Abstract
Whereas ribosomes efficiently catalyze peptide bond synthesis by most amino acids, the imino acid proline is a poor substrate for protein synthesis. Previous studies have shown that the translation factor eIF5A and its bacterial ortholog EF-P bind in the E site of the ribosome where they contact the peptidyl-tRNA in the P site and play a critical role in promoting the synthesis of polyproline peptides. Using misacylated Pro-tRNAPhe and Phe-tRNAPro, we show that the imino acid proline and not tRNAPro imposes the primary eIF5A requirement for polyproline synthesis. Though most proline analogs require eIF5A for efficient peptide synthesis, azetidine-2-caboxylic acid, a more flexible four-membered ring derivative of proline, shows relaxed eIF5A dependency, indicating that the structural rigidity of proline might contribute to the requirement for eIF5A. Finally, we examine the interplay between eIF5A and polyamines in promoting translation elongation. We show that eIF5A can obviate the polyamine requirement for general translation elongation, and that this activity is independent of the conserved hypusine modification on eIF5A. Thus, we propose that the body of eIF5A functionally substitutes for polyamines to promote general protein synthesis and that the hypusine modification on eIF5A is critically important for poor substrates like proline.
INTRODUCTION
Cellular protein synthesis is catalyzed by the ribosome with the assistance of aminoacyl-tRNAs and translation factors. The ribosome contains three aminoacyl-tRNA binding sites: the acceptor (A), peptidyl (P) and exit (E) sites. The A site binds an aminoacyl-tRNA in a codon-dependent manner. Following peptide bond formation, the A-site tRNA is translocated to the P site with concomitant movement of the now deacylated P-site tRNA to the E site. The binding of aminoacyl-tRNA to the A site is facilitated by the translation elongation factor eEF1A in eukaryotes (EF-Tu in bacteria), and translocation of the tRNAs and mRNA is promoted by the factor eEF2 (EF-G in bacteria) (reviewed in 1,2)). In addition to these two factors, a third universally conserved elongation factor was recently characterized. The factor eIF5A was first described as an initiation factor, promoting synthesis of the first peptide bond following assembly of an 80S ribosome with Met-tRNAiMet bound in the P site and base-paired with an AUG start codon on the mRNA (3–5). Subsequent studies revealed that eIF5A plays a more general role in translation elongation beyond first peptide bond synthesis (6,7, see also 8).
More recently, eIF5A and its bacterial homolog EF-P were shown to be particularly important for translation of polyproline motifs (9–11). When yeast cells expressing the temperature-sensitive eIF5A-S149P mutant were grown at a semi-permissive temperature, expression of reporter genes and yeast proteins containing runs of proline residues was impaired. Importantly, mutation of the C-terminal polyproline sequence in the yeast protein Ldb17 was sufficient to restore expression under non-permissive conditions (10). In an in vitro reconstituted assay system, as few as three consecutive proline residues was sufficient to impose a requirement for eIF5A for peptide synthesis (10). Consistent with this latter finding, ribosomes translating polyproline sequences in the absence of eIF5A were observed to stall following translation of the first two proline codons (10). Thus, in the stalled complex the nascent peptide ending with diproline would be attached to the peptidyl-tRNA with a proline codon in the A site, presumably bound to Pro-tRNAPro. While these studies clearly showed a role for eIF5A in translation of polyproline sequences, it was not clear whether the imino acid proline or the tRNAPro imposed the requirement for eIF5A.
In addition to its unique property in stimulating polyproline synthesis, eIF5A is of interest because it is the only protein known to contain the amino acid hypusine (reviewed in 8,12). The hypusine residue is formed by post-translational modification of a conserved Lys residue in eIF5A (Lys51 in yeast eIF5A encoded by HYP2). First, the enzyme deoxyhypusine synthase (Dys1 in yeast) transfers an n-butylamine moiety from the polyamine spermidine to the ε-amino group of the Lys residue forming deoxyhypusine. Second, the enzyme deoxyhypusine hydroxylase (Lia1 in yeast) hydroxylates the second methylene carbon in the appended chain to form hypusine (Nε-(4-amino-2-hydroxybutyl)lysine). The hypusine modification is essential in yeast cells (13–16) as well as for the ability of eIF5A to stimulate polyproline synthesis in vitro (10). Along with the spermidine requirement for hypusine formation, polyamines play an apparently more direct role in protein synthesis as essential components of both bacterial and eukaryotic cell-free translation systems (17–20). Further linking eIF5A and polyamines, the presence of eIF5A lowers the optimum magnesium concentration for protein synthesis in reconstituted assay systems lacking polyamines (4). Interestingly, mutations in the polyamine synthesis pathway and eIF5A promote tumorigenesis in a mouse lymphoma model, suggesting an important role for eIF5A and its hypusine modification in tumor suppression (21). However, the connection between eIF5A, polyamines and cancer is complex because, paradoxically, in other studies increased levels of polyamines and eIF5A have been linked to tumorigenesis (reviewed in 22).
Like eIF5A in eukaryotes, the archaeal aIF5A protein is post-translationally modified to contain deoxyhypusine (reviewed in 8,23). Moreover, bacterial EF-P is also post-translationally modified; however, with distinct modifications in different species (reviewed in 23). In γ-proteobacteria including Escherichia coli and Salmonella enterica, (R)-β-lysine is attached to the ε-amino group of a conserved Lys residue (corresponding to Lys51 in yeast eIF5A) and a subsequent hydroxylation generates a modification that is similar to hypusine (24–27). In other β- and γ-proteobacteria including Pseudomonas aeruginosa, the corresponding Arg residue in EF-P is glycosylated by the addition of l-rhamnose (28,29). Most recently, the corresponding Lys residue in EF-P from the Gram-positive bacterium Bacillus subtilis was found to be modified by the addition of a 5-aminopentanol moiety that closely resembles hypusine (30). The finding that both EF-P and eIF5A require a post-translational modification to stimulate polyproline synthesis highlights the unique complexity associated with proline substrates in the ribosomal peptidyl-transferase center (PTC). Recent X-ray crystallography and cryo-EM structures reveal eIF5A binding adjacent to the P-site of the yeast 80S ribosome in a position similar to where EF-P docks on the bacterial ribosome (31–33). In the eIF5A–ribosome structures, the body of eIF5A buttresses the P-site tRNA and the hypusine residue projects towards the acceptor stem of the P-site tRNA where it is proposed to reorient the proline substrates to enhance peptide bond formation (32–34). It is noteworthy that proline is an imino rather than an amino acid; however, the properties of proline that impose the apparently unique requirement for eIF5A are unknown.
In this paper, we employ in vitro peptide synthesis assays to characterize the role of eIF5A. Our studies using misacylated tRNAs as well as various proline analogs demonstrate that proline rather than tRNAPro imposes the requirement for eIF5A. Moreover, the eIF5A requirement is linked to the rigidity of proline and not its imino nature. Finally, whereas the hypusine modification is essential for polyproline synthesis, we found that the body of eIF5A functionally substituted for polyamines to stimulate general peptide synthesis. Thus, the amino acid substrates impose the requirement for eIF5A and its hypusine modification in peptide synthesis by the ribosome.
MATERIALS AND METHODS
Substrates
Proline and its analogs, α-methyl-l-proline, 3,4-dehydro-dl-proline, cis-4-hydroxy-l-proline and l-azetidine-2-carboxylic acid, as well as spermidine (catalog number S2626), spermine (S4264) and putrescine (P5780), and yeast tRNAPhe were purchased from Sigma. All DNA oligonucleotides were purchased from either Integrated DNA Technologies or Eurofins MWG Operon. The 3′-biotin-labeled RNA oligonucleotide used for affinity purification of yeast tRNAPro was also purchased from Eurofins MWG Operon; yeast tRNALys was obtained from tRNA Probes (College Station, TX, USA).
Preparation of eIF5A
Unmodified forms of His6-tagged eIF5A lacking hypusine were purified from Escherichia coli strain BL21(DE3) CodonPlus-RIL (Agilent) transformed with pC4181 (for unmodified eIF5A), pC4357 (for eIF5A K51R) or pC4358 (for eIF5A K51A) using Ni-NTA resin as described previously (10). His6-tagged hypusinated eIF5A was purified from BL21(DE3) CodonPlus-RIL transformed with the eIF5A, Dys1 (deoxyhypusine synthase), Lia1 (deoxyhypusine hydroxylase) co-expression plasmid pC4183 as described previously (10).
Preparation of translation factors and ribosomes
Preparation of translation initiation factors eIF1, eIF1A, eIF2, eIF5 and eIF5B, and translation elongation factor eEF3 was performed using published protocols (10). Native elongation factor eEF1A was purified from yeast strain YRP840 (35) using a modified version of a previously described protocol (10). Cells were grown in 4 l yeast extract peptone dextrose (YPD) medium to OD600 = 1.0, harvested and broken in 50 ml lysis buffer A (60 mM Tris–HCl [pH 7.5], 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA [pH 8.0], 10% glycerol, 1 mM DTT, 1× Complete protease inhibitor [Roche], and 0.5 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride [AEBSF, Sigma]) with glass beads using a Bead Beater (Biospec Products). Following removal of unbroken cells by centrifugation at 1855 x g for 10 min, the supernatant was clarified by centrifugation at 149 000 × g for 30 min. The supernatant was then gently mixed with 10 ml DE52 resin (Whatman, pre-equilibrated with lysis buffer A) for 1 h at 4°C. The unbound fraction containing eEF1A was isolated by pouring the mixture into a column and collecting the eluate, which was then applied to a HiTrap Q column (GE Healthcare). The unbound fraction from HiTrap Q column was applied to a HiTrap CM Sepharose column (GE Healthcare), and eEF1A was eluted with a linear gradient to 300 mM KCl. Fractions containing eEF1A were identified by SDS-PAGE, pooled, dialyzed against Storage Buffer A (20 mM Tris–HCl (pH 7.5), 40 mM KCl, 5 mM MgCl2, 10% glycerol, 1 mM DTT) and stored at –80°C.
A poly-histidine tagged version of elongation factor eEF2 was purified from yeast strain TKY675 as described previously (10) with some modifications. Cells were grown in 5 l YPD medium to OD600 = 2.0, harvested, and then suspended in lysis buffer B (50 mM potassium phosphate [pH 7.6], 300 mM KCl, 1 mM DTT, 10 mM imidazole and 1× Complete protease inhibitor EDTA-free [Roche]). The cells were broken using a bead beater, and the lysate was cleared of unbroken cells by centrifugation at 27 000 × g for 30 min. The clarified lysate was gently mixed with 1 ml Ni-NTA resin for 1 h at 4°C. The resin was then sequentially washed with 5 vol of lysis buffer B, 5 vol of lysis buffer B containing 20 mM imidazole, and then the His-tagged proteins were eluted in buffer B containing 500 mM imidazole. The eluted proteins were dialyzed against Storage Buffer B (20 mM Tris–HCl [pH 7.5], 100 mM potassium acetate, 0.1 mM magnesium acetate, 2 mM DTT and 10% glycerol) and then stored at –80°C.
Ribosomal 40S and 60S subunits were purified from Saccharomyces cerevisiae strain YRP840 based on the published protocol (52) with some modifications. Cells were grown in 10 l YPD medium to A600 = 1.0. Following cell harvest, the cell pellet was resuspended in 50 ml lysis buffer (20 mM HEPES–KOH [pH 7.4], 100 mM KOAc, 2.5 mM MgOAc, 1 mg/ml heparin, 2 mM DTT, 0.4 mM AEBSF). To break the cells a 50% volume of glass beads was added to the cell suspension. The mixture was vigorously agitated on a vortex for 1 min and then incubated on ice for 1 min. This cycle of mixing and cooling was repeated five times. Following removal of the glass beads and unbroken cells by centrifugation at ∼2560 × g, the extract was clarified by centrifugation at ∼27 000 × g and then layered (20 ml per tube) onto 3 ml sucrose cushions (20 mM HEPES–KOH [pH 7.4], 100 mM KOAc, 2.5 mM MgOAc, 0.5 M KCl, 1 M sucrose, 2 mM DTT). Ribosomes were pelleted by centrifugation at 164 000 × g in a Beckman Type70 Ti rotor. The ribosome pellet from each tube was resuspended in 1 ml Subunit Separation Buffer (20 mM HEPES–KOH [pH 7.4], 0.5 M KCl, 2 mM MgCl2, 2 mM DTT) and then stirred on ice for 1 h. To release any nascent chains associated with the ribosomes, 10 μl of 0.1 M puromycin was added to 1 ml ribosome solution, which was then sequentially incubated on ice for 15 min, at 37°C for 10 min and then on ice for 10 min. Next, 1 ml of ribosome solution was gently layered on top of 5–20% sucrose gradients, and then subjected to velocity sedimentation by centrifugation at 174 000 × g for 6 h at 4°C in a Beckman SW32 rotor. Following fractionation of the gradient, the fractions containing the 40S and 60S peaks were separately pooled and then concentrated by centrifugation at 59 000 × g for 24 h. Finally, the ribosomal subunit pellets were dissolved in buffer containing 20 mM HEPES–KOH [pH 7.4], 100 mM KOAc, 2.5 mM MgOAc, 250 mM sucrose and 2 mM DTT. The subunit concentration was calculated by measuring the A260 and using the conversion 1.0 A260 unit = 50 pmol of 40S subunits or 31 pmol of 60S subunits.
E. coli MetRS purification and aminoacylation of initiator tRNAMet
For aminoacylation of yeast initiator tRNAiMet, E. coli MetRS was purified from E. coli strain XL1 Blue (Agilent) transformed with pRA101 (36) and yeast tRNAiMet was prepared by in vitro transcription as described previously (37). Aminoacylation reactions containing 5 μM tRNAiMet, 2 mM ATP, 0.3 μM [35S]methionine (Perkin Elmer), 10 mM MgCl2 and 1 μM MetRS in 1× reaction buffer A (100 mM HEPES–KOH [pH 7.5], 10 mM KCl and 1 mM DTT) were incubated at 37°C for 30 min. In typical reactions ∼50–60% of tRNAiMet was charged with [35S]methionine.
Preparation of other aminoacyl-tRNAs
The UGG isoacceptor of tRNAPro was purified from bulk S. cerevisiae tRNA (Roche) using the biotinylated oligo 5′-CCAAAGCGAG AATCATACCA CTAGAC-BioTEG-3′ and then treated with CCA adding enzyme as described previously (10). The tRNAPhe was aminoacylated using a yeast post-ribosomal supernatant (S100, prepared from strain BY4741 as previously described (10)) as the source of phenylalanyl-tRNA synthetase (PheRS). For aminoacylation of tRNALys, the yeast lysyl-tRNA synthetase (KRS1) expression plasmid pET3a-KRS1 was introduced into the E. coli strain BL21(DE3) CodonPlus-RIL (Agilent) and KRS1 was expressed and purified as previously described (37).
Prolyl-tRNA synthetase (ProRS) was purified as described (38) with minor modifications. Transformants of E. coli strain XL1 Blue (Agilent) carrying the ProRS expression vector pQE30-yPRS (obtained from Karin Musier-Forsyth, Ohio State University) were grown in 500 ml Lysogeny Broth (LB) medium containing 100 μg/ml ampicillin at 37°C to OD600 = 0.7. Following addition of 0.1 mM IPTG, the culture was incubated at 20°C for 16 h, and then ProRS was purified as described previously (10).
For aminoacylation of tRNA, including Pro analogs (except pipecolic acid which was aminoacylated using a flexizyme), we followed our previously published protocol (10) with some modifications. In 100 μl reaction volume, 5 μM tRNA was mixed with 2 mM ATP, 0.1 mM amino acid and 1 μM tRNA synthetase in 1× reaction buffer B (40 mM Tris–HCl [pH 7.6], 10 mM magnesium acetate and 1 mM DTT) and then incubated at 30°C for 30 min. Following sequential extractions with phenol (pH 4.3) and chloroform, RNA was precipitated by addition of 10 μl 3 M sodium acetate (pH 5.3) and 3 vol ethanol. Precipitated RNA was collected by centrifugation at maximum speed in a microfuge at 4°C. The RNA pellet was resuspended in 20 μl RNase-free water, and then split into 5 μl aliquots and stored at –80°C. Charging efficiencies were typically 20–50%.
Preparation of flexizyme and misacylation of tRNA
Synthesis of phenylalanyl-cyanomethyl ester (Phe-CME), prolyl-3,5-dinitrobenzyl ester (Pro-DBE) and pipecolic-DBE was carried out as described previously (39), and the flexizymes dFx and eFx, or derivatives thereof, were prepared by in vitro transcription as described previously (40,41). As the 3′-end of yeast tRNAPro has the atypical sequence -CCCA-3′ (with C rather than A immediately preceding the CCA sequence), the 3′ end of the flexizyme eFx was mutated from –GGU-3′ to –GGG-3′ to enable base-pairing interactions with 3′ end of tRNAPro. The RNA sequence of the altered eFx is (mutated final G is underlined):
5′-GGAUCGAAAGAUUUCCGCGGCCCCGAAAGGGGAUUAGCGUUAGGG-3′.
Flexizyme catalyzed acylation of tRNA was performed as described previously (40,42–44) with minor modifications. Briefly, a mixture of 1 μl 1M HEPES–KOH (pH 7.5), 2.5 μl 200 μM flexizyme, 5 μl 100 μM tRNA and 3.5 μl RNase-free water was incubated at 95°C for 2 min and then slowly cooled at room temperature for 5 min. Following addition of 4 μl 3 M MgCl2, the mixture was incubated at room temperature for 5 min followed by incubation on ice for 3 min. To acylate the tRNA, the reaction mixture was supplemented with 4 μl 25 mM activated amino acid (dissolved in DMSO) and then incubated on ice for 2 h. Following the acylation reaction, the RNA was precipitated by adding 80 μl 0.3 M sodium acetate (pH 5.2) and 200 μl ethanol and then collected by centrifugation at 15 000 × g for 15 min at 25°C. Finally, the RNA pellet was washed in 70% ethanol, and the aminoacyl-tRNAs were resuspended in RNase-free water.
Peptide formation assay
Peptide formation assays were performed using an in vitro reconstituted system as previously described (10). To examine the polyamine effect on peptide synthesis, translation factors and ribosomes were purified and both initiation and elongation complexes were assembled without spermidine. Initiation complexes were prepared in 1× Recon Buffer A (30 mM HEPES–KOH [pH 7.5], 100 mM potassium acetate, 3 mM magnesium acetate, 2 mM DTT) as described previously (10). Final concentrations for each component are: 8 nM [35S]Met-tRNAiMet, 0.4 μM eIF2, 1 μM eIF1, 1 μM eIF1A, 0.8 μM 40S, 1 μM mRNA, 1 μM eIF5 and 0.5 μM eIF5B. Following assembly of initiation complexes, excess factors were removed by sedimenting the complexes through a 1 M sucrose cushion in 1× Recon Buffer C (30 mM HEPES–KOH [pH 7.5], 100 mM potassium acetate, 1 mM magnesium acetate, 2 mM DTT). Ribosomal complexes were resuspended in 1× Recon Buffer C, and aliquots were stored at –80°C. Peptide formation assays contained 4 nM initiation complex, 2 μM eEF1A, 1 μM eEF2, 1 μM eEF3, 1 μM aminoacyl tRNA, 1 mM GTP-Mg2+, 1 mM ATP-Mg2+ and varying amounts of polyamines in 1× Recon Buffer C. For assays with misacylated tRNAs or proline analogs, the Recon Buffer contained 1 mM spermidine. The elongation assay components were preincubated for 5 min on ice before adding the initiation complex, and then reactions were incubated at 26°C. Reactions were quenched at different times by mixing with an equivalent vol of 0.2 N KOH, and progress of peptide formation was examined by electrophoretic thin-layer chromatography (TLC) as described previously (10,45). The fractional yield of peptide products in each reaction at different times was quantified and fit using Kaleidagraph (Synergy Software) to the first-order exponential equation A[1 − exp(−kt)]. If the reaction proceeded with sigmoidal kinetics, the data were fit to the simple two-step process equation A(1 + 1/(k1 – k2)[k2 exp(–k1t) – k1 exp(–k2t)]) where A is the amplitude and k1and k2 are the observed rate constants.
RESULTS
eIF5A requirement for polyproline synthesis is imposed by proline, not by tRNAPro
Given the position of eIF5A binding on the ribosome with the body of eIF5A contacting the peptidyl-tRNA in the P site and the hypusine residue near the acceptor arm of the tRNA (32,33), the ability of eIF5A to promote polyproline synthesis could reflect specific interactions between eIF5A and tRNAPro. Alternatively, the imino acid proline is known to be a poor substrate for solvent-based and for ribosome-dependent peptide bond formation (46–49), and thus may establish the additional requirement for eIF5A. To determine whether proline or tRNAPro imposes the requirement for eIF5A, we generated misacylated Pro-tRNAPhe and Phe-tRNAPro and examined the eIF5A dependency for polyproline and polyphenylalanine synthesis using an in vitro reconstituted translation assay system.
As both proline tRNA synthetase (ProRS, 50) and phenylalanine tRNA synthetase (PheRS, 51) possess editing activities to either deacylate misacylated tRNAs or to hydrolyze non-cognate amino acid-adenylate intermediates, the canonical aminoacyl-tRNA synthetases cannot be used to generate the misacylated Pro-tRNAPhe and Phe-tRNAPro. Accordingly, flexizymes, RNA enzymes selected for the ability to aminoacylate tRNA (40,43,44) but which lack editing activity, were used to generate the misacylated tRNAs. Purified tRNAPhe (Sigma) was incubated with the dFx flexizyme and 3,5-dinitrobenzyl ester (DBE)-proline to generate Pro-tRNAPhe, and purified tRNAPro and cyanomethyl ester (CME)-phenylalanine were incubated with a modified version of the eFx flexizyme designed to base pair with the 5′-CCCA-3′ end of the acceptor arm of tRNAPro to synthesize Phe-tRNAPro (Figure 1A). The misacylated Pro-tRNAPhe and Phe-tRNAPro as well as the canonical Pro-tRNAPro and Phe-tRNAPhe, which were synthesized using the conventional synthetases, were then tested as substrates for in vitro peptide synthesis.
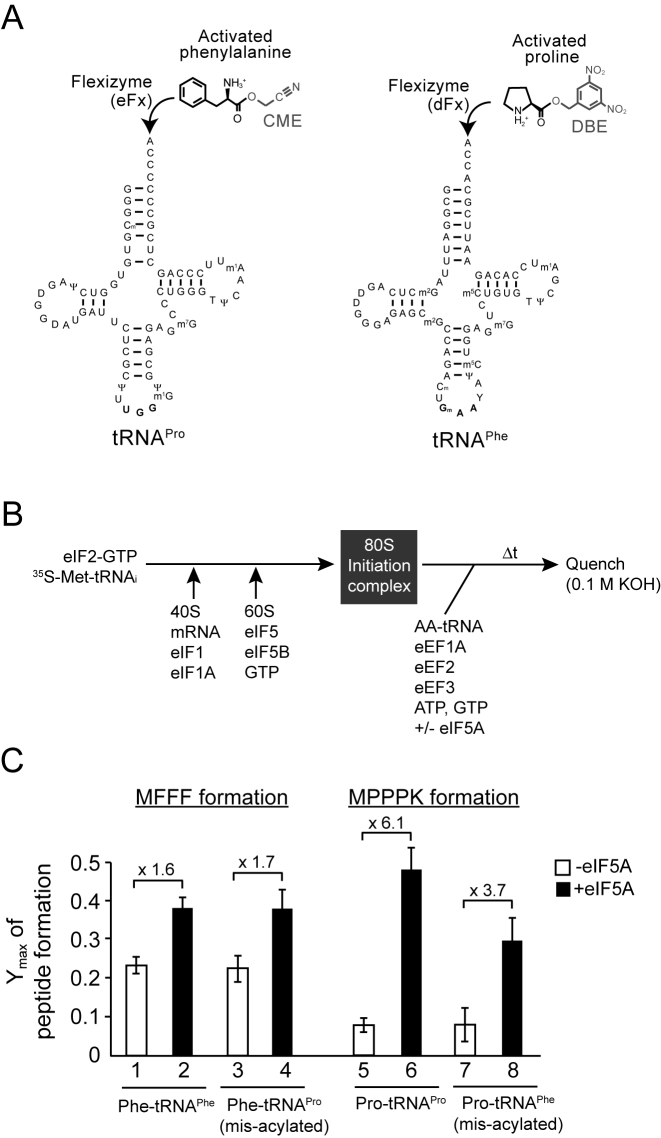
Figure 1.
Imino acid proline and not tRNAPro imposes eIF5A requirement for polyproline peptide synthesis. (A) Cloverleaf structures of yeast tRNAPro and tRNAPhe depicting their misacylation by flexizymes eFx and dFx and cyanomethyl ester (CME)-phenylalanine and 3,5-dinitrobenzyl ester (DBE)-proline, respectively. (B) Scheme for in vitro reconstituted translation elongation assay. (C) Maximum fractions (Ymax) of MFFF (lanes 1–4) and MPPPK (lanes 5–8) synthesis in elongation assays (Supplementary Figure S1) performed with the indicated canonical Pro-tRNAPro or Phe-tRNAPhe or with misacylated Pro-tRNAPhe or Phe-tRNAPro in the presence of 1 mM spermidine and in the absence (white bars) or presence (black bars) of 5 μM hypusinated eIF5A. Fold stimulation of Ymax by eIF5A is presented (above bars) for each aminoacyl-tRNA. Error bars are standard deviations (SD) from at least three independent experiments.
As depicted in Figure 1B, translation initiation complexes were assembled by first mixing [35S]Met-tRNAiMet with eIF2 and GTP to form ternary complexes, and then adding 40S ribosomal subunits, translation factors eIF1 and eIF1A, and a model unstructured mRNA encoding the peptides MPPP, MFFF, MFFFK or MPPPK. The use of the unstructured mRNA avoided the requirement to include translation factors (eIF3, eIF4A, eIF4B, eIF4E, eIF4F) that promote mRNA binding to the ribosome (52,53). Following a brief incubation to allow assembly of the 48S preinitiation complex in which the anticodon of [35S]Met–tRNAiMet is paired with the AUG start codon of the mRNA, the 60S ribosomal subunit was added along with GTP and the factors eIF5 and eIF5B. The resulting 80S initiation complex is poised with [35S]Met–tRNAiMet in the P site base-paired to the AUG start codon. Following purification by sedimentation through a sucrose cushion to remove initiation factors, translation elongation was activated by addition of the appropriate aminoacyl-tRNAs together with the delivery factor eEF1A, the ribosomal translocase eEF2, the fungal specific elongation factor eEF3, ATP and GTP in the absence or presence of saturating levels of eIF5A (5 μM). Elongation reactions were quenched by addition of base, and the released peptide products were analyzed by electrophoretic thin-layer chromatography (TLC).
To test whether tRNAPro imposes the requirement for eIF5A, 80S initiation complexes were assembled on mRNAs encoding either MFFF or MPPP and polyphenylalanine peptide synthesis was monitored in reactions containing either canonical Phe-tRNAPhe or misacylated Phe-tRNAPro, respectively. As shown in Figure 1C, in the reactions containing Phe-tRNAPhe MFFF was efficiently synthesized in the absence of eIF5A, and addition of hypusinated eIF5A resulted in an ∼1.6-fold increase in the end level of MFFF synthesis (lane 2 versus 1; see also Supplementary Figure S1A and B). Note that unless otherwise stated eIF5A will refer to the hypusinated form of the protein and this modified form of the factor was used for all experiments in Figures 1 and 2. These results in Figure 1C support our previous in vitro findings that eIF5A modestly stimulates MF, MFF and MFFF peptide synthesis (6,10). Because reaction end-points, especially for proline peptide synthesis described below, greatly vary in the presence versus absence of eIF5A, the observed reaction rates are likely impacted by competing reactions including peptidyl-tRNA drop-off from the ribosome. This phenomenon has been previously observed both in yeast (54) and bacterial (9,55) systems when monitoring the synthesis of short peptides in vitro. Given this apparent complication, analysis of the reactions will focus on end points (end levels) and not observed rates. Nearly identical yields of MFFF synthesis were obtained in the reactions containing misacylated Phe-tRNAPro using 80S complexes assembled on the mRNA encoding MPPP. Substantial MFFF peptide synthesis was observed in the absence of eIF5A (end level = 0.23) and addition of eIF5A resulted in an ∼1.7-fold increase (end level = 0.39; Figure 1C, lane 4 versus lane 3; and Supplementary Figure S1A and B). Importantly, no peptide synthesis was observed in reactions containing Phe-tRNAPro and eIF5A when the ribosome was programmed with an mRNA encoding MFFF (Supplementary Figure S1A), demonstrating that no Phe-tRNAPhe contaminated the Phe-tRNAPro preparation and that peptide synthesis by the misacylated tRNAPro is dependent on canonical decoding of the mRNA by the tRNA anticodons.
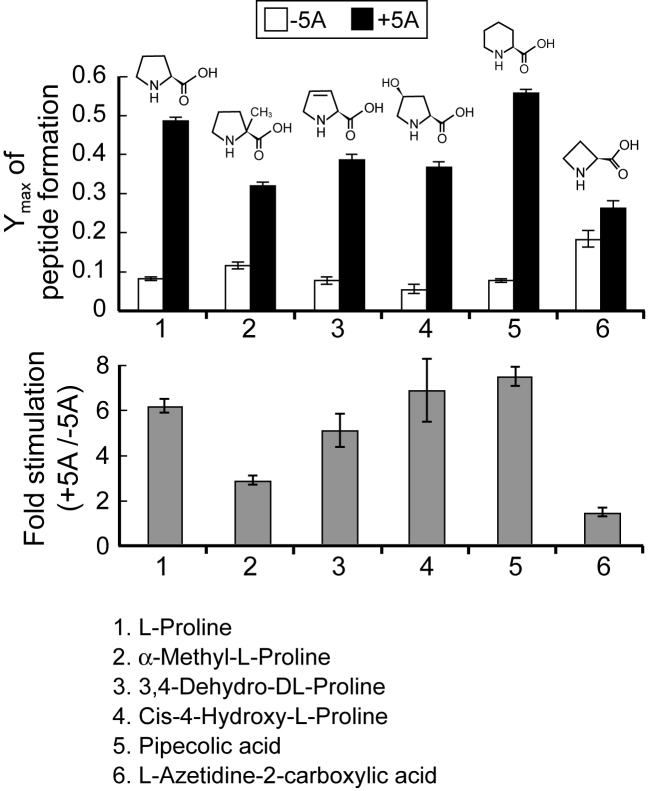
Figure 2.
eIF5A-independent synthesis of homopolymers of azetidine-2-carboxylic acid (AZC), but not of other proline analogs. Maximum fractions of MPPPK peptide synthesis in elongation assays (Supplementary Figure S2) performed with the indicated proline analogs (1, l-proline; 2, α-methyl-l-proline; 3, 3,4-dehydro-dl-proline; 4, cis-4-hydroxy-l-proline; 5, pipecolic acid; 6, l-azetidine-2-carboxylic acid) in the presence of 1 mM spermidine and in the absence (white bars) or presence (black bars) of 5 μM hypusinated eIF5A. (Bottom) Fold stimulation of Ymax by eIF5A was calculated for each reaction in the upper panel (see also Supplementary Figure S2). Error bars are SD from at least three independent experiments (A) and calculated propagated SD (B).
Next, we tested whether the imino acid proline imposes the requirement for eIF5A. 80S initiation complexes were assembled on an mRNA encoding MPPPK and polyproline synthesis was monitored in reactions containing canonical Pro-tRNAPro. In contrast to the MFF and MFFF peptides, which were well resolved during electrophoretic TLC, MPP and MPPP peptides co-migrated during TLC. Addition of a C-terminal Lys residue on the MPPPK peptide enabled resolution during TLC of the full-length tri-proline peptide from incomplete C-terminally truncated products (Supplementary Figure S1). As expected, polyproline peptide synthesis showed a strong dependence on eIF5A. In the absence of eIF5A very little MPPPK was synthesized, and addition of eIF5A stimulated polyproline peptide synthesis ∼6.1-fold (Figure 1C, lane 6 versus lane 5; and Supplementary Figure S1C and D). Similarly, when misacylated Pro-tRNAPhe was tested for MPPPK peptide synthesis using 80S initiation complexes assembled on an mRNA encoding MFFFK, polyproline synthesis was stimulated ∼3.7-fold by adding eIF5A (end level = 0.08 in the absence of eIF5A and 0.30 in the presence of eIF5A; Figure 1C, lane 8 versus lane 7, and Supplementary Figure S1C and D). In control reactions, no peptide formation was detected when misacylated Pro-tRNAPhe was used for elongation of initiation complexes assembled on mRNA encoding MPPPK (Supplementary Figure S1C), demonstrating that the observed peptide synthesis was not due to contaminating canonical Pro-tRNAPro. It is interesting to note that the end level of the MPPPK peptide formation using misacylated Pro-tRNAPhe is lower than that obtained with canonical Pro-tRNAPro (0.30 versus 0.49, respectively), suggesting that the body of tRNAPhe is less effective for eIF5A-stimulated polyproline synthesis than is the body of tRNAPro. Taken together, these data indicate that the imino acid proline and not tRNAPro imposes the primary eIF5A requirement for polyproline peptide synthesis.
In contrast to most proline analogs, azetidine-2-carboxylic acid bypasses the eIF5A requirement for synthesis of homopolymer peptides
Proline is known to be a poor substrate for both ribosome-dependent and solvent-based peptide synthesis (46–49). To gain insight into the properties of proline that impose the eIF5A requirement for peptide synthesis, the eIF5A dependency for peptide synthesis was assessed using several proline analogs. The analogs were chosen based on availability and on their differences in peptide bond flexibility relative to proline. Native yeast ProRS was used to aminoacylate tRNAPro with the analogs α-methyl-l-proline, 3,4-dehydro-dl-proline, cis-4-hydroxy-l-proline and l-azetidine-2-carboxylic acid (AZC), while the dFx flexizyme system was used to aminoacylate tRNAPro with the 6-membered ring derivative DBE-l-pipecolic acid. To monitor the eIF5A dependency for incorporation of the proline analogs into peptides, 80S initiation complexes were assembled on an mRNA encoding MPPPK and peptide synthesis was monitored in reactions containing Lys-tRNALys and the proline analogs linked to tRNAPro.
The proline analogs exhibited different rates and end levels for peptide synthesis (Figure 2 and Supplementary Figure S2). Like native proline, α-methyl-l-proline, 3,4-dehydro-dl-proline, cis-4-hydroxy-l-proline and pipecolic acid showed strong dependency on eIF5A for peptide synthesis with the end level for peptide synthesis increasing 2.9–fold (α-methyl-l-proline) to 7.4–fold (pipecolic acid) (Figure 2 and Supplementary Figure S2). Interestingly, the end level of poly-AZC peptide synthesis was ∼2–3–fold higher than polyproline synthesis in the absence of eIF5A, and addition of eIF5A only stimulated poly-AZC synthesis by ∼1.4–fold (Figure 2 and Supplementary Figure S2). This modest stimulation of poly-AZC synthesis by eIF5A is similar to the ∼1.6–fold stimulation of polyPhe synthesis by eIF5A observed in Figure 1C (lanes 1–2). As the four-membered ring derivative AZC is more flexible than proline (56) and exhibits a decreased cis to trans isomerization barrier relative to proline (56–58), the structural rigidity of proline may contribute to the requirement for eIF5A for efficient peptide synthesis.
Both hypusinated and unhypusinated eIF5A functionally substitute for polyamines to promote in vitro peptide synthesis
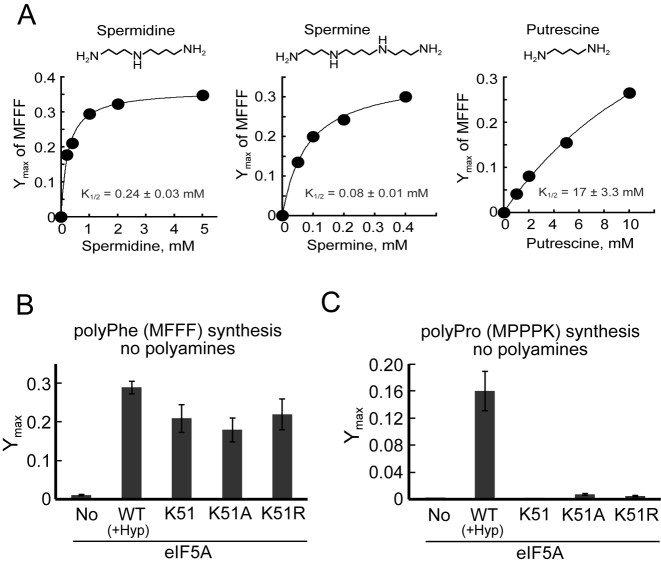
Addition of polyamines, typically spermidine, spermine or putrescine, is known to enhance the overall yield and to lower the optimum Mg2+ concentration in both bacterial and eukaryotic cell-free translation systems (17,18, see also 19,20,59–62). To examine the impact of polyamines in our reconstituted in vitro translation assays, all components of the translation assay system including translation factors, aminoacyl-tRNAs and ribosomes were prepared in buffers lacking all polyamines. To monitor general translation elongation activity, reagents prepared in the absence of polyamines were used to first assemble an 80S initiation complex on an mRNA encoding MFFF. The synthesis of the polyphenylalanine (polyPhe) peptide was then monitored in reactions containing the 80S initiation complexes plus Phe-tRNAPhe and the elongation factors eEF1A, eEF2 and eEF3 (but lacking eIF5A). The use of pre-assembled 80S initiation complexes in these assays restricted the analysis to the impact of polyamines on translation elongation and not on the initiation steps of translation. As shown in Figure 3A (and Supplemental Figure S3A), no MFFF peptide synthesis was detected in the assays lacking polyamines. Addition of spermidine, spermine or putrescine provided a dose-dependent restoration of peptide synthesis (Figure 3A). The concentration of spermidine required to reach the half-maximal end level of peptide synthesis (K1/2(endpoint)) was 0.24 (±0.03) mM. The tetraamine spermine was much more effective in stimulating peptide synthesis with K1/2(endpoint) = 0.08 (±0.01) mM. Putrescine also promoted MFFF peptide synthesis; however, the end level of peptide synthesis continuously increased without a distinct saturation up to 10 mM putrescine yielding an apparent K1/2(endpoint) = 17 (±3.3) mM (Figure 3A). Interestingly, the effectiveness of the three polyamines to stimulate peptide synthesis correlated with the number of amines in the molecule: the tetraamine spermine was more effective than the triamine spermidine, which in turn was more effective than the diamine putrescine. The limited effectiveness of putrescine to stimulate peptide synthesis raised the possibility that contaminating spermine or spermidine in the putrescine might be responsible for the observed activity. However, no spermine or spermidine was detectable during mass spectrometry analysis of the putrescine (purity >99.99%; data not shown). These results indicate that putrescine, albeit at non-physiologically high levels, can stimulate translation elongation. Despite the ability of polyamines to stimulate MFFF peptide synthesis, saturating concentrations of these polyamines were unable to stimulate polyproline peptide synthesis in reactions lacking eIF5A (data not shown). Moreover, high concentrations (>1 mM) of spermine, (but not of spermidine or putrescine up to maximum concentration tested, 5 and 10 mM, respectively), strongly inhibited the yield of MFFF peptide synthesis (data not shown).
Figure 3.
Both hypusinated and unhypusinated eIF5A substitute for polyamines to stimulate general translation. (A) Maximum fractions of MFFF peptide synthesis obtained in polyamine-deficient in vitro reconstituted translation assays supplemented with the indicated amounts of spermidine, spermine or putrescine (structures depicted above plots) were plotted and fit to the Michaelis–Menten equation. K1/2(endpoint) is the polyamine concentration at which 50% of the Ymax is obtained. (B, C) Maximum fractions of MFFF (B) and MPPPK (C) peptide synthesis obtained in polyamine-deficient in vitro reconstituted translation assays supplemented with no eIF5A (No), 5 μM wild type hypusinated eIF5A [WT(+Hyp)], or three versions of eIF5A (5 μM) lacking hypusine: wild type eIF5A (K51), eIF5A-K51A, or eIF5A-K51R, as indicated (see also Supplementary Figure S3). Error bars are SD from at least three independent experiments (B, C).
The hypusine modification on eIF5A is derived from spermidine, and we previously showed that addition of hypusinated eIF5A enhances both general (polyPhe) and polyproline peptide synthesis in reactions containing polyamines (Figure 1C, and (10)). Next, we asked if hypusinated eIF5A could stimulate translation in assays lacking polyamines. As shown in Figure 3B and Supplementary Figure S3A,B (No versus WT), wild-type hypusinated eIF5A (5 μM) readily stimulated MFFF peptide synthesis in assays lacking added polyamines. Likewise, hypusinated eIF5A (5 μM) readily stimulated polyproline peptide synthesis in assays lacking added polyamines (Figure 3C and Supplementary Figure S3C,D, No versus WT). As expected, the ability of eIF5A to stimulate polyproline synthesis in assays lacking polyamines was dependent on the hypusine modification. Unmodified eIF5A (K51) as well as the unhypusinated eIF5A-K51A and eIF5A-K51R mutants were unable to rescue polyproline peptide synthesis (Figure 3C and Supplementary Figure S3C). Interestingly, the unhypusinated forms of eIF5A (K51, K51A and K51R) at the same 5 μM concentration substituted for wild-type hypusinated eIF5A to promote MFFF peptide synthesis in reactions lacking polyamines (Figure 3B and Supplementary Figure S3A and B). Notably, the concentration of eIF5A (5 μM) used in these assays is well below the mM concentrations of polyamines required to stimulate elongation, indicating that the activity of eIF5A in these assays is not due to contaminating polyamines in our eIF5A preparations. Moreover, the level of eIF5A in the assays is physiologically relevant. We estimate the eIF5A concentration in yeast cells at 36–100 μM (data not shown), consistent with a previous report that eIF5A is present in excess of ribosomes in yeast cells and is one of the most abundant translation factors (63). Taken together, our in vitro results indicate that eIF5A might have functions beyond simply delivering hypusine to the ribosome, and that the body of eIF5A (lacking hypusine) can also stimulate protein synthesis.
DISCUSSION
The translation factor eIF5A was originally identified by its ability to stimulate methionyl-puromycin synthesis, a model assay for peptide bond formation (3–5). More recently, eIF5A was shown to function like its bacterial ortholog EF-P to promote translation of polyproline motifs (10). In light of recent structures revealing eIF5A binding to the ribosome, our biochemical data provide new insights into the function of eIF5A and the role of polyamines in protein synthesis. In the crystal structure of eIF5A bound to an 80S ribosome lacking tRNA substrates (32), the eIF5A occupies the same position as in cryo-EM images of eIF5A–80S complexes containing a P-site tRNA (33). Moreover, the position of eIF5A in these ribosomal complexes overlays with the position of EF-P bound to the bacterial 70S ribosome with Met-tRNA in the P site (31). In all three of these structures the eIF5A/EF-P is positioned to contact the acceptor stem of the P-site tRNA and to lie adjacent to the D stem–loop of the tRNA. Notably, it has been proposed that EF-P makes more intimate (direct) contact with the D stem-loop than does eIF5A (33). The structural images of eIF5A bound to the 80S ribosome are consistent with directed hydroxyl radical cleavage studies (10) that placed the N-terminal domain and hypusine residue of eIF5A near the acceptor stem and placed the C-terminal domain of eIF5A nearer to the elbow and anticodon stem of the L-shaped P-site tRNA. As described below, we propose that these interactions of eIF5A with the body of the P-site tRNA may underlie the ability of eIF5A to functionally substitute for polyamines in the stimulation of general protein synthesis.
Based on the position of eIF5A binding adjacent to the peptidyl-tRNA in the P site, it was unclear whether the requirement for eIF5A for polyproline synthesis was dictated by the tRNAPro or by the imino acid proline. Using misacylated tRNAs, we showed that Phe-tRNAPro, like Phe-tRNAPhe, was readily utilized in the synthesis of polyphenylalanine peptides in the absence of eIF5A and that addition of eIF5A provided a modest stimulation in peptide yields (Figure 1C). These results demonstrate that tRNAPro does not impose a requirement for eIF5A, and thus suggest that the imino acid proline imposes the eIF5A requirement.
Consistent with the notion that proline imposes the requirement for eIF5A, polyproline peptide synthesis directed by misacylated Pro-tRNAPhe or by canonical Pro-tRNAPro was low in the absence of eIF5A, and the peptide yield was strongly stimulated by addition of the factor (Figure 1C). These data indicate that the imino acid proline imposes the requirement for eIF5A. However, it is noteworthy that even in the presence of eIF5A the end level of polyproline synthesis directed by Pro-tRNAPhe is less than that achieved with Pro-tRNAPro (Figure 1C, lane 8 versus 6, and Supplementary Figure S1C,D). One interpretation of this result is that tRNAPro possesses features that enable it to interact more productively with eIF5A. Consistent with this hypothesis, studies with EF-P in the bacterial system identified elements in the D-arm and D-loop of tRNAPro that enabled it to function efficiently with EF-P (55). Specific residues within the 9-nt D-loop closed by two consecutive G:C base pairs in D-stem of tRNAPro were found to be important for EF-P stimulation of polyproline peptide synthesis, and substitution of these elements into heterologous tRNAs enhanced their function in EF-P stimulated polyproline peptide synthesis (55). Interestingly, the 9-nt D-loop and adjacent D-stem in yeast tRNAPro (Figure 1A) is dissimilar to bacterial tRNAPro and shows greater similarity to the D-loop of yeast tRNAPhe (Figure 1A). Thus, while our data clearly show that tRNAPro, when misacylated with Phe, does not impose a requirement for eIF5A, and that the imino acid Pro imposes an eIF5A requirement when linked to either tRNAPro or tRNAPhe, it is possible that, like EF-P, the eIF5A stimulation of peptide synthesis is dependent on elements in the P-site tRNA. Accordingly, yeast tRNAPhe may fortuitously possess structural features in common with yeast tRNAPro that enable it to functionally interact with eIF5A to stimulate peptide synthesis; however, these features are likely distinct from those identified in bacterial tRNAPro that mediate EF-P-directed peptide synthesis. At odds with this discussion, recent ribosomal profiling and biochemical experiments revealed that eIF5A has a broad role and promotes synthesis of peptides both containing and lacking proline residues (54). Accordingly, the efficient ability of eIF5A to promote peptide synthesis with both Pro-tRNAPro and Pro-tRNAPhe may indicate that, unlike EF-P, eIF5A does not recognize or interact with specific tRNA features.
While proline is known to be a poor substrate for peptide synthesis (46–49), the properties of proline that impose the requirement for eIF5A are unclear. To gain insights, we tested a collection of proline analogs to determine their dependency on eIF5A. Interestingly, all of the proline analogs tested except AZC showed the same eIF5A dependency as observed for proline (Figure 2). The pKa values of both the carboxyl and amino groups of the proline analogs did not correlate with the reactivity of the imino acids in either the presence or absence of eIF5A, indicating that the requirement for eIF5A is not imposed by the electronegativity of proline. This finding is not surprising given that the hypusine side-chain on eIF5A is not positioned to contribute to the chemistry of peptide bond formation and instead is thought to help position the substrates (32–34).
As AZC is a four-membered ring analog of proline and thus also an imino acid, the enhanced incorporation of AZC into proteins in the absence of eIF5A demonstrates that the imino nature of proline does not impose the eIF5A requirement. Rather, we suggest that the ring structure of proline may be an important determinant imposing the requirement for eIF5A. AZC peptides are more flexible than proline peptides (64,65) and the rotational barrier for converting between cis and trans peptide bonds is decreased for AZC versus proline (56,58) with AZC favoring the cis conformation (57). These differences in the physical properties of AZC versus proline point toward the structural rigidity of proline as contributing to the requirement for eIF5A. Interestingly, Doerfel et al. (66) examined the activity of proline analogs in the bacterial system with EF-P. Using kinetic analyses not yet available for the eukaryotic system, these researchers found that EF-P stimulated translation by decreasing the activation energy for peptidyl transfer primarily through a favorable entropic effect (66). This entropic role of EF-P, and by extension for eIF5A, is consistent with the structures of eIF5A on the ribosome and the position of hypusine interacting with the acceptor stem of the P-site tRNA on the ribosome (32,33). Perhaps the hypusine side chain accelerates peptide bond formation by fixing the position of the acceptor stem of the P-site tRNA for reaction with the A-site substrate. Given this apparent role of eIF5A/EF-P to enhance protein synthesis through a favorable entropy change, it is challenging to understand how the more flexible AZC accelerates the reaction in the absence of eIF5A. Moreover, it is noteworthy that in the bacterial system, synthesis of the simple tripeptides fMet-Pro-puromycin and fMet-AZC-puromycin was stimulated to similar extents by EF-P (66). However, consistent with our findings, an in vitro toe-printing assay to monitor ribosome stalling during bacterial translation revealed less stalling on a tri-AZC versus a tri-proline sequence (66). Taken together, our results indicate that the structural rigidity of proline, especially in the context of polyproline sequences, contributes to the requirement for eIF5A, and are consistent with the notion that the hypusine side chain on eIF5A promotes translation by properly positioning the P-site substrate for peptide bond formation.
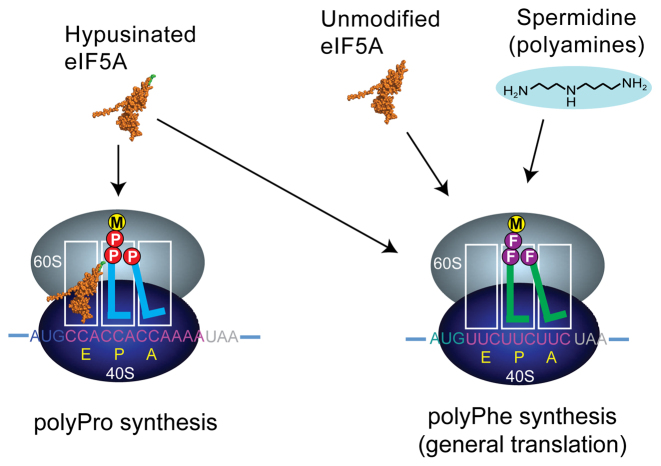
Finally, our studies provided new insights into the role of polyamines in protein synthesis. While polyamines are required for maximal translational activity in vitro, the molecular function of polyamines in protein synthesis has remained unclear. Based on their ability to bind nucleic acids and proteins, and to, in part, functionally overlap with Mg2+, polyamines are thought to perform diverse roles in RNA and DNA reactions (see 67,68). Interestingly, eIF5A was previously shown to lower the Mg2+ optimum for in vitro translation assays lacking spermidine (4), and in this work we show that eIF5A can functionally substitute for polyamines to stimulate synthesis of the generic substrate MFFF (Figure 3B). Surprisingly, this ability of eIF5A to replace polyamines was not dependent on the hypusine modification on eIF5A and thus can be attributed to the body of the factor, which is nestled beside the P-site tRNA on the ribosome (Figure 4). Accordingly, we propose two functions for eIF5A on the ribosome. First, eIF5A serves as a hypusine delivery instrument. Binding of eIF5A to the ribosome places hypusine in a position to properly orient the acceptor stem of the P-site tRNA and resolve possible geometric impediments to peptide bond synthesis incurred when diproline is linked to the P-site tRNA (see 34, and Figure 4). Second, the body of eIF5A interacts with the P-site tRNA and functionally substitutes for polyamines to stimulate general protein synthesis (Figure 4).
Figure 4.
Models of polyamine and eIF5A stimulation of general and polyproline peptide synthesis. In the absence of eIF5A polyproline synthesis (left panel) stalls with diproline bound to the P-site tRNA and Pro-tRNA in the A site of an 80S elongating ribosome. Binding of eIF5A (orange) in the E site positions the hypusine residue (green) so that it can interact with the acceptor stem of the P-site tRNA and facilitate peptide bond formation. General (polyPhe) translation (right panel) requires polyamines such as spermidine; however, both unmodified and hypusinated eIF5A can functionally substitute for polyamines to stimulate general translation. The precise binding sites for spermidine on the ribosome are not known; however, given the overlap between eIF5A and polyamine function in general translation, polyamines are proposed to interact with the P-site tRNA to stabilize its binding to the ribosome and enhance its reactivity in peptide bond formation.
Given the functional overlap between eIF5A and polyamines, it is tempting to speculate that a critical role of polyamines is to interact with the P-site tRNA and the peptidyl-transferase center (PTC) of the ribosome. Consistent with this notion, the crystal structure of free tRNAPhe revealed two spermine molecules – one bound to the anticodon stem and the other bound closer to the variable loop and elbow of the tRNA (69). Moreover, the photoreactive polyamine derivative azidobenzamidino (ABA)-spermine was found to crosslink to multiple locations in E. coli ribosomes including to 23S rRNA helices H74 and H93 near the PTC (70–72), and in a high-resolution crystal structure of the E. coli ribosome, a spermidine molecule was observed bound to helix H74 (73). Interestingly, the structural and directed hydroxyl radical probing studies of eIF5A–ribosome complexes placed the hypusine residue in close contact with 25S rRNA helices H74 and H93 in yeast ribosomes (10,32,33). Thus, eIF5A and polyamines interact with the same or overlapping sites on the ribosome. A possible insight into the function of eIF5A (or polyamines) in general translation comes from our kinetic studies of peptide synthesis. As we noted previously (10), and as also noted for the studies of EF-P stimulated peptide synthesis (55,66), addition of the factor corrects an endpoint defect that is thought to be due to peptidyl-tRNA dropoff, an effect that may be more pronounced for the short peptides synthesized in the in vitro reconstituted systems. Accordingly, binding of eIF5A, and by inference polyamines, might stabilize binding of the peptidyl-tRNA enabling higher yields of peptide synthesis.
Tying together the findings from the studies presented here, we propose that eIF5A and polyamines generally support protein synthesis by stabilizing the binding of the P-site tRNA on the ribosome. Moreover, we propose that the hypusine side chain on eIF5A contacts the acceptor arm of the P-site tRNA to help orient the attached nascent peptide for peptide bond formation with the A site residue, a process that is especially needed when structurally rigid polyproline peptides are attached to the P-site tRNA. Goals for future studies include visualizing the interaction of eIF5A, its hypusine modification, and polyamines with polyprolyl-tRNA in the P site of the ribosome in order to gain further insights into how these molecules stimulate protein synthesis.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Sarah Walker for the MetRS plasmid, Karin Musier-Forsyth for the ProRS plasmid, and Terri Kinzy for the eEF2 and eEF3 expression vectors. We thank Duck-Yeon Lee in the Biochemistry Core Facility of the NHLBI for TOF-LC/MS analysis of polyamines. We thank Anthony Schuller and Rachel Green for helpful discussions and for comments on the manuscript, and Alan Hinnebusch, Jon Lorsch and members of the Dever, Hinnebusch and Lorsch labs for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of the National Institutes of Health (NICHD) (in part); Japan Science and Technology Agency (JST) PRESTO of Molecular Technology (to T.K.); Japan Society for the Promotion of Science (JSPS) Grant-in-Aid Scientific Research (S) [26220204 to H.S.]. Funding for open access charge: Lab budget of NIH Investigator.
Conflict of interest statement. None declared.
REFERENCES
- 1. Dever T.E., Green R.. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb. Perspect. Biol. 2012; 4:a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schmeing T.M., Ramakrishnan V.. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009; 461:1234–1242. [DOI] [PubMed] [Google Scholar]
- 3. Kemper W.M., Berry K.W., Merrick W.C.. Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Bα and M2Bβ. J. Biol. Chem. 1976; 251:5551–5557. [PubMed] [Google Scholar]
- 4. Schreier M.H., Erni B., Staehelin T.. Initiation of mammalian protein synthesis. I. Purification and characterization of seven initiation factors. J. Mol. Biol. 1977; 116:727–753. [DOI] [PubMed] [Google Scholar]
- 5. Benne R., Hershey J.W.B.. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J. Biol. Chem. 1978; 253:3078–3087. [PubMed] [Google Scholar]
- 6. Saini P., Eyler D.E., Green R., Dever T.E.. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009; 459:118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gregio A.P., Cano V.P., Avaca J.S., Valentini S.R., Zanelli C.F.. eIF5A has a function in the elongation step of translation in yeast. Biochem. Biophys. Res. Commun. 2009; 380:785–790. [DOI] [PubMed] [Google Scholar]
- 8. Dever T.E., Gutierrez E., Shin B.S.. The hypusine-containing translation factor eIF5A. Crit. Rev. Biochem. Mol. Biol. 2014; 49:413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Doerfel L.K., Wohlgemuth I., Kothe C., Peske F., Urlaub H., Rodnina M.V.. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013; 339:85–88. [DOI] [PubMed] [Google Scholar]
- 10. Gutierrez E., Shin B.S., Woolstenhulme C.J., Kim J.R., Saini P., Buskirk A.R., Dever T.E.. eIF5A promotes translation of polyproline motifs. Mol. Cell. 2013; 51:35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ude S., Lassak J., Starosta A.L., Kraxenberger T., Wilson D.N., Jung K.. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013; 339:82–85. [DOI] [PubMed] [Google Scholar]
- 12. Park M.H., Nishimura K., Zanelli C.F., Valentini S.R.. Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids. 2010; 38:491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Magdolen V., Klier H., Wohl T., Klink F., Hirt H., Hauber J., Lottspeich F.. The function of the hypusine-containing proteins of yeast and other eukaryotes is well conserved. Mol. Gen. Genet. 1994; 244:646–652. [DOI] [PubMed] [Google Scholar]
- 14. Schnier J., Schwelberger H.G., Smit-McBride Z., Kang H.A., Hershey J.W.B.. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1991; 11:3105–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park M.H., Joe Y.A., Kang K.R.. Deoxyhypusine synthase activity is essential for cell viability in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1998; 273:1677–1683. [DOI] [PubMed] [Google Scholar]
- 16. Sasaki K., Abid M.R., Miyazaki M.. Deoxyhypusine synthase gene is essential for cell viability in the yeast Saccharomyces cerevisiae. FEBS Lett. 1996; 384:151–154. [DOI] [PubMed] [Google Scholar]
- 17. Atkins J.F., Lewis J.B., Anderson C.W., Gesteland R.F.. Enhanced differential synthesis of proteins in a mammalian cell-free system by addition of polyamines. J. Biol. Chem. 1975; 250:5688–5695. [PubMed] [Google Scholar]
- 18. Igarashi K., Hikami K., Sugawara K., Hirose S.. Effect of polyamines on polypeptide synthesis in rat liver cell-free system. Biochim. Biophys. Acta. 1973; 299:325–330. [DOI] [PubMed] [Google Scholar]
- 19. Igarashi K., Kashiwagi K.. Modulation of protein synthesis by polyamines. IUBMB Life. 2015; 67:160–169. [DOI] [PubMed] [Google Scholar]
- 20. Igarashi K., Sugawara K., Izumi I., Nagayama C., Hirose S.. Effect of polyamines of polyphenylalanine synthesis by Escherichia coli and rat-liver ribosomes. Eur. J. Biochem. 1974; 48:495–502. [DOI] [PubMed] [Google Scholar]
- 21. Scuoppo C., Miething C., Lindqvist L., Reyes J., Ruse C., Appelmann I., Yoon S., Krasnitz A., Teruya-Feldstein J., Pappin D. et al. A tumour suppressor network relying on the polyamine-hypusine axis. Nature. 2012; 487:244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nakanishi S., Cleveland J.L.. Targeting the polyamine-hypusine circuit for the prevention and treatment of cancer. Amino Acids. 2016; 48:2353–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lassak J., Wilson D.N., Jung K.. Stall no more at polyproline stretches with the translation elongation factors EF-P and IF-5A. Mol. Microbiol. 2016; 99:219–235. [DOI] [PubMed] [Google Scholar]
- 24. Navarre W.W., Zou S.B., Roy H., Xie J.L., Savchenko A., Singer A., Edvokimova E., Prost L.R., Kumar R., Ibba M. et al. PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica. Mol. Cell. 2010; 39:209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peil L., Starosta A.L., Virumae K., Atkinson G.C., Tenson T., Remme J., Wilson D.N.. Lys34 of translation elongation factor EF-P is hydroxylated by YfcM. Nat. Chem. Biol. 2012; 8:695–697. [DOI] [PubMed] [Google Scholar]
- 26. Roy H., Zou S.B., Bullwinkle T.J., Wolfe B.S., Gilreath M.S., Forsyth C.J., Navarre W.W., Ibba M.. The tRNA synthetase paralog PoxA modifies elongation factor-P with (R)-β-lysine. Nat. Chem. Biol. 2011; 7:667–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yanagisawa T., Sumida T., Ishii R., Takemoto C., Yokoyama S.. A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P. Nat. Struct. Mol. Biol. 2010; 17:1136–1143. [DOI] [PubMed] [Google Scholar]
- 28. Lassak J., Keilhauer E.C., Furst M., Wuichet K., Godeke J., Starosta A.L., Chen J.M., Sogaard-Andersen L., Rohr J., Wilson D.N. et al. Arginine-rhamnosylation as new strategy to activate translation elongation factor P. Nat. Chem. Biol. 2015; 11:266–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rajkovic A., Erickson S., Witzky A., Branson O.E., Seo J., Gafken P.R., Frietas M.A., Whitelegge J.P., Faull K.F., Navarre W. et al. Cyclic rhamnosylated elongation factor P establishes antibiotic resistance in Pseudomonas aeruginosa. MBio. 2015; 6:e00823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rajkovic A., Hummels K.R., Witzky A., Erickson S., Gafken P.R., Whitelegge J.P., Faull K.F., Kearns D.B., Ibba M.. Translation control of swarming proficiency in Bacillus subtilis by 5-amino-pentanolylated elongation factor P. J. Biol. Chem. 2016; 291:10976–10985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blaha G., Stanley R.E., Steitz T.A.. Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome. Science. 2009; 325:966–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Melnikov S., Mailliot J., Shin B.S., Rigger L., Yusupova G., Micura R., Dever T.E., Yusupov M.. Crystal structure of hypusine-containing translation factor eIF5A bound to a rotated eukaryotic ribosome. J. Mol. Biol. 2016; 428:3570–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schmidt C., Becker T., Heuer A., Braunger K., Shanmuganathan V., Pech M., Berninghausen O., Wilson D.N., Beckmann R.. Structure of the hypusinylated eukaryotic translation factor eIF-5A bound to the ribosome. Nucleic Acids Res. 2016; 44:1944–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Melnikov S., Mailliot J., Rigger L., Neuner S., Shin B.S., Yusupova G., Dever T.E., Micura R., Yusupov M.. Molecular insights into protein synthesis with proline residues. EMBO Rep. 2016; 17:1776–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hatfield L., Beelman C.A., Stevens A., Parker R.. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996; 16:5830–5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alexander R.W., Nordin B.E., Schimmel P.. Activation of microhelix charging by localized helix destabilization. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:12214–12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Murray J., Savva C.G., Shin B.S., Dever T.E., Ramakrishnan V., Fernandez I.S.. Structural characterization of ribosome recruitment and translocation by type IV IRES. Elife. 2016; 5:e13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. SternJohn J., Hati S., Siliciano P.G., Musier-Forsyth K.. Restoring species-specific posttransfer editing activity to a synthetase with a defunct editing domain. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:2127–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saito H., Kourouklis D., Suga H.. An in vitro evolved precursor tRNA with aminoacylation activity. EMBO J. 2001; 20:1797–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Goto Y., Katoh T., Suga H.. Flexizymes for genetic code reprogramming. Nat. Protoc. 2011; 6:779–790. [DOI] [PubMed] [Google Scholar]
- 41. Goto Y., Katoh T., Suga H.. Preparation of materials for flexizyme reactions and genetic code reprogramming. Protocol Exchange. 2011; doi:10.1038/protex.2011.209. [Google Scholar]
- 42. Kawakami T., Murakami H., Suga H.. Messenger RNA-programmed incorporation of multiple N-methyl-amino acids into linear and cyclic peptides. Chem. Biol. 2008; 15:32–42. [DOI] [PubMed] [Google Scholar]
- 43. Murakami H., Ohta A., Ashigai H., Suga H.. A highly flexible tRNA acylation method for non-natural polypeptide synthesis. Nat. Methods. 2006; 3:357–359. [DOI] [PubMed] [Google Scholar]
- 44. Terasaka N., Suga H.. Flexizymes-facilitated genetic code reprogramming leading to the discovery of drug-like peptides. Chem. Lett. 2014; 43:11–19. [Google Scholar]
- 45. Eyler D.E., Green R.. Distinct response of yeast ribosomes to a miscoding event during translation. RNA. 2011; 17:925–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Johansson M., Ieong K.W., Trobro S., Strazewski P., Aqvist J., Pavlov M.Y., Ehrenberg M.. pH-sensitivity of the ribosomal peptidyl transfer reaction dependent on the identity of the A-site aminoacyl-tRNA. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Muto H., Ito K.. Peptidyl-prolyl-tRNA at the ribosomal P-site reacts poorly with puromycin. Biochem. Biophys. Res. Commun. 2008; 366:1043–1047. [DOI] [PubMed] [Google Scholar]
- 48. Pavlov M.Y., Watts R.E., Tan Z., Cornish V.W., Ehrenberg M., Forster A.C.. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wohlgemuth I., Brenner S., Beringer M., Rodnina M.V.. Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J. Biol. Chem. 2008; 283:32229–32235. [DOI] [PubMed] [Google Scholar]
- 50. Beuning P.J., Musier-Forsyth K.. Hydrolytic editing by a class II aminoacyl-tRNA synthetase. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:8916–8920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yarus M. Phenylalanyl-tRNA synthetase and isoleucyl-tRNAPhe: a possible verification mechanism for aminoacyl-tRNA. Proc. Natl. Acad. Sci. U.S.A. 1972; 69:1915–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Algire M.A., Maag D., Savio P., Acker M.G., Tarun S.Z. Jr, Sachs A.B., Asano K., Nielsen K.H., Olsen D.S., Phan L. et al. Development and characterization of a reconstituted yeast translation initiation system. RNA. 2002; 8:382–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mitchell S.F., Walker S.E., Algire M.A., Park E.H., Hinnebusch A.G., Lorsch J.R.. The 5′-7-methylguanosine cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and to block an alternative pathway. Mol. Cell. 2010; 39:950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schuller A.P., Wu C.C.-C., Dever T.E., Green R.. eIF5A functions globally in translation elongation and termination. Mol. Cell. 2017; 66:194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Katoh T., Wohlgemuth I., Nagano M., Rodnina M.V., Suga H.. Essential structural elements in tRNAPro for EF-P-mediated alleviation of translation stalling. Nat. Commun. 2016; 7:11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jhon J.S., Kang Y.K.. Conformational preferences of proline analogues with different ring size. J. Phys. Chem. B. 2007; 111:3496–3507. [DOI] [PubMed] [Google Scholar]
- 57. Bessonov K., Vassall K.A., Harauz G.. Parameterization of the proline analogue Aze (azetidine-2-carboxylic acid) for molecular dynamics simulations and evaluation of its effect on homo-pentapeptide conformations. J. Mol. Graph. Model. 2013; 39:118–125. [DOI] [PubMed] [Google Scholar]
- 58. Kern D., Schutkowski M., Drakenberg T.. Rotational barriers of cis/trans isomerization of proline analogues and their catalysis by cyclophilin. J. Am. Chem. Soc. 1997; 119:8403–8408. [Google Scholar]
- 59. Hunter A.R., Farrell P.J., Jackson R.J., Hunt T.. The role of polyamines in cell-free protein synthesis in the wheat-germ system. Eur. J. Biochem. 1977; 75:149–157. [DOI] [PubMed] [Google Scholar]
- 60. Igarashi K., Kashiwagi K., Aoki R., Kojima M., Hirose S.. Comparative studies on the increase by polyamines of fidelity of protein synthesis in Escherichia coli and wheat germ cell-free systems. Biochem. Biophys. Res. Commun. 1979; 91:440–448. [DOI] [PubMed] [Google Scholar]
- 61. Snyder R.D., Edwards M.L.. Effects of polyamine analogs on the extent and fidelity of in vitro polypeptide synthesis. Biochem. Biophys. Res. Commun. 1991; 176:1383–1392. [DOI] [PubMed] [Google Scholar]
- 62. Takeda Y. Polyamines and protein synthesis. I. The effect of polyamines on cell free polyphenylalanine synthesis in Escherichia coli. J. Biochem. 1969; 66:345–349. [DOI] [PubMed] [Google Scholar]
- 63. Firczuk H., Kannambath S., Pahle J., Claydon A., Beynon R., Duncan J., Westerhoff H., Mendes P., McCarthy J.E.. An in vivo control map for the eukaryotic mRNA translation machinery. Mol. Syst. Biol. 2013; 9:635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zagari A., Nemethy G., Scheraga H.A.. The effect of the L-azetidine-2-carboxylic acid residue on protein conformation. II. Homopolymers and copolymers. Biopolymers. 1990; 30:961–966. [DOI] [PubMed] [Google Scholar]
- 65. Zagari A., Nemethy G., Scheraga H.A.. The effect of the L-azetidine-2-carboxylic acid residue on protein conformation. I. Conformations of the residue and of dipeptides. Biopolymers. 1990; 30:951–959. [DOI] [PubMed] [Google Scholar]
- 66. Doerfel L.K., Wohlgemuth I., Kubyshkin V., Starosta A.L., Wilson D.N., Budisa N., Rodnina M.V.. Entropic contribution of elongation factor P to proline positioning at the catalytic center of the ribosome. J. Am. Chem. Soc. 2015; 137:12997–13006. [DOI] [PubMed] [Google Scholar]
- 67. Igarashi K., Kashiwagi K.. Modulation of cellular function by polyamines. Int. J. Biochem. Cell Biol. 2010; 42:39–51. [DOI] [PubMed] [Google Scholar]
- 68. Pegg A.E. Functions of polyamines in mammals. J. Biol. Chem. 2016; 291:14904–14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Quigley G.J., Teeter M.M., Rich A.. Structural analysis of spermine and magnesium ion binding to yeast phenylalanine transfer RNA. Proc. Natl. Acad. Sci. U.S.A. 1978; 75:64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Petropoulos A.D., Xaplanteri M.A., Dinos G.P., Wilson D.N., Kalpaxis D.L.. Polyamines affect diversely the antibiotic potency: insight gained from kinetic studies of the blasticidin S and spiramycin interactions with functional ribosomes. J. Biol. Chem. 2004; 279:26518–26525. [DOI] [PubMed] [Google Scholar]
- 71. Xaplanteri M.A., Andreou A., Dinos G.P., Kalpaxis D.L.. Effect of polyamines on the inhibition of peptidyltransferase by antibiotics: revisiting the mechanism of chloramphenicol action. Nucleic Acids Res. 2003; 31:5074–5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Xaplanteri M.A., Petropoulos A.D., Dinos G.P., Kalpaxis D.L.. Localization of spermine binding sites in 23S rRNA by photoaffinity labeling: parsing the spermine contribution to ribosomal 50S subunit functions. Nucleic Acids Res. 2005; 33:2792–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Noeske J., Wasserman M.R., Terry D.S., Altman R.B., Blanchard S.C., Cate J.H.. High-resolution structure of the Escherichia coli ribosome. Nat. Struct. Mol. Biol. 2015; 22:336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.