Abstract
Environmental factors, including diet, exercise, stress, and toxins, profoundly impact disease phenotypes. This review examines how Wilson disease (WD), an autosomal recessive genetic disorder, is influenced by genetic and environmental inputs. WD is caused by mutations in the copper-transporter gene ATP7B, leading to the accumulation of copper in the liver and brain, resulting in hepatic, neurological, and psychiatric symptoms. These symptoms range in severity and can first appear anytime between early childhood and old age. Over 300 disease-causing mutations in ATP7B have been identified, but attempts to link genotype to the phenotypic presentation have yielded little insight, prompting investigators to identify alternative mechanisms, such as epigenetics, to explain the highly varied clinical presentation. Further, WD is accompanied by structural and functional abnormalities in mitochondria, potentially altering the production of metabolites that are required for epigenetic regulation of gene expression. Notably, environmental exposure affects the regulation of gene expression and mitochondrial function. We present the “multi-hit” hypothesis of WD progression, which posits that the initial hit is an environmental factor that affects fetal gene expression and epigenetic mechanisms and subsequent “hits” are environmental exposures that occur in the offspring after birth. These environmental hits and subsequent changes in epigenetic regulation may impact copper accumulation and ultimately WD phenotype. Lifestyle changes, including diet, increased physical activity, stress reduction, and toxin avoidance, might influence the presentation and course of WD, and therefore may serve as potential adjunctive or replacement therapies.
Keywords: Wilson disease, Modifier gene, Epigenetics, Mitochondria
1. Introduction
There is mounting evidence that the phenotypic expression of genetic diseases is governed by modifiable environmental inputs, including diet,1 exercise, 2,3 stress,4 and toxins.5 These factors impact disease phenotypes through several mechanisms, including alterations in mitochondrial function and epigenetic regulation of gene expression. The ability to modify the phenotype of a genetic disease through lifestyle changes is valuable, because there are often limited treatment options for such conditions. This review will discuss the evidence that the phenotype of Wilson disease (WD) is influenced by interacting genetic and epigenetic factors, affecting the regulation of gene expression and potentially WD onset and progression.
2. Clinical presentation and treatment of Wilson disease
WD is an autosomal recessive disorder that is caused by mutations in ATP7B, which encodes for a P-type ATPase that is primarily expressed in the brain and liver.6 In the liver, ATP7B traffics excess copper to the hepatocyte plasma membrane for excretion into bile and loads copper onto the ferroxidase protein ceruloplasmin. 6 WD is characterized by the accumulation of copper in the liver and brain, effecting hepatic, neurological, and psychiatric symptoms. The severity of these symptoms varies widely. Liver manifestations range from mild elevation of liver enzymes to acute liver failure and cirrhosis.7,8 The neurological effects can be similar to those of Parkinson disease and include tremors, dysarthria, and ataxia.9 The psychiatric symptoms include anxiety, depression, disinhibition, and personality changes.10
The age of onset of WD also widely varies. Liver manifestations that are related to WD have been reported to first appear in patients aged as young as 9 months and in those aged over 70 years.11,12 Although the course of the disease tends to be benign if it is diagnosed early, the response to copper chelation or zinc treatment varies and can be accompanied by many sideeffects.13,14
Two therapies commonly prescribed for WD are zinc salts and copper chelators. These can be used alone or in combination. Zinc inhibits intestinal absorption of dietary copper,15 and copper chelators bind copper in the blood and tissues for subsequent excretion in the urine. Zinc monotherapy is less effective than copper chelators in the treatment of hepatic WD.13 In a small study, zinc treatment improved the neurological symptoms in patients with WD.14 A retrospective study that compared the effects of two copper chelation therapies in 405 patients with WD found more patients discontinued D-penicillamine compared with trientine due to adverse reactions.16 Also, regardless of copper chelator, hepatic improvements were observed in more than 90% of patients, whereas 55% of patients experienced neurological improvements. In 7% of patients, neurological symptoms worsened after copper chelation therapy was initiated,16 which is believed to be attributed to over-mobilization of free copper.17
Another study utilizing brain magnetic resonance imaging (MRI) also found variable responses to D-penicillamine when combined with zinc sulfate in 50 patients with the neurological manifestation of WD.18 Based on the MRI scans, 35 patients improved, 9 remained stable, 4 declined, and 1 patient showed signs of improvement and decline. These data support the need for adjunctive or alternative therapies for patients who fail to respond adequately or experience adverse reactions to traditional treatments.
3. Genetics of Wilson disease
Recent genetic studies reported a prevalence of 1:7,026 for two mutant pathogenic ATP7B alleles,19 indicating that WD is more frequent than initially described (1:30,000).20,21 Over 500 mutations in ATP7B have been identified, 380 of which have been deemed to be disease-causing mutations.22 Most of these changes are missense mutations, small insertions and deletions in the coding region, or splice junction mutations. Whole-exon deletions, mutations in the promoter region, and multiple mutations within the same gene also occur but are less common (for a recent review 23). These various mutations decrease ATP7B levels, cause improper protein localization, and reduce substrate binding (copper, ATP) and activity.24–26
Attempts to link specific gene mutations to phenotypic outcomes have provided little explanation for the highly varied presentation of WD.27–30 Studies have found a correlation between p.H1069Q mutations, which are the most common mutations in European populations, and late onset of WD31,32—an association that Merle et. al. challenged.33 A correlation between truncated ATP7B mutations and early-onset WD has also been observed in Europeans,33–35 but other studies have not been able to corroborate these findings.36–38 The overall lack of a clear genotype-phenotype relationship has spurred researchers to investigate other mechanisms—including modifier genes and epigenetic regulation of gene expression—to account for the pleiotropic effects of WD.
4. Potential modifier genes
Modifier genes affect the expression of other genes,39 impacting disease-related phenotype, for instance, by altering expressivity—the degree to which a particular trait is expressed. ATP7B modifier genes are believed to influence tolerance to copper accumulation and copper storage capacity.40 Several modifier genes of ATP7B have been analyzed and are described briefly below. The results from these studies are often conflicting, likely due to their small cohorts and the heterogeneity of ATP7B mutations, necessitating further examination to explain the highly varied phenotypic presentation of WD.
4.1. Copper metabolism domain-containing 1 (COMMD1)
COMMD1 is a copper chaperone that interacts directly with the N-terminus of ATP7B.41 Mutations in COMMD1 cause canine copper toxicosis in Bedlington terriers.42 No exonic mutations in COMMD1 have been identified in patients with WD.43 An association between a COMMD1 polymorphism [Asn 164 (GAT/GAC)] and early onset of neurological and hepatic symptoms was reported in WD patients who were homozygous for the most common ATP7B mutation, H1069Q.43 But, subsequent studies have not been able to confirm this finding.44,45
4.2. Antioxidant 1 copper chaperone (ATOX1)
ATOX1, similar to COMMD1, is a copper chaperone that associates directly to ATP7B. By modulating the amount of copper that binds to ATP7B, ATOX1 influences the intracellularlocalization,46 posttranslational modification, and enzymatic activity of ATP7B.47 ATOX1 is also a copper-dependent transcription factor that is involved in cell proliferation.48 Despite the physical interaction betweenATOX1 and ATP7B, a study in patients with WD failed to identify any significant non-synonymous coding variations in ATOX1 beyond the expected frequencies. 44,49
4.3. X-linked inhibitor of apoptosis (XIAP)
XIAP is an anti-apoptotic protein and a potential regulator of copper-induced cell injury. The binding of copper to XIAP relieves the inhibition of caspase, thereby initiating caspase-mediated cell death in copper-loaded hepatocytes.50 XIAP might also maintain cellular copper homeostasis by promoting ubiquitination and degradation of COMMD1.51 In support of this model, experiments in Xiap-deficient mice yielded lower cellular copper levels and modestly reduced levels of COMMD1. 51 Four non-synonymous coding SNPs have been described in the coding region of XIAP, 52–53 but their function remains unknown. In 98 WD patients, 54 the frequency of 7 SNPs identified in the XIAP gene did not differ significantly from previous reports, and the statistical analysis did not reveal any correlation between each XIAP SNP and age of onset or clinical presentation (hepatic vs. neurologic vs. mixed vs. asymptomatic). 54
4.4. Patatin-like phospholipase domain-containing 3(PNPLA3)
PNPLA3is a lipase that targets triglycerides. 55 Several studies have shown that a missense mutation (I148M) in PNPLA3 increases hepatic fat deposition. 56 A hallmark of WD is hepatic fat accumulation; therefore, the prevalence of PNPLA3 mutations was investigated in 98 patients with WD.22 Multivariate logistic regression revealed the PNPLA3 G allele was an independent variable associated with moderate/severe steatosis, whereas hepatic copper content was not. 22
4.5. Apolipoprotein E (APOE)
APOE has a significant role in lipid transport. There are 3 isoforms of APOE: ε2, ε3, and ε4.57 The presence of the APOEε4 allele is associated with an increased risk of developing Alzheimer disease, whereas APOEε3/3 appears to provide moderate neuroprotection. Notably, several studies suggest that excess copper in the brain and various ATP7B genotypes are associated with the development of neurodegenerative disorders, including Alzheimer disease.58 The APOEε3/3 genotype has been associated with delayed onset of hepatic and neurological symptoms in patients with WD,59 and women with APOE ε4 present earlier with WD symptoms, particularly among ATP7BH1069Q homozygous patients.60 However, other groups have been unable to confirm these findings.61,62
4.6. Hemochromatosis (HFE)
HFE regulates circulating iron by interacting with transferrin and transferrin receptor.63 HFE mutations can increase intestinal iron uptake and are associated with hereditary hemochromatosis. Data on WD patients and in animal models of WD indicate that hepatic iron accumulation exacerbates the symptoms of WD.64–66 Two initial case reports described the concomitance of HFE and ATP7B polymorphisms and the corresponding accumulation of hepatic iron and copper.67,68 A study in 32 patients with WD from Sardinia analyzed iron and copper metabolism indices, HFE mutations, and liver biopsies.69 Patients with HFE polymorphisms had approximately 2-fold higher hepatic iron concentrations and were less responsive to anti-copper treatments compared with those who lacked them. However, other studies have failed to find an association between WD and HFE allele frequencies.70,71
4.7. 5,10-methylenetetrahydrofolate reductase (MTHFR)
MTHFR modulates homocysteine levels through folate and methionine metabolism. Mutations in MTHFR are associated with increased homocysteine levels, which can exacerbate WD symptoms and contribute to WD phenotypic variability. In patients with WD, the MTHFR 677T allele is associated with hepatic phenotype, and the 1298C allele correlates with earlier presentation. These polymorphisms are unrelated to differences in copper metabolism.72 Although hyperhomocysteinemia has not been described in WD, it is possible that homocysteine or aberrant methionine metabolism affects the WD phenotype , because homocysteine can pass the blood-brain barrier and have neurotoxic effects that are enhanced by the presence of copper.73,74 In addition, methionine metabolism is linked to DNA and histone methylation, with potential consequences for the regulation of gene expression.
4.8. Prion protein (PrP)
PrP binds to copper ion with low affinity and can affect copper metabolism,75 especially in the central nervous system, where PrP is highly expressed.76 The interaction between copper and PrP might have a protective effect on neurons.77 The prevalence of a prion gene polymorphism in codon 129is similar between patients with WD and healthy controls.78 However, the age of onset of WD and that of the neurological presentation were delayed 5 years and 7 years, respectively, in patients with homozygous methionine in codon 129 compared with subjects with at least one valine in codon 129 in the Prp gene.
4.9. Tumor protein 53 (p53)
p53regulates cell growth by inducing cell cycle arrest, apoptosis, autophagy, DNA repair, changes in metabolism, and cellular oxidative status.79 Mutations in p53in 12 patients with WD were investigated, wherein higher frequencies of G:C to T:A transversions in codon 249 and C:G to A:T transversions and C:G to T:A transitions in codon 250 were found in liver tissue compared with healthy controls.80 Patients with WD also expressed more hepatic inducible nitric oxide synthase, implicating nitric oxide as a source of increased oxidative stress in WD,80 as supported by the high levels of etheno-DNA adducts, formed by oxyradical-induced lipid peroxidation, in patients with WD. 81 Furthermore, etheno-DNA adducts in cell culture induce oxidative stress, leading to p53 mutations. 80 These data indicate that copper-induced oxidative stress leads to lipid peroxidation and consequently the formation of DNA adducts, ultimately resulting in gene mutations.
4.10. Brain-derived neurotrophic factor (BDNF)
BDNF promotes the survival and growth of neurons.82 Patients with WD more frequently harbored BDNF polymorphisms [Val/Val (−196 G/G) and −270 C/T] than healthy controls and asymptomatic WD patients.83 However, no association was identified between any BDNF polymorphism and disease presentation (hepatic vs. neurological) or age of onset of WD.
5. Overview of epigenetic regulation of gene expression
Epigenetics is a broad term that encompasses a wide range of regulatory processes that influence gene expression without altering the nucleotide sequence. Environmental factors, including diet,1 exercise,2 stress, 4 and toxins,5 alter the epigenome in humans and animal models. These modifiable elements can shape how genes are expressed and thus impact how genetic diseases, including WD, present phenotypically. The most well-understood epigenetic mechanism is DNA methylation;84 other mechanisms include histone modifications,85 small RNAs, 86 and long non-coding RNAs.87 Each mechanism is described briefly below.
Mammalian DNA methylation occurs at cytosine bases, typically in CpG sites.88 Methylation can arise in nuclear and mitochondrial DNA (mtDNA), but the mechanisms regarding mtDNA methylation are less well understood compared with that of the former. 89 The effects of DNA methylation on gene expression depend on where it takes place in the genome. Methylation of the transcription start site results in gene silencing, whereas gene body methylation correlates positively with enhanced transcription; however, this relationship is not universal.90
DNA methylation is mediated by several DNA methyltransferases (DNMTs):DNMT1, DNMT3a, and DNMT3b.DNMT1 maintains DNA methylation,91 whereas DNMT3a and DNMT3b catalyze de novo DNA methylation.92 DNA methylation depends on the availability of methyl donors, including folate, vitamin B12, methionine, betaine, and choline—the levels of which are significantly influenced by diet.93 DNA demethylation is mediated by a group of enzymes, known as ten-eleven translocation enzymes, and occurs through thymine DNA glycosylasemediated base excision repair.94
Posttranslational modifications of histones also regulate gene expression by controlling access to the underlying DNA; these modifications include acetylation, methylation, phosphorylation, ribosylation, glycosylation, and sumoylation.85 These markers can combine in myriad ways to regulate gene expression, forming the basis of the histone code.95 Histone acetylation and deacetylation enhance and repress transcription, respectively, and are catalyzed by histone acetylases (HATs) and histone deacetylases (HDACs).Histone phosphorylation and ribosylation increase transcription, whereas histone methylation, glycosylation, and sumoylation impede it.85
Non-coding RNAs also influence gene expression through posttranslational gene silencing. Small RNAs include microRNAs, small interfering RNAs, and PIWI-interacting RNAs. MicroRNAs increase the degradation of mRNA that is involved in epigenetic regulation (e.g., HDAC, DNMT3a, DNMT3b).Furthermore, epigenetic mechanisms can reciprocate, governing the expression of small RNAs.96 Long non-coding RNAs alter gene expression by interacting with chromatin-modifying proteins and control the stability and translation of complementary mRNAs.87
6. Evidence of epigenetic regulation in Wilson disease
Due to the lack of robust genotype-phenotype correlations in WD, epigenetic regulation of gene expression might be involved in disease presentation. Case studies have reported twins and siblings who have the same WD genotype presenting with disparate phenotypes— implicating factors other than genetics, including epigenetics, 97–100 in the presentation of WD.101 Animal studies in the Jackson toxic milk (tx-j) mouse model of WD support this model, 102 astx-j mice show DNA hypomethylation and altered expression of genes that are involved in DNA methylation compared with wild-type mice.103–105 Methylation reactions rely on the availability of the universal methyl donor S-adenosylmethionine (SAM), most of which is produced in the liver.106 Methionine and ATP produce SAM via methionine adenosyltransferase (MAT).SAM is then converted to S-adenosylhomocysteine (SAH) through the bi-directional enzyme S-adenosylhomocysteine hydrolase (AHCY).ACHY is a key enzyme that regulates the amount of SAM that is available for methylation reactions. Copper inhibits ACHY,107 leading to the accumulation of SAH, a reduction in SAM, the inhibition of MAT, and ultimately impaired methylation.104
Consistent with the mechanism above, our group reported that 24-week-old tx-j mice have lower hepatic ACHY protein and transcript levels, elevated SAH levels, fewer Dnmt3b transcripts and global DNA hypomethylation versus wild-type animals. Other groups have also observed reduced hepatic ACHY levels in the toxic milk mouse model.108 The mechanism of this decrease is paradoxical—ACHY is a copper-binding protein, and as expected, it is downregulated during copper deficiency.108 One possible reason for this inconsistency is the abnormal distribution of copper in hepatocytes in animal models of WD.109 It is also not clear to what degree the human WD phenotype is influenced by reduced protein levels and copper inhibition of ACHY. Betaine supplementation increases SAM and Dnmt3b levels and restores DNA methylation in the liver.104 Our group found a 17% rise in global DNA methylation, concurrent with changes in fetal hepatic gene expression, in fetal tx-j mice from dams that received choline-supplemented diets versus adequate choline in their diets; 105 notably, fetal liver bred from homozygous tx-j dams exhibited copper deficiency—not excess levels.105 Mice that are born to homozygous tx-j dams might be a useful model for studying copper deficiency-induced fatty liver.110 These data indicate that hepatic copper accumulation alters DNA methylation in a mouse model of WD whereas dietary intervention with methyl donors restoresit. Another well-established consequence of WD—mitochondrial abnormalities—can impact the availability of metabolites that are required for epigenetic regulation.
7. Copper overload, mitochondrial function, and epigenetic regulation
Mitochondria are integral to metabolism, constituting the site at which the tricarboxylic acid (TCA) cycle, electron transport chain (ETC), and β-oxidation of fatty acids take place in the cell.111 The morphology of mitochondria from patients with WD is altered, manifesting as separated outer and inner mitochondrial membranes, cristae dilations, and giant mitochondria; these changes occur in hepatocytes in early- and late-stage WD.112 Although alterations in neuronal mitochondria have not been investigated in WD patients and animal models,113 tx-j mice show neuroinflammation and develop behavioral abnormalities, 114, 115 and several animals with neuronal copper intoxication consume more oxygen, possibly indicative of mitochondrial uncoupling.116 The structural changes in hepatocyte mitochondria might be caused by copper-induced disulfide bond (thiol)formation. One study observed copper-induced thiol formation, protein crosslinking, and altered morphology in mitochondria that were isolated from the livers of Atp7b−/− mice—effects that preceded changes in oxidative phosphorylation. Furthermore, in vivo copper chelation was found to restore mitochondrial shape.117
Mitochondria are more susceptible to damage from excess copper compared with other cellular compartments due to the presence of high-affinity copper-binding partners (e.g., Cu/Zn SOD, copper chaperones, copper ligand, and glutathione). 118 Furthermore, copper inhibits key mitochondrial enzymes,119, 120 disrupting the production of metabolites that are involved in epigenetic regulation. These enzymes and metabolites are summarized below.
Pyruvate dehydrogenase converts pyruvate into acetyl-CoA, which can then enter the TCA cycle. Acetyl-CoA is required for histone acetylation by HATs.121 Patients with WD have increased blood pyruvate levels, which have been attributed to inhibition of pyruvate dehydrogenase by copper.120, 122
2-oxoglutarate dehydrogenase(aka α-ketoglutarate dehydrogenase) converts 2-oxoglutarate (2- OG) to succinyl-CoA.2-OGis a substrate for the DNA and histone demethylases TET and Jumanji C domain-containing histone demethylases.123 Copper inhibits 2-oxoglutarate dehydrogenase.119
Succinate dehydrogenase (SDH) functions in the TCA cycle and ETC. In the TCA cycle, SDH oxidizes succinate to fumarate—both of which inhibit TET and Jumanji C domain-containing histone demethylases.123 Copper inhibits SDH,124 as supported by our findings that the lower levels of hepatic SDH in tx-j mice are restored by choline and copper chelation therapy alone and in combination.103
NAD+ is a substrate for class III protein deacetylases, known as sirtuins.125 NADH is produced in the TCA cycle by malate dehydrogenase and oxidized to NAD+ by complex II in the ETC. Copper inhibits NAD+-dependent enzymes.124
Adenosine triphosphate (ATP) provides phosphate groups for histone modifications.85 Additionally, ATP is a substrate for many reactions, including generation of the universal methyl donor SAM.126 Patients with WD experience significant reductions in enzyme activity of the oxidative phosphorylation complexes I–IV and might thus have a lower capacity to produce ATP. 127
Reactive oxygen species (ROS) are by-products of mitochondrial metabolism.ROS can induce posttranslational modifications of proteins through several mechanisms, including thiol oxidation, thereby altering the conformation, enzymatic activity, and location of proteins.128 At normal concentrations, ROS are considered to be signaling molecules,129 whereas they induce DNA, lipid, and protein oxidation; membrane destruction; and depletion of NAD+ and ATP at higher concentrations.130
8. Modifiers of epigenetic regulation: Diet, exercise, stress, toxins, and sex
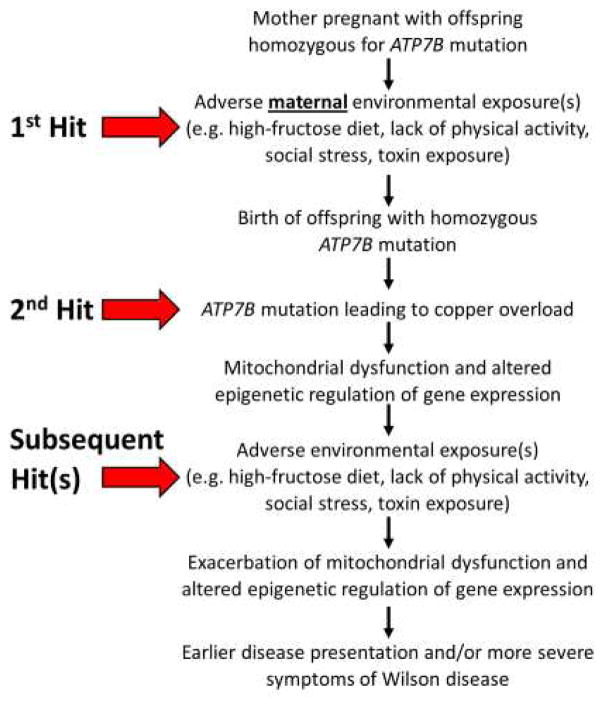
Epigenetic changes that are caused by environmental factors (e.g., diet, exercise, stress, and toxins) can blunt or exacerbate those that are induced by copper accumulation in WD, thus altering WD presentation and progression. Modifying these lifestyle factors can provide an opportunity to mitigate copper-induced changes to the epigenome in patients with WD. The multi-hit hypothesis of how copper and lifestyle factors combine to modify the presentation of WD is depicted in Fig 1. The evidence on how each of these factors impacts the epigenome is described below.
Fig. 1.
The multi-hit hypothesis of how copper and lifestyle factors combine to modify the presentation of WD
8.1. Diet
Diet influences the epigenome through such mechanisms as direct provision of methyl donors,131, 132 differences in macronutrient profile (e.g., high fat, high fructose), 1, 3, 133 and diet-derived microRNAs.134
Dietary methyl donors include vitamins (e.g.choline, folate, riboflavin, vitamin B6, vitamin B12) and amino acids (e.g.methionine, cysteine, serine, glycine).132, 135 All vitamins and methionine are considered to be essential nutrients, because the body does not produce them in adequate amounts, thus requiring them to be obtained from the diet.136 Inadequate intake of methyl donors, including choline and folate, alters DNA methylation and histone modifications.137, 138 Our group found that 24-week-old tx-j mice have hypomethylated DNA compared with wild-type mice and that DNA methylation in tx-j mice is restored to wild-type levels with 4 weeks of betaine supplementation.104 These results have not been confirmed in humans, due to the lack of studies on DNA methylation status in patients with WD.
Diets that contain excess fat or sugar have also been shown to change the epigenome. 1,3, 133 Skeletal muscle DNA methylation was assessed in 21 healthy men before and after 5 days of high-fat overfeeding (50% extra calories, 60% of kcals from a variety of fats) in a randomized crossover study. The investigators detected DNA methylation changes in >6500 genes, most of which involved genes that were related to inflammation, reproduction, and cancer.1 A study in 8-month-old male mice examined the effects of a fast food diet alone and in combination with exercise, reporting several alterations in hepatic DNA methylation and transcription after 4 months of treatment.3 The main findings were that the fastfood diet modulated lipid metabolism and that exercise prevented fast food diet-induced epigenomic changes in critical regulatory regions. These findings have implications for WD, because lipid metabolism is altered in WD patients and animal models. 139, 140
Studies in rodents have shown that high fructose consumption also affects the liver epigenome.133, 141 In rats that were provided with 20% fructose in their drinking water for 14weeks, mtDNA was hypomethylated and the promoter regions of PPARα and CTP1A were hypermethylated.133,141 We have observed downregulation of hepatic Pparα gene and protein expression in tx-j versus wild-type mice—prompting us to consider whether a high-fructose diet in WD will downregulate genes further, including Pparα.104 High-fat-, high-sugar-induced changes to the epigenome have been reversed in rodents after their return to a chow diet.142 The exact mechanisms of these diet-driven epigenetic changes are unknown, but potential mechanismsinclude fructose and a high-fat diet induced ROS production and diet-induced changes in the production and utilization of mitochondrial metabolites that are necessary for epigenetic regulation.143–146
Preliminary evidence of the presence of microRNAs in food is beginning to emerge, although it is unknown to what degree they govern mammalian gene expression.134 One study found significant amounts of milk-derived microRNAs in human blood after consumption and these microRNAs altered gene expression in kidney cell cultures and mouse liver.147
8.2. Exercise
Physical activity alters the epigenome in humans and animal models.2,3 One study subjected sedentary middle-aged men to 1 hour of exercise twice per week for 6 months and identified >2800 genes in skeletal muscle and >7600 genes in adipose tissue that were differentially methylated compared with baseline.2 After thisexercise-based intervention, most genes in the adipose tissue were hypermethylated, whereas most skeletal muscle genes were hypomethylated—indicating epigenetically regulated interorgan crosstalk. This study did not examine changes in hepatic DNA methylation or gene expression, but several animal studies suggest that there are exercise-induced changes in hepatic epigenetic regulation that are relevant to the WD phenotype (e.g., lipolysis, mitochondrial function, inflammation).
One such study determined the effects of exercise on the mouse liver epigenome and transcriptome by providing a chow diet and a running wheel to 8-month-old mice for 4 weeks and comparing them with animals without access to the wheel; the former underwent alterations in histone methylation, DNA hypermethylation, and upregulation of genes that mediate carboxylic/fatty acid biosynthesis and acute inflammatory responsesin the liver.3 Another study in mice found that 6 weeks of exercise increased hepatic levels of NAD-dependent deacetylase sirtuin-1 and cytochrome c oxidase subunit 4 isoform 1 and promoted mitochondrial remodeling.148 The mechanisms behind these exercise-induced epigenetic changes have not been determined but are likely to involve changes to the mitochondria, including greater ETC activity and mitochondrial turnover.149–150
8.3. Stress
Social stressors and stress reduction techniques, including meditation,4,151 have epigenetic effects. Mice that have been provided with environmental enrichment for 4 weeks (8 mice housed per cage, containing tunnels and a running wheel) show increased mRNA levels and altered histone trimethylation of the neurotrophic factor Bdnf compared with control mice (4 housed per cage without tunnels or running wheel).152 In contrast, mice that were exposed to an aggressor mouse for 10 minutes per day for 10 days showed downregulated hippocampal Bdnf mRNA and experienced greater histone methylation at the promoter region.153 BDNF levels might be relevant to WD, because patients with Parkinson disease—some signs of which can resemble the movement impairments that are observed in WD—have lower BDNF levels in the substantianigra,154 and serum levels of BDNF correlate negatively with motor function.155 Furthermore, certain SNPs in BDNF are more frequent in symptomatic WD patients versus healthy controls and asymptomatic WD patients.83 In humans, meditation can increase BDNF levels,156 alters histone modifications, and downregulates proinflammatory genes.151
8.4. Toxins
Mitochondria can be damaged by natural and synthetic chemicals, including environmental toxins and pharmaceutical drugs.157 Charged and lipophilic toxins are disproportionately drawn to mitochondria due to the presence of an electrochemical gradient and high lipid content.157 There are several types of mitochondrial damage, such as lower mtDNA integrity, inhibition of mitochondrial enzymes, reduced membrane potential, impaired Ca2+ transport, and promotion of apoptosis. These events can alter the production of metabolites that are required for epigenetic regulation.157 Bisphenol A, commonly found in plastic, stimulates hepatic fat accumulation by modulating DNA methylation patterns.158 Avoidance of these chemicals might be useful in the treatment of WD.
8.5. Maternal exposure
The environmental factors that influence epigenetic regulation described above also impact the developing fetus inutero. Maternal diet, 159 exercise,160 stress,4 and toxin exposure during pregnancy can change the epigenetic profile of offspring, potentially resulting in longterm effects that ultimately determine disease risk later in life.161–162 Intensive research is underway to understand the developmental origins of health and disease.163 For example, one study has investigated the effects of a maternal high-fat diet on offspring,164 finding that adult offspring from dams that are fed a high-fat diet during gestation and lactation accumulate more hepatic fat; develop deficiencies in mitochondrial enzyme complexes I–IV; and upregulate hepatic genes that are related to lipogenesis, inflammation, and oxidative stress despite being given a low-fat diet after weaning. 164 Other studies have examined the effects of maternal choline deficiency and supplementation, reporting that 1 week of maternal choline deficiency or supplementation in rats alters DNA and histone methylation in the fetal liver and brain.165, 166 Our group has observed differences in hepatic gene expression in fetal tx-j versus wild-type mice, indicating that fetal development is affected by maternal copper status.103
8.6. Sex
Sex differences have been observed in WD, wherein women tend to present more frequently with acute liver failure and neurological involvement is more common in men.13,167 This sexual dimorphism has been attributed to hormonal factors and differences in hepatic iron accumulation over time.167 However, a recent study by our group indicates that epigenetic factors contribute to the disparate presentation between sexes—in adult tx-j livers, transcript levels of genes that are related to mitochondrial function (Ndufab1, Ndufb5, Sdha, Cox5a, and Atp5j) were upregulated in female compared with male mice in response to choline and D-penicillamine alone and in combination. 103 Similarly, DNA methylation mechanisms differ between sexes.168
9. Multifaceted treatment approach
Copper-chelating agents are the main treatment option for patients with WD, but they have side effects, and the responsiveness to this therapy varies,16, 169 necessitating adjunctive or alternative therapies. Lifestyle modifications that support mitochondrial function might be valuable in the treatment of WD, because such patients develop mitochondrial structural and functional abnormalities.112,127 Improvements in or delayed impairment of mitochondrial function in WD can influence epigenetic regulation, as described in previous sections. Possible adjunctive treatment options for WD and their rationale are discussed below.
Diet
Consuming a diet that is low in sugar—especially fructose—might be more critical for patients with WD compared with the healthy population. Fructose contributes to fat accumulation in the liver and might exacerbate steatosis,170 which is commonly found in WD.170,22 The type and amount of fat consumption should also be considerations for subjects with WD. Consumption of oxidized fats can further elevate liver enzymes in patients with WD— oxidized palm oil increases plasma ALT and AST levels in rats after 18 weeks of feeding.7,171 Dietary fat composition influences mitochondrial lipid composition and function.172, 173 Brief fasting might also be useful in the treatment of WD, because fasting increases mitophagy and thus might be an effective strategy to eliminate defective mitochondria. 174 Common fasting practices include the 5:2 plan (feed for 5 days and restrict calories to <70% for 2 days each week), alternate day fasting, and restricting eating to a 12-hour window each day.175 However, few studies have examined the metabolic effects of fasting in humans; therefore, no recommendations regarding WD and fasting can be made with confidence at this time.
Dietary supplements
Supplements that support mitochondrial function—including α-lipoic acid, ubiquinol, magnesium, and B vitamins—might be useful adjuvant therapies for WD. Alpha-lipoic acid is a cofactor for 2-oxoglutarate dehydrogenase and an antioxidant.176 It has neuroprotective effects, rendering it potentially efficacious in Parkinson disease.177 One study found that the addition of the mitochondrial enzyme cofactors dihydrolipoic acid, thiamine, and pyruvate reduced copper-induced neuronal death in murine neocortical cells and that administration of thiamine to Long-Evans Cinnamon rats (a rodent model for hepatic copper accumulation) significantly increases their lifespan.178 Ames et al. 179 have written an excellent review on the use of nutrients to support mitochondrial function.
Exercise
Increased physical activity can aid in mitochondrial remodeling in the liver.148 Exercise might be particularly beneficial for WD patients with neurological manifestations, because physical activity has been shown to improve Parkinson disease symptoms; however, it is unknown what type of physical activity provides the greatest benefits.180
Stress
Stress reduction through meditation, visualization, or yoga can aid in reducing neuroinflammation and may be useful for treating patients with the neurological phenotype of WD.151, 181
Toxins
Avoidance of toxins—especially those that impair mitochondrial function—might be warranted for patients with WD. Common toxins, including those that are found in pesticides and plastics,158,182 stimulate hepatic fat accumulation and disrupt mitochondrial function.157 Copper-induced mitochondrial damage and hepatic fat accumulation might be exacerbated in the presence of chemicals with known mitochondrial toxicities.
10. Conclusion
WD is a complex illness with few treatment options. Studies are needed to evaluate the efficacy of lifestyle modifications, including diet, exercise, stress reduction, and toxin avoidance—alone and in combination with current treatments. Despite the lack of direct evidence that these lifestyle modifications improve WD, there are findings that indicate that they alter the epigenetic regulation of gene expression and mitochondrial function—both of which are disrupted in WD. Determining the mechanisms of the varied clinical presentation of WD has been challenging, but new high-throughput omics-based methods might help increase our understanding of this complex disease. Some of the studies in this review have used such technologies as metabolomics, transcriptomics, and genome-wide DNA methylation to identify additional pathways that are impacted in WD, potential therapeutic targets, and to determine treatment efficacy.
Acknowledgments
This review was supported by grant numbers R03DK099427 and R01DK104770 to V.M. from the National Institute of Diabetes and Digestive and Kidney Diseases(USA).
Footnotes
Conflict of interest: The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jacobsen SC, Brons C, Bork-Jensen J, et al. Effects of short-term high-fat overfeeding on genome-wide DNA methylation in the skeletal muscle of healthy young men. Diabetologia. 2012;55:3341–3349. doi: 10.1007/s00125-012-2717-8. [DOI] [PubMed] [Google Scholar]
- 2.Ling C, Ronn T. Epigenetic adaptation to regular exercise in humans. Drug Discov Today. 2014;19:1015–1018. doi: 10.1016/j.drudis.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Zhou D, Hlady RA, Schafer MJ, et al. High Fat Diet and Exercise Lead to a Disrupted and Pathogenic DNA Methylome in Mouse Liver. Epigenetics. 2017;12:55–69. doi: 10.1080/15592294.2016.1261239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gudsnuk K, Champagne FA. Epigenetic Influence of Stress and the Social Environment. ILAR J. 2012;53:279–288. doi: 10.1093/ilar.53.3-4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hou L, Zhang X, Wang D, Baccarelli A. Environmental chemical exposures and human epigenetics. Int J Epidemiol. 2012;41:79–105. doi: 10.1093/ije/dyr154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanzi RE, Petrukhin K, Chernov I, et al. The Wilson disease gene is a copper transporting ATPase with homology to the Menkes disease gene. Nat Genet. 1993;5:344–350. doi: 10.1038/ng1293-344. [DOI] [PubMed] [Google Scholar]
- 7.Roberts EA, Schilsky ML American Association for Study of Liver Diseases (AASLD) Diagnosis and treatment of Wilson disease: an update. Hepatology. 2008;47:2089–20111. doi: 10.1002/hep.22261. [DOI] [PubMed] [Google Scholar]
- 8.Stromeyer FW, Ishak KG. Histology of the liver in Wilson's disease: a study of 34 cases. Am J Clin Pathol. 1980;73:12–24. doi: 10.1093/ajcp/73.1.12. [DOI] [PubMed] [Google Scholar]
- 9.Taly AB, Meenakshi-Sundaram S, Sinha S, Swamy HS, Arunodaya GR. Wilson disease: description of 282 patients evaluated over 3 decades. Medicine (Baltimore) 2007;86:112–121. doi: 10.1097/MD.0b013e318045a00e. [DOI] [PubMed] [Google Scholar]
- 10.Svetel M, Potrebić A, Pekmezović T, et al. Neuropsychiatric aspects of treated Wilson's disease. Parkinsonism Relat Disord. 2009;15:772–775. doi: 10.1016/j.parkreldis.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Kim JW, Kim JH, Seo JK, et al. Genetically confirmed Wilson disease in a 9-month old boy with elevations of aminotransferases. World J Hepatol. 2013;5:156–159. doi: 10.4254/wjh.v5.i3.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ala A, Borjigin J, Rochwarger A, Schilsky M. Wilson disease in septuagenarian siblings: Raising the bar for diagnosis. Hepatology. 2005;41:668–670. doi: 10.1002/hep.20601. [DOI] [PubMed] [Google Scholar]
- 13.Weiss KH, Gotthardt DN, Klemm D, et al. Zinc monotherapy is not as effective as chelating agents in treatment of Wilson disease. Gastroenterology. 2011;140:1189–1198. e1. doi: 10.1053/j.gastro.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 14.Medici V, Trevisan CP, D'Inca R, et al. Diagnosis and management of Wilson's disease: results of a single center experience. J Clin Gastroenterol. 2006;40:936–941. doi: 10.1097/01.mcg.0000225670.91722.59. [DOI] [PubMed] [Google Scholar]
- 15.Yuzbasiyan-Gurkan V, Grider A, Nostrant T, Cousins RJ, Brewer GJ. Treatment of Wilson's disease with zinc: X. Intestinal metallothionein induction. J Lab Clin Med. 1992;120:380–386. [PubMed] [Google Scholar]
- 16.Weiss KH, Thurik F, Gotthardt DN, et al. Efficacy and safety of oral chelators in treatment of patients with Wilson disease. Clin Gastroenterol Hepatol. 2013;11:1028–1035. e1–e2. doi: 10.1016/j.cgh.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 17.Brewer GJ, Askari F, Dick RB, et al. Treatment of Wilson's disease with tetrathiomolybdate: V. Control of free copper by tetrathiomolybdate and a comparison with trientine. Transl Res. 2009;154:70–77. doi: 10.1016/j.trsl.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Sinha S, Taly AB, Prashanth LK, Ravishankar S, Arunodaya GR, Vasudev MK. Sequential MRI changes in Wilson's disease with de-coppering therapy: a study of 50 patients. Br J Radiol. 2007;80:744–749. doi: 10.1259/bjr/48911350. [DOI] [PubMed] [Google Scholar]
- 19.Coffey AJ, Durkie M, Hague S, et al. A genetic study of Wilson’s disease in the United Kingdom. Brain. 2013;136:1476–1487. doi: 10.1093/brain/awt035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheinberg IH, Sternlieb I. Wilson's disease. In: Smith LH, editor. Major Problems in International Medicine. Philadelphia, NY: W.B. Saunders Company; 1984. pp. 23–25. [Google Scholar]
- 21.Park RH, McCabe P, Fell GS, Russell RI. Wilson's disease in Scotland. Gut. 1991;32:1541–1545. doi: 10.1136/gut.32.12.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stattermayer AF, Traussnigg S, Dienes HP, et al. Hepatic steatosis in Wilson disease--Role of copper and PNPLA3 mutations. J Hepatol. 2015;63:156–163. doi: 10.1016/j.jhep.2015.01.034. [DOI] [PubMed] [Google Scholar]
- 23.Bandmann O, Weiss KH, Kaler SG. Wilson's disease and other neurological copper disorders. Lancet Neurol. 2015;14:103–113. doi: 10.1016/S1474-4422(14)70190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lutsenko S. Modifying factors and phenotypic diversity in Wilson's disease. Ann N Y Acad Sci. 2014;1315:56–63. doi: 10.1111/nyas.12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huster D, Kuhne A, Bhattacharjee A, et al. Diverse functional properties of Wilson disease ATP7B variants. Gastroenterology. 2012;142:947–956. e5. doi: 10.1053/j.gastro.2011.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braiterman LT, Murthy A, Jayakanthan S, et al. Distinct phenotype of a Wilson disease mutation reveals a novel trafficking determinant in the copper transporter ATP7B. Proc Natl Acad Sci U S A. 2014;111:E1364–E1373. doi: 10.1073/pnas.1314161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caca K, Ferenci P, Kuhn HJ, et al. High prevalence of the H1069Q mutation in East German patients with Wilson disease: rapid detection of mutations by limited sequencing and phenotype-genotype analysis. J Hepatol. 2001;35:575–581. doi: 10.1016/s0168-8278(01)00219-7. [DOI] [PubMed] [Google Scholar]
- 28.Tomić A, Dobricić V, Novaković I, et al. Mutational analysis of ATP7B gene and the genotype-phenotype correlation in patients with Wilson's disease in Serbia. Vojnosanit Pregl. 2013;70:457–462. doi: 10.2298/vsp1305457t. [DOI] [PubMed] [Google Scholar]
- 29.Mihaylova V, Todorov T, Jelev H, et al. Neurological symptoms, genotype-phenotype correlations and ethnic-specific differences in Bulgarian patients with Wilson disease. Neurologist. 2012;18:184–189. doi: 10.1097/NRL.0b013e31825cf3b7. [DOI] [PubMed] [Google Scholar]
- 30.Nicastro E, Loudianos G, Zancan L, et al. Genotype-phenotype correlation in Italian children with Wilson's disease. J Hepatol. 2009;50:555–561. doi: 10.1016/j.jhep.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 31.Gromadzka G, Schmidt HH, Genschel J, et al. p.H1069Q mutation in ATP7B and biochemical parameters of copper metabolism and clinical manifestation of Wilson's disease. Mov Disord. 2006;21:245–248. doi: 10.1002/mds.20671. [DOI] [PubMed] [Google Scholar]
- 32.Stapelbroek JM, Bollen CW, van Amstel JK, et al. The H1069Q mutation in ATP7B is associated with late and neurologic presentation in Wilson disease: results of a meta-analysis. J Hepatol. 2004;41:758–763. doi: 10.1016/j.jhep.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 33.Merle U, Weiss KH, Eisenbach C, Tuma S, Ferenci P, Stremmel W. Truncating mutations in the Wilson disease gene ATP7B are associated with very low serum ceruloplasmin oxidase activity and an early onset of Wilson disease. BMC Gastroenterol. 2010;10:8. doi: 10.1186/1471-230X-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gromadzka G, Schmidt HH, Genschel J, et al. Frameshift and nonsense mutations in the gene for ATPase7B are associated with severe impairment of copper metabolism and with an early clinical manifestation of Wilson's disease. Clin Genet. 2005;68:524–532. doi: 10.1111/j.1399-0004.2005.00528.x. [DOI] [PubMed] [Google Scholar]
- 35.Panagiotakaki E, Tzetis M, Manolaki N, et al. Genotype–phenotype correlations for a wide spectrum of mutations in the Wilson disease gene (ATP7B) Am J Med Genet A. 2004;131:168–173. doi: 10.1002/ajmg.a.30345. [DOI] [PubMed] [Google Scholar]
- 36.Nicastro E, Loudianos G, Zancan L, et al. Genotype–phenotype correlation in Italian children with Wilson’s disease. J Hepatol. 2009;50:555–561. doi: 10.1016/j.jhep.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 37.Okada T, Shiono Y, Hayashi H, et al. Mutational analysis of ATP7B and genotype-phenotype correlation in Japanese with Wilson's disease. Hum Mutat. 2000;15:454–462. doi: 10.1002/(SICI)1098-1004(200005)15:5<454::AID-HUMU7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 38.Palsson R, Jonasson JG, Kristjansson M, et al. Genotype-phenotype interactions in Wilson's disease: insight from an Icelandic mutation. Eur J Gastroenterol Hepatol. 2001;13:433–436. doi: 10.1097/00042737-200104000-00023. [DOI] [PubMed] [Google Scholar]
- 39.Nadeau JH. Modifier genes in mice and humans. Nat Rev Genet. 2001;2:165–174. doi: 10.1038/35056009. [DOI] [PubMed] [Google Scholar]
- 40.Bandmann O, Weiss KH, Kaler SG. Wilson's disease and other neurological copper disorders. Lancet Neurol. 2015;14:103–113. doi: 10.1016/S1474-4422(14)70190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tao TY, Liu F, Klomp L, Wijmenga C, Gitlin JD. The copper toxicosis gene product Murr1 directly interacts with the Wilson disease protein. J Biol Chem. 2003;278:41593–41596. doi: 10.1074/jbc.C300391200. [DOI] [PubMed] [Google Scholar]
- 42.Hardy RM, Stevens J, Band-Stowe CM. Chronic progressive hepatitis in Bedlington terriers associated with elevated liver copper concentrations. Minn Vet. 1975;15:13–24. [Google Scholar]
- 43.Stuehler B, Reichert J, Stremmel W, Schaefer M. Analysis of the human homologue of the canine copper toxicosis gene MURR1 in Wilson disease patients. J Mol Med (Berl) 2004;82:629–634. doi: 10.1007/s00109-004-0557-9. [DOI] [PubMed] [Google Scholar]
- 44.Bost M, Piguet-Lacroix G, Parant F, Wilson CM. Molecular analysis of Wilson patients: direct sequencing and MLPA analysis in the ATP7B gene and Atox1 and COMMD1 gene analysis. J Trace Elem Med Biol. 2012;26:97–101. doi: 10.1016/j.jtemb.2012.04.024. [DOI] [PubMed] [Google Scholar]
- 45.Weiss KH, Merle U, Schaefer M, Ferenci P, Fullekrug J, Stremmel W. Copper toxicosis gene MURR1 is not changed in Wilson disease patients with normal blood ceruloplasmin levels. World J Gastroenterol. 2006;12:2239–2242. doi: 10.3748/wjg.v12.i14.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DiDonato M, Hsu HF, Narindrasorasak S, Que L, Jr, Sarkar B. Copper-induced conformational changes in the N-terminal domain of the Wilson disease copper-transporting ATPase. Biochemistry. 2000;39:1890–1896. doi: 10.1021/bi992222j. [DOI] [PubMed] [Google Scholar]
- 47.Vanderwerf SM, Cooper MJ, Stetsenko IV, Lutsenko S. Copper specifically regulates intracellular phosphorylation of the Wilson's disease protein, a human copper-transporting ATPase. J Biol Chem. 2001;276:36289–36294. doi: 10.1074/jbc.M102055200. [DOI] [PubMed] [Google Scholar]
- 48.Itoh S, Kim HW, Nakagawa O, et al. Novel role of antioxidant-1 (Atox1) as a copper-dependent Transcription factor involved in cell proliferation. J Biol Chem. 2008;283:9157–9167. doi: 10.1074/jbc.M709463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simon I, Schaefer M, Reichert J, Stremmel W. Analysis of the human Atox 1 homologue in Wilson patients. World J Gastroenterol. 2008;14:2383–2387. doi: 10.3748/wjg.14.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mufti AR, Burstein E, Csomos RA, et al. XIAP Is a copper binding protein deregulated in Wilson's disease and other copper toxicosis disorders. Mol Cell. 2006;21:775–785. doi: 10.1016/j.molcel.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 51.Burstein E, Ganesh L, Dick RD, et al. A novel role for XIAP in copper homeostasis through regulation of MURR1. EMBO J. 2004;23:244–254. doi: 10.1038/sj.emboj.7600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salzer U, Hagena T, Webster DB, et al. Sequence analysis of BIRC4/XIAP in male patients with common variable immunodeficiency. Int Arch Allergy Immunol. 2008;147:147–151. doi: 10.1159/000135702. [DOI] [PubMed] [Google Scholar]
- 53.Serre D, Gurd S, Ge B, et al. Differential allelic expression in the human genome: a robust approach to identify genetic and epigenetic cis-acting mechanisms regulating gene expression. PLoS Genet. 2008;4:e1000006. doi: 10.1371/journal.pgen.1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weiss KH, Runz H, Noe B, et al. Genetic analysis of BIRC4/XIAP as a putative modifier gene of Wilson disease. J Inherit Metab Dis. 2010;(Suppl 3):S233–S240. doi: 10.1007/s10545-010-9123-5. [DOI] [PubMed] [Google Scholar]
- 55.He S, McPhaul C, Li JZ, et al. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem. 2010;285:6706–6715. doi: 10.1074/jbc.M109.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dongiovanni P, Donati B, Fares R, et al. PNPLA3 I148M polymorphism and progressive liver disease. World J Gastroenterol. 2013;19:6969–6978. doi: 10.3748/wjg.v19.i41.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wardell MR, Suckling PA, Janus ED. Genetic variation in human apolipoprotein E. J Lipid Res. 1982;23:1174–1182. [PubMed] [Google Scholar]
- 58.Squitti R, Polimanti R, Siotto M, et al. ATP7B variants as modulators of copper dyshomeostasis in Alzheimer’s disease. Neuromolecular Med. 2013;15:515–522. doi: 10.1007/s12017-013-8237-y. [DOI] [PubMed] [Google Scholar]
- 59.Schiefermeier M, Kollegger H, Madl C, et al. The impact of apolipoprotein E genotypes on age at onset of symptoms and phenotypic expression in Wilson's disease. Brain. 2000;123:585–590. doi: 10.1093/brain/123.3.585. [DOI] [PubMed] [Google Scholar]
- 60.Litwin T, Gromadzka G, Członkowska A. Apolipoprotein E gene (APOE) genotype in Wilson's disease: impact on clinical presentation. Parkinsonism Relat Disord. 2012;18:367–369. doi: 10.1016/j.parkreldis.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 61.Kocabay G, Tutuncu Y, Yilmaz H, Demir K. Impact of apolipoprotein E genotypes on phenotypic expression in Turkish patients with Wilson's disease. Scand J Gastroenterol. 2009;44:1270–1271. doi: 10.1080/00365520903225908. [DOI] [PubMed] [Google Scholar]
- 62.Gu YH, Kodama H, Du SL. Apolipoprotein E genotype analysis in Chinese Han ethnic children with Wilson's disease, with a concentration on those homozygous for R778L. Brain Dev. 2005;27:551–553. doi: 10.1016/j.braindev.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 63.Lebron JA, West AP, Jr, Bjorkman PJ. The hemochromatosis protein HFE competes with transferrin for binding to the transferrin receptor. J Mol Biol. 1999;294:239–245. doi: 10.1006/jmbi.1999.3252. [DOI] [PubMed] [Google Scholar]
- 64.Hayashi H, Yano M, Fujita Y, Wakusawa S. Compound overload of copper and iron in patients with Wilson's disease. Med Mol Morphol. 2006;39:121–126. doi: 10.1007/s00795-006-0326-7. [DOI] [PubMed] [Google Scholar]
- 65.Kato J, Kohgo Y, Sugawara N, et al. Abnormal hepatic iron accumulation in LEC rats. Jpn J Cancer Res. 1993;84:219–222. doi: 10.1111/j.1349-7006.1993.tb02859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Le A, Shibata NM, French SW, et al. Characterization of timed changes in hepatic copper concentrations, methionine metabolism, gene expression, and global DNA methylation in the Jackson toxic milk mouse model of Wilson Disease. Int J of Mol Sci. 2014;15:8004–8023. doi: 10.3390/ijms15058004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hafkemeyer P, Schupp M, Storch M, Gerok W, Haussinger D. Excessive iron storage in a patient with Wilson's disease. Clin Investig. 1994;72:134–136. doi: 10.1007/BF00184590. [DOI] [PubMed] [Google Scholar]
- 68.Walshe JM, Cox DW. Effect of treatment of Wilson's disease on natural history of haemochromatosis. Lancet. 1998;352:112–113. doi: 10.1016/S0140-6736(98)85017-4. [DOI] [PubMed] [Google Scholar]
- 69.Sorbello O, Sini M, Civolani A, Demelia L. HFE gene mutations and Wilson's disease in Sardinia. Dig Liver Dis. 2010;42:216–219. doi: 10.1016/j.dld.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 70.Erhardt A, Hoffmann A, Hefter H, Haussinger D. HFE gene mutations and iron metabolism in Wilson's disease. Liver. 2002;22:474–478. doi: 10.1034/j.1600-0676.2002.01732.x. [DOI] [PubMed] [Google Scholar]
- 71.Pfeiffenberger J, Gotthardt DN, Herrmann T, et al. Iron metabolism and the role of HFE gene polymorphisms in Wilson disease. Liver Int. 2012;32:165–170. doi: 10.1111/j.1478-3231.2011.02661.x. [DOI] [PubMed] [Google Scholar]
- 72.Gromadzka G, Rudnicka M, Chabik G, Przybyłkowski A, Członkowska A. Genetic variability in the methylenetetrahydrofolate reductase gene (MTHFR) affects clinical expression of Wilson's disease. J Hepatol. 2011;55:913–919. doi: 10.1016/j.jhep.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 73.White AR, Huang X, Jobling MF, et al. Homocysteine potentiates copper- and amyloid beta peptide-mediated toxicity in primary neuronal cultures: possible risk factors in the Alzheimer's-type neurodegenerative pathways. J Neurochem. 2001;76:1509–1520. doi: 10.1046/j.1471-4159.2001.00178.x. [DOI] [PubMed] [Google Scholar]
- 74.Linnebank M, Lutz H, Jarre E, et al. Binding of copper is a mechanism of homocysteine toxicity leading to COX deficiency and apoptosis in primary neurons, PC12 and SHSY-5Y cells. Neurobiol Dis. 2006;23:725–730. doi: 10.1016/j.nbd.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 75.Brown DR, Qin K, Herms JW, et al. The cellular prion protein binds copper in vivo. Nature. 1997;390:684–687. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]
- 76.Ford MJ, Burton LJ, Morris RJ, Hall SM. Selective expression of prion protein in peripheral tissues of the adult mouse. Neuroscience. 2002;113:177–192. doi: 10.1016/s0306-4522(02)00155-0. [DOI] [PubMed] [Google Scholar]
- 77.Gasperini L, Meneghetti E, Pastore B, Benetti F, Legname G. Prion protein and copper cooperatively protect neurons by modulating NMDA receptor through S-nitrosylation. Antioxid Redox Signal. 2015;22:772–784. doi: 10.1089/ars.2014.6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Merle U, Stremmel W, Gessner R. Influence of homozygosity for methionine at codon 129 of the human prion gene on the onset of neurological and hepatic symptoms in Wilson disease. Arch Neurol. 2006;63:982–985. doi: 10.1001/archneur.63.7.982. [DOI] [PubMed] [Google Scholar]
- 79.Brady CA, Attardi LD. p53 at a glance. J Cell Sci. 2010;123:2527–2532. doi: 10.1242/jcs.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hussain SP, Raja K, Amstad PA, et al. Increased p53 mutation load in nontumorous human liver of Wilson disease and hemochromatosis: Oxyradical overload diseases. Proc Natl Acad Sci U S A. 2000;97:12770–12775. doi: 10.1073/pnas.220416097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nair J, Carmichael PL, Fernando RC, Phillips DH, Strain AJ, Bartsch H. Lipid peroxidation-induced etheno-DNA adducts in the liver of patients with the genetic metal storage disorders Wilson's disease and primary hemochromatosis. Cancer Epidemiol Biomarkers Prev. 1998;7:435–440. [PubMed] [Google Scholar]
- 82.Binder DK, Scharfman HE. Brain-derived Neurotrophic Factor. Growth factors. 2004;22:123–131. doi: 10.1080/08977190410001723308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mirowska-Guzel D, Litwin T, Gromadzka G, Czlonkowski A, Czlonkowska A. Influence of BDNF polymorphisms on Wilson’s disease susceptibility and clinical course. Metab Brain Dis. 2013;28:447–453. doi: 10.1007/s11011-013-9399-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 85.Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol. 2013;20:259–266. doi: 10.1038/nsmb.2470. [DOI] [PubMed] [Google Scholar]
- 86.Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat struct mol biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 88.Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- 89.Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci U S A. 2011;108:3630–3635. doi: 10.1073/pnas.1012311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aran D, Toperoff G, Rosenberg M, Hellman A. Replication timing-related and gene body-specific methylation of active human genes. Hum Mol Genet. 2011;20:670–680. doi: 10.1093/hmg/ddq513. [DOI] [PubMed] [Google Scholar]
- 91.Bestor TH. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 1992;11:2611–2617. doi: 10.1002/j.1460-2075.1992.tb05326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 93.Niculescu MD, Zeisel SH. Diet, methyl donors and DNA methylation: interactions between dietary folate, methionine and choline. J Nutr. 2002;132:2333S–2335S. doi: 10.1093/jn/132.8.2333S. [DOI] [PubMed] [Google Scholar]
- 94.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 96.Sato F, Tsuchiya S, Meltzer SJ, Shimizu K. MicroRNAs and epigenetics. FEBS J. 2011;278:1598–1609. doi: 10.1111/j.1742-4658.2011.08089.x. [DOI] [PubMed] [Google Scholar]
- 97.Kegley KM, Sellers MA, Ferber MJ, Johnson MW, Joelson DW, Shrestha R. Fulminant Wilson's disease requiring liver transplantation in one monozygotic twin despite identical genetic mutation. Am J Transplant. 2010;10:1325–1329. doi: 10.1111/j.1600-6143.2010.03071.x. [DOI] [PubMed] [Google Scholar]
- 98.Członkowska A, Gromadzka G, Chabik G. Monozygotic female twins discordant for phenotype of Wilson's disease. Mov Disord. 2009;24:1066–1069. doi: 10.1002/mds.22474. [DOI] [PubMed] [Google Scholar]
- 99.Senzolo M, Loreno M, Fagiuoli S, et al. Different neurological outcome of liver transplantation for Wilson's disease in two homozygotic twins. Clin Neurol Neurosurg. 2007;109:71–75. doi: 10.1016/j.clineuro.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 100.Santhosh S, Shaji RV, Eapen CE, et al. Genotype phenotype correlation in Wilson’s disease within families-a report on four south Indian families. World J Gastroenterol. 2008;14:4672–4676. doi: 10.3748/wjg.14.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chabik G, Litwin T, Członkowska A. Concordance rates of Wilson’s disease phenotype among siblings. J Inherit Metab Dis. 2014;37:131–135. doi: 10.1007/s10545-013-9625-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Coronado V, Nanji M, Cox DW. The Jackson toxic milk mouse as a model for copper loading. Mamm Genome. 2001;12:793–795. doi: 10.1007/s00335-001-3021-y. [DOI] [PubMed] [Google Scholar]
- 103.Medici V, Kieffer DA, Shibata NM, et al. Wilson Disease: Epigenetic effects of choline supplementation on phenotype and clinical course in a mouse model. Epigenetics. 2016;11:804–818. doi: 10.1080/15592294.2016.1231289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Medici V, Shibata NM, Kharbanda KK, et al. Wilson disease: changes in methionine metabolism and inflammation affect global DNA methylation in early liver disease. Hepatology. 2013;57:555–565. doi: 10.1002/hep.26047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Medici V, Shibata NM, Kharbanda KK, et al. Maternal choline modifies fetal liver copper, gene expression, DNA methylation, and neonatal growth in the tx-j mouse model of Wilson disease. Epigenetics. 2014;9:286–296. doi: 10.4161/epi.27110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cantoni GL. The nature of the active methyl donor formed enzymatically from L-methionine and adenosinetriphosphate1,2. J Am Chem Soc. 1952;74:2942–2943. [Google Scholar]
- 107.Li M, Li Y, Chen J, et al. Copper ions inhibit S-adenosylhomocysteine hydrolase by causing dissociation of NAD+cofactor. Biochemistry. 2007;46:11451–11458. doi: 10.1021/bi700395d. [DOI] [PubMed] [Google Scholar]
- 108.Bethin KE, Cimato TR, Ettinger MJ. Copper binding to mouse liver S-adenosylhomocysteine hydrolase and the effects of copper on its levels. J Biol Chem. 1995;270:20703–20711. doi: 10.1074/jbc.270.35.20703. [DOI] [PubMed] [Google Scholar]
- 109.Ralle M, Huster D, Vogt S, et al. Wilson disease at a single cell level: intracellular copper trafficking activates compartment-specific responses in hepatocytes. J Biol Chem. 2010;285:30875–30883. doi: 10.1074/jbc.M110.114447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Aigner E, Strasser M, Haufe H, et al. A role for low hepatic copper concentrations in nonalcoholic Fatty liver disease. Am J Gastroenterol. 2010;105:1978–1985. doi: 10.1038/ajg.2010.170. [DOI] [PubMed] [Google Scholar]
- 111.Ernster L, Schatz G. Mitochondria: a historical review. J Cell Biol. 1981;91:227s–255s. doi: 10.1083/jcb.91.3.227s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sternlieb I. Mitochondrial and fatty changes in hepatocytes of patients with Wilson's disease. Gastroenterology. 1968;55:354–367. [PubMed] [Google Scholar]
- 113.Zischka H, Lichtmannegger J. Pathological mitochondrial copper overload in livers of Wilson's disease patients and related animal models. Ann N Y Acad Sci. 2014;1315:6–15. doi: 10.1111/nyas.12347. [DOI] [PubMed] [Google Scholar]
- 114.Terwel D, Loschmann YN, Schmidt HH, Scholer HR, Cantz T, Heneka MT. Neuroinflammatory and behavioural changes in the Atp7B mutant mouse model of Wilson’s disease. J Neurochem. 2011;118:105–112. doi: 10.1111/j.1471-4159.2011.07278.x. [DOI] [PubMed] [Google Scholar]
- 115.Przybyłkowski A, Gromadzka G, Wawer A, et al. Neurochemical and behavioral characteristics of toxic milk mice: an animal model of Wilson’s disease. Neurochem Res. 2013;38:2037–2045. doi: 10.1007/s11064-013-1111-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vogel FS, Kemper L. Biochemical reactions of copper within neural mitochondria, with consideration of the role of the metal in the pathogenesis of Wilson's disease. Lab Invest. 1963;12:171–179. [PubMed] [Google Scholar]
- 117.Zischka H, Lichtmannegger J, Schmitt S, et al. Liver mitochondrial membrane crosslinking and destruction in a rat model of Wilson disease. J Clin Invest. 2011;121:1508–1518. doi: 10.1172/JCI45401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zischka H, Lichtmannegger J. Pathological mitochondrial copper overload in livers of Wilson's disease patients and related animal models. Ann N Y Acad Sci. 2014;1315:6–15. doi: 10.1111/nyas.12347. [DOI] [PubMed] [Google Scholar]
- 119.Babu GR, Rao PV. Effect of copper sulphate on alpha-ketoglutarate metabolism in the digestive gland of the snail host, Lymnaealuteola. J Environ Pathol Toxicol Oncol. 1987;7:29–34. [PubMed] [Google Scholar]
- 120.Sheline CT, Choi EH, Kim-Han JS. Cofactors of mitochondrial enzymes attenuate copper-induced death in vitro and in vivo. Ann Neurol. 2002;52:195–204. doi: 10.1002/ana.10276. [DOI] [PubMed] [Google Scholar]
- 121.Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol Cell. 2006;23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 122.Walshe JM. Pyruvate metabolism in Wilson's disease. Clin Sci. 1961;20:197–203. [PubMed] [Google Scholar]
- 123.Salminen A, Kauppinen A, Hiltunen M, Kaarniranta K. Krebs cycle intermediates regulate DNA and histone methylation: epigenetic impact on the aging process. Ageing Res Rev. 2014;16:45–65. doi: 10.1016/j.arr.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 124.Heron P, Cousins K, Boyd C, Daya S. Paradoxical effects of copper and manganese on brain mitochondrial function. Life Sci. 2001;68:1575–1583. doi: 10.1016/s0024-3205(01)00948-1. [DOI] [PubMed] [Google Scholar]
- 125.Braunstein M, Rose AB, Holmes SG, et al. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 126.Mato JM, Alvarez L, Ortiz P, Pajares MA. S-adenosylmethionine synthesis: molecular mechanisms and clinical implications. Pharmacol Ther. 1997;73:265–280. doi: 10.1016/s0163-7258(96)00197-0. [DOI] [PubMed] [Google Scholar]
- 127.Gu M, Cooper JM, Butler P, et al. Oxidative-phosphorylation defects in liver of patients with Wilson's disease. Lancet. 2000;356:469–474. doi: 10.1016/s0140-6736(00)02556-3. [DOI] [PubMed] [Google Scholar]
- 128.Podrini C, Borghesan M, Greco A, Pazienza V, Mazzoccoli G, Vinciguerra M. Redox homeostasis and epigenetics in non-alcoholic fatty liver disease (NAFLD) Curr Pharm Des. 2013;19:2737–2746. doi: 10.2174/1381612811319150009. [DOI] [PubMed] [Google Scholar]
- 129.Finkel T. Signal transduction by reactive oxygen species. J Cell Biol. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zorov DB, Juhaszova M, Sollott SJ. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol Rev. 2014;94:909–950. doi: 10.1152/physrev.00026.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12:949–957. [PubMed] [Google Scholar]
- 132.Anderson OS, Sant KE, Dolinoy DC. Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J Nutr Biochem. 2012;23:853–859. doi: 10.1016/j.jnutbio.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yamazaki M, Munetsuna E, Yamada H, et al. Fructose consumption induces hypomethylation of hepatic mitochondrial DNA in rats. Life Sci. 2016;149:146–152. doi: 10.1016/j.lfs.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 134.Wagner AE, Piegholdt S, Ferraro M, Pallauf K, Rimbach G. Food derived microRNAs. Food Funct. 2015;6:714–718. doi: 10.1039/c4fo01119h. [DOI] [PubMed] [Google Scholar]
- 135.Kalhan SC, Hanson RW. Resurgence of serine: an often neglected but indispensable amino Acid. J Biol Chem. 2012;287:19786–19791. doi: 10.1074/jbc.R112.357194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Otten JJ, Hellwig JP, Meyers LD. Dietary reference intakes: the essential guide to nutrient requirements. Washington, NY: National Academies Press; 2006. [Google Scholar]
- 137.Blount BC, Mack MM, Wehr CM, et al. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci U S A. 1997;94:3290–3295. doi: 10.1073/pnas.94.7.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zeisel SH. Dietary choline deficiency causes DNA strand breaks and alters epigenetic marks on DNA and histones. Mutat Res. 2012;733:34–38. doi: 10.1016/j.mrfmmm.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Seessle J, Gohdes A, Gotthardt DN, et al. Alterations of lipid metabolism in Wilson disease. Lipids Health Dis. 2011;10:83. doi: 10.1186/1476-511X-10-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Huster D, Purnat TD, Burkhead JL, et al. High copper selectively alters lipid metabolism and cell cycle machinery in the mouse model of Wilson disease. J Biol Chem. 2007;282:8343–8355. doi: 10.1074/jbc.M607496200. [DOI] [PubMed] [Google Scholar]
- 141.Ohashi K, Munetsuna E, Yamada H, et al. High fructose consumption induces DNA methylation at PPARα and CPT1A promoter regions in the rat liver. Biochem Biophys Res Commun. 2015;468:185–189. doi: 10.1016/j.bbrc.2015.10.134. [DOI] [PubMed] [Google Scholar]
- 142.Uriarte G, Paternain L, Milagro FI, Martinez JA, Campion J. Shifting to a control diet after a high-fat, high-sucrose diet intake induces epigenetic changes in retroperitoneal adipocytes of Wistar rats. J Physiol Biochem. 2013;69:601–611. doi: 10.1007/s13105-012-0231-6. [DOI] [PubMed] [Google Scholar]
- 143.Zhang X, Zhang JH, Chen XY, et al. Reactive oxygen species-induced TXNIP drives fructose-mediated hepatic inflammation and lipid accumulation through NLRP3 inflammasome activation. Antioxid Redox Signal. 2015;22:848–870. doi: 10.1089/ars.2014.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cardoso AR, Cabral-Costa JV, Kowaltowski AJ. Effects of a high fat diet on liver mitochondria: increased ATP-sensitive K+ channel activity and reactive oxygen species generation. J Bioenerg Biomembr. 2010;42:245–253. doi: 10.1007/s10863-010-9284-9. [DOI] [PubMed] [Google Scholar]
- 145.Cannon JR, Harvison PJ, Rush GF. The effects of fructose on adenosine triphosphate depletion following mitochondrial dysfunction and lethal cell injury in isolated rat hepatocytes. Toxicol Appl Pharmacol. 1991;108:407–416. doi: 10.1016/0041-008x(91)90087-u. [DOI] [PubMed] [Google Scholar]
- 146.Satapati S, Sunny NE, Kucejova B, et al. Elevated TCA cycle function in the pathology of diet-induced hepatic insulin resistance and fatty liver. J Lipid Res. 2012;53:1080–1092. doi: 10.1194/jlr.M023382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Baier SR, Nguyen C, Xie F, Wood JR, Zempleni J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cellcultures, and mouse livers. J Nutr. 2014;144:1495–1500. doi: 10.3945/jn.114.196436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tanaka H, Iwasaki Y, Yamato H, et al. p-Cresyl sulfate induces osteoblast dysfunction through activating JNK and p38 MAPK pathways. Bone. 2013;56:347–354. doi: 10.1016/j.bone.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 149.Sun L, Shen W, Liu Z, Guan S, Liu J, Ding S. Endurance exercise causes mitochondrial and oxidative stress in rat liver: effects of a combination of mitochondrial targeting nutrients. Life Sci. 2010;86:39–44. doi: 10.1016/j.lfs.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 150.Santos-Alves E, Marques-Aleixo I, Rizo-Roca D, et al. Exercise modulates liver cellular and mitochondrial proteins related to quality control signaling. Life Sci. 2015;135:124–130. doi: 10.1016/j.lfs.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 151.Kaliman P, Alvarez-Lopez MJ, Cosin-Tomas M, Rosenkranz MA, Lutz A, Davidson RJ. Rapid changes in histone deacetylases and inflammatory gene expression in expert meditators. Psychoneuroendocrinology. 2014;40:96–107. doi: 10.1016/j.psyneuen.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kuzumaki N, Ikegami D, Tamura R, et al. Hippocampal epigenetic modification at the brain-derived neurotrophic factor gene induced by an enriched environment. Hippocampus. 2011;21:127–132. doi: 10.1002/hipo.20775. [DOI] [PubMed] [Google Scholar]
- 153.Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 154.Parain K, Murer MG, Yan Q, et al. Reduced expression of brain-derived neurotrophic factor protein in Parkinson's disease substantia nigra. Neuroreport. 1999;10:557–561. doi: 10.1097/00001756-199902250-00021. [DOI] [PubMed] [Google Scholar]
- 155.Scalzo P, Kummer A, Bretas TL, Cardoso F, Teixeira AL. Serum levels of brain-derived neurotrophic factor correlate with motor impairment in Parkinson's disease. J Neurol. 2010;257:540–545. doi: 10.1007/s00415-009-5357-2. [DOI] [PubMed] [Google Scholar]
- 156.Xiong GL, Doraiswamy PM. Does meditation enhance cognition and brain plasticity? Ann N Y Acad Sci. 2009;1172:63–69. doi: 10.1196/annals.1393.002. [DOI] [PubMed] [Google Scholar]
- 157.Meyer JN, Leung MC, Rooney JP, et al. Mitochondria as a target of environmental toxicants. Toxicol Sci. 2013;134:1–17. doi: 10.1093/toxsci/kft102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ke ZH, Pan JX, Jin LY, et al. Bisphenol a exposure may induce hepatic lipid accumulation via reprogramming the DNA methylation patterns of genes involved in lipid metabolism. Sci Rep. 2016;6:31331. doi: 10.1038/srep31331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Geraghty AA, Lindsay KL, Alberdi G, McAuliffe FM, Gibney ER. Nutrition during pregnancy impacts offspring’s epigenetic status-evidence from human and animal studies. Nutr Metab Insights. 2016;8:41–47. doi: 10.4137/NMI.S29527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Robinson AM, Bucci DJ. Maternal exercise and cognitive functions of the offspring. cognitive sciences. Cogn Sci (Hauppauge) 2012;7:187–205. [PMC free article] [PubMed] [Google Scholar]
- 161.Perera F, Herbstman J. Prenatal environmental exposures, epigenetics, and disease. Reprod Toxicol. 2011;31:363–373. doi: 10.1016/j.reprotox.2010.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Schulz LC. The Dutch Hunger winter and the developmental origins of health and disease. Proc Natl Acad Sci U S A. 2010;107:16757–16758. doi: 10.1073/pnas.1012911107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wadhwa PD, Buss C, Entringer S, Swanson JM. Developmental origins of health and disease: brief history of the approach and current focus on epigenetic mechanisms. Semin Reprod Med. 2009;27:358–368. doi: 10.1055/s-0029-1237424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bruce KD, Cagampang FR, Argenton M, et al. Maternal high-fat feeding primes steatohepatitis in adult mice offspring, involving mitochondrial dysfunction and altered lipogenesis gene expression. Hepatology. 2009;50:1796–1808. doi: 10.1002/hep.23205. [DOI] [PubMed] [Google Scholar]
- 165.Davison JM, Mellott TJ, Kovacheva VP, Blusztajn JK. Gestational choline supply regulates methylation of histone H3, expression of histonemethyltransferases G9a (Kmt1c) and Suv39h1 (Kmt1a), and DNA methylation of their genes in ratfetal liver and brain. J Biol Chem. 2009;284:1982–1989. doi: 10.1074/jbc.M807651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kovacheva VP, Mellott TJ, Davison JM, et al. Gestational choline deficiency causes global and Igf2 gene DNA hypermethylation by up-regulation of Dnmt1 expression. J Biol Chem. 2007;282:31777–31788. doi: 10.1074/jbc.M705539200. [DOI] [PubMed] [Google Scholar]
- 167.Litwin T, Gromadzka G, Członkowska A. Gender differences in Wilson's disease. J Neurol Sci. 2012;312:31–35. doi: 10.1016/j.jns.2011.08.028. [DOI] [PubMed] [Google Scholar]
- 168.Tobi EW, Lumey LH, Talens RP, et al. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009;18:4046–4053. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Rodriguez B, Burguera J, Berenguer M. Response to different therapeutic approaches in Wilson disease. A long-term follow up study. Ann Hepatol. 2012;11:907–914. [PubMed] [Google Scholar]
- 170.Stanhope KL, Schwarz JM, Keim NL, et al. Consuming fructose-sweetened, not glucose-sweetened, beverages increases visceral adiposity and lipids and decreases insulin sensitivity in overweight/obese humans. J Clin Invest. 2009;119:1322–1334. doi: 10.1172/JCI37385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Owu DU, Osim EE, Ebong PE. Serum liver enzymes profile of Wistar rats following chronic consumption of fresh or oxidized palm oil diets. Acta Tropica. 1998;69:65–73. doi: 10.1016/s0001-706x(97)00115-0. [DOI] [PubMed] [Google Scholar]
- 172.Stavrovskaya IG, Bird SS, Marur VR, et al. Dietary macronutrients modulate the fatty acyl composition of rat liver mitochondrial cardiolipins. J Lipid Res. 2013;54:2623–2635. doi: 10.1194/jlr.M036285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ramsey JJ, Harper ME, Humble SJ, et al. Influence of mitochondrial membrane fatty acid composition on proton leak and H2O2 production in liver. Comp Biochem Physiol B Biochem Mol Biol. 2005;140:99–108. doi: 10.1016/j.cbpc.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 174.Tapia PC. Sublethal mitochondrial stress with an attendant stoichiometric augmentation of reactive oxygen species may precipitate many of the beneficial alterations in cellular physiology produced by caloric restriction, intermittent fasting, exercise and dietary phytonutrients: “Mitohormesis” for health and vitality. Med Hypotheses. 2006;66:832–843. doi: 10.1016/j.mehy.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 175.Antoni R, Johnston KL, Collins AL, Robertson MD. Effects of intermittent fasting on glucose and lipid metabolism. Proc Nutr Soc. 2017;16:1–8. doi: 10.1017/S0029665116002986. [DOI] [PubMed] [Google Scholar]
- 176.Shay KP, Moreau RF, Smith EJ, Smith AR, Hagen TM. Alpha-lipoic acid as a dietary supplement: Molecular mechanisms and therapeutic potential. Biochim Biophys Acta. 2009;1790:1149–1160. doi: 10.1016/j.bbagen.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Jomova K, Vondrakova D, Lawson M, Valko M. Metals, oxidative stress and neurodegenerative disorders. Mol Cell Biochem. 2010;345:91–104. doi: 10.1007/s11010-010-0563-x. [DOI] [PubMed] [Google Scholar]
- 178.Sheline CT, Choi EH, Kim-Han JS, Dugan LL, Choi DW. Cofactors of mitochondrial enzymes attenuate copper-induced death in vitro and in vivo. Ann Neurol. 2002;52:195–204. doi: 10.1002/ana.10276. [DOI] [PubMed] [Google Scholar]
- 179.Liu J, Ames BN. Reducing mitochondrial decay with mitochondrial nutrients to delay and treat cognitive dysfunction, Alzheimer's disease, and Parkinson's disease. Nutr Neurosci. 2005;8:67–89. doi: 10.1080/10284150500047161. [DOI] [PubMed] [Google Scholar]
- 180.Goodwin VA, Richards SH, Taylor RS, Taylor AH, Campbell JL. The effectiveness of exercise interventions for people with Parkinson's disease: A systematic review and meta-analysis. Mov Disord. 2008;23:631–640. doi: 10.1002/mds.21922. [DOI] [PubMed] [Google Scholar]
- 181.Khalsa DS. Stress, meditation, and Alzheimer’s disease prevention: where the evidence stands. J Alzheimers Dis. 2015;48:1–12. doi: 10.3233/JAD-142766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Polyzos SA, Kountouras J, Deretzi G, Zavos C, Mantzoros CS. The emerging role of endocrine disruptors in pathogenesis of insulin resistance: A concept implicating nonalcoholic fatty liver disease. Curr Mol Med. 2012;12:68–82. doi: 10.2174/156652412798376161. [DOI] [PubMed] [Google Scholar]