Abstract
During embryogenesis, organisms undergo considerable cellular remodelling requiring the combined action of thousands of proteins. In the case of the well studied model Drosophila melanogaster, transcriptomic studies, most notably from the modENCODE project, have described in detail changes in gene expression at the mRNA level across development. Although such data are clearly very useful for understand how the genome is regulated during embryogenesis, it is important to understand how changes in gene expression are reflected at the level of the proteome. In this study, we describe a combination of two quantitative label free approaches, SWATH and Data Dependent Acquisition, to monitor changes in protein expression across a timecourse of Drosophila embryonic development. We demonstrate that both approaches provide robust and reproducible methods for the analysis of proteome changes. In a preliminary analysis of Drosophila embryogenesis, we identified several pathways, including the heat-shock response, nuclear protein import and energy production, that are regulated during embryo development. In some cases changes in protein expression mirrored transcript levels across development, whereas other proteins showed signatures of post-transcriptional regulation. Taken together, our pilot study provides a good platform for a more detailed exploration of the embryonic proteome.
Keywords: Early embryo development, Label-Free quantification, Mass-spectrometry, SWATH
Introduction
Embryogenesis, the process in which a zygote develops from a fertilised egg cell into a complete organism, encompasses key events such as axis specification, gastrulation, germ layer formation and organogenesis [1]. The Drosophila embryo, due to its short generation time and well established genetic tools, has proved to be a tractable model for characterising many of the molecular aspects underpinning development and a wealth of studies have provided important insights that are often transferable to other systems [2–4]. During embryogenesis, a single diploid cell, the fertilized egg, undergoes a series of rapid nuclear division over a two hour period to form the blastoderm, comprising some 6,000 cells. This is followed by further cell divisions, cell movement and cellular differentiation to generate the germ layers and specific tissues of the larva [5]. Development is a highly regulated and complex process involving hundreds of regulatory factors and characterised by the spatial and temporal expression of the majority of genes in the genome [6]; from approximately 1000 genes at the blastoderm stage to over 10,000 during subsequent stages of development and morphogenesis [6–9]. The dynamic and complex cellular remodelling that characterises embryogenesis is, to a large extent, dependent upon fine tuning of the proteome to regulate cellular processes at each stage of development. As part of the modENCODE project, RNA-Seq and tiling microarrays were used to comprehensively sample the transcriptome across embryonic development: combining these data with previous in situ hybridisation studies provides a detailed view of transcriptome dynamics [6–9]. However, changes in transcript levels or isoforms are not always reflected as variation in proteins or proteomes [10] and the relationship between the transcriptome and proteome remains poorly characterised in most species and across development. In order to study relevant changes in protein expression and abundance, we require sensitive technologies capable of interrogating the entire proteome [11].
Mass-spectrometry (MS) has emerged as a powerful tool for proteome analysis. It can be deployed to identify the protein content of a sample, measure the quantity of proteins, provide insights into protein structure and identify molecules that proteins interact with [12]. The combination of high resolution mass-spectrometry, electrospray ionisation sources, robust liquid chromatography (LC) and bioinformatics analysis has enabled the identification of thousands of proteins from several organisms [13]. Quantitative MS is the method of choice for monitoring expression changes in the proteome [12], with Data Dependent Acquisition (DDA) methods based on the fragmentation of the most abundant peptides in a sample used for peptide identification [13]. For protein quantification, methods based on stable isotopic labelling have been widely used and been shown to provide very good accuracy [13]. However, these methods cannot be applied in many experimental workflows and can be prohibitively expensive in studies involving many biological samples [14]. In parallel to stable isotopic labelling methods, quantitative Label-Free (LFQ) approaches have been developed [12–14]. The reduced cost and ease with which such approaches can be adapted to any experimental workflow in comparison to isotopic labelling methods have contributed to the growing interest in LFQ approaches. LFQ methods compare the signal intensity of a peptide between different LC-MS runs. Different LFQ strategies have been described, based on extracted ion chromatogram (XIC) of the MS1 signal of the peptides (MS1-based XICs) or XIC of the MS2 signals of the fragments (MS2-based XICs) [13]. Spectral counting, which compares the number of MSMS events for all the peptides of a protein between different LC-MS runs, has also been extensively used to monitor protein expression changes [13] LFQ approaches have been shown to be suitable for comparing protein expression levels between different conditions or to provide good approximations of absolute protein abundance, and when used in conjunction with carefully designed experiments and relevant statistical methods, they provide good alternatives to isotopic labelling methods [12–14].
As opposed to DDA approach, targeted proteomics approaches only quantify a selection of proteins of interest [15]. Methods including SRM (Selected Reaction Monitoring) [16] or PRM (Parallel Reaction Monitoring) [17] are based on the co-selection of peptides of interest and their fragments. In these acquisition modes, a peptide is selected within a defined m/z range and fragmented while all the other ions are discarded. During SRM, the ions resulting from the fragmentation step are then filtered to detect only the expected fragments from the selected precursor [16]. In PRM, all the fragments from the precursor ion are detected in a unique high resolution scan [17]. These methods enable the MS analysis to focus only on the ions, and thus peptides/proteins, of interest which results in improved sensitivity and quantification accuracy. However, only few hundreds proteins are routinely quantifiable per MS run, which does not allow for broad coverage of the proteome [18, 19]. The main application of targeted methods is therefore in hypothesis-driven experiments that require high sample numbers, to confirm results from other proteomics experiments or, due to the high sensitivity and accuracy, in biomarker characterisation [15, 20].
Recently, a new approach based on a Data Independent Acquisition mode, Sequential Window Acquisition of all THeoretical mass spectra (SWATH), has been developed [21]. In this approach, after a first MS scan, sequential wide m/z window ranges (swaths) of precursor ions are selected for fragmentation, resulting in mixed MSMS spectra. The quantification is performed through in silico SRM-like analysis. Typically, extracted ion chromatograms of the fragments of a peptide are generated from MSMS spectra and used for comparative analysis between different MS runs. A priori information has been needed for SWATH experiments. A spectral library containing spectra of the peptides as well as peptides retention times are required for this approach. This library can be built from separate DDA acquisitions [22] or directly from the SWATH data [23]. Peak areas obtained from extracted ion chromatograms of each fragment from each peptide around the predicted retention time are used to generate the quantification data. The correlation between all the fragments and the precursor, as well as the order of abundance of the fragments compared with the spectra in the library, are used to verify the identity of the precursor. Since all the precursors in the particular swath are fragmented, the SWATH strategy potentially provides information on a large fraction of the proteome and, in theory, combines the best of SRM and DDA approaches: a sensitive and accurate quantification with good proteome depth [12].
In this study, we used a combination of different Label-Free approaches, global, semi-targeted and targeted, to monitor the dynamics of the D. melanogaster embryonic proteome. We first compared the performance of SWATH and MS1-based XICs. We found that, in general, label-free quantification methods provide a robust method for the identification and quantitation of proteins between different conditions. Encouragingly, we found that the SWATH approach showed very good reproducibility. Finally, we used a combination of the different label-free quantification methods to provide, for the first time, a comprehensive analysis of the D. melanogaster proteome at different stages of embryonic development. The expression profiles of selected proteins were validated using PRM and their transcripts measured by quantitative real time polymerase chain reaction (qRT-PCR). Importantly, we observed good agreement on trends in protein expressions between all the quantitative methods we employed. Our study identified some clusters of differentially regulated proteins across embryonic development, including importins and heat-shock proteins, as well as remodelling of the global energy production systems of the embryo. This study provides new insights into the proteome dynamics during D. melanogaster embryogenesis and serve as a foundation for our ongoing functional characterisation of the developmental proteome.
Materials and Methods
Fly Lines, embryo collection and protein extraction
Flies of the sequenced D. melanogaster iso-1 strain from the Bloomington Stock Centre (y1; Gr22biso-1 Gr22diso-1 cn1 CG33964iso-1 bw1 sp1; LysCiso-1 MstProxiso-1 GstD5iso-1 Rh61) were kept on standard yeast-cornmeal media in a 12-h light-dark cycle at 25 °C and 75% relative humidity. Embryos were collected from cages on apple juice agar plates over a 16 hours period, or every 4.5 hours after egg laying and aged to produce a crude developmental time course (0-4.5h, 4.5-9h, 9-13.5h, 13.5-18h and 18-22.5h). After collection, embryos were dechorionated with 50% bleach, washed with water, frozen in liquid nitrogen and kept at -80°C. Three independent replicates were collected for each time point. Embryos were lysed in Tris 50mM pH 7.5, 4% SDS, protease inhibitor (Complete, Roche) using a dounce homogenizer (50 strokes per sample). Each sample was then boiled for 5 minutes at 95°C.
Sample preparation for mass-spectrometry analysis
In gel digestion was used for sample preparation. A detergent compatible (DC) protein assay (Bio-Rad) was used to measure protein concentration. Loading buffer (Tris 40mM pH 7.5, 2% SDS, 10% glycerol, 25 mM DTT final concentration) was added to the samples and they were boiled for 5 minutes at 95°C. Alkylation of cysteines was performed using iodoacetamide at a final concentration of 60mM for 30 min at room temperature in the dark. 100µg of protein per condition were loaded on a SDS-PAGE gel (acrylamide concentration of 4% for the stacking and 12% for the resolving). Proteins were concentrated in one band in the resolving gel (for the embryo developmental time course and to compare SWATH and DDA Label-Free quantification methods) or fully separated (for library construction). Gels were stained with colloidal coomassie and were cut and washed with a 50% acetonitrile (ACN), 25mM Ammonium Bicarbonate (AB) solution. Gel bands were dried with ACN and swollen with trypsin (Promega) in 50mM AB. 2µg or 500ng of trypsin were respectively used for single concentrated bands or bands from a full separation. Protein digestion was performed overnight at 37°C. The resulting peptides were extracted from the gel by two incubations in 10% formic acid, acetonitrile (1:1) for 15 min at 37 °C. The collected extractions were pooled with the initial digestion supernatant, dried in a Speed-Vac, and resuspended in 3% acetonitrile, 0.1% formic acid for mass-spectrometry analysis. HRM peptides (Biognosys) were added to each sample before injection on the mass spectrometer.
Data Dependent Acquisitions (DDA) Mass Spectrometry
Mass spectrometry analyses were performed on a TripleTOF 6600 mass spectrometer fitted with a Duospray ion source (AB SCIEX) and coupled to an ACQUITY UPLC System (WATERS). 5µg (for the embryo developmental time course) of sample was injected onto a MicroLC column (150 mm long x 0.3 mm inner diameter) with ChromXP C18CL, 300 Å pore size, 3 μm diameter particles (AB SCIEX). Samples were run using a 49 min gradient from 3–40% solvent B (solvent A 0.1% formic acid, 5% DMSO in water; solvent B: 0.1% formic acid, 5% DMSO in acetonitrile) at a flow rate of 5 µl/min. Data were acquired using an ion spray voltage of 5.5 kV, curtain gas at 25 psi and nebulizer gas at 10 psi. A DDA method was set up with the MS survey range set between 350 and 1250 m/z (250 ms accumulation time) followed by dependent MS/MS scans with a mass range set between 100 and 2000 m/z (100 ms accumulation time) of the 20 most intense ions in the high sensitivity mode with a 2+ to 5+ charge state. Dynamic exclusion was set for a period of 15 sec and a tolerance of 50 ppm. Rolling collision energy was used.
Analysis of Data Dependent Acquisitions (DDA) files
The DDA .wiff files were analysed with MaxQuant [24] version 1.5.2.8 software using default settings. The minimal peptide length was set to 7. Trypsin/P was used as digestion enzyme. Search criteria included carbamidomethylation of cysteine as a fixed modification, oxidation of methionine and acetyl (protein N terminus) as variable modifications. Up to two missed cleavages were allowed. The mass tolerance for the precursor was 0.07 and 0.006 Da for the first and the main searches respectively, and for the fragment ions was 50 ppm. TOF recalibration was allowed. The DDA files were searched against the D. melanogaster UniProt fasta database (July 2014, 41,773 sequences) in which the Biognosys iRT peptide sequences (11 entries) were added. The identifications were filtered to obtain FDR of 1% at the peptide and the protein level. No filter was applied to the number of peptide per protein.
For the MS1-based XICs quantification of DDA analysis, the re-quantification and match between runs modules of MaxQuant [24] were used and enabled the LFQ normalization method [25] was enabled (DDA LFQ) or not (DDA LF). Only unique peptides were used for quantification. When the LFQ normalization method was not used, the data were normalized by the median intensity in each condition. Fold change thresholds at 2 and 0.5 with an adjusted p-value inferior to 0.05 were used to consider a protein up or down-regulated. The p-values were adjusted using a Benjamini-Hochberg correction.
SWATH data acquisition
The LC gradient and TripleTOF 6600 mass spectrometer were set up as for the DDA acquisition as described above, but operated in SWATH mode using the following parameters: Acquisition of a 100-ms survey scan was followed by acquisition of 40 fragment ion spectra from 40 precursor isolation windows (swaths). The swaths were overlapping by 1 m/z and thus cover a range of 400-1250 m/z. Swath isolation windows were as follow: 399.5–422.0, 421.0–441.2, 440.2–456.6, 455.6-471.5, 470.5-484.7, 483.7-497.3, 496.3-510.0, 509.0-522.1, 521.1-533.6, 532.6-545.2, 544.2-555.6, 554.6-566.6, 565.6-577.0, 576.0-587.5, 586.5-597.4, 596.4-607.8, 606.8-618.3, 617.3-629.3, 628.3-640.3, 639.3-651.3, 650.3-663.4, 662.4-675.5, 674.5-688.7, 687.7-701.9, 700.9-715.1, 714.1-728.8, 727.8-743.1, 742.1-758.5, 757.5-774.5, 773.5-791.0, 790.0-809.1, 808.1-827.8, 826.8-848.2, 847.2-871.3, 870.3-897.1, 896.1-927.9, 926.9-965.9, 964.9-1015.4, 1014.4-1097.3, 1096.3-1249.7. The SWATH MS2 spectra were recorded with an accumulation time of 40 ms and cover 100-2000 m/z. The collision energy for each window was determined according to the calculation for a charge 2+ ion centred upon the window with a spread of 15.
SWATH analysis
For the generation of the SWATH assay library, 10 bands of a 1D gel fractionation of 100µg of a protein sample from embryos collected over a 16 hours period were acquired in Information Dependent Acquisition mode on a TripleTOF 6600 mass spectrometer (Sciex). To each peptide sample, HRM peptides (Biognosys) were added before analysis.
All the files were analysed using MaxQuant with similar parameters than for the DDA data analysis. The library was built using Spectronaut 7 from Biognosys [25], using the default settings, from the resulting combined file from the MaxQuant analysis. Spectronaut 7 (Biognosys) [26] was used to analyse the SWATH experiments. Default settings were used except for the retention time prediction type that was set to dynamic iRT with a correction factor for window of 2. Different Q-value were used to filter the reported intensity values (Q-values from 10-1 to 10-8). For the embryo development time course experiment and DDA and SWATH quantification methods comparison, a Q-value of 10-5 was used. For each proteins, the three peptides with the highest intensities were used for quantitative analysis. The data were normalized by the median protein intensity in each condition. Fold change thresholds at 2 and 0.5 with an adjusted p-value inferior to 0.05 were used to consider a protein up or down-regulated. The p-values were adjusted using a Benjamini-Hochberg correction.
Data Dependent Acquisition (DDA) and Parallel Reaction Monitoring (PRM) on a Q-Exactive mass-spectrometer
A list of peptides from DDA and SWATH analysis of the embryo developmental time course was prepared for PRM validation (at least 3 peptides per protein). An embryo sample was first run in DDA mode and analysed with MaxQuant to obtain the retention times of the peptides which were used to set up a scheduled PRM assay. Samples were trapped on a 100μm × 2cm, C18, 5μm, 100Ȧ trapping column (Acclaim PepMap 100) in µlPickUp Injection mode at 4 μL/min flow rate for 10 minutes. Samples were then loaded on a RSLC, 75μm × 50cm, nanoViper, C18, 3μm, 100Ȧ column (Acclaim, PepMap) retrofitted to an EASY-Spray source with a flow rate of 300nl/min (buffer A: HPLC H2O, 0.1% formic acid, buffer B: 100% ACN, 0.1% formic acid). A 120 minutes gradient was performed as follow; 0-3 min: 2% buffer B, 3-90 min: 2->40% buffer B, 90.3-95 min: 40%->90% buffer B, 95.3-120 min: 2% buffer B). Peptides were transferred to the gaseous phase with positive ion electrospray ionization at 2.1kV. In DDA the top 10 precursors were acquired between 400 and 1600 m/z with a 2Th (Thomson) selection window, dynamic exclusion of 30 seconds, normalised collision energy (NCE) of 25 and resolution of 70,000. For PRM, precursors were targeted in a 2Th selection window around the m/z of interest. Precursors were fragmented in HCD mode with NCE energy of 25. MS1 was performed at a 70,000 resolution, an AGC target of 3e6 and a maximum C-trap fill time of 200ms; MS/MS was performed at 35,000 resolution, an AGC target of 5e4 and a maximum C-trap fill time of 100ms. Spectra were analysed using Skyline with manual validation. Skyline quantitation data was exported to excel and the quantitation data was normalized against the TIC of the MS runs. The list of the peptides followed by PRM is given in the supplementary information (Supplementary Table 2).
Data availability
All the mass spectrometry data have been deposited to the ProteomeXchange Consortium [27] via the PRIDE partner repository with the dataset identifier PXD003178.
The results from the DDA LFQ and SWATH analysis of the time course experiment, the PRM results and the spectral library used in this study are provided in supplementary information (supplementary tables).
Total RNA Isolation, cDNA Synthesis and Quantitative Real-time PCR
For the qRT-PCR experiments, 2 biological replicates, independent from the replicates used in the MS experiments, were analysed. Total RNA was isolated using the TRIzol method (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. The quality of the extracted RNA was verified by gel electrophoresis and optical density measurements. First-strand cDNA was synthesized with the SuperScript® III Reverse Transcriptase Kit (Invitrogen) according to manufacturer instructions, the synthesized cDNAs were stored at −20°C for further qRT-PCR.
qRT-PCR was performed in the iCycler iQ system (Bio-Rad, Hercules, CA) and an IQ SYBR Green Supermix (Bio-Rad) using SensiMix™ SYBR® Hi-ROX Kit (Bioline). Data analysis was performed using comparative CT method following established protocols [28]. The rp49 (RpL32) mRNA was used as internal control and changes in mRNA levels were calculated as a fold-change relative to the value for the 0-4.5 hours sample. Primers were designed with Primer3 or retrieved from other databases [28]. Sequences of all primers used for qRT-PCR analyses are as following:
| Annotation symbol | protein | symbol | F name | Forward primer sequence | R name | Reverse primer sequence |
|---|---|---|---|---|---|---|
| CG4799 | P52295 | Pen | pen_F | CTGGCACAGATCAACAGACT | pen_R | CCTGGATCTGCTTCTGGTTAC |
| CG4916 | P23128 | me31B | me31b_F3 | GCCAAAGGACAACCGATTCAA | me31b_R3 | TCCCATCCCTTCTCGAATATACC |
| CG2637 | M9NDC3 | Fs(2)Ket | FS_F2 | TTCGTTCATCAAGCGCATCG | Fs_R2 | AAACGAAGTGCACAGATCGC |
| CG7939 | PO4359 | RpL32 | RpL32-F | AGCATACAGGCCCAAGATCG | RpL32-R3 | TGTTGTCGATACCCTTGGGC |
Results and discussion
Comparison of the performances of SWATH and DDA Label-Free quantification approaches
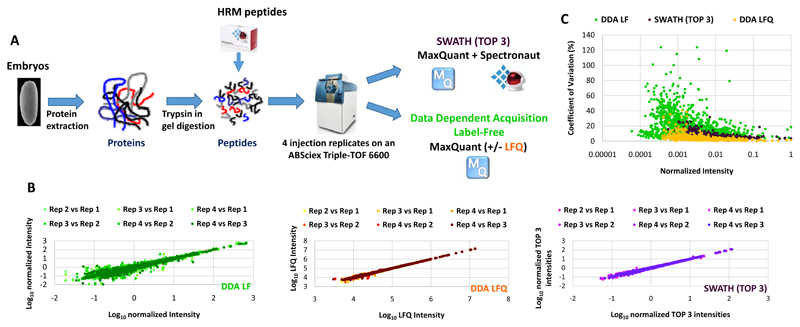
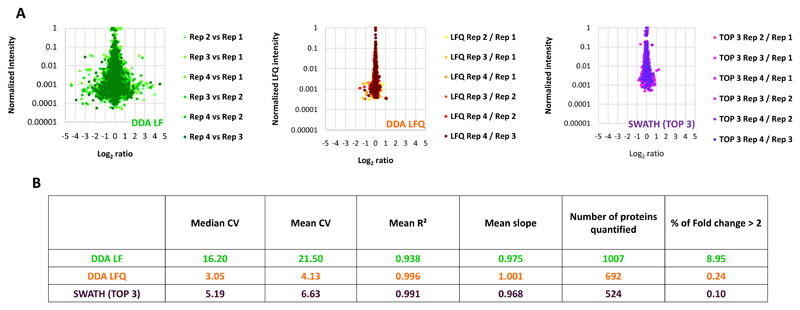
SWATH has been recently introduced as a label-free quantitative proteomic approach that can combine the advantages of DDA and targeted proteomic methods [21]. Few studies have compare SWATH quantification robustness and accuracy to other quantitative proteomic approaches [29, 30]. We first compared MS1-based XICs and SWATH label-free quantification methods (Figure 1A). Indeed, to our knowledge no comparison between the MS1-based XICs label-free approach using MaxQuant, which is one of the most used software to perform this kind of quantification, and the label-free SWATH quantification method has been performed. A protein sample prepared from 0-16hr D. melanogaster embryos and digested with trypsin was used as a standard to benchmark the different quantification methods. Four injection replicates were used to compare these quantification methods. For the MS1-based XICs, MaxQuant [24] analysis was performed with (DDA LFQ) or without (DDA LF) the LFQ normalization method [24] (Figure 1A). The LFQ normalization method developed by Cox et al [25], is a suite of algorithms used to normalize the MS1-based XICs between different LC-MS/MS runs. For the SWATH analysis, a library containing 1750 proteins was produced from a 0-16 hr embryo extract separated on a 1D SDS-PAGE gel, digested with trypsin and analysed in DDA mode on the same platform using MaxQuant and Spectronaut (Biognosys) [26]. 89% of the proteins present in the spectral library were previously identified in other Drosophila melanogaster embryo proteomics studies (ref Hughes and Gouw) (Supplementary Figure 2). Spectronaut was used to process the SWATH data and the average of the 3 highest peptide areas based on the sum of the fragment intensities were used to obtain a protein abundance index for the quantification at the protein level (Figure 1A). This approach has been shown to be suitable for label-free quantification approaches and provide a good accuracy (Rardin et al, Mol Cell Proteomics, 2015). Only the peptides of very high quality (Q-value < 1.10-5 corresponding to an FDR of 0.001%) were used for the SWATH analysis since these showed low CVs and a good run to run reproducibility at the peptide level when compared to DDA (Supplementary Figure 2). The results showed that SWATH generates more robust quantification data than DDA MS1-based XICs quantification on the same platform when the LFQ normalisation method is not used (DDA LF) (Figure 1B-1C and Figure 2A-B). Indeed, SWATH analysis provides better run to run intensity reproducibility (Figure 1B), generates lower CVs (Figure 1C) and less extreme ratios (Log2 ratios > 1 or < -1) when comparing the technical replicates (Figure 2A). In contrast, more proteins were quantified with the DDA MS1-based XICs quantification but at the cost of poor quality quantification (Figure 2B). However, when the LFQ normalization method was used (DDA LFQ), the quantification quality was greatly improved and more similar to that achieved with the SWATH analysis (Figure 1B-C and Figure 2A-B). Indeed, the observed median CVs are very close for both methods (3.05 and 5.19 for the DDA LFQ and SWATH, respectively) and both quantification approaches produce few extreme ratios (0.24 and 0.1% of Log2 ratios between the technical replicates > 1 or < -1 for the DDA LFQ and SWATH, respectively) (Figure 1B-C and Figure 2A-B). Thus, our analysis indicates that SWATH and DDA MS1-based XIC quantifications using the LFQ normalisation method in MaxQuant are robust tools for the analysis of changes in protein expression at the proteome level. Moreover, it has been shown that as these two types of quantification are performed at a different level (MS1 level for DDA MS1-based XICs and MS2 level for SWATH), they are complementary because both methods can produce errors of signal extraction and the discrepancies between the two approaches can point out this kind of problems (Rardin et al, Mol Cell Proteomics, 2015). We decided to use a combination of these two quantitative proteomics approaches to study the dynamics of the proteome of the D. melanogaster embryos during embryogenesis.
Figure 1. Comparison of Label-Free quantification methods.
A) Overall strategy for comparing SWATH and Data Dependent Acquisition Label-Free quantification methods. Protein were extracted from a Drosophila embryo sample, digested with trypsin and retention time normalisation (HRM) peptides spiked into the peptide sample before injection. Four analytical replicates were performed for SWATH and DDA acquisition modes on an ABSciex Triple-TOF 6600. The resulting files were analysed with MaxQuant with (DDA LFQ) or without (DDA LF) the LFQ module enabled or with MaxQuant and Spectronaut for the DDA and SWATH analysis, respectively. B) The Log10 intensities of the injection replicates were plotted pairwise to evaluate the run to run reproducibility of each quantification method. C) The Coefficient of variation (CV) was calculated for each protein quantified using the different quantification methods. The CVs were plotted as function of the intensities normalized to the maximal intensity.
Figure 2. Comparison of the number of extreme values generated by the different Label-Free quantification methods.
A) Ratio were calculated pairwise between the different injection replicates. Intensities, normalized to the maximal intensity, were plotted as function of the ratios for each quantification method. B) Table summarising the comparison of Label-Free quantification methods.
Monitoring protein expression during D. melanogaster embryonic development by Label-Free quantification and SWATH approaches
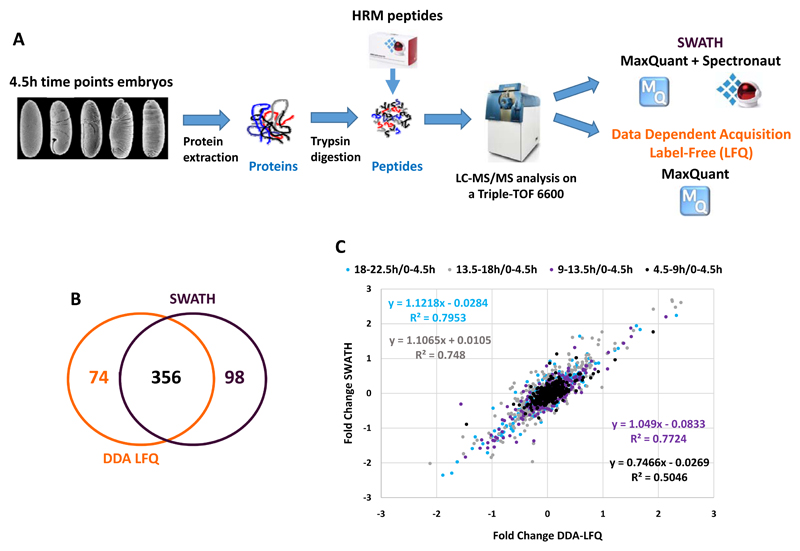
Despite its importance for all members of the Animal kingdom, very few quantitative data are available on the modulation of the proteome during embryonic development for any species [10, 31]. To begin to characterise how the proteome is regulated during embryogenesis, we used the D. melanogaster embryo as model since it is exceptionally well characterised at the genetic and genomic level [1]. Embryos were collected across development at 5 different time windows (0-4.5h, 4.5-9h, 9-13.5h, 13.5-18h and 18-22.5h) with three independent biological replicates subject to analysis in both DDA and SWATH acquisition modes (Figure 3A). The resulting files were analysed with MaxQuant using the LFQ normalization method or with MaxQuant and Spectronaut for DDA MS1-based XICs (DDA LFQ) and SWATH analysis respectively (Figure 3A). We identified over 400 proteins with each method that were quantified at all timepoints (430 with DDA LFQ and 475 with SWATH). All the timepoints were compared to the 0-4.5h timepoint. Importantly, the coefficients of variation between the biological replicates were found to be low (median CV around 10%) for both approaches, demonstrating the good quality of the data (Supplementary Figure 3). First, we observed that the protein ratios are more divergent at the later stages of embryonic development, indicating a global remodelling of the proteome across development (Supplementary Figure 4). However, the overall profile of the protein of the different stages look similar on a SDS-PAGE, suggesting that the expression of the most abundant proteins remain during embryogenesis (Supplementary Figure 5). Encouragingly, both datasets generated by DDA LFQ and SWATH resulted in similar profiles (Figure 3B).
Figure 3. Monitoring the dynamics of the proteome during embryogenesis by a combination of SWATH and DDA Label-Free quantification approaches.
A) Overall strategy to measure changes in protein expression levels across D. melanogaster embryonic development by SWATH and Data Dependent Acquisition Label-Free quantification methods. Embryos were collected for a developmental time course of five 4.5 hour timepoints. Proteins were extracted, digested with trypsin and HRM peptides spiked into the peptide sample before injection. The samples were analysed using both SWATH and DDA acquisition modes on an ABSciex Triple-TOF 6600 equipped with a microflow source. The resulting files were analysed with MaxQuant with the LFQ module (DDA LFQ) enabled or with MaxQuant and Spectronaut for the DDA and SWATH analysis, respectively. B) Venn diagram representing the overlap between the proteins identified in our DDA LFQ and SWATH analysis. C) Correlation between the ratio obtained with the DDA LFQ and SWATH analysis. The ratios measured by DDA LFQ and SWATH (4.5-9h/0-4.5h, 9-13.5h/0-4.5h, 13.5-18h/0-4.5h and 18-22.5h/0-4.5h) are plotted for each protein. The values of the fitted linear regression are reported on the graph.
Proteins regulated across D. melanogaster embryogenesis
To focus on a high confidence set of proteins we selected those that were differentially expressed at least two-fold (corrected p-value <0.05) compared to the first timepoint and found 31 proteins (15 up and 16 down) encoded by 31 genes for the DDA analysis and 64 proteins (36 up and 28 down) encoded by 64 genes for the SWATH analysis. The latter were significantly enriched (Benjamini and Hochberg corrected p= 3.7e-3) for the Gene Ontology term small molecule metabolic process, in particular multiple subunits of the ATP Synthase complex (see below). We also noticed that in both lists there was a significant enrichment for genes expressed in the embryonic/larval muscle system (Benjamini and Hochberg corrected p<1.0e3). Comparing both lists we identified 49 proteins that were common to both lists: 13 are expressed in the embryonic musculature. We elaborate on some of these commonly identified differentially expressed proteins.
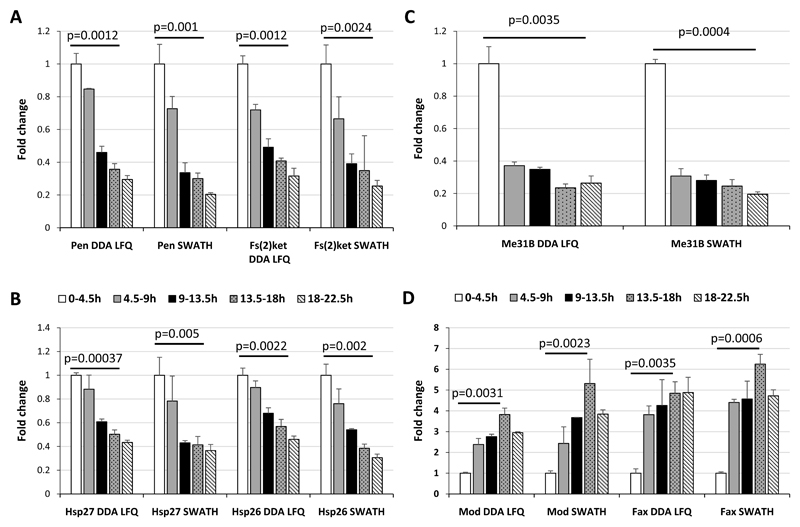
Among the differentially expressed proteins we identified, the alpha and beta subunits of the importin nuclear pore complex were down regulated over the timecourse (Pendulin and Fs(2)Ket respectively; Figure 4A). The importin heterodimer targets hundreds of proteins to the nuclear-pore complex (NPC) and facilitates their translocation across the nuclear envelope. The alpha subunit binds specifically to substrates containing either a simple or bipartite NLS (nuclear localization signal) motif [32] and the beta subunit mediates docking of the substrate to nucleoporins [32]. Both subunits of the heterodimer share very close expression profiles and our data support the view that regulation of the nuclear import system may play an important role in development [32]. An increase of the expression of Pen and Fs(2)Ket was also reported between the 2-4h and 10-12h embryonic time points using SILAC quantification (Hughes et al). The ratios measured for these proteins are very close to the ones observed in our study by DDA LFQ and SWATH for the comparison of the 0-4.5h and 9-13.5h time points (Supplementary Figure 7).
Figure 4. Proteins regulated across D. melanogaster embryonic development.
The ratios for each protein at each timepoint compared with the mean value at the 0-4.5h timepoint were calculated. The fold change obtained from the DDA Label-Free quantification across the time course were represented using histograms for: A) importin alpha (Pen) and beta (Fs(2)Ket); B) the heat-shock proteins Hsp27 and Hsp26; C) the ATP-dependent RNA helicase Me31b (Me31B); D) the DNA-binding protein Modulo (Mod) and Failed Axon Connections (Fax). Error bars represent the standard deviations between the biological replicates. Benjamini-Hochberg corrected p-values are shown for the comparison between the 0-4.5h timepoint and the 18-22.5h (A, B and C) or the 13.5-18h (D) timepoints.
We also observed decreased expression of two heat-shock proteins, Hsp26 and Hsp27, over the course of embryogenesis (Figure 4B), supporting previous studies that reported a reduction in Hsp27 expression during early development [33]. In addition, it has been shown that the subcellular localization of Hsp27 and Hsp26 varies between cellular compartments depending on developmental stages [33–35], indicating that the developmental profiles of Hsp26 and Hsp27 may be complex and suggesting that these proteins may have several roles during development. In general, our knowledge about the roles of heat-shock proteins across embryogenesis remains limited and Hsp27 and Hsp26 represent two interesting candidates for further investigations.
The DEAD-box ATP-dependent RNA helicase Me31B is involved in translation silencing through mRNA decapping [36] and was also found to decrease during embryogenesis (Figure 4C). As with the other four proteins discussed above, there is a strong maternal contribution of me31b mRNA and this appears to be reflected at the protein level [37]. Although Me31B appears to be required for normal oogenesis, the role of this protein is during embryonic development is poorly understood. We suspect that declining protein levels reflect, to a large extent, depletion of maternally contributed protein and mRNA. In support of this notion, we note that modENCODE expression profiles demonstrate that transcripts for all five of these genes dramatically decline at the onset of zygotic gene expression [6]. Interestingly, Gouw et al have also observed a decrease of the expression of Me31B at the protein level but in the very early steps of the embryo development (180-270min versus 0-90min time points).
In contrast, we observe increasing expression of the DNA-binding protein Modulo during embryogenesis (Figure 4D). Interestingly, RNA-seq data indicates relatively stable transcript levels across embryogenesis, suggesting post-transcriptional regulation. Modulo binds DNA and associates with chromatin [38] but it also contains four RNA Recognition Motifs and has been shown to provides a sequence-specific RNA binding activity as part of a nucleolar ribonucleoprotein complex [39]. It is speculated that it plays a vital role in the regulation of gene expression during Drosophila development. Due to its ability bind to nucleic acids and proteins, it may control the activity of genes critical for the process of morphogenesis of several embryonic territories via chromatin changes. Therefore, Modulo seems to be an interesting candidate to study in the context of development.
As with Modulo, the levels of the Failed Axon Connections (Fax) protein was also found to increase over embryonic development (Figure 4D) yet exhibit relatively stable transcript levels. This protein is the founding member of the conserved FAX family that, together with the Abl tyrosine kinase, is involved in axon development [40]. Mutations in fax act as dominant genetic enhancers of Abl mutant phenotypes and the Fax protein is expressed in a similar pattern to Abl in the embryonic mesoderm and axons of the central nervous system [40]. The increasing expression of Fax in the latter stages of embryonic expression is consistent with the role Fax plays in axonogenesis, since the majority of axon development occurs in the latter half of embryogenesis. In both these cases we have identified proteins encoded by transcripts that are relatively stable over embryonic development yet show significant changes in protein abundance, emphasising the need to gather more quantitative data on the proteome. Hughes et al also observed an increase of the expression of Modulo and Fax between a 2-4h and 10-12h using SILAC quantification with ratios very close to the ones measured in our study by DDA LFQ and SWATH when comparing the 0-4.5h and 9-13.5h time points (Supplementary Figure 7).
Energy production systems remodelling during D. melanogaster embryogenesis
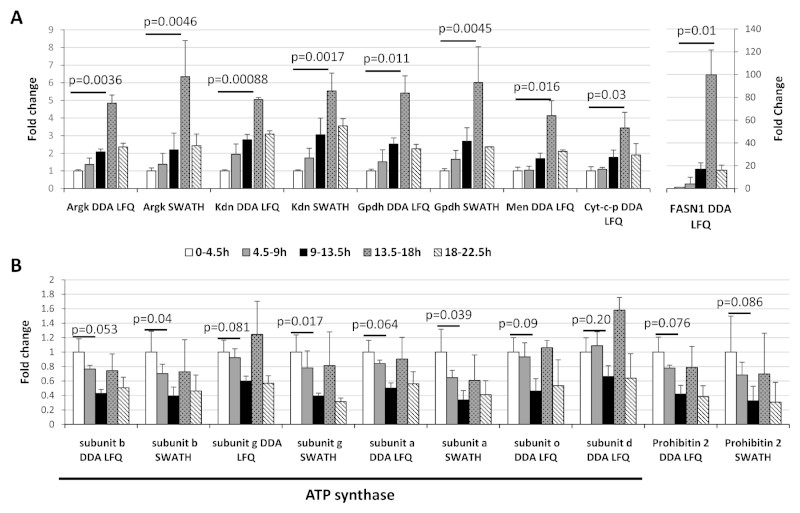
We found several enzymes involved in different energy metabolism steadily increased during embryogenesis, with peak expression at the 13.5-18h timepoint, before declining at the end of embryonic life (Figure 5). We observed this trend with proteins involved in the TCA cycle (Malic enzyme (Men) and Citrate synthase (Kdn)), amino acid metabolism (Arginine kinase (Argk)), lipid biosynthesis (Fatty acid synthase 1 (FASN1)) and electron transport chain respiration (Glycerol-3-phosphate dehydrogenase (Gpdh) and Cytochrome c proximal (Cyt-c-p)). During early development, the embryo undergoes considerable cellular remodelling which is likely to require high levels of energy and the induction of genes implicated in the main energy production pathways has already been observed at the mRNA level [41]. In contrast, subunits of the ATP synthase complex do not share the same profile (Figure 5A and B). Rather, we observed that 5 subunits (A, B, D, G and O) are all down regulated prior to peaking at 13.5-18h timepoint (Figure 5B). The profiles of some of these subunits are significant, and are similar to that of Prohibitin 2 which has been found to interact with the complex IV of the respiratory chain [42] (Figure 5B). The expression profile of the alpha subunit, representative for these results was confirmed by Parallel Reaction Monitoring (Supplementary Figure 4). These data suggests that the embryo regulates ATP production at key steps of the development by modulating the expression level of ATP synthase as well as enzymes involved in the aerobic and anaerobic energy production pathways. It is possible the regulation may be due to changes in O2 availability, for example, it is known that embryonic tracheal development is regulated by changing O2 [43]. In general, few data are available on the regulation of the metabolic pathways during embryogenesis and our new data hint at a complex interplay between metabolic pathways during development.
Figure 5. Remodelling of the energy production pathways during embryogenesis.
The ratios for different proteins involved in different metabolic pathways were calculated for each timepoint as compared to the 0-4.5h timepoint. A) The fold changes obtained from the DDA Label-Free quantification across the time course were represented using histograms for Arginine kinase (Argk), Citrate synthase (Kdn), Glycerol-3-phosphate dehydrogenase (Gpdh), Malic enzyme (Men), Cytochrome c proximal (Cyt-c-p) and the Fatty acid synthase 1 (FASN1). B) Five ATP synthase (ATPsyn) subunits and Prohibitin 2 (PHB2). Error bars represent the standard deviations between the biological replicates. Benjamini-Hochberg corrected p-values are shown for the comparison between the 0-4.5h timepoint and the 13.5-18h (A) or the 9-13.5h (B) timepoints.
Validation of protein expression changes by parallel reaction monitoring and quantitative real time PCR
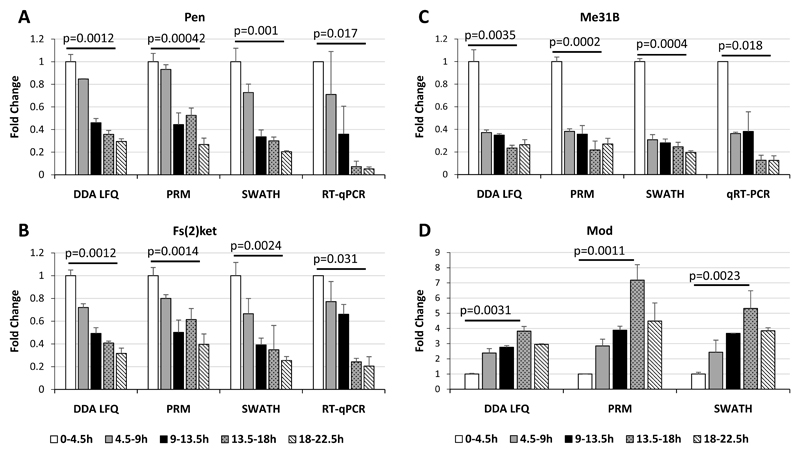
In order to validate our quantitative proteomics approaches we used parallel reaction monitoring (PRM) to measure expression changes. In the case of the alpha and beta importin subunits, Me31B and Modulo, the PRM data showed very good agreement with the DDA LFQ and SWATH data (Figure 6) We also checked mRNA expression for the importin subunits and Me31B by reverse transcriptase-quantitative PCR (Figure 6) and in all three cases saw that the trends in transcript levels mirrored the protein levels. In addition we note that the trend in transcript levels we measured were in broad agreement with the RNA-seq data from modENCODE. Taken together, these validations highlight the accuracy and reproducibility of the quantitative Label-Free methods we have used here.
Figure 6. Validation of expression profiles of a subset of proteins by PRM and qRT-PCR.
The expression profiles of: a) Pen; B) Fs(2)Ket; C) Me31B and D) Mod, were measured using Parallel Reaction Monitoring (PRM) and quantitative real time PCR (RT-qPCR). The ratios were calculated for timepoint in comparison with the 0-4.5h timepoint. Error bars represent the standard deviations between the biological replicates. Benjamini-Hochberg corrected p-values are shown for the comparison between the 0-4.5h timepoint and the 18-22.5h (A, B and C) or the 13.5-18h (D) timepoints.
Concluding remarks
In this study, we have applied Label-Free quantification methods to monitor dynamics in the D. melanogaster embryonic proteome. We compared the performance of SWATH and DDA MS1-based XICs quantification methods, showing that both approaches provide a robust way to measure variation at the proteome level. For DDA MS1-based XICs quantification we found that LFQ normalization with MaxQuant software gave the most robust performance, whereas SWATH gave comparable results without any sophisticated normalization. However, it would be interesting to apply the MaxQuant LFQ normalization algorithms to SWATH data to see if it can improve SWATH quantification quality and analytical coverage. Using a combination of DDA MS1-based XICs and SWATH quantification approaches, we found that the expression of more than 70 proteins was regulated across embryonic development. Validation of some of our results with orthogonal proteomics experiments or mRNA analysis increases our confidence in the reliability of these approaches when applied to comparative analysis of the developmental proteome. We found that proteins involved in heat-shock response, nuclear protein import or energy production pathways were differentially expressed across development, some showing expected declines due to utilisation of maternal stores, whereas others showed hallmarks of post-transcriptional regulation. We believe the preliminary work reported here is the first proteomics study covering a substantial portion of Drosophila embryogenesis and sets the scene for more detailed analysis. Although more in depth analysis of the proteome will provide additional insights, this study highlights the complexity of proteome dynamics during development and identifies some regulated biochemical pathways likely to be important. Finally, this study serves as a foundation for more detailed, higher resolution analyses to better characterise differential protein expression during embryogenesis in this well-established model system.
Supplementary Material
Significance of the study.
Lewis Wolpert is credited with the famous quote that gastrulation is the most important time in your life, highlighting the prominence of just one of the many steps in embryogenesis needed to build an organism from a single cell. Many genetic, biochemical and genomic studies have provided us with a better understanding of the role of many individual proteins play in key aspects of embryogenesis. However, in truth we have very little information on the dynamics of the wider proteome across development and this is much needed if we wish to understand how the complexity of development is controlled. In this study we use a combination of Label-Free proteomic quantification methods to monitor protein expression changes over a time course of Drosophila embryo development. We identify regulation of key pathways, such as nuclear protein import, heat-shock response and energy production systems during embryogenesis, providing new insights on proteome dynamics and a foundation for further detailed functional studies of the developmental proteome.
Acknowledgments
Funding: B.F, D.K and L.G are funded by BBSRC (Ref: BB/L002817/1). MR and JV acknowledge funding by the Wellcome Trust (RG 093735/Z/10/Z) the ERC (Starting grant 260809), M.R. is a Wellcome Trust Research Career Development and Wellcome-Beit Prize fellow.
Abbreviations
- MS
Mass-Spectrometry
- LC
Liquid Chromatography
- DDA
Data-Dependent Acquisition
- DIA
Data Independent Acquisition
- LF
Label-Free Quantification
- SWATH
Sequential Window Acquisition of all THeoretical fragment-ion
- PRM
Parallel Reaction Monitoring
- qRT-PCR
quantitative Real Time Polymerase Chain Reaction
Footnotes
The authors have declared no conflict of interest.
References
- [1].Fristrom JW. The developmental biology of Drosophila. Annu Rev Genet. 1970;4:325–346. doi: 10.1146/annurev.ge.04.120170.001545. [DOI] [PubMed] [Google Scholar]
- [2].Adams MD, Celniker SE, Holt RA, Evans CA, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- [3].An PN, Yamaguchi M, Bamba T, Fukusaki E. Metabolome analysis of Drosophila melanogaster during embryogenesis. PloS one. 2014;9:e99519. doi: 10.1371/journal.pone.0099519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Fortini ME, Skupski MP, Boguski MS, Hariharan IK. A survey of human disease gene counterparts in the Drosophila genome. J Cell Biol. 2000;150:F23–30. doi: 10.1083/jcb.150.2.f23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Weigmann K, Klapper R, Strasser T, Rickert C, et al. FlyMove--a new way to look at development of Drosophila. Trends Genet. 2003;19:310–311. doi: 10.1016/S0168-9525(03)00050-7. [DOI] [PubMed] [Google Scholar]
- [6].Graveley BR, Brooks AN, Carlson JW, Duff MO, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Manak JR, Dike S, Sementchenko V, Kapranov P, et al. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat Genet. 2006;38:1151–1158. doi: 10.1038/ng1875. [DOI] [PubMed] [Google Scholar]
- [8].Tomancak P, Beaton A, Weiszmann R, Kwan E, et al. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002;3:RESEARCH0088. doi: 10.1186/gb-2002-3-12-research0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tomancak P, Berman BP, Beaton A, Weiszmann R, et al. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2007;8:R145. doi: 10.1186/gb-2007-8-7-r145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sun L, Champion MM, Huber PW, Dovichi NJ. Proteomics of Xenopus development. Molecular human reproduction. 2015 doi: 10.1093/molehr/gav052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Artieri CG, Fraser HB. Transcript length mediates developmental timing of gene expression across Drosophila. Mol Biol Evol. 2014;31:2879–2889. doi: 10.1093/molbev/msu226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bensimon A, Heck AJ, Aebersold R. Mass spectrometry-based proteomics and network biology. Annu Rev Biochem. 2012;81:379–405. doi: 10.1146/annurev-biochem-072909-100424. [DOI] [PubMed] [Google Scholar]
- [13].Cox J, Mann M. Quantitative, high-resolution proteomics for data-driven systems biology. Annu Rev Biochem. 2011;80:273–299. doi: 10.1146/annurev-biochem-061308-093216. [DOI] [PubMed] [Google Scholar]
- [14].Bantscheff M, Lemeer S, Savitski MM, Kuster B. Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Analytical and bioanalytical chemistry. 2012;404:939–965. doi: 10.1007/s00216-012-6203-4. [DOI] [PubMed] [Google Scholar]
- [15].Ebhardt HA, Root A, Sander C, Aebersold R. Applications of targeted proteomics in systems biology and translational medicine. Proteomics. 2015;15:3193–3208. doi: 10.1002/pmic.201500004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Molecular systems biology. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gallien S, Duriez E, Crone C, Kellmann M, et al. Targeted proteomic quantification on quadrupole-orbitrap mass spectrometer. Molecular & cellular proteomics : MCP. 2012;11:1709–1723. doi: 10.1074/mcp.O112.019802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gallien S, Kim SY, Domon B. Large-Scale Targeted Proteomics Using Internal Standard Triggered-Parallel Reaction Monitoring (IS-PRM) Molecular & cellular proteomics : MCP. 2015;14:1630–1644. doi: 10.1074/mcp.O114.043968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Soste M, Hrabakova R, Wanka S, Melnik A, et al. A sentinel protein assay for simultaneously quantifying cellular processes. Nature methods. 2014;11:1045–1048. doi: 10.1038/nmeth.3101. [DOI] [PubMed] [Google Scholar]
- [20].Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nature methods. 2012;9:555–566. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- [21].Gillet LC, Navarro P, Tate S, Rost H, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Molecular & cellular proteomics : MCP. 2012;11 doi: 10.1074/mcp.O111.016717. O111 016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Schubert OT, Gillet LC, Collins BC, Navarro P, et al. Building high-quality assay libraries for targeted analysis of SWATH MS data. Nature protocols. 2015;10:426–441. doi: 10.1038/nprot.2015.015. [DOI] [PubMed] [Google Scholar]
- [23].Tsou CC, Avtonomov D, Larsen B, Tucholska M, et al. DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nature methods. 2015;12:258–264. doi: 10.1038/nmeth.3255. 257 p following 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- [25].Cox J, Hein MY, Luber CA, Paron I, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Molecular & cellular proteomics : MCP. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bruderer R, Bernhardt OM, Gandhi T, Miladinovic SM, et al. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Molecular & cellular proteomics : MCP. 2015;14:1400–1410. doi: 10.1074/mcp.M114.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Vizcaino JA, Deutsch EW, Wang R, Csordas A, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nature biotechnology. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- [29].Bourassa S, Fournier F, Nehme B, Kelly I, et al. Evaluation of iTRAQ and SWATH-MS for the Quantification of Proteins Associated with Insulin Resistance in Human Duodenal Biopsy Samples. PloS one. 2015;10:e0125934. doi: 10.1371/journal.pone.0125934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].McQueen P, Spicer V, Schellenberg J, Krokhin O, et al. Whole cell, label free protein quantitation with data independent acquisition: quantitation at the MS2 level. Proteomics. 2015;15:16–24. doi: 10.1002/pmic.201400188. [DOI] [PubMed] [Google Scholar]
- [31].Hughes CS, Foehr S, Garfield DA, Furlong EE, et al. Ultrasensitive proteome analysis using paramagnetic bead technology. Molecular systems biology. 2014;10:757. doi: 10.15252/msb.20145625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14:505–514. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- [33].Michaud S, Marin R, Tanguay RM. Regulation of heat shock gene induction and expression during Drosophila development. Cell Mol Life Sci. 1997;53:104–113. doi: 10.1007/PL00000572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Glaser RL, Wolfner MF, Lis JT. Spatial and temporal pattern of hsp26 expression during normal development. EMBO J. 1986;5:747–754. doi: 10.1002/j.1460-2075.1986.tb04277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pauli D, Tonka CH, Tissieres A, Arrigo AP. Tissue-specific expression of the heat shock protein HSP27 during Drosophila melanogaster development. J Cell Biol. 1990;111:817–828. doi: 10.1083/jcb.111.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Nishihara T, Zekri L, Braun JE, Izaurralde E. miRISC recruits decapping factors to miRNA targets to enhance their degradation. Nucleic Acids Res. 2013;41:8692–8705. doi: 10.1093/nar/gkt619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].de Valoir T, Tucker MA, Belikoff EJ, Camp LA, et al. A second maternally expressed Drosophila gene encodes a putative RNA helicase of the "DEAD box" family. Proc Natl Acad Sci U S A. 1991;88:2113–2117. doi: 10.1073/pnas.88.6.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bantignies F, Goodman RH, Smolik SM. The interaction between the coactivator dCBP and Modulo, a chromatin-associated factor, affects segmentation and melanotic tumor formation in Drosophila. Proc Natl Acad Sci U S A. 2002;99:2895–2900. doi: 10.1073/pnas.052509799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Perrin L, Romby P, Laurenti P, Berenger H, et al. The Drosophila modifier of variegation modulo gene product binds specific RNA sequences at the nucleolus and interacts with DNA and chromatin in a phosphorylation-dependent manner. J Biol Chem. 1999;274:6315–6323. doi: 10.1074/jbc.274.10.6315. [DOI] [PubMed] [Google Scholar]
- [40].Hill KK, Bedian V, Juang JL, Hoffmann FM. Genetic interactions between the Drosophila Abelson (Abl) tyrosine kinase and failed axon connections (fax), a novel protein in axon bundles. Genetics. 1995;141:595–606. doi: 10.1093/genetics/141.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tennessen JM, Baker KD, Lam G, Evans J, Thummel CS. The Drosophila estrogen-related receptor directs a metabolic switch that supports developmental growth. Cell Metab. 2011;13:139–148. doi: 10.1016/j.cmet.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Strub GM, Paillard M, Liang J, Gomez L, et al. Sphingosine-1-phosphate produced by sphingosine kinase 2 in mitochondria interacts with prohibitin 2 to regulate complex IV assembly and respiration. FASEB J. 2011;25:600–612. doi: 10.1096/fj.10-167502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Simon MC, Keith B. The role of oxygen availability in embryonic development and stem cell function. Nat Rev Mol Cell Biol. 2008;9:285–296. doi: 10.1038/nrm2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the mass spectrometry data have been deposited to the ProteomeXchange Consortium [27] via the PRIDE partner repository with the dataset identifier PXD003178.
The results from the DDA LFQ and SWATH analysis of the time course experiment, the PRM results and the spectral library used in this study are provided in supplementary information (supplementary tables).