Abstract
Physiological performance, disease and bleaching prevalence are often patchy within individual coral colonies. These responses are largely influenced by coral-associated microbes, but how the coral microbiome changes over small spatial scales has never been quantified before. We performed a high-resolution quantification of the spatial scale of microbial species turnover (β-diversity) within skeletons of boulder-forming Porites corals. We found very strong prokaryotic species turnover across spatial scales ranging from 4 mm to 2 m within individual colonies, possibly resulting from dispersal limitation and microbial interactions. The microalgal community was more homogeneously distributed, which is likely due to these photosymbionts actively boring through limestone. Our findings highlight unprecedented levels of intra-colony heterogeneity in the skeletal microbiome, which has implications for the experimental design of coral microbiome studies and for our understanding of coral resilience.
The vulnerability of corals to pathogens and climate change varies across spatial scales. Thermal tolerance, for example, varies among conspecific corals and within individual colonies depending on their associated Symbiodinium community (Rowan et al., 1997; Abrego et al., 2008). Similarly, bleaching susceptibility and diseases such as patchy necrosis are not uniformly distributed within individual colonies (Bythell et al., 2004; Shapiro et al., 2016). Coral health depends on microorganisms, but the existence and consequences of small-scale microbial structure within colonies are virtually unknown. Unveiling fine-scale variation in community composition (β-diversity) within colonies is a critical step towards understanding functional interactions between corals and microbes, and for designing sampling strategies for coral microbiome studies.
The eukaryotic and prokaryotic microorganisms living inside coral skeletons—the endolithic microbiome—have recently come into the spotlight due to their newly discovered biodiversity and potential roles in coral nutrition, nitrogen cycling, bioerosion and coral disease (Schlichter et al., 1995; Tribollet, 2008; Miller et al., 2011; Marcelino and Verbruggen, 2016; Del Campo et al., 2017; Marcelino et al., 2017). Here, we investigated the spatial distribution of prokaryotic and eukaryotic microbiota in the skeletons of individual boulder corals (genus Porites). The decrease in community similarity with distance inside the skeletons was quantified using distance–decay relationships (DDR), a measure of β-diversity and habitat heterogeneity (Nekola and White, 1999; Anderson et al., 2011; Martiny et al., 2011). We collected 90 skeleton samples from healthy, large Porites lutea (3) and P. lobata (5) colonies following a geometric progression sampling design with distances between samples ranging from 4 mm to 1.992 m (Supplementary Materials, Supplementary Figure S1 and Supplementary Tables S1 and S2). Because our goal was to characterise how community composition changes with distance, we minimised potential environmental differences by collecting each microbiome sample at equivalent positions and the same depth within colonies. Using multi-marker metabarcoding (Marcelino and Verbruggen, 2016) we characterised the prokaryotes (16S rRNA gene marker), eukaryotic microalgae and cyanobacteria (UPA and tufA markers—referred to as ‘algae’ hereafter) of each skeleton sample. We quantified within-colony β-diversity as the slope of the linear regression between (log10-transformed) pairwise community similarities and (log10-transformed) spatial distances, that is, the slope of the DDR (Supplementary Materials).
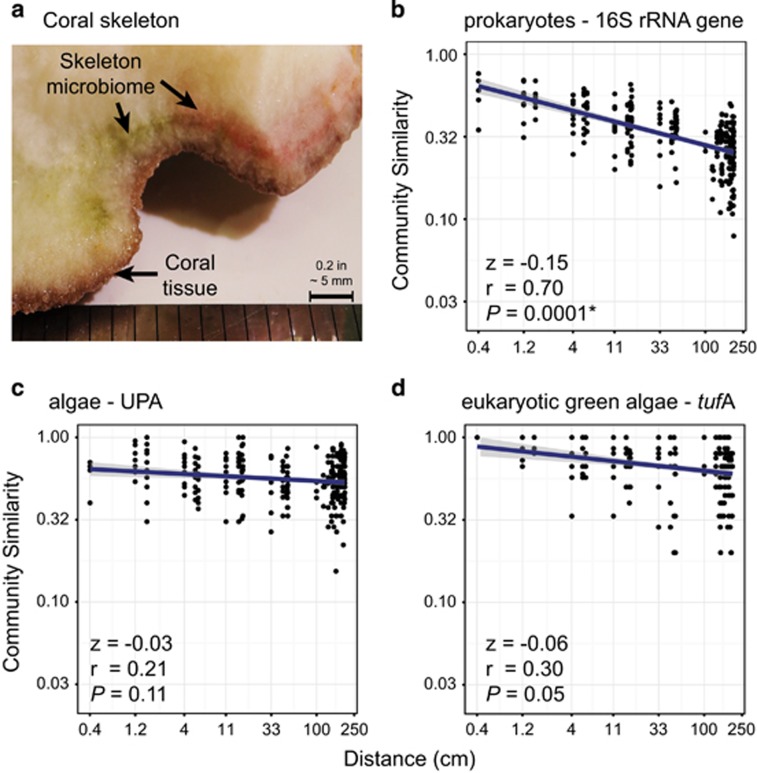
Endolithic prokaryotes showed a significant decay in community similarity with distance, suggesting a highly heterogeneous assemblage within individual colonies (Figure 1). The observed DDR slope for prokaryotes (−0.15, P=0.0001) is among the steepest ever reported for microorganisms (Supplementary Tables S3–S5). The DDR slope for bacteria in marine sediments, for example, measured at larger spatial scales, is half of the slope observed here (Zinger et al., 2014; Supplementary Table S5). In contrast, endolithic algae had a shallow DDR slope that was not significantly different from zero, suggesting they were more homogeneously distributed (Figure 1 and Supplementary Table S3). The same pattern holds when individual colonies from different locations are analysed separately, thus it is unlikely that the pattern results from environmental gradients along the colony (Supplementary Figure S2 and Supplementary Table S4). Species accumulation curves confirmed the patchy distribution of endolithic prokaryotes (Supplementary Figures S3–S5). Only a fraction of the colony’s microbial diversity was observed in each ~0.25 cm3 sample: 26.36% (±10.77 s.d.) for the 16S rRNA gene, 37.76% (±15.63 s.d.) for UPA and 51.78% (±16.67 s.d.) for tufA.
Figure 1.
Distance–decay relationships for the endolithic communities in coral skeletons at the intra-colony scale. (a) Coral fragment showing green and red patches of pigmented bacteria and algae. (b–d) Distance–decay relationships obtained with the 16S rRNA gene, UPA and tufA markers, respectively. Blue lines indicate the linear regression between (log10 transformed) spatial distance and (log10 transformed) Sørensen community similarity—that is, the slope of the DDR. Shaded areas represent the 95% confidence interval. Slope values (z), Mantel r statistics (r) and significance vales (P) are provided.
The high rate of prokaryotic species turnover may be a consequence of limited dispersal within coral colonies. Microbial motility is likely reduced in dense substrates such as limestone skeletons and aggregation of bacterial colonies may result in high β-diversity. In contrast, many species of endolithic algae actively bore their way through limestone (Tribollet, 2008) and their higher dispersal ability explains the low algal β-diversity (Figure 1). Microbial interactions and competitive sorting can result in micro-niches with different chemical properties at very small spatial scales and may also contribute to the high prokaryotic β-diversity observed here (see Nekola and White, 1999; Cordero and Datta, 2016).
The high turnover of prokaryotic species raises the question whether different lineages may fulfil similar functions in different parts of the skeleton. While amplicon data cannot answer this definitively, we did observe that a phylogeny-based distance metric produces a shallower DDR slope (Supplementary Materials and Supplementary Table S3). This suggests that while individual species have strong spatial turnover, the phylogenetic lineages to which they belong are widespread. Assuming that basic metabolic functions are conserved among related species, we can expect functional redundancy in the coral skeleton.
The ‘insurance hypothesis’ states that diversity and functional redundancy increase ecosystem stability given species’ functional complementarity and asynchronous responses to stress (Yachi and Loreau, 1999; Loreau and de Mazancourt, 2013). Extrapolating from this hypothesis, one could expect that spatial structure is associated with patchy responses to stress in the short term, but lineages with similar ecological functions will outcompete their less resilient neighbours conferring ecosystem stability in the long run. It is reasonable to expect therefore that functional redundancy and spatial structure confers an increased long-term stability for the endolithic microbiome. This is a possible mechanism for the apparently stable nature of the coral endolithic community composition, and is in agreement with the hypothesis that the skeleton serves as a stable reservoir for the coral microbiome (Marcelino et al., 2017).
This study provides the first quantitative evidence for turnover of prokaryotic species in coral skeletons at the centimetre scale. We propose that limited dispersal and non-random associations between microorganisms drive the spatial pattern of prokaryotes, while the limestone-boring nature of endolithic algae explains their low species turnover. Whether coral tissue and mucus have similarly high β-diversity still needs to be investigated, but studies indicate that heterogeneity also exist there (Rohwer et al., 2002; Hansson et al., 2009; Daniels et al., 2011). Our findings imply that multiple samples are needed for a full characterisation of a coral skeleton's microbiome and its ecological roles. The notion that microbiome structuring in corals occurs over far smaller spatial scales than previously thought should prompt us to investigate coral health and resilience at the small spatial scales where microbial interactions occur.
Acknowledgments
This research was supported by the Australian Biological Resources Study (RFL213-08), the Australian Research Council (FT110100585, DP150100705), the University of Melbourne (Botany Foundation grant to VRM and ECR grant to HV), the Albert Shimmins Fund and the Holsworth Wildlife Research Endowment. The bioinformatics aspects were supported by use of the Melbourne Bioinformatics (VLSCI) facilities (project UOM0007) and the Nectar Research Cloud, a collaborative Australian research platform supported by the National Collaborative Research Infrastructure Strategy (NCRIS). We are grateful to Zac Forsman for providing advice on DNA barcodes for coral identification, Eric Treml for advice on statistical analyses and two anonymous reviewers for their constructive comments. We thank Mike Van Keulen, Frazer McGregor, the Heron Island Research Station staff and the members of the Verbruggen lab for facilitating fieldwork.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Abrego D, Ulstrup KE, Willis BL, van Oppen MJ. (2008). Species-specific interactions between algal endosymbionts and coral hosts define their bleaching response to heat and light stress. Proc Biol Sci 275: 2273–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MJ, Crist TO, Chase JM, Vellend M, Inouye BD, Freestone AL et al. (2011). Navigating the multiple meanings of beta diversity: a roadmap for the practicing ecologist. Ecol Lett 14: 19–28. [DOI] [PubMed] [Google Scholar]
- Bythell J, Pantos O, Richardson L (2004). White plague, white band, and other ‘white’ diseases. In: Rosenberg E, Loya Y (eds). Coral Health and Disease. Springer: Berlin, Heidelberg, pp 351–365..
- Cordero OX, Datta MS. (2016). Microbial interactions and community assembly at microscales. Curr Opin Microbiol 31: 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels CA, Zeifman A, Heym K, Ritchie KB, Watson CA, Berzins I et al. (2011). Spatial heterogeneity of bacterial communities in the mucus of Montastraea annularis. Mar Ecol Prog Ser 426: 29–40. [Google Scholar]
- Del Campo J, Pombert JF, Slapeta J, Larkum A, Keeling PJ. (2017). The 'other' coral symbiont: Ostreobium diversity and distribution. ISME J 11: 296–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson L, Agis M, Maier C, Weinbauer MG. (2009). Community composition of bacteria associated with cold-water coral Madrepora oculata: within and between colony variability. Mar Ecol Prog Ser 397: 89–102. [Google Scholar]
- Loreau M, de Mazancourt C. (2013). Biodiversity and ecosystem stability: a synthesis of underlying mechanisms. Ecol Lett 16(Suppl 1): 106–115. [DOI] [PubMed] [Google Scholar]
- Marcelino VR, Verbruggen H. (2016). Multi-marker metabarcoding of coral skeletons reveals a rich microbiome and diverse evolutionary origins of endolithic algae. Sci Rep 6: 31508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelino VR, Morrow KM, van Oppen MJ, Bourne DG, Verbruggen H. (2017). Diversity and stability of coral endolithic microbial communities at a naturally high pCO2 reef. Mol Ecol(in press). Available at: http://onlinelibrary.wiley.com/doi/10.1111/mec.14268/full. [DOI] [PubMed]
- Martiny JB, Eisen JA, Penn K, Allison SD, Horner-Devine MC. (2011). Drivers of bacterial beta-diversity depend on spatial scale. Proc Natl Acad Sci USA 108: 7850–7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AW, Blackwelder P, Al-Sayegh H, Richardson LL. (2011). Fine-structural analysis of black band disease-infected coral reveals boring cyanobacteria and novel bacteria. Dis Aquat Organ 93: 179–190. [DOI] [PubMed] [Google Scholar]
- Nekola JC, White PS. (1999). The distance decay of similarity in biogeography and ecology. J Biogeogr 26: 867–878. [Google Scholar]
- Rohwer F, Seguritan V, Azam F, Knowlton N. (2002). Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser 243: 1–10. [Google Scholar]
- Rowan R, Knowlton N, Baker A, Jara J. (1997). Landscape ecology of algal symbionts creates variation in episodes of coral bleaching. Nature 388: 265–269. [DOI] [PubMed] [Google Scholar]
- Schlichter D, Zscharnack B, Krisch H. (1995). Transfer of photoassimilates from endolithic algae to coral tissue. Naturwissenschaften 82: 561–564. [Google Scholar]
- Shapiro OH, Kramarsky-Winter E, Gavish AR, Stocker R, Vardi A. (2016). A coral-on-a-chip microfluidic platform enabling live-imaging microscopy of reef-building corals. Nat Commun 7: 10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribollet A. (2008). The boring microflora in modern coral reef ecosystems: a review of its roles. In: Wisshak M, Tapanila L (eds). Current Developments in Bioerosion. Springer: Berlin, Heidelberg, pp 67–94.
- Yachi S, Loreau M. (1999). Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc Natl Acad Sci USA 96: 1463–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinger L, Boetius A, Ramette A. (2014). Bacterial taxa-area and distance-decay relationships in marine environments. Mol Ecol 23: 954–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.