Abstract
Here we define the epitopes on HA that are targeted by a group of 9 recombinant monoclonal antibodies (rmAbs) isolated from memory B cells of mice, immunized by infection with A(H1N1)pdm09 virus followed by a seasonal TIV boost. These rmAbs were all reactive against the HA1 region of HA, but display 7 distinct binding footprints, targeting each of the 4 known antigenic sites. Although the rmAbs were not broadly cross-reactive, a group showed subtype-specific cross-reactivity with the HA of A/South Carolina/1/18. Screening these rmAbs with a panel of human A(H1N1)pdm09 virus isolates indicated that naturally-occurring changes in HA could reduce rmAb binding, HI activity, and/or virus neutralization activity by rmAb, without showing changes in recognition by polyclonal antiserum. In some instances, virus neutralization was lost while both ELISA binding and HI activity were retained, demonstrating a discordance between the two serological assays traditionally used to detect antigenic drift.
Keywords: Influenza, Monoclonal antibody, Epitope mapping, Antigenic site, Virus neutralization, Antigenic drift, A/H1N1/pdm09
Introduction
Influenza viruses are common pathogens of many species, including humans, in whom seasonal influenza epidemics are a significant cause of global disease with a high annual public health and economic burden (Molinari et al., 2007). Vaccination is the most effective public health counter-measure against influenza. Antibodies play an important protective role against influenza infections, and the goal of immunization against influenza viruses is to induce a protective antibody response against the immunodominant surface protein, hemagglutinin (HA). As a small number of mutations in HA allow the virus to avoid neutralization by antibodies, influenza viruses rapidly evolve resistance to population immunity, so that influenza vaccines need to be reformulated on a regular basis. In addition to the antigenic match between vaccine and circulating virus, the protective efficacy of influenza vaccination can vary depending on the age, history of exposure and health status of the vaccinee.
HA binds to the viral receptors, sialic acids (SA), on host cells facilitating virus entry, and also brings about membrane fusion between the virus and cellular membranes that is triggered by the low pH of the endosomal compartment. Structurally, HA is composed of a globular head that includes the receptor-binding site (RBS) and previously defined antigenic sites, and a stem region that includes the membrane fusion peptide, as well as transmembrane and cytosolic regions. A number of lines of evidence, including the generation of antibody escape mutants and measurement of binding to specific mutants, have shown that protective antibodies tend to bind to specific antigenic sites on the globular head (Wan et al., 2014; Rudneva et al., 2010). For the H1 subtype of HA, the sites are termed Sa, Sb, Ca1, Ca2, and Cb (Caton et al., 1982; Yewdell and Gerhard 1981). The residues comprising these antigenic sites tend to be highly variable over time and, since mutations in these sites are well tolerated by viruses, this allows mutant viruses to arise that are capable of infecting individuals immune to the parent influenza strain. Protective antibodies can also bind to regions outside of the defined antigenic sites on the HA head (Zhu et al., 2013; Matsuzaki et al., 2014). In many cases, these regions of HA are more conserved than the antigenic sites, and antibodies targeting these regions may provide cross-reactive protection against multiple strains of influenza (Lee et al., 2014; Whittle et al., 2011; Hong et al., 2013; Krause et al., 2011).
Immunity to influenza is assessed using serological assays, such as hemagglutination inhibition (HI) or virus neutralization (VN) assays, that are correlated with protection against disease (Tsang et al., 2014; Hobson et al., 1972; Coudeville et al., 2010; Cox, 2013). These assays measure the “average” specificities and affinity of a complex mixture of antibodies. In principle, sequence analysis of the variable regions of antibodies in combination with their functional characteristics may provide more detailed information on the predicted immune response, including identification of antibodies that are present below the limit of detection of conventional assays but that have the potential for cross-protective or broadly reactive immunity.
Antibodies are comprised of a heavy and light chain. The heavy chain is the result of genomic recombination of a variable (IGHV), diversity (IGHD), and a joining (IGHJ) gene, while the light chain is composed of a variable (IGKV or IGLV) and joining (IGKJ) gene recombination. In a previous study, we analyzed the diversity and nature of the mouse antibody response against influenza HA by cloning IgH and IgL chains from individual B cells (Wilson et al., 2014). This work demonstrated that the antibody response targeting the HA of A(H1N1)pdm09 (following this particular immunization regimen) is relatively narrow, being dominated by approximately 100 heavy chain VDJ germline sequences and approximately 35 light chain VJ germline sequences. Here, we define the epitopes on HA that are targeted by a group of nine representative recombinant monoclonal antibodies (rmAbs) from this response and begin to define antigen and genomic sequence information associated with antibody recognition and neutralizing activity.
Results
rmAb recognize multiple epitopes on HA
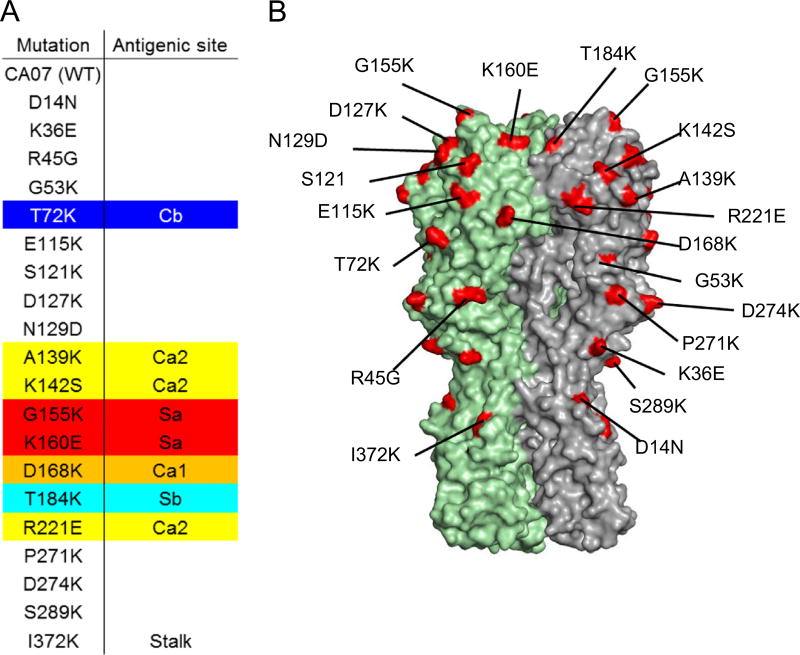
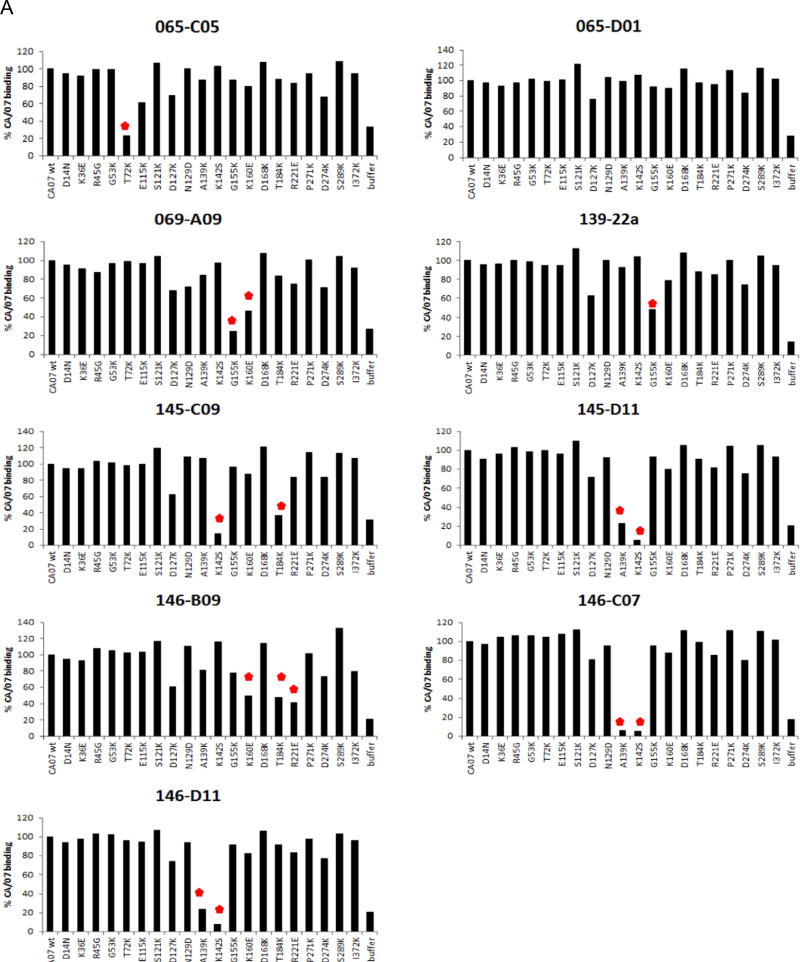
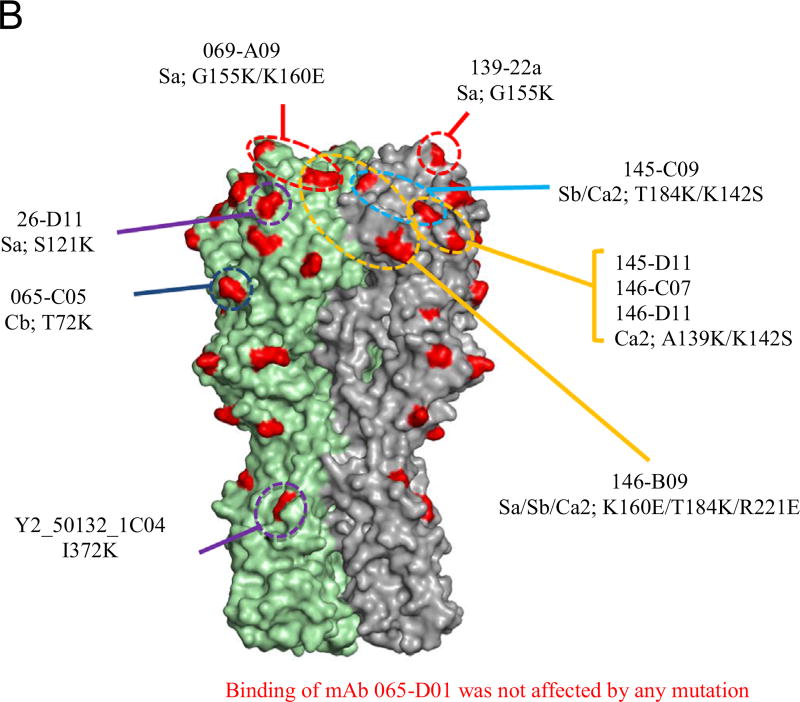
In previous work we immunized mice by sub-lethal infection with A(H1N1)pdm09 virus and boosted with TIV, leading to a robust antibody response that predominately targeted the HA of pdm09 viruses (Wilson et al., 2014). We used single-cell cloning from memory B cells to identify a number of rmAbs, including nine that bound H1N1pdm recHA with sub-nanomolar steady-state affinity (Wilson et al., 2014). These rmAb contain the most common IgH VDJ and IgK VJ rearrangements identified in that screen, and each displayed HI activity toward H1N1pdm virus (Wilson et al., 2014). To determine the binding sites of these rmAbs, we generated a panel of 20 recHAs, each containing a single point mutation within or near the defined antigenic regions of CA/07 (Fig. 1A and B), and measured binding affinity of the rmAbs to each by BLI. Control monoclonal antibodies 26-D11 and Y2_50132_1C04 (“C04”) bound with high affinity to the wild-type HA, and as expected their binding was reduced by mutations in antigenic site Sa (S121K) and in the stalk region (I372K) respectively (Fig. 2B and data not shown). For 6 of the 9 rmAbs, binding was compromised by mutation(s) in a single antigenic site, and 2 other rmAbs showed reduced binding to recHA with mutations in two (145-C09) or three (146-B09) antigenic sites (Fig. 2A and B).
Fig. 1. Epitope mapping using a recHA point mutation panel.
A panel of 20 respective single point mutations in recHA was constructed. (A) Sequence changes are shown; (B) Their locations are indicated on the 3D structure of HA.
Fig. 2. Epitope map of rmAb as determined by Biolayer Interferometry (BLI).
(A) rmAb binding affinity to each mutant HA and the percent response compared to wild-type CA/07 HA was determined. A greater than 50% reduction in binding activity was the cutoff for significance. (B) Epitopes whose mutations lead to a significant reduction in rmAb binding are indicated on the 3D structure of CA/07 HA.
Of the rmAb affected by mutations in single antigenic sites, 3 (145-D11, 146-C07 and 146-D11) were affected by the same two point mutations (A139K and K142S) within antigenic site Ca2. Binding of two other rmAbs (069-A09 and 139-22a) were reduced by Sa antigenic site mutations, G155K and K160E. 139-22a showed less reduction in binding with either mutation (Fig. 2A and B), and consistent with this observation, we have previously shown that 139-22a binds to recHA with about 100 times higher affinity than does 069-A69 (Wilson et al., 2014). Reduced HA binding to rmAb 065-C05 was observed by a T72K substitution, which is within antigenic site Cb.
Of the two rmAb affected by mutations in multiple antigenic sites, 145-C09 binding is affected by residue changes in sites Ca2 and Sb, since mutations K142S and T184K independently reduce binding. Similarly, mutations K160E (Sa), T184K (Sb), and R221E (Ca2) all reduced binding by rmAb 146-B09 and indicated that the binding footprint may span the monomer–monomer interface of the HA trimer (Fig. 2B). Although antigenic sites are defined as functionally distinct (Caton et al., 1982), anti-HA antibodies that span multiple sites (Matsuzaki et al., 2014; Tsibane et al., 2012) as well as those that cross the monomer-monomer interface (Barbey-Martin et al., 2002; Iba et al., 2014) have been described. Such antibodies might not be identified when using a classical antigenic mapping approach via mAb escape mutants, as a single mutation would probably be sufficient to avoid neutralization (Caton et al., 1982; Matsuzaki et al., 2014).
Binding of the remaining rmAb, 065-D01, was not markedly affected by any of the point mutations in the recHA panel; thus the epitope for this rmAb could not be determined in this assay (Fig. 2A). However, further analysis (see below) suggests that this rmAb binds at least partly within the Sa antigenic site.
rmAb germline gene usage and CDR3 sequence indicate constraints on the light chain
We previously sequenced and reported the germline gene analysis data for these rmAb (Wilson et al., 2014). Two rmAb in this panel (146-C07 and 146-D11) may be clonally related in that they have the same IgH and IgL germline segments, but have different somatic mutations. Both of these rmAb are equally affected by the same two residue changes in antigenic site Ca2 (Table 3). A third rmAb, 145-D11, employs a closely-related IgL V/J combination and has a similar IgL CDR3 sequence as 146-C07 and 146-D11, but has a different IgH V/D/J combination (Table 3), and this rmAb is also affected by the same two residues. Similarly, 069-A09 and 139-22a share identical IgL V/J genes and CDR3 sequence, but show distinct IgH gene usage and CDR3 sequences, and are both specific for the Sa antigenic site, further suggesting that the light chain may be important in HA antigenic targeting. However, since this panel of antibodies does not include a pair with closely-related heavy chain VDJ and CDR3 and different light chain usage, further studies are needed to determine if the light chain is particularly important in determining the antibody’s antigenic target.
Table 3.
rmAb germline gene usage and respective amino acid changes that affect CA/07 HA binding as identified by epitope mapping.
Germline gene usage was determined in previous work (Wilson et al., 2014; Reference (Wilson et al., 2014). Antigenic site Sa is shown in red, Ca1/2 in yellow, Sb in cyan, and Cb in blue.
rmAb reactivity against drifted H1N1pdm09 viruses isolated from humans
Based on the epitope profile of the panel of rmAb, we selected a group of eight H1N1pdmvirus natural isolates that contained variant residues likely to affect rmAb binding activity (Table 2). Ferret post-infection antisera are typically used to antigenically characterize influenza viruses and a reduction in HI titer of 8-fold or greater relative to the homologous titer of the vaccine strain is generally considered to be a significant antigenic difference and may warrant an update of the seasonal vaccine strain (Russell et al., 2008; Garten et al., 2009). CA/07 ferret antisera, and pooled sera from the mice used to prepare rmAb, had similar HI activity against CA/07 (vaccine strain) and Ukr/2011, Nor/2009, Mass/2011, Ind/2012 and Penn/2010 (Table 4), indicating that these viruses are antigenically similar to CA/07. HI titers against Ont/2012 were more than 8-fold lower than to CA/07 using both mouse and ferret antisera; ferret antisera also had 8-fold reduced HI titer against Par/191 while mouse sera had 4-fold reduced HI titers against Mex/2009 (Table 4). Ferret antisera MN activity paralleled that of HI activity against this virus panel, with the exception of a ≥16-fold drop in MN titer against Mex/2009 and Ind/2012 that did not correlate with the HI titer. Due to the limited amount of mouse sera, MN could not be determined with mouse sera for this panel of viruses.
Table 2.
Amino acid sequence of antigenic sites for historic H1N1 and H1N1pdm natural isolate viruses.
| Cb | Sa | Ca2 | Sa | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||||||||||||||||||
| 70 | 71 | 72 | 73 | 74 | 75 | 124 | 125 | 137 | 138 | 139 | 140 | 141 | 142 | 153 | 154 | 155 | 156 | 157 | 159 | 160 | 161 | 162 | 163 | 164 | |
| CA/07 | L | S | T | A | S | S | P | N | P | H | A | G | A | K | K | K | G | N | S | P | K | L | S | K | S |
| SC/18 | . | L | . | . | . | . | . | . | S | Y | . | . | . | S | . | . | . | S | . | . | . | . | . | . | . |
| IA/30 | . | L | . | V | . | . | . | . | . | Y | . | . | . | S | . | . | E | . | . | . | . | . | . | . | . |
| Mar/43 | . | L | S | E | R | - | . | K | S | . | . | . | K | S | E | . | D | G | . | . | N | . | N | N | . |
| NJ/76 | . | L | . | V | . | . | . | . | . | Y | . | . | . | N | E | . | . | . | . | . | . | . | . | . | . |
| USSR/77 | . | F | S | K | K | . | . | K | S | . | K | . | K | S | E | . | N | G | . | . | N | . | . | . | . |
| Tex/91 | L | F | S | K | E | . | . | . | S | . | N | . | K | S | - | . | N | G | L | . | N | V | . | . | . |
| Bris/59 | . | I | S | K | E | . | . | . | S | . | N | . | E | S | G | . | N | G | L | . | N | . | . | . | . |
| Par/2011 | P | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | D | . | . | . | . | . | . | . |
| Mex/2009 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | E | . |
| Ind/2012 | . | . | . | . | . | . | . | . | . | . | D | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| Nor/2009 | . | . | . | . | . | . | . | . | S | . | . | . | . | . | . | E | . | . | . | . | . | . | . | . | . |
| Ukr/2011 | . | . | . | . | R | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | I/L | . | . | . |
| Penn/2010 | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | N | . | . |
| Mass/2011 | . | . | . | . | . | . | . | . | . | . | . | . | E | . | . | . | . | . | . | . | . | . | N | . | . |
| Ont/2012 | . | . | . | . | . | . | . | . | . | . | . | . | . | R | . | . | E | . | . | . | . | . | . | . | . |
| Ca1 | Sb | Ca1 | Ca2 | Ca1 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|||||||||||||||||||||
| 166 | 167 | 168 | 169 | 170 | 184 | 185 | 186 | 187 | 188 | 189 | 190 | 191 | 192 | 193 | 194 | 195 | 203 | 204 | 205 | 221 | 222 | 235 | 236 | 237 | |
| CA/07 | I | N | D | K | G | T | S | A | D | Q | Q | S | L | Y | Q | N | A | S | S | R | R | D | E | P | G |
| SC/18 | V | . | N | . | . | . | G | T | . | . | . | . | . | . | . | . | . | . | . | K | . | . | . | . | . |
| IA/30 | V | . | N | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | K | . | G | . | . | . |
| Mar/43 | V | . | K | . | . | N | I | K | . | . | . | T | . | . | . | K | E | . | . | N | . | G | K | . | . |
| NJ/76 | V | . | N | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | K | . | G | . | . | . |
| USSR/77 | V | . | N | . | E | N | I | E | . | . | K | T | I | . | R | K | E | . | . | N | . | G | . | . | . |
| Tex/91 | V | . | N | K | E | N | I | G | . | . | R | A | I | . | H | T | E | . | . | H | . | . | . | . | . |
| Bris/59 | A | . | N | . | E | N | I | G | . | . | K | A | . | . | H | T | E | . | . | H | . | . | . | . | . |
| Par/2011 | . | . | . | . | . | . | T | . | G | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . |
| Mex/2009 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . |
| Ind/2012 | . | . | . | . | . | . | T | . | . | . | . | . | I | . | . | . | . | T | . | . | . | . | . | . | . |
| Nor/2009 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . |
| Ukr/2011 | . | . | . | . | . | N/T | T | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . |
| Penn/2010 | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . |
| Mass/2011 | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | T | . | . | . | G | . | . | . |
| Ont/2012 | . | . | . | . | . | . | T | . | . | . | . | R | . | . | . | . | . | T | . | . | . | G | . | . | . |
Antigenic sites are based on those determined for A/PR/8/34 by Caton et al. (1982) (Reference (Caton et al., 1982)).
Table 4.
Antisera HI and MN activity toward H1N1pdm human isolates.
| CA/07 | Ukr/2011 | Mex/2009 | Nor/2009 | Ont/2012 | Mass/2011 | Ind/2012 | Par/2011 | Penn/2010 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| +Antisera | HI | MN | HI | MN | HI | MN | HI | MN | HI | MN | HI | MN | HI | MN | HI | MN | HI | MN |
| Ferret | 256 | 512 | 512 | 128 | 256 | 32 | 128 | 128 | 4 | <4 | 512 | 1024 | 512 | <32 | 32 | 32 | 256 | 128 |
| Mouse | 160 | NT | 160 | NT | 40 | NT | 160 | NT | <20 | NT | 320 | NT | 160 | NT | 160 | NT | 160 | NT |
Data are reported as sera endpoint titer required to inhibit 4 HA units of respective virus. Day 3 post-boost sera was pooled from 5 mice and used for HI assays. Data are representative of one experiment repeated three times.
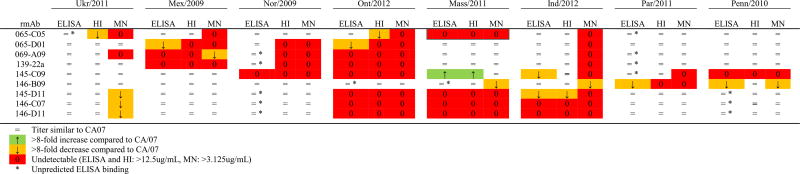
We further tested ELISA, HI and MN activity of each of the 72 possible rmAb/virus combinations, relative to the rmAb activity against CA/07 (Table 5 and Supplementary Table 1). ELISA binding analysis revealed the rmAbs mainly bound as predicted (i.e. virus containing a significant amino acid change in the identified rmAb antigenic binding site disrupted binding (Table 5)). Exceptions were 15 rmAb/virus that bound well despite apparently significant changes within the identified antigenic site (indicated by n in Table 5). In addition, one rmAb/virus pair (146-B09 vs Penn/2010) bound poorly despite the lack of an obvious change that would reduce binding, while another rmAb/virus combination (065-C05 vs Mass/2011) failed to bind despite being identical to CA/07 in the determined antigenic site. Interestingly, rmAb 145-C09, which in BLI assays showed reduced binding to recHA mutated in either Ca2 or Sb, showed enhanced reactivity (>8-fold higher titer) for Mass/2011 despite the A141E change in Ca2.
Table 5. ELISA, HI, and MN activity of rmAb’s toward natural H1N1pdm human isolates relative to A/California/07/2009.
Endpoint titers were determined by ELISA, HI and MN assays and their fold change from CA/07 was determined. Data is representative of the average of 3–4 assays. “=” indicates titer similar to CA/07, “↑” indicates >8 fold increase from CA/07, “↓” indicates >8 fold decrease from CA/07, and “0” indicates no ELISA, HI, or MN activity observed at 12500, 12500, or 3125 ng/ml, respectively.
HI activities against the H1pdm virus panel were generally consistent with ELISA binding (Table 5). In every case where ELISA binding was eliminated, no HI activity was seen, although in two cases (145-C09 vs Ind/2012 and 146-B09 vs Penn/2010 (Table 5)) HI activity was maintained in spite of reduced ELISA binding. However, in some cases, ELISA activity was maintained while HI activity was markedly decreased or lost (Table 5). Examples include the Sa-targeting rmAb 069-A09 and 139-22a, which maintained ELISA binding activity toward Nor/2009 but, as expected due to the K154E change in this virus’s Sa antigenic site, fail to show HI activity; and the Cb-specific rmAb, 065-C05, which lost most HI activity against Ukr/2011 and Ont/2012, despite equal binding based on ELISA, and in spite of the lack of mutation in antigenic site Cb of Ont/2012.
Although 6 of the viruses were antigenically identical to vaccine virus as measured by mouse and/or ferret antisera in HI assays, all 8 of the viruses showed reduced recognition by at least one rmAb. (It is important to note that these viruses are not representative of the global population of H1N1pdm09 viruses, since they were deliberately chosen to have mutations in and around the recognition sites of the rmAb). Seven of the 9 rmAb had no HI activity against Ont/2012 and an eighth showed a marked reduction in HI activity, leaving only 146-B09 capable of efficiently recognizing the virus in HI assays. This virus, which contains multiple amino acid variations across the Ca2, Sa and Sb antigenic sites, also demonstrated reduced HI titers both to ferret antisera raised against CA/07 and to our mouse sera, and is therefore expected to be less susceptible to immunity induced by the H1N1pdm09 component of current TIV. Par/2011 also showed a >8-fold reduction in HI titer with ferret antisera. However, this virus was well recognized by the mouse serum from which rmAb were prepared, and consistent with this, 8 of 9 rmAb recognized Par/2011 in HI assays. In contrast, mouse sera HI activity was reduced 4-fold toward Mex/2009, but 6 of 9 rmAb displayed HI activity toward this virus. The three rmAb that lost ELISA and HI activity with this virus included the Sa-specific as well as 065-D01, indicating that 065-D01 (which did not have a binding footprint assigned to it based on the BLI analysis) also binds at least partially within antigenic site Sa.
Virus neutralization (most commonly measured using MN assays) and HI activity have been shown to correlate with protection against influenza virus infection (McCullers and Huber, 2012; Reber and Katz, 2013). MN activity of rmAb mainly paralleled their HI activity (Table 5), with several rmAb/virus combinations showing ELISA binding while MN activity was undetectable, even though the same rmAb did neutralize other viruses in the panel. This confirms that antibodies can bind to HA in known antigenic sites without having virus-neutralizing activity and without showing HI activity. For eight rmAb/isolate combinations (i.e. 145-C09 vs Par/2011 and 069-A09 vs Ukr/2011), MN activity was lost while HI activity was still maintained, indicating that the MN assay may be more sensitive to changes in binding affinity than HI.
Cross-reactivity against historic H1N1 viruses
H1N1 viruses entered the human population in 1918, and the swine population shortly afterward. This subtype therefore includes a much wider range of sequence diversity than is represented in the H1N1pdm09 lineage tested above. We previously showed that, while none of these rmAb reacted with the recHA of A/Bris/59/07 in ELISA, three bound efficiently to recHA of SC/18 and one further bound to IA/30 (Wilson et al., 2014). We used the rmAb panel to perform HI assays on these two as well as four other H1N1 viruses that span more than 90 years of antigenic drift (1918, 1930, 1943, 1976, 1977, 1991 and 2009) and include a range of variation within antigenic sites (Table 2). For most of the virus/rmAb combinations, no HI activity was present (Table 6). The two Sa-specific antibodies (069-A09 and 139-22a) had HI activity against SC/18, as did 065-D01. 146-B09 had HI activity against NJ/76, but required a roughly 15-fold increase in antibody concentration opposed to CA/07. Although 065-D01 bound IA/30 at low concentration in our ELISA assay, HI activity was not observed, similar to the discordance between ELISA and HI noted above. rmAb 065-D01 showed the same ELISA and HI pattern as rmAb 069-A09 and 139-22a against both H1N1pdm09 panel and historical viruses, further suggesting that 065-D01 also targets antigenic site Sa but that its precise footprint is not covered by the mutations in the HA panel used for epitope mapping.
Table 6.
HI activity of rmAb toward historic H1N1 viruses.
| CA/07 | Tex/91 | USSR/77 | NJ/76 | Mar/43 | IA/30 | SC/18 | |
|---|---|---|---|---|---|---|---|
| 065-C05 | 1625 | – | – | – | – | – | – |
| 065-D01 | 98 | – | – | – | – | – | 98 |
| 069-A09 | 200 | – | – | – | – | – | 3125 |
| 139-22a | 200 | – | – | – | – | – | 98 |
| 145-C09 | 3125 | – | – | – | – | – | – |
| 145-D11 | 200 | – | – | – | – | – | – |
| 146-B09 | 98 | – | – | 1390 | – | – | – |
| 146-C07 | 200 | – | – | – | – | – | – |
| 146-D11 | 200 | – | – | – | – | – | – |
Data are reported as ng/ml required to inhibit 4 HA units of respective virus. Data are representative of one experiment repeated three times. “–” indicates titer >12500 ng/ml.
Discussion
Influenza infection or vaccination typically induces a protective, strain-specific, antibody response mainly directed against the HA glycoprotein. Current serological assays used to determine correlates of protection against influenza virus infection, including HI and MN assays, quantify the antibody response but are relatively crude measures of overall antibody reactivity. The advances in next-generation sequencing technology raise the possibility of using sequence-based repertoire analysis to rapidly assess the antibody response to influenza vaccination or infection. For this approach to be useful, antibody variable region sequence signatures must be correlated to their functional capabilities.
We have previously demonstrated that the murine immune response to HA of H1N1pdm09 is relatively restricted, originating from roughly 100 heavy V/D/J and 35 light chain V/J germ-line combinations (Wilson et al., 2014). Here we determined the partial binding footprints of a subset of high-affinity rmAb cloned from this response to identify the antigenic site(s) they recognize. Even within this small subset of high-affinity antibodies, at least seven unique binding footprints were present, targeting all four antigenic sites of HA1. Consistent with the lack of a detectable cross-reactive response in the immunized mice, no stem-reactive antibodies were identified. Humans infected with H1N1pdm09 have been shown to have detectable but rare cross-reactive antibodies; however, most of these are probably related to expansion of memory B cells originally raised against distantly-related H1N1 viruses (Wrammert et al., 2011).
This diversity in antigenic site recognition contrasts with previous observations that BALB/c and CBA/Ca mice immunized with H3N2 influenza virus (Patera et al., 1995) mainly target a single antigenic site, and that mice vaccinated against H1N1pdm09 mainly produced antibodies directed against two antigenic sites (Sa and Sb) (Rudneva et al., 2012). However, H1N1pdm09 (Matsuzaki et al., 2014) and PR/8 (Staudt and Gerhard 1983) infection or recHA from CA/04 immunization (Chen et al., 2013) generated a more diverse response that targeted multiple antigenic sites, similar to our findings. Although there could be an intrinsic difference in immunodominance between the viruses used, it is more likely that the difference between these studies is due to the immunization schedule used as well as timing of the B cell analysis.
Two of the cloned rmAb (146-D11 and 146-C07) are composed of the same IgH and IgL gene segments but encode different somatic mutations (Table 3). Unsurprisingly, both of these rmAbs showed identical patterns of binding to antigenic site Ca2 in our epitope mapping and virus panel screen, although with different affinities (Wilson et al., 2014). Interestingly, 145-D11, whose binding is also affected by the same residue changes in the Ca2 antigenic site, contains a similar IgL VJ rearrangement and CDR3 sequence but is combined with a different IgH rearrangement (Table 3). Similarly, rmAb 069-A09 and 139-22a share an IgL VJ rearrangement and CDR3 sequence but have different IgH VDJ rearrangements, and have an overlapping binding footprint within the Sa antigenic site (Table 3). Further, a pair of human derived anti-stem mAb that compete for the same HA-stem also share IgL VJ genes but use a completely different IgH VDJ rearrangement has previously been reported (Hu et al., 2013). These observations suggest that the rearrangement pattern of the light chain alone may be useful in predicting antigenic site targeting, consistent with the more limited IgL germ-line usage compared to IgH (Wilson et al., 2014). However, further examples are needed to test this possibility.
Recently, epitope mapping of H1N1pdm09 HA revealed that it is antigenically similar to PR/8, but may have a novel epitope (Matsuzaki et al., 2014). Although our study, and those of others (Rudneva et al., 2012; Chen et al., 2013; Retamal et al., 2014), did not observe this epitope, this highlights the need to further expand our understanding of the antigenicity of H1N1pdm09 HA, beyond that of the classical antigenic mapping of the HA of PR/8 (Caton et al., 1982). This will not only aid our understanding of the protective antibody response against this virus, but will also support surveillance and vaccine candidate selection efforts that monitor viral drift in the human population.
Influenza viruses typically undergo antigenic drift over time due to host immune pressure, during which time one or more antigenic sites on the HA mutate until an immune response against prior viruses is no longer completely protective. Although single amino acid mutations are capable of causing marked antigenic change (Koel et al., 2013, 2015), antigenic drift is usually associated with mutations in several amino acids located in more than one antigenic site. We examined the effect of a limited number of amino acid changes on functional recognition both by reference ferret antisera and our panel of rmAb, making use of natural isolates of H1N1pdm09. Although in most cases these natural isolates showed no evidence of antigenic drift when evaluated by HI using reference ferret serum, rmAb that were specific for individual antigenic sites lost HI ability against each of the viruses. Thus, cryptic antigenic variation can arise in a viral population, in the form of viruses are not recognized by subsets of the antibodies. It is possible that such viruses may act as intermediates for variants with changes in multiple antigenic sites that are capable of escaping immunity induced by the current vaccine strain.
Although in most cases rmAb activity could be predicted from knowledge of the sequence of HA and the antigenic targeting of the rmAb (~75% of rmAb/virus combinations), a significant number of antibodies retained binding activity in spite of significant amino acid changes in the respective HA antigenic binding site. In some cases, HI and MN activity was reduced in spite of rmAb binding, suggesting that the reduced affinity associated with the sequence changes reduced functional activity of the antibody, but in other cases normal HI and MN levels were retained. Since the virus isolates were chosen based on having biologically significant amino acid changes (e.g. Lys to Asp; Leu to Pro), these antibodies show some tolerance in their binding ability. Conversely, in two of the 72 combinations, rmAbs failed to bind despite a lack of change in the putative binding site, suggesting that either the binding footprint was incompletely mapped for these rmAbs, or that amino acid variation outside the antigenic site may alter antibody recognition, e.g. by altering the conformation of the antigenic site. Finally, in one case, an Ala to Glu change in the antigenic site was associated with an increase in binding.
In spite of these exceptions, identifying the antigenic site (s) targeted by each rmAb generally allowed us to accurately predict binding and HI reactivity toward historic H1N1 and H1N1pdm09 variant viruses, based on their HA sequence. H1 HA has undergone antigenic drift in humans and, to a lesser extent, in swine, since about 1918, when the 1918 pandemic A(H1N1) entered the human and swine populations. When comparing HI activity pattern against HA from historical H1N1 viruses circulating from 1918 to 2007, only 4 rmAb showed functional activity, with the most cross-reactivity seen with Sa-reactive rmAb against viruses with a relatively conserved Sa site. However, in other cases viruses with relatively minor or no changes in the antigenic site predicted to be involved in binding showed no HI activity. Again, this may be due to the binding footprint for the rmAb being incompletely mapped, or due to confirmation changes caused by amino acid variation outside an antigenic site.
Cross-reactivity is not all or nothing. Single site mutations can alter antibody function (ie. HI and MN activity) while still permitting antibody binding as determined by ELISA, as demonstrated by several virus/rmAb combinations that showed discordance between ELISA, HI, and MN activity. In most of these situations, epitope mapping predicted reduced antibody binding, and HI and/or MN activity was lost, but ELISA reactivity was retained. Similarly, H1N1pdm escape mutants selected in the presence of mAbs can show reduced HI activity even though antibodies can still bind in ELISA (Rudneva et al., 2012; Chen et al., 2013; Kaverin et al., 2004). In these cases, presumably the HA variant reduces rmAb binding affinity (or perhaps increases the receptor-binding affinity for sialic acids (Clarke et al., 1985; Laeeq et al., 1997; Temoltzin-Palacios and Thomas, 1994; Yewdell et al., 1986)) so that the antibody is no longer able to effectively compete with sialic acid binding, while ELISA, in the absence of competitors for binding, binding was still easily measured. Studies are underway to determine if such rmAb/virus interactions, while losing the ability to directly neutralize virus in vitro, still maintain protective effector functions in vivo (for example, via interactions with complement and antibody-dependent cell-mediated cytotoxicity (ADCC)). In any case, memory B cells expressing antibodies which still bind to antigenic variants are potential substrates for somatic hypermutation, and these (rather than naïve B cells) may be the foundation for protective antibodies against drifted variants of virus.
Characterization of infection and/or vaccine induced B cell response(s) at the monoclonal level will allow assessment of the respective protective and non-protective components of the immune response. Understanding the components of the response, and the diversity of epitopes targeted that neutralize virus, will further our understanding of vaccine efficiency. In turn, understanding the molecular nature of the neutralizing antibody/antigen interactions can aide in surveillance efforts to more quickly identify mutations, as viruses evolve in nature, that may have an impact on existing human herd immunity but which may escape identification with post-immunization reference ferret antisera.
Funding
This work was supported by the Centers for Disease Control and Prevention (CDC).
JRW received financial support for this work from the Oak Ridge Institute for Science and Education, Oak Ridge, TN.
Materials and methods
Viruses and cells
The viruses and recombinant HA (recHA) used in these experiments are summarized in Table 1, and the amino acid sequence of the virus’s antigenic sites are listed in Table 2. Virus isolate stocks were grown in 10 day old embryonated chicken eggs for 48 h at 37 °C. The sequences of all virus HA’s were confirmed before use.
Table 1.
Viruses and recombinant proteins used in this study.
| Influenza A virus | Abbreviation | Subtype | GISAID accession | Identity of HA to CA/07 (HA1) |
|---|---|---|---|---|
| A/California/07/2009 | CA/07 | A(H1N1)pdm09 | EPI177294 | – |
| A/South Carolina/1/1918 | SC/18 | A(H1N1) | EPI5571 | 86% (82%) |
| A/swine/Iowa/15/1930 | IA/30 | A(H1N1) | EPI124024 | 87% (83%) |
| A/AA/Marton/1943 | Mar/43 | A(H1N1) | EPI240837 | 82% (75%) |
| A/New Jersey/1976 | NJ/76 | A(H1N1) | EPI241033 | 91% (88%) |
| A/USSR/90/1977 | USSR/77 | A(H1N1) | EPI390455 | 80% (72%) |
| A/Texas/36/1991 | Tex/91 | A(H1N1) | EPI159432 | 79% (71%) |
| A/Mexico/5569/2009 | Mex/2009 | A(H1N1)pdm09 | EPI273882 | 99% (98%) |
| A/Norway/3206/2009 | Nor/2009 | A(H1N1)pdm09 | EPI240393 | 99% (98%) |
| A/Ukraine/130/2011 | Ukr/2011 | A(H1N1)pdm09 | EPI320158 | 98% (97%) |
| A/Pennsylvania/07/2010 | Penn/2010 | A(H1N1)pdm09 | EPI280320 | 98% (97%) |
| A/India/2005/2012 | Ind/2012 | A(H1N1)pdm09 | EPI422601 | 98% (97%) |
| A/Massachusetts/06/2011 | Mass/2011 | A(H1N1)pdm09 | EPI310002 | 98% (97%) |
| A/Paraguay/191/2011 | Par/2011 | A(H1N1)pdm09 | EPI349352 | 97% (97%) |
| A/Ontario/RV117/2012 | Ont/2012 | A(H1N1)pdm09 | EPI357951 | 97% (96%) |
The viruses and/or recombinant proteins used in this study are listed. The abbreviations by which they are referred in the text and the identity of the virus HA and HA1 domain to A/California/07/2009 is shown.
Recombinant monoclonal antibodies (rmAb)
Single-cell cloning of the rmAb used in these experiments was described previously (Wilson et al., 2014). Briefly, naïve C57BL/6 mice (Jackson) were immunized by infection with H1N1pdm virus (OH/07(H1N1pdm), antigenically identical to CA/07(H1N1pdm)), followed 21 days later by a boost with 2011/12 commercial seasonal trivalent inactivated vaccine (TIV) (Fluarix), containing HA and NA from CA/07(H1N1pdm), VIC/210(H3N2) and B/BR/60. Three days later, spleens were harvested and B cells reactive with H1N1pdm recHA were sorted onto glass slides by flow cytometry. IgH VDJ and IgL VJ gene segments were amplified and subcloned into plasmid vectors which provided human constant regions for heavy or light chain respectively, as well as a signal sequence and promoter sequences for expression in mammalian cells as previously described (Wilson et al., 2014). 293T cells were transiently transfected with single cell matching pairs of IgH and IgL expressing vectors, and supernatant was collected and concentrated to 25 µg rmAb/ml working stocks.
The nine rmAb used in these experiments are listed in Table 3; the germline gene usage and CDR3 sequences for these rmAb have been previously published (Wilson et al., 2014).
Recombinant HA cloning and transient expression
A codon-optimized cDNA encoding the ectodomain (residues 1–501) of the mature HA gene of A(H1N1) pdm09 virus CA/07 was sub-cloned into a pIEx-4 vector (EMD Millipore, MA) using the In-Fusion HD cloning system (Clontech, CA). HA mutants containing point mutations within or near known antigenic sites were generated from this wild-type pIEx-4-HA clone using the QuickChange Lightning Site-Directed Mutagenesis Kit (Agilent, CA) (Fig. 1). The point mutations were designed to induce significant size and/or charge change in surface-accessible residues. One mutation, S289K, removed a putative glycosylation addition site. All recombinant HA (recHA) proteins contained a thrombin cleavage site at the C-terminus followed by a trimerizing sequence (foldon) from the bacteriophage T4 fibritin for generating functional trimers (Yang et al., 2010), and a His-Tag to aid with subsequent assays and detection. Constructs were transiently transfected into suspension Sf9 cells (EMD Millipore, MA) using the Cellfectin II transfection reagent (Life Technologies, NY), following manufacturer’s protocols. Transfected cells were transferred into 125 ml conical flasks and maintained at 27 °C for five days in an orbital shaker/incubator (at 170 rpm). The recHAs secreted in the culture supernatant were assessed for expression by Western blot using anti-His antibody (Qiagen, CA), and applied to epitope mapping analysis without further purification.
Biolayer interferometry (BLI) assay
Binding to recHA by the cloned antibodies was measured using BLI on an Octet Red 96 instrument (Pall ForteBio, CA) according to the manufacturer’s instructions. Briefly, antibodies were diluted to 10 µg/ml in kinetics buffer (PBS containing 0.02% (v/v) Tween-20, 0.005% (v/v) sodium azide, and 100 µg/ml bovine serum albumin). recHA was coupled to anti-penta-His biosensors and antibody binding data were collected and analyzed using the system software and fitted to a 1:1 binding model. An HA1-specific mAb (26-D11; Immune Technology Corp., New York) and an HA2-specific mAb, Y2_50132_1C04 (provided by Patrick Wilson, University of Chicago), whose binding locations on the H1pdm HA were previously determined (Wrammert et al., 2011), were included in the assay as controls. Data are presented as percentages of antibody binding to mutants compared to binding to WT CA/07 (100%). A reduction of ≥50% in binding response for each mutant compared to the wild type recHA was considered significant (Throsby et al., 2008).
Hemagglutination inhibition assays
rmAbs were screened for neutralizing activity against a panel of natural H1N1pdm isolates by the hemagglutination inhibition (HI) assay as previously described (World Health Organization, 2011). Briefly two-fold serial dilutions of post immunization ferret antisera or mouse sera and three-fold serial dilutions of rmAb were mixed with equal volume of standardized influenza viruses (4 HA U/25 µl/well) for 15 min interaction. Standardized (0.5%) turkey red blood cells (50 µl) were added. After 30 min incubation at room temperature, HI titers were recorded as highest dilution of antiserum that completely inhibits hemagglutination.
ELISA assays
For detection of HA cross-reactivity, ELISA assays were performed as previously described (Wilson et al., 2014). Briefly, Costar Hi Bind plates (Corning Inc., Tewksbury, MA) were coated overnight with the appropriate virus isolate (25 HA units/well) at 4 °C. Plates were blocked for 1 h with PBS/0.1% Tween-20 (PBST) containing 1.5% BSA (blocking buffer) at room temperature. All rmAbs were serially titrated three-fold in blocking buffer and allowed to incubate with antigen-coated plates for 1 h at room temperature. After three PBST washes, wells were probed with goat anti-human (H&L)–HRP (Thermo Scientific) for 1 h at room temperature. Plates were washed three times with PBST and signal was developed with 1 step™ Turbo TMB-ELISA reagent (Thermo Scientific). Reactions were stopped with 1 N sulfuric acid and absorbance was read at 450 nm.
Microneutralization assay
MN assays were performed as described (World Health Organization, 2011). The minimum detection limit of this assay was a titer of 20 for post immunization ferret antisera.
Supplementary Material
Footnotes
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.virol.2015.08.004.
References
- Barbey-Martin C, Gigant B, Bizebard T, Calder LJ, Wharton SA, et al. An antibody that prevents the hemagglutinin low pH fusogenic transition. Virology. 2002;294:70–74. doi: 10.1006/viro.2001.1320. [DOI] [PubMed] [Google Scholar]
- Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- Chen J, Yan B, Chen Q, Yao Y, Wang H, et al. Evaluation of neutralizing efficacy of monoclonal antibodies specific for 2009 pandemic H1N1 influenza A virus in vitro and in vivo. Arch Virol. 2013 doi: 10.1007/s00705-013-1852-y. [DOI] [PubMed] [Google Scholar]
- Clarke SH, Huppi K, Ruezinsky D, Staudt L, Gerhard W, et al. Interclonal and intraclonal diversity in the antibody-response to inflienza hemagglutinin. J. Exp. Med. 1985;161:687–704. doi: 10.1084/jem.161.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudeville L, Bailleux F, Riche B, Megas F, Andre P, et al. Relationship between haemagglutination-inhibiting antibody titres and clinical protection against influenza: development and application of a bayesian random-effects model. BMC Med. Res. Methodol. 2010;10:18. doi: 10.1186/1471-2288-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RJ. Correlates of protection to influenza virus, where do we go from here? Hum. Vaccin. Immunother. 2013;9:405–408. doi: 10.4161/hv.22908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson D, Curry RL, Beare AS, Ward-Gardner A. The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J. Hyg. (London) 1972;70:767–777. doi: 10.1017/s0022172400022610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Lee PS, Hoffman RM, Zhu X, Krause JC, et al. Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J. Virol. 2013;87:12471–12480. doi: 10.1128/JVI.01388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Chen A, Miao Y, Xia S, Ling Z, et al. Fully human broadly neutralizing monoclonal antibodies against influenza A viruses generated from the memory B cells of a 2009 pandemic H1N1 influenza vaccine recipient. Virology. 2013;435:320–328. doi: 10.1016/j.virol.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba Y, Fujii Y, Ohshima N, Sumida T, Kubota-Koketsu R, et al. Conserved neutralizing epitope at globular head of hemagglutinin in H3N2 influenza viruses. J. Virol. 2014;88:7130–7144. doi: 10.1128/JVI.00420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverin NV, Rudneva IA, Ilyushina NA, Lipatov AS, Krauss S, et al. Structural differences among hemagglutinins of influenza A virus subtypes are reflected in their antigenic architecture: analysis of H9 escape mutants. J. Virol. 2004;78:240–249. doi: 10.1128/JVI.78.1.240-249.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koel BF, Burke DF, Bestebroer TM, van der Vliet S, Zondag GC, et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science. 2013;342:976–979. doi: 10.1126/science.1244730. [DOI] [PubMed] [Google Scholar]
- Koel BF, Mogling R, Chutinimitkul S, Fraaij PL, Burke DF, et al. Identification of amino acid substitutions supporting antigenic change of A (H1N1)pdm09 viruses. J. Virol. 2015 doi: 10.1128/JVI.02962-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Basler CF, et al. A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J. Virol. 2011;85:10905–10908. doi: 10.1128/JVI.00700-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laeeq S, Smith CA, Wagner SD, Thomas DB. Preferential selection of receptor-binding variants of influenza virus hemagglutinin by the neutralizing antibody repertoire of transgenic mice expressing a human immunoglobulin mu minigene. J. Virol. 1997;71:2600–2605. doi: 10.1128/jvi.71.4.2600-2605.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PS, Ohshima N, Stanfield RL, Yu W, Iba Y, et al. Receptor mimicry by antibody F045–092 facilitates universal binding to the H3 subtype of influenza virus. Nat. Commun. 2014;5:3614. doi: 10.1038/ncomms4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y, Sugawara K, Nakauchi M, Takahashi Y, Onodera T, et al. Epitope mapping of the hemagglutinin molecule of A/(H1N1)pdm09 virus by using monoclonal antibody escape mutants. J. Virol. 2014 doi: 10.1128/JVI.01381-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers JA, Huber VC. Correlates of vaccine protection from influenza and its complications. Hum. Vaccin. Immunother. 2012;8:34–44. doi: 10.4161/hv.8.1.18214. [DOI] [PubMed] [Google Scholar]
- Molinari NA, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, et al. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine. 2007;25:5086–5096. doi: 10.1016/j.vaccine.2007.03.046. [DOI] [PubMed] [Google Scholar]
- Patera AC, Graham CM, Thomas DB, Smith CA. Immunodominance with progenitor B cell diversity in the neutralizing antibody repertoire to influenza infection. Eur. J. Immunol. 1995;25:1803–1809. doi: 10.1002/eji.1830250702. [DOI] [PubMed] [Google Scholar]
- Reber A, Katz J. Immunological assessment of influenza vaccines and immune correlates of protection. Expert Rev. Vacc. 2013;12:519–536. doi: 10.1586/erv.13.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retamal M, Abed Y, Corbeil J, Boivin G. Epitope mapping of the 2009 pandemic and the A/Brisbane/59/2007 seasonal (H1N1) influenza virus hemagglutinins using monoclonal antibodies and escape mutants. J. Gen. Virol. 2014 doi: 10.1099/vir.0.067819-0. [DOI] [PubMed] [Google Scholar]
- Rudneva I, Ignatieva A, Timofeeva T, Shilov A, Kushch A, et al. Escape mutants of pandemic influenza A/H1N1 2009 virus: variations in antigenic specificity and receptor affinity of the hemagglutinin. Virus Res. 2012;166:61–67. doi: 10.1016/j.virusres.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Rudneva IA, Kushch AA, Masalova OV, Timofeeva TA, Klimova RR, et al. Antigenic epitopes in the hemagglutinin of Qinghai-type influenza H5N1 virus. Viral Immunol. 2010;23:181–187. doi: 10.1089/vim.2009.0086. [DOI] [PubMed] [Google Scholar]
- Russell CA, Jones TC, Barr IG, Cox NJ, Garten RJ, et al. Influenza vaccine strain selection and recent studies on the global migration of seasonal influenza viruses. Vaccine. 2008;26(Suppl. 4):D31–D34. doi: 10.1016/j.vaccine.2008.07.078. [DOI] [PubMed] [Google Scholar]
- Staudt LM, Gerhard W. Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. I. Significant variation in repertoire expression between individual mice. J. Exp. Med. 1983;157:687–704. doi: 10.1084/jem.157.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temoltzin-Palacios F, Thomas DB. Modulation of immunodominant sites in influenza hemagglutinin compromise antigenic variation and select receptor-binding variant viruses. J. Exp. Med. 1994;179:1719–1724. doi: 10.1084/jem.179.5.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang TK, Cauchemez S, Perera RA, Freeman G, Fang VJ, et al. Association between antibody titers and protection against influenza virus infection within households. J. Infect. Dis. 2014;210:684–692. doi: 10.1093/infdis/jiu186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsibane T, Ekiert DC, Krause JC, Martinez O, Crowe JE, Jr, et al. Influenza human monoclonal antibody 1F1 interacts with three major antigenic sites and residues mediating human receptor specificity in H1N1 viruses. PLoS Pathog. 2012;8:e1003067. doi: 10.1371/journal.ppat.1003067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Z, Ye J, Xu L, Shao H, Jin W, et al. Antigenic mapping of the hemagglutinin of an H9N2 avian influenza virus reveals novel critical amino acid positions in antigenic sites. J. Virol. 2014;88:3898–3901. doi: 10.1128/JVI.03440-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. U.S.A. 2011;108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JR, Tzeng WP, Spesock A, Music N, Guo Z, et al. Diversity of the murine antibody response targeting influenza A(H1N1pdm09) hemagglutinin. Virology. 2014;458–459:114–124. doi: 10.1016/j.virol.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza. 2011 < www.who.int/csr/disease/influenza/manual_diagnosis_surveillance_influenza/en/index.html>.
- Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J. Exp. Med. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Carney P, Stevens J. Structure and Receptor binding properties of a pandemic H1N1 virus hemagglutinin. PLoS Curr. 2010;2:RRN1152. doi: 10.1371/currents.RRN1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yewdell JW, Gerhard W. Antigenic characterization of viruses by monoclonal antibodies. Annu. Rev. Microbiol. 1981;35:185–206. doi: 10.1146/annurev.mi.35.100181.001153. [DOI] [PubMed] [Google Scholar]
- Yewdell JW, Caton AJ, Gerhard W. Selection of influenza A virus adsorptive mutants by growth in the presence of a mixture of monoclonal antihemagglutinin antibodies. J. Virol. 1986;57:623–628. doi: 10.1128/jvi.57.2.623-628.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Guo YH, Jiang T, Wang YD, Chan KH, et al. A unique and conserved neutralization epitope in H5N1 influenza viruses identified by a murine antibody against the A/goose/Guangdong/1/96 hemagglutinin. J. Virol. 2013 doi: 10.1128/JVI.01577-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.