Summary
Parental environments can influence offspring traits. However, the magnitude of the impact of parental environments on offspring molecular phenotypes is poorly understood. Here, we test the direct effects and intergenerational effects of jasmonic acid (JA) treatment, which is involved in herbivory‐induced defense signaling, on transcriptomes and metabolomes in apomictic common dandelion (Taraxacum officinale).
In a full factorial crossed design with parental and offspring JA and control treatments, we performed leaf RNA‐seq gene expression analysis, LC‐MS metabolomics and total phenolics assays in offspring plants.
Expression analysis, leveraged by a de novo assembled transcriptome, revealed an induced response to JA exposure that is consistent with known JA effects. The intergenerational effect of treatment was considerable: 307 of 858 detected JA‐responsive transcripts were affected by parental JA treatment. In terms of the numbers of metabolites affected, the magnitude of the chemical response to parental JA exposure was c. 10% of the direct JA treatment response. Transcriptome and metabolome analyses both identified the phosphatidylinositol signaling pathway as a target of intergenerational JA effects.
Our results highlight that parental environments can have substantial effects in offspring generations. Transcriptome and metabolome assays provide a basis for zooming in on the potential mechanisms of inherited JA effects.
Keywords: induced defenses, jasmonic acid (JA), LC‐MS, metabolomics, RNA‐seq, Taraxacum officinale (common dandelion), transcriptomics, transgenerational effects
Introduction
The phenotype of a plant can be affected by the environmental experiences of its direct ancestors through effects on the parents that are transmitted to the offspring. Although parental (or intergenerational) effects can be non‐adaptive, they sometimes ‘prepare’ offspring for enhanced performance when the offspring experiences similar environmental stresses to the parents (Galloway & Etterson, 2007; Holeski et al., 2012). In such cases, intergenerational effects may be evolved adaptive responses to environmental stresses, extending adaptive phenotypic plasticity across generations (Herman et al., 2014).
One area in which parental effects are thought to be particularly relevant is in plant–insect and plant–pathogen interactions. Within a single generation, priming of systemic tissue for enhanced defense has been well documented and can be induced by pathogen attack or other cues (Fu & Dong, 2013; Pieterse et al., 2014). Priming of the defense response does not constitutively activate defense responses, but results in a more rapid activation of the defense response on subsequent pathogen attack. Such effects that are induced by pathogens or herbivores can persist to offspring (Holeski, 2007) and, in some cases, effects are sustained for multiple generations (Luna et al., 2012; Rasmann et al., 2012). The underlying mechanisms are not completely understood, but mounting evidence for durable epigenetic changes in response to environmental cues (Feil & Fraga, 2012) indicates at least one possible mechanism.
Although parental environmental effects on offspring phenotypes have been shown repeatedly, there is little knowledge of the extent to which gene expression is affected by parental environment, and the limited available data to date show mixed results. Examples of transgenerational effects in plants are associated with histone modifications at defense gene promoters (Luna et al., 2012) and with small interfering RNA (siRNA) (Rasmann et al., 2012), implicating epigenetic gene regulation in offspring after parental exposure. Exposure of Arabidopsis thaliana to the bacterial elicitor flagellin has been reported to increase homologous recombination frequencies in this plant and several subsequent generations; however, whole‐transcriptome microarray analysis revealed no effects on offspring gene expression (Molinier et al., 2006). In sharp contrast, artificial leaf herbivory in Mimulus guttatus triggers gene expression changes at nearly 1000 genes in untreated offspring (Colicchio et al., 2015).
In addition to up‐ or downregulation of specific genes in offspring, variability in offspring gene expression may also be affected. Increased variability can arise as a result of variable penetrance amongst offspring individuals or of epigenetic mutations that are triggered stochastically in germline tissue in response to stress. In either scenario, the result is hypothesized to be a bet‐hedging strategy to increase levels of phenotypic variation amongst offspring, which may be adaptive when environments are variable (Levy et al., 2012; Herman et al., 2014).
Better insight into the consequences of parental environmental effects on offspring gene expression is important to understand the ecological role and evolution of phenotypic plasticity. From a practical perspective, it is also relevant to determine whether parental environments should be taken into account in the set‐up of transcriptomic studies in general, which do not always control for pre‐experiment variation. Here, we use RNA‐seq expression profiling and LC‐MS metabolomics in the apomictic common dandelion (Taraxacum officinale) to evaluate how leaf gene expression and metabolites are affected in offspring as a result of jasmonic acid (JA) treatment in the parental generation. JA is a plant signaling hormone involved in various processes, including the regulation of growth and responses to biotic and abiotic stresses (Wasternack & Hause, 2013), and plays a major role in the induction of plant chemical defenses in response to herbivory. The application of JA solutions to plants generally elicits an induction of chemical defenses that is systemic (e.g. Schenk et al., 2000; van Dam et al., 2004; De Vos et al., 2005; Tytgat et al., 2013).
Taraxacum officinale is a convenient natural model system for such studies because of its apomictic reproduction through clonal seeds (van Dijk, 2003), which permits an evaluation of transgenerational effects in the absence of genetic differences between experimental plants. In T. officinale, effects of parental JA treatment on offspring epigenetic profiles (Verhoeven et al., 2010) and on offspring resistance to caterpillar feeding (Verhoeven & van Gurp, 2012) have been reported previously, showing a potential role for epigenetically mediated parental effects on herbivore resistance in this species. In this study, we specifically aimed to: evaluate the intergenerational gene expression response, in terms of effects on gene expression means and variances, after parental JA treatment; and determine whether a parental effect of JA is associated with modified offspring leaf (secondary) chemistry including defense compounds.
Materials and Methods
Plant material and experimental design
The common dandelion, T. officinale (F.H. Wigg.), is a widespread perennial plant species which has diploid sexual and polyploid (mostly triploid) obligate apomictic variants (van Dijk, 2003). Apomixis in dandelion is through meiotic diplospory followed by parthenogenetic embryo development from unreduced egg cells and autonomous endosperm development (Koltunow, 1993), which is thought to result in seeds that are clonal copies of the heterozygous mother plant. For this study, we used a single triploid apomictic genotype (A68), an accession collected near Heteren (the Netherlands) which had been propagated for multiple generations under common glasshouse conditions before the experiment. This genotype has been studied previously in the context of parental effects (Verhoeven & van Gurp, 2012). Some genomic resources are available for T. officinale, including an expressed sequence tag (EST) database (Compositae Genome Project, compgenomics.ucdavis.edu) and a de novo assembled transcriptome based on RNA‐seq data of a different apomictic genotype than used for the current study (Ferreira de Carvalho et al., 2016), but currently no annotated reference genome has been published. For this study, we generated a new de novo assembled reference transcriptome specific for the A68 apomictic genotype (see below).
Parental generation
Eight ‘control’ and eight ‘JA’ parental treatment lineages were derived from a single A68 founder individual by subjecting plants for two subsequent generations to either JA or control treatments under common climate chamber conditions (14 h : 10 h, light : dark at 20°C : 15°C, fully randomized pots) using single‐seed descent between the generations. Exposing two subsequent generations to the same environmental stress can enhance parental effects compared with single‐generation parental exposure (Wibowo et al., 2016). Based on previous experience in dandelion (Verhoeven & van Gurp, 2012), JA treatment was applied as a 10 mM JA solution (Sigma J‐2500, dissolved in ethanol and diluted to the desired concentration with a 0.1% Triton X‐100 surfactant solution) to the upper surface of three to four fully expanded leaves. In generation 1, 0.75 ml JA solution per plant was applied to 8‐wk‐old plants; in generation 2, a total amount of 0.75 ml was applied to each plant distributed over two application treatments when plants were 5 and 7 wk old. In both generations, JA was applied during vegetative growth, c. 1 month before first flowering.
Experimental generation
For each of the G2 ‘control’ and ‘JA’ parental treatment lineages, seeds from a single seed head were weighed individually, surface sterilized (0.5% sodium hypochlorite wash) and germinated on 0.8% agar plates. After 10 d, seedlings were transplanted to individual pots and grown under climate chamber conditions as described above in fully randomized blocks. Each block contained two G3 plants from each of the eight JA (J) and control (C) parental lineages; one of these two plants received a JA treatment (JJ or CJ, depending on parental lineage) and the other a mock treatment (JC and CC) (two parental treatments × two experimental treatments × eight independent replicates = 32 plants per block; see Fig. 1 for an overview of the experimental design). JA was applied to 8‐wk‐old plants by distributing 0.25 ml of a 10 mM JA solution (see above) over the surface of two standardized leaves. Mock‐treated plants received 0.25 ml of a similar ethanol/Triton‐X solution without JA. In one block of plants, 3 h after treatment, two standardized leaves (younger than the JA‐treated leaves) were collected, discarding the latex‐rich mid‐vein, flash frozen in liquid nitrogen and stored at −80°C for subsequent RNA analysis. In a second block of 32 plants, leaf tissue was sampled in a similar way (but including the mid‐vein) 24 h after treatment for subsequent leaf chemical analysis; these samples were flash frozen in liquid nitrogen, freeze dried and stored at −80°C. Three additional blocks of 32 plants were grown for time‐series reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) gene expression analysis of a known JA early response gene (LOX2) to validate the induced JA response (see Supporting Information Notes S1). It should be noted that, for chemical and RT‐qPCR expression analysis, all available replicate plants were used but, for RNA‐seq expression analysis, only six replicates per group were used (see below).
Figure 1.
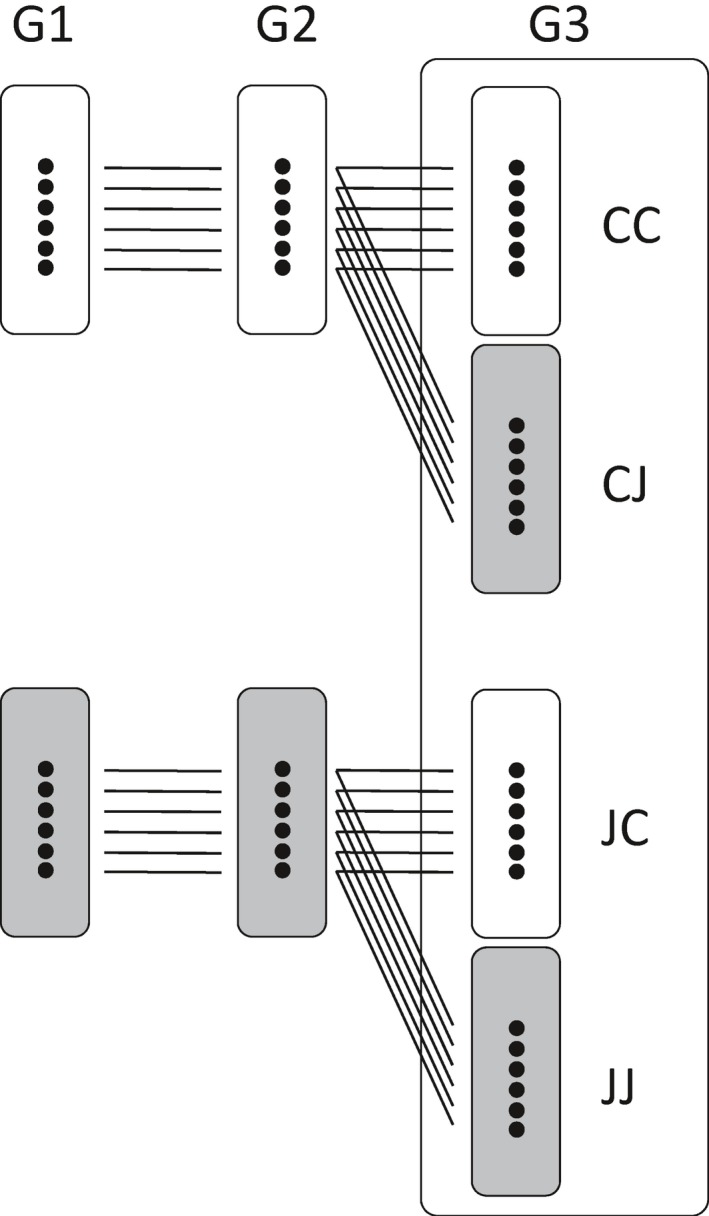
Experimental design. Common dandelion plants were exposed to jasmonic acid (JA) treatment (gray boxes) or control (white boxes). G1 and G2 indicate the ‘parental’ treatment. Transcriptome and metabolome analyses were performed in G3 plants, assessing the effects of both experimental (direct) JA treatment and parental JA treatment. The four experimental G3 groups are coded as CC, CJ, JC and JJ, where the first letter denotes the parental treatment (J = JA, C = control) and the second letter denotes the treatment of the G3 experimental plants. For chemical analyses, eight replicates per group were used but, for RNA‐seq analysis, only six replicates per group were used.
RNA‐seq expression analysis
RNA isolation
Total RNA was isolated from liquid nitrogen‐ground tissue using Trizol (Ambion, Life Technologies, Carlsbad, CA, USA) according to the manufacturer's protocol, with an additional chloroform phase separation. Quality and concentration were checked on agarose gels and on a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). For each sample, 10 μg of total RNA was DNase treated using the Turbo DNA‐free kit (Ambion, Life Technologies). Quality and concentration were checked again on agarose gels and the NanoDrop spectrophotometer, and samples were stored at −80°C until further use.
RNA‐seq library preparation
Based on RNA quality, six samples from each of the four experimental groups (CC, CJ, JC, JJ; see Fig. 1) were prepared and barcoded individually using the TruSeq RNA Sample Prep kit v.2 with 24 available barcodes from index sets A and B (Illumina, San Diego, CA, USA; cat. nos. RS‐122‐2001 and RS‐122‐2002). Before sample preparation, we added 92 synthetic ERCC RNA spike‐in control sequences (Jiang et al., 2011) (Ambion, Life Technologies, catalog number 4456739) at 50% of the manufacturer's recommended concentration. Samples were quantified (Kapa Biosystems, Wilmington, MA, USA; cat. no. KK4824) and tested using PCR primers of Ambion ERCC controls (Life Technologies, cat. no. 4456739). Two ERCC controls of low concentration (ERCC 85 and ERCC 28) and two ERCC controls of high concentration (ERCC 130 and ERCC 4) were amplified and the cycle numbers were compared. All samples showed a qualitative difference between low and high spikes. Samples were pooled and run as a single multiplexed library on the Illumina Hiseq with one lane at Florida State University (HiSeq 2000, single end 101 bp) and two lanes at Wageningen University (HiSeq 2000, paired end 101 bp). After demultiplexing, it appeared that one sample (of the CJ group) was represented by very few reads, and this sample was excluded from further analysis. The remaining samples were examined for quality using the ERCC controls. Plots of the expected concentration vs the read count for each sample (Fig. S1) and Bland–Altman (BA) plots (Bland & Altman, 1986) between samples (Fig. S2) showed high‐quality libraries, supporting quantitative interpretation of sequence read output.
De novo transcriptome assembly
Raw Fastq files were de‐multiplexed and adapters were trimmed using Fastq‐mcf (v.488 with default settings) from Ea‐utils (Aronesty, 2011), which trims adapters and filters reads based on a minimum phred score of 20. In addition, the first 10 nucleotides of all reads (both forward and reverse) were trimmed using Seqtk (https://github.com/lh3/seqtk), because this has been shown to improve the assembly of full‐length transcripts (van Gurp et al., 2013). Overlapping paired‐end reads were merged using Fastq‐join from Ea‐utils (Aronesty, 2011). De novo transcriptome assembly was performed using Trinity v.trinityrnaseq‐r2013‐02‐16 (Haas et al., 2013) using default settings. The final assembly (Dryad Digital Repository: https://doi.org/10.5061/dryad.b15tr) contained 192 951 contigs (unique Trinity comp_c_seq combinations) with minimum, median, mean and maximum lengths of 200, 809, 1107 and 17 258 bp, respectively. The contigs clustered into 77 530 putative genes (unique Trinity comp_c combinations). All 192 951 contigs were mapped to the reference proteome of eudicots (NCBI RefSeq) consisting of 1 312 075 reference proteins using uBlastx in Usearch v.6.0.307 (minimum E‐value of 1e‐5); this algorithm has similar sensitivity to NCBI Blastx, but is much faster (Edgar, 2010). Command line output parameters were set to default, except for the output format, which was set as ‘‐userfields query + target + thi + bits + raw + evalue + qlo + qhi + tlo + thi + qframe + tframe +ids + gaps + alnlen + qrow + trow + pv + ql’, in order to obtain a tabular file that was subsequently converted to an xml input file as required by Blast2go (Conesa et al., 2005). Blast2go was used to perform annotations. A maximum of 20 top Blast hits (with E‐value < 1e‐5) was retained per contig with associated gene ontology (GO) terms as determined by Blast2go. Within Blast2go, Interproscan was run, for which results were obtained for a subset of 110 016 contigs. GO terms were derived in Blast2go based on both the Blast hits as well as the Interpro results. We observed that different contigs (comp_c_seq combinations) that belonged to the same putative gene (comp_c combination) did not always produce matching annotations, which indicates that pooling contigs for an analysis at the putative gene level would introduce an unknown amount of error because of imperfect assignment of contigs to genes. Rather than working with this unknown level of uncertainty, we decided to analyze at the contig level. Although it probably carries a multiple testing penalty, this allows for more certain interpretation of the significant results.
Differential gene expression analysis
As mapping algorithms are greedy, all contigs were used as the reference for alignments. Samples were aligned using Bowtie (Langmead et al., 2009) with the following settings: –best, –tryhard, –strata, ‐a, ‐v 3 and Last (Kielbasa et al., 2011) with the ‐l 25 setting. Several normalization strategies were evaluated (Dillies et al., 2013) using BA plots of the ERCC controls. The log(RPKM) (reads per kilobase per million mapped reads) was selected as its related BA plots were the most consistent amongst all replicates (Fig. S2). Contigs were retained for quantitative analysis if they were expressed at an average of at least 10 reads per nucleotide in all four experimental groups (CC, CJ, JC, JJ) and were at least 500 nucleotides long (n = 65 827). Across all samples, this set of analyzed contigs had an average read coverage per nucleotide of 84.1 per individual sample (median 25.3). Application of the 10× coverage criterion to each of the experimental groups enables robust statistical analysis using linear models and discarded 49 658 contigs that had low expression in all of the experimental groups. This approach also excluded 11 766 contigs that showed no or low expression (< 10×) in some treatments but not in all treatments. Although we do not provide statistical evidence for treatment effects in these 11 766 contigs, this set may include contigs that are downregulated in response to treatment in one or more of the experimental groups (see Table S1 for the list of 11 766 contigs).
Normalized expression estimates were modeled using the following model: Y ij = μ + t i + εij. where i = (CC, CJ, JC, JJ) and j = (1,…, 6). ε i were assumed as ~N(0,σ2 i) (Law et al., 2014). Initial model fits with a common variance assumption did not satisfy model assumptions of residuals. The F test of the null hypothesis of homoscedastic error was rejected for 43% of the contigs at a false discovery rate (FDR) = 0.05 and 62% of the contigs at FDR = 0.20. In addition, the bayesian information criterion (BIC) for the homoscedastic model was worse than BIC for the heteroscedastic model 100% of the time. Thus, we fitted heteroscedastic models, for each contig separately, that allowed for different error variances for each experimental group. Individual contrasts were conducted to test the effect of parental JA treatment whilst controlling for the current (experimental) JA treatment (Ho: μCC − μJC = 0; Ho: μCJ − μJJ = 0) and to test the effect of the current JA treatment controlling for the parent treatment (Ho: μCC − μCJ = 0; Ho: μJJ − μJC = 0). Additional contrasts for the interaction between the parental and current JA treatment (Ho: μCC − μCJ = μJJ − μJC) and the effect of parent treatment on current JA response (Ho: μCC − μJJ = 0; Ho: μJC − μCJ = 0) were conducted. All 65 827 contrasts were simultaneously corrected for false discovery (Storey & Tibshirani, 2003). Unless otherwise specified, we consider an FDR of 0.10 to be significant. The results were qualitatively similar at FDR = 0.05 and FDR = 0.20. We selected FDR = 0.10 as a balance between type I and type II errors (Verhoeven et al., 2005). Results were merged with annotation, and enrichment tests were performed using annotation from Blast2go (Conesa et al., 2005). Because the number of differentially expressed genes was relatively low at the FDR = 0.10 significance threshold, we performed GO enrichment tests based on a significance threshold for individual transcripts at FDR = 0.20. This relaxed significance threshold results in a larger set of significant genes whilst maintaining the expected proportion of false positive results < 20%, potentially allowing for more robust enrichment analysis. Fisher's exact enrichment tests were carried out at the putative gene level, pooling for each putative gene all unique GO annotations associated with its underlying contigs and comparing the list of significant putative genes with the list of all genes analyzed.
Untargeted metabolomic profiling
LC‐MS analysis
Twenty milligrams of the freeze‐dried and finely ground leaf material were extracted with 200 μl of methanol, followed by a second extraction with 200 μl of 20% methanol containing 0.1% formic acid. Both supernatants were combined and dried in a vacuum concentrator. Pellets were re‐dissolved in 60 μl of 20% methanol with 0.1% formic acid, 5 μl of which were injected onto the analytical column. For LC‐MS analysis, a Synapt G2 mass spectrometer equipped with an Acquity UPLC (Waters, Milford, MA, USA) was used. Chromatography was performed with a flow rate of 200 μl min–1 on a Waters Acquity C18 HSS T3 column, 2.1 × 100 mm, 1.8 μm. A 10‐min gradient from 99% water to 100% methanol (both solvents with 0.1% formic acid) was used to separate the different compounds. For ionization, positive and negative electrospray ionization modes were used. The mass spectrometer was operated in MS and MSE modes in parallel with a scan range from m/z 50–2000. Extraction and alignment of the raw data were carried out using Waters MarkerLynx software.
Data analysis
Peak intensities were normalized to a total intensity of 10 000 per sample and filtered to include only mass signals present in five or more samples. To analyze differences between treatment groups, principal component analysis (PCA) and partial least squares‐discriminant analysis (PLS‐DA) on normalized data were performed in SIMCA 13.0.3. PLS‐DA models were cross‐validated with permutation tests (999 permutations). To select m/z values for further identification, we followed a two‐step approach. First, we performed orthogonal partial least squares‐discriminant analyses (OPLS‐DA) to obtain S‐plots and visually selected mass signals that showed the clearest association with JA treatment (either parental or direct JA treatment). All the (O)PLS‐DA models showed evidence of overfitting: R 2 and Q 2 of permuted data were not different from R 2 and Q 2 of real data (full model negative mode Q 2 = 0.16, CV‐ANOVA P = 0.12; positive mode Q 2 = 0.19, CV‐ANOVA P = 0.15). We therefore considered evidence from S‐plots as suggestive, but not as conclusive, for the detection of associations between mass signals and treatment.
Second, each mass signal was modeled using an ANOVA, testing the effects of direct JA treatment, parental JA treatment and the direct JA × parental JA interaction on mass signal scores. Normalized mass signals were ln‐transformed before this analysis. P values were subjected to FDR correction for multiple testing (across all P values from all model factors simultaneously) and were considered to be significant at FDR = 0.1. Only mass signals were considered for which at least three samples were present in the filtered dataset (normalized signal > 0) in each of the four experimental groups, and for which analysis of model residuals showed that residuals did not deviate significantly from a normal distribution (Shapiro–Wilk test P > 0.05).
We combined evidence from the visual OPLS‐DA S‐plots and ANOVA statistical testing approaches. We report as the subset of signals with high‐confidence treatment effects those mass signals that were identified in both approaches. The putative identification of these relevant metabolites was based on mass spectra and molecular formula.
Total phenolics assay
Total phenolics concentration was quantified using a Folin–Denis‐based protocol as described elsewhere (Engelkes et al., 2008). Briefly, freeze‐dried samples were ground to a fine powder and phenolics were extracted in 50% aqueous methanol at 90°C for 2 h. The total phenolic concentration was determined by exposing the samples to Folin–Denis reagent, and subsequently quantified spectrophotometrically at 750 nm by comparing the absorbance with a tannic acid calibration curve. Concentrations were expressed as tannic acid equivalent per gram of dried sample. All leaf samples were quantified in two independent replicates, whose phenolic content estimates were averaged for subsequent ANOVA to test for effects of JA treatment in the parental and experimental generations. The initial seed weight of experimental plants was included in the model as a covariate to correct for effects of initial size on the concentration of phenolics.
Data deposition
Dandelion leaf chemical data (LC‐MS peak intensity signals and total phenolics) and the de novo assembled dandelion transcriptome are deposited in the Dryad digital repository (https://doi.org/10.5061/dryad.b15tr).
Dandelion RNA‐seq reads are deposited in the NCBI Sequence Read Archive (BioProject accession no. PRJNA316842; samples SRS2047454–SRS2047475).
Results
Effects of direct JA exposure on gene expression
RT‐qPCR expression analysis of the JA early response gene LOX2 confirmed that the experimental JA treatment elicited a systemic response that was detectable in the tissue of leaves that had not themselves been exposed to JA (see Notes S1). In offspring of control parents, RNA‐seq analysis detected 149 contigs that were differentially expressed as a result of direct JA exposure; in offspring of JA‐treated parents, 440 contigs were differentially expressed as a result of direct JA exposure (Fig. 2a; see Table S2 for all RNA‐seq test results). Thirty‐eight contigs responded to direct JA exposure irrespective of parental treatment and, accounting for this overlap, a total of 551 unique contigs showed a direct JA effect when controlling for parental treatment. Most expression differences observed were upregulations caused by JA treatment (c. 70% of affected contigs, Table S2). The 551 contigs clustered into 244 putative genes. At a more relaxed significance threshold of FDR = 0.20, we detected a total of 1519 significant contigs, clustering into 664 putative genes, and enrichment analysis revealed that this set of 664 differentially expressed genes was significantly enriched for GO terms associated with JA responses (Table 1). Considering only the most specific GO terms, 24 terms were significantly enriched, including biological processes related to JA biosynthesis: ‘Response to wounding’, ‘Response to other organisms’, ‘Response to host immune responses’, ‘Pathogen‐associated induction of host innate immune response’ and ‘Response to JA’ (Table 1). This confirms that our RNA‐seq approach was successful in capturing the elicited JA effect.
Figure 2.
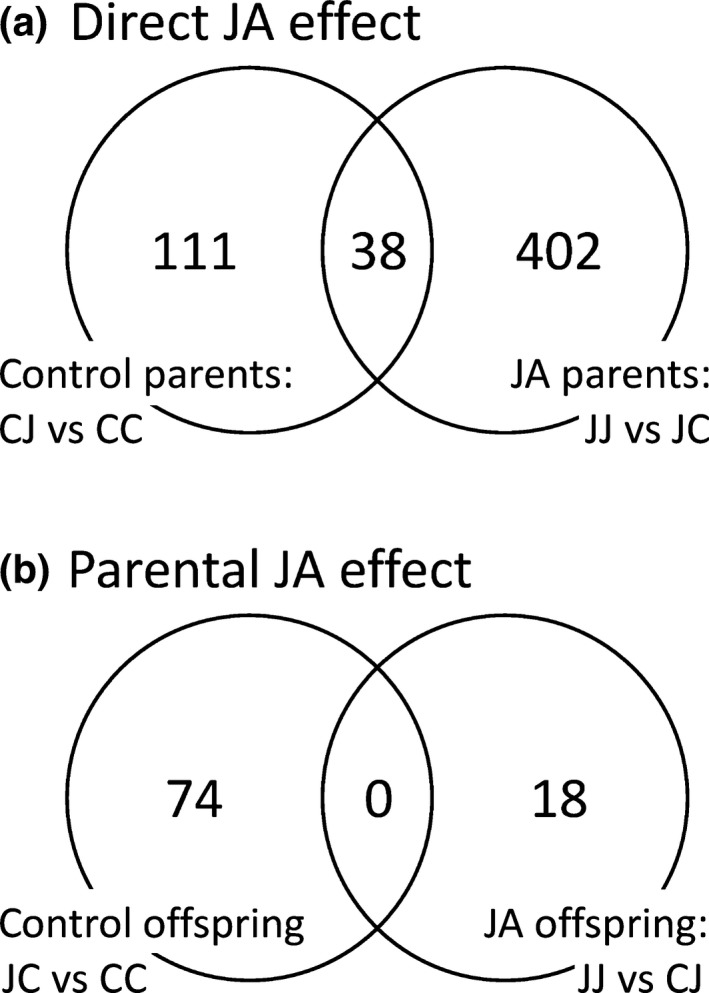
Number of differentially expressed contigs in offspring plants as a result of jasmonic acid (JA) treatment in either the offspring generation (a; direct JA effect) or parental generation (b; parental JA effect) in common dandelion. CC, CJ, JC and JJ denote the four experimental groups, in which the first letter is the parental treatment (J = JA, C = control) and the second letter is the treatment of the experimental (offspring) plants. Differential expression was tested in a priori contrasts that control for parental treatment when testing the direct JA effect, or for offspring treatment when testing the parental JA effect (significance threshold FDR = 0.1).
Table 1.
Gene ontology (GO) terms enriched amongst the jasmonic acid (JA)‐responsive genes in common dandelion (most specific GO terms only)
| GO ID | GO term | Typea | Number of genes | P valued | |
|---|---|---|---|---|---|
| Sign.b | Ref.c | ||||
| 1. Direct JA treatment: significant JJ–JC and/or CJ–CC contrasts | |||||
| GO:0009695 | Jasmonic acid biosynthetic process | P | 14 | 23 | 2.0−10 |
| GO:0009611 | Response to wounding | P | 44 | 379 | 9.2−8 |
| GO:0042401 | Cellular biogenic amine biosynthetic process | P | 17 | 78 | 7.0−7 |
| GO:0006571 | Tyrosine biosynthetic process | P | 14 | 52 | 8.0−7 |
| GO:0009094 | l‐Phenylalanine biosynthetic process | P | 14 | 52 | 8.0−7 |
| GO:0000162 | Tryptophan biosynthetic process | P | 15 | 63 | 1.2−6 |
| GO:0072329 | Monocarboxylic acid catabolic process | P | 12 | 44 | 4.4−6 |
| GO:0009423 | Chorismate biosynthetic process | P | 4 | 1 | 1.8−5 |
| GO:0006635 | Fatty acid beta‐oxidation | P | 9 | 27 | 1.9−5 |
| GO:0051707 | Response to other organism | P | 92 | 1291 | 3.6−5 |
| GO:0050660 | Flavin adenine dinucleotide binding | F | 9 | 31 | 4.6−5 |
| GO:0009821 | Alkaloid biosynthetic process | P | 6 | 11 | 5.7−5 |
| GO:0005783 | Endoplasmic reticulum | C | 51 | 609 | 7.9−5 |
| GO:0080167 | Response to karrikin | P | 29 | 294 | 2.6−4 |
| GO:0004190 | Aspartic‐type endopeptidase activity | F | 6 | 16 | 2.9−4 |
| GO:0008970 | Phosphatidylcholine 1‐acylhydrolase activity | F | 3 | 1 | 3.3−4 |
| GO:0052572 | Response to host immune response | P | 6 | 17 | 3.7−4 |
| GO:0034976 | Response to endoplasmic reticulum stress | P | 6 | 17 | 3.7−4 |
| GO:0052033 | Pathogen‐associated molecular pattern dependent induction by symbiont of host innate immune response | P | 6 | 17 | 3.7−4 |
| GO:0030433 | ER‐associated ubiquitin‐dependent protein catabolic process | P | 4 | 5 | 3.9−4 |
| GO:0005777 | Peroxisome | C | 29 | 304 | 5.5−4 |
| GO:0009415 | Response to water | P | 40 | 482 | 6.2−4 |
| GO:0001676 | Long‐chain fatty acid metabolic process | P | 4 | 6 | 6.3−4 |
| GO:0009753 | Response to jasmonic acid | P | 31 | 336 | 6.3−4 |
| 2. Parental JA treatment: significant JJ–CJ and/or JC–CC contrasts | |||||
| No significant enrichment of GO terms detected | |||||
For this GO enrichment analysis, genes were considered to be differentially expressed at an FDR = 0.20 significance threshold.
C, cellular component; F, molecular function; P, biological process.
Significant gene set for direct JA effect: 1519 significant contigs at FDR 0.20 = 664 putative genes, 526 of which with GO annotation were included in the enrichment analysis. Significant gene set for parental JA effect: 451 significant contigs at FDR 0.20 = 173 putative genes, 133 of which with GO annotation were included in the enrichment analysis.
Reference gene set consists of 11 961 analyzed and GO‐annotated putative genes.
Two‐sided Fisher's exact test; all P‐values are significant after FDR control at 0.05.
Effects of parental JA exposure on gene expression
When controlling for experimental treatment in offspring plants, 18 and 74 contigs were differentially expressed as a result of parental JA exposure in offspring groups that received JA or control treatment, respectively (Fig. 2b). Two additional contrasts, CC vs JJ and CJ vs JC, captured joint effects of direct JA treatment and parental JA treatment and were significant for 190 and 237 contigs (Table S2). In total, across the entire experimental design, 858 different contigs were differentially expressed between groups, 551 of which were detected as direct JA effects and the remaining 307 were detected only when also taking the parental treatment into account.
Hierarchical clustering of the 92 contigs that were differentially expressed in the same offspring environment as a result of different parental treatments indicates that the expression of these genes in JC plants is more similar to that in JJ plants than to that in CC plants (Fig. 3). This is consistent with direct JA effects that are sustained into the offspring generation. However, only five contigs (which clustered into three putative genes) overlapped between the genes that were significantly affected as a result of direct JA treatment and parental JA treatment (Table S2).
Figure 3.
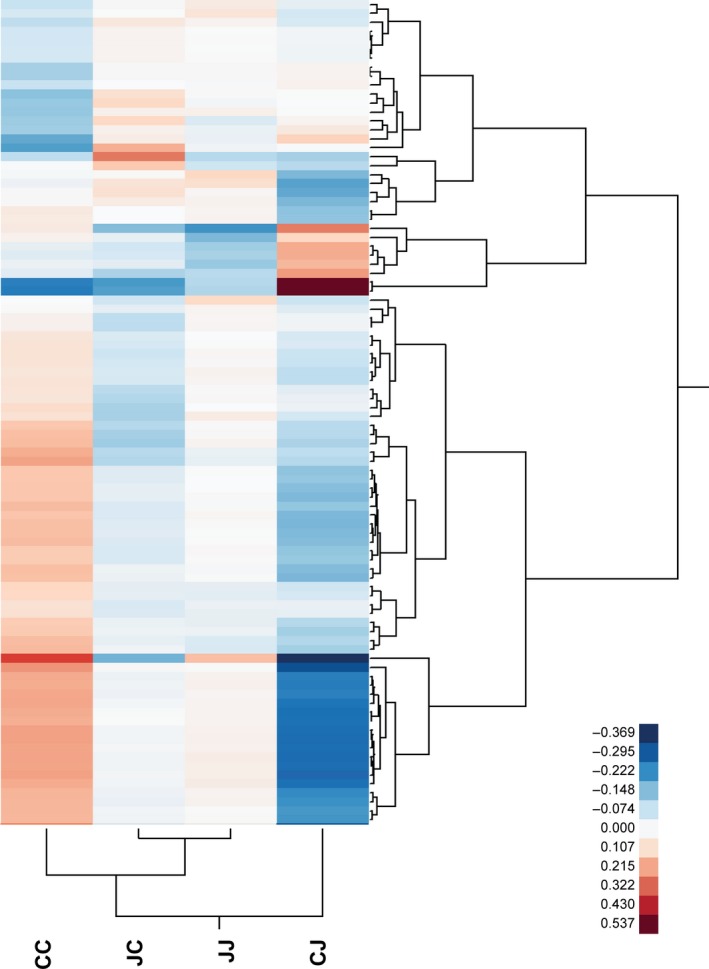
Hierarchical clustering of experimental groups and contigs based on expression scores of contigs that are differentially expressed in offspring as a result of parental jasmonic acid (JA) treatment in common dandelion. CC, CJ, JC and JJ denote the four experimental groups, in which the first letter is the parental treatment (J = JA, C = control) and the second letter is the treatment of the experimental (offspring) plants. The analysis includes 92 contigs with significant JJ–CJ contrast test and/or JC–CC contrast test (FDR = 0.1). Expression scores are log(RPKM) values averaged across replicate plants within experimental groups.
At a more relaxed significance threshold of FDR = 0.20, 451 differentially expressed contigs were detected after parental JA treatment, clustering into 173 putative genes. No significant enrichment of GO terms was observed amongst this set of 173 genes. Lack of significant GO term enrichment may be caused by the low number of significant genes. However, the list of top Blast hits for the contigs that showed a significant parental JA effect (Table S2) indicated several genes that are consistent with reported JA responses or plant defense function, such as genes associated with phosphoinositide signaling (two different inositol phosphate kinases; Sheard et al., 2010; Laxalt & Munnik, 2002), defense‐associated fatty acid epoxidation (CYP77A; Sauveplane et al., 2009), receptor‐like serine threonine kinases (often involved in pathogen recognition and defense signaling; Afzal et al., 2008), an ethylene‐responsive transcription factor (involved in pathogen defenses and the integration of hormonal signaling under stress; Müller & Munné‐Bosch, 2015) and respiratory burst oxidases (Torres & Dangl, 2005).
Effects of JA treatment on between‐replicate variation in expression
Forty‐four per cent of all analyzed contigs showed significant differences in variances between the four experimental groups (CC, CJ, JC, JJ), as indicated by a significant difference in the estimated variances (Folded F, FDR = 0.05), and all models showed an improved fit based on the BIC when the group‐specific error variances were included compared with models that assumed a common error. In these contigs with significant heteroscedasticity, almost always the CJ group had the highest variance (96.3% of contigs, see Table S2). Although there is a difference in sample size (n = 5 in the CJ group and n = 6 in the other groups), this suggests that JA treatment leads to large between‐replicate variation in gene expression 3 h after treatment. However, no trend was observed indicating that JA treatment of parents leads to increased variance in offspring gene expression. Indeed, the estimated variance of expression for the CC group was larger than the estimated variance for the JC group in the large majority of contigs with significant heteroscedasticity (Fig. 4).
Figure 4.
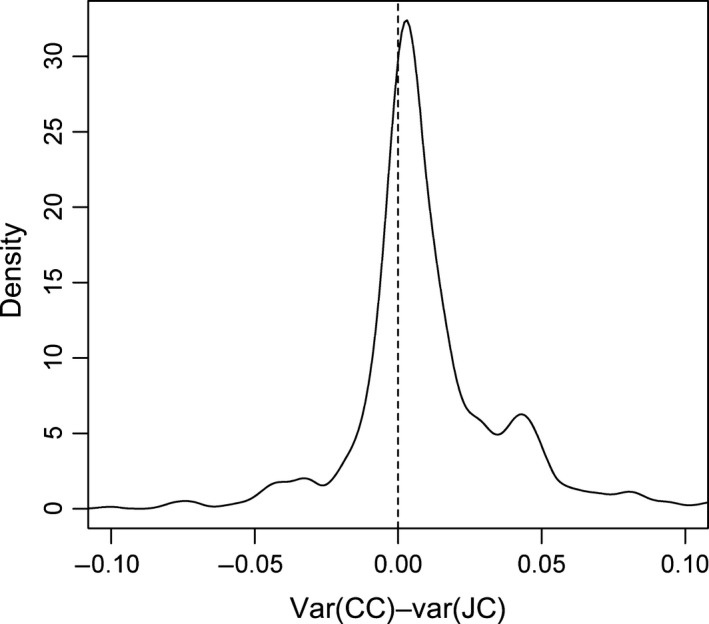
Between‐plant variance in gene expression is reduced, not increased, in common dandelion plants whose parents received jasmonic acid (JA) treatment. Density distribution of the difference in variance between CC and JC groups, based on the subset of contigs (43.8% of the total) with significant heteroscedasticity amongst the four experimental groups. CC and JC denote the experimental groups in which CC received control treatment in the parental as well as in the experimental (offspring) generation, and JC received parental JA treatment and offspring control treatment.
Effects of direct and parental JA exposure on leaf chemical composition
Untargeted LC‐MS metabolomics profiling detected, on average, 968 mass signals per sample (in total 1210 across all samples) in the negative ionization mode and an average of 5151 mass signals per sample (in total 7728 across all samples) in the positive ionization mode (data accessible at Dryad Digital Repository: https://doi.org/10.5061/dryad.b15tr). The two ion modes show a slightly different selectivity based on the propensity of a molecule to gain or lose a proton. For example, phenolic compounds are detected well in the negative mode, whereas N‐based metabolites, such as alkaloids, are generally better detected in the positive mode. In both modes, PCAs clearly separated samples based on the direct JA treatment 24 h before tissue sampling (CJ and JJ vs CC and JC, Fig. 5). An effect caused by parental JA treatment was visible: CC and JC samples clustered with only limited overlap (red vs green dots, Fig. 5). Such separation based on parental treatment was not observed in plants that received JA treatment 24 h before sampling (i.e. CJ and JJ, Fig. 5). ANOVA also indicated a strong induction of the leaf metabolome by JA treatment, where c. 16% of the tested mass signals showed a significant effect of direct JA treatment (Table 2). In addition, parental JA treatment had a significant effect (either as a main effect or in interaction with experimental treatment, Table 2) in 1.6% of mass signals.
Figure 5.
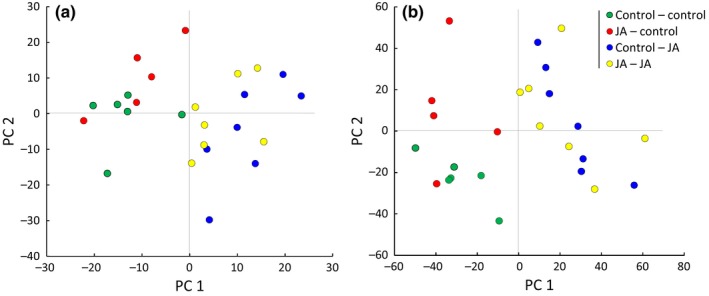
LC‐MS metabolomics analysis of leaf tissue from common dandelion plants sampled 24 h after jasmonic acid (JA) treatment (blue and yellow dots) or 24 h after control treatment (green and red dots), and whose parents had received either JA treatment (yellow and red dots) or control treatment (blue and green dots). (a) Principal component analysis (PCA) clustering based on mass signals of the LC‐MS negative ion mode; axes 1 and 2 explain 14% and 11% of the variation, respectively. (b) PCA clustering based on LC‐MS positive mode; axes 1 and 2 explain 13% and 9% of the variation, respectively.
Table 2.
Number of LC‐MS mass signals affected by direct and parental jasmonic acid (JA) treatment in common dandelion (ANOVA, significance threshold FDR = 0.1)
| No. mass signals | |
|---|---|
| Total | 8938 |
| ANOVA testeda | 2821 |
| Significant direct JA effect | 463 (16.4%) |
| Significant parental JA effect | 31 (1.10%) |
| Significant direct × parental JA interaction | 15 (0.53%) |
At least three non‐zero observations in each of the four experimental groups, and normally distributed residuals.
Based on a visual inspection of S‐plots from OPLS‐DA, we selected 33 mass signals as potentially associated with direct JA treatment and/or with parental JA treatment (Fig. S3). Of these, 16 were also significant in the ANOVA tests and six of these overlapping results could be putatively assigned to known compounds (Table S3). Based on these putative assignments, the experimental JA treatment response involved changes in linolenic acid, caftaric acid, phosphatidylglycerol and phosphatidylinositol. A response to parental JA treatment was detected in phosphatidylinositol and glycosylated malonic acid (Table S3). Mass signals that were selected in the OPLS‐DA S‐plots that showed an effect of JA, but not significant in the ANOVA test, included putative assignments to caftaric acid, chicoric acid, phosphatidylcholine and a glycosylated flavone.
When looking at a specific class of compounds with known anti‐herbivore and anti‐microbial responses, total phenolics, an effect of parental JA treatment was observed (Fig. 6). In offspring of control parents, JA treatment increased the concentration of leaf phenolics within 24 h. However, the phenolic concentration did not reach the same level on JA treatment in offspring of JA‐treated parents (Fig. 6). Although the interaction between parental and offspring JA treatment was not significant (0.05 < P < 0.1, see Fig. 6), this suggests an inhibition of JA inducibility of phenolics in offspring after parental JA treatment.
Figure 6.
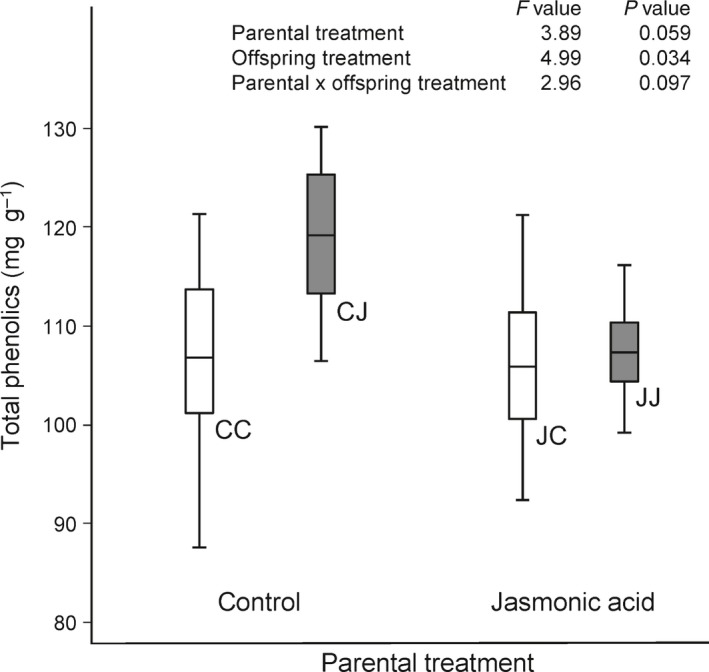
Leaf total phenolics concentration in offspring of control plants and offspring of jasmonic acid (JA)‐treated plants, 24 h after exposure to JA (dark gray box plots) or mock treatment (white box plots) in common dandelion. CC, CJ, JC and JJ denote the four experimental groups in which the first letter is the parental treatment (J = JA, C = control) and the second letter is the treatment of the experimental (offspring) plants. Boxes and whiskers denote the 25th–75th percentile and minimum–maximum observations, respectively; group mean values are indicated by the horizontal line. The inset table shows ANOVA test results from a model that also accounted for possible plant size effects as a result of differences in initial seed weight.
Discussion
Although parental environmental effects are well documented in plants, the extent to which molecular phenotypes are affected by environmental exposures in previous generations is largely unknown. Very few transcriptome‐wide evaluations have been performed and results have been ambiguous, ranging from complete absence (Molinier et al., 2006) to very widespread effects (Colicchio et al., 2015) after parental exposure to cues associated with pathogen or herbivore attack. Our study provides evidence that the inherited effect of parental exposure on molecular phenotypes can be substantial, and supports a previously noted trend that transgenerational effects of herbivore and pathogen attack may be a widespread phenomenon in plants (Holeski et al., 2012). The effect that we observed on offspring after parental JA treatment is consistent with functions related to the treatment, with both transcriptomics and metabolomics analysis converging on phosphatidylinositol signaling as a transgenerationally affected pathway. More generally, our results suggest that the interpretation of gene expression and other molecular studies needs to be mindful of effects on the seed source.
Gene expression analysis revealed a clear functional response to direct JA treatment which is consistent with known JA‐induced processes (Table 1; Notes S1). Because we putatively identified only a modest number of JA‐affected chemical compounds, a detailed pathway analysis based on the chemical JA response was not possible. However, qualitative evaluation of the compounds that were putatively identified indicated that the functional signal identified in the gene expression data was mirrored in the metabolomics data. Compounds that were JA induced included the precursor of JA biosynthesis (linolenic acid; Wasternack & Hause, 2013; consistent with the detected JA treatment effect on the JA biosynthesis pathway). Also identified were the hydrocinnamate phenolics caftaric acid and chicoric acid. These are major phenolic compounds in species from the Compositae family (Cheminat et al., 1988; Oh et al., 2009), including dandelion (Schutz et al., 2005), that are thought to function in plant defenses against pathogens and herbivores (Lee & Scagel, 2013). In dandelion, phenolic inositol esters, triterpene acetates and a sesquiterpene lactone taraxinic acid ester are important secondary metabolites in latex (Huber et al., 2015). The JA response of the hydrocinnamate phenolics in our experiment, which matches the pattern observed in total phenolics (Fig. 6), suggests that these phenolics are inducible secondary metabolites involved in herbivore defenses in dandelion.
An important result of our study is that there is also a functional signal in the inherited JA response. Several of the 40 putative genes that showed a significant parental JA effect have known functions in JA‐ or defense‐related processes. Strikingly, the gene expression and metabolomics analyses converged on one pathway that was affected by parental JA treatment: the phosphatidylinositol signaling pathway. Two of the identified genes with parental JA effect were phosphatidylinositol phosphate kinases, and one of only two parental JA effect metabolites that could be putatively assigned was phosphatidylinositol. Thus, JA treatment had a durable effect on phosphatidylinositol signaling. Phosphatidylinositol phosphate kinases are enzymes that phosphorylate the precursor inositol in a pathway that produces various phosphatidylinositol phosphates and inositol phosphates, which subsequently act as intracellular second messengers on perceiving an extracellular signal (Munnik & Testerink, 2009). Enzymes in this pathway are inducible by stress and plant hormone treatments (Lin et al., 2004), and play an important role in defense response signaling on herbivore and pathogen attack (Laxalt & Munnik, 2002; Mosblech et al., 2008; Hung et al., 2014). Phospholipid signaling is involved in various aspects of biotic defense signaling, including JA biosynthesis by affecting linoleic acid production from plasma membranes, potentiation of the COI1–JAZ complex for jasmonate recognition via a specific inositol phosphate cofactor and intracellular signaling to activate and later downregulate defense gene expression after pathogen elicitor recognition (Laxalt & Munnik, 2002; Sheard et al., 2010; Zhang & Xiao, 2015). Although the exact role of this pathway after JA treatment in dandelion remains to be determined, our congruent RNA‐seq and LC‐MS results provide a clear starting point for future work to pinpoint the (epigenetic) mechanisms that enable a JA response to persist across generations.
Genes that showed a significant effect of parental JA treatment were largely different from genes that showed an expression response on JA treatment in the experimental generation. This is counter to the idea that gene expression in offspring is limited to a sustained activity pattern in a subset of JA‐responsive genes (Bruce et al., 2007). However, this is perhaps an overly simplistic view when the timing of the JA‐induced expression response is considered. On JA application, a rapid succession of transcriptional regulatory programs unfolds with different genes being involved in the immediate, intermediate and long‐term responses (Acosta & Farmer, 2010; Wasternack & Hause, 2013). In our experiment, we tested the early expression response 3 h after JA treatment, which involves mostly different genes, and is poorly correlated with the expression response that is observed at later stages (Tytgat et al., 2013). A subset of the later stage genes may correspond to the genes that are still affected in offspring. An alternative explanation for the lack of overlap of differentially expressed genes after JA treatment vs after parental JA treatment could be related to the low statistical power to detect differentially expressed genes. If only a modest subset of genes that are affected by the treatments are recognized as statistically significant, then limited overlap between the two sets of detected genes is expected, even when many of the genes are in fact affected by both treatments.
It has been proposed that exposure to stressful environments can trigger enhanced variability amongst offspring individuals, rather than a mean shift in trait values or gene expression levels (Rapp & Wendel, 2005; Verhoeven et al., 2010; Herman et al., 2014). Enhanced variability, which is potentially mediated by an increased rate of transgenerationally stable epigenetic mutations, might reflect a bet‐hedging strategy that increases the probability of at least some progeny surviving or maintaining high fitness. In our experiment, JA treatment increased variability in gene expression 3 h after induction, which may reflect subtle timing differences in the early JA response between replicated plants. However, increased variance was not sustained into the offspring generation. By contrast, gene expression amongst offspring of JA‐treated plants showed reduced variance more often than increased variance compared with the offspring of control plants. This is perhaps a result of a conditioning of the response and a reduction in stochastic expression. Such apparent canalization of gene expression after parental JA treatment is interesting and suggests that there is some constraint on expression changes that does not exist in the control plants. Reduced variability in gene expression amongst offspring individuals might also be related to crosstalk between plant defense pathways. For instance, transgenerational priming of salicylic acid (SA)‐related plant defenses after parental treatment with SA or with other hormones or inducing agents (Luna et al., 2012; Slaughter et al., 2012) can enhance offspring expression of SA‐related genes, but, at the same time, because of crosstalk between JA and SA pathways, can suppress the activity of JA‐related defense responses (Luna et al., 2012). Thus, transgenerational activation of one pathway may result in transgenerational suppression of another pathway, and such suppression may be reflected as reduced offspring expression variability in a subset of genes.
This study revealed effects of parental treatment on the offspring transcriptome and metabolome, but the underlying mechanism of the transmission of the environmental effect between the generations remains to be elucidated. Parental environmental effects can be mediated by various mechanisms. In our experiment, parental JA treatment occurred well before flowering; thus no direct induction of the germline occurred. Possible mechanisms therefore include maternal modification of the embryonic hormone balance or inherited epigenetic effects (Boyko et al., 2007; Luna et al., 2012; Rasmann et al., 2012; Bond & Baulcombe, 2014), which is consistent with previous observations of JA‐induced heritable modification of DNA methylation patterns in dandelion (Verhoeven et al., 2010).
In conclusion, our findings demonstrate that parental environmental conditions can have long‐lasting, functional effects that are visible in the transcriptome and metabolome of offspring individuals. Our results imply that, in any gene expression study, environmental conditions should be controlled not only in the experimental generation, but also in the previous parental generation. The proportion of affected genes may be considerable, as evidenced in our study, where a single JA application in parental plants during vegetative growth, well before the induction of flowering, affected the expression of approximately one‐third of the JA‐responsive genes in offspring plants. This observation provides insight into the scope of parental environmental effects. Our results also provides a starting point for further unraveling of the underlying mechanisms that mediate transgenerational effects in plant interactions with herbivores, pathogens or parasites, where such inherited effects may be particularly common (Poulin & Thomas, 2008; Holeski et al., 2012).
Author contributions
K.J.F.V. conceived and designed the study. E.H.V. performed the experiment and the phenolics analysis. M.S. and M.M. performed the metabolomics analysis, and M.M. and K.J.F.V. performed the statistical analysis of the metabolomics data. C.O. performed RNA isolations and RT‐qPCRs. C.O. and A.M.M. carried out the RNA‐seq library preparation. T.P.v.G. and J.F.d.C. performed the de novo transcriptome assembly and annotation. A.M.M., K.J.F.V. and L.M.M. performed the analysis of differential gene expression. K.J.F.V. and L.M.M. wrote the manuscript with input from all co‐authors. All authors read and approved the final manuscript.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Observed read counts for synthetic ERCC RNA spike‐in control sequences.
Fig. S3 Orthogonal partial least squares‐discriminant analyses (OPLS‐DA) selection of jasmonic acid (JA)‐responsive mass signals.
Table S3 Putative identification of LC‐MS mass signals
Notes S1 Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) expression validation of the early jasmonic acid (JA) response candidate gene LOX2.
Fig. S2 Bland–Altman plots for within‐group pairwise comparisons based on ERCC controls.
Table S1 Contigs excluded from statistical analysis, but meeting the coverage threshold in at least one of the experimental groups
Table S2 RNA‐seq test results for differential expression analysis
Acknowledgements
We thank Tanja Bakx for help with the phenolics analysis. This work was supported by the Dutch Organization for Scientific Research (NWO VIDI grant 864.10.008 to K.J.F.V. and NWO Visiting Scholar grant 040.11.358 to L.M.M.), a KNAW Visiting Scholar grant to L.M.M. and a National Institutes of Health (NIH) grant U24 DK097209 to L.M.M.
References
- Acosta I, Farmer E. 2010. Jasmonates. The Arabidopsis book/American Society of Plant Biologists 8: e0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afzal AJ, Wood AJ, Lightfoot DA. 2008. Plant receptor‐like serine threonine kinases: roles in signaling and plant defense. Molecular Plant–Microbe Interactions 21: 507–517. [DOI] [PubMed] [Google Scholar]
- Aronesty E. 2011. ea‐utils: Command‐line tools for processing biological sequencing data. [WWW document] URL http://code.google.com/p/ea-utils. [accessed December 2012].
- Bland JM, Altman DG. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1: 307–310. [PubMed] [Google Scholar]
- Bond DM, Baulcombe DC. 2014. Small RNAs and heritable epigenetic variation in plants. Trends in Cell Biology 24: 100–107. [DOI] [PubMed] [Google Scholar]
- Boyko A, Kathiria P, Zemp FJ, Yao YL, Pogribny I, Kovalchuk I. 2007. Transgenerational changes in the genome stability and methylation in pathogen‐infected plants (Virus‐induced plant genome instability). Nucleic Acids Research 35: 1714–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce TJA, Matthes MC, Napier JA, Pickett JA. 2007. Stressful “memories” of plants: evidence and possible mechanisms. Plant Science 173: 603–608. [Google Scholar]
- Cheminat A, Zawatzky R, Becker H, Brouillard R. 1988. Caffeol conjugates from Echinacea species: structures and biological activity. Phytochemistry 27: 2787–2794. [Google Scholar]
- Colicchio JM, Monnahan PJ, Kelly JK, Hileman LC. 2015. Gene expression plasticity resulting from parental leaf damage in Mimulus guttatus . New Phytologist 205: 894–906. [DOI] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia‐Gomez JM, Terol J, Talon M, Robles M. 2005. Blast2GO: a universal tool for annotation visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- van Dam NM, Witjes L, Svatos A. 2004. Interactions between aboveground and belowground induction of glucosinolates in two wild Brassica species. New Phytologist 161: 801–810. [DOI] [PubMed] [Google Scholar]
- De Vos M, Van Oosten VR, Van Poecke RMP, Van Pelt JA, Pozo MJ, Mueller MJ, Buchala AJ, Metraux JP, Van Loon LC, Dicke M et al 2005. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Molecular Plant–Microbe Interactions 18: 923–937. [DOI] [PubMed] [Google Scholar]
- van Dijk PJ. 2003. Ecological and evolutionary opportunities of apomixis: insights from Taraxacum and Chondrilla . Philosophical Transactions of the Royal Society of London Series B: Biological Sciences 358: 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillies MA, Rau A, Aubert J, Hennequet‐Antier C, Jeanmougin M, Servant N, Keime C, Marot G, Castel D, Estelle J et al 2013. A comprehensive evaluation of normalization methods for Illumina high‐throughput RNA sequencing data analysis. Briefings in Bioinformatics 14: 671–683. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Engelkes T, Morrien E, Verhoeven KJF, Bezemer TM, Biere A, Harvey JA, McIntyre LM, Tamis WLM, van der Putten WH. 2008. Successful range‐expanding plants experience less above‐ground and below‐ground enemy impact. Nature 456: 946–948. [DOI] [PubMed] [Google Scholar]
- Feil R, Fraga MF. 2012. Epigenetics and the environment: emerging patterns and implications. Nature Reviews Genetics 13: 97–109. [DOI] [PubMed] [Google Scholar]
- Ferreira de Carvalho J, Oplaat C, Pappas N, Derks M, de Ridder D, Verhoeven KJF. 2016. Heritable gene expression differences between apomictic clone members in Taraxacum officinale: Insights into early stages of evolutionary divergence in asexual plants. BMC Genomics 17: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Dong X. 2013. Systemic acquired resistance: turning local infection into global defense. Annual Review of Plant Biology 64: 839–863. [DOI] [PubMed] [Google Scholar]
- Galloway LF, Etterson JR. 2007. Transgenerational plasticity is adaptive in the wild. Science 318: 1134–1136. [DOI] [PubMed] [Google Scholar]
- van Gurp TP, McIntyre LM, Verhoeven KJF. 2013. Consistent errors in first strand cDNA due to random hexamer mispriming. PLoS ONE 8: e85583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M et al 2013. De novo transcript sequence reconstruction from RNA‐seq using the Trinity platform for reference generation and analysis. Nature Protocols 8: 1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JJ, Spencer HG, Donohue K, Sultan SE. 2014. How stable should epigenetic modifications be? Insights from adaptive plasticity and bet hedging. Evolution 68: 632–643. [DOI] [PubMed] [Google Scholar]
- Holeski LM. 2007. Within and between generation phenotypic plasticity in trichome density of Mimulus guttatus . Journal of Evolutionary Biology 20: 2092–2100. [DOI] [PubMed] [Google Scholar]
- Holeski LM, Jander G, Agrawal AA. 2012. Transgenerational defense induction and epigenetic inheritance in plants. Trends in Ecology & Evolution 27: 618–626. [DOI] [PubMed] [Google Scholar]
- Huber M, Triebwasser‐Freese D, Reichelt M, Heiling S, Paetz C, Chandran JN, Bartram S, Schneider B, Gershenzon J, Erb M. 2015. Identification quantification spatiotemporal distribution and genetic variation of major latex secondary metabolites in the common dandelion (Taraxacum officinale agg.). Phytochemistry 115: 89–98. [DOI] [PubMed] [Google Scholar]
- Hung CY, Aspesi P, Hunter MR, Lomax AW, Perera IY. 2014. Phosphoinositide‐signaling is one component of a robust plant defense response. Frontiers in Plant Science 5: 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang LC, Schlesinger F, Davis CA, Zhang Y, Li R, H Salit M, Gingeras TR, Oliver B. 2011. Synthetic spike‐in standards for RNA‐seq experiments. Genome Research 21: 1543–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielbasa SM, Wan R, Sato K, Horton P, Frith MC. 2011. Adaptive seeds tame genomic sequence comparison. Genome Research 21: 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow AM. 1993. Apomixis: embryo sacs and embryos formed without meiosis or fertilization in ovules. Plant Cell 5: 1425–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biology 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. 2014. voom: precision weights unlock linear model analysis tools for RNA‐seq read counts. Genome Biology 15: R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxalt AM, Munnik T. 2002. Phospholipid signalling in plant defence. Current Opinion in Plant Biology 5: 332–338. [DOI] [PubMed] [Google Scholar]
- Lee J, Scagel CF. 2013. Chicoric acid: chemistry distribution and production. Frontiers in Chemistry 1: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SF, Ziv N, Siegal ML. 2012. Bet hedging in yeast by heterogeneous age‐correlated expression of a stress protectant. PLoS Biology 10: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WH, Ye R, Ma H, Xu ZH, Xue HW. 2004. DNA chip‐based expression profile analysis indicates involvement of the phosphatidylinositol signaling pathway in multiple plant responses to hormone and abiotic treatments. Cell Research 14: 34–45. [DOI] [PubMed] [Google Scholar]
- Luna E, Bruce TJA, Roberts MR, Flors V, Ton J. 2012. Next‐generation systemic acquired resistance. Plant Physiology 158: 844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J, Ries G, Zipfel C, Hohn B. 2006. Transgeneration memory of stress in plants. Nature 442: 1046–1049. [DOI] [PubMed] [Google Scholar]
- Mosblech A, Konig S, Stenzel I, Grzeganek P, Feussner I, Heilmann I. 2008. Phosphoinositide and inositolpolyphosphate signalling in defense responses of Arabidopsis thaliana challenged by mechanical wounding. Molecular Plant 1: 249–261. [DOI] [PubMed] [Google Scholar]
- Müller M, Munné‐Bosch S. 2015. Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiology 169: 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T, Testerink C. 2009. Plant phospholipid signaling: “in a nutshell”. Journal of Lipid Research 50: S260–S265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh MM, Trick HN, Rajashekar CB. 2009. Secondary metabolism and antioxidants are involved in environmental adaptation and stress tolerance in lettuce. Journal of Plant Physiology 166: 180–191. [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, Van Wees SCM, Bakker PAHM. 2014. Induced systemic resistance by beneficial microbes. Annual Review of Phytopathology 52: 347–375. [DOI] [PubMed] [Google Scholar]
- Poulin R, Thomas F. 2008. Epigenetic effects of infection on the phenotype of host offspring: parasites reaching across host generations. Oikos 117: 331–335. [Google Scholar]
- Rapp RA, Wendel JF. 2005. Epigenetics and plant evolution. New Phytologist 168: 81–91. [DOI] [PubMed] [Google Scholar]
- Rasmann S, De Vos M, Casteel CL, Tian DL, Halitschke R, Sun JY, Agrawal AA, Felton GW, Jander G. 2012. Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiology 158: 854–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauveplane V, Kandel S, Kastner P‐E, Ehlting J, Compagnon V, Werck‐Reichhart D, Pinot F. 2009. Arabidopsis thaliana CYP77A4 is the first cytochrome P450 able to catalyze the epoxidation of free fatty acids in plants. FEBS Journal 276: 719–735. [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM. 2000. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proceedings of the National Academy of Sciences, USA 97: 11655–11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz K, Kammerer DR, Carle R, Schieber A. 2005. Characterization of phenolic acids and flavonoids in dandelion (Taraxacum officinale WEB. ex WIGG.) root and herb by high‐performance liquid chromatography electrospray ionization mass spectrometry. Rapid Communications in Mass Spectrometry 19: 179–186. [DOI] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben‐Nissan G, Hinds TR, Kobayashi Y, Hsu F‐F, Sharon M, Browse J et al 2010. Jasmonate perception by inositol‐phosphate‐potentiated COI1‐JAZ co‐receptor. Nature 468: 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter A, Daniel X, Flors V, Luna E, Hohn B, Mauch‐Mani B. 2012. Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiology 158: 835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences, USA 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Dangl JL. 2005. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Current Opinion in Plant Biology 8: 397–403. [DOI] [PubMed] [Google Scholar]
- Tytgat TOG, Verhoeven KJF, Jansen JJ, Raaijmakers CE, Bakx‐Schotman T, McIntyre LM, van der Putten WH, Biere A, van Dam NM. 2013. Plants know where it hurts: root and shoot jasmonic acid induction elicit differential responses in Brassica oleracea . PLoS ONE 8: e65502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven KJF, van Gurp TP. 2012. Transgenerational effects of stress exposure on offspring phenotypes in apomictic dandelion. PLoS ONE 7: e38605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven KJF, Jansen JJ, van Dijk PJ, Biere A. 2010. Stress‐induced DNA methylation changes and their heritability in asexual dandelions. New Phytologist 185: 1108–1118. [DOI] [PubMed] [Google Scholar]
- Verhoeven KJF, Simonsen KL, McIntyre LM. 2005. Implementing false discovery rate control: increasing your power. Oikos 108: 643–647. [Google Scholar]
- Wasternack C, Hause B. 2013. Jasmonates: biosynthesis perception signal transduction and action in plant stress response growth and development. An update to the 2007 review in Annals of Botany. Annals of Botany 111: 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibowo A, Becker C, Marconi G, Durr J, Price J, Hagmann J, Papareddy R, Putra H, Kageyama J, Becker J et al 2016. Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. eLife 5: e13546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Xiao SY. 2015. Lipids in salicylic acid‐mediated defense in plants: focusing on the roles of phosphatidic acid and phosphatidylinositol 4‐phosphate. Frontiers in Plant Science 6: 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Observed read counts for synthetic ERCC RNA spike‐in control sequences.
Fig. S3 Orthogonal partial least squares‐discriminant analyses (OPLS‐DA) selection of jasmonic acid (JA)‐responsive mass signals.
Table S3 Putative identification of LC‐MS mass signals
Notes S1 Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) expression validation of the early jasmonic acid (JA) response candidate gene LOX2.
Fig. S2 Bland–Altman plots for within‐group pairwise comparisons based on ERCC controls.
Table S1 Contigs excluded from statistical analysis, but meeting the coverage threshold in at least one of the experimental groups
Table S2 RNA‐seq test results for differential expression analysis


