Abstract
Aims: Lack of Cu,Zn-superoxide dismutase (CuZnSOD) in homozygous knockout mice (Sod1−/−) leads to accelerated age-related muscle loss and weakness, but specific deletion of CuZnSOD in skeletal muscle (mSod1KO mice) or neurons (nSod1KO mice) resulted in only mild muscle functional deficits and failed to recapitulate the loss of mass and function observed in Sod1−/− mice. To dissect any underlying cross-talk between motor neurons and skeletal muscle in the degeneration in Sod1−/− mice, we characterized neuromuscular changes in the Sod1−/− model compared with mSod1KO mice and examined degenerative molecular mechanisms and pathways in peripheral nerve and skeletal muscle.
Results: In contrast to mSod1KO mice, myofiber atrophy in Sod1−/− mice was associated with increased muscle oxidative damage, neuromuscular junction degeneration, denervation, nerve demyelination, and upregulation of proteins involved in maintenance of myelin sheaths. Proteomic analyses confirmed increased proteasomal activity and adaptive stress responses in muscle of Sod1−/− mice that were absent in mSod1KO mice. Peripheral nerve from neither Sod1−/− nor mSod1KO mice showed increased oxidative damage or molecular responses to increased oxidation compared with wild type mice. Differential cysteine (Cys) labeling revealed a specific redox shift in the catalytic Cys residue of peroxiredoxin 6 (Cys47) in the peripheral nerve from Sod1−/− mice.
Innovation and Conclusion: These findings demonstrate that neuromuscular integrity, redox mechanisms, and pathways are differentially altered in nerve and muscle of Sod1−/− and mSod1KO mice. Results support the concept that impaired redox signaling, rather than oxidative damage, in peripheral nerve plays a key role in muscle loss in Sod1−/− mice and potentially sarcopenia during aging. Antioxid. Redox Signal. 28, 275–295.
Keywords: : superoxide, 20S proteasome, mitochondria, peroxiredoxins 5 and 6, myelin
Innovation.
This is the first study to compare the molecular mechanisms and pathways that occur in both skeletal muscle and peripheral nerve of Sod1−/− and mSod1KO mice in an effort to examine the relative cross-talk and role of pre- and postsynaptic changes in redox homeostasis in loss of neuromuscular integrity and function that occurs with aging. This study highlights that impaired redox signaling in peripheral nerve rather than oxidative damage appears to play a key role in altering the integrity of peripheral nerves and motor neurons and potentially age-associated muscle atrophy and functional deficits. These results are potentially clinically significant and have widespread implications for the understanding of sarcopenia during aging.
Introduction
Potential mechanisms involved in age-related muscle atrophy and weakness (sarcopenia) have been investigated through examination of homozygous Cu,Zn-superoxide dismutase (CuZnSOD) knockout mice (Sod1−/−mice) (19, 42, 53). At birth, these mice are indistinguishable from wild type (WT) mice, but by 5–8 months of age, muscles of the lower limb of Sod1−/− mice display an accelerated neuromuscular aging phenotype associated with myofiber atrophy, neurological impairments, and functional deficits that progress through adulthood.
Many features of the Sod1−/− mouse model mimic those observed in 28–30 months old WT mice (26, 42, 65) and in older humans (26, 68), including increased levels of oxidative damage (19, 26, 42, 53), a constitutive activation of redox-sensitive transcription factors (49, 66), loss of maximum contractile force (26, 42), deterioration of neuromuscular junctions (NMJs) (12, 19, 49), and deficits in mitochondrial function (19). Hence, it has been suggested that the Sod1−/− mouse model represents a useful model for the study of chronic oxidative damage in the context of neuromuscular aging, which may facilitate identification of potential mechanisms and pathways that are implicated in sarcopenia in humans (42).
Deciphering the key pathways and mechanisms underlying neuromuscular aging has been difficult due to the complex association between loss of motor units and reduction of muscle mass, which are interlinked and occur with the advance of age (27). Motor nerves and muscles are well known to have coordinated and interdependent role in maintaining a healthy neuromuscular system, specifically the viability of motor neurons is recognized to be dependent upon continued exposure to neurotrophic factors released by myofibers (18). In addition, a number of reports have also revealed that manipulations that alter NMJ integrity may induce a phenotype that closely resembles age-related muscle atrophy and weakness (6, 23, 55).
To elucidate whether the accelerated loss of muscle mass and function observed in the Sod1−/− aging model is initiated primarily by alterations in the redox status proximal or distal to the NMJ synapse, “conditional knockout models” were generated to determine whether specific CuZnSOD gene deletion targeted to skeletal muscle (mSod1KO) (70) or motor neurons (nSod1KO) (57) was sufficient to initiate the sarcopenic phenotype seen in Sod1−/− mice. In addition, a “nerve rescue” mouse model was developed in which human SOD1 was specifically expressed in neurons of Sod1−/− mice (49). This model showed no premature loss of muscle mass or function, suggesting a key role for neuronal redox status in regulation of muscle mass. Together these findings suggest that specific deletion of CuZnSOD in either skeletal muscle (70) or motor neurons (57) alone is not sufficient to induce neuromuscular degeneration and that deficits in both tissues are essential to recapitulate the atrophic phenotype observed during aging in the Sod1−/− model. Moreover, these results indicate that there is intricate muscle–nerve cross-talk that is essential for optimal skeletal muscle function.
To assess the relative role of impaired redox homeostasis in skeletal muscle and neurons in loss of skeletal muscle mass, the current study aimed to examine the molecular mechanisms and pathways in pre- and postsynaptic tissues of Sod1−/− and mSod1−/− mice using a series of biochemical, physiological, and redox proteomic techniques. Specifically, to dissect the underlying redox cross-talk between nerve and muscle in this process, we employed a proteomic approach including a differential Cys labeling step (35) to identify key regulatory redox changes and pathways that are modified in the Sod1−/− model compared with mSod1KO mice.
Results
CuZnSOD removal induces myofiber atrophy and oxidative damage in Sod1−/− but not in mSod1KO mice
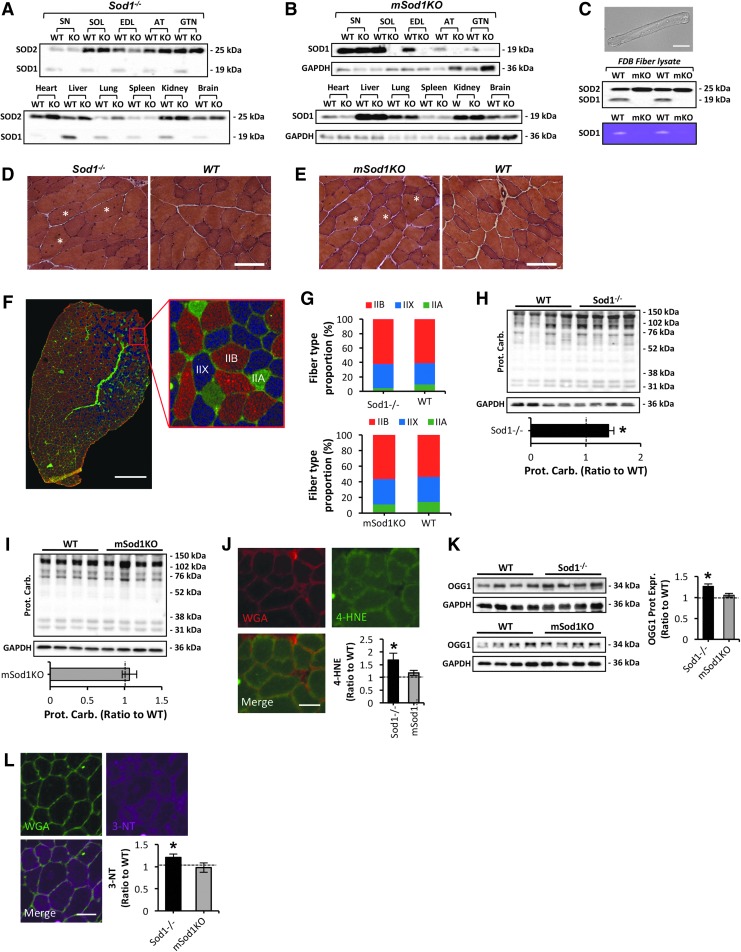
As previously reported, there was no evidence for the expression of CuZnSOD in any of the tissues from Sod1−/− mice (Fig. 1A). Loss of CuZnSOD protein expression specifically in skeletal muscle of mSod1KO mice was confirmed by immunoblotting analysis in soleus, extensor digitorum longus, anterior tibialis (AT), and gastrocnemius (GTN) muscle lysates (Fig. 1B). No change in CuZnSOD protein content in other tissues including peripheral sciatic nerve (SN), heart, liver, lung, spleen, kidney, or brain was observed (Fig. 1B). In some occasions, very faint bands depicting low expression of CuZnSOD were observed in skeletal muscle of mSod1KO mice (see GTN results, Fig. 1B). To confirm whether this was due to nonmyogenic CuZnSOD contamination, we examined CuZnSOD protein content (Fig. 1C, middle panel) and enzymatic activity (Fig. 1C, bottom panel) assessed by native gels, in a muscle fiber preparation devoid of all nonmyogenic cells, using isolated flexor digitorum brevis (FDB) muscle fibers (Fig. 1C, top panel). No expression or activity of CuZnSOD was observed in FDB lysates of mSod1KO compared with the respective WT mice, confirming specific deletion of CuZnSOD in skeletal muscle of the mSod1KO model. Body weight did not change significantly between the different mouse genotypes examined in this study; the absolute muscle and tissue masses are presented (Tables 1 and 2). There was a significant reduction in mass of AT and GTN muscles from Sod1−/− mice (42), compared with the respective WT (Sod1+/+) littermate controls (Table 1). In contrast to the Sod1−/− model, there was no evidence of atrophy in any of the muscles or tissues studied from mSod1KO mice (Table 2). Absolute mass of GTN muscle was greater in mSod1KO mice than in the respective controls (Table 2). To examine the structure of skeletal muscle, histological analysis of the AT muscle/strain was performed (Fig. 1D, E). A significant number of centrally nucleated fibers were observed in both strains (Fig. 1D, E), indicating previous cycles of degeneration and regeneration (53). Average fiber cross-sectional area (CSA) was significantly reduced in Sod1−/− mice (data not shown in detail) but not in the mSod1KO model, as previously described (53, 70). To define whether lack of CuZnSOD in Sod1−/− and mSod1KO mice altered fiber type composition, immunolabeling was performed for the four isoforms of myosin heavy chains (MHCs) on AT cryosections (Fig. 1F). As previously reported (52), AT muscle expressed MHC IIA, IIX, and IIB fibers, but there was no evidence for MHC I expression (Fig. 1F). No change in MHC isoform content was detected between muscles from either genotype compared with the respective WT controls (Fig. 1G), implying no significant changes in fiber type composition. No overt invasion of muscle by mononuclear (inflammatory) cells was seen on the histology sections from either model (Fig. 1D, E).
FIG. 1.
Characterization of muscle morphology and redox homeostasis in skeletal muscle of Sod1−/− and mSod1KO mice. (A) Representative Western blots showing CuZnSOD protein content in various tissues of Sod1−/− and the respective WT mice. (B) Representative Western blots depicting CuZnSOD protein expression in various tissues of mSod1KO and the respective WT mice. (C) Confocal image of a single isolated fiber from the FDB muscle of a WT (Sod1+/+) mouse under bright field. 20 × original magnification. Scale bar, 60 μm (top panel); protein expression of CuZnSOD and MnSOD in lysates from single isolated FDB fibers of mSod1KO and the respective WT mice (middle panel); native gel stained for SOD1 enzyme activity in lysates from single isolated FDB fibers of mSod1KO and the respective WT mice (bottom panel). (D, E) Example of hematoxylin and eosin stained transverse sections of the AT muscle from Sod1−/− (D), mSod1KO (E), and the respective WT littermate controls. White asterisks depict myofibers with central nuclei. 20 × original magnification. Scale bar, 70 μm. (F) Representative triple immunofluorescent staining of MHC isoforms; MHC IIA (green), MHC IIX (blue), and MHC IIB (red) performed on an AT cryosection obtained from a mSod1KO mouse. Right panel shows enlarged area marked by red box in the left panel to show the selective MHC isoform immunolabeling. Scale bar, 500 μm. (G) Histograms showing the fiber-type distribution of AT muscle from Sod1−/− (top panel), mSod1KO (bottom panel), and the respective WT mice. n = 3–4 mice/strain. (H, I) Western blot analysis (top panel) and quantification (bottom panel) of protein carbonyls in AT lysates of Sod1−/− (H), mSod1KO (I), and the respective WT mice. *p < 0.05 compared with values from the respective WT mice. (J) Transverse section of an AT muscle from an Sod1−/− mouse immune-labeled for 4-hydroxynonenal (4-HNE, green) protein adducts, WGA (5 μg/ml, red), to observe extracellular matrix, and a merged image as indicated and analyzed by confocal microscopy. Scale bar, 40 μm; 4-HNE content in AT skeletal muscle of Sod1−/−, mSod1KO, and the respective WT mice (bottom right panel). *p < 0.05 compared with values from the respective WT mice, n = 3 mice/strain. (K) Oxoguanine DNA glycosylase protein levels in AT skeletal muscle of Sod1−/− (top left panel), mSod1KO (bottom left panel), and the respective WT mice, and densitometric quantification of the blots (right panel). *p < 0.05 compared with values from the respective WT mice. (L) Transverse section of an AT muscle from an Sod1−/− mouse immune-labeled for 3-NT (purple) content, WGA (5 μg/ml, green), to observe extracellular matrix, and a merged image as indicated and analyzed by confocal microscopy. Scale bar, 40 μm; 3-NT content in AT skeletal muscle of Sod1−/−, mSod1KO, and the respective WT mice (bottom right panel). *p < 0.05 compared with values from the respective WT mice, n = 3 mice/strain. 3-NT, 3-nitrotyrosine; AT, anterior tibialis; EDL, extensor digitorum longus; FDB, flexor digitorum brevis; GTN, gastrocnemius; MHC, myosin heavy chain; SOL, soleus muscle; WGA, wheat germ agglutinin; WT, wild type.
Table 1.
Comparison of Tissue Weights from SOD1−/− and WT (SOD1+/+) Mice
| Tissue | WT (Sod1+/+) | Sod1−/− |
|---|---|---|
| BW (g) | 32 ± 1.2 | 30.7 ± 1.4 |
| AT (mg) | 50.1 ± 1.7 | 41.7 ± 1.1a |
| EDL (mg) | 11.4 ± 0.8 | 11.9 ± 0.7 |
| GTN (mg) | 172.5 ± 4.6 | 120.7 ± 7.6a |
| SOL (mg) | 9.6 ± 1.2 | 10.6 ± 0.4 |
| Liver (g) | 1.77 ± 0.1 | 1.85 ± 0.2 |
| Spleen (mg) | 119.4 ± 22.6 | 173.4 ± 27.3 |
| Kidney (mg) | 238 ± 26.6 | 276.5 ± 39.7 |
| Heart (mg) | 177.7 ± 7.8 | 190.9 ± 32.7 |
| Lung (mg) | 179.7 ± 26.1 | 215.3 ± 35.3 |
| Brain (mg) | 440 ± 33 | 445.3 ± 38.4 |
Values are presented as mean ± SEM.
p < 0.05 compared with values from WT mice, n = 6 mice/group.
AT, anterior tibialis; BW, body weight; EDL, extensor digitorum longus; GTN, gastrocnemius; SOL, soleus; WT, wild type.
Table 2.
Comparison of Tissue Weights from mSOD1 KO and WT Mice
| Tissue | WT | mSod1KO |
|---|---|---|
| BW (g) | 36.1 ± 3 | 36.9 ± 2.1 |
| AT (mg) | 53.3 ± 1.8 | 53.7 ± 1.5 |
| EDL (mg) | 10.9 ± 0.6 | 11.4 ± 0.6 |
| GTN (mg) | 181.7 ± 7.3 | 206 ± 8.2a |
| SOL (mg) | 8.3 ± 0.5 | 9.8 ± 0.5 |
| Liver (g) | 1.91 ± 0.2 | 1.76 ± 0.1 |
| Spleen (mg) | 88.1 ± 7.7 | 79.3 ± 5 |
| Kidney (mg) | 262.1 ± 35.9 | 242.4 ± 19.8 |
| Heart (mg) | 190 ± 17.3 | 194.5 ± 21.1 |
| Lung (mg) | 196 ± 16 | 164.1 ± 10.4 |
| Brain (mg) | 454.4 ± 7.9 | 435.8 ± 12.6 |
Values are presented as mean ± SEM.
p < 0.05 compared with values from WT mice, n = 6 mice/group.
To determine the effect of CuZnSOD ablation, in the mSod1KO and Sod1−/− models, on muscle redox homeostasis, a series of biochemical and immunohistochemical (IHC) techniques were performed. In contrast to the skeletal muscle from mSod1KO model, skeletal muscle from Sod1−/− mice showed increased protein oxidation, lipid peroxidation, and DNA damage that were apparent as elevated levels of protein carbonyls (Fig. 1H, I), 4-hydroxynonenal (4-HNE) protein adducts (Fig. 1J and Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/ars), malondialdehyde (MDA) content (Supplementary Fig. S2), and an increase in the primary enzyme responsible for excision of 7, 8-dihydro-8-oxoguanine lesions, oxoguanine DNA glycosylase (Fig. 1K), indicating an increase in oxidative DNA damage in skeletal muscle of the Sod1−/− model (20). In addition, muscle from Sod1−/− mice showed an increase in global protein nitration levels, as previously described (53), which was not evident in the mSod1KO model compared with the respective WT controls (Fig. 1L). Moreover, in contrast to the skeletal muscle of Sod1−/− mice, skeletal muscle of mSod1KO mice did not appear to alter mitochondria-mediated apoptotic pathways assessed via changes in cytochrome c cytosolic content and cleaved/procaspase-3 levels (Supplementary Fig. S3A–D). Cleaved caspase-3 was significantly elevated in the Sod1−/− model (Supplementary Fig. S3A), linked to increased release of mitochondrial cytochrome c and Smac/DIABLO proapoptotic proteins to the cytosol (Supplementary Fig. S3C). Overall, these data suggest that in contrast to Sod1−/− mice, specific ablation of CuZnSOD in mSod1KO mice does not replicate the prominent myofiber atrophy observed in Sod1−/− mice, which is associated with gross changes in muscle redox homeostasis.
NMJ degeneration and SN demyelination in Sod1−/− mice are not associated with overt changes in oxidative damage in peripheral nerve
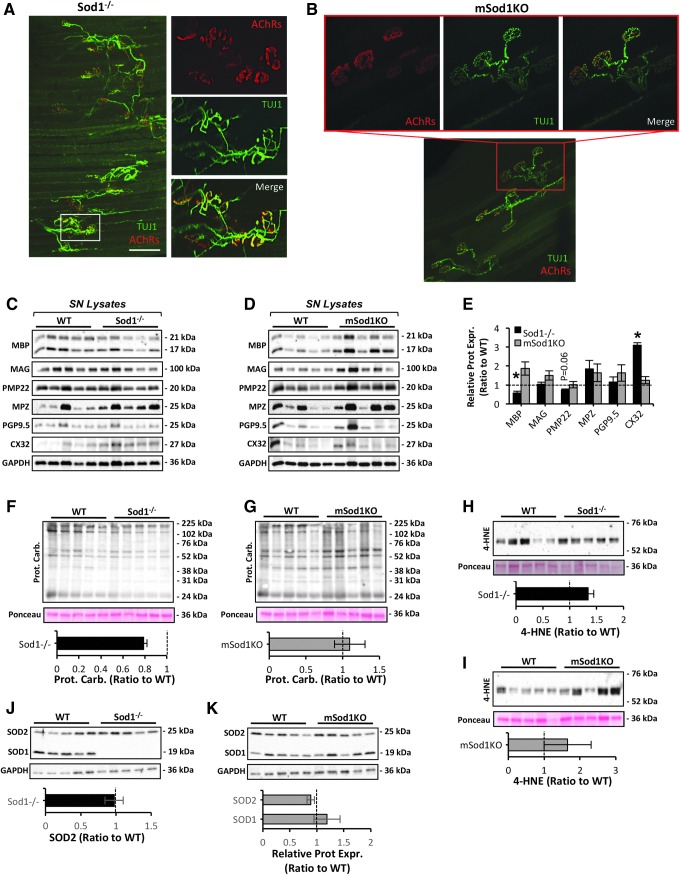
Based on previous (70) and current observations that deletion of CuZnSOD specifically from skeletal muscle does not replicate the prominent myofiber atrophy that occurs in the Sod1−/− model (Fig. 1 and Tables 1 and 2), we hypothesized that retention of CuZnSOD expression in neuronal tissue of mSod1KO mice would also preserve motor neuron integrity and function. To examine this, confocal immunofluorescence imaging of NMJs of the AT muscle was performed (Fig. 2A, B). As shown previously (19, 49), NMJs from the Sod1−/− model displayed marked structural changes as determined by immunohistology (Fig. 2A). Motor axons displayed regions of abnormal thinning, distension, and sprouting, and contrary to the pretzel-like shape of postsynaptic endplates from mSod1KO mice (Fig. 2B), endplates of Sod1−/− mice were denervated, dispersed, and extensively fragmented (Fig. 2A). To determine whether the marked structural NMJ aberrations observed in the Sod1−/− model were linked to altered peripheral nerve integrity, the contents of myelin-associated proteins involved in the formation, maturation, and turnover of myelin sheaths were examined. These included myelin basic protein (MBP), myelin-associated glycoprotein (MAG), peripheral myelin protein 22 (PMP22), myelin protein zero (MPZ), protein gene product 9.5, and connexin 32/GJB1 (CX32), which were assessed in SN lysates from the Sod1−/− and mSod1KO mouse models (Fig. 2C–E). MBP expression was significantly reduced in the Sod1−/− mice (Fig. 2C, E). Moreover, SN from the Sod1−/− model was associated with a compensatory upregulation of CX32 (also known as MYP0) and a trend toward a reduction (p = 0.06) in PMP22 levels (Fig. 2C, E). Contrary to SN from Sod1−/−, SN from mSod1KO mice showed no evidence of altered content in any of the proteins studied (Fig. 2D, E and Supplementary Fig. S4). Motor axons from SN cross sections of the Sod1−/− mouse model showed altered morphology and reduced myelin thickness, resulting in a g-ratio gain (axon diameter/myelinated fiber diameter) (Supplementary Fig. S4), also suggesting that peripheral nerve demyelination occurs in this model.
FIG. 2.
Peripheral nerve redox homeostasis and NMJ structure in Sod1−/− and mSod1KO mice. (A, B) Confocal immunofluorescence imaging of NMJs of an AT muscle from an Sod1−/− mouse (A) and mSod1KO (B) mouse. Presynaptic motor neurons immunolabeled with neuronal class III β-tubulin monoclonal antibody, a neuronal marker (green), and postsynaptic motor endplate AChRs stained with AlexaFluor 594-conjugated α-bungarotoxin (red). Small panels show enlarged area marked by a box in the larger panel. 10 × original magnification (larger panel). Scale bar, 150 μm. (C, D) Western blots of myelin-associated proteins including MBP, MAG, PMP22, MPZ, PGP9.5, and CX32 in SN lysates of Sod1−/− (C), mSod1KO (D), and the respective WT mice. (E) Densitometric analysis of the Western blots shown in (C, D). *p < 0.05 compared with values from the respective WT mice. (F, G) Western blot analysis (top panel) and quantification (bottom panel) of protein carbonyls in SN lysates of Sod1−/− (F), mSod1KO (G), and the respective WT mice. (H, I) Western blot analysis (top panel) and quantification (bottom panel) of 4-HNE protein adducts in SN lysates of Sod1−/− (H), mSod1KO (I), and the respective WT mice. (J, K) Protein expression (top panel) of the main cytosolic and mitochondrial superoxide reducing enzymes, including SOD1 and SOD2 in SN lysates of Sod1−/− (J), mSod1KO (K), and the respective WT mice; densitometric analysis of the blots (bottom panel). AChRs, acetylcholine receptors; CX32, connexin 32/GJB1; MAG, myelin-associated glycoprotein; MBP, myelin basic protein; MPZ, myelin protein zero; NMJ, neuromuscular junction; PGP9.5, protein gene product 9.5; PMP22, peripheral myelin protein 22; SN, sciatic nerve; SOD, superoxide dismutase.
To determine whether the aberrant changes in peripheral nerve/motor neuron integrity displayed in the Sod1−/− mouse model were associated with impaired redox homeostasis and elevated oxidative damage (40), we examined protein and lipid peroxidation in SN lysates of Sod1−/− and mSod1KO mice (Fig. 2F–I). Neither model showed an increase in any of the markers studied (Fig. 2F–I). To confirm the lack and/or expression of CuZnSOD in the peripheral nerve of the Sod1−/− and mSod1KO models, SOD isoforms were quantified (Fig. 2J, K). As anticipated, CuZnSOD expression was not evident in SN lysates of Sod1−/− mice (Fig. 2J), with no changes in mitochondrial superoxide scavenging system expression in the peripheral nerve of either models. Taken together, these data demonstrate that retention of CuZnSOD expression in motor neurons of the mSod1KO model prevents the degenerative alterations in motor neuron/peripheral nerve integrity that occurs in the Sod1−/− genotype, highlighting the role of motor neuron integrity in age-associated muscle atrophy, although the prominent neuromuscular degeneration seen in Sod1−/− mice was not associated with an increase in overall oxidative damage in peripheral nerve.
Global label-free proteomic analysis shows an altered redox proteome in skeletal muscle of Sod1−/− mice
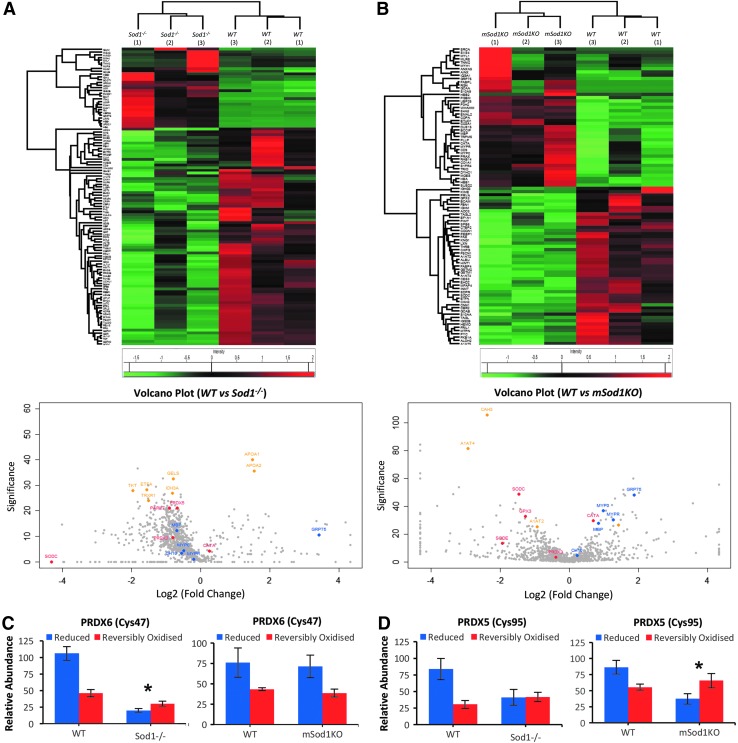
The different changes in structure and redox homeostasis observed in peripheral nerve and skeletal muscle of Sod1−/− and mSod1KO mouse models prompted examination of the molecular nerve–muscle signaling mechanisms and pathways in response to muscle-specific or global loss of CuZnSOD. Global proteomic analysis (35, 36) of skeletal muscle from each genotype was undertaken, and heatmaps representing significantly changed proteins and volcano plots highlighting proteins detected of interest are presented in Figure 3A and B. The proteomic analysis (multiscatter plots depicted in Supplementary Fig. S5A, B; identified proteins and their relative quantification are included in Supplementary Data S1 and S2) confirmed the loss of CuZnSOD (SODC) from skeletal muscle in both the Sod1−/− and mSod1KO mice (Fig. 3A, B, respectively). The label-free analysis showed a general trend for an upregulation of antioxidant proteins, including catalase (CATA), thioredoxin (THIO), and peroxiredoxins 3 and 5 (PRDX3 and PRDX5) in skeletal muscle of Sod1−/− mice (Fig. 3A). In addition, skeletal muscle of the Sod1−/− strain showed significant increases in mitochondrial metabolic and respiratory enzymes involved in energy metabolism including ATP synthase subunits alpha and beta (ATPA and ATPB), NADH dehydrogenase iron–sulfur protein 3 (NDUS3), cytochrome c oxidase subunit 2 (COX2), mitochondrial aldehyde dehydrogenase (ALDH2), Delta (3 5)–Delta (2 4)–dieonyl–CoA isomerase (ECH1), and the mitochondrial 10 kDa heat shock protein (HSP; CH10) (Fig. 3A). Analogous changes were not seen in skeletal muscle from the mSod1KO mice although significant increases in carbonic anhydrases 2 and 3 (CAH2 and CAH3) and glycoprotein alpha-1 anti-trypsin (A1AT) were observed compared with the respective WT littermate controls (Fig. 3B). In addition, mSod1KO mice exhibited a significant reduction in protein disulfide isomerase (PDIA4), a protein involved in DNA repair (EMSY), acylphosphatase (ACYP2), and the cytoplasmic glycolytic enzymes gamma-enolase (ENO2) and phosphoglucomutase-1 (PGM1). Overall, these results suggest that global loss of CuZnSOD results in an upregulation of proteins involved in both antioxidant protection and mitochondrial metabolism as a result of the altered redox homeostasis (Fig. 1) and mitochondrial dysfunction that have previously been described (19). In contrast, deletion of CuZnSOD specifically in skeletal muscle does not replicate these findings and had a predominant effect on cytoplasmic metabolism.
FIG. 3.
Global label-free proteomics of skeletal muscle from Sod1−/− and mSod1KO mice. (A, B) Heatmaps of significantly up- and downregulated proteins in skeletal muscle of Sod1−/− (A), mSod1KO (B), and the respective WT mice detected by PEAKS label-free quantification software (Significance −10log p > 20 or p < 0.01 and fold change ≥2) (top panel); volcano plots of label-free proteomic data showing changes in protein content between skeletal muscle of Sod1−/− (A), mSod1KO (B), compared with the respective WT mice (bottom panel). Proteins involved in redox regulation are highlighted in red, proteins involved in protein folding and stress response are highlighted in blue, and proteins involved in metabolism are in yellow. Proteins highlighted are alpha-1-antitrypsin 1–5 (A1AT5), acylphosphatase-2 (ACYP2), mitochondrial aldehyde dehydrogenase (ALDH2), ATP synthase subunits alpha and beta (ATPA and ATPB), carbonic anhydrase 2 and 3 (CAH2 and CAH3), catalase (CATA), heat shock protein 10 mitochondrial (CH10), cytochrome c oxidase subunit 2 (COX2), alpha crystallin B chain (CRYAB), delta (3 5)–delta (2 4)–dieonyl–CoA isomerase (ECH1), protein EMSY (EMSY), gamma enolase (ENOG), glutathione peroxidase 1 (GPX1), pyruvate kinase isozymes M1/M2 (KPYM), NADH dehydrogenase iron–sulfur protein 3 (NDUS3), protein disulfide isomerase (PDIA4), peroxiredoxins 2, 3, 5, and 6 (PRDX2, PRDX3, PRDX5, and PRDX6), phosphoglucomutase 1 (PGM1), CuZnSOD (SODC), and thioredoxin (THIO).
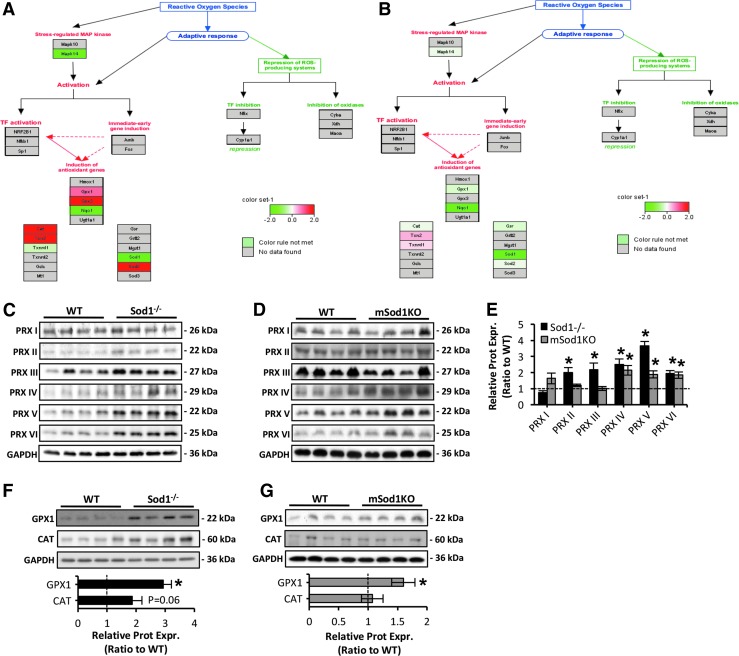
Pathway analysis of proteomic data reveals an induction of antioxidant genes in skeletal muscle of Sod1−/− mice
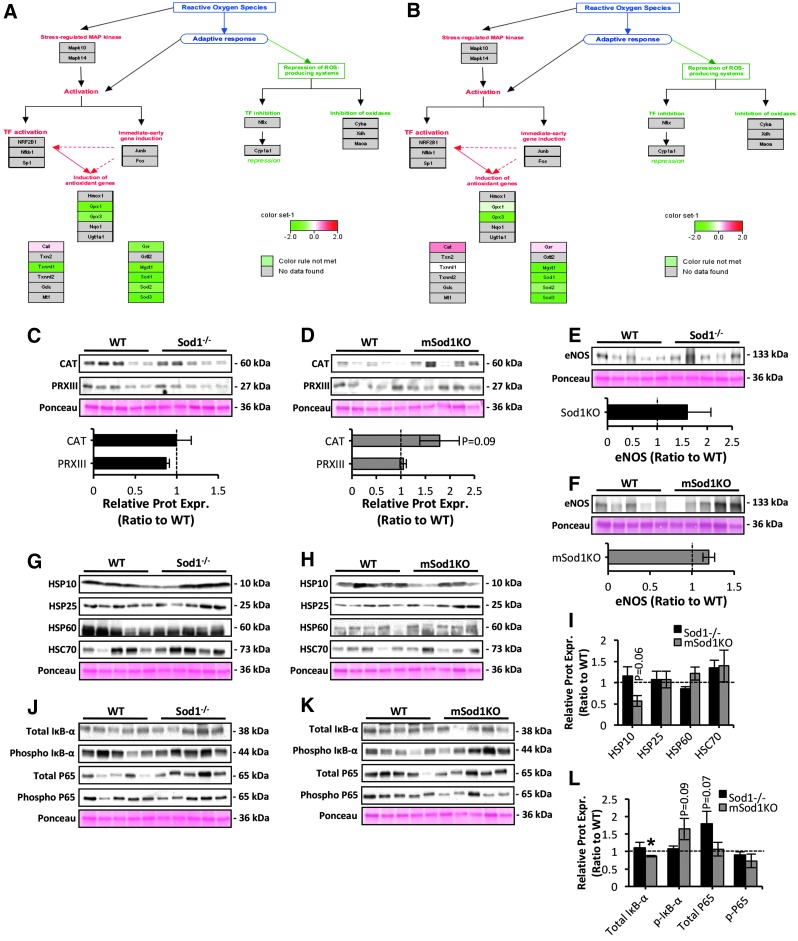
Pathway analysis of quantitative proteomic data further demonstrated an upregulated antioxidant response in skeletal muscle of Sod1−/− (Fig. 4A) with only moderate changes in mSod1KO mice (Fig. 4B). Pathway analysis indicated an induction of the main redox regulatory proteins including the mitochondrial superoxide scavenging system (MnSOD) and hydrogen peroxide (H2O2) scavenging enzymes including catalase (CAT), glutathione peroxidase (GPX), and proteins of the TRX-peroxiredoxin (PRX) system. To confirm these pathways, the protein content of regulatory enzymes for reactive oxygen and nitrogen species (RONS) including all six isoforms of the peroxiredoxin gene family (PRDX 1–6) (Fig. 4C–E), GPX1, and CAT (Fig. 4F, G) was measured. As shown in Figure 4C–G, ablation of CuZnSOD in Sod1−/− and mSod1KO mice induced an upregulation of PRXs and GPX1. Specifically, loss of CuZnSOD in skeletal muscle alone resulted in a significant increase in PRDX 4–6 (Fig. 4D, E) and GPX1 (Fig. 4G), whereas skeletal muscle from the Sod1−/− model exhibited increased contents of PRDX 2–6 (Fig. 4C, E) and GPX1 (Fig. 4F). In addition, there was a trend toward increased CAT content (p = 0.06) in skeletal muscle of the Sod1−/− mice only (Fig. 4F and confirmed by proteomics results). It is noteworthy that compensatory upregulation of all RONS regulatory proteins was greater in the Sod1−/− mice than in the mSod1KO model (Figs. 3A, B and 4C–G). Moreover, we have previously reported increased MnSOD protein content in skeletal muscle of Sod1−/− mice (53) but not in mSod1KO mice (70). The nuclear factor kappa B (NFκB) signaling pathway is known to partly regulate the expression of RONS antioxidant enzymes (47) and previous reports have indicated that in contrast to skeletal muscle of the mSod1KO model (70), skeletal muscle of Sod1−/− mice shows increased NFκB activation (49, 66). Together these findings suggest that the elevated oxidative damage seen in skeletal muscle of the Sod1−/− aging model is associated with a greater adaptive response in the antioxidant system compared with that in the mSod1KO model.
FIG. 4.
Pathway analysis of antioxidant mechanisms in skeletal muscle of Sod1−/− and mSod1KO mice. (A, B) Regulation of antioxidant proteins (as represented by Pathvisio and WikiPathways) in skeletal muscle of Sod1−/− (A) and mSod1KO mice (B). (C, D) Western blots of protein content of the PRX isoforms (PRX I–VI) in AT lysates of Sod1−/− (C), mSod1KO (D), and the respective WT mice. (E) Densitometric analysis of the Western blots shown in (C, D). *p < 0.05 compared with values from the respective WT mice. (F, G) Western blots for GPX1 and CAT protein levels (top panel) in AT skeletal muscle of Sod1−/− (F), mSod1KO (G), and the respective WT mice, and densitometric quantification of the blots (bottom panel). *p < 0.05 compared with values from the respective WT mice. CAT, catalase; GPX1, glutathione peroxidase 1; PRX, peroxiredoxin.
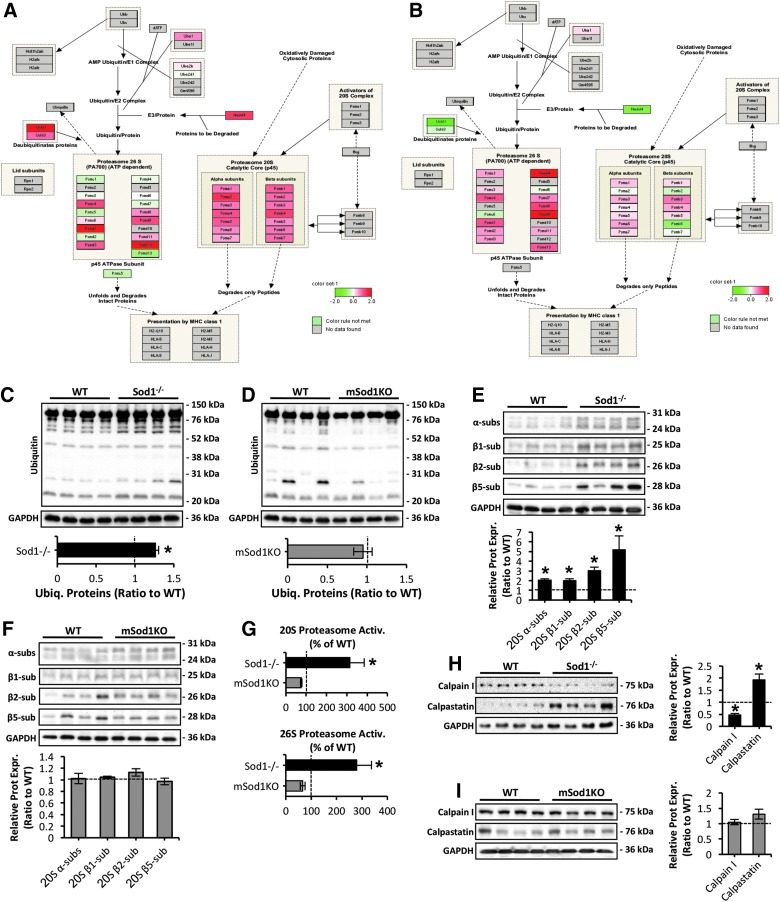
Pathway analysis of proteomic data reveals an induction of proteolytic systems in skeletal muscle of Sod1−/− mice
Previous findings have shown that proteolytic systems play a crucial role in muscle homeostasis, by the degradation of myofibrillar (56) and oxidatively modified proteins (3, 15). One of the major pathways revealed to be altered by the quantitative proteomics results due to CuZnSOD ablation was the ubiquitin–proteasome system. Analyses showed an induction of proteolytic systems in skeletal muscle of the Sod1−/− mice (Fig. 5A, B), particularly for the 20S proteasome (Fig. 5A). To confirm the pathway analysis data, we measured the level of ubiquitin-bound proteins in AT skeletal muscle of Sod1−/− and mSod1KO mice (Fig. 5C, D). Ubiquitinated proteins were significantly elevated in skeletal muscle of the Sod1−/− model (Fig. 5C) with no apparent changes in muscle of the mSod1KO mice (Fig. 5D). Moreover, elevated ubiquitination was associated with increased expression of α subunits and catalytic β1, β2, and β5 constitutive proteasomal subunits in muscle of Sod1−/− mice only (Fig. 5E, F). Proteasome regulation was further assessed by measuring the ATP-independent and ATP-stimulated degradation of the fluorogenic peptide succinyl-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin (Suc-LLVY-AMC) for 20S and 26S proteasomal activity, respectively (Fig. 5G). In contrast to muscle of the mSod1KO model, both ATP-independent (top panel) and ATP-stimulated activity (bottom panel) of the proteasome was significantly elevated in Sod1−/− mice (Fig. 5G). Next to assess the contribution of the calpain/calpastatin myofibrillar proteolytic system (61), protein levels of the Ca2+-dependent cysteine (Cys) protease calpain I and the inhibitor calpastatin (63) were assessed in muscle of Sod1−/− (Fig. 5H) and mSod1KO (Fig. 5I) genotypes. Skeletal muscle of Sod1−/− mice exhibited reduced levels of calpain I potentially due to elevated expression of the inhibitor calpastatin (Fig. 5H). Muscle-specific ablation of CuZnSOD did not alter the content of the calpain/calpastatin system (Fig. 5I). In summary, these data suggest that in contrast to the mSod1KO mice, myofiber atrophy and elevated oxidative damage to skeletal muscle seen in the Sod1−/− model are coupled to enhanced expression and proteolytic activity of the proteasome system.
FIG. 5.
Pathway analysis reveal proteasomal activation in skeletal muscle of Sod1−/− but not in mSod1KO mice. (A, B) Pathways analysis of proteasomal regulation (as represented by Pathvisio and WikiPathways) in skeletal muscle of Sod1−/− (A) and mSod1KO mice (B). (C, D) Western blots (top panel) and quantification (bottom panel) of ubiquitin-bound proteins in AT skeletal muscle of Sod1−/− (C), mSod1KO (D), and the respective WT mice. *p < 0.05 compared with values from the respective WT mice. (E, F) Western blot analysis (top panel) and quantification (bottom panel) of α, β1, β2, and β5 proteasomal subunits in AT lysates of Sod1−/− (E), mSod1KO (F), and the respective WT mice. *p < 0.05 compared with values from the respective WT mice. (G) Proteasomal activity assessed by ATP-independent (20S proteasome, top panel) and the difference between ATP-independent and ATP-stimulated (26S proteasome, bottom panel) lactacystine-sensitive degradation of the fluorogenic peptide suc-LLVY-AMC in AT lysates of Sod1−/−, mSod1KO, and the respective WT littermate controls. *p < 0.05 compared with values from the respective WT mice, n = 4 mice/strain. (H, I) Proteolytic calpain I and calpastatin protein levels (left panel) in AT skeletal muscle of Sod1−/− (H), mSod1KO (I), and the respective WT mice, and densitometric quantification of the blots (right panel). *p < 0.05 compared with values from the respective WT mice. Suc-LLVY-AMC, succinyl-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin.
Label-free proteomics and differential cysteine labeling of peripheral nerve from Sod1−/− and mSod1KO mice
Heatmaps of significantly changed proteins and volcano plots of global label-free proteomic data from the SN of Sod1−/− and mSod1KO mice are presented in Figure 6A and B (Multiscatter plots are depicted in Supplementary Fig. S6A, B; identified proteins and their relative quantification are included in Supplementary Data S3 and S4). Label-free quantitative proteomic data (Fig. 6A, B) supported the reduction in structural proteins MBP and CX32 (MYP0) in Sod1−/− mice shown in Figure 2C–E. Significant changes were also detected in redox-regulated proteins, including PRDX 5 and thioredoxin reductase 1 (TRXR1), but also metabolic enzymes known to be redox regulated (14, 33, 35, 62) including transketolase (TKT), isocitrate dehydrogenase (IDH3A), electron transport flavoprotein subunit alpha (ETFA), and the cytoskeletal regulator gelsolin (GELS) (Fig. 6A) were downregulated in SN of the Sod1−/− model. There was an upregulation of the lipid binding proteins, apolipoproteins A1 and A2 in the SN of the Sod1−/− model, which parallels the upregulation of apolipoprotein E found in the peripheral nerves of old mice (37). Peripheral nerve of the mSod1KO model showed an upregulation of catalase but a downregulation of GPX3 (Fig. 6B). A reduced protein content of CAH3 and A1AT was also seen, which contrasts with an upregulation of these proteins in skeletal muscle from this genotype (Fig. 3A, B).
FIG. 6.
Global label-free proteomics and differential cysteine labeling in peripheral nerve of Sod1−/− and mSod1KO mice. (A, B) Heatmaps of significantly up- and downregulated proteins in SN of Sod1−/− (A), mSod1KO (B), and the respective WT mice detected by PEAKS label-free quantification software (p < 0.01 and at least one unique peptide) (top panel); volcano plots of label-free proteomic data showing changes in protein content between peripheral nerve of Sod1−/− (A), mSod1KO (B), compared with the respective WT mice (bottom panel). Proteins involved in redox regulation are highlighted in red, myelin-associated proteins are highlighted in blue, and metabolic proteins are highlighted in yellow. Proteins highlighted are alpha-1 antitrypsin (A1AT), apolipoprotein A-I and A-II (APOA1 and APOA2), carbonic anhydrase 3 (CAH3), catalase (CATA), heat shock protein 10 mitochondrial (CH10), electron transport flavoprotein subunit alpha (ETFA), gelsolin (GELS), glutathione peroxidase 3 (GPX3), isocitrate dehydrogenase [NAD] alpha mitochondrial (IDH3A), myelin basic protein (MBP), myelin proteolipid protein (MYPR), myelin protein P0 (MPZ), protein DJ-1 (PARK7) peroxiredoxins 3 and 5 (PRDX3 and PRDX5), CuZnSOD (SODC), extracellular superoxide dismutase (SODE), sphingosine 1-phosphate receptor 4 (S1PR4), thioredoxin reductase 1 (TRXR1), and transketolase (TKT). (C, D) Redox state of Cys47 from PRDX 6 (C) and Cys95 from PRDX 5 (D) in peripheral nerve of Sod1−/− (left panel), mSod1KO (right panel), and their respective controls. Blue bars represent the reduced state of the Cys residue and red bars the reversibe oxidation of the Cys residue. * Indicates a significant shift (p < 0.05) in the redox state toward reversible oxidation compared with values from the respective WT mice. Cys, cysteine
Although our findings indicate no overt oxidative damage in peripheral nerve of the Sod1−/− and mSod1KO models, oxidative modification of specific proteins or local temporal alterations in reversible redox modifications may not be detected by standard approaches (37, 69), and hence we looked for more subtle changes in homeostatic redox regulation of SN tissue in both the Sod1−/− and mSod1KO models by using redox proteomic techniques to differentially label reduced and reversibly oxidized Cys residues (34, 35) (Fig. 6A–D). Although there were some minor changes to the oxidation state of specific Cys residues in key metabolic proteins, the most notable effects were found in the catalytic Cys residues of redox signaling enzymes PRDX 6 (Cys47) and 5 (Cys95) (Fig. 6C, D respectively). Figure 6C shows the relative proportion of Cys47 of PRDX 6 in the reduced (blue bars) and reversibly oxidized (red bars) state. It is clear that there was a significant shift in the redox state toward reversible oxidation of this key 1-Cys PRDX in the peripheral nerves from the Sod1−/− mice Fig. 6C, left panel that was not detected in the SN from mSod1KO mice (Fig. 6C, right panel). A representation of the redox state of Cys95 of PRDX 5 is also shown in Figure 6D. In this case, a shift in the redox state toward reversible oxidation was observed in peripheral nerves of the mSod1KO model only (Fig. 6D, right panel). It is relevant that the increased oxidation was not universal across all redox-active Cys residues detected, with some Cys residues showing a greater proportion in the reduced form, nevertheless it identifies key shifts in key regulatory redox proteins whether in overall abundance or in the oxidation state of specific Cys residues.
Pathway analysis of proteomic data reveals no overt changes in antioxidant or adaptive signaling responses in peripheral nerve of Sod1−/− and mSod1KO mice
To assess the molecular nerve signaling mechanisms and pathways in response to muscle-specific and global loss of CuZnSOD, pathways analysis of quantitative proteomic data from peripheral nerve of Sod1−/− (Fig. 7A) and mSod1KO mice (Fig. 7B) was performed. In agreement with the lack of significant changes in protein oxidation and lipid peroxidation (Fig 2F–I), neither model showed an induction in antioxidant pathways (Fig. 7A, B). To further examine the lack of changes in adaptive stress signaling responses, we assessed the protein expression of redox regulatory systems including; H2O2 scavenging enzymes including cytosolic CAT and mitochondrial PRDX 3 (Fig. 7C, D), endothelial nitric oxide synthase isoenzyme (Fig. 7E, F), and HSPs (Fig. 7G–I), all of which have a significant role in the protection against the damaging effects of increased RONS production (5, 49). Densitometric analysis of the immunoblots presented in Figure 7C–H showed no significant changes in any of the redox regulatory enzymes or protective proteins in SN lysates of Sod1−/− and mSod1KO mice (Fig. 7C–I). There were trends toward increased levels of CAT (p = 0.09) (Fig. 7D), and a reduction in mitochondrial HSP10/CH10 (p = 0.06) (Fig. 7H, I), in SN of the mSod1KO transgenic mice compared to the respective WT controls. Activation of the NFκB pathway was examined in peripheral nerves of Sod1−/− and mSod1KO genotypes (Fig. 7J–L). Contrary to increased NFκB activation in atrophic muscle of Sod1−/− strain (49, 66), peripheral nerve showed no significant changes in activation as indicated by phosphorylation of IκB-α (Phospho IκB-α), total IκB-α content (a key inhibitor of NFκB activation), or NFκB-P65 protein content (total and phosphorylated) (Fig. 7J, L). Peripheral nerve of the mSod1KO model showed some evidence of enhanced activation of NFκB signaling pathways by a significant reduction in total IκB-α content and a trend toward increased Phospho IκB-α content (p = 0.09) (Fig. 7K, L), which may partly explain the elevated CAT content in SN of mSod1KO mice (Fig. 7D). Overall, these data suggest that neuromuscular degeneration in Sod1−/− mice is not associated with adaptive stress responses in peripheral nerve but rather specific changes in oxidation state of key Cys residues in redox signaling proteins. In addition, muscle-specific ablation of CuZnSOD in the mSod1KO model appears to alter NFκB signaling pathway in peripheral nerve, indicating some redox-mediated cross-talk between skeletal muscle and motor neurons in the mSod1KO mice.
FIG. 7.
Pathway analysis of antioxidant mechanisms in peripheral nerve of Sod1−/− and mSod1KO mice. (A, B) Regulation of antioxidant proteins in pathways in peripheral nerve of Sod1−/− (A) and mSod1KO mice (B). (C, D) Western blots (top panel) of the main cytosolic and mitochondrial H2O2 reducing enzymes, including CAT and PRXIII in SN lysates of Sod1−/− (C), mSod1KO (D), and the respective WT mice; densitometric analysis of the blots (bottom panel). (E, F) Protein expression (top panel) of endothelial nitric oxide synthase (eNOS) isoform in SN lysates of Sod1−/− (E), mSod1KO (F), and the respective WT mice; densitometric analysis of the blots (bottom panel). (G, H) Protein expression of HSPs, including mitochondrial HSP10, HSP25, HSP60, and HSC70 in SN lysates of Sod1−/− (G), mSod1KO (H), and the respective WT mice. (I) Densitometric analysis of the Western blots shown in (G, H). (J, K) Western blots for total and Phospho IκB-α and P65 content (total and phosphorylated), in SN lysates of Sod1−/− (J), mSod1KO (K), and the respective WT mice. (L) Densitometric analysis of the Western blots shown in (J, K). *p < 0.05 compared with values from the respective WT mice. H2O2, hydrogen peroxide; HSPs, heat shock proteins; Phospho IκB-α, phosphorylated IκB-α; PRXIII, peroxiredoxin III.
Discussion
Substantial evidence indicates that age-related loss of skeletal muscle is linked to alterations in the oxidative status of redox-responsive proteins (34) and elevated oxidative modifications of macromolecules such as proteins, lipids, and DNA (47). Although the area remains controversial (46, 52), it has been suggested that alterations in redox homeostasis play a crucial role in sarcopenia, and we have previously reported that removal of CuZnSOD in homozygous knockout mice (Sod1−/−) induces accelerated age-related muscle loss and weakness (19, 42, 53). The atrophic phenotype displayed in the Sod1−/− model shares many features of normal aging, including a reduction in fiber CSA (19, 42) and contractile force (26, 42), a constitutive activation of redox-sensitive transcription factors (49, 66), increased levels of oxidative damage (19, 26, 42, 53), neurological impairments (12, 19, 49), and deficits in mitochondrial function (19). We have, therefore, suggested that the Sod1−/− mouse model may provide a model to investigate the mechanisms and pathways by which impaired redox signaling can contribute to age-related skeletal muscle atrophy and identify potential mechanisms and pathways that underlie sarcopenia in humans.
Deciphering the key pathways and mechanisms underlying age-related muscle atrophy and weakness has proved to be challenging due to the technical difficulties in unraveling the association between loss of motor units and reduced muscle mass that occur with advancing age (27). The symbiotic relationship between motor nerves and muscles is essential for the maintenance of a functional neuromuscular system. Several reports that have manipulated pathways involved in the maintenance of NMJ integrity and postsynaptic motor endplate acetylcholine receptors (AChRs) lead to a phenotype that closely resembles neuromuscular aging (16). These studies include enhanced degradation by the overexpression of neurotrypsin (6), depletion of the nerve-derived organizer of postsynaptic differentiation agrin (55), and a reduction in the expression of tyrosine kinase receptor B (23), a receptor for neurotrophic factors such as brain-derived neurotrophic factor and neurotrophins 4 and 5. Moreover, evidence has shown that the viability of motor neurons is recognized to be reliant on the sustained exposure to neurotrophic factors released by myofibers (18).
To unravel whether the muscle decline and weakness presented in the Sod1−/− mice are initiated by defective redox signaling within motor neurons, or skeletal muscle, conditional knockout models including mice mSod1KO (70) or nSod1KO (57) were generated together with a “nerve rescue” mouse model in which human SOD1 was specifically expressed in neurons of Sod1−/− mice (49). To directly assess the relative roles of impaired redox homeostasis in skeletal muscle or motor neurons in the loss of muscle mass and function, the focus of this study was to characterize the neuromuscular changes that occur in the Sod1−/− aging model compared with the mSod1KO strain (which show no evidence of muscle loss) and examine the molecular mechanisms and pathways that occur in both peripheral nerve and skeletal muscle. Our aim was to examine the cross-talk and relevance of pre- and postsynaptic changes in redox homeostasis on the loss of neuromuscular integrity and function that occurs with aging.
We confirmed that mice lacking CuZnSOD specifically in muscle maintain muscle mass similar to respective WT mice and do not reproduce the accelerated neuromuscular aging phenotype of the constitutive whole-body Sod1−/− strain, including the multiple biochemical and physiological changes. Ablation of CuZnSOD in skeletal muscle of mSod1KO mice induced no overt changes in oxidative damage but these mice showed some evidence for a compensatory upregulation in RONS protective enzymes. Myofiber atrophy in muscle of Sod1−/− mice was linked to elevated oxidative damage in DNA, proteins, and lipids, reflective of adaptations in stress responses to oxidants, and an increase in the protein expression and proteolytic activity of the proteasome system, previously shown to contribute to muscle wasting in inherited muscle disorders, including myopathies and muscular dystrophies (56). It is noteworthy that removal of CuZnSOD in both models resulted in many myofibers of the Sod1−/− and mSod1KO mice exhibiting centrally located nuclei, reflecting previous cycles of degeneration and regeneration (Fig. 1). The relatively minor effect seen after tissue-specific removal of CuZnSOD in skeletal muscle of mSod1KO mice supports the conclusion that compromised redox signaling within skeletal muscle is not the primary factor responsible for initiating loss of muscle mass in the Sod1−/− mice or in age-related sarcopenia. Other reports examining mice with conditional knockout of MnSOD in type IIB skeletal muscle fibers, TnIFastCreSod2fl/fl, (28) or mice with muscle-specific overexpression of rat nNOS, nNOSTg, (43, 53) where compromised redox homeostasis is predicted have also failed to produce a degenerative muscle phenotype with aging.
Consistent with our previous observations (19), the accelerated sarcopenic phenotype observed in Sod1−/− mice was accompanied with altered motor neuron integrity. Preterminal motor axons displayed regions of abnormal thinning, distension, and sprouting, and the postsynaptic endplates were denervated, dispersed, and extensively fragmented. These findings are in accord with recent reports that showed enhanced mRNA expression of AChR-α and an increase in Runx1 and GADD45α transcription factors that potentially reflect upregulation of NMJ components in response to denervation in Sod1−/− mice (49). Other data from the Sod1−/− model have shown that the denervation and NMJ fragmentation that occur in the Sod1−/− model are associated with altered electrophysiological properties, including a reduction in both the amplitude and the frequency of miniature end plate potential (mEPPs), indicating altered neurotransmission in skeletal muscle of Sod1−/− mice (49). In this study, we further report that neuromuscular aging in the Sod1−/− model is linked to altered peripheral nerve integrity. Sod1−/− mice exhibit SN demyelination and a compensatory upregulation of CX32, a neuronal protein involved in the maintenance of myelin sheaths (58). In addition, peripheral nerve of the Sod1−/− model showed diminished MBP content and a trend toward reduced levels of PMP22 neuronal protein. PMP22 is known to promote myelin development in early stage formation and has a crucial role in regulating myelin thickness and integrity (32). It has been reported that mRNA levels of both MBP and PMP22 decrease significantly in the SN of 22–24 months old male rats (38, 39) and that PMP22 rapidly declines in the degenerating distal stump after transection of SNs in adult rats (4, 22). In line with our findings, age-related changes in peripheral nerve axonal structure have previously been associated with a reduction in structural proteins and also with an increased content of proteins upregulated in brain in neurodegenerative diseases and during aging (11). Overall, these observations contribute to the understanding of the age-related alterations of the peripheral nervous system (PNS) but also help identify the role of redox-dependent proteins in the maintenance of PNS myelin sheath with aging.
Previous studies have shown that isolated mitochondria from muscle of Sod1−/− mice exhibit elevated release of H2O2 with subsequent deficits in mitochondrial function (19) and earlier studies reported that transection of the motor nerve is coupled to large and sustained increases in muscle mitochondrial RONS release (41), with an increased activation of cytosolic phospholipase A2 (cPLA2) in the denervated muscle (1, 2). cPLA2 activity controls arachidonic acid (AA) release from membrane phospholipids, and AA acts as a substrate for lipid metabolic pathways catalyzed by lipoxygenase, cyclooxygenase, and cytochrome P450 (50). It is thus reasonable to assume that Sod1−/− associated overt oxidative damage and muscle loss, potentially due to lack of appropriate innervation of the muscle, are likely secondary to PNS deficits, since these changes did not occur in the mSod1KO model, which express CuZnSOD in motor neurons and showed no evidence of altered peripheral nerve and motor neuron integrity. Further evidence to support a deficit in muscle innervation in the Sod1−/− model is provided by previous observations that direct stimulation of muscle from Sod1−/− mice elicited a consistent increase in maximum isometric force production compared with using nerve stimulation in age-matched WT littermate controls (26, 60), implying that lack of CuZnSOD in PNS of the Sod1−/− genotype results in denervation of myofibers, and a failure of neuromuscular transmission. Finally, the observation/hypothesis that age-related myofiber atrophy is not only due to effects in the muscle but is also related to processes within motor neurons that innervate skeletal muscle is supported by work from our group that has shown that neuron-specific expression of CuZnSOD in motor neurons of Sod1−/− under control of the synapsin 1 promoter, SynTgSod1−/− transgenic mice, (49) prevented NMJ degeneration and loss of muscle mass and function that occurs in the Sod1−/− mice. Overall, these data imply that alterations in redox signaling in motor neurons may play a role in initiating sarcopenia in the Sod1−/− strain.
The lack of evidence for elevated oxidative damage or adaptive stress/signaling responses in peripheral nerve of Sod1−/− mice was unexpected in light of the changes in peripheral nerve integrity seen in the Sod1−/− strain. We did not detect any apparent changes in markers of oxidative damage (protein carbonyls, 4-HNE) in peripheral nerves from either mouse model used in this study. Although we previously reported a minor increase in RONS in SN tissue from old mice, protein carbonyl levels were similar between adult and old mice (37). Previous studies of the redox-active Cys residues of proteins revealed a minor increase in reversible oxidation of specific proteins in SN of old mice compared with adult mice, including Cys47 of PRDX 6, Cys95 of PRDX 5, Cys263 of neurofilament heavy polypeptide, Cys114 of Annexin A6, Cys49 of pyruvate kinase M1/M2, and Cys254 of creatine kinase b (37). This pattern of increased oxidation did not occur across all redox-active Cys, with some Cys showing a greater proportion in the reduced form with aging (37), reflecting age-related modifications in redox signaling rather than overt increases in oxidative damage in SN tissue. Subtle changes in RONS and the subsequent redox modifications of regulatory proteins can result in altered molecular/redox signaling processes and physical function (30, 31). Of particular interest from the data presented here is the shift toward reversible oxidation in Cys47 of the 1-Cys peroxidase PRDX 6. It has previously been demonstrated that the yeast ortholog of this protein utilizes GSH to complete its catalytic cycle (45). It is noteworthy that apart from its peroxidase activity, this protein also exhibits phospholipase A2 activity that is involved in activation of NADPH oxidase 2 (13, 67). The catalytic triad responsible for its phospholipase activity 32S-140D-26H is relatively proximal to the peroxidatic Cys47 that we have detected, and it would be interesting to speculate whether a shift in its redox state would affect its phospholipase activity in the peripheral nerve of Sod1−/− mice. The atypical 2-Cys peroxiredoxin, PRDX 5, was also found to have a shift in the redox state toward reversible oxidation of its catalytic Cys95, in peripheral nerves of both Sod1−/− and mSod1KO models although the changes only reached statistical significance in mSod1KO mice. PRDX 5 has been reported in mitochondria, peroxisomes, cytosol, and nuclei, and appears to reduce H2O2, alkyl hydroperoxides, and peroxynitrite (21). Understanding how a change in the redox state of the catalytic Cys would affect redox signaling processes requires further investigation.
Proteomic analyses conducted in this study revealed an altered proteome in peripheral nerve of Sod1−/− mice, associated with downregulation of a number of metabolic enzymes that appear to be redox mediated (14, 33, 35, 62). Overall, the data presented in this study indicate that despite the observed PNS degeneration in Sod1−/− mice, peripheral nerve of this model does not exhibit altered redox homeostasis similar to that seen in the skeletal muscle that it innervates but an altered proteome and specific changes in key regulatory redox Cys residues. In addition, the results from this study and others imply that minor or transient alterations in the oxidation/reduction of key Cys may contribute to defective redox signaling in peripheral nerve with aging. Studies reported here have focused on the peripheral nerve, but it is noteworthy that the lack of any gross changes in oxidative damage in the peripheral nerve of the Sod1−/− model may not directly reflect the redox changes that occur in the cell body of the alpha motor neurons. It is possible that oxidative damage and/or disrupted redox signaling, in addition to phenotypic changes, may be more evident in the cell body of the alpha motor neurons and further studies in this area appear warranted.
Collectively, these findings demonstrate that neuromuscular integrity and redox homeostasis are differentially altered in Sod1−/− and mSod1KO mice. CuZnSOD expression in motor neurons (and potentially other cell types) of the mSod1KO model prevented the degenerative alterations in peripheral nerve integrity, increased oxidative damage, and elevated ubiquitin–proteasome regulation in skeletal muscle that occurs in the Sod1−/− genotype. These results support the concept that CuZnSOD gene deletion and altered redox regulation solely in skeletal muscle are not sufficient to initiate muscle atrophy and that impaired redox signaling in peripheral nerve rather than oxidative damage may be important in muscle loss in the Sod1−/− model and potentially sarcopenia during aging. A key role for the motor neuron is also supported by data from the “nerve rescue” mouse model wherein human SOD1 was specifically expressed in neurons of Sod1−/− mice (49), leading to complete reversal of the sarcopenic phenotype seen in Sod1−/− mice. However, the lack of a degenerative muscle phenotype in conditional knockout mice nSod1KO argues against this possibility (57). The current study suggests that impaired redox signaling associated with NMJ degeneration, denervation, and demyelination occurs in the absence of overt oxidative damage in peripheral nerve in the Sod1−/− mice, which may lead to the large increase in muscle oxidative damage seen in that model. It is relevant that we have also reported no increase in overt oxidative damage in SNs of aging WT mice (37), although large increases in oxidative damage are also seen in skeletal muscle with aging. Rather more subtle changes in the redox Cys status of specific redox-regulated proteins were seen in nerves of both Sod1−/− mice and aging WT mice, which we have argued is reflective of more subtle changes in redox signaling. It is currently unknown whether age-related myofiber atrophy and impaired function inherent with aging in humans can be attributed to deficits in motor neurons, or whether age-related deficits in skeletal muscle and/or other tissues also play a role. Overall, the models examined in this study highlight that defective redox signaling in PNS may play a key role in altering peripheral nerve integrity and potentially age-associated muscle atrophy and functional deficits.
Materials and Methods
Chemicals and reagents
Unless stated otherwise, all chemicals used in this study were obtained from Sigma Chemical Company.
Animals
All mice were generated at the University of Texas Health Science Center at San Antonio (UTHSCSA) and the Oklahoma Medical Research Foundation (OMRF). Mice were maintained under specific pathogen-free conditions and were shipped to the Biomedical Services Unit, University of Liverpool at ∼5 months of age and maintained until they were tested (mean age 9 ± 1 months). At both sites, mice were maintained under barrier conditions in microisolator cages on a 12-h dark/light cycle and were fed a CRM (P) rodent diet. Mice were euthanized by cervical dislocation, and muscles/tissues were either rapidly removed, weighed, frozen in liquid N2, and stored at −80°C, or embedded in Tissue-Tek (VWR) and rapidly frozen in nitrogen-chilled isopentane for histological analysis. All procedures were approved by the University of Liverpool Animal Welfare and Ethical Review Body. Experiments were conducted in accordance with UK Home Office guidelines under the UK Animals (Scientific Procedures) Act 1986.
Generation of constitutive whole-body Sod1 knockout (Sod1−/−) mice
Generation and characterization of the Sod1−/− mouse model have been previously described (10, 17, 42). To confirm the constitutive whole-body Sod1 deletion, several tissues were dissected and assayed for SOD1 protein expression (Fig. 1A).
Generation of muscle-specific Sod1 knockout (mSod1KO) mice
Details of the generation of the mSod1KO mouse model have been previously reported (70). To confirm the muscle-specific Sod1 deletion, a variety of tissues were dissected and assayed for SOD1 protein expression (Fig. 1B, C) and enzymatic activity (Fig. 1C).
All measures were undertaken in adult male mice, 9 ± 1 months of age. WT mice used in this study were littermate controls for either the mSod1KO or Sod1−/− mouse model. Both, mSod1KO and Sod1−/− mice were compared with their respective age-matched WT littermates.
Determination of muscle structure
Cross sections (12 μm) through the mid-belly of the AT muscle were obtained by cryosectioning at −20°C and stained with hematoxylin and eosin (Fig. 1D, E) as previously described (53). Cross sections from three to four muscles/mouse strain were examined by blinded observers to count individual fiber CSA. For these analyses, consecutive images acquired from each cryosection at 10 × magnification were merged into a single high-resolution image using Adobe photoshop CS6. Image J software was used to quantify individual fiber CSA.
IHC analysis of MHC expression
Muscle cross sections (10 μm thick) were immunolabeled for MHC isoform expression: MHC I, MHC IIA, MHC IIX, and MHC IIB using as previously described (52) (Fig. 1F).
Immunofluorescent imaging of NMJ structure
To visualize NMJs, AT muscles were fixed in 4% paraformaldehyde (PFA) for 30 min at room temperature and washed for 2 × 5 min in phosphate buffered saline (PBS). AT muscles were treated with 1% Triton X-100 in PBS for 1 h at low rocking speed, at room temperature. After permeabilization, postsynaptic motor endplate AChRs were immunolabeled with AlexaFluor 594-conjugated α-bungarotoxin (1:500; Invitrogen) for 45 min, followed by subsequent overnight staining at 4°C with AlexaFluor 488-conjugated neuron-specific class III β-tubulin antibody (1:400; Merck Millipore) in PBS supplemented with 0.2% Triton X-100. Muscles were washed for 2 × 8 min in PBS at low rocking speed at room temperature. Immunofluorescence images of the NMJs were obtained using an upright confocal microscope (Eclipse Ni-E system; Nikon). Images were captured and analyzed with the Nikon NIS-Elements imaging software. Muscles were kept in the dark at all times during staining.
Quantitative analysis of myelination
For the quantitative analysis of myelination, 1–2-cm segment of SN tissue was fixed in 4% PFA for 20 min at room temperature, followed by 2 × 5min washes in PBS and overnight incubation in 30% sucrose at 4°C. SN tissues were placed in 7.5% (w/v) porcine-derived gelatine solution supplemented with 15% (w/v) sucrose and 0.1% (w/v) sodium azide, prewarmed to 45°C, and incubated at 37°C for 3 h. Tissues were left to set at room temperature followed by overnight incubation at 4°C. SN specimens were embedded in Tissue-Tek (VWR) and rapidly frozen in nitrogen-chilled isopentane for histological analysis. Transverse cryosections of SN were performed using a cryostat (Leica) and mounted on gelatine-coated slides (FD NeuroTechnologies). Images were obtained using a Nikon C1 confocal laser scanning microscope under bright field at 20 × and 60 × magnification. To assess myelination of peripheral nerves, myelin sheath thickness (μm) and the g-ratio, which is proportional to fiber size (axon diameter/fiber diameter), were calculated using ImageJ software (44, 59).
Isolation of single mature skeletal muscle fibers
Single muscle fibers were isolated from the FDB muscle of mice as previously described (53, 54). An example of an isolated single fiber from the FDB muscle is shown in Figure 1C.
Protein extraction for redox proteomics
To identify any global redox changes, a differential Cys labeling protocol was included as reported in our recent publication (35). In brief, muscle and SN samples were homogenized directly in 50 mM ammonium bicarbonate containing 25 mM N-ethylmaleimide (d(0) NEM) pH 8. Protein lysates were prepared by centrifugation at 15,000 g for 10 min at 4°C. Excess d(0) NEM was removed using Zeba desalting columns (Thermo Scientific). One hundred micrograms of protein extract was diluted to 160 μl with 25 mM ammonium bicarbonate and denatured by the addition of 10 μl of 1% RapiGest (Waters, Manchester, United Kingdom) in 25 mM ammonium bicarbonate and incubated at 80°C for 10 min with shaking. Ten microliters of a 100 mM solution of Tris(2-carboxyethyl)phosphine hydrochloride, TCEP, was added to reduce reversibly oxidized Cys residues followed by incubation at 60°C for 10 min. Newly reduced Cys were then alkylated by addition of d(5) NEM and incubated at room temperature for 30 min. An aliquot of the samples was used at this point to check the procedure by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Proteolytic digestion was performed by incubation with trypsin overnight at 37°C. Digestion was terminated and RapiGest removed by acidification (3 μl of Trifluoroacetic acid (TFA) and incubated at 37°C for 45 min) and centrifugation (15,000 g for 15 min).
Liquid chromatography tandem mass spectrometry and label-free Mass spectrometry quantification
Samples were analyzed using an Ultimate 3000 RSLC nano system (Thermo Scientific) coupled to a QExactive mass spectrometer (Thermo Scientific). Two microliters of sample were diluted in 18 μl buffer (97% H2O, 3% MeCN, and 0.1% formic acid v/v), and 5 μl corresponding to 250 ng of protein was loaded onto the trapping column (PepMap 100, C18, 75 μm × 20 mm) using partial loop injection for 7 min at a flow rate of 4 μl/min with 0.1% (v/v) TFA. Sample was resolved on the analytical column (Easy-Spray C18 75 μm × 400 mm, 2 μm column) using gradient of 97% A (0.1% formic acid) and 3% B (99.9% acetonitrile and 0.1% formic acid) to 60% A and 40% B for 120 min at a flow rate of 300 nL/min. Data-dependent acquisition consisted of a 70,000 resolution full-scan mass spectrometry (MS) (automatic gain control [AGC] set to 106 ions with a maximum fill time of 250 ms) and the 10 most abundant peaks were selected for MS/MS using a 17,000 resolution scan (AGC set to 5 × 104 ions with a maximum fill time of 250 ms) with an ion selection window of 3 m/z and normalized collision energy of 30. Repeated selection of peptides for MS/MS was avoided by a 30 s dynamic exclusion window.
Raw spectra were converted to mascot generic files (MGFs) using Proteome Discoverer software (Thermo) and resulting MGFs were searched against the UniProt mouse sequence database using an in-house Mascot server (Matrix Science, London, United Kingdom). Search parameters used were peptide mass tolerance 10 ppm; fragment mass tolerance 0.01 Da, 1+, 2+, 3 = ions; missed cleavages 1; and instrument types ESI-TRAP. Variable modifications included in search were d(0) NEM, d(5) NEM, mon-, di-, trioxidation of Cys residues and oxidation of methionine with a false discovery rate of <1%. Label-free relative quantification was performed using PEAKS7 software (Bioinformatics Solutions, Inc., Waterloo, Canada). Label-free quantification data were imported into Perseus (64) software for generation of multiscatter plots and heatmaps. Heatmaps represent proteins that significantly change between genotypes using a −10log p-value >20 (equivalent to a p-value <0.01). The multiscatter plots (Supplementary Figs. S5 and S6) and identified proteins and their relative quantification (Supplementary Data S1–S4) are shown in Supplementary Data. Differential labeling of Cys residues allowed a targeted analysis of specific Cys residues identified as labeled with both d(0) NEM and d(5) NEM using Skyline (29), where m/z, retention times, and fragmentation spectra allowed the estimation of the d(0)/d(5) NEM or reduced:reversibly oxidized ratio for redox Cys residues from the individual parent ion intensities. Visualization of label-free proteomic data using volcano plots (maximum fold change ≤20) was performed using label-free quantification data in R studio. Pathway analysis of label-free quantitative proteomic data was performed using PathVisio (25) together with WikiPathways (24) to observe and highlight altered pathways from detected proteins.
Enzymatic activity assays
Enzymatic activity of CuZnSOD was assessed in native gels, with negative staining (Fig. 1C), as described previously (49, 51, 70). In brief, single muscle fibers isolated from the FDB muscle were homogenized in 20 mM Tris buffer (pH 7.4 at 4°C) supplemented with 0.05% Triton X-100 and protease inhibitors. After centrifugation at 15,800 g for 10 min at 4°C, protein of the supernatant was quantified using the bicinchoninic acid method following standard protocol (8). Equal amount of protein (20 μg) was separated on 10% native gel (Criterion Tris HCL precast gel; Bio-Rad) in a cold room (4°C) at 150 V for 4 h. The gel was soaked in a solution that contained nitro blue tetrazolium (NBT), riboflavin, and tetramethylethylenediamine (TEMED). In this process, TEMED reduces riboflavin, which then reduces O2, thereby forming superoxide and the superoxide reduces NBT, which forms the characteristic color of a blue formazan. Regions of SOD activity in the gel are colorless since SOD prevents the reduction of NBT. The gel image was recorded with a digital camera-imager system (Bio-Rad) and analyzed using Quantity One Software (Bio-Rad) to quantify the intensity of the regions representing CuZnSOD activity.
Proteasomal activity was quantified fluorometrically as previously described (7, 48). Skeletal muscle homogenates were lysed by three freeze–thaw cycles in ice-cold lysis buffer containing (in mM) 250 saccharose, 25 hepes, 10 MgCl2-6H2O, 1 EDTA (pH 7.8 at 4°C), supplemented with 1.6 dithiothreitol (DTT). Lysates were centrifuged at 15,100 g for 10 min at 4°C to remove nonlysed cells, membranes, and nuclei. Supernatants were incubated in proteolysis assay buffer (in mM) 225 Tris, 6 MgOAc-4H2O, 7.5 MgCl2-6H2O, and 45 KCl (pH 8.2 at 4°C), supplemented with 1.6 DTT. The fluorogenic peptide succinyl-LLVY-methylcoumarin (Suc-LLVY-AMC) (200 μM) was used as substrate to measure chymotrypsin-like activity of the proteasome. After 40 min of incubation at 37°C, release of methyl coumarin was measured using a microplate fluorometer (FLUOstar Optima; BMG) at excitation/emission wavelengths of 360/460 nm and activity calculated using free methyl coumarin as standards. To exclude other protease activities and ensure the specificity of the assay, the selective proteasome inhibitor lactacystin (20 μM final concentration) was added to the reaction mixture, and proteasomal activity was calculated as the difference between the total activity and that remaining in the presence of lactacystin and normalized to the protein concentration of each sample. For assessment of ATP-stimulated proteolysis (26S proteasome), ATP was added to 5 mM final concentration.
Immunoblotting and subcellular fractionation
Homogenates were prepared using RIPA buffer containing (in mM) 50 Tris (pH 7.4 at 4°C), 150 NaCl, and protease inhibitors. Equal amounts of protein (20−50 μg) were separated by SDS-PAGE and transferred onto nitrocellulose membranes (53). Membranes were stained with Ponceau S to ensure equivalent protein loading and transfer. Relative protein abundance was analyzed densitometrically and standardized using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or Ponceau S staining. Membranes were probed using the primary antibodies listed in Supplementary Table S1. An ECL kit was used to monitor peroxidase activity (Amersham International), and band intensities were analyzed using Quantity One Software (Bio-Rad). Mitochondrial and cytosolic fractions were obtained from skeletal muscle as previously described (9).
Measurement of protein carbonyls, 3-nitrotyrosine, 4-HNE protein adducts, and MDA content
3-Nitrotyrosine (3-NT) and 4-HNE were analyzed by IHC and immunoblotting. Cryosections (10 μm) from AT muscle were immunolabeled for 3-NT and 4-HNE, and staining intensity was measured by confocal laser scanning microscopy. Protein carbonyls, 3-NT, and MDA content in muscle lysates containing 20 μg protein were assessed with the OxiSelect protein carbonyl, nitrotyrosine, and MDA immunoblot kits (Cell Biolabs), respectively. Intensities of 2,4-dinitrophenyl-modified proteins after derivatization, 3-NT, 4-HNE protein adducts, and MDA content were quantified by densitometry.
Statistical analyses
Data are presented as mean ± SEM for each experiment. Statistical analyses for differences between groups were determined using analysis of variance followed by the post hoc least significant difference test. Single comparisons between two experimental conditions were made using an unpaired Student's t-test. Data were analyzed using SPSS 22 with p-values of <0.05 being considered statistically significant.
Supplementary Material
Abbreviations Used
- 3-NT
3-nitrotyrosine
- 4-HNE
4-hydroxynonenal
- A1AT
alpha-1 antitrypsin
- AA
arachidonic acid
- AChRs
acetylcholine receptors
- AT
anterior tibialis
- CAH2/3
carbonic anhydrases 2 and 3
- CAT
catalase
- cPLA2
cytosolic phospholipase A2
- CSA
cross-sectional area
- CX32
connexin 32/GJB1
- Cys
cysteine
- DTT
dithiothreitol
- eNOS
endothelial nitric oxide synthase
- FDB
flexor digitorum brevis
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GPX1
glutathione peroxidase 1
- GTN
gastrocnemius
- H2O2
hydrogen peroxide
- HSP
heat shock protein
- IHC
immunohistochemical
- MAG
myelin-associated glycoprotein
- MBP
myelin basic protein
- MDA
malondialdehyde
- MGFs
mascot generic files
- MHC
myosin heavy chain
- MnSOD
mitochondrial superoxide scavenging system
- MPZ
myelin protein zero
- mSod1KO mice
mice with specific CuZnSOD gene deletion targeted to skeletal muscle
- NBT
nitro blue tetrazolium
- NEM
N-ethylmaleimide
- NFκB
nuclear factor kappa B
- NMJ
neuromuscular junction
- nSod1KO mice
mice with specific CuZnSOD gene deletion targeted to motor neurons
- PBS
phosphate buffered saline
- PFA
paraformaldehyde
- PMP22
peripheral myelin protein 22
- PNS
peripheral nervous system
- PRX
peroxiredoxin
- RONS
reactive oxygen and nitrogen species
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SN
sciatic nerve
- SOD
superoxide dismutase
- Sod1−/− mice
CuZnSOD homozygous knockout mice
- Suc-LLVY-AMC
succinyl-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin
- TEMED
tetramethylethylenediamine
- TFA
Trifluoroacetic acid
- WT
wild type
Acknowledgments
We thank Dr. Betul Karademir (Marmara University, Istanbul, Turkey) for excellent technical advice and for providing the protocol used to assess proteasomal activity. This work was supported by the United States National Institute on Aging (Grant No: AG-20591). Proteomics was performed at the Centre of Proteome Research, University of Liverpool with excellent technical help from Dr. Philip Brownridge.
Author Disclosure Statement
No competing financial interests exist.
Authors Contributions
G.K.S. carried out tissue dissections, immunohistochemical analysis, confocal laser scanning microscopy, and isolation of FDB skeletal muscle fibers. G.A.N. assessed tissue weights. G.K.S., H.P., I.I.G., A.V., and G.A.N. performed morphological and histological analyses. B.M.D. and G.K.S carried out protein extraction for redox proteomics. B.M.D. performed pathway analysis, global label-free and differential Cys labeling redox proteomic analysis. G.K.S and K.E.E. carried out enzymatic activity experiments. G.K.S., H.P., and K.E.E. performed protein analysis. A.R. and H.V.R generated the transgenic mice. G.K.S., B.M.D., H.P., I.I.G., K.E., A.V., and G.A.N. were involved in data analysis. G.K.S., B.M.D., A.M., A.R., H.V.R., S.V.B., and M.J.J. designed the study. G.K.S., B.M.D., and M.J.J. wrote the article. All authors discussed the results and commented on the article.
References
- 1.Bhattacharya A, Hamilton R, Jernigan A, Zhang Y, Sabia M, Rahman MM, Li Y, Wei R, Chaudhuri A, and Van Remmen H. Genetic ablation of 12/15-lipoxygenase but not 5-lipoxygenase protects against denervation-induced muscle atrophy. Free Radic Biol Med 67: 30–40, 2014 [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharya A, Muller FL, Liu Y, Sabia M, Liang H, Song W, Jang YC, Ran Q, and Van Remmen H. Denervation induces cytosolic phospholipase A2-mediated fatty acid hydroperoxide generation by muscle mitochondria. J Biol Chem 284: 46–55, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonaldo P. and Sandri M. Cellular and molecular mechanisms of muscle atrophy. Dis Model Mech 6: 25–39, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bronstein JM. Function of tetraspan proteins in the myelin sheath. Curr Opin Neurobiol 10: 552–557, 2000 [DOI] [PubMed] [Google Scholar]
- 5.Broome CS, Kayani AC, Palomero J, Dillmann WH, Mestril R, Jackson MJ, and McArdle A. Effect of lifelong overexpression of HSP70 in skeletal muscle on age-related oxidative stress and adaptation after nondamaging contractile activity. FASEB J 20: 1549–1551, 2006 [DOI] [PubMed] [Google Scholar]
- 6.Butikofer L, Zurlinden A, Bolliger MF, Kunz B, and Sonderegger P. Destabilization of the neuromuscular junction by proteolytic cleavage of agrin results in precocious sarcopenia. FASEB J 25: 4378–4393, 2011 [DOI] [PubMed] [Google Scholar]
- 7.Catalgol B, Ziaja I, Breusing N, Jung T, Hohn A, Alpertunga B, Schroeder P, Chondrogianni N, Gonos ES, Petropoulos I, Friguet B, Klotz LO, Krutmann J, and Grune T. The proteasome is an integral part of solar ultraviolet a radiation-induced gene expression. J Biol Chem 284: 30076–30086, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cobley JN, Sakellariou GK, Owens DJ, Murray S, Waldron S, Gregson W, Fraser WD, Burniston JG, Iwanejko LA, McArdle A, Morton JP, Jackson MJ, and Close GL. Lifelong training preserves some redox-regulated adaptive responses after an acute exercise stimulus in aged human skeletal muscle. Free Radic Biol Med 70: 23–32, 2014 [DOI] [PubMed] [Google Scholar]
- 9.Dimauro I, Pearson T, Caporossi D, and Jackson MJ. A simple protocol for the subcellular fractionation of skeletal muscle cells and tissue. BMC Res Notes 5: 513, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elchuri S, Oberley TD, Qi W, Eisenstein RS, Jackson Roberts L, Van Remmen H, Epstein CJ, and Huang TT. CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene 24: 367–380, 2005 [DOI] [PubMed] [Google Scholar]
- 11.Elliott DA, Tsoi K, Holinkova S, Chan SL, Kim WS, Halliday GM, Rye KA, and Garner B. Isoform-specific proteolysis of apolipoprotein-E in the brain. Neurobiol Aging 32: 257–271, 2011 [DOI] [PubMed] [Google Scholar]
- 12.Fischer LR, Igoudjil A, Magrane J, Li Y, Hansen JM, Manfredi G, and Glass JD. SOD1 targeted to the mitochondrial intermembrane space prevents motor neuropathy in the Sod1 knockout mouse. Brain 134: 196–209, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher AB. Peroxiredoxin 6 in the repair of peroxidized cell membranes and cell signaling. Arch Biochem Biophys 617: 68–83, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia J, Han D, Sancheti H, Yap LP, Kaplowitz N, and Cadenas E. Regulation of mitochondrial glutathione redox status and protein glutathionylation by respiratory substrates. J Biol Chem 285: 39646–39654, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grune T, Reinheckel T, and Davies KJ. Degradation of oxidized proteins in K562 human hematopoietic cells by proteasome. J Biol Chem 271: 15504–15509, 1996 [DOI] [PubMed] [Google Scholar]
- 16.Hepple RT. and Rice CL. Innervation and neuromuscular control in ageing skeletal muscle. J Physiol 594: 1965–1978, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang TT, Yasunami M, Carlson EJ, Gillespie AM, Reaume AG, Hoffman EK, Chan PH, Scott RW, and Epstein CJ. Superoxide-mediated cytotoxicity in superoxide dismutase-deficient fetal fibroblasts. Arch Biochem Biophys 344: 424–432, 1997 [DOI] [PubMed] [Google Scholar]
- 18.Jackson MJ. and McArdle A. Role of reactive oxygen species in age-related neuromuscular deficits. J Physiol 594: 1979–1988, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jang YC, Lustgarten MS, Liu Y, Muller FL, Bhattacharya A, Liang H, Salmon AB, Brooks SV, Larkin L, Hayworth CR, Richardson A, and Van Remmen H. Increased superoxide in vivo accelerates age-associated muscle atrophy through mitochondrial dysfunction and neuromuscular junction degeneration. FASEB J 24: 1376–1390, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, and Barnes DE. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci U S A 96: 13300–13305, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knoops B, Goemaere J, Van der Eecken V, and Declercq JP. Peroxiredoxin 5: structure, mechanism, and function of the mammalian atypical 2-Cys peroxiredoxin. Antioxid Redox Signal 15: 817–829, 2011 [DOI] [PubMed] [Google Scholar]
- 22.Kuhn G, Lie A, Wilms S, and Muller HW. Coexpression of PMP22 gene with MBP and P0 during de novo myelination and nerve repair. Glia 8: 256–264, 1993 [DOI] [PubMed] [Google Scholar]
- 23.Kulakowski SA, Parker SD, and Personius KE. Reduced TrkB expression results in precocious age-like changes in neuromuscular structure, neurotransmission, and muscle function. J Appl Physiol 111: 844–852, 2011 [DOI] [PubMed] [Google Scholar]
- 24.Kutmon M, Riutta A, Nunes N, Hanspers K, Willighagen EL, Bohler A, Melius J, Waagmeester A, Sinha SR, Miller R, Coort SL, Cirillo E, Smeets B, Evelo CT, and Pico AR. WikiPathways: capturing the full diversity of pathway knowledge. Nucleic Acids Res 44: D488–D494, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kutmon M, van Iersel MP, Bohler A, Kelder T, Nunes N, Pico AR, and Evelo CT. PathVisio 3: an extendable pathway analysis toolbox. PLoS Comput Biol 11: e1004085, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larkin LM, Davis CS, Sims-Robinson C, Kostrominova TY, Remmen HV, Richardson A, Feldman EL, and Brooks SV. Skeletal muscle weakness due to deficiency of CuZn-superoxide dismutase is associated with loss of functional innervation. Am J Physiol Regul Integr Comp Physiol 301: R1400–R1407, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larsson JE. and Wahlstrom G. The influence of age and administration rate on the brain sensitivity to propofol in rats. Acta Anaesthesiol Scand 42: 987–994, 1998 [DOI] [PubMed] [Google Scholar]
- 28.Lustgarten MS, Jang YC, Liu Y, Qi W, Qin Y, Dahia PL, Shi Y, Bhattacharya A, Muller FL, Shimizu T, Shirasawa T, Richardson A, and Van Remmen H. MnSOD deficiency results in elevated oxidative stress and decreased mitochondrial function but does not lead to muscle atrophy during aging. Aging Cell 10: 493–505, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, and MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26: 966–968, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marino SM. and Gladyshev VN. Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. J Mol Biol 404: 902–916, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martindale JL. and Holbrook NJ. Cellular response to oxidative stress: signaling for suicide and survival. J Cell Physiol 192: 1–15, 2002 [DOI] [PubMed] [Google Scholar]
- 32.Martini R. and Schachner M. Molecular bases of myelin formation as revealed by investigations on mice deficient in glial cell surface molecules. Glia 19: 298–310, 1997 [PubMed] [Google Scholar]
- 33.McDonagh B, Pedrajas JR, Padilla CA, and Barcena JA. Thiol redox sensitivity of two key enzymes of heme biosynthesis and pentose phosphate pathways: uroporphyrinogen decarboxylase and transketolase. Oxid Med Cell Longev 2013: 932472, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonagh B, Sakellariou GK, and Jackson MJ. Application of redox proteomics to skeletal muscle aging and exercise. Biochem Soc Trans 42: 965–970, 2014 [DOI] [PubMed] [Google Scholar]
- 35.McDonagh B, Sakellariou GK, Smith NT, Brownridge P, and Jackson MJ. Differential cysteine labeling and global label-free proteomics reveals an altered metabolic state in skeletal muscle aging. J Proteome Res 13: 5008–5021, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDonagh B, Sakellariou GK, Smith NT, Brownridge P, and Jackson MJ. Redox proteomic analysis of the gastrocnemius muscle from adult and old mice. Data Brief 4: 344–348, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDonagh B, Scullion SM, Vasilaki A, Pollock N, McArdle A, and Jackson MJ. Ageing-induced changes in the redox status of peripheral motor nerves imply an effect on redox signalling rather than oxidative damage. Free Radic Biol Med 94: 27–35, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melcangi RC, Magnaghi V, Cavarretta I, Martini L, and Piva F. Age-induced decrease of glycoprotein Po and myelin basic protein gene expression in the rat sciatic nerve. Repair by steroid derivatives. Neuroscience 85: 569–578, 1998 [DOI] [PubMed] [Google Scholar]
- 39.Melcangi RC, Magnaghi V, and Martini L. Aging in peripheral nerves: regulation of myelin protein genes by steroid hormones. Prog Neurobiol 60: 291–308, 2000 [DOI] [PubMed] [Google Scholar]
- 40.Miro O, Casademont J, Casals E, Perea M, Urbano-Marquez A, Rustin P, and Cardellach F. Aging is associated with increased lipid peroxidation in human hearts, but not with mitochondrial respiratory chain enzyme defects. Cardiovasc Res 47: 624–631, 2000 [DOI] [PubMed] [Google Scholar]
- 41.Muller FL, Song W, Jang YC, Liu Y, Sabia M, Richardson A, and Van Remmen H. Denervation-induced skeletal muscle atrophy is associated with increased mitochondrial ROS production. Am J Physiol Regul Integr Comp Physiol 293: R1159–R1168, 2007 [DOI] [PubMed] [Google Scholar]
- 42.Muller FL, Song W, Liu Y, Chaudhuri A, Pieke-Dahl S, Strong R, Huang TT, Epstein CJ, Roberts LJ, 2nd, Csete M, Faulkner JA, and Van Remmen H. Absence of CuZn superoxide dismutase leads to elevated oxidative stress and acceleration of age-dependent skeletal muscle atrophy. Free Radic Biol Med 40: 1993–2004, 2006 [DOI] [PubMed] [Google Scholar]
- 43.Nguyen HX. and Tidball JG. Expression of a muscle-specific, nitric oxide synthase transgene prevents muscle membrane injury and reduces muscle inflammation during modified muscle use in mice. J Physiol 550: 347–356, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okuda T, Higashi Y, Kokame K, Tanaka C, Kondoh H, and Miyata T. Ndrg1-deficient mice exhibit a progressive demyelinating disorder of peripheral nerves. Mol Cell Biol 24: 3949–3956, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pedrajas JR, McDonagh B, Hernandez-Torres F, Miranda-Vizuete A, Gonzalez-Ojeda R, Martinez-Galisteo E, Padilla CA, and Barcena JA. Glutathione is the resolving thiol for thioredoxin peroxidase activity of 1-Cys peroxiredoxin without being consumed during the catalytic cycle. Antioxid Redox Signal 24: 115–128, 2016 [DOI] [PubMed] [Google Scholar]
- 46.Perez VI, Bokov A, Van Remmen H, Mele J, Ran Q, Ikeno Y, and Richardson A. Is the oxidative stress theory of aging dead? Biochim Biophys Acta 1790: 628–638, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Powers SK. and Jackson MJ. Exercise-induced oxidative stress: cellular mechanisms and impact on muscle force production. Physiol Rev 88: 1243–1276, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reinheckel T, Ullrich O, Sitte N, and Grune T. Differential impairment of 20S and 26S proteasome activities in human hematopoietic K562 cells during oxidative stress. Arch Biochem Biophys 377: 65–68, 2000 [DOI] [PubMed] [Google Scholar]
- 49.Sakellariou GK, Davis CS, Shi Y, Ivannikov MV, Zhang Y, Vasilaki A, Macleod GT, Richardson A, Van Remmen H, Jackson MJ, McArdle A, and Brooks SV. Neuron-specific expression of CuZnSOD prevents the loss of muscle mass and function that occurs in homozygous CuZnSOD-knockout mice. FASEB J 28: 1666–1681, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakellariou GK, Jackson MJ, and Vasilaki A. Redefining the major contributors to superoxide production in contracting skeletal muscle. The role of NAD(P)H oxidases. Free Radic Res 48: 12–29, 2014 [DOI] [PubMed] [Google Scholar]
- 51.Sakellariou GK, Pearson T, Lightfoot AP, Nye GA, Wells N, Giakoumaki II, Griffiths RD, McArdle A, and Jackson MJ. Long-term administration of the mitochondria-targeted antioxidant mitoquinone mesylate fails to attenuate age-related oxidative damage or rescue the loss of muscle mass and function associated with aging of skeletal muscle. FASEB J 30: 3771–3785, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakellariou GK, Pearson T, Lightfoot AP, Nye GA, Wells N, Giakoumaki II, Vasilaki A, Griffiths RD, Jackson MJ, and McArdle A. Mitochondrial ROS regulate oxidative damage and mitophagy but not age-related muscle fiber atrophy. Sci Rep 6: 33944, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakellariou GK, Pye D, Vasilaki A, Zibrik L, Palomero J, Kabayo T, McArdle F, Van Remmen H, Richardson A, Tidball JG, McArdle A, and Jackson MJ. Role of superoxide-nitric oxide interactions in the accelerated age-related loss of muscle mass in mice lacking Cu,Zn superoxide dismutase. Aging Cell 10: 749–760, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sakellariou GK, Vasilaki A, Palomero J, Kayani A, Zibrik L, McArdle A, and Jackson MJ. Studies of mitochondrial and nonmitochondrial sources implicate nicotinamide adenine dinucleotide phosphate oxidase(s) in the increased skeletal muscle superoxide generation that occurs during contractile activity. Antioxid Redox Signal 18: 603–621, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samuel MA, Valdez G, Tapia JC, Lichtman JW, and Sanes JR. Agrin and synaptic laminin are required to maintain adult neuromuscular junctions. PLoS One 7: e46663, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sandri M, Coletto L, Grumati P, and Bonaldo P. Misregulation of autophagy and protein degradation systems in myopathies and muscular dystrophies. J Cell Sci 126: 5325–5333, 2013 [DOI] [PubMed] [Google Scholar]
- 57.Sataranatarajan K, Qaisar R, Davis C, Sakellariou GK, Vasilaki A, Zhang Y, Liu Y, Bhaskaran S, McArdle A, Jackson M, Brooks SV, Richardson A, and Van Remmen H. Neuron specific reduction in CuZnSOD is not sufficient to initiate a full sarcopenia phenotype. Redox Biol 5: 140–148, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scherer SS, Deschenes SM, Xu YT, Grinspan JB, Fischbeck KH, and Paul DL. Connexin32 is a myelin-related protein in the PNS and CNS. J Neurosci 15: 8281–8294, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schulz A, Kyselyova A, Baader SL, Jung MJ, Zoch A, Mautner VF, Hagel C, and Morrison H. Neuronal merlin influences ERBB2 receptor expression on Schwann cells through neuregulin 1 type III signalling. Brain 137: 420–432, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi Y, Ivannikov MV, Walsh ME, Liu Y, Zhang Y, Jaramillo CA, Macleod GT, and Van Remmen H. The lack of CuZnSOD leads to impaired neurotransmitter release, neuromuscular junction destabilization and reduced muscle strength in mice. PLoS One 9: e100834, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smuder AJ, Kavazis AN, Hudson MB, Nelson WB, and Powers SK. Oxidation enhances myofibrillar protein degradation via calpain and caspase-3. Free Radic Biol Med 49: 1152–1160, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tochhawng L, Deng S, Pugalenthi G, Kumar AP, Lim KH, Yang H, Hooi SC, Goh YC, Maciver SK, Pervaiz S, and Yap CT. Gelsolin-Cu/ZnSOD interaction alters intracellular reactive oxygen species levels to promote cancer cell invasion. Oncotarget 7: 52832–52848, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Todd B, Moore D, Deivanayagam CC, Lin GD, Chattopadhyay D, Maki M, Wang KK, and Narayana SV. A structural model for the inhibition of calpain by calpastatin: crystal structures of the native domain VI of calpain and its complexes with calpastatin peptide and a small molecule inhibitor. J Mol Biol 328: 131–146, 2003 [DOI] [PubMed] [Google Scholar]
- 64.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, and Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13: 731–740, 2016 [DOI] [PubMed] [Google Scholar]
- 65.Vasilaki A, Pollock N, Giakoumaki I, Goljanek-Whysall K, Sakellariou GK, Pearson T, Kayani A, Jackson MJ, and McArdle A. The effect of lengthening contractions on neuromuscular junction structure in adult and old mice. Age (Dordr) 38: 259–272, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vasilaki A, van der Meulen JH, Larkin L, Harrison DC, Pearson T, Van Remmen H, Richardson A, Brooks SV, Jackson MJ, and McArdle A. The age-related failure of adaptive responses to contractile activity in skeletal muscle is mimicked in young mice by deletion of Cu,Zn superoxide dismutase. Aging Cell 9: 979–990, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vazquez-Medina JP, Dodia C, Weng L, Mesaros C, Blair IA, Feinstein SI, Chatterjee S, and Fisher AB. The phospholipase A2 activity of peroxiredoxin 6 modulates NADPH oxidase 2 activation via lysophosphatidic acid receptor signaling in the pulmonary endothelium and alveolar macrophages. FASEB J 30: 2885–2898, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ward RE, Boudreau RM, Caserotti P, Harris TB, Zivkovic S, Goodpaster BH, Satterfield S, Kritchevsky S, Schwartz AV, Vinik AI, Cauley JA, Newman AB, Strotmeyer ES, and Health ABCs. Sensory and motor peripheral nerve function and longitudinal changes in quadriceps strength. J Gerontol A Biol Sci Med Sci 70: 464–470, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu RF. and Terada LS. Focal oxidant and Ras signaling on the ER surface activates autophagy. Autophagy 6: 828–829, 2010 [DOI] [PubMed] [Google Scholar]
- 70.Zhang Y, Davis C, Sakellariou GK, Shi Y, Kayani AC, Pulliam D, Bhattacharya A, Richardson A, Jackson MJ, McArdle A, Brooks SV, and Van Remmen H. CuZnSOD gene deletion targeted to skeletal muscle leads to loss of contractile force but does not cause muscle atrophy in adult mice. FASEB J 27: 3536–3548, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.