Abstract
Centromeres that are essential for faithful segregation of chromosomes consist of unique DNA repeats in many eukaryotes. Although recombination is under-represented around centromeres during meiosis, little is known about recombination between centromere repeats in mitotic cells. Here, we compared spontaneous recombination that occurs between ade6B/ade6X inverted repeats integrated at centromere 1 (cen1) or at a non-centromeric ura4 locus in fission yeast. Remarkably, distinct mechanisms of homologous recombination (HR) were observed in centromere and non-centromere regions. Rad51-dependent HR that requires Rad51, Rad54 and Rad52 was predominant in the centromere, whereas Rad51-independent HR that requires Rad52 also occurred in the arm region. Crossovers between inverted repeats (i.e. inversions) were under-represented in the centromere as compared to the arm region. While heterochromatin was dispensable, Mhf1/CENP–S, Mhf2/CENP–X histone-fold proteins and Fml1/FANCM helicase were required to suppress crossovers. Furthermore, Mhf1 and Fml1 were found to prevent gross chromosomal rearrangements mediated by centromere repeats. These data uncovered the regulation of mitotic recombination between DNA repeats in centromeres and its physiological role in maintaining genome integrity.
INTRODUCTION
Repetitive elements including segmental duplication, transposons, rDNA as well as telomere and centromere repeats are prevalent in eukaryote genomes and occupy over 50% of the human genome (1,2). DNA replication problems such as fork stalling and collapse induce recombination between repeat elements (3). Non-conservative recombination such as crossover and break-induced replication between repeats give rise to gross chromosomal rearrangements (GCRs), which in turn lead to various types of genetic diseases including cancer (4–7).
Centromeres are essential for faithful segregation of chromosomes both in mitosis and meiosis. Interestingly, centromeres consist of repetitive sequences in many eukaryotes (8). Human centromeres (0.2–5 Mb) contain arrays of α-satellite repeats which comprise up to 5% of the genome. Fission yeast centromeres (40–110 kb) consist of pairs of inverted repeats (imr, dg, dh and irc) flanking the central sequence (cnt). Exchanges of entire short arms of acrocentric chromosomes, termed Robertsonian translocations, represent the most common chromosomal abnormality observed in humans (1/1000 individuals) (9). Similar translocation that occurs on the same chromosome yields isochromosomes whose arms are mirror images of each other (10). Remarkably, recombination is under-represented around centromeres to ensure faithful segregation of chromosomes at the first meiotic division (11,12); however, it remains unclear whether recombination between centromere repeats is controlled in mitotic cells.
Unique chromatin structures are formed on centromeres (13,14). In the central domain, nucleosomes containing histone H3 variant CENP–A/Cnp1 provide a platform for the assembly of the kinetochore that binds spindle microtubules. CENP–T, CENP–W, CENP–S and CENP–X histone-fold proteins are components of the constitutive centromere-associated network (CCAN) and form CENP–T–W–S–X nucleosome-like complexes that are also involved in the kinetochore assembly (15–18). CENP–S and CENP–X, also called Mhf1 and Mhf2, respectively, form (Mhf1–Mhf2)2 tetramers that preferentially bind to branched DNA and recruit Fanconi anemia (FA) complementation group M (FANCM) helicase during recombination and repair (19–22). The kinetochore chromatin that contains CENP–A and CENP–T–W–S–X is frequently flanked by heterochromatin that ensures faithful segregation of chromosomes (23,24). Interestingly, in chicken DT40 cells, heterochromatin is assembled on the centromeres that consist of DNA repeats but not on non-repetitive centromeres (25), suggesting a link between DNA repeats and heterochromatin. In pericentromeric heterochromatin domains, Suv39/Clr4 methyltransferase introduces H3K9 methylation (H3K9me) (26). A set of proteins including HP1/Swi6 specifically bind the H3K9me mark characteristic of heterochromatin and form high-order chromatin structures (27,28). Recent studies have shown that heterochromatin affects DNA damage repair and recombination to maintain genome integrity (29–31).
There are Rad51-dependent and -independent homologous recombination (HR). Rad51 forms nucleoprotein filaments on single-stranded DNA (ssDNA), catalyses strand invasion into homologous double-stranded DNA (dsDNA) and forms displacement-loop (D-loop), the resolution of which results in either crossover or non-crossover products (32). Yeast Rad52 and mammalian BRCA2 are essential for Rad51 filament formation. Rad54 binds Rad51 and facilitates early and late steps of Rad51-dependent HR (33). Rad51-dependent HR is important to maintain genome integrity, as mutations in BRCA2 elevate GCR events, resulting in predisposition to breast and ovarian cancer (34). rad51 mutations reduce gene conversion and instead increase homology-mediated GCRs (35–37). Besides its role in Rad51 filament formation, Rad52 plays a role in Rad51-independent HR. Rad52 catalyses single-strand annealing (SSA) between complementary ssDNA molecules independently of Rad51 and Rad54 (38–40). Chromosome rearrangements including deletions between tandem repeats and translocations between different chromosomes result from SSA (41–43), demonstrating that Rad51-independent HR is prone to GCR.
In previous studies, using fission yeast we have demonstrated that Rad51 and Rad54 channel spontaneous recombination between centromere inverted repeats into a non-crossover pathway, thereby limiting crossovers that result in centromere inversion or isochromosome formation (35). Here, we compared mitotic recombination between inverted repeats in centromere and in arm regions. In the centromere, Rad51, Rad54 and Rad52 were all essential for recombination. In the arm region, however, Rad51 and Rad54 were only partially required for recombination compared to Rad52. Analysis of recombinant DNA revealed that crossovers were rare in the centromere. These findings show that HR occurs in distinct ways in the context of chromatin structure. In centromeres, Rad51-dependent HR predominates and crossovers are strongly suppressed. We further showed that pericentromeric heterochromatin is not essential for the regulation of recombination; however, deletions of Mhf1/CENP–S, Mhf2/CENP–X and Fml1/FANCM increase crossovers in centromeres. mhf1 and fml1 mutations increase GCRs that are mediated by centromere repeats. Our data show that the centromere-specific regulation of recombination plays an important role to protect repetitive centromeres from homology-mediated chromosome rearrangements.
MATERIALS AND METHODS
Yeast media and strains
The fission yeast strains used in this study are detailed in Supplementary Table S1. Standard genetic procedures were used as described previously (35). Antibiotic marker genes were obtained from pFA6a-kanMX6 and pFA6a-hphMX6 (44). Yeast transformation was carried out using lithium acetate and transformants were selected on yeast extract (YE) medium supplemented with G418 (Nacalai Tesque) or hygromycin B (Nacalai Tesque) at a final concentration of 100 μg/ml. Correct integration was confirmed by polymerase chain reaction (PCR). Cells were grown on Yeast extract (YE) medium or Edinburgh minimal medium (EMM) supplemented with appropriate amino acids at a final concentration of 225 μg/ml. 5-fluoroorotic acid (5FOA; 1 mg/ml) (Apollo Science) and uracil (56 μg/ml) were added to Yeast Nitrogen Base (YNB) media containing 1.7 g/l YNB (Difco 233520, BD Biosciences), 5 g/l of ammonium sulphate and 2% glucose. Solid medium contained 1.5% agarose (Nacalai Tesque). To generate the mhf1-L78R mutant strain, the ura4+ gene was introduced at 303 bp upstream of the mhf1 coding region, after which the Ura+ cells were transformed with a 1.9 kb PCR fragment that contains the mhf1-L78R mutation and the ura4+ integration site and were selected on 5FOA plates. The integration of the mhf1-L78R mutation was confirmed by DNA sequencing. Cells were grown at 33°C, unless otherwise indicated.
Construction of cen1-Sn, ura4-Sn, ura4-Hp and ura4-Sn(cen) strains
The cen1-Sn construct was created by a series of yeast transformations essentially as described in Supplementary Figure S15 (35). To introduce the uar4+ gene at the HindIII site in imr1L, ura4-D18 mutant cells were transformed with a 2.8 kb FspI–XhoI fragment of the pTN899 plasmid and selected on EMM plates. To replace the ura4+ gene with the ade6B gene, the imr1L::ura4+ strain was transformed with a 2.6 kb DraI fragment of pTN902 and selected on 5FOA plates. To introduce the ura4+ gene at the HindIII site in imr1R, the imr1L::ade6B strain was transformed with a 2.8 kb FspI–XhoI fragment of pTN899 and selected on EMM plates. To replace the ura4+ gene with the ade6X gene, the imr1R::ura4+ strain was transformed with a 3.2 kb KpnI–SpeI fragment of pTN905 and selected on 5FOA plates.
The ura4-Sn and ura4-Hp constructs were created based on the TNF3562 strain that contains the ade6B::ura4+::ade6X construct at the ura4 locus, which was generated by transformation of a ura4-D18 mutant strain with a 6.5-kb NotI–SalI fragment of pTN480 plasmid that contains the ade6B/ade6X inverted repeats surrounding the ura4+ gene. To create the ura4-Sn construct, a 6.6-kb of the cen1-Sn central domain was amplified by PCR using cm-ade6 primer from cen1-Sn genomic DNA and introduced into TNF3562 cells. Ura– transformants were selected on 5FOA plates. To create the ura4-Hp construct, an 11.8-kb region of the cen1-Hp central domain was amplified by PCR using cm-ade6 primer from cen1-Hp genomic DNA and was used to transform TNF3562 cells.
The ura4-Sn(cen) construct was created by a series of transformations of the ura4-Sn strain as shown in Supplementary Figure S16. In vivo one-step assembly of multiple DNA fragments (45) was carried out during the transformation. To introduce pericentromeric outer repeats on one side of ura4-Sn, the ura4-Sn strain (TNF3631) was transformed with a 2.9 kb EcoO109I–SpeI fragment from pTN1015, a 14 kb EcoRI fragment from pRS140 (46) and a 5.9 kb NheI–BamHI fragment from pTN1019 that contains the ura4+ gene. Ura+ transformants were selected on EMM plates to obtain TNF4186. To eliminate ura4+, TNF4186 cells were transformed with a 1.8 kb XbaI–BamHI fragment from pTN1020 and Ura– transformants were selected on 5FOA plates to obtain TNF4226. To introduce pericentromeric repeats on the other side, TNF4226 cells were transformed with a 6.3 kb BamHI–NotI fragment from pTN1024, a 14 kb EcoRI fragment from pRS140 and a 2.4 kb BamHI–SpeI fragment from pTN1015. Ura+ transformants were then selected on EMM plates to obtain TNF4684. To suppress transcriptional silencing of the ura4+ marker, we created the ura4-Sn(sen) construct in a swi6 mutant background.
Plasmid construction
pTN899 that contains the ura4+ gene at the HindIII site in imr1 was constructed as follows: a 1.3 kb EcoRI–Eco47III fragment containing imr1 from pYC312 (47) was introduced between the EcoRI–HincII of pBluescript II KS+ (Stratagene) to form pTN886. A 1.8 kb HindIII fragment containing the ura4+ gene was introduced at the HindIII site of pTN886, yielding pTN899. A 1.9 kb DraI fragment containing the ade6B or the ade6X gene (35) was introduced at the SnaBI site, which is only 50 bp apart from the HindIII site of pTN886, forming pTN902 and pTN905, respectively.
pTN480 containing the ade6B:ura4+:ade6X construct was created as follows: a 1.9 kb DraI fragment containing ade6B was introduced between the two HindIII sites after treatment with Klenow fragment (New England Biolabs) of pTN447 that contains a 2.1 kb Sau3A-EcoRV 5′ region of ura4 on pBluescript II SK+ (Stratagene), yielding pTN478. A 1.9 kb DraI fragment containing ade6X was introduced into the HindIII site after treatment with Klenow fragment of pTN448 that contains a 2.1 kb Blp1-BamHI 3′ region of ura4 on pBluescript II SK+, yielding pTN457. A 3.3 kb Eco81I–SalI fragment containing ade6X and ura4 3′ region from pTN457 was introduced between the Eco81I-SalI of pTN478, yielding pTN480.
pTN1015 that contains ade6X and imr1 was generated as follows: pTN905 was digested with NcoI and XhoI, treated with Klenow fragment and self-ligated to delete a 1.5 kb NcoI–XhoI region containing the 3′ region of ade6X, forming pTN1011. A 1.2 kb SpeI–EcoRI fragment containing imr1 from pRS140 was introduced between the SpeI–EcoRI sites of pTN1011, yielding pTN1015.
pTN1019 that contains dh, irc1R, ura4+ and tam14+ was generated as follows: a 2.7 kb Sau3AI partially digested genome fragment containing ura4+ and tam14+ was introduced at the BamHI site of pBluescript II SK+, yielding pTN444. pTN444 was then digested with ClaI and EcoRI, treated with Klenow fragment and self-ligated to yield pNT446. A 3.8 kb genomic region containing dh and irc1R was amplified from genomic DNA by PCR using NotI-NheI-otr1R and irc1R-BlpI primers and was then digested with XbaI and BlpI. A 1.4 kb XbaI–BlpI restriction fragment of the PCR product was introduced between the XbaI-BlpI sites of pTN446 to yield pTN1016. A 2.4 kb NotI–XbaI restriction fragment of the PCR product was introduced between the NotI-XbaI sites of pTN1016 to yield pTN1019.
To generate pTN1020, pTN1016 was digested with SpeI and BlpI, treated with Klenow fragment and self-ligated to eliminate a 1.8 kb SpeI-BlpI region containing ura4+.
pTN1024 containing dh, irc1L, ura4+ and new25 was generated as follows: pTN446 was digested with BsmBI and XbaI, treated with Klenow fragment and self-ligated to yield pTN1022. A 3.7 kb region containing irc1L and dh was amplified from genomic DNA by PCR using per1-Spe and otr-Bam primers and then digested with SpeI and BamHI. A 3.6 kb SpeI–BamHI fragment was introduced between the SpeI–BamHI sites of pTN1022 to yield pTN1023. A 2.3 kb AvrII–SacI fragment containing new25 from pTN446 was introduced between the AvrII–SacI sites of pTN1023 to yield pTN1024.
Recombination between ade6B and ade6X heteroalleles
Rates of spontaneous recombination between ade6B and ade6X heteroalleles were determined as described previously (35). Yeast strains containing the cen1-Sn, cen1-Hp or ura4-Sn(cen) construct were grown on YE+A plates for 3–5 days at the indicated temperatures. EMM+A media (10 ml) was inoculated with single colonies taken from the plates and was incubated for 1–2 days. After dilution with distilled water, cultures were plated onto EMM+A and EMM+G (EMM supplemented with 50 μg/ml of guanine) plates. After 3–6 days of incubation, colonies were counted and the rate of Ade+ formation was determined by a fluctuation test using the method of medians (48). To measure the recombination rate of ura4-Sn or ura4-Hp construct, all media were supplemented with uracil.
Southern blotting to determine crossovers and non-crossovers
Chromosomal DNA was prepared as described previously (35), with some modifications. After treatment with restriction enzymes (AfeI for cen1-Sn, cen1-Hp and ura4-Sn(cen); AfeI and SmaI for ura4-Sn and ura4-Hp), DNA fragments were separated using CHEF-DRII pulse field gel electrophoresis (Bio-Rad), 0.6% agarose gel (Certified molecular biology agarose, Bio-Rad) and 0.5 × TBE buffer at 6 V/cm, switching time from 1 to 6 s, for 11–15 h. In the case of ura4-Sn and ura4-Hp, DNA fragments were also separated by standard agarose gel electrophoresis using 0.8% agarose gel (PrimGel agarose LE 1–20k, Takara) in 0.5 × TBE buffer (89 mM Tris-borate, 2 mM EDTA). After running the gel, DNA was stained with 0.2 μg/ml ethidium bromide (EtBr) and detected using the Typhoon FLA9000 (General Electric). The gel was irradiated with 300 mJ ultraviolet (UV) light and placed into an alkali solution (1.2 M NaCl, 0.4 M NaOH) for 40 min. DNA was transferred by capillary action to a nylon membrane (ClearTrans Nylon membrane 0.45 μm, Wako) in 25 mM Na-phosphate buffer (pH 6.5) and covalently attached to the membrane by UV irradiation (150 mJ). Radioactive DNA probes were prepared using a Random Primer DNA Labeling Kit Ver.2 (TaKaRa) according to the manufacturer’s protocol. A 2.8 kb HindIII–EcoRI fragment containing cnt1 prepared from pKT110, a 0.5 kb XbaI–HindIII fragment containing the new25 downstream region from pTN446 and a 1.9 kb BamHI–PstI fragment containing a 1.9 kb DraI–DraI fragment containing the ade6B gene were used as DNA templates to prepare probes 1, 2 and 3, respectively. Radioactive signals were detected by using a BAS2500 phosphoimager (Fuji film) or a Typhoon FLA9000 and were measured using Image Gauge software (Fuji film).
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) was performed as described previously (49). Immunoprecipitation was carried out using anti-Cnp1 and anti-H3 rabbit antibodies (ab1791, Abcam) attached to Dynabeads M-280 sheep anti-Rabbit IgG (Invitrogen); anti-H3K9me2 mouse antibodies (50) attached to Dynabeads M-280 sheep anti-Mouse IgG (Invitrogen); and anti-Swi6 (51), anti-Cnp20 and anti-Mhf2 rabbit antibodies attached to Dynabeads Protein A (Invitrogen). Rabbit antibodies were raised against Cnp1: NH2-MAKKSLMAEPGDPIPRPRKKRC, Cnp20: NH2-CSLMQQYLSREIAPPAIKRT and Mhf2: NH2-CLELEDLENGIAAQLALDFS peptides (Sigma). Amounts of the input and the immunoprecipitated DNA were quantified by real-time PCR using Power SYBR green PCR master mix (Applied Biosystems) in a StepOnePlus real-time PCR System (Applied Biosystems). Sequences of the primers used for real-time PCR are listed in Supplementary Table S2.
Yeast two-hybrid assay
The Matchmaker Two-Hybrid System 3 (Clontech) was used for the yeast two-hybrid (Y2H) assay according to the manufacturer’s instructions. The proteins fused to the Gal4 DNA-binding domain and those fused to the GAL4 activation domain were expressed from the pGBKT7 and pGADT7 vectors, respectively, in AH109 Saccharomyces cerevisiae cells. pBridge (Clontech) three-hybrid vectors were used instead of pGBKT7 to examine the protein–protein interaction in the presence of a third protein. Using the SMARTer RACE 5′/3′ Kit (Clontech), cDNA of mhf1 and mhf2 was produced from total RNA prepared from fission yeast cells. The full-length of Mhf1 and Mhf2 was cloned into the plasmids. The plates were incubated at 22°C.
Gross chromosomal rearrangement (GCR) assay and Pulse field gel electrophoresis
Rates of spontaneous GCR were determined by means of fluctuation tests as previously described (35,36). Pulse field gel electrophoresis (PFGE) was carried out using CHEF-DRII (Bio-Rad) under the following conditions: switching time 1800–1000 s, 2 V/cm for 45 h and then 70 s for 3 h at 10°C in 1 × TAE buffer (40 mM Tris-acetate, 1 mM EDTA) and in a 0.55% agarose gel (Certified megabase agarose gel, Bio-Rad) for broad-range PFGE; and switching time 40–70 s, 4.5 V/cm for 24 h at 10°C in 0.5 × TBE buffer and in 0.6% agarose gel (Certified megabase agarose gel, Bio-Rad) for short-range PFGE. Southern hybridization was done using Random Primer DNA Labeling Kit Ver.2 (TaKaRa) and [α-32P] dCTP (Perkin-Elmer). DNA bands were detected with a BAS2500 phosphorimager (Fuji film). For PCR analysis, DNA was recovered from agarose gel using a FastGene Gel/PCR extraction kit (Nippon Genetics).
Statistical analysis
The Fisher’s exact test and the Mann–Whitney test were performed using GraphPad Prism version 6.0g for Mac (GraphPad Software, La Jolla, CA, USA). A student’s t-test was performed using Excel (Microsoft).
RESULTS
Rad51-dependent HR predominates in centromeres
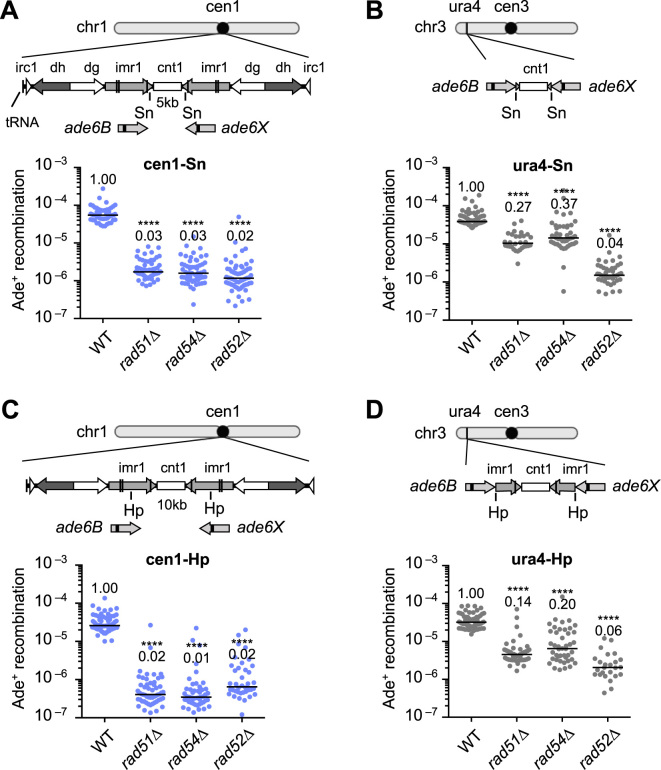
Rad51, Rad54 and Rad52 are all essential for Rad51-dependent HR, while Rad52 is also required for Rad51-independent SSA. To ascertain which type of HR occurs in centromeres, we introduced the ade6B and ade6X heteroalleles into cen1 and determined the rate of spontaneous recombination resulting in Ade+ prototroph formation (Figure 1). The cen1-Sn construct (Figure 1A) is similar to the cen1-Hp construct that we previously reported (35), but it contains the ade6B/X heteroallelles at the SnaBI (Sn) sites rather than the HpaI (Hp) sites in cen1. In the cen1-Sn construct, rad51, rad54 and rad52 deletions markedly decreased the recombination rate to the same level (Figure 1A), suggesting that Rad51-dependent HR is predominant in centromeres. To compare HR in centromere and non-centromere regions, we introduced the recombination cassette (the central region flanked by ade6B/X) into the ura4 locus on the arm region of chromosome 3 (Figure 1B). In wild-type (WT), recombination occurs at comparable rates in cen1-Sn and ura4-Sn constructs, indicating that mitotic recombination is not suppressed in centromeres. In ura4-Sn, however, rad51Δ and rad54Δ only partially decreased the recombination rate to ∼30% of the WT level, while rad52Δ severely reduced the recombination rate to 4% of the WT level, demonstrating that both Rad51-dependent HR and Rad51-independent SSA take place at the ura4 locus. To determine whether the different profiles of HR observed at the cen1 and ura4 loci are specific to the Sn sites or not, recombination rates were further assessed using cen1-Hp and ura4-Hp constructs in which ade6B/X are present in the middle of imr, HpaI (Hp) sites. Again Rad51, Rad54 and Rad52 were all essential for the recombination in cen1-Hp (Figure 1C) (35), whereas Rad51 and Rad54 were only partially required for recombination compared to Rad52 in ura4-Hp (Figure 1D). These results demonstrate that Rad51-dependent HR predominantly occurs in centromeres.
Figure 1.
Recombination between ade6B/ade6X heteroalleles in centromere and arm regions. (A) Recombination in the cen1-Sn construct. Illustrated are the central sequence cnt1 as well as the imr1, dg, dh and irc1 inverted repeats in the centromere 1 (cen1) of fission yeast. ade6B and ade6X mutant genes were integrated at the Sn sites in imr1. Spontaneous rates of Ade+ prototroph formation were determined in wild-type (WT), rad51Δ, rad54Δ and rad52Δ strains (TNF3347, 3446, 3452 and 3459, respectively). (B) Recombination in the ura4-Sn construct. From the cen1-Sn construct, the ade6B/X heteroalleles flanking the central region of cen1 were amplified and integrated at the ura4 locus. Recombination rates were determined in WT, rad51Δ, rad54Δ and rad52Δ strains (TNF3631, 3635, 3645 and 3643, respectively). (C) Recombination in the cen1-Hp construct. ade6B/X were integrated at the Hp sites in imr1 (35). Recombination rates were determined in WT, rad51Δ, rad54Δ and rad52Δ strains (TNF3144, 3257, 3286 and 3277, respectively). Recombination rates of WT, rad51Δ and rad54Δ in the cen1-Hp construct were previously published (35). (D) Recombination in the ura4-Hp construct. From the cen1-Hp construct, ade6B/X flanking the central region of cen1 were amplified and integrated at the ura4 locus. Recombination rates were determined in WT, rad51Δ, rad54Δ and rad52Δ strains (TNF3650, 3664, 3670 and 3667, respectively). Independent experimental values are shown in scatter plots and lines indicate medians. Rates relative to the WT value are indicated at the top of each column. P-values were determined by the two-tailed Mann–Whitney test. ****P < 0.0001. Sn, SnaBI; Hp, HpaI.
Crossovers are suppressed in centromeres
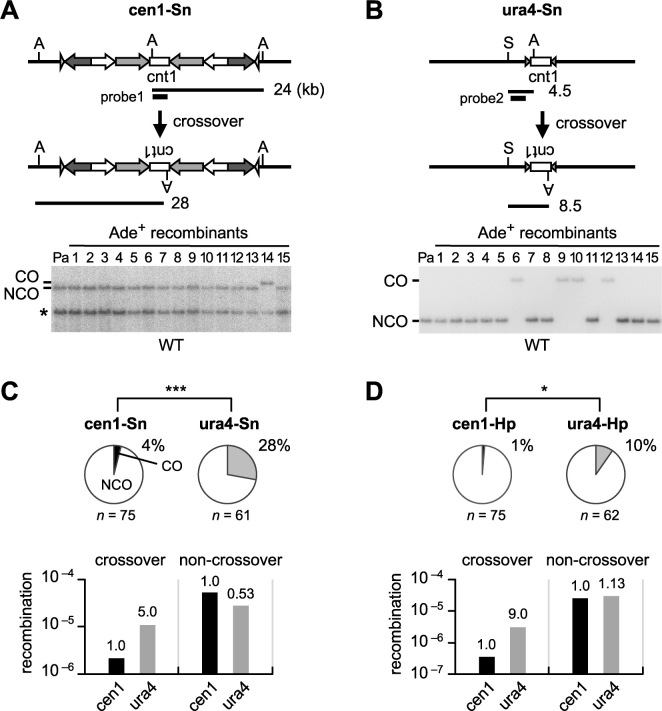
Crossover between non-allelic repeats leads to chromosome rearrangements, while non-crossover recombination retains the original configuration of chromosomes. To compare the rate of crossover between inverted repeats on the same chromatid (i.e. inversion) in centromere and arm regions (Figure 2), DNA was extracted from the parental and independent Ade+ recombinants, digested with restriction enzymes and separated by agarose gel electrophoresis. Southern hybridization was carried out to identify fragments representing crossover or non-crossover. It should be noted that crossovers that result in the isochromosome formation are not detected in this assay, as the isochromosome formation of native chromosomes is lethal in haploid cells. In cen1-Sn, only 4% of recombinants were crossovers (Figure 2A and C pie charts; Supplementary Figure S1), while in ura4-Sn, 28% of recombinants were crossovers (Figure 2B and C pie charts; Supplementary Figure S1). Net rates of crossover and non-crossover recombination were determined by multiplying recombination rates (Figure 1) by the proportion of crossovers and non-crossovers, respectively. It was revealed that crossover occurs 5-fold less frequently in cen1-Sn than in ura4-Sn, while non-crossover occurs at comparable levels in cen1-Sn and ura4-Sn (Figure 2C bar graphs). Likewise, crossover but not non-crossover was specifically suppressed by 9-fold in cen1-Hp compared to ura4-Hp (Figure 2D and Supplementary Figure S2). These findings demonstrate that crossovers between inverted repeats are suppressed in centromeres.
Figure 2.
Crossovers and non-crossovers between inverted repeats in centromere and arm regions. (A) Crossover and non-crossover recombinants produced in the cen1-Sn construct in WT (TNF3347). DNA was prepared, digested with AfeI, separated by pulse field gel electrophoresis (PFGE) (0.6% agarose, 0.5× TBE, 6 V/cm, 1–6 s switching time, for 11–15 h), transferred to a nylon membrane and subjected to Southern hybridization using probe1. An asterisk indicates the band derived from cen3. (B) Crossover and non-crossover recombinants produced in the ura4-Sn construct in WT (TNF3631). DNA was digested with AfeI and SmaI and separated by PFGE (0.6% agarose, 0.5× TBE, 6 V/cm, 1 to 5 s switching time, for 9 h). Probe2 was used for Southern hybridization. (C) Proportions of crossovers among recombinants in cen1-Sn and ura4-Sn constructs in WT are indicated in Pie charts. Net rates of crossover and non-crossover recombination are shown in bar graphs. (D) Proportions of crossovers and net rates of crossover and non-crossover recombination in cen1-Hp and ura4-Hp constructs in WT (TNF3144 and 3650, respectively). The cen1-Hp data were reported previously (35). Rates relative to the cen1 value are indicated at the top of each bar. P-values were obtained by the two-tailed Fisher’s exact test. *P < 0.05; ***P < 0.001. n, sample number; A, AfeI; S, SmaI; CO, crossover; NCO, non-crossover; Pa, parental.
Rad51-dependent HR preferentially produce non-crossovers in centromeres
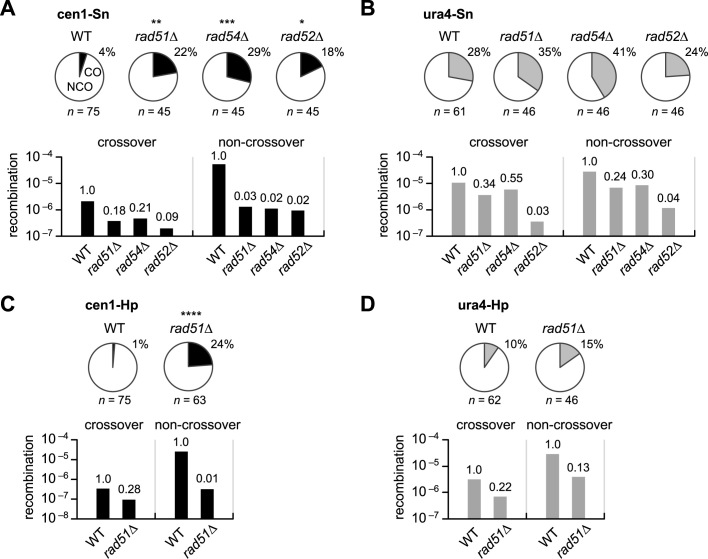
It is possible that Rad51-dependent HR has a strong preference for non-crossover recombination both in centromere and non-centromere regions and that the crossover suppression observed in centromeres is simply due to the prevalence of Rad51-dependent HR. To investigate this possibility, the crossover/non-crossover experiments were extended to the mutant strains (Figure 3). In cen1-Sn, rad51Δ, rad54Δ and rad52Δ significantly increased the proportion of crossovers (Figure 3A pie charts and Supplementary Figure S3). It should be noted that the mutations decreased the net rates of both crossover and non-crossover recombination (Figure 3A, bar graphs), although non-crossovers were dramatically decreased than crossovers. In contrast to cen1-Sn, none of the mutations significantly increased the proportion of crossovers in ura4-Sn (Figure 3B pie charts and Supplementary Figure S4). The mutations decreased crossover and non-crossover recombination to similar extents (Figure 3B bar graphs). Likewise, rad51Δ preferentially decreased non-crossover in cen1-Hp, but decreased both crossover and non-crossover to similar extents in ura4-Hp (Figure 3C and D; Supplementary Figure S5). These data show that Rad51-dependent HR has a strong preference for non-crossover recombination in centromeres.
Figure 3.
Rad51-dependent recombination preferentially promote non-crossovers in centromeres. (A) Proportions of crossovers among recombinants (pie charts) in the cen1-Sn construct in WT, rad51Δ, rad54Δ and rad52Δ strains (TNF3347, 3446, 3452 and 3459, respectively) and their net rates of crossover and non-crossover recombination (bar graphs). (B) Proportions of crossovers in the ura4-Sn construct in WT, rad51Δ, rad54Δ and rad52Δ strains (TNF3631, 3635, 3645 and 3643, respectively) and their net rates of crossover and non-crossover recombination. (C) Proportions and net rates of recombination in the cen1-Hp construct of WT and rad51Δ (TNF3144 and 3257, respectively). The proportions were published previously (35). (D) The proportions and net rates of recombination in the ura4-Hp construct of WT and rad51Δ (TNF3650 and 3664, respectively). **P < 0.01.
Pericentromeric heterochromatin does not suppress Rad51-independent SSA or crossovers
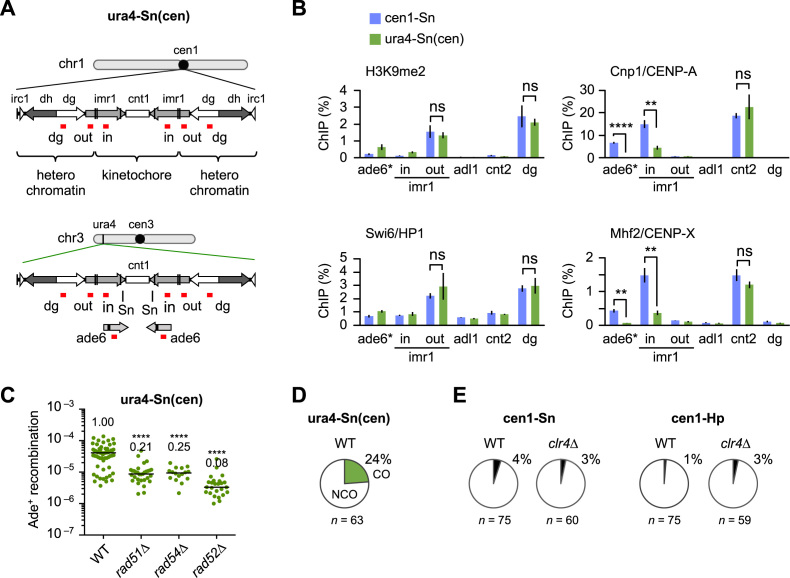
Between the cen1-Sn and ura4-Sn constructs, ade6B/X and the intervening sequence are identical but the flanking sequences are different. This raises the possibility that pericentromeric repeat regions where heterochromatin is assembled might be responsible for the specific properties of HR in centromeres. To test this, we introduced an entire region (∼40 kb) of cen1 including pericentromeric sequences into the ura4 locus (Figure 4A). This ura4-Sn(cen) construct contains the ade6B/X at the Sn site. Fragments of pericentromeric sequences are sufficient to induce the assembly of heterochromatin at an ectopic site (52). ChIP confirmed that H3K9 methylation and Swi6 binding in pericentromeric regions (imr1-out and dg) occur at similar levels in the cen1-Sn and ura4-Sn(cen) strains (Figure 4B). On the other hand, Cnp1/CENP–A and Mhf2/CENP–X were specifically bound to the central region (ade6 and imr1-in) in cen1-Sn in which ade6 is present in the original cen1. In ura4-Sn(cen), Cnp1 and Mhf2 were detected at background levels at ade6—present in the ectopic cen1—and were decreased by around half the cen1-Sn level at imr1-in. As imr1-in is present at both the original and ectopic cen1 in the ura4-Sn(cen) strain, Cnp1 and Mhf2 may not be binding to imr1-in at the ectopic site. In contrast, similar levels of Cnp1 as well as Mhf2 were observed at cnt2 of cen2 in cen1-Sn and ura4-Sn(cen) strains, showing that ectopic implantation of cen1 does not affect the formation of kinetochore chromatin on other centromeres. In contrast to heterochromatin, it seems that kinetochore chromatin is not assembled on the ectopic centromere in the ura4-Sn(cen) strain. This is probably due to the fact that di-centric chromosomes are extremely unstable in fission yeast (53). Analysis of recombination properties in ura4-Sn(cen) demonstrates that rad51Δ and rad54Δ only partially decrease recombination rates compared to rad52Δ (Figure 4C). The proportion of crossovers (24%) was comparable to that in ura4-Sn (28%) and different to that in cen1-Sn (4%, P < 0.001) (Figure 4D and Supplementary Figure S6). These data suggest that pericentromeric heterochromatin is not responsible for the regulation of recombination in the central region of centromeres. Consistent with this, a deletion of H3K9 methyltransferase Clr4 did not significantly change the proportion of crossovers in both cen1-Sn and cen1-Hp (Figure 4E and Supplementary Figure S7).
Figure 4.
Effects of pericentromere repeats on recombination. (A) Illustrated are the cen1 region on chr1 and the ectopic cen1 region introduced at the ura4 locus of chr3 in the ura4-Sn(cen) strain. Kinetochore chromatin and heterochromatin are assembled on the cen1. The positions of PCR amplification in chromatin immunoprecipitation (ChIP) analysis are shown in red. (B) Results of ChIP analysis conducted to examine H3K9me2, Swi6, Cnp1 and Mhf2 levels in the cen1-Sn and the ura4-Sn(cen) strains (TNF3347 and 4684). cnt2 and adl1 are in the centromere and arm regions of chr2, respectively. imr1-in and imr1-out are in the kinetochore and heterochromatin domains, respectively. ade6* is present in the original cen1 in cen1-Sn, while it is present only in the ectopic cen1 in ura4-Sn(cen). Data are represented as mean ± SEM from three biologically independent experiments. P-values were determined by the two-tailed student’s t-test. ns, statistically non-significant. (C) Recombination rates were determined in the ura4-Sn(cen) strain of WT, rad51Δ, rad54Δ and rad52Δ (TNF4684, 5814, 5826 and 5829, respectively). (D) Proportions of crossovers in the ura4-Sn(cen) construct in WT. (E) Proportions of crossovers in the cen1-Sn construct in WT and clr4Δ strains (TNF3347 and 3734, respectively) and in the cen1-Hp construct in WT and clr4Δ strains (TNF3144 and 3550, respectively).
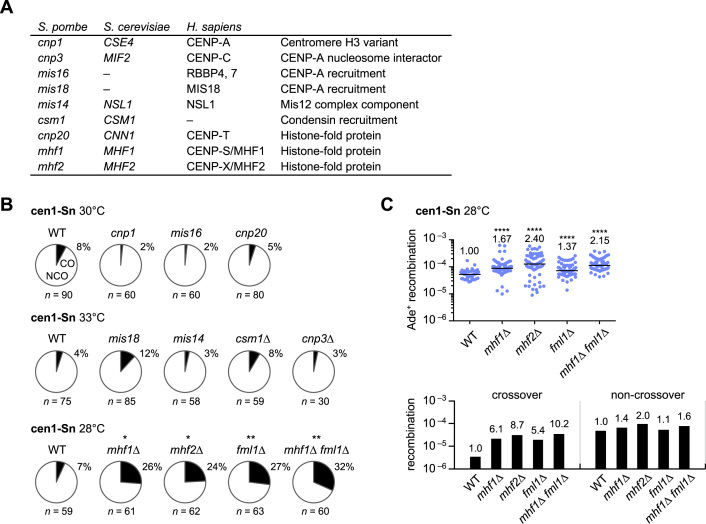
Mhf1/CENP–S, Mhf2/CENP–X and Fml1/FANCM suppress crossovers in centromeres
The above findings suggest that the factors related to the central domain rather than to pericentromeric heterochromatin regulate recombination in centromeres. To identify such factors, crossover proportions were assessed using a set of mutants listed in Figure 5A. cnp1–76 reduced Cnp1/CENP–A localization at centromeres as expected (Supplementary Figure S8) (54) but did not increase crossovers at a semipermissive temperature (Figure 5B and Supplementary Figure S9). A deletion of Cnp3/CENP–C (55) also showed no significant effects on crossovers. mis16–53 and mis18–262 impair Cnp1 localization (56), mis14–271 impairs Mis12 complexes (57) and csm1Δ reduces condensin at centromeres (58). None of these mutations significantly altered the proportion of crossovers at semipermissive temperatures (Figure 5B; the two-tailed Fisher’s exact test); however, deletions of Mhf1/CENP–S and Mhf2/CENP–X significantly increased the proportion of crossovers (Figure 5B). It is unlikely that CENP–T–W–S–X complexes (15) are responsible for the suppression of crossovers because cnp20-M447T of Cnp20/CENP–T reduced its centromere localization (Supplementary Figure S10) but did not increase crossovers (Figure 5B). Mhf1–Mhf2 recruits Fml1/FANCM to joint DNA molecules (20–22). fml1Δ increased crossovers to a similar level as mhf1Δ and mhf2Δ and it did not significantly increase the proportion of crossovers of mhf1Δ cells, demonstrating an epistatic relationship between Mhf1 and Fml1 in the crossover suppression. In contrast to rad51Δ, rad54Δ and rad52Δ (Figure 3A), mhf1Δ, mhf2Δ and fml1Δ preferentially increased the net rate of crossovers (Figure 5C). These results suggest that Mhf1/CENP–S and Mhf2/CENP–X with the aid of Fml1/FANCM helicase suppress crossovers in centromeres.
Figure 5.
Roles of the kinetochore proteins in centromere recombination. (A) List of kinetochore-related proteins examined in this study. (B) Proportions of crossovers in the mutants at their semi-permissive temperatures (30, 33 or 28°C). The cen1-Sn strain of WT, cnp1–76, mis16–53, cnp20-M447T, mis18–262, mis14–271, csm1Δ, cnp3Δ, mhf1Δ, mhf2Δ, fml1Δ and mhf1Δ fml1Δ (TNF3347, 3736, 4656, 5534, 4657, 5376, 4139, 4115, 4779, 5082, 5353 and 5128, respectively) were examined. (C) Recombination rates in the cen1-Sn strain of WT, mhf1Δ, mhf2Δ, fml1Δ and mhf1Δ fml1Δ strains at 28°C are shown in the scatter plot. Net rates of crossover and non-crossover recombination are shown in the bar graph.
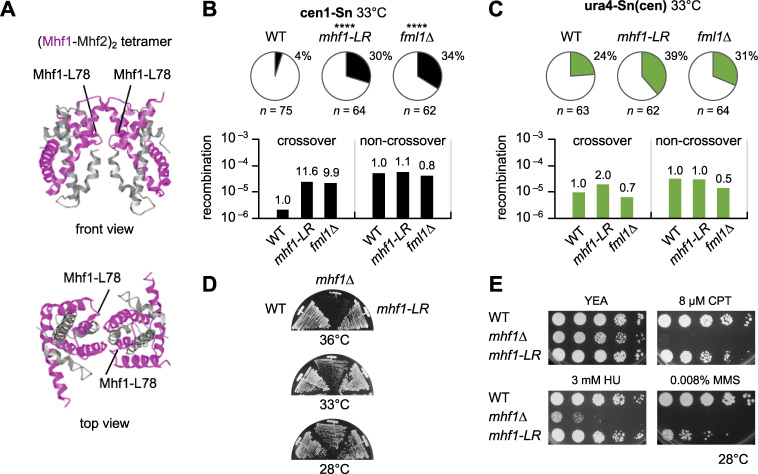
(Mhf1–Mhf2)2 tetramer formation is required for the crossover suppression in centromeres
Mhf1–Mhf2 dimers interact with each other to form (Mhf1–Mhf2)2 tetramers that preferentially bind to branched DNA (15,21,22,59). To see whether the tetramerization is important for the suppression of crossovers, the conserved leucine in the third α helix (α3) of Mhf1 that is involved in the interaction between Mhf1–Mhf2 dimers was mutated to arginine mhf1-L78R (Figure 6A). Y2H assays confirmed that mhf1-LR does not impair Mhf1–Mhf2 dimer but does abolish (Mhf1–Mhf2)2 tetramer formation (Supplementary Figure S11). In cen1-Sn, mhf1-LR—like fml1Δ—increased crossovers (Figure 6B and Supplementary Figure S12), suggesting that (Mhf1–Mhf2)2 tetramerization is required for the suppression of crossovers. Neither mhf1-LR nor fml1Δ, however, significantly increased crossovers in ura4-Sn(cen) (Figure 6C). It appears that (Mhf1–Mhf2)2 tetramers and Fml1 suppress crossovers in the context of functional centromeres.
Figure 6.
(Mhf1–Mhf2)2 tetramer formation is important for the suppression of crossovers in centromeres. (A) Ribbon diagram of the crystal structure of chicken (Mhf1–Mhf2)2 tetramers (15). The position of the conserved leucine residue in α3 helix, which corresponds to the Mhf1-L78 of fission yeast is indicated. (B) Proportions of crossovers and net rates of crossover and non-crossover recombination in the cen1-Sn strain of WT, mhf1-LR and fml1Δ (TNF3347, 5444 and 5353, respectively). (C) Proportion of crossovers and net rates of crossover and non-crossover recombination in the ura4-Sn(cen) strain of WT, mhf1-LR and fml1Δ (TNF4684, 5455 and 4806, respectively). (D) Growth of mhf1 mutants. WT, mhf1Δ and mhf1-LR strains (TNF3347, 4779 and 5444, respectively) were streaked on YE+A plates that contain adenine and were incubated at the indicated temperatures for 2 days. (E) Camptothecin (CPT), hydroxyurea (HU) and methyl methanesulphonate (MMS) sensitivities. Exponentially growing cells of WT, mhf1Δ and mhf1-LR strains (TNF3347, 4779 and 5444, respectively) were 5-fold serially diluted with distilled water and spotted on YE+A plates supplemented with the indicated concentrations of CPT, HU and MMS. Plates were incubated for 3–5 days at 28°C.
mhf1Δ cells exhibited a growth defect especially at high temperatures (19) (Figure 6D), while mhf1-LR cells grew well at all temperatures examined. A serial dilution assay showed that mhf1Δ cells are hypersensitive to camptothecin, methyl methanesulphonate and hydroxyurea, which cause replication problems and DNA damage (19,20) (Figure 6E). Compared to mhf1Δ cells, mhf1-LR cells were less sensitive to all the drugs tested. Collectively, these data suggest that (Mhf1–Mhf2)2 binding to branched DNA is required especially for the suppression of crossovers in centromeres.
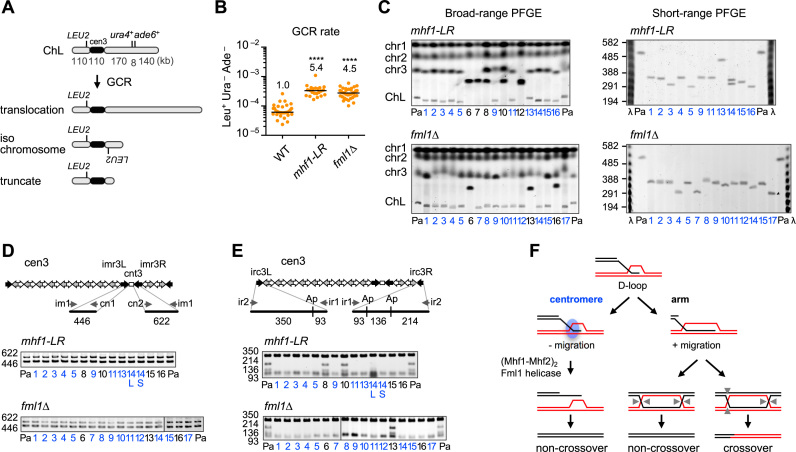
Mhf1 and Fml1 suppress gross chromosomal rearrangements in centromeres
Crossover between centromere repeats can give rise to GCRs. To see whether (Mhf1–Mhf2)2 and Fml1 suppress GCRs in centromeres, we determined the rate of spontaneous GCRs in mhf1-LR and fml1Δ strains (Figure 7A). To this aim, an extra-chromosome ChL that is derived from chr3 was used to detect otherwise lethal GCR events in haploid cells (36,60). Cells were grown in rich medium YE3S that contains leucine, uracil and adenine, and then plated onto YE plates, on which ade6– cells form red colonies. By the inspection of the red colonies using minimum medium, Leu+ Ade– Ura– clones that underwent GCRs were identified (Figure 7A) (35). A fluctuation analysis showed that both mhf1-LR and fml1Δ significantly increased the rate of GCRs compared to the WT (Figure 7B). To characterize GCR products, chromosomal DNA was prepared from the parental and independent GCR clones, separated by PFGE and stained with EtBr (Figure 7C). Large GCR products represent translocation between ChL and other chromosomes, while small products represent isochromosomes or truncates (Figure 7A). Translocations and isochromosomes have been detected at similar levels in WT (35). Broad-range PFGE revealed that more than half of the GCR products in the mhf1-LR and fml1Δ strains are small GCR products and short-range PFGE showed that the sizes of the small products match those of isochromosomes (≥300 kb; Figure 7C). The differences in size among isochromosomes is probably owing to the different copy number of dg/dh repeats. PCR analysis of GCR products that were recovered from the gel showed that both sides of cnt3–imr3 junctions were present in all the samples examined (Figure 7D); however, when the PCR product of irc3L and irc3R were digested, the restriction fragments indicative of irc3R (136 and 214 bp) were missing in all the small GCR products (Figure 7E), indicating that they are the isochromosomes whose breakpoints are present in centromere repeats. Some of the translocations were also shown to have lost irc3R in the fml1Δ mutant. These data show that (Mhf1–Mhf2)2 and Fml1 suppress GCRs that occur between centromere repeats.
Figure 7.
Mhf1 and Fml1 suppress gross chromosomal rearrangements (GCRs) in centromeres. (A) GCR assay using the extra chromosome ChL (36). GCRs associated with loss of the right arm of ChL result in Leu+ Ura– Ade–. The GCR product can be translocation, isochromosome and truncate of different lengths. (B) Spontaneous GCR rates in WT, mhf1-LR and fml1Δ strains (TNF3896, 5477 and 4813, respectively). Rates relative to WT values are indicated at the top of each column. P-values were determined by the two-tailed Mann–Whitney test. The GCR rate of WT was published previously (35). (C) Chromosomal DNA of mhf1-LR and fml1Δ were separated by broad-range and short-range PFGE and stained with ethidium bromide (EtBr) (see ‘Materials and Methods’ section). Positions of chr1, chr2, chr3 and the parental ChL are indicated on the left of the broad-range gel. Size of λ ladder (ProMega-Markers) bands are indicated on the left of the short-range gel. Pa, parental. Clone #6, 7 and 12 of mhf1-LR may have suffered complex rearrangements, resulting in two chr3 GCR products of similar sizes. Two GCR products of different sizes were detected in clone #14 of mhf1-LR, which may be due to the change in the copy number of centromere repeats. (D) PCR analysis of GCR products. PCR was carried out using ChL DNAs recovered from agarose gel using the indicated primers. cnt3–imr3 junctions were amplified and applied to standard agarose gel electrophoresis and stained with EtBr. (E) irc3L and irc3R regions were amplified and treated with ApoI. Ap, ApoI. WT data were reported previously (35). (F) Model of how (Mhf1–Mhf2)2 tetramers and Fml1 helicase suppress crossovers in centromeres. The 3′ single-stranded DNA (ssDNA) tail invades into homologous double-stranded DNA to form displacement-loops (D-loops). In the arm region, branch migration extends the length of the heteroduplex and stabilizes recombination intermediates, endonucleolytic cleavage of which results in either crossovers or non-crossovers. However, in the centromere, unidentified centromere proteins shown as a blue circle prevents branch migration, thereby stimulating (Mhf1–Mhf2)2 binding to branched DNA that recruits Fml1 helicase to dissociate D-loops, resulting in non-crossovers by synthesis-dependent strand annealing (SDSA) reactions. Arrow heads indicate endonucleotic cleavage sites of Holliday junctions.
DISCUSSION
We report here on the identification of a specific regulatory mechanism of recombination in centromeres. Rad51-dependent HR is predominant in the centromere, while both Rad51-dependent HR and Rad51-independent SSA occur in the arm region. Crossovers between inverted repeats on the same chromatid were suppressed in the centromere. Mhf1/CENP–S, Mhf2/CENP–X and Fml1/FANCM were required to suppress crossovers and GCRs in centromeres.
Since 1932 it has been observed in many organisms that meiotic recombination is under-represented around centromeres (12,61). We found that, unlike meiotic recombination, mitotic recombination was not suppressed in centromeres. Rad51, Rad54 and Rad52 were all essential for recombination at cen1, whereas Rad51 and Rad54 were only partially required compared to Rad52 at the ura4 locus. The different genetic requirements observed at cen1 and ura4 suggest that Rad51-dependent HR predominates in centromeres, while Rad51-independent SSA also occur in the arm region. Rad51-independent SSA seems specifically suppressed in centromeres, as it was observed in the arm region in budding yeast (62). In mammalian centromeres, SSA between tandem repeats causes deletions, resulting in inactivation of centromeres (63). Mammalian Rad52, which has a minor role in Rad51-dependent HR, promotes tumorigenesis (64,65). Thus, suppressing SSA in repetitive regions such as centromeres is important for maintaining genome integrity. Suppression of SSA might be owing to a lack of ssDNA, as small amounts of ssDNA-binding protein RPA has been shown to associate with centromere DNA during replication in Xenopus egg extracts (66). Post-translational modification might be also involved in the suppression of SSA. SUMOylation of Rad52 inhibits in vitro SSA activity and mutation of the SUMOylation site specifically increases deletions between tandem repeats (67–69).
Crossovers are associated with reciprocal exchange of chromosomal regions. This study showed that the proportion of crossovers between inverted repeats at cen1 is markedly lower than that of the ura4 locus. Crossovers but not non-crossovers were strongly decreased at cen1, showing a specific suppression of crossovers in centromeres. It is unlikely that the crossover suppression is simply due to the prevalence of Rad51-dependent HR. rad51 and rad54 deletions reduced non-crossovers more than crossovers at cen1, but they reduced non-crossovers and crossovers to similar extents at ura4, demonstrating a strong preference of Rad51-dependent HR for non-crossovers in centromere but not in arm regions. There are likely two distinct regulatory mechanisms behind the prevalence of Rad51-dependent HR and the crossover suppression in centromeres. Heterochromatin was assembled on pericentromeric repeats in the ura4-Sn(cen) construct; however, a high level of crossovers and Rad51-independent SSA were still observed, showing that neither an extension of homologous region nor heterochromatin assembly is sufficient to suppress crossovers and SSA at the ectopic site. Deletion of H3K9 methyltransferase Clr4 did not significantly increase the proportion of crossovers at cen1 while it slightly increased the total rate of recombination (Supplementary Figure S13), suggesting that heterochromatin assembly is not required for the specific suppression of crossovers in centromeres. Thus, it seems that the centromere proteins rather than heterochromatin factors suppress SSA and crossovers in centromeres. Surprisingly, however, Cnp1/CENP–A and its related proteins do not seem to be essential for the suppression of crossovers. A residual activity of the temperature-sensitive mutant protein might be sufficient for the crossover suppression at semipermissive temperatures. Indeed, mis18–262 increased the proportion of crossovers from 4 to 12% (Figure 5B) although the effect is statistically non-significant with the limited number of samples (P = 0.087, the two-tailed Fisher’s exact test). Thus, we do not exclude the possibility that Cnp1/CENP–A and its related proteins are involved in the crossover suppression in centromeres. Recently, using chromosome-orientation fluorescent in situ hybridization (CO-FISH) in human cells, Giunta and Fanabiki showed that CENP-A, CENP-C, CENP-T, and CENP-W protect centromere α-satellite repeats from illegitimate recombination (75), demonstrating that recombination between centromere repeats is controlled also in humans. Interestingly, they further showed that the sister chromatid exchange is increased in several cancer cell lines and during replicative senescence, suggesting an involvement of centromere instability in tumorigenesis and aging.
Mhf1/CENP–S and Mhf2/CENP–X form two different complexes: CENP–T–W–S–X (15,18,19) and (Mhf1–Mhf2)2 (15,19–22,59,70). This study found that mhf1 and mhf2 increase crossovers in centromeres. It seems that (Mhf1–Mhf2)2 rather than CENP–T–W–S–X is responsible for the crossover suppression, as cnp20-M447T did not increase crossovers but did decrease centromere localization of CENP–T. cnp20-M447T also decreased centromere localization of Mhf2 (Supplementary Figure S10), suggesting that the steady-state centromere localization of Mhf2 is dependent on CENP–T–W–S–X and it is not essential for the crossover suppression. fml1Δ decreased crossovers similar to mhf1Δ and mhf2Δ, and did not further increase crossovers in mhf1Δ cells, suggesting that Mhf1–Mhf2 and Fml1 suppress crossovers in the same pathway. In contrast to rad51, rad54 and rad52 mutations, mhf1, mhf2 and fml1 increase the net rate of crossovers, demonstrating that Mhf1–Mhf2 and Fml1 do suppress crossovers. Mhf1 and Mhf2 form Mhf1–Mhf2 heterodimers and interact with one another to form (Mhf1–Mhf2)2 tetramers that preferentially bind branched DNA and interact with FANCM helicase (15,22,59,71). mhf1-L78R did not disrupt Mhf1–Mhf2 interaction and caused only a mild defect in cell growth and mild sensitivity to DNA damaging agents, but abolished (Mhf1–Mhf2)2 tetramer formation and markedly increased crossovers at cen1. Thus, it is likely that (Mhf1–Mhf2)2 tetramers are particularly important in the crossover suppression and that Mhf1 may also have a function independent of tetramer formation. In the ura4-Sn(cen) construct, neither mhf1-LR nor fml1Δ increased crossovers significantly. It is unlikely that the heterochromatin ectopically assembled on ura4-Sn(cen) suppresses crossovers in place of Fml1, as fml1Δ did not significantly increase crossovers even in the absence of Clr4 (Supplementary Figure S14). We propose a model to explain how (Mhf1–Mhf2)2 and Fml1 suppress crossovers in centromeres (Figure 7F). In centromeres, unidentified centromere factors inhibit branch migration of joint molecules. Consistent with this, pausing of DNA replication forks has been observed at centromeres of budding yeast (72). Aberrant DNA structures such as DNA loops are accumulated in centromeres during replication in Xenopus egg extracts (66). Once branched DNA is stabilized, (Mhf1–Mhf2)2 binds it and recruits Fml1 helicase so as to disassemble joint molecules, resulting in synthesis-dependent strand annealing that generates only non-crossovers (73,74). In arm regions, branch migration extends heteroduplex, facilitating the formation of Holliday junctions, resolution of which results either in crossovers or non-crossovers. This study showed that mhf1-LR and fml1 increase the rate of GCRs whose breakpoints are present in centromere repeats. Previously, we showed that rad51Δ and rad54Δ increase the centromere GCRs (35,36). Therefore, two features of centromere recombination identified in this study: a predominance of Rad51-dependent HR and crossover suppression are important mechanisms by which homology-mediated chromosomal rearrangements is prevented in centromeres.
Supplementary Material
ACKNOWLEDGEMENTS
Some of the yeast mutant strains used in this study were obtained from the National Bio-Resource Project (NBRP), Japan. We thank Hiroshi Kimura (Tokyo Institute of Technology) for H3K9me2 antibodies, and Shiho Ogawa and Dayalini Weerasekara for comments on this manuscript.
Footnotes
Present addresses:
Yasuhiro Katahira, Department of Hematological Malignancy, Institute of Medical Science, Tokai University, 143 Shimokasuya, Isehara, Kanagawa 259-1193, Japan.
Tatsuro S. Takahashi, Department of Biology, Faculty of Science, Kyushu University, 744 Motooka, Nishi-ku, Fukuoka 819-0395, Japan.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
JSPS KAKENHI [JP21114513, JP23570212, JP26114711 to T.N.; JP23114005, JP26291072, JP16H01315 to J.N.]; Ninety-Nine Asia Student Scholarship Foundation Fellowship (to F.Z.); Japan Student Service Organization Fellowship (JASSO) (to F.Z.). Funding for open access charge: JSPS KAKENHI [JP21114513, JP23570212, JP26114711 to T.N.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Padeken J., Zeller P., Gasser S.M.. Repeat DNA in genome organization and stability. Curr. Opin. Genet. Dev. 2015; 31:12–19. [DOI] [PubMed] [Google Scholar]
- 2. de Koning A.P., Gu W., Castoe T.A., Batzer M.A., Pollock D.D.. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011; 7:e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tubbs A., Nussenzweig A.. Endogenous DNA damage as a source of genomic instability in cancer. Cell. 2017; 168:644–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Deininger P.L., Batzer M.A.. Alu repeats and human disease. Mol. Genet. Metab. 1999; 67:183–193. [DOI] [PubMed] [Google Scholar]
- 5. Campbell I.M., Gambin T., Dittwald P., Beck C.R., Shuvarikov A., Hixson P., Patel A., Gambin A., Shaw C.A., Rosenfeld J.A. et al. Human endogenous retroviral elements promote genome instability via non-allelic homologous recombination. BMC Biol. 2014; 12:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Argueso J.L., Westmoreland J., Mieczkowski P.A., Gawel M., Petes T.D., Resnick M.A.. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:11845–11850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Forsburg S.L., Shen K.F.. Centromere stability: the replication connection. Genes (Basel). 2017; 8:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aldrup-MacDonald M.E., Sullivan B.A.. The past, present, and future of human centromere genomics. Genes (Basel). 2014; 5:33–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Therman E., Susman B., Denniston C.. The nonrandom participation of human acrocentric chromosomes in Robertsonian translocations. Ann. Hum. Genet. 1989; 53:49–65. [DOI] [PubMed] [Google Scholar]
- 10. Putnam C.D., Pennaneach V., Kolodner R.D.. Saccharomyces cerevisiae as a model system to define the chromosomal instability phenotype. Mol. Cell. Biol. 2005; 25:7226–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rockmill B., Voelkel-Meiman K., Roeder G.S.. Centromere-proximal crossovers are associated with precocious separation of sister chromatids during meiosis in Saccharomyces cerevisiae. Genetics. 2006; 174:1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beadle G.W. A possible influence of the spindle fibre on crossing-over in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 1932; 18:160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Muller S., Almouzni G.. Chromatin dynamics during the cell cycle at centromeres. Nat. Rev. Genet. 2017; 18:192–208. [DOI] [PubMed] [Google Scholar]
- 14. McKinley K.L., Cheeseman I.M.. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 2016; 17:16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nishino T., Takeuchi K., Gascoigne K.E., Suzuki A., Hori T., Oyama T., Morikawa K., Cheeseman I.M., Fukagawa T.. CENP–T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell. 2012; 148:487–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hori T., Amano M., Suzuki A., Backer C.B., Welburn J.P., Dong Y., McEwen B.F., Shang W.H., Suzuki E., Okawa K. et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008; 135:1039–1052. [DOI] [PubMed] [Google Scholar]
- 17. Foltz D.R., Jansen L.E., Black B.E., Bailey A.O., Yates J.R. III, Cleveland D.W.. The human CENP–A centromeric nucleosome-associated complex. Nat. Cell Biol. 2006; 8:458–469. [DOI] [PubMed] [Google Scholar]
- 18. Amano M., Suzuki A., Hori T., Backer C., Okawa K., Cheeseman I.M., Fukagawa T.. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J. Cell Biol. 2009; 186:173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bhattacharjee S., Osman F., Feeney L., Lorenz A., Bryer C., Whitby M.C.. MHF1-2/CENP-S-X performs distinct roles in centromere metabolism and genetic recombination. Open Biol. 2013; 3:130102. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20. Yan Z., Delannoy M., Ling C., Daee D., Osman F., Muniandy P.A., Shen X., Oostra A.B., Du H., Steltenpool J. et al. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol. Cell. 2010; 37:865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singh T.R., Saro D., Ali A.M., Zheng X.F., Du C.H., Killen M.W., Sachpatzidis A., Wahengbam K., Pierce A.J., Xiong Y. et al. Mhf1–Mhf2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol. Cell. 2010; 37:879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao Q., Saro D., Sachpatzidis A., Singh T.R., Schlingman D., Zheng X.F., Mack A., Tsai M.S., Mochrie S., Regan L. et al. The MHF complex senses branched DNA by binding a pair of crossover DNA duplexes. Nat. Commun. 2014; 5:2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Allshire R.C., Ekwall K.. Epigenetic regulation of chromatin states in Schizosaccharomyces pombe. Cold Spring Harb. Perspect. Biol. 2015; 7:a018770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bloom K.S. Centromeric heterochromatin: the primordial segregation machine. Annu. Rev. Genet. 2014; 48:457–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shang W.H., Hori T., Martins N.M., Toyoda A., Misu S., Monma N., Hiratani I., Maeshima K., Ikeo K., Fujiyama A. et al. Chromosome engineering allows the efficient isolation of vertebrate neocentromeres. Dev. Cell. 2013; 24:635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rea S., Eisenhaber F., O’Carroll D., Strahl B.D., Sun Z.W., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000; 406:593–599. [DOI] [PubMed] [Google Scholar]
- 27. Nakayama J., Rice J.C., Strahl B.D., Allis C.D., Grewal S.I.. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001; 292:110–113. [DOI] [PubMed] [Google Scholar]
- 28. Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T.. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001; 410:120–124. [DOI] [PubMed] [Google Scholar]
- 29. Peters A.H., O’Carroll D., Scherthan H., Mechtler K., Sauer S., Schofer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A. et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001; 107:323–337. [DOI] [PubMed] [Google Scholar]
- 30. Chiolo I., Minoda A., Colmenares S.U., Polyzos A., Costes S.V., Karpen G.H.. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011; 144:732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zeller P., Padeken J., van Schendel R., Kalck V., Tijsterman M., Gasser S.M.. Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat. Genet. 2016; 48:1385–1395. [DOI] [PubMed] [Google Scholar]
- 32. Symington L.S., Rothstein R., Lisby M.. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics. 2014; 198:795–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ceballos S.J., Heyer W.D.. Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. Biochim. Biophys. Acta. 2011; 1809:509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yu V.P., Koehler M., Steinlein C., Schmid M., Hanakahi L.A., van Gool A.J., West S.C., Venkitaraman A.R.. Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 2000; 14:1400–1406. [PMC free article] [PubMed] [Google Scholar]
- 35. Onaka A.T., Toyofuku N., Inoue T., Okita A.K., Sagawa M., Su J., Shitanda T., Matsuyama R., Zafar F., Takahashi T.S. et al. Rad51 and Rad54 promote noncrossover recombination between centromere repeats on the same chromatid to prevent isochromosome formation. Nucleic Acids Res. 2016; 44:10744–10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nakamura K., Okamoto A., Katou Y., Yadani C., Shitanda T., Kaweeteerawat C., Takahashi T.S., Itoh T., Shirahige K., Masukata H. et al. Rad51 suppresses gross chromosomal rearrangement at centromere in Schizosaccharomyces pombe. EMBO J. 2008; 27:3036–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Putnam C.D., Hayes T.K., Kolodner R.D.. Specific pathways prevent duplication-mediated genome rearrangements. Nature. 2009; 460:984–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ivanov E.L., Sugawara N., Fishman-Lobell J., Haber J.E.. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996; 142:693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mortensen U.H., Bendixen C., Sunjevaric I., Rothstein R.. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:10729–10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kagawa W., Kurumizaka H., Ishitani R., Fukai S., Nureki O., Shibata T., Yokoyama S.. Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. Mol. Cell. 2002; 10:359–371. [DOI] [PubMed] [Google Scholar]
- 41. Pannunzio N.R., Manthey G.M., Bailis A.M.. RAD59 is required for efficient repair of simultaneous double-strand breaks resulting in translocations in Saccharomyces cerevisiae. DNA Repair (Amst). 2008; 7:788–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bhargava R., Onyango D.O., Stark J.M.. Regulation of single-strand annealing and its role in genome maintenance. Trends Genet. 2016; 32:566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Elliott B., Richardson C., Jasin M.. Chromosomal translocation mechanisms at intronic alu elements in mammalian cells. Mol. Cell. 2005; 17:885–894. [DOI] [PubMed] [Google Scholar]
- 44. Hentges P., Van Driessche B., Tafforeau L., Vandenhaute J., Carr A.M.. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast. 2005; 22:1013–1019. [DOI] [PubMed] [Google Scholar]
- 45. Gibson D.G., Benders G.A., Axelrod K.C., Zaveri J., Algire M.A., Moodie M., Montague M.G., Venter J.C., Smith H.O., Hutchison C.A. III. One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:20404–20409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chikashige Y., Kinoshita N., Nakaseko Y., Matsumoto T., Murakami S., Niwa O., Yanagida M.. Composite motifs and repeat symmetry in S. pombe centromeres: direct analysis by integration of NotI restriction sites. Cell. 1989; 57:739–751. [DOI] [PubMed] [Google Scholar]
- 47. Takahashi K., Murakami S., Chikashige Y., Funabiki H., Niwa O., Yanagida M.. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell. 1992; 3:819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lin M., Chang C.J., Green N.S.. A new method for estimating high mutation rates in cultured cells. Mutat. Res. 1996; 351:105–116. [DOI] [PubMed] [Google Scholar]
- 49. Maki K., Inoue T., Onaka A., Hashizume H., Somete N., Kobayashi Y., Murakami S., Shigaki C., Takahashi T.S., Masukata H. et al. Abundance of prereplicative complexes (Pre-RCs) facilitates recombinational repair under replication stress in fission yeast. J. Biol. Chem. 2011; 286:41701–41710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hayashi-Takanaka Y., Yamagata K., Wakayama T., Stasevich T.J., Kainuma T., Tsurimoto T., Tachibana M., Shinkai Y., Kurumizaka H., Nozaki N. et al. Tracking epigenetic histone modifications in single cells using Fab-based live endogenous modification labeling. Nucleic Acids Res. 2011; 39:6475–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nakayama J., Klar A.J., Grewal S.I.. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell. 2000; 101:307–317. [DOI] [PubMed] [Google Scholar]
- 52. Partridge J.F., Scott K.S.C., Bannister A.J., Kouzarides T., Allshire R.C.. cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr. Biol. 2002; 12:1652–1660. [DOI] [PubMed] [Google Scholar]
- 53. Sato H., Masuda F., Takayama Y., Takahashi K., Saitoh S.. Epigenetic inactivation and subsequent heterochromatinization of a centromere stabilize dicentric chromosomes. Curr. Biol. 2012; 22:658–667. [DOI] [PubMed] [Google Scholar]
- 54. Castillo A.G., Mellone B.G., Partridge J.F., Richardson W., Hamilton G.L., Allshire R.C., Pidoux A.L.. Plasticity of fission yeast CENP-A chromatin driven by relative levels of histone H3 and H4. PLoS Genet. 2007; 3:e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tanaka K., Chang H.L., Kagami A., Watanabe Y.. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev. Cell. 2009; 17:334–343. [DOI] [PubMed] [Google Scholar]
- 56. Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., Yanagida M.. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004; 118:715–729. [DOI] [PubMed] [Google Scholar]
- 57. Weir J.R., Faesen A.C., Klare K., Petrovic A., Basilico F., Fischbock J., Pentakota S., Keller J., Pesenti M.E., Pan D. et al. Insights from biochemical reconstitution into the architecture of human kinetochores. Nature. 2016; 537:249–253. [DOI] [PubMed] [Google Scholar]
- 58. Tada K., Susumu H., Sakuno T., Watanabe Y.. Condensin association with histone H2A shapes mitotic chromosomes. Nature. 2011; 474:477–483. [DOI] [PubMed] [Google Scholar]
- 59. Yang H., Zhang T., Tao Y., Wu L., Li H.T., Zhou J.Q., Zhong C., Ding J.. Saccharomyces cerevisiae MHF complex structurally resembles the histones (H3-H4)(2) heterotetramer and functions as a heterotetramer. Structure. 2012; 20:364–370. [DOI] [PubMed] [Google Scholar]
- 60. Niwa O., Matsumoto T., Yanagida M.. Construction of a mini-chromosome by deletion and its mitotic and meiotic behaviour in fission yeast. Mol. Gen. Genet. 1986; 203:397–405. [Google Scholar]
- 61. Talbert P.B., Henikoff S.. Centromeres convert but don’t cross. PLoS Biol. 2010; 8:e1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rattray A.J., Symington L.S.. Use of a chromosomal inverted repeat to demonstrate that the RAD51 and RAD52 genes of Saccharomyces cerevisiae have different roles in mitotic recombination. Genetics. 1994; 138:587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stimpson K.M., Song I.Y., Jauch A., Holtgreve-Grez H., Hayden K.E., Bridger J.M., Sullivan B.A.. Telomere disruption results in non-random formation of de novo dicentric chromosomes involving acrocentric human chromosomes. PLoS Genet. 2010; 6:e1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bhowmick R., Minocherhomji S., Hickson I.D.. RAD52 facilitates mitotic DNA synthesis following replication stress. Mol. Cell. 2016; 64:1117–1126. [DOI] [PubMed] [Google Scholar]
- 65. Sotiriou S.K., Kamileri I., Lugli N., Evangelou K., Da-Re C., Huber F., Padayachy L., Tardy S., Nicati N.L., Barriot S. et al. Mammalian RAD52 functions in break-induced replication repair of collapsed DNA replication forks. Mol. Cell. 2016; 64:1127–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Aze A., Sannino V., Soffientini P., Bachi A., Costanzo V.. Centromeric DNA replication reconstitution reveals DNA loops and ATR checkpoint suppression. Nat. Cell Biol. 2016; 18:684–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sacher M., Pfander B., Hoege C., Jentsch S.. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat. Cell Biol. 2006; 8:1284–1290. [DOI] [PubMed] [Google Scholar]
- 68. Torres-Rosell J., Sunjevaric I., De Piccoli G., Sacher M., Eckert-Boulet N., Reid R., Jentsch S., Rothstein R., Aragon L., Lisby M.. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 2007; 9:923–931. [DOI] [PubMed] [Google Scholar]
- 69. Ho J.C., Warr N.J., Shimizu H., Watts F.Z.. SUMO modification of Rad22, the Schizosaccharomyces pombe homologue of the recombination protein Rad52. Nucleic Acids Res. 2001; 29:4179–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sun W., Nandi S., Osman F., Ahn J.S., Jakovleska J., Lorenz A., Whitby M.C.. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol. Cell. 2008; 32:118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tao Y., Jin C., Li X., Qi S., Chu L., Niu L., Yao X., Teng M.. The structure of the FANCM-MHF complex reveals physical features for functional assembly. Nat. Commun. 2012; 3:782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Greenfeder S.A., Newlon C.S.. Replication forks pause at yeast centromeres. Mol. Cell. Biol. 1992; 12:4056–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Prakash R., Satory D., Dray E., Papusha A., Scheller J., Kramer W., Krejci L., Klein H., Haber J.E., Sung P. et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009; 23:67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Nassif N., Penney J., Pal S., Engels W.R., Gloor G.B.. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 1994; 14:1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Giunta S., Funabiki H.. Integrity of the human centromere DNA repeats is protected by CENP-A, CENP-C, and CENP-T. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:1928–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.