Abstract
Membrane organelles consist of both proteins and lipids. Remodeling of these membrane structures is controlled by interactions between specific proteins and lipids. Mitochondrial structure and function depend on regulated fusion and the division of both the outer and inner membranes. Here, we will discuss recent advances on the regulation of mitochondrial dynamics by two critical phospholipids, phosphatidic acid and cardiolipin. These two lipids interact with the core components of mitochondrial fusion and division (Opa1, mitofusin, and Drp1) to activate and inhibit these dynamin-related GTPases. Moreover, lipid modifying enzymes such as phospholipases and lipid phosphatases may organize local lipid composition to spatially and temporarily coordinate a balance between fusion and division to establish mitochondrial morphology.
Keywords: Dynamin-related GTPase, Mitochondrial Division, Mitochondrial Fusion, Membrane, Phospholipids, Phospholipase, Lipid Phosphatase
Mitochondrial Dynamics: Fusion and Division
Mitochondria play crucial roles in diverse cellular and physiological processes such as energy production, metabolism, intracellular signaling, cell death, development, and immune response. In these functions, mitochondria serve as a bioenergetic power plant and a dynamic signaling hub. It has become increasingly clear that these mitochondrial functions are important for human health, and defects in these processes lead to pathological consequences such as metabolic diseases, cancer, autoimmune diseases, and neurodegenerative diseases [1]. These pathologies can be caused by energy deficits and reactive oxygen species (ROS) resulting from inefficient oxidative phosphorylation. In healthy cells, ROS contribute to signal transduction, but their unregulated overproduction can damage the mitochondria themselves and other cellular components, leading to overall deficits in cellular and tissue activities. Therefore, to maintain mitochondrial, cellular, and physiological health, controlling mitochondrial function and quality is essential. During the last two decades, studies from many laboratories have demonstrated that mitochondria are highly dynamic organelles, and their dynamic behaviors are central to the maintenance of the functional competence and quality of the mitochondria [2]. There are hundreds of mitochondria in cells, and these mitochondria continuously undergo fusion and division (Fig. 1). These interactions enable mitochondria to communicate with each other and mix their contents to regulate the overall quality of the mitochondrial population as a whole. These processes, termed mitochondrial dynamics, also control the size and number of mitochondria as they grow by importing proteins from the cytosol and lipids from the endoplasmic reticulum (ER) [3].
Figure 1. Mitochondrial fusion and division.
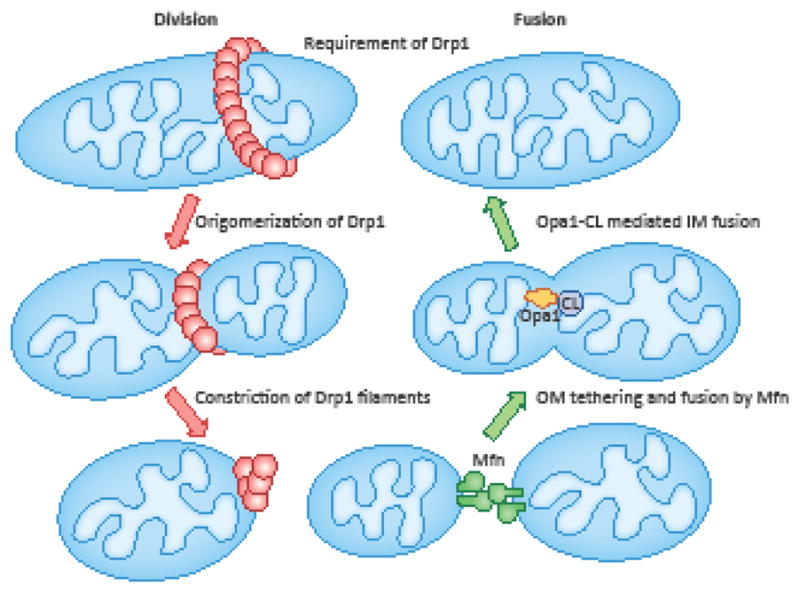
Tethering and fusion of the mitochondrial outer membrane (OM) is mediated by mitofusin (Mfn) 1 and 2. Inner membrane (IM) fusion is mediated by heterotypic interaction of Opa1 and CL. To initiate mitochondrial division, Drp1 GTPase is recruited to mitochondria via interactions with its receptor proteins located in the outer membrane (e.g., Mff, Mid49/51 and Fis1). Drp1 is then oligomerized into filaments that wrap around the mitochondria. Finally, GTP hydrolysis drives the constriction of Drp1 spirals and severs mitochondria.
Controlling the number of mitochondria in cells is critical [4]. Each mitochondrion must have at least one copy of mitochondrial DNA (mtDNA) since it encodes several essential subunits of the electron transport chain complexes. Oxidative phosphorylation depends on the components encoded by mtDNA, in addition to the majority of subunits encoded by nuclear DNA. Creating too many mitochondria by excess division would generate mitochondria that lack mtDNA and potentially compromise oxidative phosphorylation [5–7]. In contrast, overly connecting mitochondria by fusion produces mitochondria that contain too many mtDNA molecules, which can be aggregated and unable to function properly [8, 9]. In addition to the number, the size of mitochondria is also important. When mitochondria become overly enlarged due to unbalanced mitochondrial dynamics, the efficiency of mitochondrial transport into subcellular regions is decreased. This is particularly obvious in neurons, which extend long narrow axons and extensively branched dendrites. The lack of mitochondrial division, which causes the enlargement of mitochondria, blocks their distribution to axons and dendrites [10, 11]. These distribution defects result in local deficits in oxidative phosphorylation in the synapses, which require high ATP production for neural communications. In addition, excessively large mitochondria are resistant to degradation through mitophagy, likely because autophagosomes are unable to efficiently engulf them [12, 13]. As a physiological consequence of mitochondrial defects, a deficiency in mitochondrial division leads to neurodegeneration in neurons such as Purkinje cells in the cerebellum and dopaminergic neurons in the substantia nigra [14–16]. There are many other important roles for mitochondrial fusion and division [17–28], which are discussed in other excellent reviews [29–35]. In this review, we will focus on the mechanisms that control mitochondrial fusion and division with an emphasis on the emerging roles of mitochondrial phospholipids.
Critical Phospholipids for Mitochondrial Dynamics: Phosphatidic Acid and Cardiolipin
Mitochondria consist of two distinct, but physically connected membranes: the outer and inner membranes. The primary lipid components in the mitochondrial membrane are phospholipids, while sphingolipids and cholesterols are relatively uncommon compared to other membranes such as the plasma membrane [36, 37]. Two major phospholipids in the mitochondrial membranes are phosphatidylcholine (PC) and phosphatidylethanolamine, each of which contributes 30–40% of the total phospholipids in the mitochondria [38, 39]. In addition, the mitochondrial membranes contain relatively small amounts of phosphatidylglycerol, phosphatidylserine, and phosphatidic acid (PA) [38, 39]. Interestingly, levels of phosphatidylinositol vary in different organisms: they are relatively high in yeast mitochondria (~15%) and low in rat livers and plant mitochondria (~5%) [38, 39]. Cardiolipin (CL), a mitochondria-specific phospholipid with a unique structure of four acyl chains, comprises 10–15% of the total mitochondrial phospholipids [38, 39]. CL is synthesized from PA that is transported from the ER to the mitochondrial inner membrane (Fig. 2). In the inner membrane, PA is converted to CL via multiple enzymatic reactions. A fraction of CL is transported to the outer membrane (Fig. 2), likely through a CL-binding, intermembrane space-located nucleoside diphosphate kinases, NDPK-D, and/or contact sites between the outer and inner membranes and [40, 41]. In the outer membrane, CL can be converted back to PA by a mitochondria-localized phospholipase D, MitoPLD (Fig. 2) [42]. CL in the inner membrane is important for maintaining large protein complexes such as the electron transport chain complexes and the protein import machinery of the inner membrane. CL also regulates apoptosis via the activation of caspases-8 and Bax on the outer membrane and the retention of cytochrome c in the cristae of the inner membrane [43]. Furthermore, recent studies have revealed that PA and CL control mitochondrial division and fusion and coordinate the balance between these dynamic processes. We will discuss the roles of these phospholipids in mitochondrial dynamics below.
Figure 2. Biosynthesis of PA and CL.
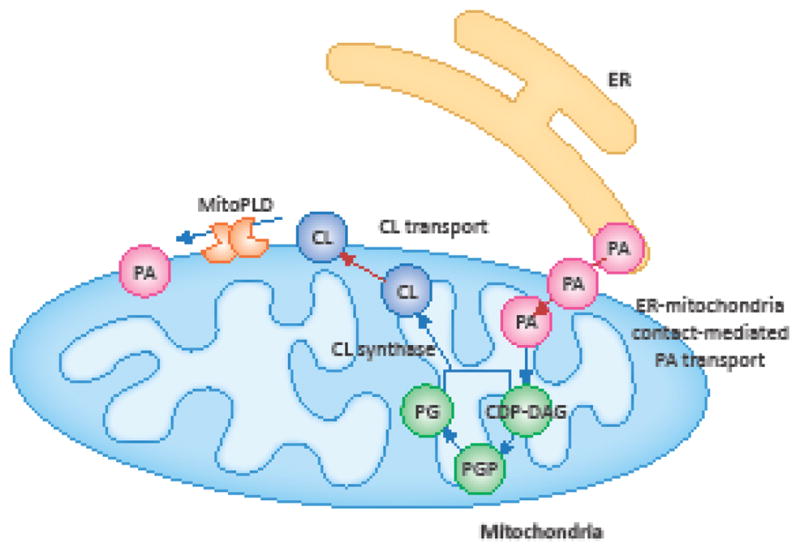
The abundance and localization of PA and CL are controlled by their synthesis, turnover and transport in mitochondria. PA is synthesized in the endoplasmic reticulum (ER) and transported to the mitochondrial outer membrane. A fraction of PA is further transported to the inner membrane, where it is converted to cytidine diphosphate diacylglycerol (CDP-DAG), phosphatidylglycerolphosphate (PGP) and then phosphatidylglycerol (PG). PG and CDP-DAP are combined to generate CL by cardiolipin synthase. In turn, a portion of the CL is exported to the outer membrane and converted to PA by MitoPLD.
Roles of Cardiolipin in Mitochondrial Fusion
As described above, CL is synthesized in the inner membrane of mitochondria. It has been shown that CL plays important roles in the fusion of the mitochondrial inner membrane through the biogenesis and assembly of a dynamin-related GTPase that mediates mitochondrial fusion (Opa1 in mammals and Mgm1 in yeast) (Fig. 1 and 3) [44–50]. A recent study also demonstrated that CL is directly involved in membrane fusion through interactions with Opa1 [51]. In addition to their role in inner membrane fusion, Opa1 and Mgm1 also facilitate outer membrane fusion, likely through interactions with outer membrane proteins such as mitofusin in mammals and Fzo1 and Ugo1 in yeast [52–56].
Figure 3. Regulation of mitochondrial fusion by PA and CL.
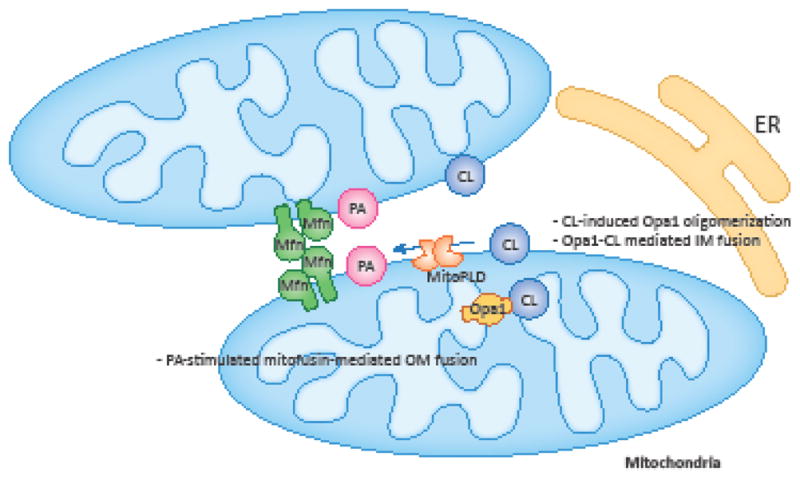
In the outer membrane, PA stimulates the mitofusin-mediated fusion. In the inner membrane, CL stimulates and mediates Opa1-mediated fusion.
A single gene encodes Opa1 and Mgm1, but multiple forms are created through alternative splicing (Opa1) and proteolytic processing (both Opa1 and Mgm1) [57]. There are two major forms, long and short. The long forms (L-Opa1/Mgm1) carry a transmembrane domain at their N-terminus that is integral to the mitochondrial inner membrane, leaving the majority of the protein in the intermembrane space. The short forms (S-Opa1/Mgm1) are produced by proteolytic cleavages of the transmembrane domain of the long forms, leaving soluble proteins in the intermembrane space. The production of these two forms is balanced to create both forms. Of these two forms, the long forms play a major role in fusion, while the short forms have been suggested to be involved in both fusion and division. A complete loss of the long forms leads to the inhibition of mitochondrial fusion.
The import of Opa1/Mgm1 into mitochondria is mediated by the translocase of the inner membrane (TIM23 complex) and presequence translocase-associated motor (PAM) [58]. CL is important for maintaining the TIM complex; decreased levels of CL lead to a partial dissociation between TIM23 complex and PAM and cause Mgm1 to be partially inserted into the inner membrane [49]. This inhibits Mgm1 processing, because the transmembrane domain that contains the cleavage site is not fully inserted into the inner membrane; instead, another alternative transmembrane domain stays in the inner membrane, creating L-Mgm1, but not S-Mgm1 [44, 59]. This incomplete biogenesis of Mgm1 results in a decrease in mitochondrial fusion. It has been suggested the role of CL in Mgm1 biogenesis is shared with another phospholipid, PE, and the decreased production of PE decreases the production of S-Mgm1 and mitochondrial fusion [47, 48]. This shared function of CL and PE in the production of S-Mgm1 may explain the observation that the production of S-Mgm1 is only modestly decreased in yeast mutants lacking CL synthase [47, 60].
CL is also involved in the assembly of Opa1 and Mgm1. The GTPase activity of dynamins and dynamin-related proteins can be stimulated by their oligomerization. Liposome-containing CL binds to purified Opa1 and Mgm1, stimulating their assembly into liposomes [45, 46, 50, 61]. The oligomerization activates S-Opa1 and S-Mgm1 to hydrolyze GTP. However, a more direct role of CL in Opa1-mediated fusion awaited discovery in a recent study in which L-Opa1 was purified and assessed for its function during fusion together with CL [51]. In this study, recombinant L-Opa1 was purified from the silk worm and reconstituted into liposomes. L-Opa1 drives the fusion of liposome-containing CL in vitro. This fusion is mediated by the heterotypic interactions of L-Opa1 and CL, but not by interactions between the L-Opa1 proteins themselves. These findings are consistent with previous observations that purified mitochondria from wild type and Opa1 knockout cells can fuse in vitro. S-Opa1 alone does not fuse liposomes, even in the presence of CL, but it does stimulate fusion when L-Opa1 is also present. Demonstrating the importance of this mechanism in cells, a cell based mitochondrial fusion assay showed that both Opa1 and CL are necessary for efficient mitochondrial fusion.
Role for CL in Mitochondrial Division
CL is also present in the outer membrane and regulates mitochondrial division mediated by another dynamin-related GTPase, Drp1, which is a cytosolic protein that is recruited to the mitochondria through interactions with integral outer membrane proteins such as Mff, Mid49, Mid51, and Fis1 (Fig. 1) [2, 62]. Drp1 constricts the mitochondria after assembly into spiral oligomers on the surface. Distinct from endocytic dynamin, which binds to phosphatidylinositol 4,5-bisphosphate, PI(4,5)P2, via a pleckstrin homology domain, Drp1 lacks a known lipid-binding domain, but binds to CL and PA. First we will discuss the role of CL in mitochondrial division. The roles PA plays in mitochondrial division, as well as fusion, are described in a later section of this review. The interaction of Drp1 with CL involves the variable domain (also called the B-insert) [63, 64]. CL has been shown to stimulate the oligomerization of Drp1 and oligomerization-induced GTP hydrolysis, similar to the effect of CL on S-Opa1 (Fig. 4) [65–67]. The oligomerization of Drp1 on liposomes induces their tubulation, which may be relevant to the formation of the mitochondrial division machinery that wraps around and constricts the mitochondrial tubules. Supporting this notion, GTP hydrolysis by Drp1 promotes the constriction of the lipid tubules formed by Drp1 oligomerization [64, 67, 68]. Interestingly, the variable domain is important for the tubulation of liposomes that contain CL. However, the removal of the variable domain stimulates the oligomerization of Drp1, but does not affect the tubulation of liposomes that lack CL [67]. These data may indicate that CL inhibits the assembly of Drp1 and the tubulation of liposomes, and that the binding of CL to the variable domain releases this inhibitory effect. While CL stimulates Drp1, Drp1 also affects the organization of CL in the membrane. Recombinant Drp1 can affect the two-dimensional distribution of CL in synthetic liposomes in a GTP-dependent manner, and it induces CL clustering [64]. Such CL-enriched lipid domains could provide for hot spots of mitochondrial division by locally activating Drp1 [64]. The CL-stimulated GTPase activity of Drp1 is synergistically enhanced in the presence of the major Drp1 receptor/effector Mff, suggesting that CL and Mff act together to potentiate mitochondrial division, and may create spatial specificity for the membrane remodeling reaction at the hot spots where these two components are concentrated [68].
Figure 4. Regulation of mitochondrial division by PA and CL.
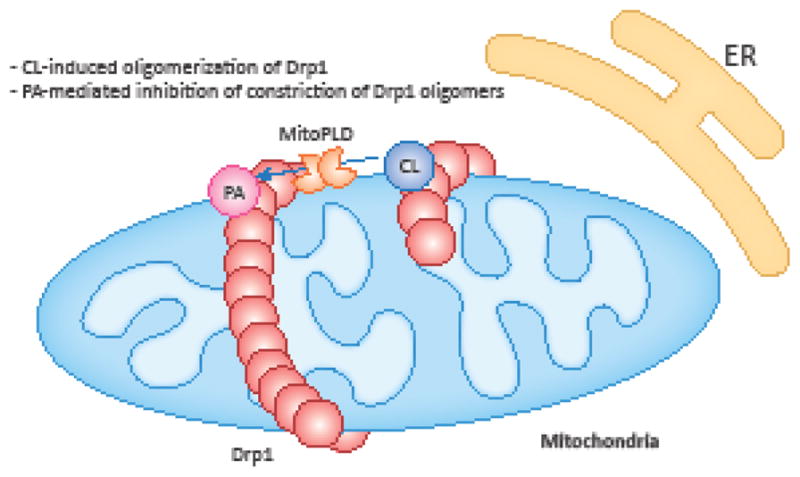
CL in the outer membrane stimulates oligomerization of Drp1 to promote mitochondrial division. In contrast, PA restrains oligomerized Drp1 to inhibit mitochondrial division.
Many in vitro biochemical and biophysical studies have demonstrated that Drp1 can induce tubulation and the constriction of liposomes, but not complete scission or small vesicle formation. A recent study has shown that the constriction mediated by Drp1 does not complete the scission reaction in cells; dynamin-2, which is known to function in the scission of endocytic vesicles from the plasma membrane, is recruited to the mitochondrial division sites and completes the mitochondrial scission [69]. These sequential fission reactions by the two dynamin proteins are consistent with their biochemical properties in terms of the diameter of the spiral oligomers. While Drp1 and its yeast homolog Dnm1 assembles into spirals with a diameter of 50–60 nm, dynamin-2 filaments can more narrowly constrict, to a 10–20 nm diameter [67, 70].
Role of PA in Mitochondrial Fusion
PA is a rare component of the mitochondrial membrane (~5%) that is predominantly produced in the ER and transferred to the outer mitochondrial membrane through an ER-mitochondrial contact site as described above (Fig. 2) [43, 71, 72]. PA is also generated from CL in the outer mitochondrial membrane by MitoPLD [42]. PA serves as both a precursor for other phospholipids as well as a signaling molecule in mitochondria as well as other membranes. Because PA lacks a headgroup, it can create a negative membrane curvature. Recent studies have shown that PA regulates both mitochondrial fusion and division in the outer membrane [73–75]. While the inner mitochondrial membrane carries Opa1 in mammals, the outer membrane has two related dynamin-related GTPases, mitofusins 1 and 2 (Fig. 1). It has been shown that mitofusins mediate the tethering of the outer membranes of two mitochondria in addition to membrane fusion (Fig. 1) [76]. Studies have shown that the PA generated by MitoPLD induces mitochondrial fusion downstream of this tethering step between two mitochondria (Fig. 3) [73, 74]. MitoPLD overexpression induced mitochondrial aggregation, suggesting mitochondria fusion, while the suppression of MitoPLD using siRNA led to mitochondrial fragmentation. Similarly, overexpression of lipin 1b, a PA phosphatase that converts PA to DAG, induces the fragmentation of mitochondria. Similar to lipin 1b, PA-PLA1 (PA-preferring phospholipase A1), which mediates the conversion of PA to lysoPA, regulates mitochondrial morphology [77]. Its overexpression fragments mitochondria, while its knockdown elongates mitochondrial tubules. In contrast to MitoPLD, both lipin 1b and PA-PLA1 are mainly located in the cytosol [73, 77]. These two lipid-modifying enzymes are likely recruited to the outer membrane for their function. These data taken together strongly suggest that PA, but not DAG or LPA, control mitochondrial morphology. It has been proposed that PA stimulates mitochondrial fusion by creating a negative curvature in the opposing mitochondrial outer membrane [78]. It is also possible that PA more directly regulates the function of mitofusin in membrane fusion. MitoPLD may not function alone in the regulation of PA in the outer membrane. MitoPLD interacts with two related outer membrane proteins, MIGA1 and MIGA2 [79]. Miga proteins (Mitoguardian) were originally identified as proteins necessary for the maintenance of photoreceptor cells in Drosophila. The mammalian homologs, MIGA1 and MIGA2, are required for mitochondrial fusion, similarly to MitoPLD. It has been suggested that MIGA1/2 stabilizes the level of MitoPLD dimerization in the outer membrane [79]. Suggesting a role for PA-regulated mitochondrial dynamics in tumorigenesis, mitochondrial division and fusion are regulated during KRAS-driven cellular transformation [80, 81], and the expression of MitoPLD is induced by the oncogenic transcriptional factor MYC [82]. Interestingly, MitoPLD is structurally similar to phosphodiesterase and exhibits endoribonuclease activity for single-stranded RNAs [83, 84] and phospholipase activity for CL [74].
Role of PA in Mitochondrial Division
In addition to mitochondrial fusion, recent studies have shown that PA also regulates mitochondrial division through interactions with Drp1 (Fig. 4) [75, 85]. The interaction of Drp1 with PA entails the PA headgroup and its acyl chains. Intriguingly, even though Drp1 is a soluble protein, it specifically recognizes the saturated acyl chains of PA and preferentially binds to saturated PA over unsaturated PA [75, 85]. These findings suggest that part of Drp1 penetrates the hydrophobic core of the lipid bilayer. Furthermore, the recognition of the headgroup and acyl chains can be mechanistically separated: Drp1 binds to liposomes that consist of both unsaturated PA and saturated PC, but not to liposomes that consist of only one or the other. This unique coincident lipid interaction inhibits Drp1 during mitochondrial division [75]. PA and the saturated acyl chains of phospholipids block Drp1 after its oligomerization onto mitochondria, and this inhibition of Drp1 makes the mitochondria resistant to the division induced by mitochondrial stress. In contrast to CL, which activates Drp1 oligomerization and GTPase activity, the interactions of Drp1 with saturated PA does not increase GTP hydrolysis. Increased levels of PA produced by MitoPLD overexpression or increased levels of saturated phospholipids produced by chemical inhibitors of stearoyl-CoA desaturase 1, which converts saturated acyl chains to unsaturated forms, induces the accumulation of Drp1 oligomers onto mitochondria without productive division, leading to the continuous assembly of Drp1 on mitochondria (Fig. 4) [75]. It has been proposed that saturated PA blocks the GTPase activity of Drp1 after oligomerization, potentially creating a primed state in the division machinery. Because CL is largely unsaturated, the PA that MitoPLD generates from CL is expected to be unsaturated. Therefore, the coincident interaction involving PA and saturated phospholipids would be physiologically relevant. In addition to MitoPLD generating PA, PA is also imported into mitochondria from the ER through ER-mitochondria contact sites [71, 86]. Since mitochondrial division often occurs at these inter-organelle contact sites and Drp1 oligomers localize to these regions [87–89], we expect that the PA imported from the ER also plays a critical role in mitochondrial division. It would be exciting to determine exactly how ER-derived PA contributes to mitochondrial division at the interfaces.
In addition to PA, Drp1 also directly interacts with the PA-producing enzyme MitoPLD in the outer membrane (Fig. 4) [75]. This protein-protein interaction suggests that PA is locally produced in the vicinity of Drp1 oligomerized on the mitochondria to create a lipid microenvironment, making this PA/saturated phospholipid-mediated regulation spatially selective and efficient (Fig. 4), as PA is transferred to the inner membrane as a precursor to other phospholipids. This potential regulatory mechanism of MitoPLD may also be important for mitochondrial fusion, since MitoPLD associates with mitofusin 1 and Opa1 [75]. As described above, PA stimulates mitofusin-mediated mitochondrial fusion, and mitofusin 1 cooperates with Opa1 to fuse mitochondria. Therefore, MitoPLD may mechanistically couple the inhibition of mitochondrial division to the stimulation of mitochondrial fusion, generating robust changes in mitochondrial morphology in response to intracellular signal transduction, the extracellular environment, and mitochondrial stress. This mechanism likely accounts for the previous observation that mitochondria have hot spots where they frequently fuse and divide.
Concluding Remarks
During the last 20 years, many protein components that mediate mitochondrial dynamics have been identified. In addition, the posttranslational modifications of these proteins in response to a variety of signals and stress have been characterized. However, there are many crucial open questions regarding the role of lipids and their regulation in the membrane remodeling processes in mitochondria (see Outstanding Questions). Because protein-lipid interactions are fundamental to membrane fusion and fission, a deeper understanding of their interaction mechanisms and the functions of these interactions is essential to elucidate a comprehensive view of the mechanism of mitochondrial dynamics. We still do not fully know about the functional microenvironments created by specific lipids and proteins in the outer and inner mitochondrial membranes. Considering the structurally defined micro-domains in mitochondria such as ER-mitochondria contact sites, outer-inner membrane contact sites, and inner membrane cristae, how the assembly and disassembly of lipid micro-domains for mitochondrial dynamics are coordinated in mitochondria in relation to these intra-mitochondrial structures would be an exciting future topic in the field.
Outstanding Questions.
What is the distribution of PA and CL in the mitochondrial outer membrane during mitochondrial fusion and division? It would be exciting to visualize PA and CL using their biosensors.
How are interactions of MitoPLD with Drp1 and mitofusin 1 and Opa1 regulated? Do these interactions contribute to the maintenance of a balance between fusion and division?
How do ER-mitochondria contact sites and outer membrane-inner membrane contact sites regulate the delivery of PA and CL to the outer membrane?
How are biosynthesis, transport and distribution of PA and CL regulated in response to intracellular and extracellular signaling and stress that affect mitochondrial dynamics?
Do mitochondrial dynamics and morphology control the production, trafficking and localization of lipids?
Trends.
Mitochondrial fusion and division play important roles in mitochondrial size, number, distribution, function, and turnover.
Cardiolipin promotes both mitochondrial division and inner membrane fusion.
Phosphatidic acid inhibits mitochondrial division and stimulates mitochondrial outer membrane fusion.
Recent studies identified phospholipases and lipid phosphatases such as MitoPLD, PA-PLA1, and lipin 1b that control the levels of cardiolipin and phosphatidic acid in mitochondria.
Lipid-modifying enzymes could provide mechanisms that coordinate mitochondrial dynamics.
Acknowledgments
We thank members of the Okamoto, Iijima, and Sesaki labs for helpful discussions. This work was supported by grants to MI (NIH GM084015, WW Smith Charitable Trust and Allegheny Health Network-Sidney Kimmel Comprehensive Cancer Center), to HS (Allegheny Health Network-Sidney Kimmel Comprehensive Cancer Center and JHU-UMD Diabetes Research Center) and to KO and HS (Osaka University International Joint Research Promotion Program).
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Picard M, et al. The rise of mitochondria in medicine. Mitochondrion. 2016;30:105–16. doi: 10.1016/j.mito.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roy M, et al. Mitochondrial division and fusion in metabolism. Current opinion in cell biology. 2015;33C:111–118. doi: 10.1016/j.ceb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horvath SE, et al. Role of membrane contact sites in protein import into mitochondria. Protein Sci. 2015;24(3):277–97. doi: 10.1002/pro.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gustafsson CM, et al. Maintenance and Expression of Mammalian Mitochondrial DNA. Annu Rev Biochem. 2016;85:133–60. doi: 10.1146/annurev-biochem-060815-014402. [DOI] [PubMed] [Google Scholar]
- 5.Hermann GJ, et al. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J Cell Biol. 1998;143(2):359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sesaki H, Jensen RE. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J Cell Biol. 1999;147(4):699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H, et al. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130(3):548–62. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 8.Ban-Ishihara R, et al. Dynamics of nucleoid structure regulated by mitochondrial fission contributes to cristae reformation and release of cytochrome c. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(29):11863–8. doi: 10.1073/pnas.1301951110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itoh K, et al. Effects of Fcj1-Mos1 and mitochondrial division on aggregation of mitochondrial DNA nucleoids and organelle morphology. Molecular biology of the cell. 2013;24(12):1842–51. doi: 10.1091/mbc.E13-03-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shields LY, et al. Dynamin-related protein 1 is required for normal mitochondrial bioenergetic and synaptic function in CA1 hippocampal neurons. Cell death & disease. 2015;6:e1725. doi: 10.1038/cddis.2015.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kageyama Y, et al. Mitochondrial division ensures the survival of postmitotic neurons by suppressing oxidative damage. The Journal of cell biology. 2012;197(4):535–51. doi: 10.1083/jcb.201110034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337(6098):1062–5. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shirihai OS, et al. How mitochondrial dynamism orchestrates mitophagy. Circulation research. 2015;116(11):1835–49. doi: 10.1161/CIRCRESAHA.116.306374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kageyama Y, et al. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. The EMBO journal. 2014;33(23):2798–813. doi: 10.15252/embj.201488658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z, et al. Mitochondrial division prevents neurodegeneration. Autophagy. 2012;8(10):1531–3. doi: 10.4161/auto.21213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berthet A, et al. Loss of mitochondrial fission depletes axonal mitochondria in midbrain dopamine neurons. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2014;34(43):14304–17. doi: 10.1523/JNEUROSCI.0930-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buck MD, et al. Mitochondrial Dynamics Controls T Cell Fate through Metabolic Programming. Cell. 2016;166(1):63–76. doi: 10.1016/j.cell.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kandimalla R, et al. Reduced Dynamin-related Protein 1 Protects Against Phosphorylated Tau-induced Mitochondrial Dysfunction and Synaptic Damage in Alzheimer’s Disease. Hum Mol Genet. 2016 doi: 10.1093/hmg/ddw312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khacho M, et al. Mitochondrial Dynamics Impacts Stem Cell Identity and Fate Decisions by Regulating a Nuclear Transcriptional Program. Cell Stem Cell. 2016;19(2):232–47. doi: 10.1016/j.stem.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 20.Manczak M, et al. Protective Effects of Reduced Dynamin-related Protein 1 Against Amyloid Beta-induced Mitochondrial Dysfunction and Synaptic Damage in Alzheimer’s Disease. Hum Mol Genet. 2016 doi: 10.1093/hmg/ddw330. [DOI] [PMC free article] [PubMed]
- 21.Santoro A, et al. DRP1 Suppresses Leptin and Glucose Sensing of POMC Neurons. Cell Metab. 2017;25(3):647–660. doi: 10.1016/j.cmet.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tezze C, et al. Age-Associated Loss of OPA1 in Muscle Impacts Muscle Mass, Metabolic Homeostasis, Systemic Inflammation, and Epithelial Senescence. Cell Metab. 2017;25(6):1374–1389. e6. doi: 10.1016/j.cmet.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahdaviani K, et al. Mfn2 deletion in brown adipose tissue protects from insulin resistance and impairs thermogenesis. EMBO Rep. 2017 doi: 10.15252/embr.201643827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pereira RO, et al. OPA1 deficiency promotes secretion of FGF21 from muscle that prevents obesity and insulin resistance. EMBO J. 2017 doi: 10.15252/embj.201696179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luchsinger LL, et al. Mitofusin 2 maintains haematopoietic stem cells with extensive lymphoid potential. Nature. 2016;529(7587):528–31. doi: 10.1038/nature16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weaver D, et al. Distribution and apoptotic function of outer membrane proteins depend on mitochondrial fusion. Mol Cell. 2014;54(5):870–8. doi: 10.1016/j.molcel.2014.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishihara T, et al. Dynamics of mitochondrial DNA nucleoids regulated by mitochondrial fission is essential for maintenance of homogeneously active mitochondria during neonatal heart development. Mol Cell Biol. 2015;35(1):211–23. doi: 10.1128/MCB.01054-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Udagawa O, et al. Mitochondrial fission factor Drp1 maintains oocyte quality via dynamic rearrangement of multiple organelles. Curr Biol. 2014;24(20):2451–8. doi: 10.1016/j.cub.2014.08.060. [DOI] [PubMed] [Google Scholar]
- 29.Flippo KH, Strack S. Mitochondrial dynamics in neuronal injury, development and plasticity. J Cell Sci. 2017;130(4):671–681. doi: 10.1242/jcs.171017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schrepfer E, Scorrano L. Mitofusins, from Mitochondria to Metabolism. Mol Cell. 2016;61(5):683–94. doi: 10.1016/j.molcel.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 31.Ishihara T, et al. Physiological roles of mitochondrial fission in cultured cells and mouse development. Ann N Y Acad Sci. 2015;1350:77–81. doi: 10.1111/nyas.12848. [DOI] [PubMed] [Google Scholar]
- 32.Serasinghe MN, Chipuk JE. Mitochondrial Fission in Human Diseases. Handb Exp Pharmacol. 2017 doi: 10.1007/164_2016_38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song M, Dorn GW., 2nd Mitoconfusion: noncanonical functioning of dynamism factors in static mitochondria of the heart. Cell metabolism. 2015;21(2):195–205. doi: 10.1016/j.cmet.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mishra P, Chan DC. Metabolic regulation of mitochondrial dynamics. J Cell Biol. 2016;212(4):379–87. doi: 10.1083/jcb.201511036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H, Chan DC. Mitochondrial Dynamics in Regulating the Unique Phenotypes of Cancer and Stem Cells. Cell Metab. 2017 doi: 10.1016/j.cmet.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mejia EM, Hatch GM. Mitochondrial phospholipids: role in mitochondrial function. J Bioenerg Biomembr. 2016;48(2):99–112. doi: 10.1007/s10863-015-9601-4. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Q, et al. Biosynthesis and roles of phospholipids in mitochondrial fusion, division and mitophagy. Cellular and molecular life sciences: CMLS. 2014 doi: 10.1007/s00018-014-1648-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vance JE. Phospholipid synthesis and transport in mammalian cells. Traffic. 2015;16(1):1–18. doi: 10.1111/tra.12230. [DOI] [PubMed] [Google Scholar]
- 39.Horvath SE, Daum G. Lipids of mitochondria. Progress in lipid research. 2013;52(4):590–614. doi: 10.1016/j.plipres.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 40.Kagan VE, et al. NDPK-D (NM23-H4)-mediated externalization of cardiolipin enables elimination of depolarized mitochondria by mitophagy. Cell Death Differ. 2016;23(7):1140–51. doi: 10.1038/cdd.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlattner U, et al. Dual function of mitochondrial Nm23-H4 protein in phosphotransfer and intermembrane lipid transfer: a cardiolipin-dependent switch. J Biol Chem. 2013;288(1):111–21. doi: 10.1074/jbc.M112.408633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nelson RK, Frohman MA. Physiological and pathophysiological roles for phospholipase D. J Lipid Res. 2015;56(12):2229–37. doi: 10.1194/jlr.R059220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osman C, et al. Making heads or tails of phospholipids in mitochondria. The Journal of cell biology. 2011;192(1):7–16. doi: 10.1083/jcb.201006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sesaki H, et al. Ups1p, a conserved intermembrane space protein, regulates mitochondrial shape and alternative topogenesis of Mgm1p. The Journal of cell biology. 2006;173(5):651–8. doi: 10.1083/jcb.200603092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ban T, et al. OPA1 disease alleles causing dominant optic atrophy have defects in cardiolipin-stimulated GTP hydrolysis and membrane tubulation. Human molecular genetics. 2010;19(11):2113–22. doi: 10.1093/hmg/ddq088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeVay RM, et al. Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J Cell Biol. 2009;186(6):793–803. doi: 10.1083/jcb.200906098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joshi AS, et al. Cardiolipin and mitochondrial phosphatidylethanolamine have overlapping functions in mitochondrial fusion in Saccharomyces cerevisiae. The Journal of biological chemistry. 2012;287(21):17589–97. doi: 10.1074/jbc.M111.330167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan EY, McQuibban GA. Phosphatidylserine decarboxylase 1 (Psd1) promotes mitochondrial fusion by regulating the biophysical properties of the mitochondrial membrane and alternative topogenesis of mitochondrial genome maintenance protein 1 (Mgm1) The Journal of biological chemistry. 2012;287(48):40131–9. doi: 10.1074/jbc.M112.399428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamura Y, et al. Ups1p and Ups2p antagonistically regulate cardiolipin metabolism in mitochondria. J Cell Biol. 2009;185(6):1029–45. doi: 10.1083/jcb.200812018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rujiviphat J, et al. Phospholipid association is essential for dynamin-related protein Mgm1 to function in mitochondrial membrane fusion. J Biol Chem. 2009;284(42):28682–6. doi: 10.1074/jbc.M109.044933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ban T, et al. Molecular basis of selective mitochondrial fusion by heterotypic action between OPA1 and cardiolipin. Nat Cell Biol. 2017 doi: 10.1038/ncb3560. [DOI] [PubMed] [Google Scholar]
- 52.Sesaki H, et al. Mgm1p, a dynamin-related GTPase, is essential for fusion of the mitochondrial outer membrane. Mol Biol Cell. 2003;14(6):2342–56. doi: 10.1091/mbc.E02-12-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sesaki H, Jensen RE. Ugo1p links the Fzo1p and Mgm1p GTPases for mitochondrial fusion. J Biol Chem. 2004;279(27):28298–303. doi: 10.1074/jbc.M401363200. [DOI] [PubMed] [Google Scholar]
- 54.Sesaki H, Jensen RE. UGO1 encodes an outer membrane protein required for mitochondrial fusion. J Cell Biol. 2001;152(6):1123–34. doi: 10.1083/jcb.152.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoppins S, et al. The soluble form of Bax regulates mitochondrial fusion via MFN2 homotypic complexes. Molecular cell. 2011;41(2):150–60. doi: 10.1016/j.molcel.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song Z, et al. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol Biol Cell. 2009;20(15):3525–32. doi: 10.1091/mbc.E09-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.MacVicar T, Langer T. OPA1 processing in cell death and disease - the long and short of it. J Cell Sci. 2016;129(12):2297–306. doi: 10.1242/jcs.159186. [DOI] [PubMed] [Google Scholar]
- 58.Schulz C, et al. Unlocking the presequence import pathway. Trends Cell Biol. 2015;25(5):265–75. doi: 10.1016/j.tcb.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 59.Herlan M, et al. Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor. J Cell Biol. 2004;165(2):167–73. doi: 10.1083/jcb.200403022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Osman C, et al. The genetic interactome of prohibitins: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. The Journal of cell biology. 2009;184(4):583–96. doi: 10.1083/jcb.200810189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meglei G, McQuibban GA. The dynamin-related protein Mgm1p assembles into oligomers and hydrolyzes GTP to function in mitochondrial membrane fusion. Biochemistry. 2009;48(8):1774–84. doi: 10.1021/bi801723d. [DOI] [PubMed] [Google Scholar]
- 62.Tamura Y, et al. SnapShot: Mitochondrial dynamics. Cell. 2011;145(7):1158, 1158 e1. doi: 10.1016/j.cell.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bustillo-Zabalbeitia I, et al. Specific interaction with cardiolipin triggers functional activation of Dynamin-Related Protein 1. PLoS One. 2014;9(7):e102738. doi: 10.1371/journal.pone.0102738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stepanyants N, et al. Cardiolipin’s propensity for phase transition and its reorganization by dynamin-related protein 1 form a basis for mitochondrial membrane fission. Mol Biol Cell. 2015;26(17):3104–16. doi: 10.1091/mbc.E15-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Macdonald PJ, et al. A dimeric equilibrium intermediate nucleates Drp1 reassembly on mitochondrial membranes for fission. Mol Biol Cell. 2014;25(12):1905–15. doi: 10.1091/mbc.E14-02-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ugarte-Uribe B, et al. Dynamin-related protein 1 (Drp1) promotes structural intermediates of membrane division. J Biol Chem. 2014;289(44):30645–56. doi: 10.1074/jbc.M114.575779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Francy CA, et al. The Mechanoenzymatic Core of Dynamin-related Protein 1 Comprises the Minimal Machinery Required for Membrane Constriction. The Journal of biological chemistry. 2015;290(18):11692–703. doi: 10.1074/jbc.M114.610881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Macdonald PJ, et al. Distinct Splice Variants of Dynamin-related Protein 1 Differentially Utilize Mitochondrial Fission Factor as an Effector of Cooperative GTPase Activity. J Biol Chem. 2016;291(1):493–507. doi: 10.1074/jbc.M115.680181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee JE, et al. Multiple dynamin family members collaborate to drive mitochondrial division. Nature. 2016 doi: 10.1038/nature20555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mears JA, et al. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat Struct Mol Biol. 18(1):20–6. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tamura Y, et al. Phospholipid transport via mitochondria. Traffic. 2014 doi: 10.1111/tra.12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murley A, Nunnari J. The Emerging Network of Mitochondria-Organelle Contacts. Mol Cell. 2016;61(5):648–53. doi: 10.1016/j.molcel.2016.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang H, et al. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Developmental cell. 2011;20(3):376–87. doi: 10.1016/j.devcel.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choi SY, et al. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat Cell Biol. 2006;8(11):1255–62. doi: 10.1038/ncb1487. [DOI] [PubMed] [Google Scholar]
- 75.Adachi Y, et al. Coincident Phosphatidic Acid Interaction Restrains Drp1 in Mitochondrial Division. Mol Cell. 2016;63(6):1034–43. doi: 10.1016/j.molcel.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cao YL, et al. MFN1 structures reveal nucleotide-triggered dimerization critical for mitochondrial fusion. Nature. 2017;542(7641):372–376. doi: 10.1038/nature21077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baba T, et al. Phosphatidic acid (PA)-preferring phospholipase A1 regulates mitochondrial dynamics. The Journal of biological chemistry. 2014;289(16):11497–511. doi: 10.1074/jbc.M113.531921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Frohman MA. Role of mitochondrial lipids in guiding fission and fusion. Journal of molecular medicine. 2015;93(3):263–9. doi: 10.1007/s00109-014-1237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y, et al. Mitoguardin Regulates Mitochondrial Fusion through MitoPLD and Is Required for Neuronal Homeostasis. Molecular cell. 2016;61(1):111–24. doi: 10.1016/j.molcel.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 80.Serasinghe MN, et al. Mitochondrial Division Is Requisite to RAS-Induced Transformation and Targeted by Oncogenic MAPK Pathway Inhibitors. Molecular cell. 2015;57(3):521–36. doi: 10.1016/j.molcel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kashatus JA, et al. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Molecular cell. 2015;57(3):537–51. doi: 10.1016/j.molcel.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.von Eyss B, et al. A MYC-Driven Change in Mitochondrial Dynamics Limits YAP/TAZ Function in Mammary Epithelial Cells and Breast Cancer. Cancer Cell. 2015;28(6):743–57. doi: 10.1016/j.ccell.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 83.Ipsaro JJ, et al. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature. 2012;491(7423):279–83. doi: 10.1038/nature11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nishimasu H, et al. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature. 2012;491(7423):284–7. doi: 10.1038/nature11509. [DOI] [PubMed] [Google Scholar]
- 85.Adachi Y, et al. An unstructured loop that is critical for interactions of the stalk domain of Drp1 with saturated phosphatidic acid. Small GTPases. 2017:1–8. doi: 10.1080/21541248.2017.1321614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kornmann B, et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325(5939):477–81. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Friedman JR, et al. ER tubules mark sites of mitochondrial division. Science. 2011;334(6054):358–62. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Korobova F, et al. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339(6118):464–7. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lewis SC, et al. ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science. 2016;353(6296):aaf5549. doi: 10.1126/science.aaf5549. [DOI] [PMC free article] [PubMed] [Google Scholar]


