Abstract
Background
Nonspecific phospholipase C (NPC), which belongs to a phospholipase C subtype, is a class of phospholipases that hydrolyzes the primary membrane phospholipids, such as phosphatidylcholine, to yield sn-1, 2-diacylglycerol and a phosphorylated head-group. NPC plays multiple physiological roles in lipid metabolism and signaling in plants. To fully understand the putative roles of NPC genes in upland cotton, we cloned NPC genes from Gossypium hirsutum and carried out structural, expression and evolutionary analysis.
Results
Eleven NPC genes were cloned from G. hirsutum, which were found on chromosomes scaffold269.1, D03, A07, D07, A08, D11, and scaffold3511_A13. All GhNPCs had typical phosphoesterase domains and have hydrolase activity that acts on ester bonds. GhNPCs were annotated as phospholipase C, which was involved in glycerophospholipid metabolism, ether lipid metabolism, and biosynthesis of secondary metabolites. These GhNPCs showed differential expression patterns in distinct plant tissues and in response to various types of stress (low-phosphate, salt, drought, and abscisic acid). They also had different types and numbers of cis-element. GhNPCs could be classified into four subfamilies. Four pairs of GhNPCs were generated by whole-genome duplication and they underwent purifying selection.
Conclusions
Our results suggested that GhNPCs are involved in regulating key abiotic stress responses and ABA signaling transduction, and they may have various functional roles for different members under complex abiotic stress conditions. Functional divergence may be the evolutionary driving force for the retention of four pairs of duplicate NPCs. Our analysis provides a solid foundation for the further functional characterization of the GhNPC gene family, and leads to potential applications in the genetic improvement of cotton cultivars.
Electronic supplementary material
The online version of this article (10.1186/s12864-017-4370-6) contains supplementary material, which is available to authorized users.
Keywords: ABA, Abiotic stress, Cloning, Evolution, Expression, Gossypium hirsutum, Nonspecific phospholipase C (NPC), Structure
Background
Phospholipases, including patatin-related phopholipase A, phospholipase A1, phospholipase A2, phospholipase C, and phospholipase D, affect not only metabolism but also intracellular signaling [1, 2]. Phospholipase C, a subfamily of the phospholipase superfamily, can be divided into phosphatidylinositol-specific phospholipase C and nonspecific phospholipase C based on respective affinities to different substrates [3, 4]. The NPCs were discovered as a type of plant phospholipid-cleaving enzyme, one that preferentially hydrolyses phosphatidylcholine (PC) but which could also act on other lipids, such as phosphatidylethanolamine (PE) [1, 2, 4].
NPC was first discovered in the pathogenic bacterium Clostridium perfringens [5]. In 1955, an NPC-like activity was identified in plant plastid fractions [6]. Since then, NPC activity was discovered successively in peanut seeds [7], rice grains [8], tomatoes [9], cultured plant cells [10], and petunia flowers [11]. However, the analysis of NPC function in plants was hindered by insufficient molecular and genetic information. Eventually, in 2005, six NPC genes were identified from the Arabidopsis genome [12]. The subsequent analyses of NPCs in Arabidopsis indicated that NPCs play important and diverse roles in various processes. For example, NPC1 is involved in the plant response to heat [13] whereas NPC3 and NPC4 are important in BL-mediated signaling in root growth [14]. NPC4 is also involved in the plant response to phosphate starvation and plays a role in auxin signaling [12, 14]. NPC4 and its derived lipids positively modulate ABA response and promote plant tolerance to drought and salt stresses [15, 16]. NPC4 also plays a role in both early and long-term responses to Al stress [17]. NPC5 and diacylglycerol promote lateral root development under mild salt stress, and the former is involved in membrane glycerolipid remodeling during phosphate limitation in leaves of Arabidopsis [18, 19]. Taken together, these results clearly indicate that Arabidopsis NPCs are involved in many biological processes.
Although NPCs have been systematically investigated in Arabidopsis, only a few physiological functions and signaling roles have been clearly demonstrated at present. Moreover, there are few studies of NPCs in other plant species. Cotton, well known for its commercial significance both as a natural textile fiber and a vegetable oil yielding cultivar, offers a remarkable model system for learning more about polyploidization, cell elongation and cell wall biosynthesis [20, 21]. To fully understand the putative roles of NPC genes in plant, a systematic analysis for the NPC gene family in cotton is necessary.
In this study, we first cloned 11 NPC genes in G. hirsutum, and then carried out analyses of gene structure, conserved domains and motifs, secondary structure, three-dimensional structure, phylogeny, chromosomal location, and gene synteny. To investigate whether the expression of the GhNPC genes show tissue specificity, quantitative real-time PCR was used to study the expression patterns of these genes in various organs. To clarify their functions in response to various types of stress, the quantitative real-time PCR was also performed under four different treatments, namely low-phosphate, salt, drought, and abscisic acid (ABA). We also conducted cis-regulatory element identification and putative functional analysis. Finally, evolutionary analysis was carried out according to the above results. The results not only broaden our insight into the roles of NPC genes in plant abiotic stress responses and signaling transduction but also lay the foundation for further functional analyses of NPCs in cotton.
Methods
Identification of GhNPCs, GaNPCs, and GrNPCs
The G. hirsutum genome sequences were downloaded from the Cotton Research Institute of Nanjing Agricultural University (http://mascotton.njau.edu.cn, Gossypium_hirsutum_v1.1) and Institute of Cotton Resrarch of CAAS (http://cgp.genomics.org.cn/page/species/index.jsp#, Gossypium_hirsutum_v1.0), respectively. The G. arboretum genome (G.arboreum_BGI-A2_v1.0) and the G. raimondii genome (G.raimondii_JGI_221_v2.1) were downloaded from the CottonGen (https://www.cottongen.org/). Then, local databases were constructed. Arabidopsis NPC sequences obtained from The Arabidopsis Information Resource (TAIR, http://www.arabidopsis.org/) were used as queries against the local databases and the National Center of Biotechnology Information (NCBI). The choice of a candidate NPC was based on the E-value (1e−5). All the obtained sequences were put into SMART (http://smart.embl-heidelberg.de/) and InterPro (http://www.ebi.ac.uk/interpro/) to confirm the existence of the phosphoesterase domain [22]. The redundant sequences were then removed.
Cloning and structural analysis of GhNPC genes
GhNPCs were amplified from the cDNA derived from the total RNA extracted from flowering stage cotton Zhong9807, cloned into the pGEM-T Easy Vector (TRANSGEN, Beijing, China), and sequenced in both directions to verify the gene sequences. The primers used for gene amplification are listed in Additional file 1: Table S1. The molecular weight (Mw) and isoelectric points (pI) of the GhNPCs were predicted by the online ExPASy proteomics server database (http://expasy.org/) [23]. The identification of exon/intron structures for each GhNPC gene was studied by comparing the sequences of CDS and genomic DNA. Diagrams of the exon-intron structures were produced by the Gene Structure Display Server 2.0 (GSDS; http://gsds.cbi.pku.edu.cn/) [24]. The conserved domains of NPC protein were examined by NCBI Conserved Domain search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and SMART (http://smart.embl-heidelberg.de/) [25], and were located with HMMSCAN (https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan) [26]. Conserved motifs of GhNPCs were analyzed by the MEME program (http://meme-suite.org/) using the following parameters: minimum motif width = 6; maximum motif width = 50; maximum motif number = 20; any number of repetitions; all of the other parameters were set at default values [27]. Furthermore, all identified motifs were annotated using the ScanProsite tool of ExPASy (http://prosite.expasy.org/scanprosite/) [28]. Secondary structures of GhNPCs were predicted using JPred4 server (http://www.compbio.dundee.ac.uk/jpred4) [29]. Three-dimensional structures were predicted using SWISS-MODEL (https://swissmodel.expasy.org/) [30]. Structure of Francisella tularensis Acid Phosphatase A (AcpA) bound to orthovanadate (2d1g.1.A) was used to build a structural model for the GhNPCs [1, 31].
Multiple sequence alignments and phylogenetic tree construction
Multiple sequence alignments of GhNPCs were performed using the software MEGA7 and tinted using DNAman [32]. To gain insights into the phylogenetic relationships between NPC in cotton and other plant species, the phylogenetic tree was constructed with the software MEGA7 that used the Maximum-likelihood (ML) method based on the WAG amino acid substitution model [32]. A bootstrap analysis was employed with 1000 replicates. The protein sequences used to generate a phylogenetic tree were from G. hirsutum (11), G. arboreum (7), G. raimondii (11), Arabidopsis thaliana (6), Glycine max (7), Oryza sativa (5), Sorghum bicolor (5), Physcomitrella patens (5), and Selaginella moellendorffii (2) (sequences are given in Additional file 2: Data Set S1).
Syntenic analysis
MCscan X was employed to identify homologous genes among G. hirsutum, G. raimondii and G. arboreum, respectively [33]. The chromosomal distribution of NPCs and syntenic blocks were drawn by Circos-0.69 (http://circos.ca/) [34]. To evaluate the selective pressure over duplicate genes, non-synonymous (K a) and synonymous (Ks) substitution were calculated using MCscan X [33].
Cis-regulatory element identification
The promoters of the GhNPCs, which were 1.5 kb upstream from the transcriptional start site, were mapped onto the G. hirsutum genome. The cis-acting regulatory elements were identified using Plant CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [35]. Among cis-acting regulatory elements that we identified, we chose abiotic stress response and phytohormone-related elements for the expression analysis.
Plant material and treatments for expression analysis
Upland cotton (G. hirsutum, Zhong9807) seeds were grown in a greenhouse at 30 °C day/25 °C night and 60−70% relative humidity, under a 14-h light/10-h dark photoperiod with a photon flux density (PFD) of 800 μmol m−2 s−1. Zhong9807 is a typically salt-tolerant variety [36]. For the expression analysis of GhNPCs in different plant tissues, we used cotton grown in soil as material. Samples of taproot, lateral root, stem, cotyledon, senescent leaf, expanded leaf, and shoot tip were collected at the six-leaf stage; bract, petal, and ovule tissues were harvested on the first day of flowering. For the expression analysis of GhNPCs under various abiotic or exogenous hormone stresses, we used cotton grown hydroponically as material. Control plants were grown in Hoagland’s solution. We transferred cotton seedlings at the six-leaf stage randomly into various stress solutions: low-phosphate (5 μM), salt (200 mM NaCl), or drought (20% PEG6000). For the hormone treatment, 200 μM of ABA was used [37]. The roots were collected at 0, 1, 2, 3, 6, 9 and 12 h. All tissues were immediately frozen in liquid nitrogen and stored at −80°C for later use.
Quantitative real-time PCR
Total RNA was extracted from the different plant tissues with EASYspin TRIzol reagent (TIANGEN, Beijing, China). The cDNA was synthesized using the PrimeScript RT Reagent Kit (TAKARA, Dalian, China), and 600 ng total RNA were used. The cDNA samples were diluted 10-fold to serve as the templates for the subsequent PCR analyses.
Real-time quantitative PCR analyses were carried out in the LightCycler® 96 System (Roche Diagnostics Corporation, Indianapolis, USA) with the SYBR Premix Ex Taq II (TAKARA, Dalian, China) in a 10-μl reaction volume, which contained 5 μl of SYBR Premix Ex Taq II, 0.4 μl of each forward and reverse primer (Additional file 3: Table S2), 0.8 μl of diluted cDNA template, and an appropriate amount of sterile double-distilled water. The conditions for amplification were as follows: 30 s at 95 °C, 40 cycles of 5 s at 95 °C, and 30 s at 60 °C. With the Histone gene (GenBank accession number NC_006639) used as the reference gene [38, 39], the expression level was calculated using the 2−ΔΔCt (ΔΔC T = (C T, GhNPCs - C T, Histone) treatments at different times - (C T, GhNPCs - C T, Histone) 0 h) method for abiotic stress treatments and the 2-ΔCt (ΔC T = C T, GhNPCs - C T, Histone) method for various tissues [40]. The entire experiment was repeated three times.
Results
Identification of GhNPCs, GaNPCs, and GrNPCs
After merging results from the above strategies, eleven NPC genes were identified from Gossypium_hirsutum_v1.1 genome and nine NPC genes were identified from Gossypium_hirsutum_v1.0 genome, respectively. While, seven NPCs were identified from G. arboretum genome and were designated as GaNPC1a, GaNPC1b, GaNPC2, GaNPC3, GaNPC4, GaNPC6a, and GaNPC6b based on Arabidopsis NPC sequences, and eleven GrNPCs were identified from G. raimondii genome and were designated as GrNPC1, GrNPC2, GrNPC3a, GrNPC3b, GrNPC3c, GrNPC4a, GrNPC4b, GrNPC6a, GrNPC6b, GrNPC6c, and GrNPC6d based on Arabidopsis NPC sequences.
Cloning and annotation of NPC genes family in cotton
Finally, a total of 11 GhNPC genes were cloned and were designated as GhNPC1a, GhNPC1b, GhNPC2a, GhNPC2b, GhNPC3a, GhNPC3b, GhNPC4, GhNPC6a, GhNPC6b, GhNPC6c, and GhNPC6d, on the basis of sequence similarities to Arabidopsis NPCs. Among them, sequences of five GhNPCs (GhNPC2b, GhNPC3a, GhNPC4, GhNPC6c, and GhNPC6d) are consistent with the Gossypium_hirsutum_v1.1 genomic data; sequences of three GhNPCs (GhNPC1b, GhNPC3b, GhNPC6a) are consistent with the Gossypium_hirsutum_v1.0 genomic data; GhNPC1a has 1 base difference with Gossypium_hirsutum_v1.0 genomic data, but the amino acid encoded has not changed; GhNPC2a has 4 base differences with Gossypium_hirsutum_v1.1 genomic data, but the amino acid encoded has not changed; GhNPC6b has 3 base differences with Gossypium_hirsutum_v1.0 genomic data, and two amino acid changes (sequences are given in Additional file 4: Data Set S2). As shown in Table 1, these GhNPC proteins varied considerably in their length, Mw, and pI. Protein length ranged from 305 to 560 amino acids (aa), with the majority (81.82%) of proteins containing 488–520 aa. Mw ranged from 34.64 to 63.80 kDa and the pI ranged from 5.18 to 6.84 pH; however, all members had pI values <7 (Table 1). Based on the genomic data, we located 11 GhNPCs on chromosomes. The GhNPC genes were unevenly distributed over the G. hirsutum genome, with chromosomes A07 and D07 each having three genes and five other chromosomes (scaffold269.1, D03, A08, D11 and scaffold3511_A13) having one gene each (Table 1 and Fig. 1).
Table 1.
The characteristics of NPC genes from G. hirsutum
| Gene name | Locus | Strand | ORF length (bp) |
Protein | Intron number |
||
|---|---|---|---|---|---|---|---|
| Length (aa) |
Mw (kDa) |
pI | |||||
| GhNPC1a | scaffold269.1:65,602-67,720 | – | 1461 | 486 | 54.52 | 6.37 | 2 |
| GhNPC1b | D03: 22,587,094-22,589,215 | – | 1461 | 486 | 54.51 | 6.37 | 2 |
| GhNPC2a | D07:17,824,914-17,829,009 | + | 1563 | 520 | 58.71 | 6.44 | 3 |
| GhNPC2b | A07:22,144,436-22,148,517 | + | 1548 | 515 | 58.13 | 6.45 | 3 |
| GhNPC3a | scaffold3511_A13:74,984-69,115 | – | 1527 | 508 | 56.96 | 5.18 | 3 |
| GhNPC3b | A08: 33,048,822-33,049,930 | – | 918 | 305 | 34.64 | 5.76 | 2 |
| GhNPC4 | D11:43,575,117-43,570,466 | – | 1683 | 560 | 63.80 | 6.38 | 2 |
| GhNPC6a | A07: 4,310,983-4,312,741 | + | 1569 | 522 | 58.04 | 6.68 | 2 |
| GhNPC6b | D07: 6,984,026-6,985,784 | + | 1569 | 522 | 58.17 | 6.84 | 2 |
| GhNPC6c | A07:5,762,622-5,765,615 | + | 1530 | 509 | 56.64 | 6.74 | 2 |
| GhNPC6d | D07:5,782,790-5,785,614 | + | 1530 | 509 | 56.80 | 6.74 | 2 |
Mw molecular weight, pI isoelectric points
Fig. 1.
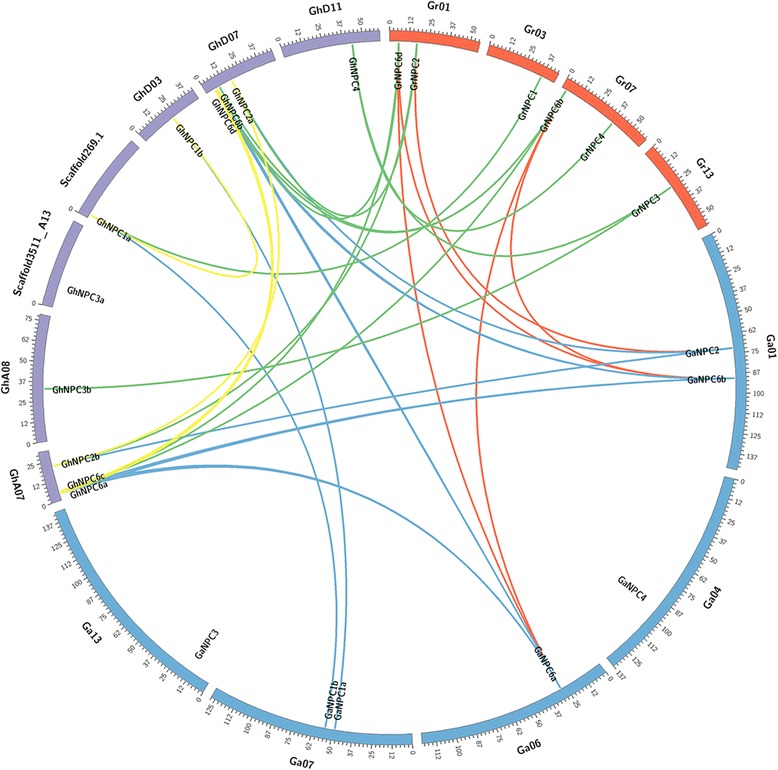
Syntenic block among NPCs from G. hirsutum, G. arboretum and G. raimondii. The chromosomes of G. hirsutum, G. arboretum and G. raimondii are indicated in purple, blue and red, respectively. Yellow lines indicate segmental duplicated pairs of GhNPCs. The putative orthologous NPCs between G.hirsutum and G. arboretum, G. hirsutum and G. raimondii, G. arboretum and G. raimondii are connected by blue, green and red lines, respectively
Structure analysis of GhNPCs
The identification of exon/intron structures for each GhNPC gene was studied by comparing the sequences of CDS and genomic DNA [24]. Most of them had two introns, though three of them (GhNPC2a, GhNPC2b, and GhNPC3a) had three introns (Fig. 2b). The GhNPCs had preserved a relatively simple constant exon-intron composition. The conserved domains of the NPC protein were examined by the NCBI Conserved Domain search, SMART and HMMSCAN [25, 26]. All of these GhNPCs contained the phosphoesterase domain (Fig. 2d), which has hydrolase activity that acts on the ester bonds. Conserved motifs of GhNPCs were analyzed using the MEME program [27]. Among the GhNPCs the number of motifs ranged from 8 to 17 (Fig. 2c). As shown in Fig. 2c, most of the GhNPC members in the same subgroup shared common motif compositions. Six motifs — motif 1, motif 2, motif 3, motif 4, motif 6, and motif 9 — were annotated as the phosphoesterase domain (Fig. 3). As shown in Fig. 2c, present in all sequences were the motifs 1, 3, 5, 7, and 9. The majority of the GhNPCs contained the motif 8 and the motif 10, with the exception of NPC3 and NPC4. However, both motif 14 and motif 15 only existed in NPC3 and NPC4. Additionally, the motif 16 and the motif 17 were unique to subfamily NPC1; motif 19 was unique to subfamily NPC2; motif 20 was unique to NPC4; motif 18 was unique to NPC6c and NPC6d. Some motifs, such as the motif 13, were shared in NPC1 and NPC6. Additional, we also predicted secondary structures using JPred4 [29], and three-dimensional structures using SWISS-MODEL [30]. As shown in Fig. 3 and Fig. 4, the structures of GhNPCs were composed by the beta sheet and several alpha helices.
Fig. 2.
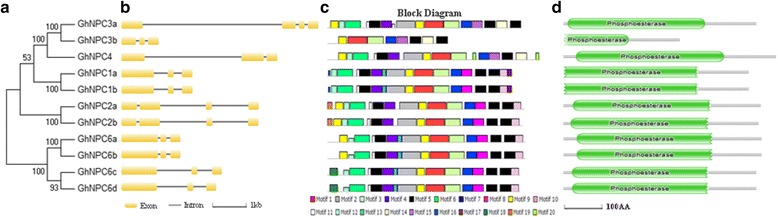
Exon-intron structure, conserved motif and conserved domain analysis of GhNPC genes. a. Phylogenetic tree of GhNPCs. Phylogenetic tree was generated using MEGA7 by the Maximum Likelihood method based on the protein sequences of GhNPCs. b. Exon-intron structures. Exons and introns of G. hirsutum are shown as yellow rounded rectangles and thin lines, respectively. c. Conserved motif analysis. Twenty motifs were identified. Different color boxes represent different types of motifs. Sequence information of twenty motifs was shown in Additional file 8: Table S5. d. Conserved domain analysis. The conserved domains of NPC protein were examined by NCBI Conserved Domain search and SMART, and were located with HMMSCAN
Fig. 3.
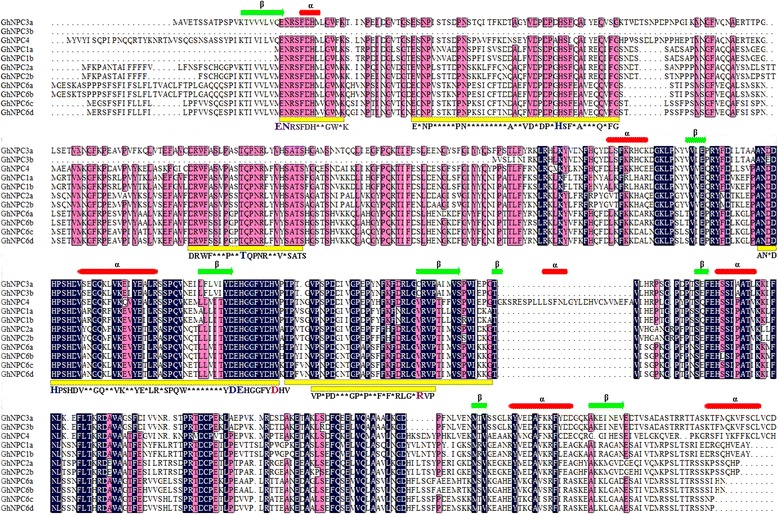
Alignments of GhNPCs. Multiple sequence alignments of GhNPCs were performed using the software MEGA7 and tinted using DNAman. Six motifs which were annotated as the phosphoesterase domain were marked in yellow. Putative active site (marked in blue) and conserved Asp-Arg ion pair (marked in purple) was indicated according to Pokotylo et al. [1]. Secondary structures of GhNPCs were predicted using JPred4 server [29]. Beta sheet were marked in green arrow and alpha helices were marked in red rectangle
Fig. 4.
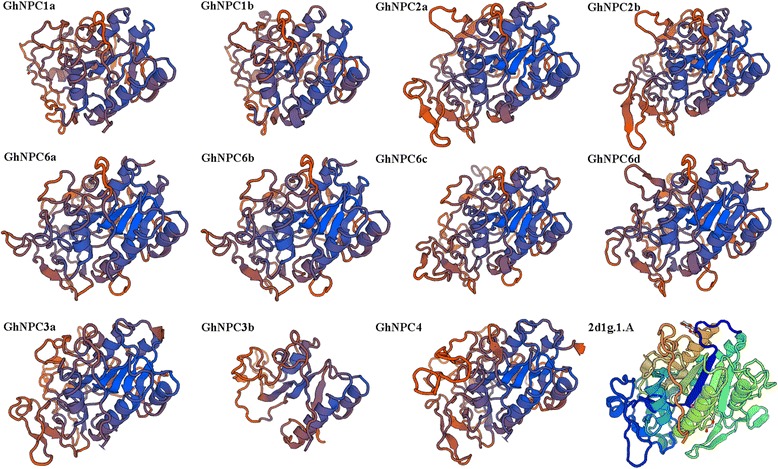
Three-dimensional structure of GhNPCs. Three-dimensional structures were predicted using SWISS-MODEL [30]. Structure of Francisella tularensis Acid Phosphatase A (AcpA) bound to orthovanadate (2d1g.1.A) are used to build a structural model for the GhNPCs [1, 31]
Phylogenetic and syntenic analysis
To gain insight into the evolution of plant NPCs, we performed a phylogenetic analysis of the NPC protein sequences known from several evolutionarily distinct plant species. In addition to its putative ancestral species, G. raimondii and G. arboreum, we also analyzed two other dicot species, Arabidopsis and soya (G. max); two monocotyledon species, rice (O. sativa) and sorghum (S. bicolor); a moss (P. patens) and a lycophyte (S. moellendorffii) (Additional file 2: Data Set S1). The phylogenetic analysis suggested that NPCs could be grouped into four clusters: I (NPC1), II (NPC2), III (NPC3, NPC4, and NPC5) and IV (NPC6) (Fig. 5). However, except for Arabidopsis, NPC5 was neither identified in the dicot soya (G. max) and cotton (G. hirsutum, G. raimondii and G. arboreum) nor in any of the other four species (O. sativa, S. bicolor, P. patens, and S. Moellendorffii). Syntenic analysis showed that four pairs of GhNPC (GhNPC1a and GhNPC1b, GhNPC2a and GhNPC2b, GhNPC6a and GhNPC6b, GhNPC6c and GhNPC6d) were segmental duplicates. GrNPCs and GaNPCs (except GaNPC3 and GaNPC4) had orthologous gene in G. hirsutum (Fig. 1). According to the results of phylogenetic and syntenic analysis, we found that GhNPC1a, GhNPC2a, GhNPC4, GhNPC6b and GhNPC6d were native to G. raimondii, while GhNPC1b, GhNPC2b, GhNPC6a and GhNPC6c were native to G. arboretum. In addition, the Ka/Ks ratio of four G. hirsutum duplicated gene pairs (GhNPC1a and GhNPC1b, GhNPC2a and GhNPC2b, GhNPC6a and GhNPC6b, GhNPC6c and GhNPC6d) were less than 1 (Additional file 5: Table S3), which suggested that they experienced purifying selection.
Fig. 5.
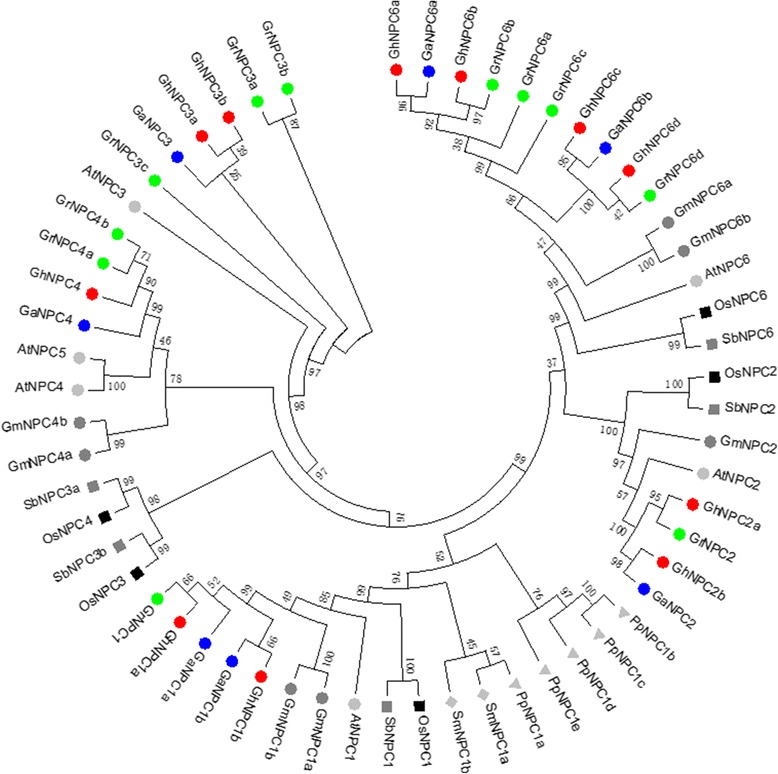
Phylogenetic tree of plant NPC. The tree was constructed using MEGA7 using the Maximum Likelihood method. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The analysis involved 59 amino acid sequences. Species abbreviations: At, Arabidopsis thaliana; Gm, Glycine max; Os, Oryza sativa; Pp, Physcomitrella patens; Sb, Sorghum bicolor; Sm, Selaginella moellendorffii; Gh, Gossypium hirsutum; Ga, G. arboretum; Gr, G. raimondii; NPC, non-specific phospholipase C
Analysis of cis-elements in the GhNPC gene promoters
To further explore gene function and regulation patterns, the cis-elements in GhNPC gene promoter sequences were studied. Regions of 1500 bp upstream from the start codons of each GhNPC gene were determined using the Plant CARE [35]. The results showed that the cis-elements could be divided into four major classes: stress-responsive, hormone-responsive, development-related, and light-responsive (Additional file 6: Excel S1). Nine stress-responsive cis-elements were identified—HSE, MBS, MBSII, CCAAT-box, LTR, TC-rich repeats, ARE, WUN-motif, and Box-W1—which reflected plant responses to heat, drought, low-temperature defense stresses, anaerobic induction, wound-responsive, and fungal elicitors (Fig. 6 and Additional file 6: Excel S1). Ten kinds of hormone-responsive cis-elements were identified, such as abscisic acid-ABA, salicylic acid-SA, methyl jasmonate-MeJA, gibberellin-GA, auxin-IAA, and ethylene (Fig. 6 and Additional file 6: Excel S1). A relatively large number of light-responsive cis-elements in GhNPC promoters were found (Additional file 6: Excel S1). As shown in Fig. 6, different GhNPCs had different types and numbers of cis-element.
Fig. 6.
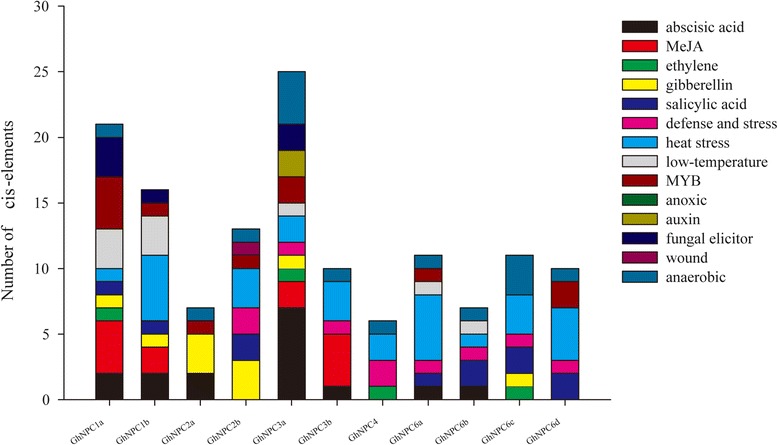
Cis-elements in the promoters of GhNPC genes that are related to stress responses. Different cis-elements with the same or similar functions are represented with the same color
Expression profiles of GhNPC genes in different plant tissues
To investigate the expression patterns of GhNPC genes in different organs and to determine their function in organ development, expression profiles were analyzed for 10 different tissues (taproot, lateral root, stem, cotyledon, senescent leaf, expanded leaf, shoot tip, bract, petal, and ovule) of the cotton cultivar ‘Zhong9807’ using real-time quantitative PCR. The mean threshold cycle (C T) of the reference gene (Histone) was 18.32. As shown in Fig. 7, GhNPC1 and GhNPC6 were ubiquitously expressed, whereas the others displayed tissue-specific expression patterns. GhNPC2a was mainly expressed in taproot and ovule. GhNPC2b was highly expressed in taproot and petal. GhNPC3a and GhNPC3b were mainly expressed in root. In addition, GhNPC4 were expressed weakly in all of the 10 tissues, though it was relatively higher in cotyledon.
Fig. 7.
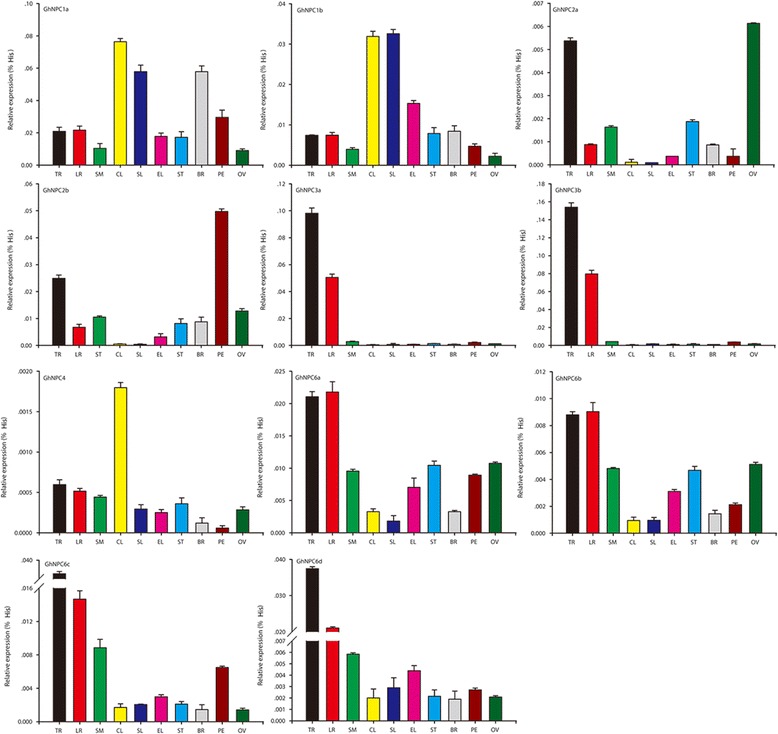
Expression of GhNPCs in various organs. The expression levels of GhNPCs were compared with Histone, wherein the expression level of Histone was defined as “1.” Note that the expression levels of different GhNPCs are scaled differently. Abbreviations: TR, taproot; LR, lateral root; SM, stem; CL, cotyledon, SL, senescent leave; EL, expanded leave; ST, shoot tip; BR, bract; PE, petal; OV, ovule. The expression level was calculated using the 2-ΔCt (ΔC T = C T, GhNPCs - C T, Histone) method [38]. The entire experiment was repeated three times
Expression patterns of GhNPC genes in response to abiotic treatments
To gain further insight into the role of the GhNPC genes under abiotic stress, we analyzed their expression profiles in response to low-phosphate, salt, drought, and ABA. As shown in Fig. 8, expression of the GhNPC genes was significantly affected by these stress treatments.
Fig. 8.
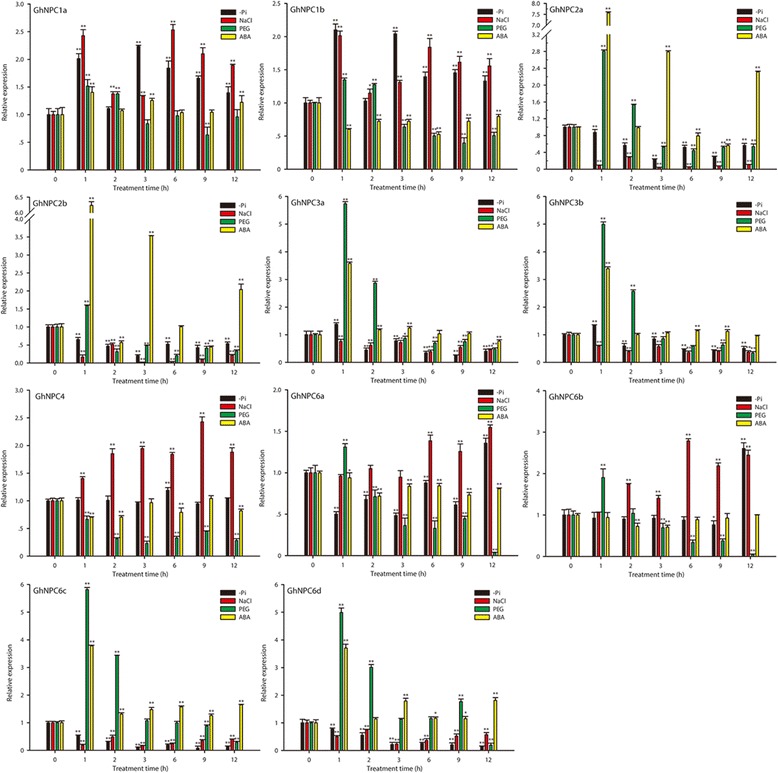
Expression of GhNPCs in root under various stresses. The expression levels of GhNPCs, which are shown in Y-axis, were compared with the control (0 h). Note that the expression levels of different GhNPCs are scaled differently. The expression level was calculated using the 2−ΔΔCt (ΔΔC T = (C T, GhNPCs - C T, Histone) treatments at different times - (C T, GhNPCs - C T, Histone) 0 h) method [38]. The entire experiment was repeated three times
GhNPC1a and GhNPC1b were up-regulated under the treatments of low-phosphate and salt, up-regulated at 1 h and 2 h but then down-regulated under the drought treatment. Under the ABA treatment, GhNPC1a was slightly up-regulated but GhNPC1b was down-regulated.
Both GhNPC2a and GhNPC2b were obviously induced at 1, 3, and 12 h under the ABA treatment, whereas they were down-regulated under the treatments of low-phosphate and salt. Under the drought treatment, both genes up-regulated at first and then down-regulated.
GhNPC3a and GhNPC3b were both down-regulated under the salt treatment. Under the treatments of low-phosphate and drought, both genes were up-regulated at first and then down-regulated. Under the ABA treatment, GhNPC3a was up-regulated at first and down-regulated at 12 h, and GhNPC3b was also up-regulated at first and had no significant differential change at 12 h. Expression of GhNPC4 was up-regulated under the salt treatment, but showed down-regulated under the drought treatment.
Expression of GhNPC6a and GhNPC6b were up-regulated at most time points under the salt treatment, yet down-regulated under the ABA treatment. By contrast, GhNPC6a and GhNPC6b were down-regulated at the beginning but up-regulated at 12 h under the low-phosphate treatment. Expression of GhNPC6c and GhNPC6d were down-regulated under both low-phosphate and salt treatments, whereas they were up-regulated at most time points under ABA treatments. GhNPC6a, GhNPC6b, GhNPC6c, and GhNPC6d were up-regulated at the beginning but then down-regulated under the drought treatment.
Discussion
Non-specific phospholipases C (NPCs) were discovered as a type of plant phospholipid-cleaving enzyme [1, 2, 4]. The six-gene family was established in Arabidopsis, and accumulating evidence suggests the involvement of Arabidopsis NPCs in abiotic stress responses as well as phytohormone activity [1, 2, 12–19]. To date, however, only a few physiological functions and signaling roles have been clearly illustrated, most of which are limited to Arabidopsis. In recent years, genome-wide identification and expression analysis have been carried out for the NPC gene family in rice and soybean [41, 42], but information of the NPC gene family in other plant species remains largely unknown. Hence, cloning, structural, functional and evolutionary analysis of the non-specific phospholipase C gene family in upland cotton appears important for pursuing further insight into the role of plant NPC in numerous biological processes.
NPC gene family in G. hirsutum
The completion of the sequencing of G. hirsutum provides great help for our research work. However, it is difficult to accurately sequence and assemble G. hirsutum genome owing to its large and complex allotetraploid genome [43]. So, the simple genome-wide identification based on genome sequencing for such complex allotetraploid G. hirsutum is not extremely accurate. There are two versions of the genome sequencing of upland cotton including Gossypium_hirsutum_v1.0 and Gossypium_hirsutum_v1.1. We identified eleven NPC genes from Gossypium_hirsutum_v1.1 genome and nine NPC genes from Gossypium_hirsutum_v1.0 genome, respectively. The nine NPC genes identified from Gossypium_hirsutum_v1.0 genome which were consistent with the result of Zhang et al. have corresponding sequences in that eleven NPC genes from Gossypium_hirsutum_v1.1 genome, but the sequences are different [44]. The number of NPC genes and the true sequences need to be verified by experiment. In this study, a total of 11 NPC genes were cloned in G. hirsutum, and five genes were consistent with the Gossypium_hirsutum_v1.1 genomic data, three genes were consistent with the Gossypium_hirsutum_v1.0 genomic data. The other three genes have 1, 3, 4 base differences with G. hirsutum genome sequencing results, respectively. The difference between exist GhNPC sequences and the G. hirsutum genome sequencing results may be due to inaccuracies in sequencing and assemble, or/and due to differences between varieties (Zhong9807 and TM-1). In addition, there are also differences in the expression levels of three corresponding genes (GhNPC1b, GhNPC3b, GhNPC6a) between our results and the paper of Zhang et al. at salt and PEG treatments. The differences in the expression levels of three corresponding genes (GhNPC1b, GhNPC3b, GhNPC6a) between our results and the paper of Zhang et al. may be because the material were taken from different tissues of different varieties at different growth stages. Our material is the root tissue of Zhong9807 at the six-leaf stage, while the material of Zhang et al. is the leaf tissue of TM-1 (the age is not applicable). Anyway, it is not comprehensive to characterize GhNPCs only using nine NPC genes identified from Gossypium_hirsutum_v1.0 genome.
Gene structure analysis showed that GhNPCs had preserved a relatively simple constant exon-intron composition. Most of them have two introns, though three of them (GhNPC2a, GhNPC2b, and GhNPC3a) have three introns (Fig. 2b). A similar phenomenon was observed in Arabidopsis, which contains 2–4 introns, and in rice, which contains 0–3 introns [42]. This suggested that plant NPC genes may have a relatively simple constant exon-intron composition. As shown in Fig. 2c, proteins with similar motif compositions were clustered in the same class. Present in all GhNPCs were the motifs 1, 3, 5, 7, and 9. Motif 1, 2, 3, 4, 6, and 9 were annotated as the phosphoesterase. Additional, four motifs (motifs 1, 3, 4, and 9) are likely important for NPC catalytic activity [1]. Thus, motif 1, motif 3, and motif 9 were crucial for NPC catalytic activity. Consistent with the previous reports, GhNPCs were also composed by the beta sheet and several alpha helices [1, 4]. The putative active site residues and the active site-stabilizing ion pairs (aspartate-arginine) of GhNPCs were conserved with AtNPCs (Fig. 3), which suggested the common catalytic reaction mechanism between AtNPCs and GhNPCs [1].
Functional analysis of the GhNPC genes
Increasing empirical evidence suggests that NPC genes could respond to abiotic stress and signaling transduction in plant. However, until now, the biological and cellular functions of most plant NPC genes remain unknown. Functional annotations showed that GhNPCs were involved in glycerophospholipid metabolism, ether lipid metabolism, and biosynthesis of secondary metabolites (Additional file 7: Table S4). The expression profile of a gene is always relative to its function. To investigate the function of the GhNPCs, we analyzed their expression profiles in different organs and in response to low-phosphate, salt, drought, and ABA. Results of real-time quantitative PCR demonstrated that GhNPCs had tissue-specific expression and were significantly altered under different abiotic stress treatments and ABA treatment.
As shown in Fig. 7, GhNPC1 and GhNPC6 were ubiquitously expressed, whereas GhNPC3a and GhNPC3b were highly expressed in root. Similar results have also been found for NPC1,NPC6 and NPC3 genes in Arabidopsis, and for which NPC2 is highly expressed in fertility organs but not in roots [15]. Meanwhile, GhNPC2a and GhNPC2b were also highly expressed in fertility organs such as petal and ovule. The main difference was that GhNPC2a and GhNPC2b were also highly expressed in taproot. In addition, NPC4 in G. hirsutum and Arabidopsis also showed different tissue specificity. The differing tissue specificity of NPC2 and NPC4 in G. hirsutum vs. Arabidopsis suggests that they might have diverse functions in these two species.
Many GhNPC genes showed great changes after treatments with low-phosphate, salt, drought, and ABA, which suggests their potential role in regulating upland cotton responses to stress abiotic conditions and in ABA signaling. Previous researcher has reported that Arabidopsis NPC1 is involved in the plant response to heat [13]. HSE, a cis-acting element involved in heat stress responsiveness, was found in the promoter of GhNPC1a and GhNPC1b (Additional file 6: Excel S1), suggesting that GhNPC1a and GhNPC1b might also play a role in upland cotton response to heat. Moreover, our results suggest that GhNPC1a and GhNPC1b were induced by low-phosphate, salt, and drought (Fig. 8), so they likely play roles in cotton response to these three types of stress. The function of the Arabidopsis NPC1 gene in response to low-phosphate, salt, and drought conditions is still not resolved, and requires further research. Previous studies revealed that Arabidopsis NPC3 is involved in the BL-mediated signaling in root growth [14]. Our study showed that both GhNPC3a and GhNPC3b were induced by low-phosphate, drought, and ABA. These results suggest that NPC3 is involved in regulating abiotic stress responses and phytohormone signaling transduction. In Arabidopsis, NPC4 plays an important role in responses to ABA and phosphate starvation, drought, and salt [12, 15, 16]. Our data shows that GhNPC4 was induced by salt, but not induced by drought, which is not completely consistent with Arabidopsis NPC4. Arabidopsis NPC2 and NPC6 have not been studied for their function yet, but we found that GhNPC2 and GhNPC6 were involved in regulating abiotic stress responses and phytohormone signaling transduction. Additionally, cis-elements play a significant role in plant stress responses [45], and many cis-elements in the promoters of the GhNPC genes were related to heat, drought, low-temperature, defense stresses, anaerobic induction, wound-responses, and fungal elicitors, ABA, SA, MeJA, GA, IAA, and ethylene (Fig. 6 and Additional file 6: Excel S1). Taken together, our results suggest that GhNPCs are involved in regulating abiotic stress responses and phytohormone signaling transduction, and that some NPCs may have diverse functions in G. hirsutum and Arabidopsis. We plan to verify the functional characteristics of the GhNPC genes in our future work.
Evolutionary analyses of the GhNPC genes
The phylogenetic analysis suggested that the entire cotton NPC gene family could be differentiated into four major classes: NPC1, NPC2, NPC6 and NPC3-5 (Fig. 5). Consistent with a previous report [1], we did not identified the NPC3–5 subfamily in the majority of species that we analyzed. In addition, P. patens and S. moellendorffii only have NPC1. It is possible that NPC1 subfamily is the ancestral NPC, which generated other NPC types, and some NPC genes had undergone species-specific evolutionary processes.
Plant genomes have undergone several rounds of whole-genome duplication (WGD) [46]. Duplicate genes are the prominent source of new genes and novel functions [47]. The allotetraploid G. hirsutum species resulted from hybridization of the two ancestral species G. arboreum and G. raimondii followed by chromosome doubling about ~1.5 million years ago (MYA) [43, 48–50]. Both G. arboreum and G. raimondii underwent 2 whole genome duplications during their evolution, one was the paleohexaploidization event common to all eudicots at ~130.8 MYA and the other was cotton-specific whole genome duplication at ~16.6 MYA [43, 50–52]. Syntenic analysis showed that four pairs of GhNPC (GhNPC1a and GhNPC1b, GhNPC2a and GhNPC2b, GhNPC6a and GhNPC6b, GhNPC6c and GhNPC6d) were segmental/WGD duplicates. Three models (pseudogenized, subfunctionalized, and neofunctionalized) have been proposed to explain why duplicated genes retained after WGD [47, 53–57]. The K a/Ks ratio of four G. hirsutum duplicated gene pairs were less than 1, which suggested that they underwent purifying selection [57]. Thus, these four duplicated genes are subfunctionalized genes and have partitioned their functions between pairs. Various functional roles may be the evolutionary driving force for the retention of these four pairs duplicate genes.
Conclusion
In this study, we first cloned 11 NPC genes in G. hirsutum and they were designated as GhNPC1a, GhNPC1b, GhNPC2a, GhNPC2b, GhNPC3a, GhNPC3b, GhNPC4, GhNPC6a, GhNPC6b, GhNPC6c, and GhNPC6d. These eleven GhNPCs were annotated as phospholipase C, which had typical phosphoesterase domains, had hydrolase activity acting on ester bonds, and were involved in glycerophospholipid metabolism, ether lipid metabolism, and biosynthesis of secondary metabolites. The GhNPCs had a relatively simple constant exon-intron composition. Motif 1, motif 3, and motif 9 were crucial for NPC catalytic activity. The backbone of GhNPCs was composed by the beta sheet and several alpha helices. Our results also suggested that GhNPCs are involved (1) in regulating key abiotic stress (low-phosphate, salt, and drought) responses and (2) in ABA signaling transduction. In addition, four pairs of GhNPCs were generated by whole-genome duplication and they underwent purifying selection. Various functional roles may be the evolutionary driving force for the retention of duplicate genes. Our analysis of the GhNPC gene family broadens our insight into the roles of NPC genes in plant abiotic stress responses and signaling transduction, provides the foundation for further functional characterization of the GhNPC gene family and for potential applications towards the genetic improvement of cotton.
Additional files
List of primers used for gene amplification. (DOC 40 kb)
The protein sequences used to generate phylogenetic tree. (DOC 54 kb)
List of primers used in quantitative real time-PCR expression analysis (DOC 39 kb)
The CDS sequences of GhNPCs (DOC 39 kb)
The K a/Ks ratio of four G. hirsutum duplicated gene pairs (DOC 30 kb)
Analysis of cis-elements in GhNPC gene promoters (XLSX 22 kb)
Functional annotations of GhNPCs. Functional annotations of GhNPCs were predicted using the Cotton Functional Genomics Database (DOC 88 kb)
Sequence information of twenty motifs (DOC 38 kb)
Acknowledgments
We thank Professor Wuwei Ye, Institute of Cotton Research, Chinese Academy of Agricultural Sciences, Anyang, Henan Province, China for providing seeds of upland cotton variety Zhong9807. We also thank Xiugui Cheng, Wen Cheng, and Wei Wang for providing advice on bioinformatics analysis.
Funding
This research was kindly supported by Shandong Natural Science Foundation of China (ZR2017MC025) and by the Genetically Modified Organisms Breeding Major Projects (Grant No. 2014ZX0800504B-003 and 2016ZX08005-004). The funding bodies were not involved in the design of the study or in any aspect of the collection, analysis and interpretation of the data or paper writing.
Availability of data and materials
The genome data supporting the conclusions of this article was downloaded from Cotton Gen (https://www.cottongen.org), Cotton Research Institute of Nanjing Agricultural University (http://mascotton.njau.edu.cn, Gossypium_hirsutum_v1.1), and Institute of Cotton Resrarch of CAAS (http://cgp.genomics.org.cn/page/species/index.jsp#, Gossypium_hirsutum_v1.0).
Abbreviations
- Aa
amino acid
- ABA
abscisic acid
- AcpA
Acid Phosphatase A
- PC
phosphatidylcholine
- CT
threshold cycle
- GA
gibberellin
- GSDS
Gene Structure Display Server
- IAA
auxin
- Ka
non-synonymous
- Ks
synonymous
- MeJA
methyl jasmonate
- ML
Maximum-likelihood
- Mw
molecular weight
- MYA
million years ago
- NCBI
National Center of Biotechnology Information
- NPC
Nonspecific phospholipase C
- PE
phosphatidylethanolamine
- PFD
photon flux density
- pI
isoelectric points
- SA
salicylic acid
- TAIR
The Arabidopsis Information Resource
- WGD
whole-genome duplication
Authors’ contributions
KZ, JS, and JZ designed all the experiments in their entirety. JS carried out the experiments. JS and YZ analyzed the data. JS wrote the manuscript. KZ and JZ reviewed the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The seeds of Zhong9807 variety used in this study were provided by Professor Wuwei Ye, Institute of Cotton Research, Chinese Academy of Agricultural Sciences, Anyang, Henan Province, China. The corresponding author of this manuscript has bilateral collaboration at laboratory level, and no permissions are needed to obtain the material. Our study complies with institutional, national, or international guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12864-017-4370-6) contains supplementary material, which is available to authorized users.
Contributor Information
Jiuling Song, Email: SongJiuling641@163.com.
Yonghe Zhou, Email: yonghezhou@163.com.
Juren Zhang, Email: jrzhang@sdu.edu.cn.
Kewei Zhang, Email: zhangkw@sdu.edu.cn.
References
- 1.Pokotylo I, Pejchar P, Potocký M, Kocourková D, Krčková Z, Ruelland E, et al. The plant non-specific phospholipase C gene family. Novel competitors in lipid signalling. Prog Lipid Res. 2013;52(1):62–79. doi: 10.1016/j.plipres.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Nakamura Y. NPC: nonspecific phospholipase Cs in plant functions. In: Wang XM, editor. Phospholipases in plant signaling. Berlin Heidelberg: Springer; 2014. pp. 55–67. [Google Scholar]
- 3.Singh A, Bhatnagar N, Pandey A, Pandey GK. Plant phospholipase C family: regulation and functional role in lipid signaling. Cell Calcium. 2015;58(2):139–146. doi: 10.1016/j.ceca.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Hong Y, Zhao J, Guo L, Kim SC, Deng X, Wang G, et al. Plant phospholipases D and C and their diverse functions in stress responses. Prog Lipid Res. 2016;62:55–74. doi: 10.1016/j.plipres.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Macfarlane MG, BCJG K. The biochemistry of bacterial toxins: the lecithinase activity of Cl. welchii toxins. Biochem J. 1941;35(8-9):884. doi: 10.1042/bj0350884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kates M. Hydrolysis of lecithin by plant plastid enzymes. Can J Biochem Physiol. 1955;33(4):575–589. doi: 10.1139/o55-072. [DOI] [PubMed] [Google Scholar]
- 7.Strauss H, Gershon ZLB, Heller M. Enzymatic hydrolysis of 1-monoacyl-SN-glycerol-3-phosphorylcholine (1-lysolecithin) by phospholipases from peanut seeds. Lipids. 1976;11(6):442–448. doi: 10.1007/BF02532834. [DOI] [PubMed] [Google Scholar]
- 8.Chrastil J, Parrish FW. Phospholipases C and D in rice grains. J Agric Food Chem. 1987;35(4):624–627. doi: 10.1021/jf00076a045. [DOI] [Google Scholar]
- 9.Rouet-Mayer MA, Valentova O, Simond-Côte E, Daussant J, Thévenot C. Critical analysis of phospholipid hydrolyzing activities in ripening tomato fruits. Study by spectrofluorimetry and high-performance liquid chromatography. Lipids. 1995;30(8):739–774. doi: 10.1007/BF02537801. [DOI] [PubMed] [Google Scholar]
- 10.Scherer GF, Paul RU, Holk A, Martinec J. Down-regulation by elicitors of phosphatidylcholine-hydrolyzing phospholipase C and up-regulation of phospholipase a in plant cells. Biochem Biophys Res Commun. 2002;293(2):766–770. doi: 10.1016/S0006-291X(02)00292-9. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura Y, Ohta H. The diacylglycerol forming pathways differ among floral organs of Petunia hybrida. FEBS Lett. 2007;581(28):5475–5479. doi: 10.1016/j.febslet.2007.10.053. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura Y, Awai K, Masuda T, Yoshioka Y, Takamiya KI, Ohta H. A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. J Biol Chem. 2005;280(9):7469–7476. doi: 10.1074/jbc.M408799200. [DOI] [PubMed] [Google Scholar]
- 13.Krčková Z, Brouzdová J, Daněk M, Kocourková D, Rainteau D, Ruelland E, et al. Arabidopsis non-specific phospholipase C1: characterization and its involvement in response to heat stress. Front Plant Sci. 2015;6:928. doi: 10.3389/fpls.2015.00928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wimalasekera R, Pejchar P, Holk A, Martinec J, Scherer GF. Plant phosphatidylcholine-hydrolyzing phospholipases C NPC3 and NPC4 with roles in root development and brassinolide signaling in Arabidopsis thaliana. Mol Plant. 2010;3(3):610–625. doi: 10.1093/mp/ssq005. [DOI] [PubMed] [Google Scholar]
- 15.Peters C, Li M, Narasimhan R, Roth M, Welti R, Wang X. Nonspecific phospholipase C NPC4 promotes responses to abscisic acid and tolerance to hyperosmotic stress in Arabidopsis. Plant Cell. 2010;22(8):2642–2659. doi: 10.1105/tpc.109.071720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kocourkova D, Krčková Z, Pejchar P, Veselková Š, Valentova O, Wimalasekera R, et al. The phosphatidylcholine-hydrolysing phospholipase C NPC4 plays a role in response of Arabidopsis roots to salt stress. J Exp Bot. 2011;62(11):3753–3763. doi: 10.1093/jxb/err039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pejchar P, Potocký M, Krčková Z, Brouzdová J, Daněk M, Martinec J. Non-specific phospholipase C4 mediates response to aluminum toxicity in Arabidopsis thaliana. Front Plant Sci. 2015;6:66. doi: 10.3389/fpls.2015.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaude N, Nakamura Y, Scheible WR, Ohta H, Dörmann P. Phospholipase C5 (NPC5) is involved in galactolipid accumulation during phosphate limitation in leaves of Arabidopsis. Plant J. 2008;56(1):28–39. doi: 10.1111/j.1365-313X.2008.03582.x. [DOI] [PubMed] [Google Scholar]
- 19.Peters C, KIM SC, Devaiah S, Li M, Wang X. Non-specific phospholipase C5 and diacylglycerol promote lateral root development under mild salt stress in Arabidopsis. Plant Cell Environ. 2014;37(9):2002–2013. doi: 10.1111/pce.12334. [DOI] [PubMed] [Google Scholar]
- 20.Yin Z, Wang J, Wang D, Fan W, Wang S, Ye W. The MAPKKK gene family in Gossypium raimondii: genome-wide identification, classification and expression analysis. Int J Mol Sci. 2013;14(9):18740–18757. doi: 10.3390/ijms140918740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emani C. Transgenic cotton for agronomical useful traits. In: Ramawat KG, Ahuja MR, editors. Fiber Plants. Switzerland: Springer International Publishing; 2016. pp. 201–216. [Google Scholar]
- 22.Zhou Y, Xu Z, Zhao K, Yang W, Cheng T, Wang J, et al. Genome-wide identification, characterization and expression analysis of the TCP gene family in Prunus mume. Front Plant Sci. 2016;7:1301. doi: 10.3389/fpls.2016.01301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasteiger E, Hoogland C, Gattiker A, Duvaud SE, Wilkins MR, Appel RD, et al. Protein identification and analysis tools on the ExPASy server. In: Walker JM, et al., editors. The proteomics protocols handbook. Totowa, NJ: Humana Press; 2005. pp. 571–607. [Google Scholar]
- 24.Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2014;31(8):1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, et al. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2014;43(D1):D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, Schreiber F, et al. HMMER web server: 2015 update. Nucleic Acids Res. 2015;43(W1):W30–W38. doi: 10.1093/nar/gkv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34(Suppl 2):369–373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, et al. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34(Suppl 2):362–365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drozdetskiy A, Cole C, Procter J, Barton GJ. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43(W1):W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42(W1):W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Felts RL, Reilly TJ, Tanner JJ. Structure of Francisella tularensis AcpA: prototype of a unique superfamily of acid phosphatases and phospholipases C. J Biol Chem. 2006;281:30289–30298. doi: 10.1074/jbc.M606391200. [DOI] [PubMed] [Google Scholar]
- 32.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Tang H, JD DB, Tan X, Li J, Wang X, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Lu X, Wang J, Wang D, Yin Z, Fan W, et al. Mining and analysis of SNP in response to salinity stress in upland cotton (Gossypium hirsutum L.) PLoS One. 2016;11(6):e0158142. doi: 10.1371/journal.pone.0158142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shah ST, Pang C, Fan S, Song M, Arain S, Yu S. Isolation and expression profiling of GhNAC transcription factor genes in cotton (Gossypium hirsutum L.) during leaf senescence and in response to stresses. Gene. 2013;531:220–234. doi: 10.1016/j.gene.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 38.Li CH, Zhu YQ, Meng YL, Wang JW, Xu KX, Zhang TZ, et al. Isolation of genes preferentially expressed in cotton fibers by cDNA filter arrays and RT-PCR. Plant Sci. 2002;163(6):1113–1120. doi: 10.1016/S0168-9452(02)00320-5. [DOI] [Google Scholar]
- 39.Zhang K, Song J, Chen X, Yin T, Liu C, Li K, et al. Expression of the Thellungiella halophila vacuolar H+-pyrophosphatase gene (TsVP) in cotton improves salinity tolerance and increases seed cotton yield in a saline field. Euphytica. 2016;211(2):231–244. doi: 10.1007/s10681-016-1733-z. [DOI] [Google Scholar]
- 40.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 41.Wang F, Deng Y, Zhou Y, Dong J, Chen H, Dong Y, et al. Genome-wide analysis and expression profiling of the phospholipase C gene family in soybean (Glycine max) PLoS One. 2015;10(9):e0138467. doi: 10.1371/journal.pone.0138467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh A, Kanwar P, Pandey A, Tyagi AK, Sopory SK, Kapoor S, et al. Comprehensive genomic analysis and expression profiling of phospholipase C gene family during abiotic stresses and development in rice. PLoS One. 2013;8(4):e62494. doi: 10.1371/journal.pone.0062494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li F, Fan G, Lu C, Xiao G, Zou C, Kohel RJ, et al. Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol. 2015;33(5):524–530. doi: 10.1038/nbt.3208. [DOI] [PubMed] [Google Scholar]
- 44.Zhang B, Wang Y, Liu JY. Genome-wide identification and characterization of phospholipase C gene family in cotton (Gossypium spp.). Sci China Life Sci. 2017:1–12. [DOI] [PubMed]
- 45.Liu Y, Wang L, Xing X, Sun L, Pan J, Kong X, et al. ZmLEA3, a multifunctional group 3 LEA protein from maize (Zea mays L.), is involved in biotic and abiotic stresses. Plant Cell Physiol. 2013;54:944–959. doi: 10.1093/pcp/pct047. [DOI] [PubMed] [Google Scholar]
- 46.Moghe GD, Shiu SH. The causes and molecular consequences of polyploidy in flowering plants. Ann N Y Acad Sci. 2014;1320(1):16–34. doi: 10.1111/nyas.12466. [DOI] [PubMed] [Google Scholar]
- 47.Zhang J. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18(6):292–298. doi: 10.1016/S0169-5347(03)00033-8. [DOI] [Google Scholar]
- 48.Wendel JF. New world tetraploid cottons contain old world cytoplasm. Proc Natl Acad Sci. 1989;86(11):4132–4136. doi: 10.1073/pnas.86.11.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wendel JF, Brubaker CL, Seelanan T. The origin and evolution of Gossypium. In: Stewart JM, Oosterhuis DM, Heitholt JJ, Mauney JR, editors. Physiology of cotton. Netherlands: Springer; 2010. pp. 1–18. [Google Scholar]
- 50.Fang L, Gong H, Hu Y, Liu C, Zhou B, Huang T, et al. Genomic insights into divergence and dual domestication of cultivated allotetraploid cottons. Genome Biol. 2017;18(1):33. doi: 10.1186/s13059-017-1167-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li F, Fan G, Wang K, Sun F, Yuan Y, Song G, et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet. 2014;46(6):567–572. doi: 10.1038/ng.2987. [DOI] [PubMed] [Google Scholar]
- 52.Llewellyn D, Showmaker KC, Shu S, Udall J, Yoo MJ, Byers R, et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton bres. Nature. 2012;492:423–427. doi: 10.1038/nature11798. [DOI] [PubMed] [Google Scholar]
- 53.Moore RC, Purugganan MD. The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol. 2005;8(2):122–128. doi: 10.1016/j.pbi.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 54.Cusack BP, Wolfe KH. When gene marriages don’t work out: divorce by subfunctionalization. Trends Genet. 2007;23(6):270–272. doi: 10.1016/j.tig.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 55.Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151(4):1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16(7):1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li S, Wang N, Ji D, Xue Z, Yu Y, Jiang Y, et al. Evolutionary and functional analysis of membrane-bound NAC transcription factor genes in soybean. Plant Physiol. 2016;172:1804–1820. doi: 10.1104/pp.16.01132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of primers used for gene amplification. (DOC 40 kb)
The protein sequences used to generate phylogenetic tree. (DOC 54 kb)
List of primers used in quantitative real time-PCR expression analysis (DOC 39 kb)
The CDS sequences of GhNPCs (DOC 39 kb)
The K a/Ks ratio of four G. hirsutum duplicated gene pairs (DOC 30 kb)
Analysis of cis-elements in GhNPC gene promoters (XLSX 22 kb)
Functional annotations of GhNPCs. Functional annotations of GhNPCs were predicted using the Cotton Functional Genomics Database (DOC 88 kb)
Sequence information of twenty motifs (DOC 38 kb)
Data Availability Statement
The genome data supporting the conclusions of this article was downloaded from Cotton Gen (https://www.cottongen.org), Cotton Research Institute of Nanjing Agricultural University (http://mascotton.njau.edu.cn, Gossypium_hirsutum_v1.1), and Institute of Cotton Resrarch of CAAS (http://cgp.genomics.org.cn/page/species/index.jsp#, Gossypium_hirsutum_v1.0).


