Abstract
The ability of the pH-Low Insertion Peptide (pHLIP) to insert into lipid membranes in a transbilayer conformation makes it an important tool for targeting acidic diseased tissues. pHLIP can also serve as a model template for thermodynamic studies of membrane insertion. We use intrinsic fluorescence and circular dichroism spectroscopy to examine the effect of replacing pHLIP’s central proline on the pH-triggered lipid-dependent conformational switching of the peptide. We find that the P20G variant (pHLIP-P20G) has a higher helical propensity than the native pHLIP (pHLIP-WT), in both water:organic solvent mixtures and in the presence of lipid bilayers. Spectral shifts of tryptophan fluorescence reveal that with both pHLIP-WT and pHLIP-P20G, the deeply penetrating interfacial form (traditionally called State II) is populated only in pure phosphocholine bilayers. The presence of either anionic lipids or phosphatidylethanolamine leads to a much shallower penetration of the peptide (referred to here as State IIS, for “shallow”). This novel state can be differentiated from soluble state by a reduction in accessibility of tryptophans to acrylamide and by FRET to vesicles doped with Dansyl-PE, but not by a spectral shift in fluorescence emission. FRET experiments indicate free energies for interfacial partitioning range from 6.2 to 6.8 kcal/mol and are marginally more favorable for pHLIP-P20G. The effective pKa for the insertion of both peptides depends on the lipid composition, but is always higher for pHLIP-P20G than for pHLIP-WT by approximately one pH unit, which corresponds to a difference of 1.3 kcal/mol in free energy of protonation favoring insertion of pHLIP-P20G.
Keywords: pH-dependent membrane insertion, tryptophan fluorescence, thermodynamics, conformational switching, transmembrane helix
Graphical abstract
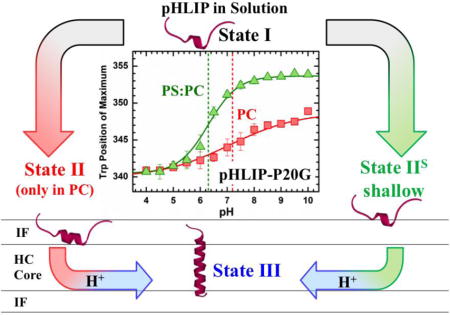
INTRODUCTION
The discovery of pHLIP (pH-Low Insertion Peptide) is a compelling example of how a basic-science study can be translated into a very useful and practical biomedical tool. While originally designed from the sequence of the C-helix of bacteriorhodopsin to study membrane protein folding [1], pHLIP is currently and most often used for imaging tumors and other acidic diseased tissues [2–9], as well as for targeted drug delivery [10–13]. The underlying mechanism of pHLIP’s pH-dependent action is associated with the protonation of Asp residues, allowing for membrane insertion at acidic pH [14–16]. Many other mechanistic and thermodynamic aspects of membrane interactions of pHLIP, however, remain the subjects of active studies.
Traditionally, membrane interactions of pHLIP are described in the context of the following three states, which were originally identified for partitioning into phosphatidylcholine (PC) bilayers [17]: unfolded and soluble state in water at neutral pH (State I), unfolded interfacial state at neutral pH (State II), and a transmembrane α-helical state at low pH (State III). Each of the states has a defined spectroscopic signature associated with the amount of secondary structure (measured by circular dichroism) and membrane penetration (blue shift of Trp fluorescence spectrum relative to that of the solution State I). In our previous study we challenged this view for bilayers of mixed lipid composition by demonstrating that the spectroscopic signature of the interfacial State II is only observed in pure PC bilayers [18].
A surprising feature of pHLIP is the apparent lack of folding upon interfacial penetration, which sets it apart from numerous membrane active peptides that undergo a partitioning-folding coupling [19–21]. To examine the effects of helical propensity on membrane interactions of pHLIP, we examine here the folding and membrane interactions of the native peptide (referred to here as the wild type, pHLIP-WT) and those of the P20G variant (pHLIP-P20G), lacking the helix-breaking proline in the middle of the hydrophobic stretch. Both pHLIP-WT and pHLIP-P20G have been previously used by Engelman and co-workers [15] to investigate the effect of acyl chain-length on membrane insertion in the context of phosphatidylcholine membranes. They have demonstrated that both membrane thickness and fluidity modulate pHLIP membrane insertion and that Pro20-to-Gly replacement resulted in a more favorable membrane insertion. By examining the pH range from 4.5 to 8, the authors concluded that the insertion pKa for pHLIP-P20G is less acidic than that of the pHLIP-WT, with the difference ranging from 0.1 to 1.0 pH units, for 22 and 14 carbon lipids, respectively. Here, we reexamine the pH-titration of both peptides into both pure POPC bilayers and bilayers containing 25% POPE or 25% POPS in POPC matrix. We find that a wider range of pH (4.5–10) is necessary to accurately characterize the insertion and that the difference for the insertion of the two peptides may be larger than reported. Our results indicate that, in all three lipid compositions, the difference is approximately 1 unit of pH and that, consistent with our previous report [18], lipid headgroups modulate the insertion of pHLIP. We introduce a revised version of the insertion scheme, which contains an alternative interfacial State IIS, to indicate a more “shallow” bilayer penetration as compared to traditional interfacial State II.
MATERIALS AND METHODS
Materials and Lipid Abbreviations
N-Hydroxybenzotriazole (HOBt), o-benzotriazol-N, N, N, N′, N′-tetramethyluronium hexafluorophosphate (HBTU), and all N-fluorenyl-9-methoxycarbonyl (Fmoc) protected L-amino acids were purchased from GL Biochem Ltd. H-Rink Amide-ChemMatrix solid support resin was purchased from PCAS BioMatrix Inc. Diisopropylehtylamine (DIEA), piperazine, N, N-dimethylformamide (DMF), dichloromethane (DCM) and trifluoroacetic acid (TFA) were from Thermo Fisher Scientific Inc. All lipids were purchased from Avanti Polar Lipids (Alabaster, AL). 1‐palmitoyl‐2‐oleoyl‐sn‐glycero‐3‐phospholcholine (POPC), 1‐palmitoyl‐2‐oleoyl‐sn‐glycero‐3‐phosphoethanolamine (POPE), 1‐palmitoyl‐2‐oleoyl‐sn‐glycero‐3‐phosphoserine (POPS).
Peptide Synthesis
pHLIP-WT (H2N-AAEQNPIYWARYADWLFTTPLLLLDLALLVDADEGG-CONH2) and pHLIP-P20G (H2N-AAEQNPIYWARYADWLFTTGLLLLDLALLVDADEGG-CONH2) were prepared by Fmoc solid-phase synthesis as described previously [10–13], using H-Rink Amide-ChemMatrix resin affording an amidated C-terminus and purified via reverse-phase high performance liquid chromatography (RP-HPLC) (Phenomenex Luna prep 10 μ 250 × 21.20 mm C8; flow rate 10mL/min; phase A: water 0.1% trifluoroacetic acid (TFA); phase B: acetonitrile 0.1% TFA; gradient 60 min from 95/5 A/B to 0/pure A/B). The purity of the peptides was determined by RP-HPLC (Agilent Zorbax Eclipse 5 μm 4.6 × 50 mm XDB-C8; flow rate 1 mL/min; phase A: water 0.01% TFA; phase B: acetonitrile 0.01% TFA; gradient 45 min from 95/5 A/B to 0/pure A/B) and their identity was confirmed via matrix-assisted laser desorption ionization time of flight (MALDI-TOF) mass spectrometry.
Preparation of lipid vesicles
Large Unilamellar Vesicles (LUV) were prepared by drying the required volume of chloroform lipid stocks under a nitrogen stream before overnight drying using high vacuum. Dried lipid films were re-suspended in 50 mM phosphate buffer (pH 8) to a final concentration of 20 mM and vortexed. For FRET measurements, 2% of Dansyl-PE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(5-dimethylamino-1-naphthalenesulfonyl) was incorporated into lipid mixture. LUV were formed by extrusion using a Mini-Extruder (Avanti Polar Lipids, Alabaster, AL) through nucleopore polycarbonate membranes of 0.1 μm pore size (Whatman, Philadelphia, PA) [22, 23]. LUV stocks were prepared in 50mM phosphate buffer, pH 8, and stored at −4°C. No lipid degradation was detected using thin layer chromatography.
Sample preparation
2 μM of pHLIP were mixed and incubated with LUV stock in 10 mM phosphate, pH 8 or 10 mM borate buffer, pH 10, resulting in molar lipid to peptide ratios of at least 500:1 (1–2 mM LUV). Membrane insertion of pHLIP was initiated by addition of the appropriate aliquots of 0.5 M acetate buffer. To avoid potential lipid degradation, the lipid-containing samples were never kept at pH 10 longer than 20 min.
Fluorescence Measurements
Fluorescence emission was measured using a SPEX Fluorolog FL3-22 steady-state fluorescence spectrometer (Jobin Yvon, Edison, NJ) equipped with double-grating excitation and emission monochromators. The measurements were made in a 2×10 mm cuvette oriented perpendicular to the excitation beam and maintained at 25 °C using a Peltier device from Quantum Northwest (Spokane, WA). For tryptophan fluorescence measurements, the excitation wavelength was 285 nm and the slits were 4 and 6 nm for emission and excitation, respectively. For acrylamide quenching, 295 nm excitation was used to reduce inner filter effects and 3 nm slits. The appropriate background spectra were subtracted in all cases. Spectral analysis was carried out using Origin 8.5 (OriginLab, MA).
Membrane partitioning
Membrane partitioning was measured by fluorescence titration of peptides with LUV containing 2% Dansyl-PE as acceptors. Interfacial partitioning is indicated by a decrease in the intensity of the donor Trp peak as it associates with the interface as a function of LUV concentration (Fig. S4). Results are plotted as the relative decrease in Trp intensity, I, versus lipid concentration, [L], and fitted as described previously using the following equation [24]:
| (Eq. 1) |
where I∞ denotes the relative intensity at infinite lipid saturation, [W] is the concentration of water (55.3 M) and Kx represents the mole fraction partitioning coefficient [25]:
| (Eq. 2) |
where [Pbil] and [Pwater] corresponds to the bulk concentrations of peptide in the bilayer and in water respectively. The calculated partitioning constant (Kx) was used to determine the free energy of interfacial partitioning (ΔGIF) using the following formula:
| (Eq. 3) |
Where R is the gas constant (1.985×10−3 kcal K−1 mol−1) and T is the experimental temperature in Kelvin (298 K). Errors in the fits were determined by support plane analysis [26] and are indicated in brackets (Fig. S5–6).
Analysis of the pH-dependent membrane insertion
The spectral data were fitted by applying nonlinear least-square analysis with the following equation [27]:
| (Eq. 4) |
Where λN and λL are the limiting values of fluorescence maximum at neutral and low pH, respectively; pKa is an apparent constant and m is a transition slope. Errors in the fits were determined by support plane analysis and are indicated in brackets (Fig. S7–8). The protonation-dependent free energy of transmembrane (TM) insertion, ΔGTMH+, was calculated using the following formula:
| (Eq. 5) |
Where R is the gas constant (1.985×10−3 kcal K−1 mol−1) and T is the experimental temperature in Kelvin (298 K).
CD experiments
CD measurements were performed using an upgraded Jasco-720 spectropolarimeter (Japan Spectroscopic Company, Tokyo). Normally 60–80 scans were recorded using a 1-mm optical path cuvette. All CD spectra were corrected for background. Percent helical folding was calculated assuming ellipticity at 222 nm corresponds only to α-helical content following the methodology proposed by Chen et al [28]:
| (Eq. 6) |
Where [θ]222 is the observed ellipticity at 222 nm, is the theoretical mean residue ellipticity for an infinitely long helical peptide (−39,500 deg cm2 dmol−1), n is the number of residues (36 in pHLIP), and k is a wavelength dependent constant (2.57 at 222 nm) [28].
RESULTS
Folding of pHLIP-WT and pHLIP-P20G in TFE
In solution, pHLIP exists as a soluble and unstructured peptide; capable of inserting into lipid bilayers as a transmembrane α-helix under mildly acidic conditions. The unstructured conformation of pHLIP in solution is maintained by electrostatic repulsion between six anionic residues throughout its sequence and two proline residues (Table 1). Several studies have shown the importance of the anionic residues on the transmembrane transition of the peptide, but little attention has been paid to the prolines. Pro20, in particular, is in the middle of the region that transitions into a transmembrane helix. To better understand the role of Pro20 on the transition of pHLIP to a transmembrane helix, we studied the folding of both pHLIP-WT and pHLIP-P20G in 2,2,2-trifluoroethanol (TFE). This organic solvent is known to induce α-helical folding of unstructured peptides [29, 30], presumably by disrupting hydrogen bonding with water, while stabilizing intramolecular hydrogen bonding [31]. As such, TFE has been traditionally used to study the α-helical folding of disordered peptides in solution and also as a simplest mimetic for thermodynamic studies of folding of membrane-active peptides [20, 32].
Table 1. pHLIP Sequences.
Both pHLIP sequences used in this study are composed of 36 amino acids total, including six anionic residues (blue). The pHLIP-P20G variant contains a Gly substitution at position Pro20 (red); located in the middle of the transmembrane region of the peptide.
| Peptide | Sequence |
|---|---|
| pHLIP-WT | AAEQNPIYWARYADWLFTTPLLLLDLALLVDADEGG |
| pHLIP-P20G | AAEQNPIYWARYADWLFTTGLLLLDLALLVDADEGG |
TFE-induced conformational changes of both pHLIP-WT and P20G were assessed by circular dichroism spectroscopy (CD). This technique allows for a simple readout of the folding transition, as it provides characteristic spectra for both unstructured and α-helical conformations. In aqueous solution (0 M TFE), pHLIP-WT showed a characteristic spectrum for an unstructured State I peptide, with a single minimum at 200 nm (Fig. 1a). Meanwhile, a spectrum with a double minimum around 208 and 222 nm, characteristic of α-helices, is obtained by substituting Pro20 for Gly. The measured molar ellipticity at 222 nm can be used to estimate the helical content of peptides as described by Chen et al. [28]. Using this method, we calculated the helical content to be 12% and 21% for pHLIP-WT and pHLIP-P20G in solution, respectively. In peptides of 36 amino acid residues in length, this amounts to only 4 and 7 folded residues in pHLIP-WT and pHLIP-P20G, respectively (Table 2). Maximum peptide helical content can be achieved by placing them in pure organic solvent, 14 M TFE. As expected, while both pHLIP variants showed α-helical spectra in TFE (Fig. 1a), pHLIP-WT is shown to remain more unstructured, with an estimated 36% helicity (13 folded residues) as opposed to 47% for pHLIP-P20G (17 folded residues).
FIGURE 1. Folding of pHLIP in TFE.
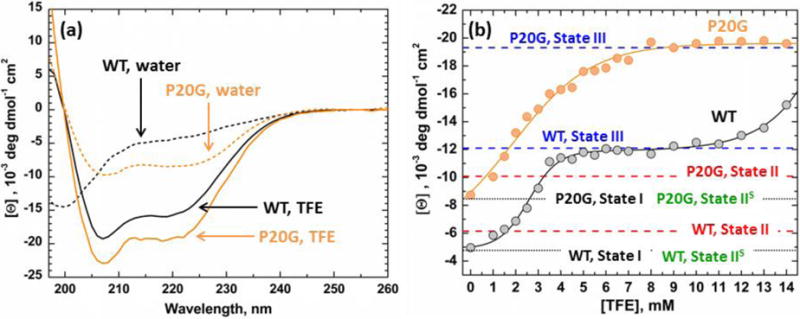
(a) Circular Dichroism spectroscopy was used to characterize the folding of pHLIP-WT (grey) and pHLIP-P20G (orange) as a function of TFE concentration. In water, both peptides start in their relatively unstructured State I (dashed lines) and transition to α-helices in higher TFE concentration (solid lines). (b) Measured molar ellipticity at 222 nm, an indication of helical content, was plotted against experimental [TFE]. As TFE concentration increases, pHLIP-P20G achieves maximum ellipticity at 9 M TFE with a sigmoidal transition. Under the same conditions pHLIP-WT undergoes a double transition with an initial plateau between 4–9 M TFE. A second transition starts after 9 M TFE and never achieves the second plateau, indicating that maximum ellipticity is not obtained. In membranes, the average ellipticity of the inserted transmembrane State III, under acidic conditions, of both peptides matched the observed plateaus in TFE (blue lines). Only a small increase in ellipticity was observed in pure POPC for each peptide State II at pH 10 (red lines). This indicates the lack of a folded interfacial form even after the P20G substitution. No change in ellipticity was observed for either peptide in non-pure POPC membranes at pH 10 (Fig. S1–3) in their respective State IIS (green lines). The black line indicates the starting ellipticity for each peptide in aqueous environment (State I).
Table 2. Helical content of pHLIP variants in LUV.
Circular Dichroism was used to measure the conformation of both pHLIP-WT and P20G in increasing TFE concentrations (Fig. S1–3). Helical fractions (fα) were calculated using estimates of maximum helicity discussed by Chen et al. [28] and described in the methods section (Eq. 6). Number of folded residues (nα) was determined using a 36 residue total for both peptides.*Number of folded residues adjusted by the fraction of peptide bound to POPC membranes, based on FRET binding data (Fig. 3)
| Sample | State | pHLIP-WT | State | pHLIP-P20G | |||
|---|---|---|---|---|---|---|---|
| fα | nα | fα | nα | ||||
| Water | pH 10 | I | 12 | 4 | I | 21 | 7 |
| pure POPC | pH 10 | II | 14 | 5 (6*) | II | 23 | 8 |
| pH 8 | II | 14 | 5 | II–III | 26 | 9 | |
| pH 4 | III | 26 | 10 | III | 48 | 17 | |
| 25POPE:75POPC | pH 10 | IIS | 12 | 4 | IIS | 18 | 7 |
| pH 8 | IIS | 12 | 4 | IIS–III | 24 | 9 | |
| pH 4 | III | 27 | 10 | III | 48 | 17 | |
| 25POPS:75POPC | pH 10 | IIS | 12 | 4 | IIS | 18 | 7 |
| pH 8 | IIS | 12 | 4 | IIS–III | 21 | 7 | |
| pH 4 | III | 27 | 10 | III | 45 | 16 | |
The folding transition of both pHLIP-WT and P20G was studied in more detail by measuring their CD at increasing TFE:water ratios and plotting the ellipticity at 222 nm vs [TFE] (Fig. 1b). This analysis revealed a large difference in the folding behavior of both pHLIP variants. As TFE concentration increases, pHLIP-P20G folds into an α-helical structure in a single sigmoidal transition, achieving maximum ellipticity at 9 M TFE (47% helicity, 17 folded residues). In contrast, pHLIP-WT exhibits two transitions: first reaching a plateau between 5–10 M TFE (27% helicity, 10 residues in helical conformation). A second transition begins above 10 M TFE and does not reach completion even in pure TFE. These results show that Pro20 is not only important in determining the maximum helical content that pHLIP can achieve, but also modulates its folding behavior in hydrophobic environments. The sequential folding observed with pHLIP-WT upon increase in TFE concentration suggests that Pro20 is breaking the helix into two independently folding segments with different helical propensities.
Folding of pHLIPs in lipid bilayer
In membranes composed purely of POPC, pHLIP-WT has been shown to form an interfacial and unstructured State II [1]. This state is characterized by a blue shift in Trp emission spectra at pH 8 from 357 nm in solution to 354 nm in the membrane interface, indicative of membrane association (Fig. S1a). This spectral change is also accompanied by a slight increase in secondary structure, measured by CD (Fig. S1b; ellipticity at 222 nm) [1]. In a previous study, however, we demonstrated extensively that this interfacial State II is not identifiable by either technique in any membrane composition tested other than pure POPC [18]. Even the presence of only 10% of a different phospholipid (i.e., phosphatidylserine, phosphatidylglycerol, phosphatidic acid, phosphatidylethanolamine and others) or cholesterol eliminated these spectral differences [18], traditionally used as the hallmarks of pHLIP’s interfacial State II.
Interfacial membrane partitioning of unstructured peptides often results in gain in secondary structure due to partitioning-folding coupling, driven by a relatively high gain in per-residue folding free energy [19, 21]. Therefore, the lack of helix formation in pHLIP during its interfacial partitioning to State II is rather unusual. To better understand the contribution of Pro20 in the peptide’s folding, we compared conformational changes of pHLIP-WT and pHLIP-P20G upon membrane addition by using CD and Trp fluorescence spectroscopy. These experiments were conducted on membranes composed either purely of POPC, or containing 25 molar percent POPS or POPE in POPC matrix.
We first measured conformational changes in both pHLIP variants under conditions known to induce formation of the interfacial State II in pHLIP-WT, namely the addition of LUV containing only POPC at pH 8. It resulted in characteristic blue shifts to its Trp spectra (357 nm to 354 nm; Fig. S1a) and small changes to its CD spectra, indicative of membrane interaction and a small gain of secondary structure (Fig. S1b, red vs black spectra). Under the same conditions (i.e., POPC LUV at pH 8), pHLIP-P20G exhibited a more pronounced change in emission blue shift (354 nm to 348 nm) and an increase in secondary structure (Fig. S1d). The larger changes in the Trp and CD spectra observed for pHLIP-P20G in POPC at pH 8 are consistent with previous reports [15], which concluded that State II for this mutant has a very high helical content. The alternative explanation would be that the pH-dependent transition from States II-to-III in pHLIP-P20G is shifted. Our data presented in the next section confirm that this is indeed the case (Fig. 4 and S9). Therefore, the standard condition for State II for pHLIP-P20G is not pH 8, as it is for the WT-pHLIP, but pH 10. (Fig. S1d red vs green spectra and Fig. 4). For this reason, all further measurements discussed in this report involving the State II were performed at pH 10.
FIGURE 4. pH-Dependent membrane insertion of pHLIP-WT (black) and pHLIP-P20G (orange) in pure POPC (a), 25POPE:75POPC (b) and 25POPS:75POPC (c), monitored by Trp fluorescence emission maximum.
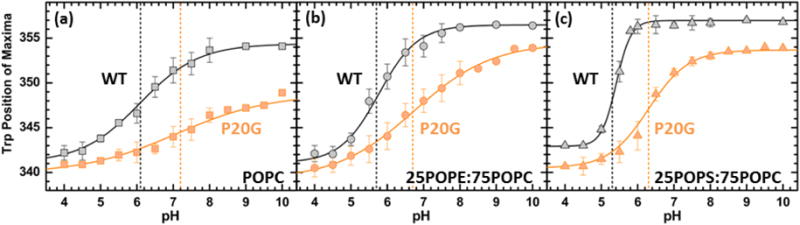
Insertion into membranes is indicated by progressive blue shifts, decreases in position of maxima, in the measured Trp emission spectra. Dashed lines indicate the pH-dependent membrane insertion pKa color coded for each pHLIP variant. The following pKa values were obtained by fitting the data to Eq. 4: (a) In POPC: WT = 6.1 [6.0;6.3] (m = 0.6 [0.5;0.7]) and P20G = 7.2 [7.0;7.4] (m = 0.4 [0.3;0.6]), (b) in 25POPE:75POPC: WT = 5.6 [5.5;5.7] (m = 0.9 [0.7;1.1]) and P20G = 6.7 [6.6;6.9] (m = 0.4 [0.3;0.5]), (c) in 25POPS:75POPC: WT = 5.4 [5.4;5.5] (m = 1.7 [1.5;2.0]) and P20G = 6.4 [6.2;6.5] (m = 0.8 [0.6;1.1]). Decrease Trp position of maxima from State I in solution, WT: 357 nm and P20G: 354 nm, in pure POPC indicates formation of State II. No changes in starting positions of maxima were observed in the pH-titrations of either 25POPE:75POPC or 25POPS:75POPC. All data points in the titrations contain error bars and are represented as the average of 3 measurements. Regardless of membrane composition, the protonation-dependent insertion of pHLIP-P20G is more favorable than that of the pHLIP-WT by 1.3 kcal/mol (equivalent of the 1 pH unit difference in pKa). Errors in the fit were calculated by support plane analysis for the protonation-dependent insertion pKa (Fig. S7) and the slope (Fig. S8).
Under acidic conditions in the presence of membranes the CD spectra of both pHLIP-WT (blue spectra in Fig. S1b, S2b, and S3b) and pHLIP-P20G (blue spectra in Fig. S1d, S2d, and S3d) contain a double minimum, characteristic of α-helices. Both peptides show conformational differences between lipid compositions consistent with our previous report [18], however, those are more pronounced in the pHLIP-P20G variant. Under acidic conditions pHLIP-P20G shows a spectral distortion below 210 nm that could be attributed to absorbance flattening artifacts (possibly due to non-uniform distribution in different vesicles). Similar variations in CD spectra of the transmembrane State III of pHLIP variants have been observed before, however, the exact reason behind it remains unclear [8, 14]. Here, we follow the example of the Engelman group [8, 14] and use the ellipticity measured at 222 nm to analyze the spectra of both pHLIP variants, which reduces the uncertainty associated with potential absorbance flattening (We emphasize that only the folding of pHLIP-P20G in State III is potentially affected, as no other samples exhibit the signs of spectral distortion).
We mapped the observed ellipticities for the different states of both peptides in the presence of membranes to their corresponding folding in TFE (Fig 1b, black vs red dashed lines). Substituting Pro20 for Gly in pHLIP allowed for higher overall interfacial folding on pure POPC bilayers (23% folded helix, 8 folded residues) compared to pHLIP-WT (14% folded helix, 6 folded residues). Spectroscopic measurements in the presence of 25POPS:75POPC and 25POPE:75POPC LUV showed no change in folding or position of Trp emission as compared to the solution State I (Fig. S2a–d, S3a–d). This absence of the expected spectroscopic signature for State II in both peptides in the presence of membranes of mixed lipid compositions is consistent with our previous pHLIP-WT results [18], and suggests that it could be a general property of the entire pHLIP family. The observed lack of a measurable blue shift in the Trp emission spectra or increase in secondary structure upon addition of LUV with mixed lipid composition at neutral to basic pH can be explained by two different possibilities: (a) pHLIP is not partitioning to these bilayers at neutral to slightly basic pH or (b) pHLIP does interact with the membrane interface but does not penetrate deep enough to cause changes in folding or to alter the environment of Trp residues. To resolve these two possibilities, we performed fluorescence quenching and Förster Resonance Energy Transfer (FRET) experiments described in subsequent sections.
The ellipticities of both peptides at 222 nm in the presence of membranes at low pH, State III, were also compared to those in TFE (Fig. 1b blue dashed lines). The ellipticities calculated for both pHLIP-WT and P20G matched their respective plateaus observed in TFE. In the case of pHLIP-WT this amounts to an average ellipticity of 26% (10 folded residues); while an average higher helical content of 46% percent (17 folded residues) was observed for pHLIP-P20G.
Fluorescence quenching with acrylamide (Identification of a new interfacial state)
Acrylamide quenching measurements were performed under conditions that allow for the formation of the interfacial State II in both pHLIP-WT and P20G and compared to their quenching in solution.
In solution, the addition of the soluble quencher acrylamide interacts with both Trp in pHLIP and reduces their observed intensity. The interaction of pHLIP with membrane interfaces, however, provides protection from acrylamide quenching, reducing its effect. Quenching constants, KSV, are obtained from the slope of intensity measurements in the absence/presence of quencher versus acrylamide concentration (Fig. 2), with higher values indicating more effective quenching. Measurements of both peptides in solution showed very similar quenching constants. We attribute the small difference between the 15.5 ± 0.1 M−1 KSV of pHLIP-WT and 14.4 ± 0.2 M−1 of pHLIP-P20G to the slightly higher protection from the higher secondary structure content of the latter (Fig. 2). The addition of LUV containing only POPC at pH 10 greatly reduce the measured KSV of both pHLIP-WT (KSV = 3.7 ± 0.1 M−1) and pHLIP-P20G (KSV = 3.1 ± 0.1 M−1) compared to solution (Fig. 2). The decrease in quenching constants observed after the addition of pure POPC LUV indicates that both pHLIP-WT and pHLIP-P20G partition to these interfaces, which is consistent with State II formation.
FIGURE 2. Stern-Volmer plots for acrylamide quenching of tryptophan fluorescence of pHLIP-WP (a) and pHLIP-P20G (b) measured at pH 10 in the absence and presence of membranes.
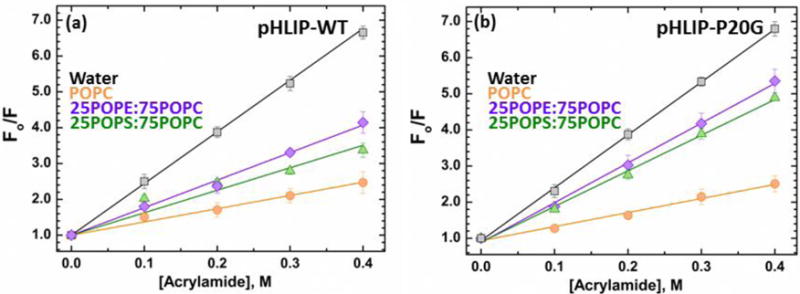
Stern-Volmer constants (KSV) were obtained from the slope of the measured intensities in the absence of quencher divided by the intensities in the presence of acrylamide vs quencher concentration. The decrease in KSV observed for both peptides in the presence of membranes compared to solution (Water KSV: pHLIP-WT = 15.1 ± 0.1 M−1, pHLIP-P20G = 14.4 ± 0.2 M−1) indicates their interaction with the membranes. The reduction in KSV is higher for either pHLIP variants in pure POPC membranes (pHLIP-WT KSV = 3.7 ± 0.1 M−1, pHLIP-P20G KSV = 3.1 ± 0.1 M−1) compared to lipids with mixed lipid compositions: 25POPE:75POPC LUV (pHLIP-WT KSV = 7.2 ± 0.2 M−1, pHLIP-P20G KSV = 9.8 ± 0.2 M−1) and 25POPS:75POPC (pHLIP-WT KSV = 6.1 ± 0.4 M−1, pHLIP-P20G KSV = 9.5 ± 0.3 M−1).
Acrylamide quenching measurements for both pHLIP variants in the presence of membranes containing either 25POPE:75POPC or 25POPS:75POPC at pH 10 also showed a decrease in their respective KSV compared to solution (Fig. 2). The observed reduction in quenching under these conditions, suggest that pHLIP is in fact partitioning to the membranes, despite being spectroscopically silent. The KSV measured for pHLIP-WT in LUV with mixed lipid compositions (7.2 ± 0.2 M−1 for 25POPE:75POPC and 6.1 ± 0.4 M−1 for 25POPS:75POPC) were higher than the one seen in POPC (3.7 ± 0.1 M−1), indicating lower protection from the membrane consistent with a more shallower interfacial location. Similarly, quenching measurements with pHLIP-P20G showed increased KSV values in 25POPE:75POPC (9.8 ± 0.3 M−1) and 25POPS:75POPC (9.5 ± 0.2 M−1) compared to pure POPC (3.1 ± 0.1 M−1). The presence of two Trp in the pHLIP constructs used in this study, however, do not allow for precise depth determination and prevent parsing out these slight differences. Future experiments using single Trp mutants will be performed to better understand the effect of interfacial properties on the depth of transmembrane pHLIP. Together, the combination of spectroscopic and quenching results indicates that pHLIP interacts with the interfaces of membranes with mixed lipid compositions without the partial folding or deeper penetration of Trp residues, characteristic of its State II in LUV containing only phosphatidylcholine. These results confirm our previous observation that the traditionally defined interfacial State II is observed only in pure PC bilayers [18]. In all mixed lipid bilayers both pHLIP-WT and pHLIP-P20G are populating a novel State IIS (for ‘shallow’ State II). This state is distinguished from State II by the lack of spectral shift of fluorescence and by lack of additional folding; and from State I by the reduction in acrylamide quenching.
Measuring the free energy for interfacial partitioning, ΔGIF
To confirm the existence of the interfacial State IIS, indirectly inferred from acrylamide experiments described above, we conducted the following membrane-binding experiments. To enhance spectroscopic response to membrane partitioning, we have doped LUV with 2% Dansyl-PE, a lipid labeled at its head group with a fluorophore that can act as a FRET acceptor for Trp fluorescence. As shown in supplemental Fig. S4, addition of such vesicles to pHLIP peptide results in a decrease of tryptophan fluorescence emission, which can be used to quantitate the partitioning. The decrease in pHLIP’s fluorescence intensity observed upon titration with Dansyl-PE-doped LUV is plotted in figure 3. The data are analyzed by fitting them to a standard partitioning equation (Eq. 1) commonly used in spectroscopic titration measurements [25]. The only distinction from more familiar cases of fluorescence increase upon partitioning (e.g., [24]), is that, in this case, we have fluorescence decrease and consequently the parameter corresponding to the ratio of intensities of completely bound peptide to that for the free peptide in solution is below one, I∞ <1. In order to explore how the variation in the latter parameter affects the precision of the determination of free energy of interfacial partitioning (ΔGIF), we performed a support-plane analysis [26]; which examines the variation of the goodness of fit (χ2 in this case) as a function of the value at which I∞ was fixed during the fit (Fig. S5). We used a cut-off of 1 standard deviation increase in χ2 over its lowest value to estimate the variation in ΔGIF that are presented in the square brackets after the most probable value of free energy below.
FIGURE 3. Membrane partitioning free energy of pHLIP-WT and P20G.
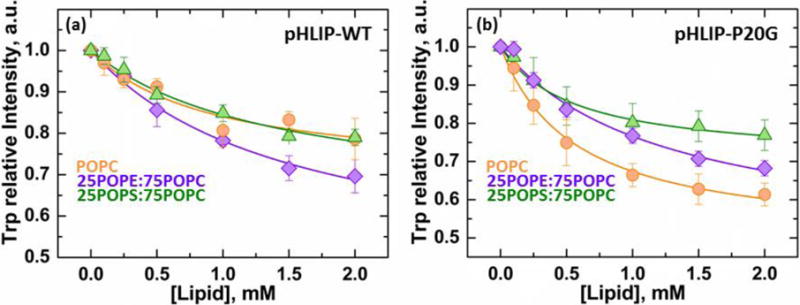
Partitioning of pHLIP to membranes was measured in duplicate by FRET between both Trp in pHLIP (donor) and 2% Dansyl-PE in LUV as acceptors (Fig. S4). The measured data was fit as explained in the methods section, and the interfacial partitioning coefficient (Kx), defined as the [Lipid] required for maximum intensity was used to calculate the partitioning free energy ΔGIF. Data is shown as the relative decrease in intensity of the Trp band as a function of lipid (acceptor) concentration for both pHLIP variants; errors were calculated by support plane analysis and indicated as a range in brackets (Fig. S5–6). (a) pHLIP-WT ΔGIF: POPC = −6.5 [−6.3; −6.7] kcal/mol (I∞ = 0.7)), 25POPE:75POPC = −6.2 [−6.1; −6.3] kcal/mol (I∞ = 0.4), 25POPS:75POPC = −6.3 [−6.1; −6.4] kcal/mol (I∞ = 0.6). (b) pHLIP-P20G ΔGIF: POPC = −6.8 [−6.7; −6.9] kcal/mol (I∞ = 0.5), 25POPE:75POPC = −6.3 [−6.2; −6.4] kcal/mol (I∞ = 0.5), 25POPS:75POPC = −6.8 [−6.8;−6.9] kcal/mol (I∞ = 0.7).
The free energy of interfacial partitioning into POPC (State II) was found to be ΔGIF = −6.8 [−6.7;−6.9] kcal/mol for pHLIP-P20G and ΔGIF = −6.5 [−6.3;−6.7] kcal/mol for pHLIP-WT (Fig.s. 3, S5). The latter corresponds well with previously reported value of −6.7 kcal/mol obtained by isothermal titration calorimetry by Reshetnyak et al. [33]. The values of ΔGIF for partitioning to the shallow interfacial State IIS, populated in mixed lipid compositions, were found to be similar to those of State II for both peptides (Fig. 3): (a) pHLIP-WT: ΔGIF = −6.3 [−6.1; −6.4] kcal/mol for 25POPS:75POPC and ΔGIF = −6.2 [−6.1; −6.3] kcal/mol for 25POPE:75POPC; (b) pHLIP-P20G: ΔGIF = −6.3 [−6.2; −6.4] kcal/mol for 25POPE:75POPC and ΔGIF = −6.8 [−6.7; −6.9] kcal/mol for 25POPS:75POPC LUV.
pH-dependent insertion into bilayers of various compositions
Under acidic conditions in the presence of membranes, pHLIP transitions into a transmembrane State III helix, regardless of membrane composition [18]. The transition to this transmembrane state is observed by increase in α-helical content (Fig. 1Sb and d, blue spectra) and by large blue shifts in Trp emission spectra from 357 to 341 nm, and from 354 to 343 nm, for pHLIP-WT and pHLIP-P20G, respectively (Fig. 1Sa and c, blue spectra). Under these conditions, CD measurements showed that the average ellipticity observed for both pHLIP variants in their transmembrane forms (Fig. S1–3, blue CD spectra) matched their respective plateaus in TFE (Fig. 1b, blue lines). The larger overall helical fraction of the transmembrane State III of pHLIP-P20G compared to that of pHLIP-WT (Table 2) indicates that the presence of Pro20 constrains the transmembrane conformation of pHLIP-WT. This difference in helicity of State III between the two peptides is about 6–7 residues and is independent of lipid composition (Table 2; pH 4).
The pH-dependent membrane insertion of both pHLIP-WT and pHLIP-P20G was characterized by measuring their Trp emission spectra in membranes containing purely POPC, 25POPE:75POPC or 25POPS:75POPC as a function of pH (Fig. 4). The pH range used in these experiments was extended to pH 10 to accommodate the formation of State II in pHLIP-P20G; which is formed under more basic conditions than pHLIP-WT (Fig. S10). The observed pH dependencies of the positions of maxima are fitted (Fig. 4a–c), as explained in the Methods section (Eq. 4), errors were calculated by support plane analysis and are shown in brackets (Fig. S7–8). Extending the pH range of the titration had no effect on the calculated insertion pKa of pHLIP-WT in pure POPC, remaining constant at 6.1 [6.0;6.3]. Meanwhile, the protonation-dependent insertion pKa of pHLIP-P20G increased to 7.2 [7.0;7.4] compared to 6.8 obtained in previous measurements performed using a shorter pH range (pH 8−4) [15]. The previously observed pKa of 6.8 for pHLIP-P20G in pure POPC by Barrera et al. [15] can still be calculated, if only the pH 4–8 range is considered (Fig. S10). Insertion of pHLIP into LUV containing mixed lipid compositions, which start their transition from the shallower interfacial State IIS, also show more favorable insertion for pHLIP-P20G. This can be appreciated in the 1.1 pH unit difference between the protonation-dependent insertion pKa of pHLIP-P20G into 25POPE:75POPC (pKa = 6.7 [6.6;6.9]) compared to pHLIP-WT (pKa = 5.6 [5.5;5.7]). A similar difference was observed between the insertion pKa of both peptides into 25POPS:75POPC membranes (pHLIP-WT: 5.4 [5.4;5.5], pHLIP-P20G: 6.4 [6.2;6.5]. Our titration measurements clearly show that interfacial properties affect the interaction and insertion of both pHLIP-WT and P20G into membranes. Not only by affecting its starting position (i.e., interfacial state), but also the protonation-dependent pKa of insertion. Under all membrane compositions tested, pHLIP-P20G showed an average insertion pKa 1 pH unit higher than those seen for pHLIP-WT (Fig. 4a–c).
Confirmation of the spectroscopically-silent State IIS by reversed fluorescence shift
In the absence of membranes, the protonation of anionic residues in acidic environments leads to the aggregation of pHLIP. This process creates a hydrophobic environment around both Trp in the interior of the aggregate. As with membrane insertion, these changes in environment can be detected spectroscopically by measuring the resulting blue shifts in the emission spectra of Trp. In solution, pHLIP-WT and pHLIP-P20G have a pKa of aggregation of 5.9 [5.9;6.0] (Fig. 5a) and 6.4 (Fig. S9), respectively. In the case of pHLIP-WT, its pH-dependent aggregation in solution can be compared to its protonation-driven insertion into membranes containing 25POPS:75POPC. In the presence of these LUV, pHLIP-WT has a pKa of 5.4 [5.4;5.5]; 0.5 pH units lower than the pKa for its aggregation in solution (Fig. 5a).
FIGURE 5. Interaction with membranes in mixed membrane composition.
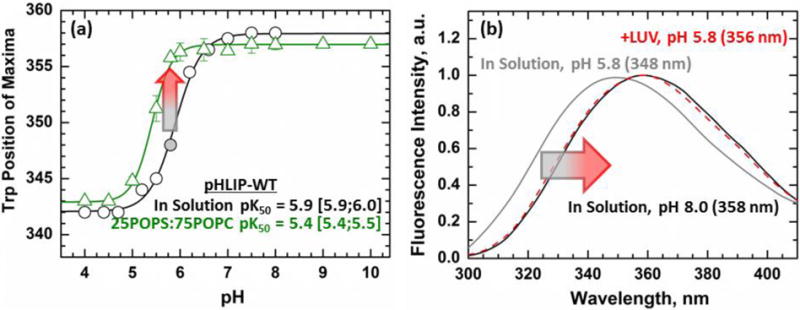
(a) Trp emission was used to measure the pH-dependent aggregation of pHLIP-WT in solution (black) and compared to the insertion of pHLIP in 25POPS:75POPC membranes (green). At pH 5.8 pHLIP-WT is in a partially aggregated form (348 nm, grey circle). At that same pH, little to no change is observed for the sample containing a peptide and 25POPS:75POPC LUV mixture (green) (b) The Trp spectra of the partial aggregate shows a large 10 nm blue shift with a position of maxima of 348 nm (grey) compared to 358 nm for the soluble peptide at pH 8 (black). Addition of membranes to this partially aggregated form results in an immediate red shift of the spectra from 348 nm (grey) to 356 nm (red).
Close to its aggregation pKa, at pH 5.8, pHLIP-WT is in a partially aggregated state in solution with a 10-nm blue shifted Trp position of maxima of 348 nm compared to 358 nm for its soluble State I (Fig. 5b, grey vs black spectra). At the same pH, pHLIP-WT in presence of 25POPS:75POPC LUV shows little to no change in Trp emission spectra from its State I in solution, with a position of maxima of 357 nm (Fig. 5a, green line), consistent with our previous report [18]. However, when 25POPS:75POPC LUV are added to partially aggregated pHLIP-WT at pH 5.8, an immediate redshift is observed in the Trp spectra to 356 nm (Fig. 5b, red spectra). For the recovery of Trp position of maxima to occur, pHLIP must be interacting with the membrane interface, which in turn causes the dissipation of the aggregates and formation of the interfacially bound state. This redshift of fluorescence provides additional confirmation of the presence of the State IIS identified by fluorescence quenching and FRET measurements.
The existence of the fluorescence red shift upon peptide binding to membranes is rather exotic and is observed for pHLIP only in a narrow pH window where solution aggregation already occurs, but full membrane insertion into State III has not yet happened. Because this window depends on a fortuitous combination of peptide properties in solution and in the membrane, this phenomenon is most pronounced for pHLIP-P20G in 25POPS:75POPC LUV. When similar measurements were made with pHLIP-WT, the difference between both spectra was found to be too small to be resolved (Fig. S9).
The tendency of pHLIP to aggregate even at moderate ionic strength (e.g., in a 50 mM phosphate buffer) is one of the challenges of using it as a targeted drug carrier [16]. Consequently, the available concentration of active/functioning pHLIP (i.e., available for binding and inserting in membranes) has always been in question. Our results indicate that, despite its tendency to aggregate in solution, pHLIP can disaggregate after binding to bilayer interfaces and become available for productive pH-mediated insertion.
DISCUSSION
The association and insertion of peptides into membranes is a complex process governed by an intricate interplay of hydrophobic and electrostatic interactions [25]. While substantial progress has been made in understanding the thermodynamics of interfacial partitioning to neutral [34] and charged bilayers [35], and interfacial folding [19, 21], the thermodynamics of transmembrane insertion present a formidable challenge due to solution aggregation of TM sequences [32, 36]. The pHLIP system bypasses this restriction by ensuring that the peptide is a monomer at (a) high pH in solution and (b) in a transmembrane state at acidic pH [17], thus making it into a useful model to study thermodynamics of membrane insertion.
Traditionally, the membrane insertion of this peptide has been described as a 3-state process [1]. In solution at neutral pH, pHLIP exists as a stable and unstructured State I peptide that can partition as a State II unstructured peptide to membrane interfaces. Under acidic conditions protonation of anionic residues allows its insertion into bilayers as a transmembrane State III α-helix. These properties make pHLIP amenable to thermodynamic membrane partitioning and insertion studies. Additionally, this peptide is of particular importance as its pH-dependent insertion into membranes is capable of directionally translocating various compounds into mildly acidic tissues. As such, it has been proposed as a targeted drug carrier into these environments, found in most solid-state tumors, inflamed tissues, and other disease conditions [2–13].
To simplify these complex systems, many thermodynamic studies involving protein-membrane interactions have been performed with membranes composed solely of phosphatidylcholine [1, 19, 36, 37]. As shown by Ladokhin and White [35], the presence of anionic lipids leads to non-additivity of free energy components and significantly complicates thermodynamic analysis. Similarly, pHLIP’s 3-state model has been confirmed only with this membrane composition [1]. The interaction of pHLIP with biological membranes, however, involves complex lipid compositions that could affect the traditional model. In our previous publication, we demonstrated that lipid headgroups can modulate the pH-dependent insertion of pHLIP and challenged the universality of the traditional 3-state model [18].
Using a combination of various fluorescence techniques, we show here that the lack of an interfacially unstructured State II observed in our previous report in lipid compositions other than pure POPC, was due to the formation of a shallower State (State IIS). Unlike the traditionally described interfacial State II in pure POPC membranes, the formation of this novel state does not result in Trp emission spectra blue shifts or measurable changes to its conformation. Instead, it is spectroscopically indistinguishable from its unstructured and soluble State I despite its association to the membrane interface. Here, we detected this new state in LUV containing mixed lipid compositions through acrylamide quenching measurements (Fig. 2), FRET between pHLIP and the acceptor-doped bilayers (Fig. 3). The identification of State IIS shows that the pH-dependent insertion of pHLIP is lipid-dependent and more complex than previously thought. In membranes composed of pure POPC, pHLIP follows the traditional 3-state pathway, passing through a deeply penetrating and relatively unstructured State II. In other lipid compositions, however, pHLIP transitions through a shallower State IIS, which is indistinguishable from its soluble State I by CD and position of Trp emission. They can be distinguished, however, by the presence of an unusual Trp red shift phenomenon that occurs when a partial pHLIP aggregate interacts and dissolves in membrane interfaces (Fig. 5). Observed under conditions where pHLIP appears to remain soluble in the presence of LUV, the measured Trp red shift shows that pHLIP-membrane interactions take place even if they are otherwise spectroscopically unresolved.
Taken together with our previous results on pHLIP-WT, we show that properties of the membrane interface have a large effect on the pH-dependent insertion of pHLIP; not only in modulating its insertion pKa, but also affecting the pathway towards insertion. Properties of the peptide itself can also affect this process. Most studies have focused on the role of the anionic Asp and Glu residues on the protonation-dependent insertion mechanism. Here, however, we inspected the role of Pro20, located in the middle of the transmembrane helix, and its role on pHLIP folding and insertion. This was done by comparing the folding of both pHLIP-WT and pHLIP-P20G variants using CD and tracking their interaction/insertion into membranes by Trp emission spectra and acrylamide quenching experiments. Substituting Pro20 for Gly had been previously observed to increase the insertion propensity of pHLIP in pure POPC membranes. We expanded this observation to membranes with different interfacial properties, previously shown to modulate pHLIP membrane insertion. Regardless of lipid composition, however, pHLIP-P20G maintained a higher insertion pKa of about 1 pH unit compared to pHLIP-WT.
Our CD measurements indicate that replacement of P20 has a profound effect on folding of pHLIP in solution and on the membrane. In water:TFE mixtures, pHLIP-WT folds in two-stage transition (Fig. 1), which is likely due to Pro20 inducing a kink and splitting the sequence into two segments with different folding propensities. Meanwhile, pHLIP-P20G folds with only a single sigmoidal transition and reaches higher helical content (Fig. 1) Similar results were observed in membranes, where pHLIP-P20G always achieved higher helical fraction than its pHLIP-WT counterpart. Interestingly, the replacement of P20 does not result in a folding interfacial state in any lipid composition. Because interfacial partitioning of unstructured peptides is thermodynamically coupled with their folding [19], the existence of unstructured States II and IIS is an unusual feature of pHLIP peptides and is possibly due to the lack of substantial hydrophobic moment.
For interfacial partitioning into vesicles of all lipid compositions, we consistently observe a marginally more favorable free energy ΔGIF for pHLIP-P20G, than that for pHLIP-WT (Fig. 3). In case of POPC (State II), this can be explained by taking in account two crucial but opposing factors: side chain hydrophobicity and free energy gain due to difference in partitioning folding coupling. For POPC, interfacial hydrophobicity scale favors the partitioning of pHLIP-WT [37]. In contrast, the difference in helix formation on the interface favors partitioning of the pHLIP-P20G by ΔGFolding ~ 0.4 kcal/mol per residue [19]. Thus, the difference in helical gain upon interfacial binding to vesicles between the two peptides (Fig. 3) is expected to make the partition of pHLIP-P20G slightly more favorable than that of pHLIP-WT. This explanation, however, cannot be automatically extended to mixed lipid compositions (State IIS), due to the lack of folding upon transfer from State I to State IIS for either peptide. It is also worth noting that interfacial hydrophobicity scale is applicable only to pure POPC and breaks down for mixed lipid compositions due to nonadditivity of various free energy components [35].
The exact reason to why the deep interfacial State II is only formed in pure PC bilayers remains puzzling, especially when considering that the presence of rather small fractions (as low as 10% [18]) of other lipids result in the formation of the shallower State IIS. One can assume that for anionic lipids such as POPS, electrostatic repulsion between charged headgroups and peptide’s anionic side chains play a role in disfavoring deeper penetration. In the case of POPE, however, this explanation is insufficient as this lipid caries no total charge. One can speculate that the reason POPE eliminated State II and converts it into State IIS might be related to ability of ethanolamine to form a network of hydrogen bonds in the interfacial region of the bilayer [38]. The exact mechanism of how this would affect the partitioning of pHLIP is not clear, however, the evidence exists that PE can engage proteins into hydrogen bonding [39]. The latter can be important in the case of unstructured peptides like pHLIP that have to satisfy their hydrogen bonds to be stable at the interface. One would expect interfacial interactions of pHLIP to become even more complex when the possibility of hydrogen bonding by headgroups in anionic phospholipids (e.g., phosphatidylserine [40, 41], phosphatidic acid [40]) is considered. The ability of pHLIP to form two different states on the interface in a lipid-dependent fashion shows the importance of membrane composition on protein-membrane interactions.
The pH-dependent insertion of pHLIP into bilayers can be coupled with the translocation of different compounds across plasma membranes. For this reason, pHLIP has been proposed as a vehicle for targeted drug delivery to mildly acidic tissues, such as tumors [2–10, 12]. Most of our knowledge of pHLIP-membrane interactions is derived from simple membrane models containing only phosphatidylcholine; however, cellular membranes are composed of many different lipid species. Our measurements using binary lipid compositions show that the protonation-dependent insertion of pHLIP into membranes is affected by interfacial properties of the membrane, modulated by lipid composition. Therefore, peculiarities of the membrane composition of the target should be taken into account for the rational design and optimization of pHLIP-derived drug delivery systems.
Our overall results showcase the importance of lipid composition on the insertion mechanism of pHLIP. Lipids headgroups were found to modulate not only the membrane partitioning, folding and insertion propensities of pHLIP peptides, but also peptide protonation in the interfacial state and the pathway (i.e., State II vs State IIS) towards insertion (Fig. 6). Thermodynamics of the transmembrane insertion of pHLIP peptides can be quantitated using the free energy of protonation associated with the transfer to State III: ΔGTMH+ =−2.3RT(pKa). Our results indicate that in each lipid composition the pKa of insertion is shifted toward neutral pH by approximately one pH unit upon P-to-G substitution (Fig. 4). This is equivalent to −1.3 kcal/mol difference in free energy in favor of pHLIP-P20G. Neither the Wimley-White octanol scale [42], nor the scale of Moon and Fleming [43] provide accurate prediction, as both suggest a more favorable insertion of the pHLIP-WT, by +1.0 and +3.2 kcal/mol, respectively. In contrast, the so-called “biological” scale [44], based on transmembrane incorporation of helical segments by a Sec61 translocon machinery, gives a rather accurate prediction of −1.5 kcal/mol for more favorable insertion of the pHLIP-P20G compared to pHLIP-WT. This somewhat surprising result (obviously pHLIP is not inserted by the translocon) may be related to the fact that the translocon sorts the hydrophobicity in the context of helical segments, for which both side-chain hydrophobicity and backbone conformation are the contributing factors. Because Pro-to-Gly replacement can affect both components, having the same helical conformational motif for both translocon-based insertion and pH-dependent insertion of pHLIP appears to be important (note that Wimley-White scale [42] and the scale of Moon and Fleming [43], utilize unfolded peptides and β-barrel proteins, respectively).
FIGURE 6. Revised scheme of membrane interaction of pHLIP.
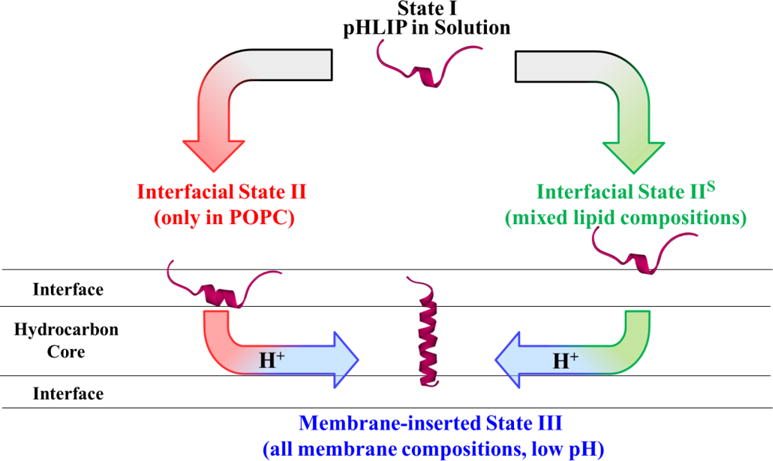
In addition to traditional Stats I, II and III [1], the scheme introduces a shallow interfacial State IIS (right). The traditionally observed State II was only observed to form on pure POPC membranes with both pHLIP-WT and pHLIP-P20G. In all other membrane compositions, pHLIP forms the newly identified State IIS. Both peptide variants transitioned to the transmembrane State III regardless of membrane composition as a function of pH. Our results presented in Fig. 3 indicate that P20-to-G replacement doesn’t affect the thermodynamics of interfacial partitioning for either State II or IIS (ΔGIF ≈−6.2÷−6.8 kcal/mol). In contrast, proline replacement led to a more favorable transmembrane insertion of pHLIP-P20G in all lipid compositions by ΔGTMH+=1.3 Kcal/mol (equivalent of 1 pH unit, Fig. 4).
The changes in side chain protonation are among the most prominent physicochemical signals capable of triggering functionally relevant structural rearrangements, especially those involved with protein-membrane interactions. For example, pH-dependent conversion of a protein structure from a water-soluble to a membrane-inserted form is a key step in many crucial processes such as cellular entry of bacterial toxins [45–50], colicins [51, 52], and viruses [53, 54], as well as membrane-mediated regulation of apoptosis by the Bcl-2 (B-cell lymphoma-2) family of proteins [55–60]. pHLIP peptides can thus serve as a relatively simple and controlled system for deciphering the physicochemical rules underlying membrane-modulated conformational switching in other systems.
Supplementary Material
Highlights.
Membrane insertion of pHLIP is modulated by lipid composition
The unstructured State II is only present in pure phosphatidylcholine membranes
A new shallow and unstructured interfacial State IIS is identified
Interfacial partitioning free energy for both peptides ranges from 6–7 kcal/mol
Protonation-dependent insertion ΔG is 1.3 kcal/mol more favorable for pHLIP-P20G than pHLIP-WT
Acknowledgments
This work was supported by the National Institute of Health (grant number R21CA181868; to D.T.) and by Lehigh University (to D.T.). We are grateful to Mahnoor Malik for her assistance with data analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors claim no conflict of interest
References
- 1.Hunt JF, Rath P, Rothschild KJ, Engelman DM. Spontaneous, pH-dependent membrane insertion of a transbilayer α-helix. Biochemistry. 1997;36:15177–15192. doi: 10.1021/bi970147b. [DOI] [PubMed] [Google Scholar]
- 2.Andreev OA, Engelman DM, Reshetnyak YK. Targeting acidic diseased tissue: New technology based on use of the pH (Low) Insertion Peptide (pHLIP) Chim Oggi. 2009;27:34–37. [PMC free article] [PubMed] [Google Scholar]
- 3.Segala J, Engelman DM, Reshetnyak YK, Andreev OA. Accurate analysis of tumor margins using a fluorescent pH Low Insertion Peptide (pHLIP) International journal of molecular sciences. 2009;10:3478–3487. doi: 10.3390/ijms10083478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vavere AL, Biddlecombe GB, Spees WM, Garbow JR, Wijesinghe D, Andreev OA, Engelman DM, Reshetnyak YK, Lewis JS. A novel technology for the imaging of acidic prostate tumors by positron emission tomography. Cancer research. 2009;69:4510–4516. doi: 10.1158/0008-5472.CAN-08-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reshetnyak YK, Yao L, Zheng S, Kuznetsov S, Engelman DM, Andreev OA. Measuring Tumor Aggressiveness and Targeting Metastatic Lesions with Fluorescent pHLIP. Mol Imaging Biol. 2010 doi: 10.1007/s11307-010-0457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andreev OA, Engelman DM, Reshetnyak YK. pH-sensitive membrane peptides (pHLIPs) as a novel class of delivery agents. Mol Membr Biol. 2010;27:341–352. doi: 10.3109/09687688.2010.509285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sosunov EA, Anyukhovsky EP, Sosunov AA, Moshnikova A, Wijesinghe D, Engelman DM, Reshetnyak YK, Andreev OA. pH (low) insertion peptide (pHLIP) targets ischemic myocardium. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:82–86. doi: 10.1073/pnas.1220038110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weerakkody D, Moshnikova A, Thakur MS, Moshnikova V, Daniels J, Engelman DM, Andreev OA, Reshetnyak YK. Family of pH (low) insertion peptides for tumor targeting. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5834–5839. doi: 10.1073/pnas.1303708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andreev OA, Engelman DM, Reshetnyak YK. Targeting diseased tissues by pHLIP insertion at low cell surface pH. Frontiers in physiology. 2014;5:97. doi: 10.3389/fphys.2014.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burns KE, Hensley H, Robinson MK, Thevenin D. Therapeutic Efficacy of a Family of pHLIP-MMAF Conjugates in Cancer Cells and Mouse Models. Molecular pharmaceutics. 2017;14:415–422. doi: 10.1021/acs.molpharmaceut.6b00847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burns KE, McCleerey TP, Thevenin D. pH-Selective Cytotoxicity of pHLIP-Antimicrobial Peptide Conjugates. Scientific reports. 2016;6:28465. doi: 10.1038/srep28465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burns KE, Thevenin D. Down-regulation of PAR1 activity with a pHLIP-based allosteric antagonist induces cancer cell death. The Biochemical journal. 2015;472:287–295. doi: 10.1042/BJ20150876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burns KE, Robinson MK, Thevenin D. Inhibition of cancer cell proliferation and breast tumor targeting of pHLIP-monomethyl auristatin E conjugates. Molecular pharmaceutics. 2015;12:1250–1258. doi: 10.1021/mp500779k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Musial-Siwek M, Karabadzhak A, Andreev OA, Reshetnyak YK, Engelman DM. Tuning the insertion properties of pHLIP. Biochimica et biophysica acta. 2010;1798:1041–1046. doi: 10.1016/j.bbamem.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrera FN, Fendos J, Engelman DM. Membrane physical properties influence transmembrane helix formation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:14422–14427. doi: 10.1073/pnas.1212665109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fendos J, Barrera FN, Engelman DM. Aspartate Embedding Depth Affects pHLIP’s Insertion pK. Biochemistry. 2013 doi: 10.1021/bi400252k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reshetnyak YK, Segala M, Andreev OA, Engelman DM. A monomeric membrane peptide that lives in three worlds: in solution, attached to, and inserted across lipid bilayers. Biophysical journal. 2007;93:2363–2372. doi: 10.1529/biophysj.107.109967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyrychenko A, Vasquez-Montes V, Ulmschneider MB, Ladokhin AS. Lipid Headgroups Modulate Membrane Insertion of pHLIP Peptide. Biophysical journal. 2015;108:791–794. doi: 10.1016/j.bpj.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ladokhin AS, White SH. Folding of amphipathic α-helices on membranes: Energetics of helix formation by melittin. J Mol Biol. 1999;285:1363–1369. doi: 10.1006/jmbi.1998.2346. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez-Vidal M, Jayasinghe S, Ladokhin AS, White SH. Folding amphipathic helices into membranes: amphiphilicity trumps hydrophobicity. J Mol Biol. 2007;370:459–470. doi: 10.1016/j.jmb.2007.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Almeida PF, Ladokhin AS, White SH. Hydrogen-bond energetics drive helix formation in membrane interfaces. Biochimica et biophysica acta. 2011 doi: 10.1016/j.bbamem.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hope MJ, Bally MB, Mayer LD, Janoff AS, Cullis PR. Generation of multilamellar and unilamellar phospholipid vesicles. Chem Phys Lipids. 1986;40:89–107. [Google Scholar]
- 23.Mayer LD, Hope MJ, Cullis PR. Vesicles of variable sizes produced by a rapid extrusion procedure. Biochim Biophys Acta. 1986;858:161–168. doi: 10.1016/0005-2736(86)90302-0. [DOI] [PubMed] [Google Scholar]
- 24.Ladokhin AS, Jayasinghe S, White SH. How to measure and analyze tryptophan fluorescence in membranes properly, and why bother? Anal Biochem. 2000;285:235–245. doi: 10.1006/abio.2000.4773. [DOI] [PubMed] [Google Scholar]
- 25.White SH, Wimley WC, Ladokhin AS, Hristova K. Protein folding in membranes: Determining the energetics of peptide-bilayer interactions. Methods Enzymol. 1998;295:62–87. doi: 10.1016/s0076-6879(98)95035-2. [DOI] [PubMed] [Google Scholar]
- 26.Montgomery DC, Peck EA. Introduction to linear regression analysis. Wiley; New York: 1982. [Google Scholar]
- 27.Scott HL, Westerfield JM, Barrera FN. Determination of the Membrane Translocation pK of the pH-Low Insertion Peptide. Biophysical journal. 2017;113:869–879. doi: 10.1016/j.bpj.2017.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen YH, Yang JT, Chau KH. Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 29.Luo PZ, Baldwin RL. Mechanism of helix induction by trifluoroethanol: A framework for extrapolating the helix-forming properties of peptides from trifluoroethanol/water mixtures back to water. Biochemistry. 1997;36:8413–8421. doi: 10.1021/bi9707133. [DOI] [PubMed] [Google Scholar]
- 30.Luo PZ, Baldwin RL. Interaction between water and polar groups of the helix backbone: An important determinant of helix propensities. Proc Natl Acad Sci USA. 1999;96:4930–4935. doi: 10.1073/pnas.96.9.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roccatano D, Colombo G, Fioroni M, Mark AE. Mechanism by which 2,2,2-trifluoroethanol/water mixtures stabilize secondary-structure formation in peptides: a molecular dynamics study. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:12179–12184. doi: 10.1073/pnas.182199699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kyrychenko A, Rodnin MV, Posokhov YO, Holt A, Pucci B, Killian JA, Ladokhin AS. Thermodynamic measurements of bilayer insertion of a single transmembrane helix chaperoned by fluorinated surfactants. J Mol Biol. 2012;416:328–334. doi: 10.1016/j.jmb.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reshetnyak YK, Andreev OA, Segala M, Markin VS, Engelman DM. Energetics of peptide (pHLIP) binding to and folding across a lipid bilayer membrane. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15340–15345. doi: 10.1073/pnas.0804746105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wimley WC, White SH. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nature Struct Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 35.Ladokhin AS, White SH. Protein chemistry at membrane interfaces: Non-additivity of electrostatic and hydrophobic interactions. J Mol Biol. 2001;309:543–552. doi: 10.1006/jmbi.2001.4684. [DOI] [PubMed] [Google Scholar]
- 36.Ladokhin AS, White SH. Interfacial folding and membrane insertion of a designed helical peptide. Biochemistry. 2004;43:5782–5791. doi: 10.1021/bi0361259. [DOI] [PubMed] [Google Scholar]
- 37.Wimley WC, Gawrisch K, Creamer TP, White SH. A direct measurement of salt-bridge solvation energies using a peptide model system: Implications for protein stability. Proc Natl Acad Sci USA. 1996;93:2985–2990. doi: 10.1073/pnas.93.7.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pink DA, McNeil S, Quinn B, Zuckermann MJ. A model of hydrogen bond formation in phosphatidylethanolamine bilayers. Biochim Biophys Acta. 1998;1368:289–305. doi: 10.1016/s0005-2736(97)00196-x. [DOI] [PubMed] [Google Scholar]
- 39.Soubias O, Teague WE, Jr, Hines KG, Mitchell DC, Gawrisch K. Contribution of membrane elastic energy to rhodopsin function. Biophysical journal. 2010;99:817–824. doi: 10.1016/j.bpj.2010.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boggs JM. Lipid intermolecular hydrogen bonding: Influence on structural organization and membrane function (BBA 85315) Biochim Biophys Acta. 1987;906:383–404. doi: 10.1016/0304-4157(87)90017-7. [DOI] [PubMed] [Google Scholar]
- 41.Slater SJ, Ho C, Taddeo FJ, Kelly MB, Stubbs CD. Contribution of hydrogen bonding to lipid-lipid interactions in membranes and the role of lipid order: Effects of cholesterol, increased phospholipid unsaturation, and ethanol. Biochemistry. 1993;32:3714–3721. doi: 10.1021/bi00065a025. [DOI] [PubMed] [Google Scholar]
- 42.Wimley WC, Creamer TP, White SH. Solvation energies of amino acid sidechains and backbone in a family of host-guest pentapeptides. Biochemistry. 1996;35:5109–5124. doi: 10.1021/bi9600153. [DOI] [PubMed] [Google Scholar]
- 43.Moon CP, Fleming KG. Side-chain hydrophobicity scale derived from transmembrane protein folding into lipid bilayers. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10174–10177. doi: 10.1073/pnas.1103979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hessa T, Kim H, Bihlmaler K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 45.Hoch DH, Romero-Mira M, Ehrlich BE, Finkelstein A, DasGupta BR, Simpson LL. Channels formed by botulinum, tetanus, and diphtheria toxins in planar lipid bilayers: Relevance to translocation of proteins. Proc Natl Acad Sci USA. 1985;82:1692–1696. doi: 10.1073/pnas.82.6.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neale EA. Moving across membranes. Nature Struct Biol. 2003;10:2–3. doi: 10.1038/nsb0103-2. [DOI] [PubMed] [Google Scholar]
- 47.Koriazova LK, Montal M. Translocation of botulinum neurotoxin light chain protease through the heavy chain channel. Nature Struct Biol. 2003;10:13–18. doi: 10.1038/nsb879. [DOI] [PubMed] [Google Scholar]
- 48.Collier RJ, Young JA. Anthrax toxin. Annu Rev Cell Dev Biol. 2003;19:45–70. doi: 10.1146/annurev.cellbio.19.111301.140655. [DOI] [PubMed] [Google Scholar]
- 49.Murphy JR. Mechanism of diphtheria toxin catalytic domain delivery to the eukaryotic cell cytosol and the cellular factors that directly participate in the process. Toxins. 2011;3:294–308. doi: 10.3390/toxins3030294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ladokhin AS. pH-triggered conformational switching along the membrane insertion pathway of the diphtheria toxin T-domain. Toxins. 2013;5:1362–1380. doi: 10.3390/toxins5081362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zakharov SD, Cramer WA. On the mechanism and pathway of colicin import across the E. Coli outer membrane. Front Biosci. 2004;9:1311–1317. doi: 10.2741/1334. [DOI] [PubMed] [Google Scholar]
- 52.Jakes KS, Cramer WA. Border crossings: colicins and transporters. Annual review of genetics. 2012;46:209–231. doi: 10.1146/annurev-genet-110711-155427. [DOI] [PubMed] [Google Scholar]
- 53.Kielian M, Rey FA. Virus membrane-fusion proteins: more than one way to make a hairpin. Nature reviews Microbiology. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neuman BW, Kiss G, Al-Mulla HM, Dokland T, Buchmeier MJ, Weikl T, Schley D. Direct observation of membrane insertion by enveloped virus matrix proteins by phosphate displacement. PloS one. 2013;8:e57916. doi: 10.1371/journal.pone.0057916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsu YT, Wolter KG, Youle RJ. Cytosol-to-membrane redistribution of Bax and Bcl-X(L) during apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:3668–3672. doi: 10.1073/pnas.94.8.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Antignani A, Youle RJ. How do Bax and Bak lead to permeabilization of the outer mitochondrial membrane? Current opinion in cell biology. 2006;18:685–689. doi: 10.1016/j.ceb.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 57.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death, Nature reviews. Molecular cell biology. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 58.Leber B, Lin J, Andrews DW. Embedded together: the life and death consequences of interaction of the Bcl-2 family with membranes. Apoptosis: an international journal on programmed cell death. 2007;12:897–911. doi: 10.1007/s10495-007-0746-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bogner C, Leber B, Andrews DW. Apoptosis: embedded in membranes. Current opinion in cell biology. 2010;22:845–851. doi: 10.1016/j.ceb.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 60.Chi X, Kale J, Leber B, Andrews DW. Regulating cell death at, on, and in membranes. Biochimica et biophysica acta. 2014;1843:2100–2113. doi: 10.1016/j.bbamcr.2014.06.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


