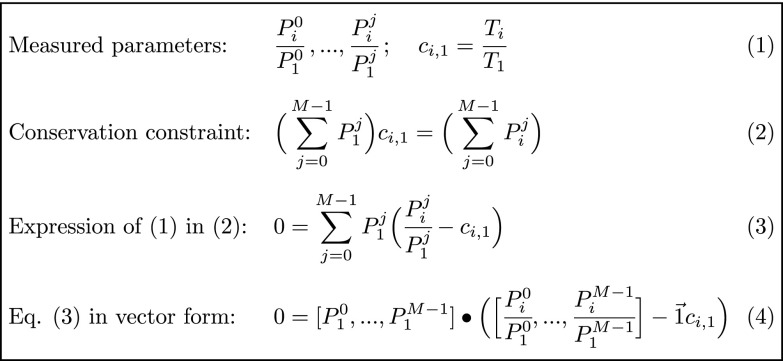Significance
Protein phosphorylation and degradation drive critical events in early embryogenesis and the cell cycle; however, comprehensive and accurate analysis of these changes is currently difficult. Using a mass-spectrometric approach, we present a quantitative view of the protein and posttranslational economy of the fertilization response in the frog egg. Protein degradation affects a small but very important class of proteins, while regulatory phosphorylation and protein release occur on a far larger scale. We have developed broadly applicable analytical methods for phosphorylation that provide absolute quantification with confidence intervals for improved interpretability of posttranslational modification analysis.
Keywords: cell cycle, polyspermy block, stoichiometry, confidence intervals, TMT
Abstract
Fertilization releases the meiotic arrest and initiates the events that prepare the egg for the ensuing developmental program. Protein degradation and phosphorylation are known to regulate protein activity during this process. However, the full extent of protein loss and phosphoregulation is still unknown. We examined absolute protein and phosphosite dynamics of the fertilization response by mass spectrometry-based proteomics in electroactivated eggs. To do this, we developed an approach for calculating the stoichiometry of phosphosites from multiplexed proteomics that is compatible with dynamic, stable, and multisite phosphorylation. Overall, the data suggest that degradation is limited to a few low-abundance proteins. However, this degradation promotes extensive dephosphorylation that occurs over a wide range of abundances during meiotic exit. We also show that eggs release a large amount of protein into the medium just after fertilization, most likely related to the blocks to polyspermy. Concomitantly, there is a substantial increase in phosphorylation likely tied to calcium-activated kinases. We identify putative degradation targets and components of the slow block to polyspermy. The analytical approaches demonstrated here are broadly applicable to studies of dynamic biological systems.
For decades, the highly synchronous events of fertilization have provided a useful system for the study of many aspects of cellular regulation, especially protein degradation and phosphorylation. The destruction of Cyclin-B and other proteins is catalyzed by the anaphase-promoting complex (APC/C), which promotes M-phase exit (1). The two activators of the APC/C are Cdc20 and Cdh1. In the egg, the cell cycle typically involves only Cdc20 (2). While the list of known Cdh1 substrates continues to grow (3), the Cdc20 target list remains small (4–6). It is of considerable interest to characterize the minimal set of Cdc20 substrates that powers the early cell cycle. Kinase activity is equally important to this regulation. Cyclin-B degradation promotes mitotic exit by inhibiting the activity of Cyclin-dependent kinase 1 (Cdk1). There is a bulk loss of phosphorylation following egg activation (7). The identities of the vast majority of these phosphoproteins remain undiscovered. However, there is a strong expectation that this regulation reflects the reversal of Cdk1 phosphorylation. Fertilization employs additional regulation not common to other cell cycles. There are timed waves of phosphorylation that correspond to the release of cortical granules just after fertilization as part of the slow block to polyspermy. This release is calcium sensitive and may reflect increases in the activity of protein kinase C (PKC) (8, 9) and CaMKII (10). An account of the secreted proteins, their function, and their upstream signaling components is limited. To investigate these unknown aspects of degradation, release, and modification of proteins at fertilization comprehensively, we employed mass spectrometry (MS) in Xenopus laevis eggs. Recent advances in multiplexed proteomics allow quantitative comparisons of multiple conditions using tandem mass tags (TMTs) (11–13). Our recent work demonstrates the power of proteomics in Xenopus for studies of early development (14–16). Although fertilization has been studied before with MS (17–20), the studies were either qualitative or did not measure phosphorylation. With an enrichment step (21–23), it is feasible to measure relative changes in a large number of phosphoproteins. However, such studies are of limited utility without measuring site stoichiometry. Relying on fold change alone for phosphorylation studies will not distinguish between 10-fold relative changes from 1% occupancy to 10% versus 10 to 100%. Several approaches are available (24–26), but none of these is able to determine occupancies of peptides measured with multiplexed MS or that have multiple phosphorylated residues. This paper introduces biological findings about fertilization and cell cycle progression as well as methods for measuring absolute stoichiometry of phosphosites that are widely applicable to MS protein modification studies.
Results
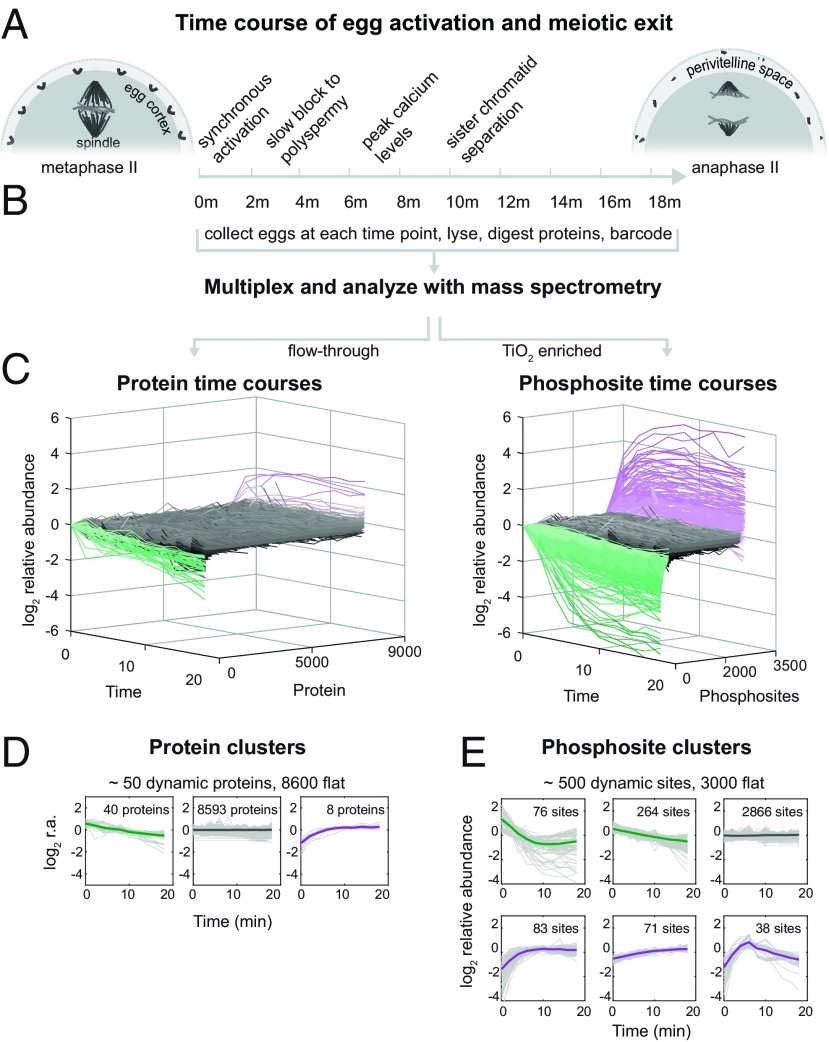
Quantitative Proteomics of Egg Activation and Meiotic Exit.
The large (1.2-mm) Xenopus laevis egg offers superb synchrony and sufficient material [30 μg of nonyolk protein per egg (27)] for proteomic and phosphoproteomic studies. To capture the progression of the rapid fertilization response (Fig. 1A), metaphase-arrested eggs were activated with electroshock and snap-frozen every 2 min in sets of 30 (Fig. 1B). We chose electrical activation over sperm addition to maximize time resolution by avoiding the asynchrony of sperm penetration through the jelly coat (28). The early morphological and molecular events to our knowledge are equivalent between fertilization and parthenogenetic activation (29, 30); hence we use the terms interchangeably here. More than 99% of eggs activate with a SD of <15 s using this approach (SI Appendix, Fig. S1 and Movies S1–S4). Time points were analyzed by MultiNotch MS3 (31). Phosphopeptides were enriched on a titanium dioxide column; the flow-through was used for protein-level MS. We multiplexed the TMT-labeled time points before enrichment to eliminate errors that arise from variation between columns. Protein-level MS was performed with four biological replicates, and phospho-level MS was performed in biological triplicate. We quantified ∼8,700 proteins (80% detected in two or more replicates) and ∼3,500 phosphosites (40% in two or more replicates) on 1,700 proteins (Fig. 1C). Less than 1% of detected proteins showed abundance changes. Forty proteins decrease in abundance over time, while eight unexpectedly show an apparent increase (Fig. 1D) (discussed below). Protein phosphosites are notably more dynamic, with ∼15% of modified residues changing (Fig. 1E). There is clear evidence for parallel dephosphorylation and phosphorylation. The phosphosite changes overwhelmingly occur on stable proteins; thus, they reflect actual changes in phosphorylation level rather than protein level.
Fig. 1.
Experimental overview. (A) Fertilization and egg activation release metaphase-arrested oocytes into anaphase and initiate the slow block to polyspermy, sister chromatid separation, remodeling of the specialized egg extracellular matrix, and inflation of the perivitelline space, among other processes. We mimic fertilization and trigger egg activation via electric shock to maximize synchrony. (B) After activation, eggs were collected every 2 min, lysed, and digested with proteases. Samples were barcoded using tandem mass tags (TMTs) and multiplexed. Protein and phosphosite dynamics were measured with mass spectrometry-based proteomics. Phosphorylated peptides were enriched using TiO2, and the column flow-through peptides were used for protein analysis. (C) Modified waterfall plots displaying the trend of every protein and phosphosite in the dataset, normalized to the first time point and ordered first by clusters (see below) and then in ascending order within each cluster. (D and E) K-means clustering to summarize dynamic classes of the protein (D) and phosphorylation time series (E) (SI Appendix, SI Methods). Bold line represents the centroid of the cluster, while the gray lines are individual time courses (normalized to the time course means).
Protein Loss Following Fertilization.
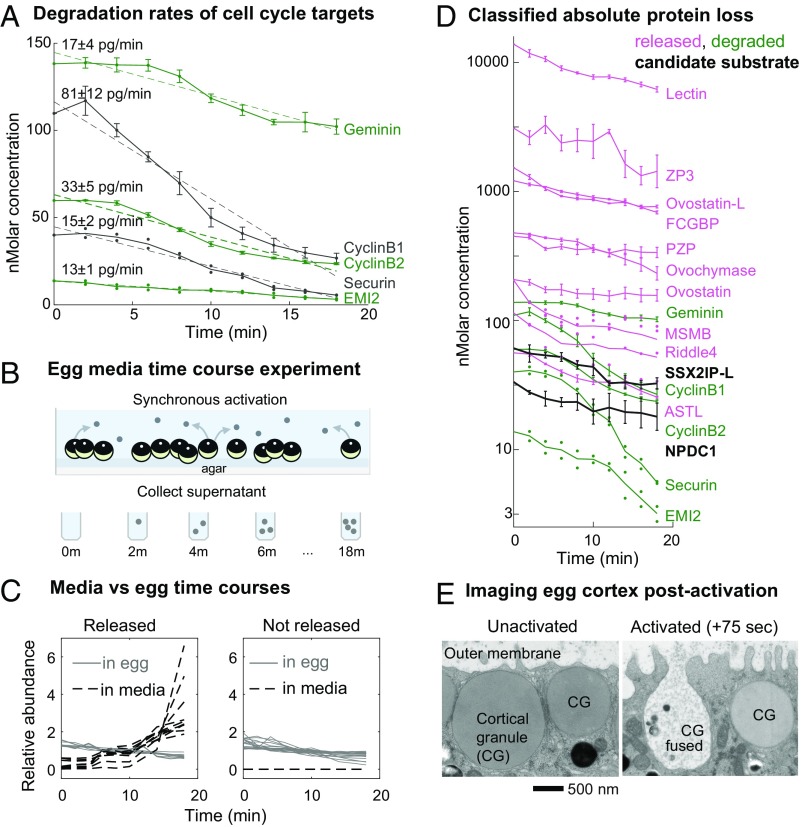
We sought to determine whether additional proteins beyond the known cell cycle targets are degraded. We identify 29 unique proteins by gene symbols that significantly decrease in abundance (Dataset S1, Tables S1 and S2). To establish statistical significance, we calculated a randomization-based false-discovery rate (FDR) (SI Appendix, Fig. S2) and set a cutoff of 1%. Specifically, we find known APC/CCdc20 substrates, including Cyclin-B1, Cyclin-B2, Geminin (32), Securin (33, 34), and the -TRCP substrate EMI2 (35) decrease significantly (Fig. 2A). Previously, we estimated the absolute abundances of ∼11,000 proteins in the frog egg (14), which allowed us to estimate absolute rates of degradation for these proteins. We see a small delay of ∼2–4 min between fertilization and initiation of degradation of the APC/C substrates. EMI2, whose destruction is required for APC/C activity, declines without delay. It is reduced by 30% 4 min after activation, when Cyclin-B1/2 and Securin begin to decline. EMI2 is stoichiometric with the least abundant components of the APC/C (14). Therefore, a ∼30% decline of EMI2 leads to a 30% increase in the maximal activity of APC/C. This activity must therefore be sufficient to initiate degradation. All observed putative APC/CCdh1 substrates, such as CDC20 (36–38), PLK1 (5), and Aurora A/B (39, 40) (SI Appendix, Fig. S3), are stable. The total loss of the known APC/CCdc20 substrates is ∼3,000 pg (or 250 nM), and they are degraded at a rate of ∼150 pg/min. From single-molecule measurements of the proteasome (41), we estimate the reactions responsible for protein degradation are approximately diffusion-limited for the reaction (SI Appendix, SI Methods). Since it is possible to competitively delay substrate degradation by adding peptide fragments of Cyclin-B (32, 42–44), the reaction in the egg may be near saturation. The overall estimated rates may be affected by the cell cycle metachrony (i.e., spatial heterogeneity) of the large Xenopus eggs. The cell cycle state should equilibrate between the top and bottom of the egg by ∼20 min given the speed of the traveling “waves” (45–47) and other previous measurements (48). Therefore, the end points we report are likely to be accurate. The half-times may be overestimated if there is substantial delay of degradation in the vegetal pole, although the exclusion of the cytoplasm by yolk may partially compensate for this phenomenon.
Fig. 2.
Protein loss occurs by degradation and release: Experiments and analysis performed to classify the mechanism of protein loss following egg activation. (A) Time series of the five proteins identified as significantly decreasing, which are known cell cycle degradation targets, plotted by their absolute changes. Error bars reflect SE (SEM) for proteins detected in at least three biological replicates. For proteins detected in fewer than three replicates, all data available are shown as points. Dashed lines represent a linear fit to the approximate zero-order kinetics (labeled with the slope and 95% confidence interval). (B) Experimental design to test for proteins released by eggs upon fertilization as an explanation for protein loss. Released proteins will increase in the media fraction over time. (C) Time series for proteins detected in both the egg and supernatant or the egg alone. These data were used to classify whether proteins were lost by release rather than degradation (by direct evidence or annotation; see text). This class comprised all but two of the significantly decreasing proteins (D) besides the known targets shown in A. (D) Time series of protein loss plotted by their absolute abundance (log10 transformed) and classified by the mechanism of loss. Error is visualized as in A. SSX2IP-L and NPDC1 homologs (black) are putative new degradation targets (see text for full criteria). (E) EM images of the cortical granules (CG) on the egg cortex. After egg activation, cortical granules fuse with the outer membrane and expel their contents.
Classifying Candidate Novel Degradation Targets.
Annotation of the decreasing proteins suggested that some candidate proteins were released from the cell rather than degraded (Dataset S1, Table S2). To test this hypothesis, we harvested the egg media in time series after egg activation (Fig. 2B). Proteins that appeared in the media and disappeared from the cell were classified as released (Fig. 2C and SI Appendix, Fig. S4). Release from the egg is the major source of protein loss rather than degradation (Fig. 2D). We found only six proteins that decreased abundance with no evidence for release (see Dataset S1, Table S3 and Methods section for full classifications and rationale). To determine whether these proteins are degraded through the ubiquitin pathway, we performed ubiquitin pulldowns with a di-glycine antibody (49) and analyzed the samples by MS. Of the six proteins that were not previously known degradation targets, two were ubiquitinated (Fig. 2D, black). A Xenopus homolog of the gene SSX2IP (sequence provided in Dataset S1, Table S1, distinguished as SSX2IP-L) is the most promising candidate for a novel APC/C substrate (Fig. 2D). SSX2IP is a centrosomal protein (50) with roles in the primary cilium (51). It is present at similar abundance to Cyclin-B (60 nM) and contains a confident D-box prediction (4). Ciliary defects occur when Cdc20 is depleted (52). SSX2IP may provide a mechanism for linking Cdc20 to the ciliary phenotypes. The other candidate is NPDC1, a neuronal protein with some association with cell cycle components (53) containing a weak D-box motif. However, it is localized to synaptic vesicles in neurons, which may suggest association with exocytosis machinery rather than degradation (although we do not detect it in the media). At the level of sensitivity in this study, the number of putative APC/CCdc20 substrates is indeed very small. All of them are below 100 nM in the egg. To estimate the sensitivity of our study, we compared the proteins identified using TMT-MS to those identified by the more sensitive (but semiquantitative) label-free approach in the frog egg (14) (SI Appendix, Fig. S5). At 50 nM, we detect 95% of the proteins with TMT-MS that we do with label-free, and still detect 50% at 10 nM. While we cannot rule out additional substrates, the data do suggest that any we did not detect are at low concentrations.
Proteins Released from the Egg.
By contrast to the relatively small mass and small number of proteins degraded after fertilization, there is a substantial reduction in cytosolic protein due to release into the medium. We detect the expected cortical granule components like an Interlectin2 homolog, as well as the proteases Ovostacin (54) and Ovochymase (55) whose function is thought to be inhibition of further sperm binding by cleaving the extracellular zona pellucida (ZP) gene family (56). We detect ZP2, 3, and 4 homologs (57) as released, which are likely to be cleaved peptide fragments that have diffused into the media (SI Appendix, Fig. S4). Therefore, at least two mechanisms are responsible for the release of protein from the interior of the egg and from its surface: exocytosis of cortical granules and proteolysis of the specialized extracellular matrix. We would not detect change for proteins that are trapped in the perivitelline space. We detect several proteins previously unknown to be released from the egg at fertilization, including several protease inhibitors. One examples is Ovostatin (AM2 homolog) (58), which was previously shown to have antitrypsin activity in the sea urchin egg (59). There are additionally a small number of other annotated protease inhibitors that decrease in the egg but are not detected in the medium. We surmise that these are released as well, rather than degraded (Dataset S1, Table S3). One example is Riddle4 (60) with known functions reported later in development (61). While the secretion of proteases is a well-recognized mechanism, the release of endogenous protease inhibitors in response to fertilization was not known. Another previously unknown released protein is a FCGBP homolog (Fig. 2D), which is similar to Zonadhesin (62, 63) that binds to the ZP proteins. It is likely extracellular and liberated by proteolysis. To compare the measured release of protein with the major morphological events of fertilization, the cell cortex of activated eggs was imaged by electron microscopy (EM) every 15 s (Fig. 2E). The release of cortical granules shows a 60-s delay, but between 75 and 90 s, ∼40% of the vesicles fuse with the outer membrane. By 10 min, the majority (∼60%) of the vesicles have fused (SI Appendix, Fig. S6). This is consistent with the generally faster loss rates of released proteins (Fig. 2D) and with previous work (9).
Observed Protein Increase Is a Phosphorylation Artifact.
A cluster of eight proteins appear to accumulate postfertilization (Fig. 1D), seven of which show a significant increase in abundance after the FDR analysis (Dataset S1, Table S4). Many of these proteins are typically stable and not expected to be synthesized during anaphase [e.g., NUP35, a nucleoporin (64)], which led us to search for an alternative explanation. Since protein trends are determined from the sum of all unmodified peptides, we examined the individual peptide trends to determine whether they all showed the same unexpected increase.
We found that these proteins did in fact have stable as well as increasing peptides (e.g., SI Appendix, Fig. S7). This discrepancy can occur from the loss of a modification on that particular peptide. The relative abundance of the unmodified peptide must increase if the modified form decreases, due to conservation. Indeed, for three of these proteins, we can directly show the loss of phosphorylation that causes the increase in the unmodified peptide (SI Appendix, Fig. S7). Overall, we have evidence that six of the seven apparently increasing proteins have phosphopeptides showing dramatic decreases in phosphorylation. Therefore, we conclude that the increasing protein trends are most likely caused by dephosphorylation rather than rapid protein synthesis. We controlled for phosphorylation artifacts in protein loss as well, and found evidence that this impacted only two proteins (Dataset S1, Table S3). We found that analyzing the data with metrics that reduce the effect of outliers (e.g., median) mitigates but does not eliminate specious trends caused by dynamic modifications. This is because multiple peptides are often affected by modifications (e.g., SI Appendix, Fig. S7C), as is the case for the majority of the proteins discussed here.
A Method for Evaluating Phosphosite Occupancy.
The phospho-MS data (Fig. 1E) show the relative abundance changes of ∼500 dynamic and ∼3,000 stable phosphorylation sites. While relative trends of phosphoforms have utility, they are often difficult to interpret. For example, a twofold increase in the occupancy of a residue from 1 to 2% appears identical to the same fold increase from 50 to 100%. These two cases can have very different functional implications; thus, we sought to calculate the phosphosite stoichiometry changes in our data (Figs. 3 and 4). It is not possible to directly determine stoichiometry from simple ratios of MS measurements without expensive spike-in standards (65). This is because the unmodified and modified forms of a peptide ionize at different efficiencies and hence the interform ratio is distorted (Fig. 4A). It is nonetheless possible to infer the absolute stoichiometry of a site by invoking the principle of mass conservation, which states that the total abundance of each peptide equals the sum of all modified and unmodified forms of the peptide (Fig. 3, Eqs. 1–3). This principle results in one constraint—a simple linear equation—for each pair of conditions in the experiment. If protein levels change, the equations must be scaled accordingly. By solving these conservation equations, we can infer the occupancy ratio from the measured relative intraform ratios of change between conditions (Fig. 3, Eq. 4). Approaches invoking mass conservation were previously reported (24, 66) for the simple case of two phosphoforms and two conditions, giving two linear equations to be solved for the stoichiometries of each form. Here, we extend the solution and broaden its utility: (i) to include cases where the number of conditions is larger than the number of observed phosphoforms; (ii) to establish a statistical measure of confidence in the results; and (iii) to include cases with multiple phosphoforms, which were not calculable by previous approaches.
Fig. 3.
The geometric relationship between the measured peptide form ratios and their unknown absolute values. Let equal the absolute abundance of peptide form j in condition i, where j = 0,1, ..., M−1, 0 = unmodified, and M is the number of forms, and where i = 1, ..., N, where N is the number of measurements. is the MS signal of form j at condition i normalized by the reference condition i = 1. is the ratio of the parent protein T between measurement (i) and measurement 1 (Eq. 1). The sum of all peptide forms is conserved or scaled by (Eq. 2). Rewriting Eq. 2 in terms of the measured parameters (1) yields Eq. 3, which is written in vector form as Eq. 4. Eq. 4 shows that the vector of absolute values is orthogonal to the M−1 dimensional subspace containing the measured ratios. When M = 2, this subspace is a line (SI Appendix, Fig. S8). For M > 2, this subspace is an M−1 dimensional plane. For overdetermined systems (N > M), this subspace can be estimated with regression (Fig. 4 and SI Appendix, SI Methods).
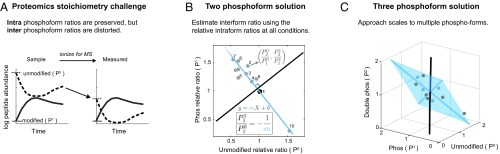
Fig. 4.
Calculation of phospho-stoichiometry from multiplexed phospho-dynamics measurements: Demonstrating a graphical approach to estimating phosphosite occupancy of multiplexed data. (A) Phosphosite occupancy cannot be directly calculated from the raw signal from the mass spectrometer because the interform ratios are distorted due to the differential ionization efficiencies of the peptide forms. This is depicted here as the unmodified form (P0) ionizing less efficiently than the modified form (P1). However, the intraform ratios are preserved (i.e., only the starting point is shifted). (B and C) Estimating the solution for the overdetermined system from multiplexed-MS data. (B) For each condition, the measured intraform ratios of change of the unmodified (P0) and single phosphorylated (P1) forms (Fig. 3) define coordinates in a 2D plane. The relative ratio is defined by the reference point, which in this case is point 1 (black point). The solution to the overdetermined system can be estimated by regression. The unknown interform P1/P0 ratio is the negative inverse of the fit line slope (i.e., is the orthogonal slope) [see Fig. 3 (Eq. 4) and SI Appendix, SI Methods and Fig. S8]. (C) For more than two P forms, the known ratios define a higher dimensional plane (e.g., a plane is fit in 3D space from the ratios of the P0, P1, and double-form P2). The plane is visualized as the blue shaded area spanned by two vectors. The solution (the black vector) is orthogonal to the plane, as in Fig. 4B. These vectors are calculated by principal-component analysis (see text). Data points are plotted in gray, and blue-shaded points are behind the plane.
Briefly, the method takes advantage of the fact that, when the number of conditions exceeds the number of possible phosphoforms, the system (Fig. 3, Eq. 4) is overdetermined (SI Appendix, Fig. S8) and the problem becomes one of multivariate linear regression. Thus, an estimated solution to the system can be determined through slope fitting, as visualized in Fig. 4B and SI Appendix, Fig. S9. This extends to fitting a hyperplane in higher dimensional space for more than two phosphoforms (Fig. 4C). The ability to incorporate many measurements under different conditions enables a more accurate calculation of occupancy than one measurement alone. The “overdetermination” of the system is key to reporting confidence intervals. We are able to estimate error in the fit coefficients using bootstrapping (67) (SI Appendix, SI Methods and Fig. S10). Solving for multisite occupancy is not possible if the number of conditions is less than the number of phosphoforms, as the system is underdetermined. Therefore, our ability to calculate multisite stoichiometries is enabled by measuring multiple conditions by multiplex-MS.
Applying Occupancy Calculation.
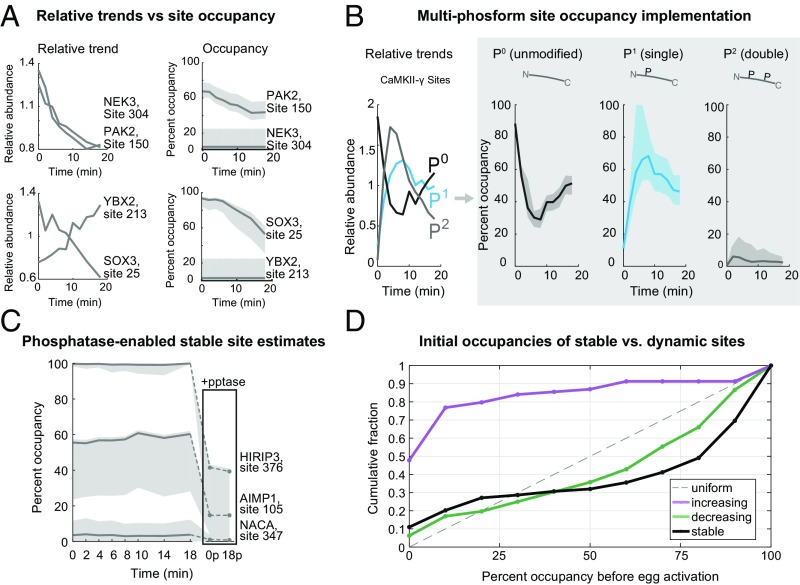
We have applied this method to the phosphorylation dynamics in the period following egg activation for sites where we detect the unmodified and modified forms. We did not scale the conservation equations in this case since the protein levels are overwhelming stable (Fig. 1 C and D). Fig. 5A shows the relative dynamics of phosphosites on the kinases NEK3 and PAK2, as well as transcription factors/nucleotide-binding proteins SOX3 and YBX2. The amount of relative change for each set is nearly identical. However, YBX2 and NEK3 sites change phosphorylation for only a small fraction of their residues, whereas the residues on SOX3 and PAK2 change substantially. These are selected examples of many important regulatory proteins with discrepancies between relative and absolute changes (SI Appendix, Fig. S11). A proof-of-principle multisite phosphorylation stoichiometry calculation is shown for a CaMKII-γ peptide (Fig. 5B) with an unmodified (P0), single-phosphorylated (P1), and double-phosphorylated (P2) form. We estimate the occupancy of the P0 form with high confidence; the confidence intervals for the P1 and P2 forms are wide but nonoverlapping, so we are able to conclude that the P1 form is more occupied than the P2. This highlights the utility of confidence intervals, especially as error increases in higher dimensional spaces. A general consideration when applying this method is that it is most effective under conditions where the relative changes are substantial. For stable sites, the minimization is unreliable and gives wide confidence intervals. To extend the use of this approach for stable sites, we repeated the fertilization time course and artificially induced dynamics with a nonspecific phosphatase treatment of two conditions (25) (SI Appendix, SI Methods and Fig. S12). We used a heat-labile phosphatase, which allowed us to inactivate the phosphatase activity for subsequent experimental steps. Three examples of calculations of stable occupancies using this method are shown in Fig. 5C. They also demonstrate the range of confidence intervals seen within the data (shown in SI Appendix, Fig. S13B). For corroboration, we used the matching phosphatase treated and untreated time points (25) to calculate occupancy using the previously reported two-condition analytical method (24). We compared these results to the occupancies derived from applying our regression-based approach to the independently measured endogenous dynamics. The results are generally consistent (Pearson coefficient = 0.8).
Fig. 5.
Occupancy of phosphorylation sites in time series postactivation. (A) Time series of two kinases (NEK3, PAK2) and two transcription factors/nucleotide binding proteins (SOX3, YBX2) demonstrating that similar amounts of relative changes can give very different occupancy changes (shaded area is the 95% confidence interval). (B) Phospho-occupancy time series of a multiphosphorylated CaMKII-γ peptide. (C) Reliable estimation of stable site occupancy is enabled by inducing dynamics with phosphatase treatment (see text; SI Appendix, Fig. S12). Examples of stoichiometry estimated with phosphatase treatment are shown here. Treated conditions are replicates of the 0- and 18-min time points (boxed). (D) Cumulative distributions of the initial phosphosite occupancies at 0 min (unactivated egg) classified by whether the sites increase, decrease, or are stable after egg activation. A uniform distribution would lie on the dotted line.
In total, we calculate confident stoichiometries (±25% occupancy of 95% confidence intervals) for ∼500 sites (15% of dataset); ∼150 of these are dynamic. With these data, we are able to compare stable versus regulated sites, which was difficult with previous approaches. For increasing, decreasing, or stable phosphosites (classified by their relative trends), we show the cumulative distribution of their occupancy at 0 min (Fig. 5D). The majority of sites that increase have a low initial occupancy. The distribution of sites that decrease is skewed toward higher occupancy, although there is more density of lower occupied sites than reported in previous studies (24, 68, 69). Interestingly, the stable site distribution is the most highly occupied class. However, the difference between the distribution of stable sites and decreasing sites is perhaps more subtle than it appears. The apparent lack of intermediate occupancies is largely a product of applying a confidence interval cutoff to the data. The cutoff improves data quality, but disproportionally filters stable sites (SI Appendix, Fig. S13).
Protein Phosphorylation Dynamics Following Egg Activation.
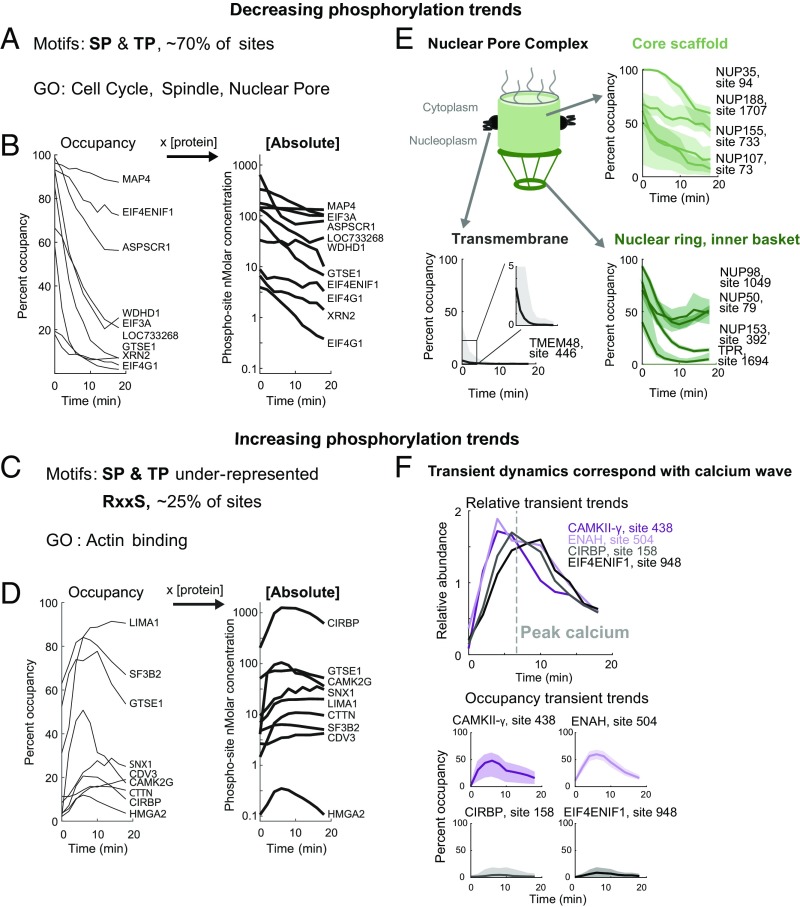
To explore the function of the regulated phosphosites, we performed motif and gene set enrichment analysis (GSEA) on the dominant trends of the dataset: increasing and decreasing stoichiometries. While the trends we detect are a subset of the total sites in the egg, their signatures are nevertheless revealing of the classes of regulation that are occurring. We were careful to exclude misleading trends that occur during multisite phosphorylation where, for example, the increase of a single form is actually the loss of a double form (SI Appendix, SI Methods and Fig. S14). Since relative trends are sufficient for motif and GSEA, we used all of the data to increase statistical power. The minimal motifs for Cdk1 and MAP kinases (S/T-P) explain ∼70% of the decreasing sites (Fig. 6A). We show several examples of the absolute dynamics for these phosphosites, calculated from our estimated site occupancies and protein concentrations (14). The majority of these show substantial loss of phosphate, which is consistent with the expected reversal of mitotic phosphorylation (Fig. 6B). The rates of the trends align with cell cycle events. The majority of proteins are dephosphorylated with a half-time of ∼10 min (some with <5 min) with minimal delay after egg activation. This matches the timing of sister chromatid separation in anaphase (30, 70) and the half-time of Cyclin-B1/2 degradation (Fig. 2A). GSEA of the dephosphorylated proteins are annotated with cell cycle, spindle, and nuclear pore functions (Fig. 6A; specific examples in SI Appendix, Fig. S15). In parallel, we also see a strong signal for increasing phosphorylation. The (S/T-P) motifs are underrepresented in this class. Instead, the minimal motif for the calcium-sensitive CaMKII is enriched, explaining 25% of the sites (Fig. 6C). Many of the increasing phosphorylation trends are fast, with half-times of 5 or ∼2 min (Fig. 6D). These are matched temporally with the calcium wave, cortical actin rearrangement (71–73), and exocytosis events (Fig. 2E). Indeed, the phosphorylated proteins are enriched for actin-binding Gene Ontology (GO) terms; all of the members of this set are also annotated as “Cell Periphery” (Dataset S1, Table S5). Given the localization of these events to the cortex, their fast rates, and the calcium sensitivity of cortical granule exocytosis (9), the increasing phosphorylation may in part promote the release of protein into the media.
Fig. 6.
Phosphosite dynamics following egg activation: Analysis of the phosphorylation trends (increasing and decreasing) following egg activation with individual examples. (A) Motif enrichment analysis (P << 0.01) results and gene set enrichment analysis (GSEA) of GO terms (P < 0.01) results for dephosphorylated proteins. (B) Time series of selected proline-directed phosphorylation plotted as occupancy and absolute phosphate dynamics (log10 transformed). All trends shown pass a 95% confidence interval width threshold of ±25%. (C) Motif analysis results as in A for proteins with increasing phosphorylation trends and GSEA results. (D) Examples of increasing phosphorylation (plotted as in B). (E) Phospho-occupancy trends showing differential dephosphorylation of nuclear pore complex (NPC) regions with confidence intervals. (F) Relative and phospho-occupancy time series of proteins showing transient trends corresponding to the peak of calcium concentrations.
Nucleoporin Dephosphorylation Corresponds to Order of NPC Assembly.
There are many intriguing vignettes in the data. One example is the differential rates of dephosphorylation of nuclear pore subcomplexes (Fig. 6E and SI Appendix, Fig. S16). During entry into mitosis, nucleoporin phosphorylation promotes nuclear pore complex (NPC) disassembly (74). The sequence of postmitotic NPC reassembly may be controlled by differential reversal of the mitotic phosphorylation (75). Our observations give some support for this hypothesis. Rates of dephosphorylation on nucleoporin phosphosites cluster by regions (76) of the NPC (Fig. 6E) (discussed further in SI Appendix, Fig. S16). These rates mostly conform to the sequence of assembly (77, 78). The fast dephosphorylation (∼2- to 5-min half-times) of inner basket and nuclear ring components (e.g., NUP153) is consistent with their early roles in NPC assembly (79). The slower dephosphorylation (∼10-min half-time) of the core scaffold components (e.g., NUP188) is consistent with their later recruitment (SI Appendix, Fig. S16). One notable exception is that while the incorporation of NUP98 is reported to be an intermediate assembly step, it is dephosphorylated rapidly (4-min half-time). Phosphorylation of NUP98 was previously shown to be rate limiting for NPC disassembly (80); it may be that its dephosphorylation is required or limiting for reassembly as well. There are two important caveats to the kinetic discrepancies: (i) nucleoporins or nucleoporin partial complexes may function outside the nuclear pore (e.g., NUP98 at the spindle) (81, 82); (ii) the nuclear envelope in the egg is packaged in structures called annulate lamellae, which also repackage dynamically after fertilization (83). Our observations may reflect this repackaging, rather than the canonical postmitotic reassembly. For example, the large relative change but small stoichiometric decrease of the transmembrane nucleoporin TMEM48 may indicate a separate pool of molecules, perhaps on the annnulate lamellae (Fig. 6E). Nevertheless, these differential rates provide information on how NPC reassembly may be regulated in the egg and by extension in other circumstances.
Ca++-Sensitive Responses Show Substantial Differences in Occupancy.
The data also offer insights into the pattern of calcium-initiated signaling. The calcium wave peaks in cytosolic concentration around 5 min postfertilization and declines thereafter (84). Fig. 6F shows a set of relative phosphorylation changes from a larger cluster (Fig. 1E) that correlates with the calcium wave. A prominent example is CaMKII-γ, which is important for egg activation in the mouse (85), although the role of its phosphorylation is unclear. Three additional proteins with phosphosites that correlate with the calcium wave are shown (Fig. 6F). While the relative changes are nearly overlapping, the stoichiometries are very different. The two translational regulators (CIRBP, EIF4ENIF1) show phosphosites that change on a small fraction (<2%) of the proteins. In contrast, over one-half of the molecules of the γ-subunit of CaMKII and ENAH (an Ena/Vasp actin-binding protein) are phosphorylated, supporting the hypothesis that the increasing phosphorylation class is related to calcium signaling and the remodeling of cortical actin. A possible explanation for the low stoichiometry of the changes on CIRBP and EIF4ENIF1 is that this site of modification plays no significant role in regulation; perhaps these sites are modified through promiscuous activity of calcium-dependent kinases (e.g., CaMKII, PKC). Alternatively, the modification may be highly localized.
Absolute Changes of Protein Phosphorylation, Degradation, and Release.
The data allow comparison of the absolute changes in the diverse yet connected processes occurring in parallel at fertilization (Table 1). We estimate the total change resulting from protein degradation as ∼300 nM (∼0.01% of the total protein mass), which is dwarfed by a nearly 50-fold higher loss from protein release [roughly consistent with older estimates (9, 86)]. The total change in phosphorylation, which occurs on a diverse set of proteins from the cell cycle machinery to calcium-sensitive kinases, is ∼7,000 nM. This number is an underestimate, as we capture only a fraction of the total dynamics. Nevertheless, this is an order of magnitude higher than the changes due to protein degradation. Unlike protein degradation, where there was a strong correlation between low abundance and instability (Fig. 2D), we see large changes in phospho-occupancy for proteins that span a 1,000-fold range of concentration (Fig. 6 B and D). This is striking evidence that phosphorylation has the capacity to change the activity of many abundant proteins within minutes. Protein degradation, even with an active E3 ligase like the APC/C, can only work so quickly on a small number of low-abundance proteins, which seem to be core to cell cycle progression.
Table 1.
Measured absolute changes
| Class of dynamic | Absolute change, nM | Fraction of proteome, % |
| Secretion | 13,000 | ∼1 |
| Protein degradation | 300 | 0.01 |
| Phosphorylation | 6,800 | 0.3 |
| Decreased | 4,200 | — |
| Increased | 2,600 | — |
Discussion
We measured the absolute protein and phosphosite dynamics at fine time resolution following fertilization. The data reveal a small number of degraded substrates and substantial protein dephosphorylation related to the cell cycle. We also detect release of protein related to the slow block to polyspermy and parallel increases in phosphorylation, which may be linked to the calcium wave. To aid in interpreting the data, we developed a method for estimating phosphosite stoichiometry using multiplexed MS.
The phosphorylation data are compatible with a recent cell cycle model, which describes a quantitative model for the cell cycle oscillator in the Xenopus egg (87). Earlier experiments demonstrated the oscillation (88, 89), but a satisfactory explanation of how it oscillated depended on several features, including a time delay (about 15 min) between activation of Cdk1 and activation of APC/C and then a very small delay in the destruction of targets once APC/C is activated (<2 min). Our measurements are strikingly consistent with fast Cdk1 inhibition, as we see immediate commencement of dephosphorylation at hundreds of proline-directed sites with no obvious delay. A discrepancy is that we see a small delay (∼2–4 min) in the degradation of APC/C substrates like Cyclin-B1/2, which is presumably required to inactivate Cdk1. The rates of these early events may be underestimated if there is sufficient metachrony of the cell cycle in the large frog egg (45, 48), but the ordering of the observations discussed here should be preserved.
The substrate specificity of the APC/C is thought to be conferred by the cofactors Cdh1 and Cdc20. We show that all detected APC/CCdh1 substrates are indeed stable in the frog egg, where Cdh1 is not expressed. The inhibition of APC/CCdh1 promotes S-phase entry (90) in the conventional cell cycle; the absence of Cdh1 and resulting stability of its substrates may assist in the bypassing of the G1 phase to S phase during the cleavage divisions in the early embryo.
All detected degradation targets (e.g., Cyclin-B1/2, EMI2) are present at low abundance, suggesting that the amount of protein degradation is small overall. However, if there is a global increase in protein synthesis at equilibrium with protein degradation, we would underestimate the flux of degradation. Protein synthesis at egg activation is reported in flies and mice (18, 91, 92), but our data show no evidence for a burst of synthesis of any of the 8,700 proteins. Our previous work also showed that new proteins comprise a small fraction of the frog embryo even hours postfertilization (15). In principle, 2 μM of new protein could accumulate in ∼20 min (∼7 μM/h, a rate measured later in development) (93). If this occurred during the first cell cycle, we would expect to identify many of these proteins given our sensitivity limits (SI Appendix, Fig. S5).
In addition to measuring the extent of protein degradation and synthesis following fertilization, our proteome-wide approach unveiled unexpected components of the slow block to polyspermy. Early studies established that protease activity is essential for this event; the inhibition of proteases leads to polyspermy (94). Paradoxically, we found that eggs release multiple endogenous protease inhibitors in response to fertilization. Although protease inhibitors were recently found in the perivitelline space of Xenopus eggs (95), their function was not addressed. There are several possible functions for released inhibitors: (i) they could prevent activity of the proteases inside the cortical granules, (ii) they might control the activity of the proteases postrelease, and (iii) they may act to block the sperm acrosome reaction (96).
Our application of quantitative proteomics to fertilization highlighted several general considerations for interpreting MS data. We concluded that phosphorylation dynamics can confuse the analysis of protein trends, giving the appearance of change in stable proteins. Dynamics in any high stoichiometric modification (e.g., acetylation) could also cause this same ambiguity. Using the median of a protein’s peptides can mitigate but will not eliminate specious trends caused by dynamic modifications. On the phosphosite level, a source of ambiguity is that a decreasing trend of a singly phosphorylated form results from an increasing multiphosphorylated form (SI Appendix, Fig. S14). This general possibility must be considered when interpreting the direction of a given phosphosite trend. This phenomenon was discussed previously (97); our data provide clear examples.
The utility of measuring phospho-occupancy is well recognized (24, 25, 66). However, current approaches are not able to estimate occupancy for multiphosphorylated peptides, stable sites, or sites measured with multiplexed proteomics. More fundamentally, these methods do not give statistical information (i.e., confidence intervals), which makes interpreting the estimates difficult. Our approach provides a unified analytical and experimental framework to address these limitations. Although we developed this approach to exploit the power of multiplexed proteomics, the advancements are compatible with other methodologies. For example, label-free phospho-proteomics typically have lower measurement precision but are conducive to high-throughput formats (98). The additional conditions, in principle, give increased statistical power with the use of the regression framework. The higher sensitivity of label-free and other techniques (99) could mitigate a limitation of this approach, which is the low efficiency of measuring both the unmodified and phosphorylated peptides. Other methods for calculating phospho-stoichiometry, such as motif-targeting proteomics (100), could also incorporate this framework. Finally, the principles of the calculation could be extended to acetylation or ubiquitination, although we expect more success if occupancies are often >1%. These approaches can increase the power of the proteomics for analysis of posttranslational modification in diverse settings.
Materials and Methods
Egg Activation Procedure.
Female X. laevis were induced with 700 U of human chorionic gonadotropin. After 14 h, eggs were harvested, washed with 1× Marc’s modified Ringer’s (MMR), de-jellied with cysteine [2% (wt/vol), pH 8.0], and then kept in dishes with agar beds to avoid the eggs sticking to the dish. Eggs were placed in open-faced gel box with a 3% agar bed in 0.1× MMR and electroshocked by applying an electric field of ∼3 V/cm for 1 s. Thirty eggs were collected every 2 min until 18 min. The excess media was removed, and samples were flash frozen in liquid nitrogen. For the phosphatase treatment experiment, the 12- and 16-min time points were replaced with phosphatase-treated replicates of the 0- and 18-min time points. Each biological replicate was performed on separate days with different frogs (termed EA, EAp2, EApR3, EAp5b, where “EA” stands for “electroactivation” and the “p” denotes an experiment where phospho-enrichment occurred). The research with X. laevis was performed under the oversight of the Harvard Medical Area Institutional Animal Care and Use Committee.
Egg Media Time Series.
Egg media was collected at the similar time points as Fig. 1B following electroactivation, except with the 0-min time point taken in triplicate and 12- and 16-min time points skipped. The media was dried to concentrate the sample and processed for MS analysis. The experiment was performed as described above with the following exceptions: (i) Eggs were placed in 0.01× MMR, to reduce the salt concentrations after dry-down. (ii) An additional ∼10 mM calcium was added to the media to assist in any wound healing caused by the electroshock. (iii) The electric field was doubled to ∼6 V/cm to account for less conductive media, as eggs did not activate at 3 V/cm. (iv) Approximately 1,000 eggs were used instead of 300 to increase the signal of protein accumulating in the media. After electroshock, the media was kept well-mixed by gently rocking the gel-box on the long axis by hand (several methods were tested by mixing with dye). Each time point was taken by extracting 2 mL of media and flash-freezing with liquid nitrogen. We replaced the volume of media removed for harvesting at each time point to keep overall volume constant throughout the experiment, and normalized afterward to account for the removal of mass in the media with each time point taken. We lyophilized the collected media samples to prevent unwanted proteolysis from the released proteases. The experiment was done in triplicate with three different clutches on the same day, and then processed in parallel as described in the MS sample preparation section below, except without the alkylation and protein precipitation steps.
General Sample Preparation for MS.
Samples were prepared essentially as previously described (14, 16). Thirty eggs per time point were snap frozen and lysed with 250 mM sucrose, 1% Nonidet P-40 substitute (Sigma), 5 mM EDTA (pH 7.2), 1 Roche Complete Mini tablet (EDTA-free), 20 mM Hepes (pH 7.2), 10 μM Combretastatin 4A, and 10 μM Cytochalasin D while frozen (14). All buffers were made with HPLC water. Eggs were lysed by pipetting up and down 40 times with a 200-μL pipette tip, vortexed at maximum speed for 10 s, incubated on ice for 10 min, and again vortexed for 10 s. Lysates were clarified by centrifugation at 4,000 relative centrifugal force (RCF) at 4 °C for 4 min in a tabletop centrifuge. The cytoplasmic and lipid layers were mixed by gentle flicking and removed from the pelleted yolk. To the lysate, Hepes (pH 7.2) was added to 100 mM, and SDS was added to 2% (wt/vol) to provide additional buffering capacity and to denature the sample. The samples were reduced with 5 mM DTT for 20 min at 60 °C. Cysteines were alkylated with 15 mM N-ethylmaleimide (NEM) for 20 min at room temperature (RT). Excess NEM was reacted with an additional 5 mM DTT at RT. Proteins were isolated by methanol/chloroform precipitation (101). The protein pellet was resuspended (∼5 mg/mL) in 6 M guanidine HCl and 50 mM 3-[4-(2-hydroxyethyl)-1-piperazinyl]propanesulfonic acid (EPPS), pH 8.5, with gentle pipetting and heated to 60 °C for 5 min. For ∼100–400 μg of protein per condition, the sample was diluted to 2 M guanidine with 5 mM EPPS, pH 8.5, and digested with LysC (Wako Chemicals) at 20 ng/μL at RT for 14 h. Next, we diluted guanidine HCl to 0.5 M with 5 mM EPPS, pH 8.5, and digested further additional LysC at 20 ng/μL for ∼15 min at RT, then added 10 ng/μL of sequencing-grade trypsin (Roche) and coincubated at 37 °C for 8 h in an incubator. For each sample, ∼100–400 μg of peptides were dried down in a SpeedVac and resuspended with 100–150 μL of 500 mM EPPS, pH 8.0, respectively. If sample was not at pH ∼8.0, an additional ∼25–50 mM HCl was added. For labeling, we added 15–50 μL of TMT stock solution (0.2 mg/40 μL of acetonitrile) to each sample and incubated at RT for 2 h (10 μg:1.5 μL peptide to TMT). Thereafter, we quenched the reaction with 10 mM hydroxylamine for 15 min. All conditions were combined, were acidified by addition of phosphoric acid to 5%, and were clarified by spinning at 21,000 RCF for 20 min. Samples were subjected to C18 solid-phase extraction (50 mg) (SepPak; Waters) to desalt and isolate peptides. To reduce sample complexity, peptides were resuspended in a 10 mM sodium carbonate buffer (pH 8.0), then fractionated by medium pH reverse-phase HPLC (Zorbax 300Extend-C18, 4.6 × 250-mm column; Agilent) using an acetonitrile gradient from 5–39%. With a flow rate of 0.8 mL/min, fractions were collected into a 96-well plate every 38 s, and then pooled into 24 fractions by combining alternating wells from each column of the plate. Each fraction was dried and resuspended in 20 μL of 1% phosphoric acid. Peptides from each fraction were desalted and extracted once more with reverse-phase purification, resuspended in 10 μL of 1% formic acid. Approximately 4 μL per fraction were analyzed by liquid chromatography (LC)-MS.
Phosphatase Treatment.
To phosphatase treat some samples without affecting the untreated conditions after multiplexing or at any subsequent steps, we made use of a thermally unstable phosphatase, which can easily be inactivated. We used the temperature-labile shrimp alkaline phosphatase (product #78390; Affymetrix). To minimize volume added to the samples, the phosphatase was concentrated to ∼4 U/μL on a 5K Amicon Ultra filter, spun at 4 °C. Since the supplied buffer contains Tris, which can interfere with TMT labeling, we exchanged this buffer with 10 mM EPPS at pH 8.0 and stored the enzyme in 50% glycerol. We see no activity loss from the buffer exchange, as assayed by p-nitrophenyl phosphate (pNPP) hydrolysis. Following completion of trypsin digestion, we allowed the temperature to cool to RT then added 10 mM MgCl, 0.1 mM EDTA, and 5 mM EPPS, pH 8.5. To the two phosphatase-treated conditions, we added enzyme at a ratio of 3:1 peptide to phosphatase units and incubated for 12 h at RT without purifying the peptides away from the proteases. The incubation is done in the presence of proteases as the dephosphorylated peptide creates a better substrate and allows for proteolysis of an otherwise missed cleavage caused by the charge of the phospho-group. Allowing for further cleavage of dephosphorylated peptides mimics the effect of an endogenous phosphatase more closely; otherwise, the forms with missed cleavages will have lower abundance in the untreated samples and confound analysis. We determined that, at RT, LysC does not cleave the phosphatase, while trypsin does but at a reduced rate. The 3:1 ratio used gives sufficient activity while avoiding peptides from the proteolysis of the phosphatase from dominating the MS signal. After incubation, samples were lyophilized to dryness, and resuspended per the TMT-MS protocol discussed above. Importantly, all samples were incubated for 5 min at 65 °C to destroy phosphatase activity in a water bath (inhibition under these conditions was confirmed beforehand with pNPP activity assays). Samples were cooled to RT before proceeding with the MS sample preparation.
Phosphopeptide Enrichment.
We chose to multiplex peptides before the enrichment column to improve the data quality. There is a trade-off of decreased yield and therefore depth, as it is not economical to label more than a few milligrams of material. We used 2.5–4 mg of TMT-labeled peptides per replicate, enriched with 5-μm titanium dioxide microspheres (GL Sciences; 5020-75000) and fractioned as previously described (102). A typical yield of 50–80 μg of peptides eluted from the column, with a median specificity of ∼80%. Additional experimental details are provided in SI Appendix.
LC-MS Analysis.
Analysis was performed essentially as previously described, with the spectra mapped to the PHROG reference database (14, 16). The main modification was the use of five-notch MS3 on the protein-level and three-notch MS3 on the phospho-level (103). Phosphorylation searches were performed with differential modification of +79.9663304104 on serine and threonine. Tyrosine was excluded as we found the majority of the identified tyrosine sites were erroneous identifications. Phosphosites were localized with in-house software based on Ascore (104). Peptides with multiple phosphosites are reported as “composite sites” and noted with a “;” delimiting each modified residue. All searches (for the protein replicates, phospho replicates, media, ubiquitin pull-downs) were mapped to a single protein map and subjected to one linear discriminator/protein sieve analysis to control the FDR.
Estimating a FDR for Protein Loss.
The full methodology is detailed and visualized in SI Appendix, SI Methods and Fig. S2. The FDR cutoff was calculated by comparing the geometric distance of the experimental data along with randomized data to an idealized degradation trend. The FDR is defined as the chance that a random trend could appear at a certain cosine distance from an idealized degradation trend.
Classification of Released Proteins and Novel APC/C Substrates.
We classified a protein as released (rather than degraded) either by direct experimental evidence, annotation, or evidence in the literature. In most cases, a protein that decreased in the egg could be clearly detected as increasing in abundance in the media. There were several cases where a protein decreased in the egg that we did not detect in the media, but these were clearly annotated as secreted proteins or were of a similar class as proteins for which we had direct evidence. For example, we infer the exocyst gene family are released into the media because vesicle trafficking and components of the exocytosis machinery were detected recently in the perivitelline space in frogs (95). To be classified as a novel APC substrate, we imposed the following criteria: the protein passes a 1% FDR threshold for decreasing abundance, had no evidence (direct or from literature) of release from the egg, no evidence of a specious trend from phosphorylation (this was established by looking for reciprocal phosphorylations, or more generally, whether all of the detected peptides behaved similarly), and was ubiquitinated.
Phospho-Stoichiometry Regression-Based Algorithm.
The full principle and methodology is included in SI Appendix. The MATLAB implementation is freely available at https://github.com/marcpresler/OccupancyMS. The total least-squares regression package (105) is adapted from File ID #31109 on MathWorks File Exchange.
Dataset S1 Guide.
All tables are provided as separate sheets in a single Excel file. Dataset S1, Table S1 shows the rank-ordered degradation candidates as unique protein IDs with protein sequence. Dataset S1, Table S2 shows the absolute loss of candidates passing a 1% FDR cutoff averaged by unique gene symbol. Dataset S1, Table S3 classifies Dataset S1, Table S2 by their mechanism of loss. Dataset S1, Table S4 shows the proteins that apparently increase in abundance. Dataset S1, Table S5 contains the GO term enrichment output. Dataset S1, Table S6 provides the raw data output for all experiments. Dataset S1, Tables S7 and S8 give the normalized protein and phosphosite data with replicates averaged. Dataset S1, Table S9 contains the phosphosite occupancy data.
SI Appendix, SI Methods.
Additional details provided exclusively in the SI Appendix, SI Methods are the EM imaging time series, data normalization, calculating absolute changes, estimating maximal degradation rates of the APC/proteasome, K-means clustering, multisite artifact correction, and motif and gene set enrichment analysis.
Supplementary Material
Acknowledgments
We thank Chris Rose and Joao Paulo for help with mass spectrometers and the S.P.G. laboratory computational team for bioinformatics support; Ying Lu, Tao Wu, Timothy J. Mitchison, and Angela H. DePace for helpful discussion; Katy Hartman, John Ingraham, and Randall W. King for insight and inspiration toward the stoichiometry calculation; and Becky Ward for critical comments on the manuscript. We thank the PRIDE team for proteomic data distribution (106). This work was supported by NIH Grants HD091846, HD073104, and GM103785 (to M.W.K.) and GM39565 (to Timothy J. Mitchison); Charles A. King Trust Postdoctoral Fellowship Program (Bank of America, N.A., Co-Trustee) and Princeton University startup funding (to M.W.); and Burroughs Wellcome Fund and Mallinckrodt Award (to A.M.K.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository (dataset identifier PXD006639).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1709207114/-/DCSupplemental.
References
- 1.Izawa D, Pines J. How APC/C-Cdc20 changes its substrate specificity in mitosis. Nat Cell Biol. 2011;13:223–233. doi: 10.1038/ncb2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tunquist BJ, Maller JL. Under arrest: Cytostatic factor (CSF)-mediated metaphase arrest in vertebrate eggs. Genes Dev. 2003;17:683–710. doi: 10.1101/gad.1071303. [DOI] [PubMed] [Google Scholar]
- 3.Singh SA, et al. Co-regulation proteomics reveals substrates and mechanisms of APC/C-dependent degradation. EMBO J. 2014;33:385–399. doi: 10.1002/embj.201385876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Z, et al. GPS-ARM: Computational analysis of the APC/C recognition motif by predicting D-boxes and KEN-boxes. PLoS One. 2012;7:e34370. doi: 10.1371/journal.pone.0034370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sivakumar S, Gorbsky GJ. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat Rev Mol Cell Biol. 2015;16:82–94. doi: 10.1038/nrm3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim HJ, et al. The G2/M regulator histone demethylase PHF8 is targeted for degradation by the anaphase-promoting complex containing CDC20. Mol Cell Biol. 2013;33:4166–4180. doi: 10.1128/MCB.00689-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karsenti E, Bravo R, Kirschner M. Phosphorylation changes associated with the early cell cycle in Xenopus eggs. Dev Biol. 1987;119:442–453. doi: 10.1016/0012-1606(87)90048-0. [DOI] [PubMed] [Google Scholar]
- 8.Tokmakov AA, Stefanov VE, Iwasaki T, Sato K, Fukami Y. Calcium signaling and meiotic exit at fertilization in Xenopus egg. Int J Mol Sci. 2014;15:18659–18676. doi: 10.3390/ijms151018659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bement WM, Capco DG. Activators of protein kinase C trigger cortical granule exocytosis, cortical contraction, and cleavage furrow formation in Xenopus laevis oocytes and eggs. J Cell Biol. 1989;108:885–892. doi: 10.1083/jcb.108.3.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markoulaki S, Matson S, Ducibella T. Fertilization stimulates long-lasting oscillations of CaMKII activity in mouse eggs. Dev Biol. 2004;272:15–25. doi: 10.1016/j.ydbio.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Thompson A, et al. Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–1904, and erratum (2006) 78:4235. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 12.McAlister GC, et al. Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal Chem. 2012;84:7469–7478. doi: 10.1021/ac301572t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Werner T, et al. High-resolution enabled TMT 8-plexing. Anal Chem. 2012;84:7188–7194. doi: 10.1021/ac301553x. [DOI] [PubMed] [Google Scholar]
- 14.Wühr M, et al. Deep proteomics of the Xenopus laevis egg using an mRNA-derived reference database. Curr Biol. 2014;24:1467–1475. doi: 10.1016/j.cub.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peshkin L, et al. On the relationship of protein and mRNA dynamics in vertebrate embryonic development. Dev Cell. 2015;35:383–394. doi: 10.1016/j.devcel.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wühr M, et al. The nuclear proteome of a vertebrate. Curr Biol. 2015;25:2663–2671. doi: 10.1016/j.cub.2015.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kronja I, et al. Quantitative proteomics reveals the dynamics of protein changes during Drosophila oocyte maturation and the oocyte-to-embryo transition. Proc Natl Acad Sci USA. 2014;111:16023–16028. doi: 10.1073/pnas.1418657111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kronja I, et al. Widespread changes in the posttranscriptional landscape at the Drosophila oocyte-to-embryo transition. Cell Rep. 2014;7:1495–1508. doi: 10.1016/j.celrep.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krauchunas AR, Horner VL, Wolfner MF. Protein phosphorylation changes reveal new candidates in the regulation of egg activation and early embryogenesis in D. melanogaster. Dev Biol. 2012;370:125–134. doi: 10.1016/j.ydbio.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo H, et al. Phosphoproteomic network analysis in the sea urchin Strongylocentrotus purpuratus reveals new candidates in egg activation. Proteomics. 2015;15:4080–4095. doi: 10.1002/pmic.201500159. [DOI] [PubMed] [Google Scholar]
- 21.Kettenbach AN, Gerber SA. Rapid and reproducible single-stage phosphopeptide enrichment of complex peptide mixtures: Application to general and phosphotyrosine-specific phosphoproteomics experiments. Anal Chem. 2011;83:7635–7644. doi: 10.1021/ac201894j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thingholm TE, Jørgensen TJ, Jensen ON, Larsen MR. Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat Protoc. 2006;1:1929–1935. doi: 10.1038/nprot.2006.185. [DOI] [PubMed] [Google Scholar]
- 23.Fíla J, Honys D. Enrichment techniques employed in phosphoproteomics. Amino Acids. 2012;43:1025–1047. doi: 10.1007/s00726-011-1111-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olsen JV, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 25.Wu R, et al. A large-scale method to measure absolute protein phosphorylation stoichiometries. Nat Methods. 2011;8:677–683. doi: 10.1038/nmeth.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curran TG, Zhang Y, Ma DJ, Sarkaria JN, White FM. MARQUIS: A multiplex method for absolute quantification of peptides and posttranslational modifications. Nat Commun. 2015;6:5924. doi: 10.1038/ncomms6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gurdon JB, Wakefield L. Microinjection of amphibian oocytes and eggs for the analysis of transcription. In: Celis JE, Graessmann A, Loyter A, editors. Microinjection and Organelle Transplantation Techniques. Academic; London: 1986. [Google Scholar]
- 28.Bernardini G, Andrietti F, Camantini M, Cosson MP. Xenopus spermatozoon: Correlation between shape and motility. Gamete Res. 1988;20:165–175. doi: 10.1002/mrd.1120200207. [DOI] [PubMed] [Google Scholar]
- 29.Gerhart J, Wu M, Kirschner M. Cell cycle dynamics of an M-phase-specific cytoplasmic factor in Xenopus laevis oocytes and eggs. J Cell Biol. 1984;98:1247–1255. doi: 10.1083/jcb.98.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ubbels GA, Hara K, Koster CH, Kirschner MW. Evidence for a functional role of the cytoskeleton in determination of the dorsoventral axis in Xenopus laevis eggs. J Embryol Exp Morphol. 1983;77:15–37. [PubMed] [Google Scholar]
- 31.McAlister GC, et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal Chem. 2014;86:7150–7158. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- 33.Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
- 34.Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 35.Hansen DV, Tung JJ, Jackson PK. CaMKII and polo-like kinase 1 sequentially phosphorylate the cytostatic factor Emi2/XErp1 to trigger its destruction and meiotic exit. Proc Natl Acad Sci USA. 2006;103:608–613. doi: 10.1073/pnas.0509549102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell. 1998;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- 37.Prinz S, Hwang ES, Visintin R, Amon A. The regulation of Cdc20 proteolysis reveals a role for APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr Biol. 1998;8:750–760. doi: 10.1016/s0960-9822(98)70298-2. [DOI] [PubMed] [Google Scholar]
- 38.Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Floyd S, Pines J, Lindon C. APC/C Cdh1 targets aurora kinase to control reorganization of the mitotic spindle at anaphase. Curr Biol. 2008;18:1649–1658. doi: 10.1016/j.cub.2008.09.058. [DOI] [PubMed] [Google Scholar]
- 40.Stewart S, Fang G. Destruction box-dependent degradation of aurora B is mediated by the anaphase-promoting complex/cyclosome and Cdh1. Cancer Res. 2005;65:8730–8735. doi: 10.1158/0008-5472.CAN-05-1500. [DOI] [PubMed] [Google Scholar]
- 41.Lu Y, Lee BH, King RW, Finley D, Kirschner MW. Substrate degradation by the proteasome: A single-molecule kinetic analysis. Science. 2015;348:1250834. doi: 10.1126/science.1250834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holloway SL, Glotzer M, King RW, Murray AW. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell. 1993;73:1393–1402. doi: 10.1016/0092-8674(93)90364-v. [DOI] [PubMed] [Google Scholar]
- 43.King RW, et al. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 44.van der Velden HM, Lohka MJ. Mitotic arrest caused by the amino terminus of Xenopus cyclin B2. Mol Cell Biol. 1993;13:1480–1488. doi: 10.1128/mcb.13.3.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang JB, Ferrell JE., Jr Mitotic trigger waves and the spatial coordination of the Xenopus cell cycle. Nature. 2013;500:603–607. doi: 10.1038/nature12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishihara K, et al. Organization of early frog embryos by chemical waves emanating from centrosomes. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130454. doi: 10.1098/rstb.2013.0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bischof J, et al. A cdk1 gradient guides surface contraction waves in oocytes. Nat Commun. 2017;8:849. doi: 10.1038/s41467-017-00979-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rankin S, Kirschner MW. The surface contraction waves of Xenopus eggs reflect the metachronous cell-cycle state of the cytoplasm. Curr Biol. 1997;7:451–454. doi: 10.1016/s0960-9822(06)00192-8. [DOI] [PubMed] [Google Scholar]
- 49.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bärenz F, et al. The centriolar satellite protein SSX2IP promotes centrosome maturation. J Cell Biol. 2013;202:81–95. doi: 10.1083/jcb.201302122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klinger M, et al. The novel centriolar satellite protein SSX2IP targets Cep290 to the ciliary transition zone. Mol Biol Cell. 2014;25:495–507. doi: 10.1091/mbc.E13-09-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang W, Wu T, Kirschner MW. The master cell cycle regulator APC-Cdc20 regulates ciliary length and disassembly of the primary cilium. Elife. 2014;3:e03083. doi: 10.7554/eLife.03083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sansal I, Dupont E, Toru D, Evrard C, Rouget P. NPDC-1, a regulator of neural cell proliferation and differentiation, interacts with E2F-1, reduces its binding to DNA and modulates its transcriptional activity. Oncogene. 2000;19:5000–5009. doi: 10.1038/sj.onc.1203843. [DOI] [PubMed] [Google Scholar]
- 54.Burkart AD, Xiong B, Baibakov B, Jiménez-Movilla M, Dean J. Ovastacin, a cortical granule protease, cleaves ZP2 in the zona pellucida to prevent polyspermy. J Cell Biol. 2012;197:37–44. doi: 10.1083/jcb.201112094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lindsay LL, Hedrick JL. Isolation and characterization of ovochymase, a chymotrypsin-like protease released during Xenopus laevis egg activation. Dev Biol. 1995;167:513–516. doi: 10.1006/dbio.1995.1045. [DOI] [PubMed] [Google Scholar]
- 56.Dunbar BS, et al. The mammalian zona pellucida: Its biochemistry, immunochemistry, molecular biology, and developmental expression. Reprod Fertil Dev. 1994;6:331–347. doi: 10.1071/rd9940331. [DOI] [PubMed] [Google Scholar]
- 57.Lindsay LL, Yang JC, Hedrick JL. Identification and characterization of a unique Xenopus laevis egg envelope component, ZPD. Dev Growth Differ. 2002;44:205–212. doi: 10.1046/j.1440-169x.2002.00635.x. [DOI] [PubMed] [Google Scholar]
- 58.Nagase H, Harris ED., Jr Ovostatin: A novel proteinase inhibitor from chicken egg white. II. Mechanism of inhibition studied with collagenase and thermolysin. J Biol Chem. 1983;258:7490–7498. [PubMed] [Google Scholar]
- 59.Yamada Y, Aketa K. Ovostatin, an endogenous trypsin inhibitor of sea urchin eggs: Purification and characterization of ovostatin from eggs of the sea urchin, Strongylocentrotus intermedius. Gamete Res. 1988;19:265–275. doi: 10.1002/mrd.1120190306. [DOI] [PubMed] [Google Scholar]
- 60.The UniProt Consortium UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lim JC, Kurihara S, Tamaki R, Mashima Y, Maéno M. Expression and localization of Rdd proteins in Xenopus embryo. Anat Cell Biol. 2014;47:18–27. doi: 10.5115/acb.2014.47.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johansson ME, Thomsson KA, Hansson GC. Proteomic analyses of the two mucus layers of the colon barrier reveal that their main component, the Muc2 mucin, is strongly bound to the Fcgbp protein. J Proteome Res. 2009;8:3549–3557. doi: 10.1021/pr9002504. [DOI] [PubMed] [Google Scholar]
- 63.Gasper J, Swanson WJ. Molecular population genetics of the gene encoding the human fertilization protein zonadhesin reveals rapid adaptive evolution. Am J Hum Genet. 2006;79:820–830. doi: 10.1086/508473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Toyama BH, et al. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 2013;154:971–982. doi: 10.1016/j.cell.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chaube R. Absolute quantitation of post-translational modifications. Front Chem. 2014;2:58. doi: 10.3389/fchem.2014.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Steen H, Jebanathirajah JA, Springer M, Kirschner MW. Stable isotope-free relative and absolute quantitation of protein phosphorylation stoichiometry by MS. Proc Natl Acad Sci USA. 2005;102:3948–3953. doi: 10.1073/pnas.0409536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shao J, Tu D. The Jackknife and Bootstrap. 1st Ed Springer; New York: 1995. [Google Scholar]
- 68.Sharma K, et al. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 2014;8:1583–1594. doi: 10.1016/j.celrep.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 69.Carpy A, et al. Absolute proteome and phosphoproteome dynamics during the cell cycle of Schizosaccharomyces pombe (fission yeast) Mol Cell Proteomics. 2014;13:1925–1936. doi: 10.1074/mcp.M113.035824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shao H, Li R, Ma C, Chen E, Liu XJ. Xenopus oocyte meiosis lacks spindle assembly checkpoint control. J Cell Biol. 2013;201:191–200. doi: 10.1083/jcb.201211041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun QY, Schatten H. Regulation of dynamic events by microfilaments during oocyte maturation and fertilization. Reproduction. 2006;131:193–205. doi: 10.1530/rep.1.00847. [DOI] [PubMed] [Google Scholar]
- 72.Bement WM, Sokac AM, Mandato CA. Four-dimensional imaging of cytoskeletal dynamics in Xenopus oocytes and eggs. Differentiation. 2003;71:518–527. doi: 10.1111/j.1432-0436.2003.07109005.x. [DOI] [PubMed] [Google Scholar]
- 73.Sokac AM, Co C, Taunton J, Bement W. Cdc42-dependent actin polymerization during compensatory endocytosis in Xenopus eggs. Nat Cell Biol. 2003;5:727–732. doi: 10.1038/ncb1025. [DOI] [PubMed] [Google Scholar]
- 74.Güttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol. 2009;10:178–191. doi: 10.1038/nrm2641. [DOI] [PubMed] [Google Scholar]
- 75.Antonin W, Ellenberg J, Dultz E. Nuclear pore complex assembly through the cell cycle: Regulation and membrane organization. FEBS Lett. 2008;582:2004–2016. doi: 10.1016/j.febslet.2008.02.067. [DOI] [PubMed] [Google Scholar]
- 76.Raices M, D’Angelo MA. Nuclear pore complex composition: A new regulator of tissue-specific and developmental functions. Nat Rev Mol Cell Biol. 2012;13:687–699. doi: 10.1038/nrm3461. [DOI] [PubMed] [Google Scholar]
- 77.Dultz E, et al. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J Cell Biol. 2008;180:857–865. doi: 10.1083/jcb.200707026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mackay DR, Ullman KS. Coordinating postmitotic nuclear pore complex assembly with abscission timing. Nucleus. 2011;2:283–288. doi: 10.4161/nucl.2.4.16189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mészáros N, et al. Nuclear pore basket proteins are tethered to the nuclear envelope and can regulate membrane curvature. Dev Cell. 2015;33:285–298. doi: 10.1016/j.devcel.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Laurell E, et al. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell. 2011;144:539–550. doi: 10.1016/j.cell.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 81.Franks TM, Hetzer MW. The role of Nup98 in transcription regulation in healthy and diseased cells. Trends Cell Biol. 2013;23:112–117. doi: 10.1016/j.tcb.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cross MK, Powers MA. Nup98 regulates bipolar spindle assembly through association with microtubules and opposition of MCAK. Mol Biol Cell. 2011;22:661–672. doi: 10.1091/mbc.E10-06-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sutovsky P, Simerly C, Hewitson L, Schatten G. Assembly of nuclear pore complexes and annulate lamellae promotes normal pronuclear development in fertilized mammalian oocytes. J Cell Sci. 1998;111:2841–2854. doi: 10.1242/jcs.111.19.2841. [DOI] [PubMed] [Google Scholar]
- 84.Nuccitelli R, Yim DL, Smart T. The sperm-induced Ca2+ wave following fertilization of the Xenopus egg requires the production of Ins(1,4,5)P3. Dev Biol. 1993;158:200–212. doi: 10.1006/dbio.1993.1179. [DOI] [PubMed] [Google Scholar]
- 85.Backs J, et al. The gamma isoform of CaM kinase II controls mouse egg activation by regulating cell cycle resumption. Proc Natl Acad Sci USA. 2010;107:81–86. doi: 10.1073/pnas.0912658106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wolf DP. The cortical response in Xenopus laevis ova. Dev Biol. 1974;40:102–115. doi: 10.1016/0012-1606(74)90112-2. [DOI] [PubMed] [Google Scholar]
- 87.Yang Q, Ferrell JE., Jr The Cdk1-APC/C cell cycle oscillator circuit functions as a time-delayed, ultrasensitive switch. Nat Cell Biol. 2013;15:519–525. doi: 10.1038/ncb2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hara K, Tydeman P, Kirschner M. A cytoplasmic clock with the same period as the division cycle in Xenopus eggs. Proc Natl Acad Sci USA. 1980;77:462–466. doi: 10.1073/pnas.77.1.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Murray AW, Kirschner MW. Dominoes and clocks: The union of two views of the cell cycle. Science. 1989;246:614–621. doi: 10.1126/science.2683077. [DOI] [PubMed] [Google Scholar]
- 90.Hsu JY, Reimann JD, Sørensen CS, Lukas J, Jackson PK. E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APC(Cdh1) Nat Cell Biol. 2002;4:358–366. doi: 10.1038/ncb785. [DOI] [PubMed] [Google Scholar]
- 91.Cascio SM, Wassarman PM. Program of early development in the mammal: Post-transcriptional control of a class of proteins synthesized by mouse oocytes and early embryos. Dev Biol. 1982;89:397–408. doi: 10.1016/0012-1606(82)90328-1. [DOI] [PubMed] [Google Scholar]
- 92.Latham KE, Garrels JI, Chang C, Solter D. Quantitative analysis of protein synthesis in mouse embryos. I. Extensive reprogramming at the one- and two-cell stages. Development. 1991;112:921–932. doi: 10.1242/dev.112.4.921. [DOI] [PubMed] [Google Scholar]
- 93.Lee G, Hynes R, Kirschner M. Temporal and spatial regulation of fibronectin in early Xenopus development. Cell. 1984;36:729–740. doi: 10.1016/0092-8674(84)90353-2. [DOI] [PubMed] [Google Scholar]
- 94.Schuel H, Schuel R. Benzohydroxamic acid induces polyspermic fertilization in the sea urchin Arbacia punctulata. Cell Biol Int Rep. 1987;11:189–196. doi: 10.1016/0309-1651(87)90025-7. [DOI] [PubMed] [Google Scholar]
- 95.Danilchik M, Tumarkin T. Exosomal trafficking in Xenopus development. Genesis. 2017;55 doi: 10.1002/dvg.23011. [DOI] [PubMed] [Google Scholar]
- 96.Penn A, Gledhill BL. Acrosomal proteolytic activity of amphibian sperm. A direct demonstration. Exp Cell Res. 1972;74:285–288. doi: 10.1016/0014-4827(72)90508-3. [DOI] [PubMed] [Google Scholar]
- 97.Solari FA, Dell’Aica M, Sickmann A, Zahedi RP. Why phosphoproteomics is still a challenge. Mol Biosyst. 2015;11:1487–1493. doi: 10.1039/c5mb00024f. [DOI] [PubMed] [Google Scholar]
- 98.Humphrey SJ, Azimifar SB, Mann M. High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat Biotechnol. 2015;33:990–995. doi: 10.1038/nbt.3327. [DOI] [PubMed] [Google Scholar]
- 99.Wühr M, et al. Accurate multiplexed proteomics at the MS2 level using the complement reporter ion cluster. Anal Chem. 2012;84:9214–9221. doi: 10.1021/ac301962s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tsai CF, et al. Large-scale determination of absolute phosphorylation stoichiometries in human cells by motif-targeting quantitative proteomics. Nat Commun. 2015;6:6622. doi: 10.1038/ncomms7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wessel D, Flügge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 102.Dephoure N, Gygi SP. A solid phase extraction-based platform for rapid phosphoproteomic analysis. Methods. 2011;54:379–386. doi: 10.1016/j.ymeth.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Erickson BK, et al. Evaluating multiplexed quantitative phosphopeptide analysis on a hybrid quadrupole mass filter/linear ion trap/orbitrap mass spectrometer. Anal Chem. 2015;87:1241–1249. doi: 10.1021/ac503934f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Beausoleil SA, Villén J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 105.Petráš I, Bednárová D. Total least squares approach to modeling: A Matlab toolbox. Acta Montan Slovaca. 2010;15:158–170. [Google Scholar]
- 106.Vizcaíno JA, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.