Abstract
Many insects rely on bacterial endosymbionts to obtain nutrients that are scarce in their highly specialized diets. The most surprising example corresponds to the endosymbiotic system found in mealybugs from subfamily Pseudococcinae in which two bacteria, the betaproteobacterium ‘Candidatus Tremblaya princeps’ and a gammaproteobacterium, maintain a nested endosymbiotic consortium. In the sister subfamily Phenacoccinae, however, a single beta-endosymbiont, ‘Candidatus Tremblaya phenacola’, has been described. In a previous study, we detected a trpB gene of gammaproteobacterial origin in ‘Ca. Tremblaya phenacola’ from two Phenacoccus species, apparently indicating an unusual case of horizontal gene transfer (HGT) in a bacterial endosymbiont. What we found by sequencing the genome of ‘Ca. Tremblaya phenacola’ PPER, single endosymbiont of Phenacoccus peruvianus, goes beyond a HGT phenomenon. It rather represents a genome fusion between a beta and a gammaproteobacterium, followed by massive rearrangements and loss of redundant genes, leading to an unprecedented evolutionary collage. Mediated by the presence of several repeated sequences, there are many possible genome arrangements, and different subgenomic sequences might coexist within the same population.
Introduction
Symbiosis has shaped the evolution of life in many ways. The huge variety of symbiotic associations described along the three domains of life is an indicator of the evolutionary relevance of this phenomenon (McFall-Ngai, 2008; Moya et al., 2008). Notably, the mutualistic relationships that animals have established with prokaryotes furnished them with new metabolic capabilities, allowing the colonization of otherwise inaccessible niches. Obligate mutualistic association with endosymbiotic bacteria (primary or P-endosymbionts) is considered a key factor for the evolutionary success of insects. Being essential for host survival and reproduction, they are fixed in the host populations (Houk and Griffiths, 1980), live inside specialized host cells (bacteriocytes) and are maternally transmitted (Buchner, 1965; Baumann, 2005). Consequently, hosts and P-endosymbionts show congruent phylogenies (Munson et al., 1991; Chen et al., 1999; Thao et al., 2000; Sauer et al., 2000; Lo et al., 2003; Moran et al., 2003; Thao and Baumann, 2004; Allen et al., 2007; Conord et al., 2008; Rosenblueth et al., 2012). Because they are highly adapted to their intracellular environment, P-endosymbionts are uncultivable. Therefore, DNA-sequencing and molecular phylogenetic analyses are frequent approaches for their characterization (Murray and Schleifer, 1994).
The study of P-endosymbionts’ genomes has revealed they share some commonalities, the most prominent being a drastic genome size reduction, with small gene sets organized in a highly compact way (Baumann, 2005; McCutcheon and Moran, 2012). Two main evolutionary mechanisms drive this genome shrinkage (Moya et al., 2008). First, living inside eukaryotic cells renders some genes unnecessary, while others become redundant with functions provided by the host. Their eventual loss has no effect on bacterial fitness and, therefore, the pressure of purifying selection over them is relaxed. Second, the strict endosymbionts’ vertical transmission reduces their effective population size, increasing the genetic-drift effects (Moran, 1996). Both factors facilitate the fixation of slightly deleterious mutations in non-essential genes, causing their inactivation and subsequent loss. Typically, this process affects genes involved in DNA repair and recombination in early association stages, further increasing the mutation rate and preventing genetic exchange by homologous recombination. In addition, the relative isolation of the endosymbiont populations usually hinders the possibility of acquiring genetic material through horizontal gene transfer (HGT). In some symbiotic systems, two or more co-primary endosymbionts complement each other to fulfill the consortium metabolic needs, leading to even more reduced genomes. The eventual acquisition of bacterial genes by the host genome might facilitate the extreme genome reduction of long-term P-endosymbionts (Sloan et al., 2014; Luan et al., 2015 and references therein).
Mealybugs (Hemiptera: Pseudococcidae) are phloem-sucking plant parasites (Hardy et al., 2008), being common pests of a wide range of plants. Like other phloem-feeding insects, they rely on endosymbionts to complement their diets. In subfamily Pseudococcinae, a nested endosymbiotic consortium has been described, in which the betaproteobacterium ‘Ca. Tremblaya princeps’ (T. princeps thereafter) harbors a gammaproteobacterium (von Dohlen et al., 2001; Kono et al., 2008; Gatehouse et al., 2012; Koga et al., 2012). Several endosymbiotic consortia from different pseudococcinae mealybugs have been partially or completely sequenced (Munson et al., 1992; Baumann et al., 2002, McCutcheon and von Dohlen, 2011; López-Madrigal et al., 2011; López-Madrigal et al., 2013; López-Madrigal et al., 2014; López-Madrigal et al., 2015; Husnik and McCutcheon, 2016; Szabó et al., 2017). These studies revealed an intricate evolutionary history involving independent acquisition and replacements of different gamma-endosymbionts by the ancestor of the extant T. princeps in different mealybug lineages. During the process, the T. princeps genomes have maintained a nearly perfect synteny conservation (McCutcheon and von Dohlen, 2011; Husnik and McCutcheon, 2016), similar to that described for Buchnera in aphids (van Ham et al., 2003), although they display several unusual genomic features: high GC content; low coding density; and presence of repeated sequences evolving under concerted evolution.
In the sister subfamily Phenacoccinae, a single beta-endosymbiont has been described, ‘Candidatus Tremblaya phenacola’ (T. phenacola thereafter; Gruwell et al., 2010). The genome sequencing of T. phenacola PAVE (Husnik et al., 2013), P-endosymbiont of Phenacoccus avenae, revealed that this endosymbiont alone can fulfill its host nutritional needs. Husnik and McCutcheon (2016) assumed that (at least) the T. phenacola PAVE gene set was present in T. princeps when the gamma-endosymbiont acquisition took place; subsequent gene losses occurred in response to this event in different T. princeps lineages, leading to endosymbiotic consortia with very few redundant metabolic functions.
In a previous work, while screening for genes involved in amino-acid biosynthesis in unexplored mealybug species, we detected what apparently represented one of the few recorded cases of HGT in a P-endosymbiont (López-Madrigal et al., 2014). Two Phenacoccus species, with a single beta-endosymbiont, had a trpB gene phylogenetically affiliated to gammaproteobacteria. To determine the magnitude of the putative HGT event, we have sequenced the genome of T. phenacola PPER. Surprisingly, while the 16S rRNA gene clearly places this strain within the T. phenacola clade, the genome is a collage of sequences of beta and gammaproteobacterial origin. The extent of HGT observed represents a unique case, both regarding the amount and function of transferred genes. Once again, bacteria–mealybugs mutualistic symbioses take us toward an unprecedented evolutionary scenario.
Materials and methods
Insect sample collection and DNA extraction
A population of P. peruvianus from an initial sample field collected in 2014 in Valencia (Spain) was reared on bougainvillea bushes in the ICBiBE facilities. Total DNA enriched in bacterial endosymbionts (TDNA) was extracted from mechanically homogenized viscera of 10–20 adult female insects, using JETFLEX Genomic DNA Purification Kit (GENOMED GmbH, Löhne, Germany).
Genome sequencing and assembly
Illumina paired-end short-insert sequencing was performed at the Sequencing facilities of the Universitat de València (see details in Supplementary Information). Reads were quality filtered using FASTX-Toolkit v0.0.14 (http://hannonlab.cshl.edu/fastx_toolkit/) and cutadapt v1.9 (Martin, 2011). Read pairs were merged using FLASH v1.2.11 (Magoc and Salzberg, 2011), and only fragments longer than 400 bp were assembled using SPAdes v3.6.2 (Bankevich et al., 2012). TBLASTX from the BLAST+ suite v2.5.0 (Camacho et al., 2009) was used to select contigs matching the T. phenacola PAVE genome. All initial paired reads were then mapped to these contigs using BBMap (Bushnell B., sourceforge.net/projects/bbmap/), with several rounds of assembly and mapping until the assembly size stopped increasing. Because the average coverage was greater than 2000 ×, one-tenth of raw reads was randomly selected and mapped against the assembly. Only 31 generated contigs with a coverage above 100 × were conserved, all ending with repeated sequences. Contigs from the original full assembly were binned according to their coverage and genomic signature (frequency of k-mers of size 4) using MyCC (Lin and Liao, 2016), confirming that the 31 contigs appear on a single bin (Supplementary Table S1).
The paired-end reads mapping to the different contigs’ edges outside repeats were used to construct a graph using Cytoscape v3.1.1 (Shannon et al., 2003) to visualize possible links between contigs. Contigs spanned entirely by repeats were merged to one of their neighbors when possible, leading to a final set of 23 contigs.
To confirm unions between contigs flanking the same repeat, primers inside bona-fide genes outside repeats were designed with Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi; Untergasser et al. 2012) (Supplementary Table S2). PCR amplifications were performed with all possible primer pair combinations (Supplementary Table S3), using 50–60 μmol of each primer per 50 μl reaction, with KAPATaq DNA Polymerase Kit (Kapa Biosystems, Wilmington, MA, USA). When needed, amplicons were cloned using pGEM-T Easy Vector System I Kit (Promega). Amplicons were ABI-sequenced using specific or vector primers T7 and SP6 (sequencing facilities, Universitat de València). Reads were quality-surveyed and assembled with Staden Package (http://staden.sourceforge.net/; Staden et al., 2000).
In addition, to confirm the existence of different rearrangements among contigs, a Nanopore sequencing experiment was performed with a MinION device (Oxford Nanopore Technologies Ltd, Oxford, UK; see details in Supplementary Information). Using megablast (Camacho et al., 2009), 3727 reads (3.97% of the obtained reads), ranging from 228–45 964 bp, were mapped against the previously assembled PPER contigs, leading to an 83 × coverage.
TBLASTX was used to identify conserved syntenic blocks between the genomes of T. phenacola PPER and PAVE. The output was parsed using custom R scripts and plotted with genoPlotR v0.8.3 (Guy et al., 2010).
Genome annotation, molecular and functional analyses
Contigs were annotated using Prokka v1.11 (Seeman, 2014), with additional manual curation in the Artemis browser (Carver et al., 2008), using BLASTX, BLASTP (Altschul et al., 1997) and Pfam (Finn et al., 2014) to correct gene boundaries, identify pseudogenes and detect missing open reading frames. Annotated open reading frames were considered functional genes if no frameshifts disrupting its coding sequence (CDS) were found and they maintained complete essential functional domains. Repeats at the contig edges were screened using ISfinder (http://www-is.biotoul.fr; Siguier et al., 2006), to identify putative remnants of insertion sequences. G+C content and codon usage were calculated using custom-made R scripts. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession MKGN00000000. The version described here is version MKGN01000000.
The functional analysis was performed by comparison with previously sequenced Tremblaya genomes (Supplementary Table S4). Additional information was retrieved from EcoCyc (Keseler et al., 2013) and Kyoto Encyclopedia of Genes and Genomes (Kanehisa et al., 2016) databases.
Taxonomic assignation of genes
Two approaches were followed to assign a taxonomic class to CDSs. The first one started with a BLASTP search against the full non-redundant database (O’Leary et al., 2016), followed by parsing of the output using MEGAN Community Edition v6.4.0 (Huson et al., 2016). The second one involved using the pipeline phylomizer (https://github.com/Gabaldonlab/phylomizer; Huerta-Cepas et al., 2014), comparing T. phenacola PPER proteins against those of T. phenacola PAVE, T. princeps PCVAL (beta-endosymbiont of Planococcus citri; López-Madrigal et al., 2011) and 10 free-living bacteria (five gammaproteobacteria and five betaproteobacteria; Supplementary Table S4). A phylogenetic tree was built for each protein and, when possible, the taxonomic origin was selected as betaproteobacteria or gammaproteobacteria. When both methods yielded the same result, or when one of them gave an inconclusive result that was not incompatible with the other one, the gene was assigned to the corresponding class. The same analysis was performed with the translated CDSs of T. phenacola PAVE and T. princeps PCVAL, as controls.
Phylogenomic analyses
OrthoMCL v2.0.9 (Li et al., 2003) was used to generate clusters of orthologous genes for the full gene sets of the aforementioned beta and gammaproteobacteria plus two alphaproteobacteria as outgroups (Supplementary Table S4). A concatenated alignment using predefined conserved proteins followed by its phylogenetic reconstruction was built with PhyloPhlAn (Segata et al., 2013). The alignments were divided in two sets, those orthologs of gammaproteobacterial origin and those of betaproteobacterial origin. Uninformative positions were excluded using trimAL v1.4 (Capella-Gutierrez et al., 2009), and the remaining positions were used to generate ML trees using RAxML 8.2.3 (Stamatakis, 2014) under the LG+G model with 100 bootstrap pseudoreplicates. In addition, each set was inserted into the already reconstructed most comprehensive tree of life (Segata et al., 2013) using PhyloPhlAn.
Results and discussion
The T. phenacola PPER genome: a puzzle with (too) many solutions
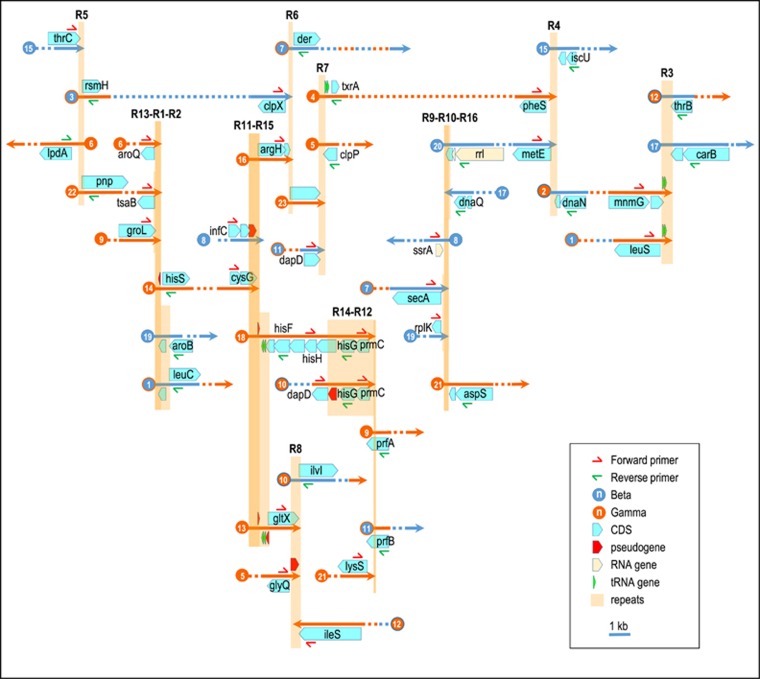
The bougainvillea mealybug P. peruvianus, similarly to other phenacoccinae mealybugs, harbors a single endosymbiont, as confirmed by FISH experiments (López-Madrigal et al., 2014), classified as the betaproteobacterium T. phenacola. However, in the same work, we detected a trpB gene phylogenetically related to gammaproteobacteria in the bacterial genome. To ascertain the magnitude of what appeared to be a HGT event, we have sequenced the complete genome of this bacterium. The unexpected presence of a large number of repeats hampered genome assembly. With the help of paired-end reads mapping to unique sequences near each contig edges (see Materials and Methods), the genome was assembled to 23 contigs (Table 1), all ending in repeated sequences ranging from 95 to 2023 bp (280 bp median size; Table 2). We determined all possible links between contigs (Supplementary Figure S1), and tested them by PCR with specific primers (Supplementary Table S2). All possible combinations gave positive results with a single exception (Supplementary Table S3), and all amplicons were sequenced to confirm the presence of single or combined repeats (Figure 1). In addition, a possible length polymorphism was detected inside contig 12. The affected region was amplified, and 10 clones were sequenced. Each clone contained an imperfect ATTGGRCAACAGTATTAGYATCCT repeat, with three to five copies of the sequence, presenting the fourth repeat a 10_11delCA, when present.
Table 1. T. phenacola PPER contigs’ features.
| Contig | Length (bp) | Unique sequence length (bp) | Coverage unique sequence | Phylogenetic affiliation | 5'-repeat | 3'-repeat |
|---|---|---|---|---|---|---|
| 1 | 14 880 | 13 581 | 209.20 | B–G | REP1.1+REP2.1 | REP3.1 |
| 2 | 14 652 | 13 822 | 278.90 | B–G | REP4.1 | REP3.2 |
| 3 | 5137 | 4719 | 257.60 | G–B | REP5.1 | REP6.1 |
| 4 | 7141 | 6581 | 275.20 | G | REP7.1 | REP4.2 |
| 5 | 5562 | 4794 | 209.50 | G | REP7.2 | REP8.1 |
| 6 | 4152 | 3565 | 240.60 | G | REP5.2c | REP1.2 |
| 7 | 13 128 | 12 667 | 229.40 | B–G–B–G–B | REP6.2 | REP9.1+REP10.1 |
| 8 | 10 425 | 9681 | 215.90 | B | REP10.2c | REP11.1 |
| 9 | 6907 | 6428 | 227.60 | G | REP12.1 | REP13.1+REP1.3 |
| 10 | 9221 | 6252 | 210.80 | B–G | REP8.2 | REP14.1+REP12.2 |
| 11 | 5390 | 5011 | 276.60 | G–B | REP12.3 | REP7.1 |
| 12 | 14 706 | 13 668 | 238.00 | B–G | REP3.3 | REP8.3c |
| 13 | 2573 | 1706 | 215.50 | G | REP11.2+REP15.1 | REP8.4 |
| 14 | 17 527 | 16 660 | 262.20 | G | REP1.4 | REP11.3 |
| 15 | 12 272 | 11 752 | 228.50 | B | REP4.3 | REP5.3 |
| 16 | 2256 | 1538 | 150.10 | G | REP11.4 | REP6.3 |
| 17 | 6528 | 5675 | 210.60 | B | REP3.4 | REP16.1c+REP10.3c |
| 18 | 6246 | 2786 | 128.00 | G | REP11.5+REP15.2 | REP14.2+REP12.4 |
| 19 | 28 817 | 27 805 | 212.40 | B | REP1.5+REP2.2 | REP9.2+REP10.4 |
| 20 | 14 257 | 13 682 | 241.90 | B | REP10.5+REP16.2 | REP4.4 |
| 21 | 10 719 | 10 404 | 198.30 | G | REP10.6 | REP12.5 |
| 22 | 4631 | 4003 | 211.50 | G | REP5.4 | REP13.2+REP1.6 |
| 23 | 1805 | 1327 | 154.60 | G | REP6.4 | REP7.4 |
Abbreviations: B, betaproteobacteria; G, gammaproteobacteria; c, complementary sequence.
Table 2. Repeats identified in the T. phenacola PPER genome.
| Repeat name | Size (bp) | Copiesa |
|---|---|---|
| REP1 | 343 | 6 |
| REP2 | 402 | 2 |
| REP3 | 554 | 4 |
| REP4 | 276 | 4 |
| REP5 | 244 | 4 |
| REP6 | 194 | 4 |
| REP7 | 284 | 4 |
| REP8 | 484 | 4 |
| REP9 | 47 | 2 |
| REP10 | 220 | 6 |
| REP11 | 524 | 5 |
| REP12 | 95 | 5 |
| REP13 | 5 | 2 |
| REP14 | 2390 | 2 |
| REP15 | 451 | 2 |
| REP16 | 76 | 2 |
Number of times annotated in the genome contigs.
Figure 1.
T. phenacola PPER genome representation illustrating all possible joins among contigs. The number of each contig is indicated in a circle at the 5′-end, and the 3′-end is indicated with an arrowhead. The contig lines are colored according to their beta (blue) or gamma (orange) origin. The vertical shadows indicate the end repeats (R1–R16) represented at scale. Genes used for PCR primers designing are shown. Dotted lines followed by an arrowhead indicate that the contig continues in other part of the Figure. When possible, the two ends of the contig are joined by dots, but not at scale. For a description of the repeats, see Table 2.
To evaluate our predicted links between more than two contigs, we performed a Nanopore sequencing experiment. Among the 286 reads mapping to more than two contigs (200 to 3, 60 to 4, 17 to 5, 6 to 6 and 3 to 7), we detected 101 out of 128 possible combinations (Supplementary Table S5; Supplementary Figure S2), thus confirming the existence of different rearrangements among contigs. The joint between contigs 18 and 11, which could not be PCR amplified, was also present.
The complete genome consists of 198 035 bp of unique sequence flanked by repeats that, altogether, amount 6589 bp. It must be noticed that not all repeats appear an even number of times (Table 2). Therefore, it is possible that different subgenomic sequences coexist within the same population. Considering that bacteriocytes contain many bacterial cells, and that endosymbionts have been described to contain many genome copies per cell (Komaki and Ishikawa, 1999; Woyke et al., 2010; van Leuven et al., 2014), it is possible that several combinations appear in a single bacteriocyte or even in a single bacterial cell. Nevertheless, according to the binning performed, the average assembly coverage and genomic signatures are quite similar for all 23 contigs (Table 1). We detected a single ribosomal cluster inside contig 20. As all cells must contain these genes to make functional ribosomes, and the mean coverage of contig 20 is close to the global mean coverage (215.9; s.d.=37.88), most contigs are expected to be present in an even number of copies in most T. phenacola PPER cells.
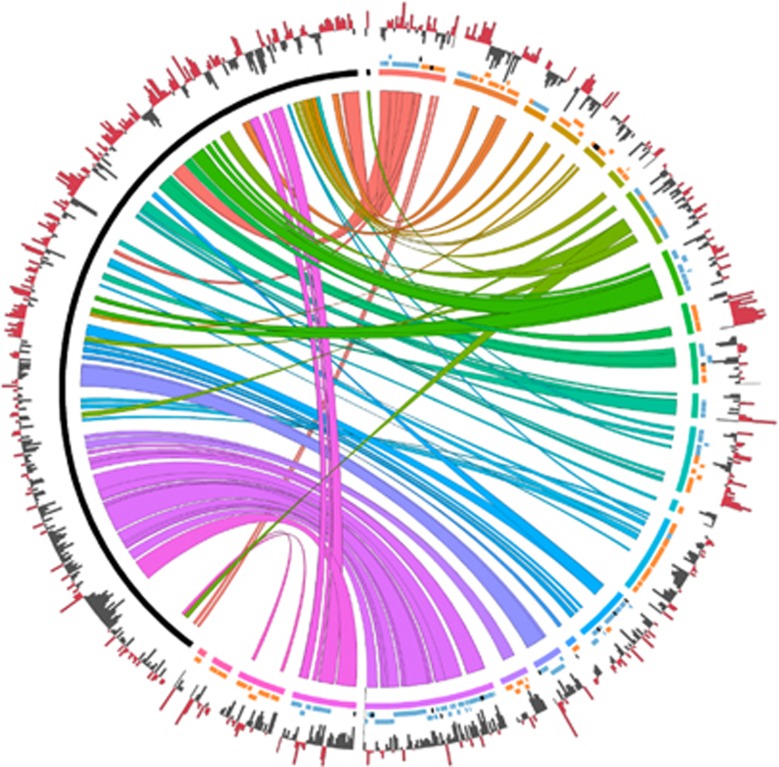
Given the impossibility to determine a single ordering for all contigs, we have chosen the one allowing the best syntenic comparison with T. phenacola PAVE (Husnik et al., 2013). The genome of T. phenacola PPER is highly rearranged (Figure 2), in contrast to the high genomic stability of all previously sequenced Tremblaya lineages, with an almost absolute synteny conservation among T. princeps strains (McCutcheon and von Dohlen, 2011; Husnik and McCutcheon, 2016), and a single inversion in T. phenacola PAVE (Husnik and McCutcheon, 2016). Chromosome rearrangements cause perturbations in GC skew, which have a deleterious impact upon the replication system (Rocha, 2004). Therefore, although bacterial chromosomes can undergo many rearrangements at the beginning of an endosymbiotic relationship (see the marked example of ‘Candidatus Sodalis pierantonius’ SOPE; Oakeson et al., 2014), long-term endosymbionts tend to present a typical GC skew, an indication that it is recovered with evolutionary time. Contrary to the PAVE genome, with a typical GC-skew pattern (Rocha, 2008), PPER genome presents a non-polarized and highly disrupted GC skew, except in the most syntenic region between both genomes, containing most ribosomal protein genes (contig 19; Supplementary Table S6), suggesting that the chimeric genomic architecture is not stabilized.
Figure 2.
Genome synteny comparison of the two T. phenacola strains already sequenced, PAVE and PPER. Bezier curves highlight regions of synteny shared between both strains. From inner to outer the rings show the PAVE chromosome and plasmid (left) in black, and the PPER contigs (right), each one in a different color; PPER genes in the forward or reverse strand are represented in different colors according to their assigned taxonomic origin, beta (blue), gamma (orange) or unassigned (black); for both genomes, the outer plots represent GC skew, with positive and negative skew depicted in red and gray, respectively.
Genome annotation, functional analysis and taxonomic assignation of the genes
The PPER genome contains 192 different CDSs, 188 with an assigned function (Supplementary Table S6). There are only four duplicated genes inside repeats (rpsU, hisG, prmC and TPPER_00169/220), and two have two homologs (infA and rlmE). As above mentioned, it possesses a single ribosomal operon and a complete set of tRNA genes for all 20 amino acids, 4 of them inside repeats. Table 3 summarizes the main genomic features compared with the strain PAVE and T. princeps PCVAL. We found no evidences of the presence of remnants of former insertion sequences.
Table 3. Main features of the Tremblaya genomes used in this study.
| Endosymbiont | T. phenacola PPER | T. phenacola PAVE | T. princeps PCVAL |
|---|---|---|---|
| Host | P. peruvianus | P. avenae | P. citri |
| Reference | This study | Husnik et al., 2013 | López-Madrigal et al., 2011 |
| Number of contigs | 23 | 1 | 1 |
| Genome size (bp) | 198 035+6589a | 171 500b | 138 931 |
| G+C (%) | 35.6 | 42.2 | 59 |
| CDS (pseudogenes) | 196 (6) | 178 (3) | 116 (19) |
| CDS coding density (%) | 90.35 | 86.3 | 71.2 |
| rRNAs | 3 | 4 | 6 |
| tRNAs (pseudogenes) | 38 | 31 | 7 (6) |
| ncRNAs | 2 | 3 | 1 |
Abbreviations: CDS, coding sequence; ncRNA, non-coding RNA.
Indicates the size of the unique sequences plus the sum of all repeats once.
Chromosome plus plasmid
The T. phenacola PPER’s gene set is similar to that of strain PAVE, with a few differences possibly reflecting an independent genome reduction process, with random loss or non-functionalization of non-essential genes, which probably will not produce remarkable differences in their functional capabilities (Supplementary Table S7). In three cases, a non-orthologous gene displacement can be invoked. Thus, the presence of pykF can compensate the absence of ndk in PPER, as pyruvate kinase has been proposed to act as a nucleoside diphosphate kinase in other endosymbionts (Gil et al., 2004); the products of hom (PPER) and thrA (PAVE) perform the same function in threonine biosynthesis, as it occurs with the products of ilvI (PPER) and ilvB (PAVE) in branched amino-acid biosynthesis (Supplementary Figure S3).
Next, we searched for the taxonomic affiliation of annotated CDSs (Supplementary Table S6). Surprisingly, only 102 CDSs appear to be of betaproteobacterial origin, but another 80 (occupying 46% of the genome) appear to belong to a gammaproteobacterium. When the taxonomic analysis was performed on the PCVAL and PAVE genomes, almost all genes were unambiguously assigned to betaproteobacteria (Supplementary Table S8). Even though two different methods were used for taxonomic assignation, eight genes could not be assigned to any taxonomic category and five gave inconsistent results. The difficulty for classifying these genes can be due to the short length of some of them, a high degree of nucleotide changes during the adaptation to the new chimeric genomic organization or the formation of chimeric genes through recombination. The lack of redundancy is remarkable: only two genes (infA and rlmE) present two copies of gamma and betaproteobacterial origin, respectively.
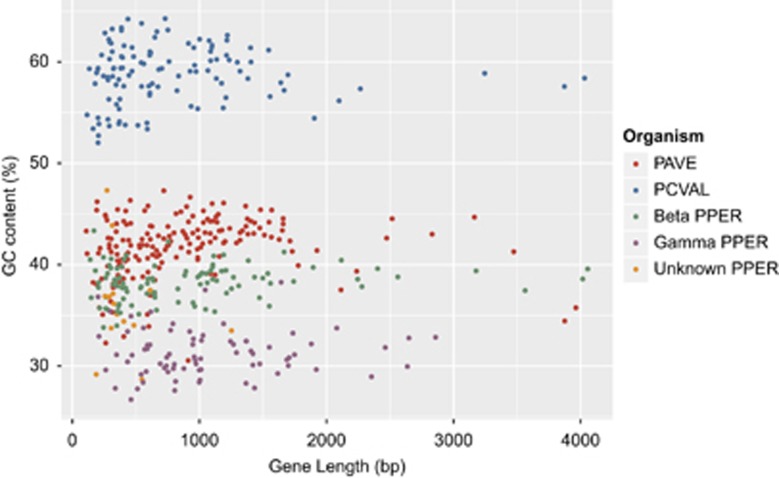
A noteworthy feature of the Tremblaya genomes (more pronounced in T. princeps) is their high G+C content compared to other endosymbionts. We have found a relationship between the taxonomic affiliation of each identified CDS and their G+C content (Figure 3). As expected, considering the general trait of beta and gammaproteobacterial genomes (Agashe and Shankar, 2014), genes with gammaproteobacterial assignation have lower G+C values. It is worth mentioning that genes not assigned to any category have a wide range in G+C content, and most of them have very short length. We also detected differences in codon usage depending on the beta or gammaproteobacterial assignation (Supplementary Figure S4).
Figure 3.
GC versus gene length in the CDS of T. phenacola PPER compared to T. phenacola PAVE and T. princeps PCVAL. The taxonomic origin of the PPER genes is indicated.
The distribution of gamma or beta genes along the genome is not random: most contigs contain only genes of one taxonomic origin, some others change the gene affiliation in the middle, and only contig 7 is completely intermixed (Figures 1 and 2; Table 1; Supplementary Table S6; Supplementary Figure S5). Remarkably, the functional distribution of genes is not random either. The transcriptional machinery and the ribosomes are of betaproteobacterial origin. This includes all subunits of the RNA polymerase and most ribosomal proteins. The difficulty to assign the phylogenetic origin of some ribosomal proteins might be related with their short length, as stated. On the contrary, aminoacyl-tRNA synthetases (not the complete set, as in other mealybugs) appear to be of gammaproteobacterial origin. The only exception is serS (a pseudogene in several T. princeps strains; Husnik and McCutcheon, 2016), which gave no clear affiliation. Except for iscSUA (involved in (Fe–S) cluster assembly), genes devoted to tRNA maturation are also of gammaproteobacterial origin. This is not surprising, as in the nested endosymbiotic consortia from pseudococcinae mealybugs, Tremblaya has retained most of its own transcriptional and translational machinery except for aminoacyl-tRNA synthetases, which must be provided by the gamma-endosymbiont. This probably indicates that extremely complex molecular machineries work better if their components share a common evolutionary origin. There are genes for ribosome maturation of both beta and gammaproteobacterial origin. In fact, rlmE, encoding an rRNA methyltransferase, is one of the few redundant genes detected, although the betaproteobacterial homolog might be degenerating, as only one taxonomy assignation method gave an unambiguous result. This could be an example of a random degeneration process affecting one of the two copies.
Although genes involved in a given function can have different evolutionary origins, those encoding proteins that need to associate to work tend to have the same one, as already noticed for ribosomal proteins. For example, all maintained subunits of the DNA polymerase (also preserved in T. princeps) are of beta origin. However, the other proteins involved in DNA replication (helicase and ligase) are of gamma origin; the first one has been preserved in all other Tremblaya genomes sequenced, while the second is absent in all of them. Genes involved in translation initiation (infA, infB and infC) and elongation (fusA and tufA) are of beta origin, although there is an additional gammaproteobacterial infA. The possibility of genes of different taxonomic origin requiring different initiation factors cannot be ruled out, although it can just be a matter of time that one of the redundant loci is randomly lost. On the other hand, genes involved in translation termination (prfA, prfB and prmC), ribosome recycling (frr) and degradation of proteins stalled during translation (smpB), as well as N-formyltransferase (fmt) and peptide deformylase (def) are of gamma origin.
As in all other mealybug endosymbionts, essential amino-acid biosynthesis appears to be the main benefit provided to the host. Remarkably, the gene set conserved for this function is quite similar to that observed in all other endosymbiotic consortia in mealybugs, including their evolutionary origin (Supplementary Figure S3; Husnik and McCutcheon, 2016; Szabó et al., 2017). Thus, as in most studied pseudococcinae mealybugs, all genes retained for the biosynthesis of methionine, threonine, isoleucine, leucine and valine, and the production of phenylalanine from chorismate are of betaproteobacterial origin, while the pathways for the production of chorismate and lysine retain the same patchwork pattern. Histidine biosynthesis is an exception, as PPER has only retained genes of gammaproteobacterial origin. The cysteine biosynthetic pathway is more complete in PPER, with all genes of gamma origin. Regarding tryptophan biosynthesis, dominated by gammaproteobacterial genes in previously analyzed mealybugs’ endosymbiotic consortia, in PPER the first step is performed by beta proteins, while the rest of the genes are of gamma origin, a similar pattern to that found in other insect’s endosymbiotic consortia (that is, Serratia/Buchnera in lachninae aphids and some Carsonella/secondary systems in psyllids; Lamelas et al., 2011; Sloan and Moran, 2012; Manzano-Marín et al., 2016). This pathway splitting at the production of the most permeable metabolite (anthranilate) has been suggested as an optimal strategy to reduce the protein cost to supply tryptophan to the whole system (Mori et al., 2016). If so, this could be an indication that the establishment of a metabolic consortium and the reduction of the corresponding genomes preceded the fusion of both organisms.
The chimeric nature of the T. phenacola PPER genome
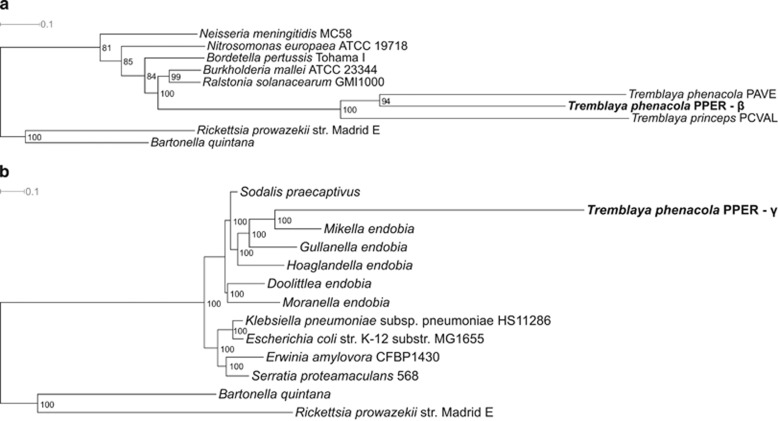
HGT has strongly influenced the evolutionary history of bacteria, as highlighted by phylogenomic reconstructions. Therefore, it is difficult to taxonomically assign a category to bacteria that have undergone extended HGT events (Comas et al., 2006). However, there are important constraints to HGT in strict intracellular bacteria. The few described examples among insects’ P-endosymbionts involve genes for the biosynthesis of a cytotoxic product (Nakabachi et al., 2013) or biotin (Nikoh et al., 2014). Nevertheless, what we have found in T. phenacola PPER goes beyond what could be considered a standard HGT event, and rather resembles the complete fusion of two genomes to form a new chimeric organism. For this reason, even though the 16S rRNA gene phylogenies clearly place this bacterium as a sister clade of T. phenacola PAVE, it is impossible to classify it as beta or gammaproteobacteria based on a global phylogenomic approach. Preliminary analyses with PhyloPhlAn, using both the tree of life (3171 microbial genomes; Segata et al., 2013) and a limited number of species (Supplementary Table S4), did not place it into the Tremblaya clade (data not shown). To get a clearer picture of the (at least) two organisms that must have merged in this chimeric genome, we performed independent phylogenomic analyses of two concatenations of the genes previously assigned as beta or gammaproteobacterial, respectively, to place the two parts of the genome in the microbial tree of life (Supplementary Figure S6) and within a phylogeny containing selected species (Figure 4). The betaproteobacterial genes place this bacterium in the T. phenacola clade (Figure 4a), with the same level of acceleration as the other Tremblaya used in the analysis. On the other hand, the gammaproteobacterial genes place it into the Sodalis-allied clade (Husnik and McCutcheon, 2016) as a sister species of ‘Candidatus Mikella endobia’, nested gamma-endosymbiont of the pseudoccinae mealybug Paracoccus marginatus (Figure 4b). In this case, the length of the PPER branch is greater compared to the other gamma-endosymbionts, probably indicating it is in the process of accommodation to the new chimeric status.
Figure 4.
Phylogenomic analysis of T. phenacola PPER. (a) Genes taxonomically affiliated to betaproteobacteria. (b) Genes taxonomically affiliated to gammaproteobacteria. The species used for the analysis are indicated in Supplementary Table S4.
How could the genomic fusion have occurred? Although HGT is uncommon in modern endosymbionts, it is an extended phenomenon in flowering-plant mitochondria (Sanchez-Puerta, 2014), derived from an ancestral α-proteobacterial endosymbiont (Andersson et al., 1998). The most notable case analyzed corresponds to Amborella trichopoda, whose mitochondrial DNA has incorporated the complete mitochondrial genomes of three green algae and one moss, plus two mitochondrial genome equivalents from other angiosperms (Rice et al., 2013). Such a high frequency of HGT has been explained by mitochondrial fusion and subsequent genomes fusion and rearrangements, mediated by homologous recombination systems (Maréchal and Brisson, 2010). Something similar might have occurred in our study case. On the basis of current evidences, the ancestor of all Tremblaya probably had a reduced genome (Husnik and McCutcheon, 2016). In the lineage driving to T. phenacola PPER, a gammaproteobacterium must have entered the consortium and, instead of replacing T. phenacola (as in the tribe Rhizoecini and genus Rastrococcus; Gruwell et al., 2010), or establishing a nested endosymbiosis (as in the T. princeps clade; reviewed by Husnik and McCutcheon, 2016), a cellular fusion event must have occurred, followed by genomic fusion. It cannot be discarded that a nested endosymbiosis preceded the cellular and genomic fusions. Because this phenomenon implies the existence of a DNA recombination machinery, the most plausible hypothesis is that such genes were present in the genome of the gammaproteobacterial donor, similarly to what has been described in P. citri (López-Madrigal et al., 2013). In fact, most mealybugs’ gamma-endosymbionts that have been completely sequenced (McCutcheon and von Dohlen, 2011; López-Madrigal et al., 2013; Husnik and McCutcheon, 2016) or screened for homologous recombination genes (López-Madrigal et al., 2015) present a more or less complete recombination machinery. Transposable elements might also facilitate a fusion process. Some authors suggest that in arthropod intracellular environments, the possibility of two bacteria co-infecting the same cell generates an ‘intracellular arena’ where distantly related bacterial lineages can exchange mobile elements (Duron, 2013). However, although insertion sequences are frequent in early endosymbiotic stages (Latorre and Manzano-Marín, 2016), they have not been identified in any sequenced mealybugs’ gamma-endosymbiont, and we did not find any indication of their former presence in T. phenacola PPER. After the fusion, the chimeric genome must have undergone massive gene loss, getting rid of almost all redundant and non-essential genes. The initial presence of homologs might have accelerated gene losses through recombination until DNA recombination genes disappeared. The remnant repeats involved in intrachromosomal recombination might have been maintained due to the loss of such genes, leading to the current, complex genome organization.
Gammaproteobacterial DNA was also found in the Madeira mealybug Phenacoccus madeirensis (López-Madrigal et al., 2014), a close relative of P. peruvianus. It is currently unclear how many phenacoccinae species carry hybrid endosymbionts or if some of them retain a pseudococcinae-like endosymbiotic consortium involving both beta (that is, T. phenacola) and gammaproteobacteria.
The study of the P. citri nested endosymbiosis showed an amazing physical and metabolic integration between two co-primary endosymbionts. In P. peruvianus, the integration goes a step beyond, reaching a physical genome integration, to produce a chimeric organism. Yet, the system is still ‘getting used’ to such a novel configuration, as it is shown by its scrambled genome, without a clear GC-skew pattern and with different possible organizations that apparently coexist in a single host. These results show the importance of HGT as a motor of bacterial evolution, also affecting bacterial endosymbionts with highly reduced genomes.
Acknowledgments
We thank Dr Antonia Soto (Universitat Politècnica de València) for biological samples supply; Dr Iñaki Comas, Galo Goig and Manuela Torres-Puente (Institut de Biomedicina de València) for help with Nanopore sequencing. This work was supported by grants BFU2015-64322-C2-1-R (co-financed by FEDER funds and Ministerio de Economía y Competitividad, Spain) and PrometeoII/2014/065 (Generalitat Valenciana, Spain). CVC is a recipient of a fellowship from the Ministerio de Economía y Competitividad (Spain).
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Agashe D, Shankar N. (2014). The evolution of bacterial DNA base composition. J Exp Zool (Mol Dev Evol) 322B: 517–528. [DOI] [PubMed] [Google Scholar]
- Allen JM, Reed DL, Perotti MA, Braig HR. (2007). Evolutionary relationships of ‘Candidatus Riesia spp.,’ endosymbiotic enterobacteriaceae living within hematophagous primate lice. Appl Environ Microbiol 73: 1659–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson SGE, Zomorodipour A, Andersson JO, Sicheritz-Pontén T, Alsmark UCM et al. (1998). The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396: 133–140. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P. (2005). Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol 59: 155–189. [DOI] [PubMed] [Google Scholar]
- Baumann L, Thao ML, Hess JM, Johnson MW, Baumann P. (2002). The genetic properties of the primary endosymbionts of mealybugs differ from those of other endosymbionts of plant sapsucking insects. Appl Environ Microbio 68: 3198–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner P. (1965) Endosymbiosis of Animals with Plant Microorganisms. Interscience: New York, NY, USA. [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T, Berriman M, Tivey A, Patel C, Bohme U, Barrell BG et al. (2008). Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24: 2672–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Li S, Aksoy S. (1999). Concordant evolution of a symbiont with its host insect species: molecular phylogeny of genus Glossina and its bacteriome-associated endosymbiont, Wigglesworthia glossinidia. J Mol Evol 48: 49–58. [DOI] [PubMed] [Google Scholar]
- Comas I, Moya A, Azad RK, Lawrence JG, González-Candelas F. (2006). The evolutionary origin of Xanthomonadales genomes and the nature of the horizontal gene transfer process. Mol Biol Evol 23: 2049–2057. [DOI] [PubMed] [Google Scholar]
- Conord C, Despres L, Vallier A, Balmand S, Miquel C, Zundel S et al. (2008). Long-term evolutionary stability of bacterial endosymbiosis in curculionoidea: additional evidence of symbiont replacement in the Dryophthoridae family. Mol Biol Evol 25: 859–868. [DOI] [PubMed] [Google Scholar]
- Duron O. (2013). Lateral transfers of insertion sequences between WolbachiaCardinium and Rickettsia bacterial endosymbionts. Heredity 2: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR et al. (2014). Pfam: the protein families database. Nucleic Acids Res 42: D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatehouse LN, Sutherland P, Forgie SA, Kaji R, Christeller JT. (2012). Molecular and histological characterization of primary (betaproteobacteria) and secondary (gammaproteobacteria) endosymbionts of three mealybug species. Appl Environ Microbiol 78: 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil R, Silva FJ, Peretó J, Moya A. (2004). Determination of the core of a minimal bacterial gene set. Microbiol Mol Biol Rev 68: 518–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruwell ME, Hardy NB, Gullan PJ, Dittmar K. (2010). Evolutionary relationships among primary endosymbionts of the mealybug subfamily Phenacoccinae (Hemiptera: Coccoidea: Pseudococcidae). Appl Environ Microbiol 76: 7521–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy L, Roat Kultima J, Andersson SGE. (2010). genoPlotR: comparative gene and genome visualization in R. Bioinformatics 26: 2334–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy NB, Gullan PJ, Hodgson CJ. (2008). A subfamily-level classification of mealybugs (Hemiptera: Pseudococcidae) based on integrated molecular and morphological data. Syst Entomol 33: 51–71. [Google Scholar]
- Lin H-H, Liao Y-C. (2016). Accurate binning of metagenomic contigs via automated clustering sequences using information of genomic signatures and marker genes. Sci Rep 6: 24175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Capella-Gutiérrez S, Pryszcz LP, Marcet-Houben M, Gabaldón T. (2014). PhylomeDB v4: zooming into the plurality of evolutionary histories of a genome. Nucleic Acids Res 42: D897–D902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houk E, Griffiths GW. (1980). Intracellular symbiotes of Homoptera. Annu Rev Entomol 25: 161–187. [Google Scholar]
- Husnik F, McCutcheon JP. (2016). Repeated replacement of an intrabacterial symbiont in the tripartite nested mealybug symbiosis. Proc Natl Acad Sci USA 113: E5416–E5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnik F, Nikoh N, Koga R, Ross L, Duncan RP, Fugie M et al. (2013). Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 153: 1567–1578. [DOI] [PubMed] [Google Scholar]
- Huson DH, Beier S, Flade I, Górska A, El-Hadidi M, Mitra S et al. (2016). MEGAN Community edition—interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput Biol 12: e1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. (2016). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44: D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Gama-Castro S, Bonavides-Martinez C et al. (2013). EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res 41: D605–D612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga R, Nikoh N, Matsuura Y, Meng X-Y, Fukatsu T. (2012). Mealybugs with distinct endosymbiotic systems living on the same host plant. FEMS Microbiol Ecol 83: 93–100. [DOI] [PubMed] [Google Scholar]
- Komaki K, Ishikawa H. (1999). Intracellular bacterial symbionts of aphids possess many genomic copies per bacterium. J Mol Evol 48: 717–722. [DOI] [PubMed] [Google Scholar]
- Kono M, Koga R, Shimada M, Fukatsu T. (2008). Infection dynamics of coexisting beta-and gammaproteobacteria in the nested endosymbiotic system of mealybugs. Appl Environ Microbiol 74: 4175–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamelas A, Gosalbes MJ, Manzano-Marín A, Peretó J, Moya A, Latorre A. (2011). Serratia symbiotica from the aphid Cinara cedri: a missing link from facultative to obligate insect endosymbiont. PLoS Genet 7: e1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre A, Manzano-Marín A. (2016). Dissecting genome reduction and trait loss in insect endosymbionts. Ann NY Acad Sci 1389: 52–75. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ Jr, Roos DS. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo N, Bandi C, Watanabe H, Nalepa C, Beninati T. (2003). Evidence for cocladogenesis between diverse dictyopteran lineages and their intracellular endosymbionts. Mol Biol Evol 20: 907–913. [DOI] [PubMed] [Google Scholar]
- López-Madrigal S, Beltrà A, Resurrección S, Soto A, Latorre A, Moya A et al. (2014). Molecular evidence for ongoing complementarity and horizontal gene transfer in endosymbiotic systems of mealybugs. Front Microbiol 5: 449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Madrigal S, Latorre A, Moya A, Gil R. (2015). The link between independent acquisition of intracellular gamma-endosymbionts and concerted evolution in Tremblaya princeps. Front Microbiol 6: 642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Madrigal S, Latorre A, Porcar M, Moya A, Gil R. (2011). Complete genome sequence of ‘Candidatus Tremblaya princeps’ strain PCVAL, an intriguing translational machine below the living-cell status. J Bacteriol 193: 5587–5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Madrigal S, Latorre A, Porcar M, Moya A, Gil R. (2013). Mealybugs nested endosymbiosis: going into the ‘matryoshka’ system in Planococcus citri in depth. BMC Microbiol 13: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan J-B, Chen W, Hasegawa DK, Simmons AM, Wintermantel WM, Ling K-S et al. (2015). Metabolic coevolution in the bacterial symbiosis of whiteflies and related plant sap-feeding insects. Genome Biol Evol 7: evv170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc T, Salzberg S. (2011). FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27: 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-Marín A, Simon JC, Latorre A. (2016). Reinventing the wheel and making it round again: evolutionary convergence in Buchnera–Serratia symbiotic consortia between the distantly related Lachninae aphids Tuberolachnus salignus and Cinara cedri. Genome Biol Evol 8: 1440–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A, Brisson N. (2010). Recombination and the maintenance of plant organelle genome stability. New Phytol 186: 299–317. [DOI] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10–12. [Google Scholar]
- McCutcheon JP, Moran NA. (2012). Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10: 13–26. [DOI] [PubMed] [Google Scholar]
- McCutcheon JP, von Dohlen CD. (2011). An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr Biol 21: 1366–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M. (2008). Are biologist in ‘future shock?’ Symbiosis integrates biology across domains. Nat Rev Microbiol 6: 789–792. [DOI] [PubMed] [Google Scholar]
- Moya A, Peretó J, Gil R, Latorre A. (2008). Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat Rev Genet 9: 218–229. [DOI] [PubMed] [Google Scholar]
- Moran NA. (1996). Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc Natl Acad Sci USA 93: 2873–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Dale C, Dunbar H, Smith WA, Ochman H. (2003). Intracellular symbionts of sharpshooters (Insecta: Hemiptera: Cicadellinae) form a distinct clade with a small genome. Environ Microbiol 5: 116–126. [DOI] [PubMed] [Google Scholar]
- Mori M, Ponce-de-León M, Peretó J, Montero F. (2016). Metabolic complementation in bacterial communities: necessary conditions and optimality. Front Microbiol 7: 1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson MA, Baumann P, Clark MA, Baumann L, Moran NA, Voegtlin DJ et al. (1991). Evidence for the establishment of aphid-eubacterium endosymbiosis in an ancestor of four aphid families. J Bacteriol 173: 6321–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson MA, Baumann P, Moran NA. (1992). Phylogenetic relationships of the endosymbionts of mealybugs (Homoptera: Pseudococcidae) based on 16S rDNA sequences. Mol Phylogenet Evol 1: 26–30. [DOI] [PubMed] [Google Scholar]
- Murray RG, Schleifer KH. (1994). Taxonomic notes: a proposal for recording the properties of putative taxa of procaryotes. Int J Syst Bacteriol 44: 174–176. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, Ueoka R, Oshima K, Tet R, Mangoni A, Gurgui M et al. (2013). Defensive bacteriome symbiont with a drastically reduced genome. Curr Biol 23: 1478–1484. [DOI] [PubMed] [Google Scholar]
- Niko N, Hosokawa T, Moriyama M, Oshima K, Hattori M, Fukatsu T. (2014). Evolutionary origin of insect–Wolbachia nutritional mutualism. Proc Natl Acad Sci USA 111: 10257–10262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakeson KF, Gil R, Clayton AL, Dunn DM, von Niederhausern AC, Hamil C et al. (2014). Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol Evol 6: 76–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44(D1): D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice DW, Alverson AJ, Richardson AO, Young GJ, Sanchez-Puerta MV, Munzinger J et al. (2013). Horizontal transfer of entire genomes via mitochondrial fusion in the angiosmerp Amborella. Science 342: 1468–1473. [DOI] [PubMed] [Google Scholar]
- Rocha E. (2004). The replication-related organization of bacterial genomes. Microbiology 150: 1609–1627. [DOI] [PubMed] [Google Scholar]
- Rocha E. (2008). The organization of the bacterial genome. Annu Rev Genet 42: 211–233. [DOI] [PubMed] [Google Scholar]
- Rosenblueth M, Sayavedra L, Sámano-Sánchez H, Roth A, Martínez-Romero E. (2012). Evolutionary relationships of flavobacterial and enterobacterial endosymbionts with their scale insect hosts (Hemiptera: Coccoidea). J Evol Biol 25: 2357–2368. [DOI] [PubMed] [Google Scholar]
- Sanchez-Puerta MV. (2014). Involvement of plastid, mitochondrial and nuclear genomes in plant-to-plant horizontal gene transfer. Acta Soc Bot Pol 83: 317–323. [Google Scholar]
- Sauer C, Stackebrandt E, Gadau J, Hölldobler B, Gross R. (2000). Systematic relationships and cospeciation of bacterial endosymbionts and their carpenter ant host species: proposal of the new taxon Candidatus Blochmannia gen. nov. Int J Syst Evol Microbiol 50: 1877–1886. [DOI] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30: 2068–2069. [DOI] [PubMed] [Google Scholar]
- Segata N, Börnigen D, Morgan XC, Huttenhower C. (2013). PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun 4: 2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34: D32–D36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Moran NA. (2012). Genome reduction and co-evolution between the primary and secondary bacterial symbionts of psyllids. Mol Biol Evol 29: 3781–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Nakabachi A, Richards S, Qu J, Murali SC, Gibbs RA et al. (2014). Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol Biol Evol 31: 857–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R, Beal KF, Bonfield JK. (2000). The Staden package, 1998. Methods Mol Biol 132: 115–130. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó G, Schulz F, Toenshoff ER, Volland J-M, Finkel OM, Belkin S et al. (2017). Convergent patterns in the evolution of mealybug symbioses involving different intrabacterial symbionts. ISME J 11: 715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao ML, Baumann P. (2004). Evolutionary relationships of primary prokaryotic endosymbionts of whiteflies and their hosts. Appl Environ Microbiol 70: 3401–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao ML, Moran NA, Abbot P, Brennan EB, Burckhardt DH, Baumann P. (2000). Cospeciation of psyllids and their primary prokaryotic endosymbionts. Appl Environ Microbiol 66: 2898–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M et al. (2012). Primer3—new capabilities and interfaces. Nucleic Acids Res 40: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ham RCHJ, Kamerbeek J, Palacios C, Rausell C, Abascal F, bastolla U et al. (2003). Reductive genome evolution in Buchnera aphidicola. Proc Natl Acad Sci USA 100: 581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leuven JT, Meister RC, Simon C, McCutcheon JP. (2014). Sympatric speciation in a bacterial endosymbiont results in two genomes with the functionality of one. Cell 158: 1270–1280. [DOI] [PubMed] [Google Scholar]
- von Dohlen CD, Kohler S, Alsop ST, McManus WR. (2001). Mealybug β-proteobacterial endosymbionts contain γ-proteobacterial symbionts. Nature 412: 433–436. [DOI] [PubMed] [Google Scholar]
- Woyke T, Tighe D, Mavromatis K, Clum A, Copeland A, Schackwitz W et al. (2010). One bacterial cell, one complete genome. PLoS One 5: e10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.