Abstract
Induced pluripotent stem cells (iPSCs), and cells derived from them, have become key tools to model biological processes, particularly in cell types that are difficult to access from living donors. We present the first map of regulatory variants in iPSC-derived neurons, based on 123 differentiations of iPSCs to a sensory neuronal fate. Gene expression was more variable across cultures than in primary dorsal root ganglion, particularly in genes related to nervous system development. Using single-cell RNA-sequencing, we found that the fraction of neuronal vs. contaminating cells was influenced by iPSC culture conditions prior to differentiation. Despite high differentiation-induced variability, using an allele-specific method we detected thousands of quantitative trait loci (QTLs) influencing gene expression, chromatin accessibility, and RNA splicing. Based on our QTLs, we estimate that recall-by-genotype studies using iPSC-derived cells will require at least 20-80 individuals to detect the effects of regulatory variants with moderately large effect sizes.
Introduction
Cellular disease models are critical for understanding the molecular mechanisms of disease and for the development of novel therapeutics. In principle, induced pluripotent stem cell (iPSC) technology enables the development of these models in any human cell type. Initial uses of iPSCs for disease modelling have focused mostly on highly penetrant, rare coding variants with large phenotypic effects1–5. However, there is growing interest in using iPSCs to model the effects of the common genetic variants of modest effect size that drive complex disease6. A key question is to what extent variability in directed differentiation is a barrier to studying the effects of common disease-associated variants in iPSC-derived cells. In addition, because cultured cells are imperfect models of primary tissues, not all common disease-associated genetic variants also alter cell phenotypes in iPSC-derived systems.
Here, we present the first large-scale study of common genetic effects in a neuronal cell type differentiated from human stem cells, iPSC-derived sensory neurons (IPSDSNs). Peripheral sensory nerve fibres innervate the skin and other organs and are brought together at the dorsal root ganglia (DRG) before synapsing with the spinal cord around the dorsal horn. The development of efficient protocols to differentiate iPSCs into nociceptive (pain-sensing) neurons7 provides the opportunity to model common genetic effects on human sensory neuron function, which may underlie individual differences in pain sensitivity and chronic pain. We investigate how power to detect common genetic effects is affected by the variability introduced by differentiation and demonstrate how initial iPSC growing conditions influence cell phenotypes in IPSDSNs. We identify quantitative trait loci (QTLs) for gene expression, RNA splicing, and chromatin accessibility and identify overlaps between molecular QTLs and common disease associations. In generating this gene regulatory map we establish effective techniques for using iPSC-derived neurons to model molecular phenotypes relevant to common diseases.
Results
Sensory neuron differentiation and characterisation
We obtained 107 IPS cell lines derived from unrelated apparently healthy individuals by the HIPSCI resource8, and followed an established small molecule protocol7 to differentiate these into sensory neurons of a nociceptor phenotype (Figure 1a , Supplementary Tables 1-3). We performed 123 differentiations; 13 using an early version of the protocol (P1) which was subsequently refined (P2) to reduce the number of differentiation failures. After QC exclusions (Supplementary Figure 1), we had gene expression data for 119 differentiations from 100 unique iPSC donors; all analyses apart from QTL calling focused on the 106 P2 protocol samples only.
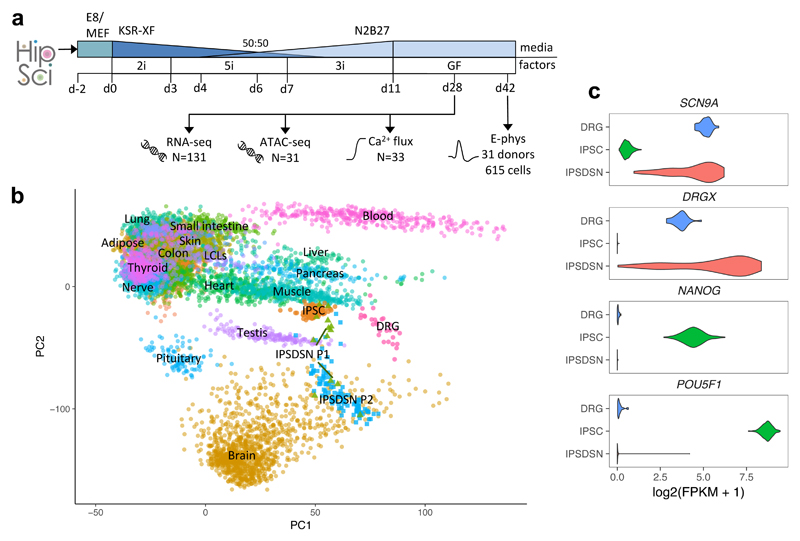
Figure 1. Characterization of molecular phenotypes in iPSC-derived sensory neurons.
(a) Schematic of IPSDSN differentiation and assays. iPSCs were received in Essential 8 (E8) medium (N=82) or on mouse embryonic fibroblasts (MEFs, N=49), and transferred to KSR-XF medium. Over 11 days, different inhibitor combinations were added (2i, 5i, 3i, see Methods), and N2B27 medium phased in, followed by transfer to growth factor medium at day 11 for neuronal maturation. (b) PCA plot projecting IPSDSN, iPSC, and DRG samples onto the first two principal components defined based on RNA-seq FPKMs in GTEx tissues. Some GTEx tissues are unlabelled due to overlapping labels. (c) Expression of sensory neuronal marker genes (SCN9A, DRGX) and key iPSC genes (NANOG, POU5F1).
We clustered our gene expression data with 239 iPSC samples from the many of same donors, 28 post-mortem DRG tissue samples from 10 donors, and 44 primary tissues from the GTEx project9 (Figure 1b). Globally, IPSDSN samples showed greatest similarity to iPSCs (gene expression correlation Spearman ρ=0.89), followed by DRG (ρ=0.84). Because different gene expression quantitation methods were used in GTEx, we cannot be certain of relative similarities between GTEx tissues and the samples we uniformly processed (IPSDSNs, iPSCs, DRG). The similarity to iPSCs may reflect lack of maturity in IPSDSNs, which is a well-recognized problem with iPSC-derived cells10–13. We also note that because the same iPSCs were differentiated to IPSDSNs, both donor genetic background and cell culture effects may contribute to the observed similarity. Despite this, key sensory neuronal marker genes were highly expressed in IPSDSNs, while pluripotency genes were not (Figure 1c). Using Ca2+ flux measurements on a subset of differentiated cultures (n=31) we confirmed that the cells consistently responded to sodium ion channel modulators veratridine and tetrodotoxin (Supplementary Figure 2). Patch-clamp electrophysiology on 616 individual neurons from 31 donors (Supplementary Figures 3,4) showed that the distribution of rheobases was comparable to those obtained from primary DRG cells, but with significant variation between donors (Supplementary Figure 5).
Quantifying differentiation variability using single-cell RNA-seq
In previous work we showed that not all individual cells express neuronal marker genes after differentiation7. Samples also appeared to differ visually in the fraction of cells with a neuronal morphology. To characterize this heterogeneity, we sequenced 177 IPSDSN cells from one individual and clustered them based on expression profiles using SC314. The data were best explained by two clusters (Figure 2a and Supplementary Figure 6), with 63% of cells forming a tight cluster expressing sensory-neuronal genes (e.g. SCN9A, CHRNB2), and the remaining 37% of cells forming a looser cluster expressing genes typical of fibroblasts (e.g. MSN, VIM). The two cell types also separated cleanly in a principal components plot (Supplementary Figure 7), indicating that the cells differentiated to distinct cell states. Gene expression in the neuronal cluster was most similar to DRG (Spearman’s ρ=0.654), followed by iPSCs (ρ=0.609) (Supplementary Figure 8) while the fibroblast-like cluster was most similar to GTEx fibroblasts (ρ=0.683), DRG (ρ=0.662), and iPSCs (ρ=0.653).
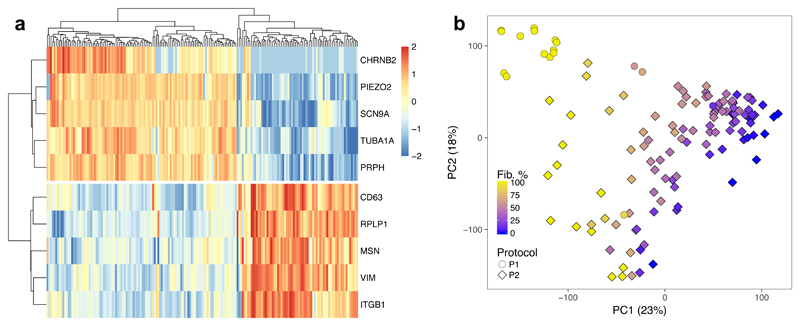
Figure 2. Single-cell sequencing of IPSDSN cells.
(a) A heatmap of RNA-seq data for ten marker genes of the two cell clusters identified by SC3. Color scale denotes normalised gene expression levels. (b) The first two principal components (PCs) of IPSDSN gene expression, with estimated fibroblast-like percentage from CIBERSORT, from samples derived using protocols 1 and 2 (P1 and P2).
Next, we used CIBERSORT15 to estimate the fraction of RNA from neuronal cells in our bulk RNA-seq samples, using the single-cell gene expression counts as signatures of neuronal or fibroblast-like expression. The estimated neuronal content correlated strongly (R2=0.75) with the first principal component of gene expression, and this corresponded well with a visual assessment of neuronal content from microscopy images (Figure 2b, Supplementary Figures 9,10). Although CIBERSORT estimated relatively high fibroblast-like content for many samples (mean 49%), a factor contributing to this may be the greater RNA content (2.3-fold greater; Supplementary Figure 11) of fibroblast-like cells. Indeed when the single-cell counts were pooled, CIBERSORT estimated the fibroblast content of this “sample” as 60%, considerably higher than the 37% of single cells in the fibroblast-like cluster. Despite this, IPSDSN samples estimated to have high fibroblast content still showed greater similarity in genome-wide gene expression with DRG than with any GTEx tissue, including fibroblast cell lines (Supplementary Figure 12).
Heterogeneity in IPSDSN gene expression
A central issue for genetic studies in iPSC-derived cells is heterogeneity of cellular phenotypes. This heterogeneity could arise from donor genetic background, effects of clonal selection and effects of the cell culture environment during reprogramming and differentiation. Genome-wide gene expression was highly correlated within lines differentiated multiple times (median Spearman ρ=0.96) and reduced slightly between IPSDSNs from different donors (median ρ=0.93) (Supplementary Figure 13). However, differentiation replicates within donor cell lines did not consistently cluster together (Supplementary Figure 14), suggesting that variability due to differentiation was at least as large as that due to donor genetic background and iPSC reprogramming together. Indeed, we observed a high degree of heterogeneity in the expression levels of some genes compared with DRG (Figure 1c and Supplementary Figure 15). These observations were independent of sample size, and were robust when comparing with DRG samples from unique donors only (Supplementary Figure 16).
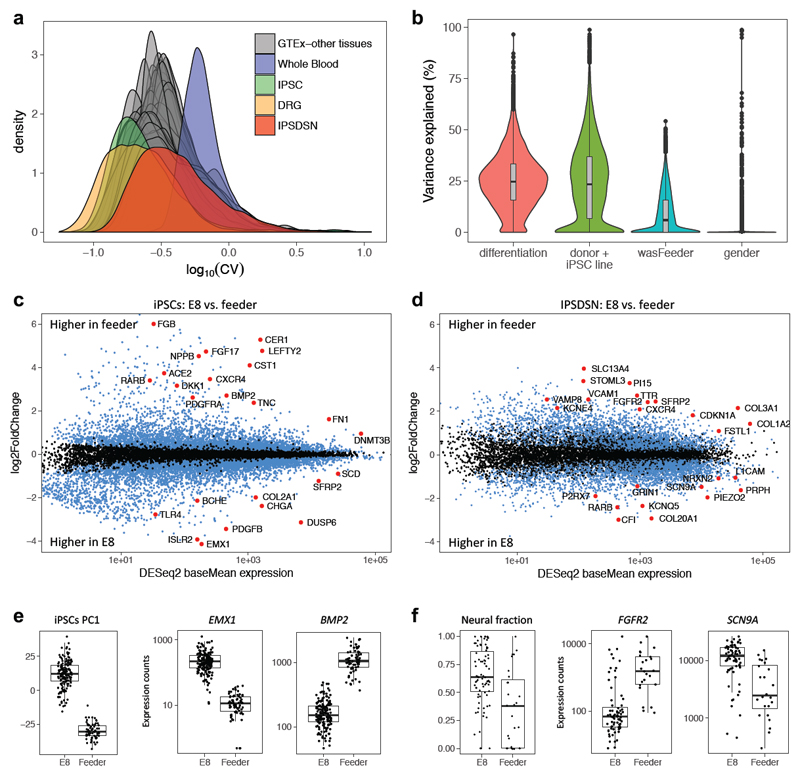
Next, we compared between-sample variability in global gene expression of IPSDSNs, measured as the coefficient of variation (CV) for each gene, to other somatic tissues and cell lines. The distribution of gene CVs in IPSDSNs (median CV=0.37) fell within the range of most GTEx tissues (Figure 3a), but was considerably higher than in DRG (median CV=0.23), indicating that IPSDSNs have greater between-sample variability in expression than the primary tissue they are intended to model. Highly variable genes in IPSDSNs were enriched for function in neuronal differentiation and development (Supplementary Table 4), whereas developmental genes were not highly variable in DRG, iPSCs, or GTEx nervous tissues (Supplementary Figure 17). Genes that were significantly upregulated between iPSCs and IPSDSNs, which includes those essential for sensory neuronal function, were also more variable than remaining genes (Supplementary Figure 18). These results highlight that expression of neuronal genes varies substantially more in IPSDSNs than in somatic nervous tissue, probably as a result of variability in differentiation. Consistent with this, variance components analysis (Figure 3b, Supplementary Figure 19) showed that more variation was explained by differentiation batch (median 24.7%) than donor/iPSC line of origin (median 23.3%), which includes both donor and reprogramming effects.
Figure 3. Gene expression variability in IPSDSNs is influenced by differentiation conditions.
(a) Density plot of the coefficient of variation of genes across samples, separately for each GTEx tissue, IPSDSN samples (n=106, P2 protocol only), iPSC (n=239), and DRG (n=28). (b) Violin plot showing, for each gene, the estimated fraction of total expression variability across samples due to differentiation batch, donor genetics or iPSC reprogramming, culture conditions (“wasFeeder”: feeder-dependent vs. E8 medium), and gender. (c) Differentially expressed genes (FDR 1%, blue and red points) between iPSC samples grown on feeders (n=68) vs. E8 medium (n=171). (d) Differentially expressed genes (FDR 1%) between IPSDSNs from feeder- (n=27) and E8-iPSCs (n=79). Neuronal differentiation genes, such as RET and L1CAM, are more highly expressed in samples from E8-iPSCs. (e) Left boxplot: global gene expression differences between feeder- and E8-iPSCs are captured in PC1. Right two boxplots: selected differentially expressed genes. (f) Left boxplot: estimated neural fraction of samples differs in IPSDSNs derived from feeder- and E8-iPSCs. Right two boxplots: selected differentially expressed genes. Boxplots show the median, 25th and 75th percentiles, with whiskers extending 1.5 times the interquartile range.
iPSC culture conditions influence cell fate
Our variance components analysis suggested that starting iPSC cell culture conditions influenced gene expression patterns in the IPSDSNs produced four weeks later (Figure 3). Of the 106 successful P2 protocol differentiations, 27 were from iPSCs maintained on mouse embryonic fibroblast (MEF) feeder cells (feeder-iPSCs), while the remaining 79 were grown in Essential 8 medium (E8-iPSCs). The first principal component (PC) of iPSC gene expression clearly differentiated feeder- and E8-iPSCs (Figure 3e), indicating that culture conditions are among the largest global effects on transcription. Similarly, PC1 of gene expression in IPSDSNs distinguished samples originating from feeder- and E8-iPSCs; moreover, IPSDSNs from E8-iPSCs had higher neuronal content (Figure 3f, 28% higher for E8-iPSCs, t-test p=1.84x10-5). A possible technical explanation for these results is that protocol implementation and batch effects changed subtly over the course of the project. However, the difference in neuronal content between IPSDSNs derived from E8 or feeder-iPSCs remained when sample derivation date was included as an explanatory covariate (linear regression p=6.5x10-4, 36% higher for E8-iPSCs, Supplementary Figure 20).
Next, we determined genes that were differentially expressed between E8- and feeder-iPSCs (Figure 3c,d). Genes upregulated in feeder-iPSCs were strongly enriched for mesenchyme development, stem cell differentiation, and Wnt and TGF-β signalling (Supplementary Tables 5-7). Notably, inhibition of TGF-β/SMAD signalling is a key step in sensory neuronal differentiation. Top differentially expressed genes include early developmental regulators EMX1 (15-fold higher in E8-iPSCs), important for specific neuronal cell fates, and BMP2 (13-fold higher in feeders), which has been shown to suppress differentiation to sensory cell fates by antagonizing Wnt/beta-catenin16 (Figure 3e). In addition, sensory neuronal markers SCN9A and TAC1 were expressed at low levels in iPSCs, with 2.2-fold and 2.9-fold higher expression in E8-iPSCs. We also considered genes differentially expressed between IPSDSNs derived from E8- and feeder-iPSCs (Figure 3d). Genes upregulated in IPSDSN samples from feeder-iPSCs were overrepresented in extracellular matrix components, pattern specification, and Wnt signalling (Supplementary Tables 8-10, Figure 3f). Genes upregulated in IPSDSN samples from E8-iPSCs were overrepresented in ion channel complexes, peripheral nervous system development, and synapse organisation. These differences likely reflect the increased neuronal content of samples from E8-iPSCs. Together these results suggest that iPSCs are primed towards different cell fates depending on the iPSC culture medium.
Since iPSC culture conditions influenced differentiation outcomes, we examined gene expression variability within subsets of IPSDSN samples. IPSDSNs differentiated from feeder-iPSCs had somewhat higher global gene expression variability, yet those from E8-iPSCs were still highly variable relative to DRG and iPSCs (Supplementary Figure 21), with neuronal and developmental gene sets enriched for highly variable genes (Supplementary Table 11).
Genetic variants influence gene expression, splicing and chromatin accessibility in sensory neurons
Using a linear model (FastQTL17), we mapped 1,403 expression quantitative trait loci (eQTLs) at FDR 10%. This number of eQTLs was lower than in GTEx tissues of comparable sample size (Supplementary Figure 22), suggesting that we may have reduced power for eQTL discovery due to variability introduced by differentiation. Using an allele-specific method18 we detected 3,778 genes with expression-modifying genetic variants, termed eGenes, at FDR 10% (Supplementary Table 12). Notably, the improvement in power from using allele-specific signals was greatest among genes with high variability across samples (Supplementary Figure 23).
We next compared our eQTLs with GTEx. When clustering tissues based on the pairwise correlation in eQTL effect sizes, IPSDSNs clustered most closely with GTEx brain tissues, while also showing elevated correlation with GTEx fibroblasts (Supplementary Figure 24). Of all 3,778 eGenes, 954 showed little or no association in GTEx v6p (Supplementary Table 13), and these included genes with known involvement in pain or neuropathies, such as SCN9A, GRIN3A, and NTRK2, suggesting that these genes have regulatory variants with IPSDSN-specific function.
Variants affecting gene splicing (sQTLs) often change either protein structure or context-dependent gene regulation, and may be more enriched for complex trait loci than are eQTLs19. Using the annotation-free method LeafCutter20 followed by FastQTL17 we discovered QTLs for 2,079 alternative splicing clusters at FDR 10% (Supplementary Table 14). Notably, only 538 (26%) of the lead variants for these splicing associations were in linkage disequilibrium (LD) r2>=0.5 with an eQTL in our dataset, indicating that these sQTLs are not merely proxies for gene-level eQTLs (or vice versa).
We collected ATAC-seq data for 31 samples21 and used this to map 6,318 chromatin accessibility QTLs (caQTLs) at FDR 10% (Supplementary Table 15). Using the LOLA Bioconductor package22 we found strong enrichment of our tissue-specific lead QTL SNPs (relative to GTEx lead SNPs) within SMARCB1 and SMARCC2 peaks (odds ratios 5.8 and 14.1; p < 5x10-5, Supplementary Tables 16,17), which are both members of the neuron-specific chromatin remodeling (nBAF) complex23. IPSDSN eQTLs and ATAC-seq peaks were enriched for ELK1 and ELK4 bidning, as well as c-Fos, a target of ELK1 and ELK4 which is widely expressed but is known to have specific functions in sensory neurons24,25 (Supplementary Table 18).
Sensory neuron QTLs overlap with complex trait loci
While we were interested in comparing our QTLs with GWAS for pain, the largest such GWAS to date included just 1,308 samples and found no associations at genome-wide significance26. We therefore considered all GWAS catalog associations with p < 5x10-8 that were in high LD (r2 > 0.8) with a QTL in our dataset, to (a) determine whether any GWAS traits are enriched overall for overlap with sensory neuron QTLs, and (b) to find individual QTLs that were strong candidates to mediate a GWAS association. Overall, IPSDSN eQTLs were enriched for overlap with GWAS catalog SNPs (p<0.001) relative to 1000 random sets of SNPs matched for minor allele frequency, distance to nearest gene, gene density, and LD27, and the overlap was consistent with that seen for eQTL studies in other tissues (Supplementary Figure 25). Although nociceptive neurons are specialized for sensing pain signals, we might expect enrichment for traits known to involve the nervous system more generally. However, among the 41 traits with at least 40 GWAS catalog associations, no trait had significantly greater overlap with our QTLs than other traits after correcting for multiple testing (Supplementary Table 19).
Across all traits, we found 156 eQTLs overlapping at least one GWAS association, and similarly 129 sQTLs and 172 caQTLs with GWAS overlap (Supplementary Tables 20-22). Among overlapping associations we found a number that relate to neuronal diseases. One striking overlap is between an eQTL for SNCA, encoding alpha synuclein, and Parkinson’s disease, for which a likely causal variant was recently identified10. The lead GWAS SNP and our lead eQTL are both in perfect LD with rs356168, in an intron of SNCA. Soldner et al. used CRISPR/Cas9 genome editing in iPSC-derived neurons to show that rs356168 alters both SNCA expression and binding of brain-specific transcription factors10. In IPSDSN cells we found that the G allele of rs356168 increased SNCA expression 1.14-fold, in line with their report of 1.06- to 1.18-fold increases in neurons and neural precursors. Despite residing in a visible ATAC-seq peak in our data, rs356168 was not a caQTL (raw p=0.22). eQTLs for SNCA are reported in GTEx (v6p), but none of the tissue lead SNPs are in LD (r2 > 0.2) with rs356168, suggesting that this SNP’s effect can be more readily detected in IPSDSNs and the frontal cortex tissue and iPSC-derived neurons studied by Soldner et al.
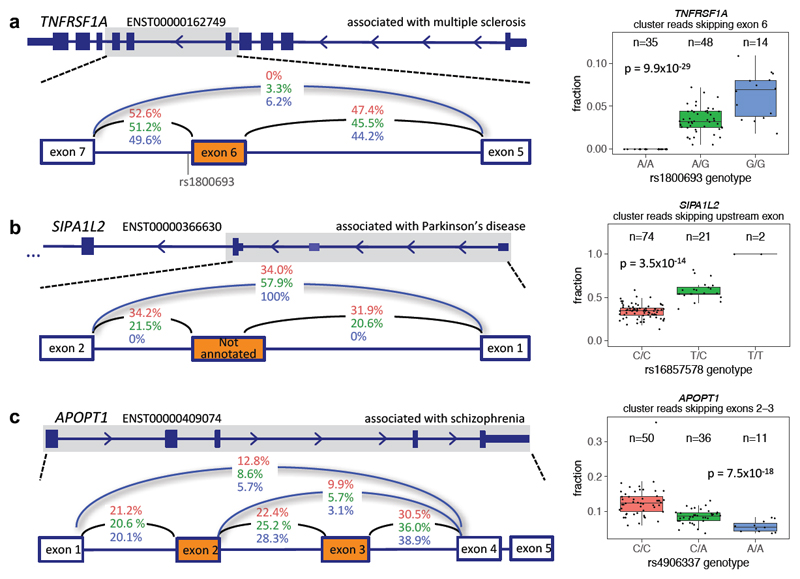
We also found multiple compelling overlaps between splice QTLs and GWAS associations (Figure 4). A strong sQTL for TNFRSF1A (p=9.9x10-29) has the same lead SNP (rs1800693) as a multiple sclerosis association. This SNP has been experimentally shown to cause skipping of exon 6, which results in a truncated, soluble form of TNFR1 that appears to reduce TNF28. TNFRSF1A is highly expressed (>15 FPKM) in both IPSDSNs and in DRG, as well as in cells of the immune system. TNF signalling has been shown to have both inflammatory and neuroprotective effects in the CNS and, despite a large body of research, the exact mechanisms and cell types responsible for the genetic risk associated with TNF receptor polymorphisms remain unclear29.
Figure 4. Splicing QTLs overlapping GWAS.
(a) An sQTL for TNFRSF1A leads to skipping of exon 6, and overlaps with a multiple sclerosis association. (b) An sQTL for SIPA1L2 leads to increased skipping of an unannotated exon between alternative promoters, and overlaps with a Parkinson’s disease association. (c) An sQTL for APOPT1 alters skipping of exons 2 and 3, and overlaps with a schizophrenia association. P values are from the FastQTL beta approximation based on 10,000 permutations. Boxplots show the median, 25th and 75th percentiles, with whiskers extending 1.5 times the interquartile range.
An sQTL for SIPA1L2 (rs16857578) is in LD with associations for both Parkinson’s disease (rs10797576, r2=0.93) and blood pressure (rs11589828, r2=0.94). An unannotated noncoding exon (chr1:232533490-232533583) between alternative SIPA1L2 promoters is included in nearly 50% of transcripts in individuals with the reference genotype, but splicing in of the exon is abolished by the variant (Figure 4b). SIPA1L2, also known as SPAR2, is a Rap GTPase-activating protein expressed in the brain and enriched at synaptic spines30 though its function is not yet clear. Interestingly, the related protein SIPA1L1 exhibits an alternative isoform with an N-terminal extension that is regulated post-translationally to influence neurite outgrowth31.
A complex sQTL for APOPT1 (rs4906337) is in near-perfect LD with a schizophrenia association (rs12887734). The splicing events involve skipping either of exon 3 only or both exons 2 and 3 (Figure 4c). At least 20 variants are in high LD (r2 > 0.9), including rs4906337, 40 bp from the exon 3 acceptor splice site, and rs2403197, 63 bp from the exon 4 donor splice site. APOPT1 is localized to mitochondria, and homozygous loss-of-function mutations lead to Cytochrome c oxidase deficiency, with affected individuals having variable motor and cognitive impairments and peripheral neuropathy32.
Recall-by-genotype studies in iPSC-derived cells will require large sample sizes
One attractive use of iPSCs is to experimentally characterise GWAS loci using a “recall-by-genotype” approach. Here, iPSC lines with specific genotypes are chosen from a large bank and differentiated into target cell types (for example, see ref.12). Our observations suggested that the cellular heterogeneity introduced by differentiation could impact the power of these studies to detect the effects of common genetic variants. Importantly, our large set of differentiations gave us accurate genome-wide estimates of effect size and expression variability in an iPSC-derived cell type. We investigated the performance of iPSC-based recall-by-genotype studies by bootstrap resampling from a stringent (FDR 1%) IPSDSN eQTL call set. For each eGene we sampled expression counts from an equal number of major and minor homozygotes for the lead SNP, sampling with replacement to achieve a specific sample size. We then estimated power as the fraction of 100 bootstrap replicates with a significant difference (p<0.05, two-tailed Wilcoxon rank sum test) in expression between homozygotes.
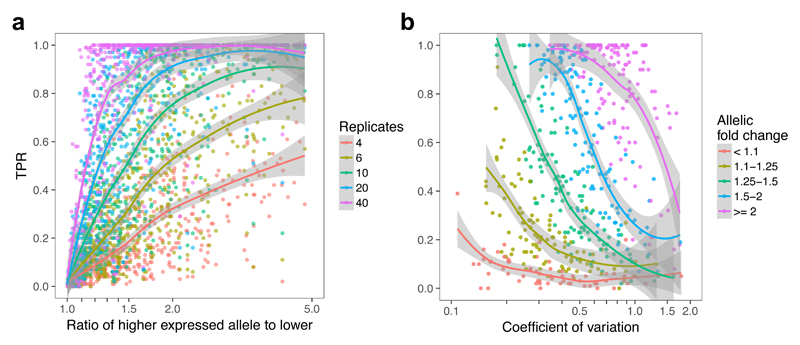
Our results illustrate important trends. First, recall-by-genotype studies in iPSC-derived cells are likely to require relatively large sample sizes, typically 20-80 unrelated individuals, for variants with a 1.5 to 2-fold allelic fold change (Figure 5a). Second, highly variable genes are more challenging, with power below 40% in a sample size of 20 for even moderately variable genes (CV 0.5 - 0.75). While expression noise will not typically be known accurately a priori, an estimate of effect size may be available from previous eQTL studies, in principle enabling estimation of the sample size required to achieve a desired power.
Figure 5. Power to detect a genetic effect in a single-variant single-gene test depends on sample size, allelic effect size, and gene expression variability.
(a) TPR as a function of allelic fold change for five different numbers of replicates (half the total sample size). (b) TPR as a function of CV for five bins of allelic fold change, with 10 samples of each genotype.
Note that these power estimates assume that a single gene is being tested, which implies a very strong prior belief in the causal gene in the region. Where multiple genes are tested, power will be further reduced. Large sample sizes will likely also be required when using genome editing to identify causal GWAS-associated variants: although genetic background can be controlled in such an experiment, differentiation noise will continue to be a major contributor to gene expression variability.
Discussion
iPSC-derived cells enable the molecular mechanisms of disease to be studied in relevant human cell types, including those which are inaccessible as primary tissue samples. Because the effect sizes of common disease-associated risk alleles tend to be small, observing their effects in cellular models is challenging10,11. In an iPSC-based system, this difficulty is compounded by variability between samples in the success of differentiation, as described for hepatocytes33, hematopoietic progenitors34, and neurons35,36.
Our study is the first that we are aware of to perform iPSC differentiation to a neuronal cell type and functionally characterise the resulting cells at scale. Sample-to-sample variability in gene expression in the iPSC-derived cells was greater than in DRGs, with highly variable genes enriched in processes relating to neuronal differentiation and development. This highlights that genes likely to be of particular interest and relevance for the function of these cells are also among the most variable, a challenge which may be broadly true of iPSC-derived cells. We detected thousands of eQTLs, sQTLs, and caQTLs in IPSDSNs, using a model that combines allele-specific and between individual differences in expression to improve power for association mapping. Some of these overlap known expression-modifying variants associated with disease, such as an eQTL for SNCA associated with Parkinson’s disease. However, for most of these disease overlaps the causal variants are not known, and await in-depth dissection of individual loci in iPSC-derived neurons.
Our study highlights the potential power of iPSC-derived cells for studying human genetic variation, but also illustrates the limitations of this approach. First, despite expressing key marker genes and exhibiting neuronal morphology and electrophysiology, IPSDSNs are transcriptionally distinct from DRG. This reflects a limitation of existing in vitro differentiation protocols, which produce cells that are not as functionally or transcriptionally mature as primary tissues. Second, our differentiations did not produce pure populations of neurons. Although we used single-cell RNA-seq to estimate neuronal content in bulk IPSDSN samples, we could not measure the purity of the resulting cultures precisely. Some of the sample-to-sample variability that we observed is likely due to this mixture of cell types, which varied across differentiations. Mature neurons can be labeled for marker genes, but are not easily sorted by automated systems, which limits the high-throughput options available for purifying neuronal populations. As a result, our QTLs do not represent those of a pure sensory neuronal cell type. For many cell types, sorting could provide one solution to the heterogeneity of differentiated cell populations.
The similarity of the fibroblast-like single cells to DRG raises the question of whether these cells are immature sensory neurons. Single-cell sequencing at multiple time points during MYOD-mediated myogenic reprogramming has suggested that some cells traverse a desired course, while others terminate at incomplete or aberrant reprogramming outcomes37. Such an approach in IPSDSNs could reveal determinants of neuronal differentiation trajectories, and may yield insights for protocol changes to improve the purity of differentiated neurons, or to specify more precise neuronal subtypes. More generally, replacing bulk RNA-seq with single-cell sequencing across many samples could enable in silico sorting of cells based on their transcriptome, and better characterisation of the sources of variation within a differentiated population of cells. Further, culturing cells from multiple donors in a pool, along with an scRNA-seq readout, could reduce differentiation-related batch effects while retaining the ability to identify donor-specific genetic effects on gene expression. These advantages suggest to us that a move towards scRNA-seq will be extremely useful in iPSC-derived cell models.
For iPSC models of common disease-associated variants to be used effectively, it is critical to know which variants exhibit a detectable cellular phenotype in vitro. We estimated the sample sizes needed to detect the effects of regulatory variants in iPSC-derived cells using a recall-by-genotype design. Power above 80% is only achieved with surprisingly large (40+) samples, even for alleles with a fold change of 1.5 to 2. Even larger samples will be needed when multiple genes in a GWAS interval are tested. These observations are consistent with a recent genome-editing experiment that required 136 differentiations in hepatocyte-like cells to discover an effect of rs12740374 on SORT1 gene expression12. Notably, the modest effect of this variant on expression in hepatocyte-like cells (1.3-fold increase) stands in contrast to the large effect of the variant (>=4-fold increase) observed previously in primary liver38. Protocol changes that Where it is possible to use a coding SNP to assess the allele-specific effect of a genome edit, as done for SNCA10, this may prove a more efficient approach to detecting causal effects of individual regulatory variants. In the future, improved differentiation protocols that show reduced variability will also enhance the ability to detect regulatory variant effects.
In summary, our catalog of QTLs reveals a large set of common variants and target genes with detectable effects in iPSC-derived neurons. These associations provide promising targets for functional studies to fine-map causal disease-associated alleles, such as by allelic replacement using CRISPR-Cas9, and our study describes the importance of considering differentiation-induced variability when planning these studies in iPSC-derived cells.
Online methods
IPS cell lines
A summary of iPSC lines used is available in Supplementary Table 2, and details of processes and assays for these iPSCs generated by the HIPSCI project are available at www.hipsci.org. Briefly, 107 human iPSCs from 103 healthy donors were obtained from HIPSCI8. Of these, 38 were initially grown in feeder-dependent medium and the remainder were grown in feeder-free E8 medium. All HIPSCI samples were collected from consented research volunteers recruited from the NIHR Cambridge BioResource (http://www.cambridgebioresource.org.uk), initially under existing ethics for iPSC derivation (REC Ref: 09/H0304/77, V2 04/01/2013), with later samples collected under a revised consent (REC Ref: 09/H0304/77, V3 15/03/2013).
Sensory neuron differentiation
All differentiations in this study were performed by a single individual, and a summary of the IPSDSN cell lines is in Supplementary Table 1. Two protocols were used, named P1 (13 differentiations) and P2 (110 differentiations). P1 protocol samples were included for QTL calling, and other analyses used P2 protocol samples exclusively. The P1 protocol (described in7) involved the addition of “2i” inhibitors (LDN193189 and SB-431542) for 5 days, followed by “5i” inhibitors (LDN193189, SB-431542, CHIR99021, DAPT, SU5402) for 6 days. When applying this protocol to a larger number of samples we observed excessive cell death prior to obtaining neural progenitors (days 9-12). We altered the protocol to make it more similar to that of Chambers et al.39, by:
using E8 rather than mTeSR1 media when maintaining iPSCs prior to differentiation;
phasing in neurobasal media beginning at day 4, and gradually increasing this to 100% by day 11;
beginning addition of inhibitors 5i at day 3 rather than day 5;
stopping addition of small molecule inhibitors LDN193189 and SB-431542 beginning at day 7 rather than day 11, referred to as “3i” for the 3 inhibitors that continued to be added.
Functional assays (Ca2+ flux, response to Veratridine) confirmed that response of the sensory neurons produced by each protocol was equivalent; however, the P2 protocol performed more consistently across cell lines and culture parameters.
P2 protocol
All reagents were from Life Technologies unless otherwise indicated. Clump-passaged iPSCs were single-cell seeded in E8 media on growth factor-reduced Matrigel (BD Biosciences) 48 hours prior to neural induction (day 0). KSR Media was prepared as 500ml DMEM-KO 130 ml Knockout Serum Replacement Xeno-Free, 1x NEAA, 1x Glutamax, 0.01 mM β-mercaptoethanol (Sigma). KSR media containing small molecule inhibitors LDN193189 (100 nM) and SB-431542 (10 µM) was added to cells from day 0 to 3 to drive anterior neuroectoderm specification. From day 3, CHIR99021 (3 µM), DAPT (10 µM) and SU5402 (10 µM) were also added to further promote neural crest phenotypes. N2B27 media was progressively phased in every two days from D4. N2B27 Media was prepared as 500 ml Neurobasal medium, 5 ml N2 supplement, 10 ml B27 supplement without vitamin A, 0.01mM β-mercaptoethanol (Sigma) and 1x Glutamax. On day 7, inhibitors LDN193189 and SB-431542 were no longer used, while CHIR99021, DAPT, and SU5402 continued to be added. On day 11 cells were reseeded at 150,000 cells/cm2 in maturation media containing N2B27 media with human-b-NGF, BDNF, NT3 and GDNF (each at 25 ng/ml). Mitomycin-C treatment (1 µg/ml) was used once at day 14 for 2 hrs to reduce the non-neuronal population. Cells were differentiated in T25 flasks for RNA and nuclei isolation, and onto coverslips and 96-well plates for electrophysiology and Ca2+-flux assays.
P1 protocol
All reagents and concentrations used were identical to the P2 protocol; the difference was timing of addition. Clump-passaged iPSCs were single-cell seeded in mTeSR1 iPSC (StemCell Technologies, Vancouver) media on growth factor-reduced Matrigel (BD Biosciences) 48 hours prior to neural induction (day 0). KSR media containing LDN193189 and SB-431542 was added to cells from day 0 to 5. From day 5, CHIR99021, DAPT and SU5402 were also added. As for the P2 protocol, cells were reseeded on day 11, and treated with Mitomycin-C on day 14.
Single-cell RNA sequencing
Full details are in the Supplementary Note. Briefly, blood-derived iPSCs from a single individual, who was not a HIPSCI donor, were differentiated to IPSDSNs in 3 batches using the P2 protocol, and were matured for 8 weeks. This differed from the bulk RNA-seq samples, which were matured for 4 weeks. Although gene expression changes are minor after 4 weeks maturation7, this difference in maturity may have influenced our estimates of neuronal content in bulk samples. Dissociated cells were prepared using a Fluidigm C1 and the Illumina Nextera XT kit, and sequenced by Illumina Nextseq500 (2x75bp). Reads were aligned to GRCh38 and Ensembl 80 transcript annotations using STAR v2.4.0d with default parameters. We excluded 9 cells expressing fewer than 20% of the ~56,000 quantified genes, and then used SC314 to cluster the remaining 177 cells based on expression counts. We examined alternative numbers of clusters from k=2 to 5 (Supplementary Figure 6). With two clusters, marker genes clearly identified one cluster (111 cells) as neuronal, whereas the other cluster (66 cells) had high expression of extracellular matrix genes reminiscent of fibroblasts. With 3 and 4 clusters, the sensory-neuronal cell cluster remained unchanged, and the fibroblast-like cluster became further subdivided. This suggests that a majority of the cells in this sample were terminally differentiated into sensory neurons, whereas the remaining cells were more heterogeneous in their gene expression.
Genotypes
We obtained imputed genotypes for all of the samples from the HIPSCI project. We used CrossMap (http://crossmap.sourceforge.net/) to convert variant coordinates from GRCh37 to GRCh38, and used bcftools (http://samtools.github.io/bcftools/) to retain only bi-allelic variants (SNPs and indels) with INFO > 0.8 and MAF > 0.05 in the 97 samples used for QTL calling.
RNA sequencing
The 131 RNA samples corresponded with 103 unique HIPSCI donors, as some samples were differentiation or RNA-extraction replicates. One sample failed in sequencing and was excluded. For QTL analyses, reads for each sample were aligned to GRCh38 and Ensembl 79 transcript annotations using STAR v2.4.0j with default parameters. Using VerifyBamID v1.1.2 40 we identified 5 mislabeled RNA samples for which the matching genotypes could be determined, as well as two samples with no matching genotypes and which were thus excluded. For comparisons among tissues, reads were aligned to the 1000 Genomes GRCh37d5 reference with Gencode v19 transcript annotations using STAR 2.5.3a.
Gene expression quantification, quality control and exclusions
Gencode Basic transcript annotations, GRCh38 release 79, were downloaded from www.gencodegenes.org. Gene expression was counted for uniquely mapping reads using featureCounts (v1.5.0)41 with options (-s 2 -p -C -D 2000 -d 25). A median of 45 million reads were generated per sample, with median 32.8 million reads (72%) uniquely mapping and assigned to genes. After excluding short RNAs and pseudogenes, we normalised expression counts for 35,033 genes using the R package cqn v5.0.242.
We determined pairwise correlation between samples using normalized counts for 14,215 expressed genes (CQN > 1) and the first five principal components of gene expression against each other. We excluded four outlier samples from subsequent analyses (Supplementary Figure 1), leaving 126 samples from 97 donors. For QTL calling, replicate BAM files from same donor were merged together using samtools.
To assess gene expression replicability, we determined the spearman correlation coefficient of CQN-normalized expression between samples across all genes for (a) extraction replicates, (b) differentiation replicates, and (c) all possible pairs of samples from different donors, and plotted the histogram of correlation coefficients in Supplementary Figure 13.
DRG samples and sequencing
Human tissue acquisition and handling was performed at Pfizer Neuroscience and Pain Research Unit in accordance with regulatory guidelines and ethical board approval. Postmortem human DRG were obtained in dissected form from Anabios or as an encapsulated sheath together with sensory/afferent axons from National Disease Research Interchange and were subsequently dissected to isolate the cell-body rich ganglion. The tissue was homogenised in QIAzol Lysis Reagent according to weight and processed according to the manufacturer's instructions for the Qiagen RNeasy Plus lipid-rich kit. RNA was prepared with the Illumina TruSeq Stranded mRNA Library Prep Kit and sequenced (2x100 bp reads) on Illumina HiSeq 2500. Reads were aligned to GRCh37 using STAR and gene counts and FPKMs obtained using featureCounts and Ensembl v75 gene annotations.
Highly variable genes in IPSDSNs and GTEx
For each of the 44 GTEx tissues, as well as IPSDSNs, DRG, and HIPSCI iPSCs, we calculated the coefficient of variation (CV) of each gene’s RPKM expression among samples of the same tissue (SMTSD in GTEx metadata). In each tissue, we subsetted the genes considered to those expressed at RPKM > 1. We plotted the distribution of CVs across all genes for each tissue as Figure 3a.
We used GeneTrail2 (https://genetrail2.bioinf.uni-sb.de) to do a gene set over-representation analysis for the top 1000 most variable genes in IPSDSNs by CV (Supplementary Table 4). Similarly, gene set over-representation analysis in E8-IPSDSN subsets was done using Genetrail2 and the top 1000 most variable genes with RPKM > 1 (Supplementary Table 11).
Variance components analysis
For Figure 3b, we selected the 106 P2 protocol IPSDSN samples after QC exclusions, and used DESeq2 to get FPKM values for each gene after size factor normalization. We included all genes with mean FPKM > 1, and input log2-transformed counts per sample into the variancePartition R package, with design formula (1|donor) + (1|differentiation) + (1|gender) + (1|wasFeeder). We plotted the distribution of variance explained for each gene across the four above factors, with unexplained variance shown as “residuals”. For Supplementary Figure 19a, we included 119 QC-passed samples, and used variancePartition as above, but with protocol in the design formula. For Supplementary Figure 19b, we used 18 samples, for which we had 3 differentiation replicates from each of 6 donor cell lines; all 6 iPSC lines were from females and had been cultured in E8 medium. We therefore included only donor and differentiation in the design formula.
Estimation of neuronal purity
We used CIBERSORT15 to estimate the fraction of RNA from neuronal cells in our bulk RNA-seq samples. We used the 14,786 genes with mean CQN expression > 0 in bulk RNA samples, and retrieved raw counts for these genes in our scRNA-seq data. We labeled the single-cell counts as “neuron” or “fibroblast-like” based on the SC3 clustering, and used these as reference samples for CIBERSORT to generate custom signature genes. We used raw expression counts for the same genes for our 126 bulk RNA-seq samples as the mixture file for CIBERSORT to use in estimating the relative fractions of neuron and fibroblast-like cell RNA.
Correlation of iPSC and IPSDSN gene expression with cell culture conditions
We selected the 106 IPSDSN samples differentiated with the P2 protocol, as well as the 87 iPSC samples these were derived from and for which we had RNA-seq data, and we used DESeq2’s variance stabilising transformation on the raw gene expression counts. We computed the first 5 principal components of gene expression separately in iPSC and IPSDSNs, and used corrplot to compute pairwise correlations among these PCs and sample metadata: gender, iPSC passage number, iPSC culture conditions (wasFeeder), iPSC PluriTest score, IPSDSN fibroblast content, and IPSDSN processing date.
We determined differentially expressed genes between feeder-iPSCs and E8-iPSCs using DESeq2, using expression counts for genes with median FPKM > 0.1 across iPSC samples (Supplementary Table 5). We removed associations driven by outliers, defined as a maximum Cook’s distance >= 5. Similarly, we determined differentially expressed genes in IPSDSNs derived from either feeder-iPSCs or E8-iPSCs (Supplementary Table 8), again for genes with median FPKM > 0.1. We used GeneTrail2 (https://genetrail2.bioinf.uni-sb.de) to do a gene set over-representation analysis for the 717 genes with expression at least 2-fold higher in feeder-iPSCs than E8-iPSCs, and similarly for the 631 genes at least 2-fold higher in E8-iPSCs (Supplementary Tables 6, 7). We did an equivalent gene set over-representation analysis for the 1,159 genes with expression at least 2-fold higher in IPSDSNs differentiated from feeder-iPSCs, and also for the 958 genes at least 2-fold higher in IPSDSNs from E8-iPSCs (Supplementary Tables 9, 10).
To determine genes upregulated on differentiation from iPSCs to IPSDSNs, we first selected the 19,658 genes with expression FPKM > 1 in at least two samples (iPSC or IPSDSN). We used DESeq2 as before, removing genes with maximum Cook’s distance > 5, identifying 4,246 differentially expressed genes at FDR 1%.
QTL calling
Full details of QTL calling are provided in the Supplementary Note. Briefly, to call cis-eQTLs, we first determined allele-specific read counts for each SNP within gene exons using GATK’s ASEReadCounter43. We used RASQUAL’s makeCovariates.R to identify 12 gene expression PCs to use as covariates. We ran RASQUAL18 with option --no-posterior-update for each of 35,033 genes, testing SNPs and indels (MAF > 0.05, INFO > 0.8) within 500 kb of the gene transcription start site. For each gene we applied Bonferroni correction to the p values based on the number of independent tests estimated by EigenMT44. We ran RASQUAL with option --random-permutation call QTLs after permuting sample labels, and noted the minimum EigenMT-corrected p value per gene. To determine the FDR for eQTL discovery at a given gene, we computed (#permuted data min pvalues < p) / (#real data min p values < p), where p is the minimum p value among SNPs for the gene in question. For QTL calling with FastQTL, we used CQN-transformed gene expression (cqn v5.0.242) with a cis-window of 500 kb, including 20 PCs as covariates. We determined tissue-specific IPSDSN genes (not in GTEx) using a protocol described for the HIPSCI project8.
To call ATAC-seq QTLs, we used featureCounts v1.5.0 to count fragments overlapping consensus ATAC-seq peaks and ASEReadCounter to count allele-specific reads at SNPs within peaks. We ran RASQUAL for each of 381,323 peaks, testing SNPs and indels within 1 kb of the center of the peak. Since >99.9% of peaks were less than 2 kb in size, this meant that we tested effectively all SNPs within peaks. We used an equivalent procedure to determine FDR as for eQTLs.
Similarity of eQTLs with GTEx
Both GTEx samples and IPSDSNs had QTLs called using FastQTL. We selected lead eQTL variants in IPSDSNs for genes with expression >= 1 FPKM. We identified effect sizes for the same variants in each GTEx tissue, where these were available. We next determined the pairwise similarity between tissues in effect sizes for these variants (in R, cor() with “pairwise.complete.obs”). IPSDSNs were a significant outlier, having lower pairwise similarity with all GTEx tissues than they had with each other, likely due to the different expression quantification methods used in the two projects. We therefore determined the relative similarity of effect sizes across tissues by Z-scaling each row of the tissue correlation matrix, and plotted the result in Supplementary Figure 24.
Motif enrichment analyses
We used the R package LOLA22 to identify enrichments in transcription factor binding sites (TFBS) and motifs. We defined three sets of loci to consider for enrichment: 1) tissue-specific eQTL SNPs with a window of 50 bp (+/- 25) around the SNP position, 2) all eQTL SNPs (50 bp window), and 3) all ATAC-seq peaks. For the QTLs we used all GTEx eQTL lead SNPs as the “universe” set against which we tested TFBS for enrichment. For this loaded GTEx eQTLs in R and used the liftOver function from rtracklayer to convert their coordinates to GRCh38. We tested for enrichment against the LOLA core database considering only ENCODE TFBS enrichments (Supplementary Tables 16 and 17). We also tested ATAC-seq peaks for enrichment relative to DNaseHS for many tissues from Sheffield45, which are available in the LOLA catalog. Motif enrichments in ATAC-seq peaks are reported in Supplementary Table 18.
Power simulations
Gene expression values were normalized to counts per million. We selected the 544 eGenes discovered by RASQUAL at FDR 1% where:
at least 10 P2-protocol samples homozygous for each allele of the lead eQTL variant,
mean expression among homozygous carriers was consistent with RASQUAL’s reported direction of effect, and
CV < 2.
For each gene we resampled the normalized expression values, with replacement, from IPSDSN samples to achieve a specified number of samples (N ∈ {4,6,10,20,40}) with each homozygous genotype. From 100 such resamplings, we defined the power to discover a given variant’s effect as the fraction of cases with p < 0.05 from a Wilcoxon rank sum test comparing expression in each genotype category. We determined the allelic fold change between genotypes using RASQUAL’s effect size (pi), as:
QTL overlap with GWAS catalog
Full details of the method to determine QTL overlap are in the Supplementary Note. Briefly, we used vcftools v0.1.1446 to compute the genotype correlation R2 across our samples between GWAS catalog variants and lead eQTL, sQTL, caQTL variants within 500 kb of each other, and retained only overlaps with R2 > 0.8. We report all overlaps with GWAS associations having p < 5x10-8 in Supplementary Tables 20-22. To determine whether our QTLs were enriched in specific GWAS catalog traits, we grouped related traits and accounted for duplicate overlaps (see Supplementary Note), and counted the number of unique GWAS-QTL overlaps for eQTLs, sQTLs, and caQTLs (Table 1). For 41 traits with at least 40 GWAS catalog associations, we then considered the binomial probability of the observed overlap, with the expected frequency being the proportion of QTL overlaps among all trait associations (6.2%). After correcting for multiple testing, no traits showed significantly greater overlap with our QTL catalog than other traits.
Table 1.
QTL associations. Columns show the number of associations and the number of unique overlaps (r2 > 0.8) between lead QTL SNPs and GWAS catalog SNPs after removing duplicates for each GWAS trait.
| Number | GWAS overlap | |
|---|---|---|
| eQTLs | 3778 | 156 |
| sQTLs | 2079 | 129 |
| ATAC QTLs | 6318 | 172 |
| Joint ATAC/eQTLs | 177 | 14 |
To test for overall enrichment of QTLs overlapping with GWAS catalog SNPs, we used vcftools to identify 1000 Genomes SNPs in LD R2 > 0.8 with a GWAS catalog SNP. We used our IPSDSN eQTL lead SNPs as input to SNPsnap (https://data.broadinstitute.org/mpg/snpsnap/), and computed 1000 random sets of SNPs matched for LD partners, MAF, gene density, and distance to nearest gene. IPSDSN eQTL lead SNPs had more overlaps (92) with GWAS catalog + R2>0.8 SNPs than did any of the matched sets (median: 58, range 37-87).
Supplementary Material
Acknowledgments
The iPSC lines were generated under the Human Induced Pluripotent Stem Cell Initiative (HIPSCI) funded by a grant from the Wellcome Trust and Medical Research Council (WT098503), supported by the Wellcome Trust (WT098051) and the NIHR/Wellcome Trust Clinical Research Facility. HIPSCI funding was used for sensory neuron RNA-sequencing. We acknowledge Life Science Technologies Corporation as the provider of Cytotune. Pfizer Neuroscience (Pfizer Ltd.) funded neuronal differentiation, functional assays, single-cell RNA-sequencing, and collection and sequencing of dorsal root ganglion samples. The authors gratefully acknowledge Natsuhiko Kumasaka for help with RASQUAL. We thank Florian Merkle for comments on the manuscript. JS gratefully acknowledges support from the Wellcome Trust for his PhD studentship.
Footnotes
URLs
OpenTargets, www.targetvalidation.org.
CIBERSORT, cibersort.stanford.edu.
ENCODE, www.encodeproject.org.
GTEx, www.gtexportal.org.
HIPSCI, www.hipsci.org.
Author contributions
JS analyzed data, and JS and DJG wrote the manuscript. SF performed all differentiations. AGu analyzed data; AGu, DJG, and PJW conceived and supervised the project. HK compared eQTLs with GTEx and identified tissue-specific eQTLs. JR and MP cultured iPSC samples. AJK performed all ATAC-seq. KA and AGon assisted with data analysis. AW performed single-cell RNA work and assisted with data analysis. RF and CLB performed RNA extraction and quantification. EI performed cell culture and Ca2+ flux assays. MB assisted with experimental design and Ca2+ flux assays. LC, SL, and AJL performed electrophysiology measurements. All authors reviewed the manuscript.
Conflicts of Interest
SF, RF, CB, AW, MB, EI, LC, SL, AJL, PJW and AGu were all employees of Pfizer at the time the experiments were performed.
Code Availability
Code used for processing and analysing data is available at https://github.com/js29/ipsdsn.
Data Availability
RNA-seq and ATAC-seq data for open access samples are deposited in the European Nucleotide Archive under accession ERP020576. These data for managed access samples are deposited in the European Genome Archive under accession EGAD00001003145. Summary statistics and gene expression counts are available at https://www.ebi.ac.uk/biostudies/studies/S-BSST16. Sample genotypes and accession numbers are available at http://www.hipsci.org/data.
References
- 1.Itzhaki I, et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 2.Liu G-H, et al. Recapitulation of premature ageing with iPSCs from Hutchinson–Gilford progeria syndrome. Nature. 2001;472:221–225. doi: 10.1038/nature09879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wainger BJ, et al. Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep. 2014;7:1–11. doi: 10.1016/j.celrep.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee G, et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao L, et al. Pharmacological reversal of a pain phenotype in iPSC-derived sensory neurons and patients with inherited erythromelalgia. Sci Transl Med. 2016;8:335ra56. doi: 10.1126/scitranslmed.aad7653. [DOI] [PubMed] [Google Scholar]
- 6.Warren CR, et al. The NextGen Genetic Association Studies Consortium: A Foray into In Vitro Population Genetics. Cell Stem Cell. 2017;20:431–433. doi: 10.1016/j.stem.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 7.Young GT, et al. Characterizing human stem cell-derived sensory neurons at the single-cell level reveals their ion channel expression and utility in pain research. Mol Ther. 2014;22:1530–43. doi: 10.1038/mt.2014.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kilpinen H, et al. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature. 2017;55160 doi: 10.1101/055160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mele M, et al. The human transcriptome across tissues and individuals. Science (80-.) 2015;348:660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soldner F, et al. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature. 2016;533:1–20. doi: 10.1038/nature17939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pashos EE, et al. Large , Diverse Population Cohorts of hiPSCs and Derived Hepatocyte-like Cells Reveal Functional Genetic Variation at Blood Lipid-Associated Loci Resource Large , Diverse Population Cohorts of hiPSCs and Derived Hepatocyte-like Cells Reveal Functional Gen. Stem Cell. 2017;20:558–570.e10. doi: 10.1016/j.stem.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warren CR, et al. Induced Pluripotent Stem Cell Differentiation Enables Functional Validation of GWAS Variants in Resource Induced Pluripotent Stem Cell Differentiation Enables Functional Validation of GWAS Variants in Metabolic Disease. Stem Cell. 2017;20:547–557.e7. doi: 10.1016/j.stem.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Sala L, Bellin M, Mummery CL. Integrating cardiomyocytes from human pluripotent stem cells in safety pharmacology: Has the time come? Br J Pharmacol. 2016;1 doi: 10.1111/bph.13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiselev VY, et al. SC3: Consensus Clustering of Single-Cell RNA-Seq Data. Nat Methods. 2017;14:483–486. doi: 10.1038/nmeth.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:1–10. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kléber M, et al. Neural crest stem cell maintenance by combinatorial Wnt and BMP signaling. J Cell Biol. 2005;169:309–320. doi: 10.1083/jcb.200411095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ongen H, Buil A, Brown AA, Dermitzakis ET, Delaneau O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics. 2016;32:1479–1485. doi: 10.1093/bioinformatics/btv722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumasaka N, Knights A, Gaffney D. Fine-mapping cellular QTLs with RASQUAL and ATAC-seq. bioRxiv. 2015;48:18788. doi: 10.1038/ng.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li YI, et al. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–4. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li YI, Knowles DA, Pritchard JK. LeafCutter: Annotation-free quantification of RNA splicing. bioRxiv. 2016:44107. doi: 10.1101/044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–8. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheffield NC, Bock C. LOLA: Enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics. 2015;32:587–589. doi: 10.1093/bioinformatics/btv612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lessard J, et al. An Essential Switch in Subunit Composition of a Chromatin Remodeling Complex during Neural Development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunt SP, Pini A, Evan G. Induction of c-fos-like protein in spinal cord neurons following sensory stimulation. Nature. 1987;328:632–634. doi: 10.1038/328632a0. [DOI] [PubMed] [Google Scholar]
- 25.Kohno T, Moore Ka, Baba H, Woolf CJ. Peripheral nerve injury alters excitatory synaptic transmission in lamina II of the rat dorsal horn. J Physiol. 2003;548:131–138. doi: 10.1113/jphysiol.2002.036186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters MJ, et al. Genome-wide association study meta-analysis of chronic widespread pain: evidence for involvement of the 5p15.2 region. Ann Rheum Dis. 2013;72:427–36. doi: 10.1136/annrheumdis-2012-201742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pers TH, Timshel P, Hirschhorn JN. SNPsnap: A Web-based tool for identification and annotation of matched SNPs. Bioinformatics. 2014;31:418–420. doi: 10.1093/bioinformatics/btu655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gregory AP, et al. TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis. Nature. 2012;488:508–11. doi: 10.1038/nature11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Probert L. TNF and its receptors in the CNS: The essential, the desirable and the deleterious effects. Neuroscience. 2015;302:2–22. doi: 10.1016/j.neuroscience.2015.06.038. [DOI] [PubMed] [Google Scholar]
- 30.Spilker C, Kreutz MR. RapGAPs in brain: Multipurpose players in neuronal Rap signalling. European Journal of Neuroscience. 2010;32:1–9. doi: 10.1111/j.1460-9568.2010.07273.x. [DOI] [PubMed] [Google Scholar]
- 31.Jordan JD, et al. Cannabinoid receptor-induced neurite outgrowth is mediated by Rap1 activation through Gαo/i-triggered proteasomal degradation of Rap1GAPII. J Biol Chem. 2005;280:11413–11421. doi: 10.1074/jbc.M411521200. [DOI] [PubMed] [Google Scholar]
- 32.Melchionda L, et al. Mutations in APOPT1, encoding a mitochondrial protein, cause cavitating leukoencephalopathy with cytochrome c oxidase deficiency. Am J Hum Genet. 2014;95:315–325. doi: 10.1016/j.ajhg.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dianat N, Steichen C, Vallier L, Weber A, Dubart-Kupperschmitt A. Human pluripotent stem cells for modelling human liver diseases and cell therapy. Curr Gene Ther. 2013;13:120–32. doi: 10.2174/1566523211313020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith BW, et al. The aryl hydrocarbon receptor directs hematopoietic progenitor cell expansion and differentiation. Blood. 2013;122:376–385. doi: 10.1182/blood-2012-11-466722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Handel AE, et al. Assessing similarity to primary tissue and cortical layer identity in induced pluripotent stem cell-derived cortical neurons through single-cell transcriptomics. Hum Mol Genet. 2016;25:989–1000. doi: 10.1093/hmg/ddv637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu B-Y, et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci U S A. 2010;107:4335–40. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cacchiarelli D, et al. Aligning single-cell developmental and reprogramming trajectories identifies molecular determinants of reprogramming outcome. 2017 doi: 10.1016/j.cels.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 38.Musunuru K, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–9. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chambers SM, et al. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat Biotechnol. 2012;30:715–720. doi: 10.1038/nbt.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jun G, et al. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am J Hum Genet. 2012;91:839–848. doi: 10.1016/j.ajhg.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao Y, Smyth GK, Shi W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 42.Hansen KD, Irizarry RA, Wu Z. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics. 2012;13:204–216. doi: 10.1093/biostatistics/kxr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castel SE, Levy-Moonshine A, Mohammadi P, Banks E, Lappalainen T. Tools and best practices for data processing in allelic expression analysis. Genome Biol. 2015;16:195. doi: 10.1186/s13059-015-0762-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davis JR, et al. An Efficient Multiple-Testing Adjustment for eQTL Studies that Accounts for Linkage Disequilibrium between Variants. Am J Hum Genet. 2016;98:216–224. doi: 10.1016/j.ajhg.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheffield NC, et al. Patterns of regulatory activity across diverse human cell types predict tissue identity, transcription factor binding, and long-range interactions. Genome Res. 2013;23:777–788. doi: 10.1101/gr.152140.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.