Abstract
Background and objectives
Genome-wide association studies identified single-nucleotide polymorphisms (SNPs) at the 17q21 locus conferring increased risk for childhood-onset asthma. Little is known about how these SNPs impact adult asthma patients. We sought to examine an adult population for associations between rs7216389 (17q21-associated SNP) and features of asthma including fractional exhaled nitric oxide (FeNO), eosinophil counts, and age of asthma onset.
Methods
Subjects were genotyped at SNP rs7216389. The geometric mean of FeNO measurements and peripheral blood eosinophil counts from 2008 to 2015 were collected. Demographics and medical history were collected including self-reported allergy diagnoses and age of asthma onset. Eosinophils, monocytes, and peripheral blood mononuclear cells (PBMCs) were isolated for the examination of ORMDL3 expression.
Results
FeNO levels from 157 genotyped subjects (31CC, 72CT, and 54TT) and peripheral eosinophil counts from 252 genotyped subjects (46CC, 122CT, and 84TT) were analyzed. In a sub-group analysis of asthma subjects, the number of attributable T alleles was associated with significantly lower age of asthma onset (P=0.03) and greater FeNO levels (geometric mean 30.0 ppb TT, 20.0 ppb CT, 20.0 ppb CC, P=0.02). In the total cohort of subjects, the T allele was associated with a higher percentage of individual eosinophil counts >200/mm3 (45% TT, 26% CT, 24% CC, P=0.005). Eosinophils expressed ORMDL3 mRNA and protein.
Conclusion
In adult subjects, the number of T alleles at SNP rs7216389 corresponds to significantly greater FeNO levels and peripheral eosinophil counts. The expression of ORMDL3 in eosinophils suggests that they may participate in mediating the asthma risk associated with the 17q21 locus.
Keywords: asthma, eosinophils, fractional exhaled nitric oxide, 17q21
Introduction
Asthma is a chronic inflammatory disease of the airways that affects 7.8% of the US population.1 A family history of allergic disease is a significant risk factor for the diagnosis of asthma. Thus, there is a great interest in identifying the genetic determinants of asthma, which in turn would lead to targeted therapies. Among these efforts to identify asthma-related genes, there have been several genome-wide association studies.2–6 One locus identified by genome-wide association studies, replicated in several different cohorts, is 17q21.6–10 Additionally, multiple single-nucleotide polymorphisms (SNPs) at the 17q21 locus are expression quantitative trait loci (eQTLs) for genes in this region, 11,12 and these SNPs are specifically associated with childhood-onset asthma.2
At one of the SNPs at the 17q21 locus, rs7216389, the TT genotype carries a 1.5 times increased risk of developing asthma.13 Other studies have shown that a combination of the risk genotype and either smoke exposure or respiratory infections in childhood further amplifies asthma development risk.8,14–17 In addition, the risk genotype at SNP rs7216389 is associated with disease severity in childhood-onset asthma.7
Long-term implications of the 17q21 genotype as children with asthma progress into adulthood are unknown. One study of adult asthmatics demonstrated decreased forced expiratory volume in 1 second (FEV1) levels with the risk allele.18 However, little else is known about the relevance of this locus in adult asthmatics. We sought to determine whether the genotype at rs7216389 is associated with differences in FeNO and peripheral blood eosinophil counts, which are two well-characterized biomarkers that are often used to distinguish asthma phenotypes and to identify responders to inhaled corticosteroid therapy.
Methods
A cohort of 255 subjects provided written informed consent to participate in the University of Wisconsin–Madison’s institutional review board-approved blood donation protocol. Individuals had physician-diagnosed asthma and/or allergic rhinitis. Individuals were between the ages of 18 and 55 years with no history of tobacco use within the past year. From 2008 to 2015, participants underwent single or repeated blood draws and fractional exhaled nitric oxide (FeNO) measurements and provided a medical history, which included self-reported ethnicity and age of asthma onset.
Genotyping
Whole blood was processed for DNA using the Gentra Puregene Blood Kit (Qiagen NV, Venlo, the Netherlands) according to the manufacturer’s protocols. SNP rs7216389 genotypes were assessed using the TaqMan SNP genotyping assay (Thermo Fisher Scientific, Waltham, MA, USA).
FeNO measurements
The NIOX MINO (Aerocrine, Stockholm, Sweden) was used for FeNO measurements which were performed according to the American Thoracic Society guidelines.19 For subjects with multiple measurements, the geometric mean was used for analysis.
Eosinophil counts
Whole blood was stained with eosin dye, and eosinophils were counted via hemacytometer. For subjects with multiple measurements, the geometric mean was used for analysis.
Cell isolation
Whole blood was centrifuged over Percoll (1.090 g/mL) to obtain the peripheral blood mononuclear cell (PBMC) fraction and the granulocyte fraction. Eosinophils were purified from the granulocyte fraction by negative selection using anti-CD16 immunomagnetic beads (MACS System; Miltenyi Biotec, San Jose, CA, USA). Blood eosinophils were >98% viable and pure. PBMCs were washed, and half the cells were collected as PBMCs and the remaining cells were used for monocyte isolation. Monocytes were isolated by negative selection using the Monocyte Isolation Kit II (Miltenyi Biotec), as per the manufacturer’s instructions.
RNA isolation
Total RNA was isolated using an RNeasy Mini Kit (Qiagen NV) according to the manufacturer’s instructions, including DNase I digestion to remove contaminating DNA. cDNA was generated using GoScript Reverse Transcriptase with random primers, nucleotides, and RNasin (Promega Corporation, Fitchburg, WI, USA). TaqMan primer and probe sets for the target gene (ORMDL3) and housekeeping/reference gene (β-actin) were obtained commercially (Thermo Fisher Scientific). Samples were run in duplicate with each reaction containing Universal Master Mix (Thermo Fisher Scientific), the primer and probe sets, and cDNA for a total reaction volume of 25 μL. Fluorescence was measured for each cycle using the StepOnePlus Real-Time PCR System (Thermo Fisher Scientific). Results were calculated using the 2−ΔCt method relative to the endogenous control and β-actin and normalized to a donor monocytes’ expression.
Western blot analysis
Whole-cell protein lysates were prepared from eosinophils or PBMCs in radioimmunoprecipitation assay buffer that included phosphatase inhibitors and complete protease inhibitors (Hoffman-La Roche Ltd., Basel, Switzerland/Thermo Fisher Scientific). Protein lysates were heated for 5 minutes at 75°C in 1× Laemmli sample buffer prior to loading, and the samples were resolved on 12% SDS-PAGE under reducing conditions. Proteins were transferred to polyvinylidene fluoride-Fl (EMD Millipore, Billerica, MA, USA), and the membranes were blocked with TBS blocking buffer (LI-COR Biosciences, Lincoln, NE, USA) +0.1% Tween for 1 hour at room temperature. Western blots were probed with rabbit anti-ORMDL3 (1:1,000; EMD Millipore) or mouse-anti-beta-actin (1:10,000; Sigma-Aldrich, St Louis, MO, USA). Blots were washed with TBS +0.05% Tween and then probed with secondary antibodies (680RD anti-mouse and 800CW anti-rabbit, 1:10,000; LI-COR Biosciences) diluted with TBS blocking buffer for 1 hour at room temperature. Western blots were washed with TBS +0.05% Tween, dried, and scanned using the Odyssey System (LI-COR Biosciences). Density measurements were captured using the Odyssey software.
Statistics
The number of measurements provided by each subject ranged from 1 to 27 for eosinophil counts and from 1 to 21 for FeNO levels. Eosinophil counts and FeNO levels for subjects providing multiple measurements were analyzed using the geometric mean of their measurements. FeNO levels and eosinophil counts were each log-transformed for analysis. Eosinophil counts were categorized as >200 or <200/mm3, which is the historical cutoff used in our laboratory to distinguish “low” and “high” eosinophil blood donors. Age of onset was square root transformed for analysis. Genotype at rs7216389 was analyzed using the additive genetic model of risk according to the number of T alleles (CC =0, CT =1, and TT =2). Eosinophil counts, FeNO levels, and age of asthma onset were each examined for associations with genotype using the linear regression. The associations between eosinophil count category and genotype were examined using the logistic regression. A two-sided P-value of <0.05 was regarded as statistically significant. Analyses were conducted using R Version 2.15 (R Foundation for Statistical Computing, Vienna, Austria).
Results
Characteristics of the 255 subjects based on rs7216389 genotype classifications (47 CC, 123 CT, and 85 TT) are listed in Table 1. There were no associations between rs7216389 genotype and FEV1, self-reported allergies, age, or gender distribution. The TT genotype exhibited a higher rate of asthma diagnosis (CC 63.0%, CT 52.5%, and TT 76.2%; P=0.09), but this was not statistically significant. Subset subject characteristics with available FeNO data are listed in Table S1.
Table 1.
Study population
| Genotype | CC | CT | TT |
|---|---|---|---|
| Population (n) | 47 | 123 | 85 |
| Age (mean) | 28.7 | 27.6 | 28.0 |
| Gender (male %) | 45.7 | 40.2 | 47.6 |
| Asthma (%) | 63.0 | 52.5 | 76.2 |
| Self-reported allergies (%) | 91.3 | 85.2 | 84.5 |
| Ethnicity (% Hispanic) | 15.2 | 23.8 | 26.2 |
| Race | 2% American Indian | 2% American Indian | 1% American Indian |
| 2% Pacific Islander | 3% Asian | 4% Asian | |
| 96% Caucasian | 4% African American | 9% African American | |
| 91% Caucasian | 86% Caucasian |
The at-risk T allele at SNP rs7216389 has been linked to early childhood-onset asthma; therefore, we examined the age of asthma onset by genotypes (Figure 1). Based on the self-reported age of asthma onset, a T allele was associated with lower age of asthma onset (CC median 12.0 years [25th–75th percentiles, 8.0–18.2], CT median 7.0 years [25th–75th percentiles, 6.0–17.0], and TT median 8.0 years [25th–75th percentiles, 4.0–15.5]; P=0.03).
Figure 1.
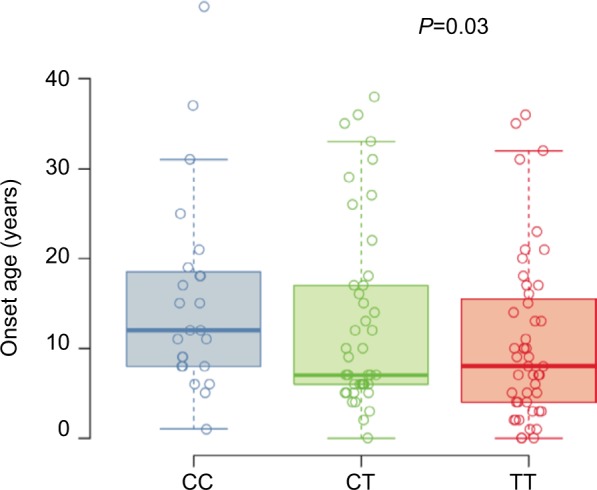
Self-reported age of asthma onset.
Notes: Box plots for the self-reported age of asthma onset in years for each subject (n=252) stratified by their genotype at SNP rs7216389, CC, CT, or TT. Boxes denote the median and 25th to 75th percentiles with whiskers extending to the farthest points within 1.5 times the box height. The P-value indicates the significance of the association between the number of T alleles and age of onset.
Abbreviation: SNP, single-nucleotide polymorphism.
FeNO levels from asthma subjects across the three genotypes are shown in Figure 2. The T allele was associated with increased FeNO levels in the subset of asthma subjects (CC 21 ppb [25th–75th percentiles, 16–33], CT 22 ppb [25th–75th percentiles, 15–35], TT 36 ppb [25th–75th percentiles, 25–48]; P=0.02). The use of inhaled corticosteroid can affect FeNO levels; therefore, we also analyzed a model of FeNO levels with both inhaled corticosteroid use and genotype terms included. The estimated genotype effect was similar in the unadjusted and adjusted analyses indicating that the relationship between FeNO and rs7216389 genotype does not differ with the use of inhaled corticosteroid.
Figure 2.
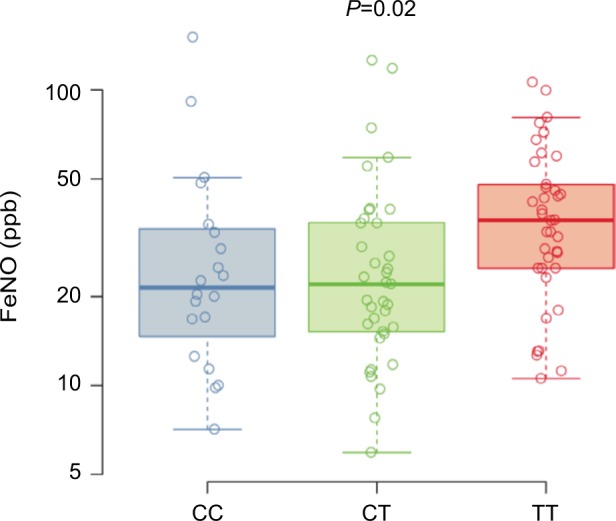
FeNO.
Notes: Box plots for the FeNO measured in ppb for the asthma subgroup (n=95) stratified by genotype at SNP rs7216389, CC, CT, and TT. Boxes denote the median and 25th to 75th percentiles with whiskers extending to the farthest points within 1.5 times the box height. The P-value indicates the significance of the association between the number of T alleles and FeNO levels.
Abbreviations: FeNO, fractional exhaled nitric oxide; SNP, single-nucleotide polymorphism.
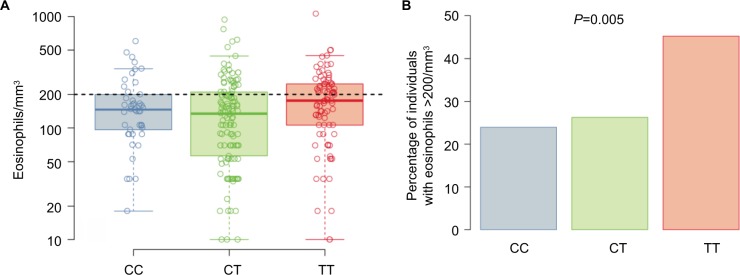
Subjects’ average (geometric mean) eosinophil counts by genotype are shown in Figure 3A. The T allele at rs7216389 was associated with a higher percentage of subjects having an average eosinophil count of >200 cells/mm3 (CC 23.9%, CT 26.2%, and TT 45.2%; P=0.005; Figure 3B). In the subset of asthma subjects, average eosinophil counts by genotype followed a similar pattern (CC 32.3%, CT 35.9%, and TT 46.9%; P=0.13) but was not statistically significant.
Figure 3.
Eosinophil counts.
Notes: (A) Box plots for the geometric mean eosinophil counts for the entire cohort (n=252) stratified by genotype at SNP rs7216389, CC, CT, and TT. Boxes denote the median and 25th to 75th percentiles with whiskers extending to the farthest points within 1.5 times the box height. The eosinophil count of 200/mm3 is shown as dashed line. (B) Percentage of subjects in entire cohort with geometric mean eosinophil count >200/mm3 stratified by genotype at SNP rs7216389, CC, CT, and TT. The P-value indicates the significance of the association between the number of T alleles and the percentage of individuals with eosinophil counts >200/mm3.
Abbreviation: SNP, single-nucleotide polymorphism.
In order to address a potential confounding influence of ethnicity/race on the genetic analyses, an additional analysis of the subset of Caucasian non-Hispanic individuals was completed. FeNO levels (Figure S1) and eosinophil counts (Figure S2) were analyzed. In this subset, the T allele remains associated with both FeNO levels (CC 20 ppb [25th–75th percentiles, 11–35], CT 19 ppb [25th–75th percentiles, 14–35], and TT 33 ppb [25th–75th percentiles, 22–46]; P=0.03; Figure S1) and eosinophil counts >200 cells/mm3 (CC 24.3%, CT 33.3%, and TT 48.1%; P=0.02; Figure S2). Therefore, our observations persisted with the exclusion of a potential ethnic/racial confounder.
In order to address the possibility that the genotype association with FeNO and blood eosinophil count is confounded by the age of asthma onset, we adjusted for the age of onset. Among subjects with asthma, there was no significant difference in the FeNO fold increase per T allele unadjusted versus adjusted (1.22 versus 1.25, P=0.73). There was also no significant difference in the eosinophil count >200 cells/mm3 odds ratio per T allele unadjusted versus adjusted (1.39 versus 1.32, P=0.29).
To determine whether eosinophils may be one of the cell types through which the 17q21 genes may be influencing asthma, we determined the expression of ORMDL3, one of the leading candidate genes at the 17q21 locus.2,20 We examined the mRNA and protein expressions of ORMDL3 in PBMCs and eosinophils. As shown in Figure 4, eosinophils expressed ORMDL3 mRNA (Figure 4A) at levels comparable to PBMCs, while ORMDL proteins (Figure 4B) were expressed, but fourfold less in total on a per cell basis. Of note, the antibody specificity in Western blots likely includes other isoforms of ORMDL and not just ORMDL3.
Figure 4.
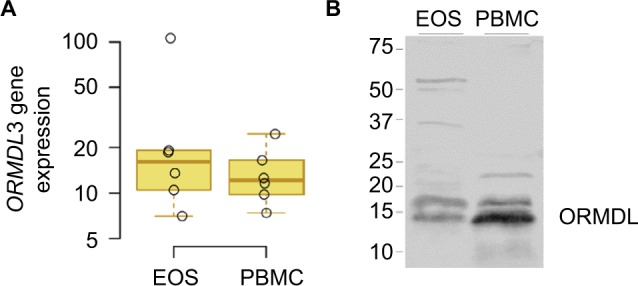
ORMDL3 expression.
Notes: (A) EOS and PBMCs were isolated from six blood donors, mRNA was purified, and gene expression was measured by Q-PCR. Box plots of ORMDL3 gene expression normalized to b-actin and shown as fold inductions relative to the control (monocyte population) from the same donor. Boxes denote the median and 25th to 75th percentiles with whiskers extending to the farthest points within 1.5 times the box height. (B) EOS and PBMC cell lysates were prepared from a representative donor and protein expression of ORMDL measured by Western blot.
Abbreviations: EOS, eosinophils; PBMCs, peripheral blood mononuclear cells.
Discussion
Very little is known about the impact of genotypic differences at the 17q21 locus on the biology of asthma in adult subjects. In our cohort, regardless of allergy diagnosis, adults with the T allele at rs7216389 were more likely to have higher FeNO levels and eosinophil counts. In addition, a greater number of T alleles in the asthma population were associated with an earlier age of asthma onset compared to those individuals with more C alleles. In order to address whether eosinophils could mediate these genotypic effects at the 17q21 locus, we examined the mRNA and protein expressions of one of the genes encoded in the locus, ORMDL3. We found that eosinophils expressed appreciable levels of ORMDL3 mRNA and ORMDL protein indicating that eosinophils may be one of the cells mediating the risk associated with the 17q21 locus. These data are consistent with several reports in the literature; however, additional analyses of large cohorts will be necessary to confirm these findings.
There are several limitations of this study. The cohort examined was not specifically recruited for this study, and thus, this study has variability in the number of blood eosinophil and FeNO observations for each subject. We chose to use a single data point for each subject based on the geometric mean of their data. There is a recall bias associated with the determination of age of asthma onset; however, we expect that the error should be similarly present in each genotype group. The asthma population was generally mild asthma patients with only 19% of subjects using controller inhaled corticosteroid therapy; thus, it is unknown whether the genotype associations with blood eosinophil counts and FeNO would be stronger in more severe disease.
In previous ethnically diverse cohort studies, several SNPs at the 17q21 locus were linked to a diagnosis of childhood asthma.2–5,9,21,22 The risk alleles at 17q21 were associated with an interaction between early infection and early onset asthma.15 More recently, 17q21 SNPs were also linked to the increased risk of wheezing upon rhinovirus exposure and the development of asthma in a pediatric population.17 However, whether 17q21 genotype has any relevance for asthma diagnosis, pathophysiology, or disease severity beyond childhood is not clear. In a European large cohort study, a 17q21 SNP exhibited strong association with childhood-onset asthma but did not show any association with adult onset asthma.20 In three cohorts separately examining association of the rs7216389 T allele with asthma severity in early-age onset asthma patients, statistically significant associations were not present; however, the combined analysis of all three cohorts exhibited a significant association between the T allele and greater odds for severe asthma. In a study of adult severe asthma patients, the odds for an rs7216389 T allele versus C allele were significantly elevated;7 however, this signal was entirely due to the severe asthma patients who had childhood onset of asthma.23 In another cohort, there was no association between the rs7216389 SNP and a diagnosis of asthma in adults;18 however, T allele was associated with both a lower FEV1 and the presence of allergic sensitization. Notably, the authors did not stratify the adult subjects based on childhood onset versus adult onset of asthma. It is intriguing to consider the possibility that the rs7216389 SNP may also be a risk factor for specific phenotypes of asthma; note that the asthma phenotype clusters from the NIH Severe Asthma Research Program identified one cluster that was notable for early age of onset, though it was also a milder form of asthma compared to the other phenotypes.24 Our results confirm that the risk allele at rs7216389 SNP has relevance in adult patients. We demonstrated that the risk allele at rs7216389 SNP is associated with greater FeNO levels and higher peripheral blood eosinophil counts. Interestingly, these observations were not restricted to the early-onset asthma patients. We did not observe an association between the rs7216389 SNP and FEV1; however, our cohort consisted of primarily mild asthma patients with essentially normal FEV1.
The elevated FeNO levels and eosinophil counts in our cohort associated with the risk alleles at SNP rs7216839 represent two biomarkers for asthma. In a study designed to identify SNPs associated with elevated FeNO levels in children, one of the SNPs (rs8069176) was within the 17q21 locus.25 Recent clinical trials with anti-IL-5 therapies have shown that clinical benefit is greatest in subjects with higher eosinophil counts.26,27 In these studies, the peripheral eosinophil count was used as an inclusion criterion to enrich the study with potential responders. Based on our report, the SNP rs7216839 represents a genotypic biomarker in asthma. We speculate that the SNP rs7216839, possibly in conjunction with other SNPs, may serve as a genotypic biomarker and predictor of response to asthma therapies such as those currently available and in development for asthma. Traditional biomarkers are subject to variability due to medication use and comorbidities. Therefore, identification of combinations of genotypic biomarkers may provide a more accurate stratification of asthma phenotypes for achieving personalized care of asthma. Further studies of large cohorts will be essential to confirm and validate these results.
Efforts to understand the mechanism of the 17q21 association with asthma have largely centered on the function of two specific genes expressed at the loci, ORMDL3 and GSDMB. Our study and others have shown that the expressions of these two genes are tightly correlated, making it difficult to identify which of these genes is directly involved in asthma pathophysiology.17,28 In asthma, whether this genotype affects the functional activity of one or multiple cell types is also unknown. Previous studies demonstrated that the risk allele at the 17q21 locus is associated with greater ex vivo rhinovirus-induced expression of ORMDL3 in PBMCs17 and increased ex vivo IL-4 and IL-13 secretions from PHA-stimulated PBMCs.29 Using an in vitro rhinovirus infection model, we recently demonstrated that increased ORMDL3 in PBMC corresponded to increased IFN-b, CXCL10, and genes associated with endoplasmic reticulum stress.30
A mouse study has shown that Ormdl3 overexpression in eosinophils induced features consistent with asthma, including spontaneous airway remodeling, increased fibrosis, mucus production, inflammation, and methacholine-induced hyper-responsiveness.31 Murine eosinophils expressed high levels of Ormdl3 mRNA and protein, and Ormdl3 was shown to have a role in eosinophil activation.32
The function of ORMDL3 appears to involve the regulation of endoplasmic reticulum-based calcium homeostasis facilitating the upregulation of the cellular unfolded protein response.33 There are also suggestions of ORMDL3 involvement in the inhibition of sphingolipid signaling, specifically sphingosine-1-phosphate. Further studies will be required to determine how these signaling events promote eosinophil production and activation.33 We speculate that the increased signaling events in eosinophils in conjunction with other cell types such as monocytes may play a significant role in mediating the 17q21 genotype-related risk and severity of asthma in adult asthmatics.
Supplementary materials
FeNO.
Notes: Box plots for the FeNO measured in ppb for the Caucasian non-Hispanic subgroup (n=120) stratified by genotype at SNP rs7216389, CC, CT, and TT. Boxes denote the median and 25th to 75th percentiles with whiskers extending to the farthest points within 1.5 times the box height. The P-value indicates the significance of the association between the number of T alleles and FeNO levels.
Abbreviations: FeNO, fractional exhaled nitric oxide; SNP, single-nucleotide polymorphism.
Percentage of donors in Caucasian non-Hispanic cohort (n=176) with geometric mean eosinophil counts >200/mm3 stratified genotype at SNP rs7216389, CC, CT, and TT. The P-value indicates the significance of the association between the number of T alleles and the percentage of individuals with eosinophil counts >200/mm3.
Abbreviation: SNP, single-nucleotide polymorphism.
Table S1.
FeNO study population
| Genotype | CC | CT | TT |
|---|---|---|---|
| Population (n) | 20 | 37 | 38 |
| Age (mean) | 31.0 | 29.0 | 30.0 |
| Gender (male %) | 70.0 | 40.5 | 55.3 |
| Self-reported allergies (%) | 95.0 | 100 | 100 |
| Ethnicity (% Hispanic) | 15.0 | 5.4 | 10.5 |
| Race | 5% Pacific Islander | 3% Asian | 5% Asian |
| 95% Caucasian | 97% Caucasian | 5% African American 90% Caucasian |
|
| FEV1% pred. (25th–75th percentile) | 88.6% (80.1–98.5) | 95.2% (85.5–109.0) | 92.7% (84.5–103.5) |
Abbreviations: FeNO, fractional exhaled nitric oxide; FEV1, forced expiratory volume in 1 second.
Acknowledgments
The authors thank the participants of the study for their dedication to asthma research. Also, we thank the nurses, research coordinators, research laboratory staff, and administrative staff of the University of Wisconsin Allergy and Asthma Research Program, including Paul Fichtinger, Gina Crisafi, Michelle Wolff, Maranda Hyde, Evelyn Falbiene, and Holly Eversoll. In addition, we would like to thank Dr Carole Ober, Dr Emma Thompson, and the laboratory staff at the University of Chicago for assistance with article editing and sample genotyping and Dr Elon Roti Roti at the University of Wisconsin–Madison for assistance with figure preparation and article editing. This study was funded by NIH P01 HL088594, NIH R21 AI122103, and NIH R21 AI121808.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Moorman J, Zahran H, Truman B, Molla M. Current asthma prevalence – United States, 2006–2008. MMWR Suppl. 2011;60(1):84–86. [PubMed] [Google Scholar]
- 2.Moffatt MF, Kabesch M, Liang L, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448(7152):470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 3.Madore A-M, Tremblay K, Hudson TJ, Laprise C. Replication of an association between 17q21 SNPs and asthma in a French-Canadian familial collection. Hum Genet. 2008;123(1):93–95. doi: 10.1007/s00439-007-0444-x. [DOI] [PubMed] [Google Scholar]
- 4.Galanter J, Choudhry S, Eng C, et al. ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am J Respir Crit Care Med. 2008;177(11):1194–1200. doi: 10.1164/rccm.200711-1644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leung TF, Sy HY, Ng MCY, et al. Asthma and atopy are associated with chromosome 17q21 markers in Chinese children. Allergy. 2009;64(4):621–628. doi: 10.1111/j.1398-9995.2008.01873.x. [DOI] [PubMed] [Google Scholar]
- 6.Li F-X, Tan J-Y, Yang X-X, Wu Y-S, Wu D, Li M. Genetic variants on 17q21 are associated with asthma in a Han Chinese population. Genet Mol Res. 2012;11(1):340–347. doi: 10.4238/2012.February.10.5. [DOI] [PubMed] [Google Scholar]
- 7.Halapi E, Gudbjartsson DF, Jonsdottir GM, et al. A sequence variant on 17q21 is associated with age at onset and severity of asthma. Eur J Hum Genet. 2010;18(8):902–908. doi: 10.1038/ejhg.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouzigon E, Corda E, Aschard H, et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. N Engl J Med. 2008;359(19):1985–1994. doi: 10.1056/NEJMoa0806604. [DOI] [PubMed] [Google Scholar]
- 9.Bisgaard H, Bønnelykke K, Sleiman PMA, et al. Chromosome 17q21 gene variants are associated with asthma and exacerbations but not atopy in early childhood. Am J Respir Crit Care Med. 2009;179(3):179–185. doi: 10.1164/rccm.200809-1436OC. [DOI] [PubMed] [Google Scholar]
- 10.Ober C, Yao T-C. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev. 2011;242(1):10–30. doi: 10.1111/j.1600-065X.2011.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hao K, Bossé Y, Nickle DC, et al. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet. 2012;8(11):e1003029. doi: 10.1371/journal.pgen.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, Hastie AT, Hawkins GA, et al. eQTL of bronchial epithelial cells and bronchial alveolar lavage deciphers GWAS-identified asthma genes. Allergy. 2015;70(10):1309–1318. doi: 10.1111/all.12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhai WH, Song CY, Huang ZG, Sha H. Correlation between the genetic polymorphism of ORMDL3 gene and asthma risk: a meta-analysis. Genet Mol Res. 2015;14(2):7101–7112. doi: 10.4238/2015.June.29.3. [DOI] [PubMed] [Google Scholar]
- 14.Flory JH, Sleiman PM, Christie JD, et al. 17q12-21 variants interact with smoke exposure as a risk factor for pediatric asthma but are equally associated with early-onset versus late-onset asthma in North Americans of European ancestry. J Allergy Clin Immunol. 2009;124(3):605–607. doi: 10.1016/j.jaci.2009.05.047. [DOI] [PubMed] [Google Scholar]
- 15.Smit LA, Bouzigon E, Pin I, et al. EGEA Cooperative Group 17q21 variants modify the association between early respiratory infections and asthma. Eur Respir J. 2010;36(1):57–64. doi: 10.1183/09031936.00154509. [DOI] [PubMed] [Google Scholar]
- 16.van der Valk RJP, Duijts L, Kerkhof M, et al. Interaction of a 17q12 variant with both fetal and infant smoke exposure in the development of childhood asthma-like symptoms. Allergy. 2012;67(6):767–774. doi: 10.1111/j.1398-9995.2012.02819.x. [DOI] [PubMed] [Google Scholar]
- 17.Calışkan M, Bochkov YA, Kreiner-Møller E, et al. Rhinovirus wheezing illness and genetic risk of childhood-onset asthma. N Engl J Med. 2013;368:1398–1407. doi: 10.1056/NEJMoa1211592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreiner-Møller E, Strachan DP, Linneberg A, Husemoen LLN, Bisgaard H, Bønnelykke K. 17q21 gene variation is not associated with asthma in adulthood. Allergy. 2015;70(1):107–114. doi: 10.1111/all.12537. [DOI] [PubMed] [Google Scholar]
- 19.American Thoracic Society; European Respiratory Society ATS/ERS recommendations for standardized procedures for the online and offline measurement of exhaled lower respiratory nitric oxide and nasal nitric oxide, 2005. Am J Respir Crit Care Med. 2005;171(8):912–930. doi: 10.1164/rccm.200406-710ST. [DOI] [PubMed] [Google Scholar]
- 20.Moffatt MF, Gut IG, Demenais F, et al. GABRIEL Consortium A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363(13):1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu H, Romieu I, Sienra-Monge J, Li H, del Rio-Navarro B, London SJ. Genetic variation in ORM1-like 3 (ORMDL3) and gasdermin-like (GSDML) and childhood asthma. J Allergy Clin Immun. 2009;123(2):S52. doi: 10.1111/j.1398-9995.2008.01912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ono JG, Worgall TS, Worgall S. 17q21 locus and ORMDL3: an increased risk for childhood asthma. Pediatr Res. 2014;75(1–2):165–170. doi: 10.1038/pr.2013.186. [DOI] [PubMed] [Google Scholar]
- 23.Binia A, Khorasani N, Bhavsar PK, et al. Chromosome 17q21 SNP and severe asthma. J Hum Genet. 2011;56(1):97–98. doi: 10.1038/jhg.2010.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore WC, Meyers DA, Wenzel SE, et al. National Heart, Lung, and Blood Institute’s Severe Asthma Research Program Identification of asthma phenotypes using cluster analysis in the severe asthma research program. Am J Respir Crit Care Med. 2010;181(4):315–323. doi: 10.1164/rccm.200906-0896OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Valk RJP, Duijts L, Timpson NJ, et al. Early Genetics & Lifecourse Epidemiology (EAGLE) Consortium Fraction of exhaled nitric oxide values in childhood are associated with 17q11.2-q12 and 17q12-q21 variants. J Allergy Clin Immunol. 2014;134(1):46–55. doi: 10.1016/j.jaci.2013.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pavord ID, Korn S, Howarth P, et al. Mepolizumab for severe eosinophilic asthma (DREAM): a multicentre, double-blind, placebo-controlled trial. Lancet. 2012;380(9842):651–659. doi: 10.1016/S0140-6736(12)60988-X. [DOI] [PubMed] [Google Scholar]
- 27.Castro M, Zangrilli J, Wechsler ME, et al. Reslizumab for inadequately controlled asthma with elevated blood eosinophil counts: results from two multicentre, parallel, double-blind, randomised, placebo-controlled, phase 3 trials. Lancet Respir Med. 2015;3(5):355–366. doi: 10.1016/S2213-2600(15)00042-9. [DOI] [PubMed] [Google Scholar]
- 28.Verlaan DJ, Berlivet S, Hunninghake GM, et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet. 2009;85(3):377–393. doi: 10.1016/j.ajhg.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schedel M, Michel S, Gaertner VD, et al. Polymorphisms related to ORMDL3 are associated with asthma susceptibility, alterations in transcriptional regulation of ORMDL3, and changes in T(H)2 cytokine levels. J Allergy Clin Immun. 2015;136(4):893. doi: 10.1016/j.jaci.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 30.Liu YP, Rajamanikham V, Baron M, et al. Association of ORMDL3 with rhinovirus-induced endoplasmic reticulum stress and type I Interferon responses in human leucocytes. Clin Exp Allergy. 2017;47(3):371–382. doi: 10.1111/cea.12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller M, Rosenthal P, Beppu A, et al. ORMDL3 transgenic mice have increased airway remodeling and airway responsiveness characteristic of asthma. J Immunol. 2014;192(8):3475–3487. doi: 10.4049/jimmunol.1303047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ha SG, Ge XN, Bahaie NS, et al. ORMDL3 promotes eosinophil trafficking and activation via regulation of integrins and CD48. Nat Commun. 2013;4:2479. doi: 10.1038/ncomms3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das S, Miller M, Broide DH. Chromosome 17q21 genes ORMDL3 and GSDMB in asthma and immune diseases. Adv Immunol. 2017;135:1–52. doi: 10.1016/bs.ai.2017.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FeNO.
Notes: Box plots for the FeNO measured in ppb for the Caucasian non-Hispanic subgroup (n=120) stratified by genotype at SNP rs7216389, CC, CT, and TT. Boxes denote the median and 25th to 75th percentiles with whiskers extending to the farthest points within 1.5 times the box height. The P-value indicates the significance of the association between the number of T alleles and FeNO levels.
Abbreviations: FeNO, fractional exhaled nitric oxide; SNP, single-nucleotide polymorphism.
Percentage of donors in Caucasian non-Hispanic cohort (n=176) with geometric mean eosinophil counts >200/mm3 stratified genotype at SNP rs7216389, CC, CT, and TT. The P-value indicates the significance of the association between the number of T alleles and the percentage of individuals with eosinophil counts >200/mm3.
Abbreviation: SNP, single-nucleotide polymorphism.
Table S1.
FeNO study population
| Genotype | CC | CT | TT |
|---|---|---|---|
| Population (n) | 20 | 37 | 38 |
| Age (mean) | 31.0 | 29.0 | 30.0 |
| Gender (male %) | 70.0 | 40.5 | 55.3 |
| Self-reported allergies (%) | 95.0 | 100 | 100 |
| Ethnicity (% Hispanic) | 15.0 | 5.4 | 10.5 |
| Race | 5% Pacific Islander | 3% Asian | 5% Asian |
| 95% Caucasian | 97% Caucasian | 5% African American 90% Caucasian |
|
| FEV1% pred. (25th–75th percentile) | 88.6% (80.1–98.5) | 95.2% (85.5–109.0) | 92.7% (84.5–103.5) |
Abbreviations: FeNO, fractional exhaled nitric oxide; FEV1, forced expiratory volume in 1 second.