Abstract
Some diets lack sufficient manganese (Mn), an essential mineral. Increasing Mn in grain by biofortification could prevent Mn deficiency, but may increase levels of the toxic element cadmium (Cd). Here, we investigated Mn in rice (Oryza sativa) grains in recombinant inbred lines (RILs) from the cross of 93–11 (low grain Mn) with PA64s (high grain Mn). Quantitative trait locus (QTL) analysis to identify loci controlling grain Mn identified a major QTL, qGMN7.1, on the short arm of chromosome 7; qGMN7.1 explained 15.6% and 22.8% of the phenotypic variation in the RIL populations grown in two distinct environments. We validated the QTL with a chromosome segment substitution line (CSSL), CSSL-qGMN7.1, in the 93–11 background harboring qGMN7.1 from PA64s. Compared to 93–11, CSSL-qGMN7.1 grain had increased Mn and decreased Cd concentrations; CSSL-qGMN7.1 roots also showed enhanced Mn uptake. Fine mapping delimited qGMN7.1 to a 49.3-kb region containing OsNRAMP5, a gene responsible for Mn and Cd uptake. Sequence variations in the OsNRAMP5 promoter caused changes in its transcript level, and in grain Mn levels. Our study thus cloned a major QTL for grain Mn concentration in rice, and identified materials for breeding rice for high Mn and low Cd concentrations in the grain.
Introduction
Manganese (Mn) is an essential trace element for plants, domestic animals, and humans1. In mammals, Mn is necessary for proper fetal development and growth and is crucial throughout the life span2,3. Humans maintain stable tissue levels of Mn and Mn is present in virtually all diets at low concentrations4. The U.S. National Research Council has established an estimated safe and adequate dietary Mn intake of 2–5 mg per day for adults5. Overt Mn deficiency diseases are extremely rare, but segments of populations from remote areas still suffer from subclinical Mn deficiency1,6. Cadmium (Cd) is a toxic heavy metal that is harmful to human health and Mn2+ and Cd2+ can be transported by the same transporters7. The international maximum limit for rice is 0.4 mg Cd·kg−1 polished rice8. As rice is the staple food for over half of the world’s population, production of rice with adequate levels of Mn and low levels of Cd would benefit human health.
Similar to other elements, Mn accumulation in rice grains occurs via root uptake, vascular transport, and transfer into the developing grain. The available Mn in the soil rhizosphere is first taken up by transporters in the plasma membrane of root cells, then loaded into the xylem, and transferred to shoots via the xylem. It is distributed to different storage compartments regulated by phloem loading and mobility, and finally remobilized from other organs, such as leaves, to the grains at the filling stage9. Many factors can affect these processes and ultimately determine the Mn level in grains, among which, genetics appears to be the dominant factor.
The use of reverse genetic approaches has uncovered several genes involved in Mn transport, translocation, or distribution in rice10–12. OsYSL2 encodes a functional iron [Fe(II)]- and Mn [Mn(II)]-nicotianamine complex transporter expressed in phloem cells and developing seeds, and OsYSL2 is required for long-distance transport of Fe and Mn10. OsNRAMP3 and OsNRAMP5 belong to the natural resistance-associated macrophage protein (NRAMP) family11,12. OsNRAMP3 is a node-localized Mn transporter that functions as a switch for regulating Mn distribution in rice11. OsNRAMP5 regulates Mn uptake and the accumulation of Mn in rice12. OsNRAMP5 also functions in iron (Fe) and Cd transport12–15. Knockout or mutation of OsNRAMP5 greatly decreased Cd uptake by the roots, resulting in a decline in the Cd concentration in the straw and grain12,13.
In the last decade, quantitative trait locus (QTL) mapping for grain Mn concentration with various mapping populations has identified a number of QTLs in rice16–24. Several major QTLs on chromosomes 3, 7, and 8 have been detected. For example, the major QTL, qMNCN-3, on chromosome 3, identified by Shen17, was also detected as qMn3 by Du et al.22, explaining 9% and 10.8% of the phenotypic variation in the two studies, respectively. qMNCN-8, on the short arm of chromosome 8, was also detected by different studies17,22,23. However, to date, no QTL associated with grain Mn concentration has been fine mapped or cloned.
The hybrid cultivar Liang-You-Pei-Jiu (LYPJ) was produced by a cross between the paternal parent 93–11 (an indica variety widely grown in China), and the maternal parent PA64s (with a mixed genetic background of indica and japonica). This pioneer super hybrid yielded 10.5 tons/ha in 200025. We previously constructed 132 LYPJ-derived core recombinant inbred lines (RILs) and re-sequenced them to establish a high-density single-nucleotide polymorphism (SNP) linkage map26. Here, we performed a QTL analysis for grain Mn concentration based on the previously developed genetic map and characterized a major QTL controlling Mn accumulation in rice grains.
Results
The qGMN7.1 from PA64s significantly increased grain Mn concentration
The RIL population from the rice super hybrid LYPJ, and the hybrid parents were grown in two different environments, Hainan (110.0 E, 18.5 N) and Hangzhou (120.0 E, 30.1 N), China. Mature seeds were harvested for determining the grain Mn concentration. The concentration of Mn in the grains was significantly different between the parents in both Hainan and Hangzhou, with concentrations in PA64s approximately 2-fold higher than in 93–11 (Fig. S1 and Table S1). The RIL population showed a wide range of phenotypic variation, in a continuous distribution (Fig. S1). Using the high-resolution SNP map, we detected 12 QTLs for grain Mn concentration distributed on all chromosomes except for chromosomes 10, 11, and 12 (Fig. S2 and Table S2). Among those QTLs, 5 were identified in the RIL populations grown in both Hainan and Hangzhou, and 8 had additive effects coming from PA64s. One major QTL with the highest LOD value, qGMN7.1, was mapped between markers SNP7-53 and SNP7-64 on the short arm of chromosome 7 and explained 15.6% and 22.8% of the phenotypic variation in the RIL populations grown in Hangzhou and Hainan, respectively (Fig. S2 and Table S2).
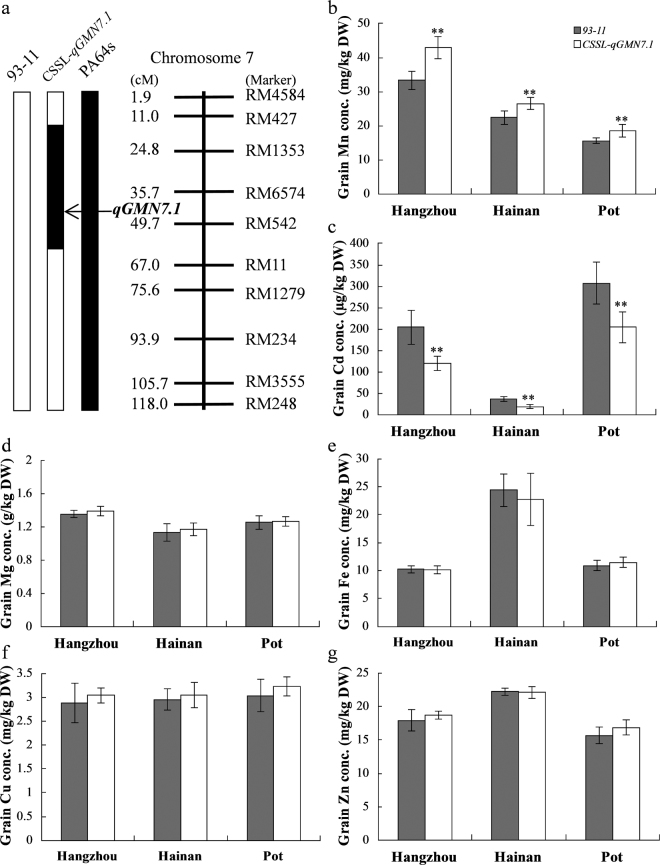
To confirm the effect of qGMN7.1 on grain Mn concentrations, we constructed a chromosome segment substitution line (CSSL), CSSL-qGMN7.1, in the 93–11 background (Fig. S3), harboring the segment from PA64s between the markers RM427 and RM11 (Fig. 1a). When 93–11 and CSSL-qGMN7.1 were cultivated in the paddy fields in Hangzhou and Hainan or grown in pots in Hangzhou (2015), the concentration of Mn in the CSSL-qGMN7.1 grain was significantly higher than that of 93–11 (Fig. 1b). Interestingly, CSSL-qGMN7.1 also showed decreased Cd concentrations in the grain, compared to 93–11 (Fig. 1c). The concentrations of other trace metals in the grains, such as magnesium (Mg), Fe, zinc (Zn), and copper (Cu), were not significantly different between CSSL-qGMN7.1 and 93–11 (Fig. 1d–g).
Figure 1.
qGMN7.1 from PA64s significantly increased grain Mn concentration. (a) Schematic graph of chromosome 7 of CSSL-qGMN7.1 and the two parents. The white and black bars represent 93–11 and PA64s alleles, respectively. The name and genetic locus of the SSR markers and the position of qGMN7.1 are indicated. (b–g) Concentrations of Mn, Cd, Mg, Fe, Cu, and Zn in the grains of 93–11 (grey) and CSSL-qGMN7.1 (white) grown in the field of Hangzhou and Hainan, China, and in pots in Hangzhou (2015). Vertical bars represent the standard deviation (n = 6). **Indicates a 1% significance level according to the t test.
Physiological characteristics of CSSL-qGMN7.1
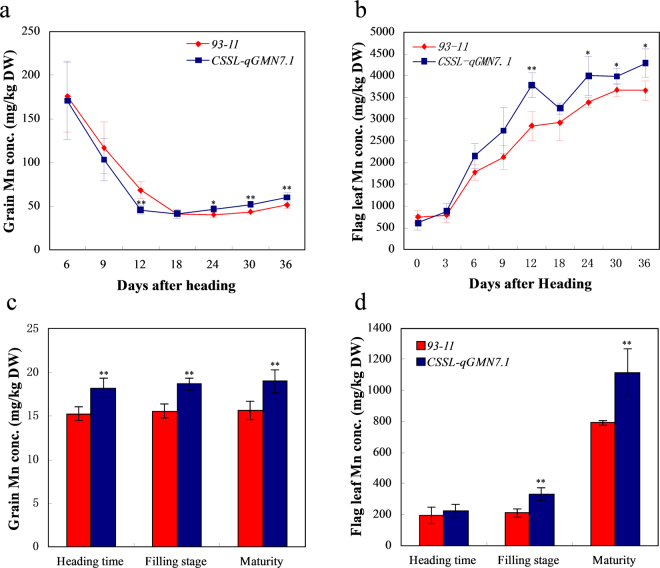
We performed a series of physiological experiments to determine the physiological mechanism underlying the increased grain Mn concentration conferred by qGMN7.1. In a time-course experiment of grain Mn accumulation, no significant difference was found between 93–11 and CSSL-qGMN7.1 at the early grain-filling stage, although both lines showed decreasing accumulation with time (Fig. 2a). At the 18th day after heading, the grain Mn concentration stabilized and then significantly increased from the 24th day after heading in CSSL-qGMN7.1 compared to 93–11. The difference remained significant through maturity (Fig. 2a). Overall, CSSL-qGMN7.1 and 93–11 had a similar pattern of Mn accumulation, and qGMN7.1 functioned during the late grain-filling stage.
Figure 2.
Comparison of time-course experiment of Mn accumulation in the grain and flag leaf between 93–11 and CSSL-qGMN7.1. (a,c) Grain Mn concentration of 93–11 (red) and CSSL-qGMN7.1 (blue). Grains were harvested at 6, 9, 12, 18, 24, 30, and 36 days after heading (a), grains were tagged and harvested until maturity after their corresponding flag leaves were sampled at the heading, grain-filling, and maturity stages (c). (b,d) Flag leaf Mn concentration of 93–11 (red) and CSSL-qGMN7.1 (blue). Flag leaves were harvested at 0, 3, 6, 9, 12, 18, 24, 30, and 36 days after heading (b), flag leaves were sampled at the heading, grain-filling, and maturity stages (d). Vertical bars represent the standard deviation (n = 4). *and **indicate a 5% and 1% significance level, respectively, according to the t test.
We also measured Mn concentrations in flag leaves at different times after heading and found that Mn increased in both CSSL-qGMN7.1 and 93–11, with higher levels in CSSL-qGMN7.1, from the late filling stage to maturity (Fig. 2b). To determine whether Mn is transferred from other organs, such as the flag leaf, into the grain at the late filling stage, we removed flag leaves and measured the effect on Mn in the grains. Removal of flag leaves at the heading and filling stages did not affect Mn accumulation in the grains (Fig. 2c, d). These results suggested that Mn is not transferred from flag leaves to the grains at the grain-filling through maturity stages.
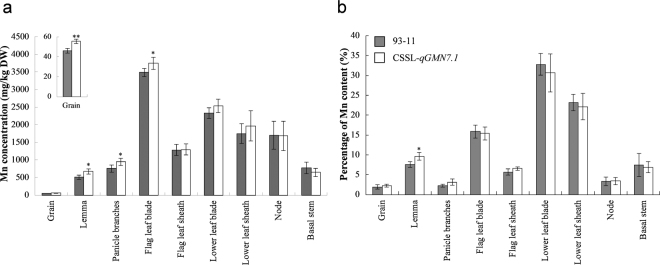
As the distribution of Mn in the aboveground parts has been reported to play an important role in grain Mn accumulation, we analyzed the Mn concentration in different organs at maturity. The highest Mn concentration was observed in flag leaf blades, with approximately 3,500 mg·kg−1 dry weight (DW), and the lowest in the grains with only about 50 mg·kg−1 DW in the rice plants (Fig. 3a). Compared to 93–11, CSSL-qGMN7.1 accumulated higher concentrations of Mn in the grains, lemma, panicle branches, and flag leaf blades. However, Mn concentrations in other leaves and stems were almost the same in both lines (Fig. 3a). An analysis of the proportion of Mn content in different organs to the whole plant showed that the proportion of grain Mn content/whole plant Mn content was similar between CSSL-qGMN7.1 and 93–11 (Fig. 3b).
Figure 3.
Mn distribution in the aboveground tissues of 93–11 and CSSL-qGMN7. (a) Mn concentration in different organs. (b) Proportion of Mn content in different organs to the Mn content in the whole plant (except roots). The aboveground tissues of 93–11 (grey) and CSSL-qGMN7.1 (white) grown in the paddy field were harvested and separated into different organs at maturity. Vertical bars represent the standard deviation (n = 4). *and **indicate a 5% and 1% significance level, respectively, according to the t test.
These results indicate that the higher Mn concentration in the grains of CSSL-qGMN7.1 did not result from greater distribution from other organs, but from higher assimilation in the roots. To confirm this, we performed a short-term (30 min) uptake experiment using intact roots of 93–11 and CSSL-qGMN7.1. The Mn uptake at 4 °C was much lower than that at 25 °C in both lines, but CSSL-qGMN7.1 exhibited higher Mn uptake than 93–11 at both temperatures (Fig. 4a). The net Mn uptake, calculated by subtracting the Mn uptake at 4 °C from that at 25 °C, was significantly greater in CSSL-qGMN7.1 than in 93–11 (Fig. 4b). Although CSSL-qGMN7.1 and 93–11 had a similar affinity for Mn, the value of Vmax for CSSL-qGMN7.1 (118.6 mg·kg−1 root DW·h−1) was significantly higher than that for 93–11 (103.4 mg·kg−1 root DW·h−1) (Fig. 4).
Figure 4.
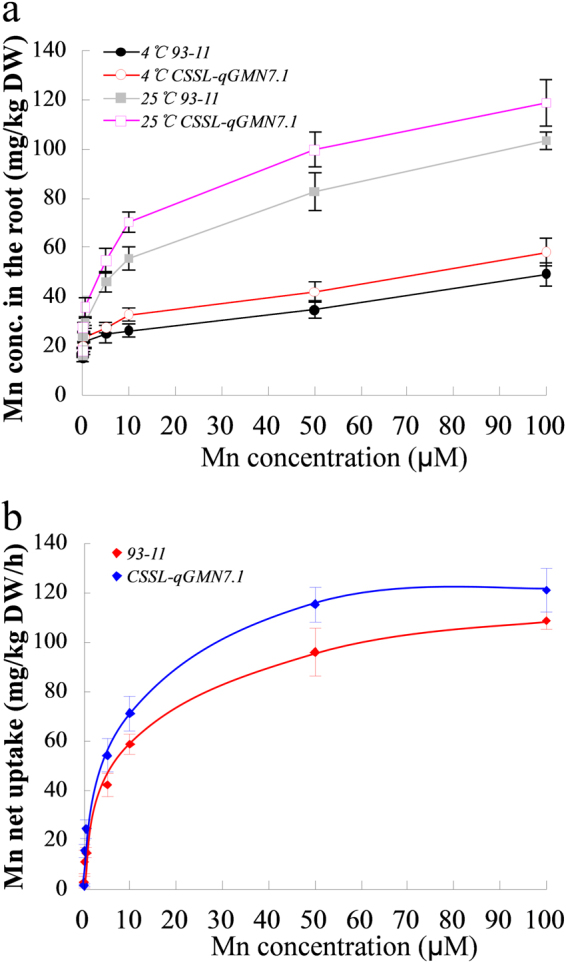
Short-term uptake of Mn by rice roots. (a) Prepared seedlings of 93–11 (closed symbols) and CSSL-qGMN7.1 (open symbols) were exposed to nutrient solutions containing various concentrations of Mn (0, 0.1, 0.5, 5, 10, 50, and 100 µM) for 30 min at 25 °C (square) or 4 °C (circle). (b) Net uptake of Mn was calculated by subtracting the uptake at 4 °C from that at 25 °C. Red rhombus, 93–11; blue rhombus, CSSL-qGMN7.1. Red and blue lines represent Michaelis-Menten curves for 93–11 and CSSL-qGMN7.1, respectively. Error bars represent standard deviation (n = 4).
Fine mapping of the qGMN7.1 to a 49.3-kb genomic region
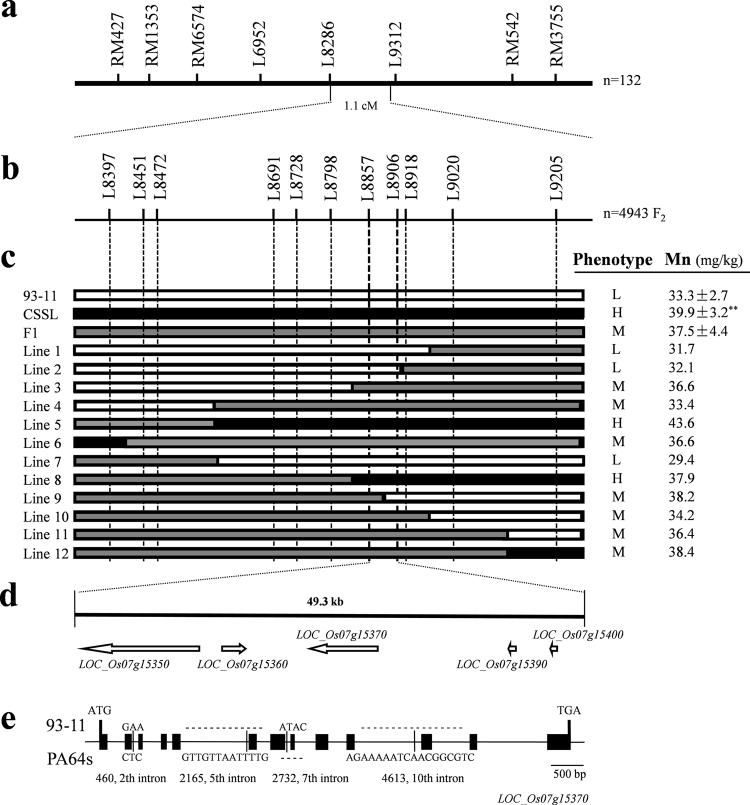
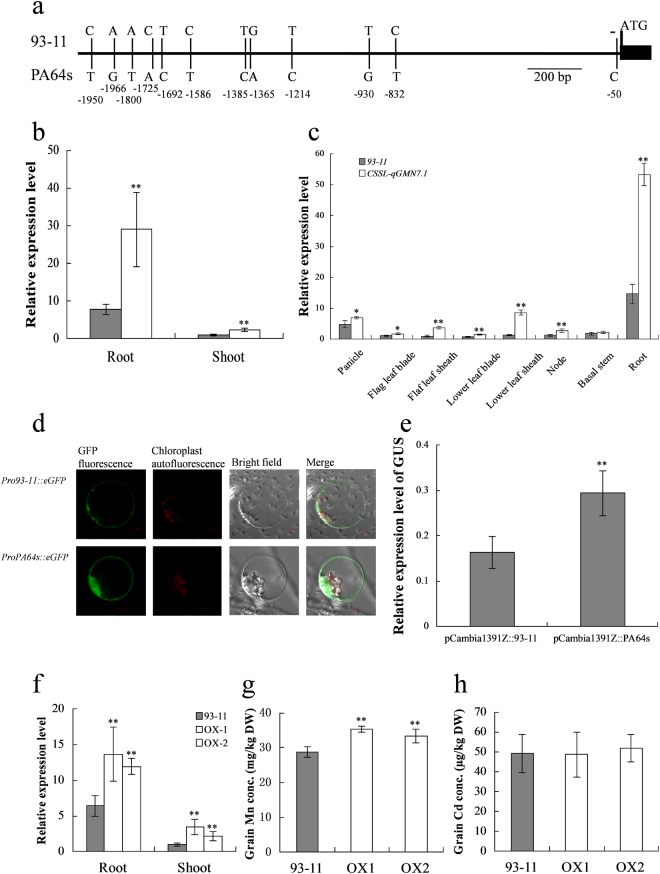
To fine map qGMN7.1, CSSL-qGMN7.1 was crossed with 93–11 to generate a segregating population. The F1 plants of this cross showed intermediate concentrations of grain Mn, indicating semi-dominant inheritance of qGMN7.1 (Fig. 5c). We planted 4,943 individuals in Hainan (2015) and screened them with markers L8286 and L9312 flanking qGMN7.1 and 12 recombinants were identified (Fig. 5). Nine newly developed insertion/deletion (InDel) markers, well distributed within the interval, were used to further genotype the 12 recombinants (Fig. 5b and Table S3). We tested the F2:3 progeny of the recombinants in Hangzhou in 2016. Grain Mn concentrations showed no significant difference among the three genotypes of recombinants Line 1, Line 2, and Line 8 (Table 1). For the other recombinant lines, qGMN7.1 segregated and significant differences in phenotypes were found in the three genotypes of qGMN7.1 (Table 1). Based on the phenotypes and genotypes of these recombinants, we delimited qGMN7.1 to a region of approximately 49.3-kb between markers L8857 and L8906 (Fig. 5c, d).
Figure 5.
Fine mapping of qGMN7.1. (a) qGMN7.1 was mapped between L8286 and L9312 based on LYPJ RILs. (b) 4,943 segregating individuals derived from a cross between 93–11 and CSSL-qGMN7.1 were used for fine mapping. (c) Genotyping and phenotyping of recombinants delimited qGMN7.1 to within a 49.3-kb region flanked by L8857 and L8906. Phenotype for grain Mn concentration: L (low), M (medium), or H (high). (d) RiceGAAS predicted five genes in the target region. (e) LOC_Os07g15370 structure and sequence variations between two parents. **Indicates a 1% significance level compared to 93–11 according to the t test (n = 6).
Table 1.
Progeny test of nine recombinants for confirmation of the fine-mapped region of qGMN7.1.
| Recombinants | Markers | Grain Mn conc. (mg/kg DW) | Grain Cd conc. (µg/kg DW) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| L8397 | L8472 | L8798 | L8857 | L8906 | L8918 | L9020 | L9205 | ||||
| Line 1 | Type 1 | 9 | 9 | 9 | 9 | 9 | 9 | P | P | 41.36 ± 2.72 | 507.34 ± 50.55 |
| Type 2 | 9 | 9 | 9 | 9 | 9 | 9 | H | H | 40.65 ± 4.03 | 467.32 ± 59.49 | |
| Type 3 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 42.00 ± 4.10 | 506.39 ± 68.26 | |
| Line 2 | Type 1 | 9 | 9 | 9 | 9 | 9 | P | P | P | 42.30 ± 3.94 | 481.06 ± 72.41 |
| Type 2 | 9 | 9 | 9 | 9 | 9 | H | H | H | 38.60 ± 3.73 | 462.25 ± 85.67 | |
| Type 3 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 40.89 ± 3.91 | 464.53 ± 83.16 | |
| Line 3 | Type 1 | 9 | 9 | 9 | 9 | P | P | P | P | 55.58 ± 4.37 | 578.19 ± 61.15 |
| Type 2 | 9 | 9 | 9 | 9 | H | H | H | H | 46.24 ± 3.86** | 620.75 ± 69.83 | |
| Type 3 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 46.87 ± 3.37** | 552.72 ± 44.53 | |
| Line 4 | Type 1 | 9 | 9 | P | P | P | P | P | P | 48.47 ± 3.99 | 508.47 ± 61.99 |
| Type 2 | 9 | 9 | H | H | H | H | H | H | 41.30 ± 4.74 | 488.22 ± 66.23 | |
| Type 3 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 38.85 ± 4.32** | 513.40 ± 55.27 | |
| Line 6 | Type 1 | P | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 46.38 ± 4.02 | 273.19 ± 53.55 |
| Type 2 | P | H | H | H | H | H | H | H | 52.80 ± 5.70 | 310.51 ± 26.32 | |
| Type 3 | P | P | P | P | P | P | P | P | 58.85 ± 5.14** | 284.98 ± 68.59 | |
| Line 8 | Type 1 | 9 | 9 | 9 | P | P | P | P | P | 53.86 ± 4.83 | 528.79 ± 63.56 |
| Type 2 | H | H | H | P | P | P | P | P | 56.57 ± 3.92 | 368.06 ± 99.61** | |
| Type 3 | P | P | P | P | P | P | P | P | 56.99 ± 5.19 | 407.64 ± 49.28** | |
| Line 9 | Type 1 | P | P | P | P | 9 | 9 | 9 | 9 | 51.05 ± 4.14 | 341.01 ± 68.95 |
| Type 2 | H | H | H | H | 9 | 9 | 9 | 9 | 46.75 ± 4.71 | 387.54 ± 62.21 | |
| Type 3 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 43.57 ± 3.64* | 499.33 ± 64.07** | |
| Line 10 | Type 1 | P | P | P | P | P | 9 | 9 | 9 | 49.71 ± 5.74 | 389.76 ± 86.87 |
| Type 2 | H | H | H | H | H | 9 | 9 | 9 | 42.39 ± 3.73* | 428.91 ± 57.30 | |
| Type 3 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 41.03 ± 2.94** | 599.43 ± 69.62** | |
| Line 12 | Type 1 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | P | 42.77 ± 3.89 | 648.47 ± 91.99 |
| Type 2 | H | H | H | H | H | H | H | P | 46.87 ± 5.39 | 488.22 ± 76.23** | |
| Type 3 | P | P | P | P | P | P | P | P | 50.89 ± 5.29* | 413.40 ± 95.27** | |
Note: Type 1, type 2, and type 3 in each panel represent the segregated genotypes of the recombinants. ‘9’, ‘P’, and ‘H’ represent the homozygote of 93–11 and PA64s, and the heterozygote of the parents, respectively. *and **indicate a 5% and 1% significance level compared to type I, respectively, according to the t test (n = 6).
We also measured the Cd concentration in the grains of the F2:3 progeny of the recombinants. Three genotypes in the recombinants Line 1 to Line 4 and Line 6 showed no significant difference in grain Cd concentration, and recombinants Line 8 to Line 10 and Line 12 exhibited segregating phenotypes (Table 1). Combined with the genetic recombination sites of these recombinants (Fig. 5c), we concluded that qGMN7.1 had little influence on Cd accumulation in the grain. Furthermore, the Cd concentration in the grain of recombinant Line 6 was about 300 µg·kg−1 DW, much lower than that of recombinant Line 1 to Line 4 (500 to 600 µg·kg−1 DW), indicating that the allele from PA64s could greatly decrease grain Cd concentration.
OsNRAMP5 is the candidate gene for the grain Mn accumulation trait
The Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/) predicted five genes in the 49.3-kb target region of qGMN7.1 (Fig. 5d): LOC_Os07g15350 encoding a transposon, LOC_Os07g15360 and LOC_Os07g15390 encoding retrotransposons, LOC_Os07g15400 encoding an expressed protein, and LOC_Os07g15370 encoding a metal transporter. Because LOC_Os07g15370 has been reported previously as OsNRAMP5, encoding a major transporter for Mn and Cd12, it was considered the most likely candidate gene for the grain Mn accumulation trait in qGMN7.1.
Sequence alignment of OsNRAMP5 between the two parents, 93–11 and PA64s, revealed no polymorphisms in the coding sequence, but 12 variations in the promoter region (Figs 5e and 6a). These sequence variations might alter the transcript levels and be responsible for the different phenotypes. Therefore, we measured the expression levels of OsNRAMP5 in 93–11 and CSSL-qGMN7.1 at different developmental stages. At the seedling stage, CSSL-qGMN7.1 had significantly higher transcript levels of OsNRAMP5 than in 93–11, with the largest difference of 3.7-fold found in the roots. Higher transcript levels were also observed in CSSL-qGMN7.1 at the booting stage, particularly in the roots (Fig. 6b, c). We then compared the promoter activity of OsNRAMP5 between 93–11 and PA64s by transient expression in rice protoplasts. The green fluorescent signals of GFP driven by the PA64s promoter were stronger than those driven by the 93–11 promoter (Figs 6d and S4), and the GUS transcript levels driven by the PA64s promoter also showed a higher level compared to that driven by the 93–11 promoter (Fig. 6e). These results suggested that the OsNRAMP5 promoter from PA64s was stronger than the OsNRAMP5 promoter from 93–11.
Figure 6.
Validation of candidate gene. (a) Sequence variations in the promoter region of OsNRAMP5 between two parents. (b,c) Comparison of the relative expression level of OsNRAMP5 between 93–11 (grey) and CSSL-qGMN7.1 (white) in roots and shoots at the seedling stage (b), and in different organs at the booting stage (c). (d,e) Promoter activity analysis. The green fluorescent signals of GFP driven by the OsNRAMP5 promoter from 93–11 and PA64s (d), the relative expression level of GUS driven by the OsNRAMP5 promoters from 93–11 and PA64s (e). (f–h) Relative expression level of OsNRAMP5 and grain Mn and Cd concentrations in 93–11 and the overexpression (OX) lines grown in pots. *and **indicate a 5% and 1% significance level, respectively, according to the t test.
To validate that OsNRAMP5 is responsible for Mn accumulation in the grain, we overexpressed it in 93–11. A significantly larger abundance of OsNRAMP5 transcript was found in roots (2 fold) and shoots (2–3 fold) of the overexpression lines than in 93–11 (Fig. 6f). The overexpression lines accumulated more Mn in the grains than 93–11 when grown in pots (Fig. 6g). However, the Cd concentration in the grains was nearly equal in 93–11 and the overexpression lines (Fig. 6h). Therefore, we concluded that OsNRAMP5 is responsible for the grain Mn accumulation trait in qGMN7.1.
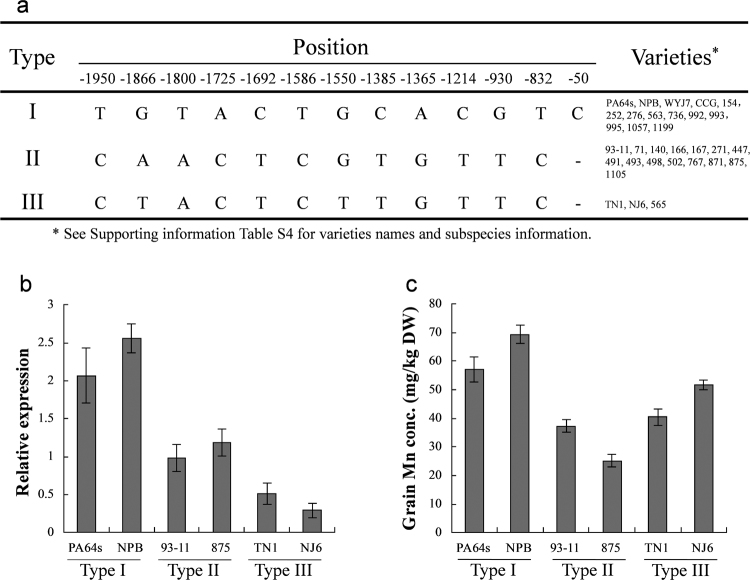
To gain further insight into the variations of the OsNRAMP5 promoters, we isolated and compared the 2-kb 5′-flanking regions of OsNRAMP5 from 30 different rice varieties. Based on the promoter sequences, three haplotypes were identified (Fig. 7a and Table S4). Among the 30 rice varieties, 13 had the same haplotype as PA64s (designated as type I) and 14 coincided with that of 93–11 (designated as type II). The promoters from varieties TN1, NJ6, and No.565 were consistent with type II, with the exception of nucleotide variations at positions −1,866 (A → T) and −1,550 (G → T) from the start codon ATG (these were designated as type III) (Fig. 7a). Compared to type I, the rice varieties containing type II and III promoters exhibited lower expression of OsNRAMP5 and lower accumulation of Mn in the grains (Fig. 7b, c).
Figure 7.
Haplotype analysis of the OsNRAMP5 promoter in different rice varieties. (a) OsNRAMP5 promoter haplotypes of rice varieties used in the study. (b) Relative expression level of OsNRAMP5 in the varieties for 3 haplotypes, the expression level was detected in the flag leaves at the grain-filling stage. (c) Grain Mn concentration in the varieties for 3 haplotypes.
Discussion
To date, hundreds of QTLs related to grain mineral elements have been identified in rice, but few have been fine mapped or cloned. In this study, we analyzed the Mn concentration in the grains of the RILs derived from the rice super hybrid LYPJ and found 12 putative QTLs in two testing environments. Among them, qGMN7.1, detected on the short arm of chromosome 7 in both environments, accounted for the largest proportion of phenotypic variation (Fig. S2 and Table S2). In the same chromosomal region, a major QTL was also detected by Ishikawa as qGMn7 using rice varieties Sasanishiki and Habataki as parents20, suggesting qGMN7.1 might be a common genetic factor for grain Mn concentrations in various rice varieties.
Previous studies showed that Mn content was significantly correlated with the contents of other mineral elements in grains, such as Mg, Fe, Zn, or Cu, suggesting that the contents of these elements might be controlled by common genes23,24. However, the concentrations of Mg, Fe, Zn, and Cu in the grains were nearly equal between CSSL-qGMN7.1 and 93–11 (Fig. 1d–g). Though the Cd concentration in the grains of CSSL-qGMN7.1 was much lower than that of 93–11, fine mapping of qGMN7.1 demonstrated that it had little influence on Cd accumulation in the grains (Table 1). These results implied that qGMN7.1 might be specialized for controlling Mn accumulation in the grains. Considering that agronomic traits could also affect the accumulation of elements in grains27,28, we investigated 9 traits of 93–11 and CSSL-qGMN7.1 and found no significant differences between them (Table S5). Therefore, CSSL-qGMN7.1 is an ideal material for rice breeding due to its improved Mn concentration and decreased Cd concentration in the grains without an accompanying loss of yield.
The role of OsNRAMP5 in controlling Mn uptake and transport has been reported in rice12–15,29. OsNRAMP5 was constitutively expressed in the roots and encodes a plasma membrane-localized protein that belongs to the natural resistance associated macrophage protein (NRAMP) family, whose members function as proton-coupled metal ion transporters that can transport Mn2+, Zn2+, Cu2+, Fe2+, Cd2+, Ni2+, Co2+, and Al3+ 12,30. OsNRAMP5 encodes a major transporter responsible for Mn uptake in rice; knockout of OsNRAMP5 resulted in a significant decline in grain Mn concentrations compared with the wild type12. Ishimaru et al. also suggested that OsNRAMP5 could play a role in Mn transport during flowering and seed development15.
Based on the RILs and the backcross population, we fine mapped qGMN7.1 to a 49.3-kb region containing OsNRAMP5 (Fig. 5d and Table 1). Although we did not find any alterations in the coding sequence of OsNRAMP5 between 93–11 and PA64s, we did find nucleotide differences in the promoter region (Figs 5e and 6a). These sequence variations lead to differences in the expression level of OsNRAMP5 and in Mn accumulation in the grains between CSSL-qGMN7.1 and 93–11 (Fig. 6b, c). Overexpression of OsNRAMP5 in the 93–11 variety increased the grain Mn concentration (Fig. 6f, g). Therefore, we inferred that the expression level of OsNRAMP5 contributed to Mn accumulation in the grains. Variations in promoter sequences commonly lead to phenotypic variation in rice31–35. The expression of OsNRAMP1 in roots was higher in high Cd-accumulating varieties (Habataki, Anjana Dhan, Jarjan) compared to low Cd-accumulating varieties (Sasanishiki, Nipponbare) due to a 400-bp deletion in the promoter region of OsNRAMP1 in the high Cd-accumulating varieties31. Consistent with previous reports, we also found that some low Mn-accumulating varieties had similar OsNRAMP5 promoter sequences as 93–11, which contains low concentrations of Mn in the grains, whereas high Mn-accumulating varieties, including Nipponbare, exhibited promoter sequences similar to PA64s, which is known for high grain Mn concentrations (Fig. 7).
Four major transport processes are involved in the accumulation of mineral elements: (1) root uptake, (2) root-to-shoot translocation by xylem flow, (3) distribution in aboveground tissues, and (4) remobilization from leaves9. In our study, Mn content in the flag leaves increased from the heading stage to maturity in the time-course experiment (Fig. 2b). Removal of the flag leaves at the heading and grain-filling stages did not affect Mn accumulation in the grains (Fig. 2c, d). Therefore, we speculated that elevation of Mn concentrations in the grain did not occur due to remobilization from the leaves. In addition, CSSL-qGMN7.1 and 93–11 showed little difference in Mn distribution (Fig. 3). However, higher Mn uptake activity was found in CSSL-qGMN7.1 compared to 93–11 (Fig. 4). Considering that OsNRAMP5 was constitutively expressed in roots (Fig. 6b, c) and that its expression was higher in CSSL-qGMN7.1 compared with 93–11, we concluded that OsNRAMP5 is responsible for the increased Mn concentrations in the grain by enhancing Mn uptake in roots.
OsNRAMP5 has also been reported to function as a Cd/Fe transporter12–14,36,37. OsNRAMP5-knockdown rice lines accumulated more Cd in the shoots, but the total Cd content was lower than in the wild-type plants36. The OsNRAMP5 knockout line lost the ability to take up Mn and Cd concurrently12, and the osnramp5 mutant exhibited decreased Cd (as well as Mn) concentrations in the straw and grain13. These studies demonstrated that the entry of Cd into rice root cells occurred via this Mn transporter, OsNRAMP5. However, in our study, CSSL-qGMN7.1 accumulated lower amounts of Cd than 93–11, contrary to the higher amounts of Mn in CSSL-qGMN7.1 compared to 93–11 (Fig. 1b, c). The recombinants that showed segregation at the qGMN7.1 region exhibited no significant differences in grain Cd concentrations (Table 1), and the overexpression lines of OsNRAMP5 did not accumulate more Cd in the grains compared with 93–11 (Fig. 6h). A possible explanation is that when plants are grown on relatively high-Mn and low-Cd soils (Hangzhou, pH = 6.04 ± 0.02; 480.35 ± 51.02 mg/kg Mn; 0.64 ± 0.12 mg/kg Cd), Cd does not readily accumulate in the grains. Additionally, an antagonistic effect may exist between Mn and Cd uptake. That is, OsNRAMP5 is mainly responsible for the transport of Mn, but not Cd when Mn is abundant. Alternatively, another locus may exist for grain Cd concentration in the substituted segments of CSSL-qGMN7.1 (Table 1), which displayed lower Cd accumulation levels when compared with 93–11.
Materials and Methods
Plant materials, and field and pot experiments
The 132 RILs of LYPJ were developed by a cross between an elite Oryza sativa ssp. indica inbred variety, 93–11, with low grain Mn concentration and an Oryza sativa ssp. indica photo-thermo-sensitive male sterile line, PA64s, with high grain Mn concentrations. The RILs and the two parents were grown in the field of CNRRI in Hangzhou (2013) and Hainan (2013), China, and the RIL population was in the F11 and F12 generation, respectively. For the determination of grain Mn, mature seeds of each line were collected from 6 plants in the middle row.
The chromosomal segment substitution line, CSSL-qGMN7.1, was selected from the advanced backcross population (BC4F2) derived from a cross of the recurrent parent 93–11 and the donor parent PA64s (Table S3). In 2015, 93–11 and CSSL-qGMN7.1 were grown in the paddy field of Hangzhou and Hainan. Both lines were also grown in pots inside a net enclosure in Hangzhou. Each pot was filled with 4 kg of sterilized paddy soil and amended with 2 mg/kg−1 CdCl2. The soil was maintained in a flooded state before heading, then kept moist until maturity.
In the paddy field of Hangzhou (2016), flag leaf blades from 93–11 and CSSL-qGMN7.1 were sampled at 0, 3, 6, 9, 12, 18, 24, 30, and 36 days after heading, and the grains were reaped at 6, 9, 12, 18, 24, 30, and 36 days after heading. At maturity, the aboveground parts of 93–11 and CSSL-qGMN7.1 were reaped and segregated as grain, lemma, panicle branches, flag leaf blade, flag leaf sheath, lower leaf blade, lower leaf sheath, node, and basal stem. In the pot experiment, the flag leaf blades were harvested at heading, filling, and maturity stages, while the grains were labeled and retained to maturity. All samples were dried at 65 °C for 3 d and then weighed before determination of Mn concentration.
Statistical analysis and QTL mapping
Statistical analysis was conducted by SAS (version 9.0). Broad-sense heritability was estimated as described by Singh and Chaudhary38. QTL analysis was performed with the MultiQTL package (www.mutiqtl.com) using the maximum likelihood interval mapping approach for the RILs. For major effect QTLs, the LOD threshold was obtained based on a permutation test (1,000 permutations, P = 0.05) for each dataset. We followed the suggestions by McCouch for the QTL nomenclature39.
Fine mapping of qGMN7.1
To fine map qGMN7.1, CSSL-qGMN7.1 was crossed with 93–11. A total of 4,943 segregating F2 individuals were grown in Hainan in 2015. Twelve recombinants were genotyped with previously developed insertion/deletion (InDel) markers supplied in Table S3. The progeny of these recombinants were grown and genotyped in Hangzhou (2016). Mature seeds were collected and prepared for mineral determination as described below.
Short-term Mn uptake experiment
To compare the transport activity for Mn between 93–11 and CSSL-qGMN7.1, we performed a short-term (30 min) uptake experiment according to a previous method12. The seedlings (28 d old) of these two lines were exposed to the nutrient solution without Mn for 1 week and then subjected to the uptake solution containing various concentrations of Mn (0.0, 0.1, 0.5, 5, 10, 50, or 100 mM) at 25 °C and 4 °C with four replicates per treatment. After 30 min, the roots were washed three times with 5 mM CaCl2 and separated from the shoots. The roots were dried at 70 °C for 3 d and used for mineral determination as described below.
Determination of metals in plant tissues
The dried samples were digested with a mixture of HNO3 (85%) and HClO4 (15%) at a gradient temperature (60 °C for 1 h, 120 °C for 1 h, 150 °C for 1 h, and up to 190 °C). The concentration of the metals in the digest solution was determined by atomic absorption spectrometry (Z-2000; Hitachi) and an inductively coupled plasma-mass spectrometer (7700X, Agilent Technologies) after dilution.
Quantitative reverse transcription PCR (qRT-PCR) analysis
Seedlings of 93–11 and CSSL-qGMN7.1 were grown in 1/2 Kimura B solution12 for 2 weeks, then the roots and shoots were harvested separately. OsNRAMP5 expression was investigated in different tissues from plants grown in the paddy field at booting stage, including root, basal stem, node, lower leaf sheath, lower leaf blade, flag leaf blade, flag leaf sheath, and panicle. Flag leaves of rice varieties were sampled at the heading stage. RNA was extracted by the Micro RNA Extraction kit (Axygen) and reverse transcribed into cDNA using a ReverTra Ace qPCR-RT kit (TOYOBA, Japan). Primers for qRT-PCR were described in a previous study12 (Table S3). Quantitative PCR was conducted on an ABI PRISM 7900HT Sequence Detector (Applied Biosystems) according to the manufacturer’s instructions. The relative expression of each transcript was obtained by comparison with the expression of rice actin1 (Table S3).
Relative promoter activity assays
The promoter fragments (2 kb) of OsNRAMP5 were amplified by PCR from the 93–11 and PA64s lines using the forward primer 5′-accatgattacgccaagcttGCGCATGTATCATTTGTTGT-3′ and the reverse primer 5′-aacgacggccagtgaattcCTCACTGCTCTCTCTCTCAA-3′, and were then cloned into the pCambia1391Z vector. The constructed pCAMBIA1391Z::93–11p and pCAMBIA1391Z::PA64sp plasmids were co-transformed with eGFP into rice protoplasts and transiently expressed40. After 12 h of incubation at 25 °C, protoplasts were collected for RNA extraction. The GUS expression level was detected by qRT-PCR with eGFP expression as the endogenous control.
The 35 S promoter of pAN580GFP was substituted with the OsNRAMP5 promoters from 93–11 and PA64s. The OsNRAMP5 promoter fragments were amplified by the forward primer 5′-aacgacggccagtgccGCGCATGTATCATTTGTTGT-3′ and the reverse primer 5′-tctagaggatccccgggtaccCTCACTGCTCTCTCTCTCAA-3′. The Pro93–11::GFP and ProPA64s::GFP plasmids were transformed into rice protoplasts and transiently expressed. After 12–16 h of incubation at 25 °C, green fluorescent signals were observed with an OLYMPUS IX71 confocal microscope.
Plasmid construction and rice transformation
The cDNA of OsNRAMP5 was amplified by PCR with the forward primer 5′-AAGGTACCATGGAGATTGAGAGAGAGAGC-3′ and the reverse primer 5′-AATCTAGACTACCTTGGGAGCGGGATGTC-3′, which include the KpnI and XbaI restriction sites, respectively. The amplified fragment was cloned into the pCAMBIA1300S vector for overexpression. The constructed vector was sequenced and introduced into 93–11 by Agrobacterium tumefaciens- (EHA105) mediated transformation. Thirteen independent transgenic plants of pCAMBIA1300S::OsNRAMP5 in the 93–11 background were obtained. Seedlings of these transgenic plants (T2 selected by hygromycin) and 93–11 were grown in 1/2 Kimura solution and transferred to pots at the four-leaf stage.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Nos 31521064 and 91535205).
Author Contributions
Z.G. and Q.Q. designed and supervised research; C.L., G.C., Y.L., K.H. and H.J. performed experiments; A.Z., B.R. and B.Z. provided technical assistances; C.L., Y.P., S.Y. and Z.G. analyzed data; C.L., G.C. and Z.G. wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-18090-7.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhenyu Gao, Email: zygao2000@hotmail.com.
Qian Qian, Email: qianqian188@hotmail.com.
References
- 1.Underwood EJ. Manganese-Trace elements in human and animal nutrition (fourth edition)-7. Quarterly Review of Biology. 1987;82:439. [Google Scholar]
- 2.Zota AR, et al. Maternal blood manganese levels and infant birth weight. Epidemiology. 2009;20:367–373. doi: 10.1097/EDE.0b013e31819b93c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wood RJ. Manganese and birth outcome. Nutr Rev. 2009;67:416–420. doi: 10.1111/j.1753-4887.2009.00214.x. [DOI] [PubMed] [Google Scholar]
- 4.Aschner JL, Aschner M. Nutritional aspects of manganese homeostasis. Mol Aspects Med. 2005;26:353–362. doi: 10.1016/j.mam.2005.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greger JL. Dietary standards for manganese: overlap between nutritional and toxicological studies. J Nutr. 1998;128:368S–371S. doi: 10.1093/jn/128.2.368S. [DOI] [PubMed] [Google Scholar]
- 6.Bjørklund G, Chartrand MS, Aaseth J. Manganese exposure and neurotoxic effects in children. Environ Res. 2017;155:380–384. doi: 10.1016/j.envres.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Hall JL, Williams LE. Transition metal transporters in plants. J Exp Bot. 2003;54:2601–2613. doi: 10.1093/jxb/erg303. [DOI] [PubMed] [Google Scholar]
- 8.Uraguchi S, Fujiwara T. Cadmium transport and tolerance in rice: perspectives for reducing grain cadmium accumulation. Rice. 2012;5:5. doi: 10.1186/1939-8433-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clemens S, Ma JF. Toxic heavy metal and metalloid accumulation in crop plants and foods. Annu Rev Plant Biol. 2016;67:489–512. doi: 10.1146/annurev-arplant-043015-112301. [DOI] [PubMed] [Google Scholar]
- 10.Ishimaru Y, et al. Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J. 2010;62:379–390. doi: 10.1111/j.1365-313X.2010.04158.x. [DOI] [PubMed] [Google Scholar]
- 11.Yamaji N, Sasaki A, Xia JX, Yokosho K, Ma JF. A node-based switch for preferential distribution of manganese in rice. Nat Commun. 2013;4:2442. doi: 10.1038/ncomms3442. [DOI] [PubMed] [Google Scholar]
- 12.Sasaki A, Yamaji N, Yokosho K, Ma JF. Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell. 2012;24:2155–2167. doi: 10.1105/tpc.112.096925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishikawa S, et al. Ion-beam irradiation, gene identification, and marker-assisted breeding in the development of low-cadmium rice. Proc Natl Acad Sci. 2012;109:19166–19171. doi: 10.1073/pnas.1211132109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishimaru Y, et al. Characterizing the role of rice NRAMP5 in Manganese, Iron and Cadmium Transport. Sci Rep. 2012;2:286. doi: 10.1038/srep00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishimaru Y, Bashir K, Nakanishi H, Nishizawa NK. OsNRAMP5, a major player for constitutive iron and manganese uptake in rice. Plant Signal Behav. 2012;7:763–766. doi: 10.4161/psb.20510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu K, et al. Quantitative trait loci controlling Cu, Ca, Zn, Mn and Fe content in rice grains. J Genetics. 2008;87:305–310. doi: 10.1007/s12041-008-0049-8. [DOI] [PubMed] [Google Scholar]
- 17.Shen XL, et al. QTL mapping for the content of five trace elements in brown rice. Molecular Plant Breeding. 2008;6:1061–1067. [Google Scholar]
- 18.Garcia-Oliveira AL, Tan L, Fu Y, Sun C. Genetic identification of quantitativetrait loci for contents of mineral nutrients in rice grain. J Integr Plant Biol. 2009;51:84–92. doi: 10.1111/j.1744-7909.2008.00730.x. [DOI] [PubMed] [Google Scholar]
- 19.Norton GJ, et al. Genetic mapping of the rice ionome in leaves and grain: identification of QTLs for 17 elements including arsenic, cadmium, iron and selenium. Plant & Soil. 2010;329:139–153. doi: 10.1007/s11104-009-0141-8. [DOI] [Google Scholar]
- 20.Ishikawa S, et al. A major quantitative trait locus for increasing cadmium-specific concentration in rice grain is located on the short arm of chromosome 7. J Exp Bot. 2010;61:923–934. doi: 10.1093/jxb/erp360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y, et al. QTL mapping for Mn, Fe, Zn and Cu contents in rice grains. Crops. 2012;6:77–81. [Google Scholar]
- 22.Du J, et al. Environmental effects on mineral accumulation in rice grains and identification of ecological specific QTLs. Environ Geochem Health. 2013;35:161–170. doi: 10.1007/s10653-012-9473-z. [DOI] [PubMed] [Google Scholar]
- 23.Zhang M, et al. Mapping and validation of quantitative trait loci associated with concentrations of 16 elements in unmilled rice grain. Theor Appl Genet. 2014;127:137–165. doi: 10.1007/s00122-013-2207-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nawaz Z, et al. Genome-wide association mapping of quantitative trait loci (QTLs) for contents of eight elements in brown rice (Oryza sativa L.) J Agric Food Chem. 2015;63:8008–8016. doi: 10.1021/acs.jafc.5b01191. [DOI] [PubMed] [Google Scholar]
- 25.Li, J. M. et al. Hybrid rice technology development: ensuring china’s food security. Ifpri Discussion Papers. 271–293 (2009).
- 26.Gao ZY, et al. Dissecting yield-associated loci in super hybrid rice by resequencing recombinant inbred lines and improving parental genomic sequences. Proc Natl Acad Sci. 2013;110:14492–14497. doi: 10.1073/pnas.1306579110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishikawa S, Ae N, Yano M. Chromosomal regions with quantitative trait loci controlling cadmium concentration in brown rice (Oryza sativa) New Phytol. 2005;168:345–350. doi: 10.1111/j.1469-8137.2005.01516.x. [DOI] [PubMed] [Google Scholar]
- 28.Anandan A, Rajiv G, Eswaran R, Prakash M. Genotypic variation and relationships between quality traits and trace elements in traditional and improved rice (Oryza sativa L.) genotypes. J Food Sci. 2011;76:H122–130. doi: 10.1111/j.1750-3841.2011.02135.x. [DOI] [PubMed] [Google Scholar]
- 29.Yang M, et al. OsNRAMP5 contributes to manganese translocation and distribution in rice shoots. J Exp Bot. 2014;65:4849–4861. doi: 10.1093/jxb/eru259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nevo Y, Nelson N. The NRAMP family of metal-ion transporters. Biochim Biophys Acta. 2006;1763:609–620. doi: 10.1016/j.bbamcr.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi R, et al. The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. J Exp Bot. 2011;62:4843–4850. doi: 10.1093/jxb/err136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, et al. Natural variation in GS5 plays an important role in regulating grainsize and yield in rice. Nat Genet. 2011;43:1266–1269. doi: 10.1038/ng.977. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, et al. Chalk5 encodes a vacuolar H(+)-translocating pyrophosphatase influencing grain chalkiness in rice. Nat Genet. 2014;46:398–404. doi: 10.1038/ng.2923. [DOI] [PubMed] [Google Scholar]
- 34.Song XJ, et al. Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc Natl Acad Sci. 2015;112:76–81. doi: 10.1073/pnas.1421127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang S, et al. The OsSPL16-GW7 regulatory module determines grain shapeand simultaneously improves rice yield and grain quality. Nat Genet. 2015;47:949–954. doi: 10.1038/ng.3352. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi R, et al. From laboratory to field: OsNRAMP5-knockdown rice is a promising candidate for Cd phytoremediation in paddy fields. PLoS One. 2014;9:e98816. doi: 10.1371/journal.pone.0098816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang L, et al. Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield. Sci Rep. 2017;7:14438. doi: 10.1038/s41598-017-14832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh, R. K. & Chaudhary, B. D. Biometrical methods in quantitative genetic analysis. Biometrical Methods in Quantitative Genetic Analysis (1977).
- 39.Mccouch SR. Gene Nomenclature System for Rice. Rice. 2008;1:72–84. doi: 10.1007/s12284-008-9004-9. [DOI] [Google Scholar]
- 40.Ren D, et al. Regulatory Role of OsMADS34 in the Determination of Glumes Fate, Grain Yield, and Quality in Rice. Front Plant Sci. 2016;7:1853. doi: 10.3389/fpls.2016.01853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.