Abstract
Ligation-based nucleic acid detection methods are primarily limited to DNA, since they exhibit poor performance on RNA. This is attributed to reduced end-joining efficiency and/or fidelity of ligases. Interestingly, chlorella virus DNA ligase (PBCV-1 DNA ligase) has recently been shown to possess high RNA-templated DNA end-joining activity; however, its fidelity has not yet been systematically evaluated. Herein, we characterized PBCV-1 ligase for its RNA-templated end-joining fidelity at single base mismatches in 3′ and 5′ DNA probe termini and found an overall limited end-joining fidelity. To improve the specificity in PBCV-1 ligase-driven RNA detection assays, we utilized structure-specific 5′ exonucleolytic activity of Thermus aquaticus DNA polymerase, used in the invader assay. In the iLock (invader padLock) probe assay, padlock probe molecules are activated prior ligation thus the base at the probe ligation junction is read twice in order to aid successful DNA ligation: first, during structure-specific invader cleavage and then during sequence-specific DNA ligation. We report two distinct iLock probe activation mechanisms and systematically evaluate the assay specificity, including single nucleotide polymorphisms on RNA, mRNA and miRNA. We show significant increase in PBCV-1 ligation fidelity in the iLock probe assay configuration for RNA detection.
INTRODUCTION
The need for accurate nucleic acid sensing and sequence determination has propelled the development of a wide range of DNA/RNA detection methods. Generally, two biochemical principles underlie specific nucleic acid detection: selective hybridization and enzyme assisted reactions. The hybridization principle is used for in situ hybridization (ISH), fluorescence ISH (FISH), high throughput microarrays, among others (1,2). Modifications to DNA and RNA hybridization assays can enhance single nucleotide polymorphism (SNP) resolution by manipulating the length of the allele/target-specific probes and the stringency of assay conditions toward a perfectly matching probe/target duplex. Despite considerable improvements in both the physicochemical properties of ISH probes and the sensing format, SNP discrimination and off-target hybridization limit sensitivity and assay throughput. Enzyme assisted methods, on the other hand, exploit inherent sensing properties of various DNA/RNA reactive enzymes, to specifically distinguish between matching or mismatching substrates (3). Both restriction endonucleases recognizing specific DNA sequence motives and structure-specific endonucleases working in a sequence independent manner are broadly used for DNA SNP analysis (4). The high nucleotide incorporation fidelity of DNA polymerases is exploited in real-time polymerase chain reaction (PCR) and next generation sequencing strategies (2). In the invader assay, 5′ flap endonucleases (FENs) (5) or 5′ exonucleolytic domains of eubacterial polymerases (6) are used to recognize and sequence-specifically cleave probe/DNA target structures (7). DNA ligation assays are based on the high end-joining fidelity of many DNA ligases (8).
A nucleic acid detection assay often combines multiple mechanisms and principles. For example, the padlock probe assay uses linear DNA oligonucleotides containing termini, that act as target specific ‘probe arm’ sequences (9). High nucleic acid detection specificity is achieved by designing target-specific arm, optimized for melting temperatures matching the optimal working temperature for the action of a high-fidelity DNA ligase. Ligase circularizes the padlock probe upon perfect matching. Circularized padlock probes can be isothermally amplified using rolling circle amplification (RCA) (10), and detected by decorating the RCA products (RCPs) with fluorophore conjugated oligonucleotides. Direct detection of RNA can be achieved with RNA hybridization methods, while enzyme assisted assay formats usually requires conversion of RNA to cDNA through reverse transcription. For example, in RNA detection reactions based on padlock probe ligation, an RNA molecule is usually converted to a cDNA copy, since the high-fidelity DNA ligases do not accept DNA/RNA heteroduplexes as efficient substrates for ligation (11,12). In the present work, we expand the available tools for direct RNA analyses by systematically exploring the favourable conditions for accurate RNA targeted padlock probe ligation and detection.
RNA templated DNA ligation activity of T4 DNA ligase has been discovered (13), and T4 DNA ligase is the only enzyme adequately studied till date, for RNA templated DNA end-joining fidelity (14,15). In fact, T4 RNA ligase 2 also allows for an efficient RNA templated end-joining, but the effect of mismatches on this enzyme's activity has only been studied for DNA templates (16). Moreover, low ligation efficiency of T4 DNA ligase on RNA (14) limits its utility as an efficient RNA genotyping enzyme, while T4 RNA ligase 2, both as native or its deletion mutant forms, has mostly been used for next-generation small RNA sequencing library preparation (17) and cloning (18). Recent discovery that PBCV-1 DNA ligase is capable of efficient RNA-templated DNA ligation (19) suggests it's use in direct RNA sensing. Indeed, a two-step method to differentiate between the let-7 miRNA family members involving ligation of short, miRNA complementary DNA probes prior to quantitative polymerase chain reaction (qPCR) amplification has been recently proposed (20). However, to our knowledge, no systematic characterization of PBCV-1 DNA ligase specificity on RNA has been done. In the present work, we evaluate PBCV-1 DNA ligase's DNA end-joining fidelity on SNP, mRNA and miRNAs. Furthermore, we propose a novel method for improving ligation specificity by combining structure-specific cleavage of probes using Thermus aquaticus (Taq) DNA polymerase together with PBCV-1 DNA ligase. We demonstrate the specificity of the method by distinguishing closely related miRNAs and SNPs in mRNA, with improved specificity.
MATERIALS AND METHODS
Oligonucleotide design
All oligonucleotides used in the PBCV-1 DNA ligase SNP discrimination fidelity experiment were purchased from IDT (Integrated DNA Technologies, Inc., Coralville, IA, USA) using standard desalting purification (Supplementary Table S1). The consensus sequence for four 25 nts long RNA templates is UCUCGCUGUCAUNCCUAUAUCCUCG (centrally located polymorphic site in the RNA target is marked as N). Padlock probes are designed such that the terminal probe arms anneal to the RNA template. All padlock probe sequences are identical, except for a discriminatory base localized at either the 3′ or the 5′ end of the probe. A reporter sequence for RCPs’ staining and detection is included in the sequence linking the probe arms, separated from probe arms with a series of ten adenines. Padlock probes were obtained as 5′ phosphorylated, to enable ligation. For miRNA detection, padlock probes were designed such that the probe arms anneal to equal halves of the target miRNA (for 22-mer let7 miRNA, both arms form 11 nt long duplexes with the miRNA (Supplementary Table S2)). The iLock probes consist of respective padlock probe sequences with an additional non-target complementary flap on the 5′ end. A synthetic wild-type KRAS RNA template, including codons 12 and 13, was ordered from IDT. Corresponding wild-type and mutated padlock probes as well as iLock probes, targeting the most common codon 12 substitutions c.34 G> A,T,C, were designed as described here previously, with the exception that the target complementary probe arms were extended to match the invader assay reaction conditions (∼51°C, Supplementary Table S3).
iLock activation and ligation
Characterization of the iLock mechanism was performed using 4:1 template excess to ensure that all matching and unmatching iLock probes participate in the activation process (10 nM RNA/miRNA/KRAS mRNA template and 2.5 nM iLock probe). Duplicate reactions were incubated in a heated-lid thermocycler at 51°C for 60 min, in a volume of 10 μl containing 1 U of Taq DNA polymerase (ThermoFisher Scientific) and 1 × Taq polymerase buffer supplied with 8 mM MgCl2. Following the iLock probe activation, 4 μl of sample volume was added to a ligation mix containing 5 U of PBCV-1 DNA ligase (SplintR, M0375S, NEB) and 1 × SplintR buffer in a final volume of 20 μl. The mixes were incubated at 37°C for 60 min. When conventional padlock probes were used, the same procedure was applied as in the initial PBCV-1 DNA ligase fidelity study, with the exception that the activation step was omitted.
RCA and detection
For RCA, 5 μl of the ligation mix was incubated with 10 nM decorator probe, 0.125 mM dNTPs, 0.2 μg/μl BSA, 250 mU of Phi29 polymerase (Monserate Biotechnology Group) and 1 × phi29 reaction buffer (Thermo Fisher) in a final volume of 25 μl at 37°C for 60 min. The polymerase was heat inactivated at 65°C for 3 min and then let to cool down to room temperature for 15 min. Fifteen microliter of the total sample was analyzed using the Aquila 400 Detection Unit (Q-linea, Uppsala). When the RCPs’ concentration was outside the instrument’s dynamic range, samples were diluted in 5 nM of the decorator probe using 1 × labeling solution (20 mM ethylenediaminetetraacetic acid, 20 mM Tris–HCl (pH 7.5), 0.05 % Tween-20 and 1 M NaCl), incubated at 65°C for 3 min, allowed to cool down to room temperature for 15 min and then counted.
qPCR
To assess if RNA templated probe activation can be combined with exponential PCR reaction to ensure more sensitive detection of RNA, templates were diluted to 50, 5 and 0.5 pM final concentration. Two invader oligonucleotides (Supplementary Table S4) were designed to form an invader structure when hybridized to a polymorphic RNA template. Invader oligonucleotide sequence was identical to 3′ and 5′ arms of 3′ T and 5′ T iLock probes respectively (Supplementary Table S1); however, the oligonucleotides were not connected with a linker and contained PCR primer binding overhangs. Invader oligonucleotides were added at 2 nM final concentration and activation reactions were conducted at 51°C for 60 min. Following probe activation, 1.5 μl of sample volume was added to a PBCV-1 ligation mix for a final volume of 15 μl. Samples were incubated at standard ligation conditions of 37°C for 60 min. As negative controls, Taq DNA polymerase, RNA template or PBCV-1 DNA ligase were individually omitted from the respective reactions. Following ligation, 1 μl of sample was transferred into qPCR mix, containing 0.1 U/μl Platinum Taq DNA polymerase (Thermo Scientific), 0.5 μM of primers (Supplementary Table S4), 0.2 mM of each dNTPs, 1 × SYBR Green, 1.5 mM MgCl2 and 1 × Platinum Taq DNA polymerase buffer in a 10 μl final volume. Theoretical final concentration of ligated qPCR template in qPCR reaction would range from 5 pM to 50 fM. qPCR reaction was performed at Mx3005P qPCR System (Agilent Genomics) and consisted of initial denaturation at 94°C for 2 min, followed by 35 cycles of 96°C for 30 s, 55°C for 30 s and 72°C for 45 s. PCR products were finally extended at 72°C for 10 min. Amplification plots and melting curves were generated using MxPro 3005P software.
PAGE
To visualize padlock/iLock probe activation and ligation, 5 μM synthetic RNA and 2.5 μM probe were processed as above. Following the ligation, 50 nM of sample was diluted in Novex® TBE (Tris-borate-EDTA)-Urea Sample Buffer (LC6876, ThermoFisher Scientific) to a final volume of 12 μl. Samples were denatured at 70°C for 3 min, placed on ice for 5 min, 10 μl was loaded onto 15 % Novex® TBE-Urea Gel (EC6885BOX, ThermoFisher Scientific) and separated in XCell SureLock™ Mini-Cell Electrophoresis System (ThemoFisher Scientific) using PowerPac Basic Power Supply (Bio-Rad) for ∼90 min at 170 V. Gels were stained using 1 × SybrGold (S11194, Invitrogen) in 1 × TBE running buffer for 15 min, followed by imaging in Gel Doc XR System (Bio-Rad).
RESULTS
RNA templated PBCV-1 DNA ligase end-joining fidelity
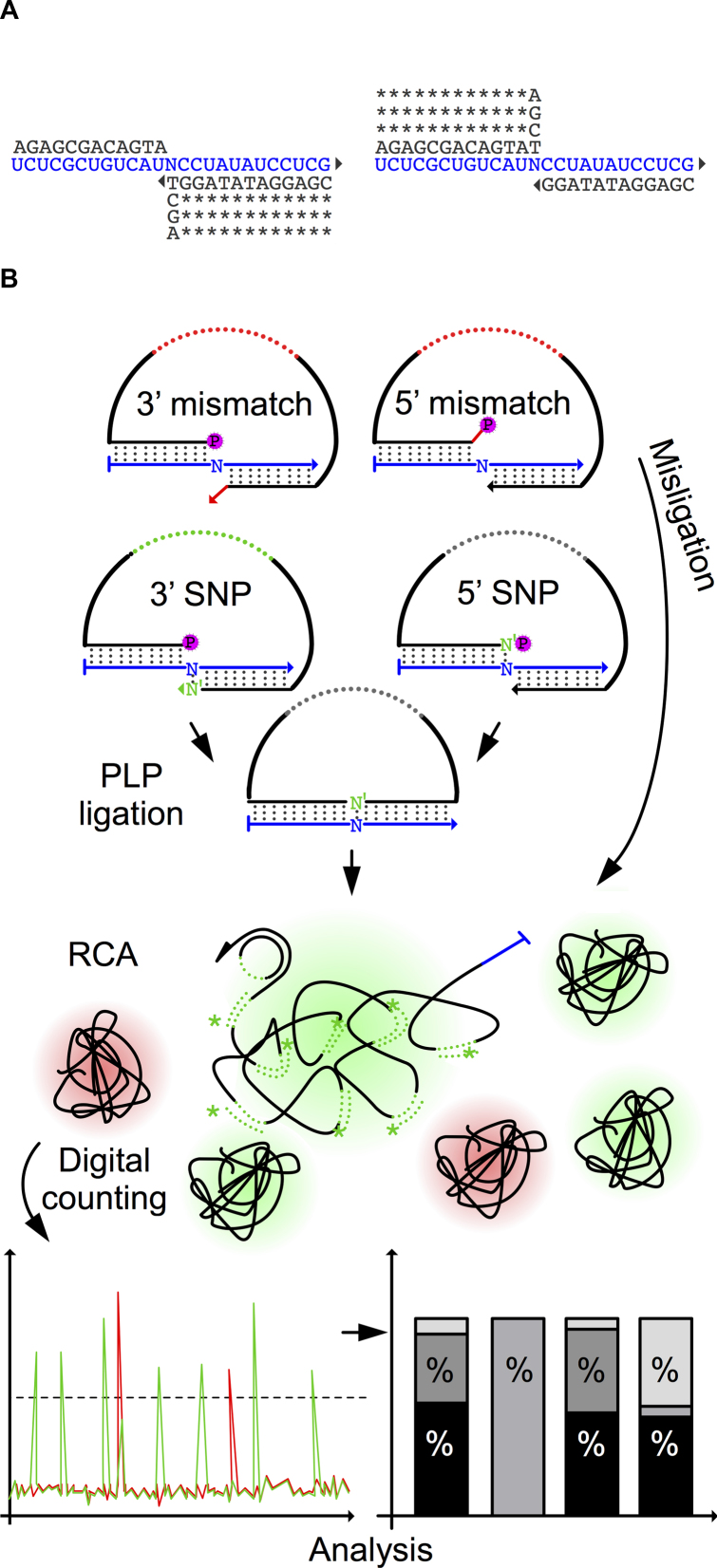
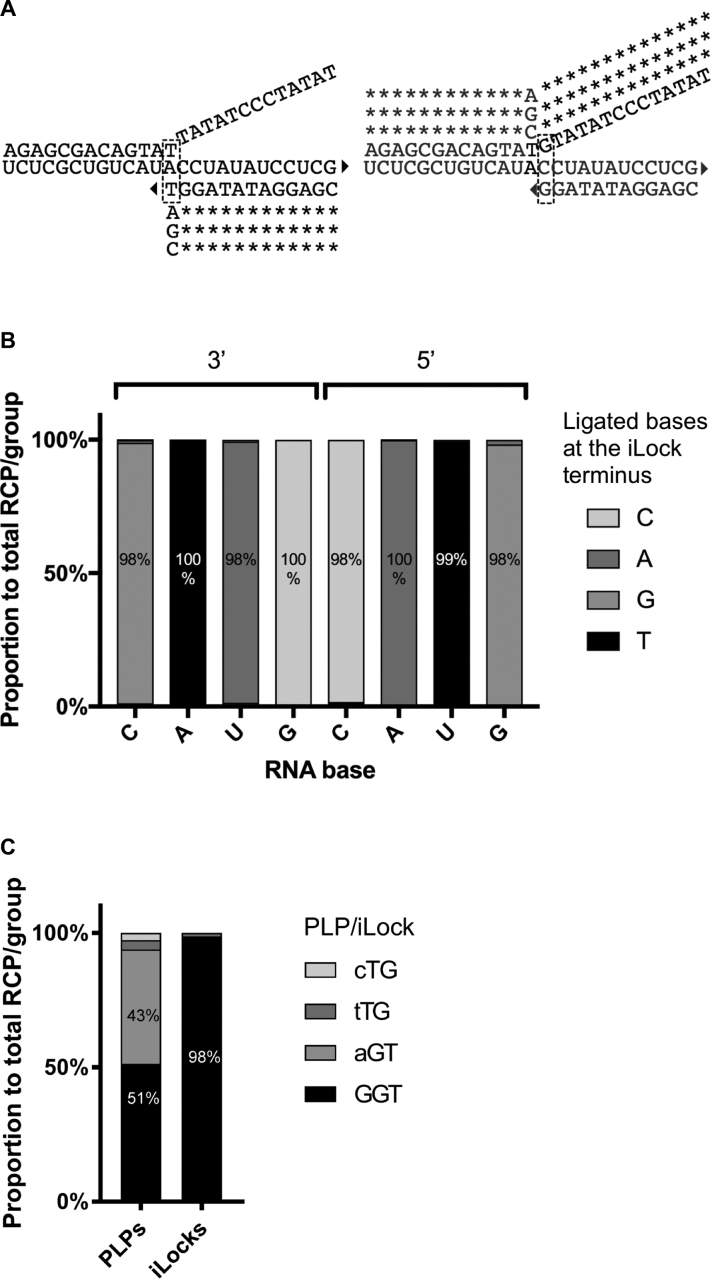
To systematically evaluate the DNA end-joining fidelity of the PBCV-1 DNA ligase on RNA substrate, we designed a DNA ligation assay using two oligonucleotides, hybridizing head-to-tail on four 25 nts long RNA templates. The four RNA templates differ in a centrally located polymorphic position, each containing one of the four bases. The RNA templates (Supplementary Table S1) have ∼50% GC content, comparable melting temperatures and have been designed to minimize secondary structures and formation of homodimers. We also designed eight identical padlock probes, except for the terminal 3′ or 5′ bases, targeting the centrally located polymorphic site in the RNA target (Figure 1A). The padlock probe arms are of comparable lengths, with a melting temperature higher than the reaction temperature of 37°C to minimize the turnover of substrates. Also, the padlock probes were designed to minimize secondary and homodimer structures formation, as in the case of RNA templates. We evaluate the ligase fidelity by amplifying circularized padlock probes (10) and then counting fluorescently labeled RCPs using digital RCA (Figure 1B) (21). The RCP count for all 32 possible SNPs, and 3′ and 5′ ends of padlock probes were recorded for each RNA base, as well as for both matching and mismatching padlock probes are presented as a percentage value (Figure 1B).
Figure 1.
PBCV-1 DNA ligase single nucleotide polymorphism (SNP) fidelity assay. (A) The structures of duplexes with mismatches at 3′ and 5′ sides of the nick. Eight PLPs were used to probe four identical RNA templates where a central base was one of the following: A, U, C or G (N). *: conserved bases; triangle: 3′ OH group; RNA template highlighted in blue. (B) All PLPs were subjected to PBCV-1 DNA ligase ligation and RCA. The generated RCPs were counted digitally and RCP numbers for both matching and mismatching PLPs are shown as a percentage.
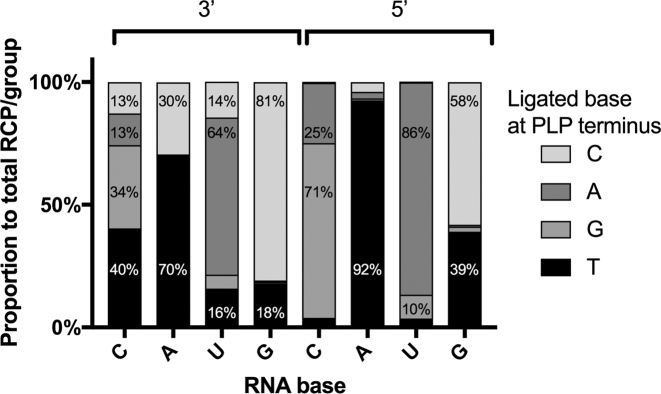
We found PBCV-1 DNA ligase as moderately accurate for certain mismatches, but highly tolerant for others. PBCV-1 DNA ligase showed the most potent ligation efficiency on terminal 3′ T on A (3′ T/A), and with similar ligation performance of 3′ T/C, 3′ A/U, 5′ A/U and 3′ C/G (Supplementary Figures S1 and S4A). Ligation fidelity, correlated to the signal for each probe, is expressed as a fraction of the total sum of RCPs in each group (for example, all 3′ probes ligated on the C template). PBCV-1 DNA ligase showed 92 % fidelity for a 5′ T/A; however, for all other mismatches, ligation fidelity was overall poor (Figure 2). In case of the C template, the enzyme was more efficient in ligating a 3′ T/C (40 %) mismatch than a matched 3′ G/C (34 %).
Figure 2.
Effects of 3′ and 5′ base mismatches on RNA templated DNA end-joining fidelity of PBCV-1 DNA ligase. For each PLP group, the contribution of each PLP in the group is presented as a percentage of the average RCPs number for all combined PLPs within a group. The average RCPs number in negative controls was subtracted from all the RCPs counts.
iLock RNA detection assay
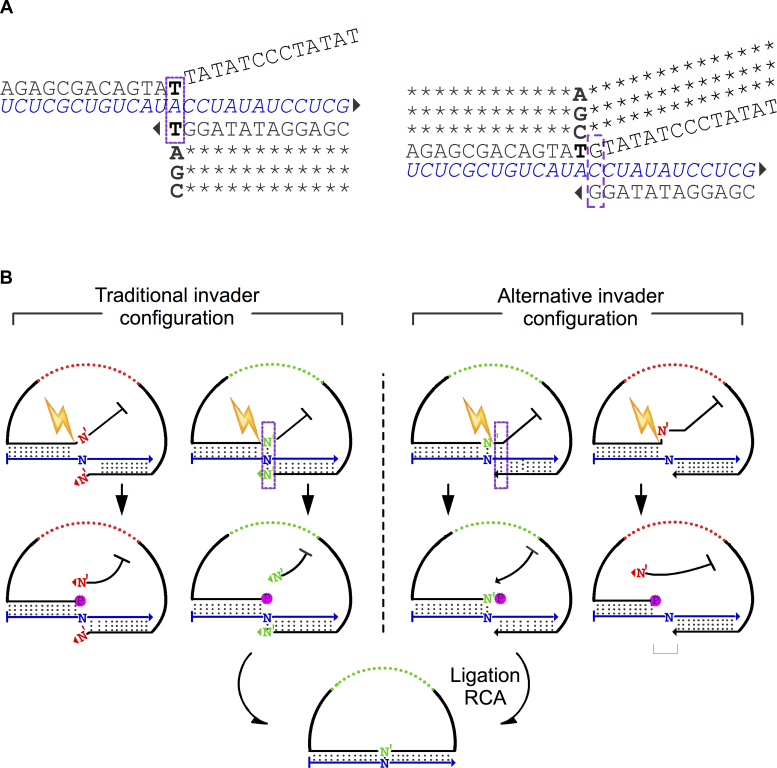
Considering the limited fidelity of PBCV-1 DNA ligase, we sought for an alternative mechanism to increase the ligation fidelity on RNA. For this, we have adopted an invasive cleavage concept into a padlock probe to form an invader padLock probe (iLock) RNA detection assay. The invader method utilizes enzymes that cleave substrates if specific sequence and structure requirements are met (6). The invasive structure recognized and cleaved by FENs and also, for example, Taq DNA polymerase, consists of a branched triple stranded structure, where two strands hybridize to the same third strand. At the site of strand migration, where one strand displaces the other, the enzyme will cleave the displaced strand at the junction, forming a nick and releasing a ‘5′ flap’. By appending a flap onto the 5′ arm of a padlock probe, we created an invasive structure upon binding to a target sequence that can be cleaved by Taq DNA polymerase. When properly hybridized to the RNA (Figure 3), iLock forms a characteristic invasive structure, where the last base of the 3′ iLock arm and a single base of the 5′ flap overlap compete for target hybridization. Mismatches in the RNA sequence at the invasive junction interfere with formation of the invasive structure, thus inhibiting cleavage and iLock activation. Depending on whether the RNA mismatch is located at the 5′ or 3′ terminus relative to the ligation junction after flap removal, two different invasive structures can be formed (Figure 3). In the traditional invader configuration, the 3′ terminal base and a single base in the 5′ flap base participate in the SNP base pairing and formation of the invasive structure. In the alternative invader structure configuration, the iLock discriminatory base is positioned 1 nt away from the invasive junction. In both scenarios, after activation through flap removal, a 5′ phosphate group is exposed and the iLock ends up having the same sequence as the corresponding padlock probe, with the discriminatory base localized on either the 3′ or the 5′ end of the iLock, respectively. At this point, the SNP is interrogated a second time, now by the PBCV-1 DNA ligase. When a mismatching iLock is hybridized to the target, the invasive structure is compromised differently depending on the invader configuration model. The traditional invader configuration results in an imperfect 3′ arm invasion, and the novel alternative approach results in the lack of a 5′ arm complementarity, both inhibiting invasive cleavage. If the iLocks are nevertheless cleaved on a mismatched target RNA, the traditional invader design will generate a ligation junction with a 3′ mismatch, while the novel, alternative approach will generate a single nucleotide gap, or less likely a 5′ mismatch (Figure 3).
Figure 3.
iLock activation using the invader principle. (A) The structures of iLock RNA binding modes according to traditional and alternative invader configurations. In a traditional configuration, the terminal 3′ iLock base displaces the first base in the flap from the invasive junction, and competes for hybridization to the SNP position. Alternatively, the discriminatory base is localized in the 5′ iLock arm, at a single base distance from the traditional invasive junction. Bold: SNP discriminatory bases; bases participating in an invader structure formation are highlighted with a purple rectangle; triangle: 3′ OH group. (B) The invader structure is formed if a matching iLock hybridizes with a template (iLocks with a green backbone). When Taq activates (spark) matching hybridized iLock in the traditional hybridization configuration, the flap is displaced, 5′P is exposed and a discriminatory base (N’) is positioned in the 3′ iLock terminus. In the alternative invader structure configuration, complementarity between iLock 5′ arm and RNA target allows for an/the (formation of) invader structure formation. After activation, phosphorylated discriminatory base (N’) is localized on the iLock 5′ terminus. When a mismatching iLock is annealed to the target, invader structure formation and iLock activation are compromised. A combination of an imperfect 3′ arm invasion (traditional approach) and a lack of 5′ arm complementarity renders the iLock probe non-functional. Misaligned iLock in the alternative approach might generate a single nucleotide gap or a 5′ mismatch, upon activation.
iLocks are activated and ligated on complementary RNA
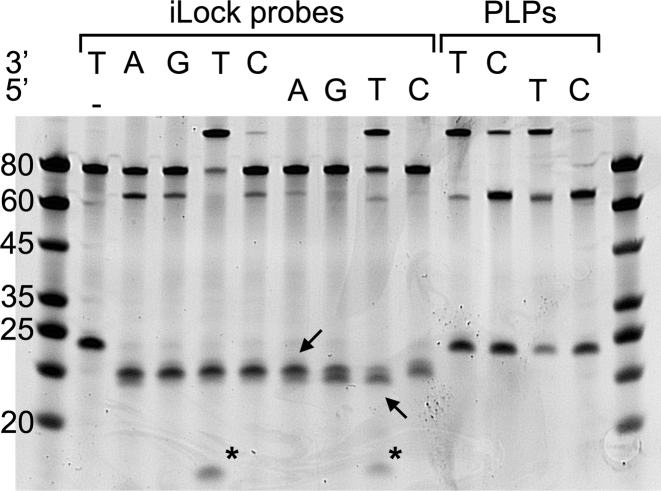
To investigate the fidelity and efficiency of iLock probe activation and ligation, the A template was queried with all possible 3′ and 5′ iLocks as well as control 3′ T, C and 5′ T, C padlock probes (Figure 4). We selected the T and C probes on an A template since the PBCV-1 DNA ligase was highly tolerant toward C/A mismatch (Figure 2). Though the standard invader assay works best at high temperatures matching the optimal temperature for endonucleolytic activity (7), we noticed efficient probe activation on RNA using Taq DNA polymerase at 37°C. In the absence of the Taq DNA polymerase, no ligation could be detected (Figure 4). In contrast, upon activation, a 5′ phosphate became available to permit iLock ligation (Figure 4). When mismatching iLocks were incubated with the RNA, a marginal activation but no ligation was seen on the polyacrylamide gel (Figure 4). Almost all matching 3′ T/A iLocks were activated by the Taq DNA polymerase, and all the activated probes were ligated (Figure 4). The ligation efficiency for the 5′ T/A iLock was lower than for the 3′ T/A, concordant with our previous experiment, when the control padlock probes were tested (Supplementary Figure S4A). For the control padlock probes, high PBCV-1 DNA ligase tolerance was again observed for the 3′ C/T mismatch. Interestingly, an observation of a potential ribonucleolytic activity of Taq DNA polymerase, removing 1–2 nts from the RNA template was noticed. Such activity has been discussed in an existing patent where DNA directed RNA template progressive hydrolysis has been shown for Taq DNA polymerase (22); however, extensive literature confirming this activity is not available.
Figure 4.
Analysis of the RNA templated iLock probe activation and ligation by polyacrylamide gelelectrophoresis (PAGE). Match and mismatch 3′/5′ iLocks (lanes 2–10) were activated and ligated on A template (lanes 3–10). 3′/5′ T and C PLPs (lanes 11–14) were ligated on A template as a reference. Inactivated iLocks are 80-nt long (lane 2). Upon activation, flap is released (visible as a faint band <20-nt long, marked with asterisks) and iLock is shortened to 70 nt. Arrows: degradation of RNA templates; -: no Taq control; DNA Ladder (lanes 1 and 15).
Characterization of nick sealing fidelity using iLock assay
After confirming that the iLock probes can successfully be activated and ligated with a high degree of fidelity, we went on to systematically test the end joining fidelity of PBCV-1 DNA ligase in the iLock assay format. We probed the four polymorphic RNA templates (Supplementary Table S1) with the eight iLocks, four for each invader mechanism (Figure 5A), using digital RCA readout for quantification. The SNP detection fidelity using PBCV-1 DNA ligase and iLocks was greatly improved for all the interrogated bases using both of the activation configurations. iLock assay showed superior specificity for matching probes, ranging from 98 % for 3′ G/C, 3′ A/U, 5′ G/C, 5′ C/G; 99 % for 5′ T/A and 100 % for remaining positions tested (Figure 5B). Compared to the PBCV-1 DNA ligase fidelity in the standard padlock probe ligation assay format, RNA detection fidelity, simultaneously exploiting properties of both the polymerase and ligase, was significantly improved. For instance, PBCV-1 DNA ligase alone showed higher ligation efficiency for mismatching 3′ T/C than 3′ G/C. In our iLock RNA detection assay, ligation fidelity for 3′ G/C was 98 %. Reaction was highly efficient for the 3′ T iLock, while the other iLocks gave less but similar amount of signal (Supplementary Figure S4B). Finally, to evaluate PBCV-1 DNA ligase genotyping fidelity on messenger RNA, we genotyped KRAS codon 12 mRNA with both padlock and iLock probes. Under standard padlock probe ligation conditions, PBCV-1 DNA ligase ligated T/G almost to the same extent as a corresponding, matching C/G padlock probe. Our result, presented in the Figure 5C, is in line with our previous observations that T/G is the most error-prone configuration for the PBCV-1 DNA ligase (Figure 2). However, using iLock probes, we could use PBCV-1 DNA ligase to successfully genotype KRAS mutation with an accuracy of over 98 % (Figure 5C).
Figure 5.
Effects of 3′ and 5′ base mismatches on RNA templated DNA end-joining fidelity of PBCV-1 DNA using iLocks. (A) Probe alignment schematics using an RNA template with base A in the central, polymorphic position. Triangle: 3′ OH group; dashed rectangle: bases participating in the invader structure formation. (B) In each iLock group, average number of RCPs was added and ligation fidelity for each iLock was is presented as a percentage within the group. (C) Wild-type KRAS codon 12 (GGT) was genotyped with a complementary and three mismatching (depicted as a lowercase SNP nucleotides in the figure legend) PLPs and iLocks. Average number of RCPs in negative controls was subtracted from all the RCPs counts.
iLock RNA assay for miRNA detection
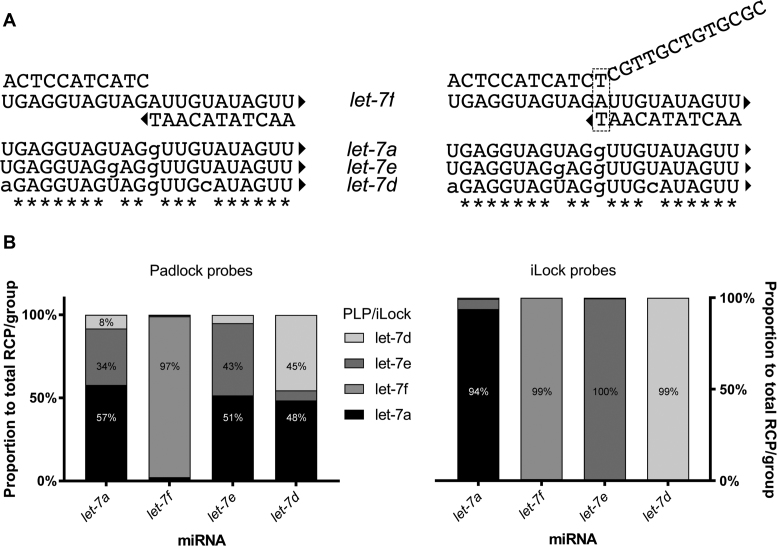
miRNA members within the same gene family often have highly related sequences. We took advantage of the diversity of let-7 miRNA family (23) to test if PBCV-1 DNA ligase can be used to distinguish highly similar RNAs. We designed padlock and iLock probes for four let-7 family members (Supplementary Table S2), with single or double nucleotide differences distributed across the 22-nt long target sequence. Each padlock and iLock probe was evaluated individually on every let-7 miRNA target. PBCV-1 DNA ligase alone, in the padlock probe assay format, could only differentiate let-7f from the other miRNAs (Figure 6B). Noteworthily, let-7f miRNA was the only miRNA having a substitution at the ligation junction. The let-7a padlock probe even generated higher signal for the let-7e and let-7d miRNA, compared to the corresponding matching probes.
Figure 6.
miRNA detection fidelity using PLP and iLock RNA detection assay. (A) Hybridization of let-7f PLP (left) and iLock (right) on let-7f miRNA. Uppercase letters: bases in the sequence corresponding to let-7f reference; lowercase: mismatches; *: conserved bases; triangle: 3′ OH group. (B) let-7 miRNA detection fidelity using PLPs (left) and iLock (right) assay. Average number of RCPs in each group was added and each probe signal is presented as a percentage. Average RCPs number in negative controls was subtracted from all the RCPs counts.
Using an initial iLock design, the specificity improved substantially, with one exception: the let-7a and let-7e iLocks showed significant crosstalk between their respective targets (Supplementary Material and Figure S1). The let-7e miRNA had, in this initial design, a mismatch to the let-7a probe’s 3 nts from the invasive junction. We investigated whether the reaction temperature or a shift of the invader junction site would improve the iLock fidelity (Supplementary Figures S2 and 3). Although, we did not observe a higher let-7a detection specificity at elevated activation temperatures, by shifting the let-7a iLock invasive junction 3 bases toward the 5′ end, all miRNAs were discriminated with high specificity (Figure 6B).
Probe activation, RNA templated ligation and PCR for sensitive RNA detection
Amplification of activated and ligated iLock probes generates individual RCP from each RNA templated probe circularization event. To investigate if the novel probe activation and RNA templated ligation reaction also can be used for other assay formats employing joining of two oligonucleotides, we designed a pair of probes forming an invader structure upon hybridization with the target that could be amplified by PCR. We monitored the ligation reaction by qPCR on a dilution series of a synthetic RNA target (Supplementary Figure S7D). No amplification products were observed when PBCV-1 DNA ligase or RNA template were omitted from the reaction. A total of 50 fM of the RNA target was detected. Both amplification- and melting curves confirmed amplification of the expected product (Supplementary Figure S7A and B).
DISCUSSION
Most enzyme-assisted RNA analysis methods are based on RNA to cDNA conversion, as most of the enzymes used in analytical methods do not work or are inefficient on RNA substrates (14). Direct and accurate detection of RNA would be desirable not only with respect to shorter assay time and protocol simplicity, but also to avoid potential bias in the cDNA synthesis step. The relatively recent discovery that the PBCV-1 DNA ligase possesses highly efficient RNA templated DNA ligation activity (19) prompted us to investigate the end joining fidelity of this enzyme for direct detection of RNA. The specificity profile of PBCV-1 DNA ligase on DNA substrates has been evaluated for a 3′ and 5′ terminal base (24). However, our ligation fidelity on RNA substrates matches the DNA templated ligation fidelity only partially. According to our data (Figure 2), ligase exhibited generally poor ligation specificity at both the 5′ and the 3′ ends of the ligation site.
To circumvent low fidelity of ligation, we have combined the invader assay principle with the padlock probe design (iLock) to evaluate if the fidelity of RNA templated DNA ligation reactions can be improved, by making use of the highly specific sequence- and structure-dependent endonucleolytic activity of Taq DNA polymerase (6). In the traditional invader configuration, iLock probe arms form a highly specific invasive structure with the RNA target, where a 3′ terminal base directly overlaps and competes to bind a single base in the 5′ flap (Figure 3). In the present work, we propose an alternative configuration of RNA binding. Combining the invader principle with high efficiency/low fidelity ligation offers many advantages. First of all, the invader method has been successfully applied in specific DNA (7) and RNA genotyping (25), as well as in miRNA detection (26). Second, iLock probes are ordered modification free, without a 5′ phosphate and without any additional arrestor oligonucleotides (preventing flap from triggering possible unspecific secondary reactions in the traditional invader assay (25)), since only the ligated iLock can generate a detectable signal. This property not only makes the assay cheaper, but also reduces the risk of template- independent ligation. Next, the RNA target base is interrogated by both by the Taq DNA polymerase and the DNA ligase and thus, effective ligation is mediated by the combined fidelities of both these enzymes.
Previous studies reported greatly reduced cleavage rate of the recombinant Taq DNA polymerase on RNA (27). In contrast, we have observed high activation and ligation activity on 25-nt long RNA templates at reaction conditions allowing for a stable probe-RNA hybridization (Figure 4). However, we have observed uncleaved iLock probes when reaction took place on relatively short miRNAs (Supplementary Figure S3). This suggests that the assay sensitivity for short RNA targets can be further improved by using RNA active recombinant enzymes or mutated variants (27). Alternatively, activation at lower temperatures (37°C) could allow for more stable probe-target hybridization or to combine Taq DNA polymerase with thermolabile ligases. Interestingly, we have observed a potential ribonucleolytic activity for Taq, also reported previously by initial invader mechanism inventors (22). While description of this activity in scientific literature is not available, inventors reported RNA hydrolysis in RNA/DNA heteroduplexes, directed by a 3′ end of the DNA strand in a heteroduplex region. According to Figure 4, target RNAs were shortened by 1, 2 or 3 nts, and this nucleolytic activity was not observed for DNA (data not shown). We have observed that RNA degradation was more pronounced at higher temperatures (data not shown), suggesting stronger degradation in less annealed heteroduplexes. As the hydrolysis potentially occurs at the 5′ end of the RNA, it did not affect PBCV-1 DNA ligation or RCA.
We have systematically investigated specificity of RNA detection in different biological contexts using novel iLock RNA detection assay, and also comparing to a traditional padlock probe ligation. To validate high RNA genotyping fidelity on biological target sequence, we have used padlock and iLock probes on frequently mutated codon 12 in KRAS mRNA (28), associated with high cancer-specific mortality rates (29). In line with the PBCV-1 DNA ligase fidelity profile (Figure 2), ligase alone could not effectively discriminate between wild-type GGT and mutant AGT codons (Figure 5C), while by using the iLock assay, only the expected probe for GGT codon gave signal.
miRNAs, due to their short length and sequence similarity, are often difficult to distinguish using traditional hybridization-based approaches. Routine methods for small RNA profiling provide excellent RNA-sequence identification. However, they involve RNA to cDNA conversion step, are strongly biased toward certain targets and might not be accurate for absolute miRNA abundance determination (30). Though high-throughput sequencing costs are declining, targeted methods offer cheaper setup and easier interpretation of results. We have investigated detection specificity of native let-7 miRNA family members using padlock and iLock probes. For traditional padlock probe ligation, only let-7f miRNA was detected with good specificity (Figure 6B). Since other padlock probes generated considerable background on other miRNAs, we concluded that enzymatic end- joining fidelity of PBCV-1 DNA ligase alone cannot be used to distinguish closely related miRNAs. In contrast, iLock probes discriminated all let-7 isoforms, however let-7a iLock probe worked least efficiently compared to other probes (melting temperature of adjusted let-7a 5′ arm was now lowered by about 15°C). Since the invader structure in the let-7a iLock was now positioned at the let-7e rU to rG mismatch, let-7a iLock probe exhibited very high fidelity on let-7e while specificity on other isoforms remained unaffected.
The iLock assay can be applied using different combinations of structure-specific endonucleases and ligases, and it can also be employed for DNA analysis. Moreover, the new ligation-based RNA sensing mechanism can be combined with exponential amplification of ligated DNA using PCR. With iLock RNA detection assay, various types of RNAs can be accurately identified, thus opening new possibilities for RNA templated DNA ligation platforms.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Malte Kühnemund, Iván Hernández-Neuta, Narayanan Srinivasan for valuable input for the project and comments on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Research Council (VR); Formas Strong Research Environment (Biobridges) [221–2011-1692]; Innovative Medicines Initiative 2 Joint Undertaking (EbolaMoDRAD) [115843]; SSF project FLU-ID [SBE13–0125]. Funding for open access charge: FLU-ID [SBE13-0125].
Conflict of interest statement. We are considering filing a patent on aspects of the described method.
REFERENCES
- 1. Yan L., Zhou J., Zheng Y., Gamson A.S., Roembke B.T., Nakayama S., Sintim H.O.. Isothermal amplified detection of DNA and RNA. Mol. Biosyst. 2014; 10:970–1003. [DOI] [PubMed] [Google Scholar]
- 2. Syvänen A.C. Accessing genetic variation: genotyping single nucleotide polymorphisms. Nat. Rev. Genet. 2001; 2:930–942. [DOI] [PubMed] [Google Scholar]
- 3. Pastinen T., Kurg A., Metspalu A., Peltonen L., Syvänen A.C.. Minisequencing: a specific tool for DNA analysis and diagnostics on oligonucleotide arrays. Genome Res. 1997; 7:606–614. [DOI] [PubMed] [Google Scholar]
- 4. Mashal R.D., Koontz J., Sklar J.. Detection of mutations by cleavage of DNA heteroduplexes with bacteriophage resolvases. Nat. Genet. 1995; 9:177–183. [DOI] [PubMed] [Google Scholar]
- 5. Kaiser M.W., Lyamicheva N., Ma W., Miller C., Neri B., Fors L., Lyamichev V.I.. A comparison of eubacterial and archaeal structure-specific 5′- exonucleases. J. Biol. Chem. 1999; 274:21387–21394. [DOI] [PubMed] [Google Scholar]
- 6. Lyamichev V., Brow M.A., Dahlberg J.E.. Structure-specific endonucleolytic cleavage of nucleic acids by eubacterial DNA polymerases. Science. 1993; 260:778–783. [DOI] [PubMed] [Google Scholar]
- 7. Lyamichev V., Mast A.L., Hall J.G., Prudent J.R., Kaiser M.W., Takova T., Kwiatkowski R.W., Sander T.J., De Arruda M., Arco D.A. et al. . Polymorphism identification and quantitative detection of genomic DNA by invasive cleavage of oligonucleotide probes. Nat. Biotechnol. 1999; 17:5–9. [DOI] [PubMed] [Google Scholar]
- 8. Landegren U., Kaiser R., Sanders J., Hood L.. A ligase-mediated gene detection technique. Science. 1988; 241:1077–1080. [DOI] [PubMed] [Google Scholar]
- 9. Nilsson M., Malmgren H., Samiotaki M., Kwiatkowski M., Chowdhary B.P., Landegren U.. Padlock probes: circularizing oligonucleotides for localized DNA detection. Science. 1994; 265:2085–2088. [DOI] [PubMed] [Google Scholar]
- 10. Banér J., Nilsson M., Mendel-Hartvig M., Landegren U.. Signal amplification of padlock probes by rolling circle replication. Nucleic acids Res. 1998; 26:5073–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Larsson C., Grundberg I., Söderberg O.. In situ detection and genotyping of individual mrna molecules. Nat. Methods. 2010; 7:395–397. [DOI] [PubMed] [Google Scholar]
- 12. Baner J., Isaksson A., Waldenström E., Jarvius J., Landegren U., Nilsson M.. Parallel gene analysis with allele-specific padlock probes and tag microarrays. Nucleic Acids Res. 2003; 31:e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fareed G.C., Wilt E.M., Richardson C., Richardsons C.C.. Enzymatic breakage and joining of deoxyribonucleic acid, VIII. Hybrids of ribo- and deoxyribonucleotide homopolymers as substrates for polynucleotide ligase of bacteriophage T4. J. Biol. Chem. 1971; 246:925–932. [PubMed] [Google Scholar]
- 14. Nilsson M., Antson D.O., Barbany G., Landegren U.. RNA-templated DNA ligation for transcript analysis. Nucleic Acids Res. 2001; 29:578–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nilsson M., Barbany G., Antson D.O., Gertow K., Landegren U.. Enhanced detection and distinction of RNA by enzymatic probe ligation. Nat. Biotechnol. 2000; 18:791–793. [DOI] [PubMed] [Google Scholar]
- 16. Chauleau M., Shuman S.. Kinetic mechanism of nick sealing by T4 RNA ligase 2 and effects of 3′-OH base mispairs and damaged base lesions. RNA. 2013; 19:1840–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P.. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001; 294:858–862. [DOI] [PubMed] [Google Scholar]
- 18. Pfeffer S., Sewer A., Lagos-Quintana M., Sheridan R., Sander C., Grässer F.A., van Dyk L.F., Ho C.K., Shuman S., Chien M. et al. . Identification of microRNAs of the herpesvirus family. Nat. Methods. 2005; 2:269–276. [DOI] [PubMed] [Google Scholar]
- 19. Lohman G.J.S., Zhang Y., Zhelkovsky A.M., Cantor E.J., Evans T.C.. Efficient DNA ligation in DNA-RNA hybrid helices by chlorella virus DNA ligase. Nucleic Acids Res. 2014; 42:1831–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jin J., Vaud S., Zhelkovsky A.M., Posfai J., McReynolds L.A.. Sensitive and specific miRNA detection method using SplintR Ligase. Nucleic Acids Res. 2016; 44:e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jarvius J., Melin J., Go J., Stenberg J., Fredriksson S., Gonzalez-rey C., Bertilsson S.. Digital quantification using amplified single- molecule detection. Nat. Methods. 2006; 3:725–727. [DOI] [PubMed] [Google Scholar]
- 22. Brow M.A.D., Lyamichev V.I., Olive D.M.. Rapid detection and identification of pathogens. 1995; Patent number: US-6, 372, 424B1 (Date granted: 2002).
- 23. Roush S., Slack F.J.. The let-7 family of microRNAs. Trends Cell Biol. 2008; 18:505–516. [DOI] [PubMed] [Google Scholar]
- 24. Sriskanda V., Shuman S.. Specificity and fidelity of strand joining by chlorella virus DNA ligase. Nucleic Acids Res. 1998; 26:3536–3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eis P.S., Olson M.C., Takova T., Curtis M.L., Olson S.M., Vener T.I., Ip H.S., Vedvik K.L., Bartholomay C.T., Allawi H.T. et al. . An invasive cleavage assay for direct quantitation of specific RNAs. Nat. Biotechnol. 2001; 19:673–676. [DOI] [PubMed] [Google Scholar]
- 26. Allawi H.T., Dahlberg J.E., Olson S., Allawi H.T., Dahlberg J.E., Olson S., Lund E., Olson M.. Quantitation of microRNAs using a modified Invader assay quantitation of microRNAs using a modified Invader assay. RNA. 2004; 10:1153–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ma W., Kaiser M.W., Schaefer J.J., Hatim T., Takova T., Neri B.P., Lyamichev V.I., Ma W., Kaiser M.W., Lyamicheva N. et al. . RNA template-dependent 5′ nuclease activity of Thermus aquaticus and Thermus thermophilus DNA polymerases. J. Biol. Chem. 2000; 275:23693–24700. [DOI] [PubMed] [Google Scholar]
- 28. Sundström M., Edlund K., Lindell M., Glimelius B., Birgisson H., Micke P., Botling J.. KRAS analysis in colorectal carcinoma: analytical aspects of pyrosequencing and allele-specific PCR in clinical practice. BMC Cancer. 2010; 10:660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Imamura Y., Morikawa T., Liao X., Lochhead P., Kuchiba A., Yamauchi M., Qian Z.R., Nishihara R., Meyerhardt J.A., Haigis K.M. et al. . Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF wild-type colorectal cancers. Clin. Cancer Res. 2012; 18:4753–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Linsen S.E.V., de Wit E., Janssens G., Heater S., Chapman L., Parkin R.K., Fritz B., Wyman S.K., de Bruijn E., Voest E.E. et al. . Limitations and possibilities of small RNA digital gene expression profiling. Nat. Methods. 2009; 6:474–476. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.