Abstract
Background & Aims
Tests to measure serum endomysial antibodies (EMA) and antibodies to tissue transglutaminase (tTG) were developed to screen for celiac disease in patients consuming gluten. However, they are commonly used to monitor patients on a gluten-free diet (GFD). We conducted a meta-analysis to assess the sensitivity and specificity of tTG IgA and EMA IgA assays in identifying patients with celiac disease who have persistent villous atrophy despite a GFD.
Methods
We searched PUBMED, EMBASE, BIOSIS, SCOPUS, clinicaltrials.gov, Science Citation Index, and Cochrane Library databases through November 2016. Inclusion criteria were studies of subjects with biopsy-confirmed celiac disease, follow-up biopsies and measurement of serum antibodies on a GFD, biopsy performed on subjects regardless of symptoms or antibody test results. Our analysis excluded subjects with refractory celiac disease, undergoing gluten challenge, or consuming a prescribed oats-containing GFD. Tests were considered to have positive or negative findings based on manufacturer cut-off values. Villous atrophy was defined as a Marsh 3 lesion or villous height:crypt depth ratio below 3.0. We constructed forest plots to determine the sensitivity and specificity of detection for individual studies. For the meta-analysis, a bivariate random effects model was used to jointly model sensitivity and specificity.
Results
Our search identified 5408 unique citations. Following review of abstracts, 442 articles were reviewed in detail. Only 26 studies (6 of tTG assays, 15 of EMA assays, and 5 of tTG and EMA assays) met our inclusion criteria. The most common reason studies were excluded from our analysis was inability to cross-tabulate histologic and serologic findings. The serum assays identified patients with persistent villous atrophy with high levels of specificity: 0.83 for the tTG IgA assay (95% CI, 0.79–0.87) and 0.91 for the EMA IgA assay (95% CI, 0.87–0.94). However, they detected villous atrophy with low levels of sensitivity: 0.50 for the tTG IgA assay (95% CI, 0.41–0.60) and 0.45 for the EMA IgA assay (95% CI, 0.34–0.57). The tests had similar levels of performance in pediatric and adult patients.
Conclusions
In a meta-analysis of patients with biopsy-confirmed celiac disease undergoing follow-up biopsy on a gluten-free diet, we found that tests for serum tTG IgA and EMA IgA levels had low sensitivity (below 50%) in detection of persistent villous atrophy. We need more-accurate non-invasive markers of mucosal damage in children and adults with celiac disease who are following a GFD.
Keywords: tissue transglutaminase antibody, endomysial antibody, monitoring and follow up diagnostics
Introduction
Serum endomysial antibodies (EMA) were first reported as a biomarker of dermatitis herpetiformis and celiac disease by Chorzelski and coworkers in 19841. Identification of tissue transglutaminase (tTG) as the autoantigen to which EMA antibodies bind2 led to the development of tTG antibody screening tests which are less labour intensive than immunofluorescent assays for EMA. Widespread availability of these tests along with increased awareness has facilitated diagnosis of celiac disease. Consequently, there is a growing population of patients with biopsy confirmed celiac disease who have been advised to follow a gluten-free diet that require follow-up care3. Although serum tTG and EMA IgA antibody tests were never intended for the routine monitoring of patients with celiac disease, this use is pervasive and advocated by several gastroenterology societies4–6.
In celiac disease, similar to other chronic intestinal conditions, such as inflammatory bowel disease, meaningful monitoring requires tools that reliably reflect mucosal health. Intestinal biopsy is the gold standard, yet serial intestinal biopsies are not obtained routinely due to their invasiveness, cost and inherent risks5. Consequently, serum tTG and/or EMA IgA tests are commonly used to monitor patients and are often interpreted clinically as reflecting mucosal damage on a gluten-free diet. These so-called “celiac antibody tests” were initially developed and validated for screening for celiac disease among untreated persons consuming a gluten-containing diet and they perform well in this context7,8. The aim of the present study is to assess whether serum tTG or EMA IgA antibody tests are useful biomarkers of villous atrophy in patients with celiac disease treated with a gluten-free diet.
Methods
Search Strategy
The following databases were searched: PubMed, Embase (OVID, 1974 to December 17 2012), Cochrane Central Register of Controlled Trials (CENTRAL, Issue 1, January 2013), Science Citation Index Expanded (ISI, 1970–2013), BIOSIS Previews (ISI, 1926 to January 17 2013), Clinical Trials.gov (137 687 studies registered; December 20 2012), and Scopus (January 28, 2013). The PubMed search strategy was developed by a librarian experienced in systematic review searching, and peer reviewed by other librarians, using the Peer Review Electronic Search Strategy (PRESS) standard9. The PubMed search was then adapted for the other databases and limited to English language and Human. The search results were updated from the date January 2013 to November 2016. All databases except Clinical Trials.gov were searched and the additional limit to articles (as appropriate) was applied. The search strategies are presented in Appendix 1 and are publically available via institutional repository10.
Inclusion criteria
Inclusion criteria were (1) subjects with a biopsy confirmed diagnosis of celiac disease; (2) follow-up biopsy on a gluten-free diet; (3) serum antibody measurement contemporaneous with biopsy (as defined by the authors); (4) biopsy of subjects with both negative and positive antibody testing regardless of symptoms; (5) sufficient data presented to enable construction of contingency tables. Subjects with refractory celiac disease, undergoing gluten challenge, or consuming a prescribed oats-containing gluten-free diet were excluded.
Records review
All identified relevant abstracts and articles were independently reviewed by two authors who selected studies based upon the inclusion criteria described above. A standardized data abstraction form was used to collect the following information: publication year; study design; geographic region of study; characteristics of study subjects (e.g., age, gender, duration of gluten-free diet at time of follow-up biopsy); and experimental methods (e.g., diet adherence assessments, serologic assays, intestinal biopsy, histology reporting). Data were abstracted and contingency tables were constructed independently by two investigators. Any discrepancies were resolved by consensus or by arbitration involving a third author.
Quality assessment of studies
Many studies were not designed to answer the question of interest and there was a wide variability in data reporting. Thus, we developed a quality assessment score based upon the four Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) domains (patient selection, index test, reference standard, and flow and timing)11. Scores ranging for 0–4 were assigned for each of 9 criteria: cohort size; handling of IgA deficiency; time between biopsy and antibody testing; patient account of diet; dietician assessment; method, location and number of biopsies; and blinding of pathologist (Table 1). For handling of IgA deficiency, studies which did not consider IgA status were scored 0. In cases where all participants had positive serology at diagnosis and/or a positive antibody test was an inclusion criteria, subjects were presumed to be IgA sufficient and a score of 4 was assigned. In cases where data were unavailable, an attempt was made to contact corresponding authors by email. Possible total scores ranged from 0–36. Studies were further categorized as low (1–12), medium (13–24) or high (25–36) quality.
Table 1.
Quality scoring matrix
| Criterion | Score | ||||
|---|---|---|---|---|---|
|
| |||||
| 0 | 1 | 2 | 3 | 4 | |
| Dietitian assessment | No/Not reported | Yes not clear what they did | Yes Dietician interview | Yes documented using scoring system | |
|
|
|||||
| Patient account of diet | None/Not reported | Self-report | Diet recall | Food record/diary | |
|
|
|||||
| Biopsy Location | Not specified | Jejunum | Distal duodenum | Duodenal bulb | Both bulb and distal duodenum |
|
|
|||||
| Number of biopsies | <2 or Not reported | 2 | 3 | 4–5 | ≥6 |
|
|
|||||
| Biopsy method | Not reported | Capsule | Capsule and/or endoscopy | Endoscopy | |
|
|
|||||
| Blinding of pathologist | No/not reported | Yes | |||
|
|
|||||
| Cohort size | <10 | 10–49 | 50–99 | 100–199 | ≥200 |
|
|
|||||
| IgA deficiency | Not reported or included IgA deficient | IgA deficient included with IgG based testing | Only IgA sufficient included or IgA deficiency excluded | ||
|
|
|||||
| Interval between antibody test and biopsy | Not reported | <26 weeks | 26 weeks – 12 weeks | 12 weeks – 1 week | <1 week |
Definitions
Antibody testing was considered to be ‘positive’ or ‘negative’ as reported in the manuscript. Where this was not apparent and/or multiple cut-offs were used, tests were classified using the manufacturer’s recommended cut-off. Subjects with ‘indeterminate’ antibody testing were excluded. Histologic Marsh classification was considered the ‘gold standard’. Villous atrophy was predefined as Marsh 3 (destructive lesions with flat mucosa)12 or, where quantitative methods were used, villous height:crypt depth ratio (Vh:CrD) < 3.0. Thus, for the primary analysis, true positives were those with positive antibody testing and villous atrophy and true negatives were those with negative antibody testing and intact villi (Marsh 0, 1 or 2 or Vh:CrD > 3). We also performed a secondary analysis of the ability to discern Marsh 0–1 from Marsh 2–3 lesions.
Statistical Analysis
Forest plots were constructed to depict the sensitivity and specificity of the individual studies. Tests of diagnostic accuracy often display considerable variation which may reflect true heterogeneity. Thus, in addition to visual assessment with the use of the forest plots, the extent of heterogeneity was estimated by the area under the prediction zone. For meta-analysis, a bivariate random effects model was used to jointly model sensitivity and specificity13,14. This approach accounts for the known negative correlation between sensitivity and specificity while a random effects model is appropriate in settings such as diagnostic testing where heterogeneity is due to variations in the study populations or procedures used.
Results are presented as a summary receiver operating characteristic (ROC) plot with sensitivity (true positive rate) on the y-axis and 1-specificity (false negative rate) on the x-axis. In addition to individual and summary points, the 95% confidence region denotes the precision of the pooled estimate of the available studies and the 95% prediction region shows the area where the next study is likely to lie, which reflects variability among studies. Statistical analysis was performed using R15 version 3.3.1 with RStudio16 version 0.99.903. All authors had access to the study data and reviewed and approved the final manuscript.
Results
Identification of studies
Initially, 9302 records were identified through the database search and brought into an EndNote database, where duplicate references were removed, resulting in 4120 records for screening. In the search update, 2378 records were identified and after duplicate references were removed, 1288 records were screened (Figure S1). A review of the titles and abstracts eliminated 4,966 articles because the articles did not include both antibody testing and histology (1247), related to diagnosis or screening (1157), were non-primary studies (1015) or case reports (927), or did not include the target population (556) or tests (64).
The remaining 442 manuscripts were reviewed in detail, and 412 were eliminated because the articles were not primary studies (70), did not include both antibody testing and histology (164), it was not possible to construct a contingency table from the data provided (101), antibody testing and/or histology was used to define study population or gluten-free diet adherence (55) or to select subjects to biopsy (6), not the target population (5) or test (2), biopsy and/or antibody testing were done but not reported (6), conflicting data in the manuscript (2) or subjects intentionally infected with hookworm (1). The 30 remaining articles underwent a detailed assessment with data extraction.
Following detailed review, two studies were excluded because biopsies were performed only on symptomatic subjects on a GFD17,18, a large epidemiologic study was excluded because there was no information about gluten-free diet adherence19 and a prospective study of 30 patients with 100% mucosal recovery, all of whom had negative EMA antibody titers at 12 months, was also excluded20. Two studies which reported results of EMA as well as tTG assays with guinea pig tTG as a substrate21,22 were excluded from analysis of tTG due to the known inferior sensitivity and specificity of guinea pig tTG compared to human tTG23.
Study and population characteristics
There were 26 studies which qualified to be included (6 tTG24–29, 15 EMA30–42, and 5 both tTG and EMA21,22,43–47; Tables 2 and 3). All but two were single center studies and most originated from Italy (10) or elsewhere in Europe (11). Remaining studies were from North America (2), Australia (2) and India (1). Six studies included pediatric subjects (4 EMA, 1 tTG, 1 both tTG and EMA). The number of eligible subjects based on inclusion criteria ranged from 12 to 945 while the actual number of subjects with complete data ranged from 11 to 390. Subject attrition from analysis ranged from 0% to 94%. Reasons for attrition were not always apparent due to the retrospective nature of many studies. Substrates for EMA IgA were typically human umbilicus or monkey esophagus. The stated duration of a gluten-free diet ranged from 2 to 600 months. Most authors reported pooled data. In cases where subjects had serial biopsies at prescribed intervals, we selected the data for the longest duration reported.
Table 2.
Characteristics of included studies of tTG IgA antibodies
| Author (year), country | Assay substrate | Age (years) | Male (%) | Study design | Total participants | Participants with complete data | GFD duration (months) | |
|---|---|---|---|---|---|---|---|---|
| Bannister (2014)24, Australia | Human | mean 7.5 range 1–16 |
43 | Retrospective cohort | Longitudinal | 150 | 1501 | median 17 range 12–149 |
| Duerksen (2010)43, Canada | Human | mean 50.5 | 14 | Prospective cohort | Cross-sectional | 22 | 21 | mean 116 range 16–600 |
| Hopper (2008)44, UK | Human | mean 53 range 21–78 |
31 | Prospective cohort | Longitudinal | 48 | 48 | minimum 12 |
| Lichtwark (2014)25, Australia | Human | mean 33 range 22–39 |
27 | Prospective cohort | Longitudinal | 16 | 11 | prescribed,12 |
| Pekki (2015)26, Finland | Human | median 45 range 15–75 |
32 | Prospective cohort | Longitudinal | 263 | 263 | prescribed, 12 |
| Raivio (2006)45, Finland | Human | median 58 range 23–82 |
33 | Prospective cohort | 91 | 91 | median108 range 12–288 |
|
| Rubio-Tapia (2010)46, USA | Human | median 47 range 18–84 |
27 | Retrospective cohort | Longitudinal | 241 | 162 | minimum 60 |
| Sharkey (2013)27, UK | Human | Adult | ND | Retrospective database | 447 | 237 | mean 11 range 2–268 |
|
| Sjöberg (2014)47, Sweden | ND | mean 8.3 range 0.8–12 |
37 | Prospective cohort | Longitudinal | 13 | 13 | mean 13 range 11–15 |
| Volta (2008)28, Italy | Human | median 35 range 14–70 |
ND | Prospective cohort | Longitudinal | 53 | 53 | prescribed,12 |
| Zanini (2012)29, Italy | Human | Adult range 16–82 |
ND | Retrospective database | Cross-sectional | 945 | 60 | median 12 |
Only 129 subjects in meta-analysis, there were 21 subjects with indeterminate results;
Non-adherent subjects (11) excluded from analysis;
GFD – Gluten-free diet; UK – United Kingdom; USA – United States of America; ND - unable to determine.
Table 3.
Characteristics of included studies of EMA IgA antibodies
| Author (year), country | Assay Substrate1 | Age (years) | Male (%) | Study Design | Total participants | Participants with complete data | GFD duration (months) | |
|---|---|---|---|---|---|---|---|---|
| Cammarota (2007)22, Italy | ND | range 19–66 | 16 | Prospective cohort | Cross-sectional | 62 | 62 | mean 11.3 range 6–14 |
| Ciacci (2002)30, Italy | Monkey | mean 34.8 | 23 | Prospective cohort | Longitudinal | 698 | 390 | mean 83 range 24–264 |
| Cuoco (1998)31, Italy | ND | adult | ND | Retrospective | Cross-sectional | 23 | 13 | prescribed, 12 |
| Dickey (2000)32, Ireland | Monkey | mean 51 range 16–18 |
26 | Prospective cohort | Longitudinal | 62 | 53 | prescribed, 12 |
| Duerksen (2010)43, Canada | Human | mean 50 | 14 | Prospective cohort | Cross-sectional | 22 | 21 | mean 116 range 16–600 |
| Hopper (2008)44, UK | Monkey | mean 52.7 range 21–78 | 31 | Prospective cohort | Longitudinal | 48 | 48 | minimum 12 |
| Kaukinen (2002)21, Finland | Human | median 49 range 22–73 | 28 | Prospective cohort | Longitudinal | 87 | 872 | median 12 range 12–216 |
| Kolacek (2004)33, Croatia | Monkey | mean 5.4 range 3–10 | ND | Prospective case series | Cross-sectional | 17 | 17 | minimum 24 |
| O’Keeffe (2001)34, Ireland | ND | mean 37 range 17–59 |
33 | Retrospective cohort | Cross-sectional | 12 | 12 | mean 36 range 2–84 |
| Raivio (2006)45, Finland | Human | median 58 range 23–82 |
33 | Prospective cohort | 91 | 91 | median108 range 12–288 |
|
| Rubio-Tapia (2010)46, USA | Monkey | median 47 range 18–84 | 27 | Retrospective cohort | Longitudinal | 241 | 124 | >60 |
| Sategna-Guidetti (1993)36, Italy | Monkey | Adult range 15–71 |
ND | Prospective cohort | Longitudinal | 19 | 14 | prescribed, 12 |
| Sategna-Guidetti (1996)37, Italy | Monkey | range 18–68 | 45 | Prospective cohort | Longitudinal | 47 | 47 | ND |
| Sategna-Guidetti (1997)35, Italy | Human & Monkey | adult | ND | Prospective cohort | Longitudinal | 47 | 47 | prescribed, 12 |
| Sjöberg (2014)47, Sweden | ND | mean 8.3 range 0.8–12 |
37 | Prospective cohort | Longitudinal | 28 | 13 | mean 13 range 11–15 |
| Troncone (1995)38, Italy | Monkey | mean 14 range 10–19 |
66 | Prospective cohort | Longitudinal | 23 | 23 | > 120 |
| Valentini (1994)39, Italy | Monkey | adult | ND | Prospective cohort | Longitudinal | 33 | 33 | mean 9.9 range 6–12 |
| Vécsei (2009)40, Austria | Monkey | median 44 range16–74 | 34 | Retrospective database | Longitudinal | 250 | 26 | > 24 |
| Vécsei (2014)41, Austria | Monkey | pediatric | ND | Prospective cohort | Cross sectional | 53 | 53 | median 26 range 12–155 |
| Yachha (2007)42, India | Monkey | mean 12 | 56 | Prospective cohort | Longitudinal | 25 | 21 | mean 44 range 14–84 |
Monkey esophagus or human umbilicus;
Non-adherent subjects (11) excluded from analysis;
ND - unable to determine; GFD – Gluten-free diet; UK – United Kingdom; USA – United States of America;
Quality assessment of studies
Cumulative quality scores ranged from 1 to 33 with studies of low (7), medium (7) and high (12) quality (Table 4). Low scores were most commonly due to failure to report the data in the categories of dietician assessment (14 studies), patient report of diet (18), blinding of pathologist (16 studies), handling of IgA deficiency (19 studies) and interval between antibody test and biopsy (17 studies). Four or more biopsies were obtained in 17 studies, with only 6 studies including duodenal bulb biopsies. Quality of reporting in the manuscript did not change significantly between older and more recent studies; however, scores were ultimately higher for more recent studies because authors were successfully contacted by email. All scores improved when authors responded (median 7, range 1 to 17). For all studies of low quality, authors could not be contacted so low ratings reflect missing data.
Table 4.
Quality scoring of included studies
| Author (year), country | Dietician assessment | Patient report of diet | Biopsy location | Number of biopsies | Biopsy method | Blinding of pathologist | Cohort Size | Handling of IgA Deficiency | Interval between lab test and biopsy | Total score |
|---|---|---|---|---|---|---|---|---|---|---|
| Bannister (2014)24, Australia | 0 | 4 | 4 | 3 | 4 | 4 | 3 | 3 [0] | 3 | 28 [25] |
| Cammarota (2007)22, Italy | 0 | 0 | 2 | 3 | 4 | 4 | 2 | 4 [0] | 0 [0] | 19 [15] |
| Ciacci (2002)30, Italy | 4 [0] | 4 [0] | 2 | 2 | 4 | 4 | 4 | 4 [0] | 3 [0] | 31 [16] |
| Cuoco (1998)31, Italy | 0 | 2 | 2 | 3 | 4 | 0 | 1 | 0 | 0 | 12 |
| Dickey (2000)32, Ireland | 2 | 0 | 2 | 2 | 4 | 4 | 2 | 4 | 0 | 20 |
| Duerksen (2010)43, Canada | 4 | 4 | 2 | 3 | 4 | 4 | 1 | 3 [0] | 4 [0] | 29 [22] |
| Hopper (2008)44, UK | 0 | 0 | 2 | 3 | 4 | 4 | 1 | 0 | 4 | 18 |
| Kaukinen (2002)21, Finland | 3 | 4 | 2 | 3 [0] | 4 | 3 [0] | 2 | 4 | 3[0] | 28 [19] |
| Kolacek (2004)33, Croatia | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| Lichtwark (2014)25, Australia | 4 | 4 | 4 | 4 | 4 | 4 | 1 | 0 | 4 | 29 |
| O’Keeffe (2001)34, Ireland | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 0 | 5 |
| Pekki (2015)26, Finland | 4 | 4 [0] | 2 | 4 | 4 | 0 [0] | 4 | 3 [0] | 4 [0] | 29 [18] |
| Raivio (2006)45, Finland | 3 [0] | 3 [0] | 2 | 3 [0] | 4 | 4 [0] | 2 | 4 | 4 [0] | 29 [12] |
| Rubio-Tapia (2010)46, USA | 4 | 3 [0] | 3 | 3 | 4 | 0 [0] | 4 | 3 [0] | 3[0] | 27 [18] |
| Sategna-Guidetti (1993)36, Italy | 0 | 0 | 2 | 0 | 2 | 0 | 1 | 0 | 0 | 5 |
| Sategna-Guidetti (1996)37, Italy | 2 | 0 | 2 | 3 | 4 | 4 | 1 | 0 | 0 | 16 |
| Sategna-Guidetti (1997)35, Italy | 0 | 0 | 2 | 0 | 4 | 0 | 1 | 0 | 0 | 7 |
| Sharkey (2013)27, UK | 3 | 4 [0] | 2 | 3 | 4 | 0 [0] | 4 | 4 | 3[0] | 27 [20] |
| Sjöberg ( 2014)47, Sweden | 2 | 4 | 2 | 1 [0] | 2 | 4 | 1 | 0 | 4 | 20 [19] |
| Troncone (1995)38, Italy | 4 | 3 | 2 | 0 [0] | 2 | 4 [0] | 1 | 4 [0] | 1 [0] | 21 [12] |
| Valentini (1994)39, Italy | 0 | 0 | 2 | 0 | 2 | 0 | 1 | 0 | 0 | 5 |
| Vécsei (2009)40, Austria | 4 [0] | 3 [0] | 4 | 4 | 4 | 0 | 4 | 4 | 4 | 31 [24] |
| Vécsei (2014)41, Austria | 4 [0] | 3 [0] | 4 | 4 | 4 | 4 | 2 | 4 | 4 | 33 [26] |
| Volta (2008)28, Italy | 3 | 3 | 4 [0] | 4 [0] | 4 [0] | 0 [0] | 2 | 3 [0] | 4 | 27 [12] |
| Yachha (2007)42, India | 0 | 0 | 2 | 3 | 4 | 0 | 1 | 0 | 1 | 11 |
| Zanini (2012)29, Italy | 2 | 0 | 2 | 3 | 4 | 0 | 1 | 0 | 3 | 15 |
Quality scores were determined by rating each of the following from 0 to 4: dietitian assessment, patient account of diet, biopsy location, number of biopsies, biopsy method, blinding of pathologist, cohort size, handling of IgA deficiency and interval between antibody test and biopsy. For a detailed description of the scoring system, please refer to Table 1. In cases where authors responded to email requests for clarification, the score is based upon scoring after qualification and initial scores are indicated in brackets. UK – United Kingdom; USA – United States of America.
Diagnostic accuracy of tTG IgA antibodies for detecting persistent villous atrophy on a GFD
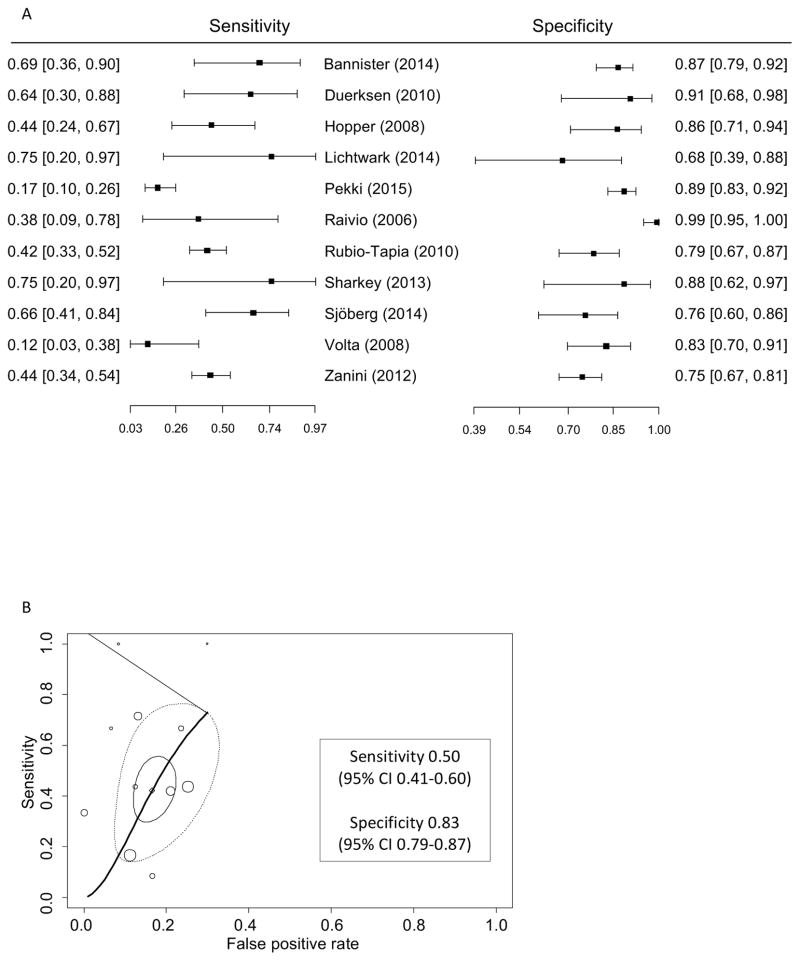
Eleven studies including 1088 patients (31% with villous atrophy) were used in the meta-analysis of the diagnostic accuracy of tTG IgA for predicting persistent villous atrophy on a gluten-free diet. Sensitivity of tTG IgA ranged from 0.12 to 0.75 and specificity ranged from 0.75 to 0.99 (Figure 2). The bivariate model point estimates were sensitivity 0.50 (95% CI 0.41–0.60) and specificity 0.83 (95% CI 0.79–0.87). The area under the summary ROC was 0.781 (Figure 1). For pediatric subjects, the area under the summary ROC was 0.879 (2 studies; 142 subjects; Table 5). The relatively large area of the 95% prediction region reflects the high heterogeneity of the included studies. Sensitivity and specificity of tTG IgA for villous atrophy in treated celiac disease did not differ with the number or location of biopsies, assay type, biopsy method or patient age (Table 5).
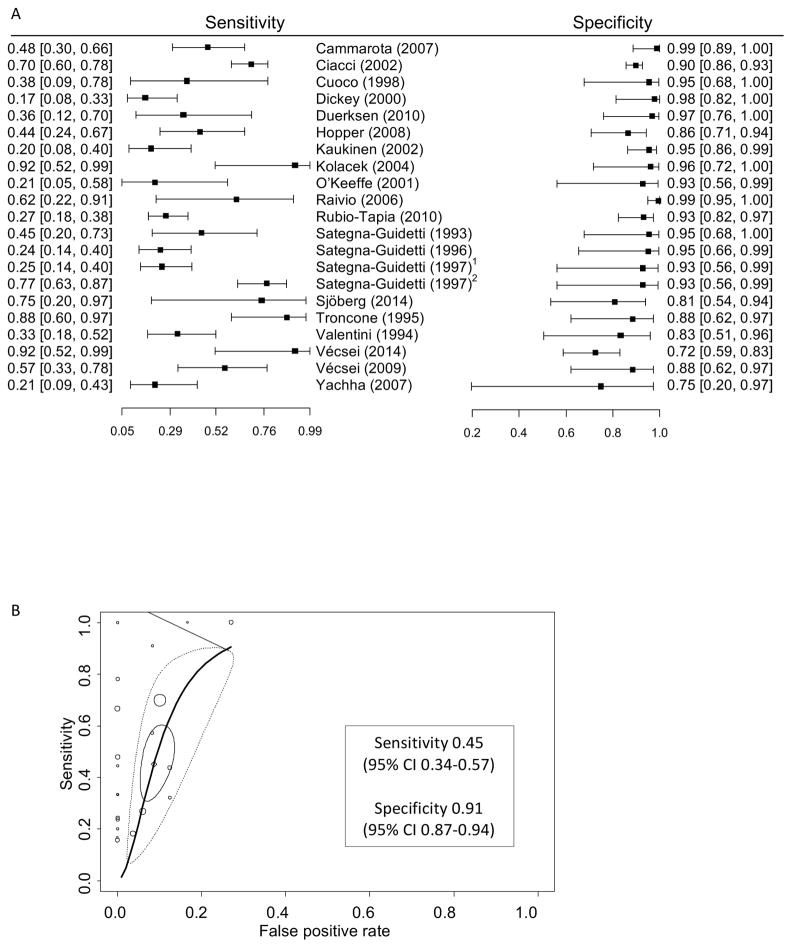
Figure 2.
Performance of EMA IgA for detection of persistent villous atrophy in patients with biopsy confirmed celiac disease on a gluten-free diet.
A. Forest plot of sensitivity and specificity of EMA IgA for persistent villous atrophy on a gluten-free diet.
B. Summary receiver operating characteristic curve of EMA IgA for persistent villous atrophy. In addition to individual and summary points, the 95% confidence region (solid circle) denotes the precision of the pooled estimate of the available studies and the 95% prediction region (dashed circle) shows the area where the next study is likely to lie, which reflects variability among studies. 1monkey, 2human.
Figure 1.
Performance of tTG IgA for detection of persistent villous atrophy in patients with biopsy confirmed celiac disease on a gluten-free diet.
A. Forest plot of sensitivity and specificity of tTG IgA for persistent villous atrophy on a gluten-free diet.
B. Summary receiver operating characteristic curve of tTG IgA for persistent villous atrophy. In addition to individual and summary points, the 95% confidence region (solid circle) denotes the precision of the pooled estimate of the available studies and the 95% prediction region (dashed circle) shows the area where the next study is likely to lie, which reflects variability among studies.
Table 5.
Sub-group analysis of factors associated with assay performance
| tTG IgA | EMA IgA | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Subgroup | Sensitivity (95% CI) | Specificity (95% CI) | AUC | Subgroup | Sensitivity (95% CI) | Specificity (95% CI) | AUC |
| All studies (n=13) | 0.42 (0.32–0.53) | 0.83 (0.79–0.87) | 0.764 | All studies (n=20)1 | 0.45 (0.34–0.57) | 0.91 (0.87–0.94) | 0.871 |
| Age | Age | ||||||
| Pediatric (n=2) | 0.70 (0.38–0.90) | 0.87 (0.80–0.91) | 0.879 | Pediatric (n=5) | 0.74 (0.35–0.94) | 0.78 (0.66–0.87) | 0.806 |
| Adult (n=9) | 0.38 (0.27–0.51) | 0.80 (0.75–0.85) | 0.720 | Adult (n=16) | 0.39 (0.50–0.71) | 0.93 (0.90–0.95) | 0.906 |
| Biopsy method | Biopsy method | ||||||
| Endoscopic (n=8) | 0.44 (0.31–0.58) | 0.81 (0.75–0.86) | 0.756 | Endoscopic (n=14) | 0.42 (0.29–0.57) | 0.92 (0.87–0.95) | 0.863 |
| Crosby capsule (n=3) | 0.27 (0.08–0.62) | 0.93 (0.71–0.99) | 0.598 | Crosby capsule (n=5) | 0.56 (0.31–0.78) | 0.89 (0.78–0.95) | 0.889 |
| Biopsy number | Biopsy number | ||||||
| < 4 biopsies (n=1) | - | - | < 4 biopsies (n=4) | 0.64 (0.25–0.90) | 0.90 (0.82–0.95) | 0.906 | |
| ≥ 4 biopsies (n=9) | 0.42 (0.27–0.58) | 0.84 (0.79–0.87) | 0.811 | ≥ 4 biopsies (n=11) | 0.37 (0.27–0.48) | 0.91 (0.85–0.95) | 0.741 |
| Biopsy location | Biopsy location | ||||||
| Bulb biopsied (n=4) | 0.55 (0.37–0.73) | 0.81 (0.73–0.86) | 0.794 | Bulb biopsied (n=3) | 0.57 (0.20–0.88) | 0.86 (0.67–0.95) | 0.836 |
| Distal duodenum (n=7) | 0.32 (0.21–0.45) | 0.83 (0.76–0.88) | 0.700 | Distal duodenum (n=10) | 0.43 (0.31–0.55) | 0.92 (0.89–0.94) | 0.906 |
| Assay substrate | |||||||
| Human umbilicus (n=4) | 0.49 (0.21–0.78) | 0.97 (0.92–0.99) | 0.966 | ||||
| Monkey esophagus (n=13) | 0.45 (0.30–0.61) | 0.90 (0.85–0.93) | 0.858 | ||||
AUC (area under the receiver operator curve);
Sategna-Guidetti (1997) included twice because both substrates were used.
Diagnostic accuracy of EMA IgA antibodies for detecting persistent villous atrophy on a GFD
Twenty studies including 1189 patients (38% with villous atrophy) were used in the meta-analysis of the diagnostic accuracy of EMA IgA for predicting persistent villous atrophy on a gluten-free diet. The sensitivity of EMA IgA ranged from 0.17 to 0.92 with a bivariate model point estimate of 0.45 (95% CI 0.34–0.57) and specificity ranged from 0.72 to 0.99 with a bivariate model point estimate of 0.91 (95% CI 0.87–0.94). The area under the summary ROC was 0.871 (Figure 2). For pediatric subjects, the area under the summary ROC was 0.806 (5 studies; 127 subjects; Table 5). Heterogeneity was less than for tTG IgA. There was one study that compared the performance of assays using human umbilical vein to monkey esophagus. The sensitivity of human tissue was significantly higher (0.77 vs 0.25), but specificity was the same (0.93)35. There was no significant difference in sensitivity or specificity of the substrates when comparing between studies (Table 5) nor was there a clear relationship between sensitivity and degree of villous atrophy (data not shown).
Diagnostic accuracy of EMA IgA and tTG IgA antibodies for detecting Marsh 2 or 3 lesions
When comparing diagnostic accuracy for Marsh 0–1 vs Marsh 2–3 lesions, there were 924,25,28,29,41,44,45,47,48 eligible studies of tTG IgA (479 patients; 25% Marsh 2–3 lesions; see supplementary figure S2). Sensitivity of tTG IgA ranged from 0.16 to 0.79 and specificity ranged from 0.68 to 0.99. The bivariate model point estimates were 0.50 (95% CI 0.32–0.69) for sensitivity and 0.86 (95% CI 0.80–0.91) for specificity for Marsh 2–3 lesions. The area under the ROC was 0.839. There were 1722,31–42,44,45,47,48 eligible studies of EMA IgA (594 patients; 47% Marsh 2–3 lesions; see supplementary figure S3). Sensitivity of EMA IgA ranged from 0.17 to 0.93 with a bivariate model point estimate of 0.44 (95% CI 0.32–0.57) and specificity ranged from 0.74 to 0.99 with a bivariate model point estimate of 0.90 (95% CI 0.84–0.94). The area under the ROC was 0.851.
Discussion
This meta-analysis demonstrates the limitations of celiac antibody testing as a surrogate marker for mucosal recovery in persons with celiac disease on a gluten-free diet. While both tTG IgA and EMA IgA had relatively high specificity for villous atrophy (tTG IgA 0.83, 95% CI 0.79–0.87; EMA IgA 0.91, 95% CI 0.87–0.94), their sensitivities are low (tTG IgA 0.50, 95% CI 0.41–0.60; EMA IgA 0.45, 95% CI 0.34–0.57). Consequently, the majority of persons with villous atrophy on a gluten-free diet had normal levels of tTG or EMA.
This contrasts with the high sensitivity (tTG IgA 85–95%; EMA IgA 80–90%) and high specificity (tTG IgA 95–99% ; EMA IgA 95–100%) of these tests in untreated celiac disease49. The vastly different test performance characteristics in the treated population may reflect decreased quantity and frequency of gluten exposure. In controlled gluten challenge studies, histologic damage precedes elevation of tTG IgA or EMA IgA antibodies by several weeks50. Thus, intermittent unsuspected gluten exposure may partially explain the high rate of false negative antibody tests. False positives were less common. This may reflect the discordance between normalization of serum antibodies and mucosal recovery following initiation of a gluten-free diet51. This difference may also relate to the dose of gluten exposure. Prior to diagnosis and initiation of a gluten-free diet, individuals may be consuming many grams of gluten per day. Many individuals on a gluten-free diet significantly reduce their gluten intake, but may continue to be exposed to trace amounts52 which may be sufficient to cause persistent mucosal damage. Overall, 38% had persistent villous atrophy and rates of recovery were higher in children (81%) than adults (62%). This is consistent with historical rates of mucosal recovery over the period studied19 which suggests that the participants in the included studies are representative of patients with celiac disease on a GFD.
Recommendations to monitor tTG and/or EMA antibodies endure because there are no other validated sensitive measures that predict mucosal recovery. Alternatives, including lactulose mannitol based intestinal permeability testing53,54, intestinal fatty acid binding protein55, and simvastatin absorption56 have been proposed, but none have been widely adopted. Novel tools to measure excretion of gluten immunogenic peptides appear promising as a marker of gluten-free diet adherence57–59, but are limited to use in trials of alternative therapies to a gluten-free diet60. Our findings highlight the need for a sensitive non-invasive biomarker that predicts mucosal recovery that can be used routinely to monitor individuals with celiac disease.
This systematic review was limited by few studies specifically designed to examine the relationship between serum antibody testing and mucosal damage in patients who are trying to follow a gluten-free diet. Many potentially eligible studies were designed to answer a specific clinical question using protocols which included antibody testing and follow-up duodenal biopsies. Frequently, results were reported in a way which precluded cross-tabulation of histologic findings and antibody test results. This was the most common reason for exclusion from this systematic review. Systematic reviews of diagnostic test accuracy do not commonly include analysis of publication bias; nevertheless, the number of studies which otherwise qualified for inclusion in which it was not possible to link serology and histology exceeded the number of included studies by a factor of four which suggests that there may be systematic underreporting. Given the number of studies involved, it was not practical to contact authors of these studies to enquire whether more detailed results were available and this would have potentially introduced additional biases.
Where possible, we included only those persons allegedly adherent to a gluten-free diet. Given the complexities of determining GFD adherence, persons with ongoing gluten exposure have likely been included. Arguably, these persons would be more likely to have concordance between mucosal findings and serum antibodies which would be expected to make the tests appear to be more, not less, accurate. Theoretically, if serologic recovery precedes histologic recovery, then inclusion of subjects who had a follow-up biopsy very soon after diagnosis might underestimate test performance. There was 1 study of tTG27 (237 patients) and 3 studies of EMA22,34,39 (total 107 patients) which included any patients who had been on a GFD for less than 11 months. Based upon the mean/median GFD duration in these studies, the number of patients involved is small and unlikely to have meaningfully influenced our findings. It is an important question whether persistent villous atrophy is related to lack of healing or if there are interspersed periods of healing and re-injury. This requires careful prospective studies with serial biopsy and close monitoring of gluten exposure and cannot be answered using the current study design.
There was considerable heterogeneity in the included studies; however, patients with varying degrees of gluten exposure who have been on a gluten-free diet for varying time periods is most reflective of the population that practitioners see in clinical practice. Similarly, participants with negative or unknown antibody test results at diagnosis could not be excluded based upon the data reported. Given a sensitivity of 85–90% for tTG IgA at diagnosis, this may have contributed to the rate of false negative serologic tests, but is unlikely to have significantly changed the findings of our meta-analysis. Comparing results from antibody tests is also complicated by use of different manufacturers, substrates and laboratories. In our analysis, neither the substrate for EMA nor the type of tTG test appeared to significantly affect performance. Some have suggested alternative cut-offs for these tests, particularly for patients on a gluten-free diet. In this study, we chose to use the manufacturer recommended cut-off as this is the threshold most likely to be used clinically. We also excluded those with “indeterminate” test results as it is unclear how to interpret these results. Ultimately, there were only 21 subjects (in a study of 150 subjects24) with indeterminate results which represents less than 2% of the total and their exclusion is unlikely to have meaningfully affected our findings.
Strengths of this study include the identification, through quality assessment scores, of problems associated with inadequate reporting of data in the celiac medical literature. There is great variability in how well authors document patient and dietician reported gluten-free diet adherence, and methodological variables, such as blinding of pathologists, location of biopsies, number of biopsies and interpretation of biopsies. Inclusion of studies from different regions of the world and EMA assays performed by different operators increases the generalizability of our findings to clinical practice. One might argue that villous atrophy is a high bar and that Marsh 1 and 2 lesions may also be of concern. Given that the celiac antibody tests suffer from low sensitivity, we chose to use a more severe lesion as the reference point because failure to detect villous atrophy is a more significant shortcoming than failure to detect less severe Marsh 1 or 2 lesions. As well, there is very poor correlation among pathologists regarding Marsh 2 lesions and much better agreement regarding Marsh 3 lesions61. In fact, Marsh 2 lesions were relatively uncommon in most studies and test performance characteristics did not improve when Marsh 2 and 3 lesions were considered. The number of studies which pooled Marsh 0 and Marsh 1 lesions precluded a meaningful analysis of Marsh 0 vs any abnormality (Marsh 1, 2, or 3).
A gluten-free diet is difficult to follow and the treatment burden is high62. Patients require ongoing and accurate feedback regarding gluten consumption as this is a measure of the effectiveness of their self-management63. Currently, silent gluten exposures typically go undetected until a repeat biopsy is performed64. Useful monitoring tools for patients with celiac disease must be sensitive to gluten ingestion, highly predictive of mucosal recovery outcomes, convenient, and affordable. A tool which is sensitive to gluten ingestion is necessary to alert the patient that self-management is inadequate. The ability to reliably predict mucosal status is even more necessary. Mucosal recovery, not elimination of gluten, is the therapeutic goal. Persistent mucosal damage is associated with bone disease65, cancer66 and possibly excess mortality19,46. Failure to detect persistent villous atrophy delays institution of dietary or behavioral modifications to reduce gluten exposure67.
Although widely available, and relatively non-invasive, serum tTG IgA and EMA IgA antibodies are poorly correlated with mucosal outcomes. Most patients with celiac disease have negative antibody tests on a gluten-free diet, even those with persistent mucosal damage. A positive test result is helpful as this has good specificity for persistent villous atrophy (tTG IgA 0.82, EMA IgA 0.91) and signals probable ongoing gluten ingestion. Such a finding should prompt dietary assessment and review with a dietitian with expertise in celiac disease. A negative antibody test is much less informative and should not be interpreted as an indicator of mucosal recovery nor as a proxy for gluten-free diet adherence. The high proportion of studies that were excluded because antibody tests were used to determine which patients to biopsy and/or to define gluten-free diet adherence suggests that such misinterpretations are common. In the absence of a non-invasive biomarker, follow-up duodenal biopsy remains the only appropriate test to assess mucosal recovery in children and adults with celiac disease.
Supplementary Material
Acknowledgments
Grant support: JAS is supported by NIH/NIDDK T32 DK 07760 and a Fellowship from the Canadian Institutes of Health Research and the Canadian Association of Gastroenterology.
The authors thank Candice Lewis, Document Delivery Supervisor at the University of Manitoba Neil John Maclean Health Sciences Library for her perseverance in successfully obtaining all required manuscripts.
Abbreviations
- EMA
anti-endomysial antibody
- GFD
gluten-free diet
- tTG
anti-tissue transglutaminase antibody
- Vh:CrD
villous height to crypt depth ratio
Footnotes
Disclosures: DAL is an employee of Takeda Pharmaceuticals. AS, DRD, CPK, SK and JAS have nothing to disclose.
Author contributions: DRD and JAS designed the study. AS designed and conducted the literature search. DRD, SK and JAS reviewed abstracts and manuscripts. SK and JAS wrote the first draft of the paper. All authors contributed to data analysis and data interpretation, and critically reviewed the manuscript for important intellectual content. JAS and SK contributed equally to this work.
References
Author names in bold designate shared co-first authorship.
- 1.Chorzelski TP, Beutner EH, Sulej J, et al. IgA anti-endomysium antibody. A new immunological marker of dermatitis herpetiformis and coeliac disease. Br J Dermatol. 1984;111:395–402. doi: 10.1111/j.1365-2133.1984.tb06601.x. [DOI] [PubMed] [Google Scholar]
- 2.Dieterich W, Ehnis T, Bauer M, et al. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat Med. 1997;3:797–801. doi: 10.1038/nm0797-797. [DOI] [PubMed] [Google Scholar]
- 3.Kelly CP, Bai JC, Liu E, et al. Advances in Diagnosis and Management of Celiac Disease. Gastroenterology. 2015;148:1175–1186. doi: 10.1053/j.gastro.2015.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kagnoff MF. AGA Institute Medical Position Statement on the Diagnosis and Management of Celiac Disease. Gastroenterology. 2006;131:1977–1980. doi: 10.1053/j.gastro.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ludvigsson JF, Bai JC, Biagi F, et al. Diagnosis and management of adult coeliac disease: guidelines from the British Society of Gastroenterology. Gut. 2014;63:1210–28. doi: 10.1136/gutjnl-2013-306578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai JC, Ciacci C, Corazza GR, et al. Celiac disease. World Gastroenterol Organ Glob Guidel. 2016:1–35. [Google Scholar]
- 7.Rostom A, Murray JA, Kagnoff MF. American Gastroenterological Association (AGA) Institute technical review on the diagnosis and management of celiac disease. Gastroenterology. 2006;131:1981–2002. doi: 10.1053/j.gastro.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Lewis NR, Scott BB. Meta-analysis: Deamidated gliadin peptide antibody and tissue transglutaminase antibody compared as screening tests for coeliac disease. Aliment Pharmacol Ther. 2010;31:73–81. doi: 10.1111/j.1365-2036.2009.04110.x. [DOI] [PubMed] [Google Scholar]
- 9.Sampson M, McGowan J, Cogo E, et al. An evidence-based practice guideline for the peer review of electronic search strategies. J Clin Epidemiol. 2009;62:944–952. doi: 10.1016/j.jclinepi.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 10.Szwajcer A, Silvester J. Systematic Review and Meta-Analysis of the Sensitivity and Specificity of TTG and EMA IgA antibodies for persistent mucosal damage in persons with celiac disease following a gluten-free diet: Literature search documentation. 2016:8. Available at: http://hdl.handle.net/1993/31937.
- 11.Whiting PF, Rutjes AWS, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–36. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 12.Marsh MN. Gluten, major histocompatibility complex, and the small intestine. A molecular and immunobiologic approach to the spectrum of gluten sensitivity (‘celiac sprue’) Gastroenterology. 1992;102:330–54. [PubMed] [Google Scholar]
- 13.Leeflang MMG. Systematic reviews and meta-analyses of diagnostic test accuracy. Clin Microbiol Infect. 2014;20:105–13. doi: 10.1111/1469-0691.12474. [DOI] [PubMed] [Google Scholar]
- 14.Reitsma JB, Glas AS, Rutjes AWS, et al. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol. 2005;58:982–90. doi: 10.1016/j.jclinepi.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 15.R Core Team. R: A Language and Environment for Statistical Computing. 2016 Available at: http://www.r-project.org.
- 16.RStudio Team. RStudio: Integrated Development for R. 2016. [Google Scholar]
- 17.Carroccio A, Brusca I, Iacono G, et al. IgA anti-actin antibodies ELISA in coeliac disease: a multicentre study. Dig Liver Dis. 2007;39:818–23. doi: 10.1016/j.dld.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Carroccio A, Ambrosiano G, Di Prima L, et al. Clinical symptoms in celiac patients on a gluten-free diet. Scand J Gastroenterol. 2008;43:1315–21. doi: 10.1080/00365520802200044. [DOI] [PubMed] [Google Scholar]
- 19.Lebwohl B, Granath F, Ekbom a, et al. Mucosal healing and mortality in coeliac disease. Aliment Pharmacol Ther. 2013;37:332–9. doi: 10.1111/apt.12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fotoulaki M, Nousia-Arvanitakis S, Augoustidou-Savvopoulou P, et al. Clinical application of immunological markers as monitoring tests in celiac disease. Dig Dis Sci. 1999;44:2133–2138. doi: 10.1023/a:1026651224985. [DOI] [PubMed] [Google Scholar]
- 21.Kaukinen K, Sulkanen S, Mäki M, et al. IgA-class transglutaminase antibodies in evaluating the efficacy of gluten-free diet in coeliac disease. Eur J Gastroenterol Hepatol. 2002;14:311–5. doi: 10.1097/00042737-200203000-00017. [DOI] [PubMed] [Google Scholar]
- 22.Cammarota G, Cuoco L, Cesaro P, et al. A highly accurate method for monitoring histological recovery in patients with celiac disease on a gluten-free diet using an endoscopic approach that avoids the need for biopsy: a double-center study. Endoscopy. 2007;39:46–51. doi: 10.1055/s-2006-945044. [DOI] [PubMed] [Google Scholar]
- 23.Wong RCW, Wilson RJ, Steele RH, et al. A comparison of 13 guinea pig and human anti-tissue transglutaminase antibody ELISA kits. J Clin Pathol. 2002;55:488–494. doi: 10.1136/jcp.55.7.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bannister EG, Cameron DJ, Ng J, et al. Can celiac serology alone be used as a marker of duodenal mucosal recovery in children with celiac disease on a gluten-free diet? Am J Gastroenterol. 2014;109:1478–83. doi: 10.1038/ajg.2014.200. [DOI] [PubMed] [Google Scholar]
- 25.Lichtwark IT, Newnham ED, Robinson SR, et al. Cognitive impairment in coeliac disease improves on a gluten-free diet and correlates with histological and serological indices of disease severity. Aliment Pharmacol Ther. 2014;40:160–170. doi: 10.1111/apt.12809. [DOI] [PubMed] [Google Scholar]
- 26.Pekki H, Kurppa K, Maki M, et al. Predictors and Significance of Incomplete Mucosal Recovery in Celiac Disease After 1 Year on a Gluten-Free Diet. Am J Gastroenterol. 2015;110:1078–1085. doi: 10.1038/ajg.2015.155. [DOI] [PubMed] [Google Scholar]
- 27.Sharkey LM, Corbett G, Currie E, et al. Optimising delivery of care in coeliac disease - comparison of the benefits of repeat biopsy and serological follow-up. Aliment Pharmacol Ther. 2013;38:1278–91. doi: 10.1111/apt.12510. [DOI] [PubMed] [Google Scholar]
- 28.Volta U, Granito A, Fiorini E, et al. Usefulness of Antibodies to Deamidated Gliadin Peptides in Celiac Disease Diagnosis and Follow-up. Dig Dis Sci. 2008;53:1582–1588. doi: 10.1007/s10620-007-0058-0. [DOI] [PubMed] [Google Scholar]
- 29.Zanini B, Magni A, Caselani F, et al. High tissue-transglutaminase antibody level predicts small intestinal villous atrophy in adult patients at high risk of celiac disease. Dig Liver Dis. 2012;44:280–285. doi: 10.1016/j.dld.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 30.Ciacci C, Cirillo M, Cavallaro R, et al. Long-Term Follow-Up of Celiac Adults on Gluten-Free Diet: Prevalence and Correlates of Intestinal Damage. Digestion. 2002;66:178–185. doi: 10.1159/000066757. [DOI] [PubMed] [Google Scholar]
- 31.Cuoco L, Cammarota G, Tursi A, et al. Disappearance of gastric mucosa-associated lymphoid tissue in coeliac patients after gluten withdrawal. Scand J Gastroenterol. 1998;33:401–5. doi: 10.1080/00365529850171035. [DOI] [PubMed] [Google Scholar]
- 32.Dickey W, Hughes DF, McMillan SA. Disappearance of endomysial antibodies in treated celiac disease does not indicate histological recovery. Am J Gastroenterol. 2000;95:712–4. doi: 10.1111/j.1572-0241.2000.01838.x. [DOI] [PubMed] [Google Scholar]
- 33.Kolacek S, Jadresin O, Petkovic I, et al. Gluten-free diet has a beneficial effect on chromosome instability in lymphocytes of children with coeliac disease. J Pediatr Gastroenterol Nutr. 2004;38:177–180. doi: 10.1097/00005176-200402000-00014. [DOI] [PubMed] [Google Scholar]
- 34.O’Keeffe J, Lynch S, Whelan a, et al. Flow cytometric measurement of intracellular migration inhibition factor and tumour necrosis factor alpha in the mucosa of patients with coeliac disease. Clin Exp Immunol. 2001;125:376–82. doi: 10.1046/j.1365-2249.2001.01594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sategna-Guidetti C, Grosso SB, Bruno M, et al. Is human umbilical cord the most suitable substrate for the detection of endomysium antibodies in the screening and follow-up of coeliac disease? Eur J Gastroenterol Hepatol. 1997;9:657–660. doi: 10.1097/00042737-199707000-00002. [DOI] [PubMed] [Google Scholar]
- 36.Sategna-Guidetti C, Pulitanó R, Grosso S, et al. Serum IgA antiendomysium antibody titers as a marker of intestinal involvement and diet compliance in adult celiac sprue. J Clin Gastroenterol. 1993;17:123–7. doi: 10.1097/00004836-199309000-00007. [DOI] [PubMed] [Google Scholar]
- 37.Sategna-Guidetti C, Grosso S, Bruno M, et al. Reliability of immunologic markers of celiac sprue in the assessment of mucosal recovery after gluten withdrawal. J Clin Gastroenterol. 1996;23:101–4. doi: 10.1097/00004836-199609000-00006. [DOI] [PubMed] [Google Scholar]
- 38.Troncone R, Mayer M, Spagnuolo F, et al. Endomysial antibodies as unreliable markers for slight dietary transgressions in adolescents with celiac disease. J Pediatr Gastroenterol Nutr. 1995;21:69–72. doi: 10.1097/00005176-199507000-00012. [DOI] [PubMed] [Google Scholar]
- 39.Valentini RA, Andreani ML, Corazza GR, et al. IgA endomysium antibody: A valuable tool in the screening of coeliac disease but not its follow-up. Ital J Gastroenterol. 1994;26:279–282. [PubMed] [Google Scholar]
- 40.Vécsei AK, Graf UB, Vogelsang H. Follow-up of adult celiac patients: which noninvasive test reflects mucosal status most reliably? Endoscopy. 2009;41:123–8. doi: 10.1055/s-0028-1103484. [DOI] [PubMed] [Google Scholar]
- 41.Vécsei E, Steinwendner S, Kogler H, et al. Follow-up of pediatric celiac disease: value of antibodies in predicting mucosal healing, a prospective cohort study. BMC Gastroenterol. 2014;14:1–20. doi: 10.1186/1471-230X-14-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yachha SK, Srivastava A, Mohindra S, et al. Effect of a gluten-free diet on growth and small-bowel histology in children with celiac disease in India. J Gastroenterol Hepatol. 2007;22:1300–5. doi: 10.1111/j.1440-1746.2007.04929.x. [DOI] [PubMed] [Google Scholar]
- 43.Duerksen DR, Wilhelm-Boyles C, Veitch R, et al. A comparison of antibody testing, permeability testing, and zonulin levels with small-bowel biopsy in celiac disease patients on a gluten-free diet. Dig Dis Sci. 2010;55:1026–31. doi: 10.1007/s10620-009-0813-5. [DOI] [PubMed] [Google Scholar]
- 44.Hopper AD, Hadjivassiliou M, Hurlstone DP, et al. What is the role of serologic testing in celiac disease? A prospective, biopsy-confirmed study with economic analysis. Clin Gastroenterol Hepatol. 2008;6:314–20. doi: 10.1016/j.cgh.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 45.Raivio T, Kaukinen K, Nemes E, et al. Self transglutaminase-based rapid coeliac disease antibody detection by a lateral flow method. Aliment Pharmacol Ther. 2006;24:147–54. doi: 10.1111/j.1365-2036.2006.02957.x. [DOI] [PubMed] [Google Scholar]
- 46.Rubio-Tapia A, Rahim MW, See JA, et al. Mucosal recovery and mortality in adults with celiac disease after treatment with a gluten-free diet. Am J Gastroenterol. 2010;105:1412–20. doi: 10.1038/ajg.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sjöberg V, Hollén E, Pietz G, et al. Noncontaminated dietary oats may hamper normalization of the intestinal immune status in childhood celiac disease. Clin Transl Gastroenterol. 2014;5:e58. doi: 10.1038/ctg.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duerksen DR, Leslie WD, Hospital SB. Positive celiac disease serology and reduced bone mineral density in adult women. Can J Gastroenterol. 2010;24:103–107. doi: 10.1155/2010/285036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rostom A, Dubé C, Cranney A, et al. The diagnostic accuracy of serologic tests for celiac disease: A systematic review. Gastroenterology. 2005;128:38–46. doi: 10.1053/j.gastro.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 50.Leffler D, Schuppan D, Pallav K, et al. Kinetics of the histological, serological and symptomatic responses to gluten challenge in adults with coeliac disease. Gut. 2012 doi: 10.1136/gutjnl-2012-302196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newnham ED, Shepherd SJ, Strauss BJ, et al. Adherence to the gluten-free diet can achieve the therapeutic goals in almost all patients with coeliac disease: A 5-year longitudinal study from diagnosis. J Gastroenterol Hepatol. 2016;31:342–9. doi: 10.1111/jgh.13060. [DOI] [PubMed] [Google Scholar]
- 52.Silvester JA, Graff LA, Rigaux L, et al. Symptomatic suspected gluten exposure is common among patients with coeliac disease on a gluten-free diet. Aliment Pharmacol Ther. 2016;44:612–9. doi: 10.1111/apt.13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duerksen DR, Wilhelm-Boyles C, Parry DM. Intestinal Permeability in Long-Term Follow-up of Patients with Celiac Disease on a Gluten-Free Diet. Dig Dis Sci. 2005;50:785–790. doi: 10.1007/s10620-005-2574-0. [DOI] [PubMed] [Google Scholar]
- 54.Vilela EG, de Abreu Ferrari MDL, de Gama Torres HO, et al. Intestinal permeability and antigliadin antibody test for monitoring adult patients with celiac disease. Dig Dis Sci. 2007;52:1304–9. doi: 10.1007/s10620-006-9511-8. [DOI] [PubMed] [Google Scholar]
- 55.Adriaanse MPM, Tack GJ, Passos VL, et al. Serum I-FABP as marker for enterocyte damage in coeliac disease and its relation to villous atrophy and circulating autoantibodies. Aliment Pharmacol Ther. 2013;37:482–90. doi: 10.1111/apt.12194. [DOI] [PubMed] [Google Scholar]
- 56.Morón B, Verma AK, Das P, et al. CYP3A4-catalyzed simvastatin metabolism as a non-invasive marker of small intestinal health in celiac disease. Am J Gastroenterol. 2013;108:1344–51. doi: 10.1038/ajg.2013.151. [DOI] [PubMed] [Google Scholar]
- 57.Comino I, Fernández-Bañares F, Esteve M, et al. Fecal Gluten Peptides Reveal Limitations of Serological Tests and Food Questionnaires for Monitoring Gluten-Free Diet in Celiac Disease Patients. Am J Gastroenterol. 2016:1–10. doi: 10.1038/ajg.2016.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Moreno ML, Cebolla Á, Muñoz-Suano A, et al. Detection of gluten immunogenic peptides in the urine of patients with coeliac disease reveals transgressions in the gluten-free diet and incomplete mucosal healing. Gut. 2015:1–8. doi: 10.1136/gutjnl-2015-310148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Comino I, Real A, Vivas S, et al. Monitoring of gluten-free diet compliance in celiac patients by assessment of gliadin 33-mer equivalent epitopes in feces. Am J Clin Nutr. 2012;95:670–7. doi: 10.3945/ajcn.111.026708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leffler D, Kupfer SS, Lebwohl B, et al. Development of Celiac Disease Therapeutics: Report of the Third Gastroenterology Regulatory Endpoints and Advancement of Therapeutics Workshop. Gastroenterology. 2016;151:407–11. doi: 10.1053/j.gastro.2016.07.025. [DOI] [PubMed] [Google Scholar]
- 61.Corazza GR, Villanacci V, Zambelli C, et al. Comparison of the interobserver reproducibility with different histologic criteria used in celiac disease. Clin Gastroenterol Hepatol. 2007;5:838–43. doi: 10.1016/j.cgh.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 62.Shah S, Akbari M, Vanga R, et al. Patient perception of treatment burden is high in celiac disease compared with other common conditions. Am J Gastroenterol. 2014;109:1304–11. doi: 10.1038/ajg.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bebb JR, Lawson A, Knight T, et al. Long-term follow-up of coeliac disease – what do coeliac patients want? Aliment Pharmacol Ther. 2006;23:827–831. doi: 10.1111/j.1365-2036.2006.02824.x. [DOI] [PubMed] [Google Scholar]
- 64.Dewar DH, Donnelly SC, McLaughlin SD, et al. Celiac disease: management of persistent symptoms in patients on a gluten-free diet. World J Gastroenterol. 2012;18:1348–56. doi: 10.3748/wjg.v18.i12.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lebwohl B, Michaëlsson K, Green PHR, et al. Persistent Mucosal Damage and Risk of Fracture in Celiac Disease. J Clin Endocrinol Metab. 2014:jc20133164. doi: 10.1210/jc.2013-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lebwohl B, Granath F, Ekbom A, et al. Mucosal healing and risk for lymphoproliferative malignancy in celiac disease: a population-based cohort study. Ann Intern Med. 2013;159:169–75. doi: 10.7326/0003-4819-159-3-201308060-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hollon JR, Cureton PA, Martin ML, et al. Trace gluten contamination may play a role in mucosal and clinical recovery in a subgroup of diet-adherent non-responsive celiac disease patients. BMC Gastroenterol. 2013;13:40. doi: 10.1186/1471-230X-13-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.