Abstract
The tumor suppressor p53 is a key regulator of apoptosis induced by various cellular stresses. p53 can induce apoptosis by two mechanisms. First, p53 acts as a transcription factor inducing and repressing pro-apoptotic and anti-apoptotic targets genes, respectively. Second, p53 is able to translocate to the mitochondria, where it interacts with BCL-2 family members to induce membrane permeabilization and cytochrome c release. p53 transcriptional activity is regulated by a set of post-translational modifications that have been well documented. However, how these modifications impact the direct mitochondrial pathway of death remain poorly understood. In this study, we focused on the role of serine 392 phosphorylation in the control of p53-dependent apoptosis. We used CRISPR/Cas9 genome editing to substitute serine 392 by a non-phosphorylatable alanine in HCT-116 colon carcinoma cells. The S392A mutant displayed normal transcriptional activity following genotoxic stress, but markedly impaired ability to localize to mitochondria. The decreased mitochondrial localization of the S392A mutant correlated with a lower ability to induce apoptosis. Confirmatory observations were made following enforced expression of the S392A p53 mutant or a phospho-mimetic S392E mutant in H1299 lung carcinoma cells. Our observations support the premise that serine 392 phosphorylation of p53 influences its mitochondrial translocation and transcription-independent apoptotic function.
The tumor suppressor p53 occupies a nodal position in various cellular pathways that preserve the integrity of the genome in response to a wide variety of stress. Linked to its ability to induce cell cycle arrest and cell death by apoptosis, the TP53 gene is found mutated in over 50% of human cancers.1 Regulation of p53 function is complex, involving interactions with numerous proteins, localization in different cell compartments and multiple post-translational modifications (PTMs) of several types, including phosphorylations.2
The C-terminal phosphorylations of p53 are less studied than those of its N-terminal region. p53 has at least four C-terminal serines (S315, S366, S378 and S392) that can be phosphorylated.3, 4, 5 Among these, S392 is highly conserved in vertebrates. The kinase(s) responsible for S392 phosphorylation still remain(s) elusive, although several candidates have been proposed.6, 7, 8, 9, 10, 11, 12 The effects of this phosphorylation on p53 activity also remain a matter of debate. S392 phosphorylation was described to increase the stability of the tumor suppressor, to enhance tetramer formation and to regulate p53 DNA-binding affinity for specific and non-specific sites.13, 14, 15 Evidences also exist that S392 phosphorylation modulates the capacity of p53 to suppress cell growth and substitution of mouse S389 to alanine promoted UV-induced skin tumors.16, 17
To study the impact of S392 phosphorylation on p53-dependent apoptosis, we used the CRISPR/Cas9n technology to substitute S392 by a non-phosphorylatable alanine in HCT-116 colon carcinoma cells. We failed to identify a marked difference in p53 stabilization or transcriptional activity between cells expressing the S392A mutant and cells harboring wt p53. Besides its transcriptional activity, p53 has a transcription-independent function in induction of cell death, based on its ability to regulate outer mitochondrial membrane (OMM) permeabilization through interactions with BCL-2 family members such as BAX, BAK, BCL-2, and BCL-XL.18, 19, 20, 21, 22, 23, 24 Notably, direct interaction of p53 with BAX and BAK triggers their oligomerization, leading to OMM permeabilization and release of apoptotic effectors such as cytochrome c into the cytosol.22, 23, 24 We found that the p53 form phosphorylated on S392 (P-S392 p53) is enriched in the mitochondrial fraction of cells exposed to a genotoxic stress. Moreover, the p53 S392A mutant displays a markedly impaired ability to translocate to the OMM and a lower ability to induce apoptosis.
Results
Generation of a HCT-116 cell line expressing the p53 S392A mutant by CRISPR/Cas9n genome edition
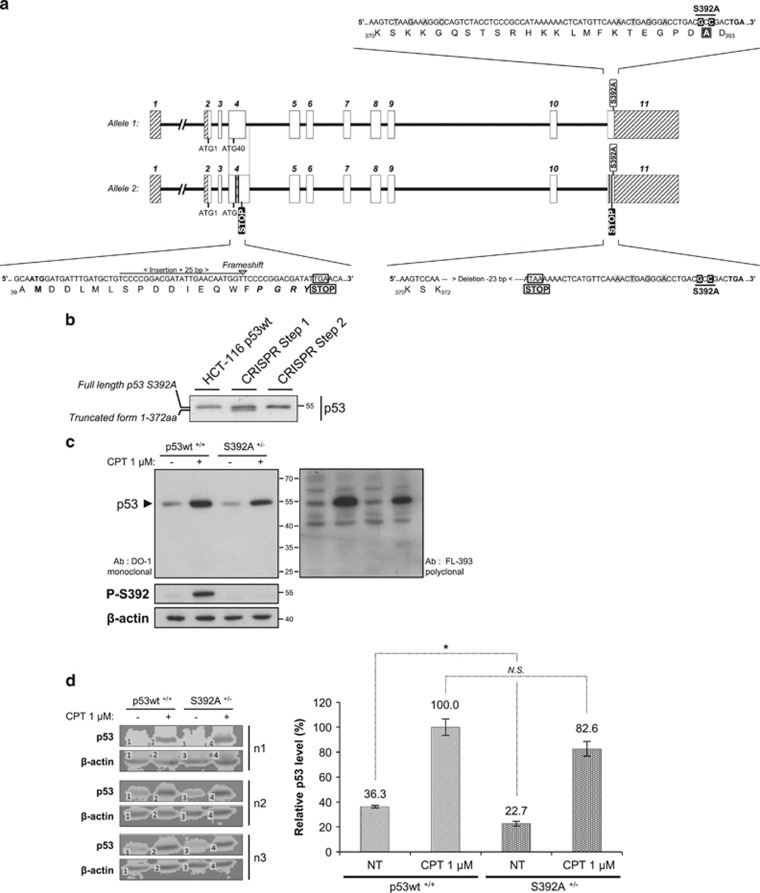
Alignment of p53 aa sequences of different vertebrate species highlights the presence of three highly conserved serines (S15, S215, and S392) (Supplementary Figure S1). The impact of S392 phosphorylation on p53 functions remains a matter of debate. To determine if S392 phosphorylation modulates p53 activity, we substituted S392 by an alanine in HCT-116 cells by CRISPR/Cas9 genome edition (Figure 1 and Supplementary Figure S2 for details).25, 26, 27 While no clone homozygous for the desired mutation could be identified, we isolated one clone with a S392A allele and an allele with a 23 bp deletion, leading to formation of a stop codon at position 373 (Figure 1a). Western blot analysis of this clone detected both full-length p53 and a shorter form, suggesting that the truncated allele (p53 1–372) was expressed (Figure 1b). As this could interfere with our observations, we performed a second round of CRISPR/Cas9n to knockout the truncated allele using sgRNAs guides targeting codon 40 corresponding to an alternative ATG start codon (Figure 1a). This procedure allowed to isolate a clone with a p53 S392A allele and an allele with a STOP codon in exon 4, expected to be a null allele (Figures 1a and b). This clone was called HCT-116 p53 S392A. We also isolated a clone with an homozygous 8 bp deletion, leading to a premature STOP codon. This clone expressed no detectable p53 protein and was considered to be p53 knockout (Supplementary Figure S3).
Figure 1.
Generation of a HCT-116 p53 S392A cell line by CRISPR/Cas9 genome edition. (a) The figure displays a schematic representation of the two p53 alleles and the modifications introduced. The TP53 gene is composed of 11 exons, the full-length p53 protein being encoded by exons 2–11. S392 is located in the last exon. The repair template (RT) was correctly introduced in the first allele (allele 1). This allele is functional and allows the expression of p53 S392A. Allele 2 is not functional. In this second allele, partial insertion of the RT with a deletion of 23 bases is observed (exon 11). This indel mutation generates a stop codon at position 373. Allele 2 also displays an insertion of 25 bp in exon 4 (after ATG40) that generates a frameshift and formation of a stop codon. Bases in grey represent the silent mutations introduced (near and in the PAM sequence areas recognized by the two sgRNAs guides). These mutations prevent that the ssODN becomes a substrate for the Cas9n enzyme. (b) Western blot analysis of p53 expression in HCT-116 p53 wt cells and after each CRISPR step. Western blot analysis after step 1 (targeting of S392) reveals two bands. They correspond to full-length p53 S392A (allele 1) and the p53 1–372 truncated form (allele 2). After CRISPR step 2 (targeting of ATG40), allele 2 is no more functional as indicated by loss of expression of the truncated 1–372 form. (c) HCT-116 cells expressing p53 wt and the S392A mutant (CRISPR/Cas9n) were treated with 1 μM CPT for 24 h. Total protein extracts were then prepared and p53 levels were assessed with the anti-p53 DO-1 monoclonal and FL-393 polyclonal antibodies. S392 phosphorylation was also measured. (d) Semi-quantitative western blot analysis for the expression of total p53 using the ScanLater western blot technology (Molecular Devices). HCT-116 p53 wt and S392A cells were treated or not (NT, no treatment) with 1 μM CPT for 24 h. Total protein extracts were prepared and detection of p53 was assessed with p53 DO-1 antibody. For the quantification of p53 levels, the signal for p53 was reported to that of β-actin. The data are means±SD from three independent experiments. Student's t-test was used to calculate significance (*P<0.05; N.S., non-significant)
To characterize the cell line, we treated p53 S392A+/− and wt+/+ cells with 1 μM camptothecin (CPT) for 24 h. Western blot analysis with monoclonal DO-1 and polyclonal FL-393 antibodies revealed that both p53 wt and S392A are induced by CPT. Moreover, no aberrant band was detected in HCT-116 S392A cells using the polyclonal antibody (Figure 1c). Since S392A cells have only one functional allele, we compared its p53 protein level with that of wt cells. We performed a semi-quantitative western blot analysis of cells treated or not with CPT. The data indicate a significant difference in basal p53 level between S392A+/− and p53+/+ cells. However, in presence of a stress, only a non-significant difference of 17% was recorded, suggesting a minimal impact of one allele knockout on p53 induction in response to a genotoxic stress (Figure 1d). Additional data on p53 mRNA level in HCT-116 KO, wt and S392A cells are displayed in Supplementary Figure S4.
HCT-116 p53 S392A cells display reduced apoptosis
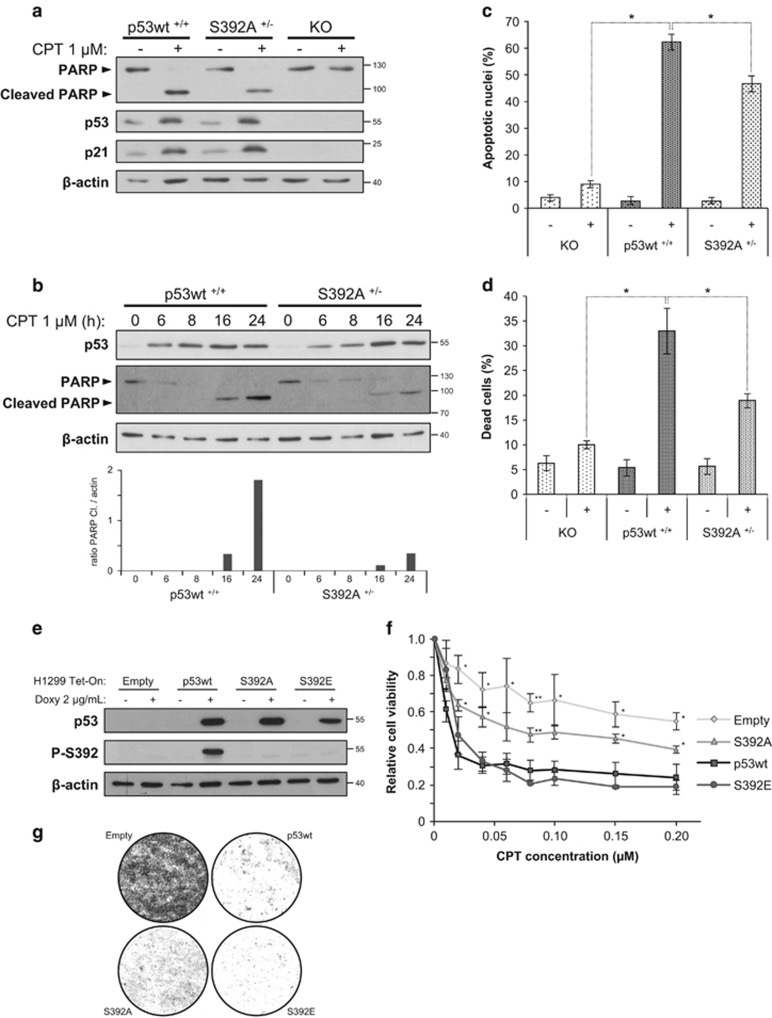
We determined if S392A p53 mutation altered induction of apoptosis after CPT exposure. HCT-116 p53 KO, wt and S392A cells were treated with 1 μM CPT for 24 h. Presence of the caspase-cleaved 89 kDa PARP fragment and expression of p21waf1 were assessed by western blot. As expected, KO cells failed to induce p21waf1 expression and PARP cleavage in response to CPT. Moreover, p21waf1 protein expression was induced similarly in p53 wt and S392 A cells, while a decreased PARP cleavage was observed in cells expressing p53 S392A (Figure 2a). A kinetic of PARP cleavage in presence of CPT was next performed, indicating a marked reduction of the level of cleaved PARP at 16 and 24 h in S392A cells (Figure 2b). We next monitored DNA fragmentation by flow cytometry analysis of the sub-G1 peak.28 We found that cells expressing S392A p53 developed significantly less apoptosis than their wt counterparts after treatment with CPT, while KO cells were markedly deficient for apoptosis induction (Figure 2c). The impact of the S392A mutation on cell viability was assessed using the trypan blue dye exclusion method. After a 24 h treatment with CPT, we recorded an approximatively two-fold reduction of cell death in S392A cells as compared to wt cells (Figure 2d).
Figure 2.
Failure to phosphorylate S392 is correlated with a decreased apoptosis in response to a genotoxic stress. (a) HCT-116 cells expressing p53 wt, the S392A mutant as well as p53 KO cells were treated or not with 1 μM CPT for 24 h. Total protein extracts were then prepared. PARP cleavage, p53, and p21Waf-1 expression were detected by western blot analysis. (b) HCT-116 cells expressing p53 wt and S392A were treated with 1 μM CPT for 0, 6, 8, 16, or 24 h. Then, total protein extracts were collected and measurement of apoptosis induction was assessed by quantification of PARP cleavage. The graph indicates the relative PARP cleavage in the different extracts based on a densitometry analysis of the band corresponding to cleaved PARP (normalization to β-actin). (c) HCT-116 cells were treated for 24 h with 1 μM CPT. After treatment, cells were collected, fixed and stained with propidium iodide. The percentage of apoptotic nuclei (sub-G1 population) was evaluated using flow cytometry. Results show the percentage of apoptotic nuclei in each experimental condition and are the means±S.D. from three independent experiments. Statistical analysis was performed using Student's t-test with Bonferroni correction (Bonferroni-corrected, *P<0.0253; CPT-treated KO versus p53 wt, P<0.001; CPT-treated S392A versus p53 wt P=0.003). (d) HCT-116 cells were treated with 1 μM CPT. After 24 h of treatment, viability was determined by the trypan blue dye exclusion method. Results show the percentage of dead cells in each experimental condition and are the means±S.D. from three independent experiments. Statistical analysis was performed using Student's t-test with Bonferroni correction (Bonferroni-corrected, *P<0.0253; KO versus p53 wt, P=0.0111; S392A versus p53 wt P=0.0251). (e) Detection of total p53 and its form phosphorylated on S392 in inducible (Tet-On) H1299 clones expressing p53 wt, S392A and S392E as well as cells transfected with the empty vector. The expression of p53 was induced with 2 μg/ml doxycycline for 24 h. (f) H1299 Tet-On clones expressing p53 wt, S392A and S392E or transfected with the empty vector were incubated for 72 h with 2 μg/ml doxycycline and increasing concentrations of CPT (from 0 to 0.2 μM). After treatments, relative cell viability was assessed by a MTS assay. The data are means±S.D. from three independent experiments. The one-way ANOVA test was used to compare independent groups (for each CPT concentration). When the ANOVA was significant, a Dunnett's test was used to compare a group of interest with p53 wt as reference (*P<0.05, **P<0.01). Exact P-values and details on the test are available in Supplementary Table S1. (g) Colony formation assay of H1299 Tet-On clones expressing p53 wt, S392A and S392E or transfected with the empty vector were treated simultaneously with 2 μg/ml of doxycycline and 1 μM of CPT for 3 days
We next investigated the consequences of the S392A mutation in p53-null H1299 cells. We developed clones with a tetracycline-inducible expression of wt p53, S392A p53, or S392E p53, in which S392 was replaced by a glutamic acid, corresponding to a phospho-mimetic mutation (Figure 2e). Expression of the different p53 forms was induced by doxycycline and cells were exposed to CPT. Proliferation was then determined using a MTS assay. Control cells without p53 were less sensitive to CPT than cells containing any of the p53 forms (Figure 2f). The S392E mutant behaved like wt p53 but the non-phosphorylable S392A mutant displayed a significantly reduced sensitivity to CPT (Figure 2f and Supplementary Table S1 for the detailed statistical analysis). A colony formation assay further confirmed that failure to phosphorylate S392 compromises p53 tumor suppressor activity (Figure 2g).
The p53 S392A mutant displays normal post-translational modifications and transcriptional activity after exposure to CPT
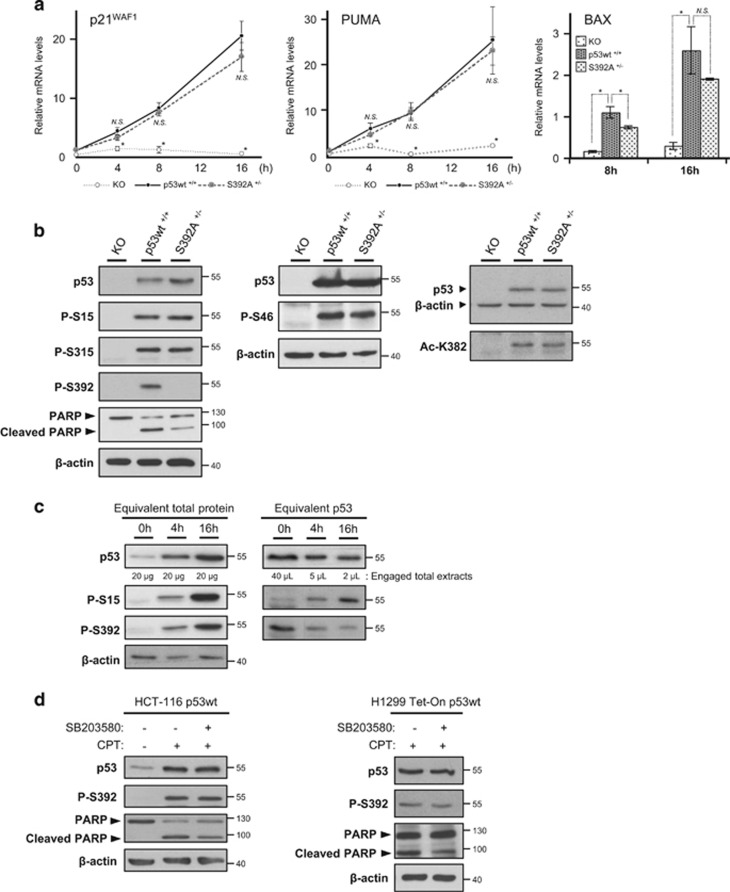
Phosphorylation of S392 was described to promote p53 binding to DNA.13, 14, 15 The impaired ability of p53 S392A to induce apoptosis could therefore be linked to an altered transcriptional activity. To test this hypothesis, we compared the transcriptional activity of S392A and wt p53 in HCT-116 cells. We determined by RT-qPCR the levels of transcripts of several target genes whose promoter have low (BAX and BBC3/PUMA), or high (CDKN1A, p21waf1) affinity for p53.29, 30 After treatments of 4, 8, and 16 h with 1 μM CPT, we observed a marked induction of the genes encoding for p21waf1 and PUMA in cells expressing p53 wt or the S392A mutant, as compared to KO cells (P<0.05). In addition, no significant difference was found between p53 wt and S392A (Figure 3a). The level of BAX mRNA was determined after 8 and 16 h of exposure to CPT. We observed a significant 32% reduction of BAX mRNA level in cells expressing p53 S392A after 8 h, but not 16 h of treatment (Figure 3a and Supplementary Table S2). A similar analysis was performed after 24 h of treatment. At this later time point, we observed no difference of RNA levels for p21waf1, BAX, PUMA, PIG3, or BCL-2 (Supplementary Figure S5). Measure of p21waf1 protein induction by western blot also suggests an equal ability of both p53 forms to transactivate CDKN1A (Figure 2a). In addition, S392A and wt HCT-116 cells showed similar levels of MDM2 after CPT exposure (Supplementary Figure S6).
Figure 3.
p53 wt and the S392A mutant display similar induction of key p53 target genes and post-translational modifications. (a) Kinetic of the expression level of genes encoding for p21waf1, PUMA and BAX by RT-qPCR in p53 KO HCT-116 cells and cells expressing p53 wt or S392A. Cells were treated with 1 μM CPT for the indicated times before RNA extraction. Values are means±S.D. of 3 independent experiments. The one-way ANOVA test was used to compare independent groups (for each CPT concentration). When the ANOVA was significant, a Dunnett's test was used to compare a group of interest with p53 wt as reference (*P<0.05). Exact P-values and details on the test are available in Supplementary table S2. (b) Western blot analysis of different post-translational modifications of p53 after a 24 h treatment of HCT-116 p53 KO, p53 wt and S392A cells with 1 μM CPT. The primary antibodies used specifically recognized the p53 forms phosphorylated on serines 15, 46, 315 and 392, and the p53 form acetylated on lysine 382. In parallel, cleavage of PARP was also assessed. (c) Western blot analysis of total p53 and its phosphorylations on S15 and S392 in HCT-116 p53 wt cells treated with 5 μM camptothecin (CPT) for 0, 4 and 16 h. In the left panel, equal amounts of proteins were loaded in each well of the gel, while in the right panel we loaded equivalent amounts of total p53 at the different time points. (d) In the left panel, HCT-116 p53 wt cells were incubated or not with 20 μM of the p38 MAPK inhibitor SB203580 1 h before addition of 10 μM CPT. After 16 h of treatment, the expression of p53 and its form phosphorylated on S392 as well as PARP cleavage were assessed by western blot analysis. In the right panel, H1299 Tet-On cells expressing p53 wt received 20 μM of SB203580 1 h before addition of 2 μg/ml doxycycline and 10 μM CPT for 16 h
Post-translational modifications play a key role in regulating various aspects of p53 function.2 S46 phosphorylation promotes transactivation of pro-apoptotic target genes like PUMA and p53AIP1.31, 32, 33, 34, 35 This modification also enhances p53 mitochondrial translocation.36 In contrast, phosphorylation of mitochondrial p53 on S315 reduces its association with BCL-2 and BCL-XL.4 Western blot analysis showed that p53 S392A and wt were similarly phosphorylated on S15, S315 and S46 in cells exposed to CPT (Figure 3b and Supplementary Figure S7). The acetylation status of the neighboring K382 was also analyzed. Indeed, acetylations regulate both p53 transcriptional activity and transcription-independent functions. For instance, acetylation of K320/373/382 favors the disruption of a Ku70-BAX complex, promoting apoptosis.37 We observed that K382 was acetylated in both wt and S392A cells (Figure 3b and Supplementary Figure S7). In addition, the ubiquitination profile of wt and S392A p53 was similar in HCT-116 cells exposed to CPT (Supplementary Figure S8).
The p53 form phosphorylated on S392 accumulates after exposure to CPT. However, the fraction of p53, which is phosphorylated on S392 decreases with time (Figure 3c). Thus, in cells exposed to CPT, the level of P-S392 p53 increases more slowly than the total p53 level. This could be due to a limiting kinase activity. The kinase(s) responsible for S392 phosphorylation in response to stress is (are) not clearly identified, but evidences suggest that CK2, CDK9, PKR or p38 MAPK are candidates.6, 7, 8, 9, 10, 11, 12 We exposed p53 wt HCT-116 cells to CPT in the presence of the p38 MAPK inhibitor SB203580 and observed no effect of the inhibitor on S392 phosphorylation while PARP cleavage was reduced (Figure 3d). We obtained similar results with H1299 p53 wt Tet-On cells treated for 4 h with or without SB203580 (Figure 3d). We tested the effect of two other molecules, TBB and Cdk9 Inhibitor II that inhibit respectively CK2 and CDK9. Both of them failed to decrease S392 phosphorylation in presence of CPT (Supplementary Figure S9).
Phosphorylation of S392 promotes p53 mitochondrial translocation during a genotoxic stress
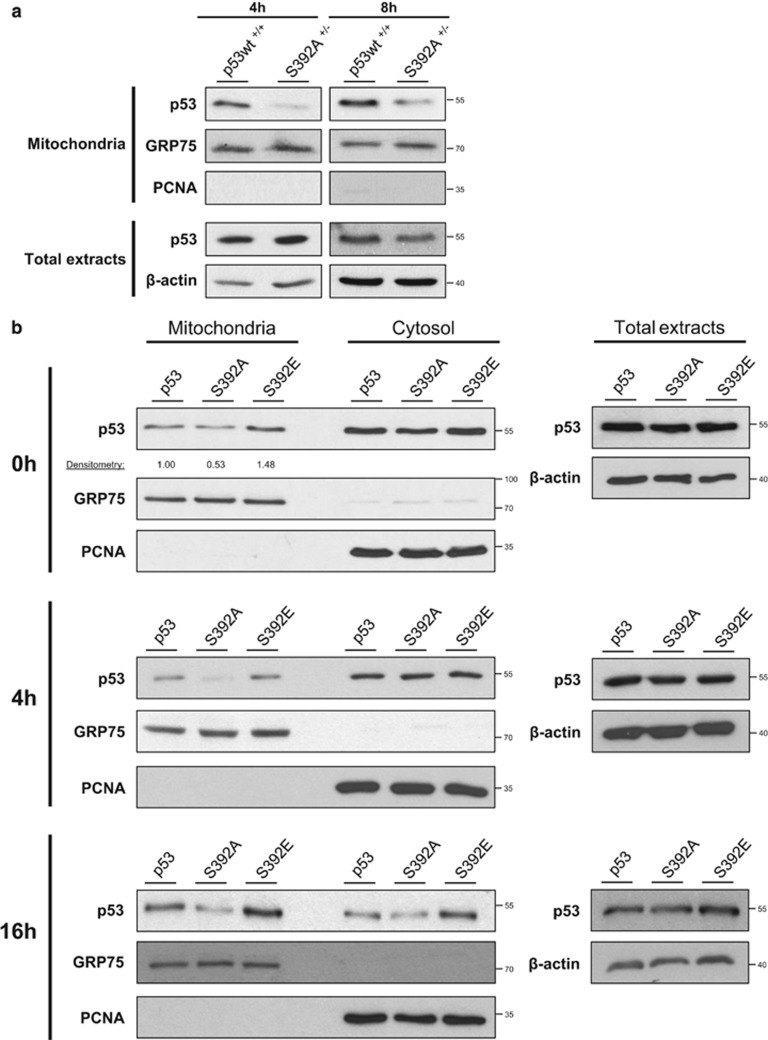
Since p53 transcriptional activity was not markedly affected by S392 phosphorylation status, we investigated the possibility that the S392A mutation disturbed p53 mitochondrial translocation. After 4 and 8 h of exposure to CPT, mitochondrial fractions were isolated from wt and S392A HCT-116 cells, and the abundance of p53 was determined by western blot. The S392A mutant displayed a markedly reduced mitochondrial translocation, suggesting that phosphorylation of S392 is an essential step for p53-mediated transcriptional-independent signaling, leading to apoptosis in stress condition (Figure 4a). We next characterized the localization of p53 wt and its mutants in the H1299 Tet-On clones. We induced p53 expression with doxycycline for 24 h before addition of CPT for 0, 4 or 16 h. In absence of stress (0 h), the fraction of p53 present in the mitochondrial fraction was higher for the S392E p53 form than for p53 wt and S392A, suggesting that S392 phosphorylation enhances mitochondrial localization (Figure 4b). After 4 or 16 h of exposure to CPT, p53 wt and the S392E mutant were abundantly present in the mitochondrial fraction of stressed cells. Comparatively, p53 S392A showed a reduced mitochondrial localization. Of note, p53 S392A is expressed at a similar level than p53 wt or S392E, and is present in the cytosolic fraction (Figure 4b). Similar data were obtained after exposure to a higher CPT concentration (10 μM rather than 1 μM) (Supplementary Figure S10).
Figure 4.
S392 phosphorylation promotes p53 mitochondrial translocation in presence of a genotoxic stress. (a) HCT-116 cells expressing p53 wt or the S392A mutant were treated with 1 μM camptothecin for 4 and 8 h. Total protein extracts as well as mitochondrial fractions were then prepared and screened for the presence of p53. GRP75 is a mitochondrial protein, while PCNA is both cytosolic and nuclear. Detection of GRP75 and PCNA was performed to control for the loading and the purity of the fractions. (b) Kinetic of the mitochondrial relocation of p53 in H1299 Tet-On clones expressing p53 wt, S392A and S392E. H1299 Tet-On clones were incubated overnight with 2 μg/ml doxycycline to induce p53 expression. The next day, cells were treated with 1 μM CPT for 0, 4 and 16 h. Total protein extracts as well mitochondrial and cytosolic fractions were prepared. The presence of p53 was characterized by western blot analysis. Detection of GRP75 and PCNA was performed to control for equal loading of the wells and purity of the fractions. For the time point 0 h, the densitometry analysis of p53 mitochondrial signals was based on the ratio of the intensities of p53 bands on GRP75 bands
The S392-phosphorylated form of p53 is enriched in the mitochondrial fraction of cells exposed to a genotoxic stress
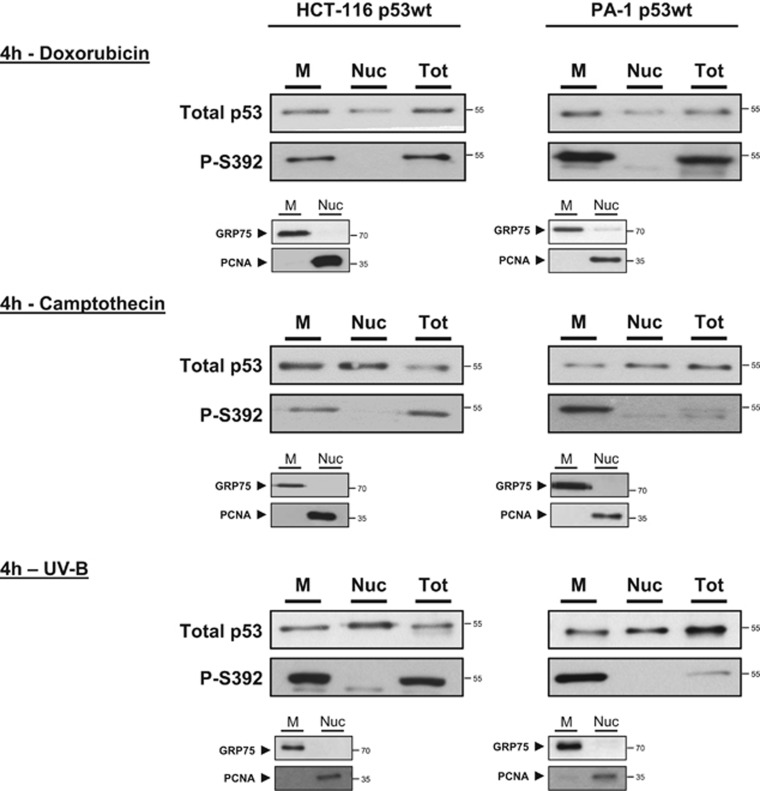
Since phosphorylation of S392 promotes p53 mitochondrial translocation, we next asked whether P-S392 p53 was enriched in the mitochondrial fraction of cells exposed to apoptotic stimuli. We induced DNA damage to p53 wt HCT-116 and PA-1 cells by exposure to CPT, doxorubicin (DOX) or UV-B. The abundance of P-S392 and total p53 was evaluated by western blot in total protein extracts as well as mitochondrial and nuclear fractions. We observed that P-S392 p53 was abundant in the mitochondrial fraction of the two cell lines while absent from the nuclei (Figure 5). We also examined additional phosphorylation and acetylation sites by western blot after a 4h treatment with DOX. No other post-translational modification was found to be enriched in the mitochondrial fraction of both cell lines (Supplementary Figure S11).
Figure 5.
The p53 form phosphorylated on S392 is enriched in the mitochondrial fraction of cells exposed to a genotoxic stress. HCT-116 and PA-1 p53 wt cells were treated for 4 h with 1 μM doxorubicin or camptothecin. Total protein extracts (Tot) as well mitochondrial (M) and nuclear (Nuc) fractions were prepared thereafter. For UV-B treatment, we used doses of 400 J/m2 and 100 J/m2 for, respectively, HCT-116 and PA-1 cells. Total protein extracts and fractions were prepared 4 h after the UV-B exposure. For western blot analysis, we loaded equal amounts of total p53 in each well of the gel. Presence of P-S392 p53 was assessed with a monoclonal antibody that specifically recognizes the p53 form phosphorylated on S392. To control for the purity of the fractions, we detected GRP75 and PCNA
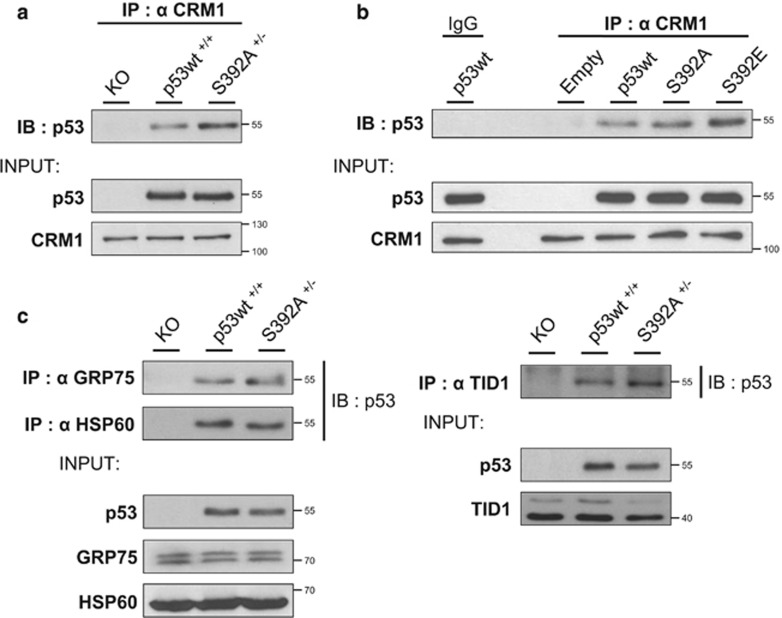
The S392A mutant is not impaired for interactions with CRM1, GRP75, HSP60 or TID1
Among the numerous interactions of p53, some regulate its mitochondrial localization. The increased mitochondrial localization of the arginine 72 p53 polymorphic variant was previously linked to its enhanced binding to the E3 ubiquitin ligase MDM2 and CRM1 (Exportin-1).19 Indeed, interactions that favor p53 nuclear export are also likely to promote its mitochondrial localization. In addition, S392 phosphorylation could impact on p53 nuclear export.38 We tested the hypothesis that the S392A mutation disturbs interaction with CRM1. HCT-116 p53 wt, KO and S392A cells were treated with CPT for 16 h. Total protein extracts were immunoprecipitated with an anti-CRM1 antibody and the interaction with p53 was detected by western blot. We found that p53 S392A was immunoprecipitated as efficiently with CRM1 as wt p53 (Figure 6a). Moreover, in H1299 cells expressing wt, S392A or S392E p53, all p53 forms interacted well with CRM1 (Figure 6b).
Figure 6.
The S392A mutant is not deficient for different interactions known to modulate p53 mitochondrial translocation. (a) HCT-116 cells expressing p53 wt or the S392A mutant as well as p53 KO cells were treated with 1 μM CPT for 16 h. Total protein extracts were then prepared and immunoprecipitated with an anti-CRM1 antibody. The interaction with p53 was detected by western blot. Inputs for p53 and CRM1 in the different extracts are also depicted. (b) H1299 Tet-On clones transfected with the empty vector as well as cells expressing p53, S392A and S392E were incubated overnight with 2 μg/ml doxycycline to induce p53 expression. The next day, cells were treated with 1 μM CPT for 4 h. Total protein extracts were next prepared and immunoprecipitated with an anti-CRM1 antibody or control IgG. Detection of p53 was assessed by western blot analysis. Inputs for p53 and CRM1 in the different extracts are shown. (c) HCT-116 cells expressing p53 wt, the S392A mutant as well as p53 KO cells were treated with 1 μM CPT for 16 h. Total protein extracts were prepared and immunoprecipitated with anti-GRP75, -HSP60 and -TID1 antibodies. The interaction with p53 was detected by western blot. The inputs for p53, GRP75, HSP60 and TID1 in the different extracts are depicted
The chaperones GRP75 (mtHSP70) and HSP60 as well as the co-chaperone TID1 are known to promote p53 mitochondrial translocation.19, 39, 40 We hypothesized that phosphorylation of S392 could facilitate p53 translocation by enhancing its interaction with one or several of these chaperones. To investigate this possibility, we performed co-immunoprecipitation experiments in HCT-116 cells expressing p53 wt or S392A. Both p53 forms similarly immunoprecipitated with each of the three chaperones, showing that phosphorylation of S392 is not essential for these interactions (Figure 6c).
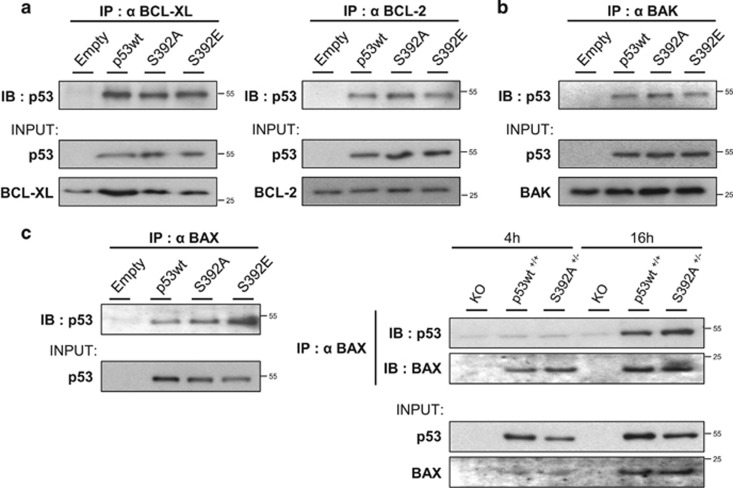
The S392A p53 mutant shows an intact ability to interact with BCL-2 family members
The tumor suppressor p53 regulates OMM permeabilization through interactions it establishes with pro- and anti-apoptotic proteins of the BCL-2 family.19, 20, 21, 22, 23, 24 In addition, members of this family attached to the OMM could act as mitochondrial anchors for p53.22, 41 The defective mitochondrial translocation of S392A p53 could thus be explained by a defective interaction with one or several BCL-2 family members.
This hypothesis was tested by co-immunoprecipitation in H1299 cells expressing wt, S392A or S392E p53. After an overnight induction of p53 with doxycycline followed by a 4 h treatment with CPT, total extracts were prepared and analyzed. Regarding the anti-apoptotic proteins BCL-2 and BCL-XL, no difference of association was observed between the three p53 forms (Figure 7a). In absence of stress, BAK is mainly localized at the OMM, but it also associates with the endoplasmic reticulum membrane.42 Moreover, in certain cell types, a fraction of BAK is cytosolic.43 We observed that the three p53 forms co-immunoprecipitated with BAK in total protein extracts from H1299 cells (Figure 7b). However, when the immunoprecipitations were performed in isolated mitochondrial fractions, S392E and wt p53 displayed a stronger interaction with BAK as compared to p53 S392A, in accordance with its defective mitochondrial translocation (Supplementary Figure S12). Similar results were obtained in HCT-116 cells exposed to CPT. In the mitochondrial fractions, less S392A p53/BAK complex than wt p53/BAK complex was present (Supplementary Figure S13). BAX is mainly cytosolic in unstressed cells.44 We observed no difference of association between S392A and wt p53 in total extracts of H1299 cells exposed to CPT for 4 h (Figure 7c, left panel). Similar data were obtained when the cells were treated for 16 h (Supplementary Figure S14). We also analyzed the BAX-p53 interaction in wt and S392A HCT-116 cells. The S392A mutant induced BAX expression like wt p53. Moreover, both p53 forms interacted well with BAX (Figure 7c, right panel). The sum of these data indicate that S392 phosphorylation status does not influence interactions of p53 with BCL-2 family members.
Figure 7.
Phosphorylation of S392 does not modulate p53 interactions with BCL-2 family members. (a) H1299 Tet-On clones transfected with the empty vector as well as cells expressing p53, S392A and S392E were incubated overnight with 2 μg/ml doxycycline to induce p53 expression. The next day, cells were treated with 1 μM CPT for 4 h. Total protein extracts were next prepared and immunoprecipitated with anti-BCL-XL and -BCL-2 antibodies. The interaction with p53 was detected by western blot. The inputs for p53, BCL-XL and BCL-2 in the different extracts are depicted. (b) Immunoprecipitation-western blot analysis of the interaction between p53 and BAK in the H1299 Tet-On clones after a 4 h treatment with CPT. The inputs for p53 and BAK are depicted. (c) Interaction of p53 with BAX in H1299 and HCT-116 cells. In the left panel, H1299 Tet-On clones transfected with the empty vector as well as cells expressing p53, S392A and S392E were incubated overnight with 2 μg/ml doxycycline to induce p53 expression. The next day, cells were treated with 1 μM CPT for 4 h. Total protein extracts were next prepared and immunoprecipitated with an anti-BAX antibody. The interaction with p53 was detected by western blot. In the right panel, HCT-116 cells expressing p53 wt, the S392A mutant as well as p53 KO cells were treated with 1 μM CPT for 4 or 16 h. Total protein extracts were prepared and immunoprecipitated with an anti-BAX antibody. The interaction with p53 was detected by western blot. The direct immunoprecipitation of BAX and the inputs for p53 and BAX in the different extracts are also depicted
Discussion
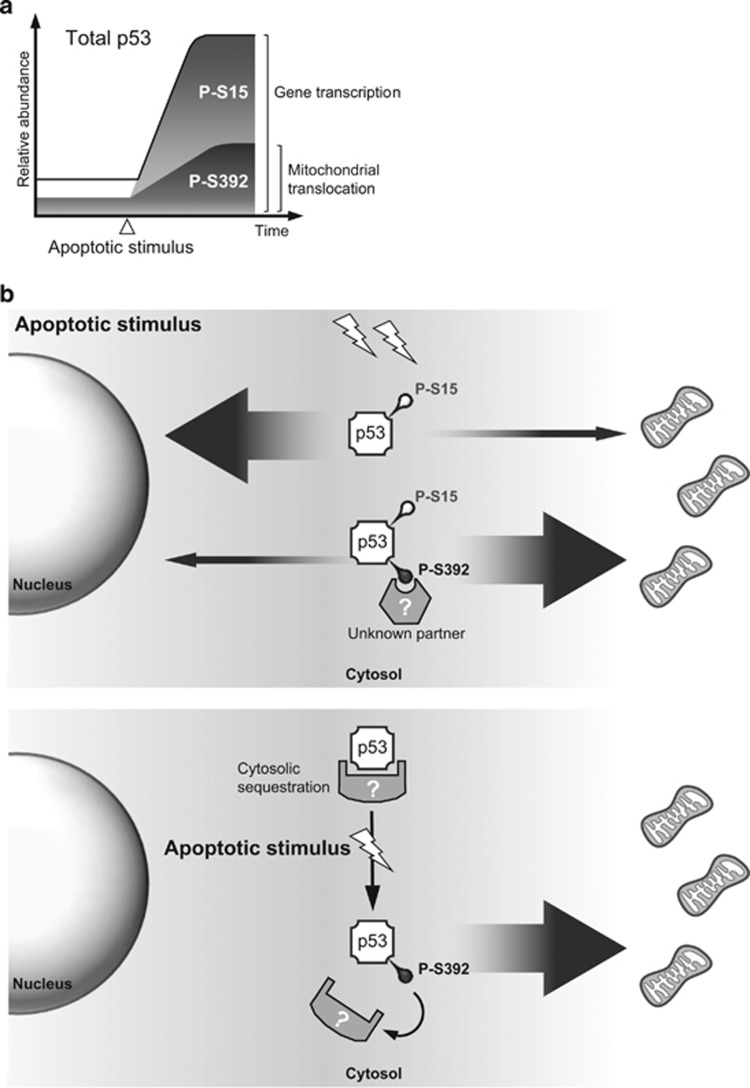
We observed that a fraction of p53 proteins are constitutively phosphorylated on S392 and that the abundance of P-S392 p53 markedly increases after exposure to a DNA damaging agent. However, the fraction of P-S392 p53 molecules decreases when p53 accumulates in stressed cells (Figure 8a). This could be explained if the responsible kinase activity is limiting or if a phosphatase activity increases in stressed cells. This is in accordance with previously published reports showing that phosphorylation of S392 is a slow process and that the short half-life of p53 in unstressed cells prevents accumulation of P-S392 p53, while in presence of a stress, the stabilization of p53 allows its accumulation.10, 11
Figure 8.
S392 phosphorylation of p53 increases in cells exposed to a genotoxic stress, promoting its mitochondrial localization and apoptosis. (a) In healthy cells, a fraction of p53 is already phosphorylated on S392. In addition, the S392 phosphorylated form of p53 becomes more abundant after exposure to a genotoxic stress, although accumulation of P-S392 p53 is poor in comparison to other phosphorylated forms such as P-S15 p53. Therefore, it is likely that, even in cells exposed to an apoptotic stimulus, only a minor fraction of p53 proteins carries a phosphorylated S392. This post-translational modification modulates the transcription-independent function of p53, P-S392 p53 being preferentially targeted to the mitochondria. Failure to phosphorylate S392 correlates with a decreased mitochondrial localization of p53 and apoptosis. (b) Hypotheses on the involvement of S392 phosphorylation in the regulation of p53 trafficking to mitochondria. In the top diagram, we postulate that p53 needs to interact with a cytosolic, yet unknown, protein partner in order to be translocated to the mitochondria and trigger OMM permeabilization; and that the interaction is facilitated by phosphorylation of S392. In the bottom diagram, we propose that in the absence of a stress, mitochondrial translocation of cytosolic p53 could be largely prevented through interaction with one or more proteins able to interact and sequester the tumor suppressor. Moreover, formation of this inhibitory complex is prevented by the increased phosphorylation of serine 392 in cells exposed to a genotoxic stress, allowing cytosolic p53 to reach the OMM to trigger apoptosis.
To investigate the impact of S392 phosphorylation on p53 function in apoptosis, we substituted S392 by a non-phosphorylatable alanine in p53 wt HCT-116 cells. We obtained cells harboring one p53 S392A allele and one p53 knockout allele. These S392A hemizygous cells express p53 protein levels similar to that of p53 wt cells after exposure to CPT. Interestingly, the p53 S392A cells displayed an enhanced resistance to the genotoxic agent CPT associated with a reduced apoptosis as assessed by PARP cleavage and DNA fragmentation. These observations were recapitulated in H1299 cells with a tetracycline-inducible expression of wt, S392A or S392E p53.
The reduced ability of S392A p53 to induce apoptosis after a genotoxic stress could result from an alteration of its transcriptional activity. We tested this hypothesis by assessing the RNA levels of different p53 target genes (p21waf1, BAX, PUMA, PIG3 and BCL-2) and observed no marked alteration. This is in line with previous observations showing that the S392A mutant (or its mouse homologous p53 S389A) was not affected in its ability to transactivate p21waf1, PUMA or BAX.45, 46 However, other reports indicate that S392 phosphorylation impacts on the formation of p53 tetramers and DNA binding.13, 14, 15 We thus cannot exclude that subtle differences exist between the transcriptional activity of wt and S392A p53.
Independently of its transcriptional activity, p53 also induces apoptosis by directly triggering OMM permeabilization. This activity involves its translocation to the mitochondria and interactions with several BCL-2 family members. How PTMs impact on p53 mitochondrial activity is still unclear. It has been evidenced that p53 ubiquitination by MDM2 favors its mitochondrial translocation.19, 47 Sykes and colleagues showed that p53 acetylation on K120 increased its ability to release BAK from MCL1.48 Moreover, a fraction of MDMX resides at the OMM and promotes the recruitment of P-S46 p53.36 We previously showed that S20 phosphorylation is influential on the p53-BAK interaction and modulates p53 mitochondrial level during late apoptosis.49 More recently, Zhang et al.50 showed that ubiquitination of cytosolic p53 on K24 by the TRAF6 E3 ligase inhibits its mitochondrial translocation and interaction with the BAK-MCL1 complex.50 We show here in two different cell models that failure to phosphorylate S392 markedly reduces p53 mitochondrial translocation after exposure to an apoptotic stimulus. We also found that P-S392 p53 is mainly mitochondrial after a genotoxic stress. Interestingly, exposure of rat RINm5F insulinoma cells to high glucose triggers mitochondrial translocation of P-S390 (homologous to human S392) p53 and presence of P-S390 p53 at the mitochondria correlates with increased apoptosis.51
Different hypothesis could explain the compromised ability of S392A p53 to localize to mitochondria. (1) It is possible that phosphorylation of S392 is required for additional PTMs necessary to promote mitochondrial translocation. However, we observed no effect of the S392 mutation on phosphorylation of S46 or S315, which respectively regulates p53 mitochondrial translocation and association with BCL-2 or BCL-XL.4, 36 (2) p53 might need to interact with a cytosolic partner in order to be transported to the OMM, as member of a protein complex. This interaction could be promoted by S392 phosphorylation (Figure 8b). We observed that S392A mutation does not disturb the interaction of p53 with GRP75, HSP60 or TID1, which are known to regulate its mitochondrial translocation.19, 39, 40 Moreover, S392 phosphorylation status does not modulate interaction of p53 with BCL-2 family members. However, it cannot be ruled out that it regulates p53 interaction with a yet unidentified carrier. (3) In another model, in unstressed cells, p53 would be sequestered in the cytosol through interaction with another protein, and this interaction would be disrupted by phosphorylation of S392 (Figure 8b).
In summary, we showed that P-S392 p53 is enriched at the mitochondria after cell exposure to genotoxic agents and that the S392A non-phosphorylatable p53 mutant displays impaired mitochondrial translocation and decreased ability to trigger apoptosis. The exact mechanism by which S392 phosphorylation controls p53 mitochondrial localization awaits further studies. A growing body of evidence now indicates that several PTMs regulate p53 proapoptotic mitochondrial activity through modulating its interaction with protein partners, as described above.4, 49, 50, 51, 52, 53 Targeting such interactions may constitute an effective strategy to trigger p53 mitochondrial localization and hence apoptosis in cancer cells.
Materials and methods
Cell culture and treatments
The p53 wt HCT-116 (human colorectal carcinoma) and PA-1 (human ovarian teratocarcinoma) cell lines were cultured in DMEM (Lonza, BE12-604 F) supplemented with 10% FBS and 1% penicillin-streptomycin (Lonza DE17-602E). The p53-null H1299 cell line (ATCC CRL-5803) was employed to derive Tet-On clones expressing wt p53 or mutants at position 392. Tet-On clones were grown in DMEM supplemented with 10% FBS, 1% penicillin-streptomycin, 800 μg/ml G418 (Santa Cruz, Dallas, TX, USA, sc-29065) and 1 μg/ml puromycin (Gibco, A1113803). All cells were tested for mycoplasma contamination. Camptothecin (Sigma-Aldrich, St Louis, MO, USA, C9911), doxorubicin (Sigma-Aldrich, D1515) and SB203580 (Cell Signaling, Danvers, MA, USA, 5633) were used as indicated in the figures. For UV-B treatment, HCT-116 cells were exposed to 400 J/m2 and PA-1 cells to 100 J/m2 (Vilber Lourmat, Marne-la-Vallée, France, RMX-3 W, France).
Tet-On system
H1299 Tet-On clones were generated by transfection of the pTet-On vector coding for the transactivator (Clontech, Mountain View, CA, USA) followed by a second transfection with the pTRE2pur-ΔMyc plasmid containing the p53 cDNA. The Myc tag was deleted from pTRE2pur-Myc vector (Clontech) using the BamHI and MluI restriction sites followed by fill-in of 5′overhangs to generate blunt ends for plasmid ligation. The wt p53 cDNA (R72 polymorphic variant) was cloned into pTRE2pur-ΔMyc using the EcoRV restriction site. To generate the p53 S392 A and S392E mutants, a site-directed mutagenesis (Agilent Technologies, Santa Clara, CA, USA) was performed on the p53 wt vector.
CRISPR/Cas9n genome editing
The protocol for genome engineering using CRISPR/Cas9 is detailed in the works of Feng Zhang and colleagues. We followed the procedure that utilizes the double nicking mediated by Cas9n D10A.25, 26 Briefly, regions of interest of the TP53 gene from HCT-116 cells were first sequenced. Sequences were analyzed with the online analysis tools available on benchling.com to identify and score CRISPR sgRNAs according to their specificity and efficiency. Sense and anti-sense oligos corresponding to the selected sgRNAs guides (See Supplementary Table S3 for the sequences) were purchased from Eurogentec (Liège, Belgium), annealed and cloned into the pSpCas9n(BB)-2A-Puro PX462 v2.0 vector (Feng Zhang – Addgene 62987). To introduce the desired mutation into the genome by homologous recombination, we used a synthetic single-stranded DNA oligonucleotide (ssODN) of 200 bases as repair template. The ssODN was purchased from Integrated DNA Technologies (Liège, Belgium) (See Supplementary Table S3 for the sequence). HCT-116 cells were transfected using Lipofectamin 2000 (Invitrogen, Carlsbad, CA, USA). For one well of a 6-well plate, the transfection was performed with 1 μg of each pX462 vector coding for the two guides, with or without 1 μl of 20 μM ssODN. The next day, the transfected cells were selected with puromycin for 48 h. After 5 days of recovery, a dilution cloning was performed.
Total protein extracts
Proteins were extracted with NP40 lysis buffer (50 mM Tris-HCl pH 8, 150 mM NaCl, 5 mM EDTA, 1% NP40) supplemented with protease (Thermofisher, Waltham, MA, USA, 78429) and phosphatase inhibitors (Phosphatase Inhibitor cocktails 2 and 3, Sigma-Aldrich P5726 and P0044).
Mitochondria isolation
Cells were scraped and centrifuged at 500 g. After two washes in PBS, the pellet was resuspended in fractionation buffer (10 mM Hepes pH 7.4, 300 mM Mannitol, 1 mM EDTA, 0.2 mM EGTA) supplemented with protease and phosphatase inhibitors. The cell suspension was lysed with a pre-chilled type B Dounce homogenizer. To remove intact cells, cell debris, lysed membrane and nuclei, the lysate was centrifuged 3 times at 500 g (10 min, 4 °C). The supernatant from the last centrifugation was collected and centrifuged at 9000 × g (10 min, 4 °C). After centrifugation, the supernatant (cytosolic fraction) was stored at −20 °C. The pellet (mitochondrial fraction) was resuspended in 100 μl of fractionation buffer and stored at −20 °C overnight for complete lysis of mitochondria.
Isolation of nuclear extracts
Cells were scraped and centrifuged at 500 g. After two washes with PBS, the pellet was resuspended in hypotonic buffer (10 mM Hepes-NaOH pH 7.9, 0.1 mM EDTA, 10 mM KCl) supplemented with 1 mM DTT as well as protease and phosphatase inhibitors. After a 15 min incubation on ice, cells were lyzed with a type B Dounce homogenizer and centrifuged at 4000 × g for 2 min at 4 °C. The pellet was washed with hypotonic buffer and after a second centrifugation, it was resuspended in hypertonic buffer (20 mM Hepes-KOH pH 7.9, 400 mM NaCl, 1 mM EDTA, 20% Glycerol) supplemented with 1 mM DTT as well as protease and phosphatase inhibitors. After a 45 min incubation on ice with brief vortexing every 5–10 min, the suspension was centrifuged at 14 000 g for 5 min at 4 °C and the supernatant (soluble nuclear fraction) was collected.
Western blot
Protein concentrations were determined using the BCA Protein Assay (Thermofisher, 23225). Equal amounts of protein were separated on SDS-polyacrylamide gels and transferred to PVDF membrane. Membranes were blocked with 5% non-fat dry milk or alternatively 5% BSA in PBS or containing 0.2% Tween-20 (PBS-T) for 30 min. After blocking, the membrane was incubated with the primary antibody 1 h at room temperature or overnight at 4 °C. Next, the membrane was washed three times with PBS-T and incubated 1 h with the appropriate peroxidase-conjugated secondary antibody (1:5,000 dilution; Dako). After three washes, the immunoreactive bands were detected using the Western Lightning Plus-ECL kit (PerkinElmer, Waltham, MA, USA). For detection of phospho-specific residues of p53, the proteins were transferred to a nitrocellulose membrane and we used TBS-T (0.1% Tween-20) during the incubation and the washes. The following primary antibodies were used: mouse anti-actin Ab (Sigma-Aldrich, A1978), mouse anti-cleaved PARP (Asp214) Ab (Cell Signaling, 9546), mouse anti-p53 (DO-1) Ab (Millipore, Billerica, MA, USA, OP43), rabbit anti-p53 (FL-393) Ab (Santa Cruz, sc-6243), mouse anti-p21 Ab (Cell Signaling, 2946), rabbit anti-p53 P-Ser15 Ab (Cell Signaling, 9284), rabbit anti-p53 P-Ser46 Ab (Cell Signaling, 2521), rabbit anti-p53 P-Ser315 Ab (Cell Signaling, 2528), rabbit anti-p53 P-Ser392 Ab (Cell Signaling, 9281), rabbit anti-p53 Acetyl-Lys382 Ab (Cell Signaling, 2525), mouse anti-PCNA Ab (Santa Cruz, sc-56), goat anti-GRP75 Ab (Santa Cruz, sc-1058), rabbit anti-HSP60 Ab (Santa Cruz, sc-1722), mouse anti-TID1 Ab (Santa Cruz, sc-18820) and rabbit anti-CRM1 Ab (Santa Cruz, sc-5595), rabbit anti-BAK (NT) Ab (Millipore, 06-536), mouse anti-Bcl-2 (Clone 7) Ab (BD Biosciences, Franklin Lakes, NJ, USA, 610539), mouse anti-Bcl-xL (7B2.5) Ab (Santa Cruz, sc-56021), rabbit Anti-Bax (NT) Ab (Millipore, ABC11). To perform semi-quantitative WB analysis, we used the ScanLater Western Blot Detection Cartridge in a SpectraMax® i3 microplate reader system (Molecular Devices, Sunnyvale, CA, USA) according to manufacturer’s instructions. After transfer of proteins to PVDF membranes and blocking, the membrane was incubated simultaneously with the anti-p53 DO-1 and anti-actin antibodies. After 3 washes, the membrane was incubated with a secondary Europium-labeled anti-mouse antibody (Molecular Devices, R8205). Immunoreactive bands were scanned using the SpectraMax i3 microplate reader system and analyzed with the integrated software.
MTS assay
Cells seeded in 96-well plates (5,000 cells/well) were treated for 72 h as described in the figures. After treatment, cells were incubated for 1 h in fresh culture medium supplemented with a solution of MTS/PMS (Promega, Madison, WI, USA; G5421) according to manufacturer’s instructions. After the incubation, global cell growth was assessed by measuring the absorbance at 490 nm directly proportional to the number of live cells.
RNA extraction and quantitative RT-PCR analysis
Cells from one 100 mm round culture dish were lysed directly into Ribozol reagent (Amresco, Solon, OH, USA). The upper aqueous phase was collected, treated with DNAse and purified with the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions. The purified RNAs were reverse transcribed using the GoScript Reverse Transcription System (Promega) with 1 μg of total RNA and a mix of 0.5 μg of random and Oligo(dT) primers. After the RT-reaction, the samples were diluted four-fold. Quantitative PCR of mRNA levels were performed in triplicate on a StepOne+ (Applied Biosystems, USA) using SYBR Green dye (with ROX dye as reference) as detection method (Thermofisher, K0221). Detection of GAPDH mRNA level was run in parallel to the genes of interest. The primers used during the quantitative RT-PCR analysis are listed in Supplementary Table S4.54, 55, 56, 57, 58
Cell viability measurement using trypan blue
Cells were treated as described in the figures. After treatment, the culture medium containing dead cells was collected and the adherent cells were trypsinized. Trypsin was neutralized with the collected medium. Cell viability was assessed directly on the cell suspension with the trypan blue dye exclusion method (Vi-CELL Cell Counter automate, Beckman, USA).
Co-immunoprecipitation
Cells were lysed in NP40 lysis buffer (50 mM Tris-HCl pH 8, 150 mM NaCl, 1% NP40). Alternatively, for CRM1 immunoprecipitation, cells were lysed in high-salt lysis buffer (50 mM Tris-HCl pH 8, 300 mM NaCl, 5 mM EDTA, 1% NP40). Buffers were supplemented with protease and phosphatase inhibitors. After lysis, 800 μl to 2 ml of buffer containing 300 μg (for CRM1 IP), 500 μg (for GRP75, HSP60 and BAK IPs) or 2 mg (for BCL-2, BCL-XL and BAX IPs) of proteins were incubated with 1 to 2 μg of the adequate antibody for 2 h to overnight at 4 °C. Protein complexes were immunoprecipitated with 20 μl of agarose protein G slurry (Pierce, 20398) for 30 min at 4 °C, followed by an incubation of 30 min at room temperature. The beads were washes four times with 700 μl of NP40 (or high-salt) lysis buffer. Protein complexes were eluted with 20 μl of 1 × Laemmli sample buffer and analyzed by western blot. Western blot detection of the primary antibody was performed with HRP Anti-Light Chain-Specific Antibodies (Jackson Laboratories, Bar Harbor, ME, USA) or with HRP anti-IgG conformation specific (TrueBlot anti-Rabbit IgG HRP and TrueBlot ULTRA anti-Mouse IgG HRP (Rockland, Pottstown, PA, USA)).
Sub-G1 peak analysis by flow cytometry
The protocol followed has been described in details by C. Riccardi and I. Nicoletti.28 Briefly, after treatment, cells were washed with PBS, resuspended in 70% EtOH and stored at −20 °C. On the day of the analysis, cells were washed with PBS and resuspended in 1 ml of DNA staining solution containing 20 μg of propidium iodide and 0.2 mg of DNase free RNase per mL of PBS. After 30 min of incubation in the dark, analysis of the sub-G1 peak was performed cells on a Guava easyCyte 6HT-2 L flow cytometer (Millipore, USA).
Colony formation assay
Cells were treated as described in the figure. After treatment, the cells were washed twice with PBS and fixed with 4% formaldehyde for 15 min. After 2 washes with PBS, cells were stained with 0.03% crystal violet for 20 min at room temperature. The stained colonies were washed twice with PBS and once with deionized water. Next, the plates were air dried.
Statistical analyses
The statistical analysis was performed using the JMP Pro v.12.1.0 software (SAS Institute, Cary, NC, USA). The statistical tests employed are described in the figure legends.
Acknowledgments
We thank Catherine Rasse (SMCS, Université catholique de Louvain, Belgium) for the statistical analyses. We thank Prof. Maureen Murphy (The Wistar Institute, Philadelphia PA, USA) for critical reading of the manuscript. The laboratory of PD is financed by the FNRS-TELEVIE (Fonds National de la Recherche Scientifique) (Brussels, Belgium) and the FSR (Fonds Spéciaux de Recherche UCL, Louvain-la-Neuve). CC is the owner of a grant from the FNRS-TELEVIE (grant Télévie N°7458713F) and FSR (Fonds Spéciaux de Recherche UCL, Louvain-la-Neuve).
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by D Green
The authors declare no conflict of interest.
Supplementary Material
References
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature 2000; 408: 307–310. [DOI] [PubMed] [Google Scholar]
- Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer 2006; 6: 909–923. [DOI] [PubMed] [Google Scholar]
- Bischoff JR, Friedman PN, Marshak DR, Prives C, Beach D. Human p53 is phosphorylated by p60-cdc2 and cyclin B-cdc2. Proc Natl Acad Sci USA 1990; 87: 4766–4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantajit D, Fan M, Duru N, Wen Y, Reed JC, Li JJ. Cyclin B1/Cdk1 phosphorylation of mitochondrial p53 induces anti-apoptotic response. PloS ONE 2010; 5: e12341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Y-H, Chung P-H, Sun T-P, Shieh S-Y. p53 C-terminal phosphorylation by CHK1 and CHK2 participates in the regulation of DNA-damage-induced c-terminal acetylation. Mol Biol Cell 2005; 16: 1684–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller D, Zeng X, Li X, Kapoor M, Iordanov MS, Taya Y et al. The p38MAPK inhibitor SB203580 alleviates ultraviolet-induced phosphorylation at serine 389 but not serine 15 and activation of p53. Biochem Biophys Res Commun 1999; 261: 464–471. [DOI] [PubMed] [Google Scholar]
- Huang C, Ma WY, Maxiner A, Sun Y, Dong Z. p38 kinase mediates UV-induced phosphorylation of p53 protein at serine 389. J Biol Chem 1999; 274: 12229–12235. [DOI] [PubMed] [Google Scholar]
- Keller DM, Lu H. p53 serine 392 phosphorylation increases after UV through induction of the assembly of the CK2.hSPT16.SSRP1 complex. J Biol Chem 2002; 277: 50206–50213. [DOI] [PubMed] [Google Scholar]
- Claudio PP, Cui J, Ghafouri M, Mariano C, White MK, Safak M et al. Cdk9 phosphorylates p53 on serine 392 independently of CKII. J Cell Physiol 2006; 208: 602–612. [DOI] [PubMed] [Google Scholar]
- Cox ML, Meek DW. Phosphorylation of serine 392 in p53 is a common and integral event during p53 induction by diverse stimuli. Cell Signal 2010; 22: 564–571. [DOI] [PubMed] [Google Scholar]
- Meek DW, Cox M. Induction and activation of the p53 pathway: a role for the protein kinase CK2? Mol Cell Biochem 2011; 356: 133–138. [DOI] [PubMed] [Google Scholar]
- Cuddihy AR, Wong AH, Tam NW, Li S, Koromilas AE. The double-stranded RNA activated protein kinase PKR physically associates with the tumor suppressor p53 protein and phosphorylates human p53 on serine 392 in vitro. Oncogene 1999; 18: 2690–2702. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K, Sakamoto H, Lewis MS, Anderson CW, Erickson JW, Appella E et al. Phosphorylation of serine 392 stabilizes the tetramer formation of tumor suppressor protein p53. Biochemistry 1997; 36: 10117–10124. [DOI] [PubMed] [Google Scholar]
- Nichols NM, Matthews KS. Human p53 phosphorylation mimic, S392E, increases nonspecific DNA affinity and thermal stability. Biochemistry 2002; 41: 170–178. [DOI] [PubMed] [Google Scholar]
- Lu H, Fisher RP, Bailey P, Levine AJ. The CDK7-cycH-p36 complex of transcription factor IIH phosphorylates p53, enhancing its sequence-specific DNA binding activity in vitro. Mol Cell Biol 1997; 17: 5923–5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne DM, Palmer RH, Meek DW. Mutation of the casein kinase II phosphorylation site abolishes the anti-proliferative activity of p53. Nucleic Acids Res 1992; 20: 5565–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruins W, Zwart E, Attardi LD, Iwakuma T, Hoogervorst EM, Beems RB et al. Increased sensitivity to UV radiation in mice with a p53 point mutation at Ser389. Mol Cell Biol 2004; 24: 8884–8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P et al. p53 has a direct apoptogenic role at the mitochondria. Mol Cell 2003; 11: 577–590. [DOI] [PubMed] [Google Scholar]
- Dumont P, JI-J Leu, Della Pietra AC 3rd, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet 2003; 33: 357–365. [DOI] [PubMed] [Google Scholar]
- Tomita Y, Marchenko N, Erster S, Nemajerova A, Dehner A, Klein C et al. WT p53, but not tumor-derived mutants, bind to Bcl2 via the DNA binding domain and induce mitochondrial permeabilization. J Biol Chem 2006; 281: 8600–8606. [DOI] [PubMed] [Google Scholar]
- Xu H, Tai J, Ye H, Kang CB, Yoon HS. The N-terminal domain of tumor suppressor p53 is involved in the molecular interaction with the anti-apoptotic protein Bcl-Xl. Biochem Biophys Res Commun 2006; 341: 938–944. [DOI] [PubMed] [Google Scholar]
- Leu JI-J, Dumont P, Hafey M, Murphy ME, George DL. Mitochondrial p53 activates Bak and causes disruption of a Bak-Mcl1 complex. Nat Cell Biol 2004; 6: 443–450. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 2004; 303: 1010–1014. [DOI] [PubMed] [Google Scholar]
- Pietsch EC, Perchiniak E, Canutescu AA, Wang G, Dunbrack RL, Murphy ME. Oligomerization of BAK by p53 utilizes conserved residues of the p53 DNA binding domain. J Biol Chem 2008; 283: 21294–21304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013; 8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin C-Y, Gootenberg JS, Konermann S, Trevino AE et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013; 154: 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 2013; 31: 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccardi C, Nicoletti I. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat Protoc 2006; 1: 1458–1461. [DOI] [PubMed] [Google Scholar]
- Weinberg RL, Veprintsev DB, Bycroft M, Fersht AR. Comparative binding of p53 to its promoter and DNA recognition elements. J Mol Biol 2005; 348: 589–596. [DOI] [PubMed] [Google Scholar]
- Szak ST, Mays D, Pietenpol JA. Kinetics of p53 Binding to Promoter Sites In Vivo. Mol Cell Biol 2001; 21: 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Dumont P, Della Pietra A, Shetler C, Murphy ME. The codon 47 polymorphism in p53 is functionally significant. J Biol Chem 2005; 280: 24245–24251. [DOI] [PubMed] [Google Scholar]
- Grison A, Mantovani F, Comel A, Agostoni E, Gustincich S, Persichetti F et al. Ser46 phosphorylation and prolyl-isomerase Pin1-mediated isomerization of p53 are key events in p53-dependent apoptosis induced by mutant huntingtin. Proc Natl Acad Sci USA 2011; 108: 17979–17984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 2000; 102: 849–862. [DOI] [PubMed] [Google Scholar]
- Taira N, Nihira K, Yamaguchi T, Miki Y, Yoshida K. DYRK2 is targeted to the nucleus and controls p53 via Ser46 phosphorylation in the apoptotic response to DNA damage. Mol Cell 2007; 25: 725–738. [DOI] [PubMed] [Google Scholar]
- Rinaldo C, Prodosmo A, Mancini F, Iacovelli S, Sacchi A, Moretti F et al. MDM2-regulated degradation of HIPK2 prevents p53Ser46 phosphorylation and DNA damage-induced apoptosis. Mol Cell 2007; 25: 739–750. [DOI] [PubMed] [Google Scholar]
- Mancini F, Di Conza G, Pellegrino M, Rinaldo C, Prodosmo A, Giglio S et al. MDM4 (MDMX) localizes at the mitochondria and facilitates the p53-mediated intrinsic-apoptotic pathway. EMBO J 2009; 28: 1926–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Woods NT, Piluso LG, Lee H-H, Chen J, Bhalla KN et al. p53 acetylation is crucial for its transcription-independent proapoptotic functions. J Biol Chem 2009; 284: 11171–11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-Y, Park B-J, Kim D-J, Kim W-H, Kim S, Oh K-S et al. Modification of serine 392 is a critical event in the regulation of p53 nuclear export and stability. FEBS Lett 2004; 572: 92–98. [DOI] [PubMed] [Google Scholar]
- Ahn BY, Trinh DLN, Zajchowski LD, Lee B, Elwi AN, Kim S-W. Tid1 is a new regulator of p53 mitochondrial translocation and apoptosis in cancer. Oncogene 2010; 29: 1155–1166. [DOI] [PubMed] [Google Scholar]
- Trinh DLN, Elwi AN, Kim S-W. Direct interaction between p53 and Tid1 proteins affects p53 mitochondrial localization and apoptosis. Oncotarget 2010; 1: 396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietsch EC, JI-J Leu, Frank A, Dumont P, George DL, Murphy ME. The tetramerization domain of p53 is required for efficient BAK oligomerization. Cancer Biol Ther 2007; 6: 1576–1583. [DOI] [PubMed] [Google Scholar]
- Scorrano L, Oakes SA, Opferman JT, Cheng EH, Sorcinelli MD, Pozzan T et al. BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science 2003; 300: 135–139. [DOI] [PubMed] [Google Scholar]
- Todt F, Cakir Z, Reichenbach F, Emschermann F, Lauterwasser J, Kaiser A et al. Differential retrotranslocation of mitochondrial Bax and Bak. EMBO J 2015; 34: 67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YT, Wolter KG, Youle RJ. Cytosol-to-membrane redistribution of Bax and Bcl-X(L) during apoptosis. Proc Natl Acad Sci USA 1997; 94: 3668–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruins W, Bruning O, Jonker MJ, Zwart E, Hoeven TV, van der, Pennings JLA et al. The absence of Ser389 phosphorylation in p53 affects the basal gene expression level of many p53-dependent genes and alters the biphasic response to UV exposure in mouse embryonic fibroblasts. Mol Cell Biol 2008; 28: 1974–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughery J, Cox M, Smith LM, Meek DW. Critical role for p53-serine 15 phosphorylation in stimulating transactivation at p53-responsive promoters. Nucleic Acids Res 2014; 42: 7666–7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchenko ND, Wolff S, Erster S, Becker K, Moll UM. Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J 2007; 26: 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes SM, Stanek TJ, Frank A, Murphy ME, McMahon SB. Acetylation of the DNA binding domain regulates transcription-independent apoptosis by p53. J Biol Chem 2009; 284: 20197–20205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrogiovanni C, Vandaudenard M, Waterschoot B, De Backer O, Dumont P. Decrease of mitochondrial p53 during late apoptosis is linked to its dephosphorylation on serine 20. Cancer Biol Ther 2015; 16: 1296–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Li C-F, Zhang L, Wu C-Y, Han L, Jin G et al. TRAF6 Restricts p53 Mitochondrial Translocation, Apoptosis, and Tumor Suppression. Mol Cell 2016; 64: 803–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-López LA, Díaz-Flores M, García-Macedo R, Ávalos-Rodríguez A, Vergara-Onofre M, Cruz M et al. High glucose induces mitochondrial p53 phosphorylation by p38 MAPK in pancreatic RINm5F cells. Mol Biol Rep 2013; 40: 4947–4958. [DOI] [PubMed] [Google Scholar]
- Speidel D. Transcription-independent p53 apoptosis: an alternative route to death. Trends Cell Biol 2010; 20: 14–24. [DOI] [PubMed] [Google Scholar]
- Marchenko ND, Moll UM. Mitochondrial death functions of p53. Mol Cell Oncol 2014; 1: e955995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Qiu W, Dudgeon C, Liu H, Huang C, Zambetti GP et al. PUMA is directly activated by NF-kappaB and contributes to TNF-alpha-induced apoptosis. Cell Death Differ 2009; 16: 1192–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim RH, Kang MK, Kim T, Yang P, Bae S, Williams DW et al. Regulation of p53 during senescence in normal human keratinocytes. Aging Cell 2015; 14: 838–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Luo J, Chen F, Yang F, Song W, Zhu A et al. BRCA1 regulates PIG3-mediated apoptosis in a p53-dependent manner. Oncotarget 2015; 6: 7608–7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capitani N, Lucherini OM, Sozzi E, Ferro M, Giommoni N, Finetti F et al. Impaired expression of p66Shc, a novel regulator of B-cell survival, in chronic lymphocytic leukemia. Blood 2010; 115: 3726–3736. [DOI] [PubMed] [Google Scholar]
- Larson Gedman A, Chen Q, Kugel Desmoulin S, Ge Y, LaFiura K, Haska CL et al. The impact of NOTCH1, FBW7 and PTEN mutations on prognosis and downstream signaling in pediatric T-cell acute lymphoblastic leukemia: a report from the Children’s Oncology Group. Leukemia 2009; 23: 1417–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.