Significance
Efforts to understand and treat fibrolamellar hepatocellular carcinoma (FL-HCC) have been confounded by a lack of models that accurately reflect the genetics and biology of the disease. Here we demonstrate that the Dnajb1–Prkaca gene fusion drives tumorigenesis in mice, and that fusion to DNAJB1 drives FL-HCC initiation more effectively than wild-type PRKACA overexpression. The requirement of the PRKACA kinase domain in tumor initiation establishes the potential utility of kinase inhibitors targeting the fusion. By identifying genetic and environmental factors that can enhance the consistency and aggressiveness of disease progression, we reveal biological characteristics of the disease and advance a robust platform for future preclinical studies.
Keywords: fibrolamellar hepatocellular carcinoma, CRISPR, mouse cancer models, protein kinase A, β-catenin
Abstract
A segmental deletion resulting in DNAJB1–PRKACA gene fusion is now recognized as the signature genetic event of fibrolamellar hepatocellular carcinoma (FL-HCC), a rare but lethal liver cancer that primarily affects adolescents and young adults. Here we implement CRISPR-Cas9 genome editing and transposon-mediated somatic gene transfer to demonstrate that expression of either the endogenous fusion protein or a chimeric cDNA leads to the formation of indolent liver tumors in mice that closely resemble human FL-HCC. Notably, overexpression of the wild-type PRKACA was unable to fully recapitulate the oncogenic activity of DNAJB1–PRKACA, implying that FL-HCC does not simply result from enhanced PRKACA expression. Tumorigenesis was significantly enhanced by genetic activation of β-catenin, an observation supported by evidence of recurrent Wnt pathway mutations in human FL-HCC, as well as treatment with the hepatotoxin 3,5-diethoxycarbonyl-1,4-dihydrocollidine, which causes tissue injury, inflammation, and fibrosis. Our study validates the DNAJB1–PRKACA fusion kinase as an oncogenic driver and candidate drug target for FL-HCC, and establishes a practical model for preclinical studies to identify strategies to treat this disease.
Fibrolamellar hepatocellular carcinoma (FL-HCC) ubiquitously harbors an ∼400-kb deletion on chromosome 19 that produces an in-frame fusion of DnaJ heat shock protein family member B1 (DNAJB1) and protein kinase cAMP-activated catalytic subunit alpha (PRKACA) (1, 2) (Fig. S1A). DNAJB1 encodes a subunit of the heat shock factor 40 (HSP40) complex, which activates the ATPase of HSP70 and serves as a molecular chaperone that can be induced by an array of environmental stresses (3). PRKACA encodes a catalytic subunit of protein kinase A (PKA), which resides in the cytoplasm in an inactive tetrameric complex with PKA C-β and two regulatory subunits of the PKA holoenzyme (4). Activation of G protein-coupled receptors leads to cAMP-dependent activation of the PKA catalytic subunits and subsequent phosphorylation of a panoply of cellular substrates (4). The crystal structure of the DNAJB1–PRKACA fusion protein shows that the catalytic site, regulatory subunit binding, and anchoring protein binding remain similar to those of the wild-type PRKACA (5).
Beyond the presence of DNAJB1–PRKACA fusions, FL-HCC tumorigenesis is poorly understood. Few, if any, other significantly recurrent mutated genes have been described (6, 7), and while broad copy-number alterations have been observed, they do not specifically implicate known oncogenes or tumor suppressors (7). Unlike liver cancer in older adults, FL-HCC is not associated with any known etiological risk factors such as alcoholism, chronic hepatitis infection, or liver flukes (8).
Currently, FL-HCC is diagnosed on the basis of histological features such as large cells with granular eosinophilic cytoplasm, vesiculated nuclei, and large nucleoli. Ultrastructural studies observe a hyperaccumulation of mitochondria and abundant endoplasmic reticulum (9). While early onset and lack of chronic liver disease are suggestive of FL-HCC, classic HCC can also occur in young patients and misdiagnosis is common (10). Given the specificity of DNAJB1–PRKACA fusion for FL-HCC, its detection will likely be decisive for correct diagnosis (2).
Surgical resection is currently the primary treatment for FL-HCC patients. Although often described as a relatively indolent disease, a high rate of recurrence represents a major clinical challenge (11), and the 5-y survival rate is 34% (12). There is no evidence of survival benefit from adjuvant chemotherapy or any systemic treatment applicable to classic HCC (13). While the unique demographics and genetics of FL-HCC suggest that these patients should be treated differently from those with HCC, there have been few clinical trials that have been tailored to this patient population (www.clinicaltrials.gov). The development of FL-HCC–specific therapies has been further hindered by the lack of genetically and biologically accurate model systems.
Mouse models have been a powerful tool for evaluating the oncogenic potential of candidate drivers and studying the biology of tumorigenesis, and as preclinical systems to test novel therapeutics (14). In this study, we employed hydrodynamic transfection combined with either CRISPR-Cas9–mediated editing of the endogenous deletion or transposon-mediated transgenesis of fusion cDNA and variants, allowing us to introduce genetic lesions in a subset of hepatocytes without the time and expense of producing germ-line genetic strains (15, 16). Using this approach, we demonstrate that the DNAJB1–PRKACA fusion is a bona fide oncogene and identify genetic and environmental factors that cooperate with the fusion event to drive aggressive disease. Our results further show that the PRKACA kinase domain is required for these effects, providing a rationale for targeting kinase activity pharmacologically. We anticipate that the models presented herein will serve as a powerful platform for future biological and preclinical studies.
Results
CRISPR-Mediated Deletion Results in the Fusion Oncogene and Drives Tumorigenesis in Vivo.
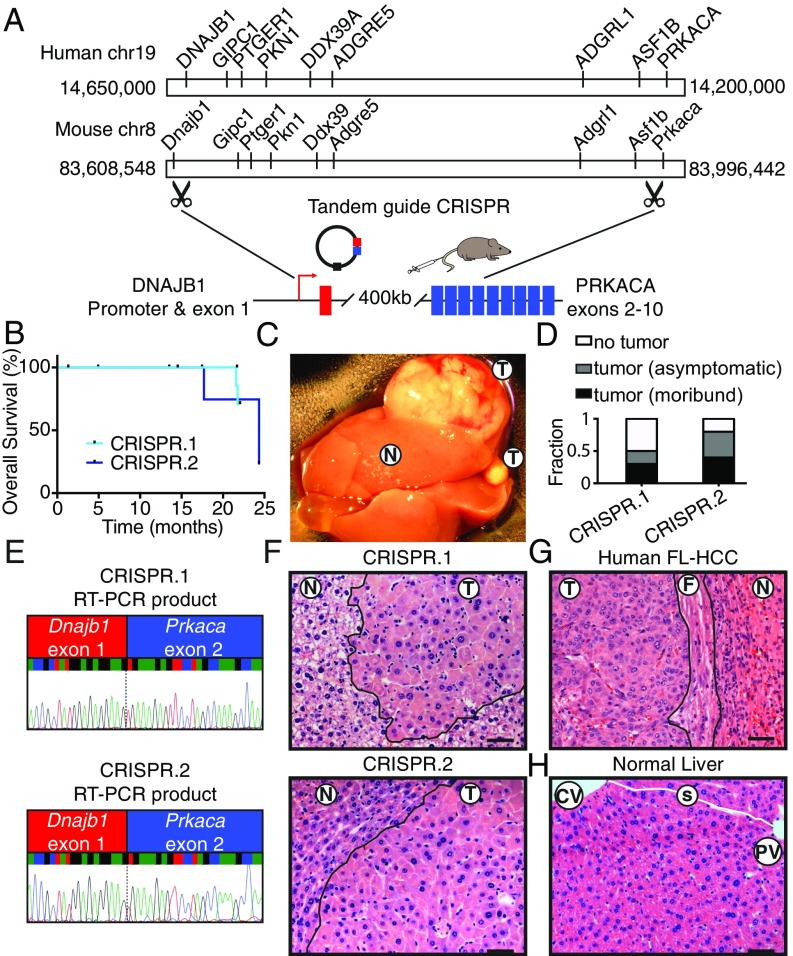
The oncogenic potential of the endogenous DNAJB1–PRKACA fusion in vivo was assessed. Coexpression of Cas9 with multiple single-guide RNAs (sgRNAs) can be used to model chromosome translocations, inversions, and deletions by generating DNA double-strand breaks at the breakpoints of chromosome rearrangements, which are subsequently joined by nonhomologous end joining (17–21). While such events are rare, an oncogenic rearrangement is expected to be positively selected in vivo. We determined whether the FL-HCC–associated rearrangement could be generated in hepatocytes of young adult mice via hydrodynamic tail-vein injection using tandem sgRNAs corresponding to the breakpoints of the disease-associated deletion in the first introns of Dnajb1 and Prkaca (Fig. 1A). Importantly, the deleted region on human chromosome 19 in FL-HCC is syntenic to a corresponding region on mouse chromosome 8. In fact, all protein-coding genes present in the human region have orthologs present in the mouse region, and are arranged in the same order (Fig. 1A).
Fig. 1.
CRISPR-mediated deletion results in a fusion oncogene and drives tumorigenesis in vivo. (A) Configuration of human chromosome 19, including DNAJB1 and PRKACA, configuration of mouse chromosome 8, including Dnajb1 and Prkaca, and schematic of an endogenous 400-kb deletion targeted by hydrodynamic injection of vector containing tandem sgRNAs targeting introns of Dnajb1 and Prkaca and Cas9. (B) Overall survival of mice injected with CRISPR.1 (n = 10) or CRISPR.2 (n = 9). (C) Macroscopic view of a tumor-bearing liver. N, adjacent normal; T, tumor. (D) Fraction of mice harvested with no detectable tumor, asymptomatic mice with histologically detectable disease (asymptomatic), or moribund mice with tumors (moribund) for each indicated genotype. (E) Sanger sequencing of chimeric transcript amplified from tumors generated by CRISPR.1 (Top) and CRISPR.2 (Bottom). (F) H&E staining of tumors generated by CRISPR.1 (Top) and CRISPR.2 (Bottom). (G) Human case of FL-HCC (F, fibrosis). (H) Normal mouse liver, where sinusoids trace from central veins to portal triads (CV, central vein; PV, portal vein) with intact sinusoids (white line; s). (Scale bars, 50 μm.)
To test the feasibility of this approach, different sgRNAs capable of targeting the first intron of Dnajb1 and Prkaca were coexpressed with Cas9 in NIH 3T3 cells or adult livers using lentiviral transduction or hydrodynamic injection, respectively (Fig. S1A), and confirmed to produce a fusion event using PCR (Fig. S1 B and C). Next, two sgRNA pairs, targeting different sequences within the same introns (herein CRISPR.1 and CRISPR.2), were introduced into the livers of adult mice and the animals were monitored over time. A subset of animals transduced with both sgRNA combinations became moribund with liver tumors 16 to 24 mo post injection (Fig. 1B). Tumor-bearing mice typically harbored disease involving multiple lobes, presumably from independent initiating events, and ranged from diffuse to macroscopically visible (Fig. 1C). In samples evaluated histologically, 2/9 mice injected with CRISPR.1, and 3/7 mice injected with CRISPR.2, died as a result of tumor burden. Additionally, nonmoribund animals that were killed harbored histological evidence of disease (annotated as “asymptomatic”) in 2/9 CRISPR.1 mice and 2/7 of CRISPR.2 mice (Fig. 1D). For both guide pairs, RT-PCR and Sanger sequencing with fusion-specific primers confirmed expression of the intended Dnajb1–Prkaca fusion oncogene in these lesions (Fig. 1E).
Histologically, the CRISPR-induced mouse tumor cells (Fig. 1F) were strikingly similar to human FL-HCC (Fig. 1G). Like human FL-HCC, the mouse liver tumors were composed of large, pleiomorphic polygonal cells with abundant eosinophilic cytoplasm, large vesicular nuclei, and prominent nucleoli. Furthermore, the lobular structure of the tumors was clearly disrupted (Fig. 1H) and distinctive cytoplasmic inclusions were observed (see below). However, unlike the human disease, the mouse tumors were not surrounded by detectable fibrosis. Supporting the robustness of these results, tumors with similar latency and histology were recapitulated with an independent Cas9-expressing vector (Fig. S1D). Thus, induction of an endogenous Dnajb1–Prkaca fusion through intrachromosomal deletion drives tumors with features of FL-HCC in mice. Independent work from others recently demonstrated an FL-HCC phenotype of lesions in mice using similar methods (22).
DNAJB1–PRKACA Fusion Drives Liver Tumorigenesis.
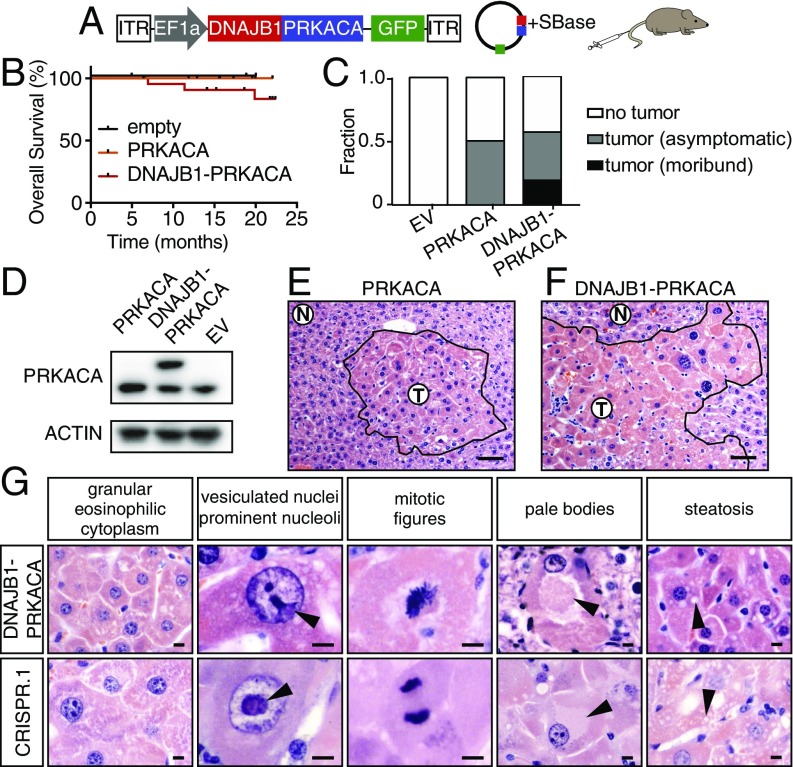
The segmental deletion that results in the DNAJB1–PRKACA gene fusion entails heterozygous loss of seven other coding genes, with unknown functional contribution. In other contexts, such deletions can contribute to tumorigenesis directly through attenuating the function of haploinsufficient tumor suppressors (23). To determine whether the DNAJB1–PRKACA fusion is sufficient to drive tumorigenesis (uncoupled from the typical genomic deletion), and whether simply the overexpression of the wild-type PRKACA gene could recapitulate this effect, we used hydrodynamic injection to deliver a transposon expressing the human DNAJB1–PRKACA fusion cDNA or a full-length wild-type PRKACA cDNA. Cotransfection of transiently expressed Sleeping Beauty transposase (SBase) with a transposon construct allows for stable integration and constitutive overexpression of the cDNA that mimics the high levels in human tumors (1) (Fig. 2A).
Fig. 2.
DNAJB1–PRKACA fusion drives liver tumorigenesis. (A) Schematic of the Sleeping Beauty transposon strategy to deliver DNAJB1–PRKACA fusion cDNA to young adult livers (ITR, inverted terminal repeat). (B) Overall survival of mice injected with cDNA encoding DNAJB1–PRKACA (n = 23), wild-type PRKACA (n = 12), or empty vector (n = 4). (C) Fraction of mice harvested with no detectable tumor, asymptomatic mice with histologically detectable disease, or moribund mice with tumors expressing empty vector (EV), wild-type PRKACA, or DNAJB1–PRKACA fusion. (D) Western blot of liver progenitor cells 4 d after viral transduction with the indicated cDNA in vitro. (E) Cluster of atypical hepatocytes in liver injected with wild-type pT3-PRKACA. (Scale bar, 50 μm.) (F) Tumor generated by pT3-DNAJB1–PRKACA. (Scale bar, 50 μm.) (G) Higher-magnification images highlighting common murine FL-HCC features: large granular, eosinophilic cytoplasm; vesiculated nuclei with prominent nucleoli (arrowheads); mitotic figures; pale bodies; and steatosis. (Scale bars, 10 μm.)
Expression and protein stability are similar between the DNAJB1–PRKACA fusion protein and full-length WT PRKACA, and overexpression of wild-type PRKACA produced some changes to hepatocyte histology but did not trigger the formation of lethal tumors (Fig. 2 B–E). However, expression of the DNAJB1–PRKACA cDNA produced tumors with similar kinetics, penetrance, and morphology as CRISPR-driven murine FL-HCC (Fig. 2 B, C, and F). Again, a spectrum of histological findings supported the similarity between these complementary methods and human FL-HCC (Figs. 1 and 2), including the presence of large tumor cells with granular, eosinophilic cytoplasm and prominent nucleoli (Fig. 2F). Mitotic figures, steatosis, and pale bodies (8) were also observed (Fig. 2G). These results imply that the DNAJB1 portion of the fusion protein contributes to disease beyond facilitating overexpression of PRKACA, and that the chromosome 19 deletion event is not required for oncogenesis.
Murine Tumors Display Structural and Molecular Features of Human FL-HCC.
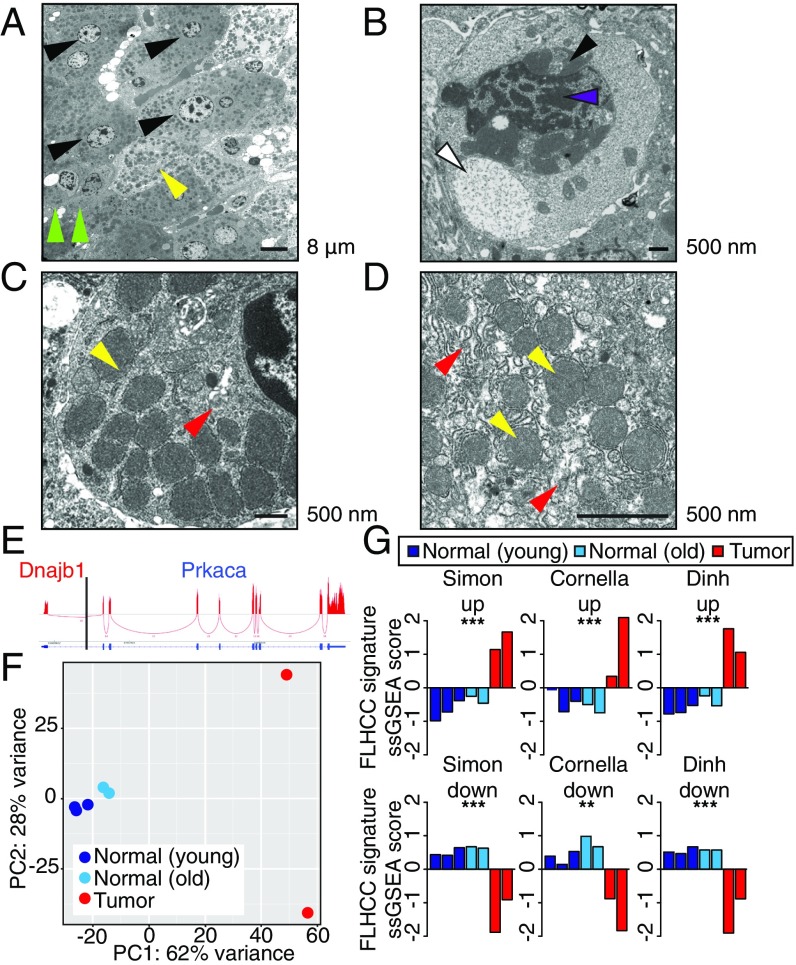
To validate that the murine model recapitulates other aspects of the human disease, we further characterized the phenotype of murine tumors. Human FL-HCC has consistently recognizable ultrastructural features that were also observed in murine FL-HCC tumors by electron microscopy (EM). Like human FL-HCC, murine tumor cells were typically larger than adjacent normal hepatocytes and contained clumped heterochromatin (Fig. 3 A and B and Fig. S2A), occasional pale bodies (Fig. 3B), and prominent nucleoli (Fig. S2 B and C). Most notably, tumor cells exhibited a marked increase in mitochondria with atypical appearance (8, 9, 24) (yellow arrowheads, Fig. 3 C and D and Fig. S2 E–H). Numerous megamitochondria were observed. The mitochondria were round to oval and homogeneous without obvious cristae and were surrounded by abundant rough endoplasmic reticulum (red arrowheads, Fig. 3 C and D and Fig. S2). Throughout the tumor, cells had moderate to severe perinuclear and cytoplasmic aggregates of lipofuscin pigment (Fig. 3B and Fig. S2I). This finding, along with the mitochondrial phenotype, could be consistent with a state of oxidative stress (25, 26).
Fig. 3.
Murine tumors display structural and molecular features of human FL-HCC. Ultrastructural analysis of a transposon-induced tumor by electron microscopy. (A) Tumor cells (black arrowheads) with numerous mitochondria (yellow) and larger cell size than compressed adjacent hepatocytes at tumor margin (green). (B) Pale body (white), perinuclear accumulation of lipofuscin (black), and heterochromatin (purple). (C) Tumor cell with abundant mitochondria (yellow) and rough endoplasmic reticulum (RER; red). (D) Abnormal mitochondria (yellow) with indistinct cristae surrounded by RER (red). (E) Sashimi plot indicating RNA-seq reads that cross the DNAJB1–PRKACA junction. (F) Principal component analysis depicting the first and second principal components (PC1 and PC2) of young control livers (dark blue), aged control livers (light blue), and tumors derived from CRISPR.1 (red). (G) Z-scored single sample gene set enrichment analysis for normal (blue) and tumor (red) samples. **P < 0.01, ***P < 0.001.
Human FL-HCC is known to often express markers of multiple lineages, including hepatic, biliary, and neuroendocrine (27, 28). The murine tumors were positive for hepatocyte markers HNF4A and HNF1A, with some cells showing reduced expression, consistent with reduced hepatocyte lineage commitment (Fig. S3 A and B). However, the murine tumors were negative for other proteins that are often expressed in FL-HCC, including biliary markers CK7 and CK19 as well as CD68 (Fig. S3 C–F), perhaps reflecting the fact that mature hepatocytes are targeted by the hydrodynamic transfection technique and thus are necessarily the cell of origin of the murine FL-HCC model, whereas the cell of origin in human FL-HCC is unknown.
To further validate the mouse model, gene expression analysis by RNA-seq (sequencing) was performed on murine tumors and control liver tissue and compared with human FL-HCC. Sequencing reads that cross the junction were observed, confirming expression of the fusion (Fig. 3G). Principal component analysis demonstrated that the vast majority of the variance between samples described the differences between tumor and normal samples (Fig. 3H). A focused analysis to investigate the similarity between mouse and human tumors was evaluated in two ways. First, single-sample gene set enrichment analysis (ssGSEA) (29), using FL-HCC expression signatures from three independent published studies (7, 10, 30), was used to confirm that differentially expressed genes in human tumors were, in aggregate, significantly enriched in our murine tumors (Fig. 3I). Second, a supervised analysis of curated functional gene sets previously reported as enriched in FL-HCC (30) was consistent with murine tumor expression data (Fig. S4A). These results provide a global analysis that classifies the murine model as FL-HCC. Overall, the murine tumors arising in the presence of the DNAJB1–PRKACA fusion show most, but not all, features of the human disease.
Molecular Profiling of Murine FL-HCC Reveals Processes Linked to Tumorigenesis.
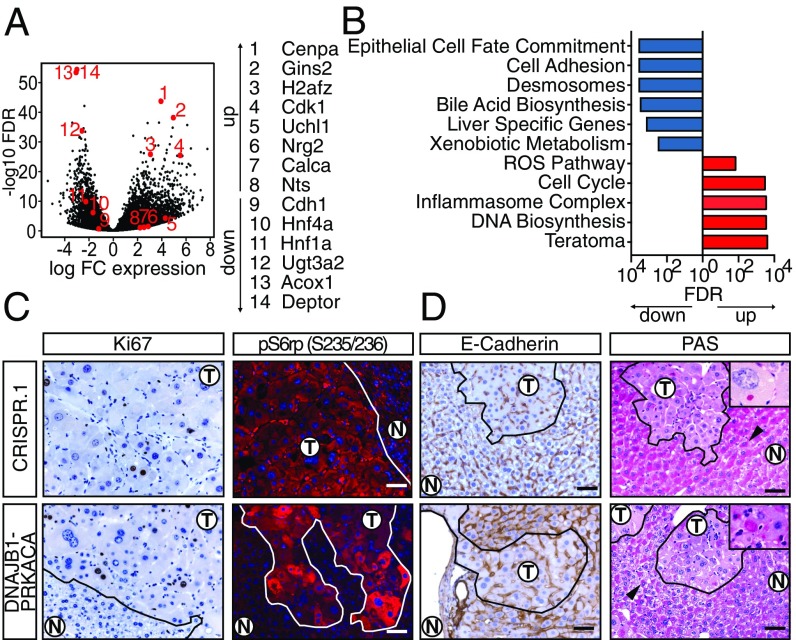
Unbiased analysis of expression data also suggested yet other biological processes that may be relevant to FL-HCC pathogenesis. A total of 5,710 genes were significantly differentially expressed between tumor and normal tissue (Fig. 4A and Dataset S1), and further global analysis by GSEA was performed (Fig. 4B and Dataset S2). The gene expression data, in agreement with human signatures, showed evidence of increased proliferation and mitogenic signaling (Fig. 4 A–C). For example, cell-cycle and DNA biosynthesis gene sets, and specifically Cdk1, Gins2, and Cenpa, were highly up-regulated in the experimental tumors (Fig. 4 D and E). Accordingly, tumors displayed an elevated Ki67 index (∼9%) compared with adjacent normal tissue (∼1%) (Fig. 4C). Activation of the PI3K pathway was indicated by down-regulation of Deptor (a negative regulator of mTORC1 also decreased in human tumors) and up-regulation of the receptor tyrosine kinase ligands Egf, Nrg2, and Ereg in both mouse and human tumors (30) (Fig. 4A and Fig. S4E). This observation was validated by immunofluorescence showing high levels of phospho-S6rp, a marker of mTOR activity that is highly expressed in most FL-HCCs but rarely in classic HCC (7, 31) (Fig. 4C).
Fig. 4.
Molecular profiling of murine FL-HCC reveals processes linked to tumorigenesis. (A) Volcano plot depicting differentially expressed genes in CRISPR-induced tumors with respect to normal liver. (B) ssGSEA analysis for select functionally annotated gene sets. (C) Ki67 IHC and p-S6rp S235/236 immunofluorescence of CRISPR-induced tumor (Top) or transposon-induced tumor (Bottom). N, normal. (D) E-cadherin (CDH1) staining and periodic acid Schiff staining. Note that adjacent normal hepatocytes are positive with asymmetrical subcellular distribution (black arrowheads). (D, Insets) PAS+ hyaline bodies. (Scale bars, 50 μm.)
Supporting previous reports that dedifferentiation is associated with DNAJB1–PRKACA–driven transformation (32), we observed down-regulation of hepatocyte lineage markers and up-regulation of some neuroendocrine markers (Fig. 4D and Fig. S4G), as has been observed in human FL-HCC (30). GSEA further showed down-regulation of epithelial cell fate commitment, bile acid biosynthesis, liver-specific genes, and xenobiotic metabolism (33), and up-regulation of a teratoma-associated gene set (Fig. 4E). Similarly, gene expression signatures defining specialized zones of hepatocytes (34) were universally lost, consistent with human data (30) (Fig. S4H).
Murine FL-HCC cells may also have defects in cell adhesion. Tumor cells displayed a decrease in Cdh1/E-cadherin expression, and cell adhesion- and desmosome-associated gene sets were down-regulated (Fig. 4 A and B). E-cadherin down-regulation was confirmed by immunohistochemistry (IHC), and EM revealed indistinct or simple tumor cell–cell junctions (Fig. S2 J and K). Loss of cell polarity was indicated by a loss of synchronized glycogen storage evident in neighboring hepatocytes, shown by largely negative periodic acid Schiff (PAS) staining (Fig. 4D). An exception to this pattern was the observation of PAS+ Mallory/hyaline bodies, which are commonly found in FL-HCC (8) and are reminiscent of a stress-induced phenotype (35) (Fig. 4D, Insets).
Murine and human tumors also showed evidence of a response to oxidative stress, as indicated by the up-regulation of enzymes involved in detoxifying reactive oxygen species (ROS) (e.g., Nqo1, Gpx3, Gpx4, and Acox1) (Fig. 4B and Fig. S4F). The accumulation of mitochondria can be driven by oxidative stress (36); up-regulation of mitochondrial-encoded transcripts (Fig. S4B) and an increase in the number of mitochondria observed by EM (Fig. 3 and Fig. S2 E–H) were also evident in both the mouse model and clinical samples. Whether or how each of these features contributes to the pathogenesis of FL-HCC remains to be determined.
WNT Pathway Cooperates with DNAJB1–PRKACA to Accelerate FL-HCC.
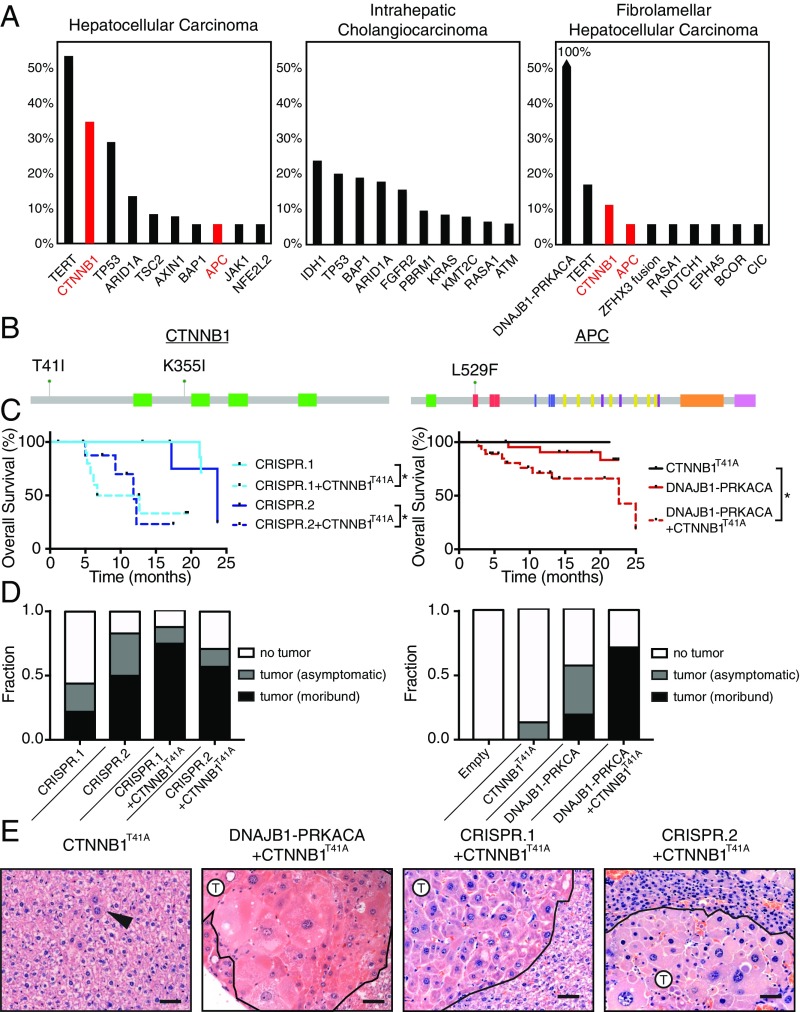
To further understand the genetic basis of FL-HCC and address the long latency of the single-hit models, we investigated additional factors that could accelerate disease. We queried the MSK-IMPACT collection of targeted sequencing data of over 18,000 cancer patients (37). Eighteen liver cancer patients (age 18 to 36) whose tumors harbored the DNAJB1–PRKACA fusion were identified (Fig. 5A). As expected, the fusion was not detected in any liver cancer patient over the age of 36 (0/414 patients) or in any nonliver cancer patient (0/18,367 patients), confirming the remarkable specificity of the DNAJB1–PRKACA fusion to liver oncogenesis (Fig. S5A). While HCC and intrahepatic cholangiocarcinoma (ICC) share several common mutations, none of these have been linked to FL-HCC. Surprisingly, we noted previously unreported recurrent mutations in the Wnt pathway in human FL-HCC (Fig. 5 A and B). The MSK-IMPACT cohort contains 3/18 (17%) samples of FL-HCC with CTNNB1 or APC mutation (age 18 to 21 y old). These cases each showed classic histological features of FL-HCC (Fig. S5C).
Fig. 5.
WNT pathway cooperates with DNAJB1–PRKACA to accelerate FL-HCC. (A) Frequency of top 10 most commonly mutated genes in hepatocellular carcinoma (n = 142), intrahepatic cholangiocarcinoma (n = 175), and fibrolamellar carcinoma (n = 18) samples from MSKCC IMPACT sequencing. (B) Domain schematic of loci of Wnt pathway mutations in FL-HCC. (C) Overall survival of mice injected with CTNNB1T41A and: CRISPR.1 (n = 10, log-rank P = 0.0042 ± CTNNB1T41A), CRISPR.2 (n = 8, P = 0.022 ± CTNNB1T41A), and DNAJB1–PRKACA (n = 30, P = 0.069 ± CTNNB1T41A, P = 0.045 ± DNAJB1–PRKACA). CRISPR.1, CRISPR.2, and DNAJB1–PRKACA alone are repeated from Figs. 1 and 2 for reference. *P < 0.05. (D) Fraction of mice harvested with no detectable tumor, asymptomatic mice with histologically detectable disease (asymptomatic), or moribund mice with tumors (moribund) for each indicated genotype. (E) H&E images depicting the histology of mice injected with CTNNB1T41A in combination with CRISPR.1, CRISPR.2, or DNAJB1–PRKACA. Arrowhead, atypical hepatocyte; T, tumor. (Scale bars, 50 μm.)
In parallel, candidate drivers of liver cancer were evaluated for their ability to synergize with DNAJB1–PRKACA to transform hepatocytes. Neither transposon-based delivery of MYC, AKTmyristoylated, NOTCHICD, YAPS127A, Fgf15, Il10, and Il18 nor CRISPR-mediated inactivation or knockout of p19ARF, Pten, Prkar1a, Rb1, Cdkn1b, and Tsc2 cooperated with DNAJB1–PRKACA (Fig. S6). However, an activated form of β-catenin uniquely cooperated with DNAJB1–PRKACA (using a transposon encoding CTNNB1T41A cDNA). Of note, CTNNB1T41A is the same allele that co-occurred with the DNAJB1–PRKACA fusion in a human primary FL-HCC and its corresponding brain metastasis (Fig. 5B and Fig. S5B). Both pairs of tandem guide CRISPRs, as well as transposon delivery of fusion cDNA, synergized with transposon delivery of stabilized β-catenin, increasing penetrance and reducing latency of the model (Fig. 5 C and D). In all cases, the histology of the resulting tumors matched the single-hit models, though some features (e.g., cell size) were more pronounced (Fig. 5E). The acceleration of the model by Wnt signaling was further validated by the combination of DNAJB1–PRKACA cDNA and disruption of Apc using CRISPR, which yielded tumors with a similar phenotype (Fig. S7). While expression of the DNAJB1–PRKACA fusion alone led to increased membranous β-catenin and phosphorylation of β-catenin at PKA phosphorylation site S675, expression of the canonical Wnt target AXIN2 was negative or weak in samples without genetic manipulation of the Wnt pathway (Fig. S7), which is corroborated by low expression of AXIN2 mRNA (Dataset S1) and the “HALLMARK_WNT_BETA_CATENIN_SIGNALING” gene set (Dataset S2) in samples driven by the fusion only. Of note, tumor cells arising in a mouse injected with the CRISPR.1 sgRNA pair and the CTNNB1T41A transposon formed tumors upon multiple rounds of s.c. transplantation into syngeneic recipients (Fig. S8). Hence, genetic lesions that activate Wnt signaling occur in the human disease and can cooperate with DNAJB1–PRKACA to accelerate FL-HCC.
Inflammatory and Fibrotic Agent 3,5-Diethoxycarbonyl-1,4-Dihydrocollidine Enhances FL-HCC Tumorigenesis.
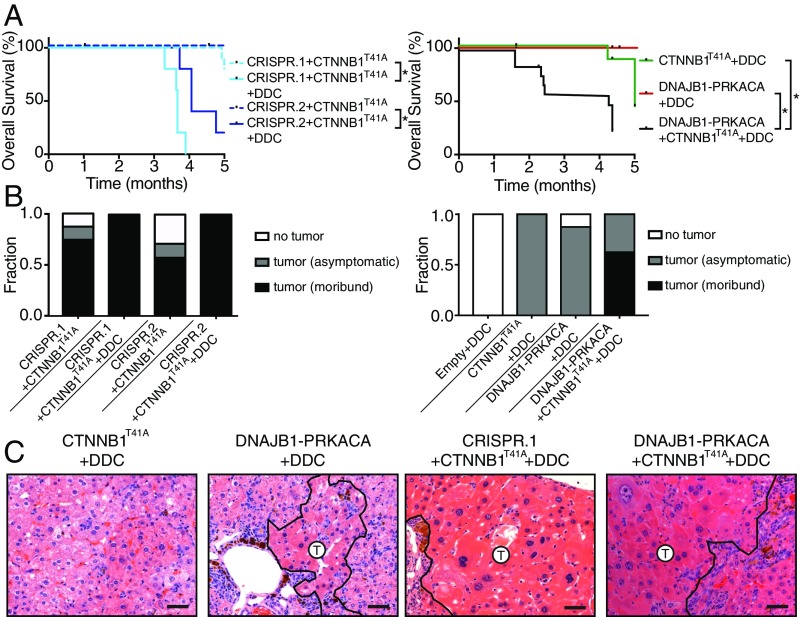
FL-HCC typically occurs in young patients who do not have the overt chronic liver diseases that often promote fibrosis and contribute to the emergence of classic HCC (8). Nevertheless, since the tumors arising in our mouse model lacked the eponymous fibrosis characteristic of human FL-HCC, we wondered whether experimental strategies to induce fibrosis might accelerate murine tumors. The hepatotoxin 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC) causes oxidative liver damage, cell death in periportal hepatocytes, atypical ductal expansion of progenitor cells, and, ultimately, fibrosis (38), and can accelerate HCC tumorigenesis by specific oncogenic events (39, 40).
Consistent with published results, mice treated with a 0.1% DDC-containing diet develop hepatomegaly, inflammation, and fibrosis with portal bridging by 8 wk of treatment but did not develop tumors (38, 41) (Fig. 6 and Fig. S9A). Although the DDC diet showed some increase in the onset of tumors following expression of mutant CTNNB1 alone, the effects on the combination of mutant CTNNB1 and DNAJB1–PRKACA were dramatic: In fact, 6-mo survival was decreased from 60 to 70% with CTNNB1T41A/DNAJB1–PRKACA to 0% observed with the same combination in DDC-treated mice (Fig. 6 A and B). The histology of the tumor cells themselves remained largely unchanged by DDC treatment but, as expected, the surrounding tissue acquired DDC-associated phenotypes associated with tissue regeneration following injury (Fig. 6C and Figs. S9B and S10). Surprisingly, the morbidity of the combination often preceded the establishment of significant fibrosis (Fig. 6C and Fig. S9). Therefore, these data suggest that one or more factors associated with the DDC-induced regenerative response can fuel murine FL-HCC. Furthermore, the combination of our non–germ-line genetic approaches and a DDC diet produces FL-HCC–like tumors at high penetrance and with a short latency.
Fig. 6.
Inflammatory and fibrotic agent DDC enhances FL-HCC tumorigenesis. (A) Overall survival of mice injected with the indicated genotypes ± DDC diet: CRISPR.1 + CTNNB1T41A (n = 5, log-rank P < 0.001 ± DDC), CRISPR.2 + CTNNB1T41A (n = 5, log-rank P < 0.001 ± DDC), and DNAJB1–PRKACA + CTNNB1T41A (n = 15, log-rank P < 0.001 ± DDC). Survival data of mice without DDC treatment repeated from Figs. 1, 2, and 5. *P < 0.05. (B) Fraction of mice harvested with no detectable tumor, asymptomatic mice with histologically detectable disease, or moribund mice with tumors for each indicated genotype with or without the DDC diet. (C) H&E images depicting the histology of mice of the indicated genotypes and fed the DDC diet. (Scale bars, 50 μm.)
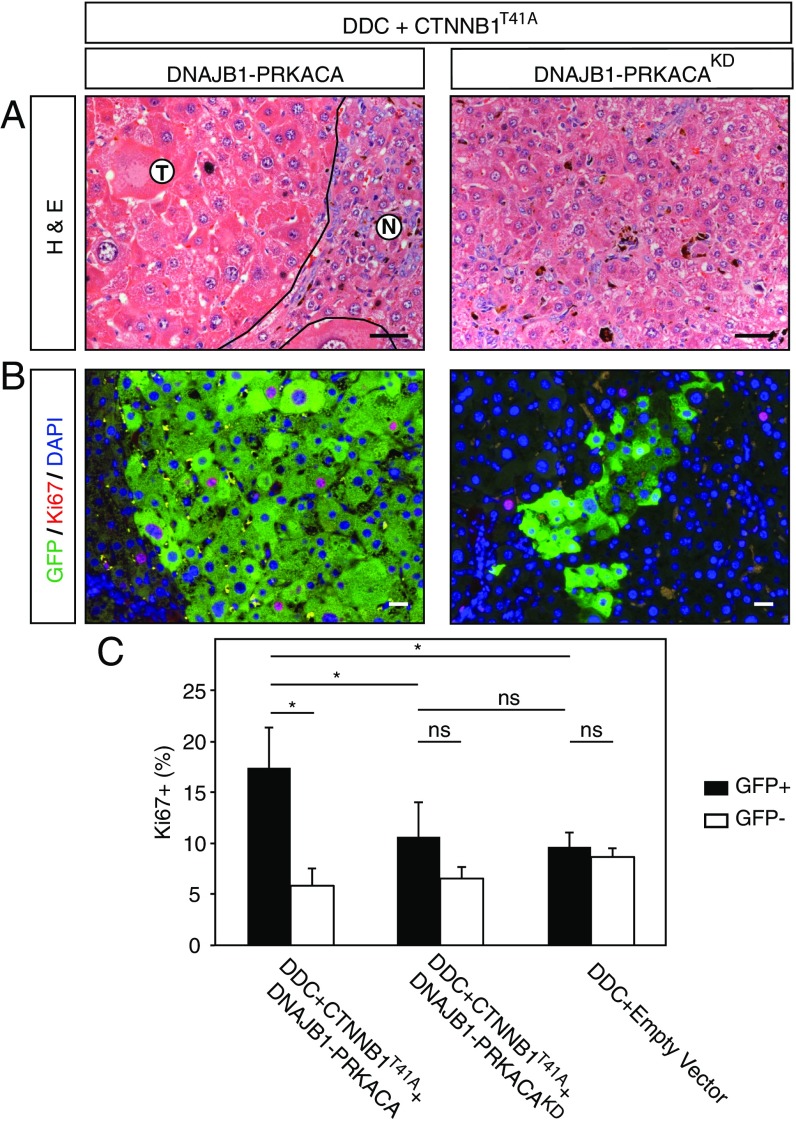
Tumorigenicity of DNAJB1–PRKACA Is Dependent on the Kinase Domain.
To illustrate the potential of a rapid and robust model of FL-HCC, we used the above methods to address whether the kinase activity of the DNAJB1–PRKACA fusion is essential for its ability to drive tumorigenesis—a prerequisite for rationalizing the use of small-molecule inhibitors targeting the PRKACA kinase for treatment of FL-HCC. To this end, we produced a kinase-dead version of the DNAJB1–PRKACA fusion harboring a mutation in the PRKACA component, equivalent to the previously described K72H mutation (42), and compared its oncogenic potential with the intact fusion cDNA when combined with DDC and CTNNB1T41A. A cohort of mice was produced and killed at 9 to 10 wk to examine the presence or absence of liver lesions. While clusters of neoplastic hepatocytes with classic FL-HCC morphology were observed in samples with an intact kinase domain, no such atypical hepatocytes were identified in samples expressing the kinase-dead fusion cDNA (Fig. 7A). Furthermore, expression of the kinase-intact fusion led to a significantly elevated Ki67-positive fraction of GFP+ cells, while the GFP+ cells in samples expressing the kinase-dead fusion cDNA did not show a significantly higher Ki76-positive fraction compared with adjacent normal liver or DDC-treated empty vector controls (Fig. 7C). Thus, the PRKACA kinase domain is required for FL-HCC tumor initiation.
Fig. 7.
Tumorigenicity of DNAJB1–PRKACA is dependent on the kinase domain. (A) H&E staining of DDC-treated mouse livers transfected with CTNNB1T41A combined with either DNAJB1–PRKACA or kinase-dead DNAJB1–PRKACA (KD; Right). (B) Immunofluorescence costaining for Ki67 (red) and GFP (green). Nuclei are counterstained with DAPI (blue). (C) Quantification of Ki67-positive cells. Error bars represent SD. Two-tailed t test, *P < 0.05. ns, not significant. (Scale bars, 50 μm.)
Discussion
Using a combination of hydrodynamic transfection, somatic genome editing, and transposon-mediated gene delivery, we demonstrate that the DNAJB1–PRKACA fusion is a bona fide oncogene that drives FL-HCC. Since tumors were produced by both the endogenous fusion and ectopic expression of the fusion cDNA, it appears that the loss of the intervening 400-kb deleted region, encompassing seven additional genes, is dispensable for tumorigenesis. The gene fusion appears to be functionally distinct from the mere overexpression of wild-type PRKACA, which is insufficient to drive tumor progression, at least when expressed in adult hepatocytes. On the other hand, the kinase domain of PRKACA is required for the FL-HCC phenotype. Importantly, this indicates that the potentially druggable enzymatic activity of the chimera is important for tumorigenesis.
Numerous features of the FL-HCC models described here mimic the human disease. Histologically, our methods generate tumors whose morphology is strikingly reminiscent of human FL-HCC. Dramatic accumulation of mitochondria appears to be a consistent consequence of DNAJB1–PRKACA activity, further linking our model to the human disease. Globally, we observe an expression signature significantly enriched with the genes differentially expressed in human FL-HCC. Nonetheless, DNAJB1–PRKACA expression in the murine hepatocytes does not appear to directly drive all molecular features of FL-HCC (fibrosis, expression of biliary markers, CD68). Additionally, while human FL-HCC often metastasizes, no metastases were observed in the mouse model. Perhaps other biological factors, or time, are needed for metastatic progression. These differences notwithstanding, the mouse model allows for insight into the phenotype of protein kinase A fusion activity in the liver in a controlled setting. We observe that induction of the fusion results in tumors with activated mTOR signaling (a druggable pathway) and detectable effects of oxidative stress (a potential vulnerability), validated by multiple previous reports describing human clinical samples (7, 30, 31, 33).
In human FL-HCC, we observed recurrent mutations that hyperactivate the Wnt pathway together with the DNAJB1–PRKACA fusion. Furthermore, genetic alteration of this pathway—but not several other oncogenes or tumor suppressors—synergized with DNAJB1–PRKACA–driven tumorigenesis in the mouse. While the basis for this genetic interaction remains to be determined, it is possible that modification of β-catenin may be one downstream output of DNAJB1–PRKACA and that stabilization of β-catenin may amplify its functional consequences. PKA has been described to regulate β-catenin through a variety of mechanisms, including direct modification via C-terminal phosphorylation (43–45); accordingly, phospho-β-catenin is elevated in human FL-HCC (46, 47). Alternatively, β-catenin may protect cells from oxidative stress imposed by DNAJB1–PRKACA (48).
DDC dramatically shortened tumor latency, unexpectedly preceding extensive fibrosis. By causing oxidative stress-induced cell death in a subset of periportal hepatocytes, DDC sets in motion a liver regenerative response that is associated with compensatory expansion of liver progenitor cells, activation of myofibroblasts, fibrosis, and massive immune cell infiltration (41) in a process supported by β-catenin expression in hepatocytes (48). Any or all of the activated and recruited cell types could contribute paracrine signals that protect and stimulate growth in the spatially separated (largely pericentral) population of hepatocytes that are transfected by hydrodynamic injection (49). While chronic liver damage is not considered to be necessary for human FL-HCC, our data raise the possibility that some environmental factor, perhaps in a susceptible population, may be relevant in the etiology of the disease. Alternatively, it is possible that β-catenin and/or DDC facilitated a change in cell state that supports or expands a susceptible progenitor cell population more prevalent in adolescents.
Animal models set a gold standard for assessing the oncogenic potential of aberrations observed in human cancer and provide experimental systems to study disease mechanisms or test novel therapeutic strategies (14). While one patient-derived xenograft of FL-HCC has been described (32), our models introduce the ability to examine the entire process of tumor initiation in immune-competent organisms. The somatic engineering methods described here involve only delivery of plasmid DNA to hepatocytes and do not require expensive and time-consuming generation or breeding of germ-line mouse strains, cell transplantation, or stable expression of Cas9. The model is easily implemented, reproducible, and genetically defined. As such, these systems are a powerful platform for further understanding the biology of FL-HCC and facilitating drug discovery for a disease that disproportionately affects young patients and has limited treatment options.
Materials and Methods
Animals and Treatments.
Female, 6- to 10-wk-old C57BL6/N mice were purchased from Envigo. All animal experiments were approved by the Memorial Sloan Kettering Cancer Center (MSKCC) Institutional Animal Care and Use Committee (protocol 11-06-011). For hydrodynamic tail-vein injection, a sterile 0.9% NaCl solution was prepared containing plasmid DNA of either 40 μg CRISPR vector or 20 μg transposon vector together with CMV-SB13 transposase (1:5 molar ratio). Mice were injected into the lateral tail vein with a total volume corresponding to 10% of body weight (typically 2 mL for a 20-g mouse) in 5 to 7 s (15, 50). DDC treatment was administered through a diet containing 0.1% DDC (Sigma-Aldrich; Envigo) until sacrifice (40). Transplants were performed by finely mincing freshly isolated tumors, suspending in 1:1 PBS:Matrigel, and injecting s.c. in a 100-μL volume.
Electron Microscopy.
Tissue was fixed in 4% glutaraldehyde and transferred to cold PBS until further processed. The tissues were post fixed in 1% osmium tetroxide in PBS. After washing in water, the tissue was stained with 2% aqueous uranyl acetate for ∼2 h at 4 °C. Tissues were dehydrated through a dilution series of acetone followed by propylene oxide and embedded in Epon. Ultrathin sections were deposited on grids and stained with uranyl acetate for 15 min and lead citrate for 5 min.
Immunohistochemistry and Immunofluorescence.
Tissue was prepared for histology by fixing in 10% buffered formalin overnight and then transferred to 70% ethanol until paraffin embedding and sectioning (IDEXX RADIL). Antigen retrieval was performed in a pressure cooker with sodium citrate buffer. The following primary antibodies were used: Ki67 (Abcam; ab16667; 1:200), p-S6rp (Cell Signaling; 2211; 1:200), E-cadherin (BD; 610181; 1:500), HNF1a (Santa Cruz; sc-6547; 1:100), HNF4a (Abcam; ab41898; 1:200), CK7 (Abcam; ab181598; 1:500), CK19 (Abcam; ab133496; 1:1,000), CD68 (MSKCC Pathology Core Facility; 5 μg/mL), IBA1 (Wako; 1:500), GFP (Abcam; ab13970; 1:200), β-catenin (BD; 610154; 1:500), p-β-catenin (Cell Signaling; 9567; 1:100), and AXIN2 (Abcam; ab32197; 1:800). Primary antibodies were incubated at 4 °C overnight in blocking buffer. Sections were incubated with anti-rabbit ImmPRESS HRP-conjugated secondary antibodies (MP7401; Vector Laboratories), and chromagen development was performed using ImmPACT DAB (SK4105; Vector Laboratories). Stained slides were counterstained with Harris hematoxylin. Images of stained sections were acquired on a Zeiss Axio Scope Imager Z.1. Raw tif files were processed using Photoshop CS5 software (Adobe Systems) to adjust white balance.
RNA-Seq.
Total RNA was isolated from frozen tissue using the RNeasy Mini Kit (Qiagen), quality control was performed on an Agilent Bioanalyzer, and 500 ng of total RNA (RNA integrity number >8) underwent polyA selection and TruSeq library preparation according to instructions provided by Illumina (TruSeq RNA Sample Prep Kit version 2) with six cycles of PCR. Single-end, 75-bp sequencing was performed at the Cold Spring Harbor Laboratory core facility. Approximately 8 million reads were acquired per sample. Resulting RNA-seq data were analyzed as described previously (23). Adaptor sequences were removed using Trimmomatic (51). RNA-seq reads were then aligned to the mouse genome (mm10) using STAR (52) with default parameters, and genome-wide transcript counting was performed using Subread to generate a count matrix (53, 54). Differential expression analysis was performed by DESeq2 (55). Genes were considered to be significantly differentially expressed if tumor/normal comparison was greater than twofold and false discovery rate (FDR)-adjusted P value was less than 0.05.
Human–murine mapping of orthologs was performed based on the Ensembl database accessed through the Biomart R/Bioconductor package (56). Human fibrolamellar HCC signatures were defined as genes with at least twofold and significant expression changes in fibrolamellar tumors with respect to normal. Human transcriptional profiling data were obtained from published studies (7, 30) and The Cancer Genome Atlas/Broad GDAC firehose using annotation from Dinh et al. (10). For comparison with human datasets and for gene set enrichment analysis, the ssGSEA method was implemented using the GSVA package within R (29). The GSVA outputs were subsequently compared across groups using the limma package (57). The C2, C3, C5, and Hallmark collections of gene sets from MSigDB version 6.0 were queried (58).
Human Tumor Sequencing Data.
The MSK-IMPACT sequencing data (37) were obtained from the MSKCC cBioPortal (59) (www.cbioportal.org). Of the 18 DNAJB1–PRKACA fusion cases, 14 were annotated as fibrolamellar HCC and 4 were annotated as HCC. We considered all DNAJB1–PRKACA–positive liver cancers as FL-HCC, given the common misdiagnosis of this rare cancer type (10), for which the presence of the DNAJB1–PRKACA fusion should be considered diagnostic (2).
Supplementary Material
Acknowledgments
We thank Francisco Sanchez-Rivera (MSKCC), Yadira Soto-Feliciano (Rockefeller University), and John P. Morris IV (MSKCC) for critical reading of the manuscript, and all members of the S.W.L. laboratory for advice and discussions. We thank Wei Luan and Leah Zamachek for technical assistance, and David Klimstra, Paul Romesser, Robert Bowman, Joana DeCampos Vidigal, and Andrea Ventura for critical insights and materials. We gratefully acknowledge the members of the Molecular Diagnostics Service in the Department of Pathology (MSKCC). This work was supported primarily by a project grant from the Starr Cancer Consortium (I10-0098) (to S.W.L. and S.M.S.). This work was also funded in part by the Marie-Josée and Henry R. Kravis Center for Molecular Oncology, National Cancer Institute Cancer Center Core Grants P30-CA008748 and NIH P01 CA013106, the Andrew McDonough B+ Foundation, and Grant UL1 TR001866 from the National Center for Advancing Translational Sciences (NCATS) National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program. E.R.K. is supported by an F31 NRSA predoctoral fellowship from the NCI/NIH under Award F31CA192835. G.L., D.R., and S.M.S. were supported by a grant from the NIH (1R56CA207929). T.B. is the William C. and Joyce O’Neil Charitable Trust Fellow and is supported by the MSKCC Single Cell Sequencing Initiative. S.W.L. is an investigator of the Howard Hughes Medical Institute and the Geoffrey Beene Chair for Cancer Biology.
Footnotes
The authors declare no conflict of interest.
Data deposition: The RNA-sequence reported in this paper has been deposited in the Sequence Read Archive (SRA), https://www.ncbi.nlm.nih.gov/bioproject (accession no. PRJNA415225).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716483114/-/DCSupplemental.
References
- 1.Honeyman JN, et al. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science. 2014;343:1010–1014. doi: 10.1126/science.1249484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham RP, et al. DNAJB1-PRKACA is specific for fibrolamellar carcinoma. Mod Pathol. 2015;28:822–829. doi: 10.1038/modpathol.2015.4. [DOI] [PubMed] [Google Scholar]
- 3.Priya S, Sharma SK, Goloubinoff P. Molecular chaperones as enzymes that catalytically unfold misfolded polypeptides. FEBS Lett. 2013;587:1981–1987. doi: 10.1016/j.febslet.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 4.Turnham RE, Scott JD. Protein kinase A catalytic subunit isoform PRKACA; history, function and physiology. Gene. 2016;577:101–108. doi: 10.1016/j.gene.2015.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung J, et al. Structural insights into mis-regulation of protein kinase A in human tumors. Proc Natl Acad Sci USA. 2015;112:1374–1379. doi: 10.1073/pnas.1424206112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darcy DG, et al. The genomic landscape of fibrolamellar hepatocellular carcinoma: Whole genome sequencing of ten patients. Oncotarget. 2015;6:755–770. doi: 10.18632/oncotarget.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornella H, et al. Unique genomic profile of fibrolamellar hepatocellular carcinoma. Gastroenterology. 2015;148:806–818.e10. doi: 10.1053/j.gastro.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torbenson M. Fibrolamellar carcinoma: 2012 update. Scientifica (Cairo) 2012;2012:743790. doi: 10.6064/2012/743790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham RP, et al. Environmental exposures as a risk factor for fibrolamellar carcinoma. Mod Pathol. 2017;30:892–896. doi: 10.1038/modpathol.2017.7. [DOI] [PubMed] [Google Scholar]
- 10.Dinh TA, et al. Comprehensive analysis of The Cancer Genome Atlas reveals a unique gene and non-coding RNA signature of fibrolamellar carcinoma. Sci Rep. 2017;7:44653. doi: 10.1038/srep44653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaseb AO, et al. Prognostic indicators and treatment outcome in 94 cases of fibrolamellar hepatocellular carcinoma. Oncology. 2013;85:197–203. doi: 10.1159/000354698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eggert T, et al. Fibrolamellar hepatocellular carcinoma in the USA, 2000–2010: A detailed report on frequency, treatment and outcome based on the Surveillance, Epidemiology, and End Results database. United European Gastroenterol J. 2013;1:351–357. doi: 10.1177/2050640613501507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riggle KM, Turnham R, Scott JD, Yeung RS, Riehle KJ. Fibrolamellar hepatocellular carcinoma: Mechanistic distinction from adult hepatocellular carcinoma. Pediatr Blood Cancer. 2016;63:1163–1167. doi: 10.1002/pbc.25970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kersten K, de Visser KE, van Miltenburg MH, Jonkers J. Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol Med. 2017;9:137–153. doi: 10.15252/emmm.201606857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tschaharganeh DF, et al. p53-dependent Nestin regulation links tumor suppression to cellular plasticity in liver cancer. Cell. 2014;158:579–592. doi: 10.1016/j.cell.2014.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawakami K, Largaespada DA, Ivics Z. Transposons as tools for functional genomics in vertebrate models. Trends Genet. 2017;33:784–801. doi: 10.1016/j.tig.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maddalo D, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cook PJ, et al. Somatic chromosomal engineering identifies BCAN-NTRK1 as a potent glioma driver and therapeutic target. Nat Commun. 2017;8:15987. doi: 10.1038/ncomms15987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han T, et al. R-Spondin chromosome rearrangements drive Wnt-dependent tumour initiation and maintenance in the intestine. Nat Commun. 2017;8:15945. doi: 10.1038/ncomms15945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun. 2014;5:3728. doi: 10.1038/ncomms4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blasco RB, et al. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep. 2014;9:1219–1227. doi: 10.1016/j.celrep.2014.10.051. [DOI] [PubMed] [Google Scholar]
- 22.Engelholm LH, et al. CRISPR/Cas9 engineering of adult mouse liver demonstrates that the Dnajb1-Prkaca gene fusion is sufficient to induce tumors resembling fibrolamellar hepatocellular carcinoma. Gastroenterology. September 15, 2017 doi: 10.1053/j.gastro.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, et al. Deletions linked to TP53 loss drive cancer through p53-independent mechanisms. Nature. 2016;531:471–475. doi: 10.1038/nature17157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Payne CM, Nagle RB, Paplanus SH, Graham AR. Fibrolamellar carcinoma of liver: A primary malignant oncocytic carcinoid? Ultrastruct Pathol. 1986;10:539–552. doi: 10.3109/01913128609007211. [DOI] [PubMed] [Google Scholar]
- 25.Sohal RS, Brunk UT. Lipofuscin as an indicator of oxidative stress and aging. Adv Exp Med Biol. 1989;266:17–26; discussion 27–29. doi: 10.1007/978-1-4899-5339-1_2. [DOI] [PubMed] [Google Scholar]
- 26.Höhn A, Sittig A, Jung T, Grimm S, Grune T. Lipofuscin is formed independently of macroautophagy and lysosomal activity in stress-induced prematurely senescent human fibroblasts. Free Radic Biol Med. 2012;53:1760–1769. doi: 10.1016/j.freeradbiomed.2012.08.591. [DOI] [PubMed] [Google Scholar]
- 27.Ward SC, et al. Fibrolamellar carcinoma of the liver exhibits immunohistochemical evidence of both hepatocyte and bile duct differentiation. Mod Pathol. 2010;23:1180–1190. doi: 10.1038/modpathol.2010.105. [DOI] [PubMed] [Google Scholar]
- 28.Ross HM, et al. Fibrolamellar carcinomas are positive for CD68. Mod Pathol. 2011;24:390–395. doi: 10.1038/modpathol.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hänzelmann S, Castelo R, Guinney J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon EP, et al. Transcriptomic characterization of fibrolamellar hepatocellular carcinoma. Proc Natl Acad Sci USA. 2015;112:E5916–E5925. doi: 10.1073/pnas.1424894112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riehle KJ, et al. mTORC1 and FGFR1 signaling in fibrolamellar hepatocellular carcinoma. Mod Pathol. 2015;28:103–110. doi: 10.1038/modpathol.2014.78. [DOI] [PubMed] [Google Scholar]
- 32.Oikawa T, et al. Model of fibrolamellar hepatocellular carcinomas reveals striking enrichment in cancer stem cells. Nat Commun. 2015;6:8070. doi: 10.1038/ncomms9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kannangai R, Vivekanandan P, Martinez-Murillo F, Choti M, Torbenson M. Fibrolamellar carcinomas show overexpression of genes in the RAS, MAPK, PIK3, and xenobiotic degradation pathways. Hum Pathol. 2007;38:639–644. doi: 10.1016/j.humpath.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 34.Halpern KB, et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature. 2017;542:352–356. doi: 10.1038/nature21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Celli R, Zhang X. Pathology of alcoholic liver disease. J Clin Transl Hepatol. 2014;2:103–109. doi: 10.14218/JCTH.2014.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo C, et al. Mitochondrial accumulation under oxidative stress is due to defects in autophagy. J Cell Biochem. 2013;114:212–219. doi: 10.1002/jcb.24356. [DOI] [PubMed] [Google Scholar]
- 37.Zehir A, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Preisegger KH, et al. Atypical ductular proliferation and its inhibition by transforming growth factor beta1 in the 3,5-diethoxycarbonyl-1,4-dihydrocollidine mouse model for chronic alcoholic liver disease. Lab Invest. 1999;79:103–109. [PubMed] [Google Scholar]
- 39.Matter MS, et al. Oncogenic driver genes and the inflammatory microenvironment dictate liver tumor phenotype. Hepatology. 2016;63:1888–1899. doi: 10.1002/hep.28487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beer S, et al. Hepatotoxin-induced changes in the adult murine liver promote MYC-induced tumorigenesis. PLoS One. 2008;3:e2493. doi: 10.1371/journal.pone.0002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fickert P, et al. A new xenobiotic-induced mouse model of sclerosing cholangitis and biliary fibrosis. Am J Pathol. 2007;171:525–536. doi: 10.2353/ajpath.2007.061133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iyer GH, Moore MJ, Taylor SS. Consequences of lysine 72 mutation on the phosphorylation and activation state of cAMP-dependent kinase. J Biol Chem. 2005;280:8800–8807. doi: 10.1074/jbc.M407586200. [DOI] [PubMed] [Google Scholar]
- 43.Kang DE, et al. Presenilin couples the paired phosphorylation of beta-catenin independent of axin: Implications for beta-catenin activation in tumorigenesis. Cell. 2002;110:751–762. doi: 10.1016/s0092-8674(02)00970-4. [DOI] [PubMed] [Google Scholar]
- 44.Hino S, Tanji C, Nakayama KI, Kikuchi A. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase stabilizes beta-catenin through inhibition of its ubiquitination. Mol Cell Biol. 2005;25:9063–9072. doi: 10.1128/MCB.25.20.9063-9072.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang X, et al. Phosphorylation and inactivation of glycogen synthase kinase 3 by protein kinase A. Proc Natl Acad Sci USA. 2000;97:11960–11965. doi: 10.1073/pnas.220413597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cieply B, Zeng G, Proverbs-Singh T, Geller DA, Monga SP. Unique phenotype of hepatocellular cancers with exon-3 mutations in beta-catenin gene. Hepatology. 2009;49:821–831. doi: 10.1002/hep.22695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Veelen W, et al. β-Catenin tyrosine 654 phosphorylation increases Wnt signalling and intestinal tumorigenesis. Gut. 2011;60:1204–1212. doi: 10.1136/gut.2010.233460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tao GZ, et al. Wnt/β-catenin signaling protects mouse liver against oxidative stress-induced apoptosis through the inhibition of forkhead transcription factor FoxO3. J Biol Chem. 2013;288:17214–17224. doi: 10.1074/jbc.M112.445965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen X, Calvisi DF. Hydrodynamic transfection for generation of novel mouse models for liver cancer research. Am J Pathol. 2014;184:912–923. doi: 10.1016/j.ajpath.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bell JB, et al. Preferential delivery of the Sleeping Beauty transposon system to livers of mice by hydrodynamic injection. Nat Protoc. 2007;2:3153–3165. doi: 10.1038/nprot.2007.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liao Y, Smyth GK, Shi W. The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao Y, Smyth GK, Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 55.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liberzon A, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cerami E, et al. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.