Abstract
Self/non-self-discrimination by the innate immune system relies on germline-encoded, non-rearranging receptors expressed by innate immune cells recognizing conserved pathogen-associated molecular patterns. The natural killer group 2D (NKG2D) receptor is a potent immune-activating receptor that binds human genome-encoded ligands, whose expression is negligible in normal tissues, but increased in stress and disease conditions for reasons that are incompletely understood. Here it is not clear how the immune system reconciles receptor binding of self-proteins with self/non-self-discrimination to avoid autoreactivity. We now report that increased expression of NKG2D ligands after virus infection depends on interferon response factors activated by the detection of viral double-stranded RNA by pattern-recognition receptors (RIG-I/MDA-5) and that NKG2D ligand up-regulation can be blocked by the expression of viral dsRNA-binding proteins. Thus, innate immunity-mediated recognition of viral nucleic acids triggers the infected cell to release interferon for NK cell recruitment and to express NKG2D ligands to become more visible to the immune system. Finally, the observation that NKG2D-ligand induction is a consequence of signaling by pattern-recognition receptors that have been selected over evolutionary time to be highly pathogen-specific explains how the risks of autoreactivity in this system are minimized.
Keywords: cellular immune response, double-stranded RNA (dsRNA), natural killer cells (NK cells), pattern recognition receptor (PRR), virus, NKG2D, NKG2D ligands, dsRNA, self/non-self-discrimination, virus infection
Introduction
Self/non-self-discrimination by the innate immune system depends on germline-encoded, non-rearranging receptors expressed by innate immune cells recognizing conserved molecular patterns specific to pathogens, such as LPS and flagellin (1). However, this paradigm does not seem to hold for natural killer (NK)2 cells, because although the majority of NK activating receptors are non-rearranging and essentially invariant, the ligands for these receptors are not pathogen-specific, but rather are encoded in the human genome (2) and, a system based on immune recognition of self, inevitably involves a risk of self-damage. The best characterized of these receptor/ligand systems is the interaction of natural killer group 2D (NKG2D) with major histocompatibility complex class I chain-related A and B (MICA/B) and UL16-binding protein (ULBP) molecules and it has been reported that inappropriate expression of NKG2D-ligands (NKG2DL) is a feature of various autoimmune diseases including rheumatoid arthritis, celiac disease, and diabetes (3). NK cell expression of inhibitory receptors able to recognize constitutively expressed self-molecules such as MHC class I is one way to control autoreactivity, but this inhibitory signaling can be overcome by activation via the NKG2D receptor after it binds NKG2DL whose expression, although negligible on healthy tissues, is up-regulated in disease (4).
The basis of this increased expression in disease is not completely understood. Prior studies using tumor cell lines, or actively proliferating normal cells, have shown that NKG2DL expression is influenced by multiple pathways, both transcriptionally and posttranscriptionally (4–8). However, the observation that inhibition of these pathways typically decreases NKG2DL expression partially, but not completely, suggests that other levels of regulation of the expression of NKG2DL likely exist. Moreover, practically all this information has been obtained from the study of tumor cells, so that even though an increase in expression of ligands for the NKG2D receptor occurs after infection with multiple pathogens (4), relatively little is known as to how NKG2DL expression is coupled to cellular processes associated with infectious disease. Histone deacetylase 3 (HDAC3) is known to be a repressor of ULBPs expression in epithelial cancer cells (9) and recently it was shown that expression of the murine cytomegalovirus protein M18, which blocks HDAC action, can also lead to expression of ULBP1 (10). These authors proposed that in uninfected cells, constitutive HDAC activity normally suppresses NKG2DL expression, but that viral HDAC inhibition relieved this block, however, this passive model of regulation is likely simplistic and NKG2DL expression probably requires the recruitment of positive elements to the promoters of these genes, as well as the relief of blocks on promoter activity (4).
Here we show that the detection by RIG-I-like receptors (RLRs) of double-stranded RNA (dsRNA) produced during viral infection of human fibroblasts induces expression of various NKG2DL and that interferon response factors (IRFs) can intervene in this process. We also provide evidence that the expression of virally encoded dsRNA-binding proteins can block the increased expression of molecules such as MICA or ULBP2. These data demonstrate that innate recognition of viral dsRNA can trigger both the secretion of type I interferons, and induce expression of NKG2DL, so that the infected cell is a better target for the cytotoxic lymphocytes recruited to the site of viral infection by those interferons. The observation that induced expression of NKG2DL is a consequence of pattern-recognition receptor (PRR) signaling also suggests that the problem of how the binding of activating NK receptors to self-proteins can be squared with self/non-self-discrimination and the avoidance of autoimmunity is solved by making expression of those ligands contingent on signaling from PRR.
Results
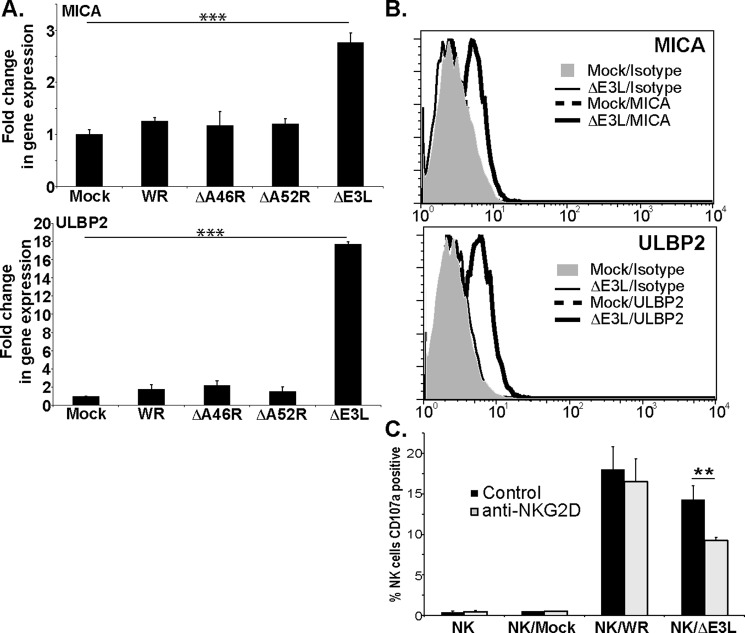
Virus infections, but not vaccinia, often increase expression of ligands for the NKG2D receptor (4, 11, 12). To test the idea that some gene product of vaccinia virus (VV) might block induced expression of NKG2DL, human fibroblasts were infected with either wild-type VV (strain WR), or mutant VVs deficient in the A46R and A52R genes that interfere with TLR signaling (13, 14) or the E3L gene, which impairs the mammalian IFN-regulated innate antiviral response (15). After overnight culture, changes in the expression of mRNA for various NKG2DL genes were analyzed by qPCR. Infection with either wild-type VV or mutants deficient in A46R and A52R did not induce changes in the levels of mRNA for NKG2DL (Fig. 1A). In contrast, infection with the ΔE3L mutant consistently led to significantly increased levels of mRNA for MICA and ULBP2. Similar observations were made after infection of another fibroblast cell line (supplemental Fig. 1A). Consistent with previous data (16, 17), infection with herpes simplex virus I, as another negative control, did not induce any modification of NKG2DL mRNA levels in either of these cell lines (supplemental Fig. 1B).
Figure 1.
Infection with vaccinia virus deficient in the E3L protein triggers a functionally relevant increase in expression of NKG2DL. A, hTert fibroblasts were infected with the indicated virus or left uninfected and cultured overnight. Total RNA was extracted and qRT-PCR was used to assay MICA and ULBP2 expression. The results are expressed as fold-change in gene expression compared with uninfected cells. The data presented are representative ±S.D. of eight experiments. B, flow cytometry analysis of MICA and ULBP2 expression in fibroblasts infected with the indicated viruses, (4 experiments with fibroblast from two different donors). C, NK cells, preincubated with either an isotype control mAb or an NKG2D-specific mAb, were cultured alone or in the presence of mock-infected, wild-type vaccinia WR-infected or VVΔE3L-infected fibroblasts at an effector:target ratio of 1:2. The graph shows the proportion of CD107a+ NK cells in the different conditions after incubation for 2 h at 37 °C (three experiments with NK cells from two different donors). Statistical significance was determined using one-way ANOVA (**, p < 0.01; ***, p < 0.001; no bars, S.D. values smaller than symbol used; no symbol means not statistically significant, p > 0.05).
These increased levels of mRNA expression were associated with increased expression of MICA and ULBP2 protein as visualized by FACS (Fig. 1B) and Western blot (supplemental Fig. S2). This increased expression of NKG2DL at the cell surface was sufficient to affect NK cell recognition of VV-infected fibroblasts, which normally depends on the NKp30, NKp44, and NKp46 receptors (11, 12), so that whereas mAb blockade of the NKG2D receptor had almost no effect on NK cell recognition of cells infected with wild-type VV, this treatment significantly reduced NK cell recognition of human fibroblasts infected with the ΔE3L virus (Fig. 1C). Although the levels of degranulation were low, these data were reproducible in experiments with NK cells from unrelated donors and confirm that up-regulation of NKG2DL on the infected cell produced by infection with this virus was functionally relevant. That NKG2D blockade reduces only partially the lysis almost certainly reflects the contribution of interactions between other NK activating receptors and the VV-infected cell.
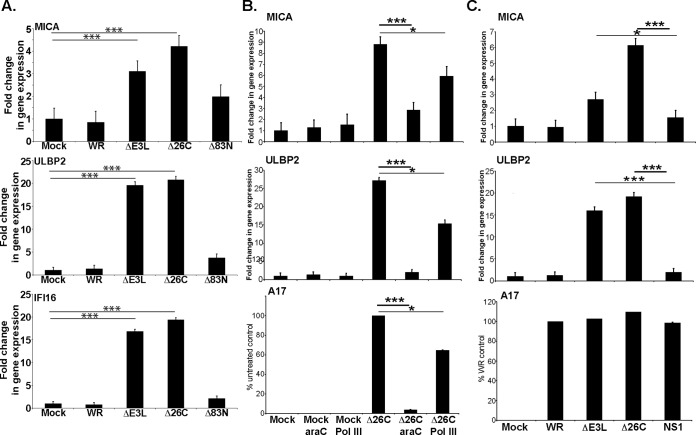
E3L contains two functional domains: an N-terminal sequence required for nuclear localization and Z-DNA-binding activity, and a C-terminal domain that sequesters dsRNA (18). Cells were infected with wild-type VV, the ΔE3L mutant, and mutant viruses lacking either the N-terminal domain (Δ83N) or the C-terminal domain (Δ26C) of E3L, and NKG2DL mRNA levels were assessed. Deletion of the 26 amino acids at the C terminus of E3L that bind dsRNA removed the ability of the E3L protein to block the increase in NKG2DL mRNA after infection (Fig. 2A), suggesting that viral dsRNA was likely a key stimulus triggering increased NKG2DL expression. Poxvirus genomes are transcribed in a temporal cascade; early genes are transcribed prior to viral DNA replication, whereas transcription of intermediate and late genes occurs after viral DNA replication and is generally thought to be the major source of viral dsRNA (19). Treatment with arabinoside C (araC) inhibits poxvirus DNA replication, and so the accumulation of viral RNAs, without affecting the synthesis of viral early mRNAs and early proteins. Culture of VVE3LΔ26C-infected fibroblasts with araC markedly reduced the increase in MICA and ULBP2 mRNA generally seen after infection with this mutant (Fig. 2B), supporting the hypothesis that RNA produced late in infection stimulates expression of NKG2DL mRNA. Treatment with an inhibitor of RNA polymerase III reduced only modestly the induction of ULBP2 mRNA, suggesting that the action of this enzyme in transcription of cytosolic DNA into RNA for detection through the RIG-I/MDA-5 pathway (20) does not play an important role in the induction of NKG2DL expression in this system. Importantly, when cells were infected with a recombinant VV lacking the E3L gene, but expressing a distinct viral dsRNA-binding protein, influenza NS1 (VVΔE3L/NS1) (21) (Fig. 2C), the reconstitution of dsRNA sequestration was sufficient to block NKG2DL induction, again suggesting that viral dsRNA is a key stimulus triggering increased expression of NKG2DL.
Figure 2.
Accumulation of dsRNA in infected cells triggers the increase in NKG2DL expression induced by viral infection. A, fibroblasts were infected with the indicated viruses and qPCR was used to assay changes in MICA, ULBP2, and IFI16 expression (3 experiments). B, fibroblasts were infected with the VVE3LΔ26C virus and then treated with either araC or the RNA Pol III inhibitor. qRT-PCR was used to assay MICA, ULBP2, and A17 expression. For MICA and ULBP2 the results are expressed as fold-change in gene expression compared with uninfected cells. For A17 the data are represented as a fraction of untreated control. The data presented are representative of four experiments with the VVE3LΔ26C virus. Similar data were obtained in experiments where VVΔE3L-infected fibroblasts were treated with these inhibitors. C, fibroblasts were infected with the indicated viruses and then qRT-PCR was used to assay MICA, ULBP2, and A17 expression, as described above. The data presented are representative of four experiments. *, p < 0.05; ***, p < 0.001.
To confirm this hypothesis in a virus-independent system, we transfected fibroblasts with cytoplasmic RNA purified from VV, VVΔE3L infected and control cells (Fig. 3). These experiments demonstrated that transfection of purified RNA is sufficient to trigger slight increases in mRNA levels for both MICA and ULBP2, but that RNA isolated from cells infected with VVΔE3L was a significantly more potent stimulus for NKG2DL expression than RNA from either VV or uninfected cells, consistent with prior data showing that the RLR system preferentially recognizes the RNA species associated with virus infection (20), but that expression of E3L suppresses dsRNA accumulation in vaccinia-infected cells (22).
Figure 3.
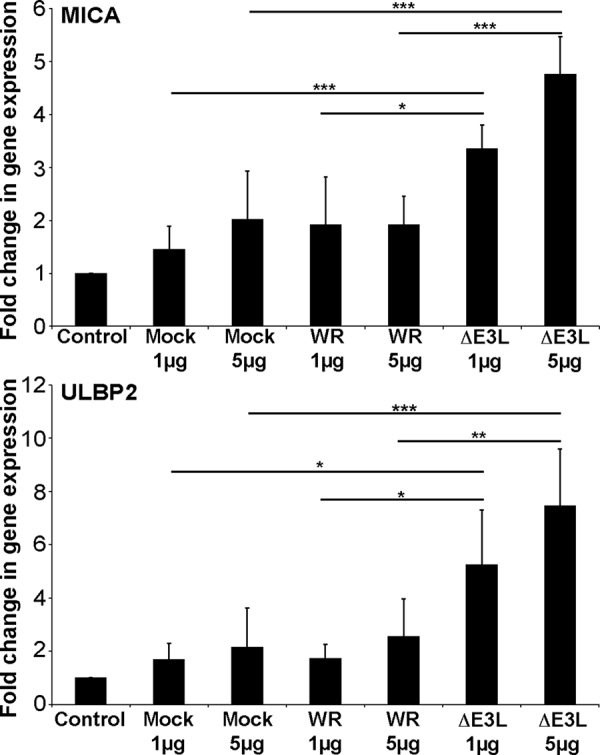
Transfection with purified viral RNA is sufficient to increase expression of MICA and ULBP2 mRNA. Fibroblasts were transfected with the indicated quantity of cytoplasmic RNA isolated from control, WR-, or VVΔE3L-infected fibroblasts. After overnight culture total RNA was extracted and qRT-PCR used to assay MICA and ULBP2 expression. The data are presented as the means of 3–5 experiments ± S.D. Statistical significance was determined using one-way ANOVA (*, p < 0.05; **, p < 0.01; ***, p < 0.001. No symbol means not statistically significant,
To test whether receptors, like RIG-I and MDA-5 that detect cytosolic viral RNA ligands or processed self-RNA (20), were involved in this increased expression of NKG2DL mRNA, fibroblast cells were prepared where signaling by both MDA-5 and RIG-I was inhibited by overexpression of the V protein of parainfluenzavirus type 5 (PIV5) (23, 24). The increase in NKG2DL RNA after VVΔE3L infection was blocked in fibroblasts expressing the PIV5 V protein (Fig. 4A). In a more direct approach shRNA-mediated silencing of the MAVS protein (supplemental Fig. S3B) that acts downstream of RIG-I and MDA-5 significantly reduced the increase in NKG2DL mRNA triggered by VVΔE3L infection (Fig. 4B). Infection of cell lines where CRISPR/Cas9 technology had been used to specifically silence either RIG-I or MDA-5 (supplemental Fig. S3C) showed that deletion of either of these RLRs singly had very little effect on the increase in expression of NKG2DL such as ULBP2 suggesting that both signaling pathways are activated after infection with the ΔE3L virus and that signaling through either RIG-I or MDA-5 is sufficient to produce an increase NKG2DL mRNA levels (Fig. 4C). Consistent with this idea, transfection of human fibroblast lines with either full-length RIG-I or MDA-5 proved to be sufficient to stimulate an increase in NKG2DL mRNA expression (Fig. 4D) in agreement with previous data showing that over-expression of wild-type RIG-I or MDA-5 could trigger increased activation of an IFN-β reporter gene (25, 26). In aggregate, these data strongly suggest that the recognition by RLRs, either RIG-I or MDA-5, of dsRNA produced during viral infection stimulates increased levels of mRNA for NKG2DL.
Figure 4.
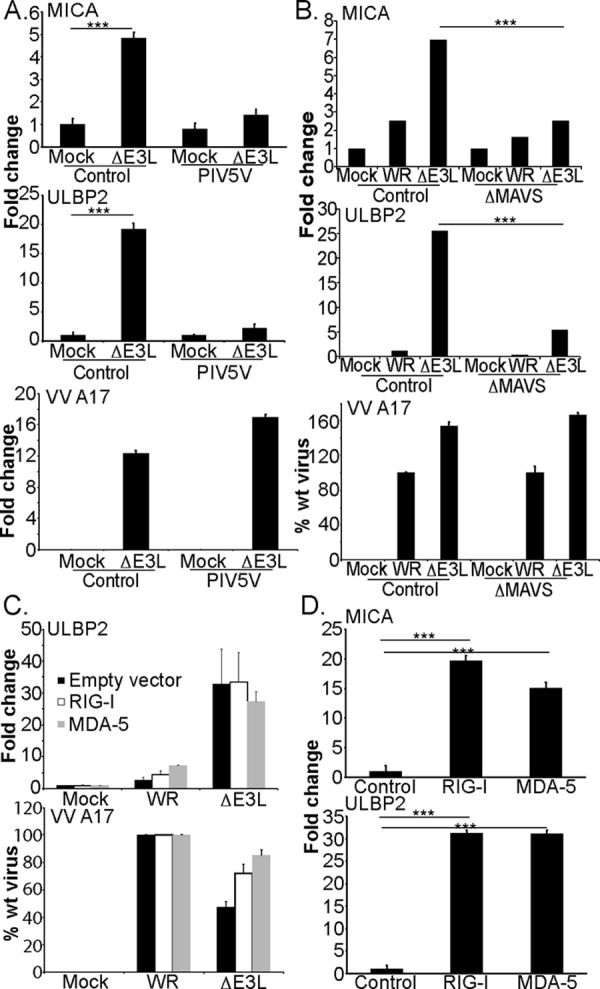
Increased NKG2DL expression depends on signaling by the RIG-I and/or MDA-5 receptors. Fibroblasts expressing the V5 gene of parainfluenza virus (A) were either left uninfected or infected with the VVΔE3L virus. qRT-PCR was used to assay MICA and ULBP2 expression (4 experiments). B, fibroblasts, transduced with lentivirus expressing either control or MAVS-specific shRNA, were infected with the indicated viruses and after overnight culture total RNA was extracted and qRT-PCR used to assay MICA and ULBP2 expression. C, fibroblasts, where expression of the indicated gene had been reduced by CRISPR/Cas9 targeting, were infected with the indicated viruses and after overnight culture total RNA was extracted and qRT-PCR used to assay ULBP2 and VVA17 expression (3 experiments). D,fibroblasts were transfected with the indicated plasmids using the 4D-Nucleofector system. After overnight culture total RNA was extracted and qRT-PCR was used to assay MICA and ULBP2 expression (3 experiments). Data are presented ± S.D. Statistical significance was determined using one-way ANOVA (***, p < 0.001; no bars, S.D. values smaller than symbol used; no symbol means not statistically significant, p > 0.05).
RLR activation could increase NKG2DL transcription either directly via cell intrinsic factors modulated by RLR signaling or indirectly via the secretion of type I interferons from the virus-infected cell, which then drive a secondary wave of transcription of hundreds of other genes. Treatment of fibroblasts with high doses of recombinant IFN-α and -β strongly induced expression of the interferon responsive gene IFI16, but no changes were noted in the expression of MICA or ULBP2 (Fig. 5A), suggesting that the up-regulation of MICA and ULBP2 triggered by dsRNA were not triggered by type I IFNs. These data were confirmed in experiments where mAb-mediated neutralization of IFN-α/β signaling reduced IFI16 induction more than 10-fold, but had no effect on the induction of MICA or ULBP2 after VVΔE3L infection (Fig. 5B).
Figure 5.
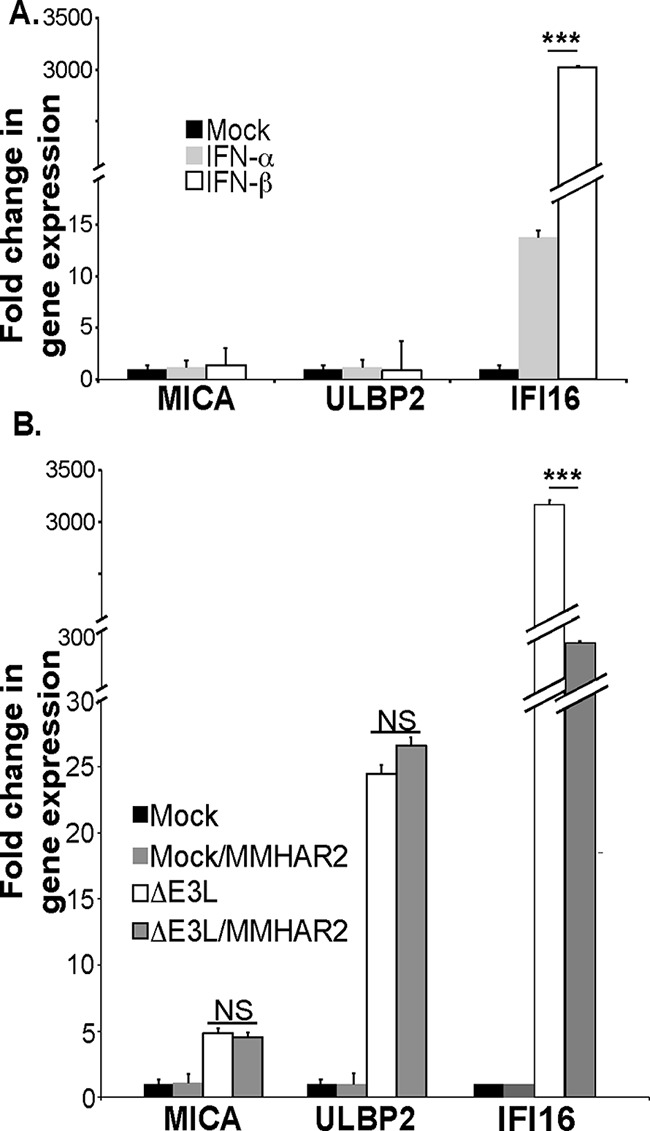
The increased NKG2DL expression by fibroblasts after viral infection does not depend on interferon α/βs. A, fibroblasts were cultured with vehicle, IFNα (1000 units/ml), or IFNβ (1000 units/ml) overnight and then qRT-PCR was used to assay MICA, ULBP2, and IFI16 expression (3 experiments). B, fibroblasts were either left uninfected or infected with the VVΔE3L virus in the presence or absence of 5 μg/ml of the MMHAR2 antibody that blocks the interaction of α/β interferons with their receptor. qRT-PCR was used to assay MICA, ULBP2, and IFI16 expression (6 experiments). ***, p < 0.001.
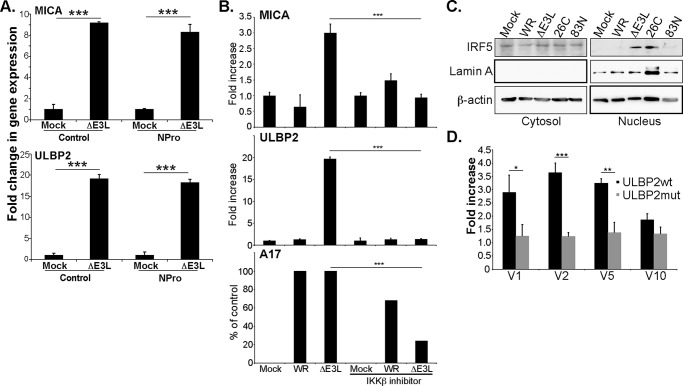
The previous data suggested that signaling downstream of RIG-I/MDA-5/MAVS led to the increase in NKG2DL mRNA. Surprisingly, transfection of cells with the NPro product of bovine viral diarrhea virus (BVDV) that drives proteasomal degradation of interferon regulatory factor 3 (27) had no effect on the induction of NKG2DL after infection with VVΔE3L (Fig. 6A). Thus, to better define the factors underlying the increased levels of NKG2DL mRNA after infection with VVΔE3L, the effects of treatment with chemical inhibitors of a wide range of pathways, including those involving ATM, ATR, PI-3 kinase, NF-κB, and TBK1/IRF3 were tested (supplemental Fig. 4). Inhibition of neither NF-κB nor the TBK1/IRF3 axis had much effect and this conclusion was supported by experiments of infection using fibroblasts where IRF3 or NF-κB expression had been specifically silenced (supplemental Fig. S5). In contrast, treatment with the IKKβ inhibitor (BI605906) blocked the increased mRNA for MICA and ULBP2 seen after VVΔE3L infection (Fig. 6B). This compound is known to block IKKβ-mediated phosphorylation of IFN regulatory factor 5 (IRF5) (28), suggesting that IRF5 could be a relevant factor in the induction of MICA and ULBP2 in this system. Consistent with this idea, infection with the VVΔE3L and 26C mutant viruses that induce increased NKG2DL mRNA also induced nuclear translocation of IRF5 (Fig. 6C). Finally, IRF5 co-expression could enhance transcription from a reporter vector expressing luciferase under the control of ∼1000 bp of ULBP2 promoter sequence (Fig. 6D), whereas having little effect on a ULBP2 promoter where a strong candidate IRF-binding site, identified using the TransFac package, was mutated to destroy the consensus sequence for IRF binding. Because various alternatively spliced forms of IRF5 have been described, four different IRF5 isoforms were co-transfected with either the wild-type or mutant ULBP2 promoter in these experiments. For three of these variants, co-expression of IRF5 triggered an ∼3-fold increase in luciferase transcription from the ULBP2 reporter vector (Fig. 6D), confirming that at least some IRF5 variants can modulate NKG2DL transcription.
Figure 6.
A role for IKKβ and IRF5 in the increased transcription of NKG2DL after VVΔE3L infection. A, fibroblasts expressing the NPro gene of BVD virus were either left uninfected or infected with the VVΔE3L virus. qRT-PCR was used to assay MICA and ULBP2 expression (4 experiments). B, fibroblasts were either left uninfected or infected with the indicated virus in the presence or absence of 5 μm BI605906 that inhibits IKKβ. qRT-PCR was used to assay MICA, ULBP2, and VVA17 expression (3 experiments). C, Western blot analysis of IRF5 expression in cytosolic and nuclear lysates of fibroblasts infected with the indicated viruses, Lamin A was used as a fractionation control (2 experiments). D, 293T cells were co-transfected with Firefly luciferase reporter plasmid containing either wt or mutant ULBP2 promoter, Renilla luciferase, and an expression vector for the indicated IRF5 isoform. After 24 h, cells were lysed and the Renilla and Firefly luciferase activities were measured by the dual luciferase assay (9 experiments). The data were normalized using the Renilla luciferase levels. Data are presented ± S.D. Statistical significance was determined using one-way ANOVA (*, p < 0.05; **, p < 0.01; ***, p < 0.001; no bars, S.D. values smaller than symbol used; no symbol means not statistically significant, p > 0.05).
Discussion
Recognition of dsRNA by RIG-I and MDA-5 during virus infection triggers an increase in NKG2DL expression by the infected cell. Thus the same stimulus that triggers interferon release for NK cell activation and recruitment to sites of infection also induces changes in the target cell surface to make it a better target for those NK cells when they arrive. This dual effect of RLR activation is reminiscent of the effects produced by TLR stimulation of macrophages and dendritic cells, where activation via these molecules triggers both the transcription of cytokine genes and the up-regulation of costimulatory molecules such as CD40 and CD86. Indeed, treatment of both human and murine macrophages with TLR ligands induces cell surface expression of NKG2DL as well as differentiation of these cells (4). These observations raise the possibility that increased NKG2DL expression could be a general downstream feature of PRR activation. The interaction between PRRs and pathogen-associated molecular patterns (PAMPs) has undergone stringent selection over evolutionary time to efficiently initiate immune responses while safeguarding self/non-self-discrimination (1). Thus the hypothesis that the expression of the NKG2DL is downstream of signals originating from PRRs resolves the apparent problem of reconciling self/non-self-discrimination with high affinity binding of a potent immune activating receptor to self-proteins. The pattern of expression of NKG2DL by different cell types infected with different pathogens would depend on the integration of 1) the output from the repertoire of PRRs expressed by that cell and activated by the PAMPs expressed by the infectious agent and 2) the effects of viral immune evasion molecules that target PRR/PAMP interactions. This model can explain the heterogeneous patterns of NKG2DL expression between different cell types under different kinds of stress.
With regard to the details of how PRR activation signals for increased NKG2DL expression our data show that inhibition of IKKβ significantly reduced the normal increase in NKG2DL mRNA. It was surprising that IRF3 knockdown did not affect the increase in MICA and ULBP2 RNA after VVΔE3L infection, because overexpression of IRF3 could enhance transcription from the ULBP2wt reporter vector (supplemental Fig. S6) and transfection of DNA has been shown to induce expression of murine NKG2DL (RAE-1) via a pathway involving IRF3 (5). However, here it is important to note that vaccinia virus is known to encode multiple gene products that act to block IRF3 activation, both in the cytosol and nucleus (29, 30). Similar considerations, VACV is well-known to express many proteins that act to restrict NF-κB activation (31, 32), may also explain why neither inhibition nor silencing of NF-κB affected the induction of NKG2DL mRNA after infection even though the canonical NF-κB pathway is generally considered the major pathway triggered by PRR activation. A prediction from these observations would be that different pathogen immune evasion proteins targeting different PRRs will markedly influence NKG2DL expression after infection; either the degree of induction of expression or the repertoire of NKG2DL affected. This hypothesis that the regulation of NKG2DL expression depends on interplay between various factors that influence PRR activation may also be a factor in the aberrant expression of NKG2DL in autoimmunity where multiple examples of mutations and polymorphisms in PRRs have been implicated in the pathogenesis of various autoimmune diseases (33). For example, it is interesting to note that not all the isoforms of IRF5 were able to up-regulate transcription from a ULBP2 luciferase reporter vector. IRF5 is a critical regulator of the immune response to infection, mediating interferon activation and apoptosis in a number of inflammatory signaling pathways (34). Moreover, variation in IRF5 has been associated with multiple autoimmune diseases, in particular SLE (35), but also with cancer (36). In most cases the bases of these associations are not fully understood, but different IRF5 splice isoforms have distinct patterns of cell type-specific expression, localization, and dissimilar functions in type I IFN gene induction in virus infection (37). Thus it does not appear unreasonable to suggest that differences in the expression of IRF5 (due either to allelic polymorphism or splicing variation) could contribute to the observed variation in expression patterns of NKG2DL. In fact, given the extensive cross-talk between the NF-κB and IRF immune signaling pathways after PRR activation, it is not surprising that increased NKG2DL expression is a common outcome after ligation of multiple PRRs, including TLRs, RLRs, and cytosolic DNA sensors (4).
More speculatively, the key roles of PRRs in inflammatory responses and the known connection between inflammation and cancer (38) suggest that PRR activation could also be a factor at least in initiating the expression of NKG2DL by tumor cells, although obviously other events including DNA damage, mutation, and dysregulated proliferation could later become key factors in the maintenance of NKG2DL expression by these tumor cells.
Materials and methods
Cells and viruses
Primary fibroblasts and the immortalized fibroblast cell lines BJ1 (Promega) and hTERT human fibroblasts (39) as well as 293T cells and baby hamster kidney cells (BHK-21) were cultured in DMEM with 10% fetal calf serum, glutamine, and antibiotics. VACV wild-type Western reserve strain (WR), ΔA46R, ΔA52R mutants (kind gifts of Prof. G. L. Smith, University of Cambridge), and ΔE3L/NS1 have been described (13, 14, 21) and were grown on BSC-40 cells. VACV mutants deleted of E3L (ΔE3L), of the first 83 N-terminal amino acids of E3L (Δ83N), or of the 26 C-terminal amino acids of E3L (Δ26C) were grown and titrated in BHK-21 cells (21).
Human NK cells (95–99% CD3-CD56+) were isolated from peripheral blood using a negative selection kit (Miltenyi Biotec) and expanded in vitro as described (11). The NK cells were washed free of IL-2 and cultured overnight prior to use.
Virus infection
Cells, synchronized by confluence arrest, were trypsinized, replated in complete medium, and allowed to attach for 1–2 h. The cells were then infected with the indicated virus and the cultures incubated overnight at 37 °C and 5% CO2. All experiments were done with virus stocks titered in baby hamster kidney (BHK) cells and equal multiplicity of infections were used in every experiment. No effects of variation in multiplicity of infection were seen over a 5–50 range.
Plasmids and transfection
Expression vectors for RIG-I and MDA-5 (40) were obtained from Addgene (plasmids 27236 and 27225). Fibroblasts were transfected using the 4D-Nucleofector system (Lonza).
Recombinant lentiviruses produced using the plasmids, pdl.PIV5/V.w3.puro and pdl.BVDV/NPro.puro (41)(gifts of Prof. R. Randall University of St. Andrews), were used to create fibroblast lines expressing the V protein of PIV5 or NPro of BVDV. IRF3 knockdown was assayed directly by Western blotting, whereas the expression of PIV5V was followed indirectly by Western blotting for STAT1, another target for PIV5V (42). Cells were used within four passages of transduction. Lentiviruses expressing shRNA to silence expression of MAVS were prepared using the oligos shown in supplemental Table S1 cloned in the pLKO.1 vector. Lentiviruses directing expression of Cas9 and guide RNAs specific for the indicated genes were prepared using the vector lentiCRISPR v2 (a gift from Feng Zhang; Addgene plasmid number 52961) (43).
Transfection with cytoplasmic RNA
Cytoplasmic RNA was isolated from mock, VACV-, and VVΔE3L-infected cells using standard protocols (44) and the RNEasy kit (Qiagen) before transfection into fibroblasts using Lipofectamine Plus.
Chemicals
Recombinant interferon-α and interferon-β were purchased from PeproTech. mAb specific for interferon-α receptor chain 2 (MMHAR-2) was purchased from Millipore. Arabinoside C (araC) was purchased from Sigma and was used at a final concentration of 40 μg/ml. The inhibitor of Pol III (ML-60218) was purchased from Millipore and used at a final concentration of 12.5 μm. The BI605906 inhibitor, which blocks phosphorylation of IRF5 by IKKβ (28), a kind gift of Prof. Philip Cohen (University of Dundee), was used at a final concentration of 5 μm. Other inhibitors were purchased from Sigma, Millipore, and Selleck Biochemicals.
RNA extraction, cDNA synthesis, and qPCR
Total RNA was reverse-transcribed using random hexamers and SuperScript II RNase (Invitrogen). Quantitative PCR analysis was carried out using SYBR Green PCR Master Mix (BD Biosciences) and the primers shown in supplemental Table S1. These reactions produced single PCR amplicons of the expected length and melting temperature, as assessed by dsDNA melting curve analysis. Data were analyzed with SDS2.2 sequence detection systems and are presented as fold-changes with respect to uninfected fibroblasts.
Cell fractionation, SDS-PAGE, and Western blot analysis
Nuclear and cytoplasmic cell lysates were prepared as described previously (45). SDS-PAGE and Western blotting were carried out as described (46). β-Actin was visualized using the mAb AC-15 (Sigma). IRF3, RIG-I, MDA-5, NF-κB p65, and STAT1 were detected using rabbit monoclonal antibodies purchased from Cell Signaling Technology. Lamin A-specific antibody was purchased from Santa Cruz Biotechnology, whereas the polyclonal antibody to IRF5 (28) was purchased from the University of Dundee, Scotland. Western blot data were quantitated using Image J software (rsb.info.nih.gov/ij/, 1997–2008).
Flow cytometry
Cells were incubated with primary antibodies specific for ULBP2 and MICA, purchased from R&D Systems (Abingdon, UK), or isotype controls, followed by PE-labeled F(ab′)2 fragments of goat anti-mouse Ig (Dako). Cell death was evaluated by the analysis of 7-aminoactinomycin D staining. Samples were analyzed using BD FACSCalibur (BD Biosciences).
Degranulation assay
Degranulation assays to quantify cell surface CD107a expression were performed as previously described (46). For blocking experiments NK cells were preincubated with 10 μg/ml of mAb anti-NKG2D (R&D systems) or an isotype control mAb.
Luciferase reporter assays
Approximately 1 kb of DNA immediately upstream of the initiation codon of ULBP2 was amplified using oligonucleotides (5′-ggtaccgggcagagcaggaacaagac-3′ and 5′-ctcgagtaaggacccagagcgctaggg-3′) and cloned into the pGL3 vector (Promega). A mutant destroying the best consensus sequence for IRF binding was prepared by QuikChange PCR mutagenesis using the oligonucleotides (5′-gggcaacaagaggctttctccgtctctaa-3′ and 5′-ttagagacggagaaagcctcttgttgccc-3′). 293T cells were transfected, using the JETPEI reagent, with a mixture of the pGL3 vector containing the ULBP2 promoter (either wild-type or mutant), a vector to express the Renilla luciferase, and either empty pcDNA3 plasmid, one of four plasmids containing FLAG-tagged IRF5 variants (47) (corresponding to the Q13568, Q13568-2, Q13568-3, and Q13568-4 isoforms described in the UniProt database), a kind gift of Dr. Pat Gaffney (Oklahoma Medical Research Foundation, Oklahoma City, OK) and IRF3 or IRF7 expression vectors. After 24 h, cells were lysed and the ratio of Renilla and Firefly luciferase activities was measured by the dual luciferase assay (Promega).
Author contributions
H. T. R. conceived and coordinated the study and wrote the paper. H. T. R. and G. E. performed and analyzed the experiments. S. G. and M. V. G. provided essential reagents and technical advice and contributed to the preparation of the figures. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Profs. Rick Randall, Mariano Esteban, Pat Gaffney, Geoff Smith, and Dr. Pablo Gastaminza for kind gifts of reagents and advice.
This work was supported by Fondo de Investigación Sanitaria (FIS) Grants PI11/00298 and PS09/00181, Ministerio de Economía y Competividad (MINECO) Grants SAF2012-32293, SAF2014-54623, SAF2014-58752-R, and SAF2015-69169-R, and Comunidad de Madrid Grant S2010/BMD-2326. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S6 and Table S1.
- NK
- natural killer
- NKG2D
- natural killer group 2D
- HDAC
- histone deacetylase
- ANOVA
- analysis of variance
- RLR
- RIG-I-like receptors
- IRF
- interferon response factor
- PRR
- pattern-recognition receptor
- VV
- vaccinia virus
- qPCR
- quantitative PCR
- PIV5
- parainfluenzavirus type 5
- BCDV
- bovine viral diarrhea virus
- PAMP
- pathogen-associated molecular pattern
- araC
- arabinoside C
- WR
- Western reserve
- BHK
- baby hamster kidney
- MICA/B
- major histocompatibility complex class I chain-related A and B
- ULBP
- UL16-binding protein
- MAVS
- mitochondrial antiviral signaling.
References
- 1. Janeway C. A. Jr., and Medzhitov R. (2002) Innate immune recognition. Annu. Rev. Immunol. 20, 197–216 [DOI] [PubMed] [Google Scholar]
- 2. Fernández-Messina L., Reyburn H. T., and Valés-Gómez M. (2012) Human NKG2D-ligands: cell biology strategies to ensure immune recognition. Front. Immunol. 3, 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Van Belle T. L., and von Herrath M. G. (2009) The role of the activating receptor NKG2D in autoimmunity. Mol. Immunol. 47, 8–11 [DOI] [PubMed] [Google Scholar]
- 4. Raulet D. H., Gasser S., Gowen B. G., Deng W., and Jung H. (2013) Regulation of ligands for the NKG2D activating receptor. Annu. Rev. Immunol. 31, 413–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lam A. R., Bert N. L., Ho S. S., Shen Y. J., Tang L. F., Xiong G. M., Croxford J. L., Koo C. X., Ishii K. J., Akira S., Raulet D. H., and Gasser S. (2014) RAE1 ligands for the NKG2D receptor are regulated by STING-dependent DNA sensor pathways in lymphoma. Cancer Res. 74, 2193–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nanbakhsh A., Pochon C., Mallavialle A., Amsellem S., Bourhis J. H., and Chouaib S. (2014) c-Myc regulates expression of NKG2D ligands ULBP1/2/3 in AML and modulates their susceptibility to NK-mediated lysis. Blood 123, 3585–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nachmani D., Gutschner T., Reches A., Diederichs S., and Mandelboim O. (2014) RNA-binding proteins regulate the expression of the immune activating ligand MICB. Nat. Commun. 5, 4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gowen B. G., Chim B., Marceau C. D., Greene T. T., Burr P., Gonzalez J. R., Hesser C. R., Dietzen P. A., Russell T., Iannello A., Coscoy L., Sentman C. L., Carette J. E., Muljo S. A., and Raulet D. H. (2015) A forward genetic screen reveals novel independent regulators of ULBP1, an activating ligand for natural killer cells. Elife 4, e08474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. López-Soto A., Folgueras A. R., Seto E., and Gonzalez S. (2009) HDAC3 represses the expression of NKG2D ligands ULBPs in epithelial tumour cells: potential implications for the immunosurveillance of cancer. Oncogene 28, 2370–2382 [DOI] [PubMed] [Google Scholar]
- 10. Greene T. T., Tokuyama M., Knudsen G. M., Kunz M., Lin J., Greninger A. L., DeFilippis V. R., DeRisi J. L., Raulet D. H., and Coscoy L. (2016) A Herpesviral induction of RAE-1 NKG2D ligand expression occurs through release of HDAC mediated repression. Elife 5, E14749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chisholm S. E., and Reyburn H. T. (2006) Recognition of vaccinia virus-infected cells by human natural killer cells depends on natural cytotoxicity receptors. J. Virol. 80, 2225–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jarahian M., Fiedler M., Cohnen A., Djandji D., Hämmerling G. J., Gati C., Cerwenka A., Turner P. C., Moyer R. W., Watzl C., Hengel H., and Momburg F. (2011) Modulation of NKp30- and NKp46-mediated natural killer cell responses by poxviral hemagglutinin. PLoS Pathog. 7, e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harte M. T., Haga I. R., Maloney G., Gray P., Reading P. C., Bartlett N. W., Smith G. L., Bowie A., and O'Neill L. A. (2003) The poxvirus protein A52R targets Toll-like receptor signaling complexes to suppress host defense. J. Exp. Med. 197, 343–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stack J., Haga I. R., Schröder M., Bartlett N. W., Maloney G., Reading P. C., Fitzgerald K. A., Smith G. L., and Bowie A. G. (2005) Vaccinia virus protein A46R targets multiple Toll-like-interleukin-1 receptor adaptors and contributes to virulence. J. Exp. Med. 201, 1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chang H. W., Watson J. C., and Jacobs B. L. (1992) The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc. Natl. Acad. Sci. U.S.A. 89, 4825–4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chisholm S. E., Howard K., Gómez M. V., and Reyburn H. T. (2007) Expression of ICP0 is sufficient to trigger natural killer cell recognition of herpes simplex virus-infected cells by natural cytotoxicity receptors. J. Infect. Dis. 195, 1160–1168 [DOI] [PubMed] [Google Scholar]
- 17. Alvarez-Breckenridge C. A., Yu J., Price R., Wojton J., Pradarelli J., Mao H., Wei M., Wang Y., He S., Hardcastle J., Fernandez S. A., Kaur B., Lawler S. E., Vivier E., Mandelboim O., Moretta A., Caligiuri M. A., and Chiocca E. A. (2012) NK cells impede glioblastoma virotherapy through NKp30 and NKp46 natural cytotoxicity receptors. Nat. Med. 18, 1827–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Perdiguero B., and Esteban M. (2009) The interferon system and vaccinia virus evasion mechanisms. J. Interferon Cytokine Res. 29, 581–598 [DOI] [PubMed] [Google Scholar]
- 19. Boone R. F., Parr R. P., and Moss B. (1979) Intermolecular duplexes formed from polyadenylylated vaccinia virus RNA. J. Virol. 30, 365–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kato H., Takahasi K., and Fujita T. (2011) RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol. Rev. 243, 91–98 [DOI] [PubMed] [Google Scholar]
- 21. Guerra S., Abaitua F., Martínez-Sobrido L., Esteban M., García-Sastre A., and Rodríguez D. (2011) Host-range restriction of vaccinia virus E3L deletion mutant can be overcome in vitro, but not in vivo, by expression of the influenza virus NS1 protein. PLoS ONE 6, e28677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu R., and Moss B. (2016) Opposing roles of double-stranded RNA effector pathways and viral defense proteins revealed with CRISPR-Cas9 knockout cell lines and vaccinia virus mutants. J. Virol. 90, 7864–7879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Andrejeva J., Childs K. S., Young D. F., Carlos T. S., Stock N., Goodbourn S., and Randall R. E. (2004) The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-β promoter. Proc. Natl. Acad. Sci. U.S.A. 101, 17264–17269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Childs K., Randall R., and Goodbourn S. (2012) Paramyxovirus V proteins interact with the RNA Helicase LGP2 to inhibit RIG-I-dependent interferon induction. J. Virol. 86, 3411–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Childs K., Stock N., Ross C., Andrejeva J., Hilton L., Skinner M., Randall R., and Goodbourn S. (2007) mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology 359, 190–200 [DOI] [PubMed] [Google Scholar]
- 26. Bamming D., and Horvath C. M. (2009) Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J. Biol. Chem. 284, 9700–9712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hilton L., Moganeradj K., Zhang G., Chen Y. H., Randall R. E., McCauley J. W., and Goodbourn S. (2006) The NPro product of bovine viral diarrhea virus inhibits DNA binding by interferon regulatory factor 3 and targets it for proteasomal degradation. J. Virol. 80, 11723–11732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lopez-Pelaez M., Lamont D. J., Peggie M., Shpiro N., Gray N. S., and Cohen P. (2014) Protein kinase IKKβ-catalyzed phosphorylation of IRF5 at Ser462 induces its dimerization and nuclear translocation in myeloid cells. Proc. Natl. Acad. Sci. U.S.A. 111, 17432–17437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Unterholzner L., Sumner R. P., Baran M., Ren H., Mansur D. S., Bourke N. M., Randow F., Smith G. L., and Bowie A. G. (2011) Vaccinia virus protein C6 is a virulence factor that binds TBK-1 adaptor proteins and inhibits activation of IRF3 and IRF7. PLoS Pathog. 7, e1002247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ferguson B. J., Benfield C. T., Ren H., Lee V. H., Frazer G. L., Strnadova P., Sumner R. P., and Smith G. L. (2013) Vaccinia virus protein N2 is a nuclear IRF3 inhibitor that promotes virulence. J. Gen. Virol. 94, 2070–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bahar M. W., Graham S. C., Chen R. A., Cooray S., Smith G. L., Stuart D. I., and Grimes J. M. (2011) How vaccinia virus has evolved to subvert the host immune response. J. Struct. Biol. 175, 127–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith G. L., Benfield C. T., Maluquer de Motes C., Mazzon M., Ember S. W., Ferguson B. J., and Sumner R. P. (2013) Vaccinia virus immune evasion: mechanisms, virulence and immunogenicity. J. Gen. Virol. 94, 2367–2392 [DOI] [PubMed] [Google Scholar]
- 33. Takeuchi O., and Akira S. (2010) Pattern recognition receptors and inflammation. Cell 140, 805–820 [DOI] [PubMed] [Google Scholar]
- 34. O'Neill L. A., and Bowie A. G. (2007) The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 7, 353–364 [DOI] [PubMed] [Google Scholar]
- 35. Cham C. M., Ko K., and Niewold T. B. (2012) Interferon regulatory factor 5 in the pathogenesis of systemic lupus erythematosus. Clin. Dev. Immunol. 2012, 780436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Uccellini L., De Giorgi V., Zhao Y., Tumaini B., Erdenebileg N., Dudley M. E., Tomei S., Bedognetti D., Ascierto M. L., Liu Q., Simon R., Kottyan L., Kaufman K. M., Harley J. B., Wang E., Rosenberg S. A., and Marincola F. M. (2012) IRF5 gene polymorphisms in melanoma. J. Transl. Med. 10, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mancl M. E., Hu G., Sangster-Guity N., Olshalsky S. L., Hoops K., Fitzgerald-Bocarsly P., Pitha P. M., Pinder K., and Barnes B. J. (2005) Two discrete promoters regulate the alternatively spliced human interferon regulatory factor-5 isoforms: multiple isoforms with distinct cell type-specific expression, localization, regulation, and function. J. Biol. Chem. 280, 21078–21090 [DOI] [PubMed] [Google Scholar]
- 38. Mantovani A., Allavena P., Sica A., and Balkwill F. (2008) Cancer-related inflammation. Nature 454, 436–444 [DOI] [PubMed] [Google Scholar]
- 39. McSharry B. P., Jones C. J., Skinner J. W., Kipling D., and Wilkinson G. W. (2001) Human telomerase reverse transcriptase-immortalized MRC-5 and HCA2 human fibroblasts are fully permissive for human cytomegalovirus. J. Gen. Virol. 82, 855–863 [DOI] [PubMed] [Google Scholar]
- 40. Rothenfusser S., Goutagny N., DiPerna G., Gong M., Monks B. G., Schoenemeyer A., Yamamoto M., Akira S., and Fitzgerald K. A. (2005) The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 175, 5260–5268 [DOI] [PubMed] [Google Scholar]
- 41. Everett R. D., Young D. F., Randall R. E., and Orr A. (2008) STAT-1- and IRF-3-dependent pathways are not essential for repression of ICP0-null mutant herpes simplex virus type 1 in human fibroblasts. J. Virol. 82, 8871–8881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Didcock L., Young D. F., Goodbourn S., and Randall R. E. (1999) The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73, 9928–9933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sanjana N. E., Shalem O., and Zhang F. (2014) Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ribaudo R., Gilman M., Kingston R. E., Choczynski P., and Sacchi N. (2001) Preparation of RNA from tissues and cells. Curr. Protoc. Immunol. Chapter 10, Unit 10.11 [DOI] [PubMed] [Google Scholar]
- 45. Valés-Gómez M., Winterhalter A., Roda-Navarro P., Zimmermann A., Boyle L., Hengel H., Brooks A., and Reyburn H. T. (2006) The human cytomegalovirus glycoprotein UL16 traffics through the plasma membrane and the nuclear envelope. Cell Microbiol. 8, 581–590 [DOI] [PubMed] [Google Scholar]
- 46. Esteso G., Luzón E., Sarmiento E., Gómez-Caro R., Steinle A., Murphy G., Carbone J., Valés-Gómez M., and Reyburn H. T. (2014) Altered microRNA expression after infection with human cytomegalovirus leads to TIMP3 downregulation and increased shedding of metalloprotease substrates, including MICA. J. Immunol. 193, 1344–1352 [DOI] [PubMed] [Google Scholar]
- 47. Wen F., Ellingson S. M., Kyogoku C., Peterson E. J., and Gaffney P. M. (2011) Exon 6 variants carried on systemic lupus erythematosus (SLE) risk haplotypes modulate IRF5 function. Autoimmunity 44, 82–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.