinonen et al., 2014; Sporn and Liby, 2012). Under basal conditions, the bZip transcription factor NRF2 binds to the negative regulator KEAP1, which directs rapid and constitutive ubiquitination and proteasomal degradation of NRF2. Under conditions of oxidative stress, key cysteines in KEAP1 are oxidatively modified to block interaction with NRF2, stabilizing the transcription factor to allow for nuclear translocation and coordination of a gene expression program that induces detoxification and metabolic enzymes to restore redox homeostasis (Gorrini et al., 2013; Mitsuishi et al., 2012). Cancers stimulate NRF2 function in multiple ways, including genetic mutations in NRF2 and KEAP1 that disrupt their interaction and are found in > 20% of non-small cell lung cancers (NSCLCs) (Berger et al., 2016; Cancer Genome Atlas Research, 2014). Despite maturation in our understanding of how NRF2 becomes activated and promotes a transcriptional program that responds to oxidative stress, the underlying molecular mechanisms by which stimulation of this pathway imparts a survival and growth advantage to cancer cells remain poorly defined. Moreover, to date, only a handful of early-stage small molecules have been identified that inhibit NRF2 function (Bollong et al., 2015; Lu et al., 2016), and as a consequence, oncogenic mutations in the KEAP1-NRF2 complex remain unactionable from a therapeutic perspective.
One amino acid residue that is particularly susceptible to oxidative modification is cysteine. Cysteines play important roles in proteins, including as nucleophiles in catalysis (Giles et al., 2003), as metal-binding residues (Pace and Weerapana, 2014), and as sites for post-translational modification (Chung et al., 2013). While low levels of ROS can stimulate cell growth, excessive ROS has damaging effects on many fundamental biochemical processes in cells, including, for instance, metabolic (Gorrini et al., 2013) and protein homeostasis (Lee et al., 2013) pathways. Activation of NRF2 in cancer cells may thus serve to protect biochemical pathways from ROS-induced functional impairments.
Cysteine residues not only constitute sites for redox regulation of protein function, but also for covalent drug development (Johnson et al., 2010; Liu et al., 2013). Both catalytic and non-catalytic cysteines in a wide range of proteins have been targeted with electrophilic small molecules to create covalent inhibitors for use as chemical probes (Ostrem et al., 2013; Johnson et al., 2010; Liu et al., 2013) and therapeutic agents, including ibrutinib, which targets Bruton’s tyrosine kinase BTK for treatment of B-cell cancers (Pan, 2008) and afatinib and AZD9291, which target mutant forms of EGFR for treatment of lung cancer (Finlay et al., 2014). Recently, our lab performed a global analysis of cysteine ligandability in human cancer cell proteomes, revealing a rich content of cysteines amenable to modification by electrophilic small molecules (Backus et al., 2016). Some of these cysteines were found in proteins that have been historically considered undruggable, such as transcription factors and adaptor proteins, suggesting that cysteine-reactive covalent chemistry may substantially expand the portion of the human proteome that can be targeted by small-molecule probes.
Here, we have applied chemical proteomics to quantitatively and globally map cysteine reactivity in KEAP1-mutant and KEAP1-WT human NSCLC cell lines, resulting in the discovery of a core group of ligandable cysteines that are regulated by NRF2 and support key metabolic and transcriptional pathways. Chief among these was a conserved cysteine (C274) in the orphan nuclear receptor NR0B1, which is a strictly NRF2-regulated protein that supports the anchorage-independent growth of KEAP1-mutant NSCLC cells. We further discovered that NR0B1 forms a multimeric transcriptional complex important for regulating a subset of the NRF2 gene expression program in NSCLC cells. Finally, we describe covalent small-molecule ligands that target C274 of NR0B1 with good selectivity in cancer cells and find that these probes disrupt NR0B1 protein complexes and alter the transcriptional output and growth of NRF2-activated cancer cells.
Results
Mapping cysteine reactivity in KEAP1-WT and KEAP1-mutant NSCLC cells
We identified several human NSCLC cell lines that contain inactivating mutations in the gene encoding KEAP1 (H2122, H460, A549 and H1792), as well as additional NSCLC lines that were wild type (WT) for this gene (H1975 and H2009) (Table S1). Small hairpin RNA (shRNA)-mediated knockdown of NRF2 in NSCLC cell lines with KEAP1 mutations, where NRF2 protein levels are stabilized (Figure S1A), substantially impaired cell proliferation in conjunction with lowering NRF2 protein content (Figures 1A, B and S1B–D). In contrast, KEAP1-WT NSCLC lines were only marginally affected by NRF2-knockdown (Figures 1A and S1E). Depletion of NRF2 in the KEAP1-mutant NSCLC line H2122 also led to a marked reduction in glutathione and a concomitant rise in cytosolic H2O2 compared to KEAP1-WT H1975 cells. (Figure S1F, G).
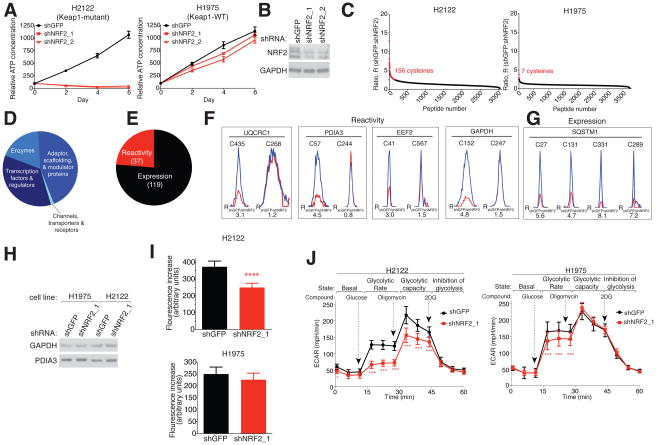
Figure 1. Chemical proteomic map of NRF2-regulated cysteines in NSCLC cells.
A) Proliferation of KEAP1-mutant (H2122) and KEAP1-WT (H1975) cells expressing shRNAs targeting NRF2 (shNRF2) or a control (shGFP), as determined by measuring intracellular ATP concentrations. Data represent mean values ± SD (n = 6/group).
B) Immunoblot of NRF2 in shNRF2- or shGFP-H2122 cells.
C) isoTOP-ABPP (R) ratios for cysteines in shNRF2- or shGFP-H2122 of -H1975 cells. Red data points mark R values ≥ 2.5, which was used as a cutoff for NRF2-dependent changes in cysteine reactivity. Average R values from n = 4–5 biological replicates per group are shown. See also Table S1.
D) Distribution of proteins harboring NRF2-regulated cysteines by functional class.
E) Distribution of NRF2-regulated cysteines reflecting changes in reactivity versus protein expression. See also Table S1.
F) Representative proteins with NRF2-regulated changes in cysteine reactivity. Representative parent mass (MS1) profiles for tryptic peptides with IA-alkyne-reactive cysteines in shNRF2- (red) and shGFP- (blue) H2122 cells. Two cysteines are shown per protein, one with altered and the other with unaltered reactivity between shNRF2- and shGFP-H2122 cells.
G) Representative MS1 profiles for cysteine-containing tryptic peptides in SQSTM1 in shNRF2- (red) and shGFP- (blue) H2122 cells (F).
H) Immunoblot of GAPDH and PDIA3 expression in shNRF2- and shGFP-H1975 and H2122 cells.
I) GAPDH activity in shNRF2- and shGFP-H2122 and -H1975 cells. Data represent mean values ± SD (n =16/group). ****p < 0.0001 for shNRF2 versus shGFP groups.
J) Glycolytic flux is impaired in shNRF2-H2122 cells. ECAR = extracellular acidification rate. Data represent mean values ± SD (n = 20–26/group) from three biological replicates. ***p < 0.001, *p < 0.05 for shNRF2 versus shGFP groups.
We next mapped cysteine reactivities in KEAP1-mutant (H2122) and KEAP1-WT (H1975) NSCLC lines following shRNA-mediated knockdown of NRF2 (shNRF2) using the isoTOP-ABPP platform, which employs a broadly reactive iodoacetamide alkyne (IA-alkyne, 1) probe for labeling, enriching, and quantifying cysteine residues in proteomes (Weerapana et al., 2010) (Figure S1H). We evaluated cells at early (24, 48 h) time points following NRF2 knockdown (Figure S1I) to minimize changes in cysteine reactivity that may have been indirectly caused by proliferation defects. We defined NRF2-regulated cysteines as those showing ≥ 2.5-fold changes in reactivity in shNRF2 cells compared to control shRNA (shGFP) cells (i.e., isoTOP-ABPP Ratio (R) ≥ 2.5 for shGFP/shNRF2) and found that 156 cysteines of > 3000 total quantified cysteines in H2122 cells satisfied this criterion (Figure 1C and Table S1). Approximately three times as many NRF2-regulated cysteines were observed on day 2 versus day 1 post-NRF2 knockdown in H2122 cells (Figure S1J), which may reflect a proportional increase in changes caused by NRF2-regulated gene/protein expression (see below). In contrast, NRF2 depletion had minimal effects on cysteine reactivity in H1975 cells (Figure 1C and Table S1). We also noted that several cysteines with prominent changes in shNRF2-H2122 cells were not detected in H1975 cells, likely reflecting that the proteins harboring these cysteines are themselves regulated by NRF2 (see below). We further evaluated changes in cysteine reactivity in NSCLC cells caused by other anti-proliferative mechanisms – specifically treatment with the general kinase inhibitor staurosporine or the EGFR inhibitor AZD9291 – neither of which caused substantive changes in cysteine reactivity in KEAP1-mutant or KEAP1-WT cells (Figures S1K–M and Table S1). These results indicate that NRF2 disruption produces specific and widespread alterations in cysteine reactivity in KEAP1-mutant NSCLC cells.
NRF2-regulated cysteines were found in proteins from many different functional classes (Figure 1D). In instances where all quantified cysteines for a given protein were altered in shNRF2-H2122 cells, we concluded that the changes reflected an alteration in protein expression. In contrast, if only one of multiple cysteines for a given protein had a substantial reduction in IA-alkyne-reactivity (R ≥ 2.5), with the other quantified cysteines remaining constant (R < 1.5), we concluded that the change was reactivity-based. We supplemented this analysis by determining changes in gene expression in shNRF2- versus shGFP-H2122 cells by RNA sequencing (RNA-seq) (Table S2), which provided an expression estimate for proteins that contained only one quantified IA-alkyne-reactive cysteine. By combining our proteomic and gene expression analysis, we determined that ~80% of all changes in cysteine reactivity reflected alterations in protein abundance following NRF2-knockdown, with the remaining ~20% identified as alterations in reactivity (Figure 1E and Table S1). Proteins harboring cysteines that underwent specific reactivity changes in shNRF2-H2122 cells were found in central pathways that include glycolysis (GAPDH), protein folding (PDIA3), protein translation (EEF2), and mitochondrial respiration (UQCRC1) (Figure 1F). An example of a protein showing expression changes in shNRF2-H2122 cells was the canonical NRF2-regulated protein SQSTM1 (Figure 1G). None of these cysteines were affected by NRF2 knockdown in H1975 cells (Figure S1M).
A recent cysteine proteomics study performed in Kras-mutated mouse pancreatic cancer organoids deleted for NRF2 expression identified several redox-regulated cysteines (Chio et al., 2016). We noted, however, a minimal overall overlap (~3%) in NRF2-regulated cysteines in our results compared to this previous study, which may reflect differences in the mode of NRF2 activation (KEAP1 mutations versus Kras/p53 mutations) tumor of origin (NSCLC versus pancreatic), species (human versus mouse), and/or method of assigning changes in cysteine reactivity (fold-change versus statistical).
The NRF2-regulated cysteines in PDIA3 (C57) and GAPDH (C152) are catalytic residues, designating them as candidate sites for NRF2 control over fundamental biochemical pathways in cancer cells. Another quantified cysteine outside of the GAPDH active site – C247 (Figure 1F) – was unaltered in reactivity by NRF2 knockdown (Figure 1F), and we confirmed that GAPDH protein expression was unaffected in shNRF2 cells by immunoblotting (Figure 1H). C152 in GAPDH is a redox-sensitive residue that is subject to S-sulphenylation (Yang et al., 2014) and S-sulfhydration (Shestov et al., 2014) and can be affected by pharmacologically induced forms of oxidative stress (Yun et al., 2015). To our knowledge, however, the regulation of C152 by NRF2 has not been previously reported. Consistent with the conserved catalytic function performed by C152, shNRF2-H2122 cells, but not shNRF2-H1975 cells, showed a significant decrease in GAPDH activity (Figure 1I). NRF2 knockdown also produced reductions in basal glycolysis and maximal glycolytic rate that were more substantial in magnitude in H2122 cells compared to H1975 cells (Figure 1J). These results thus illuminate a post-translational mechanism by which NRF2 preserves the functional state of a catalytic cysteine to support GAPDH activity and glycolysis in cancer cells.
Mapping cysteine ligandability in KEAP1-WT and KEAP1-mutant NSCLC cells
We next investigated the ligandability of cysteines in NRF2-regulated proteins by performing competitive isoTOP-ABPP of proteomes from three KEAP1-mutant (H2122, H460 and A549) and three KEAP1-WT (H1975, H2009 and H358) NSCLC lines with two electrophilic fragments – 2 and 3 (Figure 2A) – that showed broad cysteine reactivity in previous studies (Backus et al., 2016). We refer to these compounds as ‘scout’ fragments capable of providing a global portrait of covalent small molecule-cysteine interactions in native biological systems.
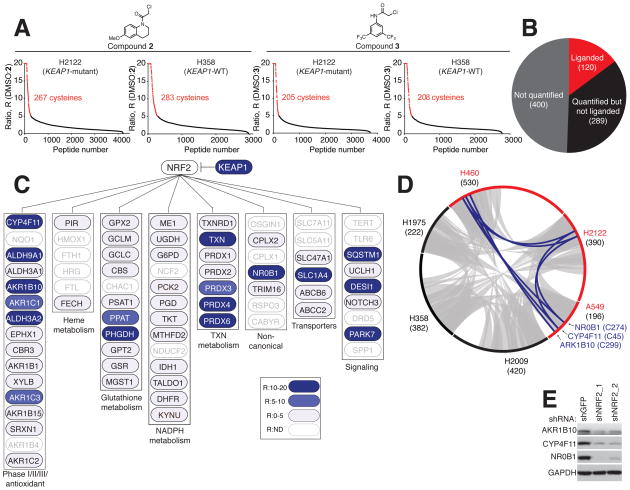
Figure 2. Cysteine ligandability mapping of KEAP1-mutant and KEAP1-WT NSCLC cells.
A) isoTOP-ABPP ratios (R values; DMSO/compound) for cysteines in H2122 cell (KEAP1-mutant) and H358 cell (KEAP1-WT) proteomes treated with DMSO or ‘scout’ fragments 2 or 3 (500 μM, 1 h). Red data points mark R values ≥ 5, which was used as a cutoff for defining liganded cysteines. Average R values from n = 3 biological replicates per group are shown. See also Table S3.
B) Pie chart of NRF2-regulated genes/proteins in NSCLC cell lines denoting the subset that contain liganded cysteines (red).
C) Cysteine ligandability map for representative NRF2 pathways. Blue marks proteins with liganded cysteines in NSCLC cells. ND, not detected.
D) Circos plot showing the overlap in liganded cysteines between KEAP1-mutant (red) and KEAP1-WT (black) NSCLC cells. Gray and blue chords represent liganded cysteines found in both KEAP1-WT and KEAP1-mutant cell lines and selectively in KEAP1-mutant cell lines, respectively. Numbers in parenthesis indicate total liganded cysteines per cell line.
E) Immunoblot of AKR1B10, CYP4F11 and NR0B1in shNRF2- and shGFP- H2122 cells.
From a total of ~9700 cysteines quantified across the proteomes of six NSCLC lines, ~1100 scout fragment-sensitive, or ‘liganded’, cysteines were identified (Figure 2A and S2A–B, and Table S3). We next overlayed this ligandability map with the fraction of proteins showing changes in cysteine reactivity and/or gene expression in shNRF2 cells (Figure S2C and Table S2), resulting in the identification of ~120 NRF2-regulated proteins with liganded cysteines (Figure 2B). These proteins populated diverse metabolic and signaling pathways known to be modulated by NRF2 (Figure 2C), but most were observed in both KEAP1-mutant and KEAP1-WT cells (Figures 2D and S2D), indicating that NRF2 influenced, but did not strictly control the expression of these proteins in NSCLCs. Opposing this general profile was a much more restricted subset of liganded proteins that were exclusive to KEAP1-mutant cells (Figures 2D and S2D). These proteins included NR0B1 (liganded at C274), CYP4F11 (liganded at C45), and AKR1B10 (liganded at C299) (Figures 2D and S2D), which we confirmed by RNA-seq and western blotting were all decreased following knockdown of NRF2 in KEAP1-mutant NSCLC cells (Figures 2E and S2E–F).
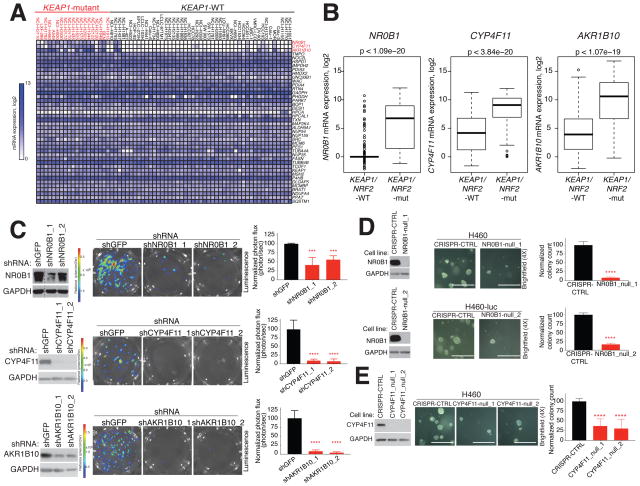
A broader survey of gene expression across > 30 NSCLC lines confirmed the remarkably restricted expression of NR0B1, CYP4F11, and AKR1B10 to KEAP1-mutant cells (Figures 3A and S3A). This expression profile was confirmed by western blotting (Figure S3B) and was also observed in primary human lung adenocarcinoma (LUAD) tumors (Figure 3B). NR0B1 and AKR1B10 have been shown to be important for the proliferation of certain cancers (Garcia-Aragoncillo et al., 2008; Kinsey et al., 2009; Oda et al., 2009; Wang et al., 2009), including KEAP1-mutant NSCLC cells (Oda et al., 2009; Wang et al., 2009). The role of CYP4F11 in cancer cell growth has not, to our knowledge, been examined. Consistent with past work, we found that shRNA knockdown of NR0B1 and AKR1B10 impaired the three-dimensional growth of H460 (Figure 3C) and H2122 cells (Figure S3C). Similar effects were observed for CYP4F11 (Figures 3C and S3C). We also found that CRISPR-mediated knockout of NR0B1 or CYP4F11 in H460 cells strongly reduced colony formation (Figures 3D–E). Efforts to generate CRISPR knockout cells lacking AKR1B10 were unsuccessful.
Figure 3. Characterization of liganded proteins selectively expressed in KEAP1-mutant NSCLC cells.
A) Heat map depicting RNAseq data in KEAP1-WT and KEAP1-mutant NSCLC cell lines for genes encoding NRF2-regulated proteins with liganded cysteines. RNAseq data obtained from (Klijn et al., 2015) (also see Figure S3A).
B) NR0B1, AKR1B10, and CYP4F11 expression in lung adenocarcinoma (LUAD) tumors grouped by NRF2/KEAP1 mutational status. Data obtained from TCGA.
C) Effect of knockdown of ARK1B10, CYP4F11, and NR0B1 on the anchorage-independent growth of H460 cells. Left: Immunoblot of the indicated proteins in H460 cells expressing the luciferase protein and indicated shRNAs. Dashed line represents a lane that was cropped from an immunoblot. Middle and right: Representative brightfield images (middle) and quantification (right) of growth of indicated cell variants in soft agar. Data represent mean values ± SD (n = 4–10/group) from at least two biological replicates. ****p < 0.0001, ***p < 0.001 for shRNAs targeting indicated genes vs shGFP.
D) and E) Effect of CRISPR-generated knockout of NR0B1 (D) or CYP4F11 (E) on the anchorage-independent growth of H460 cells. Left: Immunoblot of NR0B1 or CYP4F11 in null clones (_1 and _2) or H460 cells expressing sgRNA CRISPR-CTRL and/or luciferase (denoted as H460-luc). Middle and right: Representative brightfield images (middle) and quantification of growth of indicated cell variants. Data represent mean values ± SD (n = 6–12/group) from at least two biological replicates. ****p < 0.0001, for NR0B1- or CYP4F11-null H460 clones vs sgRNA CRISPR-CTRL H460 cells. Scale bar equals 0.75 mm.
In summary, by integrating chemical proteomic maps of scout fragment reactivity with differential expression profiles, we identified ligandable proteins selectively expressed by and important for the growth of KEAP1-mutant NSCLCs.
NR0B1 nucleates a transcriptional complex that supports the NRF2 gene network
Considering the general druggability of enzymes and their key role in managing cellular redox homeostasis, it is perhaps not surprising that enzymes like AKR1B10 and CYP4F11 comprised a major portion of the NRF2-regulated ligandability map. We also noted that most of these enzymes, as well as other NRF2-regulated genes and proteins, were expressed broadly across many human tissues (Figure S3C). NR0B1, however, stood out as a striking contrast, being an atypical orphan nuclear receptor with very limited normal tissue expression (Figure S3C). Structural studies have shown that NR0B1 possesses a very shallow pocket in place of the typical ligand-binding domain found in other nuclear receptors, indicating that NR0B1 may function as a “ligandless” adaptor or co-regulatory protein (Sablin et al., 2008). Consistent with this premise, NR0B1 acts as a transcriptional repressor of the nuclear receptors SF1 and LRH1 and supports development of Lydig and Serotoli cells in mice (Iyer and McCabe, 2004). Mutations in the NR0B1 gene lead to adrenal hypoplasia congenita (AHC) in human males (Iyer and McCabe, 2004). The biochemical and cellular functions of NR0B1 in human cancer and in particular, KEAP1-mutant cancer cells, however, remain poorly understood.
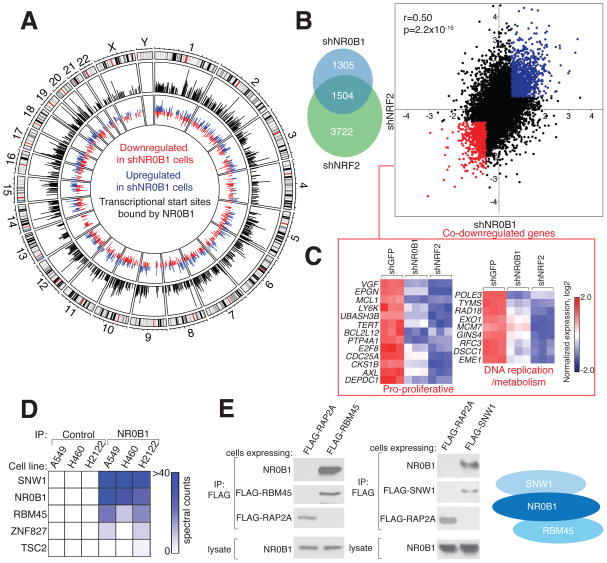
We first assessed whether NR0B1 acts as a transcriptional regulator in KEAP1-mutant NSCLC cells. RNAseq analysis identified more than >2500 genes that were substantially altered (1.5-fold) in expression in shNR0B1 H460 cells (Table S2), and ~30% of these genes were located near transcriptional start sites (TSSs) bound by NR0B1 as determined by chromatin immunoprecipitation sequencing (ChIP-seq) (Figure 4A and Table S2). These results suggest that many of the NR0B1-regulated genes in NSCLC cells are in open chromatin under direct transcriptional control of NR0B1. Unbiased functional enrichment analysis (Huang da et al., 2009) revealed an overrepresentation of cell cycle-related and pro-proliferation functions in genes reduced in expression in shNR0B1 NSCLC cells (Figure S4A and Table S2) that included, for instance, strong E2F and Myc gene signatures (Figure S4B). RNAseq analyses further revealed a substantial correlation in global gene expression changes induced by knockdown of NR0B1 or NRF2 in NSCLC cells (Figure 4B and Table S2), with > 50% of the genes with substantially altered (> 1.5 fold) expression in shNR0B1 cells showed a similar magnitude directional change in shNRF2 cells (Figure 4B). Among the most co-downregulated genes were those involved in proliferation and DNA metabolism/replication (Figure 4C), consistent with the enrichment of these terms in the NR0B1-regulated gene set (Figure S4B).
Figure 4. NR0B1 nucleates a transcriptional complex that supports the NRF2 gene-expression program.
A) Intersection between NR0B1-regulated genes and transcriptional start sites (TSSs) bound by NR0B1. Outer circle: Chromosomes with cytogenetic bands. Middle circle: Whole genome plot of mapped NR0B1 reads (black) determined by ChIP-Seq corresponding to the transcriptional start sites (TSSs) of genes differentially expressed (up- (blue) or down- (red) regulated > 1.5-fold) in shNR0B1-H460 cells compared to shGFP-H460 cells (inner circle). See also Table S2.
B) Overlap (left) and correlation (right) between genes up- (red) or down- (blue) regulated (> 1.5-fold) in shNR0B1- and shNRF2-H460 cells compared to shGFP-H460 control cells. r and p values were determined by Pearson correlation analysis.
C) Heat map depicting RNAseq data for the indicated genes in shNR0B1-, shNRF2-, or shGFP-H460 cells. Expression was normalized by row. See also Table S2.
D) Heat map representing NR0B1-interacting proteins in NSCLC cells.
E) Endogenous NR0B1 co-immunoprecipitates with FLAG-RBM45 and FLAG-SNW1, but not control protein FLAG-RAP2A, in H460 cells, as determined by immunoblotting (left); right: schematic of NR0B1 protein interactions.
Considering the established function of NR0B1 as a coregulatory protein that participates in nuclear receptor complexes (Iyer and McCabe, 2004; Sablin et al., 2008), we hypothesized that NR0B1 may interact with other proteins to regulate transcriptional pathways in KEAP1-mutant cancer cells. We expressed a FLAG epitope-tagged form of NR0B1 in KEAP1-mutant NSCLC cells, immunoprecipitated NR0B1 from these cells, and identified associated proteins by mass spectrometry (MS)-based proteomics. Eleven proteins were substantially co-enriched (> 20-fold) with NR0B1 compared to a control protein METAP2 (Figure S4C and Table S1). A subset of these proteins, including RBM45 and SNW1, were also confirmed by MS-based proteomics to interact with endogenous NR0B1 (Figure 4D). Stably expressed FLAG-SNW1 and FLAG-RBM45, but not a control protein (FLAG-RAP2A), interacted with NR0B1 in multiple NSCLC cells (Figures 4E and S4D), and both SNW1 and RBM45, like NR0B1, were localized to the nucleus of NSCLC cells (Figure S4F). SNW1 did not directly interact with RBM45 in the absence of NR0B1 (Figure S4E), indicating that NR0B1 bridges these two proteins to nucleate a multimeric protein complex (Figure 4E). While very little is known about RBM45, SNW1 has been implicated as a transcriptional activator (Zhang et al., 2003) and found to interact with multiple nuclear receptors, including NR0B1, in large-scale yeast two-hybrid assays (Albers et al., 2005). Consistent with this role and with a coordinated function for SNW1 and NR0B1 in KEAP1-mutant cancer cells, RNAi-mediated knockdown of SNW1 produced a similar set of gene expression changes to those observed in shNR0B1 cells (Figure S4G and Table S2). SNW1 knockdown also blocked the anchorage independent growth of KEAP1-mutant NSCLC cells (Figure S4H).
Covalent small molecules that disrupt NR0B1 protein interactions
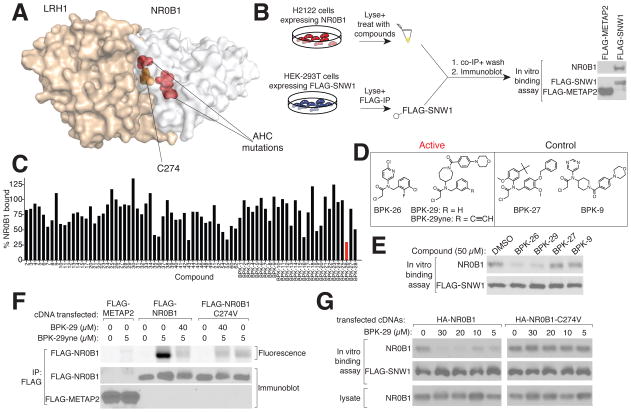
The liganded cysteine in NR0B1 – C274 – is located within a conserved “repression helix” that commonly possesses a LXXLL sequence in other nuclear receptors, but, in NR0B1, has been replaced by a PCFXXLP sequence, where the “C” is C274 (Sablin et al., 2008). Missense mutations within this general region of NR0B1 have been found to cause AHC (Suntharalingham et al., 2015) (Figure 5A), pointing to an important functional role for the repression helix. The hydrophobic residues in the repression helix of NR0B1, including C274, are solvent-exposed and appear to contribute to protein-protein interactions (Sablin et al., 2008) (Figure 5A), suggesting that ligands targeting C274 might disrupt NR0B1 protein complexes.
Figure 5. A covalent ligand targeting C274 disrupts NR0B1 protein complexes.
A) Co-crystal structure of mouse NR0B1 (white) and LRH1 (burnt orange) from (Sablin et al., 2008) highlighting the location of C274 (orange) at the protein interaction interface that is also flanked by AHC mutations: R267, V269 and L278 (red).
B) Left: Schematic for an NR0B1-SNW1 in vitro-binding assay. Right: immunoblot showing that NR0B1 interacts with SNW1, but not a control (METAP2) protein.
C) Small molecule screen of electrophilic compounds (50 μM) for disruption of binding of FLAG-SNW1 to NR0B1 as shown in (B). Percentage of NR0B1 bound to SNW1 was normalized to vehicle (DMSO). A hit compound BPK-26 is marked in red.
D) Structures of NR0B1 ligands (BPK-26 and BPK-29), clickable probe (BPK-29yne), and inactive control compounds (BPK-9 and BPK-27).
E) BPK-26 and BPK-29, but not BPK-9 and BPK-27, disrupt the in vitro interaction of FLAG-SWN1 with NR0B1.
F) BPK-29yne labels WT-NR0B1, but not an NR0B1-C274V mutant. HEK293T cells expressing the indicated proteins were treated with BPK-29 or vehicle (3 h) prior to treatment with BPK-29yne (30 min). Immunoprecipiated proteins were analyzed by in-gel fluorescence-scanning and immunoblotting.
G) BPK-29 disrupts protein interactions for NR0B1-WT, but not a NR0B1-C274V mutant. HEK293T cells expressing HA-NR0B1-WT or HA-NR0B1-C274V proteins were treated with DMSO or BPK-29, after which lysates were generated and evaluated for binding to FLAG-SNW1, as shown in (B).
We next aimed to develop a more advanced chemical probe targeting C274 of NR0B1. Using an in vitro binding assay (Figure 5B), we screened an ~80-member library of cysteine-reactive electrophilic compounds (Table S3) (Backus et al., 2016) at 50 μM for blockade of interactions between endogenous NR0B1 and recombinant FLAG-SNW1 in cell lysates (Figure 5C). Among the hits (> 50% blockade) were a series of N-disubstituted chloroacetamides (CAs), including BPK-26 (Figure 5D, E), that were selected for further investigation. The initial structure-activity relationship indicated more tolerance to substitution of the N-aryl compared to N-benzyl group of BPK-26, including a hit BPK-28 where the N-aryl group was replaced with an azepane group with only modest reductions in potency (Figure S5A). Modifications to BPK-28, including installation of a morpholine group, generated compound BPK-29 (Figure 5D) that recovered potency (Figures 5E and S5B). Both BPK-26 and BPK-29 inhibited the NR0B1-SNW1 interaction with IC50 values between 10–20 μM in vitro (Figure S5C). The initial screen also identified structurally related, inactive control compounds – BPK-9 and BPK-27 (Figure 5C, D) – that did not inhibit the NR0B1-SNW1 interaction across a tested concentration range of 1–50 μM (Figures 5E and S5C). Finally, we confirmed by LC-MS/MS analysis that BPK-26 and BPK-29 covalently modified C274 of NR0B1 (Figures S5D, E and Table S3).
We also synthesized an alkyne analogue of BKP-29 (BPK-29yne) and found that this probe labeled WT-NR0B1, but not a C274V mutant (Figure 5G), and this labeling was blocked by pre-treatment with BPK-29 in a concentration dependent manner (Figures 5G and SF5). The C274V-NR0B1 mutant maintained binding to SNW1, but this protein-protein interaction was not sensitive to BPK-26 or BPK-29, supporting that these ligands disrupt the NR0B1 protein-protein interactions by covalently modifying C274 (Figures 5G and S5G).
Cellular studies with NR0B1 ligands
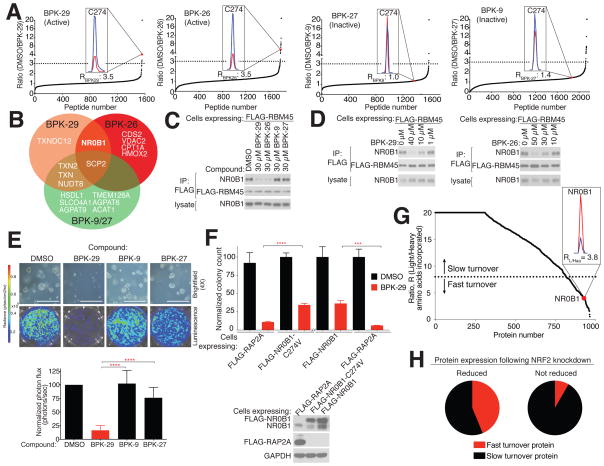
IsoTOP-ABPP confirmed the cellular engagement of C274 of NR0B1 by BPK-26 and BPK-29 in NSCLC cells (Figure 6A and Tables 1 and S3), with both compounds achieving ~70% target occupancy when tested at 40 μM for 3 h (Figures 6A and S6A). In contrast, the inactive control compounds BPK-9 and BPK-27 did not engage C274 (Figure 6A and Tables 1 and S3). Nine additional cysteines among the > 1500 total cysteines quantified by isoTOP-ABPP cross-reacted with BPK-26 and/or BPK-29 in NSCLC cell proteomes (Figures 6A, B and Tables 1 and S3), and most of these cysteines also reacted with the control compounds (Figures 6B and Tables 1 and S3). NR0B1 was the only target shared between BPK-26 and BPK-29 that did not cross-react with the control compounds (Figures 6B and Table 1). C274 was also the only cysteine in NR0B1 engaged by BPK-26 and BPK-29 among several other quantified cysteines (Figure S6B). BPK-29 displayed superior potency compared to BPK-26, achieving > 50% engagement of C274 at 5 μM in NSCLC cells (Figure S6A). We finally employed the BPK-29yne probe to further characterize the protein targets of BPK-29 in NSCLC cells following the chemical proteomic workflow outlined in Figure S6C, which verified most of the targets mapped by isoTOP-ABPP and revealed another seven proteins engaged by BPK-29, all of which also cross-reacted with the control compounds (Tables 1 and S3). Taken together, these data indicate that BPK-26 and BPK-29 substantially engage NR0B1 with good overall proteomic selectivity in KEAP1-mutant NSCLCs.
Figure 6. Characterization of NR0B1 ligands in KEAP1-mutant NSCLC cells.
A) isoTOP-ABPP of H460 cells treated with NR0B1 ligands and control compounds (40 μM, 3 h). Dashed lines designate R values ≥ 3 (DMSO/compound), which was used as a cutoff to define cysteines liganded by the indicated compounds. Insets show MS1 profiles for C274 in NR0B1 for DMSO (blue) versus compound (red) treatment. Data are from individual experiments representative of at least three biological replicates.
B) Venn diagram comparing the proteome-wide selectivity of NR0B1 ligands BPK-29 and BPK-26 and control compounds BPK-9 and BPK-27 in H460 cells as determined in (A). (See also Tables 1 and S3).
C) BPK-29 and BPK-26 block the RBM45-NR0B1 interaction in H460 cells. H460 cells stably expressing FLAG-RBM45 were incubated with indicated compounds for 3 h, whereupon FLAG immunoprecipitates were performed and analyzed by immunoblotting.
D) Concentration-dependent blockade of NR0B1 binding to FLAG-RBM45 by BPK-29 (left) and BPK-26 (right) in H460 cells. Experiments performed as described in (C).
E) Effect of BPK-29 and control compounds (5 μM) on anchorage-independent growth of KEAP1-mutant H460 cells. Representative brightfield images (top) and quantification (bottom) of cell growth. Data represent mean values ± SD (n = 4–8 per group) and are representative of two to four biological replicates; ****p < 0.0001 for BPK-29 vs BPK-9 or BPK-27. Scale bar equals 0.75 mm.
F) Ectopic expression of NR0B1-WT or NR0B1-C274V, but not control protein (RAP2A) partially and significantly rescues the impairment in anchorage-independent growth produced by BPK-29 (5.7 μM) in H460 cells expressing luciferase. Proliferation was determined by colony counting and data represent mean values ± SD (n = 3/group) and representative of at least two biological replicates; ****p < 0.0001 for FLAG-Rap2a, BPK-29 vs FLAG-NR0B1-C274, BPK-29; ***p < 0.001 for FLAG-RAP2A, BPK-29 vs FLAG-NR0B1-WT, BPK-29. Bottom: Immunoblot showing expressed NR0B1 (or RAP2A control) proteins.
G) SILAC ratio plots for light amino acid-labeled cells (pulse phase) switched into media containing heavy amino acids for 3 h (chase phase) followed by proteomic analysis. Dashed line designates R values (light/heavy) of < 8, which was used as a cutoff for fast-turnover proteins. Inset shows MS1 peak ratio for NR0B1, which is among the top 5% of fast-turnover proteins. See also Table S1.
H) Proteins regulated by NRF2 in NSCLC cells are enriched in fast-turnover proteins. Charts comparing fraction of NRF2-regulated genes (as determined by RNAseq) for which the corresponding proteins are designated as fast or slow turnover (as determined in G) further divided into groups showing reduced expression (left) or not (right) on day 1 following NRF2 knockdown (as determined by isoTOP-ABPP). See also Table S1.
Table 1.
Proteome-wide selectivity of NR0B1 ligand BPK-29.
| UniProt ID | Protein | BPK-29-competed isoTOP-ABPP analysis# | BPK-29-competed BPK-29yne analysis* | Competed residues (peptide) | Competed by control ligands BPK-9/27*,# |
|---|---|---|---|---|---|
| P51843 | NR0B1 | Yes | Yes | C274 | No |
| Q8WV74 | NUDT8 | Yes | Yes | C207 | Yes |
| P22307 | SCP2 | Yes | Yes | C94 | Yes |
| P10599 | TXN | Yes | Yes | C35 | Yes |
| Q16881 | TXNRD1^ | Yes | Yes | U648 | Yes |
| O95881 | TXNDC12 | Yes | Yes | C66 | No |
| Q99757 | TXN2 | Yes | – – | C90 | Yes |
| P00352 | ALDH1A1 | – – | Yes | – – | Yes |
| Q9BRX8 | FAM213A | – – | Yes | – – | Yes |
| Q9BVL4 | SELO^ | – – | Yes | – – | Yes |
| P78417 | GSTO1 | – – | Yes | – – | Yes |
| Q5TFE4 | NT5DC1 | – – | Yes | – – | Yes |
| Q9H7Z7 | PTGES2 | – – | Yes | – – | Yes |
Contains conserved functional (seleno)cysteine residue
Competed defined as showing R value ≥ 2.5 at 20 μM of test compound
Competed defined as showing R value ≥ 3.0 at 40 μM of test compound
– –BPK-29-competed protein or peptide not detected
We asked next asked whether BPK-26 and BPK-29 inhibited NR0B1 protein interactions in cells using two complementary systems. First, we generated KEAP1-null HEK293T cells and found that these cells show elevated expression of NR0B1 (Figure S6D). KEAP1-null HEK293T cells, or KEAP1-mutant NSCLC cells, were then engineered to stably express FLAG-tagged RMB45 or SNW1 and treated with BPK-26 and BPK-29 or inactive control compounds. In both cell systems, BPK-26 and BPK-29, but not control compounds, blocked the interactions of FLAG-tagged RMB45 or SNW1 with endogenous NR0B1 (Figures 6C and S6E–F). BPK-29 blocked NR0B1-protein interactions with better potency than BPK-26 (Figures 6D and S6G).
Based on its superior in situ activity (Figures 6D and S6A, G) and selectivity (Figure 6A, B), BPK-29 was chosen for additional biological studies. Treatment of KEAP1-mutant NSCLC cells with BPK-29 (5 μM) blocked colony formation in soft agar (Figures 6E and S7A). Control compounds BPK-9 and BPK-27 had much less of an effect (Figures 6E and S7A). Exogenous expression of WT or a C274V mutant of NR0B1 significantly, albeit partially rescued the growth inhibition caused by BPK-29 (Figure 6F). In contrast, BPK-29 (5 μM), or NR0B1 knockdown, minimally affected the anchorage-independent growth of KEAP1-WT NSCLC cells (Figure S7B–C).
BPK-29 (30 μM, 12 h) also produced some of the gene expression changes caused by shRNA-mediated disruption of NR0B1 or NRF2 in KEAP1-mutant NSCLC cells (Figure S7D, Table S2), including reductions in CRY1, DEPDC1, and CPLX2 (Figures S7E–F), which were not observed in KEAP1-WT NSCLC cells treated with BPK-29 (Figure S7E). We further confirmed that BPK-29-treated cells also showed a substantial reduction in CRY1 protein content (Figure S7G). These gene and protein expression changes were not observed in KEAP1-mutant NSCLC cells treated with control compound BPK-9 (Figures S7D–G).
In the course of studying the cellular activity of BPK-29, we were perplexed by the relatively shallow concentration-dependent change in engagement of C274 of NR0B1 compared to other targets of the compound (Figure S6A). Covalent ligands like BPK-29 engage proteins in a time-dependent manner, which led us to speculate that differences in protein turnover rate in cells could affect the maximal absolute engagement of NR0B1 by BPK-29. We accordingly performed SILAC pulse-chase chemical proteomics experiments in Keap1-mutant NSCLC cells, which revealed that NR0B1 was among a select subset of NRF2-regulated proteins that exhibit rapid turnover in NSCLC cells (Figure 6G and Table S1). These fast-turnover proteins generally corresponded to those that displayed early time point changes in protein abundance in our original isoTOP-ABPP analysis of shNRF2 cells (Figure 6H and Table S1). Similar results were obtained in KEAP1-mutant NSCLC cells treated with cycloheximide, which provided a half-life estimate for NR0B1 of ~4.8 h (Figure S7H). These findings demonstrate that NR0B1 is a short half-life protein in KEAP1-mutant NSCLC cells, possibly explaining its rapid decrease following NRF2 disruption (Table S1) and substantive, but incomplete engagement by BPK-29 in cells (Figures 6A and S6A). More generally, our chemical proteomic data highlight how NRF2 can exert strict temporal and coordinated control over proteins in cancer cells by integrated transcriptional and post-translational mechanisms.
Discussion
Oncogenic pathways alter the biochemical state of cancer cells to promote unregulated growth and survival. The extent to which genetically defined cancers depend on such biochemical alterations may create druggable vulnerabilities, an outcome that is especially relevant for oncogenes like NRF2 that have proven difficult to directly target with established therapeutic modalities. Here we have used chemical proteomics to discover proteins harboring cysteine residues that are both regulated by NRF2 and targeted by electrophilic small molecules. By using a pair of scout fragments found previously to show broad reactivity with cysteines across the human proteome (Backus et al., 2016), we simplified the search for ligandable cysteines in NSCLC lines, and we anticipate that this approach should facilitate the discovery of druggable site in other biological systems. Some of the liganded cysteines discovered in KEAP1-mutant cells, such as the catalytic nucleophiles of GADPH and PDIA3, were regulated post-translationally by NRF2, indicating that this transcription factor establishes a redox environment conducive to proper cysteine-dependent enzyme function in NSCLC cells. NRF2 disruption may also produce post-translational effects that indirectly regulate cysteine reactivity, such as changes in the structure of protein complexes or non-redox-related protein modifications (e.g., phosphorylation). GAPDH and PDIA3, as well as most of the other NRF2-regulated proteins discovered in our chemical proteomic analysis, however, are broadly expressed in human tissues, leading us to question their suitability as co-dependency targets specific to NRF2-dependent NSCLCs. The atypical orphan nuclear receptor NR0B1 stood out, however, as a notable exception, as it displayed a much more restricted expression profile that was tightly linked to the KEAP1/NRF2 mutational state of NSCLCs. NR0B1 was also among the most rapidly downregulated proteins in shNRF2 NSCLC cells, and we found this portion of the NRF2-regulated proteome was enriched in fast-turnover proteins. We speculate that coupling NRF2 transcription to rapid protein turnover provides cancer cells with a mechanism to tightly coordinate the functions of proteins like NR0B1.
Our findings furnish a model where NR0B1 promotes the expression of a specific subset of NRF2-regulated genes that are enriched in pro-proliferative functions, including genes that regulate the cell cycle and DNA synthesis. NR0B1 may serve a similar function in cancers that depend on other oncogenic transcription factors, such EWS/FLI-1-dependent Ewing’s sarcoma, where NR0B1 knockdown also suppresses cell growth and cell cycle-related gene expression (Garcia-Aragoncillo et al., 2008).
Considering that NR0B1 lacks a clear ligand-binding pocket, this protein may act primarily as a coregulatory protein to affect transcription (Iyer and McCabe, 2004). We identified multiple proteins (RBM45, SNW1) that interact with NR0B1 in KEAP1-mutant NSCLC cells and found that covalent ligands reacting with C274 of NR0B1 block these protein-protein interactions. Mutation of C274 to valine fully protected NR0B1 protein complexes from covalent ligand action, but only partially rescued the growth blockade caused by these compounds. Potential reasons for this difference include a non-equivalent function for the C274V-NR0B1 mutant in NSCLC cells and/or that the covalent ligands impair cancer cell growth by a combination of NR0B1-dependent and -independent mechanisms. Further optimization of the potency and selectivity of NR0B1 ligands should enable more advanced biological studies, where we expect that the rapid resynthesis rate of the protein will present an additional challenge for covalent ligands targeting C274. NR0B1 has been shown to display a short half-life in other cell systems (Ehrlund et al., 2009), suggesting that this property is an intrinsic feature of the protein. Nonetheless, we are emboldened by the successful development of cysteine-reactive ligands that target other proteins with short half-lives, where optimization of the pharmacokinetic properties of the covalent drugs proved critical (Thorarensen et al., 2017).
In summary, our studies have identified key transcriptional and post-translational mechanisms by which NRF2 regulates the proteome of human NSCLC cells. Recent studies have also revealed that NRF2 can control the translation machinery of mouse pancreatic tumor cells to promote the expression of key drivers of proliferation (Chio et al., 2016). While it is possible that the modulation of general cell processes, such as translation, glycolysis, and protein folding, by NRF2 may provide an expanded therapeutic window for agents that disrupt these core functions in NRF2-activated cancers, such approaches are ultimately predisposed to affect many cells in the body. Proteins like NR0B1, which show a much more restricted expression profile aligned with NRF2 activation, present a potentially attractive alternative. Further work is needed, however, to determine whether ligands that disrupt NR0B1 function can impede NRF2-dependent cancer growth in vivo, either independently or in combination with other targeted therapies. Beyond oncology, NR0B1 plays an important role in testicular and adrenal gland development, where it appears to engage in a distinct set of protein-protein interactions that involve LRH1 and SF1 (Iyer and McCabe, 2004; Sablin et al., 2008; Suntharalingham et al., 2015). These interactions, as well as mutations that cause congenital adrenal hypoplasia (Suntharalingham et al., 2015) involve the repression helix region of NR0B1 containing C274, indicating that the ligands reported herein may serve as useful chemical probes to understand NR0B1 function in mammalian development and other cell biological processes. More generally, our results underscore the value of emerging chemical proteomic methods for mapping ligandable sites in human proteins (Backus et al., 2016; Niphakis et al., 2015), in this case, unveiling a cysteine in the protein-protein interaction domain of a transcriptional regulator that is amenable to covalent inactivation by small-molecule electrophiles.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resoures and reagents should be directed to the Lead Contact, Benjamin Cravatt (cravatt@scripps.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
All cell lines were obtained from ATCC. All cells were maintained at 37 °C with 5% CO2. HEK-293T cells were grown in DMEM (Corning) supplemented with 10% fetal bovine serum (FBS, Omega Scientific), penicillin (100 U/ml), streptomycin (100 μg/ml) and L-glutamine (2 mM). H2122, H460, A549, H1975, H358, H1792, and H2009 cells were grown in RPMI-1640 (Invitrogen) supplemented as above. H2009 cells were additionally supplemented with Insulin-Transferrin-Selenium (Invitrogen). For SILAC experiments, each cell line was passaged at least six times in SILAC RPMI (Thermo), which lack L-lysine and L-arginine, and supplemented with 10% (v/v) dialyzed FBS (Gemini), penicillin, streptomycin, L-glutamine (as above), and either [13C6, 15N2]-L-lysine and [13C6, 15N4]-L-arginine (100 mg/mL each) or L-lysine and L-arginine (100 mg/mL each). Heavy and light cells were maintained in parallel and cell aliquots were frozen after six passages in SILAC media and stored in liquid N2 until needed. Whenever thawed, cells were passaged at least three times before being used in experiments.
METHODS DETAILS
Compound synthesis
Compounds 2–15, 17, 20–46, 48, 49, 51, 52, 54, 55, 60, 62 were obtained from the library of reactive electrophiles previously synthesized in (Backus et al., 2016). Compounds BPK-1, BPK-2, and SI-7 were synthesized following literature procedures (Backus et al., 2016), detailed in Synthesis of compounds section (Methods S1). Compounds BPK-1 – BPK-26, BPK-28 – BPK-34 were synthesized by WuXi AppTec Co., Ltd. The corresponding procedures for BPK-1 – BPK-34 can be found in Methods S1. All small molecule starting materials used in these syntheses were purchased from Aldrich Chemical Co., Alfa Aesar, TCI, J&K Scientific Ltd, Bide Pharmatech Ltd, Adamas-beta Ltd, Sinopharm Group Co Ltd, Shanghai Shaoyuan Co Ltd, Shanghai Arbor Chemical Co Ltd, PharmaBlock Sciences (Nanjing), Inc, Beijing Ouhechem Ltd, or BePharm Ltd. and were used without further purification.
cDNA cloning and mutagenesis
cDNAs encoding for NR0B1, SNW1, RBM45 were amplified from a cDNA pool generated from A549 cells and were subcloned into the FLAG-pRK5 or HA-pRK5 expression vectors. These cDNAs were also subcloned into the lentiviral expression vector FLAG-pLJM1 (Bar-Peled et al., 2013). The firefly luciferase gene was cloned into the lentiviral expression vector pLenti-pgk BLAST as described before (Goodwin et al., 2014). Cysteine mutants were generated using QuikChange XLII site-directed mutagenesis (Agilent), using primers containing the desired mutations. All constructs were verified by DNA sequencing.
Mammalian lentiviral shRNAs expression
Lentiviral shRNAs targeting the messenger RNA for human NR0B1, SWN1, and AKR1B10 were cloned into pLKO.1 vector at the Age1, EcoR1 sites as described in (Sarbassov et al., 2004). The sequences for these shRNAs is described below (sequences are 5′ to 3′).
shSNW1_1-fwd: CCGGGCCCGATGAAGAAGCTATTAACTCGAGTTAATAGCTTCTTCATCGGGCTTTTTG
shSNW1_1-rev: AATTCAAAAAGCCCGATGAAGAAGCTATTAACTCGAGTTAATAGCTTCTTCATCGGGC
shSNW1_2-fwd: CCGGGGTGGAGAAGATGAAATTTATCTCGAGATAAATTTCATCTTCTCCACCTTTTTG
shSNW1_2-rev: AATTCAAAAAGGTGGAGAAGATGAAATTTATCTCGAGATAAATTTCATCTTCTCCACC
shNR0B1_1-fwd: CCGGTCAAGTGCTTTCTTTCCAAATCTCGAGATTTGGAAAGAAAGCACTTGATTTTTG
shNR0B1_1-rev: AATTCAAAAATCAAGTGCTTTCTTTCCAAATCTCGAGATTTGGAAAGAAAGCACTTGA
shNR0B1_2-fwd: CCGGTCAGCAAATACTCAGTGAACACTCGAGTGTTCACTGAGTATTTGCTGATTTTTG
shNR0B1_2-rev: AATTCAAAAATCAGCAAATACTCAGTGAACACTCGAGTGTTCACTGAGTATTTGCTGA
shNR0B1_3-fwd: CCGGACAGATTCATCGAACTTAATACTCGAGTATTAAGTTCGATGAATCTGTTTTTTG
shNR0B1_3-rev: AATTCAAAAAACAGATTCATCGAACTTAATACTCGAGTATTAAGTTCGATGAATCTGT
shAKR1B10_2-fwd: CCGGCTATCTGGACGTCTATCTTATCTCGAGATAAGATAGACGTCCAGATAGTTTTTG
shAKR1B10_2-rev: AATTCAAAAACTATCTGGACGTCTATCTTATCTCGAGATAAGATAGACGTCCAGATAG
shRNA-encoding plasmids were co-transfected with ΔVPR envelope and CMV VSV-G packaging plasmids into 2.5 × 106 HEK-293T cells using the Xtremegene 9 transfection reagent (Sigma-Aldrich). Virus-containing supernatants were collected forty-eight hours after transfection and used to infect target cells in the presence of 10 μg/ml polybrene (Santa Cruz). Twenty-four hours post-infection, fresh media was added to the target cells which were allowed to recover for an additional twenty-four hours. Puromycin was then added to cells, which were analyzed immediately or on the 2nd or 3rd day after selection was added.
Generation of CRISPR-mediated knockout HEK-293T cell lines
sgRNAs targeting KEAP1 or NRF2 (described below) were designed, amplified, and cloned into transient pSpCas9-2A-Puro (Addgene, PX459). 1 × 106 HEK-293T cells were transfected with the pSpCa9-2A-Puro plasmid containing sgRNAs targeting KEAP1 or NRF2. Following puromycin selection, clonal cells were isolated by flow cytometry and analyzed for the increased or decreased expression of NRF2 by immunoblot for KEAP1-null or NRF2-null cells, respectively.
sgRNAs targeting KEAP1 (sequences are 5′ to 3′).
KEAP1-fwd: CACCGTTGGCATCATGAACGAGCTG
KEAP1-rev: AAACCAGCTCGTTCATGATGCCAAC
sgRNAs targeting NRF2 (sequences are 5′ to 3′).
NRF2-fwd: CACCGTGGAGGCAAGATATAGATCT
NRF2-rev: AAACAGATCTATATCTTGCCTCCAC
Generation of CRISPR-mediated knockout H460 cell lines
NR0B1-null or CYP4F11-null H460 cells were generated using the protocol described in (Shalem et al., 2014). In brief, sgRNAs targeting NR0B1, CYP4F11 or AKR1B10 (described below) were designed, amplified, and cloned into transient Lenti-CRISPR v2 (Addgene). Mammalian lentiviral particles harboring sgRNA-encoding plasmids were generated as described above, with the exception that the viral supernatant was concentrated with LentiX (Clontech) prior to infection of H460 cells. Following 10 days of puromycin selection, clonal cells were isolated by flow cytometry and analyzed for decreased expression of NR0B1, CYP4F11 or AKR1B10 when compared to a parental population expressing a non-targeting sgRNA (CRISPR-CTRL).
Non-targeting sgRNA (sequences are 5′ to 3′).
Lenti-CRISPRv2:sgCRISPR-CTRL-fwd: GCGAGGTATTCGGCTCCGCG
Lenti-CRISPRv2:sgCRISPR-CTRL-rev: CGCGGAGCCGAATACCTCGC
sgRNAs targeting NR0B1 (sequences are 5′ to 3′).
Lenti-CRISPRv2:sgNR0B1-fwd: GCCGCTTGCAGTTCGAGACTG
Lenti-CRISPRv2:sgNR0B1-rev: CAGTCTCGAACTGCAAGCGGC
sgRNAs targeting CYP4F11 (sequences are 5′ to 3′).
Lenti-CRISPRv2:sgCYP4F11-fwd: GTCAGCTGCTGTCGCACCCA
Lenti-CRISPRv2:sgCYP4F11-rev: TGGGTGCGACAGCAGCTGAC
sgRNAs targeting ARK1B10 (sequences are 5′ to 3′).
Lenti-CRISPRv2:sgARK1B10-1-fwd: GTGCCGATATCCTGCATCAA
Lenti-CRISPRv2:sg ARK1B10-1-rev: TTGATGCAGGATATCGGCAC
sgRNAs targeting AKR1B10 (sequences are 5′ to 3′).
Lenti-CRISPRv2: sgARK1B10-2-fwd: GTCTTTGAGAGACCCCTTGTG
Lenti-CRISPRv2: sgARK1B10-2-rev: CACAAGGGGTCTCTCAAAGAC
Mammalian lentiviral cDNA expression
Mammalian lentiviral particles harboring cDNA-encoding plasmids were generated as described above, with the exception that the viral supernatant was concentrated with LentiX (Clontech) prior to infection of target cells. Cells were allowed to recover for 24 h followed by continuous selection with puromycin.
Identification of NR0B1 interacting proteins
Confluent 15 cm dishes of A549 stably or transiently expressing FLAG-NR0B1 or FLAG-METAP2, were rinsed with ice-cold PBS and were sonicated in the presence of Chaps IP buffer (0.3% Chaps, 40mM Hepes pH 7.4, 50mM KCl, 5mM MgCl2 and EDTA-free protease inhibitors (Sigma)). Following lysis, samples were clarified by centrifugation for 10 min at 16,000 × g. FLAG-M2 beads (100 μL, 50:50 slurry) was added to the clarified supernatant and incubated for 3 h while rotating at 4°C. Beads were washed once with Chaps IP buffer and three times with Chaps IP buffer supplemented with 150 mM NaCl. Proteins were eluted with the FLAG peptide (sequence DYKDDDDK) from the FLAG-M2 beads, run on a 4–20% Tris-glycine gel (Invitrogen) and stained with InstantBlue (Expedeon). Each lane was cut into 10 pieces and in-gel trypsin (Promega) digestion was performed. The resulting digests were analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS). MS2 spectra data were extracted from the raw file using RAW Convertor (version 1.000; available at http://fields.scripps.edu/downloads.php). MS2 spectra data were searched using the ProLuCID algorithm (publicly available at http://fields.scripps.edu/downloads.php) using a reverse concatenated, non-redundant variant of the Human UniProt database (release-2012_11). Cysteine residues were searched with a static modification for carboxyamidomethylation (+57.02146) and one differential modification for oxidized methionine (+15.9949). Spectral counts for proteins from FLAG-NR0B1 immunoprecipitates were compared to spectral counts for proteins from FLAG-METAP2 immunoprecipitates across 5–6 biological replicates. Interacting proteins were classified as those proteins whose corresponding peptides were enriched by greater that 20-fold in FLAG-NR0B1 immunoprecipitates compared to FLAG-METAP2 immunoprecipitates.
For identification of endogenous NR0B1 interacting proteins, A549, H2122 or H460 cell lysates were prepared as described above. The NR0B1 (Cell Signaling Technology), RagC (Cell Signaling Technology) or GAPDH (Santa Cruz) antibodies were added to each lysate and incubated with rotation at 4°C for 1.5 h. Subsequently, protein G sepharose beads (50 μL, 50:50 slurry) were added to each sample and incubated for an additional 1.5 h. Beads were washed as described above and proteins were eluted with 8M urea at 30°C for 1 h. Proteins were reduced by treatment with DTT (10 mM for 30 min at 65 °C) and cysteines were alkylated with iodoacetamide (20 mM for 30 min at 37°C). Urea was diluted to 2M and proteins were digested with 2 μg of Trypsin (Promega). The resulting digests were analyzed by mass spectrometry as described below.
Co-transfection based interaction experiments
For transfection experiments, 4 × 106 HEK-293T cells were plated in a 10 cm dish. The next day, cells were transfected with the pRK5-based cDNA expression plasmids indicated in the figures in the following amounts. Figure S4: 25 ng FLAG-RBM45, 100 ng FLAG-NR0B1, 200 ng HA-SNW1; Figure 5 and S5: for in-vitro binding experiments: 5000 ng FLAG-SNW1; for in vitro binding experiments with transiently transfected NR0B1: 25 ng HA-NR0B1 or HA-NR0B1-C274V; for fluorescence experiments: 5000 ng Flag-NR0B1 or 5000 ng FLAG-NR0B1-C274V; Figure 5S: for site of labeling experiments, 5000 ng FLAG-NR0B1. Following transfections, cells were grown for 48 h and processed as described below.
Compound treatment for assessment of protein-protein interactions
Confluent 10 cm plates of indicated cell lines were rinsed once with warm PBS and incubated in serum/dye-free RPMI with indicated compounds or vehicle for 3 h at 37 °C. Cells were washed once ice-cold PBS and snap frozen.
Cell lysis and Immunoprecipitations
Cells were rinsed once with ice-cold PBS, and lysed by sonication in Triton IP buffer. Lysates were clarified by centrifugation at 16,000 × g for 10 min. Samples were normalized to 1 mg ml−1 and boiled following the addition of sample buffer. For FLAG- or HA-immunoprecipitations, FLAG or HA resins (30 μL, 50:50 slurry) were added to the pre-cleared lysates and incubated with rotation for 3 hours at 4°C. Following immunoprecipitation, the beads were washed once with IP buffer followed by 3 times with IP buffer containing 500mM NaCl. Loading buffer (40 μL) was added to the immunoprecipitated proteins which were subsequently denatured by boiling. Proteins were resolved by SDS-PAGE, analyzed by immunoblotting and relative band intensities were quantified using ImageJ software.
In vitro binding assay
H2122 clarified cell lysate (100 μL, 1 mg ml−1) in IP-buffer were incubated with the indicated compounds or vehicle (DMSO) for 3 hours at 4°C with rotation. Following treatment, 3 volumes of IP-buffer was added along with immobilized FLAG-SNW1 beads (30 μL, 50:50 slurry), which was incubated for an additional hour at 4°C. Beads were washed three times with IP-buffer supplemented with 500 mM NaCl. Immunoprecipitated proteins were resolved by SDS-PAGE and analyzed by immunoblotting. NR0B1 and HA-NR0B1 levels were determined by using the NR0B1 antibody (Cell Signaling). IC50 curves were determined using Prism 6 (Graphpad) software, with maximum and minimum values set at 100% NR0B1 bound 0% NR0B1 bound respectively.
Immunofluorescence
Samples were prepared as described in (Bar-Peled et al., 2013). In brief, 1 × 105 A549 cells stably expressing FLAG-RBM45 or FLAG-SNW1 were plated on poly-lysine coated glass coverslips in 12-well tissue culture plates. Forty-eight hours later, the culture media was removed and cells were fixed with 4% paraformaldehyde (Electron microscopy services). The slides were rinsed three times with PBS and cells were permeabilized with 0.05% Triton X-100 in PBS for 1 min. The slides were rinsed four times with PBS and incubated with primary antibodies in 5% normal donkey serum (Thermo) overnight at 4°C. After rinsing four times with PBS, the slides were incubated with secondary antibodies conjugated to the indicated fluorophores (Invitrogen) for 1 h at room temperature. Following an additional four washes with PBS, the slides were stained with Hoechst (Invitrogen) following the manufacturer’s protocol. Slides were mounted on glass coverslips using Prolong Gold® Antifade reagent (Invitrogen) and imaged on Zeiss LSM 780 laser scanning confocal microscope. Images were processed using ImageJ software.
Measurement of glycolytic flux
Cells were plated on poly-L-lysine coated 96-well Seahorse plates (Seahorse Biosciences) after lentiviral infection with shNRF2 or shGFP and equilibrated for 1 h in DMEM (Sigma D6030) containing 2 mM glutamine in the absence of serum and glucose. Basal extracellular acidification rate (ECAR) was then analyzed in the Seahorse XFe96 flux analyzer (Seahorse Biosciences), followed by ECAR measurements after sequential injections of 10 mM glucose, 2 μM oligomycin and 100 mM 2-deoxyglucose (2-DG).
Measurement of intracellular glutathione levels
H2122 or H1975 cells expressing shRNAs targeting a control or NRF2 were cultured in 6-well plates and total cellular glutathione content was determined using the Glutathione Assay Kit (Cayman Chemical) following the manufacturer’s protocol. Absorbance from GSH reaction with DTNB was measured using a Biotek Synergy 2 microplate reader (Biotek).
Measurement of GAPDH activity
2.5 × 105 H2122 or H1975 cells expressing shRNAs targeting a control or NRF2 were cultured in 6-well plates and GAPDH activity was determined using Ambion KDalert GAPDH Assay Kit (Fisher) following the manufacture’s protocol. This assay measures the conversion of NAD+ to NADH by GAPDH in the presence of glyceraldehyde-3-phosphate. The rate of NADH production correlated to an increase in fluorescence was measured by using a Biotek Synergy 2 microplate reader (Biotek).
Measurement of cytosolic hydrogen peroxide levels
Cytosolic hydrogen peroxide was measured using the Peroxyfluor-6 acetoxymethyl ester (PF6-AM) fluorescent probe as described in (Dickinson et al., 2011). In brief, cells were washed twice with warm PBS and incubated with 250 nM of PF6-AM in serum-free RPMI for 20 min at 37°C. Cells were allowed to recover in complete RPMI for 1 h and were subsequently harvested and resuspended in sorting buffer (PBS + 1% FBS). Flow cytometry acquisition was performed with BD FACSDiva™-driven BD™ LSR II flow cytometer (Becton, Dickinson and Company) which measured the increase in PF6-AM fluorescence. Data was analyzed with FlowJo software (Treestar Inc.)
Monolayer proliferation assay
Cells were cultured in 96-well plates at 3 × 103 cells per well in 100 μl of RPMI. At the indicated time points 50 μl of Cell Titer Glo reagent (Promega) was added to each well and the luminescence read on a Biotek Synergy 2 microplate reader (Biotek).
Anchorage-independent growth assay
Multiple NSCLC cell lines were tested for their ability to form colonies in soft agar. H460 cells were seeded at concentrations between 0.75–3.0 × 104 cells/well of a 6-well plate. H2122 cells were seeded at concentration between 4.0–6.0 × 104 cells/well. H2009 anchorage-independent growth was substantially poorer and required a higher concentration of cells (1 × 105 cells/well) to form colonies. For comparison of cells expressing indicated shRNAs cells were infected with virus-containing supernatants as described in Mammalian lentiviral shRNAs expression. 48 h after puromycin selection cells were seeded for experiment. For comparison of H460 cells expressing exogenous cDNAs, cells expressing the luciferase protein were infected with virus-containing supernatants as described in Mammalian lentiviral cDNA expression. CRISPR-mediated knockout cell lines were generated as described in Generation of CRISPR-mediated knockout H460 cell lines. An equal number of cells from each comparison group was embedded in a solution of 0.4% Noble agar solution (Difco Labs) and placed on top of hardened layer of 0.6% agar in a 6-well plate. For comparison of cells treated with indicated compound versus DMSO, compound (5 μM) or DMSO was added to cells in the 0.4% Noble agar solution before solidification. Cells were grown for 6–20 days at 37°C with 5% CO2. Fresh media (200 μL) was added every 5 days. Colony formation was determined by either counting colonies or bioluminescent imaging using an IVIS Spectrum (Caliper Life Science). Luciferin was added to cells and bioluminescence was quantified by measuring photon flux (photons/sec) using Living Image 3.2 software (Caliper Life Science) (Goodwin et al., 2014) and normalized to cells treated with vehicle (DMSO) or expressing shGFP.
qPCR analysis
2.5 × 105 cells/well of a 6-well plate were seeded the night before treatment. Cells were treated with the indicated concentrations of compound as denoted in the figure legends for 12 h. Total RNA was isolated using the RNeasy Kit (Qiagen) according to the manufacturer’s protocol. cDNA amplification was preformed using iScript Reverse Transcription Supermix kit (Bio-Rad). qPCR primer sequences were obtained from PrimerBank (https://pga.mgh.harvard.edu/primerbank/) (Wang et al., 2012) and are listed below. qPCR analysis was performed on a ABI Real Time PCR System (Applied Biosystems) with the SYBR green Mastermix (Applied Biosystems). Relative gene expression was normalized to the 18S gene.
qPCR primers (sequences are listed 5′ to 3′)
18S-fwd: CGCCGCTAGAGGTGAAATTCT
18S-rev: CGAACCTCCGACTTTCGTTCT
CRY1-fwd: CTCCTCCAATGTGGGCATCAA
CRY1-rev: CCACGAATCACAAACAGACGG
DEPDC1-fwd: ATGCGTATGATTTCCCGAATGAG
DEPDC1-rev: CACAGCATAACACACATCGAGAA
CPLX2-fwd: GAGGAGCGTAAGGCCAAGC
CPLX2-rev: CCGGGCAGGTATTTGAGCA
Gel-based competition of BPK-29yne labeling of NR0B1
4 × 106 HEK-293T cells were seeded in poly-L-lysine coated 10 cm plates and transfected the next day with 5 μg of FLAG-NR0B1, FLAG-NR0B1-C274V, or FLAG-METAP2 cDNA in a pRK5-based expression vector. 48 h after transfection, cells were treated with indicated concentrations of BPK-29 or control compound BPK-27 for 3 h at 37 °C in DMEM containing 10% FBS and supplements as described in Cell Culture. BPK-29yne (5 μM) was then added and incubated for an additional 30 min at 37 °C. FLAG immunoprecipitates were prepared as described above and following washes, the FLAG resin was resuspended in PBS (100 μL). To each sample, 12 μL of a freshly prepared “click” reagent mixture was added to conjugate the fluorophore to probe-labeled proteins. CuAAC reaction mixture consisted of TAMRA azide (1 μL of 12.5 mM stocks in DMSO, final concentration = 125 μM), 1 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP; 2 μL of fresh 50 × stock in water, final concentration = 1mM), ligand (6 μL 17× stock in DMSO:t-butanol 1:4, final concentration = 100 μM) and 1 mM CuSO4 (2 μL of 50x stock in water, final concentration = 1 mM). Upon addition of the click mixture, each reaction was immediately mixed by vortexing and then allowed to react at ambient temperature for 1 h before quenching the reactions with 100 μL of loading buffer. Samples were boiled for 5 min and proteins were resolved by SDS-PAGE (10% acrylamide), and visualized by in-gel fluorescence on a Bio-Rad ChemiDoc MP flatbed fluorescence scanner. Samples were also analyzed by immunoblotting. Recombinantly expressed FLAG-tagged protein levels were determined with the FLAG antibody (Sigma). Gel fluorescence and imaging was processed using Image Lab (v 5.2.1) software.
Measurement of NR0B1 degradation
7.5–8 × 105 H460 cells were seeded the night before per well of a 6-well plate. Cells were treated with cycloheximide (100 μg/mL) for the indicated time points. Cells were rinsed in ice-cold PBS, scraped on ice and processed for immunoblot analysis as described above. Proteins were resolved by SDS-PAGE, analyzed by immunoblotting and NR0B1 band intensities were quantified using ImageJ software and compared to a loading control (Beta-actin or GAPDH).
RNA sequencing
RNA was isolated by RNeasy Kit (Qiagen) and digested with DNase (Qiagen) from n=3 samples per condition (cells expressing shGFP, shNRF2_1, shNR0B1_1 or shSNW1_1 or treated with DMSO, 30 μM BPK-29 or 30 μM BPK-9). RNA integrity (RIN) numbers were determined using the Agilent TapeStation prior to library preparation. mRNA-seq libraries were prepared using the TruSeq RNA library preparation kit (version 2) according to the manufacturer’s instructions (Illumina). Libraries were then quantified, pooled, and sequenced by single-end 50 base pairs using the Illumina HiSeq 2500 platform at the Salk Next-Generation Sequencing Core. Raw sequencing data were demultiplexed and converted into FASTQ files using CASAVA (version 1.8.2). Libraries were sequenced at an average depth of 15 million reads per sample.
The spliced read aligner STAR (Dobin et al., 2013) was used to align sequencing reads to the human hg19 genome. Gene-level read counts were obtained based on UCSC hg19 gene annotation. DESeq2 (Love et al., 2014) was used to calculate differential gene expression based on uniquely aligned reads, and p-values were adjusted for multiple hypothesis testing with the Benjamini-Hochberg method.
ChIP-seq analysis
ChIP was conducted as previously described (Komashko et al., 2008). H460 cells were fixed in 1% formaldehyde (Sigma) for 15 minutes at 25 °C. After lysis, samples were sonicated using a biorupter sonicator (Diagenode) for 60 cycles (30 seconds per cycle/30 seconds cooling) at a high power level. Chromatin sheering was optimized to a size range of 200 to 600 bp. Chromatin (100 μg) was immunoprecipitated with the NR0B1 antibody (Cell Signaling Technology). For DNA sequencing, samples were prepared for library construction, flow cell preparation and sequencing were performed according to Illumina’s protocols. Sequencing was accomplished on Illumina HiSeq 2500 using PE 2 × 125 bp reads with over 14 million clusters per sample.
Sequencing reads were aligned to the hg19 genome using bowtie2 (Langmead and Salzberg, 2012). Peak detection was carried out using HOMER (homer.ucsd.edu), comparing the NR0B1 IP sample against a whole-cell extract (WCE) with default parameters for transcription factor-style analysis. This requires relevant peaks to be significantly enriched over WCE and the local region with an uncorrected Poisson distribution-based p-value threshold of 0.0001 and false discovery rate threshold of 0.001. These peaks were further restricted to a 2kb window around annotated transcription start sites.
Correlation analysis
For shRNA gene expression analysis data, the correlation of gene expression levels between the shNR0B1-cells and shNRF2-cells and shNR0B1-cells and shSNW1-cells was calculated using Pearson’s correlation coefficient, and a correlation analysis was performed to calculate the p-value.
Circos plot
A graphical summary of NR0B1 genome-wide effects. The inner track shows the change in gene expression following NR0B1 knockdown (red indicates an increase, blue a decrease). The middle track shows the normalized peak height of the NR0B1 ChIP. Only genes with both significantly altered expression (adjusted p-value threshold of 0.01 and 1.5-fold expression threshold) and an NR0B1 peak near a TSS are shown.
A graphical summary of liganded cysteines in KEAP1-WT and KEAP1-mutant cell lines. The outer track denotes total liganded cysteines in a given cell line (cysteines were defined as liganded if they had an average R ≥5 and were quantified in two or more replicates). Grey chords connect liganded cysteines that are found in two or more cell lines.
GSEA
GSEA (Subramanian et al., 2005) was carried out using pre-ranked lists from FDR or fold change values, setting gene set permutations to 1000 and using either c1 collection in MSigDB version 4.0 (Figure S4C) or c2 collection in MSigDB V.5.0 (Figure S7A).
Functional Gene Enrichment Analysis
Functional enrichment in gene sets was determined using the DAVID functional annotation tool (version 6.7) with “FAT” Gene Ontology terms (Huang da et al., 2009).
isoTOP-ABPP Sample preparation
Sample preparation and analysis were based on (Backus et al. 2016.) with modifications noted below.
For analysis of NR0B1 ligands or control compound reactivity, H460 cells or H460 cells expressing luciferase in a 10 cm plate were incubated with indicated compounds in serum/dye-free RPMI for 3 hours at 37 °C. Cells were washed once ice-cold PBS and lysed in 1% Triton X-100 dissolved in PBS with protease inhibitors (Sigma) by sonication. Samples were clarified by centrifugation for 10 min at 16,000 × g. Lysate was adjusted to 1.5 mg ml−1 in 500 μL.
For analysis of cysteines regulated by NRF2, H2122 or H1975 cells expressing shGFP or shNRF2 were lysed and processed as described above. Lysate was adjusted to 1.5 mg ml−1 in 500 μL.
For analysis of cysteines that change following induction of apoptosis, H2122 and H1975 cells were treated with DMSO or staurosporine (1 μM, 4 h) in full RPMI. H1975 cells were treated with DMSO or AZD9291 (1 μM, 24 h) in full RPMI. Cells were lysed as described above.
For analysis of ligandable cysteines in KEAP1-WT (H2122, H460 and A549) cells and KEAP1-mutant (H1975, H2009 (expressing the luciferase protein) and H358) cells, lysate was prepared as described in (Backus et al., 2016). Samples were treated with 500 μM of compound 2, 3 or vehicle for 1 h at room temperature.
isoTOP-ABPP IA-alkyne labeling and click chemistry
Samples were labeled for 1 h at ambient temperature with 100 μM iodoacetamide alkyne (1, IA- alkyne, 5 μL of 10 mM stock in DMSO). Samples were conjugated by copper-catalyzed azide-alkyne cycloaddition (CuAAC) to isotopically labeled, TEV-cleavable tags (TEV-tags). Heavy CuAAC reaction mixtures was added to the DMSO-treated or shGFP control samples and light CuAAC reaction mixture was added to compound-treated or shNRF2 samples. The CuAAC reaction mixture consisted of TEV tags (light or heavy, 10 μL of 5 mM stocks in DMSO, final concentration = 100 μM), 1 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP; fresh 50 × stock in water, final concentration = 1mM), ligand (17x stock in DMSO:t-butanol 1:4, final concentration = 100 μM) and 1 mM CuSO4 (50x stock in water, final concentration = 1 mM). The samples were allowed to react for 1 h at which point the samples were centrifuged (16,000 × g, 5 min, 4°C). The resulting pellets were sonicated in ice-cold methanol (500 μL) and the resuspended light- and heavy-labeled samples were then combined pairwise and centrifuged (16,000 × g, 5 min, 4°C). The pellets were solubilized in PBS containing 1.2% SDS (1 mL) with sonication and heating (5 min, 95°C) and any insoluble material was removed by an additional centrifugation step at ambient temperature (14,000 × g, 1 min).
isoTOP-ABPP streptavidin enrichment
For each sample, 100 μL of streptavidin-agarose beads slurry (Fisher) was washed in 10 mL PBS and then resuspended in 6 mL PBS (final concentration 0.2% SDS in PBS). The SDS-solubilized proteins were added to the suspension of streptavidin-agarose beads and the bead mixture was rotated for 3 h at ambient temperature. After incubation, the beads were pelleted by centrifugation (1,400 × g, 3 min) and were washed (2 × 10 mL PBS and 2 × 10 mL water).
isoTOP-ABPP trypsin and TEV digestion
The beads were transferred to eppendorf tubes with 1 mL PBS, centrifuged (1,400 × g, 3 min), and resuspended in PBS containing 6 M urea (500 μL). To this was added 10 mM DTT (25 μL of a 200 mM stock in water) and the beads were incubated at 65 °C for 15 mins. 20 mM iodoacetamide (25 μL of a 400 mM stock in water) was then added and allowed to react at 37°C for 30 mins with shaking. The bead mixture was diluted with 900 μL PBS, pelleted by centrifugation (1,400 × g, 3 min), and resuspended in PBS containing 2 M urea (200 μL). To this was added 1 mM CaCl2 (2 μL of a 200 mM stock in water) and trypsin (2 μg, Promega, sequencing grade) and the digestion was allowed to proceed overnight at 37 °C with shaking. The beads were separated from the digest with Micro Bio-Spin columns (Bio-Rad) by centrifugation (1,000 × g, 1 min), washed (2 × 1 mL PBS and 2 × 1 mL water) and then transferred to fresh eppendorf tubes with 1 mL water. The washed beads were washed once further in 140 μL TEV buffer (50 mM Tris, pH 8, 0.5 mM EDTA, 1 mM DTT) and then resuspended in 140 μL TEV buffer. 5 μL TEV protease (80 μM) was added and the reactions were rotated overnight at 29 °C. The TEV digest was separated from the beads with Micro Bio- Spin columns by centrifugation (1,400 × g, 3 min) and the beads were washed once with water (100 μL). The samples were then acidified to a final concentration of 5% (v/v) formic acid and stored at −80°C prior to analysis.
isoTOP-ABPP liquid-chromatography-mass-spectrometry (LC-MS) analysis
Samples processed for multidimensional liquid chromatography tandem mass spectrometry (MudPIT) were pressure loaded onto a 250 μm (inner diameter) fused silica capillary columns packed with C18 resin (Aqua 5 μm, Phenomenex). Samples were analyzed using an LTQVelos Orbitrap mass spectrometer (Thermo Scientific) coupled to an Agilent 1200- series quaternary pump. The peptides were eluted onto a biphasic column with a 5 μm tip (100 μm fused silica, packed with C18 (10 cm) and bulk strong cation exchange resin (3 cm, SCX, Phenomenex,)) in a 5-step MudPIT experiment, using 0%, 30%, 60%, 90%, and 100% salt bumps of 500 mM aqueous ammonium acetate and using a gradient of 5–100% buffer B in buffer A (buffer A: 95% water, 5% acetonitrile, 0.1% formic acid; buffer B: 5% water, 95% acetonitrile, 0.1% formic acid) as has been described in (Weerapana et al., 2007). Data were collected in data-dependent acquisition mode with dynamic exclusion enabled (20 s, repeat of 2). One full MS (MS1) scan (400–1800 m/z) was followed by 30 MS2 scans (ITMS) of the nth most abundant ions.
isoTOP-ABPP peptide and protein identification
The MS2 spectra data were extracted from the raw file using RAW Convertor (version 1.000; available at http://fields.scripps.edu/downloads.php). MS2 spectra data were searched using the ProLuCID algorithm (publicly available at http://fields.scripps.edu/downloads.php) using a reverse concatenated, non-redundant variant of the Human UniProt database (release-2012_11). Cysteine residues were searched with a static modification for carboxyamidomethylation (+57.02146) and up to two differential modification for either the light or heavy TEV tags or oxidized methionine (+464.28595, +470.29976, +15.9949 respectively).
MS2 spectra data were also searched using the ProLuCID algorithm using a custom database containing only selenocysteine proteins, which was generated from a reverse concatenated, nonredundant variant of the Human UniProt database (release-2012_11). In the database, selenocysteine residues (U) were replaced with cysteine (C) and were searched with a static modification for carboxyamidomethylation (+57.02146) and up to two differential modification for either the light or heavy TEV tags or oxidized methionine (+512.2304+ or +518.2442 +15.9949). Peptides were required to have at least one tryptic terminus and to contain the TEV modification. ProLuCID data was filtered through DTASelect (version 2.0) to achieve a peptide false-positive rate below 1%.
isoTOP-ABPP R value calculation and processing
The isoTOP-ABPP ratios (R values) of heavy/light for each unique peptide (DMSO/compound treated or shGFP/shNRF2) were quantified with in-house CIMAGE software (Weerapana et al., 2010) using default parameters (3 MS1 acquisitions per peak and signal to noise threshold set to 2.5). Site-specific engagement of cysteine residues was assessed by blockade of IA-alkyne probe labelling. A maximal ratio of 20 was assigned for peptides that showed a ≥95% reduction in MS1 peak area from the experimental proteome (light TEV tag) when compared to the control proteome (DMSO, shGFP; heavy TEV tag). Ratios for unique peptide sequences entries were calculated for each experiment; overlapping peptides with the same modified cysteine (for example, different charge states, MudPIT chromatographic steps or tryptic termini) were grouped together and the median ratio is reported as the final ratio (R). Additionally, ratios for peptide sequences containing multiple cysteines were grouped together. Biological replicates of the same treatment and cell line were averaged if the standard deviation was below 60% of the mean; otherwise, for cysteines with at least one R value <4 per treatment, the lowest value of the ratio set was taken. For cysteines where all R values were ≥ 4, the average was reported. The peptide ratios reported by CIMAGE were further filtered to ensure the removal or correction of low-quality ratios in each individual data set. The quality filters applied were the following: removal of half tryptic peptides; removal of peptides which were detected only once across all data sets reported herein; removal of peptides with R = 20 and only a single MS2 event triggered during the elution of the parent ion; manual annotation of all the peptides with ratios of 20, removing any peptides with low-quality elution profiles that remained after the previous curation steps.
For selenocysteines, the ratios of heavy/light for each unique peptide (DMSO/compound treated; isoTOP-ABPP ratios, R values) were quantified with in-house CIMAGE software using the default parameters described above, with the modification to allow the definition of selenocysteine (amino acid atom composition and atomic weights). Extracted ion chromatograms were manually inspected to ensure the removal of low quality ratios and false calls.
Cysteine residues were deemed to have significantly changed following NRF2 knockdown if they had R-values ≥ 2.5. Changes in cysteine reactivity were considered reactivity based if a cysteine for a given protein had an R-value ≥ 2.5 and all the remaining cysteines in that protein had R-values < 1.5. If only one cysteine was identified per protein with an R value ≥ 2.5, and if the corresponding change in the mRNA transcript was < 1.5 (shGFP/shNRF2) then that change was also considered reactivity based. Changes in cysteine reactivity were considered expression based if a cysteine for a given protein had an R-value ≥ 2.5 and all the remaining cysteines in that protein had R-values ≥ 1.5. If only one cysteine was identified per protein with an R-value ≥ 2.5, and if the corresponding change in the mRNA transcript was ≥ 1.5 (shGFP/shNRF2) then than change was also considered expression based. For datasets corresponding to changes in cysteine reactivity in H2122 cells expressing shNRF2 or shGFP at ‘Day1/2’ two replicates were taken from the ‘Day 1’ time point and three replicates were taken from the ‘Day 2 time point’ (Table S1). For datasets corresponding to changes in cysteine reactivity in H1975 cells expressing shNRF2 or shGFP at ‘Day1/2’ two replicates were taken from the ‘Day 1’ time point and two replicates were taken from the ‘Day 2 time point’ (Table S1). For datasets corresponding to changes in cysteine reactivity in H2122 cells expressing shNRF2 or shGFP at ‘Day1’ three replicates were used (Table S1). Cysteine residues were designated as expression-based changes for this experiment if following NRF2 knockdown they had R-values ≥ 2.5 and were considered unchanged if they had R-values < 1.5 (Table S1). Cysteines were considered significantly changed following staurosporine or AZD9291 treatment if they had R values ≥ 2.5.
Cysteine residues were considered liganded in vitro by electrophilic fragments (compounds 2 or 3) if they had an average R-value ≥ 5 and were quantified in at least 2 out of 3 replicates. Targets of NR0B1 ligands or control compounds were defined as those cysteine residues that had R-values ≥ 3 in more than one biological replicate following ligand treatment in cells.
Protein turnover
For analysis of protein turnover in H460 cells, confluent 10 cm plates were washed twice with warm PBS, then incubated in “heavy” RPMI for 3 h. Cells were washed once ice-cold PBS and lysed in 1% Triton 100-X dissolved in PBS with protease inhibitors (Sigma) by sonication. Lysate was adjusted to 1.5 mg ml−1 in 2 × 500 μL. Samples were processed identically to other samples (lysates were adjusted to 1.5 mg ml−1 in 2 × 500 μL), with the following modification: only isotopically light TEV tag was used. After the “click” reaction, both 2 × 500 μL were centrifuged (16,000 × g, 5 min, 4°C) and resuspended by sonication in ice-cold methanol (500 μL). Aliquots were then combined and resolubilized in PBS containing 1.2% SDS (1 mL) as detailed in isoTOP-ABPP IA-alkyne labeling and click chemistry. Samples were further processed and analyzed as detailed in: isoTOP-ABPP streptavidin enrichment, isoTOP-ABPP trypsin and TEV digestion, isoTOP-ABPP liquid-chromatography-mass-spectrometry (LC-MS) analysis, isoTOP-ABPP peptide and protein identification and isoTOP-ABPP R value calculation and processing with the following exceptions: Samples processed for protein turnover were searched with ProLuCID with mass shifts of SILAC labeled amino acids (+10.0083 R, +8.0142 K) in addition to carboxyamidomethylation modification (+57.02146) and two differential modification for either the light TEV tag or oxidize methionine (+464.28595, +15.9949 respectively). 1 peptide identification was required for each protein. ProLuCID data was filtered through DTASelect (version 2.0) to achieve a peptide false-positive rate below 1%. Ratios of light/heavy peaks were calculated using in-house CIMAGE software. Median SILAC ratios from one or more unique peptides were combined to generate R values. Proteins were required to be quantified in at least two biological replicates. The mean R values and standard deviation for multiple biological experiments were calculated from the average ratios from each replicate. Proteins were designated as rapid turnover if they had R-values ≤ 8.
ABPP-SILAC sample preparation and LC-MS analysis
Isotopically labeled H460 cell lines were generated as described above. Light and heavy cells were treated with compounds (20 μM) or DMSO, respectively, for 3 h, followed by labeling with the BPK-29yne (5 μM) for 30 min. Cells were washed once ice-cold PBS and lysed in 1% Triton 100-X dissolved in PBS with protease inhibitors (Sigma) by sonication. Lysate was adjusted to 1.5 mg ml−1 in 500 μL. Samples were conjugated by CuAAC to Biotin-PEG4-azide (5 μL of 10 mM stocks in DMSO, final concentration = 100 μM). CuAAC “click” mix contained TCEP, TBTA ligand and CuSO4 as detailed for isoTOP-ABPP sample preparation. Samples were further processed as detailed in: isoTOP-ABPP streptavidin enrichment and isoTOP-ABPP trypsin TEV digestion with the following exception: after overnight incubation at 37 °C with trypsin, tryptic digests were separated from the beads with Micro Bio-Spin columns (Bio-Rad) by centrifugation (1,000 × g, 1 min). Beads were rinsed once with water (200 μL) and combined with tryptic digests. The samples were then acidified to a final concentration of 5% (v/v) formic acid and stored at −80°C prior to analysis. Samples were processed for multidimensional liquid chromatography tandem mass spectrometry (MudPIT) as described in isoTOP-ABPP liquid-chromatography-mass-spectrometry (LC-MS) with the exception that peptides were eluted using the 5-step MudPIT protocol with conditions: 0%, 25%, 50%, 80%, and 100% salt bumps of 500 mM aqueous ammonium acetate and using a gradient of 5–100% buffer B in buffer A (buffer A: 95% water, 5% acetonitrile, 0.1% formic acid; buffer B: 5% water, 95% acetonitrile, 0.1% formic acid).
ABPP-SILAC peptide and protein identification and R value calculation and processing
The MS2 spectra data were extracted and searched using RAW Convertor and ProLuCID algorithm as described in isoTOP-ABPP peptide and protein quantification. Briefly, cysteine residues were searched with a static modification for carboxyamidomethylation (+57.02146 C). Searches also included methionine oxidation as a differential modification (+15.9949 M) and mass shifts of SILAC labeled amino acids (+10.0083 R, +8.0142 K) and no enzyme specificity. Peptides were required to have at least one tryptic terminus and unlimited missed cleavage sites. 2 peptide identifications were required for each protein. ProLuCID data was filtered through DTASelect (version 2.0) to achieve a peptide false-positive rate below 1%. Ratios of heavy/light (DMSO/test compound) peaks were calculated using in-house CIMAGE software. Median SILAC ratios from two or more unique peptides were combined to generate R values. The mean R values and standard deviation for multiple biological experiments were calculated from the average ratios from each replicate. Targets of NR0B1 ligands or control compounds were defined as those proteins that had R-values ≥2.5 in two or more biological replicates following ligand treatment in cells.
Site of labeling
For site of labeling with BPK-29, 4 × 106 HEK-293T cells were seeded in a 10 cm plate and transfected the next day with 5 μg of FLAG-NR0B1 cDNA in a pRK5-based expression vector. 48 hours after transfection, cells were treated with vehicle, BPK-29 (50 μM) in serum-free RPMI for 3 h at 37 °C. FLAG immunoprecipitates were prepared as described above in Identification of NR0B1 interacting proteins. FLAG-NR0B1 was eluted from FLAG-M2 beads with 8M urea and subjected to proteolytic digestion, whereupon tryptic peptides harboring C274 were analyzed by LC-MS/MS. The resulting mass spectra were extracted using the ProLuCID algorithm designating a variable peptide modification (+252.986 and +386.1851 for BPK-26 and BPK-29, respectively) for all cysteine residues. For site of labeling with BPK-26, HEK-293T cell lysate transfected with FLAG-NR0B1 as described above was treated with vehicle or BPK-26 (100 μM) for 3 h at 4°C. FLAG immunoprecipitates were processed for proteomic analysis as described above. See Table S3 for MS/MS spectra and fragmentation tables.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was preformed using GraphPad Prism version 6 or 7 for Mac, GraphPad Software, La Jolla California USA, www.graphad.com or the R statistical programing language (www.R-project.org). Statistical values including the exact n and statistical significance are also reported in the Figure Legends. Inhibition curves of the NR0B1-SNW1 interactions by NR0B1-ligand are fit as using log(inhibitor) vs % normalized remaining of NR0B1-SNW1 interaction and data points are plotted as the mean ± SD (n=2–5 per group). NR0B1 half-life was calculated from a one-phase exponential decay curve plotted as mean ± SD (4–10 per group). Statistical significance was defined as p< 0.05 and determined by 2-tailed Student’s t-test (Figure 1I, S1D, 3B, 3C–F, 6E–F, S3C, S4H, S7A), two-way Anova with Bonferroni post-test analysis (Figure 1J) or correlation analysis using Pearson product-moment correlation coefficient (Figures 4B, S4G).
DATA AND SOFTWARE AVAILABILITY
Data Resources
The RNA-seq data are reported in this paper has been deposited in the NCBI under GEO: GSE89569.
Supplementary Material
Acknowledgments
We thank D. Hermanson for assistance with metabolomics, C. Brenner and M. Lazear for assistance with bioinformatics, D. Sabatini for providing lentiviral vectors, K. Lamia for providing the anti-Cry antibody, and C. Chang for PF6-AM. This work was supported by the NIH (CA087660, CA132630 to B.F.C.; CA172229 to R.J.S.; CA215249-01 to L. B-P; M.M.D., CA211526; NIH-NCI CCSG: P30 014195 to Salk NGS Core Facility), the Chapman Foundation, the Helmsley Charitable Trust, the Damon Runyon Cancer Research Foundation (L.B-P.), the American Cancer Society (R.U.S), the Life Sciences Research Foundation (E.V.V.), and the NIH (K.B.M., B.D.H.).
Footnotes
Author contributions.
L.B-P. and B.F.C conceived of research and wrote the paper. L.B-P. and EKK developed methods, preformed experiments and analysed data. L.B-P. and T.A.P. prepared samples for ChIP-Seq. R.M.S and M.M.D. analyzed chemical-proteomics data. K.B.M and B.D.H designed BPK compounds, E.K.K. generated BPK compounds, and E.K.K. and E.A.V. analytically characterized BPK compounds. R.U.S helped with anchorage independent growth assays and R.J.S provided the facilities for the quantification of these assays. J.O. assisted with structural analysis. M.W.C, Z.Z. and T.-A.I. analyzed RNAseq, ChIP-seq and TCGA data. N.I., P.J, and M.M.H. assisted with identification of KEAP1-WT and KEAP1-mutant cell lines. B.P.K. and E.S. assisted with Seahorse analysis.
Competing Financial Interests
The authors declare competing financial interests. B.F.C. is a founder and advisor to Vividion Therapeutics, a biotechnology company interested in using chemical proteomic methods to develop small-molecule drugs to treat human disease. T.A.P., N.I., P.J., Z.Z. and M.M.H. are employees of Pfizer.
References
- Albers M, Kranz H, Kober I, Kaiser C, Klink M, Suckow J, Kern R, Koegl M. Automated yeast two-hybrid screening for nuclear receptor-interacting proteins. Mol Cell Proteomics. 2005;4:205–213. doi: 10.1074/mcp.M400169-MCP200. [DOI] [PubMed] [Google Scholar]
- Ashouri A, Sayin VI, Van den Eynden J, Papagiannakopoulos T, Larsson E. Pan-cancer transcriptomic analysis associates long non-coding RNAs with key mutational driver events. 13197 EP. Nature Communications. 2016 doi: 10.1038/ncomms13197. [DOI] [PMC free article] [PubMed]
- Backus KM, Correia BE, Lum KM, Forli S, Horning BD, Gonzalez-Paez GE, Chatterjee S, Lanning BR, Teijaro JR, Olson AJ, et al. Proteome-wide covalent ligand discovery in native biological systems. Nature. 2016;534:570–574. doi: 10.1038/nature18002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger AH, Brooks AN, Wu X, Shrestha Y, Chouinard C, Piccioni F, Bagul M, Kamburov A, Imielinski M, Hogstrom L, et al. High-throughput Phenotyping of Lung Cancer Somatic Mutations. Cancer Cell. 2016;30:214–228. doi: 10.1016/j.ccell.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollong MJ, Yun H, Sherwood L, Woods AK, Lairson LL, Schultz PG. A Small Molecule Inhibits Deregulated NRF2 Transcriptional Activity in Cancer. ACS Chem Biol. 2015;10:2193–2198. doi: 10.1021/acschembio.5b00448. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso R, Love R, Nilsson CL, Bergqvist S, Nowlin D, Yan J, Liu KK, Zhu J, Chen P, Deng YL, et al. Identification of Cys255 in HIF-1alpha as a novel site for development of covalent inhibitors of HIF-1alpha/ARNT PasB domain protein-protein interaction. Protein Sci. 2012;21:1885–1896. doi: 10.1002/pro.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chio II, Jafarnejad SM, Ponz-Sarvise M, Park Y, Rivera K, Palm W, Wilson J, Sangar V, Hao Y, Ohlund D, et al. NRF2 Promotes Tumor Maintenance by Modulating mRNA Translation in Pancreatic Cancer. Cell. 2016;166:963–976. doi: 10.1016/j.cell.2016.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Wang SB, Venkatraman V, Murray CI, Van Eyk JE. Cysteine oxidative posttranslational modifications: emerging regulation in the cardiovascular system. Circ Res. 2013;112:382–392. doi: 10.1161/CIRCRESAHA.112.268680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson BC, Peltier J, Stone D, Schaffer DV, Chang CJ. Nox2 redox signaling maintains essential cell populations in the brain. Nat Chem Biol. 2011;7:106–112. doi: 10.1038/nchembio.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlund A, Anthonisen EH, Gustafsson N, Venteclef N, Robertson Remen K, Damdimopoulos AE, Galeeva A, Pelto-Huikko M, Lalli E, Steffensen KR, et al. E3 ubiquitin ligase RNF31 cooperates with DAX-1 in transcriptional repression of steroidogenesis. Mol Cell Biol. 2009;29:2230–2242. doi: 10.1128/MCB.00743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay MR, Anderton M, Ashton S, Ballard P, Bethel PA, Box MR, Bradbury RH, Brown SJ, Butterworth S, Campbell A, et al. Discovery of a potent and selective EGFR inhibitor (AZD9291) of both sensitizing and T790M resistance mutations that spares the wild type form of the receptor. J Med Chem. 2014;57:8249–8267. doi: 10.1021/jm500973a. [DOI] [PubMed] [Google Scholar]
- Garcia-Aragoncillo E, Carrillo J, Lalli E, Agra N, Gomez-Lopez G, Pestana A, Alonso J. DAX1, a direct target of EWS/FLI1 oncoprotein, is a principal regulator of cell-cycle progression in Ewing’s tumor cells. Oncogene. 2008;27:6034–6043. doi: 10.1038/onc.2008.203. [DOI] [PubMed] [Google Scholar]
- Giles NM, Giles GI, Jacob C. Multiple roles of cysteine in biocatalysis. Biochem Biophys Res Commun. 2003;300:1–4. doi: 10.1016/s0006-291x(02)02770-5. [DOI] [PubMed] [Google Scholar]
- Goldstein LD, Lee J, Gnad F, Klijn C, Schaub A, Reeder J, Daemen A, Bakalarski CE, Holcomb T, Shames DS, et al. Recurrent Loss of NFE2L2 Exon 2 Is a Mechanism for Nrf2 Pathway Activation in Human Cancers. Cell Rep. 2016;16:2605–2617. doi: 10.1016/j.celrep.2016.08.010. [DOI] [PubMed] [Google Scholar]
- Goodwin JM, Svensson RU, Lou HJ, Winslow MM, Turk BE, Shaw RJ. An AMPK-independent signaling pathway downstream of the LKB1 tumor suppressor controls Snail1 and metastatic potential. Mol Cell. 2014;55:436–450. doi: 10.1016/j.molcel.2014.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nature reviews Drug discovery. 2013;12:931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- Iyer AK, McCabe ER. Molecular mechanisms of DAX1 action. Mol Genet Metab. 2004;83:60–73. doi: 10.1016/j.ymgme.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Weerapana E, Cravatt BF. Strategies for discovering and derisking covalent, irreversible enzyme inhibitors. Future Med Chem. 2010;2:949–964. doi: 10.4155/fmc.10.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost C, Nitsche C, Scholz T, Roux L, Klein CD. Promiscuity and selectivity in covalent enzyme inhibition: a systematic study of electrophilic fragments. J Med Chem. 2014;57:7590–7599. doi: 10.1021/jm5006918. [DOI] [PubMed] [Google Scholar]
- Kinsey M, Smith R, Iyer AK, McCabe ER, Lessnick SL. EWS/FLI and its downstream target NR0B1 interact directly to modulate transcription and oncogenesis in Ewing’s sarcoma. Cancer Res. 2009;69:9047–9055. doi: 10.1158/0008-5472.CAN-09-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klijn C, Durinck S, Stawiski EW, Haverty PM, Jiang Z, Liu H, Degenhardt J, Mayba O, Gnad F, Liu J, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol. 2015;33:306–312. doi: 10.1038/nbt.3080. [DOI] [PubMed] [Google Scholar]
- Komashko VM, Acevedo LG, Squazzo SL, Iyengar SS, Rabinovich A, O’Geen H, Green R, Farnham PJ. Using ChIP-chip technology to reveal common principles of transcriptional repression in normal and cancer cells. Genome Res. 2008;18:521–532. doi: 10.1101/gr.074609.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen HM, Kansanen E, Polonen P, Heinaniemi M, Levonen AL. Role of the Keap1-Nrf2 pathway in cancer. Adv Cancer Res. 2014;122:281–320. doi: 10.1016/B978-0-12-420117-0.00008-6. [DOI] [PubMed] [Google Scholar]
- Liu Q, Sabnis Y, Zhao Z, Zhang T, Buhrlage SJ, Jones LH, Gray NS. Developing irreversible inhibitors of the protein kinase cysteinome. Chem Biol. 2013;20:146–159. doi: 10.1016/j.chembiol.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu MC, Ji JA, Jiang ZY, You QD. The Keap1-Nrf2-ARE Pathway As a Potential Preventive and Therapeutic Target: An Update. Med Res Rev. 2016;36:924–963. doi: 10.1002/med.21396. [DOI] [PubMed] [Google Scholar]
- Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Niphakis MJ, Lum KM, Cognetta AB, 3rd, Correia BE, Ichu TA, Olucha J, Brown SJ, Kundu S, Piscitelli F, Rosen H, et al. A Global Map of Lipid-Binding Proteins and Their Ligandability in Cells. Cell. 2015;161:1668–1680. doi: 10.1016/j.cell.2015.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Tian T, Inoue M, Ikeda J, Qiu Y, Okumura M, Aozasa K, Morii E. Tumorigenic role of orphan nuclear receptor NR0B1 in lung adenocarcinoma. Am J Pathol. 2009;175:1235–1245. doi: 10.2353/ajpath.2009.090010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace NJ, Weerapana E. Zinc-binding cysteines: diverse functions and structural motifs. Biomolecules. 2014;4:419–434. doi: 10.3390/biom4020419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z. Bruton’s tyrosine kinase as a drug discovery target. Drug News Perspect. 2008;21:357–362. doi: 10.1358/dnp.2008.21.7.1255308. [DOI] [PubMed] [Google Scholar]
- Planken S, Behenna DC, Nair SK, Johnson TO, Nagata A, Almaden C, Bailey S, Ballard TE, Bernier L, Cheng H, et al. Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR. J Med Chem. 2017;60:3002–3019. doi: 10.1021/acs.jmedchem.6b01894. [DOI] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Sablin EP, Woods A, Krylova IN, Hwang P, Ingraham HA, Fletterick RJ. The structure of corepressor Dax-1 bound to its target nuclear receptor LRH-1. Proc Natl Acad Sci U S A. 2008;105:18390–18395. doi: 10.1073/pnas.0808936105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, Gao S, Puigserver P, Brugge JS. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestov AA, Liu X, Ser Z, Cluntun AA, Hung YP, Huang L, Kim D, Le A, Yellen G, Albeck JG, et al. Quantitative determinants of aerobic glycolysis identify flux through the enzyme GAPDH as a limiting step. Elife. 2014:3. doi: 10.7554/eLife.03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporn MB, Liby KT. NRF2 and cancer: the good, the bad and the importance of context. Nat Rev Cancer. 2012;12:564–571. doi: 10.1038/nrc3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingham JP, Buonocore F, Duncan AJ, Achermann JC. DAX-1 (NR0B1) and steroidogenic factor-1 (SF-1, NR5A1) in human disease. Best Pract Res Clin Endocrinol Metab. 2015;29:607–619. doi: 10.1016/j.beem.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorarensen A, Dowty ME, Banker ME, Juba B, Jussif J, Lin T, Vincent F, Czerwinski RM, Casimiro-Garcia A, Unwalla R, et al. Design of a Janus Kinase 3 (JAK3) Specific Inhibitor 1-((2S,5R)-5-((7H-Pyrrolo[2,3-d]pyrimidin-4-yl)amino)-2-methylpiperidin-1-yl)prop -2-en-1-one (PF-06651600) Allowing for the Interrogation of JAK3 Signaling in Humans. J Med Chem. 2017;60:1971–1993. doi: 10.1021/acs.jmedchem.6b01694. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Yan R, Luo D, Watabe K, Liao DF, Cao D. Aldo-keto reductase family 1 member B10 promotes cell survival by regulating lipid synthesis and eliminating carbonyls. J Biol Chem. 2009;284:26742–26748. doi: 10.1074/jbc.M109.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Speers AE, Cravatt BF. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)--a general method for mapping sites of probe modification in proteomes. Nat Protoc. 2007;2:1414–1425. doi: 10.1038/nprot.2007.194. [DOI] [PubMed] [Google Scholar]
- Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Gupta V, Carroll KS, Liebler DC. Site-specific mapping and quantification of protein S-sulphenylation in cells. Nat Commun. 2014;5:4776. doi: 10.1038/ncomms5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J, Mullarky E, Lu C, Bosch KN, Kavalier A, Rivera K, Roper J, Chio II, Giannopoulou EG, Rago C, et al. Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science. 2015;350:1391–1396. doi: 10.1126/science.aaa5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Dowd DR, Staal A, Gu C, Lian JB, van Wijnen AJ, Stein GS, MacDonald PN. Nuclear coactivator-62 kDa/Ski-interacting protein is a nuclear matrix-associated coactivator that may couple vitamin D receptor-mediated transcription and RNA splicing. J Biol Chem. 2003;278:35325–35336. doi: 10.1074/jbc.M305191200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.