Abstract
CRISPR–Cas (clustered regularly interspaced short palindromic repeats-CRISPR-associated) systems provide adaptive immunity against invasive nucleic acids guided by CRISPR RNAs (crRNAs) in archaea and bacteria. Type III CRISPR–Cas effector complexes show RNA cleavage and RNA-activated DNA cleavage activity, representing the only known system of dual nucleic acid interference. Here, we investigated the function of Cmr1 by genetic assays of DNA and RNA interference activity in the mutants and biochemical characterization of their mutated Cmr complexes. Three cmr1α mutants were constructed including ΔβΔ1α, Δβ1α-M1 and Δβ1α-M2 among which the last two mutants carried a double and a quadruple mutation in the first α-helix region of Cmr1α. Whereas the double mutation of Cmr1α (W58A and F59A) greatly influenced target RNA capture, the quadruple mutation almost abolished crRNA binding to Cmr1α. We found that Cmr2α-6α formed a stable core complex that is active in both RNA and DNA cleavage and that Cmr1α strongly enhances the basal activity of the core complex upon incorporation into the ribonucleoprotein complex. Therefore, Cmr1 functions as an integral activation module in III-B systems, and the unique occurrence of Cmr1 in III-B systems may reflect the adaptive evolution of type III CRISPR–Cas systems in thermophiles.
INTRODUCTION
CRISPR–Cas (clustered regularly interspaced short palindromic repeats, CRISPR- associated) systems are composed of CRISPR loci and cas gene cassettes, which code for CRISPR RNAs (crRNAs) and Cas proteins, respectively. The system mediates the small RNA-guided nucleic acid interference to defend against invasive genetic elements in archaea and bacteria (1–4). There are three distinct stages in the adaptive immunity: adaptation, crRNA biogenesis and invading nucleic acid (NA) interference. In the first stage, two nucleases (Cas1 and Cas2) are required for acquisition of new spacers in all tested bacteria and archaea, which function together with some type-specific Cas proteins (5,6). At the second step, CRISPR loci are expressed as long precursor RNAs from which mature crRNAs are generated (7). During the final interference stage, mature crRNA and Cas proteins form a ribonucleoprotein complex (crRNP) that recognizes incoming foreign DNA (or RNA) by their sequence complementarity with crRNA and targets the homologous sequence for cleavage (8).
In the latest classification scheme, CRISPR–Cas systems fall into two main classes based on the number of Cas proteins required for invading NA silencing. Whereas multiple Cas proteins are required for NA interference in class 1 CRISPR–Cas systems, only a single Cas protein is required by each of class 2 systems (2). At least six different types of CRISPR–Cas systems are known (1,2) among which only type III systems have been shown to mediate dual DNA and RNA interference and their DNA interference is transcription-dependent (9–12). Several type III ribonucleoprotein complexes (RNPs) have been isolated and characterized, and all tested RNPs show the RNA-activated DNA cleavage activity (12–16). Furthermore, the DNA shredding activity can be strongly elevated when substrate DNA is present in a large excess amount (16).
All known type III-B Cmr complexes of dual NA interference contain six Cmr proteins that form RNPs containing crRNAs of two distinct sizes, i.e. 39 nt and 45 nt in Pyrococcus furiosus Cmr (PfuCmr) and 40 nt and 46 nt in Thermus thermophilus Cmr (TthCmr) and Sulfolobus islandicus Cmr-α (SisCmr-α) (16–19). In a structural model of PfuCmr based on structures of individual Cmr proteins (20) and a near-atomic resolution of TthCmr structure solved by cryo-electron microscopy reconstructions (21), Cmr1 is an integral part of each effector complex. Nevertheless, a stable hybrid Cmr complex lacking Cmr1 has been obtained and used for determination of the first crystal structure for a type III effector complex (22). Furthermore, it has been shown that crRNA binding requires only four Cmr proteins, i.e. Cmr2–5, but target RNA binding requires the two additional proteins, Cmr1 and Cmr6 (23). In addition, Cmr complexes lacking Cmr1 are functional in RNA cleavage (22,24). Together, this has raised a question about whether Cmr1 has an essential role in III-B CRISPR–Cas systems.
We employ S. islandicus REY15A (25) as the model for investigation of CRISPR mechanisms since a comprehensive array of genetic tools has been developed for this archaeon (26). Genetic tools developed for studying CRISPR mechanisms include invader plasmid assays for DNA interference by different CRISPR–Cas systems (9,27), an artificial mini-CRISPR-based RNA interference assay (10) and in vivo site-directed gene mutagenesis using endogenous CRISPR–Cas systems (26,28). The archaeon contains a complete I-A CRISPR–Cas system and two III-B Cmr modules named Cmr-α and Cmr-β all of which have been characterized. The I-A system has been shown to mediate the Protospacer Adjacent Motif (PAM)-dependent DNA interference (27,29) and is active in adaptation of new spacers (30). The Cmr-α system confers dual DNA and RNA interference (10) and the DNA interference is transcription-dependent (9) whereas the Cmr-β system only mediates RNA interference (10).
Here, we aimed to investigate the functions of Cmr1α in the dual DNA/RNA interference by the S. islandicus Cmr-α. Mutants of cmr1α gene deletion and amino acids substitutions were conducted and employed for in vivo genetic analysis on Cmr-α function and for biochemical characterization of Cmr-α complex isolated from each mutant. We found that Cmr1α functions as an activation module in Cmr-α to strongly enhance its activity and the protein achieves the activation by strongly binding to both target RNA and crRNA.
MATERIALS AND METHODS
Strains, growth conditions and transformation of Sulfolobus
All Sulfolobus strains were derived from the original isolate S. islandicus REY15A (31). S. islandicus E233 and Δβ were reported previously (28,32). Mutants of cmr1α deletion mutant and two alanine-substitution mutations were generated by following the CRISPR-based genome-editing method recently developed in our laboratory (28). Genetic hosts and plasmids employed in this work are listed in Supplementary Tables S1 and S2, respectively. S. islandicus strains were grown at 78°C in SCV medium (basic salts plus 0.2% sucrose, 0.2% casamino acids plus 1% vitamin solution) or SCVy (SCV + 0.0025% yeast extract), with uracil supplemented to 20 μg/ml if required. Sulfolobus competent cells were prepared and transformed by electroporation as previously described (32).
Construction of plasmids
Protospacer SS1 of the lacS gene was employed for gene silencing previously using a plasmid carrying an artificial CRISPR array containing SS1 spacer (10). Genome-editing plasmids were constructed as described previously (28) using the oligonucleotides listed in Supplementary Table S3. Sulfolobus expression vector pSeSD1 (33) was employed for construction of a C-terminal 10xHis-tagged cmr6α by inserting its coding sequence into the vector at the NdeI and StuI sites. Then, the fusion gene was amplified by PCR using the primer pair of MCS-fwd and MCS-rev (Supplementary Table S3). The resulting PCR product was digested with SmaI and XhoI, and inserted into pAC10-SS1 (Supplementary Table S2) at SalI and SmaI sites, giving pAC-cmr6α-10His. All the oligonucleotides were synthesized from Tsingke (Wuhan, China) and the sequences of all plasmid constructs were verified by DNA sequencing (Tsingke, Wuhan, China).
Determination of in vivo RNA interference efficiency by qRT-PCR
Total RNAs were prepared from three transformants of each mutant harboring either pAC-SS1 or pSe-Rp (A600 = 0.3, absorbance at 600 nm) as described previously (34). 1 μg RNA was treated with DNase I (Thermo Fisher Scientific) in a total volume of 20 μl from which 2 μl treated RNA was used for generating complementary DNA (cDNA) with the RevertAid Reverse Transcriptase (Thermo Scientific) using the lacS-RT-R primer aligning to a downstream position of the lacS gene (Figure 1A, Supplementary Table S3). Two primer sets, Qtar and Qref that were designed for amplification of PCR products in the target region and in a reference region of the mRNA, respectively, were employed for PCR amplification using iTaq™ Unicersal SYBR® Green Supermix (Bio-Rad) and a CFX384 Real-Time System (Bio-Rad) with the following PCR condition: denaturing at 95°C for 5 min, 40 cycles of 95°C 15 s, 55°C 15 s and 72°C 20 s. Data analysis was performed using the comparative Ct value method (35). The amount of uncut mRNA is expressed as the ratio between the amount of SS1 RNA protospacer in pAC-SS1 transformants and that in the corresponding pSe-RP transformants.
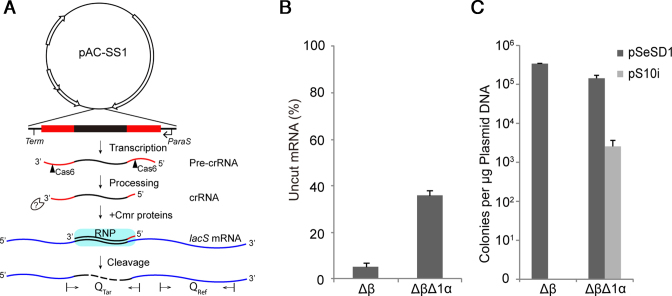
Figure 1.
Effect of cmr1α deletion on RNA and DNA interference by Cmr-α in S. islandicus. (A) Schematic of in vivo RNA interference activity assay in S. islandicus using artificial CRISPR plasmids (pAC). pAC-SS1, an artificial mini-CRISPR plasmid producing a crRNA that guides Cmr-α proteins to target the SS1 protospacer of lacS mRNAs for degradation. Two primer sets, Qtar and Qref designed for amplification of PCR products from the target region and a reference region, respectively, are indicated on the lacS mRNA. (B) Quantification of mRNAs expressed from the chromosomal lacS gene in different S. islandicus strains by qRT-PCR. Total RNAs were extracted from transformants of pAC-SS1 and pSe-RP, the latter of which a Sulfolobus artificial mini-CRISPR cloning vector (reference plasmid), and lacS mRNA levels were estimated by qRT-PCR. Error bars represent standard deviations of replicates determined for three independent transformants for each plasmid. Amounts of uncut mRNA are determined as the ratio of the amounts of SS1 RNA protospacer in pAC-SS1 transformants versus those determined for the corresponding pSe-RP transformants. (C) In vivo DNA interference activity assayed in different S. islandicus strains by an invader plasmid assay. pSeSD1—a shuttle vector for Sulfolobus, pS10i—an invader plasmid carrying a target sequence of spacer 10 in CRISPR locus 2 in S. islandicus REY15A. Δβ—S. islandicus Δβ: a reference strain that carries a deletion of the complete Cmr-β gene cassette; ΔβΔ1α—S. islandicus ΔβΔ1α: derived from S. islandicus Δβ from which the cmr1α gene was deleted. All strains were derived from the genetic host S. islandicus E233 that carries the β-glycosidase (lacS) gene.
Purification of Cmr-α crRNA ribonucleoprotein complex
S. islandicus strains (Δβ, ΔβΔ1α, Δβ1αM1 and Δβ1αM2) carrying pAC-cmr6α-10His were grown in SCVy medium, and cells were collected from at least 6 L of culture for each strain by centrifugation when A600 of the culture reached 0.7–0.8. Cell pellet was re-suspended in Buffer A (20 mM HEPES pH 7.5, 20 mM Imidazole, 250 mM NaCl) and disrupted by French press and centrifuged for 30 min at 12 000 rpm. Supernatant was loaded onto a 1 ml HisTrap HP column (GE Healthcare) pre-equilibrated with Buffer A. After washing with 15 ml of Buffer A, protein bound to the column was eluted with linear gradient of imidazole (20–500 mM) generated by mixing Buffer A and Buffer B (20 mM HEPES pH 7.5, 500 mM Imidazole, 250 mM NaCl). Sample fractions were analyzed by SDS-PAGE, and those containing Cmr-α effector complexes were pooled together, concentrated and further purified by size exclusion chromatography in Buffer C (20 mM Tris–HCl pH 8.0, 500 mM NaCl) with a Superdex 200 Hiload column (GE Healthcare). After SDS-PAGE analysis, the fractions containing the complete set of Cmr-α subunits were pooled together and concentrated. The purified Cmr-α complexes were used for further analysis.
Extraction and analysis of crRNA from the Cmr-α ribonucleoprotein complex
One hundred microliters of purified Cmr-α complexes (about 10 μg protein for each) were mixed with 600 μl Trizol agent (Sigma) and 300 μl chloroform in the indicated order. After incubation at room temperature for 5 min, the mixture was centrifuged at 12 000 rpm for 15 min. The upper phase was transferred into a new tube and precipitated by one volume of isopropanol, and RNA precipitants were recovered as a pellet by centrifugation. After washing with 1 ml of 75% ethanol pre-chilled on ice, the RNA pellet was air-dried for 30 min at room temperature, dissolved in 15 μl DEPC-H2O, and used for further analysis.
An aliquot of the purified crRNA was first 5′ labeled by 32P-γ-ATP (PerkinElmer) using T4 polynucleotide kinase (Thermo Fisher Scientific), and then separated on a large denatured polyacrylamide gel (12%, 40 cm long, containing 40% urea). The labeled RNAs were identified by exposing the gel to a phosphor screen (GE Healthcare), and scanned by using a Typhoon FLA 7000 laser scanner (GE Healthcare). RNA ladders were generated by Decade™ Marker RNA (Ambion) and labeled by 32 P with T4 polynucleotide kinase.
Labeling of DNA and RNA substrates
DNA and RNA substrates used in cleavage assays were 5′ labeled with 32P-γ-ATP using T4 polynucleotide kinase (Thermo Fisher Scientific). All nucleic acids were purified by recovering the corresponding bands from a denaturing polyacrylamide gel. DNA and RNA oligonucleotides to be used as substrate for cleavage assays were purchased from IDT, USA.
RNA cleavage and DNA cleavage assay
RNA and DNA cleavage assays were conducted as described previously (16). The reaction mixture (10 μl in total) contains 50 mM Tris–Cl (pH 7,6), 10 mM MgCl2, 5 mM DTT, and indicated amount of complex and substrates. In the DNA cleavage assay, 200 nM (unless otherwise indicated) unlabeled RNA was supplemented into the reaction mixture to activate DNA cleavage activity. The reaction was performed at 70°C and stopped at the indicated time point by the addition of 2 × RNA loading dye (New England Biolabs) and cooling on ice. Finally, the samples were heated for 5 min at 95°C and separated on denaturing polyacrylamide gels (18% polyacrylamide, 40% urea) and visualized by phosphor imaging using the Typhoon laser scanner. Amounts of RNA/DNA products and remaining NA substrates in each assay were extracted from the images using the Image Quant TL analysis software of the Typhoon scanner. The percentage was normalized by those present in reaction without Cmr-α effector complex and averaged with three times measurement.
Target RNA binding assay
The binding assay was conducted with the amounts of Cmr-α complex indicated in each assay with the binding buffer of 50 mM Tris–Cl (pH 7,6), 10 mM MgCl2, 5 mM DTT. 25 nM of 32P-5′-labeled target RNA was added to each reaction and incubated for 5 min at 70°C. Then, 2 × RNA loading dye was added to each reaction. The formation of Cmr–α-crRNA:target RNA tertiary complexes in the reactions was detected by gel electrophoresis on a 10% non-denaturing polyacrylamide gel. Radioactive signals were visualized by phosphor imaging using the Typhoon FLA 7000 laser scanner (GE Healthcare).
RESULTS
Depletion of Cmr1α by gene deletion eliminated in vivo RNA interference and strongly impaired the transcription-dependent DNA interference of Cmr-α in S. islandicus
Previous studies show that Cmr effector complexes lacking Cmr1 are either active or inactive in RNA cleavage in vitro (14,17,22,24). To investigate whether Cmr1 could have an essential function in vivo, we constructed a deletion mutant of the corresponding gene in S. islandicus REY15A. The strain carries two III-B Cmr modules named Cmr-α and Cmr-β, both of which showed RNA interference activity in a previous experiment (34). In order to eliminate any possible influence on functional analysis of Cmr1α by Cmr-β, a mutant devoid of the Cmr-β system was constructed, giving S. islandicus E233 Δβ (denoted Δβ), and further deletion of cmr1α yielded the mutant carrying Δcmr1α and Δcmr-β mutant alleles (ΔβΔ1α). The latter was suitable for genetic analysis of Cmr1α function using Δβ as a reference.
RNA interference in ΔβΔ1α was analyzed by the mini-CRISPR assay reported previously (Figure 1A) (10). The artificial CRISPR plasmid pAC-SS1 containing spacers matching the lacS protospacer 1 (SS1) was introduced into the double deletion mutant by electroporation, and the amount of uncleaved lacS mRNA in the transformants were determined by qRT-PCR with two sets of primer, Qtar and Qref. Whereas qRT-PCR with the latter set of primer estimates the total lacS mRNA level in the RNA preparations, amplification with the former set of primer reveals the level of uncut lacS mRNAs since the amplified region contains the SS1 RNA protospacer, which is to be disrupted by Cmr-α RNA cleavage in the presence of SS1 crRNA. As shown in Figure 1B, more than 90% of lacS mRNAs were cleaved in the reference strain (S. islandicus Δβ) and at least 35% mRNAs retained in ΔβΔ1α cells. These results indicated that depletion of Cmr1α by gene deletion strongly inhibited the Cmr-α RNA interference in the S. islandicus mutant.
The transcription-dependent DNA interference was evaluated by the invader plasmid assay (also called interference plasmid assay), using an invader plasmid carrying an artificial protospacer of spacer 10 of CRISPR locus 2 in a reverse orientation (pS10i). As a result, transcription of S10i on the plasmid produces a RNA protospacer that guides DNA interference by III-B systems. The same invader plasmid was employed for studying Cmr-α DNA interference in a previous work (9). As shown in Figure 1C, whereas transformation of the wild-type Cmr-α strain (Δβ) with pSeSD yielded 2 × 105 transformants/μg DNA, transformation with pS10i did not give any transformants. In comparison, whereas transformation of the double mutant with the reference plasmid showed a similar rate of transformation as for the wild-type strain, transformation of ΔβΔ1α with the invader plasmid gave ∼100-fold reduction in transformation rate (Figure 1C). These data indicated that Cmr-1α should play an important role in the Cmr-mediated DNA interference in this crenarchaeon.
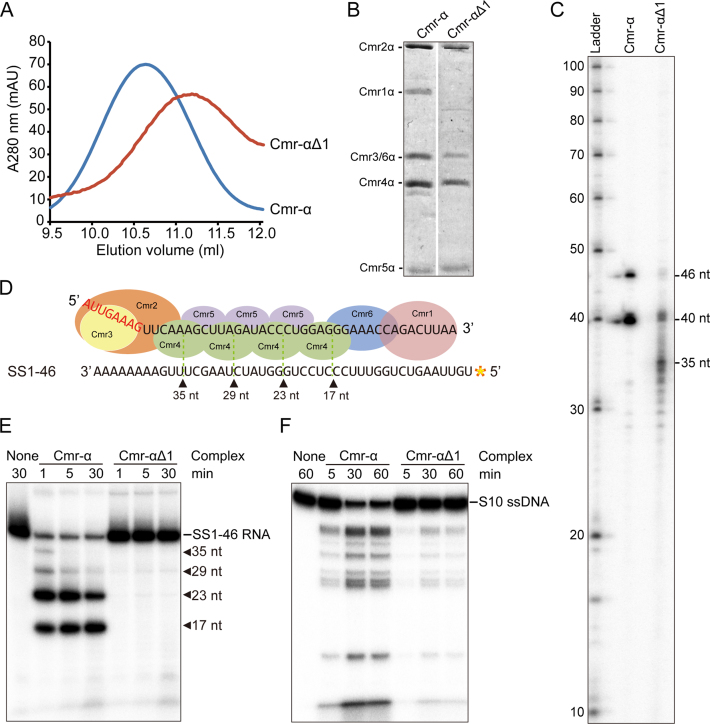
Cmr2α-6α formed a stable core nucleoprotein complex in vivo, exhibiting little residual RNA and DNA cleavage activity
Recently we have established a procedure to express a define crRNA and a His-tagged Cmr6α from pAC-cmr6α in S. islandicus, and purify a native Cmr-α RNP by His-tagged Cmr6α copurification (Supplementary Figure S1A, S1B). The same expression plasmid was introduced into S. islandicus ΔβΔ1α by electroporation, and the resulting transformants were used for isolating the Cmr-α complex by the two-step purification procedure. In gel filtration, the UV absorbance peak of the Cmr complex isolated from ΔβΔ1α appeared 0.5 ml later than that of the wild-type Cmr-α (Figure 2A, 10.5 ml versus 11 ml), suggesting that the former complex could be smaller in size than the latter. Analyzing the protein components of the fractions collected in the peak region showed that Cmr1α is indeed absent from the Cmr-α complex isolated from ΔβΔ1α (Figure 2B), indicating that five Cmr proteins (Cmr2α-6α) formed protein complex in vivo. The RNA component of the complex was then examined by RNA extraction and radio-labeling of the extracted RNA, and subsequently, denatured polyacrylamide gel analysis of the labeled products. As shown in Figure 2C, while the vast majority of crRNAs extracted from the wild-type Cmr-α complex were 40 and 46 nt in size, crRNAs present in Cmr-αΔ1 were smaller in size: majority of crRNAs appeared as a sequencing ladder from 28–35 nt, although RNA bands of 46 nt and 40 nt were still visible. These results suggested that crRNA could be more vulnerable to exonuclease degradation in the absence of Cmr1α. Nevertheless, the data showed that the effector complex formed by Cmr2α-6α contains crRNAs (Figure 2C). The ribonucleoprotein complex was designated Cmr-αΔ1 and characterized.
Figure 2.
The core Cmr-α complex lacking Cmr1 is active in nucleic acid interference. (A) Gel filtration profiles of Cmr-α (blue) and Cmr-αΔ1 (red). The latter is composed of Cmr2–6α. A280: UV absorbance at 280 nm. (B) SDS-PAGE analysis of purified Cmr-α and Cmr-αΔ1 complexes. (C) crRNAs present in Cmr-α and Cmr-αΔ1 complexes. RNAs were extracted from Cmr-α and Cmr-αΔ1, 5′-labeled with radio-active γ-32P-ATP and analyzed by denaturing PAGE. Ladder: RNA size ladder (nt). (D) Schematic of cleavage sites on SS1–46 RNA by Cmr-α complex. (E) RNA cleavage assay. 50 nM Cmr-α and Cmr-αΔ1 complexes were incubated with 25 nM labeled SS1–46 RNA for indicated time points. Then, the samples were analyzed by denaturing PAGE. (F) RNA-activated DNA cleavage assay. Each reaction contained 25 nM of labeled ssDNA, 200 nM SS1–46 RNA and 50 nM effector complex. After incubation at 70°C for 1 h, cleavage products were analyzed by denaturing PAGE.
First, we analyzed the in vitro RNA cleavage by Cmr-αΔ1 as described previously since the effector complex only contained a single type of crRNAs matching the lacS SS1 protospacer (Supplementary Table S4). First, SS1–46 RNA was radio-labeled at 5′-end, which is complementary to the spacer region of the crRNA but has mismatches to the 5′-handle sequence of cRNA (Figure 2D). Then the labeled RNA was incubated with either Cmr-α or Cmr-αΔ1 for the indicated times and analyzed by denaturing gel electrophoresis. As shown in Figure 2E, the wild-type Cmr-α cleaved majority of RNA substrate within 1 min incubation whereas Cmr-αΔ1 produced weak signals of multiple RNA cleavage products after 30 min incubation. Indeed, quantification of uncleaved RNA substrate revealed that ca. 85% of RNA substrate was retained in the Cmr-αΔ1 cleavage reaction, indicative of a low RNA cleavage activity. These data suggested that the Cmr1α subunit is very important for the backbone RNA cleavage by Cmr-α.
The RNA-activated DNA cleavage by Cmr-αΔ1 was then tested using a non-specific DNA substrate, S10 ssDNA that does not match the crRNA in the ribonucleoprotein complex (Supplementary Table S4). The reaction was set up with 200 nM SS1–46 RNA, 25 nM S10 ssDNA and 50 nM Cmr-α complexes and incubated for 60 min during which samples were taken at different time points. The reaction was stopped by addition of the loading buffer, and the cleavage products were analyzed by denaturing gel electrophoresis. As shown in Figure 2F, Cmr-α and Cmr-αΔ1 showed the same pattern of DNA cleavage. Quantification of residual DNA substrate in each reaction revealed that, while wild-type Cmr-α cleaved ∼55% of DNA substrate within 5 min, it required 60 min for Cmr-αΔ1 to cleave 58% of DNA substrate. These results indicated that Cmr-αΔ1 represents a core III-B effector complex that shows both RNA and DNA cleavage activity in vitro and transcription-dependent DNA interference in vivo.
Substitution mutation of a few well-conserved amino acids in Cmr1α exerted different effects on DNA and RNA interference by Cmr-α in S. islandicus
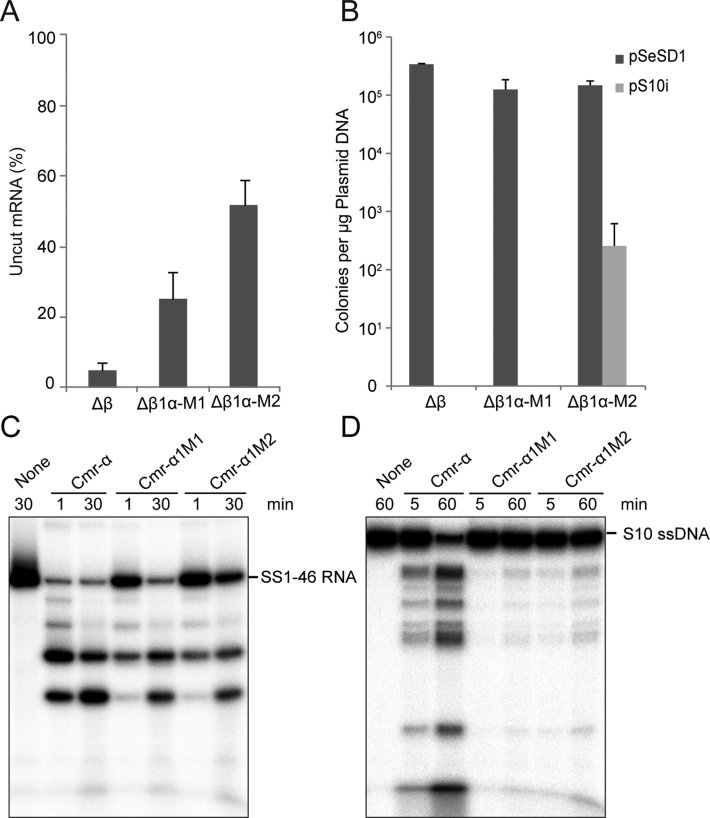
Multiple sequence alignments of a few Cmr1 proteins revealed several highly conserved amino acids in the first alpha helix of the protein (Supplementary Figures S2 and S3). Using the SWISS-MODEL web-server (36–39), a structural model was generated for the S. islandicus Cmr1α protein based on the structure of AfCmr1 (PDB id. 4l6u) (40) and PfCmr1 (PDB id. 4w8x) (20). Two groups of highly conserved amino acids were identified in the Cmr1α structural model: the first one containing two hydrophobic amino acids (W58 and F59, positions in S. islandicus Cmr1α) that were found to bind nucleotides in a previous study (20), and the other group of four invariant amino acids (I52, G54, R57 and R61) that positioned differently in the model (Supplementary Figure S3). Since the hand-like groove was predicted to bind RNA in Cmr1, each set of amino acids was subjected to substitution mutagenesis using a recently developed CRISPR-based in vivo gene mutagenesis. Transformation of S. islandicus Δβ with each genome-editing plasmid (Supplementary Table S2) yielded cmr1α mutant 1 (Δβ1α-m1) coding for Cmr1αW58AF59A and cmr1α mutant 2 (Δβ1α-m2) coding for Cmr1αI52AG54AR57AR61A (Supplementary Figure S4).
The two mutants were then employed for investigating RNA interference and transcription-dependent DNA interference together with the wild-type reference (the Δβ strain). We found that, while pAC-SS1-guided RNA interference strongly reduced lacS mRNA (95%) in the wild-type Cmr-α strain RNA interference, it reduced the target RNAs by 70% and 50% in Δβ1α-m1 and Δβ1α-m2, respectively (Figure 3A). The Δβ1α-m2 mutant was still active in RNA interference albeit at a lowered level, which showed similarity to the system present in the cmr1α deletion mutant (ΔβΔ1α, Figure 1A). These results indicated that the two hydrophobic amino acids (W58, F59) in the α1 helix could be very important for the Cmr-α RNA interference whereas the four invariant amino acids should have an essential function in RNA interference by Cmr-α (Figure 3A). Together, these data suggested that the surface groove in Cmr1 could have multiple functions in RNA interference.
Figure 3.
Influence of Cmr1α RNA-binding activity on the in vivo nucleic acid interference and in vitro RNA/DNA cleavage by the Cmr-α system. RNA interference (A) and transcription-dependent DNA interference (B) were determined for the cmr1α mutants as described in the legends of Figure 1B and C. RNA cleavage activity (C) and RNA-activated DNA cleavage activity (D) were assayed as described in the legends of Figure 2C and D. Δβ—S. islandicus Δβ, a reference strain carrying a deletion of the complete Cmr-β gene cassette; Δβ1α-M1—S. islandicus Δβ1α-M1: derived from S. islandicus Δβ with which substitution mutations of W58A and F59A were generated on the cmr1α gene. Δβ1α-M2—S. islandicus Δβ1α-M2: derived from S. islandicus Δβ with which substitution mutations of I52A, G54A, R57A and R61A were generated on the cmr1α gene. Cmr-α1M1: mutant Cmr-α complex purified from S. islandicus Δβ1α-M1 (containing Cmr1αW58A,F59A); Cmr-α1M2: mutant Cmr-α complex purified from S. islandicus Δβ1α-M2 (carrying a much reduced level of Cmr1αI52A, G54A, R57A,R61A).
We also employed the invader plasmid assay to test the transcription-dependent DNA interference by the III-B system in both cmr1α mutants. Strikingly, transformation of the wild-type Cmr-α strain and Δβ1α-m1 with the invader plasmid produced no or very few colonies whereas transformation of Δβ1α-m2 with the same plasmid gave hundreds fold higher transformation rate (Figure 3B). Nevertheless, the transformation efficiency with Δβ1α-m2 is still hundreds fold lower than that obtained with the cloning vector pSeSD, a reference plasmid (Figure 3B). These results indicated that, whereas W58A and F59A substitution in Cmr1α did not influence the transcription-dependent DNA interference by the Cmr-α system, mutation of the four invariant amino acids (I52A, G54A, R57A and R61A) strongly impaired the DNA interference activity.
Mutant Cmr-α complex exhibited distinctive RNA and DNA cleavage activity in vitro
To gain a further insight into the function of Cmr1α in antiviral immunity by Cmr-α, RNP was purified from each mutant by following the procedure described for purification of the Cmr-α complex. We found that profiles of UV absorbance in the gel filtration purification fell into two distinct groups: while the profile of the Cmr-α complex purified from the Δβ1α-m1 mutant was very similar to that of the wild-type effector complex, the UV profile of the effector complex obtained from Δβ1α-m2 resembled that of Cmr-αΔ1 (Supplementary Figure S5A). These two mutant complexes were named as Cmr-α1M1 and Cmr-α1M2, respectively, and further characterized.
SDS-PAGE analysis of Cmr-α1M1 and Cmr-α1M2 showed that a protein band corresponding to Cmr1α was present in the former but could be hardly detected in the latter (Supplementary Figure S5B). Analysis of their crRNAs revealed that crRNAs extracted from Cmr-α1M1 resembled those present in the wild-type Cmr-α whereas crRNAs present in Cmr-α1M2 resembled those isolated from Cmr-αΔ1 (Supplementary Figure S5C). These results suggested that, while W58A and F59A substitutions in Cmr1α did not affect the capability of the protein to form RNP, substitution of I52A G54A R57A and R61A strongly impaired the interaction between Cmr1α and other components of the RNP, either Cmr proteins or crRNA.
Both mutant Cmr-α effector complexes were analyzed for RNA cleavage using a radio-labeled SS1–46 RNA substrate as for Cmr-αΔ1. To better assess the cleavage activity of Cmr-α1M1 and Cmr-α1M2, 50 nM of the enzyme was used in RNA cleavage reaction for each mutant complex. Cleavage products were analyzed by denaturing gel electrophoresis and the results were shown in Figure 3C. Quantification of the cleavage products revealed that, after 1 min incubation, up to 80% of RNA substrate was cleaved by the wild-type Cmr-α whereas only 5% of RNA substrate was destructed by Cmr-α1M1 and Cmr-α1M2. Nevertheless, both mutant complexes showed RNA cleavage, and the pattern of RNA cleavage remained the same.
Both mutant effector complexes were then assayed for their RNA-activated DNA cleavage activity using a radio-labeled S10 ssDNA as the substrate. Again, 50 nM of Cmr1 mutant complex was used in each assay. As shown in Figure 3D, weaker DNA cleavage activity was observed for each mutant complex. Quantification of uncleaved DNA substrate showed that ca. 50% of ssDNA was cleaved by the WT Cmr-α in 5 min, <5% of DNA substrate was cleaved by Cmr-α1M1 and Cmr-α1M2. After incubation of 60 min, additional 40% of ssDNA was cleaved by each of the Cmr complexes. Furthermore, these results suggested that the two sets of amino acids in Cmr1α should have different roles in NA cleavage by the effector complex.
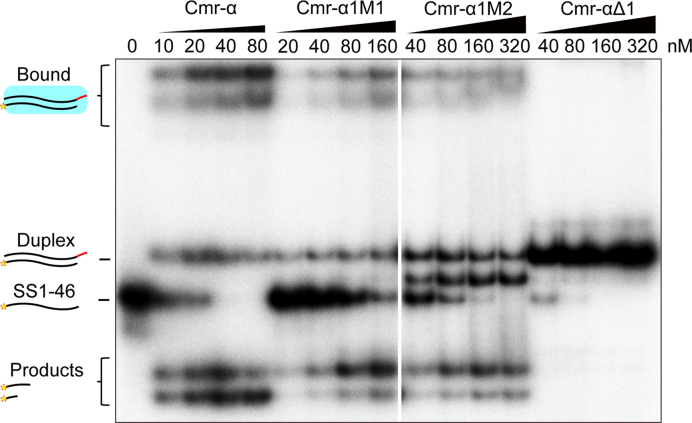
To yield an insight into whether Cmr1α could influence the RNA and DNA cleavage via RNA binding, an electrophoretic gel mobility shift assay was employed to examine the interaction between the target RNA and different types of Cmr-α effector complexes purified in this work, including Cmr-α, Cmr-αΔ1, Cmr-α1M1 and Cmr-α1M2. As shown in Figure 4, only Cmr-αΔ1 was not capable of forming any stable RNP::target RNA tertiary complexes under the experimental conditions. Furthermore, tertiary complexes formed by the remaining three RNPs showed different stabilities: The super-complexes formed by 10 nM Cmr-α were comparable to those formed by 160 nM Cmr-α1M1 and those by 320 nM Cmr-α1M2 (Figure 4). These data indicated that the first α-helix domain of Cmr1α plays a very important function in anchoring cognate target RNA in the effector complex of III-B Cmr-α system.
Figure 4.
Conserved amino acids in the Cmr1α ribonucleotide-binding groove are important for target RNA binding. Twenty-five nM of labeled SS1–46 RNA was mixed with the amounts of Cmr-α, Cmr-α1M1, Cmr-α1M2 or Cmr-αΔ1 indicated in each experiment. After incubation for 15 min, 2× loading buffer was added to each reaction. The samples were then analyzed by non-denaturing polyacrylamide gel electrophoresis. The amounts of effector complex used in the assay are: 10–80 nM Cmr-α, 20–160 nM Cmr-α1M1, 40–320 nM Cmr-α1M2 or Cmr-αΔ1. Duplex: RNA complex of crRNA and target RNA; Bound: Cmr-α-crRNA:target RNA tertiary complexes.
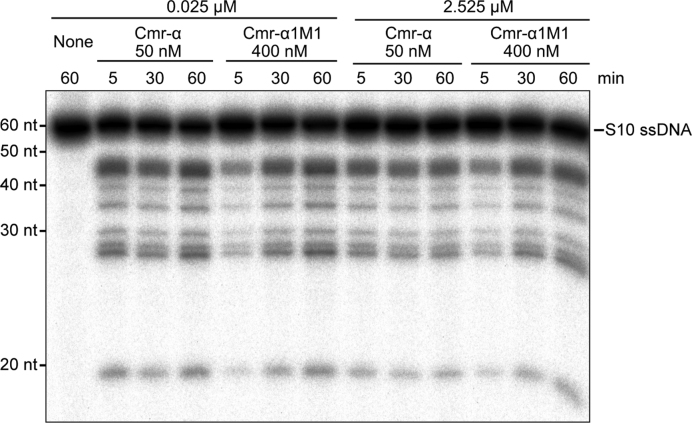
Another important feature for the S. islandicus Cmr-α system is that the effector complex is capable of mediating massive DNA destruction (16). To investigate if the activity is Cmr1α dependent, we tested whether Cmr-α1M1 could exert the massive ssDNA cleavage activity recently revealed for the wild-type Cmr-α except that 8-fold of Cmr-α1M1 was used in the assay. As shown in Figure 5, very similar patterns of DNA cleavage were observed for substrate concentrations of 100-fold difference for both the wild-type RNP and the mutant RNP. These results indicated that reduction of target RNA binding did not impair the massive ssDNA destruction by the Cmr-α system.
Figure 5.
Cmr1α-M1 does not impact the massive DNA cleavage of Cmr-α complex. Twenty-five nM labeled S10 ssDNA substrate were incubated with 50 nM Cmr-α/400 nM Cmr-α1M1 and 200 nM SS1 RNA, in the presence or absence of 2.5 μM of unlabeled S10 ssDNA as indicated. Reaction was conducted for 60 min during which samples were taken at 5, 30 and 60 min and analyzed by denaturing PAGE.
In conclusion, our genetic and biochemical studies on the S. islandicus Cmr-α system have revealed that Cmr1α is not essential for NA interference by the III-B Cmr-α system but the protein strongly facilitates the interference activity of the antiviral system and the stimulation depends on Cmr1α binding activity to both crRNA and target RNA in the effector complex.
DISCUSSION
Recently, it has been shown that III-A and III-B systems mediate transcription-dependent DNA interference in vivo whereas their effector complexes exert RNA-activated DNA cleavage in vitro (reviewed in (12)). The active sites for RNA and DNA cleavage have been identified. RNA cleavage occurs at each of the large subunits of the helical backbone in the complex (13–15,22), i.e. Cmr4 for III-B RNPs and Csm3 for III-A effector complexes whereas active sites for DNA cleavage reside on Cas10 (Cmr2/Csm1), the signature protein of type III CRISPR–Cas systems (13–16,41). The RNA-activated DNA cleavage process is subjected to a spatiotemporal regulation (15): (a) the binding of a cognate target RNA to the corresponding binary RNP complex yields a tertiary RNP complex that is an active DNase. The formation of tertiary RNP involves a conformational change that activates the DNase activity; (b) upon the target RNA cleavage and release of cleaved RNA products, the tertiary status returns to the binary status that is an inactive DNase. Furthermore, the S. islandicus Cmr-α shows massive ssDNA destruction when DNA substrate is present in a large amount (16).
Two active RNPs have been reported for several III-B Cmr systems. The smaller ones have been reconstituted with five different Cmr proteins (Cmr2–6) of P. furiosus, T. thermophilus, T. maritima and a hybrid complex (composed of PfCmr2dHD-Cmr3 and Cmr4-Cmr5-Cmr6 from A. fulgidus), which, together with the native Cmr-αΔ1 complex purified from S. islandicus in this work, are termed as the core Cmr complex. Several core Cmr complexes are reported to be active in RNA cleavage in vitro (14,17,22–24). Consistent with these observations, Cmr-αΔ1, the core complex of the S. islandicus Cmr-α system is also active in RNA cleavage. Here we have investigated in vivo RNA interference activity of the core Cmr-α system using S. islandicus ΔβΔ1α, the cmr1α deletion mutant. Initially, the mini-CRISPR-based reporter gene assay was employed to reveal in vivo RNA interference in the cmr1α deletion mutant, but it failed to reveal any activity (data not shown). Then, qRT-PCR was employed to directly analyze the quantity of mRNAs of lacS, the reporter gene. RNA interference activity was detected in these cmr1α mutants. Furthermore, the mutant (ΔβΔ1α) is active in DNA interference as revealed by invader plasmid assay, albeit at a level of 100-fold lower than the activity present in the wild-type strain. To this end, Cmr1α can be regarded as an integral activation module in the Cmr-α effector complex and functions in elevating both RNA and DNA interference by the III-B system.
Comparative studies of the wild-type Cmr-α and its three Cmr1α mutant derivative complexes have yielded important insights into the molecular mechanisms of Cmr1α activation. (a) Cmr1α stabilizes crRNA in the effector complex since crRNAs in Cmr-αΔ1 are not only shorter, but also degenerated, in contrast to the strictly 40 and 46 nt crRNAs present in Cmr-α (Figure 2C). (b) Cmr1α hosts the key interaction between crRNA and its cognate target RNA since a seed sequence identified by in vivo RNA interference assay for the S. islandicus Cmr-α system is located in 28–30 nt of crRNAs (10) interacts with Cmr1 proteins in all known Cmr structural models (18–22). (c) We have shown that alanine substitutions of W58 and F59 in Cmr1α have not influenced its capability of crRNA binding but greatly reduced the interaction of the mutated effector complex with the cognate target RNA (Figure 4). Further, it has been shown that the P. furiosus and Thermotoga maritima Cmr1 proteins do not interact with any other Cmr subunits in the absence of crRNAs (14,23). Together these results indicate that the Cmr subunit retains to III-B effector complexes by binding to crRNA and the well conserved hydrophobic amino acids of W58 and F59 play a crucial role in crRNA binding. (d) We further show that purification of Cmr-α complex from the mutant (Cmr-α1M2) carrying the quadruple mutation of I52A, G54A, R57A and R61A of Cmr1α has yielded an effector complex that is largely devoid of the mutant Cmr1α subunit (only detectable in SDS-PAGE with a large amount of sample) indicating that the interaction between Cmr-α1M2 and crRNAs has greatly impaired by the mutations. Furthermore, the Cmr-α1M2 effector complex shows a stronger affinity to the target RNA relative to Cmr-αΔ1, suggesting that target RNA binding by Cmr-α1M2 is probably not weakened by the mutation. Therefore, it has been reasoned that these amino acids are involved in crRNA binding. Since all six characterized amino acids are very well conserved among Cmr1 proteins and present in the Cmr1 ribonucleotide-binding groove identified by structural analyses (Supplementary Figures S3 and S4), this suggests that III-B CRISPR–Cas systems may share the mechanisms of crRNA and target RNA binding as well as the function of Cmr1 as the activation module in the effector complex as demonstrated for the S. islandicus Cmr-α system in this work.
Furthermore, in III-A effector complexes, there is only one subunit, i.e. Csm5, in the corresponding position of Cmr1 and Cmr6 in the III-B effector complexes. This raises at least two interesting questions: (i) what is the function of Cmr6 in Cmr complexes? (ii) Does Csm5 play the roles of both Cmr1 and Cmr6 in III-A effector complexes or III-A complexes represent a derivative of the core effector complex of III-B complexes? Conduction of direct comparisons of these immune systems will help lead to the answers of these questions. On the other hand, investigation of the diversity and distribution of CRISPR–Cas systems shows that, while III-B systems are over-represented among thermophiles, III-A systems predominate in mesophiles (2,42). In this regard, there appears to be a correlation between the Cmr1 occurrence and thermophilic life style of microorganisms, and this may reflect the adaptive evolution of type III CRISPR–Cas systems in thermophilic organisms.
To avoid self-immunity, the spatiotemporal regulation of the RNA-activated DNA cleavage by III-A and III-B RNPs should only allow the ssDNA cleavage to be active very briefly, and the DNase should be inactivated immediately after target DNA cleavage by removing target RNA. Nevertheless, it remains to be investigated as to which Cmr/Csm subunits are involved in the conformation change process and how the conformation is controlled. The present work has revealed that Cmr1 probably does not have a role in the spatiotemporal regulation in III-B CRISPR–Cas systems. As a result, core III-B complexes provide useful materials for investigation of molecular mechanisms of the RNA-activated DNA cleavage by III-B CRISPR–Cas systems such as the resolution of the crystal structure for a hybrid III-B core complex (22).
Supplementary Material
ACKNOWLEDGEMENTS
We thank members in the German CRISPR consortium and colleagues in the Archaea Centre for stimulating discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Danish Council for Independent Research [DFF-0602-02196, DFF-1323–00330]; Natural Science Foundation of China [31128011]; Scientific and Technological Self-Innovation Foundation of Huazhong Agricultural University [2014RC011]. Funding for open access charge: Huazhong Agricultural University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Mohanraju P., Makarova K.S., Zetsche B., Zhang F., Koonin E.V., van der Oost J.. Diverse evolutionary roots and mechanistic variations of the CRISPR–Cas systems. Science. 2016; 353:aad5147. [DOI] [PubMed] [Google Scholar]
- 2. Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J., Charpentier E., Haft D.H. et al. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015; 13:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barrangou R., Marraffini L.A.. CRISPR–Cas systems: prokaryotes upgrade to adaptive immunity. Mol. Cell. 2014; 54:234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wiedenheft B., Sternberg S.H., Doudna J.A.. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012; 482:331–338. [DOI] [PubMed] [Google Scholar]
- 5. Sternberg S.H., Richter H., Charpentier E., Qimron U.. Adaptation in CRISPR–Cas systems. Mol. Cell. 2016; 61:797–808. [DOI] [PubMed] [Google Scholar]
- 6. Jackson S.A., McKenzie R.E., Fagerlund R.D., Kieper S.N., Fineran P.C., Brouns S.J.. CRISPR–Cas: adapting to change. Science. 2017; 356:eaal5056. [DOI] [PubMed] [Google Scholar]
- 7. Charpentier E., Richter H., van der Oost J., White M.F.. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR–Cas adaptive immunity. FEMS Microbiol Rev. 2015; 39:428–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Plagens A., Richter H., Charpentier E., Randau L.. DNA and RNA interference mechanisms by CRISPR–Cas surveillance complexes. FEMS Microbiol. Rev. 2015; 39:442–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deng L., Garrett R.A., Shah S.A., Peng X., She Q.X.. A novel interference mechanism by a type IIIB CRISPR-Cmr module in sulfolobus. Mol. Microbiol. 2013; 87:1088–1099. [DOI] [PubMed] [Google Scholar]
- 10. Peng W., Feng M., Feng X., Liang Y.X., She Q.. An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res. 2015; 43:406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Samai P., Pyenson N., Jiang W., Goldberg G.W., Hatoum-Aslan A., Marraffini L.A.. Co-transcriptional DNA and RNA cleavage during Type III CRISPR–Cas immunity. Cell. 2015; 161:1164–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tamulaitis G., Venclovas C., Siksnys V.. Type III CRISPR–Cas immunity: major differences brushed aside. Trends Microbiol. 2017; 25:49–61. [DOI] [PubMed] [Google Scholar]
- 13. Elmore J.R., Sheppard N.F., Ramia N., Deighan T., Li H., Terns R.M., Terns M.P.. Bipartite recognition of target RNAs activates DNA cleavage by the Type III-B CRISPR–Cas system. Genes Dev. 2016; 30:447–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Estrella M.A., Kuo F.T., Bailey S.. RNA-activated DNA cleavage by the Type III-B CRISPR–Cas effector complex. Genes Dev. 2016; 30:460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kazlauskiene M., Tamulaitis G., Kostiuk G., Venclovas C., Siksnys V.. Spatiotemporal control of Type III-A CRISPR–Cas immunity: coupling DNA degradation with the target RNA recognition. Mol. Cell. 2016; 62:295–306. [DOI] [PubMed] [Google Scholar]
- 16. Han W., Li Y., Deng L., Feng M., Peng W., Hallstrom S., Zhang J., Peng N., Liang Y.X., White M.F. et al. A type III-B CRISPR–Cas effector complex mediating massive target DNA destruction. Nucleic Acids Res. 2017; 45:1983–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hale C.R., Zhao P., Olson S., Duff M.O., Graveley B.R., Wells L., Terns R.M., Terns M.P.. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009; 139:945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Staals R.H., Agari Y., Maki-Yonekura S., Zhu Y., Taylor D.W., van Duijn E., Barendregt A., Vlot M., Koehorst J.J., Sakamoto K. et al. Structure and activity of the RNA-targeting Type III-B CRISPR–Cas complex of Thermus thermophilus. Mol. Cell. 2013; 52:135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Spilman M., Cocozaki A., Hale C., Shao Y., Ramia N., Terns R., Terns M., Li H., Stagg S.. Structure of an RNA silencing complex of the CRISPR–Cas immune system. Mol. Cell. 2013; 52:146–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Benda C., Ebert J., Scheltema R.A., Schiller H.B., Baumgartner M., Bonneau F., Mann M., Conti E.. Structural model of a CRISPR RNA-silencing complex reveals the RNA-target cleavage activity in Cmr4. Mol. Cell. 2014; 56:43–54. [DOI] [PubMed] [Google Scholar]
- 21. Taylor D.W., Zhu Y., Staals R.H., Kornfeld J.E., Shinkai A., van der Oost J., Nogales E., Doudna J.A.. Structural biology. Structures of the CRISPR-Cmr complex reveal mode of RNA target positioning. Science. 2015; 348:581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Osawa T., Inanaga H., Sato C., Numata T.. Crystal structure of the CRISPR–Cas RNA silencing Cmr complex bound to a target analog. Mol. Cell. 2015; 58:418–430. [DOI] [PubMed] [Google Scholar]
- 23. Hale C.R., Cocozaki A., Li H., Terns R.M., Terns M.P.. Target RNA capture and cleavage by the Cmr type III-B CRISPR–Cas effector complex. Genes Dev. 2014; 28:2432–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhu X., Ye K.. Cmr4 is the slicer in the RNA-targeting Cmr CRISPR complex. Nucleic Acids Res. 2015; 43:1257–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo L., Brugger K., Liu C., Shah S.A., Zheng H., Zhu Y., Wang S., Lillestol R.K., Chen L., Frank J. et al. Genome analyses of Icelandic strains of Sulfolobus islandicus, model organisms for genetic and virus-host interaction studies. J. Bacteriol. 2011; 193:1672–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peng N., Han W., Li Y., Liang Y., She Q.. Genetic technologies for extremely thermophilic microorganisms of Sulfolobus, the only genetically tractable genus of crenarchaea. Sci. China Life Sci. 2017; 60:370–385. [DOI] [PubMed] [Google Scholar]
- 27. Gudbergsdottir S., Deng L., Chen Z., Jensen J.V., Jensen L.R., She Q., Garrett R.A.. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol. Microbiol. 2011; 79:35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li Y., Pan S., Zhang Y., Ren M., Feng M., Peng N., Chen L., Liang Y.X., She Q.. Harnessing Type I and Type III CRISPR–Cas systems for genome editing. Nucleic Acids Res. 2016; 44:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peng W., Li H., Hallstrom S., Peng N., Liang Y.X., She Q.. Genetic determinants of PAM-dependent DNA targeting and pre-crRNA processing in Sulfolobus islandicus. RNA Biol. 2013; 10:738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu T., Li Y., Wang X., Ye Q., Li H., Liang Y., She Q., Peng N.. Transcriptional regulator-mediated activation of adaptation genes triggers CRISPR de novo spacer acquisition. Nucleic Acids Res. 2015; 43:1044–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Contursi P., Jensen S., Aucelli T., Rossi M., Bartolucci S., She Q.. Characterization of the Sulfolobus host-SSV2 virus interaction. Extremophiles. 2006; 10:615–627. [DOI] [PubMed] [Google Scholar]
- 32. Deng L., Zhu H., Chen Z., Liang Y.X., She Q.. Unmarked gene deletion and host-vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles. 2009; 13:735–746. [DOI] [PubMed] [Google Scholar]
- 33. Peng N., Deng L., Mei Y., Jiang D., Hu Y., Awayez M., Liang Y., She Q.. A synthetic arabinose-inducible promoter confers high levels of recombinant protein expression in hyperthermophilic archaeon Sulfolobus islandicus. Appl. Environ. Microbiol. 2012; 78:5630–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ao X., Li Y., Wang F., Feng M., Lin Y., Zhao S., Liang Y., Peng N.. The Sulfolobus initiator element is an important contributor to promoter strength. J. Bacteriol. 2013; 195:5216–5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schmittgen T.D., Livak K.J.. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008; 3:1101–1108. [DOI] [PubMed] [Google Scholar]
- 36. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L. et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014; 42:W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Arnold K., Bordoli L., Kopp J., Schwede T.. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006; 22:195–201. [DOI] [PubMed] [Google Scholar]
- 38. Guex N., Peitsch M.C., Schwede T.. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis. 2009; 30(Suppl. 1):S162–S173. [DOI] [PubMed] [Google Scholar]
- 39. Kiefer F., Arnold K., Kunzli M., Bordoli L., Schwede T.. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009; 37:D387–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sun J., Jeon J.H., Shin M., Shin H.C., Oh B.H., Kim J.S.. Crystal structure and CRISPR RNA-binding site of the Cmr1 subunit of the Cmr interference complex. Acta Crystallogr. D Biol. Crystallogr. 2014; 70:535–543. [DOI] [PubMed] [Google Scholar]
- 41. Zhang J., Graham S., Tello A., Liu H., White M.F.. Multiple nucleic acid cleavage modes in divergent type III CRISPR systems. Nucleic Acids Res. 2016; 44:1789–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vestergaard G., Garrett R.A., Shah S.A.. CRISPR adaptive immune systems of Archaea. RNA Biol. 2014; 11:156–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.