Abstract
DNA primases recognize single-stranded DNA (ssDNA) sequences to synthesize RNA primers during lagging-strand replication. Arabidopsis thaliana encodes an ortholog of the DNA primase-helicase from bacteriophage T7, dubbed AtTwinkle, that localizes in chloroplasts and mitochondria. Herein, we report that AtTwinkle synthesizes RNA primers from a 5′-(G/C)GGA-3′ template sequence. Within this sequence, the underlined nucleotides are cryptic, meaning that they are essential for template recognition but are not instructional during RNA synthesis. Thus, in contrast to all primases characterized to date, the sequence recognized by AtTwinkle requires two nucleotides (5′-GA-3′) as a cryptic element. The divergent zinc finger binding domain (ZBD) of the primase module of AtTwinkle may be responsible for template sequence recognition. During oligoribonucleotide synthesis, AtTwinkle shows a strong preference for rCTP as its initial ribonucleotide and a moderate preference for rGMP or rCMP incorporation during elongation. RNA products synthetized by AtTwinkle are efficiently used as primers for plant organellar DNA polymerases. In sum, our data strongly suggest that AtTwinkle primes organellar DNA polymerases during lagging strand synthesis in plant mitochondria and chloroplast following a primase-mediated mechanism. This mechanism contrasts to lagging-strand DNA replication in metazoan mitochondria, in which transcripts synthesized by mitochondrial RNA polymerase prime mitochondrial DNA polymerase γ.
INTRODUCTION
Organelles are products of early bacterial endosymbiotic events. Phylogenetic analyses, however, reveal that the gene products responsible for transcription and replication in human and yeast mitochondria share an evolutionary origin with the replication apparatus of T-odd bacteriophages (1–3). In the T7 replisome, three bacteriophage-encoded proteins and one host protein coordinate DNA synthesis: T7 DNA polymerase (gp5), T7 primase-helicase (T7 primase-helicase or gp4), single-stranded DNA binding protein (T7 SSB or gp2.5) (4,5) and Escherichia coli′s thioredoxin. T7 primase-helicase is a modular protein that includes a primase module that synthesizes primers for the lagging-strand T7 DNA polymerase and a helicase module that processively unwinds dsDNA (4,6,7). T7 primase belongs to the family of prokaryotic primases that is composed of a RNA polymerization domain (RPD) that catalyzes ribonucleotide addition and a zinc-binding domain (ZBD) involved in ssDNA template recognition (8,9). Organellar proteins derived from T7-odd bacteriophages have undergone evolutionary changes with respect to the ancestral bacteriophage proteins (10–12). An example of these changes occurs in the metazoan replicative mitochondrial DNA helicase TWINKLE (T7 gp4-like protein with intra-mitochondrial nucleoid localization). Metazoan TWINKLE lacks the cysteines necessary for zinc coordination in the region homologous to the ZBD of T7 primase and the conserved residues in the RPD-like region required for RNA synthesis (13,14). In metazoans, transcripts generated by mitochondrial RNA polymerase are used as primers during leader and lagging strand-synthesis (14). Furthermore, fungi are devoid of a TWINKLE homologue, and short RNA transcripts synthesized by mitochondrial RNA polymerases may be used as primers (10,15–18).
Prokaryotic primases recognize a variety of template sequences. For example, bacterial primases from Staphylococcus aeurus, Aquifex aeolicus, E. coli, and primases-helicases from bacteriophages T4, T7 and SP6 recognize 5′-CTA-3′, 5′-CCC-3′, 5′-CTG-3′, 5′-GTT-3′, 5′-GTC-3′ and 5′-GCA-3′, respectively, as ssDNA template sequences (19–25). In a primase, the identity of the middle base of a minimal trinucleotide recognition sequence determines the identity of the initial or 5′ NTP, and the 5′ base determines 3′ NMP incorporation specificity (9,26). Despite the diversity in template recognition sequences, in all primases studied to date the 3′-nucleotide of the initiation triplet sequence is not transcribed. It is required, however, for template recognition, and for this reason it is called cryptic.
Seminal work predicted that TWINKLE harbors both primase and helicase activities in most eukaryotes, except in metazoans where this enzyme is predicted only have helicase acitivity (2,27). The nuclear-encoded TWINKLE homolog from the flowering plant Arabidopsis thaliana (dubbed AtTwinkle) is a 709-residue protein that is translocated into mitochondria and chloroplasts owing to its dual N-terminal targeting sequence (28–32). The module with a predicted primase activity contains the six conserved regions present in prokaryotic primases (2,27). Motif I corresponds to the ZBD necessary for template recognition (26), whereas regions II to VI are located in the RNAP domain. In vitro characterization of AtTwinkle demonstrated that this enzyme has both primase and helicase activities (28,29,32). However, its preferred ssDNA template sequence, its putative role in organellar replication, and its interactions with plant organellar DNA polymerases remain unclear.
Herein, we found that AtTwinkle requires for template recognition a ssDNA template sequence that consists of two cryptic nucleotides and that RNA products generated by AtTwinkle are used as primers for plant organellar DNA polymerases. Our data suggests that the plant organellar replisome functionally resembles the replisome of T-odd bacteriophages.
MATERIALS AND METHODS
Expression and purification of recombinant proteins
The open reading frame corresponding to the predicted processed AtTwinkle (residues 92–709) was codon optimized for bacterial expression and cloned into a pUC19 vector. Open reading frames of AtPrimase-Helicase and AtPrimase were PCR amplified using the cloning vector as a template. The primase domain of bacteriophage T7 primase-helicase (residues 1–262) was PCR amplified from genomic T7 DNA. AtPrimase-Helicase (residues 92–709) and AtPrimase (residues 92–410) gene products were subcloned into a modified pET19 vector between NdeI and BamHI restriction sites, whereas T7 primase was cloned into a pET11 vector. The modified pET19 vector adds a nine-histidine tag at the N-terminus of each construct before the cleavage site of the human rhinovirus 3C protease (PreScission protease, GE Healthcare) allowing the histidine tag to be removed by specific proteolysis. Protein expression of AtPrimase-helicase, AtPrimase, and T7 primase were carried out in an E. coli BL21(DE3) strain at 21°C in Luria Bertani medium supplemented with 0.1 mM ZnCl2. After cell cultures reached an OD600 between 0.6 to 0.8, protein expression was induced by adding 1 mM IPTG and the cell cultures were incubated for 12 h. Cell cultures were harvested by centrifugation and resuspended in 30 ml of buffer A (20 mM Tris–HCl pH 7.5 and 150 mM NaCl), and freeze-thawed three times in the presence of lysozyme (0.2 mg/ml). Clear bacterial lysate was collected by centrifugation and loaded onto a Ni-NTA column by gravity. Proteins were subsequently eluted from the column with buffer A containing 0, 50, 100, 250 or 500 mM imidazole. Protein containing fractions were dialyzed in 20 mM Tris–HCl pH 7.5, 25 mM NaCl and 10% glycerol. Proteins then were loaded in a HiTrap HP heparin column and washed with a linear gradient from 0 to 1000 mM of NaCl. Fractions containing pure protein were pooled and concentrated. Protein samples were further purified by gel filtration, using a Superdex 75 10/300 for AtPrimase and a Superdex 200 10/300 for the AtPrimase-Helicase. Both gel filtration columns were equilibrated in 20 mM Tris–HCl pH 7.5, 25 mM NaCl and 10% glycerol. T7 primase was expressed and purified as previously described (33).
To determine the molar amount of oligoribonucleotide synthesis product by T7 and Arabidopsis primase, the radioactive [α-32P]-ATP substrate and the oligonucleotide RNAs were quantified with ImageQuant. The processed intensity was subjected to the following equation, previously derived by the Patel group (34):
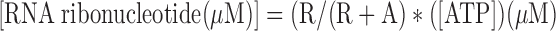 |
where R refers to the band intensity observed for each ribonucleotide, A is the intensity of the radioactively label ATP substrate and [ATP] refers to the total molar concentration of ATP (labeled and unlabeled). The molar amount of RNA products is obtained by dividing the number of ATPs in the RNA chain.
Template-directed oligoribonucleotide synthesis
Primase reactions were assayed in a buffer containing 40 mM Tris–HCl pH 7.5, 50 mM potassium glutamate, 10 mM MgCl2 and 10 mM DTT. Primase reactions contained 100 μM of the indicated NTPs and 10 μCi of [α-32P]-NTP (3,000 Ci/mM). After incubation at 30°C for 60 min, reactions were terminated by 15 mM of EDTA. Loading buffer (95% formamide, 0.1% xylene cyanol) was immediately added to the reaction and the products were separated on a 27% denaturing polyacrylamide gel containing 3 M urea. Each primase reaction contained varied amounts of recombinant protein or single-stranded DNA template as indicated in the figure legends. The ribonucleotide products were analyzed by phosphorimaging on a Storm 860 PhosphorImager.
Phylogenetic analysis and molecular modeling
The phylogenetic tree was constructed using the amino acid residue sequences of enzymes related to T7 primase-helicase. The amino acid residue sequences were aligned in Clustal Omega. This sequence alignment was used to construct a dendogram with the Neighbor-Joining method of the Molecular Evolutionary Genetic Analysis (MEGA) software. The robustness of the dendogram was assessed by a bootstrap analysis of 1000 replicates. To build the structural model of AtPrimase, its amino acid sequence (residues 98 to 402) was aligned with the corresponding amino acid sequence present in the bacteriophage T7 primase crystal structure (PDB ID: 1NUI) (8) and a homology model was constructed based on this alignment. A structural alignment was carried out using the align module of the Molecular Operating Environment (MOE). Ten models were generated and each was minimized using the CHARMM27 force field.
RESULTS
AtTwinkle is an evolutionarily conserved prokaryotic primase-helicase
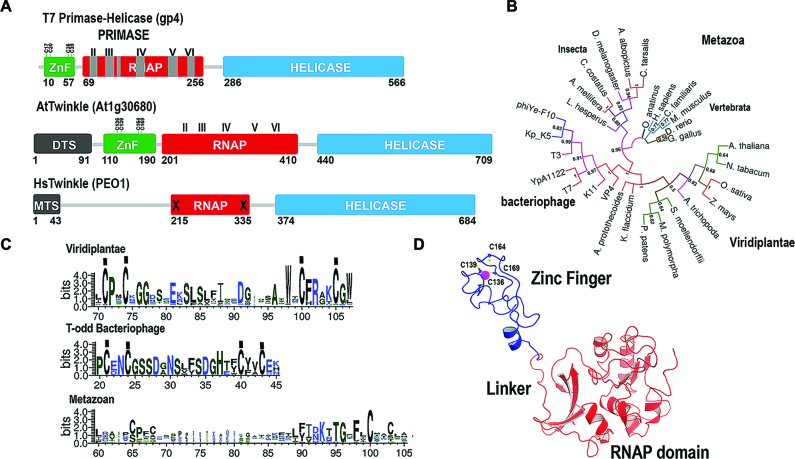
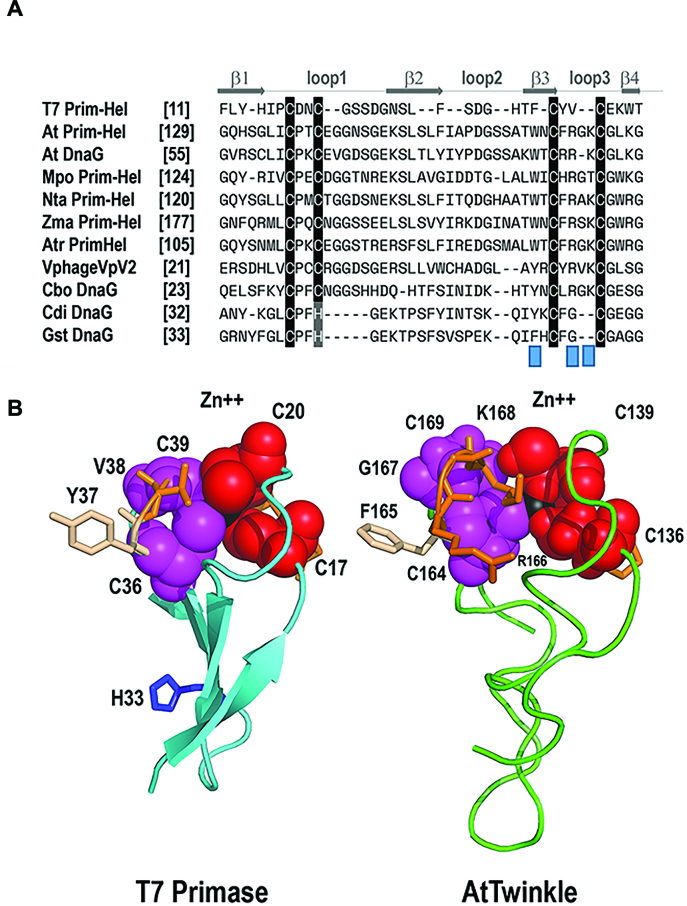
Computational analysis predicts that the nuclear-encoded AtTwinkle is processed at residue 91 during its translocation into organelles (28). The residues corresponding to the predicted processed AtTwinkle have only 22% amino acid identity with T7 DNA primase-helicase. Despite this limited amino acid identity, seminal work by Shutt and Gray (1,2) identified that green plants and algae contain the four conserved cysteines predicted to coordinate a Zn2+ atom and the catalytic amino acids for RNA polymerase and DNA helicase activities (Figure 1A and Supplementary Figures S1 and S2). A phylogenetic analysis using primase-helicases from bacteriophage, plants, and metazoans indicates that plant primase-helicases are closely related to T-odd primase-helicases (Figure 1B). Furthermore, metazoan TWINKLEs lost the cysteine residues involved in metal coordination and present amino acid substitutions of the putative catalytic residues of the RPD domain (Figure 1A and C) (2). The main differences between the conserved motifs of green plants and T-odd bacteriophage primase-helicases reside in the length and sequence of their ZBD. In green plants, the ZBD is on average 10 amino acids longer than primase-helicases from T-odd bacteriophages (Figure 1C). A homology model of the primase module of AtTwinkle, in comparison to the crystal structure of T7 primase, which includes the ZBD and RPD domains (8,35,36), predicts that the primase domain of AtTwinkle would follow similar structural arrangements to T7 primase-helicase and illustrates the possible coordination of the Zn2+ atom by the conserved cysteines in its ZBD (Figure 1D).
Figure 1.
AtTwinkle is a homolog of bacteriophage T7 primase-helicase and mitochondrial Twinkle. (A) Schematic representation of the bifunctional T7 primase-helicase in comparison to Arabidopsis and human Twinkles. T7 primase-helicase contains the six conserved motifs necessary for primase activity. Motif I is located at the ZBD and motifs II to VI in the RNAP domain. AtTwinkle conserves these motifs and contain an extra N-terminal amino acid sequence (residues 1–91) that harbors a dual organellar localization signal. Human Twinkle contains an N-terminal mitochondrial targeting sequence but lacks functional ZBD and RNAP domains. (B) Phylogenetic analysis between phage, plant and metazoan primase-helicases. The phylogenetic tree shows that plant and phage primase-helicases share a closer evolutionary relationship than plant and metazoan Twinkles. (C) Logo sequence of the amino acid region corresponding to the ZBD. The main differences between T7 Primase and AtTwinkle are the length of the ZBD that in AtTwinkle is ∼15 amino acids longer. (D) Computational homology model of the primase domain of AtPrimase-Helicase. The zinc finger is colored in blue and the RNAP domain in red. The conserved cysteines that coordinate the zinc atom are in a ball-stick representation.
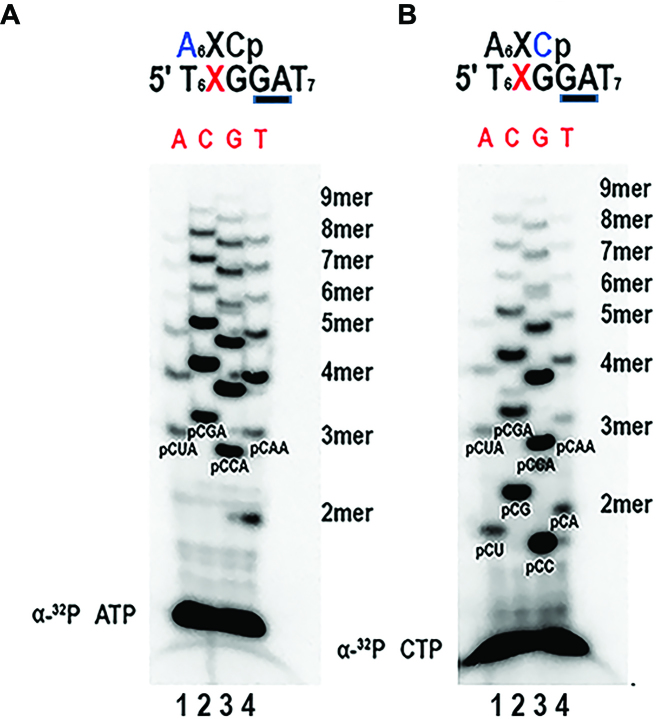
Recombinant AtTwinkle presents low activity on a T7 template recognition sequence
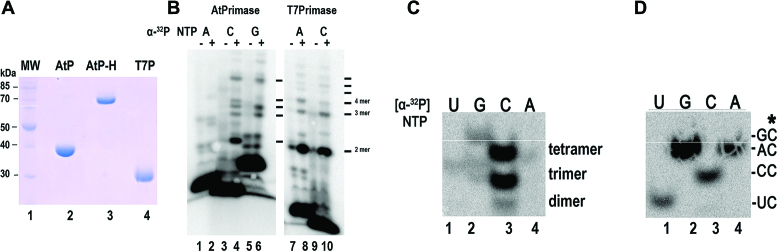
In order to biochemically elucidate the ssDNA template recognition sequence of AtTwinkle, we codon-optimized its open reading frame for bacterial protein expression. We designed a construct that encodes for the primase and helicase modules after the predicted organellar targeting sequence (residues 92 to 709) (28) and a construct containing only the primase module (residues 92–410). Herein, these protein fragments of AtTwinkle are dubbed AtPrimase-Helicase and AtPrimase respectively. Both recombinant proteins were purified to homogeneity after three chromatographic steps, with similar purity as a recombinant primase module (residues 1–262) from the T7 primase-helicase gene used as a control during this study (Figure 2A). Previous biochemical characterizations of AtTwinkle indicated that this protein is active on single-stranded M13 templates (28,29,32). To assess if our purified proteins exhibit oligoribonucleotide synthesis activity we used a random heptaoligonucleotide as a template and assayed AtPrimase and T7 primase fragments for ribonucleotide synthesis. We used a random oligonucleotide to avoid bias in the different combinations of template sequences that may occur in the fixed nucleotide sequence of the M13 template. The most abundant T7 primase product from its minimal recognition sequence 5′-GTC-3′ is the 5′-pppAC diribonucleotide (26). In this minimal recognition template, the 3′ cytidine is cryptic, the middle thymidine drives the initial binding of the 5′ ATP, and the 5′ guanosine templates for 3′ CMP incorporation (26). T7 primase generated a product that migrates with the same molecular mass when the reactions were labeled with [α-32P]-CTP or [α-32P]-ATP, suggesting that this product corresponds to the 5′-pppAC dinucleotide (Figure 2B, lanes 7–10). On the basis of a control experiment using a synthetic template containing the 5′-GTC-3′ sequence we inferred that the main T7 primase product present in the random heptamer is indeed the 5′-pppAC dinucleotide (data not shown). A similar experiment was executed with recombinant AtPrimase but with the addition of [α-32P]-CTP, [α-32P]-ATP or [α-32P]-GTP as labeled ribonucleotides. In the presence of radioactive ATP only a faint product band is observed in comparison to the control reaction without added AtPrimase (Figure 2B, lanes 1 and 2). However, in the presence of radioactively labeled CTP or GTP, AtPrimase catalyzed the synthesis of oligoribonucleotides longer than five ribonucleotides. A strong signal corresponding to the migration of a diribonucleotide is observed only when CTP is used as the radioactive label, suggesting that the identity of this product is 5′-pppCC (Figure 2B, lanes 4–8). As ATP is not efficiently incorporated by AtPrimase, a template thymidine may not be part of the template sequence recognized by AtTwinkle. On the other hand, CTP and GTP are incorporated into the RNA products. In order to directly identify the initial ribonucleotides synthesized by AtPrimase-Helicase we characterized the 5′ and 3′ ribonucleotides synthesized on a single-stranded M13 template using strategies previously developed in the study of the T7 replisome (37,38). We identified the 3′-ribonucleotide synthesized by AtPrimase-Helicase by measuring the incorporation of radioactive ribonucleotide products in individual reactions using each of the four [α-32P]-NTPs (ATP, UTP, CTP and GTP) in the presence of the other three non-radioactive NTPs and posterior phosphatase treatment. Since phosphatase removes the 5′-triphosphate the only radioactive product would correspond to the second or third phosphodiester bond. In this experiment, strong product bands were observed only in the presence of [α-32P]-CTP, indicating that CMP is the main ribonucleotide in the 3′ position (Figure 2C, lane 3). The elucidation of CMP as the preferred nucleotide in the 3′ position allows the identification of the 5′ ribonucleotide by a primer extension reaction in the presence of each of the four non-radioactively labeled NTP in the presence of [α-32P]-CTP. To our surprise, radioactively labeled products were observed in the presence of each of the four NTPs (Figure 2D), suggesting that when CTP is incorporated in the 3′ position AtTwinkle is non-selective for the identity of the 5′ ribonucleotide.
Figure 2.
Recombinant AtTwinkle is an active primase. (A) Heterologous purification of recombinant primases showing the purified proteins after three chromatographic steps. The expected theoretical molecular masses for AtPrimase, AtPrimase-Helicase and T7 Primase are 36, 70, and 28 kDa, respectively. Purified protein samples were run onto a 12.5% SDS-polyacrylamide gel and stained with Coomassie Brilliant Blue. The proteins migrate near their expected theoretical molecular mass. (B) Oligoribonucleotide synthesis in a random heptameric template using [α-32P]-ATP, [α-32P]-CTP and [α-32P]-GTP by AtPrimase in comparison to a synthesis reaction without added primase. The appearance of ribonucleotide products (lanes 4 and 6) in comparison to a T7 primase control (lanes 8 and 10) indicate that AtTwinkle is an active primase. (C) Identification of the 3′ nucleotide synthesized by AtPrimase-Helicase in M13 ssDNA template. The identity of each of the four labeled [α-32P]-NTPs is indicated. In this experiment the rest of the unlabeled rNTPs were included in the reaction. After an incubation of 30 min the products were treated with alkaline phosphatase to eliminate the label corresponding to the 5′ nucleotide. (D) Identification of the 5′ nucleotide synthesized by AtPrimase-Helicase when CTP is the preferred nucleotide in the 3′ position. In this experiment individual unlabeled NTPs were incubated in the presence of [α-32P]-CTP. The products were treated with alkaline phosphatase and run on a 27% sequencing gel.
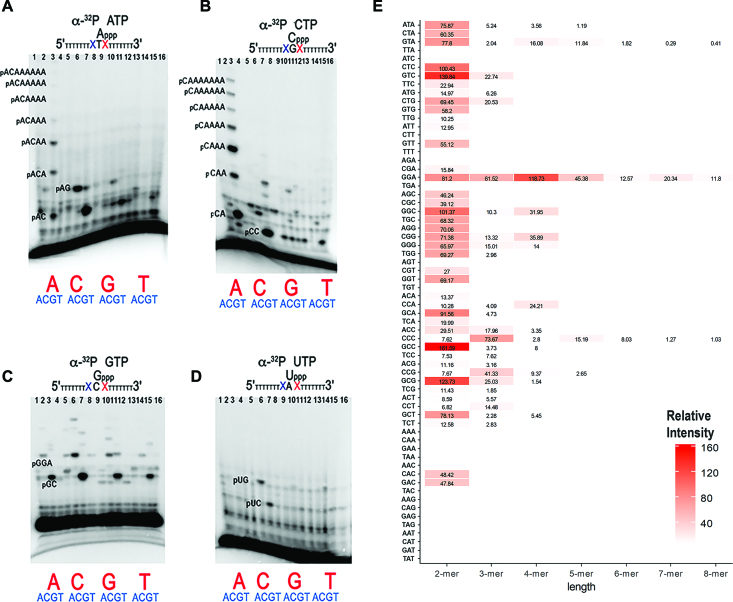
AtPrimase-Helicase synthetizes ribonucleotides longer than 4 nts in 4 out of 64 trinucleotide sequences
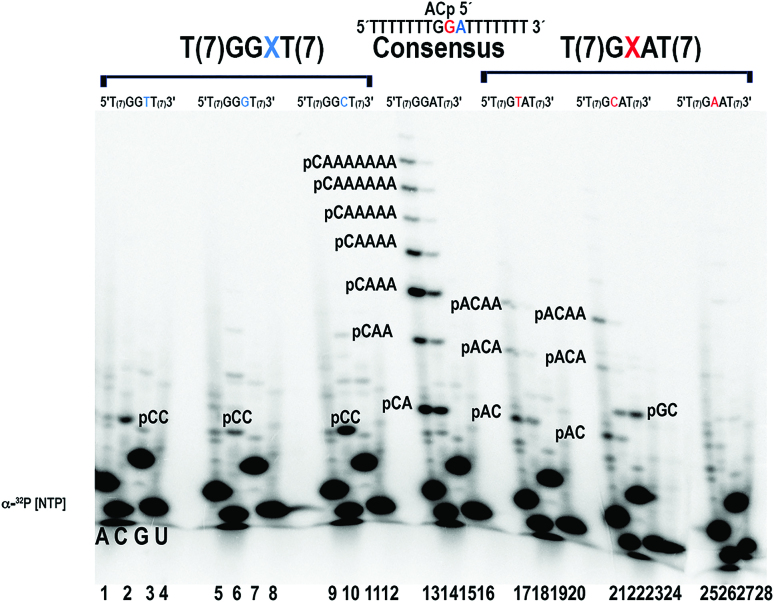
To precisely elucidate the trinucleotide recognition sequence of AtTwinkle, we used an array of 64 oligonucleotides corresponding to all possible trinucleotide combinations on primer extension reactions. We assayed oligoribonucleotide synthesis reactions labeled with either [α-32P]-ATP, [α-32P]-CTP, [α-32P]-GTP or [α-32P]-UTP using the AtPrimase-Helicase construct (Supplementary Figures S3–S6). These experiments show that AtPrimase-Helicase efficiently uses 5′-GGA-3′ as the preferred recognition sequence during oligoribonucleotide synthesis (Supplementary Figures S3 and S4, lane 11). A condensed summary of these experiments is shown in Figure 3. In this experiment, the base in middle of the random trinucleotide sequence serves as a template for the labeled [α-32P]-NTP (Figure 3, Supplementary Figure S7). This experiment shows that AtPrimase-Helicase abundantly synthesizes 2-mers in 42 out of 64 possible trinucleotide combinations. However AtPrimase-Helicase predominantly uses the template sequence 5′-GGA-3′ to synthesize products longer than four oligoribonucleotides (Figure 3B, lane 3) and to a lesser extent the 5′-GTA-3′ and 5′-CCC-3′ sequences (Figure 3A, lane 3 and Figure 3C, lane 6, Figure 3E and Supplementary Figure S7). Reaction products corresponding to 5′-pppAC (Figure 3A, lanes 3 and 7), pppCC (Figure 3B, lane 7), pppGC (Figure 3C, lane 3, 7, 11 and 15) and pppUC (Figure 3D, lane 7) diribonucleotides are abundantly observed, however these ribonucleotides are not converted into longer oligomers in the majority of the substrates (Figure 3A, lane 2, lane 8, lanes 12–14, 16).
Figure 3.
AtPrimase-Helicase is active in 4 out 64 trinucleotide sequences. Oligoribonucleotide synthesis reactions on an array of 64 possible trinucleotide combinations. The ribonucleotide complementary to the middle nucleotide dictates the identity of the radioactively labeled products. Reactions labeled with [α-32P]-ATP, [α-32P]-CTP, [α-32P]-GTP and [α-32P]-UTP, were run in panels (A), (B), (C) and (D) respectively. (E) Relative intensity of RNA products synthetized by each of the 64 trinucleotide templates by AtTwinkle. The Y-axes indicates the identity of the trinucleotide substrate and the X-ayes present the relative intensity of each product. Graphical representation of RNA synthesis indicating the percentage of each RNA product (2-mer to 8-mer) for each individual template. If the case of empty bars the intensity for the background and the reactions were equal. Oligonucleotide templates contained seven thymidines after the assayed trinucleotide sequence. Incorporation opposite those thymidines produce ribonucleotide products longer than three nucleotides. Reactions contained 5 mM of the synthetic template and 2 mM AtPrimase-Helicase.
A comparison of the migration of the RNA products observed in the 5′-GTA-3′ and 5′-GGA-3′ templates (Figure 3A, lane 3 and Figure 3B, lane 3) shows that their ribonucleotide products migrate with the same molecular mass (data not shown). This observation suggests that the 5′-GGA-3′ oligonucleotide directs the synthesis of a 5′-pppCA diribonucleotide that is further extended with adenines and that the 5′-GGA-3′ sequence contains two cryptic bases. A direct comparison between the RNA products synthesized by AtPrimase and T7 primase on templates containing 5′-GGA-3′ and 5′-GTA-3′ sequences shows that the synthesized ribonucleotides present the same migration pattern. This comparison also shows that AtTwinkle is an efficient DNA primase with comparable catalytic activity to T7 primase (Figure 4). The migration pattern of the synthesized ribonucleotides is independent of the identity of the labeled [α-32P]-NTP. This experiment further reveals that AtTwinkle recognizes two nucleotides, 5′-GA-3′, as a cryptic element and starts oligoribonucleotide synthesis with a 5′ CTP as its initial NTP.
Figure 4.
Direct comparison of the ribonucleotide synthesis activity between AtPrimase and T7 Primase. A sequencing gel showing the migration of the synthesized products by AtPrimase and T7 Primase in 5′-GGA'-3 and 5′-GTC'-3 single-stranded DNA sequences. In the case of T7 primase the lower RNA migrating product corresponds to a 5′-pppAC diribonucleotide (lanes 3 and 4) that is extended up to nine ribonucleotides according to the template thymidines. In At-Primase (lanes 1 and 2) the lower migrating product corresponds to a 5′-pppCA diribonucleotide that is extended following incorporation of AMP using the template thymidines. Reaction products were specifically labeled with [α-32P]-ATP (lanes 1 and 3) or [α-32P]-CTP (lanes 2 and 4). Primase reactions were carried out using 5 μM of the synthetic template and 1 μM of AtPrimase and 0.2 μM of T7 Primase.
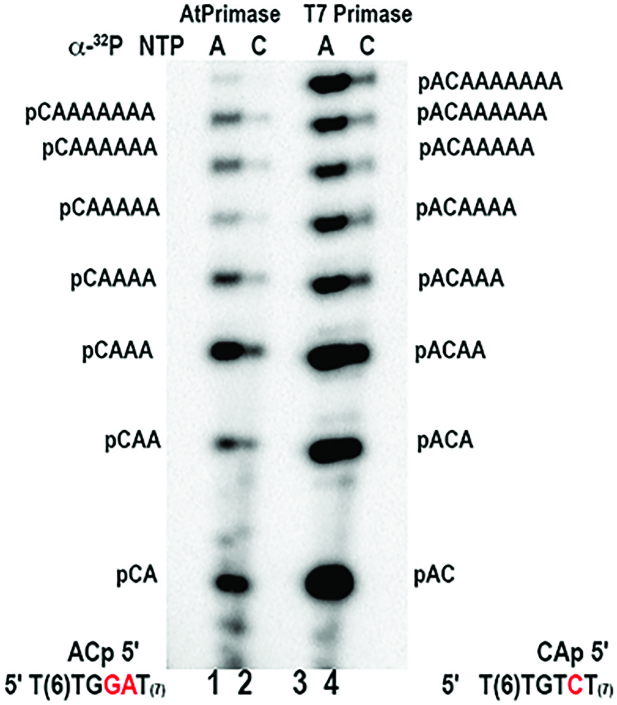
AtTwinkle recognizes two cryptic 5′-GA-3′ bases
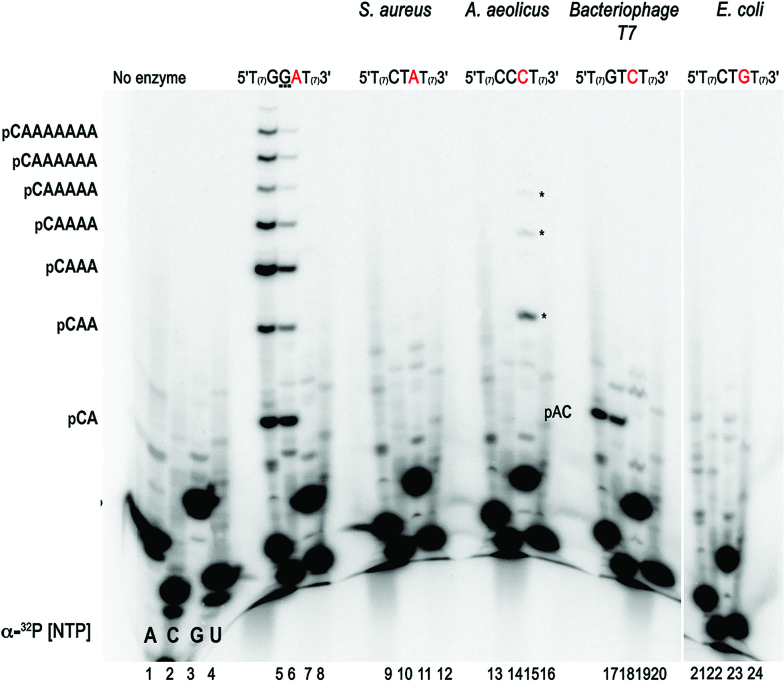
To further corroborate the unique character of the template recognized by AtTwinkle, we compared ribonucleotide synthesis by AtPrimase-Helicase using the 5′-GGA-3′ sequence and well characterized primase recognition sequences using independently labeled ribonucleotides (Figure 5, lanes 5 and 6). This experiment confirms that 5′-pppCA is the initial ribonucleotide synthesized by AtPrimase-Helicase and that it is extended up to nine ribonucleotides following insertion opposite template thymidines (Figure 5, lanes 5–8). Similar results were observed with the AtPrimase construct (data not shown). This experiment also shows that at the T7 primase recognition sequence, AtPrimase-Helicase synthesizes a 5′-AC-3′ ribonucleotide that is not further extended (Figure 5, lanes 17–20). Interestingly, oligoribonucleotide synthesis using the primase recognition sequence of A. aeolicus (5′-CCC-3′) is only observed when [α-32P]-GTP is used as a label (Figure 5, lanes 13–16). As the 5′-CCC-3′ template sequence contains seven thymidines that are instructional for adenines, the absence of a labeled RNA product in the presence of radioactive ATP indicates that poly-rG is synthesized by an iterative process. This process resembles the iterative abortive RNA synthesis by bacterial RNAPs (39,40).
Figure 5.
AtPrimase does not recognize bacterial and bacteriophage derived template sequences. Oligoribonucleotide synthesis by AtPrimase-Helicase on synthetic templates with canonical primase recognition sequences (S. aeurus, A. aeolicus, bacteriophage T7, E. coli) in comparison to the 5′-GGA'-3 sequence. RNA products were independently labeled with [α-32P]-ATP, [α-32P]-CTP, [α-32P]-GTP or [α-32P]-CTP. The identity of the RNA products from the 5′-pppCA'-3 diribonucleotide to the 5′-pppCAAAAAAA'-3 octaribonucleotide are indicated in the sequencing gel. Reactions contained 5 μM of the synthetic template and 2 μM of AtPrimase.
To further corroborate that the 5′-GGA-3′ sequence contains two cryptic bases, we generated oligonucleotides in which we systematically modified the 3′ adenine and middle guanosine and performed ribonucleotide synthesis experiments with all four labeled NTPs (Figure 6). Substitution of either ‘cryptic’ adenine or guanosine for any other base resulted in great decrease of ribonucleotide synthesis (Figure 6, lanes 1–12). In oligoribonucleotide synthesis experiments in which the 3′ adenine is modified, faint radioactive bands corresponding to products longer than 4 ribonucleotides are observed in the 5′-GGG-3′ sequence solely when CTP is used as radioactive label (Figure 6, lane 6). The absence of radioactively labeled products in reactions labeled with ATP, indicates that AtTwinkle interacts with homopolymeric 5′-CCC-3′ or 5′-GGG-3′ sequences in a process that drives iterative poly-rG or poly-rC synthesis. This experiment implies that the identity of the 3′ adenine and middle guanosine are critical for efficient ribonucleotide synthesis.
Figure 6.
Template recognition requires the cryptic 5′-GA'-3 pair. The identity of the ‘cryptic’ guanosine or adenine in the 5′-GA'-3 pair is necessary for oligoribonucleotide synthesis. Oligoribonucleotide synthesis decreases when the cryptic adenine (lanes 1–12) or guanosine (lanes 17–28) of the 5′GA'3 pair are modified with any other deoxynucleotide. Products were independently labeled with all four radioactive ribonucleotides. A control experiment with the canonical 5′-GA'-3 pair (lanes 13–16) is present and the identity of the oligoribonucleotide products are indicated.
AtTwinkle display low selectivity for 3′ ribonucleotide incorporation on the 5′ GGA 3′ sequence
Modifying the identity of the 5′-G base in the 5′-GGA-3′ sequence results in a ssDNA sequence that is a template for primer synthesis by AtTwinkle (Supplementary Figures S3 and S4, lanes 9–12). Altogether, these results argue that the identity of the 5′ rNTP during oligoribonucleotide synthesis by AtTwinkle is CTP and that two cryptic bases are required for template recognition. To test the preference for 3′ NMP incorporation we use a 5′-XGGA-3′ synthetic template sequence and systematically changed the identity of the 5′ base to all deoxynucleotides. An examination of the extension products by AtPrimase when dCMP or dGMP are in the 5′ position of the template oligonucleotide shows that the extent of ribonucleotide synthesis is approximately the same (Figure 7A and B). In comparison, ribonucleotide synthesis decreases when dAMP or dTMP are in 5′ position of the templating oligonucleotide. Thus, when ‘G’ is the base 5′ to the cryptic GA dinucleotide, AtTwinkle uses 5′-GGGA-3′, 5′-CGGA-3′3′, and to a lesser extent 5′-TGGA-3′ and 5′-AGGA-3′ as templates. This lack of selectivity for the 3′-incorporated ribonucleotide contrasts with the selectivity of T7 primase that preferentially binds CTP in the RNAP binding site (26).
Figure 7.
AtTwinkle is an efficient enzyme with non-selective incorporation for the 3′ rNTP base. AtTwinkle presents a preference for CTP as the 5′-NTP or initiating nucleotide. To assess the specificity for the 3′-rNTP base, the identify to a 5′-XGGA-‘-3 template sequence was systematically changed to all deoxynucleotides. The underlined GA corresponds to the cryptic element and X corresponds to the identity of the deoxynucleotide that templates for the incorporation of the 3′-rNMP. Oligoribonucleotides synthesis reactions where labeled with [α-32P]-ATP (A) or [α-32P]-CTP (B). The identity of the template base was either adenine (lane 1), citosine (lane 2), guanosine (lane 3) or thymidine (lane 4). The identity of the ribonucleotide products is indicated in the gel
AtTwinkle performs efficient ribonucleotide synthesis
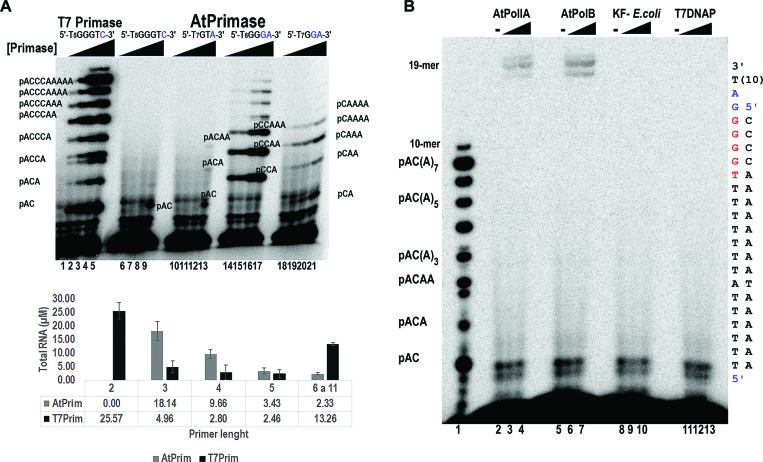
In order to directly compare the enzymatic activities of AtPrimase and T7 primase, we measured their ribonucleotide synthesis. The concentrations of AtPrimase varied from 100 to 800 nM whereas T7 primase was present at protein concentrations ranking from 6.25 to 400 nM. At a protein concentration of 6.25 nM, T7 primase synthesized approximately the same amount of a nonaribonucleotide products than AtPrimase present at a protein concentration of 800 nM on the 5′-GGGA-3′ template (Figure 8A, lanes 2 and 17). This comparison establishes that T7 primase is more active that AtPrimase. However, it is interesting to note that AtPrimase, at a concentration of 100 nM, synthesizes similar amounts of a tetraribonucleotide as 200 nM of T7 Primase (Figure 8A, lanes 3 and15). A quantification of the synthesized oligoribonucleotides indicates that AtPrimase is a highly efficient primase for the synthesis of RNA products between three to five ribonucleotides.
Figure 8.
AtPrimase efficiently synthesizes RNA primers for organellar DNA polymerases. (A) Oligoribonucleotide synthesis by the AtPrimase in 5′-GGGA-3′, 5′-GGA-3′, 5′-GTA-3′, 5′-GTC-3′ synthetic templates (lanes 6–21) in comparison to the synthesis present by T7 Primase (lanes 1–5) in a 5′-GGGTC'-3 recognition sequence. AtPrimase was present from 100 to 800 nM, whereas T7 primase was present at lower protein concentrations (from 6.25 to 400 nM). Each reaction was labeled with [α-32P]-ATP. The identity of the oligoribonucleotides is indicated in the gel. The bottom part of the figure illustrates the molar amount of oligoribonucleotide synthesis products by 100 nM of T7 and Arabidopsis primases in their preferred 5′-GGGC-3′ and 5′-GGGA-3′ ssDNA templates. To determine the molar amount of RNA, the intensity of each radioactive band was corrected with the amount of AMPs present in each ribonucleotide. (B) AtPrimase primes organellar DNA polymerases. Coupled primase-DNA polymerase reactions labeled with [α-32P]-dATP. RNA ladder generated by T7 Primase labeled with [α-32P]-ATP in the absence of DNA polymerases used as molecular weight markers. (lane 1). AtPolIA and AtPolIB, generate DNA:RNA products that correspond to the expected migration of a 19-mer in the presence of AtPrimase and unlabeled NTPs (lanes 2–4 and 5–7). In contrast the Klenow fragment of E. coli DNA polymerase I (lanes 8–10) and T7 DNA polymerase (lanes 11–13) are unable to perform coupled DNA synthesis. Coupled primase-DNA polymerase reactions were labeled with [α-32P]-dATP. AtPrim was present at 150 nM, whereas DNA polymerases were used to 75 and 150 nM. The template sequence used in the coupled primase-polymerase assay was 3′(T)10AGGGGGT(9)A(T5). Within this sequence, the AtPrimase recognition sequence is underlined
AtPrimase primes Plant Organellar DNA polymerases
Arabidopsis thaliana encodes two organellar DNA polymerases able to bypass an Apurinic/apyrimidinic (AP) site ((41) accompanying manuscript). These polymerases, AtPolIA and AtPolIB, are the only DNA polymerases located in plant organelles and therefore are the logical candidates to extend RNA primers synthesized by AtTwinkle at the lagging strand of the organellar replication fork (42). In order to elucidate the capabilities of AtTwinkle to synthesize primers for AtPolI's, we evaluated the efficiencies of AtPolIA and AtPolIB in comparison to E. coli′s Klenow fragment and T7 DNA polymerase to extend RNA primers synthesized by AtTwinkle. In this experiment, a single-stranded DNA template containing the 5′-(T)7A-(T)6GGGGA(T)9-3′ sequence was incubated with increased concentrations of AtTwinkle and the assayed DNA polymerases in the presence of [α-32P]-dATP for 1 h. A control experiment shows that at the lowest concentrations all DNA polymerases completely extend an annealed double stranded primer-template substrate in 10 min and that the single stranded oligonucleotide is not used as a template by those polymerases (data not shown). However, in the coupled primase-polymerase assay, products were observed in reactions incubated with AtPolIA and AtPolIB (Figure 8B, lanes 2–7) and not with equimolar amounts of Klenow Fragment of E. coli DNA polymerase I or T7 DNA polymerase (Figure 8B, lanes–13). Our results contrast with the primer extension observed in a coupled primase-polymerase reaction with E. coli DNA polymerase I and AtPrimase-Helicase using single-stranded M13 as a substrate (28). Although the precise concentration of E. coli DNA polymerase I was not provided in that study, RNA transcripts may be used as primers if DNA polymerases are present at high concentrations.
DISCUSSION
The organellar DNA Primase-Helicase from the plant model A. thaliana, AtTwinkle, recognizes a ssDNA sequence in which two cryptic nucleotides are necessary for efficient RNA synthesis. AtTwinkle effectively synthesizes long ribonucleotides on a trinucleotide 5′-GGA-3′ sequence (Figure 3). In which the 5′-GA-3′ pair functions as a cryptic element. Substitutions of either the cryptic 5′-G or cryptic A-3′ nucleotides and substitutions of the initial 5′-G, in the trinucleotide recognition sequence abolishes ribonucleotide synthesis (Supplementary Figures S3–S6).
In synthetic templates AtTwinkle efficiently synthesizes pppAC, pppUC, pppGC, and pppCC di-ribonucleotides, however these are not extended to longer RNA products (Figures 2C and 3). The apparent lack of discrimination for the 5′ position observed in a phosphodiesterase assay using a M13 ssDNA template may be due to the synthesis of these di-ribunucleotides (Figure 2D). In synthetic templates containing the 5′-GA-3′ cryptic sequence, AtTwinkle exhibits a strong selectivity for rCTP as the 5′ ribonucleotide. This selectivity contrasts with the relaxed selectivity for the 3′ ribonucleotide in which the identity at the 3′ elongation nucleotide can be any NTP, however CMP or GMP are preferentially incorporated (Figure 7). Our data reveals that the preferred ssDNA recognition sequences by AtTwinkle are 5′-GGGA-3′ and 5′-CGGA-3′, within these sequences the two underlined bases are cryptic nucleotides. We show that the preferred ribonucleotide products by AtTwinkle are preferentially composed of cytosines and guanines, suggesting that plant primases selectively incorporate these ribonucleotides from the A/T-rich genome of organelles (43). The preferred ribonucleotides synthesized by AtTwinkle resemble those synthesized by Aquifex aeolicus (24) suggesting that plants have selected thermodynamically stable base pairing during primer DNA synthesis. Although the stability conferred by these sequences that may be needed during primer hand-off to the replicating DNA polymerase, the selectivity for a G/C-rich recognition sequences also may play a role in determining the length of the Okazaki fragments during organellar replication.
The precise mechanisms by which a ssDNA template sequence is recognized by a primase are not fully elucidated. Biochemical and structural evidence strongly suggest that an interaction between the ZBD and the RNAP domain drives ssDNA template recognition directing the synthesis of the initial di-ribonucleotide (26,38,44–46). The ZBD residues involved in metal coordination are only composed of cysteines (Cys4 motif) in bacteriophage primases, whereas in most bacteria a histidine replaces one of these cysteines (Cys-His-Cys2 motif). The highly conserved CXXC(H)(X15–18)CXXC motif of the ZBD in prokaryotic primases folds into a zinc ribbon (47) that can be divided in two conserved CXXC(H) repeats. Within these repeats, the cysteines or histidine residues involved in Zn2+ coordination are separated by two non-conserved amino acids. The most striking difference between plant and T-odd bacteriophage primases is the variation in the residues near the conserved cysteines that coordinate the Zn2+ atom. In land plant primases, the first CXXC repeat is conserved, whereas the second CXXC repeat is substituted by a CXRXKC sequence (Figure 9A). Biochemical studies indicate that residues His33 and Lys70 located before the second CXXC repeat in T7 and Clostridium difficile primases drive ssDNA template recognition (26,48). However, the corresponding amino acids in AtTwinkle (Thr161 and Asn163) are not conserved in land plants suggesting that these residues may not be involved in ssDNA template recognition (Figure 9A and Supplementary Figure S2). A computational model of the ZBD of AtTwinkle shows that the side chains of Arg166 and Lys168 and a conserved tryptophan residue (Trp 162) (Figure 9B and Supplementary Figure S2) are in similar orientations than His33 of T7 primase and Lys70 of C. difficile primase. This substrate-accessible orientation suggests that Arg166, Lys168, and Trp162 may play a role in ssDNA template recognition. The CXRXKC sequence is also present in primases from C. botulinum and bacteriophage VpV262 (49) (Figure 9A) suggesting that if this element plays a role in template recognition, the need for two cryptic bases would not be unique to plants. Furthermore, the nuclear genome of Arabidopsis encodes for an organellar protein that only contains the primase module of AtTwinkle (At1g30660 or AtDnaG). This primase also contains a sequence that resembles the CXRXKC element (28). Although structure-function studies are needed to elucidate the mechanisms that account for the need of two cryptic bases in the ssDNA template recognition sequence by AtTwinkle, it is tempting to speculate that the amino acid differences at the ZBD with prokaryotic primase-helicases may be relevant for template selectivity.
Figure 9.
Structural suggestion for the altered template recognition by AtTwinkle. (A) Multiple sequence alignment between the ZBD of T7 primase-helicase, bacterial primases, and plant Twinkles. The fours cysteines that coordinate the Zn2+ atom in T7 primase can be divided in two CXXC elements. In land plant primases, the first CXXC elements is conserved, whereas the second one is substituted by a CXRXKC element. Protein are designated with abbreviations as follows: Mpo, Marchantia polymorpha; Nta, Nicotiana tabacum; Zma, Zea mays; Atri, Amborella trichopoda; Cbo, Clostridium botulinum; Cdi, Clostridium difficile; Gst, geobacillus stearothermophilus. (B) Computational model of the zinc finger of AtTwinkle in comparison to the crystal structure of T7-Primase. The four β-strands that correspond to the zinc finger are colored in blue. The cysteine residues CXXC element are colored in magenta and red. The side chains of the amino acids in the second CXXC element are in a ball-stick representation. The Zn2+ atom is colored in black. His33 implicated in template recognition in T7 primase is in a ball-stick representation. The computational model of AtTwinkle shows the proposed structural localization of residues R166 and K168.
Furthermore, understanding the recognition sequence of AtTwinkle is a first step toward our biochemical understanding of the plant organellar replisome. We found that RNA products synthesized by AtTwinkle are efficiently used as primers for plant organellar DNA polymerases. In the analogous bacteriophage T7 replisome, T7 DNA polymerase uses short tetra-ribonucleotide synthesized by T7 primase-helicase as primers via a ‘handoff’ mechanism. This mechanism implicates a precise interaction between primase and polymerase (50,51). It is interesting to note that AtTwinkle synthesizes abundant RNA products from three to five ribonucleotides, as these short oligoribonucleotides are easily dissociated from a DNA template. It is possible that organellar DNA primase and polymerase physically interact during primer extension. Although the precise length and identity of the oligo-ribonucleotide primers synthesized by AtTwinkle remain to be identified, our work demonstrates that AtTwinkle is a functional primase that selectively primes AtPolIs. The efficient synthesis of product of tetra-ribonucleotide or penta-ribonucleotides by AtTwinkle may be correlated with an RNA of four or five ribonucleotides that is used as a primer. It is possible that the extra recognition cryptic base provides a mechanism to generate specificity during primer synthesis and coordinate leader and lagging DNA synthesis.
Our studies strongly suggest that organellar DNA replication mechanistically resembles phage T7-replication. This process contrasts with DNA replication in metazoan mitochondria in which human mitochondrial RNAP primes human mitochondrial DNA polymerase γ. In this work, we demonstrate that a plant organellar primase-helicase, phylogenetically related to bacteriophage T7 primase-helicase, efficiently primes plant organellar DNA polymerases that are phylogenetically related to bacteria, illustrating that two enzymes with different evolutionary origins coevolved to assemble the plant organellar replisome.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Corina Diaz-Quezada for her technical support. We thank Oleg Tsodikov, Alfredo Hernandez, Teri Markow, Seung-Joo Lee and Charles Richardson for critical reading.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
CONACYT (Ciencia Basica 2015) [253757 to L.G.B.]. Funding for open access charge: CONACYT [253757]; Cinvestav-IPN.
Conflict of interest statement. None declared.
REFERENCES
- 1. Shutt T.E., Gray M.W.. Bacteriophage origins of mitochondrial replication and transcription proteins. Trends Genet. 2006; 22:90–95. [DOI] [PubMed] [Google Scholar]
- 2. Shutt T.E., Gray M.W.. Twinkle, the mitochondrial replicative DNA helicase, is widespread in the eukaryotic radiation and may also be the mitochondrial DNA primase in most eukaryotes. J. Mol. Evol. 2006; 62:588–599. [DOI] [PubMed] [Google Scholar]
- 3. Masters B.S., Stohl L.L., Clayton D.A.. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987; 51:89–99. [DOI] [PubMed] [Google Scholar]
- 4. Hamdan S.M., Richardson C.C.. Motors, switches, and contacts in the replisome. Annu. Rev. Biochem. 2009; 78:205–243. [DOI] [PubMed] [Google Scholar]
- 5. Lee S.J., Richardson C.C.. Choreography of bacteriophage T7 DNA replication. Curr. Opin. Chem. Biol. 2011; 15:580–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kulczyk A.W., Akabayov B., Lee S.J., Bostina M., Berkowitz S.A., Richardson C.C.. An interaction between DNA polymerase and helicase is essential for the high processivity of the bacteriophage T7 replisome. J. Biol. Chem. 2012; 287:39050–39060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang H., Lee S.J., Richardson C.C.. Essential protein interactions within the replisome regulate DNA replication. Cell Cycle. 2011; 10:3413–3414. [DOI] [PubMed] [Google Scholar]
- 8. Kato M., Ito T., Wagner G., Richardson C.C., Ellenberger T.. Modular architecture of the bacteriophage T7 primase couples RNA primer synthesis to DNA synthesis. Mol. Cell. 2003; 11:1349–1360. [DOI] [PubMed] [Google Scholar]
- 9. Frick D.N., Richardson C.C.. DNA primases. Annu. Rev. Biochem. 2001; 70:39–80. [DOI] [PubMed] [Google Scholar]
- 10. Sanchez-Sandoval E., Diaz-Quezada C., Velazquez G., Arroyo-Navarro L.F., Almanza-Martinez N., Trasvina-Arenas C.H., Brieba L.G.. Yeast mitochondrial RNA polymerase primes mitochondrial DNA polymerase at origins of replication and promoter sequences. Mitochondrion. 2015; 24:22–31. [DOI] [PubMed] [Google Scholar]
- 11. Cupp J.D., Nielsen B.L.. Minireview: DNA replication in plant mitochondria. Mitochondrion. 2014; 19:231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Korhonen J.A., Pham X.H., Pellegrini M., Falkenberg M.. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004; 23:2423–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Spelbrink J.N., Li F.Y., Tiranti V., Nikali K., Yuan Q.P., Tariq M., Wanrooij S., Garrido N., Comi G., Morandi L. et al. . Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 2001; 28:223–231. [DOI] [PubMed] [Google Scholar]
- 14. Wanrooij S., Fuste J.M., Farge G., Shi Y., Gustafsson C.M., Falkenberg M.. Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:11122–11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Seow F., Sato S., Janssen C.S., Riehle M.O., Mukhopadhyay A., Phillips R.S., Wilson R.J., Barrett M.P.. The plastidic DNA replication enzyme complex of Plasmodium falciparum. Mol. Biochem. Parasitol. 2005; 141:145–153. [DOI] [PubMed] [Google Scholar]
- 16. McKinney E.A., Oliveira M.T.. Replicating animal mitochondrial DNA. Genet. Mol. Biol. 2013; 36:308–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ramachandran A., Nandakumar D., Deshpande A.P., Lucas T.P., R R.B., Tang G.Q., Raney K., Yin Y.W., Patel S.S.. The yeast mitochondrial RNA polymerase and transcription factor complex catalyzes efficient priming of DNA synthesis on single-stranded DNA. J. Biol. Chem. 2016; 291:16828–16839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ling F., Hori A., Yoshitani A., Niu R., Yoshida M., Shibata T.. Din7 and Mhr1 expression levels regulate double-strand-break-induced replication and recombination of mtDNA at ori5 in yeast. Nucleic Acids Res. 2013; 41:5799–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tseng T.Y., Frick D.N., Richardson C.C.. Characterization of a novel DNA primase from the Salmonella typhimurium bacteriophage SP6. Biochemistry. 2000; 39:1643–1654. [DOI] [PubMed] [Google Scholar]
- 20. Mendelman L.V., Richardson C.C.. Requirements for primer synthesis by bacteriophage T7 63-kDa gene 4 protein. Roles of template sequence and T7 56-kDa gene 4 protein. J. Biol. Chem. 1991; 266:23240–23250. [PubMed] [Google Scholar]
- 21. Swart J.R., Griep M.A.. Primase from Escherichia coli primes single-stranded templates in the absence of single-stranded DNA-binding protein or other auxiliary proteins. Template sequence requirements based on the bacteriophage G4 complementary strand origin and Okazaki fragment initiation sites. J. Biol. Chem. 1993; 268:12970–12976. [PubMed] [Google Scholar]
- 22. Koepsell S.A., Larson M.A., Griep M.A., Hinrichs S.H.. Staphylococcus aureus helicase but not Escherichia coli helicase stimulates S. aureus primase activity and maintains initiation specificity. J. Bacteriol. 2006; 188:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koepsell S.A., Larson M.A., Frey C.A., Hinrichs S.H., Griep M.A.. Staphylococcus aureus primase has higher initiation specificity, interacts with single-stranded DNA stronger, but is less stimulated by its helicase than Escherichia coli primase. Mol. Microbiol. 2008; 68:1570–1582. [DOI] [PubMed] [Google Scholar]
- 24. Larson M.A., Bressani R., Sayood K., Corn J.E., Berger J.M., Griep M.A., Hinrichs S.H.. Hyperthermophilic Aquifex aeolicus initiates primer synthesis on a limited set of trinucleotides comprised of cytosines and guanines. Nucleic Acids Res. 2008; 36:5260–5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cha T.A., Alberts B.M.. Studies of the DNA helicase-RNA primase unit from bacteriophage T4. A trinucleotide sequence on the DNA template starts RNA primer synthesis. J. Biol. Chem. 1986; 261:7001–7010. [PubMed] [Google Scholar]
- 26. Kusakabe T., Hine A.V., Hyberts S.G., Richardson C.C.. The Cys4 zinc finger of bacteriophage T7 primase in sequence-specific single-stranded DNA recognition. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:4295–4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaguni L.S., Oliveira M.T.. Structure, function and evolution of the animal mitochondrial replicative DNA helicase. Crit. Rev. Biochem. Mol. Biol. 2016; 51:53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Diray-Arce J., Liu B., Cupp J.D., Hunt T., Nielsen B.L.. The Arabidopsis At1g30680 gene encodes a homologue to the phage T7 gp4 protein that has both DNA primase and DNA helicase activities. BMC Plant Biol. 2013; 13:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Towle-Weicksel J.B., Cao Y., Crislip L.J., Thurlow D.L., Crampton D.J.. Chimeric proteins constructed from bacteriophage T7 gp4 and a putative primase-helicase from Arabidopsis thaliana. Mol. Biol. Rep. 2014; 41:7783–7795. [DOI] [PubMed] [Google Scholar]
- 30. Carrie C., Kuhn K., Murcha M.W., Duncan O., Small I.D., O’Toole N., Whelan J.. Approaches to defining dual-targeted proteins in Arabidopsis. Plant J. 2009; 57:1128–1139. [DOI] [PubMed] [Google Scholar]
- 31. Leipe D.D., Aravind L., Grishin N.V., Koonin E.V.. The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res. 2000; 10:5–16. [PubMed] [Google Scholar]
- 32. Cao Y. A bifunctional mitochondrial DNA primase/helicase (AMH) identified in Arabidopsis thaliana: Preliminary biochemical characterization of its functions. Department of Chemistry and Biochemistry. 2012; Clark University. [Google Scholar]
- 33. Frick D.N., Baradaran K., Richardson C.C.. An N-terminal fragment of the gene 4 helicase/primase of bacteriophage T7 retains primase activity in the absence of helicase activity. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:7957–7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Deshpande A.P., Patel S.S.. Interactions of the yeast mitochondrial RNA polymerase with the +1 and +2 promoter bases dictate transcription initiation efficiency. Nucleic Acids Res. 2014; 42:11721–11732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Toth E.A., Li Y., Sawaya M.R., Cheng Y., Ellenberger T.. The crystal structure of the bifunctional primase-helicase of bacteriophage T7. Mol. Cell. 2003; 12:1113–1123. [DOI] [PubMed] [Google Scholar]
- 36. Sawaya M.R., Guo S., Tabor S., Richardson C.C., Ellenberger T.. Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7. Cell. 1999; 99:167–177. [DOI] [PubMed] [Google Scholar]
- 37. Kusakabe T., Richardson C.C.. Template recognition and ribonucleotide specificity of the DNA primase of bacteriophage T7. J. Biol. Chem. 1997; 272:5943–5951. [DOI] [PubMed] [Google Scholar]
- 38. Kusakabe T., Richardson C.C.. The role of the zinc motif in sequence recognition by DNA primases. J. Biol. Chem. 1996; 271:19563–19570. [DOI] [PubMed] [Google Scholar]
- 39. Guajardo R., Lopez P., Dreyfus M., Sousa R.. NTP concentration effects on initial transcription by T7 RNAP indicate that translocation occurs through passive sliding and reveal that divergent promoters have distinct NTP concentration requirements for productive initiation. J. Mol. Biol. 1998; 281:777–792. [DOI] [PubMed] [Google Scholar]
- 40. Pal M., Luse D.S.. The initiation-elongation transition: lateral mobility of RNA in RNA polymerase II complexes is greatly reduced at +8/+9 and absent by +23. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:5700–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Baruch-Torres N., Brieba L.G.. Plant organellar DNA polymerases are replicative and translesion DNA synthesis polymerases. Nucleic Acids Res. 2017; doi:10.1093/nar/gkx744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moriyama T., Terasawa K., Sato N.. Conservation of POPs, the plant organellar DNA polymerases, in eukaryotes. Protist. 2011; 162:177–187. [DOI] [PubMed] [Google Scholar]
- 43. Smith D.R. Updating our view of organelle genome nucleotide landscape. Front Genet. 2012; 3:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Corn J.E., Pease P.J., Hura G.L., Berger J.M.. Crosstalk between primase subunits can act to regulate primer synthesis in trans. Mol. Cell. 2005; 20:391–401. [DOI] [PubMed] [Google Scholar]
- 45. Zhou Y., Luo H., Liu Z., Yang M., Pang X., Sun F., Wang G.. Structural Insight into the specific DNA template binding to DnaG primase in bacteria. Sci. Rep. 2017; 7:659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee S.J., Zhu B., Hamdan S.M., Richardson C.C.. Mechanism of sequence-specific template binding by the DNA primase of bacteriophage T7. Nucleic Acids Res. 2010; 38:4372–4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Krishna S.S., Majumdar I., Grishin N.V.. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 2003; 31:532–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. van Eijk E., Paschalis V., Green M., Friggen A.H., Larson M.A., Spriggs K., Briggs G.S., Soultanas P., Smits W.K.. Primase is required for helicase activity and helicase alters the specificity of primase in the enteropathogen Clostridium difficile. Open Biol. 2016; 6:160272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hardies S.C., Comeau A.M., Serwer P., Suttle C.A.. The complete sequence of marine bacteriophage VpV262 infecting vibrio parahaemolyticus indicates that an ancestral component of a T7 viral supergroup is widespread in the marine environment. Virology. 2003; 310:359–371. [DOI] [PubMed] [Google Scholar]
- 50. Kato M., Frick D.N., Lee J., Tabor S., Richardson C.C., Ellenberger T.. A complex of the bacteriophage T7 primase-helicase and DNA polymerase directs primer utilization. J. Biol. Chem. 2001; 276:21809–21820. [DOI] [PubMed] [Google Scholar]
- 51. Kato M., Ito T., Wagner G., Ellenberger T.. A molecular handoff between bacteriophage T7 DNA primase and T7 DNA polymerase initiates DNA synthesis. J. Biol. Chem. 2004; 279:30554–30562. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.