Abstract
The three-dimensional (3D) structure of the intestine is a key determinant of differentiation and function; thus, preserving this architecture is an important consideration for studies of intestinal homeostasis and disease. Over the past decade, a number of systems for 3D intestinal organoid cultures have been developed and adapted to model a wide variety of biological phenomenon.
Purpose of this review
We discuss the current state of intestinal and colorectal cancer (CRC) 3D modeling, the most common methods for generating organoid cultures, and how these have yielded insights into intestinal physiology and tumor biology.
Recent findings
Organoids have been used to model numerous aspects of intestinal physiology and disease. Recent adaptations have further improved disease modeling and high-throughput therapeutic screening.
Summary
These studies show intestinal organoid models are a robust, highly tractable system which maintains many vital features of intestinal tissue, making them a pivotal step forward in the field of gastroenterology.
Keywords: Colorectal cancer, intestinal physiology, 3D culture, organoid, enteroid, model systems
Introduction
Tissue culture cell lines have proven invaluable to researchers and are readily available, easily expandable, amenable to genetic modification, and inexpensive to maintain [1]. However, as two-dimensional monolayers, cells lack complex interactions and microenvironmental cues, leading to morphologies and behaviors that can be vastly different that those observed in situ [2, 3]. Interestingly, researchers have noted that moving tissue culture cells from a 2D to a 3D culture environment and restoring interaction with the extracellular matrix (ECM) can induce dramatic effects on morphology, gene expression, differentiation, and metabolism, and thus more accurately model the native tumor state [4]. While these 3D culture techniques improved tumor modeling, it remained difficult to extrapolate results to non-diseased states since most cell lines are derived from human tumors. To this end, researchers modified 3D culture methods to allow culture of primary tissues and thus more accurately recapitulate normal tissue biology. To date, these normal ex vivo “organoid” models have been developed for a wide array of organ systems, such as the kidney, breast, and brain [5–7], which have yielded invaluable insight into organ homeostasis [8, 9].
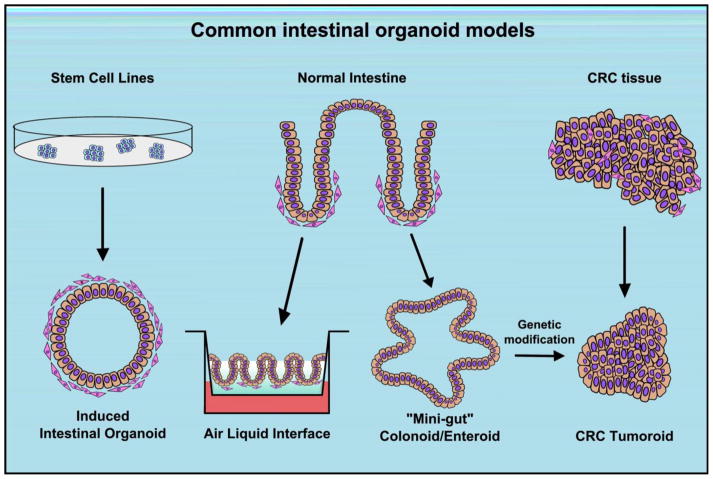
As the 3D structure of the intestine is a key regulator of differentiation and function, the ability to preserve architecture, spatial regulation, and cell polarization is crucial for studies of normal physiology [10]. Yet, long-term culture of intestinal tissue ex vivo was not described until 2009, when two groups described methods for ex vivo culture in seminal publications [11, 12]. To-date, these systems have been applied to growth of human- and mouse-derived intestinal tissues of both normal and malignant origin (Figure 1). In this review, we discuss the current state of colon and colorectal cancer (CRC) organoid culture methods and how these have yielded insights into intestinal physiology and tumor biology. Finally, we look to the future as new methodologies such xenograft growth and high-throughput screening make CRC tumor-derived organoid models viable candidates for precision tumor modeling to guide patient therapies.
Figure 1.
Overview of common methods for intestinal organoid culture
A note on nomenclature
Since 2009, methods for generating intestinal organoid cultures have been greatly expanded, with each method yielding cultures with different biologies, experimental strengths, and limitations. However, as culture varieties increase, the manner in which to describe intestinal 3D cultures has become somewhat ambiguous. In 2012, the Intestinal Stem Cell Consortium set forth nomenclature guidelines in an attempt to standardize descriptions and more readily differentiate between types of intestinal cultures [13]. Here, they reserve the term “organoid” for cultures which contain multiple cell types (particularly mesenchyme), while cultures of pure epithelial populations were designated as “enteroids” or “colonoids,” if from the small intestine or colon, respectively. While the terms “enteroid” and “colonoid” are still commonly used and aid distinction between epithelial cultures of small and large intestine, the restriction on the term “organoid” has proven somewhat cumbersome. In part, this is likely due to the fact that the term “organoid” has long been used in other organ systems to refer to all manners of 3D cultures, regardless of cell composition. Furthermore, the Clevers group refers to their purely epithelial cultures as organoids in the original 2009 publication, and have continued to do so in multiple high-impact publications over the past decade. Therefore, for the purpose of this review, we will utilize the terms “enteroid” and “colonoid” where applicable to aid distinction between small and large intestinal epithelial organoids, with “organoids” being an inclusive term for any type of 3D culture.
Ex vivo methods for culturing normal intestine
While both 2009 methods for generating intestinal organoids relied on ex vivo plating of harvested intestinal tissue, perhaps the most widely utilized method remains that described by Sato et al. [12]. Also referred to as the “mini-gut method” or “R-spondin method,” whole or partial crypts are isolated by exposure to chelating agents and then suspended in a basement membrane extract (BME) such as Matrigel. Addition of the growth factors R-spondin (WNT agonist), Noggin (a bone morphogenic protein inhibitor), and epithelial growth factor (EGF), promote stem cell expansion and cell proliferation. For the murine small intestine, these relatively simple conditions support robust cultures of pure epithelium that contain a full complement of common differentiated cell types, display a crypt-villus axis, and are amenable to serial passaging, expansion, and genetic modification. To date, this system has allowed successful culture of all segments of the small intestine (i.e., enteroids) as well as the colon (i.e., colonoids) [14–16]. However, culture conditions differ depending on the tissue source; colonoids require a higher degree of WNT3A stimulation than enteroids, and human-derived cultures require additional factors to achieve long term passage, such as nicotinamide, gastrin, the Alk4/5/7 inhibitor A-8301, and the p38 inhibitor SB202190 [17, 15]. Interestingly, both enteroid and colonoid cultures can be generated from single stem cells with minor modifications to growth factor requirements, allowing for studies of stem cell hierarchy and regenerative capabilities [12, 15, 16]. It is also notable that adult-derived mouse colonoid cultures are capable of engraftment in colonic epithelium following epithelial loss, indicating that mini-gut cultures are capable of normal intestinal function in vivo [18].
While powerful, a drawback to the mini-gut method is the lack of associated mesenchyme, which is a central regulator of the stem cell niche and differentiation in vivo. This presents a need for culture methods that preserve mesenchymal-epithelial interaction, such as the air-liquid interface (ALI) model developed by the Kuo group and published the same year as the work by Sato et al. [11]. The ALI method utilizes minced whole intestinal tissue, as opposed to isolated crypts, which are embedded in a collagen gel and then exposed to air in a transwell culture dish. Under these conditions isolated intestinal cells first form round, cystic organoids that progress to differentiated, crypt-like structures at the gel-air interface. Importantly, stromal cells such as myofibroblasts are readily apparent and closely associate with the intestinal crypts within the matrix. However, while these cultures contain all common differentiated cell types in the intestine and can be maintained in culture for over a year, proliferation is not as robust as the mini-gut model and they are unable to be propagated, making them less suited to large-scale or high-throughput experiments. Conversely, others have expanded on mini-gut-type methods by co-culturing with fibroblast feeder layers or fibroblast-conditioned media to more accurately model stem cell niche interactions [19–21].
It is also worth noting that intestinal organoids from primary tissues can be dramatically different based on whether the intestinal cells are fetal- or adult-derived. For example, in the ALI model, viability of fetal-derived cultures is higher than those derived from adult mice [11]. Using mini-gut culture methods, fetal intestine yields a predominantly spheroid population of undifferentiated intestine, termed fetal enterospheres (FEnS), which may also contain associated stromal cells [22, 23]. While these cultures are more difficult to use in studies of adult homeostasis without prior differentiation, fetal-derived mini-gut cultures are amenable to transplant and engraftment models and readily differentiate in vivo [23, 18]. Furthermore, exciting developments have been made in intestinal engineering utilizing multicellular fetal organoid units grown on polymer scaffolds [24]. While organoid units themselves are not grown ex vivo in this model, implantation into the omentum can yield functional, fully-developed intestinal tissue capable of rescuing rat models of short bowel syndrome [25].
Induced intestinal organoids
Established lines of embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) have long been used to generate differentiated tissue through timed exposure to growth and differentiation factors [26]. In 2005 it was shown that exposure to the TGFβ-like molecule, Activin A, directs stem cells to a definitive endodermal fate [27], which later serves as the basis to generate cells of mature endoderm lineages such as the pancreas and liver [28, 29]. More recently, seminal work by Spence et al. reported intestinal differentiation of endodermal cultures by concurrent exposure to fibroblast growth factor 4 and WNT3A, which formed induced human intestinal organoids (iHIOs) [30]. Interestingly, this differentiation process closely mimics early gut development and produces spheroids with a closely-associated mesenchymal layer. While early spheroid cultures are primarily undifferentiated, culture of iHIO spheroids in a mini-gut-like system allow for additional intestinal maturation and development of stem cell zones and villus-like protrusions after 1–2 months in culture. However, while these fully differentiated iHIOs can be expanded and passaged and contain cells of all intestinal lineages, drawbacks include the time necessary for maturation as well as the lack of ability to differentiate between large and small intestine.
Interestingly, ESC-derived cultures are widely considered to be more similar to fetal tissue than adult tissue and often express fetal tissue markers [30, 31]. Thus, like the fetal tissue-derived organoids, one strength of iHIOs is their ability to engraft; in fact, tissue-specific differentiation models may rely on implantation to promote tissue maturation [32]. iHIOs implanted into the mouse kidney capsule form mature intestinal epithelium around a central lumen, with the organoid giving rise to the majority of cells in the implant’s epithelium, lamina propria, muscularis mucosa, submucosa, and smooth muscle layers [33]. These organoids are also suitable to intestinal tissue engineering methods (as discussed above), and seeded organoids have been similarly grown on acellular scaffolds and implanted into the mouse omentum [34]. Indeed, the future of iHIOs in tissue engineering is bright, and currently work is underway to expand iHIO models to include other cell types important for intestinal function in order to generate fully autonomous intestinal tissue [35].
Novel uses of intestinal organoid models
The ability to preserve structure, cell-cell interactions, and differentiation in organoid cultures has permitted these methods to be widely adopted for studies of normal intestinal physiology. As proliferation is constantly maintained by an active stem cell compartment, intestinal organoid cultures are particularly well-suited for analysis of stem cell biology. Many of their earliest applications investigated mechanisms of stem cell regulation; for example, a series of elegant studies illustrated how Paneth cell secretion of WNT3A ligand maintains stem cell homeostasis [36, 37, 21, 38]. However, intestinal organoid cultures are not only suited to studies of stem cell biology. Indeed, fully-differentiated enterocytes in organoid cultures display functional glucose, peptide, and ion transporters, allowing for studies of nutrient absorption, anion and fluid secretion, and other cellular transport mechanisms [39–41]. By addition of short chain fatty acids to the culture media of intestinal mini-guts, researchers have also recently developed protocols to induce the formation of L-cells, a rare insulin-producing intestinal cell that could not previously be studied in vitro due to lack of an appropriate model system [42]. Researchers now have a novel avenue to investigate L-cell development, function, and dysregulation that may prove highly relevant to diabetes pathology. Finally, as enteroid and colonoid cultures have a pure epithelial cell pool without stromal contamination, they are easily adapted to single-cell analysis methods. In particular, recent work by Grun et al. utilizes enteroids in a single cell RNA-sequencing approach to thoroughly characterize markers of differentiation, lineage commitment, and specific cell types, and ultimately identified novel markers for rare cell linages [43]. Together, these studies and others have greatly expanded our understanding of the basic forces which drive intestinal differentiation and epithelial cell function.
Modeling intestinal disease states
While organoid technology has been widely applied to investigate normal intestinal function, others have utilized these approaches to study intestinal dysfunction and disease. For example, a number of studies have focused on cystic fibrosis (CF), a genetic disorder caused by mutation of the cystic fibrosis transmembrane conductance receptor (CFTR). While CF most notably affects the lungs, altered fluid and electrolyte homeostasis are also observed in the intestine. Interestingly, these transport-mediated phenotypes are preserved in rectal-derived cultures of CFTR mutant mice and human CF patients [39]. Because of this, patient-derived organoids are currently being utilized to determine individual drug response to CFTR potentiators and gene therapy-based treatments [44–46]. In addition to genetic disease models, organoid modeling has been applied to investigate intestinal injuries, such as radiation enteritis. These irradiation models involve both organoids generated from irradiated mice as well as direct organoid irradiation in order to identify modifiers of sensitivity, intestinal viability, stem cell function, and repair following radiation treatment [47–49].
Another interesting application of intestinal organoids is modeling bacterial and viral infections. Indeed, enteroids have been shown to support replication of rotavirus and norovirus, which provides relevant infection models as well as a means to passage viral cultures [50–53]. However, infection studies can be complicated by the fact that the apical surface, the most common site of bacterial/viral attachment, is not readily accessible with the most common 3D culture methods. To circumvent this, Wang et al. utilized ALI cultures to model infection with Clostridium difficile, a common cause of human diarrhea [54]. Others have instead opted to grow enteroids or colonoids as monolayers on transwells, which disrupts 3D architecture but preserves differentiation, allows polarization and exposes the apical surface. While this model has proven useful for studies of barrier function, transcytosis, and cell polarity, it also allows ready infection and detection of attaching and effacing lesions associated with infection [55, 56]. Conversely, intraluminal injection methods have been used to directly inoculate into the enteroid lumen, although this is a much more time consuming model and not amenable to high-throughput applications [57–59].
One drawback to organoid cultures is the lack of intestinal immune cell populations, important in both intestinal homeostasis and pathology such as inflammatory bowel disease. While phenotypes of epithelial-derived cultures from these patients have not yet been reported, intestinal organoids differentiated from hair follicle stem cells of CD patients demonstrate no differences in morphology, perhaps due to lack of immune-derived contributions [60]. However, useful insights have still been made for inflammatory diseases. The presence of a mesenchyme in iHIOs has made them useful for studies of fibrosis, a common complication of CD [61]. Furthermore, organoids can be co-cultured with immune cells such as isolated intraepithelial lymphocytes (IELs) in order to model immune/epithelial cell interactions that may influence pathology of inflammatory diseases. Indeed, these models have allowed researchers to observe direct interactions between IELs and intestinal epithelial cells and how these interactions affect T-cell differentiation, function, and dynamics [62, 63]. Others have utilized supernatants from immune cell cultures or purified cytokines to investigate mechanisms of mucosal immunology [64, 65]. Thus, it will be interesting to see if new modifications of organoid culture methods are developed to continue to adapt these models to studies of immune biology.
Development of colon cancer models
In the realm of intestinal dysfunction, perhaps the most exciting application of 3D culture methodology has been as a means to study intestinal tumor biology. While many methods have been described to-date for generation of tumor cultures, the most straightforward is likely those that start with normal intestinal organoids from inducible Apc or Ctnnb1 (i.e., β-catenin) floxed mouse lines. Subsequent delivery of Cre recombinase (or 4-hydroxytamoxifen to creER-expressing cultures) induces rapid transformation, leading to cultures resembling human tubular adenomas or undifferentiated spheroid structures in the ALI and mini-gut models, respectively [66, 67]. After transformation, these murine tumor cultures do not require exogenous R-spondin as do their normal epithelial counterparts, allowing researchers to select for pure populations of transformed tissues through growth factor depletion. Human intestinal organoid cultures can be similarly transformed by CRISPR/Cas9-mediated genome editing of the APC tumor suppressor gene [68, 69].
The more common method for establishing 3D tumor cultures is enzymatic digestion of established tumor tissue to generate tumor organoids. Cultures established from these methods will be collectively referred to as CRC “tumoroids” for the purpose of this review. In the method described by Sato et al., tumor tissue of mouse and human origin is subjected to an enzymatic digestion that generates small tumor fragments, which are then plated in Matrigel [17]. A similar protocol has been reported by Kondo et al., although cultures are placed in suspension overnight prior to plating in ECM [70]. In both cases, culture conditions select for purely epithelial structures of tumor cells which can be propagated in the matrix. Tumoroids isolated from human CRCs are highly reflective of the primary tumor and present with a range of patient-specific morphologies and corresponding histopathological features that are further maintained when xenotransplanted into immunodeficient mice [70–72]. Tumoroid cultures also show highly variable growth characteristics that are likely a result of their individual mutational profiles; recent work by Fujii et al. identifies genotype-specific growth characteristics conferred by specific mutations [71]. Finally, whole exome sequencing, copy number analysis, and mutational profiling also support high concordance between the tumoroid and primary tumor, even in late-passage tumoroid cultures maintained in culture for at least 6 months [71–73]. Together, these data suggest patient-derived tumoroids can be a powerful platform for investigating the biology of individual tumors.
Recently, tumoroid banks have been established from a wide array of CRC primary tumors which are now available to researchers [72]. However, as the majority of these tumors are early stage, microsatellite stable, and well to moderately differentiated, others have aimed to expand the available tumoroid lines to include more rare tumor subtypes, advanced tumors, and metastatic lesions. Indeed, the work by Fujii et al. establishes ideal culture methods based on patient genotype and reports growth factor modifications that permit culture of rare tumor subtypes such as sessile serrated adenomas [71]. Additionally, they report optimized protocols for culture of advanced stage colon and rectal cancers, in which tumor cell infiltration and adherence to the submucosal connective tissues confounds epithelial cell isolation by conventional methods. These modifications are then utilized to establish tumoroids from needle biopsies of diverse metastatic disease sites. Importantly, genetic analysis of these metastatic lines confirm common driver mutations with the matched primary tumor, although the metastasis-derived lineages are more invasive in immunocompromised mice in vivo [71, 74].
Modeling cancer biology
As 3D tumoroid models recapitulate many biological features of their primary tumor, a current focus has been analysis of their therapeutic potential. One exciting potential application of tumoroid technology is the ability to rapidly predict individual patient response to therapy, as patient-derived tumoroid models were recently found amenable to high-throughput drug screening assay. Indeed, studies by van de Wetering et al. describe a robotized screen in which they generated greater than 5,000 measurements of organoid-drug interactions in 19 tumor organoids and identified correlations between oncogenic mutations and response to targeted therapy [72]. Others similarly describe high-throughput drug screening using automated 384-well plates and ATP-based viability assays [75, 76]. In contrast, the current gold standard of patient tumor modeling remains patient-derived xenograft (PDX) models, in which fragments of the primary tumor are embedding ectopically in immunocompromised mice. There remain many strengths to PDX systems; for example, a recent study showed a strong correlation between the preclinical xenograft data and patient response [77]. However, PDX models remain expensive, require weeks to months for tumor growth, and are not similarly amenable to rapid high-throughput testing of combinatorial drugs [77]. Thus, tumoroid cultures may fill a unique niche in the therapeutic pipeline and pre-clinical drug development. While the reliability of ex vivo tumoroids to predict patient response to chemotherapeutics and targeted therapy has not yet been established, the Skala group recently demonstrated that 3D breast cancer tumor cultures closely predict xenograft treatment response [78]. It is also worth noting that CRC tumoroids themselves can be grown as xenografts [70, 72]. Thus, CRC tumoroids remain an intriguing option for high-throughput drug screening and predicting patient response in a time frame that will allow for direct clinical translation.
In addition to modeling aspects of individual tumor biology, tumoroids can provide a robust model for investigating CRC biology in general. Some studies have used these systems to analyze features that should be ubiquitous in cancers, such as polarity [79], while others have employed genetic modifications to study the role of particular oncogenes in a manner reminiscent of studies in 2D cell culture. Broadly, use of genetic approaches in tumoroid cultures have been described as “bottom-up” and “top-down” methods [80]. In the bottom-up approach, individual oncogenes are introduced into normal 3D cultures, allowing analysis of driver mutations without the often-complicated mutagenic backgrounds of 2D cell lines. To date, these studies have been used to model the classic multi-hit hypothesis for colorectal carcinogenesis with sequential and/or combinatorial alteration of APC, KRAS, TP53, SMAD4, and PIK3CA, with additional mutations generating increasingly aggressive tumoroid cultures [66, 68, 69]. For the top-down approach, additional alterations are added in the context of an established tumor. Many of these studies have analyzed the effects of particular genetic alterations observed in human tumors, such mutation of RNF43 or overexpression of miR-483 [81, 66]. Others have investigated particular genes of interest, such as PROX1 and BIM in the context of tumor cell survival and apoptosis resistance [82, 83]. Importantly, both bottom-up and top-down modeling approaches can take advantage of the inherent tractability of the 3D culture system, and 3D culture phenotypes can be combined with xenograft growth, high-throughput screening methods, drug treatment response, and metastasis modeling to investigate multiple aspects of tumor progression and effects on therapeutic efficacy/resistance. Indeed, we anticipate that a growing number of researchers will opt for 3D tumor culture models over standard cell lines in the coming years and that additional experimental techniques will be developed for 3D-based culture platforms.
Limitations of 3D modeling techniques
While only recently adopted by the GI field, intestinal organoid cultures have proven pivotal in extending our understanding of normal intestine, intestinal diseases, and colorectal cancer. However, as with any culture system, there remain inherent drawbacks to organoid modeling. For example, the quiescent stem cell populations which have described in vivo may not remain quiescent in culture, as sorted BM1 (+) cells can initiate organoid cultures despite being rarely active in vivo without tissue injury [84]. It remains a possibility that continuous growth factor stimulation, while yielding the ability to rapidly grow and expand organoid cultures, also alters intestinal homeostasis in a way consistent with injury and activates reserve stem cell populations. Perhaps a wider concern is that, while standard protocols have been published, a high degree of variability can occur in culture methods which may ultimately influence the results obtained across groups. One such factor is ECM composition; while Matrigel is the choice most consistent within the field, its composition is undefined and differs from lot-to-lot, potentially affecting reproducibility. The source of growth factors and their activity can also be a cause for variation, especially with the increasing use of undefined conditioned media from WNT3A, R-spondin, and Noggin-expressing cell lines.
Tumoroid establishment inherently involves sampling a small subset of a much larger tumor. Therefore, accurate patient-tumor modeling can also be confounded by intratumoral heterogeneity. As cancer is characterized by genomic instability, a single biopsy from which the tumoroid is derived is unlikely to reveal the wide range of genetic aberrations present in an entire tumor [85]. Indeed, in the tumoroid drug screenings completed by van de Wetering et al., some variation within individual organoid drug response was attributed to tumor heterogeneity. However, while not all tumor populations will be represented, it is likely that several subclones are initially present within tumoroid cultures. Unfortunately, these subclones can be preferentially selected over time, as mixtures of isolated populations of fluorescently-labeled organoids leads to progressive domination by single populations after 3–4 passages [71]. While tumor growth is similarly fluid in vivo, whether the selective pressures applied in culture select for the same clonal populations as the primary tumor has yet to be explored.
Conclusions
The goal of extended culture of primary intestinal cells has been a challenge in the field for decades. However, recent advances in organoid cultures permit study of biological phenomenon in intestinal cells in the context of normal differentiation and 3D structure. Here, we have outlined the most common methods of organoid culture for the normal and malignant intestine and have identified a subset of the recent advances that are made possible by these approaches. While limitations exist, these cultures offer advantages over tissue culture cell lines for many questions of basic biology. Furthermore, many in the field are looking ahead to perfecting high-throughput methods for patient-tumor modeling. Indeed, in an age of precision cancer therapies, it is now increasingly apparent that we also need precision cancer modeling. As the field of intestinal organoid cultures and its techniques are rapidly evolving, we look forward to many exciting and novel applications for both basic biology and translational research in the years to come.
Footnotes
Compliance with Ethics Guidelines
Conflict of Interest
Sarah P. Short, Patricia W. Costacurta, and Christopher S. Williams declare they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Masters JR. Human cancer cell lines: fact and fantasy. Nat Rev Mol Cell Biol. 2000;1(3):233–6. doi: 10.1038/35043102. [DOI] [PubMed] [Google Scholar]
- 2.Burdall SE, Hanby AM, Lansdown MR, Speirs V. Breast cancer cell lines: friend or foe? Breast Cancer Res. 2003;5(2):89–95. doi: 10.1186/bcr577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan C, Kumar C, Bohl S, Klingmueller U, Mann M. Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions. Mol Cell Proteomics. 2009;8(3):443–50. doi: 10.1074/mcp.M800258-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5(9):675–88. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 5.Qiao J, Sakurai H, Nigam SK. Branching morphogenesis independent of mesenchymal-epithelial contact in the developing kidney. Proc Natl Acad Sci U S A. 1999;96(13):7330–5. doi: 10.1073/pnas.96.13.7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simian M, Hirai Y, Navre M, Werb Z, Lochter A, Bissell MJ. The interplay of matrix metalloproteinases, morphogens and growth factors is necessary for branching of mammary epithelial cells. Development. 2001;128(16):3117–31. doi: 10.1242/dev.128.16.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3(5):519–32. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345(6194):1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 9.Shamir ER, Ewald AJ. Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nat Rev Mol Cell Biol. 2014;15(10):647–64. doi: 10.1038/nrm3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fatehullah A, Tan SH, Barker N. Organoids as an in vitro model of human development and disease. Nat Cell Biol. 2016;18(3):246–54. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 11.Ootani A, Li X, Sangiorgi E, Ho QT, Ueno H, Toda S, et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med. 2009;15(6):701–6. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–5. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 13.Stelzner M, Helmrath M, Dunn JC, Henning SJ, Houchen CW, Kuo C, et al. A nomenclature for intestinal in vitro cultures. Am J Physiol Gastrointest Liver Physiol. 2012;302(12):G1359–63. doi: 10.1152/ajpgi.00493.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuller MK, Faulk DM, Sundaram N, Shroyer NF, Henning SJ, Helmrath MA. Intestinal crypts reproducibly expand in culture. J Surg Res. 2012;178(1):48–54. doi: 10.1016/j.jss.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung P, Sato T, Merlos-Suarez A, Barriga FM, Iglesias M, Rossell D, et al. Isolation and in vitro expansion of human colonic stem cells. Nat Med. 2011;17(10):1225–7. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- ▪▪16.Mahe MM, Aihara E, Schumacher MA, Zavros Y, Montrose MH, Helmrath MA, et al. Establishment of Gastrointestinal Epithelial Organoids. Curr Protoc Mouse Biol. 2013;3(4):217–40. doi: 10.1002/9780470942390.mo130179. Standardized protocols for generating, passaging, freezing, and imaging enteroid and colonoid cultures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141(5):1762–72. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 18.Yui S, Nakamura T, Sato T, Nemoto Y, Mizutani T, Zheng X, et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5(+) stem cell. Nat Med. 2012;18(4):618–23. doi: 10.1038/nm.2695. [DOI] [PubMed] [Google Scholar]
- 19.Lahar N, Lei NY, Wang J, Jabaji Z, Tung SC, Joshi V, et al. Intestinal subepithelial myofibroblasts support in vitro and in vivo growth of human small intestinal epithelium. PLoS One. 2011;6(11):e26898. doi: 10.1371/journal.pone.0026898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gracz AD, Fuller MK, Wang F, Li L, Stelzner M, Dunn JC, et al. Brief report: CD24 and CD44 mark human intestinal epithelial cell populations with characteristics of active and facultative stem cells. Stem Cells. 2013;31(9):2024–30. doi: 10.1002/stem.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farin HF, Van Es JH, Clevers H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology. 2012;143(6):1518–29. e7. doi: 10.1053/j.gastro.2012.08.031. [DOI] [PubMed] [Google Scholar]
- 22.Mustata RC, Vasile G, Fernandez-Vallone V, Strollo S, Lefort A, Libert F, et al. Identification of Lgr5-independent spheroid-generating progenitors of the mouse fetal intestinal epithelium. Cell Rep. 2013;5(2):421–32. doi: 10.1016/j.celrep.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Fordham RP, Yui S, Hannan NR, Soendergaard C, Madgwick A, Schweiger PJ, et al. Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell. 2013;13(6):734–44. doi: 10.1016/j.stem.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi RS, Vacanti JP. Preliminary studies of tissue-engineered intestine using isolated epithelial organoid units on tubular synthetic biodegradable scaffolds. Transplant Proc. 1997;29(1–2):848–51. doi: 10.1016/s0041-1345(96)00164-9. [DOI] [PubMed] [Google Scholar]
- 25.Grikscheit TC, Siddique A, Ochoa ER, Srinivasan A, Alsberg E, Hodin RA, et al. Tissue-engineered small intestine improves recovery after massive small bowel resection. Ann Surg. 2004;240(5):748–54. doi: 10.1097/01.sla.0000143246.07277.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hynds RE, Giangreco A. Concise review: the relevance of human stem cell-derived organoid models for epithelial translational medicine. Stem Cells. 2013;31(3):417–22. doi: 10.1002/stem.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D’Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23(12):1534–41. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 28.D’Amour KA, Bang AG, Eliazer S, Kelly OG, Agulnick AD, Smart NG, et al. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24(11):1392–401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 29.Cai J, Zhao Y, Liu Y, Ye F, Song Z, Qin H, et al. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology. 2007;45(5):1229–39. doi: 10.1002/hep.21582. [DOI] [PubMed] [Google Scholar]
- 30.Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470(7332):105–9. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dye BR, Hill DR, Ferguson MA, Tsai YH, Nagy MS, Dyal R, et al. In vitro generation of human pluripotent stem cell derived lung organoids. Elife. 2015:4. doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Przyborski SA. Differentiation of human embryonic stem cells after transplantation in immune-deficient mice. Stem Cells. 2005;23(9):1242–50. doi: 10.1634/stemcells.2005-0014. [DOI] [PubMed] [Google Scholar]
- ▪33.Watson CL, Mahe MM, Munera J, Howell JC, Sundaram N, Poling HM, et al. An in vivo model of human small intestine using pluripotent stem cells. Nat Med. 2014;20(11):1310–4. doi: 10.1038/nm.3737. Transplatation of human intestinal organoids yields differentiated, functional intestinal tissue in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ▪34.Finkbeiner SR, Freeman JJ, Wieck MM, El-Nachef W, Altheim CH, Tsai YH, et al. Generation of tissue-engineered small intestine using embryonic stem cell-derived human intestinal organoids. Biol Open. 2015;4(11):1462–72. doi: 10.1242/bio.013235. Human intestinal organoids grown on scaffolds produce mature intestinal tissue and provides pivitol proof of concept for iHIO-based tissue engineering. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinagoga KL, Wells JM. Generating human intestinal tissues from pluripotent stem cells to study development and disease. EMBO J. 2015;34(9):1149–63. doi: 10.15252/embj.201490686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469(7330):415–8. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.VanDussen KL, Carulli AJ, Keeley TM, Patel SR, Puthoff BJ, Magness ST, et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139(3):488–97. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farin HF, Jordens I, Mosa MH, Basak O, Korving J, Tauriello DV, et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature. 2016;530(7590):340–3. doi: 10.1038/nature16937. [DOI] [PubMed] [Google Scholar]
- 39.Dekkers JF, Wiegerinck CL, de Jonge HR, Bronsveld I, Janssens HM, de Winter-de Groot KM, et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat Med. 2013;19(7):939–45. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- 40.Zietek T, Rath E, Haller D, Daniel H. Intestinal organoids for assessing nutrient transport, sensing and incretin secretion. Sci Rep. 2015;5:16831. doi: 10.1038/srep16831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Foulke-Abel J, In J, Yin J, Zachos NC, Kovbasnjuk O, Estes MK, et al. Human Enteroids as a Model of Upper Small Intestinal Ion Transport Physiology and Pathophysiology. Gastroenterology. 2016;150(3):638–49. e8. doi: 10.1053/j.gastro.2015.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petersen N, Reimann F, Bartfeld S, Farin HF, Ringnalda FC, Vries RG, et al. Generation of L cells in mouse and human small intestine organoids. Diabetes. 2014;63(2):410–20. doi: 10.2337/db13-0991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ▪43.Grun D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525(7568):251–5. doi: 10.1038/nature14966. Use of enteroid cultures with a single-cell RNA-sequencing approach. Enhances knowledge of intestinal differentiation pathways and identifies new markers of rare cell types. [DOI] [PubMed] [Google Scholar]
- 44.Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13(6):653–8. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 45.Vidovic D, Carlon MS, da Cunha MF, Dekkers JF, Hollenhorst MI, Bijvelds MJ, et al. rAAV-CFTRDeltaR Rescues the Cystic Fibrosis Phenotype in Human Intestinal Organoids and Cystic Fibrosis Mice. Am J Respir Crit Care Med. 2016;193(3):288–98. doi: 10.1164/rccm.201505-0914OC. [DOI] [PubMed] [Google Scholar]
- 46.Dekkers JF, Berkers G, Kruisselbrink E, Vonk A, de Jonge HR, Janssens HM, et al. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci Transl Med. 2016;8(344):344ra84. doi: 10.1126/scitranslmed.aad8278. [DOI] [PubMed] [Google Scholar]
- 47.Poindexter SV, Reddy VK, Mittal MK, Williams AM, Washington MK, Harris E, et al. Transcriptional corepressor MTG16 regulates small intestinal crypt proliferation and crypt regeneration after radiation-induced injury. Am J Physiol Gastrointest Liver Physiol. 2015;308(6):G562–71. doi: 10.1152/ajpgi.00253.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reddy VK, Short SP, Barrett CW, Mittal MK, Keating CE, Thompson JJ, et al. BVES Regulates Intestinal Stem Cell Programs and Intestinal Crypt Viability after Radiation. Stem Cells. 2016;34(6):1626–36. doi: 10.1002/stem.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilbert S, Nivarthi H, Mayhew CN, Lo YH, Noah TK, Vallance J, et al. Activated STAT5 confers resistance to intestinal injury by increasing intestinal stem cell proliferation and regeneration. Stem Cell Reports. 2015;4(2):209–25. doi: 10.1016/j.stemcr.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finkbeiner SR, Zeng XL, Utama B, Atmar RL, Shroyer NF, Estes MK. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. MBio. 2012;3(4):e00159–12. doi: 10.1128/mBio.00159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kovbasnjuk O, Zachos NC, In J, Foulke-Abel J, Ettayebi K, Hyser JM, et al. Human enteroids: preclinical models of non-inflammatory diarrhea. Stem Cell Res Ther. 2013;4(Suppl 1):S3. doi: 10.1186/scrt364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ▪52.Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, et al. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353(6306):1387–93. doi: 10.1126/science.aaf5211. Use of human enteroid cultures to maintain and study norovirus cultures. Enteroid use overcomes the prior lack of a robust norovirus cultivation system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ▪53.Saxena K, Blutt SE, Ettayebi K, Zeng XL, Broughman JR, Crawford SE, et al. Human Intestinal Enteroids: a New Model To Study Human Rotavirus Infection, Host Restriction, and Pathophysiology. J Virol. 2015;90(1):43–56. doi: 10.1128/JVI.01930-15. Patient-derived enteroids provide a highly relevant model to analyze susceptibility to rotavirus infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X, Yamamoto Y, Wilson LH, Zhang T, Howitt BE, Farrow MA, et al. Cloning and variation of ground state intestinal stem cells. Nature. 2015;522(7555):173–8. doi: 10.1038/nature14484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ▪55.Moon C, VanDussen KL, Miyoshi H, Stappenbeck TS. Development of a primary mouse intestinal epithelial cell monolayer culture system to evaluate factors that modulate IgA transcytosis. Mucosal Immunol. 2014;7(4):818–28. doi: 10.1038/mi.2013.98. Describes growth of organoids on transwell inserts to allow monolayer formation, polarization, and accessibitlity of the apical surface. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.In J, Foulke-Abel J, Zachos NC, Hansen AM, Kaper JB, Bernstein HD, et al. Enterohemorrhagic Escherichia coli reduce mucus and intermicrovillar bridges in human stem cell-derived colonoids. Cell Mol Gastroenterol Hepatol. 2016;2(1):48–62. e3. doi: 10.1016/j.jcmgh.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang YG, Wu S, Xia Y, Sun J. Salmonella-infected crypt-derived intestinal organoid culture system for host-bacterial interactions. Physiol Rep. 2014;2(9) doi: 10.14814/phy2.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Forbester JL, Goulding D, Vallier L, Hannan N, Hale C, Pickard D, et al. Interaction of Salmonella enterica Serovar Typhimurium with Intestinal Organoids Derived from Human Induced Pluripotent Stem Cells. Infect Immun. 2015;83(7):2926–34. doi: 10.1128/IAI.00161-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leslie JL, Huang S, Opp JS, Nagy MS, Kobayashi M, Young VB, et al. Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect Immun. 2015;83(1):138–45. doi: 10.1128/IAI.02561-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hohwieler M, Illing A, Hermann PC, Mayer T, Stockmann M, Perkhofer L, et al. Human pluripotent stem cell-derived acinar/ductal organoids generate human pancreas upon orthotopic transplantation and allow disease modelling. Gut. 2016 doi: 10.1136/gutjnl-2016-312423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodansky ES, Johnson LA, Huang S, Spence JR, Higgins PD. Intestinal organoids: a model of intestinal fibrosis for evaluating anti-fibrotic drugs. Exp Mol Pathol. 2015;98(3):346–51. doi: 10.1016/j.yexmp.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ▪62.Rogoz A, Reis BS, Karssemeijer RA, Mucida D. A 3-D enteroid-based model to study T-cell and epithelial cell interaction. J Immunol Methods. 2015;421:89–95. doi: 10.1016/j.jim.2015.03.014. Enteroid co-culture with T lymphocytes provides a means to study mucosal immunology and model immune/epithelial interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nozaki K, Mochizuki W, Matsumoto Y, Matsumoto T, Fukuda M, Mizutani T, et al. Co-culture with intestinal epithelial organoids allows efficient expansion and motility analysis of intraepithelial lymphocytes. J Gastroenterol. 2016;51(3):206–13. doi: 10.1007/s00535-016-1170-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Farin HF, Karthaus WR, Kujala P, Rakhshandehroo M, Schwank G, Vries RG, et al. Paneth cell extrusion and release of antimicrobial products is directly controlled by immune cell-derived IFN-gamma. J Exp Med. 2014;211(7):1393–405. doi: 10.1084/jem.20130753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lindemans CA, Calafiore M, Mertelsmann AM, O’Connor MH, Dudakov JA, Jenq RR, et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528(7583):560–4. doi: 10.1038/nature16460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ▪▪66.Li X, Nadauld L, Ootani A, Corney DC, Pai RK, Gevaert O, et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat Med. 2014;20(7):769–77. doi: 10.1038/nm.3585. Bottom-up approach in murine-derived ALI organoids to sequentially model oncogenic tranformation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sakamori R, Yu S, Zhang X, Hoffman A, Sun J, Das S, et al. CDC42 inhibition suppresses progression of incipient intestinal tumors. Cancer Res. 2014;74(19):5480–92. doi: 10.1158/0008-5472.CAN-14-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ▪▪68.Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015;21(3):256–62. doi: 10.1038/nm.3802. Bottom-up approach in human colonoids to model oncogenic tranformation. Also demonstrates feasibility of the CRISPR-Cas9 approach in intestinal organoids. [DOI] [PubMed] [Google Scholar]
- ▪69.Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521(7550):43–7. doi: 10.1038/nature14415. Bottom-up approach in iHIOs using CRISPR-Cas9 technology. [DOI] [PubMed] [Google Scholar]
- 70.Kondo J, Endo H, Okuyama H, Ishikawa O, Iishi H, Tsujii M, et al. Retaining cell-cell contact enables preparation and culture of spheroids composed of pure primary cancer cells from colorectal cancer. Proc Natl Acad Sci U S A. 2011;108(15):6235–40. doi: 10.1073/pnas.1015938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ▪▪71.Fujii M, Shimokawa M, Date S, Takano A, Matano M, Nanki K, et al. A Colorectal Tumor Organoid Library Demonstrates Progressive Loss of Niche Factor Requirements during Tumorigenesis. Cell Stem Cell. 2016;18(6):827–38. doi: 10.1016/j.stem.2016.04.003. Expansion of available tumoroid banks and updated methodologies to culture more rare subtypes, advanced tumors, and metastatic disease. Also includes genotype-phenotype analysis of growth factor requirements. [DOI] [PubMed] [Google Scholar]
- ▪▪72.van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161(4):933–45. doi: 10.1016/j.cell.2015.03.053. Generation of a large CRC tumoroid panel with mutational and histological analysis. Provides proof of concept for tumoroids as models for individual patient tumor biology and therapuetic modeling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee SH, Hong JH, Park HK, Park JS, Kim BK, Lee JY, et al. Colorectal cancer-derived tumor spheroids retain the characteristics of original tumors. Cancer Lett. 2015;367(1):34–42. doi: 10.1016/j.canlet.2015.06.024. [DOI] [PubMed] [Google Scholar]
- ▪74.Weeber F, van de Wetering M, Hoogstraat M, Dijkstra KK, Krijgsman O, Kuilman T, et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc Natl Acad Sci U S A. 2015;112(43):13308–11. doi: 10.1073/pnas.1516689112. Molecular profiling of tumoroid and matched metastasis. High genetic coorrelation supports the use of tumoroid culture to model tumor biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ▪75.Francies HE, Barthorpe A, McLaren-Douglas A, Barendt WJ, Garnett MJ. Drug Sensitivity Assays of Human Cancer Organoid Cultures. Methods Mol Biol. 2016 doi: 10.1007/7651_2016_10. Description and validation of high-throughput screening in human CRC tumoroids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boehnke K, Iversen PW, Schumacher D, Lallena MJ, Haro R, Amat J, et al. Assay Establishment and Validation of a High–Throughput Screening Platform for Three-Dimensional Patient-Derived Colon Cancer Organoid Cultures. J Biomol Screen. 2016;21(9):931–41. doi: 10.1177/1087057116650965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med. 2015;21(11):1318–25. doi: 10.1038/nm.3954. [DOI] [PubMed] [Google Scholar]
- 78.Walsh AJ, Cook RS, Sanders ME, Aurisicchio L, Ciliberto G, Arteaga CL, et al. Quantitative optical imaging of primary tumor organoid metabolism predicts drug response in breast cancer. Cancer Res. 2014;74(18):5184–94. doi: 10.1158/0008-5472.CAN-14-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Okuyama H, Kondo J, Sato Y, Endo H, Nakajima A, Piulats JM, et al. Dynamic Change of Polarity in Primary Cultured Spheroids of Human Colorectal Adenocarcinoma and Its Role in Metastasis. Am J Pathol. 2016;186(4):899–911. doi: 10.1016/j.ajpath.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 80.Neal JT, Kuo CJ. Organoids as Models for Neoplastic Transformation. Annu Rev Pathol. 2016;11:199–220. doi: 10.1146/annurev-pathol-012615-044249. [DOI] [PubMed] [Google Scholar]
- 81.Yan HH, Lai JC, Ho SL, Leung WK, Law WL, Lee JF, et al. RNF43 germline and somatic mutation in serrated neoplasia pathway and its association with BRAF mutation. Gut. 2016 doi: 10.1136/gutjnl-2016-311849. [DOI] [PubMed] [Google Scholar]
- 82.Wiener Z, Band AM, Kallio P, Hogstrom J, Hyvonen V, Kaijalainen S, et al. Oncogenic mutations in intestinal adenomas regulate Bim-mediated apoptosis induced by TGF-beta. Proc Natl Acad Sci U S A. 2014;111(21):E2229–36. doi: 10.1073/pnas.1406444111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wiener Z, Hogstrom J, Hyvonen V, Band AM, Kallio P, Holopainen T, et al. Prox1 promotes expansion of the colorectal cancer stem cell population to fuel tumor growth and ischemia resistance. Cell Rep. 2014;8(6):1943–56. doi: 10.1016/j.celrep.2014.08.034. [DOI] [PubMed] [Google Scholar]
- 84.Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A. 2012;109(2):466–71. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]