Abstract
Previous findings suggested that the motor activity of human myosin IIIA (HM3A) is influenced by phosphorylation [Kambara, T., et al. (2006) J. Biol. Chem. 281, 37291–37301]; however, how phosphorylation controls the motor activity of HM3A is obscure. In this study, we clarify the kinetic basis of the effect of phosphorylation on the ATP hydrolysis cycle of the motor domain of HM3A (huM3AMD). The affinity of human myosin IIIA for filamentous actin in the presence of ATP is more than 100-fold decreased by phosphorylation, while the maximum rate of ATP turnover is virtually unchanged. The rate of release of ADP from acto-phosphorylated huM3AMD is 6-fold greater than the overall cycle rate, and thus not a rate-determining step. The rate constant of the ATP hydrolysis step of the actin-dissociated form is markedly increased by phosphorylation by 30-fold. The dissociation constant for dissociation of the ATP-bound form of huM3AMD from actin is greatly increased by phosphorylation, and this result agrees well with the significant increase in the Kactin value of the steady-state ATPase reaction. The rate constant of the Pi off step is greater than 60 s−1, suggesting that this step does not limit the overall ATP hydrolysis cycle rate. Our kinetic model indicates that phosphorylation induces the dissociation of huM3AMD from actin during the ATP hydrolysis cycle, and this is due to the phosphorylation-dependent marked decrease in the affinity of huM3AMD· ATP for actin and the increase in the ATP hydrolysis rate of huM3AMDin the actin-dissociated state. These results suggest that the phosphorylation of myosin IIIA significantly lowers the duty ratio, which may influence the cargo transporting ability of the native form of myosin IIIA that contains the ATP-independent actin binding site in the tail.
It is known that myosin constitutes a superfamily of more than 20 classes. Class III myosins are present in photoreceptor cells and auditory hair cells (2, 3). The loss of class III myosins leads to photoreceptor degeneration in Drosophila (4, 5), and mutations in the myosin IIIA genes leads to progressive hearing loss (6). Thus, class III myosins are critical for the normal function of photoreceptors and auditory hair cells. At the molecular level, class III myosin is unique in having an N-terminal protein kinase domain (5). The protein kinase activity of this domain has been biochemically identified for Drosophila myosin III (7), Limulus myosin III (8), and human-myosin IIIA (9). The motor activity has been identified for human myosin IIIA (9), while a recombinant form of Limulus myosin III displayed no motor activity (8). There is no evidence that Drosophila myosin III has motor activity.
In vertebrates, two genes encoding class III myosin (myosin IIIA and myosin IIIB) were identified (10). So far, most studies were performed with myosin IIIA (11). Myosin IIIA is produced in retina and inner ear hair cells, and it is responsible for progressive, nonsyndromic hearing loss in humans (6). The physiological function of myosin IIIA is still unknown, but recent studies have suggested that myosin IIIA may function as a cargo carrier (12, 13). It was shown that GFP-myosin IIIA expressed in HeLa cells localizes at the tip of filopodia (12, 13), suggesting that myosin IIIA accumulates at the plus end of actin bundles. In photoreceptor cells, myosin IIIA is concentrated in the distal ends of rod and cone ellipsoids and colocalizes with the plus (distal) ends of inner segment actin filament bundles where actin forms the microvilli-like calycal processes (14). Quite recently, it was reported that myosin IIIa colocalizes with espin 1, an actin bundling protein, at stereocilia tips and transports espin 1 in filopodia (15). These results suggest that myosin IIIA can move on actin filament bundles for a long distance to be accumulated at the plus end of the actin track. Consistently, it is found that myosin IIIA is a plus end-directed motor (9).
Except for its unique N-terminal protein kinase domain, human myosin IIIA heavy chain consists of a motor domain, a neck domain, and a C-terminal tail domain, similar to other myosin superfamily members (2, 3). Following the motor domain, there is a neck/tail domain containing two proximal IQ motifs and one distal IQ motif that serve as a light chain/CaM1 binding site. There is no coiled-coil region in the tail region, suggesting that myosin III is a single-headed structure, although it may be possible that myosin III forms an oligomeric structure with its binding proteins in vivo.
For unconventional myosins, it has been thought that the class specific tail domain functions as a target protein binding domain (2, 3) or as an intramolecular inhibitor domain (16). For myosin IIIA, it was shown that the tail domain is required for localization at rod calycal processes (17). Consistently, the tail domain is required for the localization of myosin IIIA to distal filopodial tips. Interestingly, the tip of the tail can interact with actin (12). The actin binding site in the tail was identified as a DFRXXL motif that was originally identified in myosin light chain kinase (18). While the physiological importance of this ATP-independent actin binding site is unclear, it is plausible that the actin binding site at the tail serves as a tethering site and plays a role in preventing myosin IIIA from dissociating from the actin track during movement as is shown for the processive movement of kinesin (19).
The motor function of class III myosin was first identified for human myosin IIIA (9). It was shown that myosin IIIA having both kinase and motor domains exhibited actin-activated ATPase activity and actin translocating activity. Since it was also shown that the motor domain of myosin IIIA is autophosphorylated, a key question is whether autophosphorylation influences motor activity. To address this issue, Kambara et al. (1) used a myosin IIIA motor domain construct without the kinase domain, since the motor domain can be rapidly autophosphorylated in the presence of ATP during the actomyosin ATPase reaction. It was found that phosphorylation at the motor domain by the exogenous myosin IIIA kinase domain markedly increased Kactin by 500-fold, although Vmax was unchanged. This finding has raised an idea that autophosphorylation alters the ATP hydrolysis cycle pathway, thus changing the cargo transporting activity of myosin III, presumably by changing the duty ratio of the molecule.
In our study, we clarified the effect of autophosphorylation on the ATP hydrolysis mechanism of human myosin IIIA. The results suggest that phosphorylation significantly shifts myosin IIIA from the actin-associated intermediates to the actin-dissociated intermediates; thus, it spends a majority of its ATP hydrolysis cycle time at the actin-dissociated forms.
EXPERIMENTAL PROCEDURES
Materials
Restriction enzymes and modifying enzymes were purchased from New England Biolabs (Beverly, MA). Actin was prepared from rabbit skeletal muscle according to the method of Spudich and Watt (20). Phosphate-binding protein (PBP) labeled with 7-(diethylamino)-3-({[(2-maleimidyl)ethyl]amino}-carbonyl)coumarin (MDCC) was prepared as previously described (21, 22).
Myosin IIIA Expression Vector Construction and Preparation of Phosphorylated Myosin IIIA
To generate a human myosin IIIA kinase domain (huM3AKD)-expressing construct, the cDNA was truncated at Gly327 and inserted into the pFastBac1 vector (Invitrogen) containing a c-myc tag and an octahistidine tag at the C-terminus. The recombinant baculovirus was expressed in Sf9 cells, and huM3AKD protein was purified using a Ni-NTA affinity column (Qiagen, Valencia, CA). To express myosin IIIA motor domain (huM3AMD), a fragment of human myosin IIIA (Met311–Lys1058) was subcloned into modified pFastBac1 containing a C-terminal FLAG tag. The huM3AMD construct contains the motor domain but not the kinase domain and IQ motifs. Sf9 cells were infected with the recombinant virus expressing huM3AMD. The cells were lysed, and the supernatant was loaded onto an anti-FLAG chromatography column. After the mixture was washed, 2 μM huM3AKD and 1 mM ATP were added to the bead slurry and incubated in the presence of mycrocystin-LR for 30 min at 25 °C. To remove huM3AKD and ATP, the beads were washed and then phosphorylated huM3AMD was eluted with a buffer containing 10 μg/mL FLAG peptide.
Steady-State ATPase Assay
The ATPase activity was measured in 30mM HEPES (pH7.5), 30 mM KCl, 2mM MgCl2, 1 mM EGTA, 1 mM DTT, 1 μM mycrocystin-LR, 0.01 μM huM3AMD, and various concentrations of actin (0–75 μM) in the presence of an ATP regeneration system (22, 23) at 25 °C. The amount of liberated pyruvate was determined as described previously.
Kinetic Experiments
Transient kinetic experiments were performed in buffer A at 25 °C using a KinTek SF-2004 stopped-flow apparatus (KinTek Co.) equipped with a 150 W mercury–xenon arc lamp. 2′-Deoxy-mant ATP and ADP were excited by energy transfer at 280 nm, and the emission was detected through a 400 nm long pass filter (Oriel). The rates of actin binding to and dissociation from myosin IIIA were measured by monitoring the light scattering signal at 400 nm. The kinetics of phosphate release were measured using MDCC-PBP excited at 433 nm with a 436 nm band-pass filter, and the emission was measured through a 455 nm long pass filter.
Quenched-Flow Measurements
Quenched-flow measurements were performed in buffer A at 25 °C using aKinTekRQF-3 apparatus (KinTek Co.) as described previously (22).
RESULTS
Expression and Purification of Phosphorylated huM3MD
To determine the effect of autophosphorylation in the motor domain on the actin-activated ATP hydrolysis cycle, we used the kinase domain-deleted construct to avoid the potential complexity arising from phosphorylation due to the presence of the N-terminal kinase domain during the ATPase assay. We chose the construct having no IQ motifs, since the affinity of CaM light chains for human myosin III heavy chain is relatively low and the isolated myosin III construct having IQ motifs can be a mixture of myosin III with and without bound CaM light chain. To avoid the complexity arising from the heterogeneity of the molecules in terms of bound CaM, we used the huM3AMD construct shown in Figure 1A. To confirm that the elimination of the IQ domain does not have a significant effect on the ATPase activity of myosin IIIA, we examined the actin-activated ATPase activity of huM3IQ2 having two IQ motifs in addition to huM3AMD. It exhibited a Kactin and a Vmax, i.e., the actin concentration giving a half-Vmax and the maximum ATPase activity at an infinite actin concentration, respectively, similar to those of huM3MD in the presence of exogenous calmodulin (not shown). The isolated huM3AMDhad an apparent molecular mass of 90.6 kDa, and no low-molecular mass band was observed (Figure 1B). The purity of p-huM3MD was approximately 98% on the basis of densitometry analysis of SDS–PAGE gels stained with CBB, and no band for the kinase domain (calculated molecular mass of 42 kDa) was observed (Figure 1B).
Figure 1.
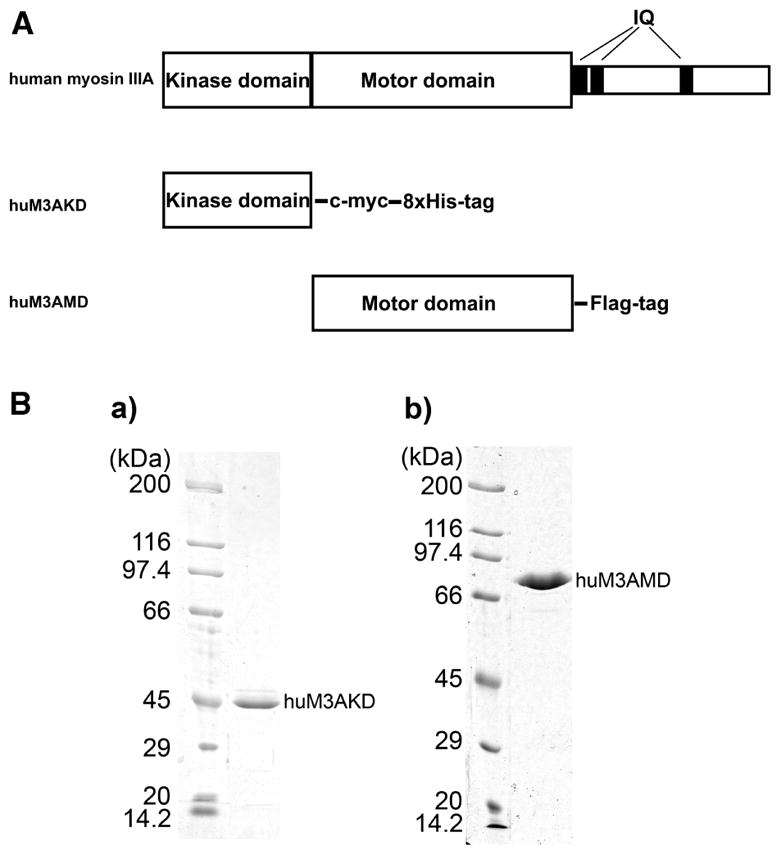
Human myosin IIIA constructs. (A) Schematic diagram of myosin IIIA constructs used in this study. The human myosin IIIA kinase domain construct (huM3AKD) contains Met1–Gly327, a C-terminal c-myc tag, and an octahistidine tag. The human myosin IIIA motor domain construct (huM3AMD) contains Met311–Lys1058 and a C-terminal FLAG tag. (B) SDS–PAGE of purified myosin IIIA constructs. huM3AKD (a) and huM3AMD (b) were expressed in Sf9 cells and purified as described in Experimental Procedures. The purified proteins were analyzed by SDS–PAGE. No CaM band is observed since the huM3AMD has no IQ motifs.
Steady-State ATPase Activity
The isolated huM3AMD was phosphorylated by the human myosin IIIA kinase domain (huM3AKD) for various amounts of time. The actin-activated ATPase activity in the presence of 5 μM actin significantly decreased by 3-fold within 2.5 min, and the activity was unchanged until 90min. Therefore, we used huM3AMD phosphorylated for 30 min in this study. The steady-state ATP activity of the phosphorylated huM3AMD as a function of actin concentration was measured (Figure 2). The basal ATPase activity (0.07 s−1) was markedly activated in the presence of actin. The solid curve is the best fit of the experimental data with the Michaelis–Menten equation and yielded Vmax and Kactin values of 1.63 s−1 and 10.3 μM, respectively. On the other hand, the Vmax and Kactin values of the unphosphorylated huM3AMD were 1.69 s−1 and <0.10 μM, respectively (not shown). A similar Kactin value for unphosphorylated myosin IIIA was also reported previously (1). Phosphorylation increased the Kactin by more than 100-fold, while it had virtually no effect on Vmax.
Figure 2.
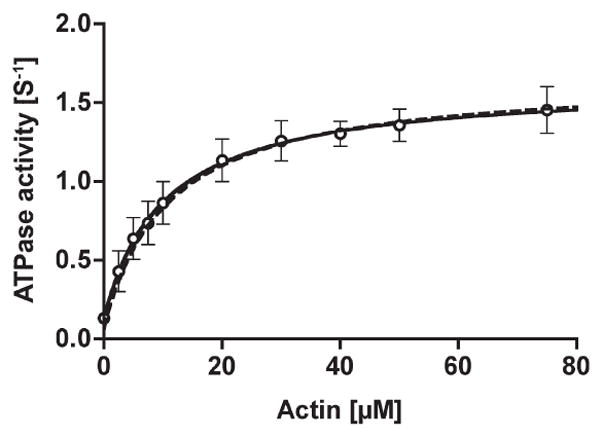
The steady-state ATPase activity of phosphorylated huM3AMD. The ATPase activity of the p-huM3AMD was measured in the presence of an ATP regeneration system and various concentrations of actin. The solid curve is the best fit to Michaelis–Menten kinetics with Vmax and Kactin values of 1.6 s−1 and 10.3 μM, respectively. The dashed line is simulated on the basis of the parameters obtained in this study (see Discussion), giving a Vmax of 1.67 s−1 and a KATPase of 10.9 μM. The error bars represent the standard error from three independent experiments.
MgATP Binding to huM3MD and acto-huM3AMD
The kinetics of ATP binding to p-huM3AMD were measured by using the fluorescent nucleotide, dmant-ATP. The rate of increase in fluorescence intensity of dmant-ATP upon binding to p-huM3AMD was fitted to double exponentials in the presence and absence of actin (Figure 3, insets). In the absence of actin, the fast phase was predominant and explained 80–90% of the total signal intensity. The rate constants of the fast and slow phases were linearly dependent on dmant-ATP concentration, yielding second-order rate constants for dmant-ATP binding (K1k+2) of 0.20 and 0.05 μM−1 s−1, respectively (Figure 3A). The rate of the reverse reaction of ATP binding obtained from the y-intercept of the fast phase in the absence of actin was 0.05 s−1. The fast phase of dmant-ATP binding to acto-p-huM3AMDwas 60–70% of the fluorescence signal. The rates of dmant-ATP binding were linearly increased with dmant-ATP concentration, and the slope of fast and slow phases gave second-order rate constants (K′1k′+2) of 0.13 and 0.03 μM−1 s−1, respectively (Figure 3B). The rate of the reverse reaction of ATP binding obtained from the y-intercept of the fast phase in the presence of actin was 0.65 s−1, suggesting that the ATP dissociation rate is increased in the presence of actin. At present, we do not understand the origin of this slow phase of the change in fluorescence intensity. The obtained K1k+2 (0.20 μM−1 s−1) and K′1k′+2 (0.13 μM−1 s−1) values for phosphorylated huM3AMD compare with values of 0.14 and 0.16 μM−1 s−1, respectively, for unphosphorylated huM3AMD previously determined (1).
Figure 3.
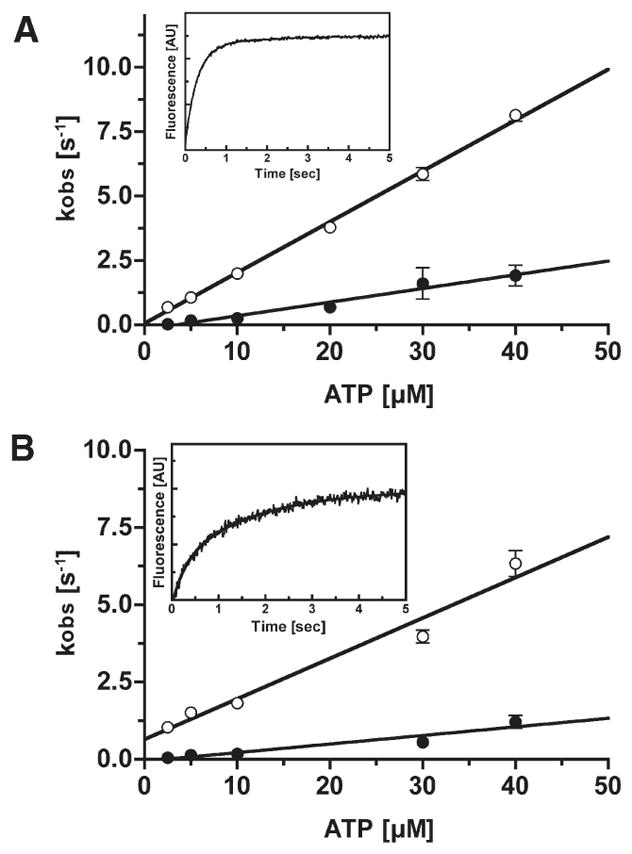
dmant-ATP binding to p-huM3AMD and acto-p-huM3AMD. (A) Rate constant of dmant-ATP binding to p-huM3AMD as a function of ATP concentration. The observed rates (kobs) were obtained by fitting the fluorescence transients to double exponentials. The apparent second-order rate constants for dmant-ATP binding to p-huM3AMDwere 0.20 μM−1 s−1 for the fast phase (predominant phase) (○) and 0.05 μM−1 s−1 for the slow phase (●). The rate of dissociation of dmant-ATP determined from the y-intercept of the fast phase is 0.05 s−1. The inset shows a typical recording of the binding of dmant-ATP (20 μM) to 0.5 μM p-huM3AMD. The solid line is the best fit to double-exponential kinetics with a fast kobs of 3.60 s−1 and a slowkobs of 0.64 s−1. (B)Rate constant of dmant-ATP binding to acto-p-huM3AMDas a function of ATP concentration. The observed rates (kobs) were obtained by fitting the fluorescence transients to double exponentials. The apparent second-order rate constants for dmant-ATP binding to acto-p-huM3AMD for the fast phase (predominant phase) (○) and the slow phase (●) are 0.13 and 0.03 μM−1 s−1, respectively. The dissociation rate of dmant-ATP determined from the y-intercept of the fast phase is 0.65 s−1. The error bars represent the standard error from three independent experiments. The inset shows a typical recording of the binding of dmant-ATP (30 μM) to 0.5 μM acto-p-huM3AMD. The solid line is the best fit to double-exponential kinetics with a fast kobs of 3.42 s−1 and a slow kobs of 0.59 s−1.
MgATP-Induced Dissociation of acto-huM3AMD
We know that myosin quickly dissociates from actin when MgATP binds. The kinetics of the MgATP-induced dissociation of acto-phosphorylated huM3AMD was monitored by measuring the change in the light scattering of acto-phosphorylated huM3MD after it was mixed with the complex with MgATP. The phosphorylated huM3AMD was mixed with 0.5 μM actin to produce acto-myosin IIIA complex, and then various concentrations of MgATP were added to the mixture. The decrease in light scattering due to the dissociation of huM3AMD from actin followed double exponentials. The rates of the fast phase (major phase) increased with MgATP concentration (Figure 4). The value was not saturated at 1.6mMMgATP, which suggested that the maximum rate constant is larger than 100 s−1. This is significantly larger than that of unphosphorylated huM3MD (1).
Figure 4.
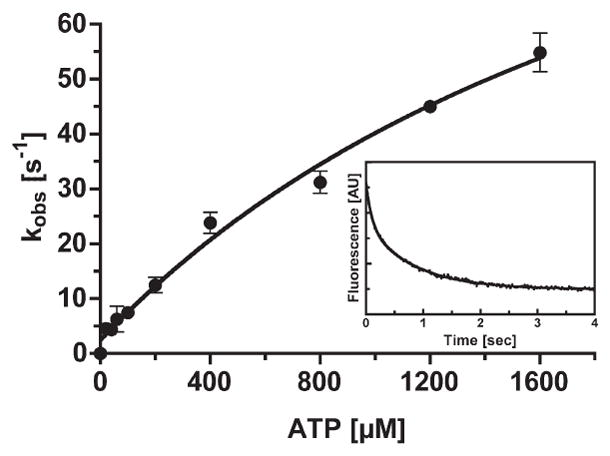
ATP-induced dissociation of acto-phosphorylated-huM3MD. The rate of ATP-induced dissociation of acto-p-huM3AMD was measured by monitoring the decrease in the light scattering signal. Dissociation rates as a function of ATP concentration are shown. The experiment was conducted in the presence of 0.1 μM actin. The error bars represent the standard error from three independent experiments. The inset shows the time course of light scattering change after 0.5 μM acto-p-huM3AMD was mixed with 100 μM MgATP. The solid line is the best fit to double-exponential kinetics with a fast kobs of 10.01 s−1 and a slow kobs of 1.19 s−1.
ATP Hydrolysis by Phosphorylated huM3AMD
The kinetics of the formation of the myosin · ADP · Pi complex were studied by using a quenched-flow apparatus. Since the ATP binding rate constant is low, we performed a multiturnover experiment with 30 μM ATP. The time course of Pi release exhibited an initial rapid phase followed by a slow linear phase (Figure 5). It has been known that the initial rapid Pi release phase (Pi burst) is due to the formation of the myosin · ADP · Pi complex, in which the bound Pi is released by acid quench. The observed Pi burst size was 0.84 mol/mol, suggesting that a significant fraction of huM3AMD is in the hydrolyzed form (M · ADP · Pi). It should be noted that the obtained Pi burst size could be underestimated, if there is some inactive myosin fraction in the sample. The ATP binding rate at 30 μM ATP was 6.0 s−1, and the rate of the reverse reaction was 0.05 s−1 (Figure 3), indicating that approximately 1%of huM3AMDis present in the ATP unbound form. On the basis of these numbers, the equilibrium constant of the hydrolysis step (K3) was estimated to be ≥6.3. The observed rate constant of the initial phase was 3.7 s−1. Since the ATP binding rate at 30 μM ATP was 6.0 s−1, the observed rate constant of the initial phase was equal to the rate constant of the hydrolysis step (k+3 + k−3). On the basis of the equilibrium constant of the hydrolysis step (K3) estimated above, k+3 and k−3 were estimated to be 3.1 and 0.6 s−1, respectively. These values are much larger than those of unphosphorylated huM3AMD (1), suggesting that phosphorylation significantly increases the rate of the ATP hydrolysis step in the actin dissociated form of huM3AMD.
Figure 5.
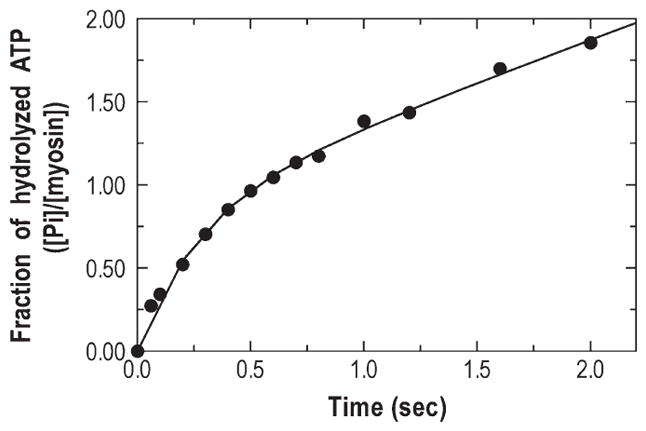
Pi burst of p-huM3MD. The quench-flow experiment was conducted after 1.4 μM p-huM3AMD and 30 μM [γ-32P]ATP were mixed and allowed to age for the indicated time. The solid line is the best fit to an initial single-exponential Pi liberation followed by the steady-state rate. The observed Pi burst size and the initial hydrolysis rate are 0.84 and 3.7 s−1, respectively.
Phosphate Release Rate
The fluorescently labeled phosphate binding protein (MDCC-PBP) was used to measure the rate of the phosphate release step, since it rapidly binds free Pi, which is recognized by the increase in fluorescence intensity. The p-huM3AMD was mixed with 60 μM MgATP, aged for 5 s to produce ATP binding and hydrolysis, and then mixed with various concentrations of actin. The observed fluorescence increase was analyzed by doule-exponential kinetics. The obtained apparent rate constants of the fast phase showed hyperbolic actin dependence due to the actin rebinding step (K9) (Figure 6). The maximum rate of release of phosphate from the acto-phosphorylated M3AMD· ADP · Pi complex (k′+4) was calculated to be 70 s−1 on the basis of the saturation curve. This value was much higher than the Vmax of the steady-state actin-activated ATPase activity. The result suggests that the Pi release step does not limit the overall ATP hydrolysis cycle rate.
Figure 6.
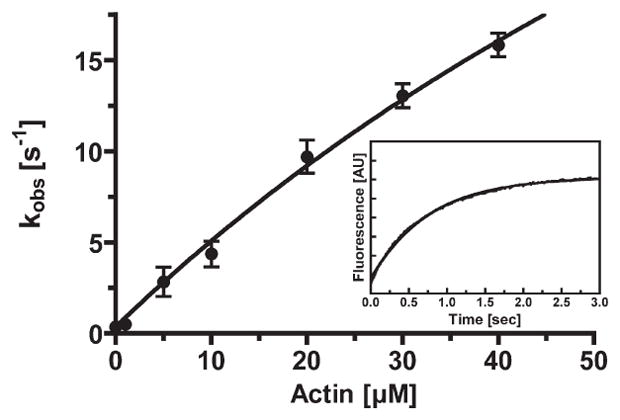
Release of phosphate from acto-p-huM3AMD. The rate of release of phosphate from huM3AMD was measured by using MDCC-PBP. p-huM3AMD (1.5 μM) was mixed with 60 μM MgATP, aged for 5 s, and then mixed with 3 μM phosphate binding protein in the presence and absence of actin. The actin concentration dependence of the rate of phosphate release is shown. The apparent rate was increased with actin concentration to yield the maximum rate of phosphate release of 70 ± 24 s−1 and a Kactin of 138 ± 58 μM. The error bars represent the standard error from three to six independent experiments. The inset shows a typical recording of the MDCC-PBP fluorescence change at 5 μM F-actin. Time course fits to double-exponential kinetics with a fast kobs of 2.0 s−1.
Kinetics of ADP Binding to and Dissociation from p-huM3MD and acto-p-huM3MD
dmant-ADP was used to determine the rate of binding of ADP to p-huM3AMD and acto-p-huM3AMD. The fluorescence intensity of dmant-ADP increased after mixing, and the time course of the increase in fluorescence intensity followed a single exponential. The observed rates increased linearly with dmant-ADP concentration in both the presence and absence of actin (Figure 7A,B). In the absence of actin, the second-order rate constant for binding of dmant-ADP to phosphorylated huM3AMD (k−5) was 0.87 μM−1 s−1 (Figure 7A). The dmant-ADP off rate constant (k+5) obtained from the y-intercept of the dmant-ADP dependence was 8.6 s−1. In the presence of actin, the second-order rate constant for dmant-ADP binding (k′ −5) was determined to be 0.56 μM−1 s−1. The dmant-ADP dissociation rate constant (k′+5) obtained from the y-intercept was 8.1 s−1. These values for the ADP off rate and ADP binding rate are similar to those obtained for unphosphorylated huM3AMD (1). The results suggest that the phosphorylation of myosin IIIA in the motor domain does not influence ADP binding to and ADP dissociation from myosin IIIA in the presence or absence of actin.
Figure 7.
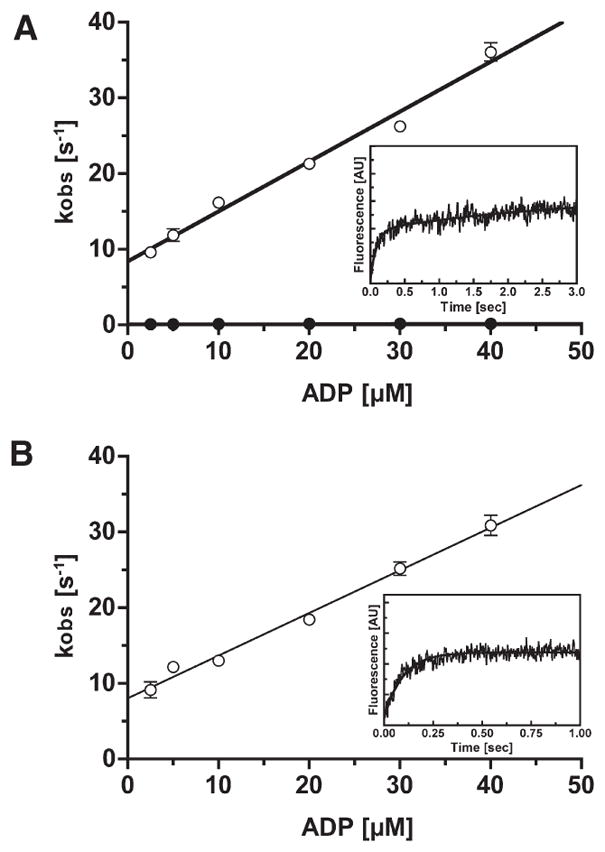
dmant-ADP binding to p-huM3AMD and acto-P-huM3AMD. (A) Rate of binding of dmant-ADP to p-huM3AMD as a function of nucleotide concentration. The observed rates (kobs) were obtained by fitting the fluorescence transients to double exponentials. The apparent second-order rate constants for binding of dmant-ADP to p-huM3AMD are 0.87 μM−1 s−1 for the fast phase (○) and 0.002 μM−1 s−1 for the slow phase (●). The dissociation rates of dmant-ADP determined from the y-intercept of the fast phase and slow phase are 8.6 and 0.12 s−1, respectively. The inset shows a typical recording of the binding of dmant-ADP (5 μM) to 0.5 μM p-huM3AMD. The solid line is the best fit to double-exponential kinetics with a fast kobs of 11.13 s−1 and a slow kobs of 0.13 s−1. (B) Rate of binding of dmant-ADP to acto-p-huM3AMD as a function of nucleotide concentration. The observed rates (kobs) were obtained by fitting the fluorescence transients to a single exponential. The apparent second-order rate constant for binding of dmant-ADP to acto-p-huM3AMDis 0.56 μM−1 s−1. The dissociation rate of dmant-ADP determined from the y-intercept is 8.1 s−1. The error bars represent the standard error from three independent experiments. For several data points, the error bars are within the circles. The inset shows a typical recording of the binding of dmant-ADP (5 μM) to 0.5 μM acto-p-huM3AMD. The solid line is the best fit to single-exponential kinetics with a kobs of 10.41 s−1.
Actin Binding to p-huM3MD
The rate of actin binding to phosphorylated huM3AMD was measured by monitoring the change in the light scattering intensity upon actin binding. The magnitude of the light scattering signal increased when actin and phosphorylated-huM3AMD were mixed, and the time course of the change followed double-exponential kinetics (Figure 8A). In the absence of ADP, the rate of the fast phase (major fraction) linearly increased with actin concentration to yield a second-order rate constant (k−6) of 0.54 μM−1 s−1 (Figure 8B). The dissociation rate constant was determined from the y-intercept to be 0.13 s−1, and the calculated dissociation constant for actin binding (K6) was 0.24 μM in the absence of ADP. This value is 4-fold higher than that obtained for unphosphorylated huM3AMD (1). The second-order rate constant for the binding of actin to phosphorylated huM3AMD· ADP was 0.14 μM−1 s−1 (Figure 8B). The dissociation rate constant for acto-phosphorylated huM3AMD· ADP obtained from the y-intercept of the actin dependence was 0.15 s−1. These values gave a dissociation constant of acto-phosphorylated huM3AMD· ADP (K10) of 1.07 μM. This value is much higher than that obtained for unphosphorylated huM3AMD (71.4 nM) previously determined.
Figure 8.
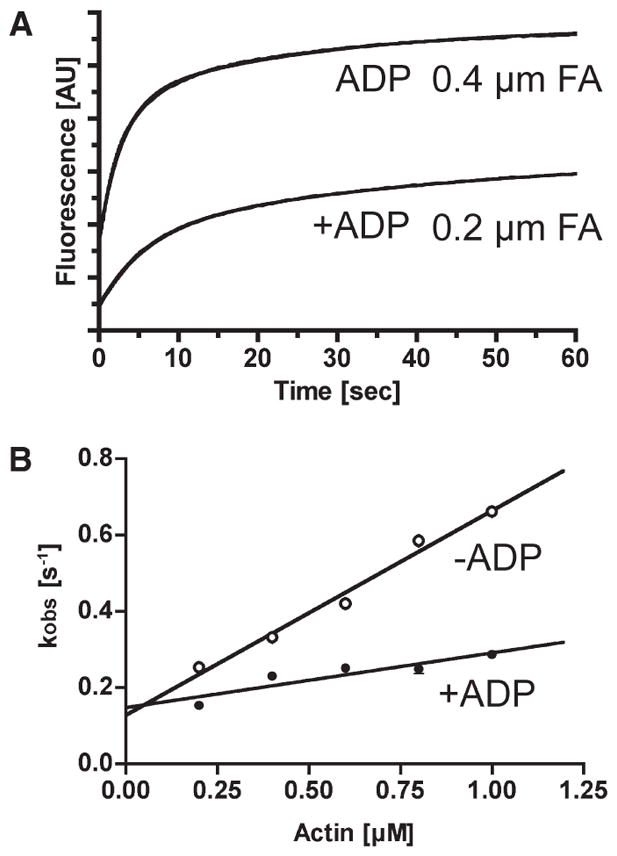
Actin binding to p-huM3AMD. (A) Typical recordings of light scattering of actin binding to 0.5 μM p-huM3AMD in the presence or absence of 0.1mMMgADP.The solid lines are the best fit to double-exponential kinetics with fast kobs values of 0.17 s−1 (0.2 μM actin with ADP) and 0.35 s−1 (0.4 μM actin without ADP). (B) The rate of binding of actin to p-huM3AMDas a function of actin concentration is shown in the presence (●) and absence (○) of 0.1mM MgADP. The fast phase (predominant) was linearly increased with actin concentration. The apparent second-order rate constant for p-huM3AMDis 0.54 μM−1 s−1, and the dissociation rate determined from the y-intercept is 0.13 s−1 in the absence of nucleotide (○). The apparent second-order rate constant for p-huM3AMD is 0.14 μM−1 s−1, and the dissociation rate determined from the y-intercept is 0.15 s−1 in the presence of nucleotide (●). The error bars represent the standard error from three independent experiments. For several data points, the error bars are within the circles.
DISCUSSION
We found previously (1) that the “M · ATP and M · ADP · P states” of unphosphorylated human myosin IIIA have much higher affinities for actin than those of other of myosin. This is reflected by the extremely low Kactin value of the acto-unphosphorylated myosin IIIA ATPase reaction. A critical finding is that phosphorylation of human myosin IIIA at the motor domain markedly increased the Kactin of the actin-activated ATPase activity (1). In this study, we clarified the effects of phosphorylation on the ATP hydrolysis cycle of acto-human myosin IIIA.
All the rate constants and the equilibrium constants obtained in this study are summarized in Table 1. Using these parameters of each elementary kinetic step of the acto-myosin III ATPase cycle, we conducted a computer simulation of the overall ATP hydrolysis cycle pathway at a saturated ATP concentration with an ATP regeneration system as a function of actin concentration. The contribution of k+4, [M], and [MD] to the overall ATPase rate was ignored for the sake of simplicity. The initial values, rates, and equilibrium constants employed in the simulation were as follows: [AM]0 = 20 nM, [AMT]0 = [MT]0 = [MDP]0 = [AMDP]0=[AMD]0=0nM, K′1k′+2[ATP]=60 s−1, K8=150 μM (rapid equilibrium), k+3=3.1 s−1, k−3=0.6 s−1 (K3=6.3), K9 = 350 μM (rapid equilibrium), k′+4 = 60 s−1, and k′+5 = 8.1 s−1. The calculated ATPase activity as a function of actin concentration is shown in Figure 2. Vmax and Kactin values calculated by this simulation were 1.67 s−1 and 10.9 μM, respectively. The calculated ATPase activities at various actin concentrations were consistent with the experimentally obtained ones, suggesting that the kinetic parameters obtained in this study are valid.
Table 1.
Kinetic Parameters of the Phosphorylated and Unphosphorylated huM3MD ATPase Cycle
| signal/method | p-myosin IIIA | up-myosin IIIAa | |
|---|---|---|---|
| steady state | |||
| Vo (s−1) | 0.07±0.02 | 0.05 | |
| Vmax (s−1) | 1.6±0.2 | 0.54 | |
| Kactin (μM) | 10.3±3.2 | 0.13 | |
| ATP binding | |||
| K1k+2 (μM−1 s−1) | mant-ATP | 0.20± 0.01 | 14 |
| k−2 (s−1) | 0.05±0.02 | 0.2 | |
| K′1k′+2 (μM−1 s−1) | mant-ATP | 0.13±0.01 | 0.16 |
| k′−2 (s−1) | 0.65±0.2 | 7.4 | |
| ATP hydrolysis | |||
| k+3 + k−3 (s−1) | quenched-flow | 3.7 ±0.4 | 0.15 |
| phosphate release | |||
| k+4obs (s−1) | MDCC-PBP | 0.36±0.01 | 0.14–0.16 |
| k′+4obs (s−1) | 70± 24 | 1.14–1.26 | |
| ADP binding | |||
| k+5 (s−1) | mant-ADP cold chase | 8.6±0.6 | 6.5 |
| k−5 (μM−1 s−1) | mant-ADP binding | 0.87±0.03 | 0.4 |
| k′+5 (s−1) | mant-ADP cold chase | 8.1±0.06 | 6.2 |
| k′−5 (μM−1 s−1) | mant-ADP binding | 0.56±0.03 | 0.58 |
| actin binding | |||
| k−6 (μM−1 s−1) | light scattering | 0.54±0.03 | 29.8 |
| k+6 (s−1) | light scattering | 0.13±0.02 | 2.3 |
| K6 (nM) | 240 | 68.8 | |
| k−8 (μM−1 s−1) | light scattering | 92.9 | |
| k+8 (s−1) | light scattering | 8.6 | |
| K8 (μM) | 0.09 | ||
| k−10 (μM−1 s−1) | light scattering | 0.14±0.02 | 23.8 |
| k+10 (s−1) | light scattering | 0.15±0.02 | 2 |
| K10 (nM) | 1071 | 71.4 | |
From ref 1.
The actin-activated ATPase cycle of phosphorylated huM3AMD can be explained as follows. The rate of formation of the A · M · ATP complex at physiological ATP concentration (several millimolar) is expected to be higher than 60 s−1, and thus not rate-determining. The off rate for dissociation of ADP from the A · M · ADP complex is more than 4-fold higher than the entire ATPase cycle rate; therefore, this step is not the rate-limiting step, although it partially explains the entire cycle rate. The off rate for dissociation of Pi from the A · M · ADP · Pi complex is significantly higher than the rate of the entire cycle. On the other hand, the ATP hydrolysis rate (k+3 + k−3) can mostly explain the overall cycle rate, and we found on the basis of the simulation that the M · ATP and M · ADP · Pi complexes are 63% of the steady-state intermediates (Figure 9).
Figure 9.
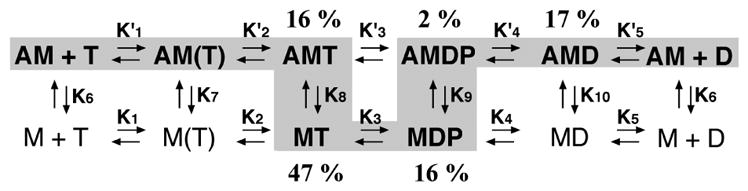
Steady-state distribution of actomyosin IIIA intermediates: A, actin; M, myosin head; T, ATP; D, ADP; P, inorganic phosphate. Note that the myosin IIIA head spends a majority of the ATP hydrolysis cycle in the weak actin binding states.
It was reported previously that the ADP off rate of myosin IIIA can be explained by two steps. This was observed for both myosin IIIA constructs with and without the kinase domain (24, 25). The slow ADP off step was identified as the rate-limiting step for the construct with the kinase domain, while it was thought to be off-pathway for the KD-deleted construct because the rate of this step was significantly slower than the overall cycle rate. In this study, we did not observe the slow ADP off step for the phosphorylated huM3MD construct, and the rate constant of the ADP off step was significantly higher than the rate of the entire cycle; therefore, the ADP off step is not the rate-determining step of phosphorylated huM3MD.
On the basis of the analysis, we found that phosphorylation influences several steps in the ATPase cycle. It was previously shown that the ATP hydrolysis rate (k+3 + k−3) of unphosphorylated huM3AMD of the actin-dissociated form is quite low and comparable with the basal ATPase cycle rate. Accordingly, we could not observe the initial Pi burst with the unphosphorylated form due to the absence of the rapid hydrolysis of ATP to form the M · ADP · Pi ternary complex (1). As shown in Figure 5, we were able to observe the initial Pi burst of phosphorylated huM3AMD. We found that phosphorylation markedly increases the ATP hydrolysis rate by several hundred-fold. The phosphorylation also markedly increases K8. This favors the actin-dissociated form of the A · M · ATP–M · ATP equilibrium. The K9 value was determied by computer simulation to be 350 nM, which is 1000-fold higher than that obtained for unphosphorylated myosin IIIA (1). These results suggest that the effect of phosphorylation is to increase K8 and K9, thus promoting the actin-dissociated intermediates.
On the basis of the computer simulation, we determined the steady-state distribution of intermediates during the acto-myosin IIIA ATPase cycle at the physiological ATP concentration (2 mM) (Figure 9). The major difference is that the predominant intermediates for phosphorylated myosin III are the actin-dissociated forms (M · ATP and M · ADP · Pi), while the actin-associated forms are the predominant intermediate for unphosphorylated myosin III (1). Therefore, we anticipate that phosphorylation induces the dissociation of human myosin IIIA from actin.
It has been shown that fish myosin IIIA without the kinase domain localizes at the tip of filopodia, while full-length myosin IIIA does not significantly localize at the tip of filopodia (17). This finding suggests that myosin IIIA can move on actin bundles within filopodia and phosphorylation attenuates the continuous movement on actin filaments in filopodia. The present finding is consistent with the previous in vivo finding and suggests that the kinase domain is responsible for phosphorylation of the motor domain, thus attenuating the continuous movement on actin bundles in filopodia.
Consistent with this view, it was reported recently that removal of the kinase domain of myosin IIIA shifts to a higher duty ratio, although this change can be due to either the change in phosphorylation level or the structural effect of deleting the kinase domain (25). It is likely that both the phosphorylation of myosin IIIA at the motor domain and the structural interaction between the kinase domain and the motor domain influence the motor properties of human myosin IIIA. Further studies are required for the mechanism of regulation of human myosin III.
In this study, we focused our effort to clarify the effect of phosphorylation of myosin IIIA in the motor domain on its motor properties. To do this, we used the motor domain construct to avoid complications due to the presence of the kinase domain and the tail. Our results clearly indicate that the phosphoryation significantly lowers the affinity for actin and the duty ratio. It is also known that the myosin IIIA tail has an ATP-independent actin binding site. We propose a model in which unphosphorylated myosin IIIA, which has an extremely high affinity for actin and a high duty ratio moves on actin filaments using the ATP-insensitive actin binding at the tail as a tether to actin filaments, thus moving processively without diffusing from the actin track (Figure 10).
Figure 10.
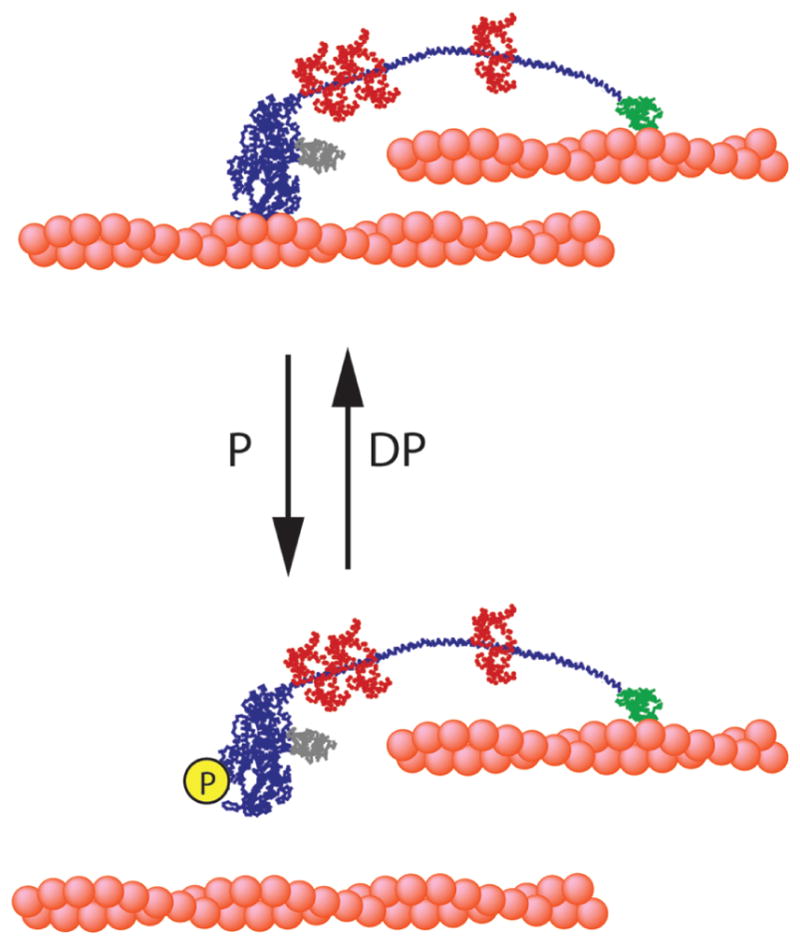
Graphical model of the possible regulation and movement of myosin IIIA: P, phosphorylation; DP, dephosphorylation; ABS, ATP-insensitive actin binding site in the tail.
Footnotes
This work was supported by National Institutes of Health Grants AR048898, AR04856, and DC006103 (to M.I.).
Abbreviations: HM3A, human myosin IIIA; huM3AMD, human myosin IIIA motor domain; PBP, phosphate binding protein; MDCC, 7-(diethylamino)-3-({[(2-maleimidyl)ethyl]amino}carbonyl)coumarin; CBB, Coomassie Brilliant Blue; dmant-, 2′-deoxy(N-methylanthraniloyl)-; CaM, calmodulin; HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; p-, phosphorylated; A, actin; M, myosin head; T, ATP; D, ADP; Pi, inorganic phosphate.
References
- 1.Kambara T, Komaba S, Ikebe M. Human myosin III is a motor having an extremely high affinity for actin. J Biol Chem. 2006;281:37291–37301. doi: 10.1074/jbc.M603823200. [DOI] [PubMed] [Google Scholar]
- 2.Sellers JR. Myosins:Adiverse superfamily. Biochim Biophys Acta. 2000;1496:3–22. doi: 10.1016/s0167-4889(00)00005-7. [DOI] [PubMed] [Google Scholar]
- 3.Berg JS, Powell BC, Cheney RE. A millennial myosin census. Mol Biol Cell. 2001;12:780–794. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsumoto H, Isono K, Pye Q, Pak WL. Gene encoding cytoskeletal proteins in Drosophila rhabdomeres. Proc Natl Acad Sci USA. 1987;84:985–989. doi: 10.1073/pnas.84.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montell C, Rubin GM. The Drosophila ninaC locus encodes two photoreceptor cell specific proteins with domains homologous to protein kinases and the myosin heavy chain head. Cell. 1988;52:757–772. doi: 10.1016/0092-8674(88)90413-8. [DOI] [PubMed] [Google Scholar]
- 6.Walsh T, Walsh V, Vreugde S, Hertzano R, Shahin H, Haika S, Lee MK, Kanaan M, King MC, Avraham KB. From flies’ eyes to our ears: Mutations in a human class III myosin cause progressive nonsyndromic hearing loss DFNB30. Proc Natl Acad Sci USA. 2002;99:7518–7523. doi: 10.1073/pnas.102091699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng KP, Kambara T, Matsuura M, Burke M, Ikebe M. Identification of myosin III as a protein kinase. Biochemistry. 1996;35:9392–9399. doi: 10.1021/bi960181a. [DOI] [PubMed] [Google Scholar]
- 8.Kempler K, Toth J, Yamashita R, Mapel G, Robinson K, Cardasis H, Stevens S, Sellers JR, Battelle BA. Loop 2 of limulus myosin III is phosphorylated by protein kinase A and autophosphorylation. Biochemistry. 2007;46:4280–4293. doi: 10.1021/bi062112u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komaba S, Inoue A, Maruta S, Hosoya H, Ikebe M. Determination of human myosin III as a motor protein having a protein kinase activity. J Biol Chem. 2003;278:21352–21360. doi: 10.1074/jbc.M300757200. [DOI] [PubMed] [Google Scholar]
- 10.Dose AC, Burnside B. A class III myosin expressed in the retina is a potential candidate for Bardet-Biedl syndrome. Genomics. 2002;79:621–624. doi: 10.1006/geno.2002.6749. [DOI] [PubMed] [Google Scholar]
- 11.Dose AC, Burnside B. Cloning and chromosomal localization of a human class III myosin. Genomics. 2000;67:333–342. doi: 10.1006/geno.2000.6256. [DOI] [PubMed] [Google Scholar]
- 12.Les Erickson F, Corsa AC, Dose AC, Burnside B. Localization of a class III myosin to filopodia tips in transfected HeLa cells requires an actin-binding site in its tail domain. Mol Biol Cell. 2003;14:4173–4180. doi: 10.1091/mbc.E02-10-0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider ME, Dose AC, Salles FT, Chang W, Erickson FL, Burnside B, Kachar B. A new compartment at stereocilia tips defined by spatial and temporal patterns of myosin IIIa expression. J Neurosci. 2006;26:10243–10252. doi: 10.1523/JNEUROSCI.2812-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dose AC, Hillman DW, Wong C, Sohlberg L, Lin-Jones J, Burnside B. Myo3A, one of two class III myosin genes expressed in vertebrate retina, is localized to the calycal processes of rod and cone photoreceptors and is expressed in the sacculus. Mol Biol Cell. 2003;14:1058–1073. doi: 10.1091/mbc.E02-06-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salles FT, Merritt RC, Jr, Manor U, Dougherty GW, Sousa AD, Moore JE, Yengo CM, Dose AC, Kachar B. Myosin IIIa boosts elongation of stereocilia by transporting espin 1 to the plus ends of actin filaments. Nat Cell Biol. 2009;11:443–450. doi: 10.1038/ncb1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li XD, Jung HS, Mabuchi K, Craig R, Ikebe M. The globular tail domain of myosin Va functions as an inhibitor of the myosin Va motor. J Biol Chem. 2006;281:21789–21798. doi: 10.1074/jbc.M602957200. [DOI] [PubMed] [Google Scholar]
- 17.Lin-Jones J, Parker E, Wu M, Dose A, Burnside B. Myosin 3A transgene expression produces abnormal actin filament bundles in transgenic Xenopus laevis rod photoreceptors. J Cell Sci. 2004;117:5825–5834. doi: 10.1242/jcs.01512. [DOI] [PubMed] [Google Scholar]
- 18.Smith L, Su X, Lin P, Zhi G, Stull JT. Identification of a novel actin binding motif in smooth muscle myosin light chain kinase. J Biol Chem. 1999;274:29433–29438. doi: 10.1074/jbc.274.41.29433. [DOI] [PubMed] [Google Scholar]
- 19.Thorn KS, Ubersax JA, Vale RD. Engineering the processive run length of the kinesin motor. J Cell Biol. 2000;151:1093–1100. doi: 10.1083/jcb.151.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 21.Brune M, Hunter JL, Corrie JE, Webb MR. Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment. 1994 doi: 10.1021/bi00193a013. [DOI] [PubMed] [Google Scholar]
- 22.Homma K, Ikebe M. Myosin X is a high duty ratio motor. J Biol Chem. 2005;280:29381–29391. doi: 10.1074/jbc.M504779200. [DOI] [PubMed] [Google Scholar]
- 23.Reynard AM, Hass LE, Jacobson DD, Boyer PD. The correlation of reaction kinetics and substrate binding with the mechanism of pyruvate kinase. J Biol Chem. 1961;236:2277–2282. [PubMed] [Google Scholar]
- 24.Dose AC, Ananthanarayanan S, Moore JE, Burnside B, Yengo CM. Kinetic mechanism of human myosin IIIA. J Biol Chem. 2007;282:216–231. doi: 10.1074/jbc.M605964200. [DOI] [PubMed] [Google Scholar]
- 25.Dose AC, Ananthanarayanan S, Moore JE, Corsa AC, Burnside B, Yengo CM. The kinase domain alters the kinetic properties of the myosin IIIA motor. Biochemistry. 2008;47:2485–2496. doi: 10.1021/bi7021574. [DOI] [PubMed] [Google Scholar]


