Abstract
The causative agent of malaria, Plasmodium, replicates inside a membrane-bound parasitophorous vacuole (PV) that shields this intracellular parasite from the cytosol of the host cell1. One common threat for intracellular pathogens is the homeostatic process of autophagy through which cells capture unwanted intracellular material for lysosomal degradation2. During the liver stage of a malaria infection, Plasmodium parasites are targeted by the autophagy machinery of the host cell and the PV membrane (PVM) becomes decorated with several autophagy markers, including LC3 (microtubule-associated protein 1 light chain 3)3,4. Here we show that Plasmodium berghei parasites infecting hepatic cells rely on the PVM transmembrane protein UIS3 to avoid elimination by host cell-mediated autophagy. We found that UIS3 binds host LC3 through a non-canonical interaction with a specialised surface on LC3 where host proteins with essential functions during autophagy also bind. UIS3 acts as a bona fide autophagy inhibitor by competing with host LC3-interacting proteins for LC3 binding. Our work identifies UIS3, one of the most promising candidates for a genetically-attenuated vaccine against malaria5, as a unique and potent mediator of autophagy evasion in Plasmodium. We propose that the protein-protein interaction between UIS3 and host LC3 represents a target for antimalarial drug development.
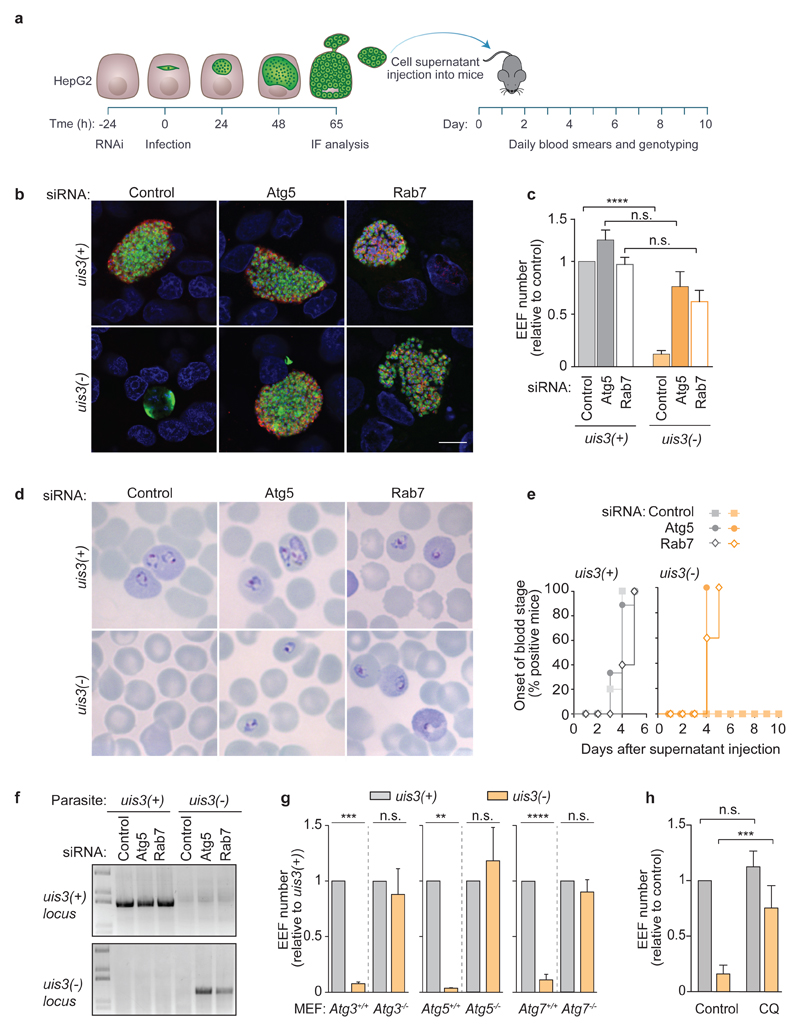
One key function of LC3 is to facilitate the delivery of autophagosomal membranes to lysosomes6. Intriguingly, most LC3-decorated hepatic parasites seem to escape this fate3,4. This implies that Plasmodium exo-erythrocytic forms (EEFs) actively disrupt the autophagic flux to avoid the deleterious effects of fusing the vacuole with lysosomes. The PVM is ideally positioned to carry out this task and several genetic studies point to the critical contribution of PVM resident proteins to parasite survival5,7,8. Nevertheless, a putative autophagy subversion activity in Plasmodium has yet to be discovered. Of the few PVM proteins identified thus far, UIS3 has the greatest impact on Plasmodium survival inside host hepatocytes5,9. Parasites lacking UIS3 (uis3(-)) infect host cells, but disappear rapidly thereafter and fail to complete development5. To determine whether UIS3 influences Plasmodium’s susceptibility to host autophagy, we allowed uis3(+) and uis3(-) P. berghei parasites to infect HepG2 cells that had been depleted of Atg5 or Rab7 to arrest the autophagic flux in early (LC3 membrane conjugation) and late (lysosomal fusion) stages, respectively10,11 (Fig. 1a, Supplementary Fig.1 a, b). Consistent with previous reports5, uis3(-) parasites were virtually undetectable in control HepG2 cells and the few hepatic schizonts observed 65 h after infection did not express MSP1, the merozoite surface marker seen in fully mature uis3(+) parasites (Fig. 1b, c, Supplementary Fig. 1c). Strikingly, depletion of Atg5 or Rab7 in HepG2 cells was sufficient to fully revert the phenotype of uis3(-) mutants to a wild-type one (Fig. 1b, c, Supplementary Fig. 1c). With the autophagy flux of the host cell arrested, uis3(-) parasites behaved as their uis3(+) counterparts with respect to schizont abundance as well as MSP1 expression (Fig. 1b, c, Supplementary Fig. 1c). Parasites that complete the liver stage successfully induce the release of blood-infective merozoite-filled sacs (merosomes) from infected cells12. To test the viability of the uis3(-) progeny released by HepG2-infected cells, we harvested the cell culture supernatants (HepG2 SN) 65 h after infection, inoculated them into mice, and monitored the onset of blood-stage infection by collecting blood samples daily (Fig. 1a). HepG2 SN from Atg5 or Rab7-depleted cells infected with uis3(-) mutants caused patent blood-stage infections within 4 to 5 days of mice inoculation, similar to mice inoculated with HepG2 SN derived from uis3(+)-infected cells (Fig. 1d-f). In stark contrast, HepG2 SN collected from control uis3(-) infected cells did not cause infection in mice (Fig. 1d-f). Thus, loss of autophagy during the liver stage fully restores uis3(-) infectivity. Additionally, the survival rates of uis3(-) and uis3(+) parasites in Atg3, Atg5, or Atg7 knockout mouse embryonic fibroblasts (MEFs) were also identical, whereas few uis3(-) mutants survived in wild-type MEFs (Fig. 1g). To obtain additional evidence that uis3(-) parasites are removed by host autophagy, newly invaded hepatoma cells were treated with chloroquine (CQ), a lysosomotropic alkalizing agent that impairs the lysosomal degradation of autophagic cargo. CQ treatment, like genetic impairment of autophagy, rescued uis3(-) mutants (Fig. 1h). On rare occasions, we were able to observe uis3(-) parasites that exhibited distinctive signs of having undergone fusion with LAMP1-positive lysosomes, most prominently, the presence of LAMP1 within the vacuole and loss of the PVM marker, UIS4 (Supplementary Fig. 1 d). Thus, we conclude that uis3(-) mutants normally fail to establish liver stage infections because they are promptly eliminated by host autophagy and that the critical function of UIS3 is to protect Plasmodium from this innate defense mechanism.
Figure 1. Plasmodium UIS3 protects liver-stage parasites from host autophagy.
a, Timeline of HepG2 infection and cell supernatant (HepG2 SN) transfer into C57BL/6 mice. b, c, HepG2 cells treated with siRNAs to silence Atg5 or Rab7 expression and infected with uis3(+) or uis3(-) parasites were fixed 65 h after infection and immunostained with anti-MSP1 (red), anti-PbHsp70 (green), and Hoechst (blue). Scale bar, 10 μm. Differentiated progeny are shown surrounded by a ring of MSP1 (b). The numbers of EEFs were quantified by microscopy (c). The bars represent the mean + SEM of pooled replicates from two (Atg5, Rab7) to four (Control) independent experiments normalised to the control. d, Representative images of Giemsa-stained blood smears from C57BL/6 mice 10 days after receiving the indicated HepG2 SN. e, Onset of blood stage parasitemia following inoculation of C57BL/6 mice with the indicated HepG2 SN. Animals that received control uis3(-) HepG2 SN remained parasite free up to 30 days after inoculation. The data represents the % of mice (out of 5 to 10; two independent experiments) with positive blood smears. f, Genotype of blood stage parasites harvested 10 days after mice inoculation with the indicated HepG2 SN. Wild-type and knockout uis3 genomic loci were amplified with specific primer sets. One representative mouse from each experimental group is shown. g, Control (Atg3+/+, Atg5+/+, Atg7+/+) and autophagy-deficient MEFs (Atg3-/-, Atg5-/-, and Atg7-/-) were infected with uis3(+) or uis3(-) parasites, fixed at 48 h, and labeled with anti-PbHsp70 and Hoechst. EEF numbers were quantified by microscopy and normalised to the uis3(+) control of each cell line. The bars represent the mean + SEM of five independent experiments. h, Huh7 cells treated with chloroquine (CQ) from 1 h to 24 h after infection with uis3(+) or uis3(-) parasites were immunostained with anti-PbUIS4, anti-PbHsp70, and Hoechst. The numbers of EEFs were quantified by microscopy in three independent experiments. Statistical significance was assessed using non-parametric two-tailed Mann-Whitney test. n.s. P > 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
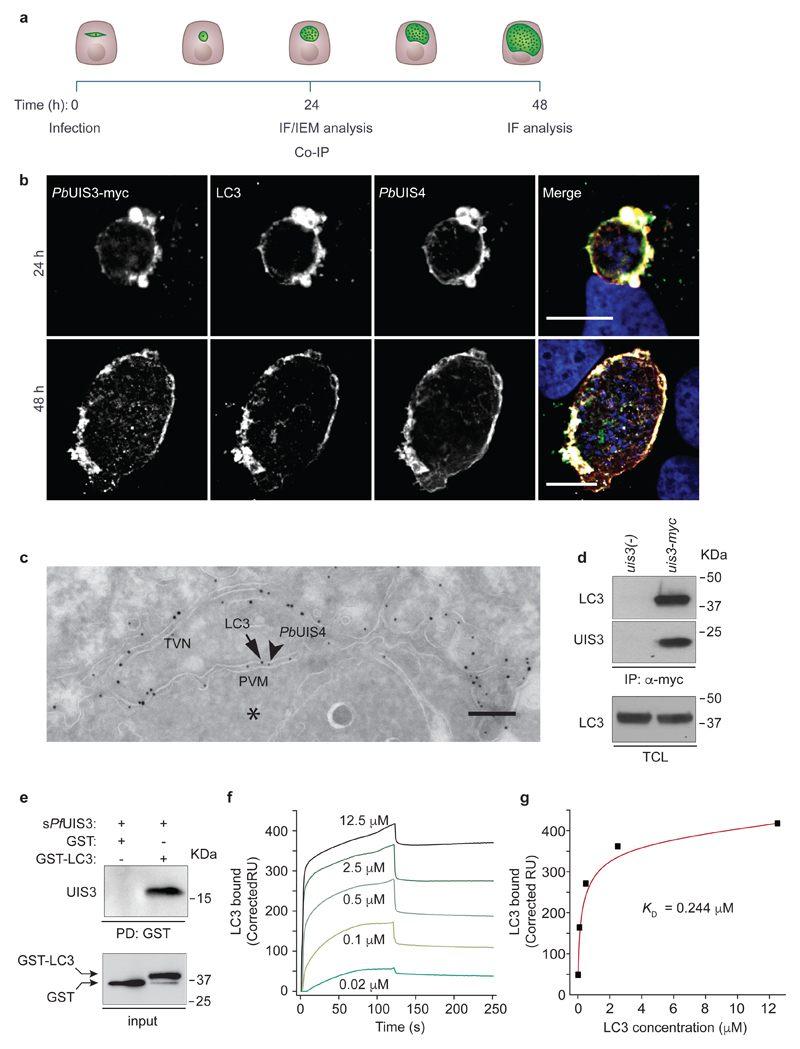
Next, we aimed to elucidate how UIS3 interferes with host autophagy. To that end, we complemented uis3(-) parasites with the UIS3 gene fused C-terminally to c-Myc (this parasite line will be referred to as uis3-myc) (Supplementary Fig. 2). Immunofluorescence analysis of liver stage schizonts, revealed that UIS3-myc co-localised with host LC3 and UIS4 to the PVM (Fig. 2a, b, Supplementary Fig. 3a, b). Of note, immuno-electron microscopy showed LC3 in direct association with the PVM, with no evidence of autophagosomal double membranes – a hallmark of canonical autophagy – forming around the PV (Fig. 2c). Prompted by these observations, we asked whether UIS3 might form a molecular complex with LC3. We allowed uis3-myc parasites to infect HeLa cells stably expressing GFP-LC3 and, 24 h after infection, proceeded to immunoprecipitate UIS3-myc. Strikingly, GFP-LC3 was found to co-immunoprecipitate with UIS3 (Fig. 2d). We further confirmed this association in P. berghei infected mouse primary hepatocytes (Supplementary Fig. 3c), thus ruling out the possibility of a potential artefact linked to GFP-LC3 over-expression. LC3 immunoprecipitation could not be detected in cells infected with uis3(-) mutants, which do not express UIS3 (Fig. 2d, Supplementary Fig. 3c). Next, we used recombinant UIS3 and LC3 to test whether the two proteins directly associate. Full-length UIS3 comprises a signal peptide, a short intravacuolar N-terminal domain, a transmembrane anchor, and a C-terminal domain that spans residues 83 to 229 and is predicted to be exposed to the host cytosol. Previous studies reported that the C-terminal domain of P. falciparum UIS3 is prone to dimerisation, but undergoes proteolytic cleavage in solution yielding a stable and soluble monomeric fragment consisting of residues 130 to 229 (hereafter referred to as soluble UIS3, sPfUIS3)13. Using recombinant GST-LC3, we were able to pull down recombinant His-tagged sPfUIS3 (Fig. 2e). The dissociation constant (KD) for the complex measured by surface plasmon resonance was of 0.244 μM (Fig. 2f, g). Despite being co-localised with both UIS3 and UIS4 on the PVM, LC3 was shown to co-immunoprecipitate with sPbUIS3, but not with the corresponding C-terminal domain of PbUIS4 (Supplementary Fig. 3d). We also did not observe co-immunoprecipitation of L-FABP with recombinant sPfUIS3 (Supplementary Fig. 3e), in line with previous work suggesting that the reported interaction between the C-terminus of P. yoelii UIS3 and mouse L-FAPB14 might be species specific15. Collectively, these data demonstrate that the distal C-terminal subdomain of UIS3, spanning amino acids 130-229, binds to LC3 directly. While our data establish that UIS3 forms a molecular complex with LC3, the association of LC3 with the PVM was found to be host-driven – two core components of the LC3 conjugation system, Atg3 and Atg5, were shown to be essential – and independent of UIS3 (Supplementary Fig. 4a-d). In contrast, neither ULK1, the AMPK-regulated autophagy-initiating kinase, nor the autophagy receptors p62 and NDP52, all previously implicated in xenophagy16, were required (Supplementary Fig. 4e-g). This is consistent with recent reports highlighting the role of Atg5 and the lack of involvement of the autophagy-initiation complex in targeting LC3 to the PVM3,17. LC3 association with the PVM was also unaffected by ROS scavengers (Supplementary Fig. 4h, i), implying that the host response against Plasmodium is mechanistically unrelated to LC3-associated phagocytosis (LAP). LAP has been described as a microbicidal pathway wherein elements of the LC3 conjugation system transfer LC3 in a ROS-dependent manner to pathogen-containing phagosomes18. Intriguingly, the pathway leading to Plasmodium targeting by autophagy appears to be inhibited by ULK1. Indeed, we observed significantly higher levels of PVM-associated LC3 upon knockdown of ULK1 expression (Supplementary Fig. 4f). These results confirm the unconventional nature of the host autophagy response against Plasmodium and imply that the association of LC3 to the PVM precedes the UIS3-LC3 interaction.
Figure 2. Plasmodium UIS3 binds to host LC3.
a, Timeline of in vitro infection and analysis. b, Representative confocal images (two independent experiments) of Huh7 cells infected with P. berghei expressing UIS3-myc (uis3-myc). Cells were fixed 24 h (top) and 48 h (bottom) after infection and immunostained with anti-myc (white), anti-UIS4 (red), anti-LC3 (green), and Hoescht (blue). Scale bar, 10 μm. c, Representative IEM micrograph of the host-parasite interface in a Huh7 cell 24 h after infection with P. berghei. PbUIS4 and host LC3 are shown labeled with 12 nm (arrowheads) and 18 nm (arrows) gold particles, respectively. The EEF cytosol is marked with an asterisk (*). The PbUIS4-positive structures correspond to the PVM and tubular-vesicular network (TVN) of the parasite. Scale bar, 200 nm. Seventeen randomly selected cells were analysed. d, Lysates of GFP-LC3 HeLa cells infected with uis3-myc or uis3(-) P. berghei were immunoprecipitated with anti-myc. Analysis of the precipitates was done by western-blot with anti-LC3 and anti-myc antibodies (top and middle panels). Total cell lysates (TCL) were immunoblotted with anti-LC3 (bottom panel). e, Purified GST or GST-LC3 were used as bait to pull-down recombinant His-tagged sPfUIS3. Pull-down fractions were immunoblotted with anti-GST and anti-His antibodies. One of four independent experiments is shown (d, e). f, Binding of GST-LC3 to sPfUIS3 immobilised on a sensor chip was analysed by surface plasmon resonance (SPR). The indicated concentrations of GST-LC3 were perfused over the sPfUIS3-coated sensor. Normalized response sensorgrams were calculated by subtracting the unspecific binding contribution of the same molar concentrations of GST alone. g, SPR response units at equilibrium plotted as a function of LC3 concentration. The dissociation constant of the interaction, KD, is shown.
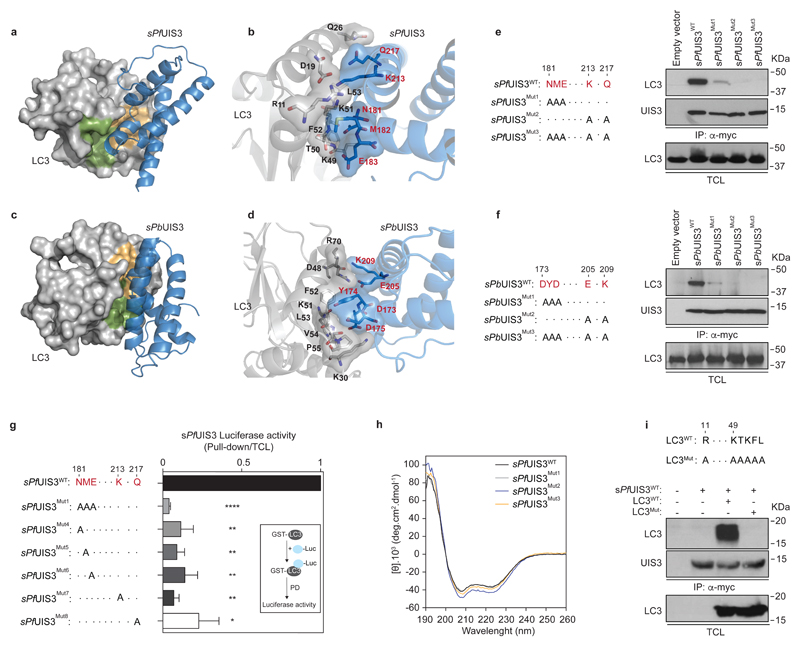
Whereas LC3 binding partners typically contain an LC3-Interacting Region (LIR) with the consensus sequence W/F-x-x-I/L/V19,20, UIS3 lacks this canonical LIR (Supplementary Fig. 5a). To gain further insight on the specific mode of binding of UIS3 to LC3, we modelled the three-dimensional structure of the putative complex between sPfUIS3 or sPbUIS3 and LC3 using a molecular docking algorithm and the published X-ray structures of sPfUIS313 and LC3B21 (Fig. 3a-d, and Supplementary Fig. 5). Our in silico analysis predicted that the interaction is likely to be driven by a flexible turn between helices α2 and α3 of UIS3 (N181, M182, and E183 in P. falciparum UIS3) and the β2 strand of LC3 (Fig. 3a, b). The complex appears to be additionally stabilised by polar interactions involving K213 and Q217 (Fig. 3b). In the P. berghei complex, the residues located in equivalent positions are D173, Y174, and D175, with E205 and K209 stabilising the complex through electrostatic interactions (Fig. 3c, d). In order to validate these predictions, we substituted the above amino acids for alanine to measure the effect of the substitutions on UIS3 binding to LC3. Simultaneous or individual mutations on all five residues abolished the interaction of sPfUIS3 with LC3 (Fig. 3e, g). Likewise, alanine substitutions of the equivalent positions in sPbUIS3 abrogated binding to LC3 almost completely (Fig. 3f). Of note, the circular dichroism (CD) spectra of recombinant sPfUIS3 proteins harboring the above mutations did not reveal any significant effect of the amino acid substitutions on protein conformation (Fig. 3h). Detailed analysis of the predicted LC3-UIS3 complexes from P. berghei and P. falciparum (Fig. 3a-d, Supplementary Fig. 5d, e) indicated that the LC3 surface where UIS3 putatively binds coincides with the surface recognised by autophagy proteins containing canonical LIR motifs, such as p6219,20,22. Mutations of key residues in this LIR-binding LC3 surface to alanine abolished the interaction between sPfUIS3 and LC3 (Fig. 3i), as predicted. Thus, although UIS3 lacks a canonical LIR, our data demonstrate that the LIR-binding surface of LC3 can accommodate this atypical interaction.
Figure 3. Non-canonical binding of Plasmodium UIS3 to the LIR pocket of LC3.
a-d, Representations of the putative complex between the soluble domain of UIS3 and LC3 obtained by molecular docking. The LC3 areas depicted in green and orange represent the two hydrophobic pockets responsible for the recognition of LIR motifs and overlap significantly with the surface recognised by UIS3 (see Supplementary Fig. 5d, e). All key interacting amino acids are highlighted in black (LC3) and red (UIS3). The P. falciparum (a, b) and P. berghei complexes (c, d) are shown. e, f, Lysates of GFP-LC3 HeLa cells expressing the indicated sPfUIS3 (e) or sPbUIS3 (f) myc-tagged proteins (amino acid substitutions shown on the left) were immunoprecipitated with anti-myc. Precipitates were analysed by western-blot with anti-LC3 (top panels) and anti-myc (middle panels). TCL were immunoblotted with anti-LC3 (bottom panels). g, Relative contribution of single amino acid substitutions (shown on the left) to the binding of luciferase-tagged sPfUIS3 to recombinant GST-LC3. The inset shows the outline of the pulldown assay. The average + SEM of two independent experiments is shown. h, Circular dichroism spectra of sPfUIS3 recombinant proteins (wild-type and the indicated mutants). Each spectrum is the average of 10 technical replicates, corrected for the buffer background. i, Immunoprecipitation of myc-tagged sPfUIS3WTfrom HeLa cell lysates co-expressing the indicated HA-tagged LC3 proteins (amino acid substitutions shown on top). Precipitates were probed with anti-HA (top panel) and anti-myc (middle panel). The bottom panel shows the levels of LC3 in TLC. One representative immunoblot of at least three independent experiments is shown (e, f, i). Statistical significance was assessed using unpaired two-tailed t test. *P < 0,05; **P < 0.01; ****P < 0.0001.
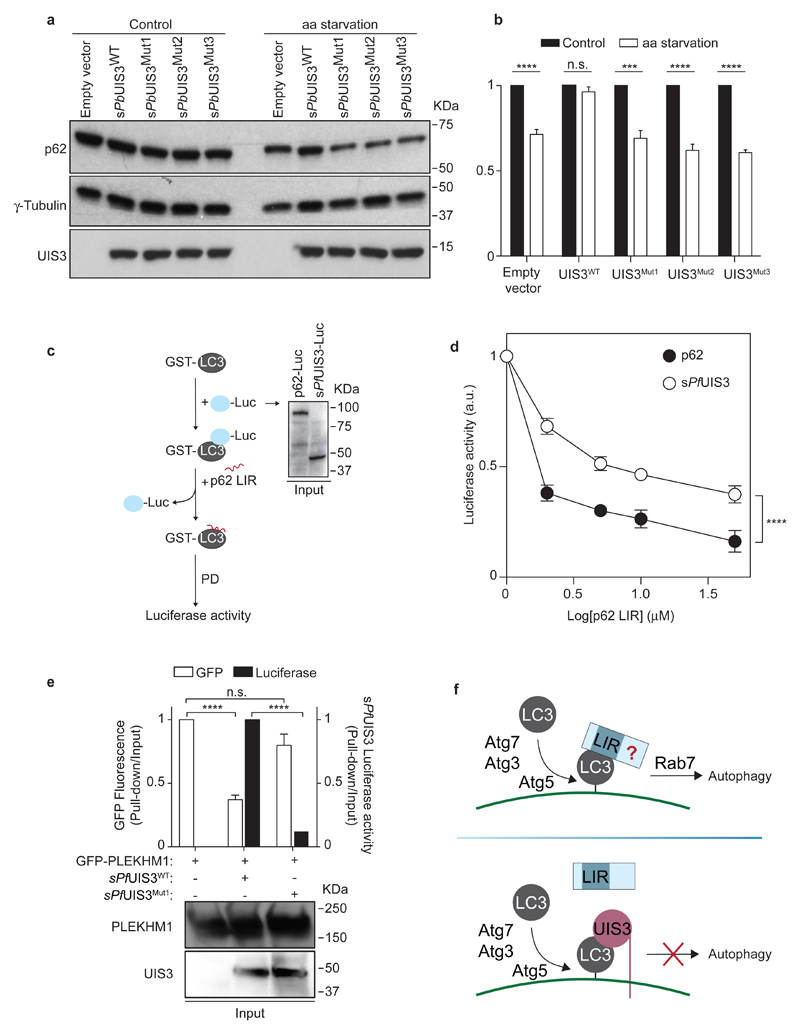
Since UIS3 and canonical LIR motifs interact with the same region of LC3, we next hypothesised that UIS3 might interfere with the binding of LIR-containing proteins to LC3, thus interrupting the autophagic flux. To examine whether the UIS3-LC3 interaction represents a physiological autophagy subversion mechanism, we quantified the impact of heterologous UIS3 expression on the autophagic flux of HeLa cells. As a readout for autophagy, we monitored p62 degradation in response to amino acid starvation23,24. Strikingly, sPbUIS3WT inhibited the degradation of p62 induced by amino acid depletion, whereas UIS3 mutants had no effect (Fig. 4a, b). The data demonstrates that binding of UIS3 to LC3 disrupts the autophagic flux, hinting at the possibility that UIS3 functions as an autophagy inhibitor, through a hindrance effect on the LC3 surface. To explore this hypothesis, we setup a LUMIER-based competition assay25 in which sPfUIS3 or p62 were complexed to GST-LC3 and subsequently incubated with increasing concentrations of the p62 LIR22 (Fig. 4c). We found that higher concentrations of p62 LIR were necessary to disrupt the UIS3-LC3 complex compared to p62-LC3 (Fig. 4d). This is consistent with the lower KD measured for the UIS3-LC3 interaction (Fig. 2g) compared to similar measurements for canonical LIR sequences26. To determine how pervasive the UIS3 hindrance effect was, we examined the impact of sPfUIS3 on the association between the LIR-containing Rab7 effector, PLEKHM127, and recombinant GST-LC3. When sPfUIS3WT and PLEKHM1 were co-expressed, LC3 pulldowns were significantly depleted of PLEKHM1 and enriched in UIS3, whereas mutations that reduced the affinity of UIS3 for LC3 restored the PLEKHM1-LC3 association (Fig. 4e). Taken together, these observations not only demonstrate that Plasmodium UIS3 antagonizes LIR-mediated LC3 interactions by binding to a conserved LIR-interacting surface on LC3, but they also confirm UIS3 as a bona fide autophagy inhibitor.
Figure 4. UIS3 acts as a bona fide autophagy inhibitor by competing with host LC3-interacting proteins for LC3 binding.
a, Western-blot analysis of p62 levels in HeLa cells expressing the indicated sPbUIS3 proteins and kept for 8 h in complete (control) or amino acid-depleted medium (aa starvation). Whole cell lysates were analysed by western-blot for p62, tubulin, and UIS3-myc. Immunoblots representative of five independent experiments are shown. b, The levels of p62 before and after aa starvation were calculated as the ratio of p62 to tubulin. The bars represent the mean + SEM of five independent experiments. c, Outline of LUMIER competition assay. The assay measures the amount of p62 LIR peptide necessary to displace luciferase-tagged p62 or sPfUIS3 from their complexes with recombinant GST-LC3. The inset shows the input levels (detected with anti-Renilla luciferase) of luciferase-tagged p62 and sPfUIS3 in one (of three) representative experiment. d, Binding of luciferase-tagged p62 or sPfUIS3 to recombinant GST-LC3 in the presence of the indicated concentrations of p62 LIR. The mean + SEM of three independent experiments is shown. For the data points corresponding to 2 and 5 µM of p62 LIR, n = 2. e, Recombinant GST-LC3 was used to pulldown extracts of HEK 293T cells co-transfected with the indicated plasmids. The ratio of PLEKHM1 and sPfUIS3 in the pulldown versus input was calculated from the GFP and luminescence signals of each fraction. The bottom panels show the levels of PLEKHM1 and UIS3 in the input extract. The bars represent the average + SEM of three independent experiments. Statistical significance was assessed using unpaired two-tailed t test (b, e) and two-way ANOVA (d): n.s. P > 0.05; ***P < 0.001; ****P < 0.0001. f, Summary of the observed effects of Plasmodium UIS3 on host autophagy. Antiplasmodial autophagy depends on the LC3 conjugation machinery and on lysosome-associated Rab7, with unidentified LIR-motif proteins mediating one or several steps leading to lysosomal fusion. The C-terminus of UIS3 binds to the LIR-docking site on LC3, rendering the site inaccessible for autophagy-promoting interactions.
In this study, we show that Plasmodium parasites escape autophagy by engaging host LC3 in an inhibitory interaction with the cytosolic domain of the PVM transmembrane protein UIS3. Our observations demonstrate that UIS3 competitively inhibits the binding of LIR-containing host proteins to the LIR-docking surface of LC3. This strongly suggests that the UIS3-mediated protection against antiplasmodial autophagy is a direct consequence of LC3 being sequestered into UIS3-LC3 complexes on the PVM (Fig. 4f, Supplementary Fig. 6). Although UIS3 is necessary to avert the potential deleterious effects of the association of LC3 to the PVM, whether Plasmodium succeeds in evading autophagy probably depends on how much of the PVM-associated LC3 is bound by UIS3. This, in turn, is determined by the levels of UIS3 expression and the robustness of the antiplasmodial response, both of which may diverge significantly between individual parasites and hosts, and could explain the observed vulnerability of P. berghei parasites to autophagy in previous reports3,4.
Evidence that IFN-γ enhances LC3 recruitment onto the PVM of P. vivax, causing parasite elimination28, supports the idea that host autophagy may be chemically enhanced to suppress the protective effect of UIS3 expression. Repurposing autophagy-inducing drugs for malaria prophylaxis might thus be a safe and cost-effective way of breaking the transmission cycle in endemic areas. Whether clinically available autophagy inducers activate the non-canonical autophagy pathway targeting Plasmodium in hepatic cells remains to be determined. Of note, neither metformin nor rapamycin, two of the best-known autophagy-inducing drugs, affect the number of infected cells in vitro or in vivo3,8,29, supporting our observations that antiplasmodial autophagy is mechanistically distinct from the AMPK- and mTOR-regulated canonical autophagy pathway.
Recent reports underscored the risk of breakthrough infections in mice immunised with a uis3(-) P. berghei ANKA whole-organism vaccine30. The data presented here suggest that host genetic or metabolic factors compromising the autophagy response might be the cause of such breakthrough infections. Although insufficiently safe on its own, UIS3 deletion is likely to improve the safety of any genetically-attenuated whole-organism vaccine against malaria by enhancing the susceptibility of hepatic parasites to host autophagy. Additionally, with the spread of drug resistance casting a shadow over malaria eradication efforts, the protein-protein interaction between UIS3 and LC3 constitutes a promising new target for tailored pharmacological interventions aiming at controlling Plasmodium liver infection and disease.
Methods
Chemicals
RPMI 1640, DMEM, EBSS, and other cell culture reagents were purchased from Gibco Invitrogen. All chemicals were from Sigma-Aldrich, except specified otherwise. Lipofectamine (Invitrogen) was used for transfection of HepG2 cells. siRNAs were transfected using Lipofectamine RNAiMAX in OptiMEM (Invitrogen).
Cell Lines
Huh7, HepG2, and HEK 293T (ATCC) cells were cultured under standard conditions in RPMI 1640 (Huh7) or DMEM (HepG2 and HEK 293T) medium supplemented with 10% FCS, 1% non-essential aminoacids (RPMI 1640), 1% glutamine, 1% penicillin/streptomycin, and 1% Hepes. Mouse embryonic fibroblasts (MEFs) were obtained from the Riken BioResource Center (Atg5+/+ (RCB2710) and Atg5-/- (RCB2711)31; M. Komatsu, Tokyo Metropolitan Institute of Medical Science (Atg3+/+ and Atg3-/-)32, and T. Finkel, NIH National Heart, Lung, and Blood Institute (Atg7+/+ and Atg7-/-)33. HeLa cells expressing GFP-LC3 were a kind gift from T. Hyman, MPI-CBG. MEFs and HeLa cells were maintained in DMEM supplemented as before. MEFs were validated through qRT-PCR and HeLa cells were tested for the expression of GFP-LC3 by Western blot. All cell lines were routinely tested for mycoplasma contamination.
Mice
All mice used in this study were C57BL/6J males, between 6 to 8 weeks of age, housed in the animal facility of the Instituto de Medicina Molecular in Lisbon. All protocols were approved by the internal animal care committee of the Instituto de Medicina Molecular and were performed according to national and European regulations.
Mouse primary hepatocytes
Mouse primary hepatocytes were isolated using a modified two-step perfusion protocol followed by a Percoll purification step34,35. Briefly, mice were sacrificed by CO2 inhalation and immediately processed for cannulation of the portal vein using a 26-gauge needle. Next, the inferior vena cava (IVC) was cut to allow fluid to drain. Liver perfusion medium (LPM) was perfused at 8–9 mL/min for 10 min, followed by liver digestion medium (LDM) also at a rate of 8–9 mL/min for 10 min. Intermittent clamping of the IVC (3-s clamp every 30 s) was performed during LDM perfusion to improve tissue digestion. After digestion, the liver was excised and the cells were liberated by tearing and shaking of the liver with forceps. The cell suspension was then sequentially filtered through a 100-μm and a 70-μm cell strainer and spun at 50g for 3 min. The cell pellet was resuspended in Williams's Medium E with 10% of fetal calf serum (FBS) and carefully overlaid on a 60% Percoll solution (1:1). The cell suspension was fractionated by centrifugation at 750g for 20 min, without break, at 20 °C. Viable hepatocytes deposited in the pellet were washed with Williams's Medium E with 10% FBS, spun at 50g for 3 min and resuspended in complete Williams's Medium E (supplemented with 4% FBS and 1% penicillin/streptomycin). Viability and yield were assessed by trypan blue staining. Viable hepatocytes were plated on collagen-coated 24-well plates.
RNAi
HepG2 cells were reverse-transfected with target-specific SMARTpool siRNA pools, as previously described36. Non-targeting SMARTpool siRNA was used as a negative control (Dharmacon, GE Healthcare). The efficiency of knockdown was assessed 48 h after transfection by qRT-PCR using kits from Applied Biosystems. Gene expression levels were normalised against those of hypoxanthine guanine phosphoribosyltransferase (Hprt1). The primer pairs used for qRT-PCR reactions are listed in Table S3.
Parasite lines
Plasmodium berghei sporozoites were obtained through dissection of the salivary glands of infected female Anopheles stephensi mosquitoes bred at the Instituto de Medicina Molecular. The following parasite lines were used: GFP-expressing P. berghei ANKA (259cl2, Leiden Malaria Research Group), uis3(-)5, uis3(+), and uis3-myc P. berghei NK65 (generated for this study).
Plasmodium transfection
P. berghei uis3-myc and uis3(+) were obtained by complementing the uis3(-) parasite line via single cross-over recombination as described in Supplementary Fig. 2. Transfection was done by electroporation of purified schizonts as previously described37. Parasites were selected with WR99210 (kindly provided by Jacobus Pharmaceuticals, Princeton, NJ, USA) for 4 consecutive days, harvested on day 10 post transfection, and genotyped by PCR with the primers listed in Table S2.
Infections
Hepatoma cells, MEFs, and mouse primary hepatocytes were plated on 24-well culture plates at a density of 65.000, 50.000, and 120.000 cells/well, respectively. Cells were infected with freshly dissected P. berghei sporozoites (30.000/well) 1 day after seeding and processed for analysis at 24 h or 48 h. siRNA-treated HepG2 cells were infected with uis3(+) or uis3(-) sporozoites 36 h after siRNA transfection and cultured for another 65 h in standard medium supplemented with 0.3 % Fungizone. At 65 h, the entire volume of cell culture medium (HepG2 SN) in each well was harvested and immediately inoculated into mice (1 well for each animal). The onset of blood stage parasitemia was monitored through Giemsa-stained thin blood smears by two unblinded independent observers. The smears were analysed daily from day 1 through day 10 after inoculation, when most animals suffered from hyperparasitemia and had to be sacrificed. On day 10, parasite genomic DNA was extracted from the blood of each experimental animal for parasite genotyping. The primer pairs used to probe the uis3 wild-type and knockout genomic loci are listed in Table S2. For ROS scavenging experiments, Huh7 cells were incubated for 2 h in complete medium supplemented with 20 mM N-acetylcysteine (NAC), before sporozoite addition. Chloroquine (50 µM) was added to hepatoma cells 1 h after sporozoite infection.
Plasmids
Plasmids used in this study are listed in Table S1. All Plasmodium sequences (except single point mutants) and human LC3B were chemically synthesised and subcloned into pCMV-Myc-N or pCMV-HA-N expression vectors (Clontech) using the EcoRI and KpnI restriction sites (GeneArt, Invitrogen). The Renilla luciferase sequence was chemically synthesised (GeneArt, Invitrogen) and subcloned into pCMV-MYC-sPfUIS3 using the EcoRI restriction site. Single point mutants of sPfUIS3 were obtained by site-directed mutagenesis using the primers listed in Table S4 and the QuickChange II site-directed mutagenesis kit (Agilent Technologies), following the manufacturer’s instructions. For production of His6-sPfUIS3 (WT and mutants), the soluble domain of P. falciparum UIS3 was amplified from pCMV-MYC-sPfUIS3 (forward primer: 5’CATGGGATCCATGGAGCAGAAGCTG3’; reverse primer: 5’CCGCGGTCTCGAGTCAGTTCTCTTCCT3) and cloned into pET28a (Novagen). All plasmids were verified by sequencing. Plasmodium sequences were optimised for expression in mammalian cells.
Immunofluorescence
Cells plated on glass coverslips were fixed in 4% paraformaldehyde (ChemCruz) for 10 min at room temperature (RT) and permeabilised/blocked with a solution of 0,2% saponin-1% BSA. For immunostaining, samples were incubated with primary antibodies diluted in blocking solution (2 h, RT), washed with PBS, incubated for 1 h at RT with AlexaFluor-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) and Hoechst 33342 (Invitrogen), and washed again. The coverslips were then mounted on microscope slides with Fluoromount (SouthernBiotech). The following primary antibodies were used for confocal microscopy: LC3 (mouse monoclonal, MBL, M152-3), c-Myc (mouse monoclonal, MBL, M192-3), PbHSP70 (2E638), PbUIS429 (goat polyclonal, SicGen, AB0042-200), LAMP1 (rabbit polyclonal, Sigma, L1418), and PbMSP129 (rabbit polyclonal, GenScript, custom-made). All images were acquired on Zeiss confocal microscopes (LSM 510 META or LSM 710) and processed in ImageJ. For co-localisation analysis, the region of interest (ROI) corresponding to the PVM was defined by thresholding the UIS4 signal, after which the Manders coefficients M1 and M2 within the ROI were calculated using the Colocalisation Threshold plugin from ImageJ. For infection quantification by microscopy, parasites were detected with anti-PbHSP70 (2E6) and anti-PbMSP1 and imaged on a Zeiss Axiovert 200M wide-field microscope equipped with an automated stage. All images (42 per coverslip) were processed and analysed using ImageJ.
Immuno-EM
Huh7 cells were fixed 24 h after infection with 4% paraformaldehyde - 0.05% glutaraldehyde in 100 mM piperazine-N,N[prime]-bis(2-ethanesulfonic acid) (PIPES)-0.5 mM MgCl2 (pH 7.2) for 1 h at 4°C and processed as described previously39. Sections were stained with antibodies against human LC3 (mouse monoclonal, MBL, M152-3) and UIS4 (goat polyclonal, SicGen, AB0042-200), followed by gold-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories), and viewed on a JEOL 1200 EX transmission electron microscope (JEOL).
Molecular Docking of UIS3-LC3 complexes
Homology models of the UIS3 soluble domains belonging to different Plasmodium species and human LC3 were built with the Phyre2 server (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index) using the crystal structures of P. falciparum UIS3 (PDB code: 2VWA) and LC3B (PDB code: 2ZJD) as reference models. The quality of the homology models obtained was improved by atomic coordinate refinement using a two-step atomic-level energy refinement protocol implemented in ModRefiner software40. Experimental and homology-derived protein coordinates were then used to construct models of the human and mouse UIS3-LC3 complexes by flexible molecular docking with the Swarmdock algorithm41. LC3 proteins were considered as receptors and UIS3 as ligands. For each complex, an initial set of 100 ligand starting positions was generated in the space around the receptor, being further optimized by energy minimization. The top 10 docking solutions were ranked by increasing free energy, and submitted to an additional energy minimization cycle by conjugated gradient during 1000 steps with GROMACS42, in order to avoid potential steric clashes between contact atoms. Visual inspection and cross comparison between the docking results obtained for the human and mouse complex showed a conserved pattern of molecular orientations for the top 5 docking solutions. Consensus residues involved in UIS3-LC3 interactions in the best docking solution were determined using CONS-COCOMAPS application43 and graphically represented with the LigPlot+ software44.
Co-Immunoprecipitation
HeLa cells expressing GFP-LC3 were transfected with plasmids encoding c-Myc or HA fusion proteins (Table S1) on 10 cm dishes and harvested 48 h after transfection in lysis buffer - 0.5 % NP-40, 150 mM NaCl, 10 mM Tris-Cl pH 7.4, 0.5 mM EDTA, supplemented with Complete protease inhibitor and PhosStop phosphatase inhibitor cocktails (Roche). On some experiments, HeLa cells were co-transfected with both myc-UIS3 and HA-tagged LC3. c-Myc- and HA-tagged proteins were purified from total cell lysates using the c-Myc tagged protein mild purification kit and the HA tagged protein purification kit, respectively, according to the manufacturer’s instructions (MBL). GFP-LC3 was immunoprecipitated from total cell lysates using GFP-Trap® coupled to agarose beads (Chromotek). For immunoprecipitation of parasite-encoded UIS3-myc from infected cells, HeLa cells or mouse primary hepatocytes were lysed 24 h after infection with uis3-myc P. berghei and total cell lysates were processed as described above. Co-immunoprecipitated proteins and total cell lysates were analysed by Western blot using antibodies against LC3 (rabbit polyclonal, MBL, PM036), and c-Myc (mouse monoclonal, MBL, M192-3) or HA tags (mouse monoclonal, Biolegend, 901503) to detect UIS3 or UIS4, followed by HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories). Immunoblots were developed using Luminata Crescendo Western HRP substrate reagent (Merck Millipore).
Expression and purification of recombinant proteins
Escherichia coli (E. coli) BL21 cells were transformed with GST-His-tagged LC3 or GST-His bacterial expression vectors. After overnight culture in LB, cells were diluted 1/100 in fresh medium to an OD ~ 0.7 and incubated with 0.3 mM IPTG for 5 h to induce the expression of the recombinant proteins. All cultures were performed at 37°C with vigorous shaking. For expression of His-tagged sPfUIS3WT, E. coli BL21 C43 bacteria were used. Cells grown overnight in LB were diluted 1/100 in fresh medium to an OD ~1.2. Recombinant protein expression was induced by the addition 0.3 mM IPTG and overnight culture at 25 °C. His-tagged sPfUIS3 mutants were expressed in E. coli BL21 bacteria. Overnight cell cultures were diluted 1/100 in LB supplemented with NZY auto-induction reagent (NZYTech). Recombinant His-tagged proteins were initially purified by metal chelating affinity chromatography using His-Trap Ni-containing columns (GE Healthcare) connected to an AKTA Explorer chromatographic system (GE Healthcare). The His-trap column (1 mL bed volume) was equilibrated with sodium phosphate buffer pH 6.8 containing 1 M NaCl and 25 mM imidazole before protein injection. His-tagged proteins were eluted by a linear gradient of imidazole from 25-500 mM in the same phosphate buffer. Fractions containing protein were collected and pooled, and further polished by size exclusion chromatography purification using a Sephadex S-200 column (GE Healthcare) eluted with PBS. Fractions containing protein were analysed by SDS-PAGE, concentrated by ultrafiltration and stored at -80°C in PBS buffer containing 15% glycerol.
Interaction between recombinant proteins
Recombinant sPfUIS3 (5 µg) was incubated with recombinant GST-His6-LC3 (10 µg) or with GST-His6 (5 µg), pre-adsorbed to glutathione beads in lysis buffer (150 mM NaCl, 20 mM Tris–HCl (pH 7.4), 0.1% Triton-X100, 5% glycerol, 5 mM EDTA and Complete protease inhibitors (Roche)). After washing, the complex was eluted in lysis buffer supplemented with 10 mM reduced L-glutathione (Sigma-Aldrich) and analysed by western-blot with antibodies against LC3 (rabbit polyclonal, MBL, PM036) and c-Myc (mouse monoclonal, MBL, M192-3).
Surface plasmon resonance
Recombinant sPfUIS3 was immobilized on a Biacore CM5 chip surface following standard protocols provided by the manufacturer (GE-Healthcare). In brief, recombinant sPfUIS3 was chemically immobilized via amine coupling to the free carboxyl groups on the CM5 chip through standard NHS/EDC procedures. Recombinant GST-His-LC3 dissolved in PBS buffer was injected over the immobilized sPfUIS3 at various concentrations to generate affinity sensorgrams that were collected in a single-cycle mode with no regeneration between sample injections. Unspecific binding was monitored by injecting recombinant GST-His protein over the sPfUIS3-coated chip under the same conditions. Normalized response sensorgrams were calculated by subtracting the unspecific binding contribution of the same molar concentrations of GST protein alone. All biosensor data processing and analysis and KD calculation were performed using the Biacore T100 Evaluation Software (GE Healthcare version 2.01). For KD calculation, a monovalent interaction model was assumed, with both proteins interacting with 1:1 stoichiometry.
Circular Dichroism
Recombinant His-tagged sPfUIS3 proteins (wild type and mutants) used for Circular Dichroism were purified using a modification of the already described protocol, where the elution buffer of the size exclusion chromatography was substituted by 50 mM sodium phosphate, 200 mM NaF, pH 7.5. CD spectra of sPfUIS3 variants were collected between 190 and 260 nm in a JASCO J-815 spectropolarimeter (Tokyo, Japan) using 1 mm quartz cells from Hellma Analytics (Müllheim, Germany). Experiments were performed at 25° C. Each CD spectrum corresponds to the average of 10 technical replicates, corrected for the buffer background contribution. Mean molar residue ellipticity values, [θ], were determined through the following relationship45:
where θ is the observed ellipticity, N is the number of amino acid residues in each protein, l is the quartz cell optical path length and c is the protein molar concentration.
Autophagy induction and p62 degradation assay
HeLa cells were changed from normal growth medium to EBSS to induce amino acid starvation-dependent autophagy. After 7 h, cells were harvested in Laemmli buffer and analysed by Western blot with antibodies against p62 (rabbit polyclonal, Sigma-Aldrich, P0067) and gamma-tubulin (mouse monoclonal, Sigma-Aldrich, T5326). The levels of p62 were measured by quantifying the ratio of p62 to tubulin signals with ImageJ.
LUMIER competition assay
HEK 293T cells transiently transfected with Luc-p62 or Luc-sPfUIS3 expressing plasmids (Table S1) were lysed 24 h post-transfection in LUMIER lysis buffer (150 mM NaCl, 20 mM Tris–HCl (pH 7.4), 0.1% Triton-X100, 5% glycerol, 5 mM EDTA and Complete protease inhibitors (Roche))25. The Renilla luciferase activity associated with each cell lysate was assessed using the Renilla Luciferase Assay System (Promega). Equivalent saturating amounts of luciferase-tagged p62 or sPfUIS3 cell lysates were incubated with recombinant GST-His6-LC3B (500 ng), pre-adsorbed to glutathione beads. After washing away unbound protein, the precipitates were incubated with increasing concentrations of synthetic p62 LIR peptide (SGGDDDWTHLSSK) for 1 h with rotation, followed by elution in lysis buffer supplemented with 10 mM reduced L-glutathione (Sigma-Aldrich). The Renilla luciferase activity associated with LC3 after elution was quantified as described above using Tecan’s Infinite 200M multiplate reader. The expression of luciferase-tagged p62 or sPfUIS3 was additionally detected with an anti-Renilla luciferase antibody (Abcam).
LUMIER/GFP assay
HEK 293T cells transiently transfected with PLEKHM1-GFP46 and co-transfected with empty plasmid, Luc-sPfUIS3WT, or Luc-sPfUIS3Mut1 (Table S1) were lysed 24 h post-transfection in LUMIER lysis buffer (150 mM NaCl, 20 mM Tris–HCl (pH 7.4), 0.1% Triton-X100, 5% glycerol, 5 mM EDTA and Complete protease inhibitors (Roche)). The cell lysates were incubated with recombinant GST-His6-LC3B (500 ng), pre-adsorbed to glutathione beads, for 2h at 4 °C, with rotation. After washing away unbound proteins, pulldowns were eluted in lysis buffer supplemented with 10 mM reduced L-glutathione. GFP (excitation 420 nm, emission 520 nm) and Renilla luciferase activity associated with LC3 in each experimental condition were quantified using Tecan’s Infinite 200M multiplate reader.
Statistical analysis
Data are expressed as means + SEM. Statistically significant differences between two different groups were determined using non-parametric two-tailed-Mann-Whitney test, unpaired two-tailed t test, or two-way ANOVA, as indicated. P values < 0.05 were considered statistically significant. Significances are represented in the figures as follows: n.s. P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ***P < 0.0001. All tests were carried out in GraphPad Prism. The experiments were not randomised. Sample sizes on mice experiments were chosen on the basis of historical data, no statistical methods were used to predetermine sample size.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information.
Supplementary Material
Acknowledgments
We would like to thank Masaaki Komatsu (Tokyo Metropolitan Institute of Medical Science) for providing Atg3 MEFs, Tony Finkel (NIH National Heart, Lung, and Blood Institute) for the Atg7 MEFs, Felix Randow (MRC laboratory of Molecular Biology, Cambridge, UK) for the gift of p62-luciferase and GST-LC3 expression plasmids, Jacobus Pharmaceuticals for the WR99210 compound, Ana Parreira for producing P. berghei-infected Anopheles mosquitoes, Sofia Marques and Ksenija Slavic for their help with parasite cloning. This work was supported by grants from the European Research Council (ERC-2012-StG_311502 to M.M.M.), Fundação para a Ciência e Tecnologia (EXCL/IMI-MIC/0056/2012 to M.M.M. and PTDC/IMI-MICC/1568/2012 to G.G.C.), and Institut Mérieux (MRG_20052016 to M.M.M). E.R. was the recipient of EMBO (ALTF 949-2008) and FCT (SFRH/BPD/68709/2010) fellowships. L.R. is the recipient of FCT fellowship SFRH/BPD/111323/2015. G.G.C. was sponsored by Marie Curie (PIEF-GA-2009-235864) and FCT (SFRH/BPD/74151/2010) fellowships. L.M.S. was supported by the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement N. 242095 (EVIMalaR). V.Z.L. was sponsored by EMBO (ALTF 357-2009) and FCT (BPD-81953-2011) fellowships. I. M. V. and J. M.-V were supported by NIH (1F32A11042-5 021) and FCT (SFRH/BD/52226/2013) fellowships, respectively.
Footnotes
Author contribution E.R. and M.M.M. conceived and led the study and wrote the manuscript. E.R, L.R., G.G.C., J.M.V. performed the experiments, acquired the data, performed data analysis, and interpreted results. Molecular docking, protein purification, and SPR analysis were performed by F.J.E. L.M.S., L.R., I.M.V., V.Z.L. conducted animal work. W.B. performed electron microscopy analysis. Circular dichroism was performed by T.N.F. G.R.M. constructed plasmids for parasite transfections. All authors read and approved the final manuscript.
References
- 1.Liehl P, Zuzarte-Luis V, Mota MM. Unveiling the pathogen behind the vacuole. Nat Rev Microbiol. 2015;13:589–98. doi: 10.1038/nrmicro3504. [DOI] [PubMed] [Google Scholar]
- 2.Gomes LC, Dikic I. Autophagy in Antimicrobial Immunity. Mol Cell. 2014;54:224–33. doi: 10.1016/j.molcel.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 3.Prado M, et al. Long-term live imaging reveals cytosolic immune responses of host hepatocytes against Plasmodium infection and parasite escape mechanisms. Autophagy. 2015;11:1561–79. doi: 10.1080/15548627.2015.1067361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thieleke-Matos C, et al. Host cell autophagy contributes to Plasmodium liver development. Cell Microbiol. 2016;18:437–50. doi: 10.1111/cmi.12524. [DOI] [PubMed] [Google Scholar]
- 5.Mueller A-K, Labaied M, Kappe SHI, Matuschewski K. Genetically modified Plasmodium parasites as a protective experimental malaria vaccine. Nature. 2005;433:164–167. doi: 10.1038/nature03188. [DOI] [PubMed] [Google Scholar]
- 6.Shen H-M, Mizushima N. At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy. Trends Biochem Sci. 2014;39:61–71. doi: 10.1016/j.tibs.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Mueller A-K, et al. Plasmodium liver stage developmental arrest by depletion of a protein at the parasite-host interface. Proc Natl Acad Sci U S A. 2005;102:3022–7. doi: 10.1073/pnas.0408442102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanson KK, et al. Torins are potent antimalarials that block replenishment of Plasmodium liver stage parasitophorous vacuole membrane proteins. Proc Natl Acad Sci U S A. 2013;110:E2838–E2847. doi: 10.1073/pnas.1306097110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spielmann T, Montagna GN, Hecht L, Matuschewski K. Molecular make-up of the Plasmodium parasitophorous vacuolar membrane. Int J Med Microbiol. 2012;302:179–86. doi: 10.1016/j.ijmm.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 10.Mizushima N, et al. Dissection of Autophagosome Formation using Apg5-deficient Mouse Embryonic Stem Cells. J Cell Biol. 2001;152:657–667. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganley IG, Wong P-M, Gammoh N, Jiang X. Distinct Autophagosomal-Lysosomal Fusion Mechanism Revealed by Thapsigargin-Induced Autophagy Arrest. Mol Cell. 2011;42:731–43. doi: 10.1016/j.molcel.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sturm A, et al. Alteration of the Parasite Plasma Membrane and the Parasitophorous Vacuole Membrane during Exo-Erythrocytic Development of Malaria Parasites. Protist. 2009;160:51–63. doi: 10.1016/j.protis.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Sharma A, Yogavel M, Akhouri RR, Gill J, Sharma A. Crystal Structure of Soluble Domain of Malaria Sporozoite Protein UIS3 in Complex with Lipid. J Biol Chem. 2008;283:24077–88. doi: 10.1074/jbc.M801946200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikolajczak SA, Jacobs-Lorena V, MacKellar DC, Camargo N, Kappe SHI. L-FABP is a critical host factor for successful malaria liver stage development. Int J Parasitol. 2007;37:483–489. doi: 10.1016/j.ijpara.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Favretto F, Assfalg M, Molinari H, D’Onofrio M. Evidence from NMR interaction studies challenges the hypothesis of direct lipid transfer from L-FABP to malaria sporozoite protein UIS3. Protein Sci. 2013;22:133–138. doi: 10.1002/pro.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farré J-C, Subramani S. Mechanistic insights into selective autophagy pathways: lessons from yeast. Nat Rev Mol Cell Biol. 2016;17:537–52. doi: 10.1038/nrm.2016.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wacker R, et al. LC3-association with the parasitophorous vacuole membrane of Plasmodium berghei liver stages follows a noncanonical autophagy pathway. Cell Microbiol. 2017:1–13. doi: 10.1111/cmi.12754. [DOI] [PubMed] [Google Scholar]
- 18.Martinez J, et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol. 2015;17:893–906. doi: 10.1038/ncb3192. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 2010;584:1379–1385. doi: 10.1016/j.febslet.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 20.Birgisdottir ÅB, Lamark T, Johansen T. The LIR motif - crucial for selective autophagy. J Cell Sci. 2013;126:3237–47. doi: 10.1242/jcs.126128. [DOI] [PubMed] [Google Scholar]
- 21.Ichimura Y, et al. Structural Basis for Sorting Mechanism of p62 in Selective Autophagy. J Biol Chem. 2008;283:22847–57. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 22.Pankiv S, et al. p62/SQSTM1 Binds Directly to Atg8/LC3 to Facilitate Degradation of Ubiquitinated Protein Aggregates by Autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 23.Mizushima N, Yoshimori T, Levine B. Methods in Mammalian Autophagy Research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klionsky D, Abdalla F, Abeliovich H. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrios-Rodiles M, et al. High-Throughput Mapping of a Dynamic Signaling Network in Mammalian Cells. Science (80-.) 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- 26.Zaffagnini G, Martens S. Mechanisms of Selective Autophagy. J Mol Biol. 2016;428:1714–24. doi: 10.1016/j.jmb.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McEwan DG, et al. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell. 2015;57:39–54. doi: 10.1016/j.molcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 28.Boonhok R, et al. LAP-like process as an immune mechanism downstream of IFN-γ in control of the human malaria Plasmodium vivax liver stage. Proc Natl Acad Sci U S A. 2016;113:E3519–28. doi: 10.1073/pnas.1525606113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruivo MTG, et al. Host AMPK Is a Modulator of Plasmodium Liver Infection. Cell Rep. 2016;16:2539–2545. doi: 10.1016/j.celrep.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar H, et al. Protective efficacy and safety of liver stage attenuated malaria parasites. Sci Rep. 2016;6:26824. doi: 10.1038/srep26824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1–5. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 32.Sou Y, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–75. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee IH, et al. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science. 2012;336:225–8. doi: 10.1126/science.1218395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W, et al. PCB 126 and other dioxin-like PCBs specifically suppress hepatic PEPCK expression via the aryl hydrocarbon receptor. PLoS One. 2012;7:e37103. doi: 10.1371/journal.pone.0037103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonçalves LA, Vigário AM, Penha-Gonçalves C. Improved isolation of murine hepatocytes for in vitro malaria liver stage studies. Malar J. 2007;6:169. doi: 10.1186/1475-2875-6-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Itoe MAA, et al. Host Cell Phosphatidylcholine Is a Key Mediator of Malaria Parasite Survival during Liver Stage Infection. Cell Host Microbe. 2014;16:778–786. doi: 10.1016/j.chom.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janse CJ, Ramesar J, Waters AP. High-efficiency transfection and drug selection of genetically transformed blood stages of the rodent malaria parasite Plasmodium berghei. Nat Protoc. 2006;1:346–56. doi: 10.1038/nprot.2006.53. [DOI] [PubMed] [Google Scholar]
- 38.Tsuji M, Mattei D, Nussenzweig RS, Eichinger D, Zavala F. Demonstration of heat-shock protein 70 in the sporozoite stage of malaria parasites. Parasitol Res. 1994;80:16–21. doi: 10.1007/BF00932618. [DOI] [PubMed] [Google Scholar]
- 39.Fentress SJ, et al. Phosphorylation of immunity-related GTPases by a toxoplasma gondii-secreted kinase promotes macrophage survival and virulence. Cell Host Microbe. 2010;8:484–495. doi: 10.1016/j.chom.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu D, Zhang Y. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys J. 2011;101:2525–34. doi: 10.1016/j.bpj.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torchala M, Moal IH, Chaleil RAG, Fernandez-Recio J, Bates Pa. SwarmDock: a server for flexible protein-protein docking. Bioinformatics. 2013;29:807–9. doi: 10.1093/bioinformatics/btt038. [DOI] [PubMed] [Google Scholar]
- 42.Pronk S, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–54. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vangone A, Oliva R, Cavallo L. CONS-COCOMAPS : a novel tool to measure and visualize the conservation of inter-residue contacts in multiple docking solutions. BMC Bioinformatics. 2012;13:S19. doi: 10.1186/1471-2105-13-S4-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laskowski RA, Swindells MB. LigPlot + : Multiple Ligand-Protein Interaction Diagrams for Drug Discovery. J Chem Inf Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 45.Kelly SM, Jess TJ, Price NC. How to study proteins by circular dichroism. Biochim Biophys Acta - Proteins Proteomics. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 46.Van Wesenbeeck L, et al. Involvement of PLEKHM1 in osteoclastic vesicular transport and osteopetrosis in incisors absent rats and humans. J Clin Invest. 2007;117:919–930. doi: 10.1172/JCI30328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information.