Abstract
Recent single-gene-based surveys of deep continental aquifers demonstrated the widespread occurrence of archaea related to Candidatus Methanoperedens nitroreducens (ANME-2d) known to mediate anaerobic oxidation of methane (AOM). However, it is unclear whether ANME-2d mediates AOM in the deep continental biosphere. In this study, we found the dominance of ANME-2d in groundwater enriched in sulfate and methane from a 300-m deep underground borehole in granitic rock. A near-complete genome of one representative species of the ANME-2d obtained from the underground borehole has most of functional genes required for AOM and assimilatory sulfate reduction. The genome of the subsurface ANME-2d is different from those of other members of ANME-2d by lacking functional genes encoding nitrate and nitrite reductases and multiheme cytochromes. In addition, the subsurface ANME-2d genome contains a membrane-bound NiFe hydrogenase gene putatively involved in respiratory H2 oxidation, which is different from those of other methanotrophic archaea. Short-term incubation of microbial cells collected from the granitic groundwater with 13C-labeled methane also demonstrates that AOM is linked to microbial sulfate reduction. Given the prominence of granitic continental crust and sulfate and methane in terrestrial subsurface fluids, we conclude that AOM may be widespread in the deep continental biosphere.
Introduction
The deep continental biosphere has been estimated to harbor up to 19% of Earth’s total biomass (Whitman et al., 1998; McMahon and Parnell, 2014). Single-gene analyses of microbial communities thriving in geographically and geochemically distinct aquifer systems show site-specific microbial community profiles (Chivian et al., 2008; Flynn et al., 2013; Pedersen et al., 2014; Bomberg et al., 2015; Ino et al., 2016). The microbial variability likely reflects the complex organic and inorganic constituents in geological formations and variable groundwater sources, including meteoric water, fossil seawater and brine (Griebler and Lueders, 2009). In the deep continental biosphere, residual organic matter anciently buried in deep geological formations and/or recently recharged from shallow groundwater and H2 derived from the fermentation of organic matter and rock–water interactions are known to serve as energy sources (Chivian et al., 2008; Wu et al., 2015; Bagnoud et al., 2016). Recently, 13C-depleted carbonate minerals in deep granite fractures are found to suggest that methane might serve as an alternative energy source in the deep continental biosphere (Drake et al., 2015). The widespread occurrence of deep microbial communities energetically dependent on methane is also supported by the increasing number of deep aquifers colonized by archaea taxonomically grouped within a subtype 2d of anaerobic methanotrophic archaea (ANME-2d), regardless of depth, locality, rock type and groundwater source (Gihring et al., 2006; Flynn et al., 2013; Bomberg et al., 2015). ANME-2d comprises phylogenetically diverse members of the Methanosarcinales represented by Candidatus Methanoperedens nitroreducens (Haroon et al., 2013). By genome-resolved metagenomic analysis and stable carbon and nitrogen isotope labeling experiments, Ca. M. nitroreducens has demonstrated nitrate-dependent anaerobic oxidation of methane (AOM) in a nitrate-amended bioreactor containing a mixture of freshwater sediment and anaerobic wastewater sludge (Haroon et al., 2013). However, it is unclear whether ANME-2d can perform nitrate-dependent AOM in deep continental groundwaters containing low concentrations of nitrate. Alternatively, these archaea may couple AOM to the reduction of electron acceptors such as sulfate (Timmers et al., 2015), metal oxides and humics (Ettwig et al., 2016), given the high availability of methane and the alternative electron acceptors in deep continental groundwaters (Lovley and Chapelle, 1995).
To investigate microbiologically mediated AOM in a deep subsurface granitic environment, we combined genome-resolved metagenomic analysis with laboratory incubation experiments. We focused on a granitic system because this igneous rock type represents a significant fraction of terrestrial subsurface environments, covering at least 15% of the land area (Leopold et al., 2012). Our study was conducted at the Mizunami Underground Research Laboratory (URL) in central Japan. The URL provides access to sufficient groundwater needed to acquire enough biomass for metagenomics and metabolic investigations, as demonstrated by previous studies using underground facilities (Chivian et al., 2008; Wu et al., 2015; Bagnoud et al., 2016). The granitic bedrock of the URL has both highly fractured and sparsely fractured domains (hereafter referred to as HFDB and SFDB, respectively). At a depth of 300 m below the surface, we drilled horizontal boreholes into the HFDB and SFDB with outflowing groundwater in 2009, an approach that minimized contamination (Figures 1a and b; Suzuki et al., 2014; Iwatsuki et al., 2015; Ino et al., 2016). The evidence supporting the involvement of ANME-2d in AOM was deduced from (1) the correlated abundance of ANME-2d phylotypes with hydrologic and geochemical features, (2) the recovery of a near-complete genome from the subsurface ANME-2d from the site showing evidence of a complete reverse methanogenesis pathway but interestingly lacking nitrate reductase and large multiheme cytochromes and (3) tracer studies with 13CH4 showing sulfate-dependent AOM in the granitic groundwater.
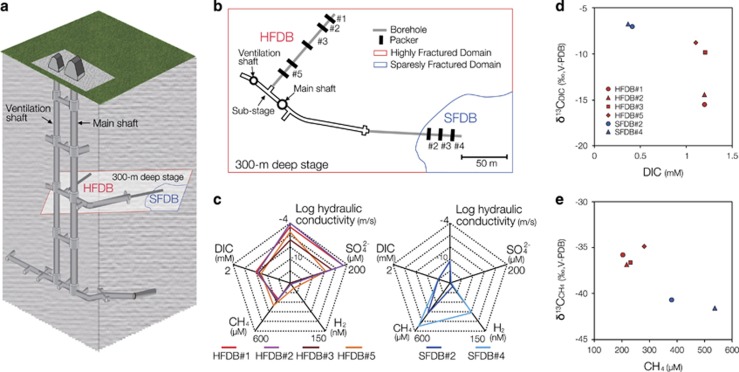
Figure 1.
Hydrological and geochemical characters of the Mizunami underground research laboratory (URL). (a) The layout of the Mizunami URL showing investigated boreholes. (b) The horizontal configuration of a 300-m deep stage showing the spatial relation with rock fracture densities and the investigated boreholes. (c) Pentadiagrams with the hydraulic conductivity and the concentrations of SO42−, H2, CH4 and DIC in groundwater from HFDB and SFDB in 2012. (d and e) Plots of stable carbon isotopic compositions and concentrations of DIC and CH4 in the HFDB and SFDB in 2012.
Material and methods
Study site
The Mizumani URL is located in the Gifu prefecture in central Japan. The Mizunami URL consists of a main shaft, a ventilation shaft, access tunnels at 300 and 500 m below ground level (mbgl) and substages (Figure 1a). Cretaceous Toki granite is overlain by the Tertiary sedimentary rocks at ~100–200 mbgl around the Mizunami URL (Yuguchi et al., 2013).
Drilling and hydraulic tests
The lengths of horizontal boreholes named the HFDB and SFDB were ~100 m. The HFDB and SFDB were located at a depth of 300 mbgl and previously referred to as 09MI20 and 09MI21, respectively (Suzuki et al., 2014; Iwatsuki et al., 2015; Ino et al., 2016). The boreholes were drilled mostly with outflowing groundwater. In case of the lack of outflowing groundwater during drilling, outflowing groundwater or local drinking water stored in water tanks was pumped into the borehole during drilling. A fluorescence tracer was added to drilling fluid stored in water tanks. Uranine and amino G acid were added to drilling fluid at 0.2±0.02 and 5±0.5 mg l−1 to drill the HFDB and SFDB, respectively. To evaluate drilling fluid contamination, the concentrations of uranine and amino G acid in collected groundwater samples were measured on site. As the concentrations of uranine and amino G acid were below detection limits (<0.001 and <0.003 mg l−1, respectively; Table 1) in groundwater samples used for biogeochemical and microbiological investigations, the contamination from drilling fluid appears to be negligible. To measure hydraulic conductivity (k: m s−1), hydraulic tests were performed. The hydraulic pressure and flow rate were determined by enclosing with double-ended packers. Then k was calculated by the methods of Hvorslev, 1951and Cooper and Jacob (1946) in the case of low and high specific storage coefficients, respectively. Shortly after hydraulic tests, multi-packer systems were installed into the HFDB and SFDB to divide the boreholes into six and four intervals, respectively. We investigated the packer intervals #1, #2, #3 and #5 of the HFDB and the packer intervals #2, #3 and #4 of the SFDB.
Table 1. Hydrogeochemical properties of granitic groundwater from the Mizunami URL.
| Sampling year |
HFDB#1 |
HFDB#2 |
HFDB#3 |
HFDB#5 |
SFDB#2 |
SFDB#4 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2012 | 2013 | 2012 | 2013 | 2012 | 2013 | 2012 | 2013 | 2011 | 2012 | 2013 | 2011 | 2012 | |
| Hydraulic conductivity (m s−1) | 3.0 × 10−5 | 1.0 × 10−4 | 2.3 × 10−7 | 3.9 × 10−6 | 4.56 × 10−11 | 4.96 × 10−11 | |||||||
| Distance from the tunnel (m) | 96.1–102.0a | 84.7–95.2a | 58.7–83.8a | 19.4–33.9a | 67.1–77.1a | 89.0–103.0a | |||||||
| Internal volume (l) | 9 | 13 | 31 | 18 | 18 | 58 | |||||||
| Outflow rate (ml min−1) | 1350 | 1380 | 1430 | 1240 | 80 | 20 | |||||||
| Amino G acid (mg l−1) | 0.003 | 0.002 | 0.003 | 0.002 | 0.003 | 0.002 | 0.002 | 0.002 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| Uranine (mg l−1) | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| pH | 8.7b | 8.6 | 8.7b | 8.6 | 8.7b | 8.5 | 8.6b | 8.4 | 9c | 8.9c | 8.9 | 9c | 8.9c |
| Temp. (°C) | 20b | 23.5 | 20.3b | 23.5 | 20b | 23.5 | 20.2b | 23.3 | 22.3c | 24.3c | 23.7 | 22.4c | 24.1c |
| DO (mg l−1) | <0.02b | <0.02 | <0.02b | <0.02 | <0.02b | <0.02 | <0.02b | <0.02 | <0.02c | <0.02c | <0.02 | <0.02c | <0.02c |
| Na+ (mM) | 3.6b | 3.4 | 3.5b | 3.4 | 3.7b | 3.9 | 4.3b | 4.4 | 5.3c | 5.2c | 5.3 | 5.6c | 5.6c |
| K+ (μM) | 10.2b | 10.2 | 10.2b | 10.2 | 12.8b | 12.8 | 17.9b | 17.9 | 10.2c | 10.2c | 10.2 | 10.2c | 10.2c |
| Ca2+ (mM) | 0.2b | 0.2 | 0.2b | 0.2 | 0.3b | 0.3 | 0.4b | 0.4 | 0.5c | 0.5c | 0.5 | 0.6c | 0.5c |
| Mg2+ (μM) | <4.1b | <4.1 | <4.1b | <4.1 | 12.3b | 16.5 | 28.8b | 32.9 | <4.1c | <4.1c | <4.1 | <4.1c | <4.1c |
| Cl− (mM) | 2.0b | 2 | 1.8b | 2 | 2.2b | 2.7 | 3.3b | 3.7 | 5.5c | 5.3c | 5.4 | 6c | 5.8c |
| NO3− (μM) | <0.8b | <0.8 | <0.8b | <0.8 | <0.8b | <0.8 | <0.8b | <0.8 | <0.8c | <0.8c | <0.8 | <0.8c | <0.8c |
| NO2− (μM) | <1.1b | <1.1 | <1.1b | <1.1 | <1.1b | <1.1 | <1.1b | <1.1 | <1.1c | <1.1c | <1.1 | <1.1c | <1.1c |
| SO42− (μM) | 188b | 135 | 188b | 135 | 146b | 104 | 125b | 90.6 | 7.3c | 4.2c | 3.1 | 4.2c | 4.2c |
| Fe2+ (μM) | <0.15b | <0.15 | <0.15b | <0.15 | <0.15b | <0.15 | <0.15b | <0.15 | <0.15c | <0.15c | <0.15 | <0.15c | <0.15c |
| HS− (μM) | 21.9b | 12.5 | 21.9b | 6.3 | 9.4b | 6.3 | 6.3b | 3.1 | <3c | <3c | 6.3 | <3c | <3c |
| NH4+ (μM) | <11.2b | <11.2 | <11.2b | <11.2 | <11.2b | <11.2 | <11.2b | <11.2 | <11.2c | <11.2c | <11.2 | <11.2c | <11.2c |
| DIC (mM) | 1.2b | 1.1 | 1.2b | 1.2 | 1.2b | 1.1 | 1.1b | 1 | 0.46c | 0.41c | 0.41 | 0.41c | 0.4c |
| DOC (μM) | 44.2b | 41.7 | 52.5b | 41.7 | 48.3b | 50 | 37.5b | <41.7 | 40.3c | 18.3c | <41.7 | 34.3c | 49.2c |
| Acetate (μM) | <2.7b | <2.7 | <2.7b | <2.7 | <2.7b | <2.7 | <2.7b | <2.7 | <2.7c | <2.7c | <2.7 | <2.7c | <2.7c |
| H2 (nM) | 3.1b | 4.3 | 2.4b | 8.2 | 2.1b | 3.4 | 15.7b | 3.7 | 1.4c | 12.2c | 9.4 | 135c | 93.1c |
| CH4 (μM) | 203b | 225 | 221b | 220 | 225b | 290 | 282b | 315 | 361c | 380c | 476 | 438c | 537c |
| C2H6 (nM) | 9b | NM | 9b | NM | 11b | NM | 15b | NM | 80c | 230c | NM | 130c | 390c |
| δ13CCH4 (‰VPDB) | −35.8b | −40.2 | −36.8b | −40.8 | −36.8b | −41.6 | −35.0b | −38.5 | −41.3 | −40.7 | −43.4 | −42 | −41.6 |
| δ13CDIC (‰VPDB) | −15.6b | −13 | −14.6b | −13.1 | −9.8b | −10.7 | −8.9b | −11.9 | −7 | −7.1 | −7.2 | −8.5 | −7.1 |
| Total cell numbers (cells ml−1) | 0.7±0.3 × 104 | 4.4±0.4 × 104 | 1.1±0.1 × 104 | 4.3±0.3 × 104 | 0.7±0.1 × 104 | 5.8±0.3 × 104 | 1.6±0.1 × 104 | 3.2±0.3 × 104 | 2.8±0.7 × 104 c | 0.39±0.1 × 104 c | 2.3±0.2 × 104 | 5.8±0.2 × 104 c | 1.9±0.2 × 104 c |
Abbreviations: DO, dissolved oxygen; DOC, dissolved organic carbon; DIC, dissolved inorganic carbon; HFDB, highly fractured domain; NM, not measured; SFDB, sparsely fractured domain; VPDB, Vienna Pee Dee Belemnite.
Geochemical data from the HFDB in 2012 and the SFDB in 2011 and 2012 have been previously published (Suzuki et al., 2014; Ino et al., 2016), except for the stable isotopic compositions of DIC and CH4 in the SFDB in 2011 and 2012. Total cell numbers in groundwater are also listed.
Referred from Iwatsuki et al. (2015).
Referred from Suzuki et al. (2014).
Referred from Ino et al. (2016).
Geochemical analyses
Methods for groundwater sampling and geochemical analyses of dissolved aqueous species and gaseous compounds have been described previously (Suzuki et al., 2014; Ino et al., 2016). Before collecting groundwater from the boreholes, at least three interval volumes of groundwater were discarded, because groundwater in borehole intervals might be geochemically and microbiologically different from that in surrounding rock fractures. Briefly, multi-parameter electrodes and probes (Ocean Seven 305; Idronaut, Brugherio, Italy) were used for measuring pH and temperature on site. The Indigo Carmine method with an absorptiometer (DR2800; HACH, Loveland, CO, USA) was used for measuring the concentration of dissolved oxygen on site. A subsample was collected through a 0.45-μm membrane filter with polypropylene housing and stored at 4 °C to perform following analysis in the laboratory. The concentrations of uranine and amino G acid were determined using a fluorescence spectrophotometer (RF1500; Shimadzu, Kyoto, Japan). Ion chromatography (ICS-1000; Dionex, Sunnyvale, CA, USA) was used to measure the concentrations of ionic species (Na+, K+, Ca2+, Mg2+, NH4+, Cl−, NO3−, NO2− and SO42−). Ferrous iron reagent powder pillows and hydrogen sulfide reagents (HACH) and a spectrophotometer (UVmini-1240; Shimadzu) were used to measure concentrations of Fe2+ and HS− in filtered groundwater samples on site. Fe2+ and HS− measurements were performed by following the manufacturer’s instructions (HACH). To measure the concentration of dissolved inorganic carbon (DIC), a combustion carbon analyzer (TOC-VCSH; Shimadzu) was used. For dissolved organic carbon (DOC), thermocatalytic oxidation and MC-NDIR detection (multi N/C 3100; Analytik Jena Japan, Kanagawa, Japan) were used. A high performance liquid chromatography coupled to a conductivity detector (Prominence; Shimadzu) was used to measure the concentration of acetate. Ammonia-concentrated SrCl2 solution was added to filtered groundwater immediately after sampling to precipitate DIC, including HCO3− and CO32− as SrCO3. After the SrCO3 was converted to CO2 through a reaction with phosphoric acid in a vacuum line, the carbon isotopic composition of DIC was determined by an isotope ratio mass spectrometer (Micromass Dual inlet OPTIMA, Waters Corporation, Milford, MA, USA).
Groundwater was collected directly into a preevacuated glass vial (65 ml volume, Nichiden-Rika Glass, Kobe, Japan) sealed with a butyl gray rubber for the analysis of dissolved CH4, C2H6 and H2 and the carbon isotopic of compositions of CH4. Degassed HgCl2-saturated solutions were added to vials for analyzing the concentrations and carbon isotopic compositions of CH4 and C2H6 using a Finnigan MAT 252 isotope ratio mass spectrometer (Thermo Finnigan, Bremen Germany) (Tsunogai et al., 2000). The concentration of H2 was determined immediately after sampling using a trace reduction gas detector (TRA-1; Round Science, Kyoto, Japan) based on the mercury oxide to mercury vapor conversion (Konno et al., 2006). The carbon isotopic compositions of DIC and methane were expressed as δ13C (per mille, ‰) against carbonate from Vienna Pee Dee Belemnite (VPDB).
Microscopic observations
For measuring total cell numbers in groundwater samples, we conducted direct counting as previously described (Konno et al., 2013; Ino et al., 2016). Briefly, 3 ml of groundwater was fixed by 3.7% formaldehyde at neutral pH and filtered through a 0.22-μm pore size, 25-mm diameter black polycarbonate filter (Advantec, Tokyo, Japan). After the filter was incubated for 5 min at 25 °C in 1 × TAE buffer containing 1 × SYBR Green I (TaKaRa, Tokyo, Japan) to stain microbial cells, the filter was observed under an epifluorescence microscope (Olympus BX51, Olympus, Tokyo, Japan) equipped with an Olympus DP70 digital camera (Olympus). We examined three filters per sample and 50 fields of view on a single filter or >300 cells per sample.
DNA extraction
Groundwater was filtered on a 0.22-μm pore size filter (type GVWP; Millipore, Billerica, MA, USA) and subjected to DNA extraction as described previously (Kouduka et al., 2012; Konno et al., 2013; Ino et al., 2016). The filter was incubated at 65 °C for 30 min in 150 μl of alkaline solution (pH 13.5, 75 μl of 0.5 N NaOH and 75 μl of TE buffer, including 10 mM Tris-HCl and 1 mM EDTA). The supernatant after centrifugation at 5000 g for 30 s at room temperature was neutralized with 750 μl of TE buffer and 150 μl of 1 M Tris-HCl (pH 6.5). The DNA was concentrated by ethanol precipitation from the DNA-bearing solutions (pH 7.0–7.5) and the DNA precipitation was dissolved in 50 μl of TE buffer (pH 8.0).
Pyrosequencing and quality control
For PCR amplification of 16S rRNA gene sequences from extracted DNA, the primers Uni530F and Uni907R with adaptor sequences were used (Nunoura et al., 2012). The forward primer Uni530F was tagged with 8-mer oligonucleotides to obtain sequences from multiple samples in a single run (Hamady et al., 2008). An initial denaturation at 96 °C for 3 min, 35 cycles of denaturation at 96 °C for 30 s, annealing at 54 °C for 45 s and extension at 72 °C for 45 s and a final extension at 72 °C for 5 min were conducted. The PCR products were subjected to electrophoresis on 1.5% agarose gels and purified by the MinElute Gel Extraction Kit (Qiagen, Valencia, CA, USA). Concentrations of the purified DNA were determined using the Quant-iT dsDNA HS Assay Kit and the Qubit fluorometer (Invitrogen, Grand Island, NY, USA) and then adjusted to 5 ng μl−1. To amplify DNA library beads for the 454 GS Junior System sequencer (Roche Applied Science, Penzberg, Germany), emulsion PCR was conducted by the GS Junior Titanium emPCR Kit Lib-L (Roche Applied Science). Amplified DNA fragments were sequenced following the manufacturer’s instructions (Roche Applied Science).
All quality control processing was performed in the Mothur program (Schloss et al., 2009). Sequences having more than one ambiguous base and/or a base below a quality score of 25 were removed in addition to sequences <250 base pairs (bp) and >500 bp. After a non-redundant set of sequences was obtained, the unique sequences were aligned to the SILVA reference alignment version 123 (http://www.mothur.org/wiki/Silva_reference_files). Chimeric sequences were detected and eliminated by ChimeraSlayer (Haas et al., 2011) in the Mothur program. The raw sequence reads were deposited to the DNA Data Bank of Japan (DDBJ) Sequences Read Archive with the accession number DRA004963.
Illumina sequencing by synthesis and quality control
16S rRNA gene sequences were also analyzed for groundwater collected from the HFDB#1 in 2015, which was used for the activity measurements of AOM. We employed a two-step tailed PCR approach to construct the paired-end libraries. In the first PCR, the prokaryote-universal primers Uni530F and Uni907R were concatenated with an internal adapter sequence. An initial denaturation at 96 °C for 3 min, 35 cycles of denaturation at 96 °C for 30 s, annealing at 54 °C for 5 min and extension at 72 °C for 45 s and a final extension at 72 °C for 5 min were conducted. The first PCR products were purified using ExoSAP-IT reaction (Affymetrix, Santa Clara, CA, USA). In the second PCR, all amplicons were reamplified using a set of Truseq adaptor sequences and 7-mer index sequences (Siddique and Unterseher, 2016). An initial denaturation at 96 °C for 3 min, 20 cycles of denaturation at 96 °C for 25 s, annealing at 65 °C for 45 s and extension at 72 °C for 45 s and a final extension at 72 °C for 7 min were conducted. The second PCR products were subjected to electrophoresis on 1.5% agarose gels and purified using the MinElute Gel Extraction Kit (Qiagen). Concentrations of the purified second PCR products were measured using the Quant-iT dsDNA HS Assay Kit and the Qubit fluorometer (Invitrogen) and then adjusted to 35.4 ng μl−1. The purified second PCR products were mixed and used as templates for pair-end sequencing by MiSeq Genome Analyzer using MiSeq Reagent Nano Kit v2 with 500 cycles following the manufacturer’s instructions (Illumina, San Diego, CA, USA). Raw reads complementary in forward and reverse directions were assembled using the Mother program (Schloss et al., 2009). Sequences including at least one ambiguous base and/or >450 bp were excluded. All of raw reads were deposited to the DDBJ Sequences Read Archive with the accession number DRA005131.
Library construction and Sanger sequencing for mcrA and dsrA genes
PCR using LA Taq polymerase (TaKaRa) and Ampdirect Plus (Shimadzu) was performed by a primer set of ME1 and ME2 (Hales et al., 1996) for mcrA gene sequences and two primer sets of DSR1F and 4DSR (Wagner et al., 1998) and DSR1F and 1334R (Santillano et al., 2010) for dsrA gene sequences. The primers ME1 and ME2 have mismatches in target sequences of newly found methanogen groups named Bathyarchaeaota and Verstraetearchaeota (Supplementary Table S1). The mcrA gene was analyzed for groundwater collected from the HFDB#1 in 2012 and 2015 and the SFDB#2 in 2012 and 2015. dsrA gene sequences were analyzed for groundwater collected from the HFDB#1 in 2012. Thermal cycling with 35 cycles of denaturation at 96 °C for 20 s, annealing at 55 °C (mcrA) or 55 °C (both of the dsrA primer sets) for 45 s and extension at 72 °C for 60 s were conducted. A PCR reaction mixture contained 0.1 μM of each oligonucleotide primer and 0.1 ng μl−1 of the DNA template. Both PCR products were subjected to 1.5% agarose gel electrophoresis and purified using MinElute Gel Extraction Kit (Qiagen). Using the TOPO TA Cloning Kit (Invitrogen), the purified PCR product was cloned into vector pCR2.1 and transformed into ECOS competent Escherichia coli DH5α (Nippon gene, Tokyo, Japan). Plasmid DNA was sequenced with the BigDye Terminator v3.1 Kit (Applied Biosystems, Foster City, CA, USA) and the vector M13 primers. The mcrA and dsrA gene sequences were deposited to DDBJ under the accession numbers LC177179 to LC177185 and LC269242 to LC269250.
Phylogenetic analysis
For grouping 16S rRNA gene sequences with >97% similarity, the farthest neighbor clustering algorithm in the Mothur program was used (Schloss et al., 2009). Rarefaction analysis was performed for the grouped 16S rRNA gene sequences using the vegan package (Oksanen et al., 2016) in R (R Development, Core Team, 2013). The Greengenes 16S rRNA reference data set released in August 2013 (http://greengenes.lbl.gov/Download/Sequence_Data/) and the Needleman–Wunsch algorithm in the Mothur program (Schloss et al., 2009) were used to align the recovered 16S rRNA gene sequences. Representative mcrA and dsrA gene sequences were aligned with closely related mcrA and dsrA gene sequences using the ClustalW program and the ambiguous nucleotide positions were rectified manually. Phylogenetic affiliations of operational taxonomic units were determined using the neighbor-joining method in the ARB software package (Ludwig et al., 2004). By using BLASTn (Altschul et al., 1990), closely related 16S rRNA, mcrA and dsrA gene sequences were obtained to construct neighbor-joining trees by the distance matrix method in the ARB software package (Ludwig et al., 2004). Bootstrap analysis was performed with 1000 replicates for trees based on 16S rRNA, mcrA and dsrA gene sequences.
Clustering and heatmap analysis of relative abundance of taxonomic groups
To clarify the abundance similarity of taxonomic groups among groundwater samples, a heatmap displaying the abundance orders of taxonomic groups in HFDB and SFDB was created by the gplot package (Warnes, 2016) in R (R Development, Core Team, 2013). A dendrogram was also constructed by hierarchical clustering based on the abundance of taxonomic groups among groundwater samples. The heatmap display and the dendogram were based on 16S rRNA gene sequences obtained by pyrosequencing.
Correlation analysis for environmental factors and microbial community structures
Principal correspondence analysis with 10 geochemical variables was performed by R (R Development, Core Team, 2013) for groundwater samples listed in Table 1. As principal correspondence analysis identified sulfate, hydrogen and methane to be representative of geochemical parameters and important substrates for microbial metabolisms, canonical correspondence analysis was performed to show correlation between the three variables and overall microbial communities and specific taxonomic groups using the vegan package (Oksanen et al., 2016) in R (R Development, Core Team, 2013). Nitrate was excluded because the concentrations were under detection limit. Monte Carlo Test based on 499 replicates was performed using the ade4 package (Dray et al., 2015) in R to evaluate the hypothesis that environmental factors affect the taxa distribution. When a P-value is <0.05, the correlation becomes statistically significant.
Thermodynamic calculations
In situ Gibbs reaction energies (ΔG) of biogeochemical reactions likely mediated in groundwater were calculated by the following equation:
ΔGoin situ refers to the Gibbs standard reaction energy under in situ temperature and pressure conditions, R refers to the ideal gas constant, T (K) refers to groundwater temperature and Q refers to the activity quotient for a reaction. The ΔGoin situ was calculated by the SUPCRT92 software package (Johnson et al., 1992) and all activities and fugacities were calculated by the Geochemist’s Workbench software package (Bethke, 1992).
Genome reconstructions and metabolic potential estimations
For metagenomic analysis, 35 liters of HFDB#1 groundwater was filtered on a 0.22 m membrane filter (Millipore filter code: GVWP) for metagenomic analysis. DNA extraction was performed using the Extrap Soil DNA Kit Plus ver. 2 (Nippon Steel and Sumikin Eco-Tech Corporation, Tokyo, Japan). In all, 150 bp paired-end sequencing with a 550 bp insert size was conducted with an Illumina HiSeq by Hokkaido System Science Co., Ltd (Sapporo, Japan). Raw shotgun sequencing reads trimmed by Sickle (Joshi and Fass, 2011) were subjected to de novo assembling using IDBA-UD (Peng et al., 2012) and mapped to >1000-bp scaffolds using Bowtie2 (Langmead et al., 2009). More than 1000-bp scaffolds were initially binned by GC content, read coverage and phylogenetic profile within the binning interface of ggkbase (http://ggkbase.berkeley.edu/). The refinement of candidate bins was performed using emergent self-organizing maps constructed from tetranucleotide frequencies and the coverage of the scaffolds (Dick et al., 2009). Resulting bins were evaluated for accuracy and completeness by CheckM version 1.06 based on a set of 188 euryarchaeal conserved single-copy phylogenetic marker genes (Parks et al., 2015). Open reading frames were predicted for all >1000- bp scaffolds with Prodigal (Hyatt et al., 2012). Functional genes in all open reading frames were annotated by BLAST searches (Ludwig et al., 2004) against databases, including Uniref100 (Suzek et al., 2007), Uniprot (Magrane and UniProt Consortium, 2011) and KEGG (Kanehisa et al., 2012). For selected functional genes, closely related sequences and their source organisms were initially accessed using NCBI Protein BLAST Program against NCBI Reference Sequence Database (RefSeq; Tatusova et al., 2014). For the phylogenetic relationship, the ARB software package (Ludwig et al., 2004) was used to align amino-acid sequences using Muscle (Edgar, 2004) and to construct a neighbor-joining tree with bootstrap values with 1000 replications. A draft genome and a contig sequence were deposited to DDBJ BioProject under accession numbers PRJDB5105, LC180219 and LC269251.
Stable isotope labeling experiments
Microbial cells were collected on 0.22-μm pore-size membrane filters (type GPWP; Millipore) in pressure-resistant stainless filter holders (Millipore) by filtering 325 and 150 liters of groundwater from the HFDB#1 and SFDB#2 in 2015, respectively. Microbial cells were stored under in situ hydraulic pressure conditions within 1 day and transferred to an anoxic glove box filled with Ar, in which microbial cells were concentrated in 12 ml of anoxic filter-sterilized SFDB#2 groundwater. Then 1 ml of the cell suspension was diluted with 13 ml of the anoxic filter-sterilized SFDB#2 groundwater and amended with ~70 μM of Na sulfate (Wako, Tokyo, Japan) in a 26-ml glass vial. Six and two vials were amended without and with 20 mM Na molybdate (Sigma-Aldrich, St Louis, MO, USA), a well-established inhibitor of sulfate reduction (Compeau and Bartha, 1985). To remove trace amounts of O2 from the vials closed with butyl rubber stoppers and aluminum caps, five cycles of evacuation and Ar purging were performed (a final pressure of 1.7 atm). In the anoxic globe box, 2.5 ml 12C methane (GL Sciences, Tokyo, Japan) and 1 ml 13C-labeled methane (CLM-3590; Cambridge Isotope Laboratories, Andover, MA, USA) were added into the vials by using a syringe with a needle (δ13CCH4=+35 000‰). The concentration of dissolved O2 was confirmed to be <10 p.p.b. using a fiber-optic oxygen meter (MICROX TX3-TRACE; PreSens, Regensburg, Germany). One vial without molybdate was autoclaved as a negative control. During the incubation, the concentration of H2 was measured by gas chromatography (GC-2010 Plus and BID-2010 Plus; Shimadzu). After 2-week incubation at room temperature in the dark, the cell suspension was filtered, and then the filtrate was measured for the concentration of HS− as described above. The filtrate was transferred into a vacuumed blood-collection tube without being exposed to air for measuring the carbon isotopic composition of DIC (δ13CDIC). By using MICAL3c (Ishimura et al., 2008), the carbon isotopic composition was determined for DIC as described previously (Miyajima et al., 1995).
Results and discussion
Hydrogeochemical and microbiological crosslinkages
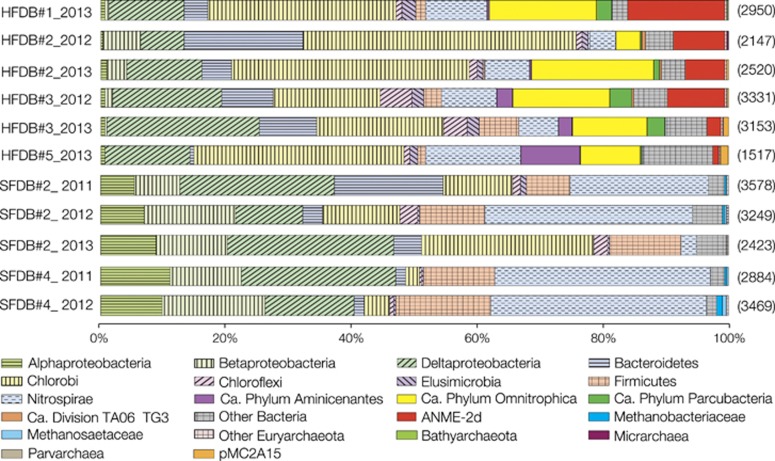
Samples were collected for groundwater chemical analysis and microbiological characterization from the HFDB in 2012–2013 and from the SFDB in 2011–2013. The two fractured domains have distinct hydrogeochemical features (Figure 1c and Table 1): the HFDB has high hydraulic conductivity, with fluids enriched in sulfate (90.6–188 μM) and DIC (1–1.2 mM), whereas the SFDB is characterized by low hydraulic conductivity and relatively high concentrations of methane (361–537 μM) and molecular hydrogen (H2; 1.4–135 nM). Stable isotopic compositions of methane and DIC were also distinct between the two sites: 12C enrichment appears to be correlated with high concentrations of DIC in the HFDB and methane in the SFDB, respectively (Figures 1d and e). As biogeochemical profiles are shaped by microbial metabolisms in the deep subsurface (Chapelle, 2001), 16S rRNA gene sequence analysis was conducted to profile the HFDB and SFDB microbial communities (Figure 2). Although the numbers of high-quality reads ranged from ~1500 to ~3500, nearly saturated rarefaction curves suggest that the biodiversity is sufficiently covered by pyrosequencing (Supplementary Figure S1). Bacteria of the Candidate Phyla Parcubacteria and Omnitrophica (formerly known as OD1 and OP3) were abundant in the HFDB, whereas α-, β- and δ-Proteobacteria, Chlorobi, Firmicutes and Nitrospira were abundant in both borehole communities. Similarly, the dominant archaea were distinct between the two sites: primarily within ANME-2d in the HFDB samples, whereas the methanogen families Methanosaetaceae, Methanospirillaceae, Methanomassiliicoccaceae and Methanobacteriaceae in the SFDB.
Figure 2.
Multiple-year taxonomic profiles of granitic groundwater from the HFDB and SFDB. Band graphs showing the distributions of taxonomic groups ranging from family to phylum. Bands without patterns indicate taxonomic groups distributed either in the HFDB or in the SFDB. Numbers in parentheses on the right indicate the numbers of high-quality reads in samples. Microbiological data from the SFDB in 2011 and 2012 have been previously published (Ino et al., 2016), whereas those from the HFDB in 2012 and 2013 and the SFDB in 2013 were obtained in this study.
As methyl-CoM reductase (mcr) is a key enzyme gene for anaerobic methanotrophy and methanogenesis, mcrA (encoding subunit A gene of mcr) gene sequences from the both sites were analyzed. mcrA gene sequences from the HFDB were all affiliated within ANME-2d, whereas those from the SFDB were affiliated within the families Methanosaetaceae and Methanobacteriaceae (Figure 3). The separate occurrence of ANME-2d and methanogen lineages is consistent between 16S rRNA and mcrA gene sequences. Given that the in situ activities of AOM and methanogenesis likely produce 12C-enriched DIC and methane in the HFDB and SFDB, respectively (Whiticar, 1999), the predicted metabolism by microorganisms recovered in the diversity surveys is consistent with the in situ geochemical signatures of the HFDB and SFDB (Figures 1d and e).
Figure 3.
Phylogenetic relationships of euryarchaeal sequences obtained by single-gene and metagenomic analyses. Neighbor-joining trees of the 16S rRNA (left) and the mcrA (right) gene sequences were constructed with bootstrap values with >50% and >75% indicated by open and solid circles, respectively. HFDB and SFDB sequences obtained by pyrosequencing and Sanger sequencing are shown in red and blue, respectively. Pink sequences were obtained by Illmina sequencing by synthesis from the HFDB#1 groundwater for the activity measurements of AOM. ANME-2d sequences in the left tree are represented by >30 pyrosequencing reads and >90 reads obtained by Illmina sequencing by synthesis. Bold sequences are represented by >100 reads for the 16S rRNA gene and >10 clonal sequences for the mcrA gene. Underlined sequences were obtained by metagenomic analysis. Scale bars represent 0.05 and 0.1 expected changes per nucleotide position. Note that 16S rRNA gene sequence of the subsurface ANME-2d reconstructed in this study was not obtained owing to the assemblage failure.
As shown in 16S rRNA and mcrA gene trees (Figure 3), ANME-2d sequences found in the HFDB were phylogenetically distinct from nitrate-reducing Ca. M. nitroreducens and more closely related to those found in the terrestrial subsurface where sulfate-dependent AOM has been suspected, based on the occurrence of ANME-2d in the sulfate–methane transition zone (Flynn et al., 2013; Bomberg et al., 2015). Consistent with the possible importance of sulfate-dependent AOM, the level of nitrate in the HFDB groundwater was low and sulfate concentrations were elevated at ~200 μM (Table 1). Further, the ANME-2d and sulfate abundances were correlated, based on multivariate function analyses (Supplementary Figure S2).
Genome-enabled metabolic reconstructions
To investigate the genomic profile of the subsurface ANME-2d, metagenomic sequencing was performed for HFDB#1 groundwater collected in 2014. All mcrA gene sequences obtained by metagenomic sequencing were affiliated within ANME-2d, which suggests that ANME and methanogene lineages not detected by PCR amplification with the primers are not substantially involved in AOM in the granitic groundwater. A 99% complete ANME-2d genome (based on euryarchaeal single copy gene content) was reconstructed (Supplementary Table S2). The 2.04 Mbp genome is smaller than those of ANME-1 (3.40 Mbp; Meyerdierks et al., 2010) and ANME-2a (3.40 Mbp; Wang et al., 2014), both of which conduct sulfate-dependent AOM, and the nitrate-reducing ANME-2d (3.20–3.74 Mbp; Haroon et al., 2013; Arshad et al., 2015). Despite this, the newly reconstructed ANME-2d genome contains nearly the full set of genes for methane oxidation (Figure 4), homologous genes of which are present in the nitrate-reducing ANME-2d genomes (Supplementary Table S3). As the ANME-2d clade contains uncultured methanogen members of the cluster ZC-1 (accession numbers of EU275986 and EU155958 in Figure 3) known to mediate hydrogenotrophic and methylotrophic pathways for methanogenesis (Zhang et al., 2008), key methanogenic genes were investigated. The subsurface ANME-2d genome was found to lack substrate-specific methyl transferases for methylotrophic methanogenesis (Krzycki, 2004), an electron bifurcating complex that supplies reduced ferredoxin for hydrogenotrophic methanogenesis (Kaster et al., 2011) and acetate kinase and phosphotransacetylase involved in aceticlastic methanogenesis (Jetten et al., 1992). Although the possibility of hitherto unknown mechanisms involved in methanogenesis is not excluded, it is unlikely that the subsurface ANME-2d is capable of producing methane.
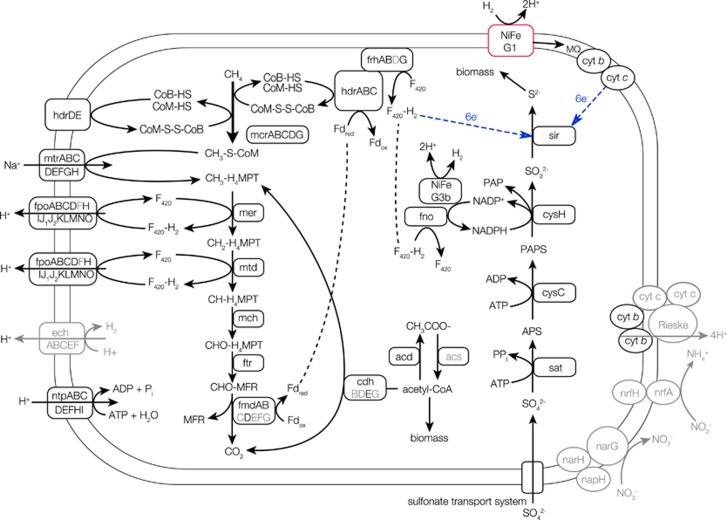
Figure 4.
Key carbon, sulfur and hydrogen metabolisms putatively inferred from the subsurface ANME-2d genome. Arrows show the directions of reaction pathways with compounds and enzymes involved in the reaction pathways as well as ion and electron transports. A gene not found in the other ANME genomes (ANME-1, ANME-2a and nitrate-reducing ANME-2d genomes) but in the subsurface ANME-2d genome is highlighted in red, whereas genes not found in subsurface ANME-2d genome but in nitrate-reducing ANME-2d genomes are shown in gray. Blue arrows show the electron transport hypothesized based on the homology of the Ni-Fe hydrogenases Group 1 gene sequence and that of the sulfite reductase gene sequence. Full names of genes are listed in Supplementary Table S3.
In sharp contrast to the genomes of nitrate-reducing ANME-2d, genes involved in nitrate/nitrite reduction were absent in the subsurface ANME-2d genome. Thus we conclude that AOM mediated by the subsurface ANME-2d is not coupled to nitrate/nitrite reduction. Genes encoding multiheme c-type cytochromes are numerous in archaea affiliated within ANME-1 (Wegener et al., 2015) and ANME-2 (McGlynn et al., 2015). The multiheme c-type cytochromes are involved in direct electron transfers to bacterial syntrophic partners and to solid Fe(III) and Mn(IV) oxides. AOM coupled to the reduction of solid Fe(III) and Mn(IV) oxides was also demonstrated for the nitrate-reducing ANME-2d with many genes encoding multiheme c-type cytochromes (Ettwig et al., 2016). Only one locus encoding multiheme c-type cytochromes was found in the subsurface ANME-2d genome (Supplementary Table S3). This makes sense, given the low availability of Fe(III) and Mn(IV) due to abiotic reduction of any oxidized compounds by hydrogen sulfide.
Nearly all genes involved in assimilatory sulfate reduction were also present in the draft genome of the subsurface ANME-2d and homologous to those found in the nitrate-reducing ANME-2d genomes (Figure 4 and Supplementary Table S3). The draft genome contains all genes involved in assimilatory sulfate reduction except for the 3′-phosphoadenylyl sulfate (PAPS) reductase (Meyerdierks et al., 2010). As the assimilation of sulfur is energetically expensive via the pathway for assimilatory sulfate reduction and feasible from hydrogen sulfides in sulfidic habitats, the alternative role of genes normally involved in assimilatory sulfate reduction has been suspected to be in dissimilatory sulfate reduction in ANME-1 (Meyerdierks et al., 2010). The additional energy required for assimilatory sulfate reduction is the generation of nicotinamide adenine dinucleotide phosphate (NADPH) to reduce PAPS to sulfite (Madigan et al., 2012). Based on bioenergetic calculations from the Mizunami groundwater (Supplementary Table S4), AOM coupled to H2-oxidation-based sulfate reduction is exothermic, even if the additional energy consumption for the cytosolic conversion of NADP to NADPH is accounted (ΔG=18 kJ mol−1; Schuchmann and Müller, 2014). Furthermore, the cytosolic Ni-Fe hydrogenase Group 3b (NiFe hydrogenase G3b) found in the subsurface ANME-2d genome is known to catalyze the bidirectional reactions between H2 and 2H+ (Wrighton et al., 2012) and to reduce NADP to NADPH (van Haaster et al., 2008).
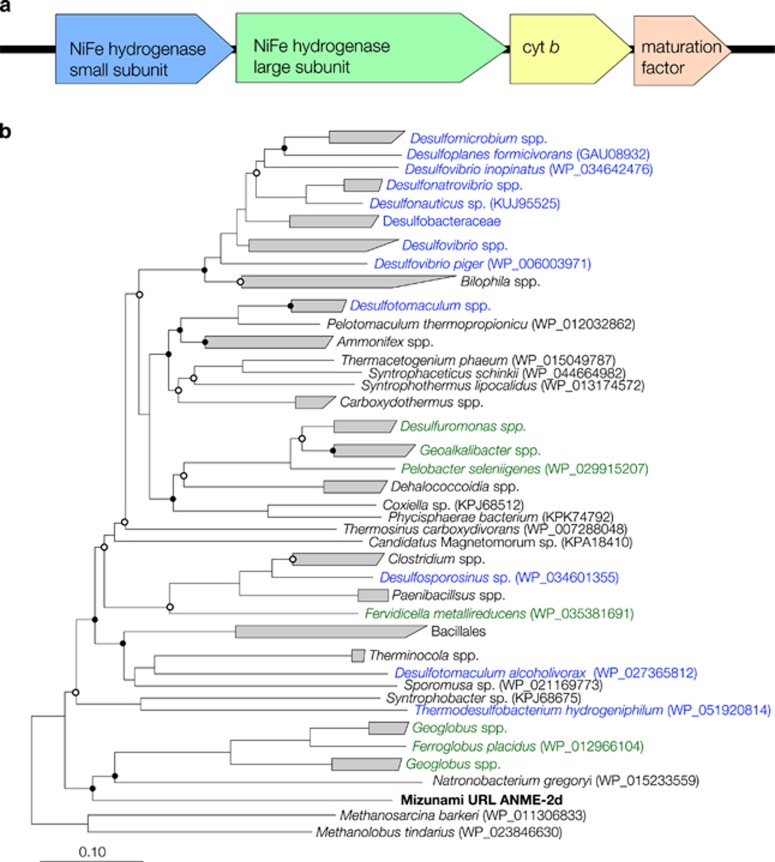
Based on metal-binding motifs and the gene organization (Greening et al., 2016), a membrane-bound H2-uptake NiFe hydrogenase Group 1 (NiFe hydrogenase G1) was identified in the subsurface ANME-2d genome (Figure 5 and Supplementary Table S5). It is noted that other ANME lineages previously examined by metagenomics analysis lack membrane-bound hydrogenase genes putatively involved in the electron transport chain (Meyerdierks et al., 2010; Haroon et al., 2013; Wang et al., 2014; Arshad et al., 2015). The NiFe hydrogenase G1 is known to oxidize H2, linked to electron transport and used for ATP generation from the proton motive force (Greening et al., 2016). Phylogenetic analysis of amino-acid sequences of the NiFe hydrogenase G1 gene of the subsurface ANME-2d studied here revealed a close relationship to those of thermophilic archaea of the genera Ferroglobus and Geoglobus known to mediate H2 oxidation coupled to Fe(III) reduction (Figure 6). In case of Geoglobus acetivorans, the NiFe hydrogenase G1 is inferred to perform respiratory H2 oxidation linked to Fe(III) reduction via quinone reduction (Mardanov et al., 2015). As the subsurface ANME-2d genome contained genes for menaquinone synthesis (Supplementary Table S3), it is likely that the subsurface ANME-2d might use the NiFe hydrogenase Group 1 for respiratory H2 oxidation via menaquinone reduction. The NiFe hydrogenase G1 is also involved in the electron transfer from H2 to methanophenazine for methanogenesis (Greening et al., 2016). However, the NiFe hydrogenase Group 1 gene sequences found in closely related methanogens such as Methanolobus tindarius and Methanosaricina barkeri were distantly related to that in the subsurface ANME-2d. It is therefore unlikely that the NiFe hydrogenase G1 is involved in methanogenesis. In the nitrate-reducing ANME-2d genomes, energy-conserving hydrogenase (ech) genes are present to oxidize H2 and translocate H+ for ATP synthesis (Figure 5; Greening et al., 2016). As the subsurface ANME-2d genome lacked the ech genes, the NiFe hydrogenase G1 might be important for producing the transmembrane H+ gradient for ATP synthesis. A sulfite reductase gene found in the subsurface ANME-2d is homologous but distantly related to the sulfite reductase and coenzyme F420 hydrogenase genes of Methanosarcina spp. (42–43% amino-acid similarities). Thus six electrons for the reduction of sulfite to HS− are hypothesized to be transported from coenzyme F420 or from the NiFe hydrogenase G1 via menaquinone and an unknown oxidoreductase complex (Figure 4).
Figure 5.
Genetic characteristics of the Ni-Fe hydrogenases Group 1 in the subsurface ANME-2d genome. (a) Gene organization surrounding a large subunit with Ni-Fe biding motifs. (b) Neighbor-joining tree of Ni-Fe hydrogenase genes based on deduced amino-acid sequences. Bootstrap values with >50% and >75% are indicated by open and solid circles, respectively. Blue and green taxa are known as sulfate and Fe(III) reducers, respectively.
Figure 6.
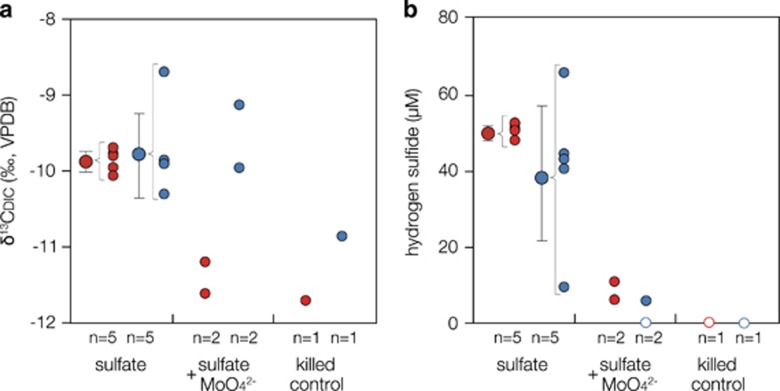
End point measurements of δ13CDIC and hydrogen sulfide production during stable isotope labeling experiments. Two-week incubations of SFDB#2 groundwater amended with ~400 μM 13C-labeled methane, ~60 nM H2 and ~70 μM sulfate were conducted with alive and killed cells from HFDB#1 and SFDB#2. To verify sulfate-dependent AOM, molybdate, an inhibitor of sulfate reduction, was added. Filled circles indicate (a) the stable carbon isotopic composition of DIC and (b) the concentrations of hydrogen sulfides. Open circles indicate that the products were undetected. Larger filled circles with error bars represent the means and s.d. of five replications. Numbers of replication (n) and experimental conditions are shown below the horizontal axis.
Sulfate-dependent AOM activities
To test for AOM coupled to sulfate reduction in the HFDB, microbial cells were incubated in sulfate-depleted SFDB#2 groundwater (pH=~9) amended with ~400 μM 13C-labeled methane, ~60 nM H2 and ~70 μM sulfate. To detect AOM activities from subsurface microbial communities within a short period, the biomass was concentrated from 1.8 × 104 cells ml−1 in the original HFDB#1 groundwater to ~1 × 107 cells ml−1 in incubation vials. It was confirmed by Illumina sequencing that the original HFDB#1 groundwater was dominated by the subsurface ANME-2d in 2015 (Supplementary Figure S3). Production of 13C-enriched DIC and hydrogen sulfide was detected during the 2-week incubation (Figures 6a and b) and the dependency of AOM on sulfate reduction was indicated by the lack of both 13C-enriched DIC and hydrogen sulfide during the incubation with molybdate, a well-established inhibitor for microbial sulfate reduction. The inhibition of AOM by molybdate also excludes the possibility of trace methane oxidation mediated in reverse by methanogens. This clarification is necessary, because the ZC-1 methanogens are phylogenetically classified within ANME-2d (Zhang et al., 2008). To further support this inference, the same incubation experiments were conducted for SFDB#2 groundwater colonized by methanogens (Figure 3). The original biomass (1.8 × 104 cells ml−1) was also enriched for incubation (~1 × 107 cells ml−1). As shown in Figures 6a and b, 13C-enriched DIC and hydrogen sulfide were both produced regardless of the inhibition of sulfate reduction with molybdate. Thus it is likely that the 13C-enriched DIC in SFDB#2 groundwater is produced by methanogens via the reverse methanogenesis pathway and that the 13C enrichment in HFDB#1 groundwater is linked to AOM coupled to sulfate reduction rather than DIC consumption by methanogenesis.
As the oxidation of 13C-labeled methane (δ13CCH4=+35 000‰) produces DIC with a δ13CDIC value of +34 000‰, given that anaerobic methanotrophs are known to preferentially oxidize 12CH4 over 13CH4 with a fractionation factor of 20‰ (Whiticar, 1999). The 1:1 production of DIC and hydrogen sulfide from AOM coupled sulfate reduction (~50 μM; Figure 6) leads to the +3800‰ enrichment of δ13CDIC from an initial cell suspension containing 400 μM DIC with a δ13CDIC value of −12‰ (Figure 6). During the short-term incubation experiments, the +2‰ enrichment was observed, which is most likely explained by the production of DIC from the oxidation of naturally occurring DOC with a δ13CDOC range around −30‰ (Hasegawa et al., 2010). To clarify the contribution of AOM to the production of hydrogen sulfide, the dilution of 13C-labeled DIC produced from AOM (δ13CDIC=+34 000‰) by DIC (δ13CDIC=−30‰) produced from the oxidation of naturally occurring organic matter (δ13CDOC=−30‰) was quantified. The stoichiometric ratio of sulfate reduction to DIC production was set to 1:1 for AOM and 1:2 for organic matter oxidation (LaRowe et al., 2008). To obtain a final suspension with a δ13CDIC value of −10‰ (Figure 6), the mass balance was achieved when AOM and organic matter oxidation produce 0.08 and 99.84 μM DIC, respectively. Although the contribution of sulfate reduction coupled to AOM is small, organic matter oxidation might be overestimated based on the fact that naturally occurring organic matter and microbial cells were highly concentrated for the short-term incubation experiments. To avoid the overestimation of organic matter oxidation, the 1-month incubation was performed without the concentration of cells. However, it was also confirmed by Illumina sequencing that the subsurface ANME-2d sequences were not obtained after the 1-month incubation, which likely resulted from the lysis of the subsurface ANME-2d cells under artificial groundwater conditions (data not shown).
The in situ rate of AOM calculated from this experiment was 3.7 nM year−1, which is similar to that of microbial sulfate reduction previously estimated by geochemical modeling of the same aquifer (1–5 nM year−1; Suzuki et al., 2014). However, this rate of AOM is much slower than those estimated from deeply buried marine sediments, anoxic marine water columns and lake sediments and water (>370 nM year−1; Knittel and Boetius, 2009 and references therein). As AOM activity is suppressed under H2-enriched conditions (LaRowe et al., 2008), the relatively slow rate of AOM might be explained by the experimental concentration of H2 (60 nM) higher than the in situ concentration (<5 nM; Table 1). It was technically impossible to reproduce the in situ H2 concentration in the short-term incubation experiments, because the original stock of 13C-labeled methane contained a trace amount of H2 as impurity. The production of 13C-enriched DIC could be caused by dissimilatory CO2 reduction coupled to methanogenesis and acetogenesis and the assimilatory CO2 fixation (Hayes, 2001). To discriminate the isotopic effects caused by microbial DIC consumption and AOM, the control incubation of HFDB#1 groundwater without CH4 is considered to be necessary. However, the omission of CH4 might stimulate microbial DIC consumption, thereby producing 13C-enriched DIC. In addition, the difference in hydrogen sulfide production appears to be within analytical errors, given the small contribution of AOM to hydrogen sulfide produced during the incubation. Thus it is difficult to measure the rate and mass balance of AOM coupled to sulfate reduction in complex microbial processes concurring in the granitic groundwater.
AOM depending on syntrophic sulfate reducers?
From the metabolic potential of the subsurface ANME-2d reconstructed by metagenomics analysis, the independent capability of AOM coupled to sulfate reduction is suggested. This notion is also supported by the lack of cell aggregates in collected groundwater samples, when microbial cells were observed by fluorescence microscopy for the total cell count. However, the biofilm formed in rock fractures is considered to constituent the substantial biomass in the deep continental biosphere (Whitman et al., 1998; McMahon and Parnell, 2014). Thus the possibility that groundwater microbes are dispersed after the detachment from the fracture surface is not excluded. As molybdate inhibits assimilatory and dissimilatory sulfate reduction pathways (Reuveny, 1977), it is necessary to perform the incubation experiments amended with inhibitors specific to assimilatory sulfate reduction such as 3′-phosphoadenosine-5′-phosphate, 5′-adenosinemonophosphate (5′-AMP) and 2′-AMP (Setya et al., 1996).
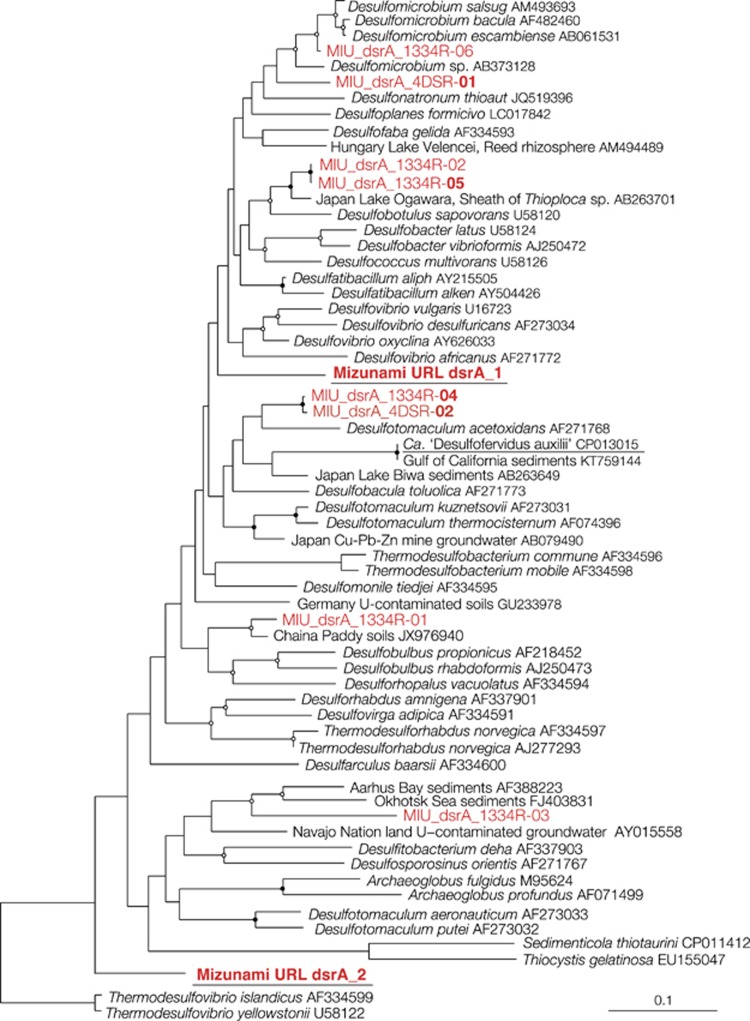
Alternatively, AOM coupled to sulfate reduction is mediated by the subsurface ANME-2d in the syntrophic association with sulfate-reducing bacteria. Previously known syntrophic partners affiliated within sulfate-reducing bacterial lineages (Knittel and Boetius, 2009) such as Desulfosarcina/Desulfococcus (ANME-1 and -2a/b/c), Desulfobacteriaceae (ANME-2d) and Desulfobulbus (ANME-3) were undetected or proportionally uncorrelated with the subsurface ANME-2d, based on the 16S rRNA gene sequence analysis (Supplementary Figure S4). The lack of the syntrophic relationship might be reasonable, given that the known syntrophic partners are revealed from marine sediments. A novel syntrophic relationship is suggested from highly correlated bacterial lineages, including δ-Proteobacteria BPC076, Nirospirae LCP-6 and Omnitrophica (Supplementary Figure S4). To link taxonomic classification based on 16S rRNA gene sequences to the sulfate-reducing physiology, single-gene analysis of dissimilatory sulfite reductase (dsr), a key enzyme for dissimilatory sulfate reduction, was performed with two primer sets targeting different regions of dsrA (encoding subunit A gene of dsr). As shown in Figure 7, dsrA gene sequences closely related to δ-proteobacterial sulfate reducers such as Desulfonatonum spp. and Desulfobulbus spp. and Desulfobotulus sapovorans were obtained. Other dsrA gene sequences were distantly related to known sulfate reducers. To further correlate the phylogenetic affiliations based on 16S rRNA and dsrA gene sequences, draft genomes that contained 16S rRNA and dsrA gene sequences were investigated for the HFDB#1 groundwater collected in 2014. None of the draft genomes contained both dsrA gene and 16S rRNA gene sequences, and two dsrA gene sequences were obtained by metagenomic sequencing. One of the dsrA gene sequences obtained by metagenomics analysis was distantly related to those obtained by single-gene analysis and Desulfotomaculum acetoxidans of Firmicutes, while the other dsrA gene sequence was also distantly related to those of Thermodesulfovibrio spp. of Nitroapirae (Figure 7). The syntrophic association needs to be clarified by fluorescence in situ hybridization analysis with probes specific to the subsurface ANME-2d and the putative sulfate reducers, which are likely aggregated on the rock surface.
Figure 7.
Neighbor-joining tree based on the amino-acid sequences of dsrA gene. The tree was constructed with bootstrap values with >50% and >75% indicated by open and solid circles, respectively. HFDB sequences obtained by single-gene and metagenomics analyses are shown in red. dsrA gene sequences obtained by single-gene analysis using two primer sets are distinguished by names with the reverse primers 4DSR and 1334R. Bold sequences are represented by >5 clonal sequences. Underlined sequences were obtained by metagenomic analysis. Scale bars represent 0.1 expected changes per amino-acid position.
Significance for the deep continental biosphere
As granitic rocks are formed deep within the continental crust by cooling of intruded magma, the organic content is extremely low. H2 production by low temperature processes is negligible in granitic rocks, given the low content of olivine and pyroxene minerals that can produce H2 during serpentinization reactions (McCollom and Bach, 2009). As abiotic methane from magmatic processes is ubiquitous in granitic rocks long after the formation, methane could serve as a major energy source in the deep granitic environment (Etiope and Sherwood Lollar, 2013; Kietäväinen and Purkamo, 2015). Geochemical analysis of deep granitic aquifers around the globe has clarified that sulfate is widely available at >1000 m depths (Gascoyne and Thomas, 1997; Pedersen, 2001; Metcalfe et al., 2006; Laaksoharju et al., 2008; Sahl et al., 2008). A few exceptions exist including granitic rocks with hydraulic conductivities <~10−10 m s−1 at the Mizunami URL (SFDB#2 and #4) and at the Olkiluoto site in western Finland (Vaittinen et al., 2009), where the scarcity of fractures appears to limit the existence of microbial life. The widespread occurrence of 13C-depleted carbonate minerals in granite fractures in the Scandinavian shield appears to be resulted from the past AOM activities coupled to sulfate reduction (Drake et al., 2015). Thus we conclude that sulfate-dependent AOM demonstrated in HFDB is potentially widespread in the deep continental biosphere and could strongly impact the global biomass production and the hydrogen, carbon and sulfur cycles.
Acknowledgments
We thank the members of the Geological Isolation Research Group of AIST and Crystalline Environment Research Group of JAEA. We also thank Y Nakamura and T Tonooka for their technical assistant. Comments from three anonymous reviewers and discussions with SE McGlynn for ANME genomes substantially improved our manuscript. Main part of this research project has been conducted as the regulatory supporting research funded by the Secretariat of Nuclear Regulation Authority (Secretariat of NRA), Japan. This work was partly supported by JSPS KAKENHI Grant Number JP26287133. As part of ‘The project for validating assessment methodology in geological disposal system’, this study was funded by the Ministry of Economy, Trade and Industry of Japan.
Author contributions
Drilling, hydraulic tests and geochemical analyses were performed by KI, UK, MK, AH, YT, KI, AF, TI, TM, DK, UT and YS. Microbial community analyses were conducted by KI, UT, MK, KY, AH, AF and YS. Multivariate function analyses were performed by KI and MS. Thermodynamic calculation was performed by YS. KI, TI and YS performed stable isotope labeling experiments. Metagenomic analysis was performed by AWH, YA, SK, BCT, JFB and YS. KI, AWH, BCT, JFB and YS wrote the manuscript with contributions of all co-authors.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Arshad A, Speth DR, de Graaf RM, Op den Camp HJM, Jetten MSM, Welte CU. (2015). A metagenomics-based metabolic model of nitrate-dependent anaerobic oxidation of methane by Methanoperedens-like archaea. Front Microbiol 6: 1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnoud A, Chourey K, Hettich RL, De Bruijn I, Andersson AF, Leupin OX et al. (2016). Reconstructing a hydrogen-driven microbial metabolic network in Opalinus Clay rock. Nat Commun 7: 12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke C. (1992) The Geochemist's Workbench: A Users Guide to Rxn, Act2, Tact, React, and Gtplot. Hydrogeology program, University of Illinois: Urbana, IL, USA. [Google Scholar]
- Bomberg M, Nyyssönen M, Pitkänen P, Lehtinen A, Itävaara M. (2015). Active microbial communities inhabit sulphate-methane interphase in deep bedrock fracture fluids in Olkiluoto, Finland. Biomed Res Int 2015: 979530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapelle FH. (2001) Ground-Water Microbiology and Geochemistry. Wiley: New York, USA. [Google Scholar]
- Chivian D, Brodie EL, Alm EJ, Culley DE, Dehal PS, DeSantis TZ et al. (2008). Environmental genomics reveals a single-species ecosystem deep within Earth. Science 322: 275–278. [DOI] [PubMed] [Google Scholar]
- Compeau GC, Bartha R. (1985). Sulfate-reducing bacteria: principal methylators of mercury in anoxic estuarine sediment. Appl Environ Microbiol 50: 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper HH, Jacob CE. (1946). A generalized graphical method for evaluating formation constants and summarizing well-field history. Eos Trans AGU 27: 526–534. [Google Scholar]
- Dick GJ, Andersson AF, Baker BJ, Simmons SL, Thomas BC, Yelton AP et al. (2009). Community-wide analysis of microbial genome sequence signatures. Genome Biol 10: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake H, Åström ME, Heim C, Broman C, Åström J, Whitehouse M et al. (2015). Extreme 13C depletion of carbonates formed during oxidation of biogenic methane in fractured granite. Nat Commun 6: 7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray S, Dufour AB, Thioulouse J. (2015). ade 4: analysis of ecological data: exploratory and Euclidean methods in environmental sciences. R package version 1.7-2. http://www.r-project.org/web/packages/ade4/.
- Edgar RC. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etiope G, Sherwood Lollar B. (2013). Abiotic methane on Earth. Rev Geophys 51: 276–299. [Google Scholar]
- Ettwig KF, Zhu B, Speth D, Keltjens JT, Jetten MS, Kartal B. (2016). Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc Natl Acad Sci 113: 12792–12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn TM, Sanford RA, Ryu H, Bethke CM, Levine AD, Ashbolt NJ et al. (2013). Functional microbial diversity explains groundwater chemistry in a pristine aquifer. BMC Microbiol 13: 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne M, Thomas DA. (1997). Impact of blasting on groundwater composition in a fracture in Canada’s Underground Research Laboratory. J Geophys Res Sol Ea 102: 573–584. [Google Scholar]
- Gihring TM, Moser DP, Lin LH, Davidson M, Onstott TC, Morgan L et al. (2006). The distribution of microbial taxa in the subsurface water of the Kalahari Shield, South Africa. Geomicrobiol J 23: 415–430. [Google Scholar]
- Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB et al. (2016). Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J 10: 761–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griebler C, Lueders T. (2009). Microbial biodiversity in groundwater ecosystems. Freshw Biol 54: 649–677. [Google Scholar]
- Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21: 494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales BA, Edwards C, Ritchie DA, Hall G, Pickup RW, Saunders JR. (1996). Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl Environ Microbiol 62: 668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. (2008). Error-correcting barcoded primers allow hundreds of samples to be pyrosequenced in multiplex. Nat Methods 5: 235–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P et al. (2013). Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500: 567–570. [DOI] [PubMed] [Google Scholar]
- Hasegawa T, Nakata K, Tomioka Y, Goto K, Kashiwaya K, Hama K et al. (2010) Research and Development on Groundwater Dating (Part 12) -APPLICATION on Groundwater Dating Using 4He and 14C in Tono Area. Central Research Institute of Electric Power Industry report, N10001, Chiba, Japan. [Google Scholar]
- Hayes JM. (2001). Fractionation of carbon and hydrogen isotopes in biosynthetic processes. Rev Mineral Geochem 43: 225–277. [Google Scholar]
- Hvorslev MJ. (1951). Time lag and soil permeability in ground-water observations. US Army Corps Eng Water Exp Stn Bull 36: 50. [Google Scholar]
- Hyatt D, LoCascio PF, Hauser LJ, Uberbacher EC. (2012). Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics 28: 2223–2230. [DOI] [PubMed] [Google Scholar]
- Ino K, Konno U, Kouduka M, Hirota A, Togo YS, Fukuda A et al. (2016). Deep microbial life in high-quality granitic groundwater from geochemically and geographically distinct underground boreholes. Environ Microbiol Rep 8: 285–294. [DOI] [PubMed] [Google Scholar]
- Ishimura T, Tsunogai U, Nakagawa F. (2008). Grain-scale heterogeneities in the stable carbon and oxygen isotopic compositions of the international standard calcite materials (NBS 19, NBS 17, IAEA-CO-1, and IAEA-CO-8). Rapid Commun Mass Spectrom 22: 1925–1932. [DOI] [PubMed] [Google Scholar]
- Iwatsuki T, Hagiwara H, Ohmori K, Munemoto T, Onoe H. (2015). Hydrochemical disturbances measured in groundwater during the construction and operation of a large-scale underground facility in deep crystalline rock in Japan. Environ Earth Sci 74: 3041–3057. [Google Scholar]
- Jetten MS, Stams AJ, Zehnder AJ. (1992). Methanogenesis from acetate: a comparison of the acetate metabolism in Methanothrix soehngenii and Methanosarcina spp. FEMS Microbiol Rev 8: 181–197. [Google Scholar]
- Johnson JW, Oelkers EH, Helgeson HC. (1992). SUPCRT92: A software package for calculating the standard molal thermodynamic properties of minerals, gases, aqueous species, and reactions from 1 to 5000 bar and 0 to 1000 °C. Comput Geosci 17: 899–947. [Google Scholar]
- Joshi NA, Fass JN. (2011), Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files. https://github.com/najoshi/sickle.
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. (2012). KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40: D109–D114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaster AK, Moll J, Parey K, Thauer RK. (2011). Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc Natl Acad Sci 108: 2981–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kietäväinen R, Purkamo L. (2015). The origin, source, and cycling of methane in deep crystalline rock biosphere. Front Microbiol 6: 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knittel K, Boetius A. (2009). Anaerobic oxidation of methane: progress with an unknown process. Annu Rev Microbiol 63: 311–334. [DOI] [PubMed] [Google Scholar]
- Konno U, Tsunogai U, Nakagawa F, Nakaseama M, Ishibashi J, Nunoura T et al. (2006). Liquid CO2 venting on the seafloor: Yonaguni Knoll IV hydrothermal system, Okinawa Trough. Geophys Res Lett 33: L16607. [Google Scholar]
- Konno U, Kouduka M, Komatsu DD, Ishii K, Fukuda A, Tsunogai U et al. (2013). Novel microbial populations in deep granitic groundwater from Grimsel Test Site, Switzerland. Microb Ecol 65: 626–637. [DOI] [PubMed] [Google Scholar]
- Kouduka M, Suko T, Morono Y, Inagaki F, Ito K, Suzuki Y. (2012). A new DNA extraction method by controlled alkaline treatments from consolidated subsurface sediments. FEMS Mcrobiol Lett 326: 47–54. [DOI] [PubMed] [Google Scholar]
- Krzycki JA. (2004). Function of genetically encoded pyrrolysine in corrinoid-dependent methylamine methyltransferases. Curr Opin Chem Biol 8: 484–491. [DOI] [PubMed] [Google Scholar]
- Laaksoharju M, Smellie J, Tullborg EL, Gimeno M, Molinero J, Gurban I et al. (2008). Hydrogeochemical evaluation and modelling performed within the Swedish site investigation programme. Appl Geochem 23: 1761–1795. [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRowe DE, Dale AW, Regnier P. (2008). A thermodynamic analysis of the anaerobic oxidation of methane in marine sediments. Geobiology 6: 436–449. [DOI] [PubMed] [Google Scholar]
- Leopold LB, Wolman MG, Miller JP. (2012) Fluvial Processes in Geomorphology. Courier Corporation: Massachusetts, USA. [Google Scholar]
- Lovley DR, Chapelle FH. (1995). Deep subsurface microbial processes. Rev Geophys 33: 365–381. [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan MT, Martingo JM, Stahl DA, Clark DP. (2012) Brock Biology of Microorganisms 13th edn Pearson Education: San Fransisco, CA, USA. [Google Scholar]
- Magrane M, UniProt Consortium. (2011) UniProt Knowledgebase: a hub of integrated protein data Database (Oxford) 2011: bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardanov AV, Slododkina GB, Slobodkin AI, Beletsky AV, Gavrilov SN, Kublanov IV et al. (2015). The Geoglobus acetivorans genome: Fe (III) reduction, acetate utilization, autotrophic growth, and degradation of aromatic compounds in a hyperthermophilic archaeon. Appl Environ Microbiol 81: 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollom TM, Bach W. (2009). Thermodynamic constraints on hydrogen generation during serpentinization of ultramafic rocks. Geochim Cosmochim Acta 73: 856–875. [Google Scholar]
- McGlynn SE, Chadwick GL, Kempes CP, Orphan VJ. (2015). Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526: 531–535. [DOI] [PubMed] [Google Scholar]
- McMahon S, Parnell J. (2014). Weighing the deep continental biosphere. FEMS Microbiol Ecol 87: 113–120. [DOI] [PubMed] [Google Scholar]
- Metcalfe R, Takase H, Sasao E, Ota K, Iwatsuki T, Arthur RC et al. (2006). A system model for the origin and evolution of the Tono Uranium Deposit, Japan. Geochem Explor Envir Anal 6: 13–31. [Google Scholar]
- Meyerdierks A, Kube M, Kostadlnov I, Teellng H, Glöckner FO, Relnhardt R et al. (2010). Metagenome and mRNA expression analyses of anaerobic methanotrophic archaea of the ANME-1 group. Environ Microbiol 12: 422–439. [DOI] [PubMed] [Google Scholar]
- Miyajima T, Yamada Y, Hanba YT, Yoshii K, Koitabashi T, Wada E. (1995). Determining the stable isotope ratio of total dissolved inorganic carbon in lake water by GC/C/IRMS. Limnol Oceanogr 40: 994–1000. [Google Scholar]
- Nunoura T, Takaki Y, Kazama H, Hirai M, Ashi J, Imachi H et al. (2012). Microbial diversity in deep-sea methane seep sediments presented by SSU rRNA gene tag sequencing. Microb Environ 27: 382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D et al. (2016), Package ‘vegan’. http://www.r-project.org, http://vegan.r-forge.r-project.org/.
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25: 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K. (2001) Diversity and activity of microorganisms in deep igneous rock aquifers of the Fennoscandian Shield. In: Fredrickson JK, Fletcher M (eds). Subsurface Microbiology and Biogeochemistry. Wiley-Liss Inc: New York, USA, pp 97–139. [Google Scholar]
- Pedersen K, Bengtsson AF, Edlund JS, Eriksson LC. (2014). Sulphate-controlled diversity of subterranean microbial communities over depth in deep groundwater with opposing gradients of sulphate and methane. Geomicrobiol J 31: 617–631. [Google Scholar]
- Peng Y, Leung HCM, Yiu SM, Chin FYL. (2012). IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28: 1420–1428. [DOI] [PubMed] [Google Scholar]
- R Core Team. (2013) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. http://www.r-project.org.
- Reuveny Z. (1977). Derepression of ATP sulfurylase by the sulfate analogs molybdate and selenate in cultured tobacco cells. Proc Natl Acad Sci 74: 619–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl JW, Schmidt R, Swanner ED, Mandernack KW, Templeton AS, Kieft TL et al. (2008). Subsurface microbial diversity in deep-granitic-fracture water in Colorado. Appl Environ Microbiol 74: 143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santillano D, Boetius A, Ramette A. (2010). Improved dsrA-based terminal restriction fragment length polymorphism analysis of sulfate-reducing bacteria. Appl Environ Microbiol 76: 5308–5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchmann K, Müller V. (2014). Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria. Nat Rev Microbiol 12: 809–821. [DOI] [PubMed] [Google Scholar]
- Setya A, Murillo M, Leustek T. (1996). Sulfate reduction in higher plants: molecular evidence for a novel 5′-adenylylsulfate reductase. Proc Natl Acad Sci 93: 13383–13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique AB, Unterseher M. (2016). A cost-effective and efficient strategy for Illumina sequencing of fungal communities: a case study of beech endophytes identified elevation as main explanatory factor for diversity and community composition. Fungal Ecol 20: 175–185. [Google Scholar]
- Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. (2007). UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23: 1282–1288. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Konno U, Fukuda A, Komatsu DD, Hirota A, Watanabe K et al. (2014). Biogeochemical signals from deep microbial life in terrestrial crust. PLoS ONE 9: e113063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova T, Ciufo S, Fedorov B, O'Neill K, Tolstoy I. (2014). RefSeq microbial 565 genomes database: new representation and annotation strategy. Nucleic Acids Res 566: D553–D559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers PH, Suarez-Zuluaga DA, van Rossem M, Diender M, Stams AJ, Plugge CM. (2015). Anaerobic oxidation of methane associated with sulfate reduction in a natural freshwater gas source. ISME J 10: 1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunogai U, Yoshida N, Ishibashi J, Gamo T. (2000). Carbon isotopic distribution of methane in deep-sea hydrothermal plume, Myojin Knoll Caldera, Izu-Bonin arc: implications for microbial methane oxidation in the oceans and applications to heat flux estimation. Geochim Cosmochim Acta 64: 2439–2452. [Google Scholar]
- Vaittinen T, Ahokas H, Nummela J, Paulamäki S. (2009) Hydrogeological Structure Model of the Olkiluoto Site: Update in 2008. Working Report 2009-15. Posiva Oy: Eurajoki, Finland. [Google Scholar]
- van Haaster DJ, Silva PJ, Hagedoorn PL, Jongejan JA, Hagen WR. (2008). Reinvestigation of the steady-state kinetics and physiological function of the soluble NiFe-hydrogenase I of Pyrococcus furiosus. J Bacteriol 190: 1584–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FP, Zhang Y, Chen Y, He Y, Qi J, Hinrichs KU et al. (2014). Methanotrophic archaea possessing diverging methane-oxidizing and electron-transporting pathways. ISME J 8: 1069–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, Roger AJ, Flax JL, Brusseau GA, Stahl DA. (1998). Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol 180: 2975–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes MGR. (2016), Package ‘gplots’ http://cran.r-project.org/web/packages/gplots/.
- Wegener G, Krukenberg V, Riedel D, Tegetmeyer HE, Boetius A. (2015). Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 526: 587–590. [DOI] [PubMed] [Google Scholar]
- Whiticar MJ. (1999). Carbon and hydrogen isotope systematics of bacterial formation and oxidation of methane. Chem Geol 161: 291–314. [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. (1998). Prokaryotes: the unseen majority. Proc Natl Acad Sci USA 95: 6578–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton KC, Thomas BC, Sharon I, Miller CS, Castelle CJ, VerBerkmoes NC et al. (2012). Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337: 1661–1665. [DOI] [PubMed] [Google Scholar]
- Wu X, Holmfeldt K, Hubalek V, Lundin D, Åström M, Bertilsson S et al. (2015). Microbial metagenomes from three aquifers in the Fennoscandian shield terrestrial deep biosphere reveal metabolic partitioning among populations. ISME J 10: 1192–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuguchi T, Tsuruta T, Hama K, Nishiyama T. (2013). The spatial variation of initial 87Sr/86 Sr ratios in the Toki granite, Central Japan: Implications for the intrusion and cooling processes of a granitic pluton. J Miner Petrol Sci 108: 1–12. [Google Scholar]
- Zhang G, Tian J, Jiang NA, Guo X, Wang Y, Dong X. (2008). Methanogen community in Zoige wetland of Tibetan plateau and phenotypic characterization of a dominant uncultured methanogen cluster ZC-I. Environ Microbiol 10: 1850–1860. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.