Significance
Inflammasomes are cytosolic multiprotein complexes that initiate innate immune responses to microbial infection. Inflammasome specificity is determined by cytosolic innate immune sensors, including nucleotide-binding domain, leucine-rich repeat-containing family, apoptosis inhibitory proteins (NAIPs). In mice, which encode seven different NAIPs, individual NAIPs recognize specific components of the structurally related bacterial type III secretion system (T3SS) and flagellar apparatus. Humans encode a single functional NAIP, raising the question of whether human NAIP recognizes the same repertoire of bacterial ligands. Here, we find that, in contrast to the ligand specificity exhibited by the murine NAIPs, the single human NAIP broadly detects multiple T3SS and flagellin proteins. These findings provide a basis for understanding the mechanisms underlying human-specific innate immune responses against bacterial infection.
Keywords: inflammasome, NAIP, NLRC4, flagellin, type III secretion system
Abstract
Inflammasomes are cytosolic multiprotein complexes that initiate host defense against bacterial pathogens by activating caspase-1–dependent cytokine secretion and cell death. In mice, specific nucleotide-binding domain, leucine-rich repeat-containing family, apoptosis inhibitory proteins (NAIPs) activate the nucleotide-binding domain, leucine-rich repeat-containing family, CARD domain-containing protein 4 (NLRC4) inflammasome upon sensing components of the type III secretion system (T3SS) and flagellar apparatus. NAIP1 recognizes the T3SS needle protein, NAIP2 recognizes the T3SS inner rod protein, and NAIP5 and NAIP6 recognize flagellin. In contrast, humans encode a single functional NAIP, raising the question of whether human NAIP senses one or multiple bacterial ligands. Previous studies found that human NAIP detects both flagellin and the T3SS needle protein and suggested that the ability to detect both ligands was achieved by multiple isoforms encoded by the single human NAIP gene. Here, we show that human NAIP also senses the Salmonella Typhimurium T3SS inner rod protein PrgJ and that T3SS inner rod proteins from multiple bacterial species are also detected. Furthermore, we show that a single human NAIP isoform is capable of sensing the T3SS inner rod, needle, and flagellin. Our findings indicate that, in contrast to murine NAIPs, promiscuous recognition of multiple bacterial ligands is conferred by a single human NAIP.
In response to pathogenic bacteria, the innate immune system is required for inflammatory responses that promote host defense. Host defense is initiated by the engagement of pattern recognition receptors (PRRs) by pathogen-associated molecular patterns (1). Cytosolic PRRs detect pathogens that introduce products into host cells as a consequence of bacterial virulence activities, such as specialized secretion systems. A subset of cytosolic PRRs, termed the nucleotide-binding domain, leucine-rich repeat-containing (NLR) family, is composed of 23 members in humans and 34 members in mice (2, 3). A subfamily of NLRs, known as nucleotide-binding domain, leucine-rich repeat-containing family, apoptosis inhibitory proteins (NAIPs), recognizes bacterial proteins that are translocated into the host cell by Gram-negative bacteria. One such pathogen is Salmonella, which uses a virulence-associated type III secretion system (T3SS) to inject effector proteins into the host cell cytosol that promote bacterial invasion and survival (4). These secretion systems also translocate structurally related components of the T3SS or closely related flagellar apparatus, enabling cytosolic detection of bacteria by NAIPs (5). In mice, ligands for four of seven distinct NAIPs are known: NAIP1 recognizes the T3SS needle protein, NAIP2 recognizes the T3SS inner rod protein, and both NAIP5 and NAIP6 recognize flagellin (6–11). Upon binding their cognate ligands, the NAIPs recruit the adaptor nucleotide-binding domain, leucine-rich repeat-containing family, CARD domain-containing protein 4 (NLRC4) (12–14). The resulting NAIP/NLRC4 inflammasome then recruits and activates caspase-1 (15). Active caspase-1 mediates processing and secretion of IL-1 family cytokines and a proinflammatory cell death termed pyroptosis (16–18), which promote antimicrobial functions critical for controlling bacterial infection (19–22). This inflammasome also plays a protective role in mouse models of colitis-associated colorectal cancer and may be a useful strategy in tumor immunotherapy (23, 24). However, the NLRC4 inflammasome can cause sepsis-like disease after antibiotic disruption of the microbiota, and activating NLRC4 mutations cause an autoinflammatory syndrome in humans (25–29). Defining the mechanisms of human NAIP sensing of bacterial ligands may, therefore, provide insight into therapeutic approaches for diverse infectious and autoinflammatory diseases.
Unlike in mice, the human NAIP locus has a number of pseudogenes and gene duplications and has retained a single functional copy of the full-length NAIP gene (30, 31). Initial studies with human monocytic cell lines suggested that human NAIP could only sense the T3SS needle protein (7–9). However, a recent study found that flagellin also triggers NAIP inflammasome activation in primary human macrophages and indicated that detection of flagellin was mediated by an alternate splice isoform of NAIP (32). These findings suggested that, in humans, specificity for different bacterial ligands is encoded by distinct splicing variants of the single NAIP gene.
Here, we show that, in addition to the T3SS needle protein and flagellin, primary human macrophages also mount NAIP inflammasome responses against T3SS inner rod proteins from multiple bacterial pathogens. In addition, our data show that the Salmonella Typhimurium SPI-2 T3SS inner rod protein, SsaI, which is required for intracellular bacterial replication, does not activate the inflammasome in human macrophages, suggesting that intracellular Salmonella evades NAIP recognition in both humans and mice. Intriguingly, we find that a single human NAIP isoform is sufficient for NLRC4 inflammasome responses to the T3SS needle, inner rod, and flagellin. Overall, our findings suggest that, unlike mice, which express multiple NAIPs that each possesses exquisite ligand specificity, the single human NAIP has evolved to broadly recognize multiple bacterial ligands. These findings provide important insight into distinct mechanisms of innate immune sensing of Gram-negative bacteria by mice and humans.
Results
Salmonella Typhimurium Induces Flagellin-Independent Inflammasome Responses in Primary Human Macrophages.
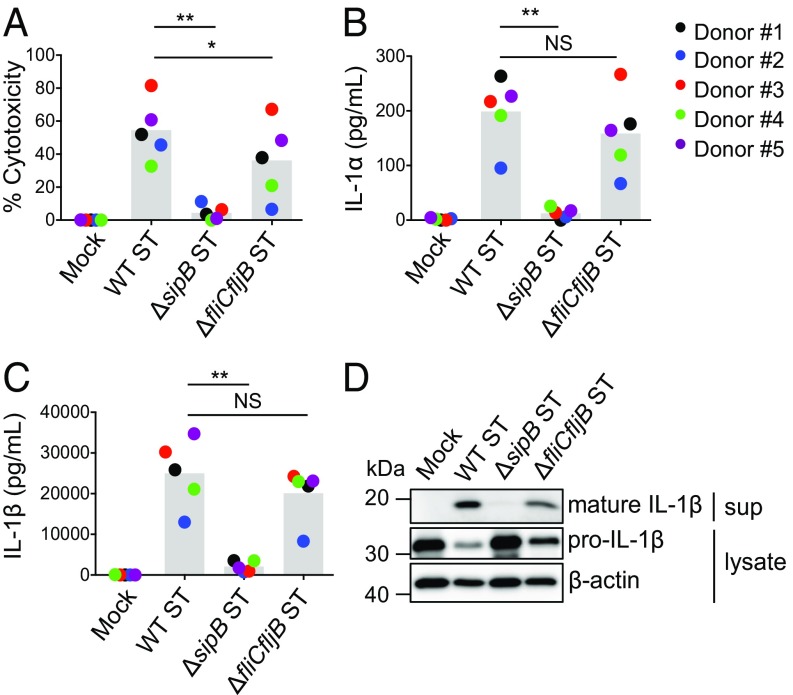
In murine macrophages, the NAIPs induce inflammasome activation on direct recognition of proteins from the T3SS and the structurally related flagellar apparatus. The relative contribution of these components to the inflammasome response in human macrophages is still unclear. Thus, we examined cell death as well as secretion of IL-1α and IL-1β after infection of human monocyte-derived macrophages (hMDMs) with WT, SPI-1 T3SS-deficient (ΔsipB), or flagellin-deficient (ΔfliCfljB) Salmonella Typhimurium strains. Compared with WT Salmonella-infected macrophages, ΔfliCfljB Salmonella-infected macrophages exhibited a slight but not statistically significant decrease in inflammasome activation as measured by IL-1α and IL-1β secretion, IL-1β processing, and cell death (Fig. 1). In contrast, inflammasome activation was abrogated in ΔsipB-infected macrophages (Fig. 1 A–C). Immunoblot analysis indicated no defect in pro–IL-1β production in ΔsipB-infected hMDMs, but inflammasome-mediated cleavage of pro–IL-1β into its active form was not observed (Fig. 1D). These results suggest that Salmonella infection of primary human macrophages induces robust flagellin-independent inflammasome activation that requires the SPI-1 T3SS.
Fig. 1.
Salmonella Typhimurium induces T3SS-dependent, flagellin-independent inflammasome responses in primary human macrophages. hMDMs were primed with LPS for 3 h and treated with PBS (mock), WT Salmonella (WT ST), ΔsipB ST, or ΔfliCfljB ST at a multiplicity of infection of 20 for 4 h. (A) Cell death (percentage cytotoxicity) was measured by lactate dehydrogenase release assay and normalized to mock infected cells. (B and C) IL-1α and IL-1β supernatant levels were measured by ELISA. (D) Immunoblot analysis was performed on supernatants (sup) for mature IL-1β and on lysates for pro–IL-1β and β-actin as a loading control (representative of two donors). Each data point represents the mean of triplicate infected wells for each of five different human donors. Shaded bars represent the overall mean of the donors. NS, not significant. *P < 0.05 by paired t test; **P < 0.01 by paired t test.
Salmonella Typhimurium T3SS Inner Rod Protein PrgJ Activates the Inflammasome in Primary Human Macrophages.
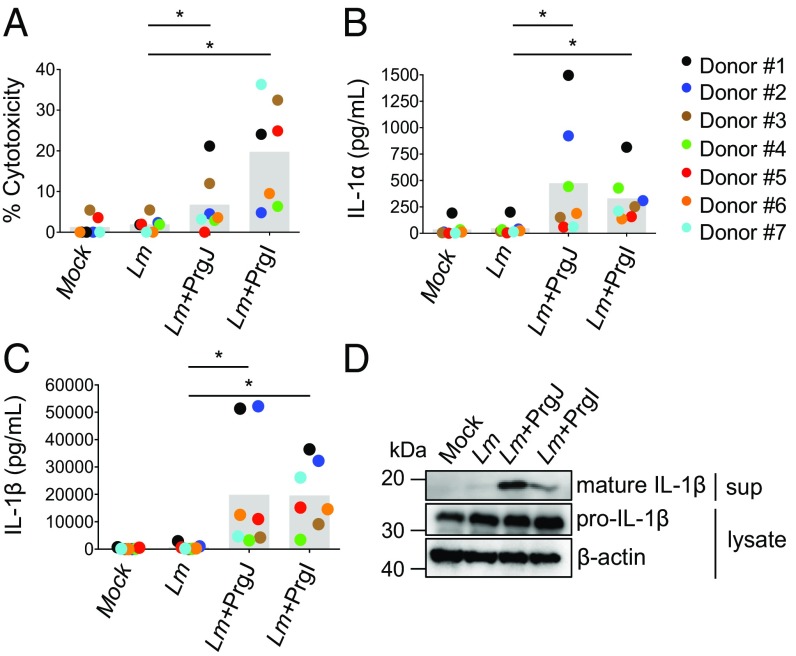
Previous studies using immortalized human monocytic cell lines found that the NAIP inflammasome could be activated by the T3SS needle protein but not flagellin or the T3SS inner rod (7–9). However, another study found that NAIP played a role in restricting the intracellular replication of flagellated bacteria (33). Recently, it was shown that flagellin can activate the NAIP inflammasome in primary hMDMs (32). As our data suggested that there is a robust flagellin-independent, T3SS-dependent inflammasome response to Salmonella, we sought to determine whether, in addition to the T3SS needle protein PrgI, the T3SS inner rod protein PrgJ could induce inflammasome activation in primary hMDMs. We utilized the Gram-positive pathogen Listeria monocytogenes, which does not encode a T3SS apparatus, to directly deliver PrgJ or PrgI into host cells (34). After infection, Listeria uses the pore-forming toxin Listeriolysin O (LLO) to escape into the cytosol, where it expresses the protein ActA on the bacterial surface to polymerize actin (35, 36). We utilized strains that ectopically express PrgJ or PrgI translationally fused to the N terminus of ActA and under control of the actA promoter. This approach of delivering flagellin into the host cell cytosol robustly activates the mouse NAIP5 inflammasome (34). Indeed, as expected, hMDMs infected with Listeria expressing PrgI induced robust IL-1α and IL-1β secretion, IL-1β processing, and cell death above that of WT Listeria-infected cells (Fig. 2). Surprisingly, infection with PrgJ-expressing Listeria also induced robust IL-1α and IL-1β release, IL-1β processing, and cell death (Fig. 2). Importantly, cytosolic access was required for inflammasome activation, as PrgJ-expressing Listeria lacking hly, the gene encoding LLO, did not induce IL-1β secretion (Fig. S1).
Fig. 2.
L. monocytogenes-mediated delivery of the T3SS inner rod protein PrgJ activates the inflammasome in primary human macrophages. hMDMs were primed with Pam3CSK4 for 3 h and infected with PBS (mock), WT Listeria (Lm), or Lm strains expressing PrgJ and PrgI at a multiplicity of infection of five for 16 h. (A) Cell death (percentage cytotoxicity) was measured by lactate dehydrogenase release assay and normalized to mock infected cells. (B and C) IL-1α and IL-1β supernatant levels were measured by ELISA. (D) Immunoblot analysis of supernatants (sup) for mature IL-1β and lysates for pro—IL-1β and β-actin as a loading control (representative of two donors). Each data point represents the mean of triplicate infected wells for each of seven different human donors. Shaded bars represent the overall mean of the donors. *P < 0.05 by paired Wilcoxon signed rank test.
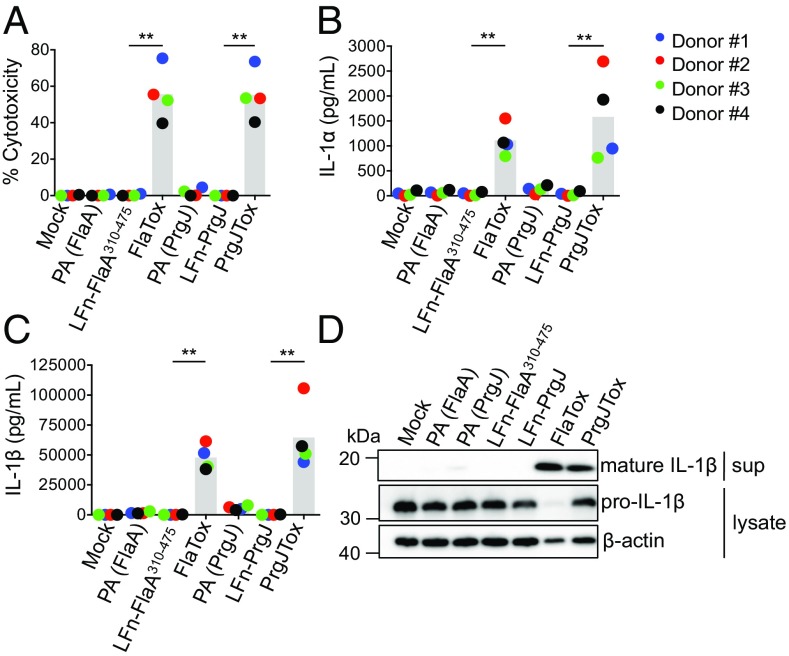
To determine whether PrgJ alone could induce inflammasome activation independently of bacterial infection, we used an anthrax toxin-based delivery system (9, 11, 37). In this system, bacterial ligands are translationally fused to the N-terminal domain of Bacillus anthracis lethal factor (LFn). The LFn domain enables ligand translocation into the host cell cytosol through a membrane channel formed by the anthrax protective antigen (PA) protein. We used a translational fusion of LFn and PrgJ (LFn-PrgJ) as well as LFn fused to flagellin as a positive control for NAIP inflammasome activation. To avoid potential confounding effects of TLR5 detection of flagellin, we used a truncated Legionella pneumophila flagellin that lacks the TLR5-activating region but retains the C-terminal 166 amino acids detected by murine NAIP5 (38, 39). In agreement with previous findings (32), hMDMs treated with PA+LFn-FlaA310–475 (referred to as FlaTox) induced robust inflammasome activation as measured by significantly increased IL-1α and IL-1β cytokine release, IL-1β processing, and cell death (Fig. 3). Treatment with PA+LFn-PrgJ (referred to as PrgJTox) also induced robust IL-1α and IL-1β cytokine secretion, IL-1β processing, and cell death (Fig. 3). In contrast, treatment with PA, LFn-FlaA310–475, or LFn-PrgJ alone did not activate, indicating that FlaA and PrgJ induce inflammasome activation only when delivered into the host cell cytosol via PA. Altogether, these results show that primary human macrophages undergo inflammasome activation on cytosolic sensing of the Salmonella Typhimurium T3SS inner rod protein.
Fig. 3.
Anthrax toxin-mediated delivery of the T3SS inner rod protein PrgJ induces robust inflammasome activation in primary human macrophages. hMDMs were primed with Pam3CSK4 for 4 h and treated with PA alone, LFn-FlaA310–475 alone, LFn-PrgJ alone, PA+LFn-FlaA310–475 (FlaTox), or PA+LFn-PrgJ (PrgJTox) for 16 h. (A) Cell death (percentage cytotoxicity) was measured by lactate dehydrogenase release assay and normalized to mock infected cells. (B and C) IL-1α and IL-1β supernatant level were measured by ELISA. (D) Immunoblot analysis of supernatants (sup) for mature IL-1β and lysates for pro–IL-1β and β-actin as a loading control (representative of two donors). Each data point represents the mean of triplicate infected wells for each of four different human donors. Shaded bars represent the overall mean of the donors. **P < 0.01 by paired t test.
Human NAIP Is Required for Maximal Inflammasome Responses to the T3SS Inner Rod Protein PrgJ.
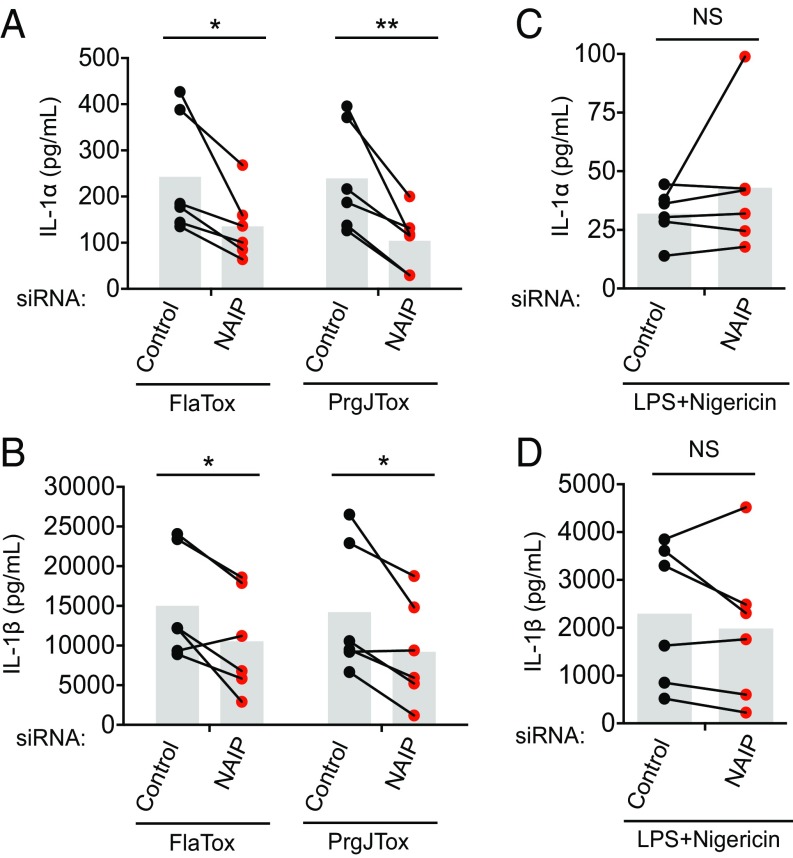
Human NAIP is required for inflammasome responses to flagellin and the T3SS needle protein (7, 9, 32). To test whether NAIP is also necessary for detecting PrgJ, we used siRNAs to silence NAIP in primary hMDMs (Fig. S2). As expected (32), anti-NAIP siRNA treatment resulted in significantly decreased IL-1α and IL-1β secretion after FlaTox administration compared with control siRNA treatment. Anti-NAIP siRNA treatment also led to significantly decreased IL-1α and IL-1β secretion in response to PrgJTox administration relative to control siRNA-treated cells, suggesting that NAIP is required for maximal inflammasome responses to PrgJ (Fig. 4 A and B). Importantly, siRNA-mediated silencing of NAIP did not significantly affect inflammasome responses to LPS+Nigericin, which specifically activates the NLRP3 inflammasome and does not engage NAIP (40) (Fig. 4 C and D). These results indicate that NAIP is required for maximal inflammasome responses to the T3SS inner rod.
Fig. 4.
Human NAIP is required for maximal inflammasome responses to flagellin and the T3SS inner rod protein PrgJ. Primary hMDMs were transfected with control siRNA or siRNA against NAIP for 48 h and primed with Pam3CSK4 for 4 h. (A and B) Cells were treated with PA+LFn-PrgJ (PrgJTox) or PA+LFn-FlaA310–475 (FlaTox) for 5 h. (C and D) Cells were treated with LPS+Nigericin for 5 h. IL-1α and IL-1β supernatant levels were measured by ELISA. Each data point represents the mean of triplicate infected wells for six different human donors. Shaded bars represent the overall mean of the donors. NS, not significant. *P < 0.05 by paired t test; **P < 0.01 by paired t test.
T3SS Inner Rod Proteins from Other Bacterial Species Induce Inflammasome Activation in Human Macrophages.
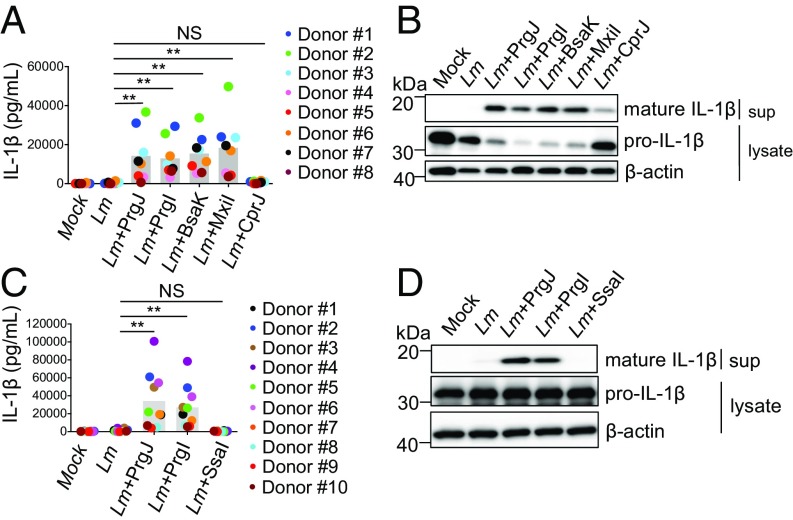
As T3SS inner rod proteins from multiple bacterial species activate the mouse NAIP2 inflammasome (41), we next examined whether other bacterial T3SS inner rod homologs similarly activate human cells. We engineered Listeria strains expressing the T3SS inner rod proteins from Burkholderia thailandensis (BsaK), Shigella flexneri (MxiI), and Chromobacterium violaceum (CprJ). In agreement with previous findings (41), mouse macrophages infected with Listeria expressing these inner rod homologs robustly secreted IL-1β (Fig. S3). hMDMs infected with Listeria expressing PrgI, PrgJ, BsaK, and MxiI also resulted in robust IL-1β secretion and processing well above that of WT Listeria-infected cells (Fig. 5 A and B). In contrast, CprJ-expressing Listeria induced relatively low levels of IL-1β secretion and processing. These findings show that human macrophages broadly detect and activate the inflammasome in response to T3SS inner rod proteins from multiple bacterial species.
Fig. 5.
T3SS inner rod proteins from multiple bacterial species activate the human inflammasome, whereas the Salmonella Typhimurium SPI-2 T3SS inner rod protein, SsaI, evades immune recognition. hMDMs were primed with Pam3CSK4 for 3 h and infected with WT Listeria (Lm) or strains ectopically expressing PrgJ, PrgI, BsaK MxiI, CprJ, or SsaI at a multiplicity of infection of five for 16 h. Cells were treated with PBS for the mock control. (A and C) IL-1β supernatant levels were measured by ELISA. (B and D) Immunoblot analysis of supernatants (sup) for mature IL-1β and lysates for pro–IL-1β and β-actin as a loading control. Each data point represents the mean of triplicate infected wells for each of 8–10 different human donors. Shaded bars represent the overall mean of the donors. NS, not significant. **P < 0.01 by paired Wilcoxon signed rank test.
Salmonella Typhimurium SPI-2 T3SS Inner Rod Protein SsaI Evades Immune Detection by Human Macrophages.
Salmonella Typhimurium uses two different T3SSs, termed SPI-1 and SPI-2. The SPI-1 T3SS plays a role in bacterial invasion, whereas the SPI-2 T3SS is required for intracellular survival and replication (42–44), suggesting a need to evade host recognition of the SPI-2 T3SS. Indeed, while the SPI-1 T3SS inner rod protein, PrgJ, robustly activates the mouse NAIP2 inflammasome, the SPI-2 T3SS inner rod protein, SsaI, evades detection (41). We, therefore, asked whether SsaI also evades human NAIP by expressing SsaI in Listeria. Consistent with previous findings (41), mouse macrophages infected with Listeria expressing SsaI secreted negligible levels of IL-1β (Fig. S3). Infection of hMDMs with SsaI-expressing Listeria also resulted in negligible IL-1β secretion and cleavage compared with infection with Listeria expressing PrgJ or PrgI (Fig. 5 C and D). These data suggest that the SPI-2 T3SS inner rod protein SsaI has evolved to evade NAIP recognition in both mice and humans.
The THP-1 Monocytic Cell Line Undergoes Inflammasome Activation in Response to T3SS Inner Rod and Flagellin Proteins.
Previous studies using the U937 and THP-1 monocytic cell lines found that anthrax toxin-mediated delivery of flagellin or inner rod proteins did not induce inflammasome activation (7, 9). Transfection of purified PrgJ protein into these cells also failed to activate the inflammasome (8). In contrast, recent findings and the data presented here show that hMDMs mount robust inflammasome responses to flagellin (32) and the T3SS inner rod. A previously proposed explanation for these discrepant findings is that distinct NAIP splicing isoforms possess differing ligand specificities and that primary human macrophages and immortalized cells express differing levels of particular isoforms (32). Alternatively, human NAIP may recognize all three bacterial ligands regardless of isoform type. As THP-1 cells express lower levels of NAIP and NLRC4 than primary macrophages (32), the method of ligand delivery or specific bacterial proteins previously used may not have been sufficient for inflammasome activation in this cell type. Previous studies utilized the C. violaceum inner rod protein CprJ (7, 9), which we found to be a poor inflammasome activator in hMDMs relative to other T3SS inner rod homologs (Fig. 5 A and B). Another study used transfection-based delivery of PrgJ protein (8), which is likely not as efficient at delivering proteins into host cells as the anthrax toxin system.
Thus, we next tested whether THP-1 cells activate inflammasome responses to PrgJ delivered via Listeria or the anthrax toxin system. Although PrgI-expressing Listeria induced IL-1α and IL-1β secretion in THP-1 cells, PrgJ-expressing Listeria failed to do so (Fig. S4), despite robustly activating hMDMs (Fig. 2). In contrast, THP-1 cells treated with PrgJTox robustly secreted IL-1α and IL-1β (Fig. 6 A and B). Consistent with previous findings (7, 9), anthrax toxin-mediated delivery of full-length flagellin failed to activate THP-1 cells (Fig. S5). Intriguingly, anthrax toxin-mediated delivery of a truncated version of flagellin robustly triggered IL-1α and IL-1β secretion (Fig. 6 A and B), likely because of more efficient delivery of truncated flagellin. These data show that THP-1 cells are capable of detecting the T3SS needle, inner rod, and flagellin but are less responsive than hMDMs, as the type of bacterial ligand and route of delivery influence the extent of inflammasome activation.
Fig. 6.
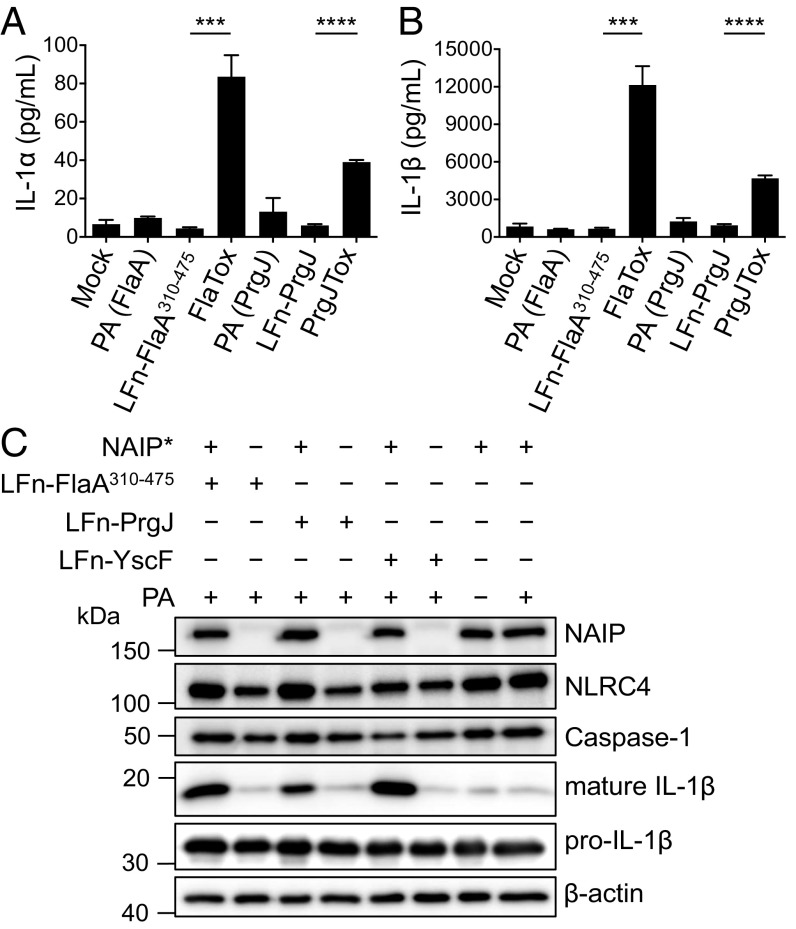
A single NAIP isoform is sufficient for inflammasome responses to flagellin, the T3SS inner rod protein, and the T3SS needle protein. (A and B) THP-1 cells were primed with Pam3CSK4 and treated with PA alone, LFn-FlaA310–475 alone, LFn-PrgJ alone, PA+LFn-FlaA310–475 (FlaTox), or PA+LFn-PrgJ (PrgJTox) for 5 h. IL-1α and IL-1β supernatant levels were measured by ELISA. Bar graphs display the mean ± SD of triplicate wells. Representative of three independent experiments. ***P < 0.001 by unpaired t test; ****P < 0.0001 by unpaired t test. (C) HEK293 cells were transfected with expression vectors encoding NLRC4, caspase-1, and IL-1β. Where indicated, cells were also transfected with vectors encoding NAIP* (+) or empty vector control (−). After 18 h, cells were treated with PA+LFn-PrgJ, PA+LFn-FlaA310–475, PA+LFn-YscF, or PA alone for 9 h. Immunoblot analysis was performed on cell lysates for mature and pro–IL-1β, NAIP*, NLRC4, caspase-1, and β-actin as a loading control. Representative of three independent experiments.
A Single NAIP Isoform Mediates Inflammasome Responses to T3SS Needle, Inner Rod, and Flagellin Proteins.
Our data show that both THP-1 cells and hMDMs recognize T3SS needle, inner rod, and flagellin and that NAIP contributes to ligand detection. We next sought to understand how a single human NAIP gene could confer recognition of all three ligands in contrast to mice, which utilize distinct NAIPs to recognize each ligand. Interestingly, studies in which chimeric mouse NAIPs were generated to define the ligand specificity domain identified a chimeric mouse NAIP capable of recognizing multiple ligands (45), suggesting the possibility that human NAIP might function as a broad receptor. Human monocytic cell lines express lower levels than hMDMs of a particular full-length NAIP splicing isoform (termed NAIP*) that enables sensing of flagellin (32). We, therefore, sought to test whether a single NAIP isoform possesses specificity for a given bacterial ligand or is capable of detecting all three bacterial ligands. We ectopically expressed the NAIP* isoform previously shown to recognize flagellin (32) along with other NLRC4 inflammasome components in HEK293 cells and then used the anthrax toxin system to deliver bacterial ligands into these cells. As expected, HEK293 cells expressing the NAIP* isoform robustly processed IL-1β in response to flagellin (Fig. 6C). Unexpectedly, however, delivery of PrgJ or the Burkholderia T3SS needle protein (YscF) also induced robust IL-1β processing (Fig. 6C). Critically, inflammasome activation by FlaA, PrgJ, and YscF required NAIP, as delivery of bacterial ligands into cells only expressing NLRC4, caspase-1, and IL-1β did not result in IL-1β processing. Inflammasome activation also required delivery of the bacterial ligands, as untreated cells or PA treatment alone did not process IL-1β. Altogether, these data indicate that a single human NAIP isoform is sufficient to mediate inflammasome responses to the T3SS needle, inner rod, and flagellin proteins.
Discussion
Our data show that, like murine cells, human macrophages sense multiple bacterial ligands from the T3SS and flagellar apparatus. In addition to the T3SS needle and flagellin, T3SS inner rod proteins from multiple bacterial species activate the human NAIP inflammasome. Furthermore, a single human NAIP isoform can mediate inflammasome responses to all three bacterial proteins in contrast to mouse NAIPs, which are highly selective for recognition of individual flagellin or T3SS proteins (6–11). Consistent with our findings, a recent study found that the Pseudomonas aeruginosa T3SS inner rod also activates the human NAIP inflammasome (46). The region of murine NAIPs that confers ligand specificity has been mapped to an internal region composed of several nucleotide-binding domain (NBD)-associated α-helical domains (45). This region has evolved under positive selection in both rodents and primates (45), suggesting that this domain mediates ligand detection in human NAIP as well. How NAIP achieves broad recognition of multiple ligands and whether NAIP binds these ligands with similar or differing affinities or binding kinetics are unclear. The T3SS inner rod, needle, and flagellin proteins exhibit low sequence conservation but have some structural conservation, as the T3SS is thought to have evolved from the flagellar apparatus (47). Thus, human NAIP may recognize structural elements common to all three ligands. It will be of interest to determine whether NAIP detection of these three ligands is functionally redundant or distinct in the initiation of antimicrobial activities.
Our study raises intriguing questions about the evolution of the NAIP/NLRC4 inflammasome. It is likely that a single NAIP progenitor was present in the last common ancestor of primates and rodents (48). In mice, there has been an expansion of NAIP genes as a consequence of several gene duplication events (49); interestingly, the murine NAIPs are specialists, as they each recognize only one of three bacterial proteins derived from the evolutionarily related T3SS and flagellar apparatus. In contrast, the single human NAIP is a generalist, as it is capable of functionally detecting all three bacterial proteins. The promiscuity displayed by human NAIP may provide a selective advantage, as it may be more difficult for pathogens to simultaneously evade recognition of all three ligands by human NAIP.
Promiscuous ligand recognition may be a general strategy used by the innate immune system to diversify protein functionality as a means of promoting responses against different pathogenic stimuli. For example, the natural killer (NK) activating receptor NKG2D broadly recognizes several MHC class I-related proteins in contrast to other NK receptors, which typically recognize a single ligand. The ability of NKG2D to recognize a broad array of stress-inducible host ligands may provide an evolutionary advantage against viruses that use mechanisms to down-regulate NKG2D ligands as well as rapidly evolving cancers (50). Furthermore, the TLR sorting adaptor TIRAP promiscuously detects multiple lipids, which diversify subcellular sites of TLR signaling and thus, enable responses to both extracellular and endosomal pathogens (51). However, one possible tradeoff with a more promiscuous mode of sensing is that human NAIP may possess weaker affinities or altered binding kinetics for its bacterial ligands and hence, decreased signaling potency. In contrast, a given mouse NAIP may possess higher affinity or half-life in binding its particular ligand and thus, confer heightened immune responses. Indeed, compared with mouse macrophages, human macrophages do not seem to be as responsive to cytosolic flagellin, as they are more permissive for intracellular replication of flagellated bacteria (33). While the precise basis for this difference is unknown, one possibility is that human NAIP detects flagellin with lower affinity or altered binding kinetics than mouse NAIP5.
It will be of interest to examine how coevolution with Gram-negative bacteria shaped the NAIP genes in humans and other mammals and whether pathogens have evolved strategies for evading human NAIP. Functional NAIP copy number varies among human populations, and increased copy number has been postulated to confer a selective advantage in antibacterial defense (52). Studies in mice have shown that inappropriate activation of the NAIP/NLRC4 inflammasome can lead to lethal systemic inflammation resembling sepsis (29, 37). Moreover, gain-of-function mutations in human NLRC4 result in pathologic enterocolitis and autoinflammation (25–28). Perhaps gain-of-function mutations in human NAIP confer similar pathological outcomes.
Our results provide insight into human NAIP detection of bacterial proteins from the T3SS and flagellar apparatus. The data presented here provide an important basis for elucidating the mechanisms underlying human NAIP inflammasome responses to bacterial infection, which could prove crucial to understanding how the NAIP/NLRC4 inflammasome contributes to human health and disease.
Materials and Methods
Ethics Statement.
All studies involving hMDMs were performed in compliance with the requirements of the US Department of Health and Human Services and the principles expressed in the Declaration of Helsinki. Samples obtained from the University of Pennsylvania Human Immunology Core are considered to be a secondary use of deidentified human specimens and are exempt via Title 55 Part 46, Subpart A of 46.101 (b) of the Code of Federal Regulations. All experiments performed with mouse bone marrow-derived macrophages were done so in accordance with the Animal Welfare Act and the recommendations in Guide for the Care and Use of Laboratory Animals of the NIH (53). The Institutional Animal Care and Use Committee of the University of Pennsylvania approved all procedures (protocol 804928).
Bacterial Strains and Growth Conditions.
Salmonella enterica serovar Typhimurium WT, ΔsipB (54), and ΔfliCfljB (55) isogenic strains on the SL1344 background were used. Three hours before infection, Salmonella was diluted into Luria–Bertani broth containing 300 mM NaCl and grown for 3 h standing at 37 °C to induce SPI-1 expression (56). L. monocytogenes WT and isogenic strains on the 10403S background were cultured in brain heart infusion medium (34). Listeria strains encoding heterologous bacterial ligands (L. pneumophila FlaA, Salmonella Typhimurium PrgJ, and Salmonella Typhimurium PrgI) translationally fused to the truncated N terminus of ActA and under the control of the actA promoter were used (34). The pPL2 vector encoding PrgJ was introduced into Δhly Listeria as previously described (34, 57). Listeria strains expressing Salmonella Typhimurium SsaI, B. thailandensis BsaK, S. flexeri MxiI, and C. violaceum CprJ were constructed using codon-optimized gene fragments (IDT) cloned into the pPL2 vector and introduced into Listeria as previously described (34, 57).
Cellular Assays.
Purified human monocytes from deidentified healthy human donors were obtained from the University of Pennsylvania Human Immunology Core. Monocytes were cultured in RPMI supplemented with 10% (vol/vol) heat-inactivated FBS, 2 mM l-glutamine, 100 IU/mL penicillin, 100 µg/mL streptomycin, and 50 ng/mL recombinant human M-CSF (Gemini Bio-Products) for 6 d to promote differentiation into hMDMs. One day before infection, adherent hMDMs were replated in media with 25 ng/mL human M-CSF lacking antibiotics at 1.0 × 105 cells per well in a 48-well plate. Pam3CSK4 (100 ng/mL) and LPS (500 ng/mL) pretreatments, bacterial infections, anthrax toxin-mediated delivery of bacterial ligands, siRNA experiments, cytotoxicity assays, ELISA, immunoblot analyses, qRT-PCR analyses, HEK293 inflammasome reconstitution assays, and statistical analyses were performed as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Russell Vance and Daniel Portnoy for helpful discussions, protocols, and reagents, and Randilea Nichols, Isabella Rauch, and Jeannette Tenthorey for providing anthrax toxin-based reagents. We also thank Igor Brodsky and members of the Brodsky laboratory and the laboratory of S.S. for helpful discussions and critical reading of the manuscript and Cierra Casson and Janet Yu for hMDM experimental protocols. We thank the Human Immunology Core of the Penn Center for AIDS Research and the Abramson Cancer Center for providing purified primary human monocytes. This work is supported, in part, by NIH National Cancer Institute Grants R01CA188034 (to J.-D.S.) and P30-CA016520, NIH National Institute of General Medical Sciences Grants T32GM07229 (to V.M.R.R.) and R25GM071745, NIH National Institute of Allergy and Infectious Diseases Grants R01AI118861 (to S.S.) and R01AI123243 (to S.S.), National Science Foundation Graduate Fellowship DGE-1321851 (to V.M.R.R.), a College Alumni Society Board of Managers and Presidents Undergraduate Research Grant (to I.J.S.), a College Alumni Society Pincus–Magaziner Family Undergraduate Research Grant (to B.M.Y.), and a Burroughs–Wellcome Fund Investigators in the Pathogenesis of Infectious Diseases Award (to S.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710433114/-/DCSupplemental.
References
- 1.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Chen G, Shaw MH, Kim YG, Nuñez G. NOD-like receptors: Role in innate immunity and inflammatory disease. Annu Rev Pathol. 2009;4:365–398. doi: 10.1146/annurev.pathol.4.110807.092239. [DOI] [PubMed] [Google Scholar]
- 3.von Moltke J, Ayres JS, Kofoed EM, Chavarría-Smith J, Vance RE. Recognition of bacteria by inflammasomes. Annu Rev Immunol. 2013;31:73–106. doi: 10.1146/annurev-immunol-032712-095944. [DOI] [PubMed] [Google Scholar]
- 4.Galán JE, Lara-Tejero M, Marlovits TC, Wagner S. Bacterial type III secretion systems: Specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol. 2014;68:415–438. doi: 10.1146/annurev-micro-092412-155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun YH, Rolán HG, Tsolis RM. Injection of flagellin into the host cell cytosol by Salmonella enterica serotype Typhimurium. J Biol Chem. 2007;282:33897–33901. doi: 10.1074/jbc.C700181200. [DOI] [PubMed] [Google Scholar]
- 6.Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477:592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J, Zhao Y, Shi J, Shao F. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc Natl Acad Sci USA. 2013;110:14408–14413. doi: 10.1073/pnas.1306376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rayamajhi M, Zak DE, Chavarria-Smith J, Vance RE, Miao EA. Cutting edge: Mouse NAIP1 detects the type III secretion system needle protein. J Immunol. 2013;191:3986–3989. doi: 10.4049/jimmunol.1301549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Y, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y, et al. Genetic functions of the NAIP family of inflammasome receptors for bacterial ligands in mice. J Exp Med. 2016;213:647–656. doi: 10.1084/jem.20160006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rauch I, et al. NAIP proteins are required for cytosolic detection of specific bacterial ligands in vivo. J Exp Med. 2016;213:657–665. doi: 10.1084/jem.20151809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L, et al. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science. 2015;350:404–409. doi: 10.1126/science.aac5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Z, et al. Structural and biochemical basis for induced self-propagation of NLRC4. Science. 2015;350:399–404. doi: 10.1126/science.aac5489. [DOI] [PubMed] [Google Scholar]
- 14.Diebolder CA, Halff EF, Koster AJ, Huizinga EG, Koning RI. Cryoelectron tomography of the NAIP5/NLRC4 inflammasome: Implications for NLR activation. Structure. 2015;23:2349–2357. doi: 10.1016/j.str.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 15.Martinon F, Burns K, Tschopp J. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 16.Li P, et al. Mice deficient in IL-1 beta-converting enzyme are defective in production of mature IL-1 beta and resistant to endotoxic shock. Cell. 1995;80:401–411. doi: 10.1016/0092-8674(95)90490-5. [DOI] [PubMed] [Google Scholar]
- 17.Kuida K, et al. Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme. Science. 1995;267:2000–2003. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- 18.Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: Host cell death and inflammation. Nat Rev Microbiol. 2009;7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miao EA, et al. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol. 2010;11:1136–1142. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jorgensen I, Zhang Y, Krantz BA, Miao EA. Pyroptosis triggers pore-induced intracellular traps (PITs) that capture bacteria and lead to their clearance by efferocytosis. J Exp Med. 2016;213:2113–2128. doi: 10.1084/jem.20151613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rauch I, et al. NAIP-NLRC4 inflammasomes coordinate intestinal epithelial cell expulsion with eicosanoid and IL-18 release via activation of caspase-1 and -8. Immunity. 2017;46:649–659. doi: 10.1016/j.immuni.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sellin ME, et al. Epithelium-intrinsic NAIP/NLRC4 inflammasome drives infected enterocyte expulsion to restrict Salmonella replication in the intestinal mucosa. Cell Host Microbe. 2014;16:237–248. doi: 10.1016/j.chom.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 23.Lin KH, et al. Carboxyl-terminal fusion of E7 into flagellin shifts TLR5 activation to NLRC4/NAIP5 activation and induces TLR5-independent anti-tumor immunity. Sci Rep. 2016;6:24199. doi: 10.1038/srep24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garaude J, Kent A, van Rooijen N, Blander JM. Simultaneous targeting of toll- and nod-like receptors induces effective tumor-specific immune responses. Sci Transl Med. 2012;4:120ra16. doi: 10.1126/scitranslmed.3002868. [DOI] [PubMed] [Google Scholar]
- 25.Kitamura A, Sasaki Y, Abe T, Kano H, Yasutomo K. An inherited mutation in NLRC4 causes autoinflammation in human and mice. J Exp Med. 2014;211:2385–2396. doi: 10.1084/jem.20141091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romberg N, et al. Mutation of NLRC4 causes a syndrome of enterocolitis and autoinflammation. Nat Genet. 2014;46:1135–1139. doi: 10.1038/ng.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Canna SW, et al. Life-threatening NLRC4-associated hyperinflammation successfully treated with IL-18 inhibition. J Allergy Clin Immunol. 2017;139:1698–1701. doi: 10.1016/j.jaci.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Canna SW, et al. An activating NLRC4 inflammasome mutation causes autoinflammation with recurrent macrophage activation syndrome. Nat Genet. 2014;46:1140–1146. doi: 10.1038/ng.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ayres JS, Trinidad NJ, Vance RE. Lethal inflammasome activation by a multidrug-resistant pathobiont upon antibiotic disruption of the microbiota. Nat Med. 2012;18:799–806. doi: 10.1038/nm.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romanish MT, Lock WM, van de Lagemaat LN, Dunn CA, Mager DL. Repeated recruitment of LTR retrotransposons as promoters by the anti-apoptotic locus NAIP during mammalian evolution. PLoS Genet. 2007;3:e10. doi: 10.1371/journal.pgen.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romanish MT, Nakamura H, Lai CB, Wang Y, Mager DL. A novel protein isoform of the multicopy human NAIP gene derives from intragenic Alu SINE promoters. PLoS One. 2009;4:e5761. doi: 10.1371/journal.pone.0005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kortmann J, Brubaker SW, Monack DM. Cutting edge: Inflammasome activation in primary human macrophages is dependent on flagellin. J Immunol. 2015;195:815–819. doi: 10.4049/jimmunol.1403100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vinzing M, et al. NAIP and Ipaf control Legionella pneumophila replication in human cells. J Immunol. 2008;180:6808–6815. doi: 10.4049/jimmunol.180.10.6808. [DOI] [PubMed] [Google Scholar]
- 34.Sauer JD, et al. Listeria monocytogenes engineered to activate the Nlrc4 inflammasome are severely attenuated and are poor inducers of protective immunity. Proc Natl Acad Sci USA. 2011;108:12419–12424. doi: 10.1073/pnas.1019041108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaillard JL, Berche P, Mounier J, Richard S, Sansonetti P. In vitro model of penetration and intracellular growth of Listeria monocytogenes in the human enterocyte-like cell line Caco-2. Infect Immun. 1987;55:2822–2829. doi: 10.1128/iai.55.11.2822-2829.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tilney LG, Portnoy DA. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J Cell Biol. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Moltke J, et al. Rapid induction of inflammatory lipid mediators by the inflammasome in vivo. Nature. 2012;490:107–111. doi: 10.1038/nature11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lightfield KL, et al. Differential requirements for NAIP5 in activation of the NLRC4 inflammasome. Infect Immun. 2011;79:1606–1614. doi: 10.1128/IAI.01187-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lightfield KL, et al. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat Immunol. 2008;9:1171–1178. doi: 10.1038/ni.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mariathasan S, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 41.Miao EA, et al. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc Natl Acad Sci USA. 2010;107:3076–3080. doi: 10.1073/pnas.0913087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.LaRock DL, Chaudhary A, Miller SI. Salmonellae interactions with host processes. Nat Rev Microbiol. 2015;13:191–205. doi: 10.1038/nrmicro3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galán JE. Salmonella interactions with host cells: Type III secretion at work. Annu Rev Cell Dev Biol. 2001;17:53–86. doi: 10.1146/annurev.cellbio.17.1.53. [DOI] [PubMed] [Google Scholar]
- 44.Figueira R, Holden DW. Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiology. 2012;158:1147–1161. doi: 10.1099/mic.0.058115-0. [DOI] [PubMed] [Google Scholar]
- 45.Tenthorey JL, Kofoed EM, Daugherty MD, Malik HS, Vance RE. Molecular basis for specific recognition of bacterial ligands by NAIP/NLRC4 inflammasomes. Mol Cell. 2014;54:17–29. doi: 10.1016/j.molcel.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grandjean T, et al. The human NAIP-NLRC4-inflammasome senses the Pseudomonas aeruginosa T3SS inner-rod protein. Int Immunol. 2017;29:377–384. doi: 10.1093/intimm/dxx047. [DOI] [PubMed] [Google Scholar]
- 47.Saier MH., Jr Evolution of bacterial type III protein secretion systems. Trends Microbiol. 2004;12:113–115. doi: 10.1016/j.tim.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 48.Growney JD, Scharf JM, Kunkel LM, Dietrich WF. Evolutionary divergence of the mouse and human Lgn1/SMA repeat structures. Genomics. 2000;64:62–81. doi: 10.1006/geno.1999.6111. [DOI] [PubMed] [Google Scholar]
- 49.Endrizzi MG, Hadinoto V, Growney JD, Miller W, Dietrich WF. Genomic sequence analysis of the mouse Naip gene array. Genome Res. 2000;10:1095–1102. doi: 10.1101/gr.10.8.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eagle RA, Trowsdale J. Promiscuity and the single receptor: NKG2D. Nat Rev Immunol. 2007;7:737–744. doi: 10.1038/nri2144. [DOI] [PubMed] [Google Scholar]
- 51.Bonham KS, et al. A promiscuous lipid-binding protein diversifies the subcellular sites of toll-like receptor signal transduction. Cell. 2014;156:705–716. doi: 10.1016/j.cell.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boniotto M, et al. Population variation in NAIP functional copy number confers increased cell death upon Legionella pneumophila infection. Hum Immunol. 2012;73:196–200. doi: 10.1016/j.humimm.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 53.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]
- 54.Lawley TD, et al. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2006;2:e11. doi: 10.1371/journal.ppat.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wynosky-Dolfi MA, et al. Oxidative metabolism enables Salmonella evasion of the NLRP3 inflammasome. J Exp Med. 2014;211:653–668. doi: 10.1084/jem.20130627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee CA, Falkow S. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc Natl Acad Sci USA. 1990;87:4304–4308. doi: 10.1073/pnas.87.11.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lauer P, Chow MY, Loessner MJ, Portnoy DA, Calendar R. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J Bacteriol. 2002;184:4177–4186. doi: 10.1128/JB.184.15.4177-4186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.