Abstract
Long non-coding RNAs (lncRNAs) play a pivotal role in tumorigenesis, exemplified by the recent finding that lncRNA maternally expressed gene 3 (MEG3) inhibits tumor growth in a p53-dependent manner. Acute myeloid leukemia (AML) is the most common malignant myeloid disorder in adults, and TP53 mutations or loss are frequently detected in patients with therapy-related AML or AML with complex karyotype. Here, we reveal that MEG3 is significantly downregulated in AML and suppresses leukemogenesis not only in a p53-dependent, but also a p53-independent manner. In addition, MEG3 is proven to be transcriptionally activated by Wilms’ tumor 1 (WT1), dysregulation of which by epigenetic silencing or mutations is causally involved in AML. Therefore MEG3 is identified as a novel target of the WT1 molecule. Ten–eleven translocation-2 (TET2) mutations frequently occur in AML and significantly promote leukemogenesis of this disorder. In our study, TET2, acting as a cofactor of WT1, increases MEG3 expression. Taken together, our work demonstrates that TET2 dysregulated WT1-MEG3 axis significantly promotes AML leukemogenesis, paving a new avenue for diagnosis and treatment of AML patients.
Introduction
Acute myeloid leukemia (AML) is the most common malignant myeloid disorder in adults, which is a heterogeneous clonal disorder of hematopoietic progenitor cells, with diverse biological, phenotypic and prognostic behaviors, and strikingly different outcomes to standard therapy.1, 2 In order to improve outcome in AML, current efforts in basic and clinical research focus on the genetic characterization of AML in the hopes of furthering our understanding of AML pathogenesis and discovering new targeted therapies that will eventually lead to an increase in the cure rate.2, 3 Long non-coding RNAs (lncRNAs), defined as transcripts longer than 200 nucleotides,4 affect various cellular functions such as gene regulation, genomic imprinting, RNA maturation and translation.5, 6 Moreover, multiple lines of evidence link dysregulation of lncRNAs to diverse human diseases, especially cancer. In the process of carcinogenesis, certain lncRNAs have been attributed to oncogenic and/or tumor suppressor roles.5
Maternally expressed gene 3 (MEG3) is a myeloid-related lncRNA that has been shown to act as tumor suppressor in solid tumors.7, 8 Previous data have demonstrated that MEG3 is capable of increasing the protein level of the tumor suppressor p53 and enhancing p53 binding to its target promoters.9 In consequence, it is very evident that MEG3 can inhibit tumorigenesis through a p53-dependent manner. However, TP53 mutations can result in loss of wild-type tumor-suppressing p53 function in diverse types of human cancer, including AML.10 MEG3 has been implicated in the regulation of the RB pathway and thus of cell proliferation,11, 12 implying a possible role in a p53-independent pathway. The RB protein can repress gene transcription by directly binding to the transactivation domain of E2F in a complex on the promoters of the E2F target genes including DNMT3A.13 Accordingly, RB is involved in transcriptional regulation of DNMT3A gene in cancer cells,13 however, the detailed role of the RB-DNMT3A pathway remains to be fully elucidated.
The Wilms’ tumor 1 (WT1) gene, located on chromosome 11p13, encodes a transcriptional regulator that is capable of activating or repressing gene transcription.14, 15 The precise role of WT1 in hematopoiesis and its contribution to leukemogenesis are open for speculation. It is reported that WT1 mutations are associated with an extremely poor outcome,15 and they can lead to progression of leukemia by conferring drug resistance.16, 17 Dysregulation of the WT1 gene by epigenetic modifications or mutations might promote leukemic cell proliferation and impair differentiation.18, 19 However, the role of WT1 in regulating cancer-related gene expression remains largely unknown, especially whether WT1 controls MEG3 activity has yet to be explored.
The ten–eleven translocation (TET) family proteins TET1, TET2 and TET3 constitute a novel family of dioxygenases, whose functions are to demethylate DNA sequence by converting 5-methylcytosine to 5-hydroxymethylcytosine.20 Pathologically, TET2 is frequently mutated in hematopoietic malignancies of the myeloid lineage, particularly in AML.21 Most recently, WT1 is found to physically interact with TET2 and recruit it to the target genes of WT1,22 suggesting that TET2 may be involved in the transcriptional activity of WT1 in AML.
In this study, we demonstrate that inactivation of MEG3 promotes AML leukemogenesis in a p53-dependent as well as a p53-independent mode. Further analyses show that WT1 specifically binds to the MEG3 promoter and activates its transcription, and thereby, inducing a reduction of its downstream molecule MDM2. We further investigate the possible association between TET2 and WT1 in AML, and the results suggest that TET2 acts as a cofactor of WT1 to promote MEG3 transcription. Given the importance of the WT1-MEG3 axis in suppressing tumor growth, our findings suggest that targeting this axis may represent a novel approach for effective AML treatment.
Materials and methods
More detailed information on materials and methods can be found in the Supplementary Information.
Patient and tissue samples
Forty-two AML patient samples were analyzed at the time of diagnosis in this study. AML patients were classified according to French–American–British. All patients gave their written informed consent. The study has been approved by the Ethics Committee of the Institute. Mononuclear cells were isolated by density gradient centrifugation using Lymphoprep, and cryopreserved. In addition, 15 potential donors for allogeneic bone marrow transplantation were used as normal controls. Highly enriched human CD34+ cells (>90%) were derived from bone marrow mononuclear cells using MiniMACS (Miltenyi Biotech, Bergisch Gladbach, Germany) following the manufacturer’s instructions. Confirmation of bone marrow-derived CD34+ cells phenotype and purity was assessed by immunophenotypic analysis using CD34-FITC (BD Biosciences, San Diego, CA, USA) coupled with flow cytometry. All patients’ samples and controls were provided by the Second Affiliated Hospital of Dalian Medical University.
Statistical analysis
Student’s t-test (two-tailed), t-test with Welch's correction, Log-rank (Mantel–Cox) test, F-test were performed to analyze the data using GraphPad Prism 6.0 software (GraphPad software Inc., San Diego, CA, USA). P-values <0.05 were considered statistically significant. *P<0.05; **P<0.01; ***P<0.001.
Results
LncRNA MEG3 is downregulated in AML
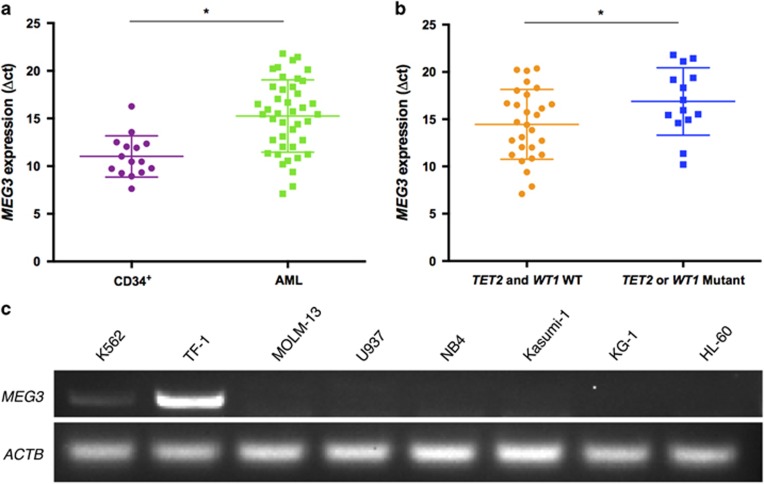
To gain insights into the patterns of activation of MEG3 gene in the AML pathogenesis, we examined its expression in normal CD34+ bone marrow cells and AML patient samples with different WT1 or TET2 mutation status. Given some variations were usually not regarded as true missense mutations (including P29R, I1762V, V218M, L1721W and H1778R) in TET2,23 only nonsense (4/42, 9.5%) and frameshift mutations (9/42, 21.4%) were used in our analysis. WT1 mutations (frameshift) were detected in two patients (2/42, 4.8% Supplementary Tables 1 and 2). Real-time quantitative PCR results indicated that MEG3 was robustly expressed in bone marrow CD34+ cells, significantly downregulated in all AML samples, particularly in the WT1- or TET2-mutant AML subtypes (Figures 1a and b).
Figure 1.
Expression of lncRNA MEG3 in AML patients and cell lines. (a) RT-qPCR analysis of MEG3 RNA expression in bone marrow of 42 samples from AML patients and CD34+ cells derived from 15 potential donors for allogeneic bone marrow transplantation. (b) RT-qPCR analysis of MEG3 RNA expression in bone marrow from AML patients with/without WT1 or TET2 mutations. (c) PCR analysis of MEG3 RNA expression in AML cell lines. *P<0.05. RT-qPCR, real-time quantitative PCR.
For subsequent cell model experiments, we also investigated the genetic backgrounds (Supplementary Tables 1 and 2) and p53 levels (Supplementary Figure 1A) of eight representative cell lines derived from AML (K562, TF-1, MOLM-13, U937, NB4, Kasumi-1, KG-1 and HL-60) and the correlations between WT1, TET2 mutation status/expression levels and MEG3 inactivation in these cells (Supplementary Figure 1B). As expected, MEG3 displayed negligible expression in the WT1-mutant AML cell lines (U937) relative to WT1- and TET2-wild-type AML cells (Figure 1c; Supplementary Figure 1B). Taken together, these findings demonstrate that dysregulation of MEG3 expression definitely correlates with WT1 or TET2 mutations, which thus probably plays an important role in AML pathogenesis.
MEG3 suppresses tumor growth through a p53-dependent pathway
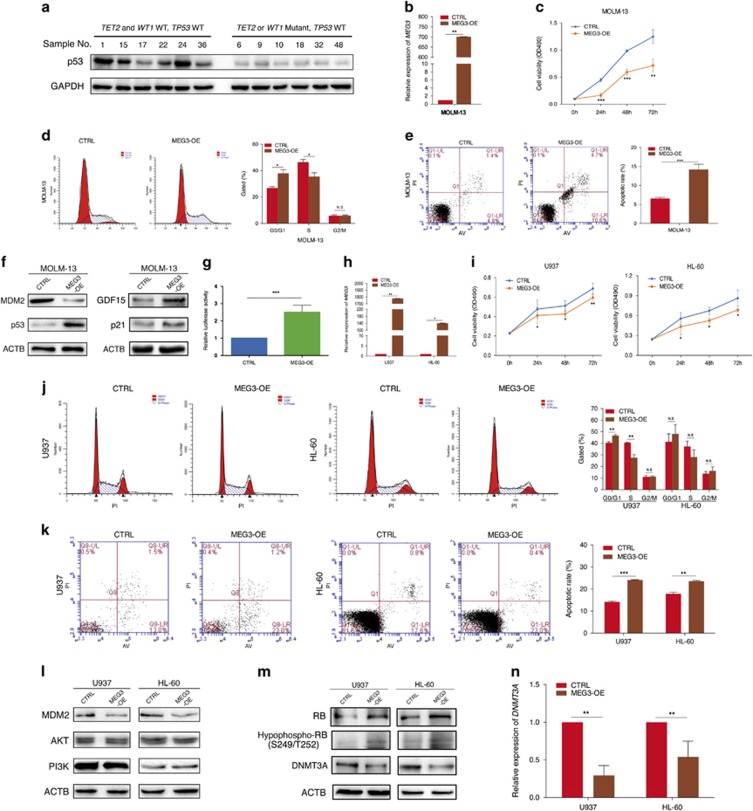
MEG3 has been widely demonstrated to exert its biological functions via p53 signaling.24, 25, 26 We reasoned that MEG3 should module AML pathogenesis to some extent in a similar way. On the basis of the data of genetic status of AML samples (Supplementary Tables 1 and 2), we discovered that p53 protein levels were dramatically reduced in samples with TET2 or WT1 mutations (Figure 2a). Then we examined the regulatory roles of MEG3 in cellular phenotypes and key signaling molecules in TP53wt AML cell line. Overexpression of MEG3 in MOLM-13 (WT1wt, TET2wtandTP53wt) cell line was found to significantly suppress cell proliferation and induce G0/G1 cell cycle arrest and apoptosis when compared with controls (Figures 2b–e). In addition, MEG3 was found to positively regulate the expression of p53 and p53 target genes (including p21 and GDF15, but has no effect on BAX, NOXA and PUMA), and negatively regulate MDM2 expression (Figure 2f; Supplementary Figure 2A). Furthermore, luciferase reporter assays confirmed the p53 transcriptional activity in the MOLM-13 cells (Figure 2g). Intriguingly, forced expression of MEG3 had no obvious effect on PI3K, AKT, RB and hypophosphorylated RB (S249/T252) expression levels (Supplementary Figure 2B).
Figure 2.
Effect of MEG3 on cell proliferation and apoptosis in vitro. (a) Western blotting analysis of p53 protein levels in samples with/without TET2 or WT1 mutations. (b) RT-qPCR analysis of MEG3 RNA expression in MOLM-13 cell line. (c) MTT assay of the proliferation of MOLM-13 cell line. (d) The bar chart represented the percentage of cells in G0/G1, S or G2/M phase, as indicated. (e) The apoptotic rates of cells were detected by flow cytometry. (f) Western blotting analysis of MDM2, p53, GDF15 and p21 after pCDH-MEG3 and control transfection. (g) Induction of TP53 promoter activity by MEG3 in MOLM-13 cell line. (h) RT-qPCR analysis of MEG3 RNA expression in U937 and HL-60 cell lines. (i) MTT assay of the proliferation of U937 and HL-60 cell lines. (j) The bar chart represented the percentage of cells in G0/G1, S or G2/M phase, as indicated. (k) The apoptotic rates of cells were detected by flow cytometry. (l, m) Western blotting analysis of MDM2, AKT, PI3K, RB, hypophospho-RB (S249/T252) and DNMT3A after pCDH-MEG3 and control transfection in U937 and HL-60 cell lines. (n) RT-qPCR analysis of DNMT3A mRNA expression in U937 and HL-60 cell lines. Results shown were from three independent experiments. ACTB or GAPDH protein was used as an internal control for western blotting analysis. *P<0.05; **P<0.01; ***P<0.001. NS, not significant; RT-qPCR, real-time quantitative PCR.
MEG3 suppresses tumor growth through a p53-independent pathway
Given that TP53 mutations or loss are frequently detected in patients with therapy-related AML or AML with complex karyotype,27, 28 we therefore next explored whether MEG3 plays a role as tumor suppressor in AML cell lines absent of TP53. As in the TP53wt AML cell line, stable overexpression of MEG3 in the two TP53mut AML cell lines U937 (WT1mut, TET2wt) and HL-60 (WT1wt, TET2wt) significantly suppressed cell proliferation, induced G0/G1 cell cycle arrest and apoptosis when compared to controls (Figures 2h–k). In contrast, knockdown of MEG3 in K562 (WT1wt, TET2wt) and TF-1 (WT1wt, TET2wt) cell lines resulted in a significant increase in cell proliferation, a failure to restrict cell cycle progression and induce apoptosis (Supplementary Figures 2C–F). These results suggest that MEG3 also suppresses tumor growth through a p53-independent pathway.
Subsequently, we interrogated the molecular mechanism by which MEG3 negatively regulates cell proliferation in a p53-independent manner by examining the expression of MDM2, PI3K, AKT and RB. Gain- or loss-of-function experiments in the above selected AML cell lines revealed that MEG3 negatively regulates MDM2 at the protein expression level (Figure 2l; Supplementary Figure 2G). Although the changes in AKT and PI3K expression were not obvious, RB and hypophosphorylated RB (S249/T252) protein levels correlated positively with MEG3 abundances (Figures 2l and m; Supplementary Figures 2G and H).
Extensive studies have suggested that RB functions as a classic tumor suppressor and inhibits cell proliferation mainly by negatively regulating the transcription of certain genes that are required for cell cycle progression. Depletion of MDM2 increases the total RB and the active form of hypophosphorylated RB at S249/T252.13 RB lacks a DNA-binding domain and is tethered to promoters through its interaction with other sequence-specific transcription factors such as members of the E2F family of proteins. RB binds to the transcription activation domain of E2F and blocks its activity. More importantly, the RB–E2F complex also actively represses transcription on promoters that contain E2F sites.29 Indeed, DNMT3A is one of genes whose promoters contain E2F sites. To examine whether MEG3 affects the expression of DNMT3A by promoting hypophosphorylation of RB at S249/T252, we determined the expression levels of DNMT3A in AML cells after silencing or overexpressing MEG3. The results showed that both mRNA and protein expression levels of DNMT3A negatively correlated with MEG3 abundances (Figures 2m and n; Supplementary Figures 2H and I). Taken together, these data demonstrate that DNMT3A is a MEG3 downstream gene and that MEG3 probably plays crucial roles in inhibiting tumor growth by downregulating DNMT3A via the MDM2/RB signaling pathway in AML cells in the absence of p53.
MEG3 inhibits AML leukemogenesis in vivo
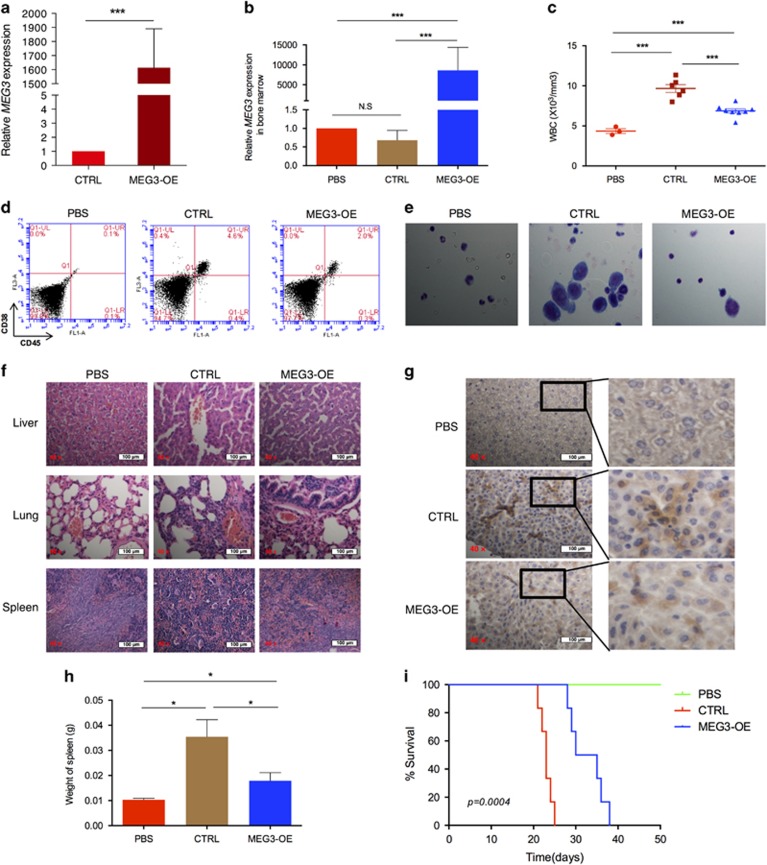
To investigate whether MEG3 affects AML leukemogenesis in vivo, two types of mouse models were established. We first examined the effects of MEG3 on the robust engraftment of AML in NOD-SCID mice. U937 cells transfected with pCDH-MEG3 (MEG3-OE) or control vector (CTRL; Figure 3a) were injected into 6–8-week-old mice. Four weeks after tail injection, the mice transplanted with U937-CTRL showed higher frequencies of serious paralysis and accumulation of urine in bladders than those with U937-MEG3-OE (Supplementary Figure 3A). After U937-MEG3-OE injection, MEG3 was highly expressed in the peripheral blood and bone marrow, and the number of white blood cells decreased dramatically (Figures 3b and c; Supplementary Figure 3B). Bone marrow assay showed that a significant decrease in CD45+/CD38+ cells in mice injected with U937-MEG3-OE, indicating that MEG3 inhibits the percentage of engraftment (Figure 3d). A large amount of monocelluar cells with megakaryocytes was observed in AML mice (Figure 3e). Leukemic infiltration was also observed in multiple organs after robust NOD-SCID engraftment of numerous AML cells (Figures 3f and g; Supplementary Figure 3C). We also detected the more serious splenomegaly in mice injected with U937-CTRL compared with U937-MEG3-OE (Figure 3h; Supplementary Figure 3D). Furthermore, our data reveal that MEG3 expression prolongs the survival of AML mice (Figure 3i).
Figure 3.
MEG3 inhibits AML leukemogenesis in vivo. (a) Relative MEG3 RNA expression in U937 cells stably transfected with CTRL and MEG3-OE. (b) Relative MEG3 RNA expression in bone marrow of PBS (n=3), U937-CTRL (n=6) and U937-MEG3-OE (n=8) at 4 weeks. (c) Peripheral blood WBC count at 4 weeks. (d) CD45+ and CD38+ immunophenotype of PBS, U937-CTRL and U937-MEG3-OE treated for 4 weeks. (e) Bone marrow morphology in PBS, U937-CTRL and U937-MEG3-OE. Scale bar, 10 μm. (f) H&E of liver, lung and spleen of PBS, U937-CTRL and U937-MEG3-OE at 4 weeks. Scale bars represented 100 μm. (g) IHC of liver with CD45 antibody of PBS, U937-CTRL and U937-MEG3-OE at 4 weeks. (h) The weight of spleen of mice treated with PBS, U937-CTRL and U937-MEG3-OE. (i) Kaplan–Meier survival curve of PBS, U937-CTRL and U937-MEG3-OE. *P<0.05; ***P<0.001. H&E, hematoxylin and eosin; IHC, immunohistochemistry; NS, not significant; PBS, phosphate-buffered saline; WBC, white blood cells.
We next injected U937 cells transfected with pCDH-MEG3 or control into female athymic nude (nu/nu) mice. Twenty-one days after the injection, we observed that the tumors formed in the U937-MEG3-OE group were substantially smaller compared to those from the U937-CTRL group (Supplementary Figures 3E and F). The average tumor weights were markedly lower in the U937-MEG3-OE group than the control group at the end of the experiment (Supplementary Figure 3G). As predicted, the expression levels of MEG3 in tumor tissues of the U937-MEG3-OE group were significantly higher than those of the control group (Supplementary Figure 3H). Moreover, hematoxylin and eosin and immunohistochemistry showed that proliferating cell nuclear antigen levels in tumor tissues of the U937-MEG3-OE group exhibited decreased positivity for proliferating cell nuclear antigen than in those of the U937-CTRL group (Supplementary Figures 3I and J). Taken together, these results suggest that MEG3 limits the proliferation of AML cells in vivo.
MEG3 is transcriptionally regulated by WT1 in AML
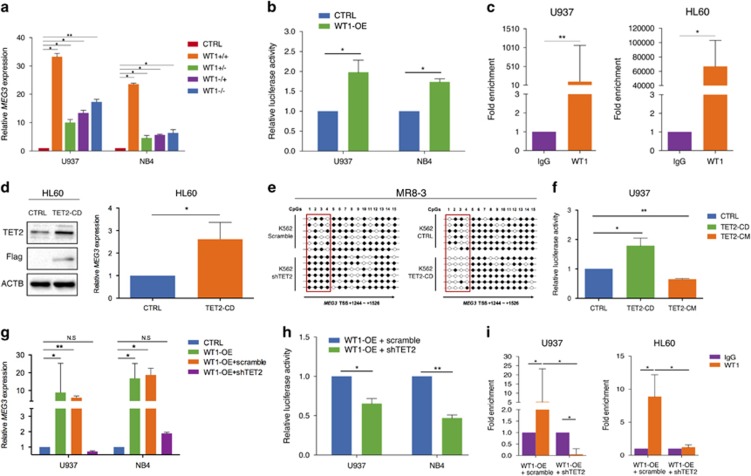
WT1 mutations or inactivation have been implicated in the leukemogenesis of AML.30 The correlation between WT1 and MEG3 expression in clinical samples and AML cell lines therefore led us to hypothesize that the WT1 transcription factor regulates MEG3 expression. To test this conjecture, we performed in silico analysis31 on the MEG3 promoter sequence and located a binding site of WT1 in this region (Supplementary Figure 4A). Relatively low-expression levels of WT1 were detected in U937 and NB4 cells (Supplementary Figures 4B and C), but a significant increase in MEG3 expression levels was observed following the transfection of wt WT1 into the two cell lines (Figure 4a). In contrast, WT1 depletion dramatically decreased MEG3 expression in KG-1 cells (WT1wt, TET2wt) (Supplementary Figures 4D and E). To examine whether WT1 can activate the transcription of MEG3, we constructed a WT1 luciferase reporter by inserting the sequence of MEG3 promoter region into the pGL4 vector. As shown in Figure 4b, compared with controls, the luciferase reporter assays revealed that overexpression of WT1 induced the transcriptional activation of MEG3, whereas knockdown of WT1 significantly reduced its transcription activity (Supplementary Figures 4F). To verify the prediction for the WT1 binding site, we performed chromatin immunoprecipitation assay in U937 and HL-60 cell lines using WT1-specific antibody after transfection of wt WT1 into the two cell lines. The chromatin immunoprecipitation results confirmed the direct binding of WT1 on the endogenous MEG3 promoter (Figure 4c). These findings provide compelling evidence that WT1 binds to MEG3 promoter and thus activates MEG3 transcription.
Figure 4.
WT1 interacts with TET2 to co-regulate MEG3 expression in AML cells. (a) RT-qPCR analysis of MEG3 RNA expression after transfected with four major WT1 splicing variants (+5/+KTS, +5/−KTS, −5/+KTS, −5/−KTS) in U937 and NB4 cell lines. (b) Induction of MEG3 promoter activity by WT1 in U937 and NB4 cell lines. (c) The WT1 binding at the promoter regions of MEG3 was assessed by ChIP analysis. (d) HL-60 cell line was transfected with a Flag-TET2CD construct, western blotting analysis of TET2 and Flag, RT-qPCR analysis of MEG3 RNA expression. (e) A representative methylation pattern of the CpGs in K562 cells after bisulfite treatment. Each line represented one PCR product, and six PCR products were shown for each sample. (f) Induction of MEG3 promoter activity by TET2CD or TET2CM in U937 cells. (g) WT1 was transiently overexpressed either singly or with shRNA against TET2 in U937 and NB4 cells, RT-qPCR was examined for the RNA expression of MEG3. (h) Induction of MEG3 promoter activity by WT1 either singly or with shRNA against TET2 in U937 and NB4 cell lines. (i) WT1 was transiently overexpressed either singly or shRNA against TET2 in U937 and HL-60 cells. The WT1 binding at the promoter regions of MEG3 was assessed by ChIP analysis. Representative images of three independent experiments were shown. ACTB protein was used as an internal control for western blotting analysis.*P<0.05; **P<0.01. •, methylated CpG; ○, unmethylated CpG; ChIP, chromatin immunoprecipitation; NS, not significant; RT-qPCR, real-time quantitative PCR; shRNA, short hairpin RNA.
MEG3 expression is associated with its first intron methylation mediated by TET2
Previous study has demonstrated that MEG3 gene expression is under epigenetic control, and aberrant CpG methylation occurs in AML patients.32 Downregulation of MEG3 in our TET2-mutant AML subtype samples implies that TET2 is likely to modulate MEG3 expression. We found that overexpression of TET2 CD domain (TET2CD) that contains both the Cys-rich and the DSBH domains could lead to the upregulation of MEG3 expression (Figure 4d), whereas depletion of TET2 resulted in the downregulation of MEG3 (Supplementary Figures 5A and B). Furthermore, we confirmed that the catalytic domain was required for the effect of TET2 on activating MEG3 expression, as the TET2 catalytic inactive mutant (TET2CM) could not upregulate MEG3 expression (Supplementary Figures 5C and D). DNA methylation is a common mechanism for promoter repression, and it also contributes to inactivation of enhancer elements usually existing in regions outside of promoters, particularly in introns. It is also noteworthy that expression of the MEG3-DLK1 locus may be regulated by differentially methylated regions (DMRs). Therefore, the methylation status of multiple cis elements (including promoter, enhancer, DMR and the first intron) of MEG3 gene was assayed to determine whether TET2 affects the expression of MEG3 through the epigenetic mechanism. Interestingly, after knockdown or overexpression of TET2 in K562 cells, no apparent changes in the DNA methylation levels were detected in the promoter, enhancer and DMR regions of MEG3, but the significant changes were observed in the first intron (Figure 4e; Supplementary Figures 6A and B). The same results were observed in HL-60 cell line (Supplementary Figure 6C). In agreement, the luciferase reporter assays showed that overexpression of TET2CD, but not the TET2CM, induced the transcriptional activity of the MEG3 promoter in U937 cells (Figure 4f). Collectively, these results suggest that TET2 can activate MEG3 transcription, which is associated with the ability of TET2 to mediate the MEG3 first intron DNA methylation.
The transactivation activity of WT1 on MEG3 is modulated by TET2 in AML
As described above, both WT1 and TET2 affect the transcriptional activity of MEG3. Both of them are found to be mutated, which are believed to play key roles in AML leukemogenesis. However, their mutations are mutually exclusive, indicating they may function in the same pathway to suppress AML. Moreover, WT1 has recently been demonstrated to physically bind and recruit TET2 to its target genes.22 This led us to propose that TET2 may serve as a cofactor of WT1 in its regulation of MEG3 transcription, as TET2 lacks a DNA-binding domain. To validate this idea, we first determined the WT1–TET2 association by coupled immunoprecipitation and western blotting (IP-western) in NB4 and 293 T-cell lines. As shown in Supplementary Figure 7A, we demonstrated that the WT1–TET2 interaction could readily be detected in the two cell lines. In addition, WT1 was transiently expressed either alone or with shTET2 in U937 and NB4 cell lines. The cells only transfected with the WT1 vector alone expressed higher MEG3 levels than those transfected with both the WT1 and shTET2 vectors (Figure 4g; Supplementary Figures 7B and C). To elucidate the influence of the WT1 and TET2 interaction on the transcriptional activity of MEG3, the luciferase assay was carried out to test the activity of MEG3 promoter. Figure 4h showed that cells transfected only with the WT1 vector exhibited higher luciferase activity than those transfected simultaneously with the WT1 and shTET2 vectors. Moreover, silencing of WT1 expression with short hairpin RNA together with transfection with TET2CD vector in KG-1 cells resulted in higher luciferase activities compared with WT1 short hairpin RNA vector only (Supplementary Figure 7D). Subsequently, chromatin immunoprecipitation and real-time quantitative PCR assays were used to further explore whether regulation of MEG3 promoter by WT1 requires TET2. Consistently, WT1 was found to be less enriched on the MEG3 promoter in cells simultaneously transfected with WT1 and shTET2 vectors than those transfected only with WT1 vector (Figure 4i). Taken together, these results suggest that TET2 serves as a cofactor for WT1 in the regulation of MEG3 transcriptional activation in AML cells.
Discussion
Mounting evidence has supported the notion that lncRNAs are involved in tumorigenesis.7 LncRNA MEG3, silenced in various types of cancer, is commonly regarded as a tumor suppressor. Although MEG3 low expression and promoter hypermethylation have been found to be markers of poor prognosis in AML patients,33 the functional consequence of MEG3 downregulation and underlying mechanisms involved have remained elusive. In this study, we show that MEG3 suppresses AML leukemogenesis through both p53-dependent and p53-independent pathways. More importantly, our work reveals MEG3 as a novel target of WT1 and that TET2 interacts with WT1 to co-regulate MEG3 transcriptional activation.
Our works show that ectopic expression of MEG3 suppresses cell proliferation and induces G0/G1 cell cycle arrest in AML, consistent with previous observations in other solid tumors.24, 25, 26, 34 The inhibitory roles of MEG3 in AML leukemogenesis are further verified in two independent in vivo mouse models. It is well-known that MEG3 protects the p53 from degradation, and thus inhibits tumor growth in a p53-dependent fashion.35 Our experiments extend our understanding of the molecular events leading to AML leukemogenesis, where decreased expression of MEG3 significantly restricts the increase of the tumor suppressor p53 protein.34, 35, 36 These observations often further support for a common tumor suppressive mechanism, whereby MEG3 functions as a tumor suppressor, at least partly via the activation of p53 in multiple cancers.
MEG3 is capable of inducing MDM2 degradation, thereby stabilizing the p53 protein. However, p53 is often lost or mutated in patients including therapy-related AML or AML with complex karyotype.27, 28 It has been demonstrated that MDM2 contributes to tumor initiation and progression even when p53 is no longer active.37 Our data confirm that MEG3 inhibits AML cell growth under genetic backgrounds of TP53 mutation, deletion or depletion, by decreasing MDM2 protein level. We discover that MDM2 functionally interacts with its target RB in AML. MEG3 promotes MDM2 degradation, and eventually increases the total RB level and the active form of hypophosphorylated RB.38, 39 After activation, RB interacts with transcription factor E2F to negatively regulate expression of their target genes. We uncover that DNMT3A, a very important target of E2F complex, is markedly downregulated by MEG3. Given the crucial role of DNMT3A in AML, it is most probably that MEG3 inhibits tumor growth through RB-DNMT3A pathway. Apart from the p53-dependent pathway, our results unveil a novel p53-independent mechanism by which MEG3 exerts its tumor-suppressive function in AML.
Currently, the observations that WT1 is expressed in the majority of AML cases, and that it is mutated in a proportion of AMLs have led to a number of studies to decipher the mechanistic role of WT1.30 The WT1 mutations clustering in exons 7 and 9 are correlated with lower complete remission rates, higher relapse rates, shorter disease-free survival and overall survival.15, 19 The specific genes that are regulated by WT1 and become deregulated by WT1 mutation remain an area of intense investigation.30 Our findings that WT1 positively modulates the transcription of MEG3 provide an explanation for the mechanism of leukemogenesis in WT1-mutant AML.
It is long known that promoter hypermethylation is a common mechanism contributing to transcriptional repression.40 Moreover, DNA methylation has been found to correlate negatively with enhancer activity and gene expression, possibly by interfering with transcription factor binding.41 These functionally relevant binding sites for transcription factors, however, usually exist in regions outside of promoters, particularly in introns.42 Gene expression is usually regulated by the methylation status of DNA cis elements such as promoters, enhancers and introns.43 It is also noteworthy that expression of the MEG3-DLK1 locus may be regulated by DMRs.44, 45 Our results show that the promoter, enhancer, DMR and the first intron of MEG3 display aberrant methylation, consistent with others findings.32, 46 However, only the first intron displays significant change in methylation levels upon TET2 expression, meaning that the methylation of this region might be in a dynamic state. Differential methylation at intronic enhancers has been previously reported to affect gene expression.47, 48, 49 Our results show that the induction of MEG3 expression by TET2 is associated with methylation of its first intron. Collectively, these results suggest the presence of putative regulatory elements in the first intron of MEG3 and MEG3 expression may thus be repressed by hypermethylation as a result of lack of functional TET2, contributing to MEG3 epigenetic silencing occurred specifically in the TET2-mutant AML subtype.
TET2 is able to inhibit leukemogenesis, but it is frequently mutated in AML. TET2 is found to be mutated in a mutually exclusive manner with WT1, indicating that they both may suppress AML through the same pathway. Most recently, WT1 has been demonstrated to physically interact with TET2 and recruit it to the target genes of WT1,22 implying that TET2 may be involved in modulating the transcriptional activity of WT1. As expected, our data confirm that WT1 interacts directly with TET2, and we further demonstrate that TET2 serves as a cofactor of WT1 to activate MEG3 expression. Not surprisingly, the mutation of TET2 substantially diminishes the transaction activity of WT1 on MEG3. Taken together, these results suggest there exists a linear TET2-WT1-MEG3 axis in the suppression of AML leukemogenesis, and therefore dysfunction of this axis; such mutations to either TET2 or WT1 may be a key contributor to AML. Evidently, our study uncovers a novel mechanism by which WT1–TET2 plays a critical role through the regulation of MEG3 gene expression.
In summary, our work demonstrates that the loss of lncRNA MEG3 leads to AML leukemogenesis, indicating that its overexpression can suppress AML development via both p53-dependent and p53-independent pathways. Furthermore, the discovery that WT1 cooperates with TET2 to upregulate MEG3 expression places TET2-WT1-MEG3 signaling axis as a central tumor suppressive pathway in AML, therefore emphasizing the potentials of this axis in AML diagnosis and therapy.
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China (Nos. 81372713, 81672497, 81570124, 81270606, 81402048 and 81372714), Liaoning Provincial Natural Science Foundation of China (No. 2014023010), the Reformation Project in the Key Clinical Departments of Provincial Hospitals on Construction of Diagnosis and Treatment Capacity in Liaoning Province (LNCCC-A02-2015), Medical Research Council UK (No. MR/N012097/1), Cancer Research UK (No. A12011) and Breast Cancer Now (Nos. 2012MayPR070, 2012NovPhD016). We would like to thank Dr Wei Cheng (ICSC Core Facility, Dalian Medical University) for the material support.
Author contributions
Conception and design by YL, JL, YY, JF, XW, QL, JY and BJ; development of methodology by YL, JL and JF; acquisition of data by YL, JL, YY, JF, YH, SH, LY, JX, DH, BM, DZ, YW, YZ, KY, BZ, PC; analysis and interpretation of data (for example, statistical analysis, flow cytometry interpretation) by YL, JL, JF; writing, review and/or revision of the manuscript by YL, JL, YY, JF, XW, EW-FL, QL, JY and BJ; administrative, technical or material support: BJ, JY, QL and XW; study supervision: BJ, JY, QL and XW.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
The authors declare no conflict of interest.
Supplementary Material
References
- Diaz-Beya M, Brunet S, Nomdedeu J, Pratcorona M, Cordeiro A, Gallardo D et al. The lincRNA HOTAIRM1, located in the HOXA genomic region, is expressed in acute myeloid leukemia, impacts prognosis in patients in the intermediate-risk cytogenetic category, and is associated with a distinctive microRNA signature. Oncotarget 2015; 6: 31613–31627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estey E, Dohner H. Acute myeloid leukaemia. Lancet 2006; 368: 1894–1907. [DOI] [PubMed] [Google Scholar]
- Hao SF, Shao ZH. HOTAIR is upregulated in acute myeloid leukemia and that indicates a poor prognosis. Int J Clin Exp Pathol 2015; 8: 7223–7228. [PMC free article] [PubMed] [Google Scholar]
- Volders PJ, Verheggen K, Menschaert G, Vandepoele K, Martens L, Vandesompele J et al. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res 2015; 43 (Database issue): D174–D180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjari M, Khoshnevisan A, Shin YK. Long non-coding RNAs in hematologic malignancies: road to translational research. Front Genet 2013; 4: 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 2015; 47: 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Malave NI, Rao DS. Long noncoding RNAs in hematopoietic malignancies. Brief Funct Genomics 2016; 15: 227–238. [DOI] [PubMed] [Google Scholar]
- Zhang X, Rice K, Wang Y, Chen W, Zhong Y, Nakayama Y et al. Maternally expressed gene 3 (MEG3 noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology 2010; 151: 939–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J et al. Long noncoding RNA MEG3 interacts with p53 protein and regulates partial p53 target genes in hepatoma cells. PLoS One 2015; 10: e0139790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol 2013; 15: 2–8. [DOI] [PubMed] [Google Scholar]
- Benetatos L, Vartholomatos G, Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int J Cancer 2011; 129: 773–779. [DOI] [PubMed] [Google Scholar]
- Zhang X, Gejman R, Mahta A, Zhong Y, Rice KA, Zhou Y et al. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res 2010; 70: 2350–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YA, Lin RK, Tsai YT, Hsu HS, Yang YC, Chen CY et al. MDM2 overexpression deregulates the transcriptional control of RB/E2F leading to DNA methyltransferase 3A overexpression in lung cancer. Clin Cancer Res 2012; 18: 4325–4333. [DOI] [PubMed] [Google Scholar]
- Call KM, Glaser T, Ito CY, Buckler AJ, Pelletier J, Haber DA et al. Isolation and characterization of a zinc finger polypeptide gene at the human chromosome 11 Wilms' tumor locus. Cell 1990; 60: 509–520. [DOI] [PubMed] [Google Scholar]
- Paschka P, Marcucci G, Ruppert AS, Whitman SP, Mrozek K, Maharry K et al. Wilms' tumor 1 gene mutations independently predict poor outcome in adults with cytogenetically normal acute myeloid leukemia: a cancer and leukemia group B study. J Clin Oncol 2008; 26: 4595–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyvold CG, Stentoft J, Braendstrup K, Melsvik D, Moestrup SK, Juhl-Christensen C et al. Wilms' tumor 1 mutation accumulated during therapy in acute myeloid leukemia: biological and clinical implications. Leukemia 2006; 20: 2051–2054. [DOI] [PubMed] [Google Scholar]
- Summers K, Stevens J, Kakkas I, Smith M, Smith LL, Macdougall F et al. Wilms' tumour 1 mutations are associated with FLT3-ITD and failure of standard induction chemotherapy in patients with normal karyotype AML. Leukemia 2007; 21: 550–551, author reply 552. [DOI] [PubMed] [Google Scholar]
- Yang L, Han Y, Suarez Saiz F, Minden MD. A tumor suppressor and oncogene: the WT1 story. Leukemia 2007; 21: 868–876. [DOI] [PubMed] [Google Scholar]
- Krauth MT, Alpermann T, Bacher U, Eder C, Dicker F, Ulke M et al. WT1 mutations are secondary events in AML, show varying frequencies and impact on prognosis between genetic subgroups. Leukemia 2015; 29: 660–667. [DOI] [PubMed] [Google Scholar]
- Pan F, Weeks O, Yang FC, Xu M. The TET2 interactors and their links to hematological malignancies. IUBMB Life 2015; 67: 438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A et al. Mutation in TET2 in myeloid cancers. N Engl J Med 2009; 360: 2289–2301. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xiao M, Chen X, Chen L, Xu Y, Lv L et al. WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol Cell 2015; 57: 662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou WC, Chou SC, Liu CY, Chen CY, Hou HA, Kuo YY et al. TET2 mutation is an unfavorable prognostic factor in acute myeloid leukemia patients with intermediate-risk cytogenetics. Blood 2011; 118: 3803–3810. [DOI] [PubMed] [Google Scholar]
- Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ et al. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer 2013; 13: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia LF, Wei SB, Gan YH, Guo Y, Gong K, Mitchelson K et al. Expression, regulation and roles of miR-26a and MEG3 in tongue squamous cell carcinoma. Int J Cancer 2014; 135: 2282–2293. [DOI] [PubMed] [Google Scholar]
- Chunharojrith P, Nakayama Y, Jiang X, Kery RE, Ma J, De La Hoz Ulloa CS et al. Tumor suppression by MEG3 lncRNA in a human pituitary tumor derived cell line. Mol Cell Endocrinol 2015; 416: 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen-Bjergaard J, Christiansen DH, Desta F, Andersen MK. Alternative genetic pathways and cooperating genetic abnormalities in the pathogenesis of therapy-related myelodysplasia and acute myeloid leukemia. Leukemia 2006; 20: 1943–1949. [DOI] [PubMed] [Google Scholar]
- Haferlach C, Dicker F, Herholz H, Schnittger S, Kern W, Haferlach T. Mutations of the TP53 gene in acute myeloid leukemia are strongly associated with a complex aberrant karyotype. Leukemia 2008; 22: 1539–1541. [DOI] [PubMed] [Google Scholar]
- Zhang HS, Dean DC. Rb-mediated chromatin structure regulation and transcriptional repression. Oncogene 2001; 20: 3134–3138. [DOI] [PubMed] [Google Scholar]
- Rampal R, Figueroa ME. Wilms tumor 1 mutations in the pathogenesis of acute myeloid leukemia. Haematologica 2016; 101: 672–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 2014; 158: 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury H, Suarez-Saiz F, Wu S, Minden MD. An upstream insulator regulates DLK1 imprinting in AML. Blood 2010; 115: 2260–2263. [DOI] [PubMed] [Google Scholar]
- Alvarez-Dominguez JR, Hu W, Gromatzky AA, Lodish HF. Long noncoding RNAs during normal and malignant hematopoiesis. Int J Hematol 2014; 99: 531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Ren Z, Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem 2012; 113: 1868–1874. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem 2007; 282: 24731–24742. [DOI] [PubMed] [Google Scholar]
- Sun M, Xia R, Jin F, Xu T, Liu Z, De W et al. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol 2014; 35: 1065–1073. [DOI] [PubMed] [Google Scholar]
- Wienken M, Dickmanns A, Nemajerova A, Kramer D, Najafova Z, Weiss M et al. MDM2 associates with polycomb repressor complex 2 and enhances stemness-promoting chromatin modifications independent of p53. Mol Cell 2016; 61: 68–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li M, Wang H, Agrawal S, Zhang R. Antisense therapy targeting MDM2 oncogene in prostate cancer: effects on proliferation, apoptosis, multiple gene expression, and chemotherapy. Proc Natl Acad Sci USA 2003; 100: 11636–11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Sheng G, Warner BW. Epidermal growth factor-induced rapid retinoblastoma phosphorylation at Ser780 and Ser795 is mediated by ERK1/2 in small intestine epithelial cells. J Biol Chem 2005; 280: 35992–35998. [DOI] [PubMed] [Google Scholar]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 2003; 349: 2042–2054. [DOI] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP. Methylation-induced repression–belts, braces, and chromatin. Cell 1999; 99: 451–454. [DOI] [PubMed] [Google Scholar]
- Farnham PJ. Insights from genomic profiling of transcription factors. Nat Rev Genet 2009; 10: 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou S, Lee HM, Qin H, Li JW, Gao Z, Liu X et al. Whole-genome bisulfite sequencing of multiple individuals reveals complementary roles of promoter and gene body methylation in transcriptional regulation. Genome Biol 2014; 15: 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astuti D, Latif F, Wagner K, Gentle D, Cooper WN, Catchpoole D et al. Epigenetic alteration at the DLK1-GTL2 imprinted domain in human neoplasia: analysis of neuroblastoma, phaeochromocytoma and Wilms' tumour. Br J Cancer 2005; 92: 1574–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami M, O'Sullivan MJ, Green AJ, Watabe Y, Arisaka O, Masawa N et al. The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genet 2010; 6: e1000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetatos L, Hatzimichael E, Dasoula A, Dranitsaris G, Tsiara S, Syrrou M et al. CpG methylation analysis of the MEG3 and SNRPN imprinted genes in acute myeloid leukemia and myelodysplastic syndromes. Leuk Res 2010; 34: 148–153. [DOI] [PubMed] [Google Scholar]
- Unoki M, Nakamura Y. Methylation at CpG islands in intron 1 of EGR2 confers enhancer-like activity. FEBS Lett 2003; 554: 67–72. [DOI] [PubMed] [Google Scholar]
- Hoivik EA, Bjanesoy TE, Mai O, Okamoto S, Minokoshi Y, Shima Y et al. DNA methylation of intronic enhancers directs tissue-specific expression of steroidogenic factor 1/adrenal 4 binding protein (SF-1/Ad4BP). Endocrinology 2011; 152: 2100–2112. [DOI] [PubMed] [Google Scholar]
- Verhulst EC, Mateman AC, Zwier MV, Caro SP, Verhoeven KJ, van Oers K. Evidence from pyrosequencing indicates that natural variation in animal personality is associated with DRD4 DNA methylation. Mol Ecol 2016; 25: 1801–1811. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.