Abstract
Quorum sensing, in which bacteria communities use signaling molecules for inter- and intracellular communication, has been intensively studied in recent decades. In order to fabricate highly sensitive easy-to-handle point of care biosensors that detect quorum sensing molecules, we have developed, as is reported here, reporter bacteria loaded alginate–methacrylate (alginate-MA) hydrogel beads. The alginate-MA beads, which were obtained by electrostatic extrusion, were reinforced by photo-cross-linking to increase stability and thereby to reduce bacteria leaching. In these beads the genetically engineered fluorescent reporter bacterium Escherichia coli pTetR-LasR-pLuxR-GFP (E. coli pLuxR-GFP) was encapsulated, which responds to the autoinducer N-(3-oxododecanoyl)homoserine lactone secreted by Pseudomonas aeruginosa. After encapsulation in alginate-MA hydrogel beads with diameters in the range of 100–300 μm that were produced by an electrostatic extrusion method and rapid photo-cross-linking, the E. coli pLuxR-GFP were found to possess a high degree of viability and sensing activity. The encapsulated bacteria could proliferate inside the hydrogel beads, when exposed to bacteria culture medium. In media containing the autoinducer N-(3-oxododecanoyl)homoserine lactone, the encapsulated reporter bacteria responded with a strong fluorescence signal due to an increased green fluorescent protein (GFP) expression. A prototype dipstick type sensor developed here underlines the potential of encapsulation of viable and functional reporter bacteria inside reinforced alginate–methacrylate hydrogel beads for whole cell sensors for bacteria detection.
Keywords: alginate−methacrylate, microbead, electrostatic extrusion, bioencapsulation, whole cell biosensor, quorum sensing
Introduction
Biosensors are generally defined as integrated analytical devices, which convert biological recognition events related to environmental changes or target analytes into a measurable signal.1 Biosensors offer rapid and on-site/point-of-care detection and have been widely used for monitoring environment pollutants, chemical substances, traces of toxins, and hormones in the field of environmental monitoring, food safety, health care, and biomedicine. In this context also biological or biologically derived sensing elements, such as enzymes, antibodies, DNA, and whole cells, have been used as biosensors.2 Among them, living cells and especially genetically engineered microorganisms have shown advantages as biosensors. After genetic modification, the microorganism can produce reporter proteins under a promoter’s transcriptional control in a dose-dependent fashion in response to the presence of target compounds.3 Thereby, whole-cell biosensors can provide a desirable signal output with very high sensitivity and selectivity. Whole-cell biosensors are a good alternative to enzyme- or antibody-based biosensors, since microbial cells can be grown rapidly at low cost, require little maintenance, and exhibit high selectivity.
Traditionally, whole bacteria cell biosensors are optimized to produce electric currents, heat, chromogens, or luminescence or fluorescence signals transcriptionally regulated via promotors that are naturally activated when exposed to chemicals or environmental physical stresses.4 Quorum sensing, in which bacteria use small signaling molecules called autoinducers to regulate their gene expression in response to e.g. fluctuations of bacteria population density, is an attractive mechanism for whole cell sensing. Gram-negative bacteria mainly use N-acylhomoserine lactones (AHLs), while Gram-positive bacteria often use oligopeptides for the population-dependent interbacteria communication.5 Quorum sensing is involved in several physiological processes, such as symbiosis, production of antibiotics, motility, conjugation, and biofilm formation. In particular, the adherence of bacteria and their formation of biofilms possess great impact in ecology, medicine, and industry. For example, the biofilm formation of the Gram-negative bacterium Pseudomonas aeruginosa (P. aeruginosa) and the accompanying increase in antibiotic resistance may cause acute and chronic infections.6 Thus, the detection of early stages of biofilm formation is an important target for point of care bacteria growth monitoring.
Poh et al. have reported on a genetically engineered Escherichia coli (E. coli) carrying a synthetic genetic system, which comprises quorum sensing, killing, and lysing devices, that enables E. coli to sense and eradicate pathogenic P. aeruginosa strains.7 The E. coli pTetR-LasR-pLuxR-GFP (E. coli pLuxR-GFP) exhibiting an AHL sensing device has been verified to possess an optimal sensing activity approximately in the concentration range of AHLs secreted by P. aeruginosa.7 In order to maintain both viability and activity of these engineered E. coli pLuxR-GFP in otherwise detrimental environmental conditions (e.g., pH, heat, and toxic compounds) and to separate the sensor bacteria from the surrounding media for easy handling, bacteria were, as reported here, encapsulated into hydrogel microbeads to form a robust biosensing element.
Related hydrogel-based capsules and beads, composed of water-swollen three-dimensionally cross-linked polymeric networks, have been extensively studied for the encapsulation of living cells to retain cell viability and functionality.8,9 Alginate, as an anionic polysaccharide with a linear copolymer composed of 1 → 4 linked β-d-mannuronic acid (M) and α-l-guluronic acid (G),10 can be cross-linked via the interaction between the carboxylic acid groups of the alginate and divalent ions like Ca2+. The corresponding rapid and nondetrimental cell encapsulation process has made alginate the most commonly used material for bioencapsulation. For instance, viable lactic acid bacteria11 and Lactobacillus plantarum(12) have been embedded in alginate beads. However, the ionic cross-linking is easily broken by cationic scavengers, such as the nongelling cations sodium, or chelators, such as citrate. Hence, as-prepared beads may suffer from insufficient stability, poor mechanical properties, and noncontrollable permeability, the modification of alginate systems has attracted considerable attention. Several covalent cross-linking strategies, including photo-cross-linking, have been utilized instead of ionic cross-linking;13 e.g., alginate functionalized with 2-aminoethyl methacrylate14 or with methacrylate groups was polymerized by photo-cross-linking.15 Up to date, most of the studies are aim for eukaryotic cells encapsulation.
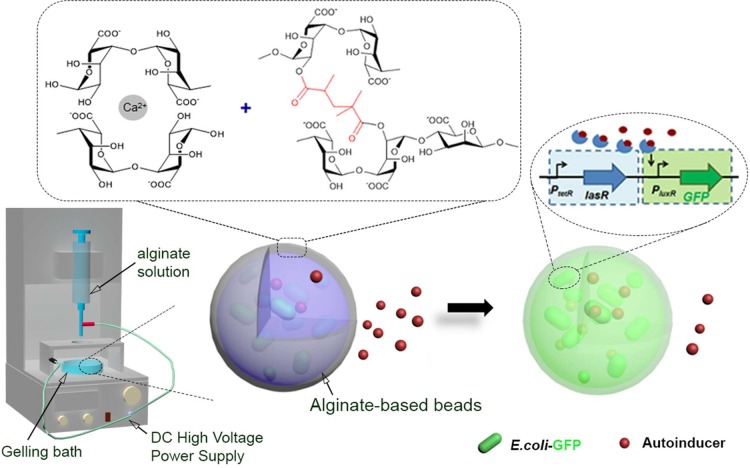
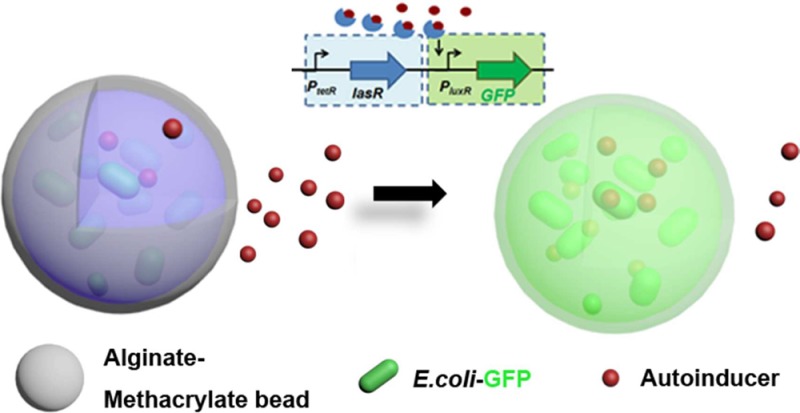
Here we report on the development of reporter E. coli pLuxR-GFP loaded alginate-based hydrogel microbeads for the detection of bacteria via their secreted autoinducers that were reinforced by photo-cross-linking to prevent bacteria leaching (Figure 1). Apart from the systematic investigation of the impact of ionic and covalent cross-linking on the swelling properties, the viability, bacteria escape, and functionality of the encapsulated reporter E. coli pLuxR-GFP were investigated, in particular in sensing N-(3-oxododecanoyl)homoserine lactone (3OC12HSL), an autoinducer secreted by P. aeruginosa, via the autoinducer-triggered expression of green fluorescent protein (GFP).
Figure 1.
Schematic of reporter bacterium E. coli pTetR-LasR-pLuxR-GFP7 loaded alginate-based hydrogel microbeads and the autoinducer-triggered expression of green fluorescent protein (right). The alginate-based hydrogel beads were fabricated using the electrostatic extrusion method (left: schematic of the electrostatic extrusion setup used for microscale bead formation). Ionic cross-linking and photo-cross-linking were involved in bead formation and reinforcement, respectively.
Experimental Section
Materials
Alginic acid sodium salt, glycidyl methacrylate, triethylamine, tetrabutylammonium bromide, calcium chloride hexahydrate, sodium chloride, 2-hydroxy-4′-(2-hydroxyethoxy)-2-methylpropiophenone (Irgacure 2959), hexamethyldisiloxane, Trizma base, Fluoresceinamine isomer I, N-(3-(dimethylamino)propyl)-N′-ethylcarbodiimide hydrochloride (EDC), N-hydroxysulfosuccinimide sodium salt (Sulfo-NHS), 2-(N-morpholino)ethanesulfonic acid (MES) hydrate, and N-(3-oxododecanoyl)homoserine lactone were purchased from Sigma-Aldrich and used without further purification. Hydrochloric acid was purchased from Fisher Chemical. N-Dodecanoyl-l-homoserine lactone-3-hydrazone fluorescein was purchased from Biomol. Luria-Bertani medium (LB broth) and LB agar were purchased from Carl Roth (Germany). Throughout the whole study, Milli-Q water drawn from a Millipore Direct Q8 system with Millimark Express 40 filter (Merck, Germany) was used.
Alginate–Methacrylate Synthesis
Photopolymerizable methacrylate groups were added to alginate to yield alginate–methacrylate (alginate-MA) via glycidyl methacrylate reaction. Briefly, 1.00 g of alginic acid sodium salt was dissolved in 100 mL of Milli-Q water to form a 1% w/v solution at 25 °C; 2.2 mL of triethylamine, 4.4 mL of glycidyl methacrylate, and 2.2 g of tetrabutylammonium bromide were added sequentially and carefully mixed before the next component was added. The reaction was conducted at 25 °C for 24 h, followed by 1 h reaction at 60 °C. Then the clear solution was precipitated in an excess amount of acetone (20 times the volume of reaction solution) twice to remove excess reactants.
Synthesis of Fluorescein-Labeled Alginate–Methacrylate
10 mg of alginate-MA was dissolved in 10 mL of MES buffer (50 mM MES, 50 mM NaCl, pH 6.5) to form a 0.1 wt % alginate-MA solution. 0.25 mM EDC and 0.25 mM Sulfo-NHS were added, and the solution was stirred at 25 °C for 2 h before adding 0.25 mM fluoresceinamine. The reaction mixture was stirred at 25 °C for 18 h. Then the solution was transferred to dialysis membranes (molecular weight cutoff: 3500 Da, Spectrum Laboratories) and dialyzed against Milli-Q water at 25 °C for 3 days (9 times water exchange) in the dark. Finally, the fluorescein-labeled alginate solution was freeze-dried and kept at 4 °C in the dark before use.
Hydrogel Microbead Formation
0.2 g of alginate-MA was suspended in 10 mL of Tris buffer (10 mM Tris, pH 8.5) in a septum-capped glass vial at 25 °C and shaken until it was fully dissolved to form a 2 wt % alginate-MA solution. Then 0.01 g of the photoinitiator Irgacure 2959 was added. This solution was transferred into a syringe (1 mL, B. Braun Melsungen AG, Germany). Care was taken to avoid the formation of air bubbles. Then the solution in the syringe was extruded through a 27G blunt end needle (B. Braun Melsungen AG, Germany, o.d. = 0.4 mm, i.d. = 0.2 mm) into 100 mM CaCl2 gelling solution that contained 0.1 wt % Irgacure in Tris buffer using a home-built syringe pump. An electric potential between 1.0 and 6.5 kV was applied by a high voltage unit (HCN14-6 500, FuG Elektronik GmbH) with the needle connected to the positively charged electrode. For alginate-MA beads, the beads in the gelling solution were exposed to UV irradiation (CL-1000 series UV cross-linker, with CL-1000L Model 365 nm UV tubes, 5 × 8 W, UVP, UK) for 5 min, and after UV irradiation the beads were kept in gelling solution for another 10 min hardening. For alginate beads, the beads were kept in gelling solution for 15 min to harden. The diameters of at least 50 randomly chosen microbeads were measured using Zen imaging software from the images taken with an optical microscope (Primo Vert light microscope, Carl Zeiss, Germany). Fluorescence microscopy images of fluorescein-labeled alginate-MA beads in CaCl2 solution were taken with an Axiovert 135 fluorescence microscope (Carl Zeiss, Germany). The average diameters and standard deviations were calculated from the measured data. Alginate beads were prepared using the same procedure as described above for alginate-MA except no Irgacure 2959 was used for alginate beads.
Hydrogel Microbead Swelling Test
The swelling behavior was investigated by measuring the size of the microbeads after various incubation times in different solutions. Alginate and alginate-MA beads were immersed in 0.9 wt % NaCl, 2 mM, 20 mM, and 100 mM CaCl2 as well as in 5 mM and 50 mM sodium citrate solution, respectively. The diameters of at least 50 randomly chosen microbeads were measured using optical microscopy after different time periods, and the average diameters and standard deviations were calculated. The swelling ratio (SR) is defined as
| 1 |
and was calculated by determining the mean value of microbead diameter at time t, Dt, according to
| 2 |
Here D0 denotes the mean value of microbead diameter at time 0, Vt is the calculated volume of microbeads at time t, and V0 is the calculated volume of microbeads at time 0.
Hydrogel Microbead Stability Tests
The hydrogel microbeads were immersed in 0.9 wt % NaCl or 100 mM CaCl2 solution and then kept at 37 °C for 1, 3, 7, 14, or 35 days. After each time interval, the microbeads were filtered using cell strainers (mesh size 40 μm, Corning Incorporated, USA), washed with Milli-Q water, dried at 37 °C, and weighted. The average values of three measurements were taken for each sample, and the maintain weight percentage at time t is defined as follows:
| 3 |
where Wt is the dry weight of degraded microbeads at time t and W0 is the dry weight of microbeads at time 0.
Hydrogel Microbead Permeability Tests
The permeability of the microbeads was studied by measuring the diffusion of N-dodecanoyl-l-homoserine lactone-3-hydrazone-fluorescein (FITC-AHL) into the hydrogel beads. 0.3 mg of alginate-based beads in 1 mL of CaCl2 Tris buffer (100 mM) was filtered using cell strainers, and excess water was absorbed using filter paper before they were immersed into 1 mL of FITC-AHL Tris solution (1.0 × 10–5 mol/L) for 1, 2, 3, 5, or 10 min. The FITC-AHL concentration of the supernatant was monitored. Beads were filtered and immersed into 1 mL of 50 mM sodium citrate solution to release FITC-AHL inside beads. The concentration of the released FITC-AHL was determined using a microplate reader (Tecan SAFIRE, Tecan, Switzerland) with a black flat-bottom 96-well plate (Greiner Bio-one, Germany). Fluorescence intensity vs concentration standard curves of FITC-AHL in Tris buffer and 50 mM sodium citrate solution were studied and plotted.
Microencapsulation of E. coli PLuxR-GFP
All cells involved in encapsulation and characterization experiments were Escherichia coli TOP10 pTetR-LasR-pLuxR-GFP7 (E. coli pLuxR-GFP as abbreviation) without further genetic modification. The glycerol stock was stored at −80 °C. Stocks were streaked and diluted onto LB agar plates containing 100 μg/mL ampicillin. Plates were incubated overnight at 37 °C to form colonies and stored at 4 °C for less than 1 month. A single colony of E. coli pLuxR-GFP was cultivated in 5 mL of LB broth supplemented with ampicillin (100 μg/mL) for 16 h in a shaking incubator (MaxQ 4000 benchtop orbital shaker, Thermo Scientific, USA) at 37 °C and 200 rpm to reach the exponential growth phase (OD600 ≈ 0.5). 1 mL cell suspension was pelleted in a sterilized Eppendorf snap-cap microcentrifuge tube by centrifugation for 10 min at 5000g (microcentrifuge, Micro Star 17, VWR, USA), and the supernatant was discarded. The solutions were sterilized using 0.22 μm Millex-GS filter (Carl Roth, Germany). The cells were resuspended in 1 mL of 2 wt % alginate solution or 1 mL of 2 wt % alginate-MA solution with 0.1 wt % Irgacure 2959. The bacteria–polymer suspension was transferred into a 1 mL syringe and extruded at electric potential difference of 6.0 kV through a 27G blunt end needle into a 100 mM CaCl2 gelling solution with or without 0.1 wt % Irgacure 2959. The formed microbeads were rinsed with sterile Tris buffer (10 mM Tris, pH 8.5) before being transferred to LB broth, followed by culturing in a shaking incubator at 37 °C and 200 rpm. The microbead diameters were measured using optical microscopy.
Encapsulation Efficiency
The encapsulation efficiency was calculated as follows:
| 4 |
where Nbead is the number of bacteria in the hydrogel beads (without UV irradiation), and Nprecursor is the number of bacteria before encapsulation; both are expressed as colony-forming unit (cfu)/mL. The number of bacteria in the hydrogel beads was evaluated as follows: The hydrogel beads were first separated from the gelling bath solution via a cell strainer and washed with excess 100 mM CaCl2 six times to remove free bacteria. Then the hydrogel beads were disrupted and dissolved using a sterile 50 mM sodium citrate solution for 15 min with gentle shaking. The solution obtained was serially diluted in Ringer’s solution, and the number of bacteria was evaluated by counting the colonies on LB agar plates after incubation at 37 °C for 24 h.
Bacteria Live/Dead Staining
After separating the microbeads from the culture media via filtration (Cell strainer, 40 μm mesh, Corning), 200 μL of fresh LB broth with 20 μg/mL propidium iodide (PI) and 100 μg/mL fluorescein diacetate (FDA) were added to resuspend the beads. After incubation in the dark at 25 °C for 5 min, the beads were filtered using the 40 μm mesh filter and washed with fresh LB broth to remove excess dye. The labeled samples embedded on a microscope slide (VWR, Germany) with a marking film (depth 0.25 mm, Carl Roth, Germany) and sealed with a glass cover (Menzel-Gläser, Germany) and Rotiseal (Carl Roth, Germany) to avoid evaporation were analyzed under an inverted fluorescence microscope (Axiovert 135, Carl Zeiss, Germany). The number of live and dead bacteria was counted using ImageJ.
Determination of Bacteria Density in the Beads
The growth of bacteria inside the microbeads over time was evaluated. The 0.3 mg alginate-based beads were suspended in 8 mL of LB medium and incubated at 37 °C. After 2, 4, 6, and 24 h culture, the number of bacteria inside beads was evaluated as above-described via counting the number of colonies formed on LB agar plates (plate count method). The data shown are the mean of three independent experiments. Bacteria viability in the hydrogel beads during storage at 4 °C was investigated via live/dead staining and CFU tests.
Leaching Tests and Proliferation of Encapsulated Bacteria
Alginate and alginate-MA beads were separated from suspension via filtration (Cell strainer, 40 μm mesh, Corning) and washed with excess 20 mM CaCl2 solution at least 4 times to remove nonencapsulated bacteria. The washing solution was collected and poured on agar plates. The agar plates were incubated at 37 °C for 24 h. The number of bacteria in the washing solution was evaluated by counting the colonies on LB agar plates. The alginate and alginate-MA beads were resuspended in fresh LB medium and stored at 37 and 4 °C, respectively. The bead concentration was about 9000 beads/mL. For storage at 37 °C, the suspension was separated from beads via filtration (Cell strainer, 40 μm mesh, Corning) after 2, 4, and 6 h incubation. The bead-free suspension was serially diluted in Ringer’s solution, and the number of bacteria was evaluated by counting the colonies on LB agar plates after incubation at 37 °C for 24 h. For storage at 4 °C, the number of bacteria in the suspension was evaluated as described above after 1, 3, 14, and 35 days incubation.
Scanning Electron Microscopy (SEM) Study
Alginate-MA beads were fixated using glutaraldehyde (1.5%) and paraformaldehyde (3%) in DPBS (Dulbecco’s phosphate-buffered saline with calcium and magnesium, Life Technologies, 1× dilution) for 2 h at 4 °C. Then the beads were dehydrated in an ethanol/water mixture at ethanol concentrations of 30%, 50%, 70%, 80%, 90%, and 95% for 15 min each and 100% for three times 15 min each. Dehydrated samples were then submerged completely in distilled hexamethyldisiloxane (HMDS) twice for 10 min followed by air-drying. Samples were sputter-coated with a thin layer of gold (∼10 nm thickness) and images obtained using a field emission scanning electron microscope (ZEISS ultra-55, Germany).
Characterization of E. coli PLuxR-GFP Sensing in Alginate-Based Beads via AHL-Assay
The fluorescence intensity due to the GFP expression was monitored in the presence of 1.0 × 10–5, 1.0 × 10–6, and 1.0 × 10–7 mol/L N-(3-oxododecanoyl)homoserine lactone. Planktonic bacteria and bacteria encapsulated inside the alginate-based beads were mixed with LB broth and transferred in triplicate aliquots of 200 μL for induction with 3OC12HSL into a transparent, flat-bottom 96-well plate. The plate was incubated in a shaking incubator at 37 °C and 200 rpm. After every 30 min, the OD600 and the fluorescence intensity were recorded using a microplate reader. The OD600 and the fluorescence intensity of LB medium were used as background.
Fabrication of Dipstick Type Biosensor
A dipstick type biosensor was fabricated by entrapping reporter bacteria loaded alginate-MA beads into a transparent poly(ethylene glycol) (PEG)-based hydrogel film (30 mm × 10 mm × 5 mm). After the beads had formed in the gelling bath, as described in Hydrogel Microbead Formation section, the beads were filtered and resuspended in LB medium with 0.1% Irgacure 2959 and 10 wt % poly(ethylene glycol) diacrylate (Mn= 575 g/mol, Sigma-Aldrich). Then the bead–precursor system was exposed to UV irradiation for 5 min. The formed hydrogels were cut with scalpel to the desired dimensions.
Results and Discussion
Alginate-MA Hydrogel Beads
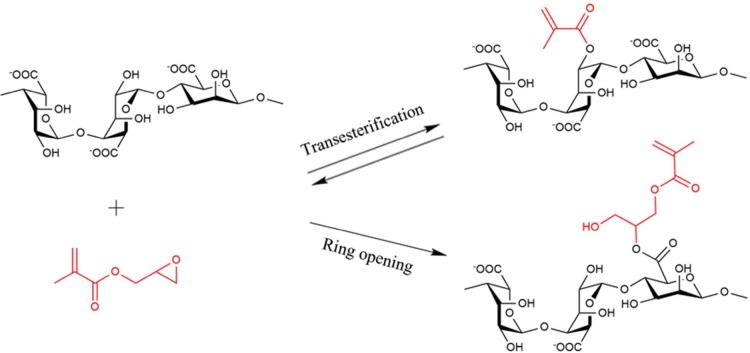
Photo-cross-linkable alginate (alginate-MA) was prepared using glycidyl methacylate, as shown in Figure 2, according to the method described by Leach et al.16 Ionic cross-linking, due to the rapid reaction speed, was used in electrostatic extrusion process for beads formation, while photo-cross-linking, due to the irreversible covalent bonding, was used to increase the stability of beads (Figure 1). The photo-cross-linking accelerates the stabilization of the fabricated alginate beads under mild conditions compared to the known thermal route reported by Wang et al. for the encapsulation of eukaryotic cells.15 An excess of glycidyl methacrylate with respect to alginate was used because of the poor solubility and hydrolysis in aqueous medium. Two reactions occurred, including a reversible transesterification through the primary group and an irreversible ring-opening conjugation through the carboxylic acid group.17,18
Figure 2.
Methacrylation of alginate by reaction with glycidyl methacrylate. The two competing pathways of transesterification and ring-opening may occur.
Because of the short reaction time used here, both ring-opening and transesterification reactions occurred. The formation of the products in the alginate-MA reaction was confirmed by 1H NMR spectroscopy (Supporting Information part 1). Resonances at 5.65, 6.09, and 1.83 ppm verified the presence of methylene and methyl protons (CHaHb=C(CH3)−) coupled to the grafted methacrylate. The degree of substitution is defined as the amount of methacrylate groups per guluronic acid repeat unit and was calculated from the relative integrals of the signals for the methacrylate protons and the protons of guluronic acid (4.94 and 4.34 ppm).13,14,19,20 The degree of substitution was calculated as 10%. A higher degree of substitution (∼25%) can be achieved by increasing the ratio of glycidyl methacrylate and alginate, reaction time, or by using dimethylformamide as a cosolvent (data not shown).
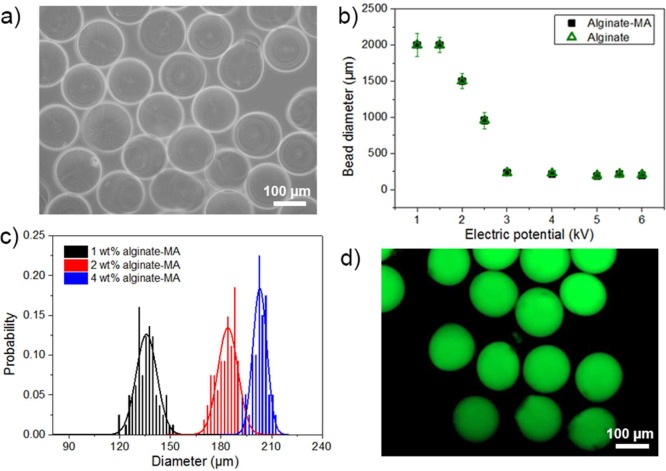
Electrostatic extrusion, which is conducted under mild conditions without using high temperatures or organic solvents,21,22 was used for microscale beads formation. As shown in Figure 1, a syringe pump was used to deliver the polymer solution to the end of a needle. An electric potential was applied between the needle and the gelling solution. The generated electric field causes mobile ions in the polymer solution to accumulate near the surface of the pendant meniscus. At sufficiently high electric fields, the electrostatic stress overcomes the capillary tension, and a jet of drops is formed to expel some portion of the surface charge (Rayleigh limit).23 An optical microscopy image of 2 wt % alginate-MA beads formed at electric potential of 6.0 kV is shown in Figure 3a. The effect of the applied voltage was investigated by a comparison of the bead sizes. Figure 3b shows that the bead diameter decreased from 2.3 mm to less than 200 μm with increasing the applied electric potential U up to the critical electric potential, Uc. Uc is characterized by surface tension of the polymer solution at U = 0 and the internal diameter of the needle.24 By increasing the applied electric potential from 3.0 to 6.0 kV, the shape of formed hydrogel beads became more regular and uniform (the coefficient of variance decreased from 20% to 5%, Supporting Information part 2). Photochemically cross-linked alginate-MA beads had similar diameter distributions as neat alginate beads formed under the same conditions, which indicates that the substitution of alginate with methacrylic acid as well as the polymerization did not influence the bead formation markedly. At an electric potential of 6.0 kV, 2% w/v alginate-MA formed hydrogel beads exhibit an average diameter around 184 μm and 3.2% coefficient of variance (Figure 3a), and the circularity 4πA/P2 was 0.92 ± 0.02 (where A represents the projected bead area and P the bead perimeter observed by optical microscopy). With increasing polymer concentration, the bead diameter was observed to increase. For 1 and 4 wt % alginate-MA, the average diameters were 140 and 200 μm, respectively (Figure 3c). The circularity 4πA/P2 calculated from the optical microscopy images was 0.90 ± 0.03 for 1 wt % alginate-MA beads and 0.95 ± 0.04 for 4 wt % alginate-MA beads.
Figure 3.
(a) Optical microscopy image of 2 wt % alginate-MA beads at electric potential of 6.0 kV. (b) Plot of bead diameters for 2 wt % alginate and alginate-MA for different applied electric potential; the data are presented as arithmetic mean ± standard deviation calculated from at least three replicates. (c) Bead diameter distribution of 1, 2, and 4 wt % alginate-MA at electric potential of 6.0 kV. (d) Fluorescence microscopy image of 2 wt % fluorescein-labeled alginate-MA beads; beads were formed at electric potential of 6.0 kV.
Alginate-MA was also fluorescently labeled by conjugation with fluoresceinamine for polymer distribution characterization. The bead size distribution was first investigated using fluorescence microscopy. Fluorescently labeled alginate beads possess a similar size distribution and shape as nonlabeled beads (Figure 3d and Supporting Information part 3), which indicates that the comparatively low labeling ratio had insignificant influence on the bead formation process. The distribution of labeled alginate-MA in the beads was examined with confocal laser scanning microscopy. Images were obtained by scanning through a section of the beads. As shown in Supporting Information part 4, in the absence of nongelling ions, the fluorescence intensity of the polymer at the surface was about 20% ± 5% higher than that in the center. This is in accordance with previously reported observations for alginate beads.25−27 This gradient is attributed to the rapid and virtually irreversible gelling characterized by strong binding of the carboxylate groups of the alginate with the cross-linking Ca2+ ions and therefore a gradient in cross-linking density. A higher polymer concentration leads to a higher local concentration of pendant methacrylic acid groups for photopolymerization and therefore ultimately higher cross-linking density.
Swelling and Stability of Hydrogel Beads
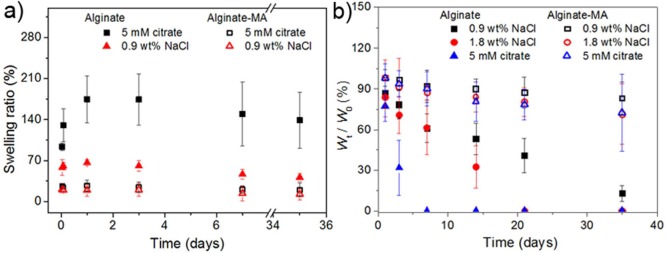
Since the alginate gelling via Ca2+ can be reversed by removing the divalent cations, the hydrogel network is destabilized with time. To investigate this effect, the swelling ratios of alginate-based beads in sodium citrate (5 mM) and sodium chloride solution (0.9 wt %) were evaluated for 35 days. From the circularity calculated from optical microscopy images of beads and SEM images of dried alginate-MA beads (Supporting Information part 5), the volume swelling ratios of beads were calculated based on the assumption that beads were spheres. Alginate and alginate-MA beads swelled, but remained intact, since such low ion concentrations only slightly weaken the polymer network.
Figure 4a shows that the alginate beads swelled fast for the first 2 h and reached equilibrium within 24 h in both citrate and sodium chloride solution. The equilibrium swelling ratio (ESR) of alginate beads was 170% ± 60% and 65% ± 10% for 5 mM citrate and 0.9 wt % NaCl, respectively. Conversely, alginate-MA beads reached swelling equilibrium in 2 h with an ESR of ∼20% for both solutions. For long periods of incubation, the swelling ratio of the beads slightly decreased, which may be due to the dissolution of some lose polymer chains. The behavior of alginate-based beads in solutions with different calcium concentration is shown in Supporting Information part 6.
Figure 4.
(a) Swelling ratio of alginate and alginate-MA beads in 5 mM sodium citrate and 0.9 wt % NaCl solution vs time. The data are presented as arithmetic mean ± standard deviation calculated from at least 50 randomly chosen microbeads from microscopy images. (b) Ratio between dried bead weight at time t and dried bead weight at time 0 in 0.9 wt %, 1.8 wt %, and 5 mM citrate solution. The solution was changed at every measured time point. The data are presented as arithmetic mean ± standard deviation calculated from at least three technical replicates.
Hydrogel beads swelled in 2 mM CaCl2 but shrank at higher concentration (20 and 100 mM) after immersion for 1 day. The shrinking in volume may be due to the rearrangement of alginate chains to form a more rigid network and the osmotic pressure. Alginate beads showed higher ESR in chelation (citrate) or nongelling ions (Na+) solution than in CaCl2 solutions of different concentration. These observations indicate that the properties of ionically cross-linked alginate beads are influenced by the incubation medium. By contrast, alginate-MA beads exhibited limited swelling with swelling ratios not exceeding 30% in 0.9 wt % NaCl, 5 mM sodium citrate, and CaCl2 solutions, which indicates that the swelling properties are mainly governed by the covalent cross-linking.
The stability of the hydrogel beads was studied gravimetrically (Figure 4b). After each measurement, fresh incubation media was added. After changing 5 mM sodium citrate solution 3 times, or 1.8 wt % NaCl solution 5 times, or 0.9 wt % NaCl solution 6 times, the alginate beads were totally dissolved. By contrast, the cross-linked alginate-MA beads remained partially stable even after 6 times media changes independent of the medium. These data are evidence in support of a significant reinforcement of the hydrogel by the photochemically introduced cross-links. The increased cross-link density is thus directly translated to a reduced swelling ratio and increase stability.
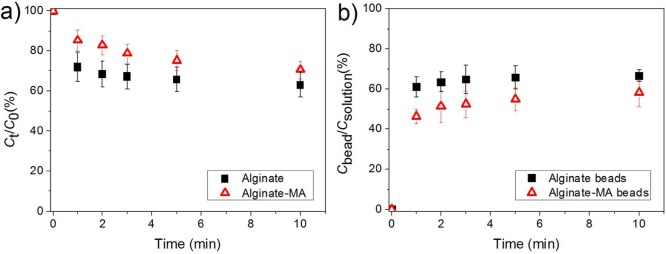
Permeability of Hydrogel Beads for AHLs
The uptake of AHL autoinducers into the alginate-based hydrogel beads was studied by investigating the uptake of a model molecule N-dodecanoyl-l-homoserine lactone-3-hydrazone-fluorescein (FITC-AHL). Figure 5 shows that the concentration of FITC-AHL in the solution decreased and the concentration of FITC-AHL in the beads increased already within 1 min and reached a plateau (diffusion equilibrium) after 2 min, which indicated that FITC-AHL could penetrate into both alginate and alginate-MA beads within minutes. The rapid uptake of FITC-AHL indicates the reinforcement of alginate beads has no significant influence on the permeability of the alginate-based beads for AHL autoinducers. The measured FITC-AHL concentration in the beads was around 50–60% of the concentration in the solution, which may be attributed to the loss of FITC-AHL during the beads separation process.
Figure 5.
Plots of (a) normalized FITC-AHL concentration in the solution against time and (b) ratio between FITC-AHL concentration inside the beads and in the surrounding solution over time; the same volume of alginate or alginate-MA beads was immersed in the FITC-AHL solution. The data are presented as arithmetic mean ± standard deviation calculated from at least three replicates.
Bacteria Encapsulation, Proliferation, and Storage
To test the feasibility of alginate-MA beads used for fabricating biosensor arrays, E. coli pLuxR-GFP, used as model reporter bacteria, was compartmentalized into hydrogel beads. The number of viable bacteria inside beads after encapsulation was investigated first by calculating the encapsulation efficiency. The encapsulation efficiency has been reported before to rely on many factors, including bacteria strain, polymer type, polymer concentration, and especially the encapsulation method.11 The encapsulation efficiency of E. coli pLuxR-GFP into 2 wt % alginate-based beads via electrostatic extrusion method used here was calculated as 76 ± 14% (Supporting Information part 9) by performing the plate count method, which is in the encapsulation efficiency range (from 55% to 83%) reported by Corbo et al.28Figure 6a shows that the beads with bacteria inside have uniform shape and sizes, and the bacteria were homogeneously distributed inside the hydrogel beads. The average diameter slightly increased to 185 μm compared to beads without embedded bacteria, which may be due to the increase of the viscosity of the solution during electrojetting.
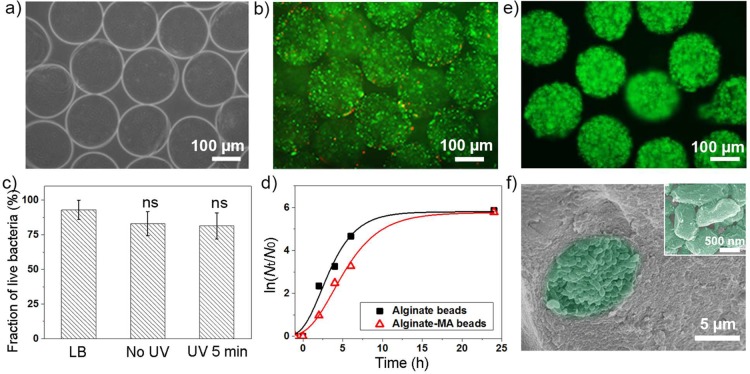
Figure 6.
(a) Optical microscopy image of bacteria loaded alginate-MA beads. (b) Live/dead staining of bacteria inside beads after 5 min UV irradiation, 0.1 wt % Irgacure 2959; green staining indicates viable bacteria, red staining indicates dead bacteria. (c) Fraction of live E. coli pLuxR-GFP before encapsulation, after encapsulation without UV irradiation, and after 5 min UV irradiation; the data are presented as arithmetic mean ± standard deviation calculated from at least three biological replicates, one-way ANOVA. (d) Natural logarithm plot of normalized E. coli pLuxR-GFP population density inside alginate and alginate-MA beads incubated in LB broth. (e) FDA staining of live bacteria entrapped in alginate-MA beads after 24 h incubation in LB media at 37 °C. (f) SEM image of E. coli pLuxR-GFP cluster inside alginate-MA bead; inset is a high-magnification image of entrapped E. coli pLuxR-GFP.
The UV damage on bacteria viability was further studied. Since it is known that short wavelength UV light may damage bacterial DNA, depending on the exposure wavelength and exposure time,29,30 a wavelength of 365 nm was chosen for the photo-cross-linking. Irgacure 2959 was used as initiator because it has been reported to exhibit relatively low cytotoxicity and has been used for cell encapsulation and biomedical applications before.31,32 5 min UV irradiation time and 0.1 wt % Irgacure 2959 were determined as best conditions for the whole study to preserve bacteria vitality. By recording an E. coli pLuxR-GFP growth curve based on OD600 kinetic measurements after UV irradiation, it turned out that E. coli pLuxR-GFP had a longer lag time after UV irradiation, while the generation time was constant (about 25 min) compared to bacteria not exposed to UV irradiation (Supporting Information part 8).
Hydrogel beads containing bacteria could be distinguished from nonbacteria or nonproliferating bacteria beads based on the difference in light scattering properties (Figure 6a). The time-lapse images of alginate-MA beads are presented in Supporting Information part 10. The light scattering properties changed over time, and small clusters were formed, which indicates the entrapped bacteria proliferated inside beads. A live/dead cell assay was used for viability analysis of bacteria after encapsulation and subsequent UV irradiation. In this assay, FDA stains live bacteria in green, while PI can only penetrate the membrane of dead cells and thus stains dead cells in red (Figure 6b). The average percentage of live bacteria in LB media before encapsulation was 93%, while the average ratio of viable bacteria inside the MA-beads without UV irradiation was 83% (Figure 6c). After 5 min UV irradiation, the fraction of viable cells was 81 ± 10%. These results indicate that the low concentration of initiator, the short UV irradiation time, and the long UV wavelength used possess low impact on bacteria viability. It cannot be excluded that the possible elongation of the cell membrane during extrusion has an additional minor effect on the cell viability.33 To check the dynamics of E. coli pLuxR-GFP growth in alginate-based beads, beads containing approximately 300 cells at the beginning of cultivation were incubated in LB medium at 37 °C, and the bacteria population density was recorded over time using a standard plate counting method after bead lysis via citrate. Figure 6d indicates that the growth curves of bacteria in hydrogel beads were similar to those of planktonic bacteria in bulk liquid media, including the exponential-growth phase and the saturation phase. The generation time calculated from maximum growth rate was about 50 min, which was longer than the generation time of planktonic bacteria in bulk LB broth (∼25 min), which might result from the limitation of space, nutrition, oxygen, etc., inside the beads. After 24 h incubation in LB medium at 37 °C, the live E. coli pLuxR-GFP entrapped in alginate-MA beads were stained with a green fluorescent dye (Figure 6e). Strong green fluorescence intensity suggests the large number of live bacteria inside the beads. SEM images of E. coli pLuxR-GFP inside alginate-MA beads showed that the bacteria formed aggregates of cells (Figure 6f), which may be because the segregation of divided cells was confined in the hydrogel networks.
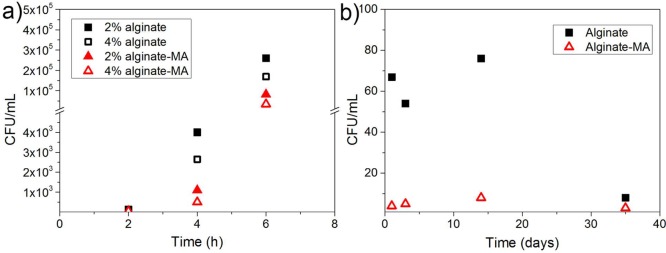
The leaching of E. coli pLuxR-GFP from the alginate-based beads was evaluated by determining the bacteria population density in the suspension of beads at 37 and 4 °C incubation. Before incubation, the free bacteria attached on the bead surface were removed by continuous washing with excess amount of 20 mM CaCl2 solution. The number of bacteria in the washing solution was determined via counting the number of colonies formed on LB agar plates (Table S3 in Supporting Information part 11). After incubation at 37 °C (Figure 7a), the free bacteria in the suspension may originate from two sources: one is the leakage or escape of encapsulated bacteria from alginate-based beads, and the other one is the proliferation of planktonic bacteria that were not initially removed in the LB medium. With increasing concentration of alginate, the number of bacteria in the suspension was reduced. For beads with the same concentration of polymer, the concentration of planktonic bacteria in the alginate bead suspension was higher than that of alginate-MA bead suspension. For storage at 4 °C (Figure 7b), the number of free bacteria in alginate-MA bead suspension was around 10 times lower than that of the alginate bead suspension during the time period investigated. The number of bacteria in the suspension dropped down after 35 days incubation, likely due to the decrease of bacteria viability during storage (see below). All these results confirm that the reinforcement by rapid photo-cross-linking indeed reduced the leakage of encapsulated bacteria significantly.
Figure 7.
E. coli pLuxR-GFP population density in alginate and alginate-MA bead suspensions vs storage time for storage at (a) 37 °C and (b) 4 °C.
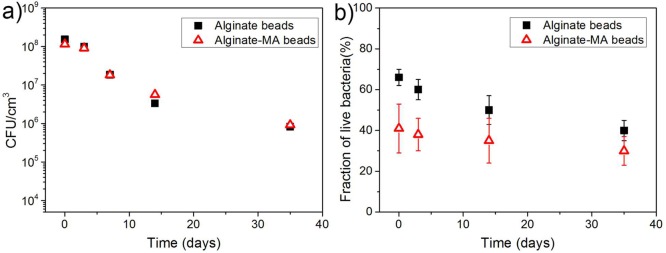
The viability of E. coli pLuxR-GFP inside alginate-based beads during storage at 4 °C in LB medium was investigated via a standard plate counting method and the FDA/PI live/dead cell assay. As shown in Figure 8a, the initial bacterial concentration was around 1 × 108 cfu/cm3 for both alginate and alginate–methacrylate beads. The bacterial concentration inside alginate-based beads decreased and showed a tailing effect with a residual bacteria concentration of 1 × 106 cfu/cm3 after 35 days; after 5–6 days the bacteria concentration was one-half of its initial value. The live cell fraction from live/dead cell staining was shown in Figure 8b. The decrease of live cell fraction for both alginate-based beads during storage is consistence with the results from plate counting method. Both methods proved that a portion of bacteria kept viability after 35 days storage.
Figure 8.
(a) Bacteria number inside alginate-based beads during 4 °C storage determined via plate count method. (b) Fraction of live bacteria inside alginate-based beads during storage at 4 °C determined via live/dead staining; the data are presented as arithmetic mean ± standard deviation calculated from at least three biological replicates.
Characterization of E. coli PLuxR-GFP Encapsulated in Alginate-Based Beads
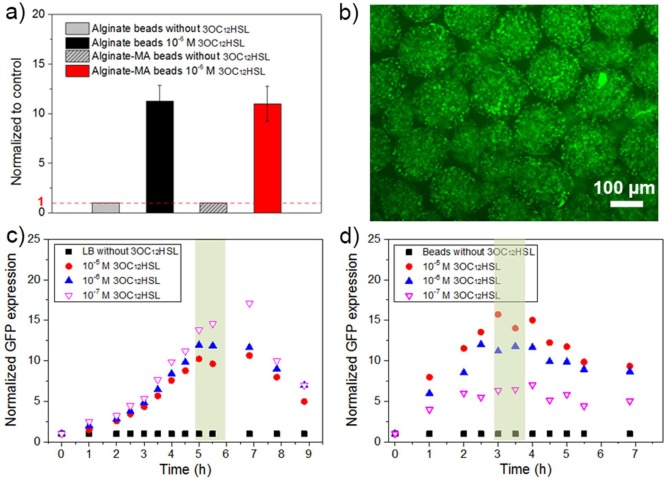
To investigate the response of encapsulated E. coli pLuxR-GFP to autoinducers, 3OC12HSL, as one of the autoinducers used by P. aeruginosa, was added to the bead suspension. The fluorescence intensity of E. coli pLuxR-GFP entrapped in alginate and alginate-MA beads with and without exposition to 3OC12HSL was recorded. In the presence of 1.0 × 10–6 mol/L 3OC12HSL, the fluorescence intensity was 11 ± 1 and 11 ± 2 times higher than in the absence of autoinducers for alginate beads and alginate-MA beads, respectively (Figure 9a). This result shows that the sensing performance of the AHL-reporter strain tested here encapsulated in the alginate-MA matrix is at least as good as in the unmodified alginate matrix. A fluorescence microscopy image of alginate-MA beads is presented in Figure 9b. The entrapped E. coli pLuxR-GFP exhibited a strong green fluorescence in the presence of 3OC12HSL, which indicates the entrapped bacteria sense and respond to 3OC12HSL in surrounding environment. To quantify the autoinducer-response, the fluorescence intensity and the population density (OD600) of bacteria in LB medium and inside alginate-based hydrogel beads were recorded over time with/without exposition to 3OC12HSL. The fluorescence intensity per cell (fluorescence intensity divided by OD600), as a relative measure for GFP expression, was normalized to control cultures (not exposed to 3OC12HSL). As shown in Figure 9c, a sharp increase in fluorescence intensity was observed for bacteria in both LB medium and alginate-MA beads in the presence of 1.0 × 10–5, 1.0 × 10–6, and 1.0 × 10–7 mol/L 3OC12HSL. For bacteria in LB medium, the GFP expression increased when exposed to 3OC12HSL and reached a maximum after 4–5 h incubation with all 3OC12HSL concentrations tested. The fluorescence intensity per cell was around 12 times higher than without induction. Exemplary data after 4 h of incubation from one out of three repeated experiments are shown in Figure S11. When bacteria were encapsulated into alginate-based beads and exposed to 3OC12HSL, the fluorescence intensity per cell increased with time and reached a maximum (6–13 times higher than without induction) around 3 h culture (Figure 9d), so the maximum signal developed even faster than in LB-Medium. Overall, the AHL concentration tested here is in the range of the extracellular concentration of 3OC12HSL (1.0 × 10–6–1.0 × 10–7 mol/L) present close to the site of P. aeruginosa infections, as reported earlier.7,34,35 This implies that the alginate-MA encapsulated E. coli pLuxR-GFP sensing device would be sensitive enough to detect the 3OC12HSL natively produced by P. aeruginosa.
Figure 9.
(a) Fluorescence intensity (normalized to that of alginate-based beads in the absence of autoinducers) of E. coli pLuxR-GFP in alginate-based beads exposed to 1.0 × 10–6 mol/L 3OC12HSL. (b) Fluorescence microscopy image of beads loaded with E. coli pLuxR-GFP in the presence of 1.0 × 10–6 mol/L 3OC12HSL after 3 h incubation. Normalized GFP expression of E. coli pLuxR-GFP in the presence of 1.0 × 10–5, 1.0 × 10–6, and 1.0 × 10–7 mol/L 3OC12HSL (fluorescence intensity divided by OD600 and normalized to that of E. coli pLuxR-GFP culture in the absence of 3OC12HSL which was set to “1” as a reference) over time: (c) in LB medium; (d) inside of alginate-MA beads.
To demonstrate the potential of the system investigated here, a dipstick type biosensor was fabricated by dispersing E. coli pLuxR-GFP loaded alginate-MA beads in a poly(ethylene glycol)-based hydrogel film (30 mm × 10 mm × 5 mm), as shown in Figure 10. The entrapped E. coli pLuxR-GFP in the dipstick type biosensor proliferated and responded to 1.0 × 10–6 mol/L 3OC12HSL with a strong fluorescence signal (Figure 10c).
Figure 10.
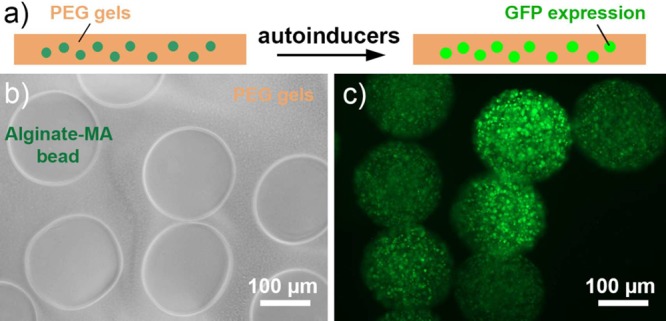
(a) Schematic diagram of dipstick type biosensor. (b) Optical microscopy image of alginate-MA beads loaded with E. coli pLuxR-GFP that were dispersed in PEG-based hydrogels (time 0). (c) Fluorescence microscopy image of dipstick type biosensor in the presence of 1.0 × 10–6 mol/L 3OC12HSL after 3 h incubation.
Conclusion
In summary, the alginate–methacrylate, obtained from reaction with glycidyl methacrylate, was used to fabricate hydrogel beads with controllable diameters in the range of 100–300 μm via an electrostatic extrusion method. The combination of ionic cross-linking and photo-cross-linking affords the formation of stable and robust alginate-based microbeads with decreased swelling ratio, increased stability, and good permeability of dye-labeled autoinducers. The AHL sensing E. coli pLuxR-GFP, as model bacteria, was encapsulated into alginate-MA hydrogel beads with an encapsulation efficiency of 76 ± 14%. The encapsulated bacteria inside the hydrogel beads kept their high viability and proliferated inside the beads with a maximum generation time of 50 min, when exposed to bacteria culture medium. The increased bead stability leads to a 10 times decrease in bacteria leaching from the beads, while the entrapped bacteria kept high viability during storage at 4 °C. Upon exposure to 3OC12HSL in the concentration range of 1.0 × 10–5–1.0 × 10–7 mol/L, the entrapped bacteria exhibited 6–13 times increased fluorescence intensity due to GFP expression as a response to the autoinducer trigger after 3 h. These results and the feasibility test with a prototype dipstick sensor show that the alginate-MA hydrogel beads with encapsulated engineered bacteria possess considerable potential to be used as whole sensors for P. aeruginosa detection.
Acknowledgments
The authors thank Dipl.-Ing. Gregor Schulte for his invaluable technical support and Dr. Yvonne Voss for stimulating discussions.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsami.7b07166.
1H NMR spectra, optical microscopy images of alginate-MA beads, fluorescein-labeled alginate-MA synthesis and the alginate distribution inside beads investigation, SEM image of alginate-MA bead, hydrogel swelling in CaCl2 solutions and 50 mM sodium citrate, FITC-AHL standard curves in Tris buffer and citrate solution, E. coli pLuxR-GFP growth curve and fitting, encapsulation efficiency, time-lapse images of bacteria proliferation inside alginate-MA beads, scheme of the washing process and bacteria density in beads washing solution, fluorescence intensity and OD600 of E. coli pLuxR-GFP in the presence of 3OC12HSL (PDF)
The authors acknowledge funding received for the project from the funding program of the Federal Ministry of Economics and Technology in Germany, “Central Innovation Programme SME” (ZIM, KF2589507SK4), as well as financial support from the EU (ERC project ASMIDIAS, grant no. 279202), the Deutsche Forschungsgemeinschaft (DFG grant no. INST 221/87-1 FUGG), and the University of Siegen.
The authors declare no competing financial interest.
Supplementary Material
References
- Lei Y.; Chen W.; Mulchandani A. Microbial Biosensors. Anal. Chim. Acta 2006, 568, 200–210. 10.1016/j.aca.2005.11.065. [DOI] [PubMed] [Google Scholar]
- Park M.; Tsai S. L.; Chen W. Microbial Biosensors: Engineered Microorganisms as the Sensing Machinery. Sensors 2013, 13, 5777–5795. 10.3390/s130505777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikova E. G.; Shumkova E. S.; Shumkov M. S. Whole-cell Bacterial Biosensors for the Detection of Aromatic Hydrocarbons and Their Chlorinated Derivatives. Appl. Biochem. Microbiol. 2016, 52, 347–357. 10.1134/S0003683816040128. [DOI] [PubMed] [Google Scholar]
- He W.; Yuan S.; Zhong W. H.; Siddikee M. A.; Dai C. C. Application of Genetically Engineered Microbial Whole-cell Biosensors for Combined Chemosensing. Appl. Microbiol. Biotechnol. 2016, 100, 1109–1119. 10.1007/s00253-015-7160-6. [DOI] [PubMed] [Google Scholar]
- Hawver L. A.; Jung S. A.; Ng W.-L. Specificity and Complexity in Bacterial Quorum-sensing Systems. FEMS Microbial. Rev. 2016, 40, 738–752. 10.1093/femsre/fuw014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flickinger S. T.; Copeland M. F.; Downes E. M.; Braasch A. T.; Tuson H. H.; Eun Y.-J.; Weibel D. B. Quorum Sensing Between Pseudomonas Aeruginosa Biofilms Accelerates Cell Growth. J. Am. Chem. Soc. 2011, 133, 5966–5975. 10.1021/ja111131f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeidi N.; Wong C. K.; Lo T. M.; Nguyen H. X.; Ling H.; Leong S. S.; Poh C. L.; Chang M. W. Engineering Microbes to Sense and Eradicate Pseudomonas Aeruginosa, a Human Pathogen. Mol. Syst. Biol. 2011, 7, 521–531. 10.1038/msb.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos P.; Lazarjani H. A.; Poncelet D.; Faas M. M. Polymers in Cell Encapsulation from an Enveloped Cell Perspective. Adv. Drug Delivery Rev. 2014, 67–68, 15–34. 10.1016/j.addr.2013.11.005. [DOI] [PubMed] [Google Scholar]
- Selimović Š.; Oh J.; Bae H.; Dokmeci M.; Khademhosseini A. Microscale Strategies for Generating Cell-Encapsulating Hydrogels. Polymers 2012, 4, 1554–1579. 10.3390/polym4031554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. Y.; Mooney D. J. Alginate: Properties and Biomedical Applications. Prog. Polym. Sci. 2012, 37, 106–126. 10.1016/j.progpolymsci.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbo M. R.; Bevilacqua A.; Speranza B.; Di Maggio B.; Gallo M.; Sinigaglia M. Use of Alginate Beads as Carriers for Lactic Acid Bacteria in a Structured System and Preliminary Validation in a Meat Product. Meat Sci. 2016, 111, 198–203. 10.1016/j.meatsci.2015.10.005. [DOI] [PubMed] [Google Scholar]
- Corbo M. R.; Bevilacqua A.; Gallo M.; Speranza B.; Sinigaglia M. Immobilization and Microencapsulation of Lactobacillus Plantarum: Performances and in vivo Applications. Innovative Food Sci. Emerging Technol. 2013, 18, 196–201. 10.1016/j.ifset.2012.12.004. [DOI] [Google Scholar]
- Mahou R.; Borcard F.; Crivelli V.; Montanari E.; Passemard S.; Noverraz F.; Gerber-Lemaire S.; Bühler L.; Wandrey C. Tuning the Properties of Hydrogel Microspheres by Adding Chemical Cross-linking Functionality to Sodium Alginate. Chem. Mater. 2015, 27, 4380–4389. 10.1021/acs.chemmater.5b01098. [DOI] [Google Scholar]
- Jeon O.; Bouhadir K. H.; Mansour J. M.; Alsberg E. Photocrosslinked Alginate Hydrogels with Tunable Biodegradation Rates and Mechanical Properties. Biomaterials 2009, 30, 2724–2734. 10.1016/j.biomaterials.2009.01.034. [DOI] [PubMed] [Google Scholar]
- Wang X.; Hao T.; Qu J.; Wang C.; Chen H. Synthesis of Thermal Polymerizable Alginate-GMA Hydrogel for Cell Encapsulation. J. Nanomater. 2015, 2015, 1–8. 10.1155/2015/970619. [DOI] [Google Scholar]
- Baier Leach J.; Bivens K. A.; Patrick C. W. Jr.; Schmidt C. E. Photocrosslinked Hyaluronic Acid Hydrogels: Natural, Biodegradable Tissue Engineering Scaffolds. Biotechnol. Bioeng. 2003, 82, 578–589. 10.1002/bit.10605. [DOI] [PubMed] [Google Scholar]
- Li Q.; Wang D.-A.; Elisseeff J. H. Heterogeneous-Phase Reaction of Glycidyl Methacrylate and Chondroitin Sulfate: Mechanism of Ring-Opening-Transesterification Competition. Macromolecules 2003, 36, 2556–2562. 10.1021/ma021190w. [DOI] [Google Scholar]
- Bencherif S. A.; Srinivasan A.; Horkay F.; Hollinger J. O.; Matyjaszewski K.; Washburn N. R. Influence of the Degree of Methacrylation on Hyaluronic Acid Hydrogels Properties. Biomaterials 2008, 29, 1739–1749. 10.1016/j.biomaterials.2007.11.047. [DOI] [PubMed] [Google Scholar]
- Llanes F.; Sauriol F.; Morin F. G.; Perlin A. S. An Examination of Sodium Alginate from Sargassum by NMR Spectroscopy. Can. J. Chem. 1997, 75, 585–590. 10.1139/v97-069. [DOI] [Google Scholar]
- Fertah M.; Belfkira A.; Dahmane E. m.; Taourirte M.; Brouillette F. Extraction and Characterization of Sodium Alginate from Moroccan Laminaria Digitata Brown Seaweed. Arabian J. Chem. 2017, 10, S3707–S3714. 10.1016/j.arabjc.2014.05.003. [DOI] [Google Scholar]
- Balanč B.; Kalušević A.; Drvenica I.; Coelho M. T.; Djordjević V.; Alves V. D.; Sousa I.; Moldão-Martins M.; Rakić V.; Nedović V.; Bugarski B. Calcium-Alginate-Inulin Microbeads as Carriers for Aqueous Carqueja Extract. J. Food Sci. 2016, 81, E65–E75. 10.1111/1750-3841.13167. [DOI] [PubMed] [Google Scholar]
- Gasperini L.; Mano J. F.; Reis R. L. Natural Polymers for the Microencapsulation of Cells. J. R. Soc., Interface 2014, 11, 20140817. 10.1098/rsif.2014.0817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-U.; Hardy M.; Kang S. J.; Barton K.; Adair K.; Mukhopadhyay D. k.; Lee C. Y.; Strano M. S.; Alleyne A. G.; Georgiadis J. G.; Ferreira P. M.; Rogers J. A. High-Resolution Electrohydrodynamic Jet Printing. Nat. Mater. 2007, 6, 782–789. 10.1038/nmat1974. [DOI] [PubMed] [Google Scholar]
- Poncelet D.; Neufeld R. J.; Goosen M. F. A.; Burgarski B.; Babak V. Formation of Microgel Beads by Electric Dispersion of Polymer Solutions. AIChE J. 1999, 45, 2018–2023. 10.1002/aic.690450918. [DOI] [Google Scholar]
- Morch Y. A.; Donati I.; Strand B. L.; Skjak-Braek G. Effect of Ca2+, Ba2+, and Sr2+ on Alginate Microbeads. Biomacromolecules 2006, 7, 1471–1480. 10.1021/bm060010d. [DOI] [PubMed] [Google Scholar]
- Strand B. L.; Morch Y. A.; Espevik T.; Skjak-Braek G. Visualization of Alginate-poly-L-lysine-alginate Microcapsules by Confocal Laser Scanning Microscopy. Biotechnol. Bioeng. 2003, 82, 386–394. 10.1002/bit.10577. [DOI] [PubMed] [Google Scholar]
- Thu B.; Gåserød O.; Paus D.; Mikkelsen A.; Skjåk-Bræk G.; Toffanin R.; Vittur F.; Rizzo R. Inhomogeneous Alginate Gel Spheres: An Assessment of the Polymer Gradients by Synchrotron Radiation-induced X-ray Emission, Magnetic Resonance Microimaging, and Mathematical Modeling. Biopolymers 2000, 53, 60–71. . [DOI] [PubMed] [Google Scholar]
- Corbo M. R.; Bevilacqua A.; Sinigaglia M. Shelf Life of Alginate Beads Containing Lactobacilli and Bifidobacteria: Characterisation of Microspheres Containing Lactobacillus Delbrueckii subsp. Bulgaricus. Int. J. Food Sci. Technol. 2011, 46, 2212–2217. 10.1111/j.1365-2621.2011.02739.x. [DOI] [Google Scholar]
- Mori M.; Hamamoto A.; Takahashi A.; Nakano M.; Wakikawa N.; Tachibana S.; Ikehara T.; Nakaya Y.; Akutagawa M.; Kinouchi Y. Development of a new Water Sterilization Device with a 365 nm UV-LED. Med. Biol. Eng. Comput. 2007, 45, 1237–1241. 10.1007/s11517-007-0263-1. [DOI] [PubMed] [Google Scholar]
- Blatchley E. R.; Dumoutier N.; Halaby T. N.; Levi Y.; Laîne J. M. Bacterial Responses to Ultraviolet Irradiation. Water Sci. Technol. 2001, 43, 179–186. [PubMed] [Google Scholar]
- Fairbanks B. D.; Schwartz M. P.; Bowman C. N.; Anseth K. S. Photoinitiated Polymerization of PEG-diacrylate with Lithium Phenyl-2,4,6-trimethylbenzoylphosphinate: Polymerization Rate and Cytocompatibility. Biomaterials 2009, 30, 6702–6707. 10.1016/j.biomaterials.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorovich N. E.; Oudshoorn M. H.; van Geemen D.; Hennink W. E.; Alblas J.; Dhert W. J. A. The Effect of Photopolymerization on Stem Cells Embedded in Hydrogels. Biomaterials 2009, 30, 344–353. 10.1016/j.biomaterials.2008.09.037. [DOI] [PubMed] [Google Scholar]
- Nguyen D. K.; Son Y. M.; Lee N. E. Hydrogel Encapsulation of Cells in Core-Shell Microcapsules for Cell Delivery. Adv. Healthcare Mater. 2015, 4, 1537–1544. 10.1002/adhm.201500133. [DOI] [PubMed] [Google Scholar]
- Pearson J. P.; Passador L.; Iglewski B. H.; Greenberg E. P. A Second N-acylhomoserine Lactone Signal Produced by Pseudomonas Aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 1995, 92, 1490–1494. 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton T. S.; De Nys R.; Netting A.; Kumar N.; Hentzer M.; Givskov M.; Kjelleberg S. A Novel and Sensitive Method for the Quantification of N-3-oxoacyl Homoserine Lactones Using Gas Chromatography–mass Spectrometry: Application to a Model Bacterial Biofilm. Environ. Microbiol. 2000, 2, 530–541. 10.1046/j.1462-2920.2000.00136.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.