Abstract
FoxO1 and FoxO3a (collectively FoxO1/3a) proteins regulate a wide array of cellular processes, including hepatic gluconeogenesis. Phosphorylation of FoxO1/3a is a key event that determines its subcellular location and transcriptional activity. During glucose synthesis, the activity of FoxO1/3a is negatively regulated by Akt-mediated phosphorylation, which leads to the cytoplasmic retention of FoxO1/3a. However, the nuclear phosphatase that directly regulates FoxO1/3a remains to be identified. In this study, we discovered a nuclear phosphatase, SCP4/CTDSPL2 (SCP4), that dephosphorylated FoxO1/3a and promoted FoxO1/3a transcription activity. We found that SCP4 enhanced the transcription of FoxO1/3a target genes encoding PEPCK1 and G6PC, key enzymes in hepatic gluconeogenesis. Ectopic expression of SCP4 increased, while knockdown of SCP4 inhibited, glucose production. Moreover, we demonstrated that gene ablation of SCP4 led to hypoglycemia in neonatal mice. Consistent with the positive role of SCP4 in gluconeogenesis, expression of SCP4 was regulated under pathophysiological conditions. SCP4 expression was induced by glucose deprivation in vitro and in vivo and was elevated in obese mice caused by genetic (Avy) and dietary (high-fat) changes. Thus, our findings provided experimental evidence that SCP4 regulates hepatic gluconeogenesis and could serve as a potential target for the prevention and treatment of diet-induced glucose intolerance and type 2 diabetes.
Introduction
Gluconeogenesis represents a major metabolic process that maintains the blood glucose level within critical limits. Elevated hepatic gluconeogenesis is crucial in the development of obesity and type 2 diabetes. Understanding the molecular mechanisms underlying the regulation of hepatic gluconeogenesis will facilitate the discovery of potential target for the prevention and treatment of diet-induced glucose intolerance and type 2 diabetes. Glucose-6-phosphatase and PEPCK1 are the two key gluconeogenic enzymes that control glucose homeostasis (1,2). There are multiple transcription factors regulating the expression of these enzymes in liver (3–10). Among them, FoxO1/3a proteins are the first identified transcription factors (11).
FoxO1/3a proteins belong to the forkhead box O subfamily of transcription factors that regulate multiple cellular functions from proliferation, metabolism, life span, and stress resistance to gluconeogenesis (12–14). FoxO1/3a promote gluconeogenesis through activating transcription of genes encoding glucose-6-phosphatase and PEPCK (9,15). FoxO1/3a are phosphoproteins (16,17). The phosphorylation status of FoxO1/3a, which is mainly regulated by insulin signaling pathway during glucose homeostasis, determines the cellular location and transcriptional activity of FoxO1/3a (18). In liver, insulin suppresses hepatic gluconeogenesis through the activation of Akt (19). Activated Akt subsequently phosphorylates FoxO1/3a at multiple sites including Ser256 on FoxO1 and Ser253 on FoxO3a, and the phosphorylated form of FoxO1/3a possesses higher binding affinity with chaperone protein 14-3-3ζ for the cytoplasmic retention and nuclear export of FoxO1/3a (20). As a result, gluconeogenesis is inhibited.
As the counterpart of protein kinases, protein phosphatases play important roles in maintaining the balanced phosphorylation level of target proteins. However, the phosphatases that control the balanced FoxO1/3a phosphorylation are largely unknown. PP2A and a dual-specific phosphatase (DUSP6; MKP3) were reported to be the cytosolic phosphatases for FoxO1 and enhance hepatic gluconeogenesis (21–23). Because Akt is present in both cytosol and nucleus, the following question remains: is there a nuclear phosphatase that antagonizes the action of Akt though dephosphorylating FoxO1/3a in the nucleus? In our effort to identify such a nuclear phosphatase for FoxO1/3a, we performed functional phosphatase library screening and identified SCP4/CTDSPL2 (SCP4 hereafter), a member of the short COOH-terminal phosphatase (SCP) family, as a novel nuclear phosphatase of FoxO1/3a. We further investigated the function of SCP4 in regulating hepatic gluconeogenesis. We found that SCP4 could dephosphorylate FoxO1/3a in vivo and directly in vitro. SCP4-induced dephosphorylation of FoxO1/3a led to the nuclear retention of FoxO1/3a and enhanced expression of gluconeogenic genes. More importantly, depletion of SCP4 caused a reduced glucose production in HepG2 cells and hypoglycemia in mice. Furthermore, wild-type (WT) mice under fasting conditions or obese mice caused by either genetic (Avy) or dietary (high-fat) changes had increased SCP4 expression in liver. These results illustrate the critical function of SCP4 in regulating FoxO1/3a activity and gluconeogenesis.
Research Design and Methods
Animals
SCP4 knockout mice were generated using an SCP4 gene-trap construct (University of California Davis) in which a lacZ cDNA was inserted into the intron 2 of SCP4 genomic DNA. Mice with heterozygous SCP4 deletion were interbred to generate SCP4 homozygous deletion. For high-fat diet experiments, 6-week-old male C57BL/6 were fed on standard chow diet or high-fat diet (41% energy from fat; no. 112734; Dyets) for 5 weeks. Livers of 6-month-old lean control or obese Agouti (Avy) mice were also used for experiments as specified in the text.
Plasmids
cDNAs encoding SCP4–WT, SCP4-DN (SCP4 catalytic inactive mutant Asp-295-Asn), FoxO1, and FoxO3a were cloned into mammalian expression vectors (pRK5; Genentech). The PEPCK1 and G6PC promoter fragments (∼1.5 kb) were obtained by PCR from the genomic DNA of C57BL/6 mouse embryonic fibroblasts (MEFs) and cloned into pGL3 basic luciferase reporter plasmid to generate pPEPCK1-luc and pG6PC-luc. The 14-3-3ζ plasmid was provided by Dr. Dihua Yu from MD Anderson Cancer Center. The SCP4-specific short hairpin RNA (shSCP4) construct was described previously (24).
Antibodies and Reagents
Antibodies against SCP4, FoxO1, phosphorylated (p-)FoxO1, FoxO3a, p-FoxO3a, Akt, p-S473–Akt, LC-3 I/III, GFP, and histone H3 were purchased from Cell Signaling Technology. Antibodies against α-tubulin, β-actin, GAPDH, FLAG tag, and HA tag were from Sigma-Aldrich. Antibodies against MYC tag and Lamin A/C were from Santa Cruz Biotechnology. Active Akt1 protein was purchased from EMD Millipore.
Cell Culture and Transfection
HEK293T, NIH3T3, HeLa, and HepG2 cells were cultured in DMEM with 10% FBS. Cells were transfected with Lipofectamine 2000 transfection reagent (Invitrogen).
Immunoprecipitation and Immunoblotting Analysis
Cells were transfected with expression plasmids and harvested 24–48 h after transfection. Coimmunoprecipitation (co-IP) and immunoblotting (IB) were carried out as described (24).
In Vitro Dephosphorylation Assay
FLAG-FoxO1/3a proteins were obtained from in vitro translation using TNT Transcription/Translation System (Promega) and phosphorylated by active Akt1 to generate p-FoxO1/3a. GST fusion proteins of SCP4-WT and D295N mutant were purified from Escherichia coli by Glutathione Sepharose 4B beads. p-FoxO1/3a and SCP4 proteins were incubated in the in vitro phosphatase reaction buffer (40 mmol/L Tris-HCl, pH 8.0, 20 mmol/L KCl, 30 mmol/L MgCl2, and 2 mmol/L dithiothreitol) at 37°C for 1 h. Phosphorylation level of FoxO1/3a was analyzed by IB.
In Vitro Protein Binding Assay
Purified GST-SCP4 protein absorbed to Glutathione Sepharose 4B beads was incubated with in vitro translated FoxO1/3a for 2 h in the binding buffer (0.1% Nonidet P-40, 120 mmol/L NaCl, 50 mmol/L Tris-HCl, pH 7.5, and 2 mmol/L EDTA). SCP4 and FoxO1/3a association was determined by IB.
Luciferase Reporter Assay
Cells were cotransfected with expression plasmids and reporter plasmids. At 36 h after transfection, cells were harvested, and luciferase activity was measured as described previously (25).
Immunofluorescence
Cells grown on coverslips were fixed with 4% formaldehyde for 10 min, permeabilized with 0.5% Triton X-100 for 15 min, and blocked with 5% BSA/PBS for 1 h. Cells were then probed with primary antibody followed by incubation with fluorescence-conjugated secondary antibody and visualized under Zeiss Axioplan II microscope (Carl Zeiss).
Quantitative RT-PCR
Total RNAs were isolated using TRIzol Reagent (Invitrogen). Quantitative RT-PCR (qRT-PCR) was performed using Power SYBR Green PCR Master Mix (Applied Biosystems). The sequences of qRT-PCR primers used in this study are: human (h)-PEPCK1 (5′-ACGGCTCTGAGGAGGAGAAT-3′ and 5′-GGGTGACGATAACCGTCTTG-3′); h-G6PC (5′-CCTCAGGAATGCCTTCTACG-3′ and 5′-CCAAAACCCACCAGTATGGA-3′); h–glycogen synthase 1 (Gys1) (5′-GAGGATGAATTCGACCTGGA-3′ and 5′-CCCCACCAGGAAGTAGTTGT-3′); h/mouse (m)-18s (5′-ATTGACGGAAGGGCACCACC-3′ and 5′-GCCAGAGTCTGTTCGTTATC-3′); h/m-SCP4 (5′-GCTGGTAGTTATGAAATGACAAAT-3′ and 5′-GTTGAAGACAGGCGAAAATATGG-3′); m-Insulin (5′-TGGAGGCTCTCTACCTGGTG-3′ and 5′-GGTCTGAAGGTCACCTGCTC-3′); m-Glucagon (5′-ACTTTGTGGCTGGATTGCTT-3′ and 5′-GTGAGTGGCGTTTGTCTTCA-3′); m-Gys1 (5′-TTGGAAGACTGGGAGGATGA-3′ and 5′-CATTCATCCCCTGTCACCTT-3′); m-Gys2 (5′-GCTGAAAGACTCCCTGTGGA-3′ and 5′-AATTCTCTCCCCACTCATCG-3′); m-Pyg1 (5′-GATCCGTACACAGCAGCACT-3′ and 5′-CTCATCGCAGGCATTTTGTA-3′); m-FoxO1 (5′-ACGAGTGGATGGTGAAGAGC-3′ and 5′-TGCTGTGAAGGGACAGATTG-3′); m-FoxO3a (5′-ACAAACGGCTCACTTTGTCC-3′ and 5′-CTGTGCAGGGACAGGTTGT-3′); m-CRTC2 (5′-CTTCGAGGAGGTGATGATGG-3′ and 5′-AACATTGGGCAGAGAACCAC-3′); m-HNF4α (5′-AAATGTGCAGGTGTTGACCA-3′ and5′-CTCGAGGCTCCGTAGTGTTT-3′); m-G6PC (5′-TCGGAGACTGGTTCAACCTC-3′ and 5′-ACAGGTGACAGGGAACTGCT-3′); and m-PEPCK1 (5′-TGTCGGAAGAGGACTTTGAGA-3′ and 5′-CACATAGGGCGAGTCTGTCA-3′).
Cell Fractionation
HepG2 cell pellet (5 × 106) was suspended in 300 μL buffer A (250 mmol/L sucrose, 20 mmol/L HEPES, pH 7.4, 10 mmol/L KCl, 1.5 mmol/L MgCl2, and 1 mmol/L EDTA) for 10 min. The nuclear pellet was obtained by centrifugation at 1,200g for 5 min with the supernatant as cytoplasmic fraction. The nuclear pellet was washed twice with buffer A, resuspended in buffer B (20 mmol/L HEPES, pH 7.9, 1.5 mmol/L MgCl2, 300 mmol/L NaCl, and 0.2 mmol/L EDTA), sonicated briefly till sample was no longer viscous, and used as nuclear fraction.
Glucose Production Assay
HepG2 cells were washed with PBS and incubated in glucose-free and phenol red–free DMEM containing 20 mmol/L sodium lactate and 2 mmol/L sodium pyruvate for 13 h. A total of 100 mmol/L glucagon was added to culture medium 3 h before sample collection. The culture medium was then collected, and glucose level was measured by Amplex Red Glucose/Glucose Oxidase Assay Kit (Invitrogen). Glucose level was normalized with cellular protein concentration.
Blood Glucose Measurement
Blood was collected from the tail of mice. Blood glucose concentration was measured by Contour Blood Glucose Meter (Ascensia Diabetes Care).
Glucose and Pyruvate Rescue Injection
Embryonic day (E)18.5 embryos were obtained by caesarean section and injected with 20 μL of 10% glucose or 20 μL of 10% pyruvate every 6–8 h until end point.
Periodic Acid Schiff Staining
Paraffin sections of liver were oxidized with 0.5% periodic acid solution and stained with Schiff reagent following the manufacturer’s instruction (periodic acid Schiff kit; Sigma-Aldrich).
Fasting/Refeeding Test
Eight C57BL/6 mice were randomly grouped and subjected to standard chow diet, fasted, or fasted followed by refeeding, as indicated in the text. Livers from these mice were collected for protein and RNA extraction.
Results
SCP4 Is a FoxO1/3a Phosphatase
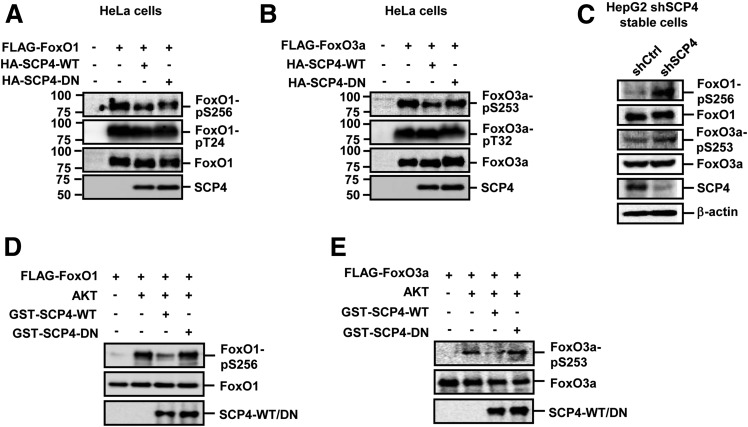
In an effort to identify FoxO1/3a phosphatase(s), we screened a phosphatase library of 40 protein serine/threonine phosphatases (PS/TPs) including 18 protein phosphatases Mg2+- or Mn2+-dependent, 13 phosphoprotein phosphatases, 5 small COOH-terminal serine/threonine phosphatases (FCP/SCPs), and 4 DUSPs for their ability to reduce the phosphorylation level of Ser256 in FoxO1 and Ser253 in FoxO3a. The PS/TP cDNA library was previously constructed and reported by our laboratory (26). Of 40 PS/TPs, SCP4, a member of FCP/SCP family, could reduce the phosphorylation level of FoxO1/3a (data not shown). This SCP4 effect was confirmed in HeLa cells in which expression plasmids for SCP4 and FoxO1 or FoxO3a were cotransfected. We have also generated an SCP4 mutant (SCP4-DN) in which the amino acid residue D295 was mutated to N295, which resulted in the loss of SCP4 phosphatase activity. As shown in Fig. 1A and B, expression of SCP4, but not the SCP4-DN mutant, decreased the phosphorylation level of FoxO1-Ser256 and FoxO3a-Ser253. As a specificity control, phosphorylation levels of FoxO1-Thr24 and FoxO3a-Thr32 were also examined. We found that SCP4 had no effects on Thr24/Thr32 phosphorylation (Fig. 1A and B). Therefore, SCP4 specifically regulates FoxO1/3a Ser256/253 phosphorylation, and this SCP4 effect requires its phosphatase activity. Conversely, efficient knockdown of SCP4 expression, which was achieved by stable expression of shSCP4 (Fig. 1C), increased the phosphorylation levels of FoxO1/3a in HepG2 cells (Fig. 1C). As controls, depletion of SCP4 had no effect on the total protein levels of FoxO1/3a. These results suggest that SCP4 regulates FoxO1/3a phosphorylation under physiological conditions.
Figure 1.
SCP4 dephosphorylates p-FoxO1/3a in vivo and in vitro. A: SCP4 dephosphorylates p-FoxO1. HeLa cells were cotransfected with FLAG-FoxO1 and HA–SCP4-WT or HA–SCP4-DN. Levels of FoxO1–p-S256, FoxO1–p-T24, total FoxO1, and SCP4 were determined by IB. B: SCP4 dephosphorylates p-FoxO3a. HeLa cells were cotransfected with FLAG-FoxO3a and HA–SCP4-WT or HA–SCP4-DN. Levels of FoxO3a–p-S253, FoxO3/a–p-T32, total FoxO3a, and SCP4 were determined by IB. C: Knockdown of SCP4 increases endogenous p-FoxO1 and p-FoxO3a levels. Cells lysates of shSCP4 and control HepG2 cells were subjected to IB to examine protein levels of FoxO1–p-S256, FoxO1, FoxO3a–p-S253, FoxO3a, and SCP4. β-Actin blot was used as a loading control. D: SCP4 directly dephosphorylates p-FoxO1 in vitro. SCP4-WT and SCP4-DN were purified as GST fusion protein from E. coli and absorbed to Glutathione Sepharose 4B beads. p-FoxO1 was obtained from in vitro translation of FLAG-FoxO3a, phosphorylated by active Akt1 protein, and immunoprecipitated by anti-FLAG antibody conjugated on agarose beads. p-FoxO1 was mixed with GST–SCP4-WT or GST–SCP4-DN in the phosphatase reaction buffer. Levels of FoxO1–p-S256 were determined by IB. E: SCP4 directly dephosphorylates p-FoxO3a in vitro. SCP4-WT, SCP4-DN, and p-FoxO3a were obtained and assayed as described in D. Levels of FoxO3a–p-S253 were determined by IB.
To distinguish whether the effect of SCP4 on FoxO1/3a phosphorylation is direct or through another kinase or phosphatase, we performed an in vitro dephosphorylation assay. As the enzyme, recombinant SCP4 or SCP4-DN proteins were purified as GST-fusion proteins. As the substrate, p-FoxO1/3a was produced by first incubating in vitro translated FLAG-FoxO1/3a with active recombinant Akt1 and then being immunopurified by anti-FLAG antibody mobilized agarose beads. In the in vitro dephosphorylation assays, SCP4, but not SCP4-DN, could decrease the levels of FoxO1–p-Ser256 and FoxO3a–p-Ser253, indicating the direct phosphatase activity of SCP4 toward phospho-FoxO1/3a (Fig. 1D and E).
SCP4 Directly Interacts With FoxO1/3a
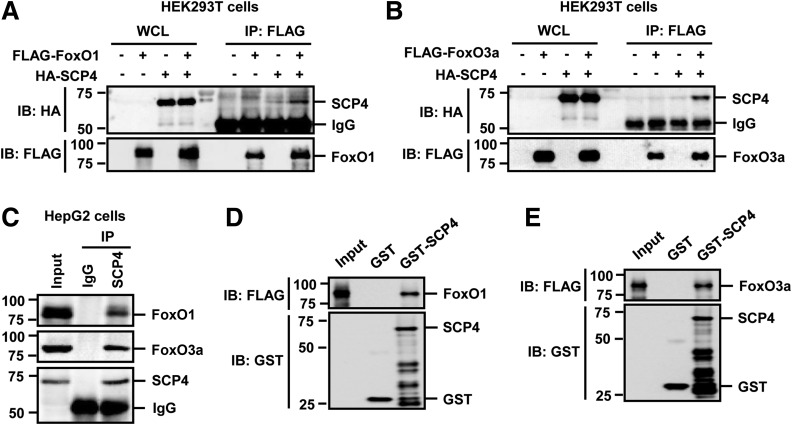
Because a physical interaction is generally present between an enzyme and its substrates, we performed a co-IP experiment to examine the association between SCP4 and FoxO1/3a. FLAG-tagged FoxO1 or FoxO3a was cotransfected with HA-tagged SCP4, and an anti-FLAG antibody was used to IP FoxO1/3a. The coprecipitation of SCP4 was clearly detected by anti-HA IB (Fig. 2A and B). Moreover, the SCP4–FoxO1/3a interaction was detected with both proteins at the endogenous level in HepG2 cells (Fig. 2C). The interaction between SCP4 and FoxO1/3a was also direct as shown in an in vitro binding assay in which GST-SCP4 fusion protein could directly bind to in vitro translated FoxO1 (Fig. 2D) and FoxO3a (Fig. 2E). Taken together, we have identified SCP4 as a FoxO1/3a phosphatase that directly interacts with and dephosphorylates FoxO1/3a.
Figure 2.
SCP4 physically interacts with FoxO1/3a. A: SCP4 co-IP with FoxO1. HEK293T cells were transfected with FLAG-FoxO1 and HA-SCP4. Co-IP was carried out by using an anti-FLAG antibody. Levels of indicated proteins in whole cell lysates (WCL) and IP products were analyzed by IB. B: SCP4 co-IP with FoxO3a. HEK293T cell transfection, co-IP, and IB were done as described in A. C: SCP4 interacts with FoxO1/3a at endogenous levels in vivo. Cell lysates of HepG2 cells were IP with an anti-SCP4 antibody or control IgG antibody. The immunocomplexes and input were analyzed by IB with indicated antibodies. D: SCP4 directly binds to FoxO1 in vitro. In vitro binding assay was carried out with purified GST-SCP4 absorbed on Glutathione Sepharose beads and in vitro translated FoxO1. IB was carried out with indicated antibodies. E: SCP4 directly binds to FoxO3a in vitro. The in vitro binding assay and IB were carried out as described in D.
SCP4 Attenuates FoxO1/3a Nuclear Export
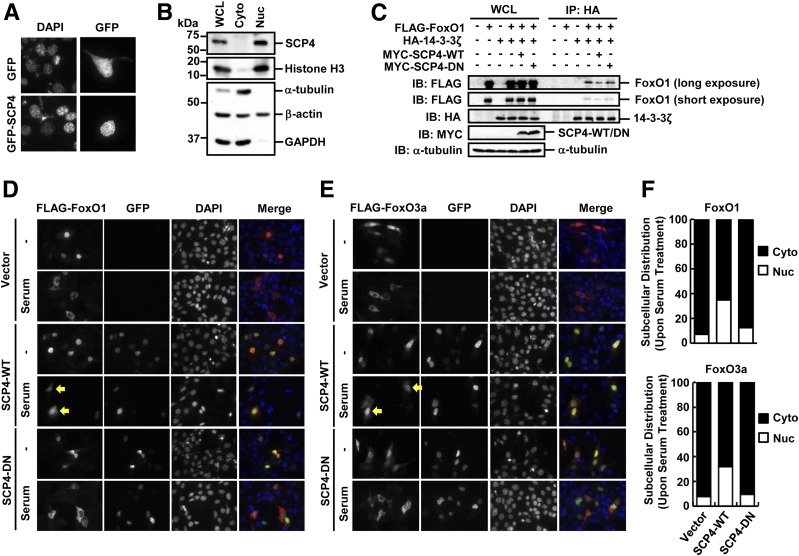
Akt-mediated phosphorylation of FoxO1/3a plays a critical role in regulating FoxO1/3a transcription activity, as phosphorylation of FoxO1/3a retains FoxO1/3a in the cytoplasm and represses FoxO1/3a-mediated transcription (20). Akt is localized in both the cytoplasm and nucleus. It prevents the nuclear import and promotes the nuclear export of FoxO1/3a (16–18). Along the same line, dephosphorylation of FoxO1/3a by phosphatases should counteract Akt’s action. After having established the role of SCP4 as a FoxO1/3a phosphatase, we were wondering in which subcellular compartment SCP4 exerts its function. To answer this, we examined the subcellular localization of SCP4. We found that transiently expressed GFP-tagged SCP4 was mainly localized in the nuclei of NIH3T3 cells (Fig. 3A). Furthermore, cell fractionation approach showed that endogenous SCP4 was only detected in the nuclear fraction (Fig. 3B), indicating that SCP4 is a nuclear protein and dephosphorylates FoxO1/3a in the nucleus.
Figure 3.
SCP4 is a nuclear protein and inhibits nuclear export of FoxO1/3a. A: NIH3T3 cells were transiently transfected with GFP-SCP4 or control GFP plasmids. The GFP and GFP-SCP4 proteins were visualized by fluorescence microscope. B: SCP4 is localized in the nucleus. Subcellular fractionation analysis was carried out in HepG2 cells. Histone H3 was used as a nuclear (Nuc) marker, and α-tubulin and GAPDH were used as cytoplasmic (Cyto) fraction markers. C: SCP4 attenuates the binding of FoxO1 to 14-3-3ζ. Co-IP experiments between FoxO1 and 14-3-3ζ were performed in the presence or absence of SCP4 coexpression. HA antibody was used to immunoprecipitate 14-3-3ζ, and the coprecipitation of FoxO1 was determined by IB with FLAG antibody. D: SCP4 blocks the nuclear export of FoxO1. NIH3T3 cells were transfected with FLAG-FoxO1 and GFP–SCP4-WT or DN mutant. After serum starvation or serum addition, cells were fixed and immunostained with anti-FLAG (red, for FoxO1) and anti-GFP (green, for SCP4) antibodies. Yellow arrows indicate the nuclear retention of FoxO1 by SCP4-WT. DNA was stained with DAPI. The subcellular location of FoxO1 after serum addition was counted in SCP4-WT– or SCP4-DN–expressing cells. E: SCP4 blocks the nuclear export of FoxO3a. Experiments were carried out in NIH3T3 cells as described in D. Yellow arrows indicate the nuclear retention of FoxO3a by SCP4-WT. F: Quantification of FoxO1 and FoxO3a subcellular distribution. Percentage of nuclear (Nuc) or cytosolic (Cyto) FoxO1 to total FoxO1-positive cells is calculated and shown in the top panel, whereas that of FoxO3a is shown in the bottom panel. WCL, whole cell lysate.
The p-Ser256 in FoxO1 and p-Ser253 in FoxO3a provide the docking sites for 14-3-3ζ binding and help the cytoplasmic retention/degradation of FoxO1/3a (27,28). If SCP4 dephosphorylates FoxO1/3a in the nucleus, we expected that SCP4 would enhance the nuclear accumulation of FoxO1/3a and attenuate the interaction between FoxO1/3a and 14-3-3ζ. Indeed, co-IP experiments demonstrated a decreased interaction between FoxO1/3a and 14-3-3ζ in the presence of overexpression of SCP4-WT (Fig. 3C and data not shown). Consistently, immunofluorescence staining in NIH3T3 cells showed that SCP4 increased the nuclear localization of FoxO1/3a (Fig. 3D–F). In control cells, FoxO1/3a were accumulated in the nucleus under serum starvation and translocated into the cytoplasm with the addition of serum. However, ectopic expression of SCP4, but not SCP4-DN, blocked this serum-induced nuclear export of FoxO1/3a.
SCP4 Enhances FoxO1/3a Transcription Activity and Promotes Glucose Production
Because FoxO1/3a are transcriptional factors that regulate multiple signaling pathways (12–14), we focused our study of SCP4 regulation of FoxO1/3a during hepatic gluconeogenesis. PEPCK1 and G6PC are the two key enzymes in gluconeogenesis (1,2) and direct targets of FoxO1/3a (9,15). FoxO1/3a (nonphosphorylated form) activates the transcription of PEPCK1 and G6PC, which in turn promotes hepatic glucose synthesis. In contrast, insulin signal activates Akt kinase activity, which phosphorylates FoxO1/3a and represses the function of FoxO1/3a (9,15–17,20).
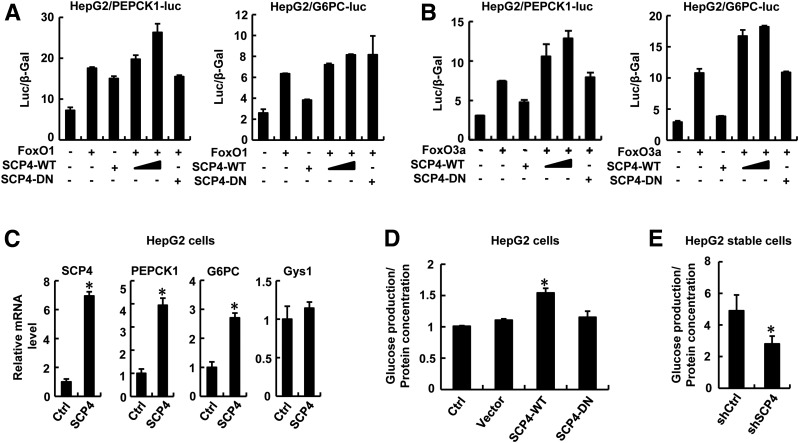
We reasoned that as a phosphatase for FoxO1/3a, SCP4 would enhance the transcription of PEPCK1 and G6PC and thus promote glucose production. To determine the effect of SCP4 on PEPCK1 and G6PC transcription, we first constructed two luciferase reporters, PEPCK1-luc and G6PC-luc, which contained the FoxO binding sites in PEPCK1 or G6PC promoter, respectively, and analyzed the effects of FoxO1/3a and SCP4 on reporter expression. As shown in Fig. 4A and B, FoxO1 or FoxO3a activated PEPCK1 and G6PC promoters. Coexpression of SCP4, but not SCP4-DN, further enhanced the FoxO-induced reporter activity. Next, qRT-PCR analysis showed that stable expression of SCP4 in HepG2 cells enabled a marked increase in the mRNA levels of PEPCK1 and G6PC (Fig. 4C). On the contrary, SCP4 failed to enhance the transcription of Gys1, an enzyme for glycogen synthesis (Fig. 4C).
Figure 4.
SCP4 enhances FoxO1/3a transcription activity and promotes glucose production. A: SCP4 potentiates FoxO1 in activating the promoters of FoxO target genes. HepG2 cells were transfected with PEPCK1-luc or G6PC-luc reporter plasmid together with FoxO1, SCP4-WT, or SCP4-DN, and cell lysates were collected for luciferase assays. B: SCP4 potentiates FoxO3a in activating the promoters of FoxO target genes. Cell transfection, luciferase assay, and data analysis were done as described in A, except that FoxO3a instead of FoxO1 was used in the experiments. C: SCP4 enhances the expression of gluconeogenic genes but not glycogen synthesis genes. In SCP4-overexpressing HepG2 cells, the transcript levels of PEPCK1, G6PC, and Gys1 were measured by qRT-PCR. Measurement of SCP4 RNA level was also shown as a sample control. D: Overexpression of SCP4 promotes glucose production. Culture media from SCP4-WT, SCP4-DN, or control (Ctrl) vector–transfected HepG2 cells were collected for glucose production assays. E: SCP4 depletion decreases glucose production. Culture media of HepG2 cells expressing shSCP4 or shCtrl were collected for glucose production assays. All data are shown as means ± SEM; n = 3. *P < 0.05.
We further directly examined the effect of SCP4 on the glucose level in HepG2 cells. In consistence with the role of SCP4 in controlling PEPCK1 and G6PC transcription, SCP4 enhanced glucose production (Fig. 4D), whereas depletion of SCP4 by shSCP4 decreased glucose production (Fig. 4E). Taken together, these data support the notion that SCP4 enhances the transcription activity of FoxO1/3a and promotes glucose synthesis.
Neonatal SCP4-Null Mice Display Hypoglycemia
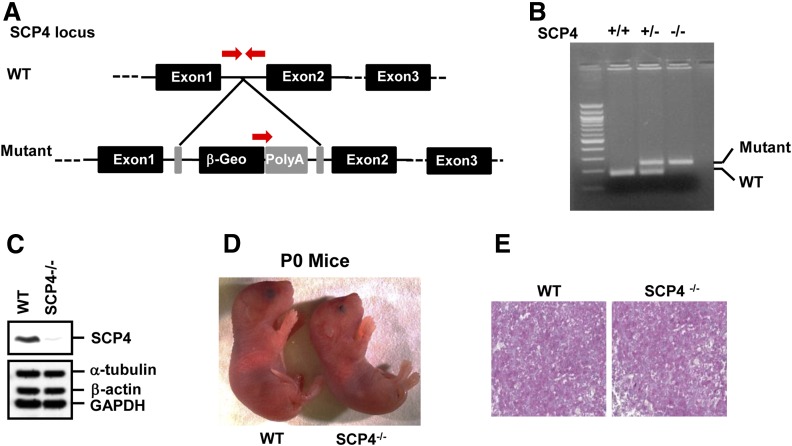
We further investigated the physiological function of SCP4 in regulating hepatic gluconeogenesis by using an SCP4 knockout mouse model. SCP4 knockout mice were generated with commercially available SCP4 gene-trap construct in which a lacZ cDNA was inserted into the intron 2 of the SCP4 locus (Fig. 5A). The homozygous deletion of the SCP4 gene and absence of SCP4 expression were confirmed by PCR (Fig. 5B) and IB (Fig. 5C), respectively. SCP4−/− mice died within 24 h after birth. There was no apparent gross phenotypic abnormality in SCP4−/− neonates, except they were slightly smaller compared with their WT littermates (Fig. 5D). Because SCP4 regulates glucose production at cellular level (Fig. 4), we examined the blood glucose and glycogen levels in the neonates immediately after birth. WT and SCP4−/− littermates exhibited comparable blood glucose level (data not shown). No significant difference in the hepatic glycogen level was observed between SCP4−/− and WT controls (Fig. 5E), suggesting that the glycogen storage was also normal in SCP4−/− mice at the perinatal stage. This is not surprising considering that the blood glucose level in perinatal pups is mainly maintained by the mothers, and the neonates can sustain their blood glucose level for the first 15–30 min with the maternally derived glucose (29). After the stop of transplacental nutrient supply, neonates have to compensate with the cessation of nutrient until supply can be restored with milk (30). During this starvation period, glycogenolysis and gluconeogenesis are important to support the glucose homeostasis (29).
Figure 5.
Generation and characterization of SCP4 knockout mice. A: Generation of SCP4 knockout mice. Constructs of the WT allele and the disrupted allele are shown. Exons are represented by black boxes. Arrows represent the PCR primers used for genotyping. B: PCR genotyping of mouse tail DNA. Tail DNA was extracted from F2 littermates of heterozygote crosses and subjected to PCR using 5′ primers specific for the WT or knockout allele and a common 3′ primer as shown in A. C: SCP4 protein is undetectable in SCP−/− MEFs. MEF lysates isolated from E13.5 WT and SCP4−/− embryos were subjected to IB with indicated antibodies. D: Images of WT and SCP4−/− mice at birth (P0). SCP4−/− mouse was smaller than WT littermate (left). E: Liver glycogen storage is not affected by SCP4 gene deletion. The E18.5 liver sections of WT and SCP4−/− mice were stained with periodic acid Schiff to determine glycogen levels.
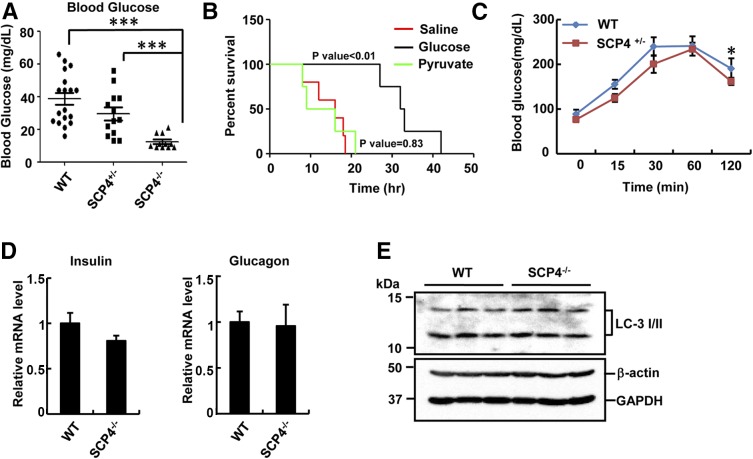
To mimic the nutrient cessation, we subjected the neonates (19 of SCP4 WT, 13 of heterozygous SCP4+/− and 12 of homozygous SCP4−/−) from caesarean delivery to starvation for 12 h, and their blood glucose levels were measured. Consistent with the promoting role of SCP4 in glucose production, blood glucose levels in SCP4−/− neonates were significantly lower than those in their WT littermates (Fig. 6A). Injection of glucose to SCP4−/− neonates could extend their life up to 42 h, indicative of a partial rescue by external glucose (Fig. 6B). Moreover, saline (control) or pyruvate, an upstream substrate for glucose synthesis, did not rescue the lethality of SCP4−/− mice (Fig. 6B). These in vivo data further confirmed our findings from in vitro studies that SCP4 plays critical role in maintaining glucose level. In supporting this, we also observed lower gluconeogenesis rate after 48-h starvation in 6-week-old SCP4 heterozygous mice compared with that in WT littermates (Fig. 6C).
Figure 6.
SCP4−/− neonates display hypoglycemia. A: Blood glucose levels are decreased in SCP4−/− mice. E18.5 WT (n = 19), SCP4+/− (n = 13), and SCP4−/− (n = 12) mice were starved for 12 h. Blood from animal tails was collected for glucose measurement. B: Glucose but not pyruvate injection can extend the survival time of SCP4−/− mice. SCP4−/− mice at E18.5 were injected with saline alone (n = 5), glucose-containing saline (n = 4), or pyruvate-containing saline (n = 4) every 6–8 h. The survival status of mice was monitored every hour. C: Gluconeogenesis rate is decreased in adult SCP4+/− mice. Six-week-old heterozygous SCP4+/− and WT littermates were starved for 48 h and then injected with sodium pyruvate. Their blood glucose levels were measured at indicated time points. D: There is no significant difference in the mRNA levels of insulin and glucagon between SCP4−/− and WT littermates. Total RNAs were extracted from the pancreas of E18.5 SCP4−/− and WT mice after 12-h starvation and subjected to qRT-PCR to examine the expression levels of insulin and glucagon. Data are shown as means ± SEM; n = 6. E: Autophagy is not affected in SCP4−/− mice. Autophagy marker LC-3 I/II was determined by IB in the liver lysates of E18.5 SCP4−/− and WT mice after 12 h starvation. *P < 0.05; ***P < 0.001.
To exclude the possibility that SCP4 deletion causes abnormal insulin or glucagon secretion to affect glucose level, we examined the insulin and glucagon expression in pancreas by qRT-PCR. No significant difference was observed between SCP4−/− and WT littermates (Fig. 6D). Because autophagy is important for the fasting neonates to generate substrates (e.g., glycogenic amino acids essential for gluconeogenesis) (31), we also investigated the process of autophagy to exclude the possibility of the substrate shortage during gluconeogenesis. Immunoblotting analysis of autophagy marker LC-3 showed that autophagy was normal in the liver of SCP4−/− mice (Fig. 6E). Thus, we believe that the lower glucose level in SCP4−/− mice is the result of decreased glucose synthesis.
PEPCK1 and G6PC Expression Are Downregulated in SCP4−/− Mouse Livers
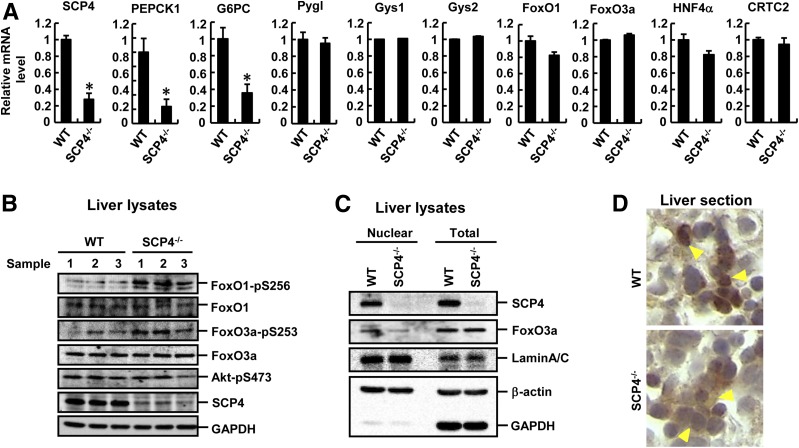
To directly demonstrate that hepatic glucose synthesis is attenuated in SCP4−/− mice, we used qRT-PCR to examine the expression level of enzymes essential for glucose homeostasis. We found that the expression of glycogenolysis- and glycogenesis-related genes (e.g., glycogen phosphorylase liver form [Pygl] and Gys1/2) was not affected in SCP4−/− mice, but rather the expression of the glucose synthesis genes PEPCK1 and G6PC was decreased (Fig. 7A). We also examined the expression levels of FoxO1, FoxO3a, HNF4α, and CRTC2, the upstream transcriptional factors for PEPCK1 and G6PC, and found that their expression was comparable between SCP4−/− and WT littermates (Fig. 7A). These observations led to a conclusion that ablation of SCP4 gene attenuated the hepatic glucose production mainly because of the decreased PEPCK1 and G6PC expression. Thus, SCP4 is an important regulator in gluconeogenesis in vivo.
Figure 7.
Ablation of SCP4 attenuates the nuclear localization of FoxO1/3a and downregulates the expression of PEPCK1 and G6PC. A: Transcription of gluconeogenic genes is diminished in SCP4−/− mice. Total RNAs were extracted from the livers of E18.5 WT and SCP4−/− mice after 12-h starvation and subjected to qRT-PCR to examine the expression levels of blood glucose homeostasis–related genes, glycogen phosphorylase liver form (Pygl), Gys1, Gys2, PEPCK1, and G6PC. The mRNA levels of upstream regulators FoxO1, FoxO3a, HNF4α, and CRTC2 were also examined. The mRNA level of SCP4 was also shown as a sample control. Data are shown as means ± SEM; n = 6. B: FoxO1–p-S256 and FoxO3a–p-S253 levels are higher in the livers of SCP4−/− mice than that in WT littermates. Liver lysates from E18.5 WT (n = 3) and SCP4−/− mice (n = 3) after 12-h starvation were collected and subjected to IB with indicated antibodies. C: Nuclear FoxO3a is reduced in SCP4−/− mouse livers. Subcellular distribution of FoxO3a was analyzed by cell fractionation assays in liver samples collected as described in B. Lamin A/C, GAPDH, and β-actin were used as subcellular markers. D: Nuclear FoxO3a is reduced in SCP4−/− mice. Immunohistochemical staining of FoxO3a was carried out in the paraffin sections of livers collected as described in B. FoxO3a nuclear staining signal was weaker in SCP4−/− than that in WT, as indicated by yellow arrowheads. *P < 0.05.
Consistent with SCP4-catalyzed dephosphorylation of FoxO1/3a observed in cell-based assays (Fig. 1), the phosphorylation levels of FoxO1/3a were markedly higher in the livers of SCP4−/− mice than that in WT littermates (Fig. 7B). This increased FoxO1/3a phosphorylation was not caused by increased Akt activity, as the level of active Akt (Akt–p-S473) was comparable between SCP4−/− and WT littermates (Fig. 7B). When examining the subcellular localization of FoxO proteins in SCP4−/− mice by fractionation assays and immunohistochemistry staining, we found that the level of nuclear FoxO1/3a was lower in the livers of SCP4−/− mice than in WT littermates (Fig. 7C and D), confirming that the nuclear localization of FoxO1/3a was impaired, and the subsequent glucose synthesis was attenuated in SCP4−/− mice.
SCP4 Expression Is Regulated by Metabolic States
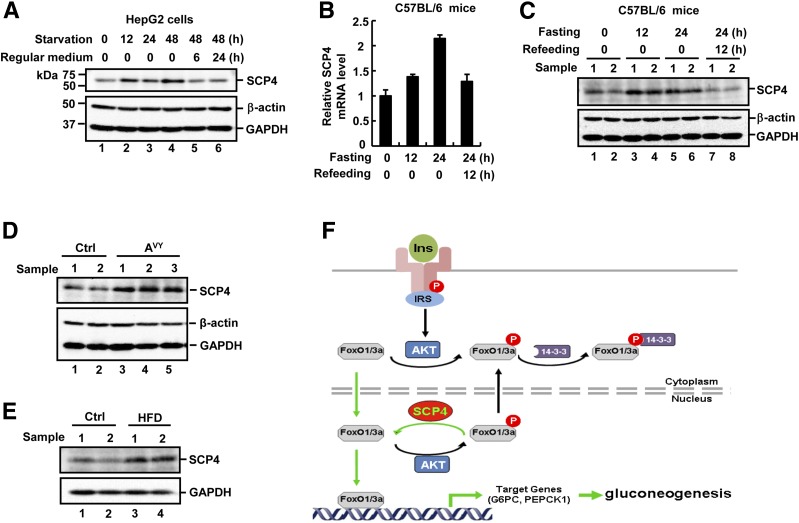
If SCP4 is a critical regulator of glucose production, is SCP4 itself regulated during glucose homeostasis? To answer this, we examined the expression of SCP4 under glucose deprivation condition. In HepG2 cells, we found that the protein level of SCP4 was increased during glucose deprivation for 12, 24, and 48 h, but decreased to the basal level after glucose refeeding for 6 or 24 h (Fig. 8A). This suggests that SCP4 expression is regulated by metabolic conditions and, as a result, regulates the glucose homeostasis. Similar regulation in the SCP4 level was also observed in vivo in mouse livers. Specifically, we examined whether SCP4 expression was changed during fasting–feeding cycles. C57BL/6 mice were grouped and subjected to regular diet feeding, fasted for 12 or 24 h, or fasted for 24 h followed by refeeding for 12 h. SCP4 mRNA and protein levels were examined from the livers of these mice by qRT-PCR or IB. Our results showed that the SCP4 mRNA (Fig. 8B) and protein (Fig. 8C) levels were increased during starvation and decreased to the basal level after refeeding, which corresponded to the need of glucose production.
Figure 8.
SCP4 expression is regulated under pathophysiological conditions. A: Glucose withdrawal induces SCP4 expression in HepG2 cells. HepG2 cells were cultured in glucose-free medium for indicated time or cultured for 48 h and then returned to regular medium for 6 or 24 h. Cell lysates were analyzed by IB with indicated antibodies. B: Fasting/refeeding regulates the mRNA level of SCP4 in mouse livers. C57BL/6 mice were fasted for indicated time or fasted for 24 h followed by refeeding for 12 h. Total RNAs were extracted from the livers of these mice and subjected to qRT-PCR. C: Fasting/refeeding regulates the protein level of SCP4 in mouse livers. Fasting/refeeding of C57BL/6 mice and liver sample collection were performed as described in B. The liver lysates were subjected to IB. D: SCP4 expression is increased in Agouti (AVY) mice. The protein level of SCP4 in the livers of 6-month-old lean control (Ctrl) and obese AVY mice was examined by IB. E: High-fat feeding increases SCP4 expression. C57BL/6 mice were fed with either standard chow diet (Ctrl) or high-fat diet (HFD) for 5 weeks. The protein level of SCP4 in the mouse livers was determined by IB. F: A working model for SCP4 enhancing FoxO1/3a activation. Activation of the insulin–Akt pathway promotes cytoplasmic accumulation of FoxO1/3a either by retaining FoxO1/3a in the cytoplasm or facilitating FoxO1/3a nuclear export. In the nucleus, SCP4 dephosphorylates p-FoxO1/3a, promotes nuclear retention of FoxO1/3a, and enhances the expression of FoxO1/3a target genes, including G6PC and PEPCK1, leading to gluconeogenesis.
More significantly, we investigated the regulation of SCP4 expression under pathophysiological conditions such as in Agouti (Avy) gene-induced obese mice or in mice that were subject to high-fat feeding for 5 weeks. We found that hepatic expression of SCP4 was elevated in obese mice caused by either genetic (Avy) (Fig. 8D) or dietary (high-fat) changes (Fig. 8E). This is in agreement with the role that SCP4 plays in glucose production and the known association between obesity and diabetes with increased hepatic gluconeogenesis.
Discussion
The elevated hepatic gluconeogenesis is crucial for the development of obesity and type 2 diabetes. It is a major contributing factor to hyperglycemia in both types 1 and 2 diabetes. Thus, understanding how gluconeogenesis is regulated will shed light on the prevention and cure of hyperglycemia. Transcription factors FoxO1/3a play critical roles in inducing the expression of gluconeogenic genes (12,14). Thus, elucidation of molecular mechanisms underlying the regulation of FoxO1/3a activity will contribute to the development of effective therapeutic intervention.
FoxO1/3a activity is regulated by phosphorylation (17,18). In liver, the insulin/Akt signaling pathway inactivates FoxO1/3a by phosphorylating FoxO1/3a to suppress glucose synthesis (19). To counteract this, PP2A, a PS/TP, and DUSP6 (MKP3), a dual-specific phosphatase, have been reported to serve as cytosolic phosphatases for FoxO and enhance hepatic gluconeogenesis (21–23). Because Akt resides in both cytosol and nucleus, the question remains if nuclear FoxO1/3a phosphatases exist. Our current study reported the first identification of a nuclear FoxO1/3a phosphatase to regulate FoxO1/3a subcellular location and transcription activity. SCP4 is exclusively nuclear localized and regulates FoxO nuclear retention. In the nucleus, SCP4 dephosphorylates p-FoxO1/3a and enhances the expression of FoxO1/3a target genes such as G6PC and PEPCK1, leading to gluconeogenesis (Fig. 8F). Therefore, we reason that although phosphatases PP2A and DUSP6 may facilitate dephosphorylation of FoxO1/3a in the cytoplasm (and thus possible nuclear import and functions), SCP4 acts to retain an active FoxO1/3a on chromatin and potentiate the activities of FoxO1/3a proteins.
SCP4 belongs to the family of small FCP/SCPs. This family includes FCP1 that dephosphorylates small COOH-terminal domain of RNA polymerase II and SCP1, SCP2, and SCP3, which exhibit various functions such as in epithelial to mesenchymal transition, transforming growth factor-β and bone morphogenetic protein (BMP) signaling, and neuronal genes expression (32–38). There are scarcely any studies on the function and regulation of SCP4. We have previously found that SCP4 dephosphorylates BMP-activated Smad1/5/8 to inhibit BMP-induced osteogenic differentiation (24) and that SCP4 enhances FoxO transcription activity and promotes cellular proteolysis and muscle wasting in chronic kidney disease mouse model (39). A recent report showed that SCP4 exhibited Ser5-preferential phosphatase activity toward RNA polymerase II in vitro (40). Whether these other substrates of SCP4 (e.g., Smad1/5/8) participate in the regulation of the SCP4-regulated glucose production awaits further investigation. Nonetheless, our present findings that SCP4 enhances the expression of gluconeogenic genes and promotes hepatic glucose synthesis assign a new function to SCP4. Our data from both in vitro cell-based assays and in vivo mouse models strongly support a notion that SCP4 is a critical regulator in glucose homeostasis through its direct dephosphorylation of p-Ser256 on FoxO1 and p-Ser253 on FoxO3a.
In further support of this, we found that the expression of SCP4 is regulated under pathophysiological conditions. Hepatic expression of SCP4 was elevated in obese mice caused by either genetic (Avy) or dietary (high-fat) change, consistent with the known association between obesity and diabetes with increased hepatic gluconeogenesis. Based on our studies, we could reason that small-molecule interference with SCP4 expression or activity can prevent FoxO1/3a activation and subsequent gluconeogenesis. Thus, our identification of SCP4 as a FoxO1/3a phosphatase expands the function of this family of phosphatases to regulating hepatic glucose synthesis and presents a potential target for the prevention and treatment of diet-induced obesity and type 2 diabetes.
Article Information
Acknowledgments. The authors thank Dr. Dihua Yu (MD Anderson Cancer Center) for the 14-3-3ζ construct and the members of the Feng and Lin laboratories for helpful discussion and technical assistance.
Funding. This study was partly supported by grants from the National Natural Science Foundation of China (91540205, 31571447, and 31090360 to X.-H.F.), Ministry of Science and Technology of the People’s Republic of China (2012CB966600 to X.-H.F.), U.S. Department of Defense (DAMD W81XWH-15-1-0650 to X.-H.F.), National Institutes of Health (R21-CA-209007 to X.-H.F., R01-DK-073932 to X.L., R01-AR-063668 to Z.H., and R01-HL-102314 and R01-HL-123953 to J.Ch.), American Heart Association Grant-in-Aid (20500033 to J.Ch.), U.S. Department of Agriculture (CRIS 3092-5-001-059 to Q.T.), and the Fundamental Research Funds for the Central Universities (to Q.T.).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. J.Ca., Y.Y., Z.Z., and X.L. performed the experiments. Q.T., J.Ch., X.-H.F., and X.L. designed the study. Z.H., Q.T., and X.C. provided technical support. Q.T., X.H.-F., and X.L. analyzed and interpreted data. J.Ca., Y.Y., X.-H.F., and X.L. wrote the manuscript. X.L. conceived the study. X.L. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
See accompanying article, p. 23.
References
- 1.Shelly LL, Lei KJ, Pan CJ, et al. . Isolation of the gene for murine glucose-6-phosphatase, the enzyme deficient in glycogen storage disease type 1A. J Biol Chem 1993;268:21482–21485 [PubMed] [Google Scholar]
- 2.Nordlie RC, Lardy HA. Mammalian liver phosphoneolpyruvate carboxykinase activities. J Biol Chem 1963;238:2259–2263 [PubMed] [Google Scholar]
- 3.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol 2001;2:599–609 [DOI] [PubMed] [Google Scholar]
- 4.Park EA, Roesler WJ, Liu J, et al. . The role of the CCAAT/enhancer-binding protein in the transcriptional regulation of the gene for phosphoenolpyruvate carboxykinase (GTP). Mol Cell Biol 1990;10:6264–6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhee J, Inoue Y, Yoon JC, et al. . Regulation of hepatic fasting response by PPARgamma coactivator-1alpha (PGC-1): requirement for hepatocyte nuclear factor 4alpha in gluconeogenesis. Proc Natl Acad Sci U S A 2003;100:4012–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durham SK, Suwanichkul A, Scheimann AO, et al. . FKHR binds the insulin response element in the insulin-like growth factor binding protein-1 promoter. Endocrinology 1999;140:3140–3146 [DOI] [PubMed] [Google Scholar]
- 7.Koo SH, Flechner L, Qi L, et al. . The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature 2005;437:1109–1111 [DOI] [PubMed] [Google Scholar]
- 8.Yang J, Croniger CM, Lekstrom-Himes J, et al. . Metabolic response of mice to a postnatal ablation of CCAAT/enhancer-binding protein alpha. J Biol Chem 2005;280:38689–38699 [DOI] [PubMed] [Google Scholar]
- 9.Vander Kooi BT, Streeper RS, Svitek CA, Oeser JK, Powell DR, O’Brien RM. The three insulin response sequences in the glucose-6-phosphatase catalytic subunit gene promoter are functionally distinct. J Biol Chem 2003;278:11782–11793 [DOI] [PubMed] [Google Scholar]
- 10.Schmoll D, Walker KS, Alessi DR, et al. . Regulation of glucose-6-phosphatase gene expression by protein kinase Balpha and the forkhead transcription factor FKHR. Evidence for insulin response unit-dependent and -independent effects of insulin on promoter activity. J Biol Chem 2000;275:36324–36333 [DOI] [PubMed] [Google Scholar]
- 11.Ogg S, Paradis S, Gottlieb S, et al. . The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 1997;389:994–999 [DOI] [PubMed] [Google Scholar]
- 12.Barthel A, Schmoll D, Unterman TG. FoxO proteins in insulin action and metabolism. Trends Endocrinol Metab 2005;16:183–189 [DOI] [PubMed] [Google Scholar]
- 13.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene 2005;24:7410–7425 [DOI] [PubMed] [Google Scholar]
- 14.Kousteni S. FoxO1, the transcriptional chief of staff of energy metabolism. Bone 2012;50:437–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakravarty K, Cassuto H, Reshef L, Hanson RW. Factors that control the tissue-specific transcription of the gene for phosphoenolpyruvate carboxykinase-C. Crit Rev Biochem Mol Biol 2005;40:129–154 [DOI] [PubMed] [Google Scholar]
- 16.Biggs WH 3rd, Meisenhelder J, Hunter T, Cavenee WK, Arden KC. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci U S A 1999;96:7421–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunet A, Bonni A, Zigmond MJ, et al. . Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 1999;96:857–868 [DOI] [PubMed] [Google Scholar]
- 18.Kops GJ, de Ruiter ND, De Vries-Smits AM, Powell DR, Bos JL, Burgering BM. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature 1999;398:630–634 [DOI] [PubMed] [Google Scholar]
- 19.Garofalo RS, Orena SJ, Rafidi K, et al. . Severe diabetes, age-dependent loss of adipose tissue, and mild growth deficiency in mice lacking Akt2/PKB beta. J Clin Invest 2003;112:197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Der Heide LP, Hoekman MF, Smidt MP. The ins and outs of FoxO shuttling: mechanisms of FoxO translocation and transcriptional regulation. Biochem J 2004;380:297–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan L, Lavin VA, Moser LR, Cui Q, Kanies C, Yang E. PP2A regulates the pro-apoptotic activity of FOXO1. J Biol Chem 2008;283:7411–7420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Z, Jiao P, Huang X, et al. . MAPK phosphatase-3 promotes hepatic gluconeogenesis through dephosphorylation of forkhead box O1 in mice. J Clin Invest 2010;120:3901–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu H, Yang Q, Shen M, et al. . Dual specificity MAPK phosphatase 3 activates PEPCK gene transcription and increases gluconeogenesis in rat hepatoma cells. J Biol Chem 2005;280:36013–36018 [DOI] [PubMed] [Google Scholar]
- 24.Zhao Y, Xiao M, Sun B, et al. . C-terminal domain (CTD) small phosphatase-like 2 modulates the canonical bone morphogenetic protein (BMP) signaling and mesenchymal differentiation via Smad dephosphorylation. J Biol Chem 2014;289:26441–26450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai F, Lin X, Chang C, Feng XH. Nuclear export of Smad2 and Smad3 by RanBP3 facilitates termination of TGF-beta signaling. Dev Cell 2009;16:345–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin X, Duan X, Liang YY, et al. . PPM1A functions as a Smad phosphatase to terminate TGFbeta signaling. Cell 2006;125:915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehtinen MK, Yuan Z, Boag PR, et al. . A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell 2006;125:987–1001 [DOI] [PubMed] [Google Scholar]
- 28.Yuan Z, Lehtinen MK, Merlo P, Villén J, Gygi S, Bonni A. Regulation of neuronal cell death by MST1-FOXO1 signaling. J Biol Chem 2009;284:11285–11292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darlington GJ. Molecular mechanisms of liver development and differentiation. Curr Opin Cell Biol 1999;11:678–682 [DOI] [PubMed] [Google Scholar]
- 30.Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy in glucose homeostasis. Pathol Res Pract 2006;202:631–638 [DOI] [PubMed] [Google Scholar]
- 31.Kuma A, Hatano M, Matsui M, et al. . The role of autophagy during the early neonatal starvation period. Nature 2004;432:1032–1036 [DOI] [PubMed] [Google Scholar]
- 32.Cho H, Kim TK, Mancebo H, Lane WS, Flores O, Reinberg D. A protein phosphatase functions to recycle RNA polymerase II. Genes Dev 1999;13:1540–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palancade B, Dubois MF, Dahmus ME, Bensaude O. Transcription-independent RNA polymerase II dephosphorylation by the FCP1 carboxy-terminal domain phosphatase in Xenopus laevis early embryos. Mol Cell Biol 2001;21:6359–6368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeo M, Lin PS, Dahmus ME, Gill GN. A novel RNA polymerase II C-terminal domain phosphatase that preferentially dephosphorylates serine 5. J Biol Chem 2003;278:26078–26085 [DOI] [PubMed] [Google Scholar]
- 35.Yeo M, Lee SK, Lee B, Ruiz EC, Pfaff SL, Gill GN. Small CTD phosphatases function in silencing neuronal gene expression. Science 2005;307:596–600 [DOI] [PubMed] [Google Scholar]
- 36.Sapkota G, Knockaert M, Alarcón C, Montalvo E, Brivanlou AH, Massagué J. Dephosphorylation of the linker regions of Smad1 and Smad2/3 by small C-terminal domain phosphatases has distinct outcomes for bone morphogenetic protein and transforming growth factor-beta pathways. J Biol Chem 2006;281:40412–40419 [DOI] [PubMed] [Google Scholar]
- 37.Wrighton KH, Willis D, Long J, Liu F, Lin X, Feng XH. Small C-terminal domain phosphatases dephosphorylate the regulatory linker regions of Smad2 and Smad3 to enhance transforming growth factor-beta signaling. J Biol Chem 2006;281:38365–38375 [DOI] [PubMed] [Google Scholar]
- 38.Wu Y, Evers BM, Zhou BP. Small C-terminal domain phosphatase enhances snail activity through dephosphorylation. J Biol Chem 2009;284:640–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X, Yu R, Sun L, et al. . The nuclear phosphatase SCP4 regulates FoxO transcription factors during muscle wasting in chronic kidney disease. Kidney Int 2017;92:336–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wani S, Sugita A, Ohkuma Y, Hirose Y. Human SCP4 is a chromatin-associated CTD phosphatase and exhibits the dynamic translocation during erythroid differentiation. J Biochem 2016;160:111–120 [DOI] [PubMed] [Google Scholar]