Abstract
Southeast (SE) Asia is 1 of the most biodiverse regions in the world, and it holds approximately 20% of all mammal species. Despite this, the majority of SE Asia's genetic diversity is still poorly characterized. The growing interest in using environmental DNA to assess and monitor SE Asian species, in particular threatened mammals—has created the urgent need to expand the available reference database of mitochondrial barcode and complete mitogenome sequences. We have partially addressed this need by generating 72 new mitogenome sequences reconstructed from DNA isolated from a range of historical and modern tissue samples. Approximately 55 gigabases of raw sequence were generated. From this data, we assembled 72 complete mitogenome sequences, with an average depth of coverage of ×102.9 and ×55.2 for modern samples and historical samples, respectively. This dataset represents 52 species, of which 30 species had no previous mitogenome data available. The mitogenomes were geotagged to their sampling location, where known, to display a detailed geographical distribution of the species. Our new database of 52 taxa will strongly enhance the utility of environmental DNA approaches for monitoring mammals in SE Asia as it greatly increases the likelihoods that identification of metabarcoding sequencing reads can be assigned to reference sequences. This magnifies the confidence in species detections and thus allows more robust surveys and monitoring programmes of SE Asia's threatened mammal biodiversity. The extensive collections of historical samples from SE Asia in western and SE Asian museums should serve as additional valuable material to further enrich this reference database.
Keywords: invertebrate-derived (iDNA), metabarcoding, GenBank, taxonomic assignment
Data Description
Context
Southeast (SE) Asia is 1 of the most biodiverse regions in the world, hosting ∼20% of mammal species, but it is experiencing rapid deforestation for agriculture and development. To assess the ecological consequences of land use change, there is growing interest in using environmental DNA to monitor mammal populations, particularly threatened taxa that often underpin conservation policies [1–4]. Yet current efforts are hampered by the lack of a reference database of mitochondrial barcodes and complete mitogenome sequences. Currently there are 922 mammalian mitogenomes available in Genbank. Unfortunately, most are not tagged by location/origin. Data mining through manual screening of each mitogenomes resulted in 174 terrestrial mammal species that are typical to SE Asia. In this work, 30 novel species are added, contributing to ∼17% expansion of the current SE Asia mammal mitogenome database.
DNA extraction
Genomic DNA was extracted from different sample types of 72 small mammals, comprising 52 species, listed in Table 1 and Table 2. DNA from modern tissue and blood samples was isolated using the Qiagen DNeasy extraction kit (Qiagen, Hilden, Germany, [QIAGEN, RRID:SCR_008539]) or Invitek DNA extraction kit (Invitek GmbH, Berlin, Germany), as per standard protocols following the manufacturer's guidelines. Historical samples obtained from the Zoological Museum, Natural History Museum of Denmark, and University of Copenhagen (ZM, KU) were treated differently according to type of tissue (Additional file 1a), while at the German Primate Center, DNA extraction from museum specimens followed Liedigk et al. (2015) [5] using the Gen-IAL First All Tissue Kit (Gen-IAL, Troisdorf, Germany). Complete details of sample information are provided in Additional file 2.
Table 1:
List of mitogenomes assembled in this work that supplement preexisting mitogenome references already available in GenBank
| No. | GenBank ID | Common name | Genus | Species | Assembly size | Locality | Source | Sample date of collection | Data by |
|---|---|---|---|---|---|---|---|---|---|
| 1 | KY117537 | Hog deer | Axis | porcinus | 16 402 | CPH Zoo | ZM, KU | 21/8/1912 | F.M.S./F.P. |
| 2 | KY117538 | Pallas's squirrel | Callosciurus | erythraeus | 16 656 | Bangkok, Thailand | ZM, KU | 25/5/1969 | F.M.S./F.P. |
| 3 | KX265095 | Bay cat | Catopuma | badia | 16 960 | Sabah, Malaysia | National Museum Scotland | 20/04/2000 | P.R.P. |
| 4 | KX224524 | Asiatic golden cat | Catopuma | temminckii | 16 960 | Thailand | American Museum of National History, New York. | 10/10/1927 | P.R.P. |
| 5 | KY117545 | Sumatran rhino | Dicerorhinus | sumatrensis | 16 466 | Sumatra, Indonesia | Naturalis, Leiden, The Netherlands | 1880 | R.M. |
| 6 | KY117546 | Least pygmy squirrel | Exilisciurus | exilis | 16 637 | Indonesia | ROM | 16/06/1993 | F.M.S./F.P. |
| 7 | KY117548 | Hose's mongoose | Herpestes | javanicus | 16 340 | Java, Indonesia | ZM, KU | 12/3/1947 | F.M.S./F.P. |
| 8 | KY117550 | Three-striped ground Squirrel | Lariscus | indsignis | 16 399 | Maybe Malaysia | ZM, KU | Unknown | F.M.S./F.P. |
| 9 | KY117592 | Black crested macaque | Macaca | nigra | 16 558 | Captive | Gettorf Zoo, Germany | 18/07/2000 | C.R. |
| 10 | KY117593 | Northern pig-tailed macaque | Macaca | leonina | 16 554 | Captive | Ludwig-Maximilans-University Munich, Germany | 6/3/1995 | C.R. |
| 11 | KY117594 | Southern pig-tailed macaque | Macaca | nemestrina | 16 531 | Peninsular Malaysia | National Museum Scotland, Edinburgh, UK | Unknown | C.R. |
| 12 | KT288227 | Marbled cat | Pardofelis | marmorata | 17 218 | Sumatra, Indonesia | National Archaeological Museum of the Netherlands, Leiden | 30/08/1930 | P.R.P. |
| 13 | KY117602 | Sumatra surili | Presbytis | melalophos | 16 558 | Captive | Howletts Wild Animal Park, UK | 23/7/1999 | C.R. |
| 14 | KR135743 | Flat-headed cat | Prionailurus | planiceps | 17 704 | Sabah, Malaysia | Sabah Wildlife Department | 25/04/2000 | P.R.P. |
| 15 | KY117580 | Malayan field rat | Rattus | tiomanicus | 16 415 | SPF Bidor, Perak, Malaysia | FRIM | 12/2/2011 | F.M.S./F.P. |
| 16 | KY117579 | Malayan field rat | Rattus | tiomanicus | 16 312 | Indonesia | ROM | 01/06/1993 | F.M.S./F.P. |
| 17 | KY117581 | Malayan field rat | Rattus | tiomanicus | 16 305 | Hutan Simpan Chikus, Tapah Perak, Malaysia | FRIM | 13/1/2011 | F.M.S./F.P. |
| 19 | KY117582 | Black giant squirrel | Ratufa | bicolor peninsulae | 16 600 | Bang Nara, Malakka, Thailand | ZM, KU | 3/12/1932 | F.M.S./F.P. |
| 18 | KY117574 | Javan rhino | Rhinoceros | sondaicus | 16 417 | Java, Indonesia | Copenhagen Natural History Museum | Unknown | R.M. |
| 20 | KY117575 | Javan rusa | Rusa | timorensis | 16 437 | Toeloeng Agoeng, West Java, Indonesia | Naturalis, Leiden, The Netherlands | Unknown | R.M. |
| 21 | KY117576 | Indian sambar deer | Rusa | unicolor dejeani | 16 437 | Mentawai, Indonesia | Naturalis, Leiden, The Netherlands | Unknown | R.M. |
| 22 | KY117599 | Western purple-faced langur | Semnopithecus | vetulus | 16 545 | Captive | Belfast Zoo, UK | 9/11/1998 | C.R. |
| 23 | KY117589 | Malayan tapir | Tapirus | indicus | 16 794 | Captive | Copenhagen Zoo | 11/1/2015 | F.M.S./F.P. |
| 24 | KY117598 | Silvered langur | Trachypithecus | cristatus | 16 551 | North Sumatra, Indonesia | Bavarian State Collection Munich, Germany | 1911 | C.R. |
FRIM: Forest Research Institute, Malaysia; ROM: Royal Ontario Museum; ZM, KU: Zoological Museum, University of Copenhagen.
Table 2:
List of mitogenomes assembled in this work that have no previous complete mitogenome reference available in GenBank
| No. | GenBank ID | Common name | Genus | Species | Assembly size | Locality | Source | Sample date of collection | Data by |
|---|---|---|---|---|---|---|---|---|---|
| 1 | KY117536 | Asian small-clawed otter | Aonyx | cinereus | 16 153 | Captive | Copenhagen Zoo | 08/08/11 | F.M.S./F.P. |
| 2 | KY117535 | Asian small-clawed otter | Aonyx | cinereus | 16 153 | Sarawak, Malaysia | British Museum of Natural History, London | 25/8/2010 | F.M.S./F.P. |
| 3 | KY117560 | Binturong | Arctictis | binturong | 17 067 | Unknown | Tierpark, Berlin | 29/11/2010 | P.R.P. |
| 4 | KY117541 | Plantain squirrel | Callosciurus | notatus | 16 582 | Hutan Bidor, Perak, Malaysia | FRIM | 11/2/2011 | F.M.S./F.P. |
| 5 | KY117542 | Plantain squirrel | Callosciurus | notatus | 16 599 | East Kalimantan, Indonesia | ROM | 03/06/1993 | F.M.S./F.P. |
| 6 | KY117543 | Prevost's squirrel | Callosciurus | prevostii | 16 674 | East Kalimantan, Indonesia | ROM | 15/06/1993 | F.M.S./F.P. |
| 7 | KY117540 | Variable squirrel | Callosciurus | finlaysonii frandseni | 15 747 | Koh Chang, Thailand | ZM, KU | 14/1/1900 | F.M.S./F.P. |
| 8 | KY117539 | Variable squirrel | Callosciurus | finlaysonii | 16 489 | Central Thailand | ZM, KU | 2/2/1928 | F.M.S./F.P. |
| 9 | KY117544 | Sunda otter civet | Cynogale | bennetti | 15 784 | Borneo | British Museum of Natural History, London | 25/8/2010 | F.M.S./F.P. |
| 10 | KY117549 | Greater mouse deer | Tragulus | napu | 15 778 | Bang Nara, Thailand | ZM, KU | 11/10/1931 | F.M.S./F.P. |
| 11 | KY117552 | Long-tailed giant rat | Leopaldamys | sabanus | 15 973 | G. Telapak Buruk, Negeri Sembilan, Malaysia | FRIM | 24/2/2010 | F.M.S./F.P. |
| 12 | KY117553 | Long-tailed giant rat | Leopaldamys | sabanus | 15 972 | Teluk Segadas, P. Pangkor, Perak, Malaysia | FRIM | 19/3/2010 | F.M.S./F.P. |
| 13 | KY117554 | Long-tailed giant rat | Leopaldamys | sabanus | 15 974 | Hutan Simpan Temengor, Gerik Perak, Malaysia | FRIM | 23/1/2014 | F.M.S./F.P. |
| 14 | KY117555 | Long-tailed giant rat | Leopaldamys | sabanus | 15 972 | Hutan Simpan Lenggor, Kluang, Johor, Malaysia | FRIM | 19/2/2014 | F.M.S./F.P. |
| 15 | KY117551 | Long-tailed giant rat | Leopaldamys | sabanus | 15 974 | Malaysia | ROM | 28/05/1993 | F.M.S./F.P. |
| 16 | KY117556 | Hairy-nosed otter | Lutra | sumatrana | 16 580 | Bang Nara, Thailand | ZM, KU | 1/4/1939 | F.M.S./F.P. |
| 17 | KY117557 | Smooth-coated otter | Lutrogale | perspicillata | 16 042 | Melaka, Malaysia | British Museum of Natural History, London | 25/8/2010 | F.M.S./F.P. |
| 18 | KY117558 | Smooth-coated otter | Lutrogale | perspicillata | 16 041 | Bang Nara, Thailand | ZM, KU | 24/1/1933 | F.M.S./F.P. |
| 19 | KY117591 | Moor macaque | Macaca | maura | 16 563 | Captive | Hannover Zoo, Germany | 20/8/1998 | C.R. |
| 20 | KY117564 | Rajah/brown spiny rat | Maxomys | rajah | 16 200 | Indonesia | ROM | 06/06/1993 | F.M.S./F.P. |
| 21 | KY117562 | Rajah/brown spiny rat | Maxomys | rajah | 16 296 | Teluk Segadas, P. Pangkor, Perak, Malaysia | FRIM | 19/3/2010 | F.M.S./F.P. |
| 22 | KY117563 | Rajah/brown spiny rat | Maxomys | rajah | 16 296 | Pasir Bogak, P.Pangkor, Perak, Malaysia | FRIM | 18/3/2010 | F.M.S./F.P. |
| 23 | KY117567 | Red spiny rat | Maxomys | surifer | 16 286 | 50 ha, Pasoh, Negeri Sembilan, Malaysia | FRIM | 12/6/2008 | F.M.S./F.P. |
| 24 | KY117566 | Red spiny rat | Maxomys | surifer | 16 290 | Indonesia | ROM | 21/05/1993 | F.M.S./F.P. |
| 25 | KY117565 | Red spiny rat | Maxomys | surifer | 16 286 | Malaysia | ROM | 17/05/2013 | F.M.S./F.P. |
| 26 | KY117570 | Whitehead's spiny rat | Maxomys | whiteheadi | 16 316 | Hutan Simpan Bikam, Perak, Malaysia | FRIM | 12/2/2011 | F.M.S./F.P. |
| 27 | KY117571 | Whitehead's spiny rat | Maxomys | whiteheadi | 16 316 | Keruing Trail, FRIM, Kepong, Selangor, Malaysia | FRIM | 13/3/2013 | F.M.S./F.P. |
| 28 | KY117568 | Whitehead's spiny rat | Maxomys | whiteheadi | 16 287 | Hutan Simpan Bikam, Perak, Malaysia | FRIM | 12/2/2011 | F.M.S./F.P. |
| 29 | KY117569 | Whitehead's spiny rat | Maxomys | whiteheadi | 16 429 | Bukit Tapah, Perak, Malaysia | FRIM | 23/3/2011 | F.M.S./F.P. |
| 30 | KY052142 | Indian muntjac | Muntiacus | muntjak | 16 354 | West Java, Indonesia | Vienna NHM | 1858 | R.M. |
| 31 | KY117559 | Bornean yellow muntjac | Muntiacus | atherodes | 16 354 | Koemai, West Borneo | Bonn NHM | 1938 | R.M. |
| 32 | KY117573 | Dark-tailed tree rat | Niviventer | cremoriventer | 16 322 | Track 5 (G.Inas), Kedah, Malaysia | FRIM | 5/11/2009 | F.M.S./F.P. |
| 33 | KY117572 | Dark-tailed tree rat | Niviventer | cremoriventer | 16 234 | Malaysia | ROM | 17/05/2013 | F.M.S./F.P. |
| 34 | KY117600 | Grizzled leaf monkey | Presbytis | comata comata | 16 551 | Captive | Howletts Wild Animal Park, UK | 23/12/1999 | C.R. |
| 35 | KY117601 | Mitred leaf monkey | Presbytis | mitrata | 16 555 | Captive | Howletts Wild Animal Park, UK | 12/11/1998 | C.R. |
| 36 | KX857784 | Leopard cat | Prionailurus | bengalensis | 16 989 | Thailand | American Museum of National History, New York. | 25/02/1924 | P.R.P. |
| 37 | KY117578 | Annandale's sundaic rat | Rattus | annandalei | 16 297 | Hutan Simpan Bikam, Perak, Malaysia | FRIM | 12/2/2011 | F.M.S./F.P. |
| 38 | KY117577 | Annandale's sundaic rat | Rattus | annandalei | 16 301 | Hutan Simpan Bikam, Perak, Malaysia | FRIM | 11/2/2011 | F.M.S./F.P. |
| 39 | KY117583 | Mountain giant sunda rat | Sundamys | infraluteus | 16 297 | Malaysia | ROM | 18/05/2013 | F.M.S./F.P. |
| 40 | KY117585 | Müller's giant sunda rat | Sundamys | meulleri | 16 326 | Track 1 (G.Inas), Kedah, Malaysia | FRIM | 5/11/2009 | F.M.S./F.P. |
| 41 | KY117584 | Müller's giant sunda rat | Sundamys | meulleri | 16 304 | Malaysia | ROM | 01/06/2013 | F.M.S./F.P. |
| 42 | KY117586 | Brooke's squirrel | Sundasciurus | brookei | 16 417 | East Kalimantan, Indonesia | ROM | 13/06/1993 | F.M.S./F.P. |
| 43 | KY117587 | Low's squirrel | Sundasciurus | lowii | 16 307 | East Kalimantan, Indonesia | ROM | 06/06/1993 | F.M.S./F.P. |
| 44 | KY117588 | Low's squirrel | Sundasciurus | sp | 16 458 | East Kalimantan, Indonesia | ROM | 21/06/1993 | F.M.S./F.P. |
| 45 | KY117595 | Phayre's langur | Trachypithecus | phayrei phayrei | 16 548 | South West Myanmar | Natural History Museum Berlin, Germany | Unknown | C.R. |
| 46 | KY117596 | East Javan ebony langur | Trachypithecus | auratus | 16 552 | Captive | Bristol Zoo, UK | 26/10/2010 | C.R. |
| 47 | KY117597 | West Javan ebony langur | Trachypithecus | mauritius | 16 554 | West Java, Indonesia | Naturalis Leiden; Netherlands | Unknown | C.R. |
| 48 | KY117590 | Long-tailed porcupine | Trichys | fasciculata | 16 328 | Borneo | ZM, KU | 5/10/1894 | F.M.S./F.P. |
FRIM: Forest Research Institute, Malaysia; NHM: Natural History Museum; ROM: Royal Ontario Museum; ZM, KU: Zoological Museum, University of Copenhagen.
Data Validation and Quality Control
Mitogenome sequencing, assembly, and annotation
Mitogenomes were generated using several approaches. In Copenhagen, author F.M.S. constructed Illumina shotgun libraries with insert sizes ranging between 50 and 400 bp. To construct libraries, DNA was sheared to the target size range using Bioruptor® XL (Diagenode, USA [Diagenode, RRID:SCR_014807]) and converted into an Illumina-compatible sequencing library using the NEBNext E6070 Kit (New England Biolabs, UK). The libraries were polymerase chain reaction (PCR) amplified with index primers and purified using Qiaquick columns (Qiagen, Hilden, Germany) according to the manufacturer's instruction (Additional file 1b). Multiple libraries were combined together into 3 pools, normalized to 10 nM, and sequenced across 3 lanes of Illumina HiSeq 2500 using SR100 bp chemistry. In Berlin and Goettingen, mitogenomes were generated by authors P.R.P. and C.R. using overlapping PCR products using long-range PCR (Additional file 1c) followed by library construction and MiSeq sequencing, or Sanger sequencing as described in Patel, Förster, and Kitchener (2016) [6] and Liedigk et al. (2015), Roos et al. (2011), and Liedigk et al. (2014) [5, 7, 8], respectively. Author R.M.’s mitogenomes were done using methods outlined in Fortes and Paijmans (2015) [9]. Further details about laboratory methods are described in Additional file 1.
Raw reads for F.M.S. samples were assembled independently by authors F.M.S. and F.P. using 2 different approaches, then compared for consistency. Author F.M.S. trimmed the reads for sequencing adapters, low-quality stretches, and leading/tailing Ns using AdapterRemoval 1.2 (AdapterRemoval, RRID:SCR_011834) [10]. The mitochondrial genome was reconstructed with MITObim v. 1.8 [11] using the reference mitogenome of the closest species available in GenBank as the seed reference (Additional file 2). In order to obtain the mapping statistics of the samples, we ran PALEOMIX v. 1.2.6 [12] with default parameters where reads shorter than 25 bp after trimming were discarded. The trimmed reads were aligned against the newly assembled mitogenome generated by MITObim using Burrows–Wheeler Aligner [13]. Alignments showing low-quality scores and PCR duplicates were further removed using the MarkDuplicates program from Picard tools, and reads were locally realigned around small insertions and deletions (indels) to improve overall genome quality using the IndelRealigner tool from the Genome Analysis Toolkit (GATK, RRID:SCR_001876) [14]. In contrast, author F.P. inputted the trimmed reads into mitoMaker [15], which performs a de novo and reference-based assembly using SOAPdenovoTrans v. 1.03 (SOAPdenovo-Trans, RRID:SCR_013268) [16] and MITObim v. 1.7 [11]. Post-assembly, the F.M.S. and F.P. mitogenomes were manually compared for consistency by F.M.S. to generate the final consensus sequences. These assemblies were automatically annotated using tRNAscan-SE v. 1.4 (tRNAscan-SE, RRID:SCR_010835) [17] and Basic Local Alignment Search Tool v. 2.2.29 (NCBI BLAST, RRID:SCR_004870) [18] using the mitochondrial genomes found in the National Center for Biotechnology Information Reference Sequence Database (RefSeq, RRID:SCR_003496) [19] as references.
For the mitogenome constructed by author R.M., Illumina sequence reads were de-multiplexed according to the respective indexes with the Illumina software bcl2fastq v. 2.17 (Illumina, San Diego, CA, USA), and adapters were clipped from the sequence reads with the software cutadapt v. 1.3 [20]. Quality trimming was done through a sliding window approach (10 bp; Q20), and all reads shorter than 20 bp were removed from the analyses. Mitogenome references from target or closely related species were used for mapping of the sequencing reads. Aligned reads were de-duplicated using MarkDuplicates from Picard-tools v. 1.106 (Picard, RRID:SCR_006525) [21]. VariantCalling was carried out using Samtools v. 1.1 (SAMTOOLS, RRID:SCR_002105) [13] and Bcftools v. 1.2 (SAMtools/BCFtools, RRID:SCR_005227) [22]. For each sample, GATK [14] variant calling output files were further filtered to have a minimum read coverage ≥ ×3, and variants were only called when the corresponding base was represented by ≥50%; otherwise this position was “N”-masked.
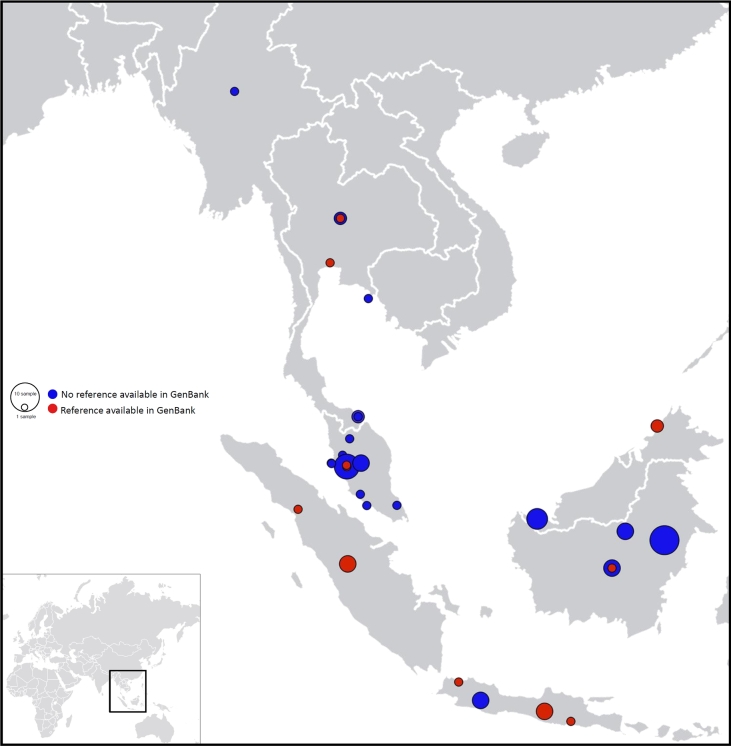
Numbers of raw reads generated for each sample and mapping statistics for all 72 mitogenome assemblies are shown in Additional file 2. Sanger sequenced mitogenomes were checked with 4Peaks 1.8 (4Peaks, RRID:SCR_000015) [23], assembled with SeaView 4.5.4 [24], and annotated with DOGMA [25]. All mitogenomes were checked manually by eye to identify possible errors caused by insertion and deletions in Tablet [26]. The final mitochondrial genomes have been uploaded to GenBank (accession numbers are provided in Tables 1 and 2). The details of all new mitogenomes assembled in this work are given in Tables 1 and 2. Mitogenomes (60 samples) with known localities were geotagged and mapped to display their geographical distribution (Fig. 1).
Figure 1:
Geographical distribution of mitogenomes assembled in this work (60 mitogenomes with known locality).
Phylogenetic analysis
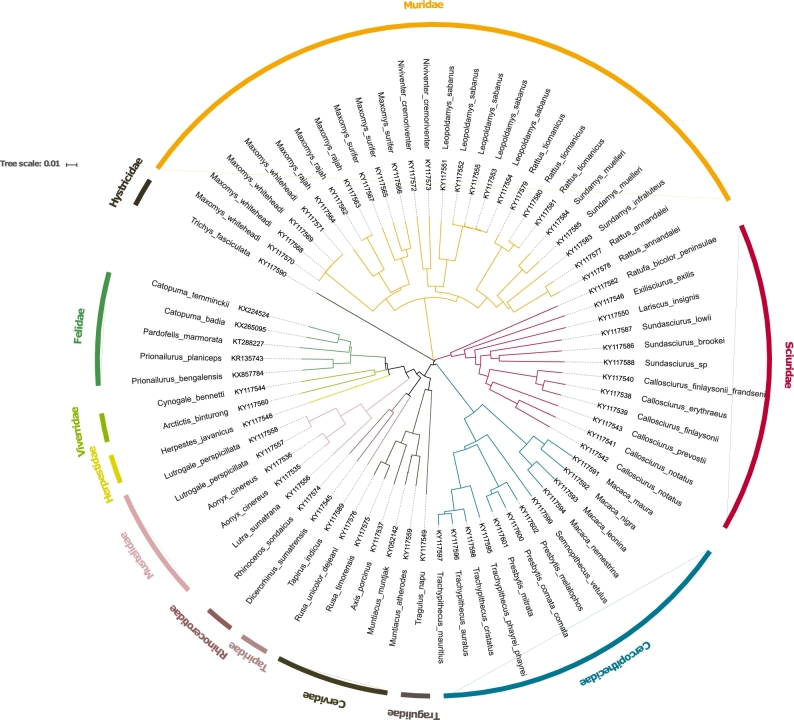
All the sequenced mitogenomes were aligned using MAFFT v. 7.158b (MAFFT, RRID:SCR_011811) [27] using the E-INS-i option (Additional file 3). Randomized Axelerated Maximum Likelihood (RAxML) v. 8.0.26 (RAxML, RRID:SCR_006086) [28] was used to perform the phylogenetic analysis with a GTR+GAMMA model of nucleotide substitution. To obtain node support, we used 100 bootstrap pseudo-replicates (Fig. 2). The newick file is provided as Additional file 4.
Figure 2:
Phylogenetic tree of mitogenomes assembled in this work.
Re-use Potential
We anticipate that the now-expanded mitogenome reference dataset for SE Asian mammals will provide benefits for a number of research areas. First, it should enhance the power of environmental DNA and other metabarcoding/barcoding approaches that relate to the identification of SE Asian mammals by conferring the ability to identify more species to the species level. This in turn has practical applications for those monitoring SE Asia's threatened mammal biodiversity, combatting trade in mammal species and so on. Second, the data will also have relevance to phylogenetic and population studies based on mtDNA data, which will be of use as we investigate the evolutionary history of this biodiversity hotspot.
Availability of supporting data
Raw shotgun data are deposited in the SRA under bioproject number PRJNA361218 and are available in the GigaScience repository, GigaDB [29]. Details of the method to support this work can be found in protocols.io [30].
Additional files
1. Additional file 1: DNA extraction of historical samples, library construction, and primer information
2. Additional file 2: Sample information sheet of mitogenomes assembled in this work
3. Additional file 3: Alignment of mitogenomes assembled in this work
4. Additional file 4: Newick file for phylogenetic tree
Abbreviations
BLAST: Basic Local Alignment Search Tool; bp: base pair; GATK: Genome Analysis Toolkit; MAFFT: Multiple Alignment using Fast Fourier Transform; NCBI RefSeq: National Center for Biotechnology Information Reference Sequence Database; PCR: polymerase chain reaction; RAxML: Randomized Axelerated Maximum Likelihood; SE: southeast.
Competing interests
The authors declare that they have no competing interests.
Funding
This project was funded by the Malaysian Government (F.M.S.), Lundbeck Foundation grant R52–5062 (M.T.P.G.), Leibniz-Association grant SAW-2013-IZW-2 (J.F.), the German Federal Ministry of Education and Research grant BMBF FKZ: 01LN1301A (A.W.), and the German Primate Center (C.R.).
Author contributions
F.M.S., A.W., J.F., and M.T.P.G. conceived the project. F.M.S., M.H.S.S., M.S.S., M.S.A., R.M., P.R.P., C.R., B.K.L., and S.J.R. collected the samples and extracted the genomic DNA. F.M.S., R.M., P.R.P., and C.R. constructed the libraries and did sequencing. F.M.S., J.R.M., F.P., S.L., P.R.P., R.M., D.L., and C.R. assembled the mitogenomes and performed mitogenome analysis. F.M.S., S.L., P.R.P., and M.T.P.G. wrote the article. All authors discussed the project and data. All authors read and approved the final manuscript.
Supplementary Material
Acknowledgements
We thank the Danish National High-throughput Sequencing Centre for assistance in generating Illumina data. We also thank the staff of the zoos in Hannover, Copenhagen, Gettorf, Belfast, Bristol, and Howletts Wild Animal Park; the staff of the Ludwig-Maximilans-University Munich, the Bavarian State Collection Munich, the National Museums Scotland, Edinburgh, the Natural History Museum Berlin, and Naturalis Leiden; and the Zoological Museum, Natural History Museum of Denmark, University of Copenhagen for providing samples for this work. B.K.L. gratefully acknowledges the Indonesian and Malaysian government authorities for issuing scientific permits.
References
- 1. Bohmann K, Evans A, Gilbert MTP et al. Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol Evol 2014;29:358–67. [DOI] [PubMed] [Google Scholar]
- 2. Lee P, Gan HM, Clements GR et al. Field calibration of blowfly-derived DNA against traditional methods for assessing mammal diversity in tropical forests 1. Genome 2016;59:1008–22. [DOI] [PubMed] [Google Scholar]
- 3. Schnell IB, Sollmann R, Calvignac-Spencer S et al. iDNA from terrestrial haematophagous leeches as a wildlife surveying and monitoring tool–prospects, pitfalls and avenues to be developed. Front Zool 2015;12:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schnell IB, Thomsen PF, Wilkinson N et al. Screening mammal biodiversity using DNA from leeches. Curr Biol 2012;22:R262–3. [DOI] [PubMed] [Google Scholar]
- 5. Liedigk R, Kolleck J, BöKer KO et al. Mitogenomic phylogeny of the common long-tailed macaque (Macaca fascicularis fascicularis). BMC Genomics 2015;16:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Patel RP, Förster DW, Kitchener AC. Two species of Southeast Asian cats in the genus Catopuma with diverging histories: an island endemic forest specialist and a widespread habitat generalist. Open Science 2016;3:160350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roos C, Zinner D, Kubatko LS et al. Nuclear versus mitochondrial DNA: evidence for hybridization in colobine monkeys. BMC Evol Biol 2011;11:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liedigk R, Roos C, Brameier M et al. Mitogenomics of the Old World monkey tribe Papionini. BMC Evol Biol 2014;14:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fortes GG, Paijmans JLA. Analysis of whole mitogenomes from ancient samples. Methods Mol Biol 2015;1347:179–95. [DOI] [PubMed] [Google Scholar]
- 10. Lindgreen S. AdapterRemoval: easy cleaning of next generation sequencing reads. BMC Res Notes 2012;5:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hahn C, Bachmann L, Chevreux B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads–a baiting and iterative mapping approach. Nucleic Acids Res 2013;41(13):e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schubert M, Ermini L, Sarkissian CD et al. Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat Protoc 2014;9:1056–82. [DOI] [PubMed] [Google Scholar]
- 13. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mckenna A, Hanna M, Banks E et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. mitoMaker https://sourceforge.net/projects/mitomaker/. Accessed 26 May 2017.
- 16. Xie Y, Wu G, Tang J et al. SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 2014;30:1660–6. [DOI] [PubMed] [Google Scholar]
- 17. Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997;25:955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Camacho C, Coulouris G, Avagyan V et al. BLAST+: architecture and applications. BMC Bioinformatics 2009;10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. RefSeq: NCBI Reference Sequence Database. https://www.ncbi.nlm.nih.gov/refseq. Accessed 26 May 2017. [Google Scholar]
- 20. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 2011;17:10–12. [Google Scholar]
- 21. picard https://agithub.com/broadinstitute/picard. Accessed 20 July 2017.
- 22. bcftools. https://github.com/samtools/bcftools. Accessed 26 May 2017. [Google Scholar]
- 23. Nucleobytes: software for science. http://nucleobytes.com. Accessed 26 May 2017. [Google Scholar]
- 24. Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 2010;27:221–4. [DOI] [PubMed] [Google Scholar]
- 25. Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004;20:3252–5. [DOI] [PubMed] [Google Scholar]
- 26. Milne I, Stephen G, Bayer M et al. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform 2013;14(2):193–202. [DOI] [PubMed] [Google Scholar]
- 27. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013;30:772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014;30:1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Salleh FM, Ramos-Madrigal J, Penaloza F et al. Supporting data for “An expanded mammal mitogenome dataset from Southeast Asia.” GigaScience Database 2017. http://dx.doi.org/10.5524/100313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Salleh FM, Ramos-Madrigal J, Penaloza F et al. An expanded mammal mitogenome dataset from Southeast Asia. protocols.io 2017. http://dx.doi.org/10.17504/protocols.io.im6cc9e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.