SUMMARY
Eliciting broadly neutralizing antibody (bNAb) responses against HIV-1 is a major goal for a prophylactic HIV-1 vaccine. One approach is to design immunogens based on known broadly neutralizing epitopes. Here we report the design and synthesis of an HIV-1 glycopeptide immunogen derived from the V3 domain. We performed glycopeptide epitope mapping to determine the minimal glycopeptide sequence as the epitope of V3-glycan specific bNAbs PGT128 and 10–1074. We further constructed a self-adjuvant three-component immunogen that consists of a 33-mer V3 glycopeptide epitope, a universal T-helper epitope P30, and a lipopeptide (Pam3CSK4) that serves as a ligand of Toll-like receptor 2. Rabbit immunization revealed that the synthetic self-adjuvant glycopeptide could elicit substantial glycan dependent antibodies that exhibited broader recognition of HIV-1 gp120s than the non-glycosylated V3 peptide. These results suggest that the self-adjuvant synthetic glycopeptides can serve as an important component to elicit glycan-specific antibodies in HIV vaccine design.
Keywords: HIV Vaccine, Broadly Neutralizing Antibody, Glycopeptide, Glycoprotein, N-Glycan, Chemoenzymatic Synthesis, Synthetic Immunogen, Adjuvant, Neutralizing Epitope
In Brief
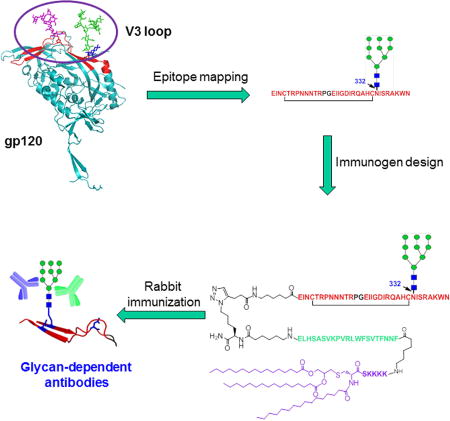
Cai et al. designed and synthesized a three-component, self-adjuvanting vaccine carrying a minimal HIV-1 V3 glycopeptide epitope. Rabbit immunization showed that the glycopeptide immunogen could elicit substantial glycan-dependent antibodies that exhibited broad recognition of HIV-1 gp120s across clades.
INTRODUCTION
The HIV-1 envelope glycoprotein (Env) is responsible for virus entry into host cells by mediating binding to receptors and is the sole target for HIV vaccine design (Wyatt and Sodroski, 1998). A key feature of the Env is the heavily glycosylation, including about 90 N-linked glycans that cover most of the Env trimer surface (Stewart-Jones et al., 2016, Zhang et al., 2004). These N-linked glycans are created by the host cell and are recognized as immunologically “self” by the human immune system. Therefore these N-glycans form a dense “glycan shield” and help the Env successfully evade antibody neutralization (Wei et al., 2003). To a large degree, this evasion is responsible for the difficulty in developing an effective HIV vaccine. Nevertheless, broadly neutralizing activity against the HIV Env has been found in 5–25% of donor sera after 2–3 years of infection, suggesting the possibility to elicit neutralizing activities by vaccination (Scheid et al., 2011, Stamatatos et al., 2009). Broadly neutralizing antibodies (bNAbs) have been isolated from infected individuals and have been shown to target several different epitopes on Env (Huang et al., 2012, Walker et al., 2011, Walker et al., 2009, Corti and Lanzavecchia, 2013, Kwong and Mascola, 2012). Interestingly, most of the recently discovered bNAbs are found to be glycan-dependent antibodies. The most potent neutralizers of these bNAbs appear to be from the PGT121–130 group that target and penetrate the glycan shield to recognize both glycans and the protein surface of the V3 region of gp120 (Walker et al., 2011, Kong et al., 2013, Mouquet et al., 2012). Structural studies have revealed that most of the PGT-series bNAbs recognize the high-mannose patch centered on the N-glycan at the N332 site immediately adjacent to the C-terminal base of the V3 loop, suggesting the N332 high-mannose N-glycan is highly accessible and vulnerable to the human immune system (Kong et al., 2013, Mouquet et al., 2012, Walker et al., 2011, Pejchal et al., 2011, Julien et al., 2013, Sok et al., 2014). However, elicitation of glycan-dependent antibodies that are cross-reactive to HIV-1 Env through active immunization turned out to be a challenging task (Saunders et al., 2017, Horiya et al., 2014). Several previous immunization studies with synthetic glycoconjugates raised carbohydrate-specific antibody responses in rabbits, but the antibodies showed poor recognition of, or were not cross-reactive to, HIV-1 gp120s (Astronomo et al., 2010; Ni et al., 2006, Joyce et al., 2008). Recently, an immunization study has shown that repetitive vaccination in macaques over a 4-year period with a high-mannose glycoform of HIV-1 Env is able to induce V3 glycan-dependent antibodies, but the antibodies neutralize only the HIV-1 pseudoviruses carrying high-mannose N-glycans (Saunders et al., 2017). In addition, it should be pointed out that many Env related immunogens also elicit antibody responses to non-neutralizing epitopes that would likely dilute the immune responses to those desired neutralizing epitopes (Williams et al., 2015).
To focus the immune response on the neutralizing epitope, an alternative strategy is to design appropriate immunogens based on the minimal neutralizing epitope of the glycan-dependent bNAbs (Pejchal et al., 2011, Alam et al., 2013, Fernandez-Tejada et al., 2015, Amin et al., 2013, Horiya et al., 2014). For that purpose, a crucial first step is to characterize those glycopeptide neutralizing epitopes. We have recently reported the chemoenzymatic synthesis and antibody binding analysis of homogeneous V3 glycopeptides derived from HIV-1 JR-FL strain, which identifies a 33-mer mini-V3 glycopeptide carrying a high-mannose N-glycan at the N332 site as the epitope of bNAbs PGT128 and 10–1074, and the mini-V3 glycopeptide carrying a sialylated complex type N-glycan at the N301 site as a possible epitope for PGT121, which lay a foundation for a design of a glycopeptide-based vaccine (Orwenyo et al., 2017). Independently, Haynes and coworkers have recently reported that a 30-mer synthetic mini-V3 glycopeptide derived from the JR-FL strain, when formulated in the Toll-like receptor 4 agonist GLA-SE adjuvant, is able to induce V3-glycan targeted antibodies in rhesus macaques, suggesting the synthetic V3 glycopeptide could mimic the V3 glycan specific epitopes of bNAbs (Alam et al., 2017). Nevertheless, the monomeric V3 glycopeptide used in the immunization study lacks T-helper epitope and is found to be weakly immunogenic and to require high-dose and long repeated immunization to raise glycan-specific antibodies, as shown by the experimental data (Alam et al., 2017). In this paper, we describe a rational design of HIV-1 immunogen based on the V3 glycan-dependent neutralizing epitope. We performed glycopeptide epitope mapping through synthesis and antibody binding analysis of truncated glycopeptides, which indicated that a 33-mer V3 glycopeptide appeared to be the suitable epitope mimic of the V3-glycan specific bNAbs PGT128 and 10–1074. We further designed and synthesized a self-adjuvant three-component immunogen that consists of the 33-mer V3 glycopeptide epitope carrying a high-mannose glycan at the N332 site, an universal T-helper epitope P30 (Cai et al., 2013a), and a lipopeptide (Pam3CSK4) that is a ligand of Toll-like receptor 2 for stimulating immune response (Ingale et al., 2007). We found that the synthetic three-component glycopeptide immunogen, when formulated into liposomes and administered to rabbits without any additional adjuvants, was able to elicit glycan-dependent antibodies in a relatively short period of time. Moreover, sera antibodies induced by the glycopeptide immunogen exhibited broad recognition of several HIV-1 gp120s across clades.
RESULTS
Probing the minimal neutralizing epitopes through glycopeptide epitope mapping
Synthesis of complex glycopeptides carrying natural N-glycans is a challenging work (Gamblin et al., 2009, Schmaltz et al., 2011, Unverzagt and Kajihara, 2013, Danby and Withers, 2016). Here we chose to synthesize the V3 glycopeptides using a chemoenzymatic method, which we have recently developed for homogeneous N-glycoprotein synthesis (Wang and Amin, 2014, Amin et al., 2013, Orwenyo et al., 2017). Briefly, an N-acetylglucosamine (GlcNAc) moiety is installed at the predetermined glycosylation site during automated solid-phase peptide synthesis (SPPS) to provide a GlcNAc-peptide as an acceptor, and then a synthetic high-mannose glycan, in the form of activated glycan oxazoline, is transferred to the GlcNAc moiety by the endoglycosidase mutant, EndoA-N171A, to provide a homogeneous glycopeptide with natural glycosidic linkages (Toonstra et al., 2016, Huang et al., 2009). Recently we have used this strategy to synthesize V1V2 glycopeptides for characterizing the glycan specificity of bNAb PG9 and PG16 (Amin et al., 2013).
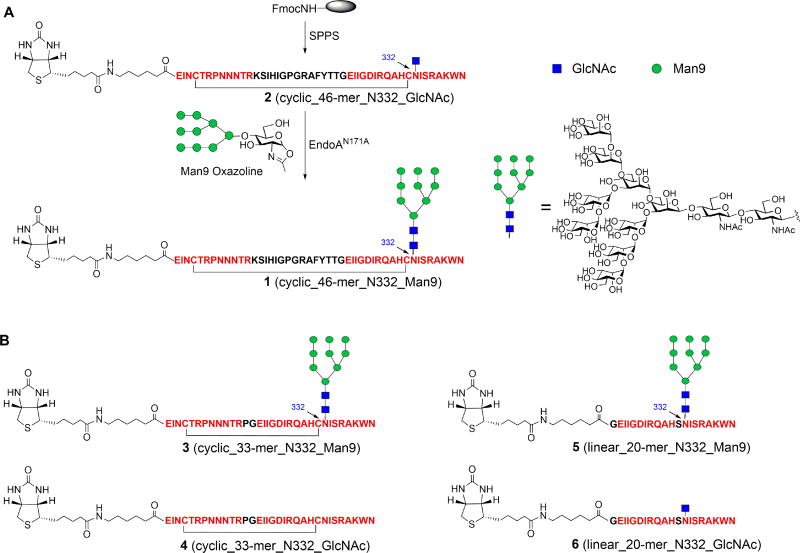
HIV-1 V3 domain typically consists of 35–40 amino acids with a disulfide bond between C296 and C331 to form a loop structure (Huang et al., 2005). We selected an extended V3 domain sequence (aa 293–339, HXB2 numbering) from the HIV-1 B-clade JR-FL strain, and focused on the N332 site, which is highly conserved to carry a high-mannose glycan (Sok et al., 2014). We first synthesized the 46-mer cyclic full-length V3 high-mannose glycopeptide 1 by transferring the high-mannose glycan with EndoA-N171A to the precursor GlcNAc-peptide 2, which was obtained by SPPS (Figure 1A). A biotin tag was introduced at the N-terminus to facilitate site-specific immobilization for binding analysis. To determine the minimal peptide requirement to recapitulate the neutralizing epitope, we also synthesized the truncated V3 sequence, including a 33-mer cyclic mini-V3 sequence corresponding to the residues 293–304 and 321–339 with a replacement of the tip residues (304–320) by a Pro-Gly (PG) dipeptide insert (Orwenyo et al., 2017), and a 20-mer linear mini-V3 sequence corresponding to the residues 320–339 with a replacement of the C331 by a Ser (Figure 1B). The high-mannose glycopeptides (3 and 5) were synthesized in excellent yields by the similar chemoenzymatic method used for the synthesis of 46-mer glycopeptide 1 (Figure S1 and S2). We also synthesized the corresponding aglycone peptide (4) and the GlcNAc-peptides (2 and 6). The purity and identify of these glycopeptides were characterized by analytical HPLC and ESI-MS (see STAR Method).
Figure 1. Synthesis of gp120 V3 glycopeptide carrying N-glycan by chemoenzymatic method.
A) Synthesis of 46-mer V3 glycopeptide. B) Structure of truncated 33-mer and 20-mer V3 peptides.
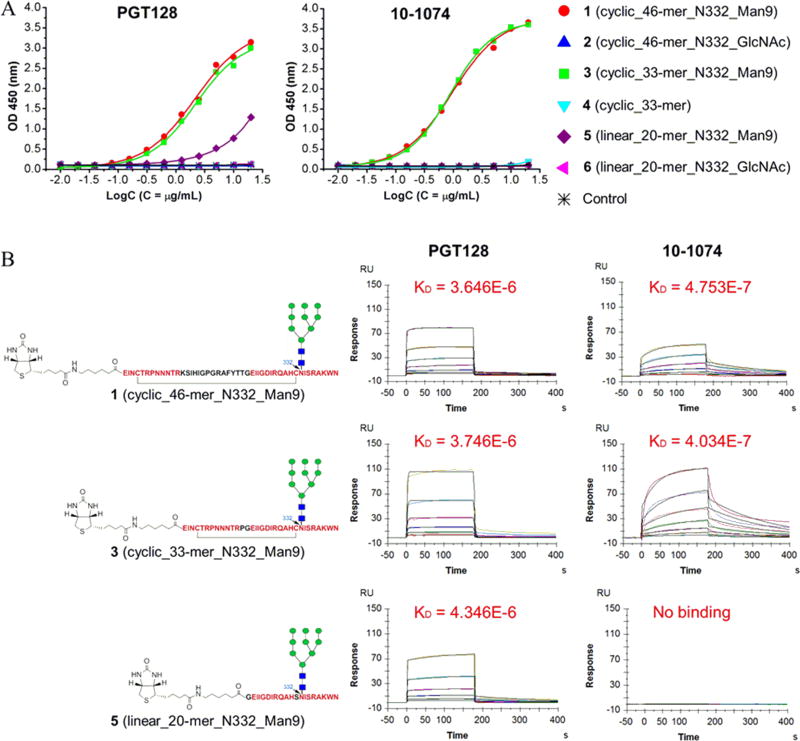
Previous structural studies have revealed that the two bNAbs PGT128 and 10–1074 bind to the base of the gp120 V3 loop with a contact of the GDIR motif and interact with the N332 high-mannose glycan (Kong et al., 2013, Gristick et al., 2016, Pejchal et al., 2011, Mouquet et al., 2012, Sok et al., 2016). However, the requirement of minimal peptide domain to recapitulate the neutralizing epitope remains to be further characterized. For this purpose, we first evaluated the binding of the synthetic V3 glycopeptides to PGT128 and 10–1074 by enzyme-linked immunosorbent assay (ELISA). As shown in Figure 2A, both PGT128 and 10–1074 showed strong binding to the 46-mer high-mannose glycopeptide 1 and 33-mer high-mannose glycopeptide 3. The binding profiles for PGT128 or 10–1074 between the two glycopeptides (1 and 3) appear to be similar, implying comparable binding affinity to the 46-mer and 33-mer high-mannose V3 glycopeptides. Thus, the tip sequence of the V3 domain does not enhance or reduce the affinity for the binding to bNAbs. Interestingly, PGT128 also showed binding to the 20-mer high-mannose glycopeptide 5 in spite of a reduced affinity, while 10–1074 did not show any binding to this linear shorter glycopeptide. Both PGT128 and 10–1074 did not bind to the 33-mer aglycone peptide 4 and 46-mer GlcNAc-peptide 2 in ELISA assay. We also performed binding analysis using the surface plasmon resonance (SPR) technology. PGT128 displayed comparable binding affinity to 46-mer high-mannose glycopeptide 1 (KD = 3.65 µM) and 33-mer high-mannose glycopeptide 3 (KD = 3.75 µM), but decreased binding affinity to 20-mer high-mannose glycopeptide 5 (KD = 4.35 µM). For 10–1074, the binding affinity to the 33-mer high-mannose glycopeptide 3 (KD = 0.40 µM) was about the same as that of the 46-mer high-mannose glycopeptide 1 (KD = 0.48 µM). Similar to ELISA results, no binding of 10–1074 to the 20-mer high-mannose glycopeptide 5 was observed in the SPR analysis. Taken together, ELISA and SPR binding results suggest that the cyclic 46-mer and 33-mer high-mannose V3 glycopeptides could equally recapitulate the neutralizing epitopes of PGT128 and 10–1074, while the 20-mer high-mannose glycopeptide showed significantly reduced affinity for antibody 10–1074. Previous immunizations with HIV Env glycoproteins have been reported to elicit strain specific neutralizing antibodies primarily against the V3 tip (Javaherian et al., 1990, Gao et al., 2005, Moody et al., 2015). Therefore, the 33-mer high-mannose V3 glycopeptide, in which the highly variable and immune-predominant tip sequence (aa 304–319) was deleted, would be more suitable for HIV vaccine design by focusing the immune response to the conserved neutralizing epitope.
Figure 2. Binding of the synthetic V3 glycopeptides to the bNAb PGT128 and 10–1074.
A) ELISA analysis of the binding of the synthetic V3 glycopeptides to PGT128 and 10–1074. Streptavidin was coated on the microtiter plates and biotinylated V3 glycopeptide (2 µg/mL) were immobilized. PGT128 and 10–1074 IgGs were titrated with two-fold serial dilutions starting from 20 µg/mL; B) SPR binding of the V3 high-mannose glycopeptides to PGT128 and 10–1074. The biotinylated V3 glycopeptides were immobilized on CM5 chips coated with neutravidin. PGT128 and 10–1074 IgGs were flowed through as analytes. The sensorgrams were recorded with two-serial dilutions starting at the highest concentration of 500 nM. Data were fit with a 1:1 Langmuir binding model (fitting was shown in black line). KD was given in M.
Glycopeptide immunogen design and synthesis
Based on the binding results, we selected the cyclic 33-mer V3 glycopeptide carrying a high-mannose glycan at the N332 site as a key component for vaccine design and immunization studies. We decided to use the three-component vaccine strategy for immunogen design, which has been used previously for anti-tumor vaccine development (Ingale et al., 2007, Lakshminarayanan et al., 2012, Wilkinson et al., 2011, Cai et al., 2013b, Pett et al., 2017). Briefly, the V3 glycopeptide carrying a high-mannose glycan at the N332 site, a T helper epitope peptide P30 derived from tetanus toxoid (Cai et al., 2013a) and the toll-like receptor ligand lipopeptide Pam3CSK4 (Ingale et al., 2007) were covalently conjugated together. The T helper epitope peptide could activate T cells to enhance the antibody response, while the TLR ligand lipopeptide could serve as a self-adjuvanting immune stimulant by stimulating B lymphocytes and macrophages. Finally, the amphiphilic lipopeptide-glycopeptide could form a stable liposome to efficiently present the V3 glycopeptide to antigen-presenting cells (Ingale et al., 2007, Wilkinson et al., 2011). Thus, the designed three-component immunogen is expected to be superior for mounting a much stronger immune response than the plain mini-V3 glycopeptide used recently in the rhesus macaques immunization study (Alam et al., 2017).
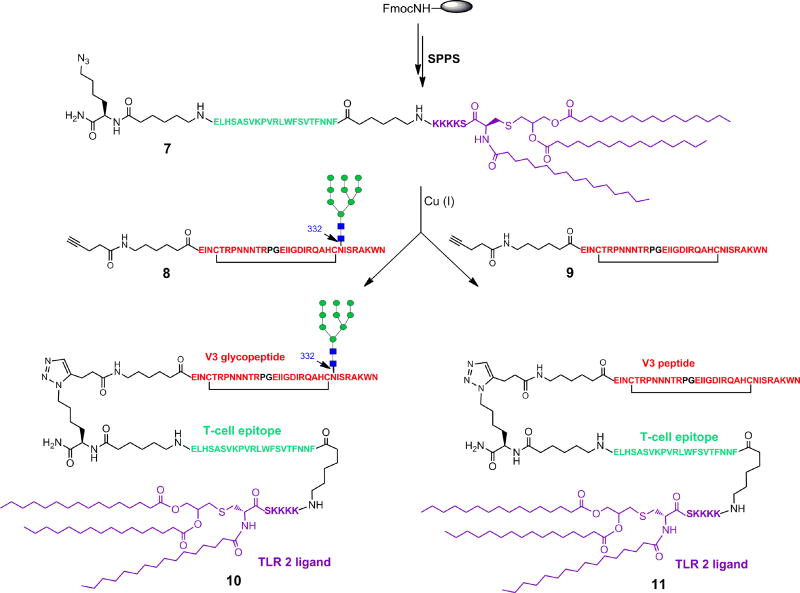
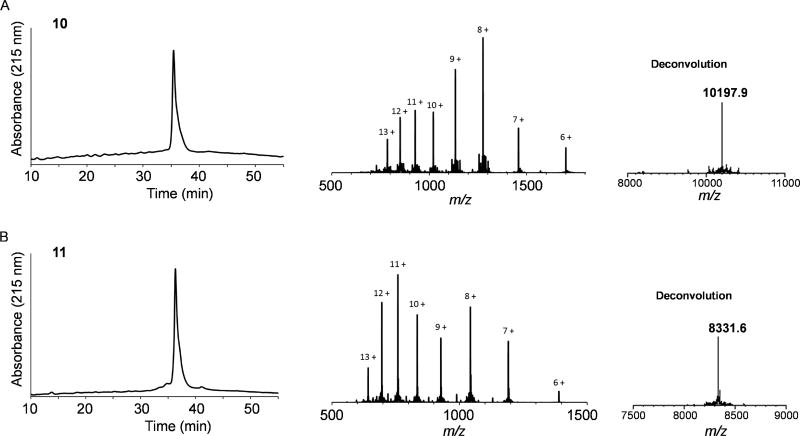
Several methods were reported to synthesize the MUC1 three-component anti-tumor vaccines, including the liposome-mediated native chemical ligation (Ingale et al., 2007), thioester ligation (Cai et al., 2013b) and linear synthesis strategy (Thompson et al., 2015). Here we applied copper (I)-catalyzed alkyne-azide 3 + 2 cycloaddition (click chemistry) to assembly the lipopeptide and HIV V3 glycopeptide (Figure 3). First, the T cell epitope peptide P30 and the Pam3CSK4 lipopeptide were synthesized by stepwise SPPS using Fmoc chemistry to give the lipopeptide 7 with excellent yield. The purity and identity of lipopeptide 7 were confirmed by HPLC and ESI-MS analysis (See STAR Method). A Lys(N3) residue was placed at the C-terminus of the P30 peptide to allow site-specific ligation to V3 glycopeptide. On the other hand, the cyclic 33-mer high-mannose V3 glycopeptide 8 carrying an alkyne moiety at the N-terminus was synthesized using the chemoenzymatic method (Figure S3). Then the V3 high-mannose glycopeptide 8 was ligated to the lipopeptide 7 to afford the desired three-component glycopeptide immunogen 10 by click chemistry (Figure 3). To study the influence of N332 high-mannose glycan on the immunogenicity of V3 domain, we also synthesized the aglycone V3 peptide 9 (Figure S3) and ligated to the lipopeptide 7 to afford the aglycone peptide three-component immunogen 11. The purity and identity of the synthetic three-component immunogens 10 and 11 were confirmed by HPLC and ESI-MS analysis (Figure 4).
Figure 3. Synthesis of three-component immunogen 10 and 11.
Alkyne V3 glycopeptide 8 was prepared by the chemoenzymatic method. The final click reaction was performed in DMF using copper (I) acetate as the catalysis. The compounds carrying the Pam3CSK4 were purified by RP-HPLC on a polar CN column.
Figure 4. HPLC and ESI-MS analysis of the synthetic three-component glycopeptide/peptide immunogens.
A) The three-component glycopeptide immunogen (10); B) The three-component non-glycosylated peptide immunogen (11).
Immunogenicity of synthetic HIV immunogens
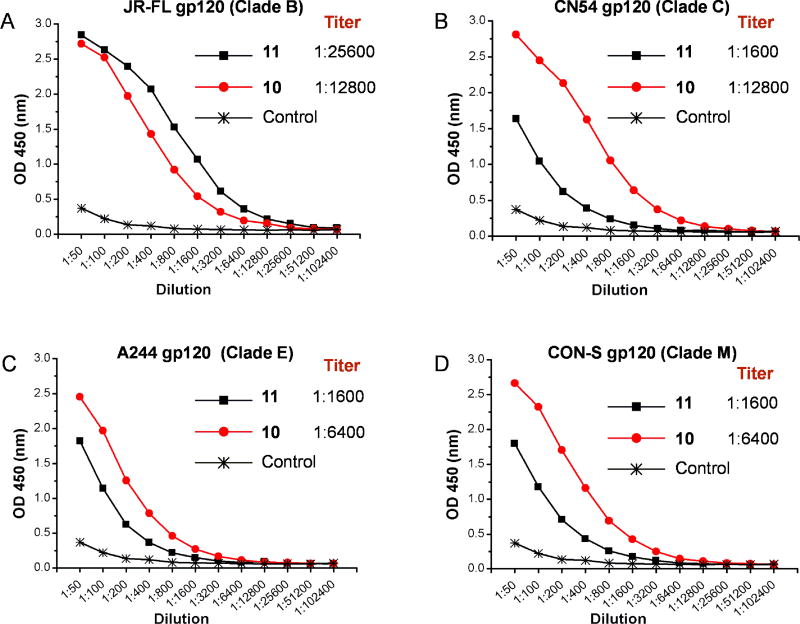
Without additional adjuvants, the synthetic three-component immunogen 10 and 11 were incorporated into liposome following a reported procedure (Ingale et al., 2007). We immunized three New Zealand white rabbits with each immunogen in a dose of 50 µg via intramuscular and subcutaneous injections. The priming and two boosting injections were performed at 21 day intervals and bleeds were taken 7 days later. We combined the antisera from the three immunized rabbits and first evaluated the binding to gp120s from several different strains. The antisera induced by both immunogens 10 and 11 showed substantial binding to the self JR-FL gp120 (Figure 5A). The titer for the antisera induced by the aglycone immunogen 11 was about two folds higher than that of the glycopeptide immunogen 10 against the homologous gp120. However, antisera induced by peptide immunogen 11 only weakly recognize the gp120s of CN54, A244 and CON-S strains, while the antisera induced by the glycopeptide immunogen 10 showed substantial recognition to all the heterologous HIV-1 gp120s tested with comparable titers as observed for the JR-FL gp120 (Figure 5B–D). These results indicated that while peptide immunogen 11 induced strain specific antibody response, the glycopeptide immunogen 10 could induce antibodies with broad binding activities against gp120s across strains and clades. Many recently isolated bNAbs with broad neutralizing activities were found to target N-glycans (Walker et al., 2011, Kong et al., 2013, Mouquet et al., 2012). The broad recognition of the antisera to gp120s across strains and clades implied that the glycopeptide immunogen 10 might elicit antibodies target the common N332 high-mannose glycan.
Figure 5. ELISA binding of the rabbit antisera to gp120s.
A) with JR-FL gp120; B) with CN54 gp120; C) with A244 gp120; D) with CON-S gp120. Gp120s were coated on the microtiter plates (2 µg/mL). The rabbit antisera induced by the immunogen 10 and 11 were titrated with two-fold serial dilutions starting from 1:50 dilution. Titer (endpoint titer) was defined as the last highest dilution gives an optical density of 0.1 or greater than the control.
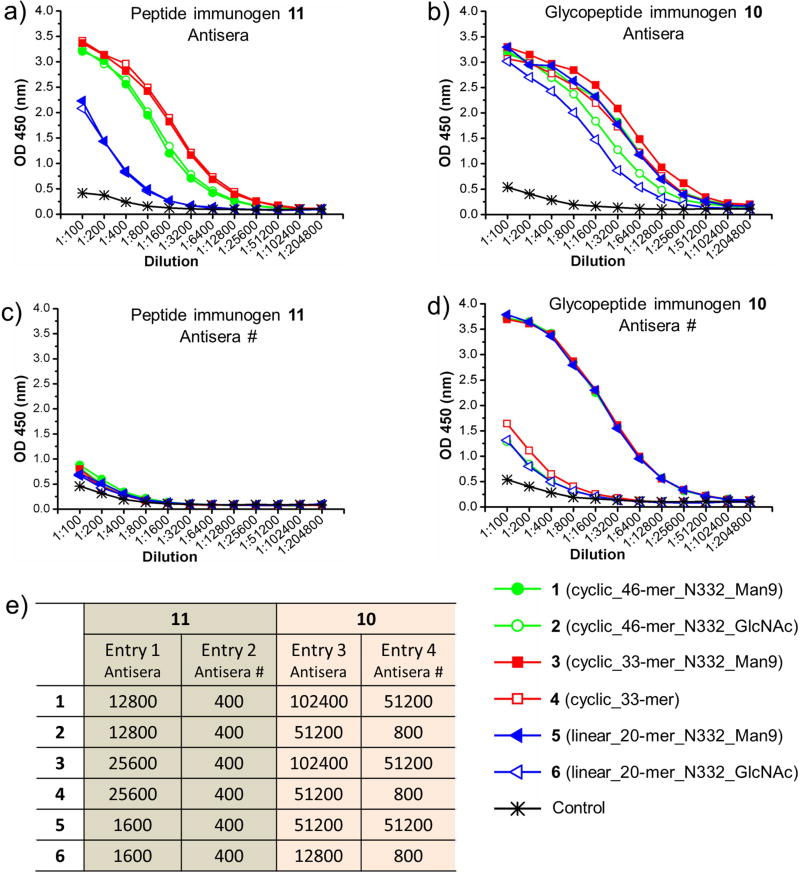
We next performed ELISA binding of the antisera to the synthetic V3 peptides/glycopeptides 1–6. The antisera induced by the peptide immunogen 11 showed strong binding to the 33-mer cyclic V3 aglycone peptide 4 (titer: 25 600), and a similar binding profile was observed to the high-mannose glycopeptide 3 with identical titer (Figure 6A). The binding of the antisera to the 46-mer cyclic V3 GlcNAc-peptide 2 and high-mannose glycopeptide 1 were comparable to the 33-mer V3 peptides, but the antisera showed significantly decreased binding to the 20-mer V3 high-mannose glycopeptide 5 and GlcNAc-peptide 6. These observations suggested that the antibodies induced by peptide immunogen 11 mainly targeted the V3 region corresponding to residues 292–303. Antibodies induced by the high-mannose glycopeptide immunogen 10 showed strong binding to all of the V3 peptides 1–6 (Figure 6B). Interestingly, the antisera displayed preference to the high-mannose glycopeptides over the corresponding GlcNAc-peptide and the aglycone peptide. The titer was two folds higher for the 46-mer and 33-mer high-mannose V3 glycopeptides (1 and 3) than the 46-mer GlcNAc-peptide 2 and 33-mer aglycone peptide 4. However, the titer was about 4 folds higher to the 20-mer high-mannose V3 glycopeptide 5 than the GlcNAc-peptide 6. More interestingly, the antisera induced by the glycopeptide immunogen 10 showed enhanced binding to the V3 peptides 1–6 than the antibodies induced by peptide immunogen 11. The binding enhancement was greater to the high-mannose glycopeptides (1, 3 and 5) than to the corresponding GlcNAc-peptides or aglycone peptide (2, 4 and 6), as reflected by the endpoint titers. The binding of the antibodies induced by peptide immunogen 11 was very weak to the 20-mer high-mannose glycopeptide 5 and GlcNAc-peptide 6 (Figure 6A). However, antibodies induced by glycopeptide immunogen 10 showed enhanced binding to the 20-mer V3 glycopeptides (Figure 6B). These results indicated that glycopeptide immunogen 10 was able to raise N332-glycan dependent antibodies. We also performed ELISA binding to a synthetic high-mannose glycan (Man9-Asn-Biotin), which was synthesized following our recently described procedure (Orwenyo et al., 2017). Similar to most of the V3 glycan dependent bNAbs, the antibodies induced by glycopeptide immunogen 10 did not bind to the high-mannose glycan alone (Figure S4), suggesting that the antibody responses were raised to target the unique glycopeptide moieties, instead of the polypeptide or the glycan components alone.
Figure 6. ELISA analysis of the binding of the antisera raised by three-component immunogens (10 and 11) to the synthetic V3 glycopeptides 1–6 (coating antigens).
A) the antisera induced by the three-component peptide immunogen 11; B) the antisera induced by the three-component glycopeptide immunogen 10; C) the antisera induced by the peptide immunogen (11) that were pretreated with magnetic beads carrying V3 peptide 4 to depress the V3 peptide-specific antibodies; D) the antisera induced by the glycopeptide immunogen (10) that were pretreated with magnetic beads carrying V3 peptide 4 to depress the V3 peptide-specific antibodies; E) Endpoint titers of the ELISA binding between the rabbit antisera and V3 glycopeptides, which were derived from A–D).
To further confirm that glycan dependent antibodies were induced by glycopeptide immunogen 10, we pretreated the antisera prior to ELISA analysis by incubating with magnetic beads carrying aglycone V3 peptide 4, which could remove the peptide specific antibodies in the sera. After this pretreatment, the binding of the antisera induced by peptide immunogen 11 to all of the V3 peptides and glycopeptide (1–6) was almost eliminated (Figure 6C). Strikingly, after pretreated with the polypeptide (4), the antisera induced by glycopeptide immunogen 10 still exhibited strong binding to high-mannose glycopeptides (1, 3 and 5), but the binding to the GlcNAc-peptide or aglycone peptide (2, 4 and 6) was significantly reduced (Figure 6D). Interestingly, the pretreated antisera showed similar ELISA binding profiles and identical titers (51200) to the 46-mer, 33-mer and 20-mer V3 high-mannose glycopeptides (2, 4 and 6). Compared to the untreated antisera, the titers of the pretreated antisera to the 46-mer and 33-mer high-mannose glycopeptides (1 and 3) was reduced to half, while the titer of the pretreated antisera to the 20-mer high-mannose glycopeptide 5 was identical to that of the untreated antisera (Figure 6E, Entry 3 and 4). These data suggest that substantial glycan dependent antibodies were raised by the synthetic glycopeptide immunogen (10), which recognize the unique structures presented by the glycopeptide moiety.
The neutralizing activity of the antisera was assessed against tier 1 and tier 2 HIV-1 viruses via a TZM-bl cell-based neutralization assay (Sarzotti-Kelsoe et al., 2014). Our preliminary assay indicated that the antisera induced by both peptide immunogen 11 and glycopeptide immunogen 10 did not show neutralizing activities (data not shown). This result is similar to a recent immunization study with a synthetic mini-V3 glycopeptide in rhesus macaque, showing that a V3 glycopeptide mixed with an adjuvant could raise glycan dependent antibodies using a dose escalation protocol over a one-year period, but the antibodies failed to neutralize HIV-1 viruses (Alam et al., 2017). As demonstrated in another recent immunization study, repetitive vaccination with a high-mannose glycoform of HIV-1 Env over a 4-year period resulted in induction of V3-glycan specific antibodies (Saunders et al., 2017). The antibodies showed neutralizing activities but they were neutralizing only against pseudoviruses carrying high density of high-mannose N-glycans.
DISCUSSION
Development of broadly neutralizing antibodies by active immunization remains a challenging task in HIV-1 vaccine research. Among others, both the immunogen design and the immunization protocols should be optimized in order to develop affinity maturation and/or to elicit sufficient levels of somatic hypermutation (SHM), which is a common observation among bNAbs (Sok et al., 2013, Klein et al., 2013, Wu et al., 2011, Haynes and Burton, 2017). The intrinsic high-mannose patch centered on the N332 high-mannose glycan adjacent to the V3 domain is a distinguished neutralizing epitope targeted by many broad and potent bNAbs (Kong et al., 2013, Walker et al., 2011). A recent study showed that a Man9-V3 glycopeptide could be used to isolate a potent bNAb (DH270.6) from HIV-infected individual CH765, and was also used to isolate glycan-dependent antibody after vaccination with the Man9-V3 glycopeptide in rhesus macaques (Alam et al., 2017). V3 glycopeptide could also prime precursors of V3-glycan B cell lineages to clonally expand (Bonsignori et al., 2017). These findings indicate V3 glycopeptides could recapitulate the neutralizing epitope of bNAbs. However, the V3 glycopeptide alone is not a potent immunogen (Alam et al., 2017). The glycopeptide alone lacked a built-in T-helper and thus required a very high-dose (50–500 µg of the glycopeptide) and repetitive immunization over a one-year period to raise glycan-dependent antibodies, even with the co-injection of the Toll-like receptor 4 agonist GLA-SE (glucopyranosyl lipid adjuvant-stable emulsion) adjuvant. Rationally designed immunogens are required to enhance the immune responses. In the present study, we designed and synthesized a novel three-component immunogen that the identified the 33-mer cyclic V3 glycopeptide carrying a high-mannose N-glycan at the N332 site, a T helper epitope peptide (P30), and a TLR2 receptor ligand (Pam3CSK4). Our immunization study in rabbits demonstrated that the three-component glycopeptide-based immunogen was significantly immunogenic and was able to raise substantial glycan-dependent antibody responses within a relatively short period of immunization with a much lower dose. Interestingly, a clear difference in the immunogenicity between the glycopeptide-based (10) and the non-glycosylated peptide-based immunogen (11) was observed. While the glycopeptide-based immunogen (10) raised substantial glycan-dependent antibodies that bind to an array of HIV-1 gp120 envelope glycoproteins across clades, the non-glycosylated peptide-based immunogen elicited antibodies that could bind tightly to the specific strain (JR-FL), but only had low affinity for those HIV-1 gp120s from different strains. Thus, our experimental data suggest that the V3 glycopeptide-based immunogen could have greater potential than the non-glycosylated polypeptide based immunogen in eliciting broadly reactive antibody responses targeting the shared glycopeptide motifs on the Env from different HIV-1 strains. Notably, previous immunization studies have shown that synthetic oligomannose-containing glycoconjugate immunogens, designed to mimic the 2G12 epitope, were able to raise carbohydrate-dependent antibodies, but the antibodies induced were not cross-reactive with HIV-1 envelope glycoprotein gp120, even a number of high-mannose N-glycans are present on the viral envelope (Astronomo et al., 2008, Astronomo et al., 2010).
It should be pointed out that the antisera induced by the three-component V3 glycopeptide immunogen in the current preliminary immunization study failed to neutralize HIV-1 viruses, probably due to the lack of somatic mutation of the antibodies and/or the low titers of antibody responses. Nevertheless, the ability to elicit V3 glycan-dependent antibodies cross-reactive to heterologous HIV-1 gp120s by a well-defined glycopeptide immunogen within a short period of immunization represents an important discovery, as few immunogens have been able to raise glycan-dependent antibodies cross-reactive to various HIV-1 gp120s (Haynes and Burton, 2017). The three-component V3 glycopeptide immunogen described in the present study appears to be superior to the weakly immunogenic synthetic V3 glycopeptides that lacks a T helper epitope (Alam et al., 2017), and to the unusual high-mannose Env trimer glycoform that contains various non-neutralizing epitopes (Saunders et al., 2017) in eliciting V3 glycan-dependent antibodies. Future studies should be directed to further characterizing the glycan-dependent antibodies raised by the glycopeptide immunogen and to the assessment of the immunogenicity of the synthetic three-component glycopeptide immunogen in non-human primate models, either alone or in combination with other vaccine candidates such as the Env trimer and related constructs, to evaluate if the glycopeptide immunogen can focus the immune response to the glycopeptide epitopes to raise broadly HIV-neutralizing antibodies.
SIGNIFICANCE
A class of recently discovered bNAbs, including the PGT 128 and 10–107, appear to target the intrinsic high-mannose patch centered on the N332 high-mannose glycan and a conserved peptide segment at the base of the V3 loop. However, elicitation of such glycan-dependent neutralizing antibodies through active immunization has yet to be achieved. As described in this paper, we first performed glycopeptide epitope mapping to identify the minimal glycopeptide sequence recognized by PGT128 and 10–1074. Then we incorporated the identified mini-V3 glycopeptide epitope in the design of a novel three-component self-adjuvanting immunogen that consists of the V3 glycopeptide epitope, a T-helper peptide P30, and the TLR2 ligand lipopeptide Pam3CSK4. We demonstrate that the three-component glycopeptide immunogen could elicit substantial glycan-dependent antibodies in rabbits within only two months with a relatively low dose immunization. The serum antibodies induced by the glycopeptide immunogen showed broad recognition to several gp120s across clades, while the serum antibodies induced by the non-glycosylated peptide immunogen elicited mainly strain-specific antibody responses. Our results demonstrated that the N-332 high-mannose N-glycan was immunogenic and vulnerable to the immune system. While the antibodies raised in the short-term immunization regimen failed to neutralize HIV-1, raising substantial glycan-dependent, gp120-reactive antibodies by a well-defined glycopeptide immunogen represents an important step toward the goal of stimulating glycan-dependent broadly neutralizing antibody responses in HIV vaccine research. Our experimental data suggest that the three-component V3 glycopeptide immunogen could mimic the neutralizing epitopes of those glycan-specific bNAbs and holds the promise to potentially raise glycan-dependent, broadly neutralizing antibody responses in an appropriate immunization regimen.
STAR METHOD
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Lai-Xi Wang (wang518umd.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Rabbit immunizations were carried out by Spring Valley Laboratories, Inc., which is licensed by the United States Department of Agriculture (APHIS registration number, 51-R-051) and has been accredited fully and continuously by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALACi). All procedures were performed according to the NIH Guide for the Care and Use of Laboratory Animals. All New Zealand White rabbits were female and 8 weeks old when the experiments started.
METHOD DETAILS
General Procedures
Analytical reverse-phase HPLC was carried out on a Waters 626 HPLC system equipped with a dual absorbance UV detector. All the V3 peptides and glycopeptides were run on a C18 column (YMC-Triart C18, 4.6 × 250 mm, 5 µm) at a flow rate of 1 mL/min using a linear gradient of 15–45% MeCN containing 0.1% TFA over 30 min. The lipopeptides were run on a CN column (YMC-Pack CN, 4.6 × 250 mm, 5 µm) using a linear gradient of 20–70% MeCN containing 0.1% TFA over 50 min. ESI-MS spectra were measured on a Micromass ZQ-4000 single-quadrupole mass spectrometer. For the V3 peptides and glycopeptides, preparative reverse-phase HPLC was carried out on a Waters 600 HPLC system equipped with a dual absorbance UV detector using a C18 column (Waters XBridge, Prep Shield RP 10 × 250 mm, 5µm) at a flow rate of 4 mL/min. The preparative HPLC purification of the lipopeptides was performed on a CN column (YMC-Pack CN, 10 × 250 mm, 5 µm).
HIV V3 Peptide Synthesis
All Fmoc-protected amino acid building blocks and coupling reagents were purchased from Bachem. Abbreviations: TBTU = N,N,N′,N′-Tetramethyl-O-(benzotriazol-1-yl)uronium tetrafluoroborate; DIPEA = Diisopropylethylamine; DMF = Dimethylformide; HOBt = Hydroxybenzotriazole; DCM = Dichloromethane; TFA = Trifluoroacetic acid; DMSO = Dimethyl sulfoxide.
V3 peptide synthesis was performed under microwave synthesis conditions using a CEM Liberty Blue microwave peptide synthesizer. Synthesis was based on Fmoc chemistry using PAL-PEG-PS resin (0.18 mmol/g) on a 0.1 mmol scale. Couplings were performed using 6 equiv. of Fmoc-protected amino acids, 6 equiv. of TBTU and 12 equiv. of DIPEA in DMF. The couplings were carried out at 45 °C for 20 min with a repeat of the coupling cycle for the longer full-length V3 peptides. The glycosyl amino acid Fmoc-(Ac3GlcNAc)-Asn-OH was introduced at the N332 sites and Fmoc deprotection was carried out with 20% piperidine in DMF containing 0.1 M HOBt. Fmoc-ε-Acp-OH was extended at the N-terminus as spacer and then Biotin or 4-pentynoic acid was coupled at the N-terminus to install the corresponding biotin or alkyne group. The resin was washed with DMF (3×) and DCM (3×) then cleavage was carried out using cocktail R (TFA/Thioanisole/Ethanedithiol/Anisole = 90/5/3/2). The resin was then filtered and the obtained solution was added to cold diethyl ether for peptide precipitation. The crude peptide was dissolved in glacial acetic acid and then lyophilized. For the 46-mer and 33-mer V3 glycopeptides, cyclization was performed in a 20% aqueous DMSO solution. De-O-acetylation of the GlcNAc moiety was then performed in a 5% aqueous hydrazine solution. The crude peptides were purified on preparative RP-HPLC to afford the desired peptides.
GlcNAc-peptide (2) (cyclic_46-mer_N332_GlcNAc)
ESI-MS: Calcd., M = 5764.46; found (m/z): 721.49 [M + 8 H]8+, 824.42 [M + 7 H]7+, 961.65 [M + 6 H]6+. RP-HPLC retention time, tR = 23.8 min.
GlcNAc-peptide (3a) (cyclic_33-mer_N332_GlcNAc)
ESI-MS: Calcd., M = 4333.86; found (m/z): 722.86 [M + 6 H]6+, 867.23 [M + 5 H]5+, 1083.78 [M + 4 H]4+, 1444.70 [M + 3 H]3+. RP-HPLC retention time, tR = 21.1 min.
Peptide (4) (cyclic_33-mer)
ESI-MS: Calcd., M = 4128.64; found (m/z): 688.99 [M + 6 H]6+, 826.59 [M + 5 H]5+, 1032.98 [M + 4 H]4+, 1377.61 [M + 3 H]3+. RP-HPLC retention time, tR = 20.9 min.
GlcNAc-peptide 6 (linear_20-mer_N332_GlcNAc)
ESI-MS: Calcd., M = 2807.15; found (m/z): 702.62 [M + 4 H]4+, 936.49 [M + 3 H]3+, 1404.23 [M + 2 H]2+. RP-HPLC retention time, tR = 22.5 min.
GlcNAc-peptide (8a)
ESI-MS: Calcd., M = 4185.62; found (m/z): 698.52 [M + 6 H]6+, 838.02 [M + 5 H]5+, 1047.27 [M + 4 H]4+, 1396.03 [M + 3 H]3+. RP-HPLC retention time, tR = 21.1 min.
Peptide (9)
ESI-MS: Calcd., M = 3982.43; found (m/z): 664.67 [M + 6 H]6+, 797.40 [M + 5 H]5+, 996.50 [M + 4 H]4+. RP-HPLC retention time, tR = 21.4 min.
Chemoenzymatic Synthesis of Homogeneous V3-Glycopeptides
Glycopeptide 1 (cyclic_46-mer_N332_Man9)
Glycopeptide 2 (1.5 mg, 0.26 µmol) was incubated at 30 °C together with Man9-oxazoline (2.16 mg, 1.3 µmol) and Endo-AN171A (40 µg) in phosphate buffer (100 mM, p H 7, 100 µL). Upon completion of the enzymatic reaction after 2 h, as indicated by monitoring with RP-HPLC analysis, the reaction was quenched using 0.1% aq. TFA. The product was purified by RP-HPLC to give glycopeptide 1 as a white powder (1.56 mg, 81%). ESI MS: calcd. M = 7426.91; found (m/z): 929.31 [M + 8 H]8+, 1061.92[M + 7 H]7+, 1238.75 [M + 6 H]6+. RP-HPLC retention time, tR = 23.5 min.
Glycopeptide 3 (cyclic_33-mer_N332_Man9)
Glycopeptide 3a (2 mg, 0.46 µmol), Man9-oxazoline 2 (4.6 mg, 2.76 µmol) and Endo-AN171A (80 µg) were incubated as described for glycopeptide 1 to give glycopeptide 3 as a white powder (2.46 mg, 89%). ESI-MS: Calcd., M =5994.34; found(m/z): 889.12 [M + 7 H]7+, 999.95 [M + 6 H]6+, 1199.95 [M + 5 H]5+. RP-HPLC retention time, tR = 20.1 min.
Glycopeptide 5 (linear_20-mer_N332_Man9)
Glycopeptide 6 (1.3 mg, 0.46 µmol), Man9-oxazoline (4.6 mg, 2.76 µmol) and Endo-AN171A (80 µg) were incubated as described for glycopeptide 1 to give glycopeptide 5 as a white powder (1.9 mg, 90%). ESI-MS: Calcd., M = 4469.60; found(m/z): 1118.25 [M + 4 H]4+, 1490.67 [M + 5 H]5+. RP-HPLC retention time, tR = 19.7 min.
Glycopeptide 8
Glycopeptide 8a (1.9 mg, 0.46 µmol), Man9-oxazoline (4.6 mg, 2.76 µmol) and Endo-AN171A (80 µg) in phosphate buffer (100 mM, pH 7, 200 µl) were incubated as described for glycopeptide 1 to give glycopeptide 8 a white powder (2.33 mg, 87%). ESI-MS: Calcd., M = 5848.08; found (m/z): 975.61 [M + 6 H]6+, 1170.53 [M + 5 H]5+, 1462.91 [M + 4 H]4+. RP-HPLC retention time, tR = 21.0 min.
Synthesis of Lipopeptide 7
Lipopeptide 7 was synthesized using a similar procedure for the V3 peptide synthesis on the automated peptide synthesizer with a 0.1 mmol scale. Briefly, the Lys-N3 was first coupled to the resin, and then Fmoc-ε-Acp-OH was attached. The P30 sequence was coupled stepwise followed by Fmoc-ε-Acp-OH coupling. Then the SKKKK sequence was coupled and finally the Pam3Cys was coupled. The resin was washed and cleaved using cocktail R. The crude peptide was purified by RP-HPLC on a CN column to give the purified 7 (200 mg, yield 46%). ESI-MS: Calcd., M = 4347.70; found (m/z): 725.94 [M + 6 H]6+, 870.92 [M + 5 H]5+, 1088.40 [M + 4 H]4+, 1450.86 [M + 3 H]3+. RP-HPLC retention time, tR = 38.7 min.
Synthesis of Three-component Immunogens 10 and 11
Lipopeptide 7 (1.5 mg, 0.35 µmol), V3 alkyne glycopeptide 8 (2.5 mg, 0.43 µmol) and CuOAc (4 µg, 0.03µmol) were dissolved in 100 µL DMF. Then the mixture was incubated at 40 °C for 18 hours. The reaction mixture was diluted by 1 mL water and then lyophilized. The crude product was purified by RP-HPLC on a CN column to give the desired immunogen 10 (2.8 mg, 78%). ESI-MS: Calcd., M = 10200.61; found (m/z): 785.48 [M + 13 H]13+, 850.79 [M + 12 H]12+, 928.04 [M + 11 H]11+, 1020.77 [M + 10 H]10+, 1134.13 [M + 9 H]9+, 1275.70 [M + 8 H]8+, 1457.80 [M + 7 H]7+, 1700.70 [M + 6 H]6+, 2040.57 [M + 5 H]5+. RP-HPLC retention time, tR = 35.4 min.
The three-component immunogen 11 was prepared from alkyne peptide 9 (1.7 mg, 0.43 µmol) and lipopeptide 7 (1.5 mg, 0.35 µmol) using the same procedure as described for the preparation of 10. After purification, the desired immunogen 11 (2.5 mg, 86%) was obtained. ESI-MS: Calcd., M = 8329.69; found (m/z): 641.98 [M + 13 H]13+, 695.31 [M + 12 H]12+, 758.43 [M + 11 H]11+, 834.17 [M + 10 H]10+, 926.74 [M + 9 H]9+, 1042.47 [M + 8 H]8+, 1191.35 [M + 7 H]7+, 1389.65 [M + 6 H]6+, 1667.49 [M + 5 H]5+. RP-HPLC retention time, tR = 36.3 min.
ELISA Analysis
The 96-well ELISA microtiter plates were coated with 5 µg/mL Neutravidin in PBS (100 µL/well) and incubated at 4 °C overnight. The plates were washed with PBS/0.05% Tween-20 and blocked with 2% sodium caseinate (w/v) in PBS at room temperature for 1 h. After washing three times, 2 µg/mL of the biotinylated glycopeptide dissolved in 1% casein PBS (100 µL/well) was added and incubated at 37 °C for 1 h. Controls were set up by adding PBS without V3 glycopeptide to the wells. Then plates were washed three times and titrated against 1:2 serial dilutions of monoclonal antibodies (10–1074 and PGT128) or rabbit antisera in 1% sodium caseinate. The plates were incubated at 37 °C for 1 h. After washing three times, a solution (100 µl/well) of 1:3000 diluted horseradish peroxidase (HRP)-conjugated goat anti-human IgG (H + L) antibody or horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (H + L) antibody in 1% PBS was added to the plates. The plates were incubated for 1 h at 37 °C. After washing three times, a solution of 3, 3’, 5, 5’-tetramethylbenidine (TMB) was added. Color was allowed to develop for 5 min, and then quenched by adding a solution of 1 M H3PO4. The readout was measured at a wavelength of 450 nm. A modified procedure was used for the ELISA binding to the gp120s. The gp120 was diluted in PBS (2 µg/mL) and coated on the plate (100 µL/well), then following the above mentioned procedures to complete the experiments.
SPR Measurement
The SPR binding was performed on a BIAcore T200 system (GE Healthcare) at 25 °C. Biotinylated V3 glycopeptides were immobilized on neutravidin-coated CM5 sensor chips in HBS-P buffer (10 mM HEPES, 150 mM NaCl, P20 surfactant 0.05% v/v, pH 7.4) until 200 response unit (RU) was achieved. IgGs of PGT128 and 10–1074 were injected individually over four cells at two-fold increasing concentration in HBS-P buffer with a flow rate of 40 µL/min for 180 sec. Then HBS-P buffer was injected for 210s to allow for dissociation. Regeneration was performed by injection of 3M MgCl2 with a flow rate of 50 µL/min for 3 min followed by injection of HBS-P buffer with a flow rate of 50 µL/min for 5 min. Data processing was carried out using the BIAcore T200 evaluation software to subtract appropriate blank references and to fit the sensorgrams globally using a 1:1 Langmuir binding model to obtain the apparent kinetic parameters.
Rabbits Immunization
The synthetic multi-component immunogen 10 and 11 were incorporated into liposome following a reported procedure (Ingale et al., 2007). New Zealand White rabbits in a group of 3 were immunized via intramuscular and subcutaneous injections containing 50 µg of 10 or 11. After priming, three booster injections were given at interval of 21 days, and bleeds were taken 7 days postimmunization. The combined sera from each group were used for ELISA analysis.
Antisera Pretreatment to Subtract Peptide Specific Antibodies
To subtract the peptide specific antibodies in the antisera, Streptavidin magnetic beads (1 mg, New England BioLabs) were incubated with the synthetic 33er V3 peptide 4 (0.5 mg/ml, 500 µL) at 37 °C for 30 min. The beads were washed three times with PBS buffer. Then 1 mL antisera induced by immunogen 10 and 11 were added and incubated with the beads for 30 min at 37 °C. The supernatant (Antisera #) was separated by applying magnet and used for ELISA analysis.
QUANTIFICATION AND STATISTICAL ANALYSIS
SPR data processing was carried out using the BIAcore T200 evaluation software to subtract appropriate blank references and to fit the sensorgrams globally with a 1:1 Langmuir binding model. Rabbit antisera elicited by the same immunogen were mixed for immunological evaluation. ELISA data were presented as means of triplicates. PGT128 and 10–1074 ELISA data were processed by fitting a sigmoid.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PGT128 | Prof. Pamela Bjorkman Lab | N/A |
| 10-1074 | Prof. Pamela Bjorkman Lab | N/A |
| Horseradish peroxidase-conjugated goat anti-human IgG (H + L) antibody | SeraCare Life Sciences (Maryland) | 04-10-06 |
| Horseradish peroxidase-conjugated goat anti-rabbit IgG (H + L) antibody | SeraCare Life Sciences (Maryland) | 04-15-06 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Fmoc-PAL-PEG-PS resin | Thermo Fisher Scientific | GEN913383 |
| Fmoc-Asn(Ac3GlcNAc)-OH | Toonstra et al., 2016 | N/A |
| Man9GlcNAc | Huang et al., 2009 | N/A |
| EndoAN171A | Huang et al., 2009 | N/A |
| PamsCys-OH | Sigma-Aldrich | 670820 |
| D(+)-Biotin | Acros Organic | 230090010 |
| 4-pentynoic acid | Acros Organic | 213730010 |
| Fmoc-6-aminohexanoic acid | ChemPep Inc | 180102 |
| Fmoc-Lys(N3)-OH | ChemPep Inc | 101227 |
| N,N-Dimethylformamide | Fisher Chemical | BP1160-4 |
| Dichloromethane | Thermo Fisher Scientific | GEN902017 |
| Diethyl ether | Thermo Fisher Scientific | 615080010 |
| DMSO | Thermo Fisher Scientific | 610420010 |
| Trifluoroacetic acid | Acros Organics | 293811000 |
| Piperidine | Alfa Aesar | A12442 |
| Acetonitrile | Fisher Chemical | A998-4 |
| Copper (I) acetate | Sigma-Aldrich | 403342 |
| TMB 1-Component Microwell Peroxodase Substrate | SeraCare Life Sciences (Maryland) | 53-00-02 |
| Sodium caseinate | Sigma-Aldrich | C8654 |
| Magnesium chloride | Sigma-Aldrich | M8266 |
| Tween 20 | Bio-Rad | 1706531 |
| HBS-P buffer 10× | GE healthcare | BR100671 |
| Neutravidin protein | Thermo Fisher Scientific | 31000 |
| Critical Commercial Assays | ||
| Amine coupling kit | GE healthcare | BR100050 |
| Experimental Models: Organisms/Strains | ||
| New Zealand White Rabbit | Spring Valley Laboratories | N/A |
| Software and Algorithms | ||
| Xcalibur v2.0.7 SP1 | Thermo Fisher Scientific | N/A |
| BIAcore T200 evaluation software | GE healthcare | N/A |
| MagTran deconvolution software | Gundry Research Group | |
| Other | ||
| Sensor chip CM5 | GE healthcare | BR100012 |
| 96-well ELISA plate | Santa Cruz Biotechnology | SC-204463 |
| Streptavidin magnetic beads | New England BioLabs | S1420S |
Highlights.
A cyclic 33-mer V3 glycopeptide was identified as a mimic of neutralizing epitopes
A novel three-component glycopeptide immunogen was constructed via click chemistry
The glycopeptide immunogen induced substantial glycan-dependent antibody response
The glycan-dependent antibodies showed recognition of HIV-1 gp120 across clades
Acknowledgments
We thank other members of the Wang lab for technical assistance and discussions. We also thank Prof. Pamela Bjorkman and her lab members for kindly providing PGT128 and 10–1074 antibodies. The gp120s were obtained from the NIH AIDS reagent program. This work was supported by the National Institutes of Health (NIH grant R01AI113896).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
H.C. and L.-X.W. conceived and designed the project. H.C. planed and performed the experiments. J.O. helped with the peptide synthesis. J.G. and Y.Q. expressed the enzyme. R.Z. helped with the ELISA. C.C.L. and D.C.M. performed and analyzed the neutralizing experiments. H.C. and L.-X.W. wrote the manuscript. All the authors revised the manuscript.
References
- Alam SM, Aussedat B, Vohra Y, Ryan Meyerhoff R, Cale EM, Walkowicz WE, Radakovich NA, Anasti K, Armand L, Parks R, et al. Mimicry of an HIV broadly neutralizing antibody epitope with a synthetic glycopeptide. Sci. Transl. Med. 2017;9:eaai7521. doi: 10.1126/scitranslmed.aai7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam SM, Dennison SM, Aussedat B, Vohra Y, Park PK, Fernandez-Tejada A, Stewart S, Jaeger FH, Anasti K, Blinn JH, et al. Recognition of synthetic glycopeptides by HIV-1 broadly neutralizing antibodies and their unmutated ancestors. Proc. Natl. Acad. Sci. USA. 2013;110:18214–18219. doi: 10.1073/pnas.1317855110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin MN, Mclellan JS, Huang W, Orwenyo J, Burton DR, Koff WC, Kwong PD, Wang LX. Synthetic glycopeptides reveal the glycan specificity of HIV-neutralizing antibodies. Nat. Chem. Biol. 2013;9:521–526. doi: 10.1038/nchembio.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astronomo RD, Lee HK, Scanlan CN, Pantophlet R, Huang CY, Wilson IA, Blixt O, Dwek RA, Wong CH, Burton DR. A glycoconjugate antigen based on the recognition motif of a broadly neutralizing human immunodeficiency virus antibody, 2G12, is immunogenic but elicits antibodies unable to bind to the self glycans of gp120. J. Virol. 2008;82:6359–6368. doi: 10.1128/JVI.00293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astronomo RD, Kaltgrad E, Udit AK, Wang SK, Doores KJ, Huang CY, Pantophlet R, Paulson JC, Wong CH, Finn MG, Burton DR. Defining Criteria for Oligomannose Immunogens for HIV Using Icosahedral Virus Capsid Scaffolds. Chem. Biol. 2010;17:357–370. doi: 10.1016/j.chembiol.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Kreider EF, Fera D, Meyerhoff RR, Bradley T, Wiehe K, Alam SM, Aussedat B, Walkowicz WE, Hwang KK, et al. Staged induction of HIV-1 glycan-dependent broadly neutralizing antibodies. Science Translational Medicine. 2017;9:eaai7514. doi: 10.1126/scitranslmed.aai7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Chen MS, Sun ZY, Zhao YF, Kunz H, Li YM. Self-adjuvanting synthetic antitumor vaccines from MUC1 glycopeptides conjugated to T-cell epitopes from tetanus toxoid. Angew. Chem. Int. Ed. 2013a;52:6106–6110. doi: 10.1002/anie.201300390. [DOI] [PubMed] [Google Scholar]
- Cai H, Sun ZY, Huang ZH, Shi L, Zhao YF, Kunz H, Li YM. Fully synthetic self-adjuvanting thioether-conjugated glycopeptide-lipopeptide antitumor vaccines for the induction of complement-dependent cytotoxicity against tumor cells. Chem. Eur. J. 2013b;19:1962–1970. doi: 10.1002/chem.201203709. [DOI] [PubMed] [Google Scholar]
- Corti D, Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu. Rev. Immunol. 2013;31:705–742. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- Danby PM, Withers SG. Advances in Enzymatic Glycoside Synthesis. ACS Chem. Biol. 2016;11:1784–1794. doi: 10.1021/acschembio.6b00340. [DOI] [PubMed] [Google Scholar]
- Fernandez-Tejada A, Haynes BF, Danishefsky SJ. Designing synthetic vaccines for HIV. Expert. Rev. Vaccines. 2015;14:815–831. doi: 10.1586/14760584.2015.1027690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamblin DP, Scanlan EM, Davis BG. Glycoprotein synthesis: an update. Chem. Rev. 2009;109:131–163. doi: 10.1021/cr078291i. [DOI] [PubMed] [Google Scholar]
- Gao F, Weaver EA, Lu Z, Li Y, Liao HX, Ma B, Alam SM, Scearce RM, Sutherland LL, Yu JS, et al. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group m consensus envelope glycoprotein. J. Virol. 2005;79:1154–1163. doi: 10.1128/JVI.79.2.1154-1163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gristick HB, Von Boehmer L, West AP, JR, Schamber M, Gazumyan A, Golijanin J, Seaman MS, Fatkenheuer G, Klein F, Nussenzweig MC, et al. Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat. Struct. Mol. Biol. 2016;23:906–915. doi: 10.1038/nsmb.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Burton DR. Developing an HIV vaccine. What are the paths and obstacles to a practical vaccine? Science. 2017;355:1129–1130. doi: 10.1126/science.aan0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiya S, Macpherson IS, Krauss IJ. Recent strategies targeting HIV glycans in vaccine design. Nat. Chem. Biol. 2014;10:990–999. doi: 10.1038/nchembio.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, et al. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Li C, Li B, Umekawa M, Yamamoto K, Zhang X, Wang LX. Glycosynthases enable a highly efficient chemoenzymatic synthesis of N-glycoproteins carrying intact natural N-glycans. J. Am. Chem. Soc. 2009;131:2214–2223. doi: 10.1021/ja8074677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingale S, Awolfert M, Gaekwad J, Buskas T, Boons GJ. Robust immune responses elicited by a fully synthetic three-component vaccine. Nat. Chem. Biol. 2007;3:663–667. doi: 10.1038/nchembio.2007.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javaherian K, Langlois AJ, Larosa GJ, Profy AT, Bolognesi DP, Herlihy WC, Putney SD, Matthews TJ. Broadly neutralizing antibodies elicited by the hypervariable neutralizing determinant of HIV-1. Science. 1990;250:1590–1593. doi: 10.1126/science.1703322. [DOI] [PubMed] [Google Scholar]
- Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce JG, Krauss IJ, Song HC, Opalka DW, Grimm KM, Nahas DD, Esser MT, Hrin R, Feng M, Dudkin VY, Chastain M, Shiver JW, Danishefsky SJ. An oligosaccharide-based HIV-1 2G12 mimotope vaccine induces carbohydrate-specific antibodies that fail to neutralize HIV-1 virions. Proc. Natl. Acad. Sci. USA. 2008;105:15684–15689. doi: 10.1073/pnas.0807837105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu RB, Fu BZ, et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013;153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Lee JH, Doores KJ, Murin CD, Julien JP, Mcbride R, Liu Y, Marozsan A, Cupo A, Klasse, et al. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat. Struct. Mol. Biol. 2013;20:796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshminarayanan V, Thompson P, Wolfert MA, Buskas T, Bradley JM, Pathangey LB, Madsen CS, Cohen PA, Gendler SJ, Boons GJ. Immune recognition of tumor-associated mucin MUC1 is achieved by a fully synthetic aberrantly glycosylated MUC1 tripartite vaccine. Proc. Nat.l Acad. Sci. USA. 2012;109:261–266. doi: 10.1073/pnas.1115166109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody MA, Gao F, Gurley TC, Amos JD, Kumar A, Hora B, Marshall DJ, Whitesides JF, Xia SM, Parks R, et al. Strain-Specific V3 and CD4 Binding Site Autologous HIV-1 Neutralizing Antibodies Select Neutralization-Resistant Viruses. Cell Host Microbe. 2015;18:354–362. doi: 10.1016/j.chom.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PN, Spencer DI, Seaman MS, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc. Natl. Acad. Sci. USA. 2012;109:E3268–3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Song H, Wang Y, Stamatos NM, Wang LX. Toward a carbohydrate-based HIV-1 vaccine: Synthesis and immunological studies of oligomannose-containing glycoconjugates. Bioconjug. Chem. 2006;17:493–500. doi: 10.1021/bc0502816. [DOI] [PubMed] [Google Scholar]
- Orwenyo J, Cai H, Giddens J, Amin MN, Toonstra C, Wang LX. Systematic Synthesis and Binding Study of HIV V3 Glycopeptides Reveal the Fine Epitopes of Several Broadly Neutralizing Antibodies. ACS Chem. Biol. 2017;12:1566–1575. doi: 10.1021/acschembio.7b00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pett C, Cai H, Liu J, Palitzsch B, Schorlemer M, Hartmann S, Stergiou N, Lu M, Kunz H, Schmitt E, et al. Microarray Analysis of Antibodies Induced with Synthetic Antitumor Vaccines: Specificity against Diverse Mucin Core Structures. Chem. Eur. J. 2017;23:3875–3884. doi: 10.1002/chem.201603921. [DOI] [PubMed] [Google Scholar]
- Sarzotti-Kelsoe M, Bailer RT, Turk E, Lin CI, Bilska M, Greene KM, Gao H, Todd CA, Ozaki DA, Seaman MS, et al. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J. Immunol. Methods. 2014;409:131–146. doi: 10.1016/j.jim.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders KO, Nicely NI, Wiehe K, Bonsignori M, Meyerhoff RR, Parks R, Walkowicz WE, Aussedat B, Wu NR, Cai F, et al. Vaccine Elicitation of High Mannose-Dependent Neutralizing Antibodies against the V3-Glycan Broadly Neutralizing Epitope in Nonhuman Primates. Cell Rep. 2017;18:2175–2188. doi: 10.1016/j.celrep.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TYK, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. Sequence and Structural Convergence of Broad and Potent HIV Antibodies That Mimic CD4 Binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaltz RM, Hanson SR, Wong CH. Enzymes in the synthesis of glycoconjugates. Chem. Rev. 2011;111:4259–4307. doi: 10.1021/cr200113w. [DOI] [PubMed] [Google Scholar]
- Sok D, Doores KJ, Briney B, Le KM, Saye-Francisco KL, Ramos A, Kulp DW, Julien JP, Menis S, Wickramasinghe L, et al. Promiscuous Glycan Site Recognition by Antibodies to the High-Mannose Patch of gp120 Broadens Neutralization of HIV. Sci. Transl. Med. 2014;6:236ra63. doi: 10.1126/scitranslmed.3008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D, Laserson U, Laserson J, Liu Y, Vigneault F, Julien JP, Briney B, Ramos A, Saye KF, Le K, et al. The effects of somatic hypermutation on neutralization and binding in the PGT121 family of broadly neutralizing HIV antibodies. PLoS Pathog. 2013;9:e1003754. doi: 10.1371/journal.ppat.1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D, Pauthner M, Briney B, Lee JH, Saye-Francisco KL, Hsueh J, Ramos A, Le KM, Jones M, Jardine JG, Bastidas R, et al. A Prominent Site of Antibody Vulnerability on HIV Envelope Incorporates a Motif Associated with CCR5 Binding and Its Camouflaging Glycans. Immunity. 2016;45:31–45. doi: 10.1016/j.immuni.2016.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatatos L, Morris L, Burton DR, Mascola JR. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat. Med. 2009;15:866–870. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens AJ, et al. Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell. 2016;165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson P, Lakshminarayanan V, Supekar NT, Bradley JM, Cohen PA, Wolfert MA, Gendler SJ, Boons GJ. Linear synthesis and immunological properties of a fully synthetic vaccine candidate containing a sialylated MUC1 glycopeptide. Chem. Commun. (Camb) 2015;51:10214–10217. doi: 10.1039/c5cc02199e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toonstra C, Amin MN, Wang LX. Site-Selective Chemoenzymatic Glycosylation of an HIV-1 Polypeptide Antigen with Two Distinct N-Glycans via an Orthogonal Protecting Group Strategy. J. Org. Chem. 2016;81:6176–6185. doi: 10.1021/acs.joc.6b01044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unverzagt C, Kajihara Y. Chemical assembly of N-glycoproteins: a refined toolbox to address a ubiquitous posttranslational modification. Chem. Soc. Rev. 2013;42:4408–4420. doi: 10.1039/c3cs35485g. [DOI] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Jujien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LX, Amin MN. Chemical and chemoenzymatic synthesis of glycoproteins for deciphering functions. Chem. Biol. 2014;21:51–66. doi: 10.1016/j.chembiol.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Wilkinson BL, Day S, Malins LR, Apostolopoulos V, Payne RJ. Self-adjuvanting multicomponent cancer vaccine candidates combining per-glycosylated MUC1 glycopeptides and the Toll-like receptor 2 agonist Pam3CysSer. Angew. Chem. Int. Ed. 2011;50:1635–1639. doi: 10.1002/anie.201006115. [DOI] [PubMed] [Google Scholar]
- Williams WB, Liao HX, Moody MA, Kepler TB, Alam SM, Gao F, Wiehe K, Trama AM, Jones K, Zhang R, et al. HIV-1 VACCINES. Diversion of HIV-1 vaccine-induced immunity by gp41-microbiota cross-reactive antibodies. Science. 2015;349:aab1253. doi: 10.1126/science.aab1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, Mckee K, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- Zhang M, Gaschen B, Blay W, Foley B, Haigwood N, Kuiken C, Korber B. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology. 2004;14:1229–1246. doi: 10.1093/glycob/cwh106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.