Significance
Most proteases catalyze peptide bond hydrolysis of substrate proteins in aqueous environments. Intramembrane proteases (IPs) are unusual, cleaving substrates in hydrophobic cellular membranes. IPs regulate many processes that impact health, but potential benefits of manipulating IP activities remain elusive due to insufficient knowledge about how IPs interact with substrates. We report experimental and modeling results that illuminate how intramembrane metalloprotease SpoIVFB interacts with its substrate Pro-σK. A 26-residue linker between two domains of SpoIVFB is crucial, perhaps allowing an ATP-induced conformational change to position Pro-σK for cleavage. SpoIVFB and Pro-σK are broadly conserved in endospore-forming bacteria. Endospores are highly resistant cells that promote persistence of some important human pathogens. The work may lead to new strategies to control endospore formation.
Keywords: intramembrane protease, regulated intramembrane proteolysis, Bacillus subtilis, sporulation, SpoIVFB
Abstract
Intramembrane proteases (IPs) cleave membrane-associated substrates in nearly all organisms and regulate diverse processes. A better understanding of how these enzymes interact with their substrates is necessary for rational design of IP modulators. We show that interaction of Bacillus subtilis IP SpoIVFB with its substrate Pro-σK depends on particular residues in the interdomain linker of SpoIVFB. The linker plus either the N-terminal membrane domain or the C-terminal cystathione-β-synthase (CBS) domain of SpoIVFB was sufficient for the interaction but not for cleavage of Pro-σK. Chemical cross-linking and mass spectrometry of purified, inactive SpoIVFB–Pro-σK complex indicated residues of the two proteins in proximity. A structural model of the complex was built via partial homology and by using constraints based on cross-linking data. In the model, the Proregion of Pro-σK loops into the membrane domain of SpoIVFB, and the rest of Pro-σK interacts extensively with the linker and the CBS domain of SpoIVFB. The extensive interaction is proposed to allow coordination between ATP binding by the CBS domain and Pro-σK cleavage by the membrane domain.
Intramembrane proteases (IPs) are membrane-embedded proteases that cleave their substrates within a transmembrane segment (TMS) or near the membrane surface, releasing from the membrane a fragment of the substrate (1). The released fragments impact a wide variety of processes in organisms from bacteria to humans, and there are four known families of IPs: metalloproteases that include SpoIVFB and human site-2 protease (S2P), aspartyl proteases such as presenilins and signal peptide peptidases, serine proteases known as rhomboids, and the glutamyl protease Rce1 (2, 3). Understanding how IPs interact with substrates is key to modulating IP activity, with many potential benefits. However, difficulties in studying enzyme–substrate complexes that function in biological membranes have impeded progress.
In Bacillus subtilis, SpoIVFB cleaves Pro-σK during the process of endospore formation (4). Both proteins are broadly conserved in endospore-forming Bacillus and Clostridium species (5), including many that cause disease or provide benefits (6, 7). Understanding how SpoIVFB interacts with Pro-σK could inform strategies to inhibit or enhance sporulation. More broadly, SpoIVFB is a member of the S2P family (8), named after human S2P, which is involved in regulation of cholesterol biosynthesis and responses to endoplasmic reticulum stress and viral infection (9, 10). The S2P family also includes enzymes in nonspore-forming bacteria that mediate stress responses and enhance pathogenicity (4, 11), and plant enzymes involved in chloroplast development and likely in stress responses (12). Hence, studies of SpoIVFB have far-reaching implications. Only one group of potential therapeutics targeting this family of enzymes has been reported (13). Design of additional modulators could be facilitated by knowledge about how these enzymes interact with their substrates, but no structure of an S2P family member or any other IP in complex with a native substrate has been reported, although structures of rhomboid–peptide inhibitor complexes may aid drug design (14, 15).
Mutational analysis suggested SpoIVFB is a metalloprotease (16, 17), and this was confirmed by reconstituting cleavage of Pro-σK by SpoIVFB with purified proteins (18). The reaction depended not only on zinc, but on ATP, which was shown to bind to the cystathione-β-synthase (CBS) domain of SpoIVFB. CBS domains in a variety of proteins undergo a conformational change upon ligand binding to regulate protein activity in response to cellular energy levels or ion availability (19, 20). It has been proposed that the CBS domain of SpoIVFB senses a change in ATP concentration during B. subtilis endospore formation, regulating cleavage of Pro-σK (4, 18). A C-terminal fragment of SpoIVFB including the CBS domain binds to Pro-σK (18). Here, we show that binding relies on a segment that precedes the CBS domain and links it to the N-terminal membrane domain of SpoIVFB.
Cleavage of Pro-σK by SpoIVFB can be reconstituted by coexpressing the two proteins in Escherichia coli (21). This approach facilitated mutational studies that showed the C-terminal half of Pro-σK is dispensable for cleavage (22) and revealed the preferences of SpoIVFB for residues in Pro-σK(1-127) (i.e., the N-terminal half) near the cleavage site (23). [Note: Pro-σK(1-127) has been called Pro-σK(1-126) previously (18, 21–26), but mass spectrometry data suggest that the first of two adjacent potential start codons is used (18, 26), adding one residue at the N-terminal end. We adopt the revised numbering of Pro-σK herein.] E. coli coexpression was also used in disulfide cross-linking experiments with single-Cys versions of Pro-σK(1-127) and catalytically inactive SpoIVFB, which showed that residues near the active site and in two predicted loops of SpoIVFB could be cross-linked with residues near the cleavage site in Pro-σK(1-127) (25). The loop predictions were based on a model of SpoIVFB that was made using the crystal structure of the membrane domain of an archaeal homolog (27). The cross-linking results supported the model of SpoIVFB and provided initial insight into its interaction with Pro-σK(1-127) (25). Biochemical approaches showed that the SpoIVFB–Pro-σK(1-127) complex can be solubilized from E. coli membranes with mild detergents and purified (26). Disulfide cross-linking of purified complex containing single-Cys versions of the two proteins suggested that it resembles the complex formed in vivo. Ion mobility-mass spectrometry analysis resulted in an observed mass consistent with a 4:2 SpoIVFB–Pro-σK(1-127) complex. Here, we take advantage of these advances to further characterize the complex and use all of the available data to build a molecular model.
Results
The Interdomain Linker of SpoIVFB Is Important for Interaction with Pro-σK.
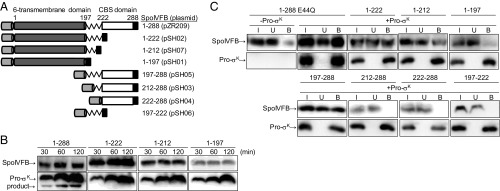
A homology model of SpoIVFB (25) suggested that the N-terminal six-transmembrane domain (residues 1–196) containing the protease active site is connected to the C-terminal CBS domain (residues 223–288) by a 26-residue linker (residues 197–222) (Fig. 1A). To test whether the N-terminal domain of SpoIVFB interacts with Pro-σK(1-127), we engineered three C-terminal deletions of SpoIVFB. The deletions removed the CBS domain but left the entire linker (1–222), the N-terminal half of the linker (1–212), or none of the linker (1–197) (Fig. 1A). The deletions were made in the context of TM-SpoIVFB, which has an N-terminal TMS from rabbit cytochrome P450 2B4 (designated TM) (28) that enhances its accumulation upon expression in E. coli, and a C-terminal FLAG2 tag for detection by immunoblot (18). None of the deletion variants cleaved Pro-σK(1-127)-His6 (designated Pro-σK) upon coexpression in E. coli, whereas full-length TM-SpoIVFB (designated 1–288) cleaved Pro-σK as expected (18) (Fig. 1B). We nevertheless tested whether the deletion derivatives could interact with Pro-σK using pull-down assays. Full-length catalytically inactive 1–288 E44Q served as a control and, as expected (26), more than half of this protein copurified with Pro-σK in the bound fraction (Fig. 1C). Interestingly, 1–222 and, to a lesser extent, 1–212 copurified with Pro-σK, but very little of 1–197 was bound (Fig. 1C), showing the importance of the N-terminal half of the linker for interaction with Pro-σK.
Fig. 1.
Derivatives of SpoIVFB and assays for cleavage of Pro-σK and interaction with it. (A) Diagram of TM-SpoIVFB (designated 1–288) and its deletion derivatives (plasmids in parenthesis). CBS domain, white; FLAG2; black; interdomain linker, jagged line; 6-transmembrane domain, dark gray; TM, light gray. Numbers above indicate SpoIVFB residues. (B) Cleavage assays. Full-length 1–288 or the indicated deletion derivative was coexpressed with Pro-σK(1-127)-His6 (designated Pro-σK) (pZR12) in E. coli, and samples collected at the indicated times after induction were subjected to immunoblot analysis with anti-FLAG (Top) or anti-His (Bottom) antibodies to detect the indicated protein, including the cleavage product. (C) Interaction with Pro-σK. Catalytically inactive 1–288 E44Q (pYZ68) or the indicated deletion derivative was expressed with (+) or without (−) Pro-σK in E. coli for 2 h, and samples were subjected to pull-down assays. Input (I), unbound (U), and bound (B) fractions were subjected to immunoblot analysis as in B. The control without Pro-σK is shown for 1–288 E44Q in C, and for the other deletion derivatives in SI Appendix, Fig. S1.
To test the importance of the linker for interaction with Pro-σK in the context of the CBS domain, we engineered three deletions spanning the six-transmembrane catalytic domain of SpoIVFB but leaving the entire linker (197–288), the C-terminal half of the linker (212–288), or none of the linker (222–288) (Fig. 1A). All three proteins retained TM to promote membrane association. Only 197–288 copurified with Pro-σK in pull-down assays (Fig. 1C), confirming the importance of the N-terminal half of the linker for interaction with Pro-σK. However, the linker alone is insufficient for the interaction since 197–222 (Fig. 1A) failed to interact with Pro-σK (Fig. 1C). Therefore, our results demonstrate that the SpoIVFB linker is necessary but not sufficient for interaction with Pro-σK. Residues 197–212 of the linker appear to be important for the interaction, but either the N- or C-terminal domain of SpoIVFB is also required, and both domains are necessary for SpoIVFB to cleave Pro-σK.
Alanine Substitutions in the SpoIVFB Linker Impair Protease Activity and Interaction with Pro-σK.
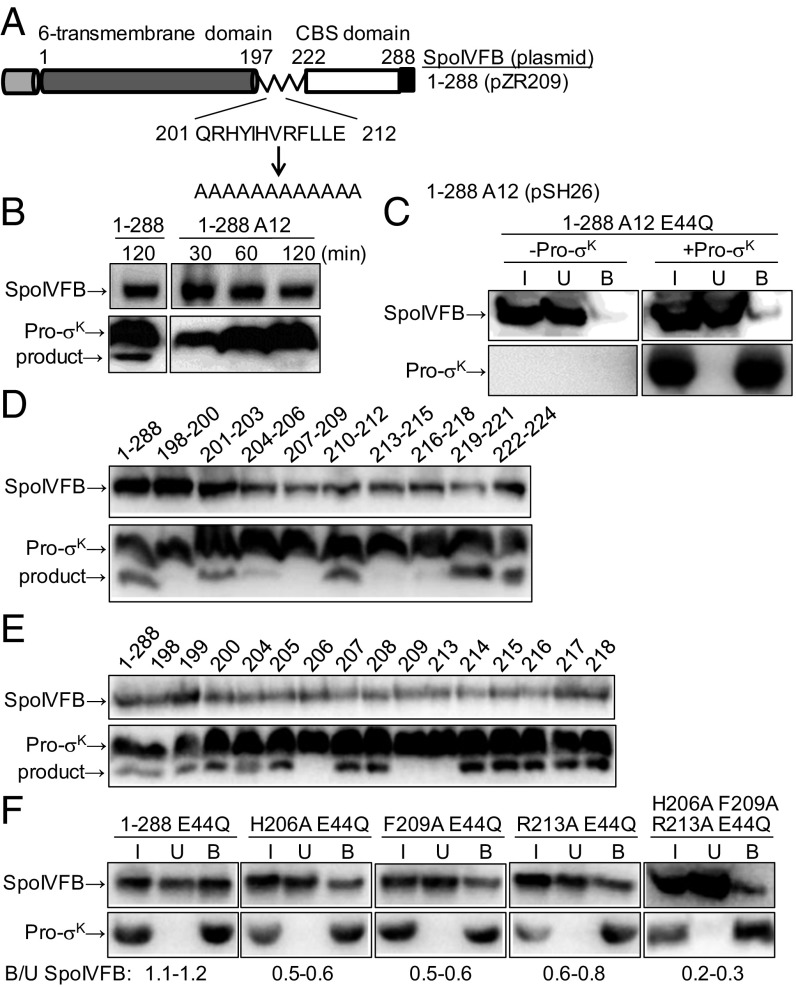
Our deletion analysis identified residues 197–212 of the SpoIVFB linker as important for interaction with Pro-σK. This region of SpoIVFB corresponds to a region of variable structure in Methanocaldococcus jannaschii S2P (mjS2P), which crystallized as a dimer in two distinct conformations (27). In the “closed” conformation, the active site is proposed to be inaccessible to the substrate, and the variable region lacks secondary structure. In the “open” conformation, TMSs 1 and 6 are farther apart, which is proposed to allow substrate access to the active site, and the variable region is α-helical, extending the TMS 6 α-helix. Based on this model of substrate gating, we reasoned that replacing the region of SpoIVFB corresponding to the structurally variable region of mjS2P, with a stretch of Ala residues which favor α-helix formation, should be compatible with the open conformation and allow Pro-σK to be cleaved, unless side chains of SpoIVFB residues in this region are important (e.g., for interaction with Pro-σK). Therefore, we replaced residues 201–212 of TM-SpoIVFB (i.e., 1–288) with Ala residues, creating 1–288 A12 (Fig. 2A). During expression in E. coli and purification, accumulation of 1–288 A12 and elution from size exclusion chromatography was similar to that of 1–288 (SI Appendix, Fig. S2A), suggesting similar in vivo stability and in vitro size and shape. Also, the purified proteins exhibited similar far-UV circular dichroism spectra (SI Appendix, Fig. S2B), indicative of similar secondary structure, which was primarily α-helical. However, in contrast to 1–288, 1–288 A12 failed to cleave Pro-σK (Fig. 2B). Although 1–288 A12 was inactive, we introduced the E44Q substitution so the protein would be comparable to 1–288 E44Q, which interacts strongly with Pro-σK in pull-down assays (Fig. 1C). In contrast, 1–288 A12 E44Q interacted weakly with Pro-σK (Fig. 2C). These results show that one or more side chains of SpoIVFB residues 201–212 are crucial for protease activity and the interaction with Pro-σK.
Fig. 2.
Effects of alanine substitutions in the SpoIVFB interdomain linker. (A) Diagram of TM-SpoIVFB (designated 1–288) emphasizing the replacement of residues 201–212 in the linker with 12 Ala residues to create 1–288 A12. See Fig. 1 for additional explanation. (B) Cleavage assays. SpoIVFB 1–288 or 1–288 A12 was coexpressed with Pro-σK(1-127)-His6 (designated Pro-σK) (pZR12) in E. coli, and samples collected at the indicated times after induction were subjected to immunoblot analysis with anti-FLAG (Top) or anti-His (Bottom) antibodies to detect the indicated protein, including the cleavage product. (C) Interaction with Pro-σK. SpoIVFB 1–288 A12 E44Q (pSH27) was expressed with (+) or without (−) Pro-σK in E. coli for 2 h, and samples were subjected to pull-down assays. Input (I), unbound (U), and bound (B) fractions were subjected to immunoblot analysis as in B. (D) Cleavage assays of triple-Ala substitutions. SpoIVFB 1–288 or its derivative with the indicated three residues changed to Ala (pSH08-pSH16) was coexpressed with Pro-σK in E. coli for 2 h, and samples were subjected to immunoblot analysis as in B. (E) Cleavage assays of single-Ala substitutions. As in B, except with 1–288 or its derivative with the indicated residue changed to Ala (pDP74, pDP75, pSH28-pSH54). (F) Interaction with Pro-σK. As in C, except with 1–288 E44Q (pYZ68) or its derivative with the indicated residue(s) changed to Ala (pSH55-pSH58). The range of ratios of bound/unbound (B/U) SpoIVFB is shown at the bottom.
To screen for important residues in the SpoIVFB linker, we made triple-Ala substitutions in the region from residue 198 to 224. We included the C-terminal half of the linker in the analysis because SpoIVFB 1–212 exhibited less interaction with Pro-σK than 1–222 (Fig. 1C). Five of the substitutions resulted in little or no cleavage of Pro-σK upon coexpression in E. coli (Fig. 2D). Each of the 15 implicated residues was changed to Ala individually. Three substitutions resulted in no cleavage of Pro-σK; H206A, F209A, and R213A (Fig. 2E). Each of the three proteins accumulated well in E. coli, indicating that diminished stability did not account for the lack of cleavage, and suggesting the proteins are folded. We conclude that the side chains of H206, F209, and R213 are required for SpoIVFB activity in E. coli. Other side chains also appear to be important, especially among residues 198–200 and 216–218 for which triple-Ala substitutions abolished cleavage (Fig. 2D), but single-Ala substitutions at these positions had little or no effect on activity (Fig. 2E), so we focused on the role of residues 206, 209, and 213.
We tested whether the three SpoIVFB residues are important for its interaction with Pro-σK by introducing H206A, F209A, and R213A substitutions individually and in combination into SpoIVFB 1–288 E44Q and performing pull-down assays. Each single-Ala substitution reduced the interaction, and the triple-Ala substitution reduced the interaction to a greater extent (Fig. 2F). All four variants accumulated well in E. coli (Fig. 2F, input samples), indicating that diminished stability did not account for reduced interaction with Pro-σK, and suggesting the proteins are folded. We conclude that the side chains of SpoIVFB residues H206, F209, and R213 are important for its interaction with Pro-σK upon coexpression in E. coli. The side chains might directly contact Pro-σK and/or they might subtly influence the structure of SpoIVFB in a way that perturbs its interaction with Pro-σK. We note that none of the three side chains is absolutely essential for the interaction in pull-down assays (Fig. 2F), but each is essential for SpoIVFB to cleave Pro-σK (Fig. 2E). Clearly, the pull-down assay can detect interactions that are not productive for cleavage, as was also observed in our analysis of SpoIVFB deletion variants (Fig. 1).
Certain Substitutions in the Linker Destabilize SpoIVFB During Sporulation.
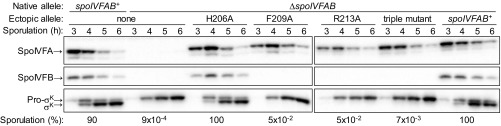
The effects of Ala substitutions in the SpoIVFB linker were assessed by ectopically integrating mutant versions of the spoIVFAB operon (i.e., the cotranscribed spoIVFA and spoIVFB genes) into the chromosome of B. subtilis deleted for the endogenous spoIVFAB operon, and inducing sporulation by nutrient deprivation. SpoIVFA accumulated in all cases, although its level appeared to be slightly lower in the strain with the R213A substitution in SpoIVFB (Fig. 3). Surprisingly, the F209A and R213A single-Ala substitutions, and the H206A F209A R213A triple-Ala substitution, resulted in little or no accumulation of SpoIVFB in sporulating B. subtilis (Fig. 3), despite the ability of these proteins to accumulate in E. coli (Fig. 2 E and F). The failure of these proteins to accumulate normally in B. subtilis can explain the apparent lack of Pro-σK cleavage and greatly reduced sporulation (Fig. 3). In contrast, the H206A substitution resulted in normal SpoIVFB accumulation in B. subtilis and allowed normal cleavage of Pro-σK and sporulation. Hence, H206 is crucial for SpoIVFB activity in E. coli (Fig. 2E), but not in sporulating B. subtilis (Fig. 3). Apparently, the reduced interaction between SpoIVFB H206A and Pro-σK observed in E. coli (Fig. 2F) either does not occur or is inconsequential in B. subtilis.
Fig. 3.
Effect of substitutions in the SpoIVFB interdomain linker during B. subtilis sporulation. B. subtilis with a wild-type spoIVFAB operon or a deletion of it at the native site, and either nothing, a spoIVFAB operon with the indicated substitution in SpoIVFB, or a wild-type spoIVFAB operon at the ectopic amyE site, were induced to sporulate, and samples collected at the indicated times postinduction were subjected to immunoblot analysis with antibodies against SpoIVFA (Top), SpoIVFB (Middle), or Pro-σK (Bottom). Sporulation was measured at 24 h after induction.
Although the H206A, F209A, and R213A substitutions in SpoIVFB appeared to reduce the interaction with Pro-σK to similar extents, based on pull-down assays using E. coli extracts (Fig. 2F), we reasoned this might not be the case in B. subtilis and could explain differential stability of the SpoIVFB variants if interaction with Pro-σK is important for SpoIVFB accumulation during sporulation. However, two B. subtilis sigK mutants that fail to accumulate Pro-σK (29) both showed normal levels of SpoIVFB during sporulation (SI Appendix, Fig. S3), demonstrating that interaction with Pro-σK is unnecessary to stabilize SpoIVFB.
To possibly identify substitutions in the SpoIVFB linker that preserve stability in B. subtilis but impair interaction with Pro-σK, we aligned the linker sequences of 136 SpoIVFB orthologs. Residues 208–210 (RFL) of B. subtilis are conserved in ≥90% of the orthologs from closely related species, and R213 is conserved in ≥80% of these orthologs (representative Bacilli are shown in SI Appendix, Fig. S4), correlating with instability of the F209A and R213A variants (Fig. 3). In contrast, H206 is conserved in ≤50% of these orthologs (SI Appendix, Fig. S4), correlating with no effect of the H206A substitution (Fig. 3). Residues at the corresponding positions in more distantly related orthologs from Clostridial species differed considerably from Bacilli (SI Appendix, Fig. S4). We used conserved differences to guide creation of substitutions in the B. subtilis SpoIVFB linker, but we were unable to identify any that impaired activity without also impairing accumulation (SI Appendix, Fig. S5). Interestingly, conservative L210I and R213K substitutions reduced accumulation of SpoIVFB in B. subtilis.
Altogether, we found three residues in the linker (F209, L210, and R213) that are important for SpoIVFB stability during B. subtilis sporulation. These residues are highly conserved in orthologs from closely related species (SI Appendix, Fig. S4). Substitutions at two of these positions (F209A and R213A) destabilize SpoIVFB in sporulating B. subtilis (Fig. 3) but not in growing E. coli (Fig. 2 E and F), perhaps due to differences in expression, proteolysis, and/or interaction partners (Discussion). The F209A and R213A substitutions in SpoIVFB impair interaction with Pro-σK in E. coli (Fig. 2F). We conclude that F209 and R213 of the SpoIVFB linker are crucial both for stability and for interaction with Pro-σK.
Chemical Cross-Linking of a SpoIVFB–Pro-σK Complex Provides Insight into Their Interaction.
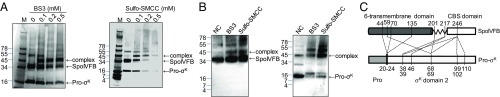
To gain further insight into the role of the SpoIVFB linker and more broadly into the interaction of SpoIVFB with Pro-σK, we purified a complex of catalytically inactive SpoIVFB-TEV-FLAG2 E44Q with Pro-σK(1-127)-His6 from E. coli engineered to coexpress the two proteins as described recently (26) (SI Appendix, Fig. S6) and subjected it to chemical cross-linking followed by protease digestion and mass spectrometry analysis to identify regions of proximity between the proteins. Incubation of the complex with the homobifunctional amine-to-amine cross-linker bis(sulfosuccinimidyl)suberate (BS3) resulted in formation of a species that migrated during gel electrophoresis at the position expected for a 1:1 complex of SpoIVFB and Pro-σK, and similar results were obtained with the heterobifunctional amine-to-sulfhydryl cross-linker sulfosuccinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate (Sulfo-SMCC) (Fig. 4A). The identity of the apparent 1:1 complexes was verified by immunoblots that detected the FLAG tag on SpoIVFB and the His tag on Pro-σK (Fig. 4B). Each cross-linked 1:1 complex was excised from the gel and digested with trypsin to produce peptides for mass spectrometry analysis. Additional analyses were carried out on 1:1 complexes obtained with other cross-linkers (SI Appendix, Table S1). One analysis relied on SpoIVFB with a Q201C substitution to provide an additional Cys residue near the N-terminal end of the linker. In total, 47 cross-linked peptides were identified. Eliminating the redundancies due to identification with more than one cross-linker and/or the same two residues being cross-linked, the number of unique cross-links identified was 34, including 9 that resulted from interchain cross-linking between SpoIVFB and Pro-σK, 16 intrachain cross-links within SpoIVFB, and 9 within Pro-σK (SI Appendix, Table S2). One SpoIVFB intrachain cross-link (between K10 and K223; SI Appendix, Table S1) was omitted from SI Appendix, Table S2 because it was not used to build the model of the SpoIVFB–Pro-σK complex presented below. All of the other chemical cross-links, plus disulfide cross-links reported previously (25), were used to derive constraints for modeling. Fig. 4C provides a cartoon representation of the interchain cross-links used. Importantly, the cross-links establish the orientation between the two proteins in the complex.
Fig. 4.
Cross-linking of a SpoIVFB–Pro-σK complex. (A) SDS/PAGE after chemical cross-linking. The purified complex was incubated with the indicated cross-linkers at the indicated concentrations, or with no cross-linker as a control, and samples including protein markers (M, molecular mass in kilodaltons at Left) were subjected to SDS/PAGE followed by Coomassie blue staining. Position of 1:1 complex of SpoIVFB and Pro-σK is indicated. (B) Immunoblot analysis after chemical cross-linking. The purified complex was incubated with the indicated cross-linkers at 0.1 mM, or with no cross-linker (NC) as a control, and samples were subjected to immunoblot analysis with anti-FLAG (Left) or anti-His (Right) antibodies to detect SpoIVFB or Pro-σK, respectively. Positions of protein markers and 1:1 complex of SpoIVFB and Pro-σK are indicated. (C) Summary of interchain cross-links used in modeling. Cys substitutions for residues 44, 70, and 135 of SpoIVFB formed disulfide cross-links with Cys substitutions for some of the residues spanning from at least residues 20–24 of Pro-σK (25). The other dashed lines depict the chemical cross-links listed in SI Appendix, Table S2. Pro-σK(1-127), including the Prosequence (1–21) and σK domain 2 (22-127), is depicted (on a different scale than SpoIVFB) since Pro-σK(1-127) was used in all of the cross-linking experiments.
A Model of the SpoIVFB–Pro-σK Complex.
A tetrameric model of SpoIVFB with two bound Pro-σK (i.e., residues 1–127) was built in stages using a combination of homology modeling and simulation-based assembly contingent on constraints derived from cross-linking experiments (see SI Appendix for details). Briefly, the N-terminal membrane domain of SpoIVFB was modeled using the structure of mjS2P (27), with two subunits of the SpoIVFB tetramer in the open conformation (chains A and C) presumed to bind Pro-σK (chains X and Y) and two subunits in the closed conformation (chains B and D) presumed unable to bind Pro-σK, consistent with the observed 4:2 SpoIVFB–Pro-σK complex (26). The C-terminal CBS domain of SpoIVFB was modeled using the Thermotoga maritima TM0935 CBS tetramer structure (30). The SpoIVFB linker and the Proregion of Pro-σK were modeled as flexible chains, and most of the rest of Pro-σK was modeled using the Thermus aquaticus SigA domain 2 structure (31). Constraints from cross-linking experiments were used to refine an initial model via molecular dynamics simulations of a coarse-grained representation (only Cα positions) in the presence of various restraint potentials (SI Appendix, Table S3).
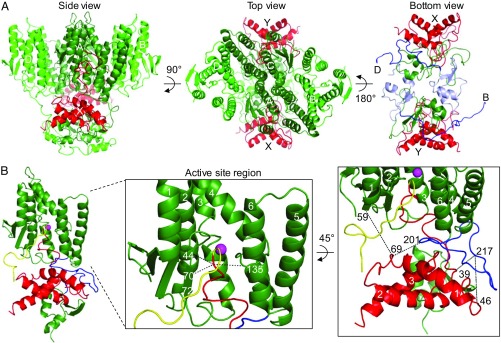
General features of the model are illustrated in Fig. 5A. The side view shows the membrane domains of the SpoIVFB tetramer at the top and the CBS domains at the bottom. This view faces the A chain (dark green), which interacts with Pro-σK chain X (red). The top view emphasizes the membrane domains of the SpoIVFB A and C chains (dark green) packed together in the center and the B and D chains (light green) packed along the sides. In this view, the SpoIVFB C chain can be seen interacting with the Pro-σK Y chain. This arrangement of SpoIVFB membrane domains is compatible with the CBS domain tetramer that resulted from the modeling template chosen. Other templates exist but are less compatible with a tetramer of membrane domains on one side of the disk-like CBS domain tetramer. The bottom view shows only the SpoIVFB linkers and CBS domains, and the two Pro-σK. In this view, the linkers (blue) and CBS domains (light blue) of chains B and D can be seen stretching across the bottom of the complex, whereas the linkers and CBS domains of chains A and C (dark green) interact with Pro-σK as described below.
Fig. 5.
Model of the SpoIVFB–Pro-σK complex. (A) SpoIVFB tetramer with two bound Pro-σK(1-127). The side view shows the SpoIVFB membrane domains at the top and the CBS domains at the bottom, facing the A chain (dark green), which interacts with the X chain of Pro-σK (red). The membrane domains of the D and B chains of SpoIVFB (light green) are on the Left and Right, respectively, and the C chain (dark green) interacting with the Y chain of Pro-σK (red) is at the back. The top view allows the arrangement of the SpoIVFB membrane domains to be seen, as well as both Pro-σK chains, which are labeled. The bottom view shows both Pro-σK chains, and only the interdomain linkers and CBS domains of SpoIVFB (for clarity). The points of attachment to the membrane domains of the D and B chains are labeled, and the linkers (blue) and CBS domains (light blue) of these chains are colored for emphasis. (B) SpoIVFB chain C bound to Pro-σK chain Y. At Left, the view is similar to that of SpoIVFB chain A bound to Pro-σK chain X in A, except the SpoIVFB linker (blue) and the Proregion (yellow) of Pro-σK are colored for emphasis, and the zinc ion (magenta) is shown. In the enlarged view of the active site region, TMSs 1–6 and residues 44, 70, and 135 of SpoIVFB are labeled. Residue 72 is also labeled to indicate the orientation of the predicted β-strand spanning residues 69–72 of SpoIVFB. At Right, the enlarged view shows residues involved in chemical cross-links (dashed lines). Pro-σK α-helices are numbered 1–3.
Fig. 5B illustrates features of the interaction between SpoIVFB chain C and Pro-σK chain Y in the model. The Proregion (yellow) of Pro-σK loops into the SpoIVFB membrane domain with a zinc ion (magenta) at its active site based on homology (27). The enlarged view of the active site region shows the locations of three SpoIVFB residues within disulfide cross-linking distance of residues near the cleavage site in Pro-σK (dashed lines connect to Cα of the residue following the cleavage site) (25). A second enlarged view emphasizes some of the residues involved in interchain chemical cross-links. The others, involving SpoIVFB C246, are illustrated in SI Appendix, Fig. S7. The cross-linking data and the model derived from it predict extensive interactions between SpoIVFB and Pro-σK that may explain how ATP binding by the SpoIVFB CBS domain promotes Pro-σK cleavage (Discussion).
Discussion
Our results show that the SpoIVFB linker plays a crucial role in Pro-σK cleavage. Substitutions in the linker eliminated cleavage of coexpressed Pro-σK in E. coli and impaired interaction between the proteins. Linker substitutions, including some conservative ones, destabilized SpoIVFB in sporulating B. subtilis. Cross-linking data and partial homology allowed a model of the SpoIVFB–Pro-σK complex to be built. The model predicts extensive interactions between Pro-σK and the SpoIVFB linker and CBS domain. We propose that these interactions allow ATP binding by the CBS domain to induce a conformational change that positions Pro-σK for cleavage by the SpoIVFB membrane domain.
Our deletion analysis provides evidence that the SpoIVFB linker coordinates functions of the membrane and CBS domains. Neither domain alone bound to Pro-σK, but the linker allowed either domain to bind Pro-σK nearly as well as full-length SpoIVFB, yet only full-length SpoIVFB cleaved Pro-σK (Fig. 1).
The effects of Ala substitutions in the SpoIVFB linker revealed three residues (H206, F209, R213) essential for cleavage of coexpressed Pro-σK in E. coli and important for binding to Pro-σK (Fig. 2). Partial binding, but elimination of cleavage, implies a subtle but important change in the interaction. The three residues are in a region of the SpoIVFB linker predicted by our model to be in close proximity to residues 43–51 that precede helix 1 of Pro-σK (SI Appendix, Fig. S8). Side chains of residues in the two regions may directly interact, although the Ala substitutions could subtly change SpoIVFB structure and indirectly perturb other interactions with Pro-σK. Disulfide cross-linking of single-Cys versions of SpoIVFB and Pro-σK, both in E. coli (25) and in vitro (26) in the presence or absence of ATP, should further elucidate how the SpoIVFB linker mediates ATP-dependent coordination between the CBS and membrane domains.
The effects of SpoIVFB linker substitutions were surprising in B. subtilis. The H206A change that eliminated Pro-σK cleavage in E. coli (Fig. 2D) was inconsequential during sporulation (Fig. 3). This difference is reminiscent of the effects of an F18A substitution in Pro-σK, which strongly impaired cleavage in E. coli but not in sporulating B. subtilis (23). We speculate that differences in membrane composition may account for these observations. Another surprise was the instability of the SpoIVFB F209A and R213A variants in B. subtilis (Fig. 3), despite their accumulation in E. coli (Fig. 2 D and E). The variants may have altered interactions with SpoIVFA and/or BofA, two proteins that form a complex with SpoIVFB and inhibit its protease activity, and strongly influence SpoIVFB accumulation during sporulation (32, 33). Topological studies in E. coli suggest that SpoIVFA and BofA help TMS 3 and 4 of SpoIVFB insert in the inner membrane (32). SpoIVFB appeared to be active at 30 °C but inactive at 37 °C in the absence of SpoIVFA and BofA (32, 34). Intriguingly, conservative substitutions in the SpoIVFB linker (V207M, L221V) permitted activity at 37 °C in the absence of SpoIVFA (32, 34), suggesting that the linker affects the thermostability of SpoIVFB in sporulating B. subtilis. We tested whether any of our SpoIVFB variants that failed to accumulate at 37 °C would exhibit a difference at 30 °C, but none did. Our results add to the evidence that the SpoIVFB linker profoundly impacts stability of the protein during sporulation, but a molecular explanation remains elusive. Our results show that coexpression with Pro-σK in E. coli allows at least some of these SpoIVFB variants to accumulate, but there are many possible reasons (e.g., differences in expression level, cellular proteases, and/or potential interaction partners such as SpoIVFA and BofA).
We used chemical cross-linking and mass spectrometry to gain further insight into the interaction between SpoIVFB and Pro-σK. Cross-links between the two proteins established their orientation in the complex, and cross-links within each protein supported predictions based on homology models of the SpoIVFB CBS domain and the Pro-σK domain 2 (SI Appendix, Tables S2 and S3). Cross-links within the SpoIVFB membrane domain were not observed, presumably due to the positions of Lys and Cys residues, the positions of trypsin cleavage sites, and/or poor ionization of hydrophobic peptides.
Our chemical cross-linking data were combined with disulfide cross-linking results reported previously (25) to constrain a model of the SpoIVFB–Pro-σK complex. In the model, the prosequence of Pro-σK loops into the active site of SpoIVFB (Fig. 5B). This allows several residues around the cleavage site in Pro-σK to be in proximity to residues E44, V70, and P135 in the active site region of SpoIVFB. Even so, the distance between Cα atoms of some residue pairs in the model exceeds the maximum theoretical distance for a disulfide cross-link (SI Appendix, Table S3) that was observed experimentally (25). This could reflect dynamic interaction between the two proteins and/or a multistep binding process, as has been proposed to explain the results of disulfide cross-linking experiments involving the E. coli IP RseP and its substrate RseA (35). Mutational studies have shown that several residues in the prosequence of Pro-σK are important for cleavage to occur (22, 23), but residues 1–17 are unconstrained by the available cross-linking data (SI Appendix, Table S3), so this aspect of the model needs further work. Likewise, residues 30–37 of Pro-σK are unconstrained by the cross-linking data, but the model predicts proximity of this unstructured region to several regions of SpoIVFB (SI Appendix, Fig. S9), which can now be tested.
The model predicts extensive interactions between Pro-σK and the SpoIVFB linker and CBS domain. Lysine residues 38, 39, 46, 68, and 69 of Pro-σK formed cross-links (Fig. 4C and SI Appendix, Table S2) that constrain this region to be in proximity to the linker (Fig. 5B and SI Appendix, Fig. S8) and CBS domain (SI Appendix, Fig. S7) of SpoIVFB. Among the Pro-σK residues that formed cross-links, residues 68 and 69 are predicted by the model to be in close proximity (5–10 Å) to SpoIVFB linker residue 201, and Pro-σK residue 39 is predicted to be in close proximity (5–8 Å) to SpoIVFB linker residue 217 (Fig. 5B and SI Appendix, Table S3). More broadly, the model predicts that Pro-σK residues 28–69 pack against the SpoIVFB linker (SI Appendix, Figs. S8–S10). The three SpoIVFB linker residues 206, 209, and 213, shown by Ala substitutions to be essential for cleavage of coexpressed Pro-σK in E. coli and important for binding to Pro-σK (Fig. 2), are predicted to be closest (7 Å between Cα residues) to Pro-σK residues 43, 51, and 46, respectively (SI Appendix, Fig. S8). The region spanning residues 38–69 of Pro-σK may form one α-helix (residues 51–65, helix 1 of σK domain 2, as shown in the figures) or two α-helices (residues 38–46 and 51–69). Pro-σK residues 38 and 69 are predicted to be within 7–13 Å of SpoIVFB CBS domain residue 246 (SI Appendix, Fig. S7 and Table S3) with which cross-links were observed (Fig. 4C and SI Appendix, Table S2), and the model predicts even closer proximity between residues 96–99 in the loop connecting α-helices 2 and 3 of σK domain 2 and several residues in a region of the CBS domain spanning residues 228–246 (SI Appendix, Fig. S11). This aspect of the model is supported by cross-links between residue 246 in the SpoIVFB CBS domain and residues 99, 102, and 110 of Pro-σK (Fig. 4C and SI Appendix, Fig. S7 and Tables S2 and S3).
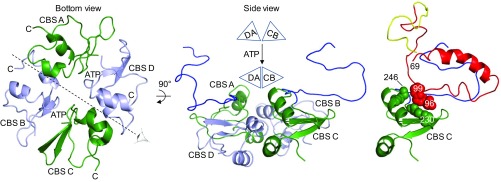
We propose that packing of Pro-σK residues 28–69 and 96–99 against the SpoIVFB linker (SI Appendix, Fig. S10) and CBS domain (SI Appendix, Fig. S11), respectively, allow ATP-induced movement of the CBS domain tetramer to position Pro-σK for cleavage. CBS domains adopt a variety of arrangements in different proteins (19, 36). SpoIVFB is tetrameric with one CBS domain per monomer (18, 26). In our model, the four CBS domains are arranged as an “antiparallel CBS module” (36) in which the A and D chain CBS domain pair is head-to-tail relative to the C and B chain CBS domain pair (Fig. 6, bottom view). The linkers of the D and B chains stretch across the bottom of the CBS module (Fig. 5A, bottom view) and are unavailable for interaction with Pro-σK. In contrast, the linkers of the A and C chains are on the same side of the CBS module as the membrane domains and interact with Pro-σK (Fig. 5B and SI Appendix, Figs. S8 and S10). Since ligand binding can induce bending of antiparallel CBS modules (36–38), we propose that ATP binding to the SpoIVFB CBS module induces a conformational change. The SpoIVFB CBS module would have two ATP-binding sites, one in the A and D chain CBS domain pair and one in the C and B chain CBS domain pair (Fig. 6, bottom view). Binding of ATP to both sites would cause movement of the CBS domain pairs relative to each other, as depicted at the top of the side view in Fig. 6. This type of “bent-to-flat” transition upon ligand binding to antiparallel CBS modules has been inferred from crystal structures of the apo- and AMP-bound forms of CBSX2 from Arabidopsis thaliana (36, 37). We propose that a similar transition upon binding of ATP to the SpoIVFB CBS module positions Pro-σK for productive interaction with the SpoIVFB active site, due to extensive interactions between Pro-σK and the SpoIVFB linker and CBS domain (Fig. 6, Right).
Fig. 6.
How ATP-induced movement of the SpoIVFB CBS domain tetramer might position Pro-σK for cleavage. (Left) The bottom view is the same as in Fig. 5A except the interdomain linkers and the Pro-σK molecules have been removed. CBS domains are labeled according to their chain, and each C terminus is indicated to highlight the antiparallel arrangement. The eye and dashed line indicate the axis of the side view shown in Middle. In the side view, the A and D chain CBS pair is on the left with CBS D in front. The pair is represented as a triangle labeled DA in the schematic above. The C and B chain CBS pair (represented as triangle CB above) is on the right with CBS C in front. The A and C chain linkers are in blue. The schematic illustrates ATP-induced movement of the CBS domain pairs, which would cause the A and C chain linkers to move. Right shows the interaction of Pro-σK chain Y with the linker and CBS domain of chain C. The interaction is proposed to allow movement of the C and B chain CBS pair to position Pro-σK for cleavage. Residues 70–95 and 100 to the C terminus of Pro-σK have been removed so that interaction of residues 28–69 with the SpoIVFB linker can be seen (see also SI Appendix, Fig. S10), and residues 96–99 of Pro-σK (red space-filling, main chain only) as well as residues 228–230, 241, and 246 of CBS C (green space-filling, main chain only) are shown to highlight their interaction (see also SI Appendix, Fig. S11).
Productive interaction of Pro-σK with the SpoIVFB active site also likely involves a membrane-reentrant loop that connects TMS 2 and TMS 3 (25). This loop is homologous to a region of E. coli RseP named the membrane-reentrant β-loop (MRE β-loop), a part of which has been proposed to form a β-strand that acts like the edge strand of many metalloproteases (39). The edge strand binds to a substrate in an extended conformation through β-strand addition and presents the extended substrate to catalytic residues at the metalloprotease active site (40, 41). In our model, residues 69–72 of the SpoIVFB C chain are depicted as a β-strand (Fig. 5B, Middle), which could act as the edge strand. E71 in the putative edge strand is one of five glutamate residues in the region spanning SpoIVFB residues 71–83. Diverse S2P family members have at least two residues with a negatively charged side chain in the corresponding region, and at least two of the five glutamate residues in SpoIVFB were required for activity (25). It was proposed that two glutamate residues interact electrostatically with K24 of Pro-σK, which had been shown to be important for cleavage by SpoIVFB (23). In our model, K24 of Pro-σK is near E82 and E83 of SpoIVFB; however, considerable flexibility would be required for E71, E73, or E74 of SpoIVFB to interact with K24 of Pro-σK. More work will be necessary to understand the roles of the MRE β-loop and the adjoining negatively charged region of S2P family members.
A short loop predicted to interrupt SpoIVFB TMS 4 also likely helps position Pro-σK in the active site region (25). This loop is part of the NX2PX4DG motif conserved in all S2P family members (8). The aspartate residue of the motif coordinates the zinc ion, along with two histidine residues in another TMS (27). The short loop of E. coli RseP is involved in substrate binding (35). Ala substitutions for N129 and D137 at the ends of the SpoIVFB short loop impair cleavage of Pro-σK (16, 17). A P132A substitution in the loop destabilized SpoIVFB (16), similar to the results we observed for F209A and R213A substitutions in the linker (Fig. 3). In our model, the side chains of N129 and D137 project toward the zinc ion at the active site, while P132 and P135 constrain the backbone of the loop so that the side chains of W134, P135, and L136 form a hydrophobic face of the active site pocket, opposite the putative edge strand of the MRE β-loop. Perhaps the hydrophobic face facilitates β-strand addition to the edge strand and/or stabilizes the β-strand in an extended conformation after addition. Residues 16–23 (LVFLVSYV) of Pro-σK are mostly hydrophobic, as are residues adjoining the cleavage site in other substrates of S2P family members since these enzymes cleave TMSs. The role of the predicted short loop of SpoIVFB and other S2P family members warrants further exploration.
Interestingly, residues 15–25 of the Pro-σK Y chain, which span the cleavage site (between residues 21 and 22), resemble an MRE β-loop in which residues 15–19 and 22–25 could form β-strands, although they are not depicted as β-strands in our model (Fig. 5B, Middle). Nevertheless, both regions are in an extended conformation compatible with β-strand addition to the putative edge strand. In this respect, Pro-σK appears to differ from most other IP substrates, which have a TMS that is presumed to be α-helical. For such substrates, α-helix–destabilizing residues typically facilitate substrate cleavage (42–48). In contrast, helix-destabilizing residues near the cleavage site are not crucial for SpoIVFB to cleave Pro-σK (25). A variety of experiments indicate that the prosequence of Pro-σK is necessary, and the first 28–32 residues are sufficient, for Pro-σK to associate peripherally with membranes (18, 22, 23, 26, 49), consistent with the notion that residues 15–25 form an MRE β-loop rather than part of an α-helical TMS. This apparent difference between Pro-σK and most other IP substrates might explain the need for an ATP-induced conformational change in SpoIVFB to position its substrate for cleavage, which in turn necessitates the extensive interactions between SpoIVFB and Pro-σK implied by our model. The extensive interactions constitute a large exosite (i.e., region outside the active site) on SpoIVFB for binding of Pro-σK. Other IPs appear to have exosites (50–52), and both IPs and their substrates are proposed to undergo conformational changes that position substrates for cleavage (48), but much more work is needed to discern general principles of intramembrane proteolysis from principles that apply more narrowly to particular types of IPs and from characteristics unique to an individual IP.
Our results provide unprecedented insight into the interaction of an IP with its substrate. Our model of the SpoIVFB–Pro-σK complex makes numerous predictions that can be tested. Obviously, structural analysis of the complex is a worthwhile goal. Also, the effects of the inhibitory proteins SpoIVFA and BofA on the interaction of SpoIVFB with Pro-σK need to be investigated. It is unknown whether Pro-σK can interact with the ternary SpoIVFB–SpoIVFA–BofA complex. The extensive interaction between SpoIVFB and Pro-σK predicted by our model raises the issue of product release after cleavage. Loss of interactions with the prosequence and perhaps a conformational change upon dissociation of ATP may be sufficient, or additional factors such as the membrane environment may be required. Since σK, but not Pro-σK, binds to core RNA polymerase (49), binding of the released product to core RNA polymerase likely ensures that it does not compete with substrate for binding to SpoIVFB. SpoIVFB and RseP have CBS and PDZ domains, respectively (8), and represent the largest subfamilies of S2Ps. It remains to be seen how well insights from studies of these model bacterial IPs translate to other S2Ps and to strategies for manipulating their activity. Some aspects of the interaction between B. subtilis SpoIVFB and Pro-σK can likely inform work on orthologs in related bacteria. Many of these bacteria cause disease or provide benefits, and persist by forming endospores (6, 7). Since cleavage of Pro-σK by SpoIVFB is a conserved and crucial event for endospore formation, a long-term potential implication of our work is the facilitation of efforts to inhibit or enhance sporulation.
Methods
Plasmids and Primers.
The plasmids and primers used in this study are described in SI Appendix, Tables S4 and S5, respectively. Site-directed mutagenesis was performed using the QuikChange kit (Stratagene). Genes subjected to site-directed mutagenesis or cloned after PCR were verified by DNA sequencing.
Proteolytic Cleavage Assay in E. coli.
Two plasmids with different antibiotic resistance genes and designed to produce Pro-σK(1-127)-His6 as the substrate or TM-SpoIVFB-FLAG2 (or a derivative) under control of a T7 RNA polymerase promoter were cotransformed into E. coli strain BL21(DE3), grown in Luria–Bertani (LB) medium supplemented with kanamycin sulfate (50 μg/mL) and ampicillin (100 μg/mL) at 37 °C, induced with 1 mM IPTG for 2 h, and equivalent amounts of cells were subjected to immunoblot analysis with chemiluminescence detection (21, 53). SeeBlue Plus2 Prestained Standard (Invitrogen) was used to judge migration of protein species. Antibodies that recognize His6 (penta-His; Qiagen) or FLAG (Sigma) were used at 1:5000 or 1:10000 dilution, respectively.
Cobalt Affinity Purification (Pull-Down Assays).
E. coli cotransformed with two plasmids were grown in 500 mL of LB medium and induced as described above. Cells were harvested, lysed, and centrifuged at low speed as described (26) except 20 mL of lysis buffer was used. The supernatant was treated with 1% n-decyl-β-d-maltoside (DM) (Anatrace) for 1 h at 4 °C to solubilize membrane proteins then centrifuged at 150,000 × g for 1 h at 12 °C. The supernatant was designated the input sample, and 100 μL was saved for immunoblot analysis. The rest was mixed with 0.5 mL of Talon superflow metal affinity resin (Clontech) that had been equilibrated with PBS containing 0.1% DM, 5 mM 2-mercaptoethanol, and 10% glycerol. The mixture was rotated for 1 h at room temperature. The cobalt resin was sedimented by centrifugation at 708 × g for 2 min at 4 °C. The supernatant was the unbound sample. The resin was washed three times with 5 mL of PBS containing 150 mM NaCl and 10% glycerol, then once with 5 mL of PBS containing 150 mM NaCl, 10% glycerol, 0.1% DM, and 40 mM imidazole, each time rotating the mixture briefly and sedimenting the resin as above. The resin was mixed with 10 mL of PBS containing 150 mM NaCl, 10% glycerol, and 0.1% DM, resulting in the bound sample. The samples (75 μL) were added to 25 μL of 4× sample buffer (100 mM Tris·HCl pH 6.8, 8% SDS, 40% glycerol, 400 mM DTT, and 0.06% bromophenol blue), boiled 3 min, and subjected to immunoblot analysis as described above. A representative result from at least two biological replicates is shown. Signal intensities were quantified using Image Lab Software (Bio-Rad) for two replicates, and the range of ratios of bound/unbound SpoIVFB is reported in Fig. 2F.
Purification and CD of SpoIVFB and SpoIVFB A12.
E. coli transformed with a plasmid to produce TM-SpoIVFB-FLAG2-His6 or its A12 derivative were grown in 4 L of LB medium supplemented with ampicillin (200 mg/mL) and induced as described above. Proteins were purified as described in SI Appendix. Far-UV CD measurements were made using an Applied Photophysics Chirascan Spectropolarimeter. The spectra were acquired for proteins purified as described above and diluted to a concentration of 3 μM in PBS containing 10% glycerol and 0.3% DM. The spectra were recorded with 0.4-s adaptive integration time and 1-nm bandwidth at 25 °C in a 1-mm path-length cell. Each spectrum shown is the average of four scans.
Sporulation and Immunoblot Analysis.
B. subtilis strain PY79 (54) with a wild-type spoIVFAB operon or its derivative BSL51 (55) in which the spoIVFAB operon was replaced with a chloramphenicol resistance gene served as controls. BSL51 was transformed with pDR18a or a derivative, and transformants were selected on LB agar containing spectinomycin (100 μg/mL) and chloramphenicol (5 μg/mL) (56). The plasmids were derived from pLD30, which permits gene replacement of amyE in the chromosome, as identified by loss of amylase activity on 1% potato starch medium with Gram’s iodine solution (56). Sporulation was induced by growing cells in the absence of antibiotics and resuspension of cells in SM medium (56). Samples (0.5 mL) collected at the indicated times after induction were centrifuged (12,000 × g), whole-cell extracts were prepared as described previously for E. coli (21) except samples were incubated at 55 °C for 5 min rather than boiling for 3 min, and proteins were subjected to immunoblot analysis (53). SeeBlue Plus2 Prestained Standard (Invitrogen) was used to judge migration of protein species. Antibodies that recognize SpoIVFA (53) or Pro-σK (29) were used at 1:3000 dilution. Antibodies that recognize SpoIVFB were affinity-purified using the antigen (17) coupled to Affigel-10 (Bio-Rad Laboratories) according to the manufacturer’s instructions. The column (1 mL) was washed with 10 mL of PBS, 10 mL of PBS containing 500 mM NaCl and 0.1% Tween-20, 10 mL of 0.2× PBS, 10 mL of 100 mM glycine (pH 2.5), and 20 mL of PBS. The column material was rotated with 10 mL of antiserum overnight at 4 °C. The column was washed as described above (first three washes) and eluted with 10 mL of 100 mM glycine (pH 2.5) collecting 1-mL fractions directly into 2 M Tris to neutralize immediately. The first fraction contained affinity-purified antibodies and was dialyzed against PBS containing 50% glycerol. The antibodies were used at 1:5000 dilution. Sporulation was measured at 24 h after induction (56).
Purification of the SpoIVFB–Pro-σK Complex.
E. coli strain BL21(DE3) transformed with pYZ42 to produce Pro-σK(1-127)-His6, and catalytically inactive SpoIVFB-TEV-FLAG2 E44Q was grown in a fermentor as described (26), induced with 1 mM IPTG for 4 h, and the complex was purified as described in SI Appendix.
Chemical Cross-Linking.
Cross-linkers were purchased from Thermo Scientific. Purified SpoIVFB–Pro-σK complex (∼5 µM each protein) was mixed with cross-linker (0.1–0.5 mM) in a 100-μL reaction with PBS containing 150 mM NaCl, 5% glycerol, and 0.02% DDM for 1 h at 25 °C, and quenched by adding 2 μL of 1 M Tris, pH 7.5. Samples were subjected to SDS/PAGE on 4–20% Mini-PROTEAN TGX gels (Bio-Rad), the gels were stained with Coomassie blue, and cross-linked products were excised.
Digestion of Cross-Linked Products and Peptide Mass Analysis.
Cross-linked products were digested in-gel, and the resulting peptides were purified as described in SI Appendix. Eluted peptides were analyzed on a ThermoFisher Q-Exactive mass spectrometer as described in SI Appendix. MS raw files were analyzed using the Mascot search algorithm (57) within the Mascot Distiller package, and cross-linked peptides were determined using the StavroX software package (58) as described in SI Appendix.
Supplementary Material
Acknowledgments
We thank Lisa Lapidus for use of the spectropolarimeter and David Rudner for pDR18a and advice on purification and use of SpoIVFB antibodies. The in-gel protease digestion and peptide mass analysis were conducted at the Michigan State University Proteomics Facility. This research was supported by National Institutes of Health Grants R01 GM43585 (to L.K.) and R01 GM084953 (to M.F.), and by Michigan State University AgBioResearch.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711467114/-/DCSupplemental.
References
- 1.Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: A control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 2.Urban S. Mechanisms and cellular functions of intramembrane proteases. Biochim Biophys Acta. 2013;1828:2797–2800. doi: 10.1016/j.bbamem.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Manolaridis I, et al. Mechanism of farnesylated CAAX protein processing by the intramembrane protease Rce1. Nature. 2013;504:301–305. doi: 10.1038/nature12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kroos L, Akiyama Y. Biochemical and structural insights into intramembrane metalloprotease mechanisms. Biochim Biophys Acta. 2013;1828:2873–2885. doi: 10.1016/j.bbamem.2013.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galperin MY, et al. Genomic determinants of sporulation in Bacilli and Clostridia: Towards the minimal set of sporulation-specific genes. Environ Microbiol. 2012;14:2870–2890. doi: 10.1111/j.1462-2920.2012.02841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Hinai MA, Jones SW, Papoutsakis ET. The Clostridium sporulation programs: Diversity and preservation of endospore differentiation. Microbiol Mol Biol Rev. 2015;79:19–37. doi: 10.1128/MMBR.00025-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Checinska A, Paszczynski A, Burbank M. Bacillus and other spore-forming genera: Variations in responses and mechanisms for survival. Annu Rev Food Sci Technol. 2015;6:351–369. doi: 10.1146/annurev-food-030713-092332. [DOI] [PubMed] [Google Scholar]
- 8.Kinch LN, Ginalski K, Grishin NV. Site-2 protease regulated intramembrane proteolysis: Sequence homologs suggest an ancient signaling cascade. Protein Sci. 2006;15:84–93. doi: 10.1110/ps.051766506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rawson RB. The site-2 protease. Biochim Biophys Acta. 2013;1828:2801–2807. doi: 10.1016/j.bbamem.2013.03.031. [DOI] [PubMed] [Google Scholar]
- 10.Ye J. Roles of regulated intramembrane proteolysis in virus infection and antiviral immunity. Biochim Biophys Acta. 2013;1828:2926–2932. doi: 10.1016/j.bbamem.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider JS, Glickman MS. Function of site-2 proteases in bacteria and bacterial pathogens. Biochim Biophys Acta. 2013;1828:2808–2814. doi: 10.1016/j.bbamem.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adam Z. Emerging roles for diverse intramembrane proteases in plant biology. Biochim Biophys Acta. 2013;1828:2933–2936. doi: 10.1016/j.bbamem.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 13.Guan M, Su L, Yuan YC, Li H, Chow WA. Nelfinavir and nelfinavir analogs block site-2 protease cleavage to inhibit castration-resistant prostate cancer. Sci Rep. 2015;5:9698. doi: 10.1038/srep09698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zoll S, et al. Substrate binding and specificity of rhomboid intramembrane protease revealed by substrate-peptide complex structures. EMBO J. 2014;33:2408–2421. doi: 10.15252/embj.201489367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho S, Dickey SW, Urban S. Crystal structures and inhibition kinetics reveal a two-stage catalytic mechanism with drug design implications for rhomboid proteolysis. Mol Cell. 2016;61:329–340. doi: 10.1016/j.molcel.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudner DZ, Fawcett P, Losick R. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc Natl Acad Sci USA. 1999;96:14765–14770. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu Y-TN, Kroos L. Evidence that SpoIVFB is a novel type of membrane metalloprotease governing intercompartmental communication during Bacillus subtilis sporulation. J Bacteriol. 2000;182:3305–3309. doi: 10.1128/jb.182.11.3305-3309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou R, Cusumano C, Sui D, Garavito RM, Kroos L. Intramembrane proteolytic cleavage of a membrane-tethered transcription factor by a metalloprotease depends on ATP. Proc Natl Acad Sci USA. 2009;106:16174–16179. doi: 10.1073/pnas.0901455106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baykov AA, Tuominen HK, Lahti R. The CBS domain: A protein module with an emerging prominent role in regulation. ACS Chem Biol. 2011;6:1156–1163. doi: 10.1021/cb200231c. [DOI] [PubMed] [Google Scholar]
- 20.Scott JW, et al. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J Clin Invest. 2004;113:274–284. doi: 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou R, Kroos L. BofA protein inhibits intramembrane proteolysis of pro-σK in an intercompartmental signaling pathway during Bacillus subtilis sporulation. Proc Natl Acad Sci USA. 2004;101:6385–6390. doi: 10.1073/pnas.0307709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prince H, Zhou R, Kroos L. Substrate requirements for regulated intramembrane proteolysis of Bacillus subtilis pro-σK. J Bacteriol. 2005;187:961–971. doi: 10.1128/JB.187.3.961-971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou R, Chen K, Xiang X, Gu L, Kroos L. Features of Pro-σK important for cleavage by SpoIVFB, an intramembrane metalloprotease. J Bacteriol. 2013;195:2793–2806. doi: 10.1128/JB.00229-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou R, Kroos L. Serine proteases from two cell types target different components of a complex that governs regulated intramembrane proteolysis of pro-σK during Bacillus subtilis development. Mol Microbiol. 2005;58:835–846. doi: 10.1111/j.1365-2958.2005.04870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Luethy PM, Zhou R, Kroos L. Residues in conserved loops of intramembrane metalloprotease SpoIVFB interact with residues near the cleavage site in pro-σK. J Bacteriol. 2013;195:4936–4946. doi: 10.1128/JB.00807-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, et al. Complex formed between intramembrane metalloprotease SpoIVFB and its substrate, Pro-σK. J Biol Chem. 2016;291:10347–10362. doi: 10.1074/jbc.M116.715508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng L, et al. Structure of a site-2 protease family intramembrane metalloprotease. Science. 2007;318:1608–1612. doi: 10.1126/science.1150755. [DOI] [PubMed] [Google Scholar]
- 28.Saribas AS, Gruenke L, Waskell L. Overexpression and purification of the membrane-bound cytochrome P450 2B4. Protein Expr Purif. 2001;21:303–309. doi: 10.1006/prep.2000.1377. [DOI] [PubMed] [Google Scholar]
- 29.Lu S, Halberg R, Kroos L. Processing of the mother-cell σ factor, σ K, may depend on events occurring in the forespore during Bacillus subtilis development. Proc Natl Acad Sci USA. 1990;87:9722–9726. doi: 10.1073/pnas.87.24.9722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller MD, et al. Crystal structure of a tandem cystathionine-β-synthase (CBS) domain protein (TM0935) from Thermotoga maritima at 1.87 A resolution. Proteins. 2004;57:213–217. doi: 10.1002/prot.20024. [DOI] [PubMed] [Google Scholar]
- 31.Feklistov A, Darst SA. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase σ subunit. Cell. 2011;147:1257–1269. doi: 10.1016/j.cell.2011.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green DH, Cutting SM. Membrane topology of the Bacillus subtilis pro-σK processing complex. J Bacteriol. 2000;182:278–285. doi: 10.1128/jb.182.2.278-285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rudner DZ, Losick R. A sporulation membrane protein tethers the pro-σK processing enzyme to its inhibitor and dictates its subcellular localization. Genes Dev. 2002;16:1007–1018. doi: 10.1101/gad.977702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cutting S, Roels S, Losick R. Sporulation operon spoIVF and the characterization of mutations that uncouple mother-cell from forespore gene expression in Bacillus subtilis. J Mol Biol. 1991;221:1237–1256. doi: 10.1016/0022-2836(91)90931-u. [DOI] [PubMed] [Google Scholar]
- 35.Koide K, Ito K, Akiyama Y. Substrate recognition and binding by RseP, an Escherichia coli intramembrane protease. J Biol Chem. 2008;283:9562–9570. doi: 10.1074/jbc.M709984200. [DOI] [PubMed] [Google Scholar]
- 36.Ereño-Orbea J, Oyenarte I, Martínez-Cruz LA. CBS domains: Ligand binding sites and conformational variability. Arch Biochem Biophys. 2013;540:70–81. doi: 10.1016/j.abb.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 37.Jeong BC, Park SH, Yoo KS, Shin JS, Song HK. Change in single cystathionine β-synthase domain-containing protein from a bent to flat conformation upon adenosine monophosphate binding. J Struct Biol. 2013;183:40–46. doi: 10.1016/j.jsb.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 38.Labesse G, et al. MgATP regulates allostery and fiber formation in IMPDHs. Structure. 2013;21:975–985. doi: 10.1016/j.str.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 39.Akiyama K, et al. Roles of the membrane-reentrant β-hairpin-like loop of RseP protease in selective substrate cleavage. Elife. 2015;4:e08928. doi: 10.7554/eLife.08928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stöcker W, Bode W. Structural features of a superfamily of zinc-endopeptidases: The metzincins. Curr Opin Struct Biol. 1995;5:383–390. doi: 10.1016/0959-440x(95)80101-4. [DOI] [PubMed] [Google Scholar]
- 41.Langklotz S, Baumann U, Narberhaus F. Structure and function of the bacterial AAA protease FtsH. Biochim Biophys Acta. 2012;1823:40–48. doi: 10.1016/j.bbamcr.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 42.Ye J, Davé UP, Grishin NV, Goldstein JL, Brown MS. Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by site-2 protease. Proc Natl Acad Sci USA. 2000;97:5123–5128. doi: 10.1073/pnas.97.10.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemberg MK, Martoglio B. Requirements for signal peptide peptidase-catalyzed intramembrane proteolysis. Mol Cell. 2002;10:735–744. doi: 10.1016/s1097-2765(02)00655-x. [DOI] [PubMed] [Google Scholar]
- 44.Urban S, Freeman M. Substrate specificity of rhomboid intramembrane proteases is governed by helix-breaking residues in the substrate transmembrane domain. Mol Cell. 2003;11:1425–1434. doi: 10.1016/s1097-2765(03)00181-3. [DOI] [PubMed] [Google Scholar]
- 45.Akiyama Y, Kanehara K, Ito K. RseP (YaeL), an Escherichia coli RIP protease, cleaves transmembrane sequences. EMBO J. 2004;23:4434–4442. doi: 10.1038/sj.emboj.7600449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akiyama Y, Maegawa S. Sequence features of substrates required for cleavage by GlpG, an Escherichia coli rhomboid protease. Mol Microbiol. 2007;64:1028–1037. doi: 10.1111/j.1365-2958.2007.05715.x. [DOI] [PubMed] [Google Scholar]
- 47.Sato T, et al. A helix-to-coil transition at the ε-cut site in the transmembrane dimer of the amyloid precursor protein is required for proteolysis. Proc Natl Acad Sci USA. 2009;106:1421–1426. doi: 10.1073/pnas.0812261106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langosch D, Scharnagl C, Steiner H, Lemberg MK. Understanding intramembrane proteolysis: From protein dynamics to reaction kinetics. Trends Biochem Sci. 2015;40:318–327. doi: 10.1016/j.tibs.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 49.Zhang B, Hofmeister A, Kroos L. The prosequence of pro-σK promotes membrane association and inhibits RNA polymerase core binding. J Bacteriol. 1998;180:2434–2441. doi: 10.1128/jb.180.9.2434-2441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arutyunova E, et al. Allosteric regulation of rhomboid intramembrane proteolysis. EMBO J. 2014;33:1869–1881. doi: 10.15252/embj.201488149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fukumori A, Steiner H. Substrate recruitment of γ-secretase and mechanism of clinical presenilin mutations revealed by photoaffinity mapping. EMBO J. 2016;35:1628–1643. doi: 10.15252/embj.201694151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kornilova AY, Bihel F, Das C, Wolfe MS. The initial substrate-binding site of γ-secretase is located on presenilin near the active site. Proc Natl Acad Sci USA. 2005;102:3230–3235. doi: 10.1073/pnas.0407640102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kroos L, Yu YT, Mills D, Ferguson-Miller S. Forespore signaling is necessary for pro-σK processing during Bacillus subtilis sporulation despite the loss of SpoIVFA upon translational arrest. J Bacteriol. 2002;184:5393–5401. doi: 10.1128/JB.184.19.5393-5401.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Youngman P, Perkins JB, Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984;12:1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]
- 55.Lu S, Kroos L. Overproducing the Bacillus subtilis mother cell sigma factor precursor, Pro-σK, uncouples σK-dependent gene expression from dependence on intercompartmental communication. J Bacteriol. 1994;176:3936–3943. doi: 10.1128/jb.176.13.3936-3943.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cutting SM, Horn PBV. Genetic analysis. In: Harwood CR, Cutting SM, editors. Molecular Biological Methods for Bacillus. John Wiley & Sons; New York: 1990. pp. 27–74. [Google Scholar]
- 57.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 58.Götze M, et al. StavroX–A software for analyzing crosslinked products in protein interaction studies. J Am Soc Mass Spectrom. 2012;23:76–87. doi: 10.1007/s13361-011-0261-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.