Abstract
The biological antagonism between the signaling proteins Numb and Notch has been implicated in the regulation of many developmental processes, especially in asymmetric cell division. Mechanistic studies show that Numb inactivates Notch via endocytosis and proteasomal degradation that directly reduce Notch protein levels at the cell surface. However, some aspects of how Numb antagonizes Notch remain unclear. Here, we report a novel mechanism in which Numb acts as a Notch antagonist by controlling the intracellular destination and stability of the Notch ligand Delta-like 4 (Dll4) through a postendocytic-sorting process. We observed that Numb/Numblike knockdown increases the stability and cell-surface accumulation of Dll4. Further study indicated that Numb acts as a sorting switch to control the postendocytic trafficking of Dll4. Of note, the Numb/Numblike knockdown decreased Dll4 delivery to the lysosome, while increasing the recycling of Dll4 to the plasma membrane. Moreover, we demonstrate that this enrichment of Dll4 at the cell surface within Numb/Numblike knockdown cells could activate Notch signaling in neighboring cells. We also provide evidence that Numb negatively controls the Dll4 plasma membrane recycling through a well-documented recycling regulator protein AP1. In conclusion, our study has uncovered a molecular mechanism whereby Numb regulates the endocytic trafficking of the Notch ligand Dll4. Our findings provide a new perspective on how Numb counteracts Notch signaling and sheds additional critical insights into the antagonistic relationship between Numb and Notch signaling.
Keywords: cell signaling, endocytosis, intracellular trafficking, Notch pathway, protein degradation, Delta-like 4, Numb
Introduction
Asymmetric cell division is a fundamental means to generate cell type diversity in metazoa (1). Numb as the first identified intrinsic cue determines asymmetric cell division of Drosophila neuroblast and sensory organ precursor (SOP)4 cells (2–5). During asymmetric cell division, Numb protein is asymmetrically localized and preferentially distributed into only one of the two daughter cells, thus generating distinct progeny (3). Numb is evolutionarily conserved. There are two mammalian homologues, Numb and Numblike, and at least four major isoforms of mammalian Numb through alternative splicing (6, 7), presumably with redundant but distinct subcellular localization and functions in different physiological and pathological processes (8–11). Both Numb and Numblike are required for asymmetric cell division, although the interpretation is different due to the functional complexity of different isoforms of Numb and cell-type heterogeneity (10, 12–15).
Genetic studies in Drosophila have shown that Numb functions as a negative regulator of Notch to determine cell fate during the development of external sensory organs and specific neurons of the peripheral and central nervous system (5, 16). Within asymmetric cell division of Drosophila SOP, the cell receiving high levels of Numb suppresses Notch signaling, whereas the cell with low levels of Numb maintains Notch activity (17). Moreover, Numb and Notch double mutants exhibit only Notch mutant phenotypes, thus postulating that Numb acts by inhibiting Notch to create asymmetry (5, 17). This antagonistic relationship between Numb and Notch has also been observed in all Numb-dependent asymmetric cell divisions in Drosophila (5, 17–20). In mammalian cells, Numb homologues appear to function in a conserved fashion. During mouse neural development, mammalian Numb takes part in the asymmetric division of precursor cells and determines the distinct cell fates of daughter cells through inhibiting Notch signaling (21–23).
Although Numb is well known to be a negative regulator of Notch, the mechanism by which Numb negatively regulates Notch is not fully uncovered. In Drosophila, Sanpodo is proposed to be a key player in Numb-dependent Notch suppression during asymmetric cell division. Sanpodo encodes a four-pass transmembrane protein that functions at the cell membrane to promote Notch signaling (24, 25). Numb interferes with Sanpodo function by preventing membrane localization of Sanpodo and thereby antagonizing Notch signaling (25–27). Further studies showed that Numb controls Sanpodo endocytosis and recycling through interaction with AP2 and AP1, respectively (28, 29). However, there is no Sanpodo in mammals, implying that the molecular mechanism is not conserved between flies and mammals. Several studies suggest that mammalian Numb antagonizes Notch by promoting its ubiquitination and thereby facilitating Notch degradation. It is found that mammalian Numb interacts with Itch, one of the E3-type ubiquitin ligases of Notch1, and recruits Itch to the Notch1 receptor to promote ubiquitination of Notch1, thus leading to decreased stability of Notch1 intracellular domain and reducing Notch1 activity (30, 31).
In addition to Notch itself, the asymmetric cell division is also controlled by the Notch ligand, Delta. An E3-type ubiquitin ligase of Delta, Neuralized, is found to be unequally segregated during asymmetric cell division, thus leading to asymmetric ubiquitination and internalization of Delta (32–34). Moreover, Delta is observed to be trafficked differently in a pair of daughter cells during Drosophila SOP asymmetric division (35). These studies highlight the role of Notch ligand in creation of asymmetry, although the molecular mechanism remains uncovered. In this study, we provide evidence that mammalian Numb regulates Notch signaling by controlling postendocytic trafficking of the Notch ligand Dll4, a mammalian homologue of the Drosophila Delta, which is the major ligand in endothelium and has important functions in vascular development. Our data show that Numb acts as a sorting switch to control the postendocytic trafficking of Dll4, thus balancing the Dll4 cell-surface recycling and lysosomal degradation. By this way, Numb regulates the cell-surface amount of Dll4 thereby controlling Notch signaling. In addition, we demonstrate that Numb controls Dll4 recycling through a well-known recycling regulator protein AP1. Collectively, our study provides a novel mechanism to elucidate the antagonism between Numb and Notch signaling.
Results
Numb/Numblike knockdown increases Dll4 protein expression
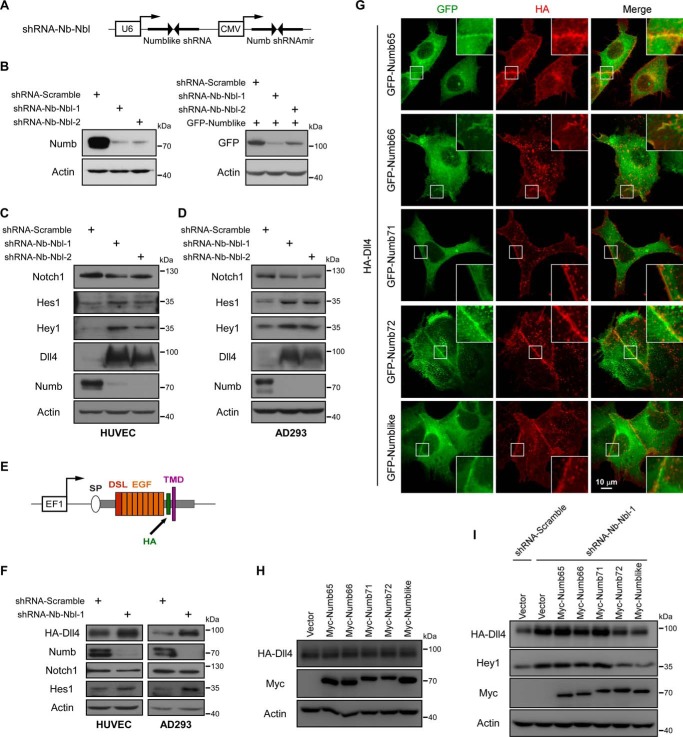
To characterize the detailed mechanisms of Numb/Numblike that control Notch signaling, we generated two short hairpin RNAs (shRNAs) to simultaneously knock down Numb and Numblike expression in human umbilical vein endothelial cells (HUVECs) (Fig. 1A). As shown, expression of either of the two shRNAs could significantly reduce the endogenous Numb expression (Fig. 1B, left panel). Because of the lack of reliable antibodies to recognize the endogenous Numblike protein, we examined the Numblike knocking down efficiency of these two shRNAs in GFP-Numblike–expressing cells. As observed, Numblike expression indicated by the GFP-Numblike fusion protein was also greatly suppressed by using these two different shRNAs (Fig. 1B, right panel). Next, the effect of Numb/Numblike knocking down on the Notch1 signaling pathway was examined by using these shRNAs. As expected, the expression of Notch1 downstream effector proteins Hes1 and Hey1 was noticeably elevated by knocking down Numb and Numblike (Fig. 1C). Surprisingly, Numb/Numblike knocking down had only marginal effect on Notch1 receptor expression but remarkably increased the expression of Dll4, a Notch1 ligand in HUVECs (Fig. 1C). These observations were also detected in AD293 cells (Fig. 1D), suggesting that this effect was not cell type–dependent. Previous studies have shown that Notch1 activation can in turn increase the Dll4 transcription as a feed-forward response (36). To address whether the increasing expression of Dll4 by knocking down of Numb/Numblike resulted from such a feed-forward regulation, we further established stable cell lines expressing HA-Dll4 in both HUVECs and AD293 cells. As the HA-Dll4 expression is driven by EF1, an exogenous promoter (graphic illustration in Fig. 1E), the Notch activation is unable to transactivate the transcription of the HA-Dll4. Thus, if the elevated Dll4 expression in Numb/Numblike knockdown cells is caused by Notch1-dependent transactivation, this effect could be abolished in these established stable cell lines. However, Numb/Numblike knockdown also induced Notch activation and remarkably increased HA-Dll4 (Fig. 1F). Of note, we also examined the Dll4 expression pattern in both Numb and Numblike single knockdown cells. Our results show that single depletion of either Numb or Numblike had no observable effect on Dll4 expression and its downstream Notch1 activation, indicating potential redundant roles of Numb and Numblike in controlling of Dll4-Notch1 signaling (supplemental Fig. S1). Based on these data, we propose a novel mechanism that Numb/Numblike can directly regulate the Notch ligand, but not only Notch itself, to control Notch signaling. Consistent with this hypothesis, we observed the co-localization of Dll4 and various isoforms of Numb at the plasma membrane in AD293 cells (Fig. 1G).
Figure 1.
Numb/Numblike knockdown increases Dll4 protein expression. A, schematic representation of Numb and Numblike double knockdown construct. From a single construct, two shRNAs (against Numb and Numblike) were expressed from the CMV and U6 promoter, respectively. B, Western blot shows transfection of the indicated constructs efficiently knocked down Numb (left panel) and Numblike (right panel) expression. C and D, Western blot analysis of expression of Notch signaling pathway components (Notch1, Hes1, Hey1, and Dll4), in HUVEC (C) and AD293 (D) cells transfected with Numb/Numblike knockdown shRNAs. E, schematic representation of HA-Dll4 expression construct driven by EF1 promoter, HA tag was inserted into Dll4 extracellular domain. SP, signal peptide; DSL, Delta/Serrate/Lag-2 domain; EGF, epidermal growth factor repeats; HA, HA tag; TMD, transmembrane domain. F, HUVEC and AD293 cell lines stably expressing HA-Dll4 (HUVEC-HA-Dll4 and AD293-HA-Dll4) were transfected with indicated shRNAs. Cells were then lysed to detect Notch signaling pathway components by Western blot analysis. G, AD293 cells co-transfected with HA-Dll4 and various GFP-tagged Numb isoforms were stained with anti-HA antibodies. The insets show enlarged views of the areas indicated by white boxes. H, AD293-HA-Dll4 cells transfected with various Myc-tagged Numb isoforms were lysed and then subjected to immunoblotting using indicated antibodies. I, control and Numb/Numblike knockdown cells (AD293-HA-Dll4) transfected with various Myc-tagged Numb isoforms were analyzed by immunoblotting using indicated antibodies.
Previous studies reported that different Numb isoforms have redundant but distinct functions in various cellular processes (8, 10, 11). Therefore, we asked whether different Numb isoforms contribute equally to Dll4 regulation. As Numb/Numblike knockdown hugely up-regulates the Dll4 expression, we expected that Numb overexpression would reversely modulate the Dll4 expression. To address this issue, various Numb isoforms were separately overexpressed within AD293 cells in which HA-Dll4 was stably expressed. To our surprise, expression of any of the Numb isoforms produced no obvious effect on HA-Dll4 expression (Fig. 1H), suggesting the complexity of the underlying mechanisms. Meanwhile, we also performed rescue experiments. As shown, re-expression of Numb72 and Numblike, but not other Numb isoforms in Numb/Numblike knockdown cells, significantly reversed the Numb/Numblike depletion-induced Dll4 up-regulation and followed Notch1 signaling activation (Fig. 1I), indicating that Numb regulates Dll4-Notch1 signaling in an isoform-specific manner.
Numb/Numblike knockdown impairs the lysosomal targeting of Dll4
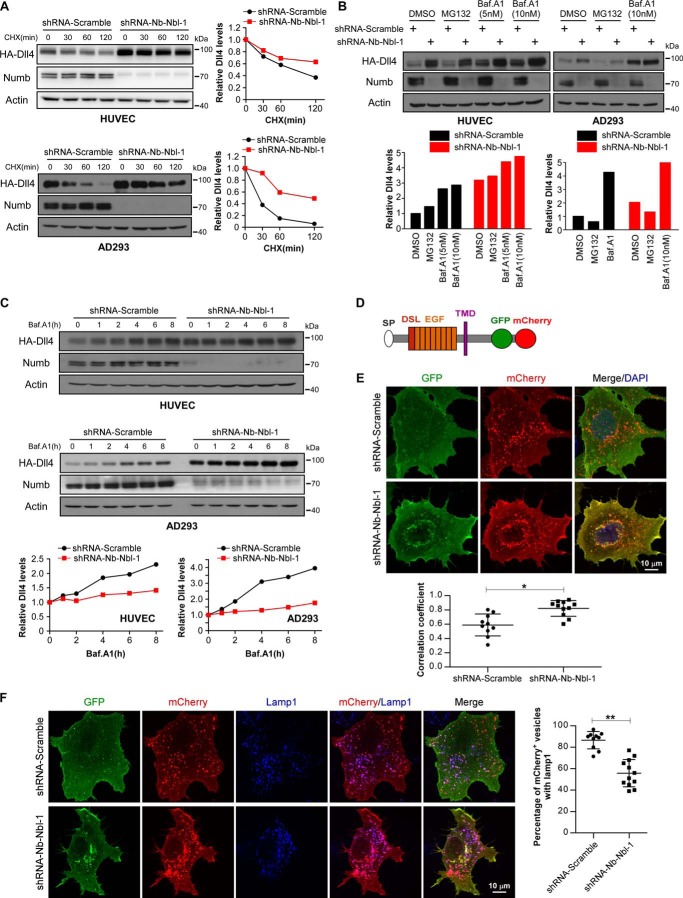
We next explored the underlying mechanism by which Numb/Numblike controls the Dll4 protein level. As Numb/Numblike knockdown strikingly increased the protein level of Dll4, we proposed that Numb/Numblike might control the Dll4 protein degradation. To address this issue, the half-life of HA-Dll4 protein was examined in both HUVECs and AD293 cells. We used cycloheximide to inhibit the translation of Dll4, and we examined the Dll4 protein level at different time points after the cycloheximide treatment. Our data showed that the half-life of the HA-Dll4 protein in Numb/Numblike knockdown cells was obviously longer than that in control cells, suggesting that the degradation rate of HA-Dll4 was reduced by Numb/Numblike knockdown (Fig. 2A). As known, mammalian proteins are degraded either through proteasome or through lysosome. Thus, we further explored which degradation pathway was responsible for the reduced turnover of Dll4 in Numb/Numblike knockdown cells. MG132 and bafilomycin A1 were used to block the proteasome and lysosome function, respectively. As shown, MG132 treatment showed no obvious effect on HA-Dll4 protein levels in both control and Numb/Numblike knockdown cells (Fig. 2B). In contrast, lysosome inhibition by bafilomycin A1 significantly increased the protein level of HA-Dll4 in both control and Numb/Numblike knockdown cells (Fig. 2B). Further quantification results show that the bafalomycin A1 treatment had a more striking effect in control cells than in Numb/Numblike knockdown cells, indicating that Numb/Numblike knockdown may partially suppress the lysosome-mediated degradation. To confirm this hypothesis, we further examined the HA-Dll4 expression in response to various times of bafalomycin A1 treatment in control and Numb/Numblike knockdown cells. As shown, the HA-Dll4 protein level in control cells increased dramatically with the increase of drug incubation time. However, the HA-Dll4 protein in Numb/Numblike knockdown cells showed a much less time-dependent response to the bafalomycin A1 treatment regardless that these cells have much more Dll4 protein (Fig. 2C). These data suggest that the Dll4 protein level in Numb/Numblike knockdown cells may have been partially saturated due to the partial inhibition of lysosome-mediated degradation. These results indicate that Numb/Numblike control the lysosome-dependent Dll4 degradation.
Figure 2.
Loss of Numb/Numblike impairs the lysosome-targeted degradation of Dll4. A, HUVEC-HA-Dll4 and AD293-HA-Dll4 cells expressing the indicated shRNAs were treated with cycloheximide (CHX, 100 μg/ml) for 0, 30, 60, and 120 min, respectively. Immunoblotting was performed as indicated. Quantification of HA-Dll4 protein levels in control and Numb/Numblike knockdown cells are shown in the right panel, and values are expressed relative to time 0 min (normalized to actin). B, expression of HA-Dll4 was analyzed by immunoblotting in control and Numb/Numblike knockdown cells (HUVEC-HA-Dll4 and AD293-HA-Dll4) treated with the indicated drugs. HA-Dll4 protein levels were quantified and shown in the bottom panel, and values are expressed relative to that in DMSO-treated control cells (normalized to actin). Baf.A1, bafilomycin A1. C, HUVEC-HA-Dll4 and AD293-HA-Dll4 cells expressing the indicated shRNAs were treated with 10 nm bafilomycin A1 for different times. Cells were harvested and subjected to immunoblotting as indicated. Quantification of HA-Dll4 levels are shown in the bottom panels, and values are expressed relative to time 0 h (normalized to actin). D, schematic representation of Dll4-GFP-mCherry expression construct, GFP and mCherry was sequentially fused to the C terminus of Dll4. E, GFP and mCherry fluorescence signals in control and Numb/Numblike knockdown AD293 cells transiently expressing Dll4-GFP-mCherry. Pearson's correlation coefficients calculated for co-localization of GFP and mCherry are plotted in the bottom panel. (n = 10 or 11 cells for each condition, means ± S.D.) *, p < 0.05. F, lamp1 immunostaining of control and Numb/Numblike knockdown AD293 cells transiently transfected with Dll4-GFP-mCherry. The percentages of mCherry+ vesicles that co-localize with lamp1 were quantified and shown in the right panel. (n = 10 or 12 cells for each condition, means ± S.D.) **, p < 0.01.
To determine whether Numb/Numblike knockdown prevents Dll4 from reaching lysosomes, we generated a tandem-tagged Dll4-GFP-mCherry probe (Fig. 2D). This probe can dissect whether a Dll4 molecule is delivered to lysosome, based on the distinct chemical properties of GFP and mCherry fluorophores. Under non-lysosomal and near-neutral pH conditions, both GFP and mCherry fluoresce. However, the low pH in the lumen of the lysosome quenches the GFP signal but not the mCherry signal. Using this approach, we found a large population of GFP−/mCherry+ dots in control cells, indicating efficient Dll4 degradation through lysosomes. In comparison, more GFP+/mCherry+ dots were observed in Numb/Numblike knockdown cells (Fig. 2E), indicating less degradation of Dll4 through lysosomes in these cells. There are two possible explanations for these results. The first is that Numb/Numblike knockdown blocks the delivery of Dll4 toward lysosomes. The second is that Numb/Numblike knockdown disrupts the lysosome function. To address these two possibilities, we then performed immunofluorescence staining to analyze the lysosome localization of Dll4-GFP-mCherry in control and Numb/Numblike knockdown cells. In control cells, the mCherry signal showed high level of co-localization with the lysosome marker protein Lamp1 (Fig. 2F, upper panels). However, Numb/Numblike knockdown drastically reduced this co-localization (Fig. 2F, lower panels). Together, these results indicate that loss of Numb/Numblike impairs the delivery of Dll4 toward lysosome for degradation.
Numb/Numblike silencing increases the cell-surface presentation of Dll4
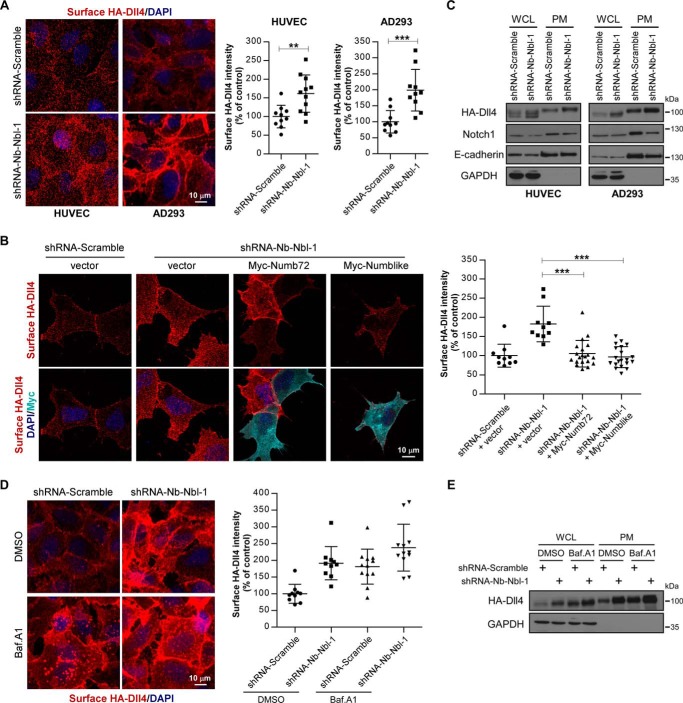
Considering the well-documented function of Numb/Numblike in endocytic trafficking, we next asked whether Numb/Numblike controls Dll4 in an endocytic manner. For this purpose, we first examined the cell-surface localization of Dll4 in control and Numb/Numblike knockdown cells. Using immunofluorescence staining, we found that Numb/Numblike knockdown clearly increased the amount of cell-surface localized HA-Dll4 in both HUVECs and AD293 cells (Fig. 3A). In addition, this phenotype in Numb/Numblike knockdown cells can be reversed by re-expression of Numb72 or Numblike (Fig. 3B). Further surface biotinylation assays showed that Numb/Numblike knockdown increased the plasma membrane accumulation of Dll4, accompanied by the decrease of Notch1 on the cell surface (Fig. 3C), suggesting that Numb/Numblike knockdown activated the Notch1 signaling by promoting the cell-surface recruitment of Dll4. We then asked whether this increased cell-surface presentation of Dll4 upon Numb/Numblike knockdown is due to the failure of Dll4 degradation through lysosomes. To address this issue, we used bafilomycin A1 to block lysosome function and then examined the cell-surface localization of HA-Dll4 in control and Numb/Numblike knockdown cells. In control cells, cell-surface localization of HA-Dll4 was strikingly increased upon bafilomycin A1 treatment (Fig. 3D). In Numb/Numblike knockdown cells, bafilomycin A1 treatment only slightly increased the cell-surface presentation of HA-Dll4 (Fig. 3D). We also exploited surface biotinylation assays to confirm this result. Consistently, either bafilomycin A1 treatment or Numb/Numblike knockdown could dramatically increase the cell-surface presentation of Dll4 (Fig. 3E). To be noted, bafilomycin A1 treatment and Numb/Numblike knockdown did not produce marked addictive effect on Dll4 cell-surface recruitment (Fig. 3, D and E). These results indicate that the excessive cell-surface accumulation of Dll4 in Numb/Numblike knockdown cells is most likely due to the failure of degradation of Dll4 via lysosomes, further confirming that Numb/Numblike plays a promoting role for Dll4 to deliver toward lysosome for degradation.
Figure 3.
Numb/Numblike depletion increases cell-surface accumulation of Dll4. A, HUVEC-HA-Dll4 and AD293-HA-Dll4 cells treated with the indicated shRNAs were fixed and stained for surface HA-Dll4. Quantification of surface HA immunofluorescence intensity are shown in the right panel. Each data point is expressed as a percentage relative to that in control cells. (n = 10 or 11 microscopic fields for each condition, means ± S.D.) **, p < 0.01; ***, p < 0.001. B, control and Numb/Numblike knockdown cells (AD293-HA-Dll4) transfected with various constructs indicated in the panel were stained for surface HA-Dll4 and Myc. Surface HA immunofluorescence intensity was quantified and shown in the right panel. Each data point is expressed as a percentage relative to that in control cells. (n = 10 or 20 cells for each condition, means ± S.D.) ***, p < 0.001. C, HUVEC-HA-Dll4 and AD293-HA-Dll4 cells treated with indicated shRNAs were biotinylated for 1 h at 4 °C. The biotinylated cell-surface molecules were precipitated and analyzed by immunoblotting as indicated. WCL, whole-cell lysate; PM, plasma membrane. D, control and Numb/Numblike knockdown cells (AD293-HA-Dll4) treated with bafilomycin A1 were stained for surface HA-Dll4. Quantification of surface HA immunofluorescence intensity are shown in the right panel. Each data point is expressed as a percentage relative to that in DMSO-treated control cells. (n = 10 or 12 microscopic fields for each condition, means ± S.D.) E, control and Numb/Numblike knockdown cells (AD293-HA-Dll4) were treated with bafilomycin A1 and subsequently biotinylated at 4 °C. The biotinylated cell-surface HA-Dll4 was precipitated and analyzed by immunoblotting.
Silencing of Numb/Numblike does not affect Dll4 endocytosis but increases Dll4 recycling
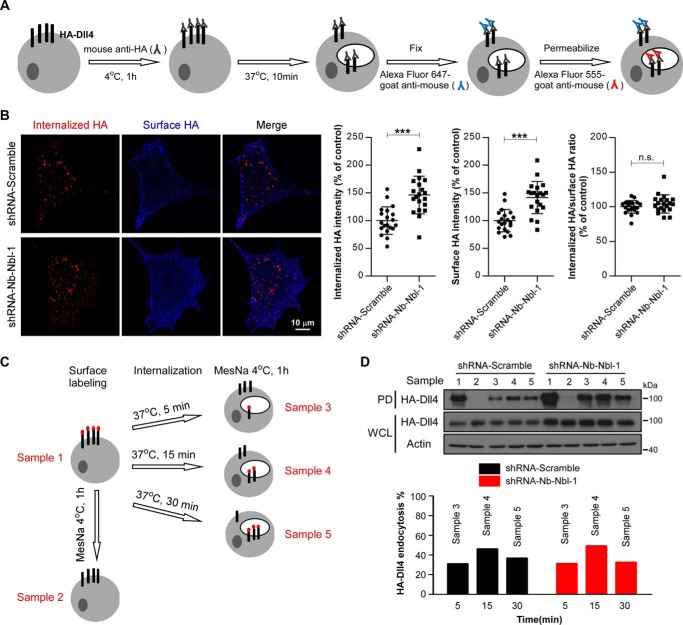
Next, we examined whether Numb/Numblike knockdown has effect on Dll4 endocytosis by using an antibody internalization assay in AD293 cells stably expressing HA-Dll4. We used anti-HA tag antibodies to label the surface HA-Dll4 on ice and allowed bound antibodies to uptake together with HA-Dll4 for 10 min at 37 °C. Then, excess amounts of secondary antibodies conjugated with Alexa Fluor 647 were added to bind the HA antibodies that remain on the cell surface. The internalized HA-Dll4 labeled by HA antibodies were finally examined by immunofluorescence staining using secondary antibodies conjugated with Alexa Fluor 555. The complete procedure of the assay is illustrated in Fig. 4A. Regardless that Numb/Numblike knockdown increased both the cell-surface and internalized Dll4, the ratio between internalized Dll4 and surface-sustained Dll4 is comparable between control and Numb/Numblike knockdown cells (Fig. 4B), suggesting that the Numb/Numblike knockdown has no effect on Dll4 endocytosis. This result was further confirmed by using biotin-labeled internalization assays. The detailed procedure is shown as a graph in Fig. 4C. Similarly, although Numb/Numblike knockdown caused a significant increase in amount of both cell-surface and internalized Dll4, the endocytic rate of Dll4 was almost the same between control and Numb/Numblike knockdown cells (Fig. 4D). Together, these results indicate that Numb/Numblike knockdown does not affect Dll4 endocytosis.
Figure 4.
Loss of Numb/Numblike has no effect on Dll4 endocytosis. A, schematic representation of antibody feeding assay for assessment of Dll4 internalization (see “Experimental procedures” for details). B, representative images showing double-label immunostaining of control and Numb/Numblike knockdown cells (AD293-HA-Dll4) for internalized HA-Dll4 and surface-remaining HA-Dll4. Quantification of the fluorescence intensity and the internalization index (fluorescence intensity of internalized HA/fluorescence intensity of surface HA) are shown in the right panel. Each data point is expressed as a percentage relative to that in control cells. (n = 20 cells for each condition, means ± S.D.) ***, p < 0.001. n.s. indicates no significance between two groups. C, schematic representation of surface biotinylation-stripping analysis to assess the rate of endocytosis (see “Experimental procedures” for details). Sample numbers 1–5 are indicated as lanes in D. D, internalization of biotinylated HA-Dll4 detected with NeutrAvidin pulldown (PD). The following lanes indicate total surface-biotinylated HA-Dll4 (lane 1), efficiency of biotin stripping (lane 2), HA-Dll4 internalization for 5 min (lane 3), 15 min (lane 4), and 30 min (lane 5). Immunoblots of whole-cell lysate (WCL) for HA-Dll4 are shown. Quantification of internalization of HA-Dll4 in control and Numb/Numblike knockdown cells (AD293-HA-Dll4) are shown in the bottom panel; values are expressed as a percentage relative to the sample 1 (normalized to actin).
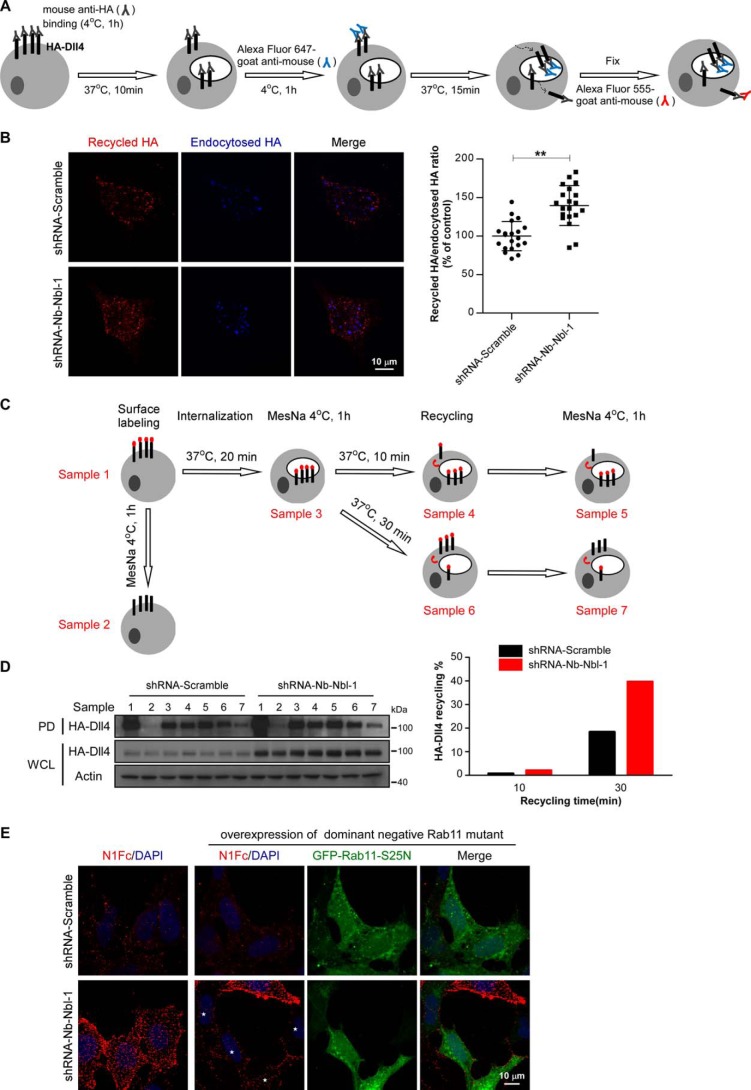
We then performed recycling assays to evaluate the potential effect of Numb/Numblike knockdown on the Dll4 recycling process. In brief, AD293 cells stably expressing HA-Dll4 were incubated with mouse anti-HA antibodies at 4 °C for 1 h to label the surface Dll4, and then the labeled Dll4 were allowed to internalize at 37 °C for 10 min. Thereafter, the non-internalized Dll4 were covered using Alexa Fluor 647-conjugated anti-mouse IgG antibodies, while the internalized Dll4 were allowed to recycle to the plasma membrane after an additional 15 min of incubation at 37 °C. Finally, the recycled Dll4 were detected using Alexa Fluor 555-conjugated anti-mouse IgG antibodies (Fig. 5A). Using this method, we examined the recycling rate of Dll4 in control and Numb/Numblike knockdown cells. As shown, we observed higher rate of Dll4 recycling in Numb/Numblike knockdown cells as compared with control cells (Fig. 5B). This result was further confirmed by using a cell-surface labeling and stripping protocol (Fig. 5C). Briefly, cell-surface Dll4 was first labeled with biotin at 4 °C. After 20 min of incubation at 37 °C, the biotin that bound with the remaining Dll4 from the cell surface was removed using MesNa. The cells were further incubated at 37 °C for 10 or 30 min for Dll4 recycling. At the end of each incubation, one-half of the cells were harvested directly, and the other half were pretreated with MesNa before harvesting. The recycle rate of Dll4 was determined by calculating the reduced amount of Dll4 biotinylation by the second MesNa treatment. In our assays, the obvious Dll4 recycling was only observed after 30 min of recycling (Fig. 5D). In control cells, ∼20% of internalized HA-Dll4 was recycled back to the cell surface after 30 min of recycling, whereas ∼40% of internalized biotinylated HA-Dll4 was detected to recycle back to the plasma membrane during the same period in Numb/Numblike knockdown cells (Fig. 5D). These results suggest that Numb/Numblike knockdown increases the Dll4 recycling. In Drosophila SOP cells, Delta is recycled through Rab11-positive endosomes (35), and then we examined whether the increased Dll4 recycling upon Numb/Numblike knockdown is also routed through Rab11. Toward this end, we expressed a dominant-negative form of Rab11, GFP-Rab11 S25N, in both control and Numb/Numblike knockdown AD293 cells. A purified Notch1 extracellular domain protein fused to the Fc region of IgG (N1Fc) can bind with the cell-surface Delta and thus was used to examine the cell-surface localized Dll4. As shown, expression of GFP-Rab11 S25N significantly abolished the increase of Dll4 recycling in Numb/Numblike knockdown cells, indicating that the increase of Dll4 recycling acts through Rab11-positive endosomes (Fig. 5E).
Figure 5.
Numb/Numblike knockdown increases Dll4 recycling. A, schematic representation of antibody feeding assay for assessment of Dll4 recycling (see “Experimental procedures” for details). B, representative images showing double-label immunostaining of control and Numb/Numblike knockdown cells (AD293-HA-Dll4) for recycled HA-Dll4 and endocytosed HA-Dll4. Quantification of the recycling index (fluorescence intensity of recycled HA/fluorescence intensity of endocytosed HA) is shown in the right panel. Each data point is expressed as a percentage relative to that in control cells. (n = 20 cells for each condition, means ± S.D.) **, p < 0.01. C, schematic representation of surface biotinylation-stripping analysis to assess the rate of recycling (see “Experimental procedures” for details). Sample numbers 1–7 are indicated as lanes in D. D, recycling of biotinylated HA-Dll4 detected with NeutrAvidin pulldown (PD). The following lanes indicate total surface biotinylated HA-Dll4 (lane 1), efficiency of biotin stripping (lane 2), HA-Dll4 internalization for 20 min (lane 3), HA-Dll4 recycling for 10 min (lane 4 versus lane 5), and HA-Dll4 recycling for 30 min (lane 6 versus lane 7). Immunoblots of whole-cell lysate (WCL) for HA-Dll4 are shown. Quantification of recycling of HA-Dll4 in control and Numb/Numblike knockdown cells (AD293-HA-Dll4) are shown in the right panel; values are expressed as a percentage relative to sample 3 (normalized to actin). E, representative images showing binding of soluble N1Fc to control and Numb/Numblike knockdown cells (AD293-HA-Dll4) transiently transfected with or without GFP-Rab11-S25N. Asterisks indicate transfection positive cells.
Taken together, our data indicate that Numb/Numblike act as a sorting switch to control the lysosomal targeting versus recycling of Dll4. In the presence of Numb/Numblike, the Dll4 prefers to go to lysosomes for degradation, thus reducing Dll4 recycling back to the cell surface and then attenuating the Dll-Notch signaling. In the absence of Numb/Numblike, Dll4 prefers to be recycled back to the plasma membrane instead of going to the lysosome for degradation, thereby promoting more recruitment of Dll4 on the plasma membrane and by this way activating more Notch receptors on the cell surface.
Numb/Numblike knockdown impairs the association of Dll4 with AP1γ1
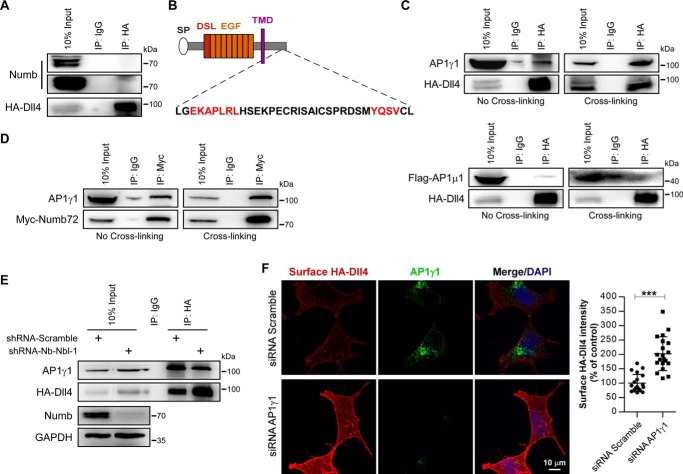
We next investigated the molecular mechanism whereby Numb controls the postendocytic sorting of Dll4 protein. We first examined the potential association between Numb and Dll4. By using co-immunoprecipitation, however, we did not find any detectable physical interaction between Dll4 and Numb (Fig. 6A). A previous study has shown that Numb can bind to adaptor protein 1γ1 subunit (AP1γ1) and acts in concert with AP1 to control the endocytic recycling of Sanpodo in Drosophila SOP cells (29). We then wondered whether Numb controls the Dll4 postendocytic trafficking by a similar mechanism. By analyzing the amino acid sequence of Dll4, two conserved AP1-binding sites were predicted based on a previous report (37). Among them, the EKAPLRL is supposed to bind with AP1γ1 subunit, and the YQSV is predicted to bind with AP1μ1 subunit (Fig. 6B). Using co-immunoprecipitation with or without cross-linking, we then proved that Dll4 can physically interact with AP1γ1 but not with AP1μ1 (Fig. 6C). Meanwhile, we also detected close association between Numb and AP1γ1 (Fig. 6D). Based on these data, we proposed that Numb might control Dll4 postendocytic trafficking through AP1γ1. Indeed, we found that silencing of Numb/Numblike clearly reduced the association between Dll4 and AP1γ1 (Fig. 6E). Consistent with this finding, siRNA-mediated knockdown of AP1γ1 also led to increased presentation of Dll4 on the cell surface (Fig. 6F), which completely resembles the phenotype of Numb/Numblike knockdown cells (Fig. 3A). Together, these data indicate that Numb negatively controls the plasma membrane recycling of Dll4 by facilitating its association with another recycling negative regulator protein AP1.
Figure 6.
Numb/Numblike knockdown impairs the interaction between Dll4 and AP1γ1. A, co-immunoprecipitation of Numb and HA-Dll4 from AD293-HA-Dll4 cell lysates. B, Dll4 contains (DE)XXXL(LI)-related motif and YXXØ motif in the cytoplasmic tail. C, AD293-HA-Dll4 cells transfected with or without FLAG-tagged AP1μ1 were treated with or without cross-linking. The lysates were then harvested and subjected to immunoprecipitation (IP) and immunoblotting as indicated. D, AD293 cells transfected with Myc-Numb72 were treated with or without cross-linking and then lysed for co-immunoprecipitation as indicated. E, control and Numb/Numblike knockdown cells (AD293-HA-Dll4) treated with cross-linking were lysed and subjected to immunoprecipitation and immunoblotting as indicated. F, AD293-HA-Dll4 cells transfected with indicated siRNAs were stained for surface HA-Dll4 and AP1γ1. Surface HA immunofluorescence intensity was quantified and shown in the right panel. Each data point is expressed as a percentage relative to that in control cells. (n = 20 cells for each condition, means ± S.D.) ***, p < 0.001.
Numb/Numblike knockdown activates Notch signaling via increasing cell-surface recruitment of Dll4
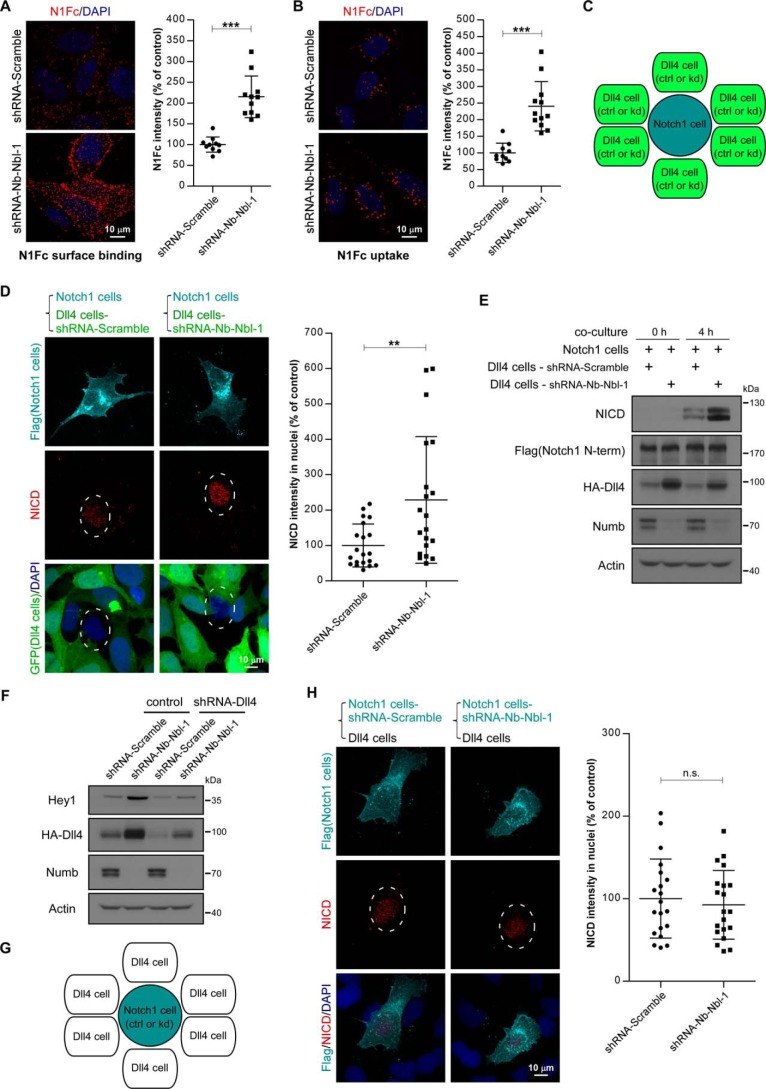
We have shown that Numb/Numblike knockdown increased the cell-surface accumulation of Dll4, and we then tested whether the cell surface–accumulated Dll4 in Numb/Numblike knockdown cells has ligand activity to promote Notch signaling. For this purpose, we first used N1Fc to test the Notch receptor binding ability of the Dll4. As shown, a significant increase of cell surface–bound N1Fc was observed in Numb/Numblike knockdown cells as compared with that in control cells (Fig. 7A). Moreover, these Dll4-bound N1Fc proteins could be internalized into cells in both control and Numb/Numblike knockdown cells (Fig. 7B). These results suggest that the cell-surface accumulated Dll4 could form a ligand–receptor complex with the Notch receptor.
Figure 7.
Numb/Numblike knockdown activates Notch signaling in neighbor cells via promoting cell-surface recruitment of Dll4. A and B, representative images showing binding (A) and uptake (B) of soluble N1Fc by AD293-HA-Dll4 cells treated with the indicated shRNAs. Quantifications of N1Fc immunofluorescence intensity are shown in A and B, respectively. Each data point is expressed as a percentage relative to that in control cells. (n = 10–12 microscopic fields for each condition, means ± S.D.) ***, p < 0.001. C, schematic representation of co-culture assay for assessment of Notch activity as shown in D and E. Notch1 cells were co-cultured with control or Numb/Numblike knockdown Dll4 cells (GFP-positive) at a ratio of 1:10. Notch1 cells refer to AD293 cells stably expressing FLAG-Notch1, and Dll4 cells refer to AD293-HA-Dll4 cells. D, representative images showing double immunostaining of co-cultures for FLAG and NICD. The nuclei of Notch1 cells are encircled by dashed line. Intensity of NICD immunofluorescence in nuclei of Notch1 cells is quantified and shown in the right panel. Each data point represents integrated intensity per cell and is expressed as a percentage relative to that in Notch1 cells co-cultured with control Dll4 cells. (n = 20 cells). **, p < 0.01. E, Notch1 cells were co-cultured with control or Numb/Numblike knockdown Dll4 cells for 4 h. Notch activation was analyzed by immunoblotting as indicated. F, AD293 cells stably expressing HA-Dll4 were transfected with indicated shRNAs. Transfection-positive cells were selected by using 1 μg/ml puromycin and subjected to immunoblotting using the indicated antibodies. G, schematic representation of co-culture assay as shown in H. Control or Numb/Numblike knockdown Notch1 cells were co-cultured with Dll4 cells. H, representative images showing double immunostaining of co-cultures for FLAG and NICD. The nuclei of Notch1 cells are encircled by a dashed line. Quantification of NICD intensity in nuclei of Notch1 cells is shown in the right panel. Each data point represents integrated intensity per cell and is expressed as a percentage relative to that in control Notch1 cells. (n = 20 cells.) n.s. indicates no significance between two groups.
Next, we performed a cell co-culture experiment to examine the Notch activation capacity of these aberrant accumulated Dll4 in Numb/Numb knockdown cells. Cells expressing FLAG-tagged Notch1 were co-cultured with control or Numb/Numblike knockdown cells that stably expressed Dll4 at a ratio of 1:10. HA-Dll4-expressing cells were labeled by GFP (graphic illustration in Fig. 7C). After 4 h of incubation, cells were immunostained with NICD antibodies to detect the Notch1 activity. As indicated, Numb/Numblike knockdown cells showed stronger ability to activate Notch signaling in their neighbor cells as compared with control cells (Fig. 7D). This observation was further confirmed using biochemical analysis (Fig. 7E). These data demonstrate that Numb/Numblike can suppress the Notch activity in neighbor cells by decreasing the cell-surface amount of Notch ligand. To further verify the significance of this novel mechanism in Numb-mediated Notch signaling regulation, we next examined to what extent the Numb knockdown-induced Notch activation is Dll4-dependent. For this purpose, we generated an shRNA construct to knock down Dll4 expression. As indicated, transfection of AD293 cells with this shRNA construct could suppress the HA-Dll4 expression efficiently in either control or Numb/Numblike knockdown cells. In the cells stably expressing HA-Dll4, Numb/Numblike knockdown significantly activated Notch signaling. However, in Dll4-depleted cells, this effect was strikingly inhibited, indicating that Notch1 signaling activation by Numb/Numblike silencing is mainly dependent on Dll4 (Fig. 7F).
Because Numb has been well documented to antagonize Notch1 signaling by directly promoting Notch1 receptor degradation (30, 31), we then also tested the requirement of Numb/Numblike in signal-receiving cells for Dll4-induced Notch activation. For this purpose, wild-type AD293 cells stably expressing HA-Dll4 were co-cultured with FLAG-Notch1-expressing cells with or without Numb/Numblike deletion (graphic illustration in Fig. 7G). After 4 h of co-culture, the Notch activity was detected by immunofluorescence staining of NICD accumulated in the nuclei. Surprisingly, we found that Notch1 activation in signal-receiving cells was almost same regardless whether Numb/Numblike was depleted or not (Fig. 7H), suggesting that the Dll4-induced Notch1 activation in signal-receiving cells is independent on Numb/Numblike. Together, our data suggest that Numb, at least in some cases, controls Notch signaling mainly through regulating its ligand but not the Notch receptor itself.
Discussion
The function of Numb as an inhibitor of Notch signaling in asymmetric cell division has been well documented (5, 6, 16, 17). However, how Numb antagonizes the Notch activity remains an open question. A long-standing model is that Numb inactivates Notch signaling by altering the Notch receptor intracellular trafficking. In Drosophila, Numb prevents Notch by inhibiting Notch receptor recycling through Sanpodo and AP1, leading to reduced plasma membrane localized Notch and thus attenuating Notch activity (29, 38). In mammalian cells, Numb was found to bind with the Notch intracellular domain and to facilitate Notch1 postendocytic trafficking toward the late endosome for degradation (31). Another link between Numb and Notch was provided by the observation that mammalian Numb interacts with the E3-type ubiquitin ligase Itch, which, in cooperation with Numb, triggers ubiquitination and degradation of the Notch intracellular domain (30). These studies suggest a Notch receptor-dependent way for Numb to antagonize Notch signaling. In this study, we provide novel evidence to show that Numb can also regulate the intracellular trafficking of Notch ligand, and in this way Numb prevents Notch signaling in a Notch ligand-dependent fashion. In the absence of Numb, Dll4, one of the Notch ligands, became highly stable and largely enriched at the cell surface, consequently leading to aberrant activation of Notch signaling. Furthermore, we show that Numb/Numblike knockdown impairs the lysosome targeting of Dll4 while facilitating its cell-surface recycling through Rab11-positive endosomes. These results provide evidence for the first time that Numb controls the postendocytic trafficking of the Notch ligand to inhibit Notch signaling in a ligand-dependent manner. Considering the existence of multiple types of Notch receptor ligands, including Delta, Serrate, and LAG-2, whether distinct types of Notch ligand are all controlled by Numb through a similar mechanism needs to be elucidated in the future.
Numb is an endocytic protein and participates in clathrin-dependent and -independent endocytic trafficking of a number of key molecules such as Notch, EGF receptor, transferrin, integrin, N-cadherin, E-cadherin, L1, etc. (23, 30, 39–43). Our previous study identified a general mechanism that Numb regulates signal sorting by acting as a docking regulator for homotypic fusion of endocytic vesicles (11). In asymmetric cell division, Numb suppresses Notch activity either by facilitating its lysosomal-dependent degradation or reducing its recycling to plasma (44). Numb also antagonizes the Notch pathway via facilitating the endocytosis of Drosophila Sanpodo, which is a membrane protein and is required for Notch activation, by interacting with a crucial recycling regulator protein AP1 (29). Here, we show that Numb controls the postendocytic sorting of the Notch ligand, Dll4, as it does for the Notch receptor. Numb knockdown significantly decreases the postendocytic trafficking of Dll4 toward the lysosome, thereby protecting Dll4 from degradation. Moreover, in the absence of Numb, Rab11-dependent recycling of Dll4 is increased, leading to a huge enrichment of Dll4 on the cell surface. Additionally, we provide evidence that Numb controls the Dll4 recycling through a well-documented recycling regulator protein AP1. These findings suggest that Numb might suppress Notch activity via regulating endocytic trafficking of either the Notch receptor or its ligand or both of them by a similar mechanism in response to different cellular signals.
Besides Notch itself, Delta is also proposed to be a key player in asymmetric cell division. During the asymmetric division of Drosophila SOP, Delta is trafficked differently in pIIa and pIIb cells. In the latter, it is delivered into Rab11-positive recycling endosomes, thus being recycled to plasma membrane. In contrast, in pIIa cells, Rab11-endosomes do not form, and thus Delta cannot be recycled back to the plasma instead of being degraded through the lysosome (35). By this mechanism, expression of Delta at the plasma membrane is skewed toward the pIIb cell, thus ensuring the asymmetric activation of Notch signaling in a pair of daughter cells. Numb, as a critical cell fate determinant, is also demonstrated to be a negative regulator of endocytic recycling. It was shown that Numb interacts physically with EHD family proteins that are involved in endocytic recycling (40, 45). This negative role of Numb on endocytic recycling is also extended to the mechanism of its counteraction over Notch. In Drosophila, Numb inhibits the recycling of Sanpodo and thus reduces the membrane-localized Notch (29, 38). In C2C12 myoblastic cells, overexpression of Numb promotes sorting of Notch1 through late endosomes for degradation, whereas deletion of Numb increases Notch1 recycling (31). In this study, we found that Numb is also a negative regulator of endocytic recycling of Delta. Numb deletion facilitates the Dll4 recycling to plasma while blocking its delivery toward the lysosome for degradation. Moreover, we provide evidence that Numb negatively controls the Dll4 plasma membrane recycling by facilitating its association with another recycling negative regulator protein AP1. These findings provide a novel mechanism that Numb, as an endocytic regulator, can also antagonize Notch via controlling intracellular trafficking of Notch ligand.
Taken together, our study uncovers a novel function of Numb in the regulation of Notch ligand endocytic trafficking, which provides a new perspective to elucidate the role of Numb in counteraction of Notch signaling. Because Numb can regulate the intracellular trafficking of both Notch receptor and its ligand, it is interesting to investigate how the two ways connect with each other to antagonize Notch signaling in the future.
Experimental procedures
Antibodies, reagents, and constructs
The antibodies used for immunoblotting and immunofluorescence were as follows: rabbit anti-Numb (Cell Signaling, 2756); rabbit anti-Notch1 (Cell Signaling, 3608); goat anti-Dll4 (R&D Systems, AF1389); rabbit anti-Hes1 (Santa Cruz Biotechnology, sc-25392); rabbit anti-Hey1 (ProteinTech, 19929-1-AP); rabbit anti-cleaved Notch1 (NICD, Cell Signaling, 4147); mouse anti-AP1γ1 (Sigma, A4200); mouse anti-HA (Covance, MMS-101P); rabbit anti-HA (Cell Signaling, 3724); mouse anti-Myc (Covance, MMS-150P); mouse anti-FLAG (PROSPEC, ANT-146); rabbit anti-E-cadherin (Cell Signaling, 4065); mouse anti-GFP (NeuroMab, 75-132); mouse anti-GAPDH (ProteinTech, 60004-1-Ig); mouse anti-β-actin (Santa Cruz Biotechnology, sc-47778); and rabbit anti-Lamp1 (Sigma, L1418). Peroxidase-conjugated donkey anti-goat (705-035-003), donkey anti-mouse (715-035-150), and goat anti-rabbit IgG (111-035-003) were from Jackson ImmunoResearch. Alexa Fluor 555- or Alexa Fluor 647-conjugated secondary antibodies to rabbit or mouse IgG (A21429, A21424, A21245, and A21236) were from Invitrogen. Reagents used were cycloheximide (Sigma), MG132 (Sigma), and bafilomycin A1 (Merck Millipore).
The coding region of human Dll4 was amplified from HUVEC cDNA by standard PCR and cloned into the pcDNA3.1 vector with an HA tag or a GFP-mCherry double tag. The resulting plasmids (HA-Dll4 and Dll4-GFP-mCherry) encode full-length human Dll4 inserted with four tandem HA epitopes between Pro-528 and Trp-529 of Dll4, or fused with GFP-mCherry at the C terminus. The coding region of human Notch1 was amplified from HUVEC cDNA and cloned into pcDNA3.1 vector with a FLAG tag. The resulting construct (FLAG-Notch1) encodes full-length human Notch1 inserted with four tandem FLAG epitopes immediately downstream of the signal peptide. HA-Dll4 and FLAG-Notch1 were subcloned into CD532A (replacing puromycin resistance gene with blasticidin resistance gene; System Biosciences) lentivirus vector. The plasmids encoding full-length mouse Numb and Numblike with a GFP or Myc tag have been previously described (11). The GFP-Rab11-S25N and FLAG-AP1μ1 expression constructs were purchased from Addgene and OriGene, respectively. pGIPZ-lentiviral shRNAmir vectors targeting human Numb (RHS4430-98843207; RHS4430-99160958) and pLKO.1-lentiviral shRNA vectors targeting human Numblike (RHS3979-97052327; RHS3979-97052329) were purchased from Open Biosystems. To generate Numb and Numblike double knockdown construct, the U6 promoter-driven shRNA expression cassette from pLKO.1 vector was cloned into pGIPZ vector in the XbaI site immediately upstream of the CMV promoter. To knock down human Dll4, pLKO.1-shRNA-Dll4 was constructed. The targeted sequence is as follows: CTACTATGGAGACAACTGCTC. All vectors were confirmed by DNA sequencing. The double-stranded siRNA targeting AP1γ1 was synthesized by RiboBio. The sequence is as follows: ACCGAAUUAAGAAAGUGGU.
Cell culture and transfection
HUVEC line was purchased from ATCC, and AD293 cell line was a kind gift from Prof. Xianming Mo (Sichuan University, China). Both cell lines were maintained in DMEM (Invitrogen) supplemented with 10% FBS (HyClone). Transfection of specific plasmids was performed using MegaTran 1.0 (OriGene) according to the manufacturer's instructions. siRNA was transfected using Lipofectamine 2000. Transfected cells were collected and analyzed by Western blotting or immunostaining.
Lentiviral transduction and generation of stable cell lines
The recombinant lentiviruses expressing HA-Dll4, FLAG-Notch1, shRNA-Nb-Nbl-1, or shRNA-Nb-Nbl-2 were produced by transfecting one of the expression plasmids together with helper plasmids (psPAX2 and pMD2.G) into HEK293T cells. Viral supernatants were collected 48 h post-transfection and were used to infect cells.
To generate stable HUVEC and AD293 cell lines expressing HA-Dll4 or FLAG-Notch1, cells were infected for 12 h with corresponding lentiviral supernatants. After 48 h of infection, cells were subjected to blasticidin (10 μg/ml, Sigma) selection for 2 weeks, and then at least five cell clones were isolated for further study. To generate Numb and Numblike double knockdown stable cell lines, cells were infected with lentivirus and selected by puromycin (1 μg/ml, Sigma) for 2 weeks. The stable cell pools were subsequently used for further analysis.
Immunostaining, immunoblotting, and biotinylation
Immunostaining was performed as described previously (11, 46). In brief, cells seeded on coverslips were fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.2% Triton X-100, and blocked in blocking buffer (PBS, 3% BSA, 2% goat serum) for 30 min. After blocking, the cells were incubated with the indicated primary antibodies for 2 h at room temperature, followed by incubation with fluorescence-labeled secondary antibodies for 1 h at room temperature. For surface staining of HA-Dll4, cells were fixed and stained with anti-HA antibodies under non-permeabilizing conditions. Images were acquired by using a Leica SD AF confocal system with a ×63 (NA 1.4) objective. Fluorescence intensity quantification and co-localization analysis were performed using Imaris software (Bitplane). All compared images were acquired under identical parameters.
Immunoblotting was performed as follows. Cells were lysed with RIPA buffer supplemented with a protease and phosphatase inhibitor mixture (Pierce). After determination of protein concentration, equal amounts of lysate were loaded and separated by SDS-PAGE. Proteins on the gels were transferred to PVDF membranes and detected with antibodies indicated in the specific experiments. Immunoblots were visualized with ECL reagent (Millipore) and quantitated using ImageJ software.
Biotinylation analysis of cell-surface molecules was performed using the Cell Surface Protein Isolation Kit (Pierce, 89881). Briefly, the cells were incubated in PBS containing 0.5 mg/ml Sulfo-NHS-Biotin for 1 h at 4 °C and lysed with RIPA buffer plus protease inhibitors. The biotinylated cell-surface molecules were precipitated with NeutrAvidin-agarose resin and analyzed by immunoblotting.
Formaldehyde cross-linking and immunoprecipitation
To obtain formaldehyde solution, 2% paraformaldehyde (Sigma) was dissolved in PBS for 2 h at ∼80 °C, and then filtered through 0.45-μm PVDF membrane (Millipore). The solution was stored in the dark at 4 °C.
For formaldehyde cross-linking, as described previously (47), cells were washed and collected, then suspended in formaldehyde solution, and incubated with agitation for 10 min at room temperature. The reaction was quenched with 1.25 m glycine in PBS. After that, the supernatant was discarded, and cells were collected for further immunoprecipitation experiments.
For immunoprecipitation, cells were lysed with IP lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40) plus protease inhibitors, and equal amounts of total protein were incubated with specific antibodies overnight at 4 °C with rotation. Protein A/G-agarose beads (Santa Cruz Biotechnology) were then added for another 2 h. Then the beads were washed and re-suspended in 2× Laemmli Buffer, followed by heating for 5 min at 95 °C. Samples were analyzed by SDS-PAGE and immunoblotting.
Internalization assay
Antibody feeding-based internalization assay was modified from previously reported methods (39, 48). Cells expressing HA-Dll4, seeded on coverslips, were incubated with mouse anti-HA antibodies for 1 h at 4 °C. Following washing twice to remove the unbound antibodies, the cells were further incubated at 37 °C for 10 min. The cells were then fixed immediately, and surface-remaining antibody-labeled HA-Dll4 was saturated by incubation with Alexa Fluor 647-conjugated secondary antibodies. The cells were then permeabilized with 0.2% Triton X-100, and internalized antibody-labeled HA-Dll4 was stained with Alexa Fluor 555-conjugated secondary antibodies. Cells were imaged using a Leica SD AF confocal system as described above. The total fluorescence intensity per cell was measured using Imaris software.
Biotinylation-based internalization assay was performed as described previously (49). Briefly, cells were labeled with Sulfo-NHS-Biotin as described above and incubated at 37 °C for various times for initiation of internalization. After incubation, the remaining biotinylated surface proteins were de-biotinylated by washing cells with MesNa stripping buffer for 1 h at 4 °C. As shown in Fig. 4C, samples 1–5, were collected to determine the level of surface biotinylation (sample 1), efficiency of stripping (sample 2), and level of internalization for various times (samples 3–5). Cells were lysed with RIPA buffer supplemented with protease inhibitors. Internalized biotinylated HA-Dll4 was precipitated with NeutrAvidin-agarose resin and detected by immunoblotting. The level of endocytosis of HA-Dll4 (samples 3–5) was determined as the % relative to total cell-surface biotinylation (sample 1), and all samples were normalized to the pixel densities of actin.
Recycling assay
Antibody feeding-based recycling assay was modified from a previously reported method (29). Cells were incubated with mouse anti-HA antibodies at 4 °C and then brought to 37 °C to allow for internalization of antibody-bound HA-Dll4, as described above. Then cells were incubated with an excess amount of Alexa Fluor 647-conjugated anti-mouse IgG antibodies at 4 °C for 1 h to block the antibodies remaining at the cell surface. Cells were next incubated at 37 °C for 15 min to allow HA-Dll4 recycling back to the cell surface. Subsequently, cells were fixed and stained with Alexa Fluor 555-conjugated anti-mouse IgG antibodies under non-permeabilizing conditions. Images were acquired, and fluorescence intensity was quantified as described above.
For the biotinylation-based recycling assay, surface-labeled HA-Dll4 was allowed to be internalized for 20 min at 37 °C followed by a first stripping of surface label with MeSNa buffer as described for the internalization assay. Cells were then washed and returned to 37 °C for the indicated time points to chase internalized HA-Dll4 back to the cell surface. Cells were then subjected to a second round of stripping to remove the biotin from recycled HA-Dll4. As shown in Fig. 5C, samples 1–7 were collected to determine the level of surface biotinylation (sample 1), efficiency of stripping (sample 2), level of internalization for 20 min (sample 3), level of recycled HA-Dll4 for 10 min (sample 4 versus sample 5), and level of recycled HA-Dll4 for 30 min (sample 6 versus sample 7). As described above, cells were lysed with RIPA buffer, and biotin-labeled HA-Dll4 was analyzed by immunoblotting. The level of recycling was determined as the % difference between sample 4 and sample 5 (10 min) and between sample 6 and sample 7 (30 min), respectively, relative to total internalized HA-Dll4 (sample 3). All samples were normalized to the pixel densities of actin.
N1Fc binding and uptake
Soluble N1Fc binding and uptake assays were performed as described previously (50) with some modifications. N1Fc (1 μg/ml; R&D Systems) dissolved in Hanks' balanced salt solution was pre-clustered for 6 h at 4 °C with unconjugated rabbit anti-mouse Fc (1:2000; Jackson ImmunoResearch) to generate N1Fc-binding buffer. For N1Fc binding, cells expressing HA-Dll4 were plated on coverslips and incubated for 1 h at 4 °C with N1Fc-binding buffer. After washing with cold PBS, cells were fixed and stained with Alexa Fluor 555-conjugated anti-rabbit IgG antibodies. For N1Fc uptake, cells were incubated with N1Fc-binding buffer at 4 °C as described above, and then shifted to 37 °C for 20 min to allow for the uptake of pre-clustered N1Fc. Subsequently, the cells were fixed and incubated with an excess amount of unconjugated anti-rabbit IgG antibodies to block the N1Fc remaining on the cell surface. The cells were permeabilized with 0.2% Triton X-100. Internalized N1Fc was then visualized with Alexa Fluor 555-conjugated anti-rabbit IgG antibodies. Images were acquired, and fluorescence intensity was quantified as described above.
Co-culture assay
AD293 cells stably expressing Dll4 (AD293-HA-Dll4) were seeded at a density of 1 × 105 cells/well on collagen-coated coverslips in 24-well plates and cultured overnight. The next day, AD293 cells stably expressing Notch1 (AD293-FLAG-Notch1) were seeded into these wells at a density of 1 × 104 cells/well. Cell co-culture was continued for 4 h. Then co-cultures were fixed and stained with anti-FLAG and anti-NICD antibodies. Images were acquired, and fluorescence intensity was quantified as described above. The co-cultures described above were also lysed for immunoblotting analysis.
Statistical analysis
Statistical analysis was performed using Student's t test. Results shown are the means ± S.D. from at least three independent experiments. p < 0.05 was considered statistically significant.
Author contributions
H. C. L. and H. S. L. conceived, designed, and supervised the project. X. S. and Z. D. designed and performed most biochemical and cell biological experiments. M. Z., K. L., H. S., and X. L. participated in cell culture, Western blotting, and immunofluorescence staining. J. C. generated vector constructions. Y. Z. and Y. H. provided materials. X. S., Z. D., H. S. L., and H. C. L. discussed the data. X. S., Z. D., and H. C. L. prepared the manuscript.
Supplementary Material
This work was supported by Grant 2014CB964602 from the Ministry of Science and Technology of the People′s Republic of China, Grants 31601174, 31671397, and 81702952 from the National Natural Science Foundation of China, Grant 2016A030312006 from Natural Science Foundation of Guangdong Province, and Grant JCYJ20160229204338907 from Shenzhen Science and Technology Program. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Fig. S1.
- SOP
- sensory organ precursor
- HUVEC
- human umbilical vein endothelial cell.
References
- 1. Knoblich J. A. (2008) Mechanisms of asymmetric stem cell division. Cell 132, 583–597 [DOI] [PubMed] [Google Scholar]
- 2. Uemura T., Shepherd S., Ackerman L., Jan L. Y., and Jan Y. N. (1989) numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell 58, 349–360 [DOI] [PubMed] [Google Scholar]
- 3. Knoblich J. A., Jan L. Y., and Jan Y. N. (1995) Asymmetric segregation of Numb and Prospero during cell division. Nature 377, 624–627 [DOI] [PubMed] [Google Scholar]
- 4. Rhyu M. S., Jan L. Y., and Jan Y. N. (1994) Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 76, 477–491 [DOI] [PubMed] [Google Scholar]
- 5. Spana E. P., and Doe C. Q. (1996) Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron 17, 21–26 [DOI] [PubMed] [Google Scholar]
- 6. Gulino A., Di Marcotullio L., and Screpanti I. (2010) The multiple functions of Numb. Exp. Cell Res. 316, 900–906 [DOI] [PubMed] [Google Scholar]
- 7. Dho S. E., French M. B., Woods S. A., and McGlade C. J. (1999) Characterization of four mammalian numb protein isoforms. Identification of cytoplasmic and membrane-associated variants of the phosphotyrosine binding domain. J. Biol. Chem. 274, 33097–33104 [DOI] [PubMed] [Google Scholar]
- 8. Verdi J. M., Bashirullah A., Goldhawk D. E., Kubu C. J., Jamali M., Meakin S. O., and Lipshitz H. D. (1999) Distinct human NUMB isoforms regulate differentiation vs. proliferation in the neuronal lineage. Proc. Natl. Acad. Sci. U.S.A. 96, 10472–10476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pece S., Confalonieri S., R Romano P., and Di Fiore P. P. (2011) NUMB-ing down cancer by more than just a NOTCH. Biochim. Biophys. Acta 1815, 26–43 [DOI] [PubMed] [Google Scholar]
- 10. Kyriazis G. A., Wei Z., Vandermey M., Jo D. G., Xin O., Mattson M. P., and Chan S. L. (2008) Numb endocytic adapter proteins regulate the transport and processing of the amyloid precursor protein in an isoform-dependent manner: implications for Alzheimer disease pathogenesis. J. Biol. Chem. 283, 25492–25502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shao X., Liu Y., Yu Q., Ding Z., Qian W., Zhang L., Zhang J., Jiang N., Gui L., Xu Z., Hong Y., Ma Y., Wei Y., Liu X., Jiang C., Zhu M., Li H., and Li H. (2016) Numb regulates vesicular docking for homotypic fusion of early endosomes via membrane recruitment of Mon1b. Cell Res. 26, 593–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Petersen P. H., Zou K., Hwang J. K., Jan Y. N., and Zhong W. (2002) Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature 419, 929–934 [DOI] [PubMed] [Google Scholar]
- 13. Petersen P. H., Zou K., Krauss S., and Zhong W. (2004) Continuing role for mouse Numb and Numbl in maintaining progenitor cells during cortical neurogenesis. Nat. Neurosci. 7, 803–811 [DOI] [PubMed] [Google Scholar]
- 14. Yan B. (2010) Numb–from flies to humans. Brain Dev. 32, 293–298 [DOI] [PubMed] [Google Scholar]
- 15. Bani-Yaghoub M., Kubu C. J., Cowling R., Rochira J., Nikopoulos G. N., Bellum S., and Verdi J. M. (2007) A switch in numb isoforms is a critical step in cortical development. Dev. Dyn. 236, 696–705 [DOI] [PubMed] [Google Scholar]
- 16. Frise E., Knoblich J. A., Younger-Shepherd S., Jan L. Y., and Jan Y. N. (1996) The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interaction in sensory organ lineage. Proc. Natl. Acad. Sci. U.S.A. 93, 11925–11932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guo M., Jan L. Y., and Jan Y. N. (1996) Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron 17, 27–41 [DOI] [PubMed] [Google Scholar]
- 18. Buescher M., Yeo S. L., Udolph G., Zavortink M., Yang X., Tear G., and Chia W. (1998) Binary sibling neuronal cell fate decisions in the Drosophila embryonic central nervous system are nonstochastic and require inscuteable-mediated asymmetry of ganglion mother cells. Genes Dev. 12, 1858–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lear B. C., Skeath J. B., and Patel N. H. (1999) Neural cell fate in rca1 and cycA mutants: the roles of intrinsic and extrinsic factors in asymmetric division in the Drosophila central nervous system. Mech. Dev. 88, 207–219 [DOI] [PubMed] [Google Scholar]
- 20. Ruiz Gómez M., and Bate M. (1997) Segregation of myogenic lineages in Drosophila requires numb. Development 124, 4857–4866 [DOI] [PubMed] [Google Scholar]
- 21. Shen Q., Zhong W., Jan Y. N., and Temple S. (2002) Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development 129, 4843–4853 [DOI] [PubMed] [Google Scholar]
- 22. Li H. S., Wang D., Shen Q., Schonemann M. D., Gorski J. A., Jones K. R., Temple S., Jan L. Y., and Jan Y. N. (2003) Inactivation of Numb and Numblike in embryonic dorsal forebrain impairs neurogenesis and disrupts cortical morphogenesis. Neuron 40, 1105–1118 [DOI] [PubMed] [Google Scholar]
- 23. Rasin M. R., Gazula V. R., Breunig J. J., Kwan K. Y., Johnson M. B., Liu-Chen S., Li H. S., Jan L. Y., Jan Y. N., Rakic P., and Sestan N. (2007) Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat. Neurosci. 10, 819–827 [DOI] [PubMed] [Google Scholar]
- 24. Babaoglan A. B., O'Connor-Giles K. M., Mistry H., Schickedanz A., Wilson B. A., and Skeath J. B. (2009) Sanpodo: a context-dependent activator and inhibitor of Notch signaling during asymmetric divisions. Development 136, 4089–4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. O'Connor-Giles K. M., and Skeath J. B. (2003) Numb inhibits membrane localization of Sanpodo, a four-pass transmembrane protein, to promote asymmetric divisions in Drosophila. Dev. Cell 5, 231–243 [DOI] [PubMed] [Google Scholar]
- 26. Skeath J. B., and Doe C. Q. (1998) Sanpodo and Notch act in opposition to Numb to distinguish sibling neuron fates in the Drosophila CNS. Development 125, 1857–1865 [DOI] [PubMed] [Google Scholar]
- 27. Cayouette M., and Raff M. (2002) Asymmetric segregation of Numb: a mechanism for neural specification from Drosophila to mammals. Nat. Neurosci. 5, 1265–1269 [DOI] [PubMed] [Google Scholar]
- 28. Hutterer A., and Knoblich J. A. (2005) Numb and α-adaptin regulate Sanpodo endocytosis to specify cell fate in Drosophila external sensory organs. EMBO Rep. 6, 836–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cotton M., Benhra N., and Le Borgne R. (2013) Numb inhibits the recycling of Sanpodo in Drosophila sensory organ precursor. Curr. Biol. 23, 581–587 [DOI] [PubMed] [Google Scholar]
- 30. McGill M. A., and McGlade C. J. (2003) Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J. Biol. Chem. 278, 23196–23203 [DOI] [PubMed] [Google Scholar]
- 31. McGill M. A., Dho S. E., Weinmaster G., and McGlade C. J. (2009) Numb regulates post-endocytic trafficking and degradation of Notch1. J. Biol. Chem. 284, 26427–26438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pavlopoulos E., Pitsouli C., Klueg K. M., Muskavitch M. A., Moschonas N. K., and Delidakis C. (2001) Neuralized encodes a peripheral membrane protein involved in delta signaling and endocytosis. Dev. Cell 1, 807–816 [DOI] [PubMed] [Google Scholar]
- 33. Lai E. C., Deblandre G. A., Kintner C., and Rubin G. M. (2001) Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of Delta. Dev. Cell 1, 783–794 [DOI] [PubMed] [Google Scholar]
- 34. Le Borgne R., and Schweisguth F. (2003) Unequal segregation of neuralized biases Notch activation during asymmetric cell division. Dev. Cell 5, 139–148 [DOI] [PubMed] [Google Scholar]
- 35. Emery G., Hutterer A., Berdnik D., Mayer B., Wirtz-Peitz F., Gaitan M. G., and Knoblich J. A. (2005) Asymmetric Rab11 endosomes regulate delta recycling and specify cell fate in the Drosophila nervous system. Cell 122, 763–773 [DOI] [PubMed] [Google Scholar]
- 36. Caolo V., van den Akker N. M., Verbruggen S., Donners M. M., Swennen G., Schulten H., Waltenberger J., Post M. J., and Molin D. G. (2010) Feed-forward signaling by membrane-bound ligand receptor circuit the case of notch Delta-like 4 ligand in endothelial cells. J. Biol. Chem. 285, 40681–40689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Park S. Y., and Guo X. (2014) Adaptor protein complexes and intracellular transport. Biosci. Rep. 34, e00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Couturier L., Mazouni K., and Schweisguth F. (2013) Numb localizes at endosomes and controls the endosomal sorting of Notch after asymmetric division in Drosophila. Curr. Biol. 23, 588–593 [DOI] [PubMed] [Google Scholar]
- 39. Nishimura T., and Kaibuchi K. (2007) Numb controls integrin endocytosis for directional cell migration with aPKC and PAR-3. Dev. Cell 13, 15–28 [DOI] [PubMed] [Google Scholar]
- 40. Santolini E., Puri C., Salcini A. E., Gagliani M. C., Pelicci P. G., Tacchetti C., and Di Fiore P. P. (2000) Numb is an endocytic protein. J. Cell Biol. 151, 1345–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nishimura T., Fukata Y., Kato K., Yamaguchi T., Matsuura Y., Kamiguchi H., and Kaibuchi K. (2003) CRMP-2 regulates polarized Numb-mediated endocytosis for axon growth. Nat. Cell Biol. 5, 819–826 [DOI] [PubMed] [Google Scholar]
- 42. Sorensen E. B., and Conner S. D. (2008) AAK1 regulates Numb function at an early step in clathrin-mediated endocytosis. Traffic 9, 1791–1800 [DOI] [PubMed] [Google Scholar]
- 43. Zhao C., Guo H., Li J., Myint T., Pittman W., Yang L., Zhong W., Schwartz R. J., Schwarz J. J., Singer H. A., Tallquist M. D., and Wu M. (2014) Numb family proteins are essential for cardiac morphogenesis and progenitor differentiation. Development 141, 281–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Johnson S. A., Zitserman D., and Roegiers F. (2016) Numb regulates the balance between Notch recycling and late-endosome targeting in Drosophila neural progenitor cells. Mol. Biol. Cell 27, 2857–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smith C. A., Dho S. E., Donaldson J., Tepass U., and McGlade C. J. (2004) The cell fate determinant numb interacts with EHD/Rme-1 family proteins and has a role in endocytic recycling. Mol. Biol. Cell 15, 3698–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee S. H., Simonetta A., and Sheng M. (2004) Subunit rules governing the sorting of internalized AMPA receptors in hippocampal neurons. Neuron 43, 221–236 [DOI] [PubMed] [Google Scholar]
- 47. Klockenbusch C., and Kast J. (2010) Optimization of formaldehyde cross-linking for protein interaction analysis of non-tagged integrin β1. J. Biomed. Biotechnol. 2010, 927585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li Z., Jo J., Jia J. M., Lo S. C., Whitcomb D. J., Jiao S., Cho K., and Sheng M. (2010) Caspase-3 activation via mitochondria is required for long-term depression and AMPA receptor internalization. Cell 141, 859–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shergill B., Meloty-Kapella L., Musse A. A., Weinmaster G., and Botvinick E. (2012) Optical tweezers studies on Notch: single-molecule interaction strength is independent of ligand endocytosis. Dev. Cell 22, 1313–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Meloty-Kapella L., Shergill B., Kuon J., Botvinick E., and Weinmaster G. (2012) Notch ligand endocytosis generates mechanical pulling force dependent on dynamin, epsins, and actin. Dev. Cell 22, 1299–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.