Abstract
Sporadic avian to human transmission of highly pathogenic avian influenza (HPAI) A(H5N1) viruses necessitates the analysis of currently circulating and evolving clades to assess their potential risk. Following the spread and sustained circulation of clade 2 viruses across multiple continents, numerous subclades and genotypes have been described. To better understand the pathogenesis associated with the continued diversification of clade 2 A(H5N1) influenza viruses, we investigated the relative virulence of eleven human and poultry isolates collected from 2006 to 2013 by determining their ability to cause disease in the ferret model. Numerous clade 2 viruses, including a clade 2.2 avian isolate, a 2.2.2.1 human isolate, and two 2.2.1 human isolates, were found to be of low virulence in the ferret model, though lethality was detected following infection with one 2.2.1 human isolate. In contrast, three of six clade 2.3.2.1 avian isolates tested led to severe disease and death among infected ferrets. Clade 2.3.2.1b and 2.3.2.1c isolates, but not 2.3.2.1a isolates, were associated with ferret lethality. All A(H5N1) viruses replicated efficiently in the respiratory tract of ferrets regardless of their virulence and lethality. However, lethal isolates were characterized by systemic viral dissemination, including detection in the brain and enhanced histopathology in lung tissues. The finding of disparate virulence phenotypes between clade 2 A(H5N1) viruses, notably differences between subclades of 2.3.2.1 viruses, suggests there are distinct molecular determinants present within the established subclades, the identification of which will assist in molecular-based surveillance and public health efforts against A(H5N1) viruses.
Keywords: Pathogenesis, Ferrets, Influenza H5N1
1. Introduction
Since the re-emergence of A(H5N1) highly pathogenic avian influenza (HPAI) viruses in 2003, this subtype has spread and become endemic in bird populations in over 70 countries of Asia, Europe, the Middle East and Africa. Active A(H5N1) virus outbreaks in poultry continue to occur in Bangladesh, Cambodia, China, Egypt, India, Indonesia, Republic of Korea, Vietnam, and several countries in West Africa (WHO, 2016a). To date, avian-to-human transmission of A(H5N1) viruses has resulted in > 850 human cases, causing death in > 450 of them; the majority of human cases and fatalities have been reported in Egypt followed by Indonesia and Vietnam (WHO, 2016b). Human infection has primarily occurred via close contact with infected poultry although limited human-to-human transmission has been reported (Abdel-Ghafar et al., 2008). The continued evolution of A(H5N1) viruses poses a direct threat to public health should mutation allow for increased zoonotic transmission events or sustained human-to-human transmission (Rejmanek et al., 2015).
Established surveillance and analysis of currently circulating viruses in poultry have continually tracked and identified genetically and antigenically distinct A(H5N1) lineages (Creanga et al., 2013; Gerloff et al., 2014; Thor et al., 2015). While genetic analysis of the hemagglutinin (HA) gene of these lineages has resulted in the classification of isolates into multiple clades and subclades, only viruses from clades 0, 1, 2, and 7 have crossed the species barrier to cause disease and death in humans (Abdel-Ghafar et al., 2008). The persistent geographic spread and genetic evolution of select clades of A(H5N1) viruses, specifically those within the 2.2 and 2.3 clusters, have required further classification into third and fourth-tiered clades (Creanga et al., 2013; WHO/OIE/FAO, 2014; Younan et al., 2013). In Egypt, only clade 2.2 viruses have been found to circulate in poultry with all confirmed human infections between 2009 and 2016 caused by 2.2.1 and 2.2.1.2 viruses (Refaey et al., 2015; Younan et al., 2013). In Vietnam, based on the available surveillance data, clade 1 viruses were gradually replaced by clades 2.3.2 and 2.3.4 viruses beginning as early as 2005. Clade 2.3.2.1c and clade 2.3.4.4 (H5N6) have subsequently become dominant throughout much of the country since 2009 (Creanga et al., 2013; Nguyen et al., 2016). The continued HA evolution of clade 2.3.2.1 viruses has resulted in three subclade classifications as of 2012 termed clade 2.3.2.1a, 2.3.2.1b and 2.3.2.1c (WHO/OIE/FAO, 2014). In China, ongoing circulation of clade 2.3.2.1c and 2.3.4.4 viruses in poultry has also been a concern due to the expanding diversity of these viruses and sporadic detection of human infections, many of which were fatal (WHO, 2016a).
Here, we use the ferret model to evaluate the virulence of a panel of clade 2 HPAI A(H5N1) viruses isolated from humans or avian species from 2006 to 2013. Ferrets are an indispensable small animal model used to assess the pathogenicity of influenza A viruses as they are naturally susceptible to influenza virus infection, present with signs of infection that closely mimic those found in human infections, and have a similar profile of sialic acid moieties of the viral receptors that are found in the human respiratory tract (Belser et al., 2011b). Our analysis of eleven clade 2 A(H5N1) avian and human isolates revealed distinct virulence profiles in the ferret model. Our results demonstrate that isolates from clade 2.2, 2.2.1, and 2.2.2.1 are generally of low virulence and not lethal in ferrets, whereas clade 2.3.2.1 display a more diverse phenotype, with 2.3.2.1a viruses possessing reduced virulence compared with 2.3.2.1b and 2.3.2.1c. As A(H5N1) influenza viruses continue to evolve, a detailed analysis of the actively emerging 2.3.2.1 clade compared to other 2.2 clades could identify clade and subclade specific molecular determinants of virulence.
2. Materials and methods
2.1. Viruses
A(H5N1) viruses used in this study are listed in Table 1. The stock viruses were grown using the same substrate as that from previous passages, either eggs or cell cultures. Viruses were propagated in Madin-Darby Canine Kidney (MDCK) cells (EG/2321, EG/3072, BnSw/HK/10, BD/5487, Dk/VN/672, Dk/VN/1206, Dk/VN/1232) or the allantoic cavity of ten-day-old embryonating hens’ eggs at 37 °C for 24–26 h (Ck/KR/06, EG/4935, Dk/VN/0004, Dk/VN/2848) as previously described (Maines et al., 2005, 2009). Pooled allantoic fluid or cell supernatant was clarified by centrifugation and aliquots were stored at −70 °C until use. Stocks were titrated in MDCK cells using standard plaque assay methods for determination of plaque forming unit (PFU) titer (Zeng et al., 2007). A 50% egg infectious dose (EID50/mL) for egg grown stocks was determined using standard methods (Reed and Muench, 1938). All experiments were performed in biosafety level 3 containment laboratories with enhancements as required by the Animal and Plant Health Inspection Service (US Department of Agriculture) and the National Select Agent Program (Department of Health and Human Services) (Chosewood and Wilson, 2009).
Table 1.
A(H5N1) viruses used in this study.
| Virus | Name | Cladea | Host | Description | Reference |
|---|---|---|---|---|---|
| A/Chicken/Korea/IS/06 | Ck/KR/06 | 2.2 | Avian | outbreak | (Lee et al., 2008) |
| A/Egypt/2321-NAMRU3/07 | EG/2321 | 2.2.1 | Human | recovered | (Younan et al., 2013) |
| A/Egypt/4935-NAMRU3/09 | EG/4935 | 2.2.1 | Human | recovered | (Younan et al., 2013) |
| A/Egypt/N03072/10 | EG/3072 | 2.2.1 | Human | fatal | (Younan et al., 2013) |
| A/Bangladesh/5487/11 | BD/5487 | 2.2.2.1 | Human | recovered | (Gerloff et al., 2014) |
| A/BarnSwallow/HongKong/D10–1161/10 | BnSw/HK/10 | 2.3.2.1b | Avian | detection | (WHO, 2011) |
| A/Duck/Vietnam/NCVD-672/11 | Dk/VN/672 | 2.3.2.1b | Avian | outbreak | (Creanga et al., 2013) |
| A/Duck/Vietnam/NCVD-1206/12 | Dk/VN/1206 | 2.3.2.1a | Avian | outbreak | (Creanga et al., 2013) |
| A/Duck/Vietnam/NCVD-1232/12 | Dk/VN/1232 | 2.3.2.1a | Avian | outbreak | (Creanga et al., 2013) |
| A/Duck/Vietnam/NCVD-0004/13 | Dk/VN/0004 | 2.3.2.1c | Avian | outbreak | (Nguyen et al., 2016) |
| A/Duck/Vietnam/NVCD-2848/13 | Dk/VN/2848 | 2.3.2.1c | Avian | outbreak | (Nguyen et al., 2016) |
Clade subdivisions described in (WHO/OIE/FAO, 2014).
2.2. Ferret pathogenesis experiments
Male Fitch ferrets (Triple F Farms, Sayre, PA), 6–12 months of age and serologically negative by hemagglutination inhibition assay for currently circulating influenza viruses, were used in this study. Ferrets were housed for the duration of each experiment in a Duo-Flo BioClean environmental enclosure (Lab Products, Seaford, DE). Animal research was conducted under the guidance of the Centers for Disease Control and Prevention's Institutional Animal Care and Use Committee in an Association for Assessment and Accreditation of Laboratory Animal Care International-accredited animal facility. Ferrets were anesthetized prior to procedures with an intramuscular injection of ketamine hydrochloride cocktail (Gustin et al., 2011). Six ferrets per experiment were intranasally (i.n.) administered 106 or 107 PFU or EID50 of virus diluted in PBS in a 1 mL volume. Ferrets were monitored daily for clinical signs of infection and nasal wash (NW) samples were collected on alternate days post-inoculation (p.i.) for virus titration in eggs as previously described (Maines et al., 2005). Ferrets that lost more than 25% total body weight or displayed signs of neurological dysfunction were euthanized. Three days p.i., three ferrets from each virus infected group were euthanized and tissues were collected for virus titration as previously described (Maines et al., 2005). The following tissues were aseptically collected in this order: intestines, spleen, kidneys, liver, trachea, lungs, brain-posterior, brain-anterior, olfactory bulb, and nasal turbinates. Lungs were also collected for histopathological examination. Statistics between titers obtained from high or low virulence viruses was performed by Student's t-test.
2.3. Histopathology and immunohistochemistry (IHC)
Necropsies were performed on three ferrets from each group on day 3 p.i. Three 5-mm lung sections of each lung lobe were collected for histopathology. Lung sections were fixed by submersion in 10% neutral buffered formalin, routinely processed, and embedded in paraffin. Sections were made at 5 µm and were stained with hematoxylin and eosin (HE). A duplicate 5-µm section was immunohistochemically stained for detection of influenza A virus nucleoprotein as described previously (Perkins and Swayne, 2001).
2.4. Virus sequencing and molecular analysis
Following virus isolation, allantoic fluid was harvested and viral RNA was isolated with the Qiagen RNeasy extraction kit (Qiagen) or Magna Pure LC system (Roche). Influenza viral RNA was amplified using the Access Quick one-step RT-PCR kit (Promega) and A(H5N1)-specific primers (sequence available upon request). The 400–600 bp amplicons were sequenced by an Applied Biosystem 3730xl system using cycle sequencing dye terminator chemistry (Life Technologies). Full-length open reading frames with at least triple coverage were generated using Sequencer 5.0 (Gene Codes). Genome sequences can be found in the GISAID and/or GenBank sequence databases. A(H5N1) virus clades were identified for each virus using the LABEL tool (Shepard et al., 2014). Full length coding sequences were aligned with related reference viruses using the Muscle algorithm (Edgar, 2004). For molecular analysis of the viral protein sequences, we used the full-length ORF sequences of NA and the internal genes starting with the ATG codon, whereas the analysis of the HA proteins was performed using sequences of the mature HA protein with signal peptide removed.
3. Results
3.1. Characteristics of A(H5N1) influenza viruses tested in this study
A selection of HPAI A(H5N1) viruses representing clades 2.2, 2.2.1, 2.2.2.1, and 2.3.2.1 isolated from either human or avian hosts were evaluated for their relative virulence in ferrets (Table 1). Of the clade 2.2, 2.2.1, and 2.2.2.1 viruses tested, all but one (Ck/KR/06) were isolated from human cases of infection. All 2.3.2.1 viruses were isolated from infected poultry. Clade 2.3.2.1 is subdivided into clade 2.3.2.1a (A/Hubei/1/2010-like), 2.3.2.1b (A/Barn Swallow/Hong Kong/D10-1161/10-like), and 2.3.2.1c (A/Hong Kong/6841/2010-like) viruses (WHO/OIE/FAO, 2014). All viruses replicated to high titer in the allantoic cavities of 10-d old embryonating eggs or Madin-Darby canine kidney (MDCK) cells, in excess of 108.5 EID50/mL and 107.5 PFU/mL, respectively.
3.2. Pathogenicity of A(H5N1) viruses in ferrets
Infection with HPAI A(H5N1) viruses bearing potential genetic indicators of pathogenicity can cause mild to severe disease in mammalian models (Herfst et al., 2012; Imai et al., 2012; Maines et al., 2005; Suguitan et al., 2012; Thor et al., 2015). To evaluate the relative virulence of selected clade 2 A(H5N1) viruses, ferrets were inoculated i.n. with 106–107 PFU or EID50 of each virus, a dose shown to consistently infect ferrets with this subtype (Table 2) (Maines et al., 2005). The relative virulence of each of the eleven A(H5N1) viruses in ferrets was classified as either high or low virulence based on > 50% (high) or < 50% (low) mortality during the acute phase of infection (Table 2).
Table 2.
Clinical signs and symptoms of A(H5N1) virus infection in ferrets.
| Virus | Clade | % Wt lossa | Feverb | Nasalc | Diarrheac | Neurod | Lethalitye | MDTf | Virulenceg |
|---|---|---|---|---|---|---|---|---|---|
| Ck/KR/06 | 2.2 | 9.4 (2–6) | 2.3 (1) | 2/3 | 0/3 | 0/3 | 0/3 | – | Low |
| EG/2321 | 2.2.1 | 20.5 (8) | 2.2 (1) | 0/3 | 1/3 | 0/3 | 2/3 | 8.0 | High |
| EG/4935 | 2.2.1 | 8.5 (3–8) | 2.3 (2) | 0/3 | 1/3 | 0/3 | 0/3 | – | Low |
| EG/3072 | 2.2.1 | 10.5 (4–12) | 1.8 (2) | 0/3 | 0/3 | 0/3 | 0/3 | – | Low |
| BD/5487 | 2.2.2.1 | 8.0 (2–5) | 2.0 (1–2) | 2/3 | 0/3 | 0/3 | 0/3 | – | Low |
| BnSw/HK/10 | 2.3.2.1b | 19.0 (6–7) | 2.4 (1) | 3/3 | 3/3 | 1/3 | 3/3 | 6.3 | High |
| Dk/VN/672 | 2.3.2.1b | 18.1 (6–7) | 2.2 (1) | 3/3 | 1/3 | 2/3 | 3/3 | 6.7 | High |
| Dk/VN/1206 | 2.3.2.1a | 15.1 (4–7) | 1.4 (1) | 3/3 | 1/3 | 0/3 | 1/3 | 7.0 | Low |
| Dk/VN/1232 | 2.3.2.1a | 10.4 (8–10) | 1.6 (1) | 2/3 | 0/3 | 0/3 | 0/3 | – | Low |
| Dk/VN/0004 | 2.3.2.1c | 24.9 (7) | 2.8 (1) | 2/3 | 3/3 | 1/3 | 3/3 | 7.0 | High |
| Dk/VN/2848 | 2.3.2.1c | 13.9 (3–10) | 2.4 (1) | 0/3 | 0/3 | 0/3 | 0/3 | – | Low |
Percent mean maximum weight loss. Day range p.i. of maximum weight loss in parentheses.
Mean maximum fever reported in degrees centigrade. Day range p.i. of maximumtemperature rise over baseline (37.3–39.2 °C) in parentheses.
Incidence of nasal discharge or diarrhea through day 14 p.i., reported as number of ferrets with symptoms/total number of ferrets.
Number of ferrets which exhibited neurological symptoms/total number of ferrets. All ferrets exhibiting neurological symptoms were humanely euthanized.
Number of ferrets required euthanasia due to severe disease through day 14 p.i./total number of ferrets.
Mean day of death among all ferrets which required euthanasia due to severe disease through day 14 p.i.
Viruses are defined as possessing high virulence when > 50% lethality is observed following infection.
Among the five viruses tested from clades 2.2 (including 2.2.1 and 2.2.2.1), only one isolated from 2007 (EG/2321) was found to be lethal and of high virulence in ferrets (Table 2). Ferrets inoculated with EG/2321 virus exhibited high fever ( > 2 °C above baseline) and severe morbidity (mean weight loss > 20%), warranting euthanasia of two of three ferrets by day 8 p.i. In contrast, reduced morbidity was observed following inoculation with other clade 2.2 viruses, with weight loss ranging from 8% to 10.5% through day 12 p.i. (Table 2). Nasal discharge and diarrhea were detected sporadically in approximately 25% of ferrets inoculated with these viruses.
Generally, we found the virulence of clade 2.3.2.1 viruses to be separated based on the group in which they have been classified. Both clade 2.3.2.1a viruses tested (Dk/VN/1206, Dk/VN/1232) exhibited transient fevers approximately 1.5 °C above baseline and moderate morbidity (mean weight loss approximately 10–15%). One ferret inoculated with Dk/VN/1206 virus was euthanized day 7 p.i. due to severe diarrhea with a suspected secondary bacterial infection; the virus remains classified as low virulence due to < 50% overall lethality. Inoculation of ferrets with clade 2.3.2.1b viruses (BnSw/HK/10, Dk/VN/672) resulted in pronounced morbidity, fever, persistent nasal discharge, and diarrhea, among other clinical signs of infection. The two 2.3.2.1b viruses tested were 100% lethal by day 7 p.i., with neurological complications necessitating euthanasia detected in half of all inoculated ferrets. In contrast, the two clade 2.3.2.1c viruses tested (Dk/VN/0004, Dk/VN/2848) exhibited divergent mammalian phenotypes. Dk/VN/0004 virus was found to possess high virulence in the ferret, with infected ferrets displaying severe morbidity and 100% lethality by day 7 p.i. However, Dk/VN/2848 virus exhibited a low virulence phenotype in ferrets, with moderate weight loss being the only clinical sign of infection. Dk/VN/2848 virus differed from Dk/VN/0004 virus by 8 amino acid changes in the 3 polymerase genes combined, however, none of these were identified in known markers of virulence. Three additional amino acid changes in the HA protein at positions 124, 163 and 188 were also identified. One of these, T188I, was previously found to increase binding to α2,6-linked sialic acid receptors (Yang et al., 2007). These data demonstrate that clade 2.3.2.1 A(H5N1) viruses cause varying degrees of disease ranging from mild clinical symptoms, similar to those seen with a seasonal influenza virus infection, to pronounced disease and death.
3.3. Replication of A(H5N1) viruses in ferrets
To assess the efficiency, kinetics, and spread of virus replication in ferrets, we first analyzed nasal wash samples collected from A(H5N1) virus-inoculated ferrets on alternate days 1–7 p.i. (Fig. 1). All viruses examined replicated to high peak titers ( > 104 EID50/mL) in nasal wash specimens, regardless of their clade or subclade, with generally comparable kinetics and magnitude between viruses of high or low virulence in the ferret model (peak mean NW titers between 104.1–106.5 EID50/mL and 104.8–105.5 EID50/mL for low and high virulence viruses, respectively). Viral clearance was evident by day 7 p.i. as titers dropped in many of ferrets to below the limit of detection (101.5 EID50/mL). Among low virulence A(H5N1) viruses, the clade 2.2 (Ck/KR/06) and 2.3.2.1a (Dk/VN/1232) viruses achieved the highest titers (≥105.2 EID50/mL) in NW specimens through day 3 p.i. With regard to clade 2.3.2.1 viruses tested, 2.3.2.1b viruses peaked at ≥104.6 EID50/mL on days 3 or 5 p.i. (both exhibiting high virulence), while 2.3.2.1a viruses peaked at ≥104.3 EID50/mL on days 1 or 3 p.i. (both exhibiting low virulence). Similarly, the clade 2.3.2.1c low virulence (Dk/VN/2848) virus peaked on day 1 p.i. (104.4 EID50/mL) and the 2.3.2.1c high virulence virus (Dk/VN/0004) peaked on day 3 p.i. (105.2 EID50/mL). Although virus replication of the 2.3.2.1 clade viruses was not always higher among those exhibiting high virulence, peak shedding was consistently observed later (3 or 5 days p.i.) compared with the low virulence viruses of the subclade that peaked on 1 or 3 days p.i. (Fig. 1).
Fig. 1. Mean viral titer in nasal washes of ferrets inoculated with HPAI A(H5N1) influenza viruses.
Three ferrets per group were inoculated with 106 or 107 PFU or EID50 of virus. Nasal washes were collected on alternate days p.i. Mean titers for each group are shown as log10 EID50/mL + standard deviation (SD) from three ferrets per group unless otherwise specified. Due to virus lethality, titers day 7 p.i. reflect one (BnSw/HK/10) or two (Dk/VN/672) ferrets only. The limit of detection was 1.5 log10 EID50/mL.
Previous studies have used the presence or absence of replication of A(H5N1) viruses in extrapulmonary tissues as a measure of pathogenicity (Belser and Tumpey, 2013; Maines et al., 2005). Here, we analyzed the level of virus replication in ferret tissues day 3 p.i. to assess the virulence of each virus. As expected, all viruses tested replicated efficiently throughout the respiratory tissues of ferrets (nasal turbinates [NasTur], trachea, lung; Fig. 2). Highly virulent and lethal A(H5N1) viruses achieved higher mean titers (NasTur 106.4, trachea 104.8, lung106 EID50/mL) than non-lethal viruses (NasTur 105.7, trachea 103.2, lung104.5 EID50/mL) in all respiratory tissues assayed; however the only significant difference between high and low virulent viruses was found in lung tissues (p < 0.02).
Fig. 2. Viral dissemination of A(H5N1) influenza viruses in ferrets.
Three ferrets per group were inoculated with 106 or 107 PFU or EID50 of virus and tissues were collected 3 days p.i. for virus titration. Mean titers for each group are shown as log10 EID50/g or mL + SD. Tissues below our limit of detection (1.5 log10 EID50/g or mL) were assigned a value of 1.5. ND, sample was not collected and not titered.
To investigate systemic spread of infection, we analyzed samples taken from brain (olfactory bulb, anterior, and posterior), spleen, and intestine tissues to quantitate virus amount (Fig. 2). Virus was detected in brain tissues analyzed for most viruses studied, demonstrating extrapulmonary spread of both lethal and non-lethal viruses. Given their proximity to upper respiratory tract tissues, the highest titers were unsurprisingly found in the olfactory bulbs (mean titer of 103.9 and 103.1 EID50/g for lethal and non-lethal viruses, respectively). Furthermore, mean titers in the olfactory bulb were higher in 2.3.2.1 viruses compared with other clade 2 viruses tested (103.8 and 102.9 EID50/g, respectively), though these differences were not statistically significant. Infectious virus in both anterior and posterior sections of the brain was detected among A(H5N1) viruses of both high and low virulence. Virus replication in spleen tissue was observed in two of three ferrets infected with either Dk/VN/672 or Ck/KR/06 viruses. Similarly, infectious virus in the intestines was only sporadically detected overall, though interestingly, virus was present in ferrets inoculated with both 2.3.2.1b viruses (one of three ferrets for BnSw/HK/10, two of three ferrets for Dk/VN/672). Infectious virus was not detected from peripheral blood nor was systemic spread to the kidneys or liver detected from any viruses tested in this study (data not shown). In summary, all viruses that were analyzed showed measureable replication throughout the respiratory tract as well as varying levels of replication in extrapulmonary tissues. Generally, lethal viruses tended to replicate to higher titers in each tissue compared to non-lethal viruses, though strain-specific differences were also apparent.
3.4. Histopathologic evaluation of lung tissue samples of A(H5N1) infected ferrets
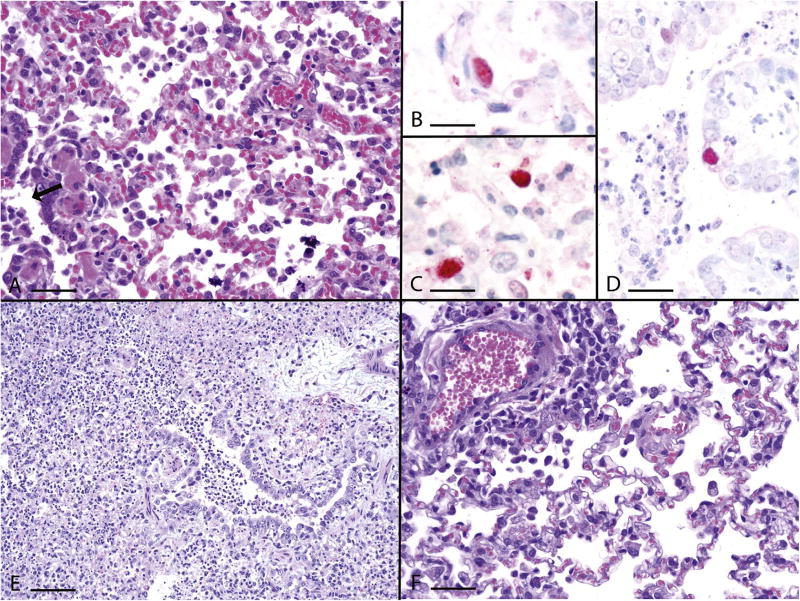
In accord with strain-specific differences observed with viral replication kinetics, magnitude, and duration, histological lesions similarly varied with individual ferrets, different viruses and different lung sections. Typically, bronchointerstitial pneumonia was identified, affecting primarily the bronchioles, especially terminal bronchioles, and generally sparing the bronchi (Fig. 3A). These bronchioles had inflammatory infiltrates of macrophages and granulocytes with accompanied degeneration to necrosis of bronchiolar epithelial cells, and peribronchiolar edema and mononuclear cell cuffs. The areas supplied by these bronchioles had alveolitis with luminal inflammatory cells, predominantly macrophages but also granulocytes, type II pneumocyte hyperplasia, some luminal fibrin and inflammatory cell infiltrates into the septal wall. Virus was localized by immunohistochemistry most commonly to the alveolar epithelium (Fig. 3B) and macrophages (Fig. 3C), but occasionally to the bronchiolar epithelium (Fig. 3D). The most severe lung lesions were noted following infection with BnSw/HK/10 virus. The bronchointerstitial pneumonia was severe and diffuse in multiple BnSw/HK/10 virus-infected lung sections and was accompanied by multiple foci of necrotizing alveolitis and alveolar hemorrhage (Fig. 3E). Lung lesions associated with Dk/VN/672 and Dk/VN/1206 virus infection were similar to BnSw/HK/10 virus, but Dk/VN/672 virus-infected lungs had fewer foci of necrotizing alveolitis while Dk/VN/1206 lacked necrotizing alveolitis and had milder bronchiolitis. In contrast, lungs from ferrets infected with Dk/VN/1232 and BD/5487 viruses had mild to moderate interstitial pneumonia in a few lung sections with minimal to mild bronchiolar lesions (Fig. 3F). In summary, clade 2.3.2.1 viruses were capable of causing pronounced pathology in ferrets, though strain-specific differences were detected. Within this clade, 2.3.2.1b viruses possessed the most severe histopathological changes.
Fig. 3. Histopathologic evaluation of ferrets infected with A(H5N1) influenza viruses.
Representative photomicrographs of hematoxylin and eosin [A, E, F] and immunohistochemically [B-D] stained lung sections from influenza virus-infected ferrets collected day 3 p.i. (A) Moderate bronchointerstitial pneumonia with predominant histiocytic alveolitis and mild bronchiolitis (arrow), Dk/VN/1206, bar =40 µm. (B) Viral antigen in type II pneumocyte, BnSw/HK/10, bar =20 µm. (C) Viral antigen in alveolar macrophages, BnSw/HK/10, bar =20 µm. (D) Viral antigen in bronchiolar epithelium, BD/5487, bar =35 µm. (E) Severe diffuse bronchointerstitial pneumonia with prominent necrotizing alveolitis and severe bronchiolitis, BnSw/HK/10, bar =80 µm. (F) Mild alveolitis with perivascular cuffing with mononuclear inflammatory cells, Dk/VN/1232, bar =40 µm.
3.5. Molecular correlates of viral pathogenicity phenotypes in mammals
An analysis of the protein sequences of the viruses included in this study reveal a myriad of amino acid changes/substitutions that have in some instances been attributed to a specific phenotype, while the effects of others remain unknown (Tables 3, 4). Amino acid differences can be observed when comparing key molecular markers (627 K, PB1-F2, HA and NS1 PDZ) among the group of six H5N1 viruses in the 2.3.2.1 subclades and viruses in clades 2.2, 2.2.1 and 2.2.2 (Table 3). These differences are not sufficient to explain the increased viral pathogenicity observed in ferrets because these amino acid changes also occur among the non-lethal H5N1 viruses. By comparing amino acid similarities among the highly virulent H5N1 viruses (EG/2321, BnSw/HK/10 and Dk/VN/672), the polybasic HA cleavage site motif, the amino acid deletions in NA and NS1, known molecular markers of virulence (CDC, 2012), were common markers among these three H5N1 viruses. However, these amino acid substitutions can be also found in the viruses of low pathogenicity, indicating that additional unknown molecular markers for virulence exist for these H5N1 viruses.
Table 3.
Potential genetic indicators of pathogenicity or human host preference.
| PB2 | PB1-F2 | HA | NA
|
NS1
|
||||
|---|---|---|---|---|---|---|---|---|
| Virus | Clade | 627Ka | #aab | # polybasic aa | antiviral sensitivityc | Δ aa 49–68d | PDZ domain ESEV | Δ aad 80–84 |
| Ck/KR/06 | 2.2 | Y | 90 | 6 | Y | Y | N | Y |
| EG/2321 | 2.2.1 | Y | 90 | 6 | Y | Y | N | Y |
| EG/4935 | 2.2.1 | Y | 90 | 6 | Y | Y | Y | Y |
| EG/3072 | 2.2.1 | Y | 90 | 6 | Y | Y | Y | Y |
| BD/5487 | 2.2.2.1 | Y | 90 | 6 | Y | Y | N | Y |
| BnSw/HK/10 | 2.3.2.1b | N | 57 | 6 | Y | Y | Y | Y |
| Dk/VN/672 | 2.3.2.1b | N | 57 | 6 | Y | Y | Y | Y |
| Dk/VN/1206 | 2.3.2.1a | N | 0 | 5 | Y | Y | Y | Y |
| Dk/VN/1232 | 2.3.2.1a | N | 90 | 5 | Y | Y | Y | Y |
| Dk/VN/0004 | 2.3.2.1c | N | 57 | 5 | Y | Y | Y | Y |
| Dk/VN/2848 | 2.3.2.1c | N | 57 | 5 | Y | Y | Y | Y |
Indicates presence of Lysine at position 627.
#aa: Number of amino acids indicating protein length
Molecular markers of antiviral resistance to oseltamivir, peramivir and zanamivir (CDC, 2012).
Δaa: Contains a deletion of amino acids at the positions shown.
Table 4.
HA amino acids reported to increase binding to α2–6 linked sialic acid receptors.
| Virus | Clade | D94N | S133A | S155N | D183Ga | T188I | K189R | P235S | E75G/ S123Pb |
L128del I151T |
S133A T188Ib |
154–6 glyc |
165–7 glyc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ck/KR/06 | 2.2 | N | S | D | D | T | R | P | E/S | S/I | S/T | Y | Y |
| EG/2321 | 2.2.1 | D | S | N | D | I | R | S | E/S | S/I | S/I | Y | Y |
| EG/4935 | 2.2.1 | N | S | N | D | T | R | S | E/S | del/T | S/T | N | Y |
| EG/3072 | 2.2.1 | N | S | N | D | T | R | S | E/S | del/T | S/T | N | Y |
| BD/5487 | 2.2.2.1 | N | S | D | D | T | K | P | E/S | S/I | S/T | Y | Y |
| BnSw/HK | 2.3.2.1b | N | A | N | D | T | R | P | E/P | L/L | A/T | N | Y |
| Dk/VN672 | 2.3.2.1b | N | A | N | D | I | R | P | E/P | L/L | A/I | N | N |
| Dk/VN1206 | 2.3.2.1a | N | A | N | N | T | R | P | E/S | L/I | A/T | N | Y |
| Dk/VN1232 | 2.3.2.1a | N | A | N | D | T | K | P | E/S | L/I | A/T | N | Y |
| Dk/VN0004 | 2.3.2.1c | N | A | N | D | T | R | P | E/S | S/I | A/T | N | Y |
| Dk/VN2848 | 2.3.2.1c | N | A | N | D | I | R | P | E/S | S/I | A/I | N | Y |
Marker not present but one or more viruses possesses a different amino acid at this position.
This combination of amino acids is necessary for binding, but some viruses only possess one.
Indicates the presence (Y) or absence (N) of glycosylation at these positions.
The six viruses in the 2.3.2.1 clade used in this study span subclades 2.3.2.1a to 2.3.2.1c; each virus contains genetic features that are either commonly shared within the viruses in the same clade or unique to each virus. Both clade 2.3.2.1b viruses (BnSw/HK/10 and Dk/VN/672) caused 100% mortality in ferrets and these two viruses are very similar to each other in amino acid composition, with a total of 99.7% similarity. Conversely, the two clade 2.3.2.1a viruses (Dk/VN/1206 and Dk/VN/1232), which did not cause ferret lethality, share genome similarity of approximately 97%, due to amino acid differences that are mostly concentrated in NA (13 aa), NS1 (9 aa), and PB1-F2 (Table 3). The main molecular differences that separates the three subclade viruses (2.3.2.1a, b, and c) from each other is found in the HA: 15 amino acids in HA1 and 5 in HA2. Although these three groups remain highly conserved at HA amino acid residues previously implicated in enhanced virulence, changes at positions 183, 188 and 189 were identified among the three subclade viruses (Table 4).
4. Discussion
HPAI A(H5N1) viruses continue to pose a public health and potential pandemic threat. While the majority of HPAI A(H5N1) influenza virus infections of poultry and humans have remained largely in China, Southeast Asia, the Middle East, and Africa, sustained human to human transmission of these viruses could result in a global pandemic. To more clearly understand the potential for divergent A(H5N1) influenza viruses to cause disease in mammals, we compared the virulence of representative clade 2 A(H5N1) viruses in the ferret animal model. With one exception, all clade 2.2, 2.2.1, and 2.2.2.1 viruses isolated from either avian or human hosts tested in this study were non-lethal and of low virulence. Our results demonstrate that clade 2.3.2.1 viruses confer a virulence phenotype depending on their genetic subgroup. Among the viruses tested here, clade 2.3.2.1a viruses possessed low virulence in the ferret model, clade 2.3.2.1b possessed high virulence, with both high and low virulence phenotypes present in 2.3.2.1 c. A single isolate, EG/2321 (clade 2.2.1) virus was also found to be of high virulence and resulted in mortality in 2 of 3 ferrets.
Despite that only 4 of 11 A(H5N1) viruses showed a high-virulence phenotype in ferrets, all viruses replicated to high titers in the respiratory tract and, with few exceptions, all viruses were capable of limited replication in brain tissues examined. Further systemic spread of these viruses was observed only sporadically in intestinal and spleen tissues, in agreement with A(H5N1) viruses from other clades (Maines et al., 2005; Yen et al., 2007). We also performed complete blood counts on acute and endpoint blood samples, as highly pathogenic A(H5N1) viruses have been shown to elicit lymphopenia following virus infection in the ferret model (Belser et al., 2011a). In summary, we found that inoculation with A(H5N1) viruses of high virulence caused significant shifts in the circulating lymphocyte, neutrophil and monocyte populations, in particular lymphopenia was most prominent during the first week of infection (data not shown). The mean percentage of lymphocytes in peripheral blood in ferrets infected with EG/2321, BnSw/HK/10, and Dk/VN/672 viruses dropped from 67–31% by day 3 p.i., persisting at < 35% by day 7 p.i.. These results are comparable to the highly virulent clade 1 A(H5N1) virus, A/Vietnam/1203/04 (Belser et al., 2011a; Maines et al., 2005) and further underscores the severe disease caused by some A(H5N1) viruses in the ferret model.
Throughout the years, several studies have been performed to assess the pathogenicity of A(H5N1) viruses using mammalian animal models (reviewed in Belser and Tumpey, 2013), linking the outcomes of infection to molecular genetic factor(s) contained in these viruses. A distinct molecular characteristic of HPAI A(H5N1) viruses that is known to play role in pathogenicity is the stretch of polybasic amino acids in the HA cleavage site (Hatta et al., 2001; Steinhauer, 1999); all A(H5N1) viruses tested in this study possess this major molecular marker of HPAI viruses. Other identified markers include amino acid substitutions in PB2 (627 K and 701N) and deletions in NA and NS1 (Hatta et al., 2001; Li et al., 2009; Matsuoka et al., 2009; Steel et al., 2009b). With respect to PB2, the amino acids 627 K and 701N are critical molecular markers of A(H5N1) virus pathogenicity in mammals (Hatta et al., 2007; Steel et al., 2009a). The 627 K mutation is one of the changes that produced a A(H5N1) transmissible in mammalian model (Herfst et al., 2012; Imai et al., 2012; Russell et al., 2012). The viruses from 2.2, 2.2.1 and 2.2.2.1 clades do possess 627 K, which is a common feature of the Qinghai Lake viruses (Chen et al., 2006), however, of the five viruses in this study that contain 627 K, only one caused lethality in ferrets (EG/2321). 627 K and 701N are not present in the clade 2.3.2.1 viruses and as such cannot explain the high pathogenicity/lethality properties of these viruses, in particular, the 2.3.2.1b viruses. This data agrees with Watanabe et al., who stated that the 627 K residue alone is not sufficient to produce a high pathogenicity phenotype in mammalian models (Watanabe et al., 2012). Alternatively, residues 147 T, 339 T and 588 T, in PB2 have been shown to be involved in the virulence of A(H5N1) viruses (Fan et al., 2014). These residues are present in all 2.3.2.1 viruses used in this study. Additional polygenic amino acids located in other viral proteins provided evidence that multiple genetic factors can modulate the virus pathogenicity in the mammalian host (Belser and Tumpey, 2013).
The HA is the viral protein with the largest number of amino acid differences separating clade 2.3.2.1a, b and c viruses. Although many of these differences are not mapped to known regions of significance, some have been reported to be associated with virus binding to 2,6 sialic acids (CDC, 2012). Importantly, several virus strains in clade 2.2.1 have been shown to contain amino acids in HA that increase binding avidity to cell receptors containing α2,6 sialic acids (Watanabe et al., 2012). One of the molecular changes in the HA among these viruses that contributes to this increase in binding is the 154–156 loss of glycosylation, which also occurs in the viruses of 2.3.2 and 2.3.2.1 clades. Interestingly, this is one of the changes that produced a transmissible A(H5N1) virus in a mammalian model (Herfst et al., 2012; Imai et al., 2012; Russell et al., 2012). Other notable changes in the viruses studied here include one substitution, G323I, an amino acid adjacent to the cleavage site only found in the highly lethal 2.3.2.1b viruses with unknown effects, and a deletion of one polybasic amino acid in the HA cleavage site among 2.3.2.1a viruses which may be associated with a decrease in pathogenicity (Zhang et al., 2012). Because A(H5N1) variants continue to emerge from avian reservoirs, it is likely that additional markers of pathogenicity will be identified. Such molecular markers of virulence have increasingly become important features utilized in the detection and prediction of potentially pathogenic and lethal viruses.
Ferrets have been widely used as a model for influenza virus pathogenesis as well as for immunity studies (Belser et al., 2011b). It is interesting to note that among the eleven HPAI A(H5N1) viruses characterized in this study, the majority of them did not cause lethal infection in ferrets. However, as A(H5N1) viruses continue to diversify in avian species (WHO/OIE/FAO, 2014), sustained surveillance, monitoring, and assessment of these viruses is needed to examine the relative risk posed by these viruses to human health. Mammalian pathotyping of HPAI A(H5N1) influenza virus isolates permits the identification of distinct molecular determinants of virulence, which are subsequently used for public health guidance. Future experiments studying the precise contribution of individual molecular changes present in clade 2.3.2.1 viruses as they compare to other viruses from clade 2 will provide valuable information in this regard.
Acknowledgments
We thank the University of Hong Kong, Egypt Ministry of Health and Population, the Naval Medical Research Unit 3, the Bangladesh Ministry of Health, the International Centre for Diarrhoeal Disease Research, Bangladesh, and the Vietnam Department of Animal Health, National Centre of Veterinary Diagnostics, and Regional Animal Health Offices for the collection and facilitating access to the viruses used. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the funding agency.
References
- Abdel-Ghafar AN, Chotpitayasunondh T, Gao Z, Hayden FG, Nguyen DH, de Jong MD, Naghdaliyev A, Peiris JS, Shindo N, Soeroso S, Uyeki TM. Update on avian influenza A A(H5N1) virus infection in humans. N. Engl. J. Med. 2008;358:261–273. doi: 10.1056/NEJMra0707279. [DOI] [PubMed] [Google Scholar]
- Belser JA, Tumpey TM. A(H5N1) pathogenesis studies in mammalian models. Virus Res. 2013;178:168–185. doi: 10.1016/j.virusres.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser JA, Gustin KM, Maines TR, Blau DM, Zaki SR, Katz JM, Tumpey TM. Pathogenesis and transmission of triple-reassortant swine H1N1 influenza viruses isolated before the 2009 H1N1 pandemic. J. Virol. 2011a;85:1563–1572. doi: 10.1128/JVI.02231-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser JA, Katz JM, Tumpey TM. The ferret as a model organism to study influenza A virus infection. Dis. Models Mech. 2011b;4:575–579. doi: 10.1242/dmm.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. A(H5N1) Genetic Changes Inventory: A Tool for International Surveillance. CDC; Atlanta: 2012. A(H5N1) Genetic Changes Inventory: a Tool for International Surveillance. [Google Scholar]
- Chen H, Li Y, Li Z, Shi J, Shinya K, Deng G, Qi Q, Tian G, Fan S, Zhao H, Sun Y, Kawaoka Y. Properties and dissemination of A(H5N1) viruses isolated during an influenza outbreak in migratory waterfowl in western China. J. Virol. 2006;80:5976–5983. doi: 10.1128/JVI.00110-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chosewood LC, Wilson DE. Centers for Disease Control and Prevention (U.S.), National Institutes of Health (U.S.). Biosafety in microbiological and biomedical laboratories. 5. U.S. Dept. of Health and Human Services, Public Health Service, Centers for Disease Control and Prevention, National Institutes of Health; Washington, D.C.: 2009. [Google Scholar]
- Creanga A, Thi Nguyen D, Gerloff N, Thi Do H, Balish A, Dang Nguyen H, Jang Y, Thi Dam V, Thor S, Jones J, Simpson N, Shu B, Emery S, Berman L, Nguyen HT, Bryant JE, Lindstrom S, Klimov A, Donis RO, Davis CT, Nguyen T. Emergence of multiple clade 2.3.2.1 influenza A (H5N1) virus subgroups in Vietnam and detection of novel reassortants. Virology. 2013;444:12–20. doi: 10.1016/j.virol.2013.06.005. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S, Hatta M, Kim JH, Halfmann P, Imai M, Macken CA, Le MQ, Nguyen T, Neumann G, Kawaoka Y. Novel residues in avian influenza virus PB2 protein affect virulence in mammalian hosts. Nat. Commun. 2014;5:5021. doi: 10.1038/ncomms6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerloff NA, Khan SU, Balish A, Shanta IS, Simpson N, Berman L, Haider N, Poh MK, Islam A, Gurley E, Hasnat MA, Dey T, Shu B, Emery S, Lindstrom S, Haque A, Klimov A, Villanueva J, Rahman M, Azziz-Baumgartner E, Ziaur Rahman M, Luby SP, Zeidner N, Donis RO, Sturm-Ramirez K, Davis CT. Multiple reassortment events among highly pathogenic avian influenza A(H5N1) viruses detected in Bangladesh. Virology. 2014;450–451:297–307. doi: 10.1016/j.virol.2013.12.023. [DOI] [PubMed] [Google Scholar]
- Gustin KM, Maines TR, Belser JA, van Hoeven N, Lu X, Dong L, Isakova-Sivak I, Chen LM, Voeten JT, Heldens JG, van den Bosch H, Cox NJ, Tumpey TM, Klimov AI, Rudenko L, Donis RO, Katz JM. Comparative immunogenicity and cross-clade protective efficacy of mammalian cell-grown inactivated and live attenuated A(H5N1) reassortant vaccines in ferrets. J. Infect. Dis. 2011;204:1491–1499. doi: 10.1093/infdis/jir596. [DOI] [PubMed] [Google Scholar]
- Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong A(H5N1) influenza A viruses. Science. 2001;293:1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- Hatta M, Hatta Y, Kim JH, Watanabe S, Shinya K, Nguyen T, Lien PS, Le QM, Kawaoka Y. Growth of A(H5N1) influenza A viruses in the upper respiratory tracts of mice. PLoS Pathog. 2007;3:1374–1379. doi: 10.1371/journal.ppat.0030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Airborne transmission of influenza A(H5N1) virus between ferrets. Science. 2012;336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Choi YK, Kim YJ, Song MS, Jeong OM, Lee EK, Jeon WJ, Jeong W, Joh SJ, Choi KS, Her M, Kim MC, Kim A, Kim MJ, Ho Lee E, Oh TG, Moon HJ, Yoo DW, Kim JH, Sung MH, Poo H, Kwon JH, Kim CJ. Highly pathogenic avian influenza virus (H5N1) in domestic poultry and relationship with migratory birds, South Korea. Emerg. Infect. Dis. 2008;14:487–490. doi: 10.3201/eid1403.070767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ishaq M, Prudence M, Xi X, Hu T, Liu Q, Guo D. Single mutation at the amino acid position 627 of PB2 that leads to increased virulence of an A(H5N1) avian influenza virus during adaptation in mice can be compensated by multiple mutations at other sites of PB2. Virus Res. 2009;144:123–129. doi: 10.1016/j.virusres.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Maines TR, Lu XH, Erb SM, Edwards L, Guarner J, Greer PW, Nguyen DC, Szretter KJ, Chen LM, Thawatsupha P, Chittaganpitch M, Waicharoen S, Nguyen DT, Nguyen T, Nguyen HH, Kim JH, Hoang LT, Kang C, Phuong LS, Lim W, Zaki S, Donis RO, Cox NJ, Katz JM, Tumpey TM. Avian influenza A(H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J. Virol. 2005;79:11788–11800. doi: 10.1128/JVI.79.18.11788-11800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines TR, Jayaraman A, Belser JA, Wadford DA, Pappas C, Zeng H, Gustin KM, Pearce MB, Viswanathan K, Shriver ZH, Raman R, Cox NJ, Sasisekharan R, Katz JM, Tumpey TM. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science. 2009;325:484–487. doi: 10.1126/science.1177238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y, Swayne DE, Thomas C, Rameix-Welti MA, Naffakh N, Warnes C, Altholtz M, Donis R, Subbarao K. Neuraminidase stalk length and additional glycosylation of the hemagglutinin influence the virulence of influenza A(H5N1) viruses for mice. J. Virol. 2009;83:4704–4708. doi: 10.1128/JVI.01987-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DT, Jang Y, Nguyen TD, Jones J, Shepard SS, Yang H, Gerloff N, Fabrizio T, Nguyen LV, Inui K, Yang G, Creanga A, Wang L, Mai DT, Thor S, Stevens J, To TL, Wentworth DE, Nguyen T, Pham DV, Bryant JE, Davis CT. Shifting clade distribution, reassortment, and emergence of new subtypes of highly pathogenic avian influenza A(H5) viruses collected in Vietnamese poultry from 2012 to 2015. J. Virol. 2017 doi: 10.1128/JVI.01708-16. (in press) http://jvi.asm.org/content/early/recent. [DOI] [PMC free article] [PubMed]
- Perkins LE, Swayne DE. Pathobiology of A/chicken/Hong Kong/220/97 A(H5N1) avian influenza virus in seven gallinaceous species. Vet. Pathol. 2001;38:149–164. doi: 10.1354/vp.38-2-149. [DOI] [PubMed] [Google Scholar]
- Reed LJ, Muench HA. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 1938;27:493–497. [Google Scholar]
- Refaey S, Azziz-Baumgartner E, Amin MM, Fahim M, Roguski K, Elaziz HA, Iuliano AD, Salah N, Uyeki TM, Lindstrom S, Davis CT, Eid A, Genedy M, Kandeel A. Increased number of human cases of influenza virus A(H5N1) Infection, Egypt, 2014–15. Emerg. Infect. Dis. 2015;21:2171–2173. doi: 10.3201/eid2112.150885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejmanek D, Hosseini PR, Mazet JA, Daszak P, Goldstein T. Evolutionary dynamics and global diversity of influenza a virus. J. Virol. 2015;89:10993–11001. doi: 10.1128/JVI.01573-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell CA, Fonville JM, Brown AE, Burke DF, Smith DL, James SL, Herfst S, van Boheemen S, Linster M, Schrauwen EJ, Katzelnick L, Mosterin A, Kuiken T, Maher E, Neumann G, Osterhaus AD, Kawaoka Y, Fouchier RA, Smith DJ. The potential for respiratory droplet-transmissible A(H5N1) influenza virus to evolve in a mammalian host. Science. 2012;336:1541–1547. doi: 10.1126/science.1222526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard SS, Davis CT, Bahl J, Rivailler P, York IA, Donis RO. LABEL: fast and accurate lineage assignment with assessment of A(H5N1) and H9N2 influenza A hemagglutinins. PloS One. 2014;9:e86921. doi: 10.1371/journal.pone.0086921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel J, Lowen AC, Mubareka S, Palese P. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 2009a;5:e1000252. doi: 10.1371/journal.ppat.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel J, Lowen AC, Pena L, Angel M, Solorzano A, Albrecht R, Perez DR, Garcia-Sastre A, Palese P. Live attenuated influenza viruses containing NS1 truncations as vaccine candidates against A(H5N1) highly pathogenic avian influenza. J. Virol. 2009b;83:1742–1753. doi: 10.1128/JVI.01920-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer DA. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology. 1999;258:1–20. doi: 10.1006/viro.1999.9716. [DOI] [PubMed] [Google Scholar]
- Suguitan AL, Jr, Matsuoka Y, Lau YF, Santos CP, Vogel L, Cheng LI, Orandle M, Subbarao K. The multibasic cleavage site of the hemagglutinin of highly pathogenic A/Vietnam/1203/2004 (A(H5N1)) avian influenza virus acts as a virulence factor in a host-specific manner in mammals. J. Virol. 2012;86:2706–2714. doi: 10.1128/JVI.05546-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thor SW, Nguyen H, Balish A, Hoang AN, Gustin KM, Nhung PT, Jones J, Thu NN, Davis W, Ngoc TN, Jang Y, Sleeman K, Villanueva J, Kile J, Gubareva LV, Lindstrom S, Tumpey TM, Davis CT, Long NT. Detection and characterization of clade 1 reassortant A(H5N1) viruses isolated from human cases in vietnam during 2013. PloS One. 2015;10:e0133867. doi: 10.1371/journal.pone.0133867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Ibrahim MS, Ellakany HF, Kawashita N, Daidoji T, Takagi T, Yasunaga T, Nakaya T, Ikuta K. Antigenic analysis of highly pathogenic avian influenza virus A(H5N1) sublineages co-circulating in Egypt. J. Gen. Virol. 2012;93:2215–2226. doi: 10.1099/vir.0.044032-0. [DOI] [PubMed] [Google Scholar]
- WHO. Antigenic and genetic characteristics of influenza A(A(H5N1)) and influenza A(H9N2) viruses for the development of candidate vaccine viruses for pandemic preparedness. 2011 [PubMed]
- WHO. Antigenic and genetic characteristics of zoonotic influenza viruses and development of candidate vaccine viruses for pandemic preparedness. 2016a [PubMed]
- WHO. Cumulative number of confirmed human cases for avian influenza A(A(H5N1)) reported to WHO, 2003–2016 2016b
- WHO/OIE/FAO. Revised and updated nomenclature for highly pathogenic avian influenza A(H5N1) viruses. Influenza Other Respir. Virus. 2014;8:384–388. doi: 10.1111/irv.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZY, Wei CJ, Kong WP, Wu L, Xu L, Smith DF, Nabel GJ. Immunization by avian H5 influenza hemagglutinin mutants with altered receptor binding specificity. Science. 2007;317:825–828. doi: 10.1126/science.1135165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HL, Lipatov AS, Ilyushina NA, Govorkova EA, Franks J, Yilmaz N, Douglas A, Hay A, Krauss S, Rehg JE, Hoffmann E, Webster RG. Inefficient transmission of A(H5N1) influenza viruses in a ferret contact model. J. Virol. 2007;81:6890–6898. doi: 10.1128/JVI.00170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younan M, Poh MK, Elassal E, Davis T, Rivailler P, Balish AL, Simpson N, Jones J, Deyde V, Loughlin R, Perry I, Gubareva L, ElBadry MA, Truelove S, Gaynor AM, Mohareb E, Amin M, Cornelius C, Pimentel G, Earhart K, Naguib A, Abdelghani AS, Refaey S, Klimov AI, Donis RO, Kandeel A. Microevolution of highly pathogenic avian influenza A(H5N1) viruses isolated from humans, Egypt, 2007–2011. Emerg. Infect. Dis. 2013;19:43–50. doi: 10.3201/eid1901.121080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Goldsmith C, Thawatsupha P, Chittaganpitch M, Waicharoen S, Zaki S, Tumpey TM, Katz JM. Highly pathogenic avian influenza A(H5N1) viruses elicit an attenuated type i interferon response in polarized human bronchial epithelial cells. J. Virol. 2007;81:12439–12449. doi: 10.1128/JVI.01134-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Chen D, Ward MP, Jiang Q. Transmissibility of the highly pathogenic avian influenza virus, subtype A(H5N1) in domestic poultry: a spatiotemporal estimation at the global scale. Geospatial Health. 2012;7:135–143. doi: 10.4081/gh.2012.112. [DOI] [PubMed] [Google Scholar]