The Caenorhabditis elegans germline is syncytial but its formation is not completely understood. During embryogenesis, the germline precursor blastomere does not complete cytokinesis and maintains a stable cytoplasmic bridge between the two primordial germ cells that is enriched in actomyosin contractility regulators.
Abstract
Stable cytoplasmic bridges arise from failed cytokinesis, the last step of cell division, and are a key feature of syncytial architectures in the germline of most metazoans. Whereas the Caenorhabditis elegans germline is syncytial, its formation remains poorly understood. We found that the germline precursor blastomere, P4, fails cytokinesis, leaving a stable cytoplasmic bridge between the two daughter cells, Z2 and Z3. Depletion of several regulators of actomyosin contractility resulted in a regression of the membrane partition between Z2 and Z3, indicating that they are required to stabilize the cytoplasmic bridge. Epistatic analysis revealed a pathway in which Rho regulators promote accumulation of the noncannonical anillin ANI-2 at the stable cytoplasmic bridge, which in turns promotes the accumulation of the nonmuscle myosin II NMY-2 and the midbody component CYK-7 at the bridge, in part by limiting the accumulation of canonical anillin ANI-1. Our results uncover key steps in C. elegans germline formation and define a set of conserved regulators that are enriched at the primordial germ cell cytoplasmic bridge to ensure its stability during embryonic development.
INTRODUCTION
Cytokinesis, the last step of cell division in which two daughter cells are physically separated, is a highly coordinated event (reviewed in Green et al., 2012; D’Avino et al., 2015). In Caenorhabditis elegans and other metazoa, cytokinesis initiates during anaphase, when antiparallel microtubules in the mitotic spindle midzone organize signaling to the cell cortex to specify the future site of ingression, in part by recruitment of the centralspindlin complex (Wheatley and Wang, 1996; Mishima et al., 2002; Oegema and Hyman, 2006). This leads to the cortical recruitment and activation of the small GTPase Rho to the site of ingression, where it orchestrates the assembly and furrowing of a contractile ring composed of several proteins including actin filaments, nonmuscle myosin II, septins, and the actomyosin scaffold protein anillin (Green et al., 2012; D’Avino et al., 2015). Ingression of the contractile ring compacts the central portion of the mitotic spindle and generates a transient intercellular bridge termed midbody (Mullins and Biesele, 1977). As furrow constriction nears completion, the contractile ring becomes progressively tighter and matures into the midbody ring, a dense structure that forms around the center of the midbody. Midbody ring formation blocks the cytoplasmic exchange between the two daughter cells, effectively resulting in cytoplasmic isolation (Agromayor and Martin-Serrano, 2013; Green et al., 2013). Cytokinesis completes with cellular abscission, when recruitment of the endosomal sorting complexes required for transport (ESCRT) proteins and microtubule depolymerization promote membrane scission and midbody ring release, generating two physically separated daughter cells (Morita et al., 2007; Schiel et al., 2011; Carlton et al., 2012; Green et al., 2013). In the early C. elegans embryo, however, ESCRT proteins were reported to be required for membrane removal at the end of cytokinesis but otherwise dispensable for membrane scission and midbody ring release (Green et al., 2013; Konig et al., 2017).
All cytokinetic steps are closely regulated to avoid cytokinetic failure, cellular binucleation, and subsequent aneuploidy. In certain contexts, however, such as the germline of many organisms across phyla, controlled cytokinesis failures take place during development, progressively leading to the formation of a cluster of joined cells that share cytoplasm through stable intercellular bridges, thus forming a syncytium (Fawcett et al., 1959; Greenbaum et al., 2011; Haglund et al., 2011). This has been well studied during Drosophila germline cyst development, where intercellular bridge stabilization following incomplete cytokinesis was proposed to lead to formation of the ring canals that connect germ cells (Robinson et al., 1994; Ong and Tan, 2010). Although their organization varies between male and female animals, ring canals are enriched in contractility regulators, often derived from the proteins present in the contractile ring, such as actin, septins, the kinesin Pavarotti (orthologue of C. elegans ZEN-4), and anillin (reviewed in Greenbaum et al., 2011). The protein Cindr, implicated in both complete and incomplete cytokineses, interacts with anillin at arrested cleavage furrows and its disappearance during gametogenesis coincides with germ cell differentiation (Haglund et al., 2010; Eikenes et al., 2013). The mechanism responsible for intercellular bridge formation after controlled cytokinesis failure was studied in detail in mouse spermatocytes, in which the germ cell–specific protein TEX14 binds to the midbody ring protein CEP55 and inhibits recruitment of the ESCRT I protein TSG-101, thus effectively blocking the completion of abscission (Greenbaum et al., 2007; Iwamori et al., 2010). Whether blocking abscission is a conserved feature of germline formation is unclear.
The C. elegans adult germline is organized as a syncytium in which each germ cell possesses a stable intercellular bridge that connects it to a central core of common cytoplasm, known as the rachis (Hubbard and Greenstein, 2005). The ring that stabilizes each germ cell intercellular bridge is enriched in regulators of actomyosin contractility that are also found in cytokinetic rings, such as actin, the nonmuscle myosin II NMY-2, the centralspindlin subunits CYK-4 and ZEN-4, as well as two anillin proteins, ANI-1 and ANI-2 (Maddox et al., 2005; Zhou et al., 2013; Amini et al., 2014). In Drosophila and cultured cells, anillin was shown to function as an adaptor protein that scaffolds contractility regulators to promote cytokinetic ring positioning and constriction (Oegema et al., 2000; Hickson and O’Farrell, 2008; Piekny and Glotzer, 2008). Anillin is proposed to achieve this in part by binding to the plasma membrane via its C-terminal Pleckstrin homology (PH) and C2 domains (Oegema et al., 2000; Liu et al., 2012; Sun et al., 2015), and interacting with actomyosin contractility regulators, such as RhoA, MgcRacGAP/RacGAP50, and Ect2, through its actin, myosin, and anillin homology (AH) domains (Straight et al., 2005; D’Avino et al., 2008; Gregory et al., 2008; Piekny and Glotzer, 2008; Frenette et al., 2012). Disruption of anillin interaction with its partners in these systems leads to cytokinesis failure (Oegema et al., 2000; Straight et al., 2005; Kechad et al., 2012). In addition to canonical ANI-1, C. elegans stable germ cell intercellular bridges are enriched in the shorter, noncanonical anillin protein ANI-2, which lacks N-terminal putative actin- and myosin-binding domains, but retains the C-terminal AH and PH domains (Maddox et al., 2005; Amini et al., 2014). Animals depleted of ANI-2 display a disorganized germline made of incomplete partitions comprising multiple nuclei (Green et al., 2011; Amini et al., 2014), indicating a role for ANI-2 in the organization of stable germ cell intercellular bridges and adult gonad architecture.
How the syncytial organization of the germline arises during C. elegans development, and especially whether it involves some degree of cytokinesis incompletion, is not known. All C. elegans germ cells originate from a single germline precursor blastomere, termed P4, which is born after successive asymmetric divisions during embryogenesis (Wang and Seydoux, 2013). Around the 100-cell stage, P4 divides into the two primordial germ cells (PGCs), Z2 and Z3, which remain mitotically quiescent throughout embryogenesis and initiate proliferation after hatching, at the mid-L1 larval stage, to give rise to the entire syncytial germline (Hirsh et al., 1976; Sulston et al., 1983). We have previously shown that, whereas ANI-1 is present in all cells, ANI-2 is specifically found in the P4 blastomere during embryogenesis, localizes to the nascent furrow during P4 division, and is enriched at the midbody ring between Z2 and Z3 during cytokinesis (Amini et al., 2014). Here we demonstrate that the P4 blastomere initiates cytokinesis but does not complete it, resulting in a stable cytoplasmic bridge between the two PGCs. We show that the accumulation of several contractility regulators at the cytoplasmic bridge, including the two C. elegans anillin proteins, is essential to ensure its stability throughout embryogenesis and may contribute to the formation of a fully organized and functional adult germline.
RESULTS
The C. elegans germline precursor blastomere does not undergo abscission
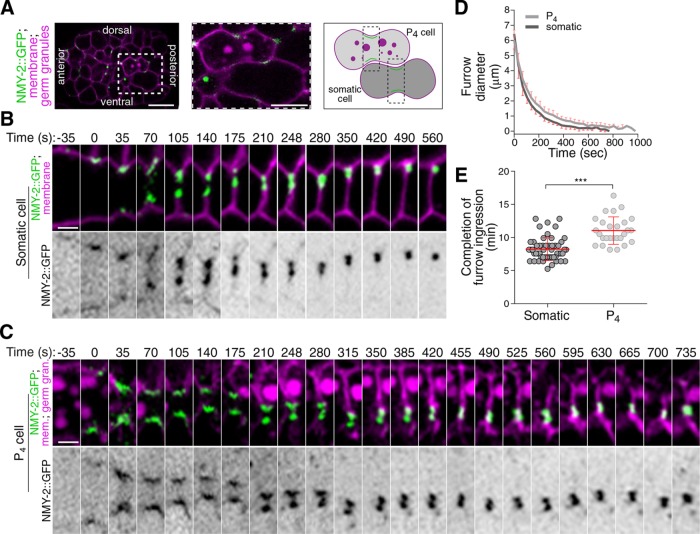
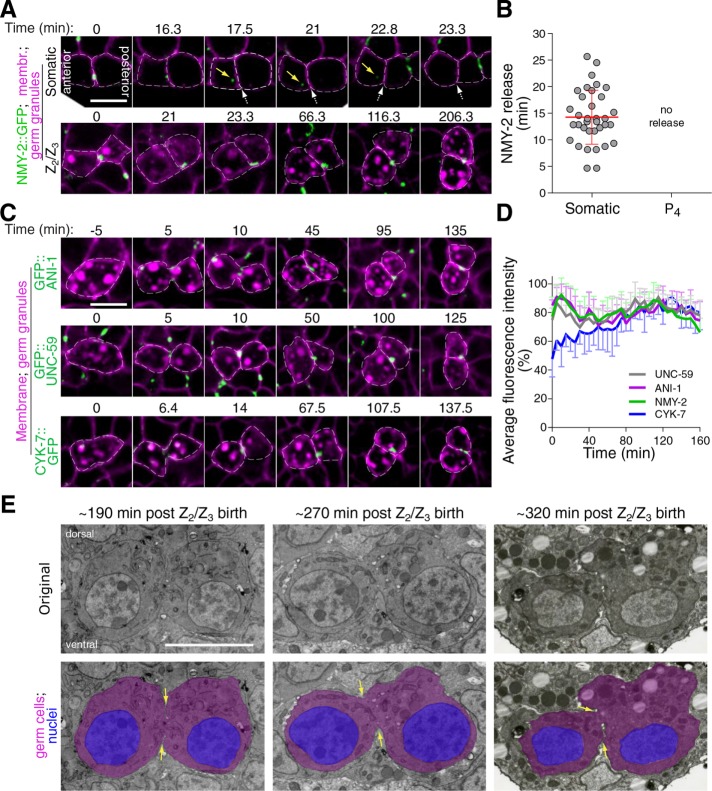
Incomplete cytokinesis is a common feature of germline development in many animals and can promote its syncytial organization. As the C. elegans germline is syncytial, we asked whether the germline precursor blastomere P4 undergoes incomplete cytokinesis during embryogenesis. This was done by monitoring the localization of GFP-tagged nonmuscle myosin II (NMY-2::GFP) in embryos coexpressing fluorescently tagged markers for the plasma membrane (TagRFP fused to the PH domain of PLCδ, hereafter TagRFP::PH) and germ cells (PGL-1::RFP), and comparing events occurring in the dividing P4 blastomere with those of neighboring somatic cells (Figure 1, A–C, and Supplemental Figure S1). Cytokinesis was previously shown to broadly occur in four successive phases: contractile ring assembly, contractile ring ingression, cytoplasmic isolation, and midbody ring release (Green et al., 2012, 2013; D’Avino et al., 2015; Konig et al., 2017). We found that the first phases of cytokinesis were normal in the P4 blastomere and that it underwent proper completion of furrow ingression in all embryos (n = 27; Figure 1C and Supplemental Movie S1), although ingression dynamics were slightly but significantly delayed compared with somatic cells (Figure 1, D and E). Interestingly, however, measuring the timing of midbody ring-associated NMY-2::GFP release from the interstitial membrane separating daughter cells revealed striking differences between P4 and somatic cells (Supplemental Figure S1, D–F). In somatic cells, we found that the midbody ring took on an average 14.3 ± 5.1 min to dissociate from the interstitial membrane separating the two sister cells (n = 35; Figure 2, A and B). In sharp contrast, the NMY-2::GFP signal remained present at the PGC interstitial boundary, even up to 200 min after the completion of furrow ingression (n = 30; Figure 2, A and B, and Supplemental Movie S1), and its intensity was stable over this entire period (Figure 2D). Similar results were obtained upon monitoring the localization of GFP-tagged versions of the canonical anillin ANI-1, the septin UNC-59, and the midbody-associated protein CYK-7 (Figure 2, C and D). Although movement of the developing embryo precluded us from monitoring NMY-2::GFP dynamics after this time, immunofluorescence analysis on fixed specimens revealed that NMY-2 is continuously enriched between the two PGCs during all remaining stages of embryogenesis (Supplemental Figure S2E). Accordingly, the monitoring of Z2 and Z3 by transmission electron microscopy in embryos undergoing epithelial morphogenesis indicated a clear cytoplasmic bridge connecting the two cells (Figure 2E), in agreement with previously reported evidence of this structure in embryos and L1 larvae (Sulston et al., 1983; Lee et al., 2017). These results indicate that although the first phases of cytokinesis occur normally in the C. elegans P4 blastomere, it fails to undergo abscission and midbody ring release at the end of cytokinesis, resulting in the two PGCs maintaining a stable cytoplasmic bridge enriched in several contractility regulators.
FIGURE 1:
The dividing P4 blastomere undergoes normal furrow ingression. (A) Confocal images (maximum intensity projection of three consecutive Z planes) of a control embryo expressing NMY-2::GFP (green), a membrane marker (TagRFP::PH; magenta), and a germ granule marker (PGL-1::RFP; magenta). The region delineated by the white dashed square is magnified by fourfold in the inset and shows a somatic cell and a P4 cell upon initiation of cytokinetic furrow ingression. The schematic illustration depicts the region of interest for monitoring furrow progression. (B, C) Time-lapse confocal images of the division plane in a somatic (B) and P4 (C) cell from a control embryo expressing the markers described in A. Time 0 (seconds) corresponds to the onset of cytokinetic furrow initiation. (D, E) Quantification of cytokinetic furrow diameter (D) and duration of cytokinetic furrow ingression (E) from the onset of furrow initiation to the completion of furrow ingression in wild-type somatic (n = 54; dark gray) and P4 (n = 27; light gray) cells. ***: p < 0.001. Scale bar for embryo in A, 10 μm; scale bar for insets in B and C, 2 μm.
FIGURE 2:
The PGCs maintain a stable cytoplasmic bridge enriched in contractility regulators. (A) Midsection confocal time-lapse images of sister somatic cells (top panels) and PGCs (bottom panels) from a control embryo expressing NMY-2::GFP (green), a membrane marker (TagRFP::PH; magenta), and a germ granule marker (PGL-1::RFP; magenta). Time 0 corresponds to completion of furrow ingression. Yellow arrows point to the midbody ring-associated NMY-2 focus upon release in the cytoplasm and white dashed arrows indicate the position in the membrane from which the midbody ring was released (top panels only). (B) Quantification of the time of midbody ring release relative to the time of completion of furrow ingression in control somatic and germ cell siblings. The red bar represents average ± SD. (C) Midsection confocal time-lapse images of control germ cells expressing membrane and germ granule markers (both in magenta) and one of three midbody ring markers (GFP::ANI-1, GFP::UNC-59, or CYK-7::GFP; green). Time 0 corresponds to completion of furrow ingression. (D) Quantification of the average fluorescence intensity (± SD) of the unreleased NMY-2::GFP, GFP::ANI-1, GFP::UNC-59, and CYK-7::GFP foci at the stable cytoplasmic bridge relative to the time of completion of furrow ingression in control PGCs. (E) Selected transmission electron microscopy images from 60-nm-thick sections of PGCs of wild-type embryos fixed 350 min (left panel), 430 min (middle panel), or 480 min (right panel) after first division. Time on the figure represents an estimate from the time of completion of furrow ingression in P4. Overlay images with colors show the cytoplasmic bridge (yellow arrows) connecting the cells’ cytoplasm (magenta), with nuclei in blue. All scale bars represent 5 μm.
Contractility regulators stabilize the cytoplasmic bridge between the two PGCs
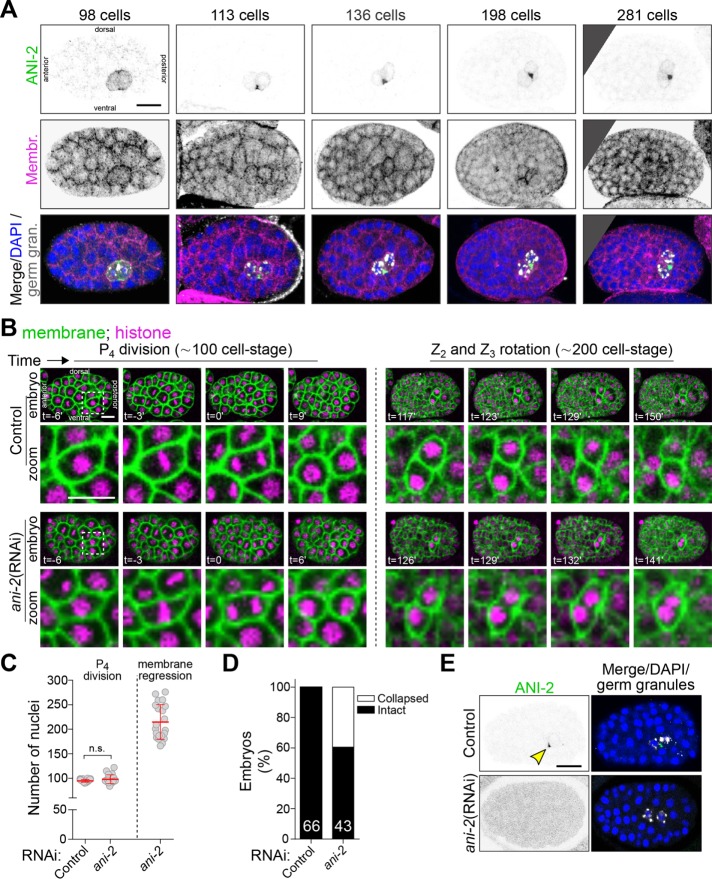
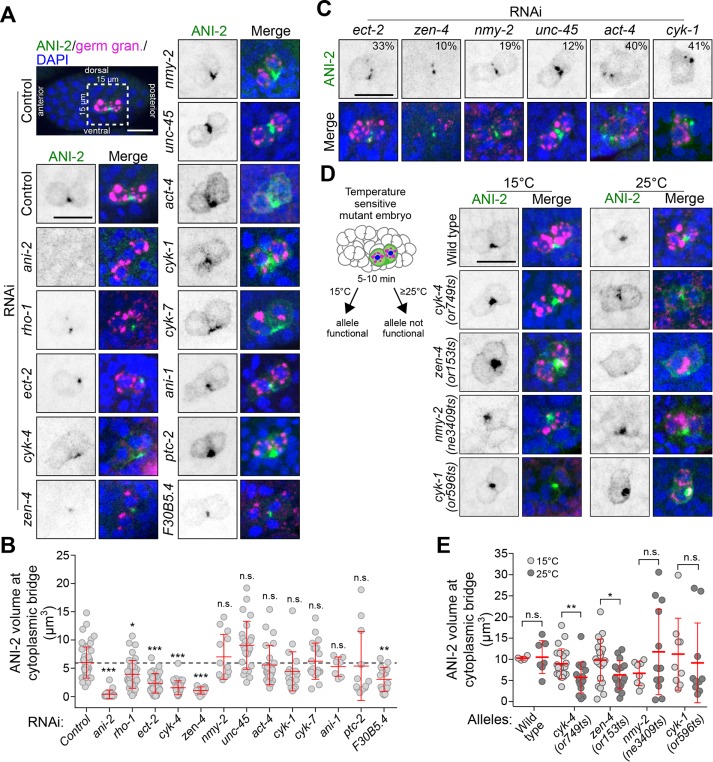
We sought to identify regulators of germ cell cytokinesis and/or PGC formation. We first asked whether the short anillin protein ANI-2 plays a role in this process, as ANI-2 possesses many properties that would make it a likely candidate: it is present specifically in the P4 blastomere during embryogenesis, it is required for intercellular bridge stability in the C. elegans adult germline, and it was proposed to function as a negative regulator of contractility (Chartier et al., 2011; Amini et al., 2014; Pacquelet et al., 2015). Using immunofluorescence, we found that ANI-2 accumulated at the cytoplasmic bridge between Z2 and Z3 and remained visible as a focus between the two PGCs up to the onset of epithelial morphogenesis at the ∼350-cell stage (Figure 3A), similar to other contractility regulators (Figure 2C and Supplemental Figure S2). To determine whether this embryonic pool of ANI-2 regulates PGC formation, we used RNA interference (RNAi) to deplete ANI-2 from animals expressing fluorescent markers enabling the visualization of plasma membrane (mNeonGreen::PH) and cell nuclei (mCherry::H2B; Figure 3B). Live imaging of ANI-2–depleted embryos revealed that they proceeded through early development normally and that, as in controls, P4 divided at the ∼100-cell stage (Figure 3, B and C). Interestingly, however, the membrane partition separating Z2 and Z3 eventually regressed in 49% of ANI-2-depleted embryos (n = 43), on average 137 ± 23 min after the membrane had visibly completed its ingression (Figure 3, B and D, and Supplemental Movie S2). Regression occurred on average at the 215 ± 36–cell stage (Figure 3C), near the end of gastrulation, when Z2 and Z3 typically undergo rotation in the long axis of the embryo (Chisholm and Hardin, 2005). RNAi-mediated depletion of ANI-2 was effective as judged by the strong decrease in ANI-2 signal in immunofluorescence experiments (Figure 3E). No membrane regression was observed in somatic cells, in which ANI-2 is undetectable. These results indicate that ANI-2 is not essential for cytokinetic furrow ingression or abscission incompletion in the P4 germline precursor blastomere but is required to stabilize the cytoplasmic bridge between the two PGCs during embryogenesis.
FIGURE 3:
Depletion of ANI-2 causes membrane collapse between the two PGCs. (A) Confocal images (maximum intensity projections of five to seven consecutive Z planes) of control embryos between the ∼100- and ∼300-cell stages expressing a membrane marker (GFP::PH), and fixed and stained with antibodies against GFP (magenta), ANI-2 (green), germ granules (white), and DAPI (blue). (B) Confocal images (maximum intensity projections of two consecutive z-stacks) of control and ani-2(RNAi) embryos expressing membrane (mNG::PH; green) and histone (mCherry::H2B; magenta) markers during birth of the two PGCs. The region delineated by the white dashed square around P4 is magnified by 3.5-fold in the inset. Bottom left, time relative to furrow initiation. (C) Quantification of the number of nuclei upon P4 division and membrane collapse in control and ani-2(RNAi) embryos. The red bars represent average ± SD. n.s. = p > 0.05. (D) Percentage of collapsed partitions between the two PGCs in control and ani-2(RNAi) embryos. The sample size (n) is indicated within each bar. (E) Midsection confocal images of control and ani-2(RNAi) embryos fixed and stained with antibodies against ANI-2 (green), germ granules (white), and DAPI (blue). Yellow arrowhead points to the ANI-2 focus in the control. All scale bars represent 10 μm.
We next sought to identify other regulators of PGC cytoplasmic bridge stabilization among genes whose depletion was previously reported to phenocopy the loss of ANI-2 function in the adult germline and/or genes encoding cytokinetic regulators (22 genes; see Supplemental Table S2 for a complete list; Green et al., 2011). We used partial RNAi-mediated depletion conditions for each gene, as the complete depletion of many of these would cause a failure in the first embryonic division, thus precluding a study of Z2 and Z3 later in embryogenesis. For each gene tested, our depletion regime resulted in a small number (<10%) of binucleated somatic cells and a high percentage of embryonic lethality, but yet allowed the division of P4 and birth of the two PGCs (Supplemental Figure S3A; see Materials and Methods for details).
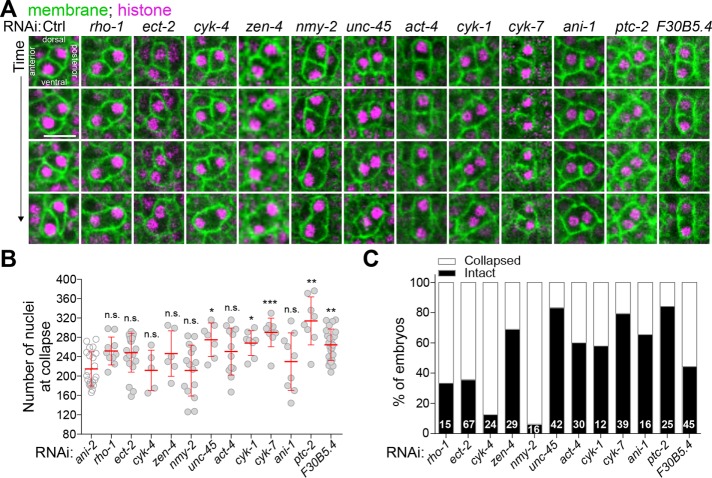
Live imaging of embryos depleted of the different genes of interest revealed that in all cases in which the initial steps of P4 blastomere cytokinesis were not compromised, P4 completed cytokinetic furrow ingression at the ∼100-cell stage, as in control. However, we observed a variable proportion of embryos in which the membrane between Z2 and Z3 had regressed after depletion of 12 of the 22 genes, a phenotype similar to that observed after ANI-2 depletion (Figure 4A). These genes encode the small GTPase Rho-1 and its guanine-nucleotide exchange factor ECT-2, the two centralspindlin subunits CYK-4 and ZEN-4 (Mishima et al., 2002), the nonmuscle myosin II NMY-2, the myosin chaperone UNC-45 (Kachur et al., 2008), the actin subunit ACT-4, the formin CYK-1, the canonical anillin ANI-1, the midbody-associated component CYK-7 (Green et al., 2011), the noncanonical Patched receptor PTC-2 (Kuwabara et al., 2000), and F30B5.4, a putative homologue of the mammalian oxidative stress-induced growth inhibitors, OSGIN1/2 (Huynh et al., 2001). Notably, the construct enabling RNAi depletion of ACT-4 could target other genes encoding actin subunits, including act-2 which is known to be expressed in early embryos (Willis et al., 2006), and thus likely reveals a general role for actin as opposed to a specific requirement for ACT-4 in this process. Most PGC membranes collapsed near the embryonic stage where collapse was observed in ANI-2-depleted embryos, on average 131 ± 44 min after the membrane had visibly completed its ingression, and some collapse events occurred at even later stages (Figure 4B). This situation is different from that of the few somatic cells that also became binucleated in these depleted embryos, which either failed membrane ingression altogether or underwent membrane collapse within 10 min after the start of ingression. This strongly suggests that the membrane collapse events observed in PGCs are not due to a general cytokinetic failure. The proportion of embryos that displayed a membrane regression phenotype varied depending on the gene depleted (Figure 4C). These variations could reflect different requirements for these proteins in regulating membrane stability but are more likely a consequence of partial RNAi-mediated depletions in each case. Insufficient depletion could also account for the absence of membrane collapse in 10 of the 22 regulators tested, although expected phenotypes were observed in each case, including defects in embryonic viability (Supplemental Figure S3A) and cytokinesis (Supplemental Figure S3B). Taken together, these results indicate that several contractility regulators are needed to stabilize the cytoplasmic bridge between the two PGCs during C. elegans embryogenesis.
FIGURE 4:
Several contractility regulators are required to stabilize the PGC cytoplasmic bridge. (A) Midsection confocal time-lapse images of PGCs from embryos expressing membrane (mNG::PH; green) and histone (mCherry::H2B; magenta) markers. Animals were differentially depleted of contractility regulators by RNAi (see Supplemental Table S2) and stability of the PGC cytoplasmic bridge was assessed by monitoring membrane signal. Scale bar, 10 μm. (B, C) Quantification of the number of nuclei at membrane collapse (B) and percentage of embryos undergoing membrane collapse (C) for each condition. The sample size (n) is indicated within each bar. The red bars represent average ± SD.
The Rho pathway promotes ANI-2 localization at the stable PGC cytoplasmic bridge
To better understand the mechanism of PGC cytoplasmic bridge stabilization, we next asked whether the regulators of bridge stability function within the same pathway by monitoring their reciprocal localization. ANI-2 accumulates specifically in the P4 blastomere (Amini et al., 2014) and thus we first determined which regulators of PGC cytoplasmic bridge stability function upstream of ANI-2 in this process. Because we were unable to generate animals producing embryos that express ANI-2 fused to a fluorescent marker, ANI-2 accumulation was scored in fixed specimens by immunofluorescence using specific antibodies and its amounts were determined by measuring the volume of the ANI-2 focus between Z2 and Z3 (Supplemental Figure S3, C and D; see Materials and Methods for details). In live embryos expressing CYK-7::GFP (another stable cytoplasmic bridge protein; see below), we found that the intensity and volume of the CYK-7::GFP fluorescence signal are directly proportional (compare Figure 2D and Supplemental Figure S2B). This method therefore provides a reliable measurement of ANI-2 accumulation at the cytoplasmic bridge.
Hermaphrodite animals were partially or strongly depleted of the different proteins of interest by RNAi (as above) and the volume of the ANI-2 focus between Z2 and Z3 was measured in embryos containing 150–250 nuclei, the stage at which the membrane between PGCs receded in ani-2(RNAi) embryos. As expected, depleting ANI-2 by RNAi resulted in a strong reduction of the ANI-2 signal and focus volume compared with control (Figure 5, A and B), providing a comparative baseline for this assay. Among the proteins required to maintain a stable PGC cytoplasmic bridge, we found that depletion of five of them resulted in a significant decrease in the volume of the ANI-2 focus: the two centralspindlin subunits CYK-4 and ZEN-4, RHO-1, ECT-2, and F30B5.4 (Figure 5, A and B). Mispositioning of the ANI-2 focus at cortical regions other than the Z2/Z3 interstitial boundary was also observed in ∼30% of embryos depleted of RHO-1 or CYK-4, and ∼7% of ECT-2–depleted embryos. These results suggested that ANI-2 could function downstream from Rho signaling in this process. To test this hypothesis, we analyzed the localization of CYK-4::GFP in embryos depleted of ANI-2 and other contractility regulators. We found that CYK-4::GFP colocalizes with ANI-2 at the stable PGC cytoplasmic bridge in control embryos, and that CYK-4::GFP accumulation at the bridge was not perturbed by depletion of ANI-2 or any of the other contractility regulators tested herein (Supplemental Figure S4, A and B). The ANI-2 focus volume in these CYK-4::GFP embryos was similar to that previously measured for each condition, thus serving as control for the efficacy of each RNAi depletion. These results indicate that Rho pathway regulators and F30B5.4 function upstream of ANI-2 to promote its localization at the stable PGC cytoplasmic bridge.
FIGURE 5:
Rho pathway regulators promote ANI-2 accumulation at the stable PGC cytoplasmic bridge. (A, C, D) Confocal images (maximum intensity projections of five to seven consecutive Z planes) of PGCs in 150- to 250-cell-stage embryos, fixed and stained with antibodies against ANI-2 (green), germ granules (magenta), and DAPI (blue). The insets depict PGCs from embryos after RNAi depletion of the genes indicated (A, C) or PGCs from embryos bearing fast-acting, temperature-sensitive alleles of the genes indicated and acquired after continuous growth at permissive temperature (15°C) or after a 5- to 10-min upshift at restrictive temperature (25°C), as depicted in the schematic (D). In C, the number (%) represents the proportion of embryos displaying fragmentation of the ANI-2 focus. Scale bars, 10 μm. (B, E) Quantification of the focus volume of ANI-2 at the PGC cytoplasmic bridge in embryos depleted of the indicated genes by RNAi (B) or in embryos bearing the temperature-sensitive alleles of the indicated genes and maintained at permissive (15°C) or restrictive (25°C) temperature (E). The red bars represent average ± SD. *: p < 0.05; **: p < 0.01; ***: p < 0.001; n.s.: p > 0.05.
In addition, depletion of ECT-2, ZEN-4, NMY-2, UNC-45, ACT-4, and CYK-1 lead to a variable proportion of embryos displaying fragmentation of the ANI-2 signal into multiple smaller foci (Figure 5C and Supplemental Figure S3G). The combined volume of these fragmented foci was indistinguishable from that of control embryos in which fragmentation was not observed. These defects in ANI-2 organization may be a consequence of Z2/Z3 interstitial membrane collapse, because they are more frequent in conditions that induce the highest frequency of collapse. Depletion of ANI-1, CYK-7, PTC-2, or any of the 10 proteins that did not cause membrane collapse had no effect on the accumulation of ANI-2 at the cytoplasmic bridge or its organization into a proper focus (Figure 5, A and B, and Supplemental Figure S3, E and F). These results indicate that most proteins whose depletion phenocopies that of ANI-2 in PGCs regulate the accumulation and/or organization of ANI-2 at the stable PGC cytoplasmic bridge.
To validate the results obtained by RNAi depletion, we took advantage of available mutants bearing fast-acting, temperature-sensitive (ts) alleles for cyk-4, zen-4, nmy-2, and cyk-1 (Davies et al., 2014). These mutant alleles provided us with the means to rapidly inactivate gene products after the birth of the two PGCs and thus exclude the possible confounding effects of early phenotypic alterations that remain a constant drawback of RNAi-mediated depletions. Embryos were upshifted to the restrictive temperature for 5–10 min before fixation and staining (Figure 5D; see Materials and Methods), and analysis of the ANI-2 focus volume was performed on fixed 150- to 250-cell-stage embryos, as above. In all mutant backgrounds, most embryos contained a few binucleated somatic cells, thus validating that the temperature upshift caused gene product inactivation without otherwise resulting in a gross membrane disorganization phenotype. While temperature upshift did not change the volume of the ANI-2 focus in wild-type embryos, the same treatment caused a significant reduction of the ANI-2 focus volume in both cyk-4(ts) and zen-4(ts) mutant embryos, similar to what had been observed after RNAi depletion of these two genes (Figure 5, D and E). Upshifting nmy-2(ts) and cyk-1(ts) embryos did not decrease the ANI-2 focus volume, consistent with RNAi results (Figure 5, D and E). Together, these results are in agreement with those obtained by RNAi and indicate that genes encoding several contractility regulators, and particularly members of the Rho pathway, function with ANI-2 to regulate cytoplasmic bridge stability between the two C. elegans PGCs.
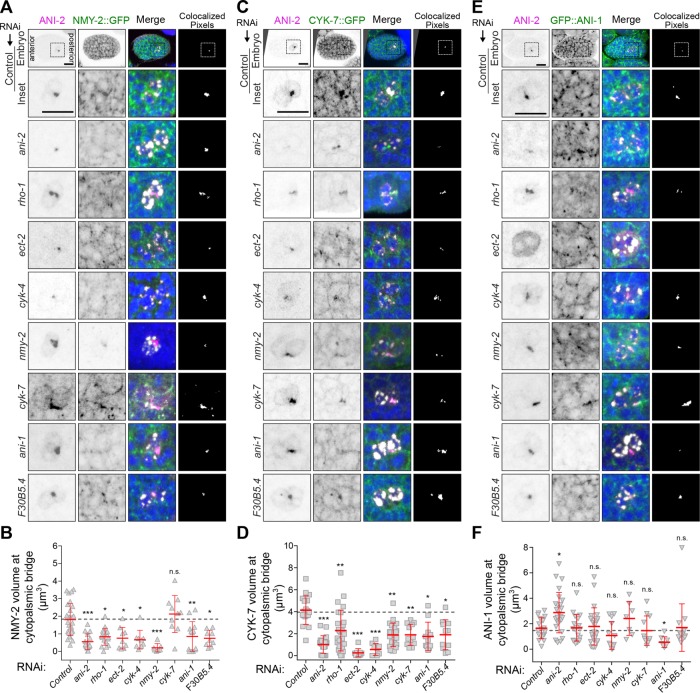
ANI-2 regulates the localization of NMY-2, CYK-7, and ANI-1 at the stable PGC cytoplasmic bridge
Although Rho pathway regulators promote the maintenance of ANI-2 at the stable PGC cytoplasmic bridge, several contractility regulators affect the stability of this bridge without impacting ANI-2 localization, raising the possibility that they could function downstream from ANI-2. To assess this, we looked at the localization of NMY-2 and CYK-7, two contractility regulators that colocalize with ANI-2 at the stable cytoplasmic bridge (Figure 6, A and C, and Supplemental Figure S2, C and E). We found that depletion of ANI-2 by RNAi significantly reduced the focus volume of both NMY-2::GFP and CYK-7::GFP at the stable cytoplasmic bridge between Z2 and Z3 (Figure 6, A–D). Similar results were obtained after RNAi depletion of the proteins required to localize ANI-2 at the stable bridge (Figure 6, A–D), consistent with those acting upstream of ANI-2 in the pathway. Interestingly, although depletion of NMY-2 caused a reduction of CYK-7 volume at the stable bridge (Figure 6, C and D), depleting CYK-7 did not affect NMY-2 localization (Figure 6, A and B), suggesting that NMY-2 acts upstream of CYK-7 in this pathway. Again, comonitoring of the ANI-2 focus volume in these embryos gave results similar to those previously measured for each condition and served as reference for the efficacy of RNAi depletion. These results suggest that ANI-2 promotes the localization of NMY-2 at stable PGC cytoplasmic bridges and that, in turn, this enables the localization of CYK-7 at this structure.
FIGURE 6:
ANI-2 regulates the localization of NMY-2, CYK-7, and ANI-1 at the stable PGC cytoplasmic bridge. (A, C, E) Confocal images (maximum intensity projections of five to seven consecutive Z planes) of PGCs in embryos at the 150- to 250-cell-stage expressing either NMY-2::GFP (A), CYK-7::GFP (C), or GFP::ANI-1 (E) and fixed and stained with antibodies against GFP (green), ANI-2 (magenta), germ granules (white), and DAPI (blue). Embryos were depleted of the genes indicated by RNAi. Scale bar, 10 μm. (B, D, F) Quantification of the focus volume of NMY-2::GFP (B), CYK-7::GFP (D), or GFP::ANI-1 (F) at the PGC cytoplasmic bridge in embryos depleted of the indicated genes by RNAi. The red bars represent average ± SD. *: p < 0.05; **: p < 0.01; ***: p < 0.001; n.s.: p > 0.05.
We and others previously demonstrated that the two C. elegans anillin proteins, the canonical ANI-1 and the short ANI-2, function opposite to each other in regulation of actomyosin contractility, in both the germline and the one-cell-stage embryo (Chartier et al., 2011; Amini et al., 2014; Pacquelet et al., 2015; Rehain-Bell et al., 2017). We therefore asked whether this antagonistic relationship was likewise required for PGC cytoplasmic bridge stability. We found that although ANI-1 is required to maintain a stable partition between the two PGCs (Figure 4), it is not required for CYK-4 or ANI-2 localization at the stable PGC cytoplasmic bridge (Figure 5, A and B, and Supplemental Figure S4, A and B), suggesting that it acts downstream from ANI-2 in this pathway. Depletion of ANI-1 caused a decrease of both NMY-2::GFP and CYK-7::GFP focus volumes at the stable bridge, indicating that ANI-1 acts upstream of these two regulators in the pathway (Figure 6, A–D). Consistent with this, we found that GFP::ANI-1 accumulates at the stable PGC cytoplasmic bridge and that its accumulation is independent of NMY-2 and CYK-7 (Figure 6, E and F). Interestingly, depletion of ANI-2 resulted in an increase in the GFP::ANI-1 focus volume at the stable bridge, indicating that ANI-2 acts to limit the amount of ANI-1 at this structure (Figure 6F). Strikingly, however, depleting Rho pathway regulators (CYK-4, ZEN-4, RHO-1, ECT-2) or F30B5.4, which are required to promote ANI-2 accumulation at the stable bridge (Figure 5, A and B), had no effect on GFP::ANI-1 accumulation (Figure 6, E and F). These results suggest that Rho pathway activity promotes the localization of both ANI-1 and ANI-2 at the stable PGC cytoplasmic bridge and that ANI-2 acts locally to limit the amount of ANI-1 at this structure, while actively promoting the accumulation of NMY-2 and CYK-7.
DISCUSSION
In this study, we show that the embryonic germline precursor cell P4 does not complete cytokinesis and that the two PGCs, Z2 and Z3, remain connected via a stable cytoplasmic bridge. We find that several actomyosin contractility regulators, including the canonical anillin ANI-1 and the short anillin ANI-2, remain stably enriched between the two PGCs and that their depletion results in a collapse of the membrane partition, contrary to what is observed in neighboring somatic cells. Our results thus support a model in which Rho pathway regulators promote the loading of anillin proteins at the stable PGC cytoplasmic bridge that is formed after P4 cytokinesis, which in turn promote the maintenance of NMY-2 and CYK-7 at the cytoplasmic bridge to ensure its stability (Figure 7).
FIGURE 7:
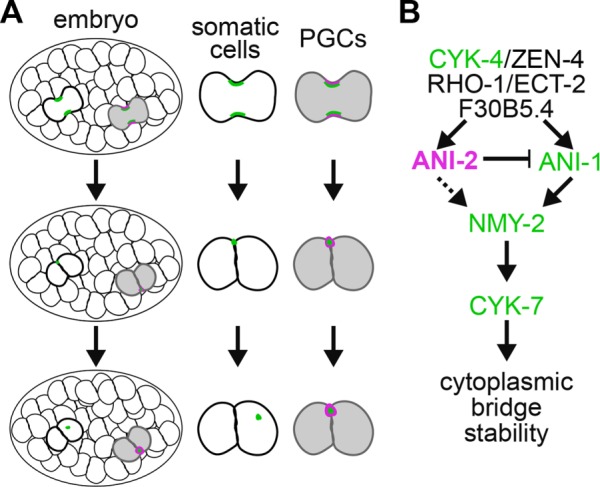
Regulation of PGC cytoplasmic bridge stability by actomyosin contractility regulators. (A) While somatic cells undergo proper midbody ring release (green) in wild-type embryos, the P4 blastomere does not complete cytokinesis and stably maintains several contractility regulators, including ANI-2 (magenta), at the PGC cytoplasmic bridge. (B) Working model depicting the proposed functional requirements for several contractility regulators in stabilizing the cytoplasmic bridge between the two PGCs during C. elegans embryogenesis. ANI-2 (magenta), ANI-1, CYK-4, NMY-2, and CYK-7 (green) all stably localize to the cytoplasmic bridge. Although ANI-2 functions upstream of ANI-1 to limit its accumulation at the stable cytoplasmic bridge, it is unclear whether it regulates NMY-2 and CYK-7 localization directly (dashed line) or through ANI-1. The epistatic relationship between regulators acting upstream of ANI-2 is unknown. See the main text for details.
Our results suggest that the centralspindlin complex proteins CYK-4 and ZEN-4 are at the top of this hierarchy because depletion of either of them results in a membrane collapse and no other regulator perturbs the localization of CYK-4 at the stable cytoplasmic bridge. This contrasts with a previous study proposing that ANI-2 acts upstream of CYK-4 to regulate the stability of intercellular bridges in adult C. elegans germ cells (Zhou et al., 2013). This apparent discrepancy could originate from differences in RNAi depletion regimes, which were longer for depletions done in adult animals and may have led to cumulative defects and more severe germline disorganization. Alternatively, it could derive from a differential requirement of contractility regulators at the stable intercellular bridge of embryonic and adult germ cells, in which the stable bridge is unlikely to directly arise from a cytokinetic ring. The requirement of Rho regulators in ANI-2 localization at the cytoplasmic bridge is consistent with ANI-2 containing C-terminal domains that, in anillin proteins of other species, were reported to bind to RhoA/RHO-1, MgcRacGAP/CYK-4, and the plasma membrane (Maddox et al., 2005; D’Avino et al., 2008; Gregory et al., 2008; Liu et al., 2012). Depleting F30B5.4 results in similar defects as RHO-1 or ECT-2 depletion; however, we currently do not know whether this regulator impinges on Rho signaling itself or where it acts between CYK-4 and ANI-2. Likewise, we do not know precisely where or how the other regulators of cytoplasmic bridge stability herein identified (UNC-45, PTC-2, CYK-1, ACT-4/actin) act in this pathway, although work in C. elegans and other systems allows us to speculate that the myosin chaperone UNC-45 works with NMY-2 (Kachur et al., 2008) and that the formin CYK-1 and actin ACT-4 function together, downstream from Rho and partly in parallel of the other regulators (Green et al., 2012; D’Avino et al., 2015).
Previous studies carried out in the one-cell-stage embryo and adult gonad revealed that the canonical C. elegans anillin ANI-1 and the short anillin ANI-2 have opposing activities on actomyosin contractility (Chartier et al., 2011; Amini et al., 2014; Pacquelet et al., 2015; Rehain-Bell et al., 2017). Our work here reveals that both ANI-1 and ANI-2 are required to localize NMY-2 and CYK-7 at the PGC cytoplasmic bridge and maintain its stability. The N-terminal region of ANI-1 contains putative actin- and myosin-binding domains that could enable ANI-1 to bind NMY-2 directly and thus mediate NMY-2 localization. Although these domains are not readily identified in ANI-2 (Maddox et al., 2005), we cannot exclude the possibility that ANI-2 could also directly interact with NMY-2 to affect its localization. However, whereas ANI-1 depletion has no noticeable effect on ANI-2 localization, depletion of ANI-2 results in an increase in ANI-1 accumulation at the stable cytoplasmic bridge, suggesting that ANI-2 acts upstream of ANI-1 in this cellular context. Another possibility is that ANI-2 exerts its effect on NMY-2 entirely through ANI-1 recruitment at the stable cytoplasmic bridge, a scenario in which both a decrease or an increase in ANI-1 levels would impede NMY-2 localization. In adult animals, ANI-2 was recently reported to act downstream of ANI-1 to limit the accumulation of NMY-2 at germline intercellular bridges (Rehain-Bell et al., 2017), suggesting that the two anillin proteins display differences in their scaffolding activities depending on the context. Interestingly, ANI-1 accumulation at the stable bridge did not vary upon depletion of the upstream Rho pathway regulators that are required for ANI-2 accumulation, suggesting that they promote the localization of ANI-1 at the stable cytoplasmic bridge independently of their activity on ANI-2 and that ANI-1 accumulation at the stable bridge is balanced by these two opposing activities. A Rho-dependent requirement for ANI-1 recruitment is consistent with the previously described function for Rho regulators acting upstream of anillin and enabling its cortical recruitment in other systems (Hickson and O’Farrell, 2008; Piekny and Glotzer, 2008). Alternatively, more complex molecular interactions between Rho regulators and C. elegans anillin proteins may impinge on their localization dynamics at the stable PGC cytoplasmic bridge.
We find that the presence of actomyosin contractility regulators at the stable cytoplasmic bridge is required around the 200-cell-stage to prevent its collapse. This coincides with a reorganization of embryonic tissues during which the two PGCs initiate a rotation along the antero-posterior axis of the embryo (see Figures 2 and 5). Although the exact role of this rotation event is unclear, it is conceivable that the forces enabling rotation of the two PGCs cause mechanical stress, and the presence of the short anillin ANI-2 at the stable bridge may be required to compensate for this stress and prevent membrane regression. We previously showed that the presence of ANI-2 at stable intercellular bridges in adult germ cells enables them to sustain deformation and that adult ani-2(-) mutant animals also display a severe membrane collapse defect that begins to manifest itself when oogenic cytoplasmic flows initiate in the gonad, an event that we postulated causes stress on the intercellular bridges (Amini et al., 2014). A balanced regulation between short and long anillin isoforms at cytoplasmic bridges may therefore be a selected molecular mechanism to ensure that they remain stable across a range of conditions that impose mechanical stress and deformation.
Finally, our findings indicate that the P4 blastomere does not complete abscission and leaves a stable cytoplasmic bridge between Z2 and Z3 that is still present in L1 larvae (Lee et al., 2017). Using photoconversion of a caged fluorescent dye, we previously demonstrated that there is little or no cytoplasmic exchange between the two PGCs soon after their birth (Amini et al., 2014). This suggests that the stable PGC cytoplasmic bridge is initially very small or physically blocked, perhaps by the midbody itself, which was previously shown to act as a diffusion barrier in other cell types (Sanger et al., 1985; Guizetti et al., 2011; Green et al., 2013). Germ cell abscission failure has been characterized in other organisms such as mouse and Drosophila and has been shown to promote germ cell syncytial organization (Hime et al., 1996; Greenbaum et al., 2007; Greenbaum et al., 2011). Several contractility regulators that we show localize to the stable PGC cytoplasmic bridge during embryogenesis are also found enriched at stable intercellular bridges connecting germ cells to the rachis in adult C. elegans animals (Zhou et al., 2013; Amini et al., 2014; Rehain-Bell et al., 2017), suggesting a functional relationship between the two developmentally distinct structures. Every adult germ cell in the C. elegans gonad appears to be connected to the rachis by an intercellular bridge, however, making it unlikely that this adult structure would directly arise from the single stable bridge connecting the two PGCs in embryos. Whether the stable PGC cytoplasmic bridge is required later for syncytial organization will require further investigation.
MATERIALS AND METHODS
C. elegans strain maintenance
Animals were cultured as previously described (Brenner, 1974) and grown at 20°C, with the exception of animals bearing temperature-sensitive alleles, which were maintained at 15°C. For all experiments, worms were synchronized at the L1 larval stage by hatching embryos in M9 buffer (0.022 mM KH2PO4, 0.042 mM Na2HPO4, 0.086 mM NaCl, 1 mM MgSO4) after sodium hypochlorite treatment (1.2% NaOCl, 250 mM KOH) and allowed to grow on OP50 bacteria to the desired developmental stage. Strains used in this study are listed in Supplemental Table S1.
RNAi-mediated depletions
To prepare RNAi plates, stationary-phase cultures of HT115 bacteria transformed with individual clones from the Ahringer library (all clones were verified by sequencing; see Supplemental Table S2 for details; Kamath et al., 2003) were diluted 1:60 and grown at 37°C for ∼2.5–3 h, to an OD600 of 0.4–0.6 (log phase). IPTG (1 mM) was added to the cultures before seeding onto nematode growth medium plates containing 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and 100 µg/ml carbenicillin, to a final concentration of 2 mM. RNAi plates were left to dry for 24 h. Adult animals were added to plates and depletion time courses were performed by feeding each RNAi clone in Supplemental Table S2 and monitoring phenotypes every 2 h for 48 h to find conditions in which a protein of interest was depleted enough to see an effect on embryonic viability (∼50% hatching failure), cytokinesis (as observed by binucleation of some somatic cells) and still allow the division of P4 into Z2 and Z3 (Supplemental Figure S3A). In each experiment, an RNAi clone containing an empty vector was used as control.
RNAi clones targeting the genes rho-1, ptc-1, and ptc-2 could not be recovered from the Ahringer library and were thus constructed by amplifying the cDNA of each gene by PCR (using the specific primer pairs designed in Sonnichsen et al. (2005), cloning the PCR product in vector L4440 and transforming ligated products into Escherichia coli strain HT115. All clones were verified by sequencing. Primer pairs were as follows: rho-1 forward: 5′-AAAAAGATATCATCGTCTGCGTCCACTCTCT-3′, rho-1 reverse: 5′-AAGGCTCGAGCTCGGCTGAAATTTCCAAAA-3′; ptc-1 forward: 5′-AAGAAAGATCTGATCGAATCTGCTGGTTGTG-3′, ptc-1 reverse: 5′-AAAAAGCTTCATTTTTCGAGAGAGCTGGC- 3′; ptc-2 forward: 5′-AAGAAAGATCTGAGATTCAAGCAGAGCCTGG-3′, ptc-2 reverse: 5′AAAAAGCTTGAGCACAGAATGATCGCAGA-3′.
Live imaging
Embryos were obtained by cutting open gravid hermaphrodites in M9 buffer using two 25-gauge needles and mounted on a poly-l-lysine–coated coverslip. The coverslip was flipped on a 3% agarose pad on a glass slide and sealed with valap (vaseline:lanolin:paraffin, 1:1:1). To image P4 cytokinesis and midbody release, two to four cell-stage embryos were manually sorted before mounting and incubated at 20°C for 5 h until the embryos had reached the desired developmental stage (i.e., P4 before division). Images were acquired with a 2 × 2- or 3 × 3-binned Axiocam 506 camera (Zeiss) mounted on a Zeiss Observer Z1 inverted microscope equipped with a Yokogawa CSU-X1 spinning disk confocal head and illuminated with 488 nm and 561 nm laser lines controlled by Zen software (Zeiss). Embryos were visualized with either a 40×/1.3 NA or a 63×/1.4 NA Plan Apochromat oil objective and z-stacks (0.5 µm or 0.75 μm) sectioning the entire embryo were acquired every 35 s for 2 h (Figures 1, A–E, and 2, A and B) or every 5 min for 5 h (Figure 2, C and D).
To visualize cytoplasmic bridge integrity, embryos were mounted as above and images were acquired with a Nikon A1R laser-scanning confocal microscope and illuminated in single-track mode with 488 nm and 561 nm laser lines controlled by NIS Elements 4.2 software (Nikon). Embryos were visualized with an Apo 40×/1.25 NA water-immersion objective and z-stacks (0.75 µm) sectioning the entire embryo were acquired every 3 min for 3–5 h. Images were processed and analyzed from the original files using ImageJ software.
Indirect immunofluorescence
Following RNAi depletions, embryos were collected by hypochlorite treatment, washed in M9 buffer, and deposited in 15 µl of M9 on a 14 × 14-mm patterned Cel-Line slide (Thermo Fisher Scientific) coated with 0.1% poly-l-lysine. A coverslip was placed on top of the embryos and the slide immediately put on a prechilled metal block on dry ice for 30 min. The coverslip was flipped to crack the eggshell and the slide immediately immersed in −20°C methanol for 20 min. The slide was allowed to dry and the sample was rehydrated in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4) for 5 min and then thrice with PBS containing 0.05% Tween 20 (PBST) for 5 min each, and 30 min in blocking buffer (PBST, 10% donkey serum). To diminish background due to nonspecific antibody interactions, primary and secondary antibodies were added sequentially. Samples were first incubated overnight at 4°C in 25 µl of PBST with primary antibodies (rabbit anti-ANI-2 [1:1000; Maddox et al., 2005] and mouse anti-P granules [clone OIC1D4, 1:300; Strome, 1986]). After three washes of 5 min in PBST, secondary antibodies (Alexa Fluor 568–coupled donkey–anti-rabbit and Alexa Fluor 647–coupled donkey–anti-mouse [1:500 each; Invitrogen]) were applied for 90 min at room temperature followed by three washes in PBST. Samples were then incubated for 90 min at room temperature in 25 µl of PBST with goat anti-GFP antibodies (1:1000; Rockland), followed by three PBST washes and incubation with Alexa Fluor 488–coupled donkey–anti-goat for another 90 min at room temperature. Samples were first washed for 20 min with PBST containing DAPI (1 µg/ml) and two additional PBST washed. Samples were mounted in Prolong Gold (Invitrogen) and allowed to dry overnight at room temperature before being imaged. Images were acquired with a Nikon A1R laser-scanning confocal microscope and illuminated in single-track mode with 405, 488, 561, and 640 nm laser lines controlled by NIS Elements 4.2 software (Nikon). Embryos were visualized with a 63×/1.4 NA Plan Apochromat oil objective and z-stacks (0.5 µm) sectioning the entire embryo were acquired. Images were processed and analyzed from the original files using ImageJ software.
Temperature-sensitive mutant embryos were processed as above except for a few differences in how they were initially collected and processed. They were first collected and washed in solutions at 15°C before being divided into two populations. Each population was suspended in M9 buffer at either 15°C (permissive, control) or 30°C (restrictive) and placed for 5 min in 15 and 25°C incubators, respectively. Embryos were collected by centrifugation (for 1.5 min) at the permissive or restrictive temperature and immediately processed as above for freeze cracking, fixation, staining, and imaging. Hence, the total treatment time at restrictive temperature was for a minimum of 5 min (initial incubation) and maximum of 10 min (subsequent processing time). This experimental setting enabled us to bypass essential gene requirements during P4 cytokinesis and monitor phenotypic consequences into PGCs that were born and developed at permissive temperature.
Quantitative image analysis
To determine the number of nuclei per embryo, confocal z-stacks were 3D reconstructed using Imaris software (version 7.6.5) to obtain voxel signals for an entire embryo. Using the “objects” tool, nuclei were defined and counted as objects in the DAPI (immunofluorescence) or mCherry::H2B (live) channel with a diameter of 2.4 ± 0.6 µm (depending on the embryonic stage).
The timings of completion of furrow ingression and midbody ring release during cytokinesis were determined from time-lapse images using ImageJ software. For each movie, average projection images of three consecutive Z planes were generated and fluorescence intensities of the membrane marker and tagged contractility proteins was measured along a 3-pixel-wide line drawn parallel (furrow ingression) or perpendicular (midbody release) to the cleavage furrow (see Supplemental Figure S1). Fluorescence background was measured in a different region of the cell and subtracted from the furrow measurements. Completion of furrow ingression was ascribed as the first time point when two ingressing peaks of fluorescence intensity coalesced into a single peak (apparent membrane closure). Midbody ring release was ascribed as the first time point when the peak of fluorescence intensity for a tagged contractility regulator became distant by more than 0.4 μm from the membrane marker at the cell boundary. Cytoplasmic bridge collapse was ascribed as the first time point when the continuous membrane fluorescence signal between the two PGCs separated into two distinct peaks of intensity (apparent membrane collapse).
The focus volume of proteins that stably localize between Z2 and Z3 was done on images from fixed or live samples using Imaris software. For each set of experiments, the control (RNAi) condition was used to set up the baseline values to do the 3D reconstruction of the cytoplasmic bridge foci as an isosurface (see Supplemental Figure S3C), using the “surface tool.” First, the intensity of the 3D rendering of the fluorescence signal (either ANI-2 or GFP) was adjusted to get an appropriate signal-to-noise ratio to visualize the focus between Z2 and Z3. Then using the “surface” tool, a cubic region was drawn around the Z2 and Z3 cells (identified using the germ granules signal) and an isosurface was automatically generated (using a surface “grain size” set to 0.1 µm) to obtain the 3D reconstruction. The isosurface was then adjusted manually (by changing the threshold values, which were kept thereafter for all foci quantified in that set of experiments) so both the rendering and the reconstruction would fit into one shape. Then, a filter was applied to exclude from the 3D reconstruction all 3D renderings having 10 voxels or less. Once reconstructed, the values given for the reconstruction’s volume (µm3) were used as markers of ANI-2 or GFP-tagged protein accumulation between Z2 and Z3.
Transmission electron microscopy
Wild-type embryos were prepared by two different methods for electron microscopy, with very similar outcomes. In one instance, isolated eggs were treated with chitinase to dissolve the eggshell (Wolf et al., 1983) and then fixed in buffered osmium tetroxide and potassium ferrocyanide at room temperature for 45 min, buffer rinsed, poststained in tannic acid, before dehydration and embedding in plastic resin (Durbin, 1987). In the second instance, isolated embryos were mounted on a glass slide in an agarose pad that was soaked in buffered glutaraldehyde fixative, before a laser was used to cut several holes in the eggshell. The glass slide was then moved to a humidified chamber for several hours to allow for fixation, followed by buffer rinses and a postfixation in buffered osmium tetroxide and potassium ferrocyanide as above. After further rinses, the embryo was poststained with uranyl acetate before dehydration and plastic resin embedment. All embryos were then thin sectioned using a diamond knife, and serial sections collected on Formvar-coated slot grids, before electron microscopy. Details are available at www.wormatlas.org/EMmethods/Laserhole.htm.
Statistical analysis
GraphPad software was used for graphing and all statistical analyses. When assumptions of normality and equal variance were met for the data analyzed (Figures 1 – 3), a parametric test of statistical significance between samples was performed by applying a Student’s t test (one comparison) or a one-way analysis of variance (ANOVA) followed by a post hoc Dunnett’s test (two or more comparisons). In cases when these assumptions failed to be met (Figures 4 – 6), nonparametric tests were used (Kruskal–Wallis with Dunn’s as post hoc for multiple comparisons). In all cases a two-tailed p value smaller than 0.05 was considered significant. All results are expressed as average ± SD unless otherwise indicated. Sample size (n) is given in each figure panel or legend. All results shown are representative of at least three independent biological replicates for each condition, except for Figure 4 (n ≥ 2). In all figures, p values are represented as follows: *p < 0.05, **p < 0.01, ***p < 0.001, and n.s. (not significant) is p > 0.05.
Supplementary Material
Acknowledgments
We thank Jim Priess, Julie Canman, Karen Oegema, Michael Glotzer, Abby Gerhold, and Amy Maddox for strains and reagents and Nicolas Chartier and Gilles Hickson for comments on the manuscript. We are also grateful to Christian Charbonneau of the Institute for Research in Immunology and Cancer’s (IRIC’s) Bio-imaging Facility for technical assistance and all members of the Labbé laboratory for helpful discussions. We thank Jonathan Hodgkin for his help in sending TEM prints and negatives from the Medical Research Council Laboratory of Molecular Biology laboratory of Sydney Brenner to the Hall laboratory for long-term curation and Carolyn Norris for help with image acquisitions. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440). E.G. is a research fellow of the Fonds de la Recherche du Québec-Santé (FRQ-S). R.A. is a research fellow of the Natural Sciences and Engineering Research Council of Canada (NSERC). This study was supported by NIH Grant No. 010943 to D.H.H. and NSERC Grant No. 434942 to J.-C.L. IRIC is supported in part by the Canada Foundation for Innovation and the FRQ-S.
Abbreviations used:
- ESCRT
endosomal sorting complexes required for transport
- GFP
green fluorescent protein
- PGC
primordial germ cell
- RNAi
RNA interference.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-08-0502) on October 26, 2017.
REFERENCES
- Agromayor M, Martin-Serrano J. Knowing when to cut and run: mechanisms that control cytokinetic abscission. Trends Cell Biol. 2013;23:433–441. doi: 10.1016/j.tcb.2013.04.006. [DOI] [PubMed] [Google Scholar]
- Amini R, Goupil E, Labella S, Zetka M, Maddox AS, Labbé JC, Chartier NT. C. elegans Anillin proteins regulate intercellular bridge stability and germline syncytial organization. J Cell Biol. 2014;206:129–143. doi: 10.1083/jcb.201310117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Caballe A, Agromayor M, Kloc M, Martin-Serrano J. ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C. Science. 2012;336:220–225. doi: 10.1126/science.1217180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier NT, Salazar Ospina DP, Benkemoun L, Mayer M, Grill SW, Maddox AS, Labbé JC. PAR-4/LKB1 mobilizes nonmuscle myosin through anillin to regulate C. elegans embryonic polarization and cytokinesis. Curr Biol. 2011;21:259–269. doi: 10.1016/j.cub.2011.01.010. [DOI] [PubMed] [Google Scholar]
- Chisholm AD, Hardin J. Epidermal morphogenesis. WormBook. 2005:1–22. doi: 10.1895/wormbook.1.35.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies T, Jordan SN, Chand V, Sees JA, Laband K, Carvalho AX, Shirasu-Hiza M, Kovar DR, Dumont J, Canman JC. High-resolution temporal analysis reveals a functional timeline for the molecular regulation of cytokinesis. Dev Cell. 2014;30:209–223. doi: 10.1016/j.devcel.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avino PP, Giansanti MG, Petronczki M. Cytokinesis in animal cells. Cold Spring Harb Perspect Biol. 2015;7:a015834. doi: 10.1101/cshperspect.a015834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avino PP, Takeda T, Capalbo L, Zhang W, Lilley KS, Laue ED, Glover DM. Interaction between Anillin and RacGAP50C connects the actomyosin contractile ring with spindle microtubules at the cell division site. J Cell Sci. 2008;121:1151–1158. doi: 10.1242/jcs.026716. [DOI] [PubMed] [Google Scholar]
- Durbin RM. Studies on the Development and Organisation of the Nervous System of Caenorhabditis elegans. Doctoral Thesis. Cambridge, UK: University of Cambridge; 1987. [Google Scholar]
- Eikenes AH, Brech A, Stenmark H, Haglund K. Spatiotemporal control of Cindr at ring canals during incomplete cytokinesis in the Drosophila male germline. Dev Biol. 2013;377:9–20. doi: 10.1016/j.ydbio.2013.02.021. [DOI] [PubMed] [Google Scholar]
- Fawcett DW, Ito S, Slautterback D. The occurrence of intercellular bridges in groups of cells exhibiting synchronous differentiation. J Biophys Biochem Cytol. 1959;5:453–460. doi: 10.1083/jcb.5.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenette P, Haines E, Loloyan M, Kinal M, Pakarian P, Piekny A. An anillin-Ect2 complex stabilizes central spindle microtubules at the cortex during cytokinesis. PLoS One. 2012;7:e34888. doi: 10.1371/journal.pone.0034888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RA, Kao HL, Audhya A, Arur S, Mayers JR, Fridolfsson HN, Schulman M, Schloissnig S, Niessen S, Laband K, et al. A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell. 2011;145:470–482. doi: 10.1016/j.cell.2011.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RA, Mayers JR, Wang S, Lewellyn L, Desai A, Audhya A, Oegema K. The midbody ring scaffolds the abscission machinery in the absence of midbody microtubules. J Cell Biol. 2013;203:505–520. doi: 10.1083/jcb.201306036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RA, Paluch E, Oegema K. Cytokinesis in animal cells. Annu Rev Cell Dev Biol. 2012;28:29–58. doi: 10.1146/annurev-cellbio-101011-155718. [DOI] [PubMed] [Google Scholar]
- Greenbaum MP, Iwamori T, Buchold GM, Matzuk MM. Germ cell intercellular bridges. Cold Spring Harb Perspect Biol. 2011;3:a005850. doi: 10.1101/cshperspect.a005850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum MP, Ma L, Matzuk MM. Conversion of midbodies into germ cell intercellular bridges. Dev Biol. 2007;305:389–396. doi: 10.1016/j.ydbio.2007.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory SL, Ebrahimi S, Milverton J, Jones WM, Bejsovec A, Saint R. Cell division requires a direct link between microtubule-bound RacGAP and Anillin in the contractile ring. Curr Biol. 2008;18:25–29. doi: 10.1016/j.cub.2007.11.050. [DOI] [PubMed] [Google Scholar]
- Guizetti J, Schermelleh L, Mantler J, Maar S, Poser I, Leonhardt H, Muller-Reichert T, Gerlich DW. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science. 2011;331:1616–1620. doi: 10.1126/science.1201847. [DOI] [PubMed] [Google Scholar]
- Haglund K, Nezis IP, Lemus D, Grabbe C, Wesche J, Liestol K, Dikic I, Palmer R, Stenmark H. Cindr interacts with anillin to control cytokinesis in Drosophila melanogaster. Curr Biol. 2010;20:944–950. doi: 10.1016/j.cub.2010.03.068. [DOI] [PubMed] [Google Scholar]
- Haglund K, Nezis IP, Stenmark H. Structure and functions of stable intercellular bridges formed by incomplete cytokinesis during development. Commun Integr Biol. 2011;4:1–9. doi: 10.4161/cib.4.1.13550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson GR, O’Farrell PH. Rho-dependent control of anillin behavior during cytokinesis. J Cell Biol. 2008;180:285–294. doi: 10.1083/jcb.200709005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hime GR, Brill JA, Fuller MT. Assembly of ring canals in the male germ line from structural components of the contractile ring. J Cell Sci. 1996;109(Pt 12):2779–2788. doi: 10.1242/jcs.109.12.2779. [DOI] [PubMed] [Google Scholar]
- Hirsh D, Oppenheim D, Klass M. Development of the reproductive system of Caenorhabditis elegans. Dev Biol. 1976;49:200–219. doi: 10.1016/0012-1606(76)90267-0. [DOI] [PubMed] [Google Scholar]
- Hubbard EJ, Greenstein D. Introduction to the germ line. WormBook. 2005:1–4. doi: 10.1895/wormbook.1.18.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh H, Ng CY, Ong CK, Lim KB, Chan TW. Cloning and characterization of a novel pregnancy-induced growth inhibitor in mammary gland. Endocrinology. 2001;142:3607–3615. doi: 10.1210/endo.142.8.8297. [DOI] [PubMed] [Google Scholar]
- Iwamori T, Iwamori N, Ma L, Edson MA, Greenbaum MP, Matzuk MM. TEX14 interacts with CEP55 to block cell abscission. Mol Cell Biol. 2010;30:2280–2292. doi: 10.1128/MCB.01392-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachur TM, Audhya A, Pilgrim DB. UNC-45 is required for NMY-2 contractile function in early embryonic polarity establishment and germline cellularization in C. elegans. Dev Biol. 2008;314:287–299. doi: 10.1016/j.ydbio.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kechad A, Jananji S, Ruella Y, Hickson GR. Anillin acts as a bifunctional linker coordinating midbody ring biogenesis during cytokinesis. Curr Biol. 2012;22:197–203. doi: 10.1016/j.cub.2011.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Frankel EB, Audhya A, Muller-Reichert T. Membrane remodeling during embryonic abscission in Caenorhabditis elegans. J Cell Biol. 2017;216:1277–1286. doi: 10.1083/jcb.201607030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara PE, Lee MH, Schedl T, Jefferis GS. A C. elegans patched gene, ptc-1, functions in germ-line cytokinesis. Genes Dev. 2000;14:1933–1944. [PMC free article] [PubMed] [Google Scholar]
- Lee K-Y, Green RA, Gutierrez E, Gomez-Cavazos JS, Kolotuev I, Wang S, Desai A, Groisman A, Oegema K. CYK-4 functions independently of its centralspindlin partner ZEN-4 to cellularize oocytes in germline syncytia. bioRxiv. 2017:196279. doi: 10.7554/eLife.36919. https://doi.org/10.1101/196279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Fairn GD, Ceccarelli DF, Sicheri F, Wilde A. Cleavage furrow organization requires PIP(2)-mediated recruitment of anillin. Curr Biol. 2012;22:64–69. doi: 10.1016/j.cub.2011.11.040. [DOI] [PubMed] [Google Scholar]
- Maddox AS, Habermann B, Desai A, Oegema K. Distinct roles for two C. elegans anillins in the gonad and early embryo. Development. 2005;132:2837–2848. doi: 10.1242/dev.01828. [DOI] [PubMed] [Google Scholar]
- Mishima M, Kaitna S, Glotzer M. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev Cell. 2002;2:41–54. doi: 10.1016/s1534-5807(01)00110-1. [DOI] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins JM, Biesele JJ. Terminal phase of cytokinesis in D-98s cells. J Cell Biol. 1977;73:672–684. doi: 10.1083/jcb.73.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K, Hyman AA. Cell division. WormBook. 2006:1–40. doi: 10.1895/wormbook.1.72.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K, Savoian MS, Mitchison TJ, Field CM. Functional analysis of a human homologue of the Drosophila actin binding protein anillin suggests a role in cytokinesis. J Cell Biol. 2000;150:539–552. doi: 10.1083/jcb.150.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S, Tan C. Germline cyst formation and incomplete cytokinesis during Drosophila melanogaster oogenesis. Dev Biol. 2010;337:84–98. doi: 10.1016/j.ydbio.2009.10.018. [DOI] [PubMed] [Google Scholar]
- Pacquelet A, Uhart P, Tassan JP, Michaux G. PAR-4 and anillin regulate myosin to coordinate spindle and furrow position during asymmetric division. J Cell Biol. 2015;210:1085–1099. doi: 10.1083/jcb.201503006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekny AJ, Glotzer M. Anillin is a scaffold protein that links RhoA, actin, and myosin during cytokinesis. Curr Biol. 2008;18:30–36. doi: 10.1016/j.cub.2007.11.068. [DOI] [PubMed] [Google Scholar]
- Rehain-Bell K, Love A, Werner ME, MacLeod I, Yates JR, 3rd, Maddox AS. A sterile 20 family kinase and its co-factor CCM-3 regulate contractile ring proteins on germline intercellular bridges. Curr Biol. 2017;27:860–867. doi: 10.1016/j.cub.2017.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DN, Cant K, Cooley L. Morphogenesis of Drosophila ovarian ring canals. Development. 1994;120:2015–2025. doi: 10.1242/dev.120.7.2015. [DOI] [PubMed] [Google Scholar]
- Sanger JM, Pochapin MB, Sanger JW. Midbody sealing after cytokinesis in embryos of the sea urchin Arabacia punctulata. Cell Tissue Res. 1985;240:287–292. doi: 10.1007/BF00222337. [DOI] [PubMed] [Google Scholar]
- Schiel JA, Park K, Morphew MK, Reid E, Hoenger A, Prekeris R. Endocytic membrane fusion and buckling-induced microtubule severing mediate cell abscission. J Cell Sci. 2011;124:1411–1424. doi: 10.1242/jcs.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- Straight AF, Field CM, Mitchison TJ. Anillin binds nonmuscle myosin II and regulates the contractile ring. Mol Biol Cell. 2005;16:193–201. doi: 10.1091/mbc.E04-08-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S. Asymmetric movements of cytoplasmic components in Caenorhabditis elegans zygotes. J Embryol Exp Morphol. 1986;97(suppl):15–29. [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Sun L, Guan R, Lee IJ, Liu Y, Chen M, Wang J, Wu JQ, Chen Z. Mechanistic insights into the anchorage of the contractile ring by anillin and Mid1. Dev Cell. 2015;33:413–426. doi: 10.1016/j.devcel.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JT, Seydoux G. Germ cell specification. Adv Exp Med Biol. 2013;757:17–39. doi: 10.1007/978-1-4614-4015-4_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley SP, Wang Y. Midzone microtubule bundles are continuously required for cytokinesis in cultured epithelial cells. J Cell Biol. 1996;135:981–989. doi: 10.1083/jcb.135.4.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis JH, Munro E, Lyczak R, Bowerman B. Conditional dominant mutations in the Caenorhabditis elegans gene act-2 identify cytoplasmic and muscle roles for a redundant actin isoform. Mol Biol Cell. 2006;17:1051–1064. doi: 10.1091/mbc.E05-09-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf N, Priess J, Hirsh D. Segregation of germline granules in early embryos of Caenorhabditis elegans: an electron microscopic analysis. J Embryol Exp Morphol. 1983;73:297–306. [PubMed] [Google Scholar]
- Zhou K, Rolls MM, Hanna-Rose W. A postmitotic function and distinct localization mechanism for centralspindlin at a stable intercellular bridge. Dev Biol. 2013;376:13–22. doi: 10.1016/j.ydbio.2013.01.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.