In this study, Wang et al. report that SET1B/COMPASS has a cytoplasmic variant with a novel catalytic-independent function. They identified ADIPOR1 signaling as a key target of SET1B/COMPASS, and their findings demonstrate a cytoplasmic function for a member of the COMPASS family in fatty acid metabolism.
Keywords: chromatin, gene expression, transcription
Abstract
Mutations and translocations within the COMPASS (complex of proteins associated with Set1) family of histone lysine methyltransferases are associated with a large number of human diseases, including cancer. Here we report that SET1B/COMPASS, which is essential for cell survival, surprisingly has a cytoplasmic variant. SET1B, but not its SET domain, is critical for maintaining cell viability, indicating a novel catalytic-independent role of SET1B/COMPASS. Loss of SET1B or its unique cytoplasmic-interacting protein, BOD1, leads to up-regulation of expression of numerous genes modulating fatty acid metabolism, including ADIPOR1 (adiponectin receptor 1), COX7C, SDC4, and COQ7. Our detailed molecular studies identify ADIPOR1 signaling, which is inactivated in both obesity and human cancers, as a key target of SET1B/COMPASS. Collectively, our study reveals a cytoplasmic function for a member of the COMPASS family, which could be harnessed for therapeutic regulation of signaling in human diseases, including cancer.
Misregulation of histones modifiers has emerged as a common therapeutic target for the treatment of different human diseases (Piunti and Shilatifard 2016). Although the physiological importance of protein lysine methylation remained obscure for many years, several protein lysine methyltransferases (KMTs) as well as lysine demethylases (KDMs) have now been identified, and their physiological significance has begun to be elucidated. Molecular perturbation of KMTs and KDMs has been demonstrated as a common mechanism underlying tumorigenesis (Hamamoto et al. 2004; Kotake et al. 2007; Mazur et al. 2014).
All known KMTs that associate with chromatin contain a catalytic SET domain, with the exception of Dot1, which harbors a unique enzymatic domain. Based on the primary amino acid sequence architecture and substrate specificity, KMTs can be divided into six subfamilies: KMT1–6 (Mohan et al. 2012). SET1A, SET1B, and MLL1–4 belong to the KMT2 family, which is capable of monomethylating, dimethylating, and trimethylating histone H3 Lys4 (H3K4), and this histone mark is considered to be involved in positive regulation of gene transcription (Kouzarides 2007; Mohan et al. 2012; Shilatifard 2012). This family of enzymes was biochemically isolated initially from yeast within a macromolecular complex and named COMPASS (complex of proteins associated with Set1) (Miller et al. 2001; Krogan et al. 2002; Shilatifard 2012). COMPASS was demonstrated to be highly conserved from yeast to humans, with one COMPASS family member in yeast and six KMT2/COMPASS members in mammals (Shilatifard 2012; Piunti and Shilatifard 2016). MLL1 and MLL2/COMPASS contain the tumor suppressor factor Menin within their complexes, while MLL3 and MLL4/COMPASS are associated with PTIP, PA1, NCOA6, and UTX (Cho et al. 2007; Shilatifard 2012). Interestingly, SET1A and SET1B are more similar to yeast SET1/COMPASS, and all of the yeast subunits have corresponding mammalian counterparts (Wu et al. 2008). Although both SET1A and SET1B share the same subunit composition, global reduction of H3K4 methylation levels is observed only in SET1A but not in SET1B knockout embryonic stem cells (ESCs) and embryos. Overexpression of SET1B fails to rescue the ESC proliferation defects induced by the loss of SET1A, suggesting that SET1B may have a function distinct from that of its homolog, SET1A (Bledau et al. 2014). SET1A/COMPASS has been shown to regulate wnt target genes and control tumor growth of colorectal cancer cells (Salz et al. 2014; Cao et al. 2017; Sze et al. 2017). However, the function of SET1B remains unclear.
Here, we report the existence of a cytoplasmic form of SET1B/COMPASS, which is essential for maintaining tumor cell viability and promotes cell growth. We further uncovered the catalytic-independent function of the SET1B/BOD1 complex in mediating the cellular metabolic program. Our study provides novel insights showing that targeting the SET1B/COMPASS pathway is a potential therapeutic strategy for different human diseases, such as diabetes and cancer.
Results
A cytoplasmic version of COMPASS
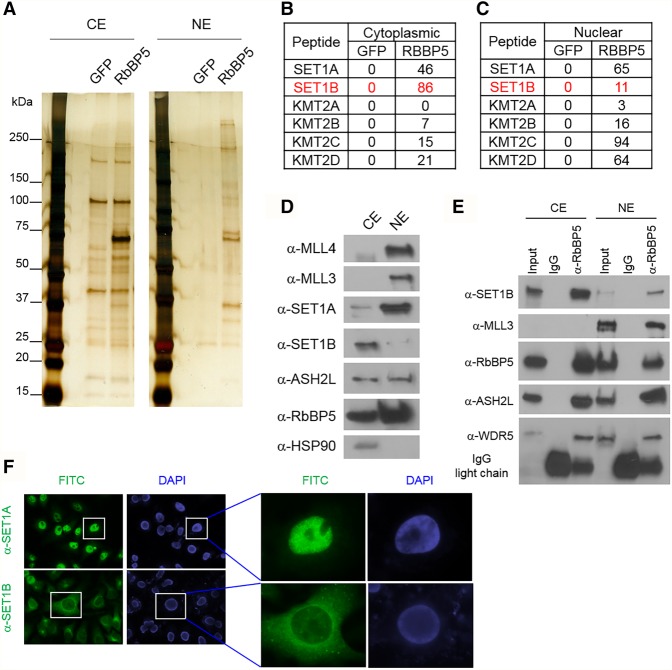
Emerging evidence uncovered the novel functions of nuclear multiprotein complexes in the cytoplasm, such as the PRC2 complex (Bodega et al. 2017) and the BAP1 complex (Bononi et al. 2017). To investigate whether the COMPASS complex exists in the cytoplasm of mammalian cells, we purified RBBP5, a common subunit of all COMPASS complexes in mammals, from both the cytoplasm and nucleus (Fig. 1A) in MCF7 cells. Mass spectrometric analysis with the purified material reveals that the majority of SET1B/COMPASS is found within the cytoplasmic fraction (Fig. 1B,C). Western blot analysis further confirms our mass spectrometric data by demonstrating that SET1B is confined mostly to the cytoplasm in these cells (Fig. 1D). In order to investigate whether cytoplasmic SET1B can function within COMPASS as a KMT, we immunoprecipitated members of the COMPASS family from cytoplasmic and nuclear extracts and analyzed their endogenous interactions with SET1B. As shown in Figure 1E, the cytoplasmic SET1B stably interacts with the WARD complex, which stimulates for COMPASS catalytic activity, while MLL3 only forms a complex with the WARD complex in the nucleus. To further confirm the subcellular localization of SET1B, we generated antibodies specific for SET1B immunofluorescence (IF) staining. To determine the specificity of the antibodies, we first tested the COMPASS-specific antibodies by performing IF in HeLa cells transfected with nontargeting shRNA or SET1B shRNA. Surprisingly, we found that in HeLa cells, most of the SET1B staining is localized in the cytoplasm, while SET1A is localized consistently in the nucleus (Fig. 1F; Supplemental Fig. S1A). In order to exclude the possibility that there may be mutations in the SET1B gene in HeLa cells that lead to its mislocalization to the cytoplasm, we transfected a Flag-tagged full-length SET1B cDNA into HEK293T cells and performed IF with anti-Flag (Supplemental Fig. S1B). Consistent with the IF experiments in HeLa cells, we found that the majority of the IF signal is localized in the cytoplasm. Moreover, the cellular localization of SET1B is the same in normal human epithelial cells and in MCF7 cells (Supplemental Fig. S1C,D).
Figure 1.
A cytoplasmic version of the COMPASS complex. (A) MCF7 cells were stably transfected with Flag-tagged RBBP5. COMPASS purified from either cytoplasmic or nuclear extracts was further subjected to mass spectrometric analysis. (B,C) The numbers of peptides from the COMPASS family that purified with RBBP5 or GFP are shown. (D) The cytoplasmic and nuclear proteins were extracted from MCF7 cells, and the endogenous levels of SET1A, SET1B, MLL3, MLL4, ASH2L, and RBBP5 were detected by Western blot analysis. HSP90 was used as the cytoplasmic fractionation marker. n = 3. (E) Immunoprecipitation was performed using anti-RBBP5 antibody from either cytoplasmic or nuclear extracts. SET1B, MLL3, ASH2L, RBBP5, and WDR5 protein levels were detected in the immunoprecipitates by Western blot analysis. (CE) Cytoplasmic extract; (NE) nuclear extract. n = 2. (F) IF was performed in HeLa cells with antibodies specific for SET1A or SET1B. Magnifications: left panel, 10×; right panel 40×. n = 4.
SET1B, but not its SET domain, maintains cell viability
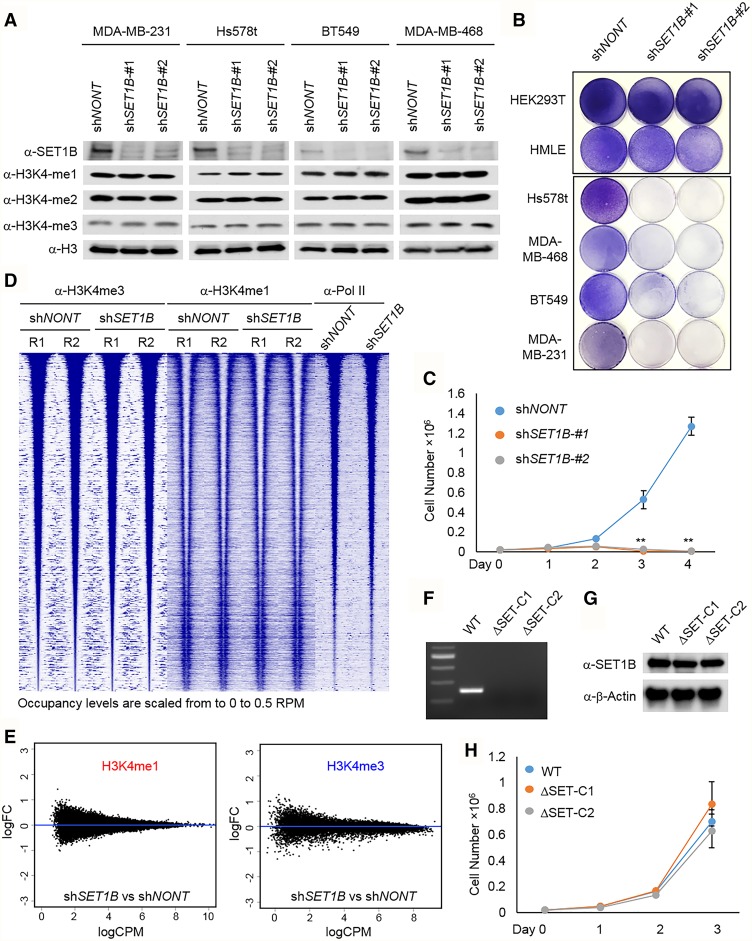
To investigate the function of cytoplasmic SET1B/COMPASS, we knocked down SET1B with two different shRNAs in multiple cell lines (Fig. 2A). The specificity of the shRNAs was tested by quantitative PCR (qPCR) (Supplemental Fig. S2A). Consistent with findings reported in mouse ESCs (Bledau et al. 2014), depletion of SET1B does not affect bulk histone H3K4 methylation (Fig. 2A). Interestingly, as exhibited by the colony formation (Fig. 2B) and cell counting (Fig. 2C) assays, we found that multiple cancer cell lines are not able to survive upon SET1B depletion. To determine that the effects observed are specific to SET1B, we also knocked down SET1A levels in five of the cell lines and assessed cell survival. After confirming the knockdown efficiency of SET1A shRNA by Western blot (Supplemental Fig. S2B), we found that, unlike SET1B knockdown effects, reducing SET1A levels does not significantly alter cell survival (Supplemental Fig. S2C). In vitro studies with SET1B/COMPASS suggested that this complex functions similarly to its yeast homolog, Set1/COMPASS, and is capable of monomethylating, dimethylating, and trimethylating histone H3K4 (Shilatifard 2012). To test whether the loss of SET1B in vivo affects subgroup histone H3K4 trimethylation (H3K4me3) on chromatin, we performed ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) analysis with anti-H3K4me1 and anti-H3K4me3 in MDA-MB-231 shCtrl and MDA-MB-231 shSET1B cells. Surprisingly, H3K4me1 and H3K4me3 were not significantly altered after knocking down SET1B (Fig. 2D,E; Supplemental Fig. S2D,E), indicating that SET1B may function independently of its catalytic domain. To further clarify whether SET1B controls cell viability in a catalytic activity-dependent or -independent manner, we knocked out the catalytic domain (SET domain) of the SET1B gene by CRISPR–Cas9 (Supplemental Fig. S2F) in MDA-MB-231 cells, which is the cell line most sensitive to SET1B depletion (Fig. 2B,C). We performed PCR genotyping and RNA sequencing (RNA-seq) as well as cDNA Sanger sequencing to confirm that the SET domain of SET1B is completely deleted in MDA-MB-231 cells (Fig. 2F; Supplemental Fig. S2G,H), while the protein level of SET1B remained stable (Fig. 2G). As expected, the cell growth assay demonstrated that removal of the SET domain of SET1B does not affect cell growth (Fig. 2H).
Figure 2.
SET1B, but not its SET domain, maintains cell viability. (A) SET1B was knocked down in cells from four cancer lines: LM2, Hs578t, MDA-MB-468, and BT549. Whole-cell lysates were extracted, and the protein levels of SET1B, H3K4me1, H3K4me2, and H3K4me3 were detected by Western blotting. Total histone H3 was used as the internal control. (B) The cell lines used in A as well as HEK293T cells and human normal epithelial cells (HMLEs) were infected with lentiviruses expressing two distinct SET1B shRNAs (shSET1B#1 and shSET1B#2). Nontargeting shRNA (shNONT) was used as a control. After puromycin selection for 48 h, 4 × 104 living cells were seeded in six-well plates and grown for 1 wk before crystal violet staining. (C) MDA-MB-231 cells were transduced by SET1B shRNA and selected with puromycin for 48 h. The cell proliferation ability was determined by cell counting. (D) The occupancies of H3K4me3, H3K4me1, and RNA polymerase II with and without shSET1B are plotted in ±5-kb windows with respect to 17,959 H3K4me3 peak centers and sorted by decreasing H3K4me3 shNONT-R1 occupancy. Two replicates were used. (E) The MA plot shows the differential occupancy of H3K4me1 and H3K4me3 under H3K4me1 and H3K4me3 peaks (shSET1B vs. shNONT), respectively. (M-values) The log of the ratio of level counts for each gene between two samples; (A-values) the average level counts for each gene across the two samples. (F) Genotyping was performed by PCR with genomic DNA extracted from parental and SET1B knockout MDA-MB-231 cells. (G) SET1B protein was detected by Western blot with whole-cell lysate extracted from SET1B wild-type and knockout cells. (H) The growth ability of SET1B wild-type and knockout cells was determined by cell counting.
BOD1 is a cytoplasmic-specific subunit of SET1B/COMPASS
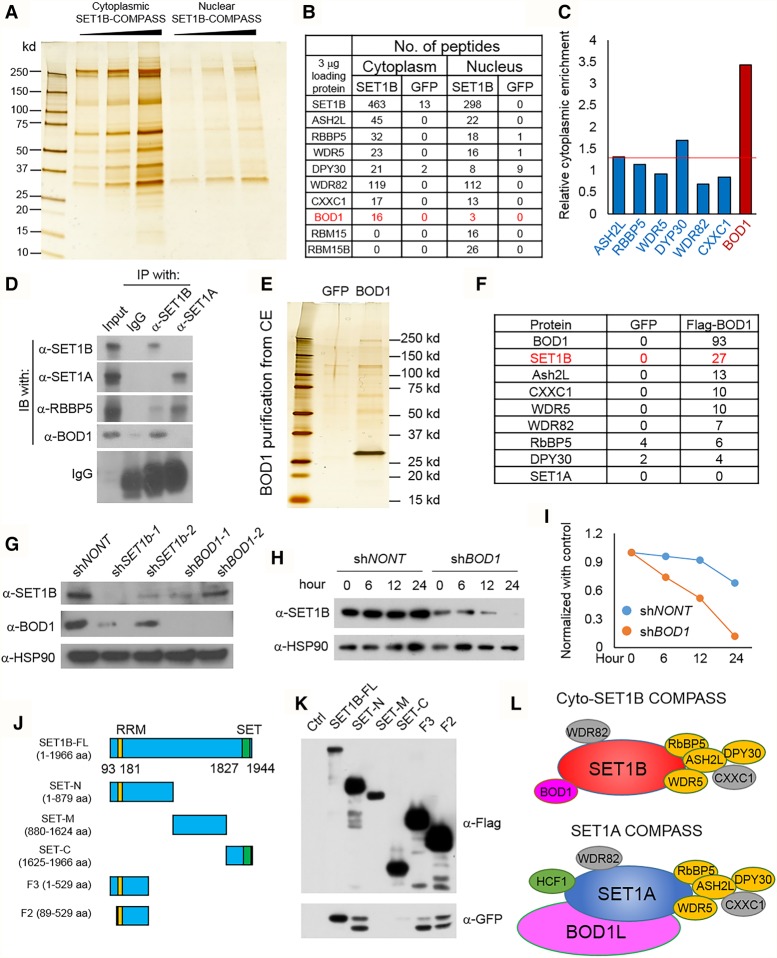
To fully characterize the interactome of SET1B/COMPASS and apply this information to identify its molecular targets, we infected MDA-MB-231 cells with retrovirus expressing Flag-tagged human full-length SET1B cDNA. After G418 selection, we obtained pooled clones of MDA-MB-231 cells stably expressing full-length SET1B. The Flag-tagged SET1B was then purified from either cytoplasmic or nuclear extracts (Fig. 3A). Silver staining confirmed that more SET1B is purified from the cytoplasmic than from the nuclear extracts of MDA-MB-231-Flag-SET1B-expressing cells. Using mass spectrometric methods, we found that SET1B forms a COMPASS-like complex with all of the core COMPASS subunits in both the cytoplasm and nucleus (Fig. 3B). However, we found the BOD1 protein, which is homologous to Cps15 in yeast Set1/COMPASS, to be largely enriched in the cytoplasmic purification of SET1B/COMPASS (Fig. 3C). Consistently, BOD1 was also found in the cytoplasmic immunoprecipitates of Flag-RBBP5 (Supplemental Fig. S3A). We also purified BOD1L1, which is the homolog of BOD1 in mammals, from 293T cells. As shown in Supplemental Figure S3B, by mass spectrometric analysis, we found that BOD1L1 firmly interacts with SET1A but not SET1B. We performed additional Western blot analyses to confirm the distribution of BOD1 and RBM15 in both cytoplasmic and nuclear purified SET1B/COMPASS. As shown in Supplemental Figure S3C, ASH2L is detected in both cytoplasmic and nuclear SET1B/COMPASS. However, RBM15/RBM15B, which contain the nuclear localization signal (NLS) in their C termini, could be detected only with nuclear SET1B purifications. We immunoprecipitated SET1A and SET1B to confirm the specific interaction between endogenous SET1B and the BOD1 protein (Fig. 3D). We also generated stable cells expressing ectopic BOD1 and purified Flag-tagged BOD1 protein from the cytoplasm (Fig. 3E). As shown in Figure 3F, the entire SET1B/COMPASS was identified by mass spectrometric analysis as the top interacting proteins in this purification.
Figure 3.
BOD1 is a cytoplasmic-specific subunit of SET1B/COMPASS. (A) MDA-MB-231 cells were stably infected with retroviruses expressing Flag-SET1B. Silver staining shows the Flag-tagged SET1B protein purified with M2 beads from either cytoplasmic extracts or nuclear extracts (equal volumes of eluted protein). (B) The mass spectrometry (MS) analysis with equal amounts of the interacting proteins of SET1B isolated from cytoplasmic and nuclear extracts are shown (equal amounts). (C) The results from B were normalized further with the spec count of the SET1B protein, and the relative cytoplasmic enrichment of the protein is shown. The cytoplasmic enrichment is presented as the ratio between cytoplasmic subunit/cytoplasmic SET1B and nuclear subunit/nuclear SET1B. (D) The endogen SET1A and SET1B proteins were immunoprecipitated, and the BOD1 protein in the immunoprecipitates was detected by Western blot. The common subunit of SET1A and SET1B COMPASS, RBBP5, was used as a control. n = 2. (E) Flag-BOD1 was purified from cytoplasmic extracts of MDA-MB-231-Flag-BOD1 cells and analyzed by silver staining. MS identified interacting proteins with Flag-BOD1. (F) Peptide numbers of subunits from SET1B COMPASS are shown. Cells stably expressing GFP were used as the negative control. (G) SET1B and BOD1 were knocked down with two distinct shRNAs in MDA-MB-231 cells, and the protein levels of SET1B and BOD1 were detected by Western blotting. HSP90 was used as the internal control. n = 3. (H) MDA-MB-231 cells were transfected with shBOD1 and selected by puromycin for 48 h. The cells were further treated with cycloheximide (CHX) for different times, and the protein level of SET1B was determined by Western blot. (I) The degradation ratios of SET1B in shNONT and shBOD1 cells were quantified by ImageJ. (J) A schematic diagram of SET1B full-length cDNA and truncated derivatives, each with a Flag tag fused to its N terminus, is shown. (K) Plasmids expressing the SET1B cDNA derivatives shown in J were transiently transfected into HEK293T cells together with GFP-BOD1 for 24 h. The Flag-tagged SET1B truncations were then purified, and the interacting GFP-BOD1 was detected by Western blotting with anti-GFP. n = 4. (L) Cartoon depictions of cyto-SET1B COMPASS and SET1A COMPASS. (Orange) WARD complex, which contains WDR5, ASH2L, RBBP5, and DPY30; (gray) SET1A/B COMPASS complex-specific subunits WDR82 and CXXC1; (pink) cyto-SET1B COMPASS-specific subunit BOD1.
BOD1 stabilizes and functions with SET1B
Our biochemical studies demonstrated that the loss of SET1B destabilized BOD1 and vice versa (Fig. 3G). Depletion of BOD1 dramatically accelerated the degradation of the SET1B protein (Fig. 3H,I). To investigate whether BOD1 contributes to SET1B's function, we knocked down BOD1 with two individual shRNAs in MDA-MB-231 cells. We found that, similar to SET1B depletion, loss of BOD1 alters cell morphology (Supplemental Fig. S3D) and decreases cell growth (Supplemental Fig. S3E). These results suggest that BOD1 functions as a part of COMPASS with SET1B in these cells in the cytoplasm. To identify common downstream targets of BOD1 and SET1B, we performed RNA-seq analyses in MDA-MB-231 cells infected with a lentivirus expressing either nontargeting shRNA or two individual BOD1 or SET1B shRNAs. As shown in Supplemental Figure S3, F and G, more mRNAs were up-regulated than down-regulated in BOD1-depleted cells, a finding similar to that with SET1B knockdown. Supporting a functional role for BOD1 and SET1B within a cytoplasmic complex, nearly half of the genes that are regulated by SET1B are also regulated by BOD1 (Supplemental Fig. S3F). We mapped the interaction region between BOD1 and SET1B and found that BOD1 interacts with the N terminus of SET1B, consistent with the mapped interaction between Cps15 and Set1 in yeast (Fig. 3J,K; Takahashi et al. 2011). Based on these observations, we conclude that the majority of SET1B/COMPASS is found with BOD1 in the cytoplasm, while SET1A/COMPASS is associated with BOD1L in the nucleus (Fig. 3L).
Loss of SET1B or BOD1 induces the activation of the adiponectin receptor 1 (AdipoR1) signaling pathway
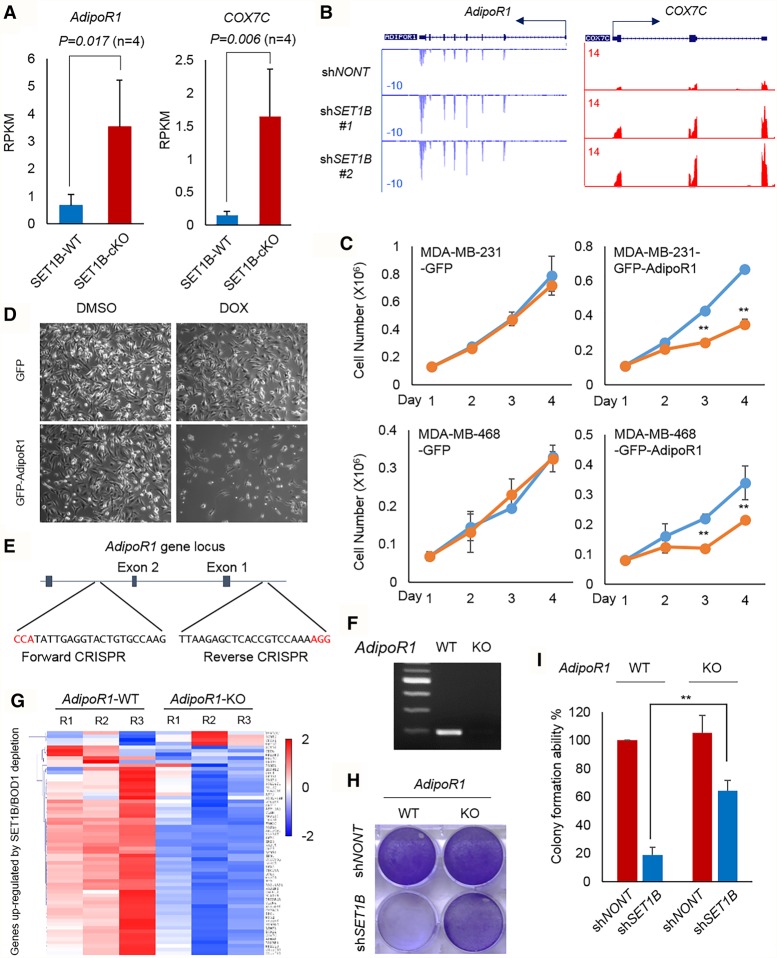
By comparing the gene expression profile between SET1B and nontargeting shRNAs, we noticed that depletion of SET1B significantly increases the expression of genes involved in cell metabolism, such as ADIPOR1, PRKAR2A, COX7C, SDC4, and COQ7 (Supplemental Fig. S4A,B). Depletion of SET1B leads to lipid accumulation in human breast cancer cells (Supplemental Fig. 4C), which is consistent with recent studies reporting that SET1B is involved in lipid metabolism (Brici et al. 2017; Han et al. 2017). We also detected that the mitochondrial trifunctional protein (HADHA/B) is copurified with BOD1 in the cytoplasm (Supplemental Fig. 4D). We further validated the interaction between SET1B/COMPASS and HADHA by immunoprecipitation (Supplemental Fig. 4E) and found that SET1B knockdown results in remarkably reduced HADHA protein levels, indicating that the cytoplasmic SET1B complex is essential for the stability of the mitochondrial trifunctional protein (Supplemental Fig. 4F). These data together support the function of SET1B involvement in mammalian metabolic processes. In addition, we noticed from our RNA-seq data that many SET1B downstream genes are involved in the adiponectin pathway. Adiponectin, which is secreted by fat tissue, directly interacts with ADIPOR1 or ADIPOR2 and further stimulates the phosphorylation of AMPK through recruiting APPL1 to the cell membrane. PPARGC1A is subsequently stimulated by pAMPK, translocates into the nucleus, and transcribes a subset of mitochondria biogenesis genes such as COX7C (Bremer et al. 2015). Consistent with what we observed in human cells, a recent study reported that conditional knockout of SET1B in mouse ovary tissue dramatically leads to lipid accumulation by changing the biosynthesis process and lipid metabolism (Brici et al. 2017). We retrieved and reanalyzed the RNA-seq data from SET1B+/+ and SET1B−/− ovary tissue (Brici et al. 2017) and found that ADIPOR1 and COX7C are also elevated threefold to fourfold in SET1B−/− tissue (Fig. 4A), which is consistent with our results (Fig. 4B). Given similar data from independent studies, we hypothesized that AdiporR1 signaling may be relevant to SET1B function. We subsequently generated multiple cell lines expressing inducible GFP or GFP-ADIPOR1 under the regulation of doxycycline (Supplemental Fig. S4G). After doxycycline treatment, we found that GFP-ADIPOR1 significantly suppresses the cell growth of both MDA-MB-231 and MDA-MB-468 cells (Fig. 4C,D). To investigate whether AdipoR1 mediates cell death induced by SET1B depletion, we designed a pair of CRISPR–Cas9 targeting the promoter region containing exons 1 and 2 of the AdipoR1 gene in MDA-MB-231 cells (Fig. 4E). According to PCR genotyping and RNA-seq results, the transcripts of AdipoR1 in the cells are completely silenced (Fig. 4F; Supplemental Fig. S4H). We next performed RNA-seq in AdipoR1 wild-type and knockout cells (Fig. 4G). Interestingly, we found that the majority of SET1B downstream genes is rescued in AdipoR1 knockout cells (Fig. 4G). Consistently, knockout of AdipoR1 largely rescued cell death induced by loss of SET1B protein (Fig. 4H,I)
Figure 4.
Loss of SET1B or BOD1 induces the activation of ADIPOR1 signaling. (A) The RNA-seq result was retrieved and analyzed in SET1B wild-type and conditional knockout ovary tissue (Brici et al. 2017). The expression level of AdipoR1 and COX7C in SET1B wild-type and SET1B knockout tissues is shown. n = 4. (B) Representative RNA-seq tracks of ADIPOR1 and COX7C in MDA-MB-231 cells infected with shNONT and shSET1B virus are shown. (C) MDA-MB-231 and BT549 cells were infected with lentiviruses inducibly expressing either GFP or GFP-ADIPOR1. After neomycin selection, the stably transfected cells were treated with DMSO or doxycycline (DOX). The proliferation ability of GFP- or GFP-ADIPOR1-inducible cell lines was determined by cell counting. n = 3. (**) P < 0.01, Student's t-test was used for statistical analysis. Error bars represent SD. (D) The cell morphology of MDA-MB-231 cells inducibly expressing GFP or GFP-AdipoR1 is shown. (E) Design of the CRIPSR–Cas9 targeting ADIPOR1 gene locus. (F) PCR was performed for genotyping of AdipoR1 wild-type and deleted cells. (G) RNA-seq was performed with AdipoR1 wild-type and knockout cells. The heat map shows the expression of SET1B downstream genes in AdipoR1 wild-type and knockout cells. (H) SET1B was further knocked down in ADIPOR1 wild-type and ADIPOR1 knockout cells, respectively. After puromycin selection for 48 h, 1 × 104 cells were seeded in a six-well plate and grown for 1 wk before crystal violet staining. (I) Quantification of crystal violet staining was determined as described in the Materials and Methods. n = 4. (**) P < 0.01, Student's t-test was used for statistical analysis. Error bars represent SD.
Targeting AdipoR1 signaling in triple-negative breast cancer
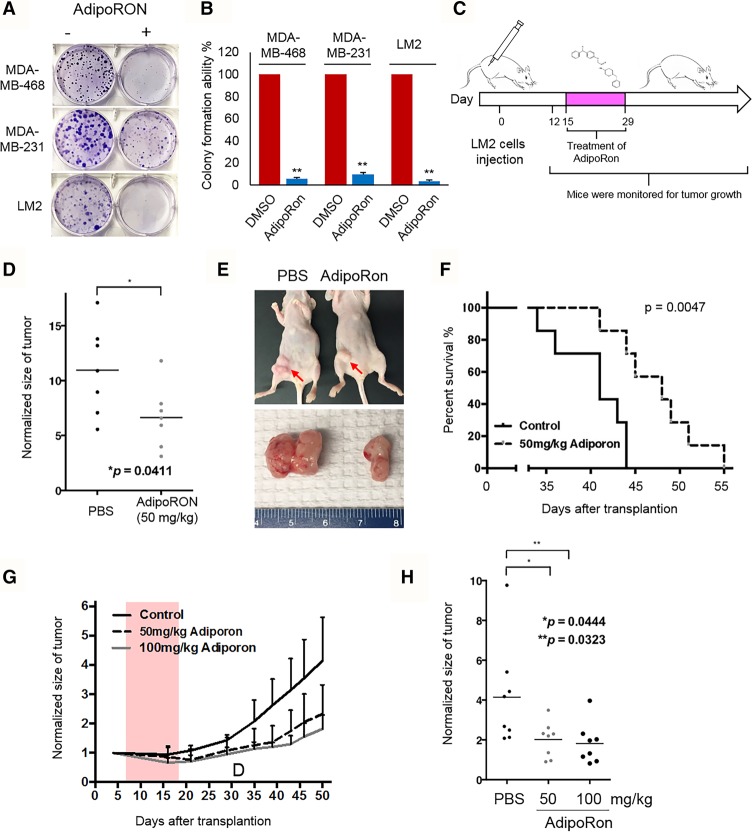
Given our findings thus far, we considered treating triple-negative breast cancer cells with one of the newly identified AdipoR1 agonists, AdipoRon, which has been reported to have promising results in insulin-resistant type 2 diabetes and longevity in obese diabetic mouse models (Okada-Iwabu et al. 2013). Interestingly, we found that treatment with AdipoRon significantly inhibited the colony formation ability of multiple triple-negative breast cancer lines (Fig. 5A,B). In order to investigate AdipoRon's potential tumor-suppressive effects in vivo, we injected LM2 cells into the mammary glands of nude mice and started the oral treatment of 50 mg/kg AdipoRon 12 d after transplantation (Fig. 5C). Tumor size was measured two times per week. As shown in Figure 5, D–F, and consistent with the in vitro studies, treatment with AdipoRon significantly reduced tumor size and increased animal survival. We also confirmed the drug effects of AdipoRon by intraperitoneal injection and observed results (Fig. 5G,H) similar to those of the oral treatment. Taken together, these results provide a novel therapeutic strategy for clinical treatment of human triple-negative breast cancer.
Figure 5.
Targeting AdipoR1 signaling in SET1B-expressing triple-negative breast cancer. (A) MDA-MB-231, MDA-MB-468, and LM2 cells were seeded in six-well plates at a concentration of 200 cells per well. The cells were treated with either DMSO or 10 µM AdipoRON for 3 wk, and the colonies were stained with crystal violet. (B) Quantification of crystal violet staining was determined as described in the Materials and Methods. n = 4. (**) P < 0.01, Student's t-test was used for statistical analysis. Error bars represent SD. (C) Flow chart of the animal experiments in D–F. (D) LM2 cells (4 × 106 cells) were inoculated into the fat pads of nude mice. Twelve days after injection, when the tumors reached 100 mm3, mice were divided randomly into two groups. They were treated 10 times with either 50 mg/kg PBS or 50 mg/kg AdipoRon, and the tumor size (D,E) and animal survival (F) were monitored. Tumor growth was measured 2 wk after inoculation. n = 7. (*) P < 0.05, Student's t-test was used for statistical analysis. Log-rank test was used to determine differences between the survival of each group. (G) LM2 cells (4 × 106 cells) were inoculated into the fat pads of nude mice. Twelve days after injection, when the tumors reached 100 mm3, mice were divided randomly into three groups. They were treated 10 times with either 50 mg/kg PBS or 100 mg/kg AdipoRon, and the tumor size was monitored. (H) Tumor growth was measured 2 wk after inoculation. n = 8. (**) P < 0.01; (*) P < 0.05, Student's t-test was used for statistical analysis.
Discussion
In this study, we report the biochemical and molecular identification of a novel SET1B/COMPASS as a cytoplasmic complex and its potential function in cells. Specifically, we demonstrated the following: (1) Purification of SET1B and subsequent proteomic analyses identified the SET1B complex as the only COMPASS family member residing predominantly in the cytoplasm, with BOD1 as its unique cytoplasmic subunit. (2) Depletion of SET1B does not affect histone H3K4 methylation. (3) SET1B is essential for cell viability in a catalytic activity-independent manner. (4) Depletion of SET1B/BOD1 leads to increased expression of several metabolic genes, such as AdiopR1. (5) Knockout of the AdipoR1 gene by CRISPR–Cas9 largely rescues cell death induced by SET1B depletion. (6) Use of AdipoR1 agonists (AdipoRon) in both culture and animal model systems of breast cancer results in significantly reduced cell growth and tumor size and increased animal survival, pointing to a role for the SET1B/COMPSS pathway in triple-negative breast cancer therapy.
SET1B was initially identified based on database analyses for proteins related to yeast Set1 (Lee et al. 2007). Human SET1A and SET1B proteins share 35% and 37% identity with yeast Set1, respectively, and the two proteins are 85% identical and 97% similar throughout the catalytic SET and post-SET domains. Both SET1A and SET1B could methylate histone H3K4 in vitro; however, in mouse ESCs, SET1A was found to be the major H3K4 methyltransferase, while loss of SET1B did not affect H3K4 methylation levels (Bledau et al. 2014). Consistent with these results, our ChIP-seq analysis in MDA-MB-231 cells did not detect obvious changes in H3K4me3 levels after depletion of SET1B.
Several KMTs have been found to localize in the cytoplasm and catalyze lysine methylation. For instance, SMYD3 was first characterized as a histone H3K4 methyltransferase in vitro (Hamamoto et al. 2004), and later studies demonstrated that SMYD3 lacks a canonical NLS and functions as an oncogene in different types of cancers by methylating MAP3K2 at K260 in the cytoplasm (Mazur et al. 2014). Consistent with SET1B depletion not affecting H3K4 methylation, we found that the majority of SET1B is present in the cytoplasm.
In our previous studies, we identified BOD1 (Cps15 or SHG1) as a component of COMPASS in yeast that had no obvious effect on H3K4 methylation levels (Miller et al. 2001; Krogan et al. 2002). In mammalian cells, BOD1 was found to function as a small kinetochore-associated protein required for mitotic chromosome congression (Porter et al. 2007) and fine-tune PP2A phosphatase activity at the kinetochore to ensure efficient chromosome congression and maintenance of chromatid cohesion (Porter et al. 2013). These findings could explain the cell growth defects that we observed after BOD1 depletion. Furthermore, our findings that half of the genes regulated by BOD1 are also regulated by SET1B and that BOD1 depletion induced the same morphological changes in cells as SET1B depletion indicate that the cytoplasmic SET1B/BOD1 complex could be involved in mitotic chromosome congression as well as maintenance of chromatid cohesion.
In agreement with our findings, recent studies are consistent with SET1B being involved in metabolism and mediates gene expression in metabolic signaling. For instance, knockout of SET1B in mouse ovary tissue resulted in increased expression of many metabolic genes, leading to defective oocyte-to-embryo transition (Brici et al. 2017). In Caenorhabditis elegans, knockout of SET2, which is SET1 in mammals, also led to elevated gene expression involved in the biosynthesis process and lipid metabolism and downstream changes in fatty acid oxidation (Han et al. 2017). Our study provides the first evidence that loss of SET1B in human cells results in the accumulation of lipid droplets and changes of expression of numerous metabolic genes such as ADIPOR1, COX7C, SDC4, and COQ7. As a cytoplasmic complex, we further identified that the cytoplasmic SET1B/BOD1 complex interacts with the mitochondrial trifunctional protein (HADHA/B), which is essential for fatty acid oxidation that breaks down fats and converts them to energy. Depletion of SET1B remarkably decreases HADHA protein levels (Supplemental Fig. 4E), indicating that the cytoplasmic SET1B complex is essential for the stability of the mitochondrial trifunctional protein. Our future work will focus on the detailed mechanism by which cytoplasmic SET1B/COMPASS regulates HADHA/B function and stability.
Adiponectin, an adipose tissue-derived hormone, has been studied extensively for the past decade because of its anti-inflammatory, anti-atherogenic, and anti-diabetic properties (Kershaw and Flier 2004). Three adiponectin receptors have been identified: ADIPOR1, ADIPOR2, and T-cadherin, with ADIPOR1 being ubiquitously expressed and having the highest binding affinity to adiponectin (Yamauchi et al. 2003). In our current study, we further proved that knockout of the ADIPOR1 gene by CRISPR–Cas9 could largely rescue the effects caused by SET1B knockdown. AdipoR1 knockout cells are also less sensitive to SET1B depletion-induced cell death. These data suggest that AdipoR1 may play a central role in the response to SET1B depletion. Therefore, the cells that are sensitive to SET1B depletion should also be sensitive to activation of AdipoR1 signaling. Recent studies identified a small molecule named AdipoRon that is a specific agonist for adiponectin receptors with low toxicity in vivo (Okada-Iwabu et al. 2013). We found that this small molecule developed for type 2 diabetes also has a dramatic anti-tumor effect in vitro at a lower concentration (10 µM) than the plasma concentration (11.6 µM) of mice treated with 50 mg/kg AdipoRon (Okada-Iwabu et al. 2013). In LM2 breast cancer cell transplanted nude mice, 12 treatments with 50 mg/kg AdipoRon significantly reduced tumor growth and significantly increased life span. Taken together, our findings reveal a new function of SET1B/COMPASS in the cytoplasm and provide a novel therapeutic strategy for targeting SET1B/BOD1/AdipoR1 signaling in human cancer.
Materials and methods
Antibodies
HSP90 (sc-7947) and GFP (sc-9996) antibodies were purchased from Santa Cruz Biotechnology, M2 Flag (F3165) antibody was purchased from Sigma, RBBP5 (A300-109A) antibody was from Bethyl Laboratories, and ASH2L (no. 5091) and WDR5 (no. 13105) antibodies were purchased from Cell Signaling. The anti-SET1A polyclonal antibody was generated with a peptide corresponding to human SET1A amino acids 1240–1444, and anti-SET1B polyclonal antibody was generated with the peptide corresponding to human SET1B amino acids 1407–1627. Anti-MLL3 was generated against human MLL3 amino acids 1–200, and anti-MLL4 was generated as described before.
Cell lines and RNAi
MCF7, T47D, BT549, Hs578t, and MDA-MB-468 cells were obtained from American Type Culture Collection and maintained with DMEM (Gibco) containing 10% FBS (Sigma). LM2 cells (derived from MDA-MB-231 cells) were kindly provided by Dr. Yibin Kang. The cells were infected with lentivirus containing shRNAs in the presence of 4 μg/ml polybrene (Sigma) for 24 h in DMEM supplemented with 10% FBS. The infected cells were selected with 2 μg/mL puromycin for an extra 48 h before harvest. The shRNA constructs were purchased from Sigma. The clone IDs for SET1B are TRCN0000237963 (shSET1B-#1) and TRCN0000237965 (shSET1B-#2); clone IDs for BOD1 are TRCN0000168741 and TRCN0000167057. The nontargeting (shCtrl) shRNA construct (SHC002) was purchased from Sigma.
ChIP-seq
For each ChIP assay, 5 × 107 cells were used, and the assays were performed as described previously. ChIP-seq libraries were prepared with Illumina's TruSeq DNA sample preparation kit.
Crystal violet staining
Crystal violet staining was performed as described previously (Wang et al. 2014). Briefly, the cells were stained with 0.5% crystal violet staining solution and incubated for 20 min at room temperature on a bench rocker with a frequency of 20 oscillations per minute. The plate was washed four times in a stream of tap water followed by air-drying for 2 h at room temperature. Next, 200 µL of methanol was added to each well, the plate was incubated with its lid on for 20 min at room temperature on a bench rocker with a frequency of 20 oscillations per minute, and the optical density of each well was measured at 570 nm (OD570) with a plate reader.
Mass spectrometry (MS)
For Orbitrap Fusion Tribrid MS analysis, the tryptic peptides were purified with Pierce C18 spin columns (Thermo Scientific). Three micrograms of each fraction was loaded onto an autosampler with a Thermo Easy nLC 1000 UPLC pump onto a vented Acclaim Pepmap 100 (75 µm × 2 cm) nanoViper trap column coupled to a 3-µm nanoViper analytical column at 100 Å, C18, 0.075 mm, and 500 mm (Thermo, 164570) with a stainless steel emitter tip assembled on the Nanospray Flex ion source with a spray voltage of 2000 V. Buffer A contained 94.785% H2O with 5% ACN and 0.125% FA, and buffer B contained 99.875% ACN with 0.125% FA. The chromatographic run was for 2 h in total with the following profile: 0%–7% for 3 min, 10% for 3 min, 25% for 80 min, 33% for 20 min, 50% for 3 min, 95% for 3 min, and again 95% for 8 min. Additional MS parameters were as follows: ion transfer tube temperature 300°C, Easy-IC internal mass calibration, default charge state 2, and cycle time 3 sec. Detector type was set to Orbitrap, with 60-K resolution, wide quad isolation, mass range normal, scan range 300–1500 (m/z), maximum injection time 50 msec, AGC target 200,000, 1 microscan, S-lens RF level 60, without source fragmentation, and centroid data type. MIPS was set as on, and included charge states were 2–6 (reject unassigned). Dynamic exclusion was enabled with n = 1 for 30- and 45-sec exclusion duration at 10 ppm for high and low. Precursor selection was as follows: decision most intense, top 20, isolation window 1.6, automatic normal scan range, first mass 110, collision energy 30%, CID, ion trap detector type, OT resolution 30 K, rapid IT scan rate, maximum injection time 75 msec, AGC target 10,000, Q of 0.25, and inject ions for all available parallelizable times.
Tandem mass spectra analysis
Spectrum raw files from samples were extracted into ms1 and ms2 files using the in-house program RawXtractor or RawConverter (http://fields.scripps.edu/downloads.php; He et al. 2015), and the tandem mass spectra were searched against the UniProt human protein database and matched to sequences using the ProLuCID/SEQUEST algorithm (ProLuCID version 3.1) (Eng et al. 1994; Xu et al. 2015) with peptide mass tolerance of 50 ppm for precursor ions and 600 ppm for fragment ions. The search space included all fully and half-tryptic peptide candidates that fell within the mass tolerance window with no miscleavage constraint, assembled and filtered with DTASelect2 (version 2.1.3) (Tabb et al. 2002; Cociorva et al. 2007) through Integrated Proteomics Pipeline (IP2 version 3, Integrated Proteomics Applications, Inc.; http://www.integratedproteomics.com). To estimate peptide probabilities and false discovery rates (FDRs) accurately, we used a target/decoy database containing the reversed sequences of all of the proteins appended to the target database (Elias and Gygi 2007). Each protein identified was required to have a minimum of one peptide of minimal length of six amino acid residues and within 10 ppm of the expected m/z. However, this peptide had to be an excellent match with a FDR <0.001 and at least one excellent peptide match. After the peptide/spectrum matches (PSMs) were filtered, we estimated that the protein FDRs were ≤1% for each sample analysis.
ChIP-seq analysis
ChIP-seq reads were aligned to the human genome (University of California at Santa Cruz [UCSC] hg19) for MDA-MB-231 cells using Bowtie version 1.0.0 (Langmead et al. 2009). Only uniquely mapping reads with up to two mismatches within the entire length of the read were considered for further analysis. The resulting reads were extended to 150 bases toward the interior of the sequenced fragment and normalized to total reads aligned (reads per million). Peak detection for H3K4me1, H3K4me3, and RNA polymerase II were done with MACS (model-based analysis of ChIP-seq) version 1.4.2 using default parameters. Gene annotations from Ensembl release 72 were used.
RNA-seq analysis
Total RNA was extracted from Trizol according to the manufacturer's instructions. Next, total RNA was treated with DNase I and cleaned with the Qiagen RNeasy minikit. Five-hundred nanograms of RNA was used for library preparation with TruSeq stranded total RNA with the Ribo-Zero Gold kit (Illumina, RS-123-2201). The sequenced reads were aligned to the human genome (UCSC hg19) with TopHat version 2.0.9 (Trapnell et al. 2009). To evaluate differential gene expression, gene count tables were constructed using Ensembl gene annotations and used as input into edgeR 3.0.8 (Robinson et al. 2010). Adjusted P-values were computed using the Benjamini-Hochburg method. Protein-coding genes, long noncoding RNA, and pseudogenes with adjusted P-values of <0.01 were used for the downstream analysis with Metascape. P-values for Venn diagrams were performed with the hypergeometric test.
Animal experiments
All animal work was performed in accordance with protocols approved by The Center for Comparative Medicine (CCM) of Northwestern University. Athymic nude mice at 5–6 wk old were used for xenograft experiments. For the tumor growth assay, 4 × 106 breast cancer cells were inoculated into the fat pads of nude mice. Tumor growth was monitored every other day 2 wk after inoculation. For treatment assay, 4 × 106 breast cancer cells were inoculated into the fat pads of nude mice. Twelve days after injection, when the tumor reached 100 mm3, mice were divided randomly into two groups. They were treated 10 times with either 50 mg/kg PBS or 50 mg/kg AdipoRon, and the tumor size and animal survival were monitored. Tumor growth was measured 2 wk after inoculation
Data availability
Next-generation sequencing data sets have been deposited at Gene Expression Omnibus with accession number GSE86855.
Supplementary Material
Acknowledgments
MDA-MB-231, LM2 cells were kindly provided by Dr. Yibin Kang. We thank Dr. Edwin Smith for critical reading of the manuscript, and Laura Shilatifard for editorial assistance. We are also grateful to Chelsee Strojny in J.N.S.’s laboratory for technical assistance for mass spectrometric studies. This work was partially supported by Northwestern University Feinberg School of Medicine. J.N.S. is supported by R00 DC-013805 and the Chicago Biomedical Consortium with support from the Searle Funds at the Chicago Community Trust. L.W. was supported by the Training Program in Signal Transduction and Cancer (T32 CA070085-19). The COMPASS studies in A.S.’s laboratory are supported by a generous grant from the Mary Kay Foundation and an Outstanding Investigator Award grant from the National Cancer Institute (R35CA197569) to A.S.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.306092.117.
References
- Bledau AS, Schmidt K, Neumann K, Hill U, Ciotta G, Gupta A, Torres DC, Fu J, Kranz A, Stewart AF, et al. 2014. The H3K4 methyltransferase Setd1a is first required at the epiblast stage, whereas Setd1b becomes essential after gastrulation. Development 141: 1022–1035. [DOI] [PubMed] [Google Scholar]
- Bodega B, Marasca F, Ranzani V, Cherubini A, Della Valle F, Neguembor MV, Wassef M, Zippo A, Lanzuolo C, Pagani M, et al. 2017. A cytosolic Ezh1 isoform modulates a PRC2-Ezh1 epigenetic adaptive response in postmitotic cells. Nat Struct Mol Biol 24: 444–452. [DOI] [PubMed] [Google Scholar]
- Bononi A, Giorgi C, Patergnani S, Larson D, Verbruggen K, Tanji M, Pellegrini L, Signorato V, Olivetto F, Pastorino S, et al. 2017. BAP1 regulates IP3R3-mediated Ca2+ flux to mitochondria suppressing cell transformation. Nature 546: 549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer K, Kocha KM, Snider T, Moyes CD. 2015. Sensing and responding to energetic stress: the role of the AMPK–PGC1α–NRF1 axis in control of mitochondrial biogenesis in fish. Comp Biochem Physiol B Biochem Mol Biol 199: 4–12. [DOI] [PubMed] [Google Scholar]
- Brici D, Zhang QY, Reinhardt S, Dahl A, Hartmann H, Schmidt K, Goveas N, Huang JH, Gahurova L, Kelsey G, et al. 2017. Setd1b, encoding a histone 3 lysine 4 methyltransferase, is a maternal effect gene required for the oogenic gene expression program. Development 144: 2806–2817. [DOI] [PubMed] [Google Scholar]
- Cao KX, Collings CK, Marshall SA, Morgan MA, Rendleman EJ, Wang L, Sze CC, Sun TJ, Bartom ET, Shilatifard A. 2017. SET1A/COMPASS and shadow enhancers in the regulation of homeotic gene expression. Gene Dev 31: 787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, et al. 2007. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem 282: 20395–20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cociorva D, Tabb DL, Yates JR. 2007. Validation of tandem mass spectrometry database search results using DTASelect. Curr Protoc Bioinformatics 16: 13.4.1–13.4.14. [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. 2007. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4: 207–214. [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5: 976–989. [DOI] [PubMed] [Google Scholar]
- Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, Yagyu R, Nakamura Y. 2004. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol 6: 731–740. [DOI] [PubMed] [Google Scholar]
- Han S, Schroeder EA, Silva-Garcia CG, Hebestreit K, Mair WB, Brunet A. 2017. Mono-unsaturated fatty acids link H3K4me3 modifiers to C. elegans lifespan. Nature 544: 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Diedrich J, Chu YY, Yates JR III. 2015. Extracting accurate precursor information for tandem mass spectra by RawConverter. Anal Chem 87: 11361–11367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershaw EE, Flier JS. 2004. Adipose tissue as an endocrine organ. J Clin Endocr Metab 89: 2548–2556. [DOI] [PubMed] [Google Scholar]
- Kotake Y, Cao R, Viatour P, Sage J, Zhang Y, Xiong Y. 2007. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4α tumor suppressor gene. Genes Dev 21: 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell 128: 693–705. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A. 2002. COMPASS, a histone H3 (lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem 277: 10753–10755. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Tate CM, You JS, Skalnik DG. 2007. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J Biol Chem 282: 13419–13428. [DOI] [PubMed] [Google Scholar]
- Mazur PK, Reynoird N, Khatri P, Jansen PW, Wilkinson AW, Liu S, Barbash O, Van Aller GS, Huddleston M, Dhanak D, et al. 2014. SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature 510: 283–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. 2001. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci 98: 12902–12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Herz HM, Shilatifard A. 2012. SnapShot: histone lysine methylase complexes. Cell 149: 498–498.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada-Iwabu M, Yamauchi T, Iwabu M, Honma T, Hamagami K, Matsuda K, Yamaguchi M, Tanabe H, Kimura-Someya T, Shirouzu M, et al. 2013. A small-molecule AdipoR agonist for type 2 diabetes and short life in obesity. Nature 503: 493–499. [DOI] [PubMed] [Google Scholar]
- Piunti A, Shilatifard A. 2016. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 352: aad9780. [DOI] [PubMed] [Google Scholar]
- Porter IM, McClelland SE, Khoudoli GA, Hunter CJ, Andersen JS, McAinsh AD, Blow JJ, Swedlow JR. 2007. Bod1, a novel kinetochore protein required for chromosome biorientation. J Cell Biol 179: 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter IM, Schleicher K, Porter M, Swedlow JR. 2013. Bod1 regulates protein phosphatase 2A at mitotic kinetochores. Nat Commun 4: 2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salz T, Li G, Kaye F, Zhou L, Qiu Y, Huang S. 2014. hSETD1A regulates Wnt target genes and controls tumor growth of colorectal cancer cells. Cancer Res 74: 775–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. 2012. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 81: 65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze CC, Cao K, Collings CK, Marshall SA, Rendleman EJ, Ozark PA, Chen FX, Morgan MA, Wang L, Shilatifard A. 2017. Histone H3K4 methylation-dependent and -independent functions of Set1A/COMPASS in embryonic stem cell self-renewal and differentiation. Genes Dev 31: 1732–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR III. 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res 1: 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi YH, Westfield GH, Oleskie AN, Trievel RC, Shilatifard A, Skiniotis G. 2011. Structural analysis of the core COMPASS family of histone H3K4 methylases from yeast to human. Proc Natl Acad Sci 108: 20526–20531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhao ZB, Meyer MB, Saha S, Yu MG, Guo AL, Wisinski KB, Huang W, Cai WB, Pike JW, et al. 2014. CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis. Cancer Cell 25: 21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A. 2008. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol 28: 7337–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Park SK, Venable JD, Wohlschlegel JA, Diedrich JK, Cociorva D, Lu B, Liao L, Hewel J, Han X, et al. 2015. ProLuCID: an improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J Proteomics 129: 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T, Kamon J, Ito Y, Tsuchida A, Yokomizo T, Kita S, Sugiyama T, Miyagishi M, Hara K, Tsunoda M, et al. 2003. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature 423: 762–769. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Next-generation sequencing data sets have been deposited at Gene Expression Omnibus with accession number GSE86855.