Abstract
UHRF1 is a key mediator of inheritance of epigenetic DNA methylation patterns during cell division and is a putative target for cancer therapy. Recent studies indicate that interdomain interactions critically influence UHRF1's chromatin-binding properties, including allosteric regulation of its histone binding. Here, using an integrative approach that combines small angle X-ray scattering, NMR spectroscopy, and molecular dynamics simulations, we characterized the dynamics of the tandem tudor domain–plant homeodomain (TTD–PHD) histone reader module, including its 20-residue interdomain linker. We found that the apo TTD–PHD module in solution comprises a dynamic ensemble of conformers, approximately half of which are compact conformations, with the linker lying in the TTD peptide–binding groove. These compact conformations are amenable to cooperative, high-affinity histone binding. In the remaining conformations, the linker position was in flux, and the reader adopted both extended and compact states. Using a small-molecule fragment screening approach, we identified a compound, 4-benzylpiperidine-1-carboximidamide, that binds to the TTD groove, competes with linker binding, and promotes open TTD–PHD conformations that are less efficient at H3K9me3 binding. Our work reveals a mechanism by which the dynamic TTD–PHD module can be allosterically targeted with small molecules to modulate its histone reader function for therapeutic or experimental purposes.
Keywords: drug screening, epigenetics, molecular dynamics, molecular modeling, nuclear magnetic resonance (NMR), protein dynamic
Introduction
Epigenetic memory of cell identity requires the faithful propagation of DNA methylation patterns through cell division and is dependent on the function of UHRF1 (ubiquitin-like containing RING and PHD fingers 1). UHRF1 is a pentadomain protein that is targeted to hemimethylated DNA and repressive histone H3K9me3 modification states, where it recruits DNMT1 for the methylation of cytosine residues on daughter strands during DNA replication (for recent reviews of UHRF1, see Refs. 1–3). Dysregulation of DNA methylation is a hallmark of many cancers, and UHRF1 has been suggested as a target for anti-cancer therapy (2, 4). It is overexpressed in multiple cancer lines (5, 6) and thought to play a critical role in the down-regulation of tumor suppressor proteins (2). Importantly, the ability of UHRF1 to maintain DNA methylation patterns is dependent on its H3K9me3-binding function.
Hemimethylated DNA is recognized by the SRA domain (SET and RING-associated), whereas H3K9me3 marked chromatin is recognized by the plant homeodomain (PHD)3 and tandem tudor domain (TTD), which are connected by a 20-residue linker to form the TTD–PHD histone reader module (see Fig. 1A). The PHD in isolation recognizes the unmodified N terminus of H3 (7, 8), whereas the TTD binds to the H3K9me3 mark (9). The crystal structure of H3-bound TTD–PHD (10, 11) shows that the two domains can bind to their target sites cooperatively. This coordinate binding requires the linker to be bound to a peptide-binding groove formed at the interface of the TTDN/TTDC subdomains; disruption of linker-groove contacts prevents high-affinity binding of TTD–PHD to H3K9me3. One putative mechanism to disrupt multivalent binding is by phosphorylation of the linker at Ser298 by PKA or PIM1 (10, 12, 13). It is unclear to what degree cooperative histone interaction occurs in the context of full-length UHRF1. Some studies indicate that the module may adopt histone-binding states that are solely PHD– or TTD–mediated (14, 15).
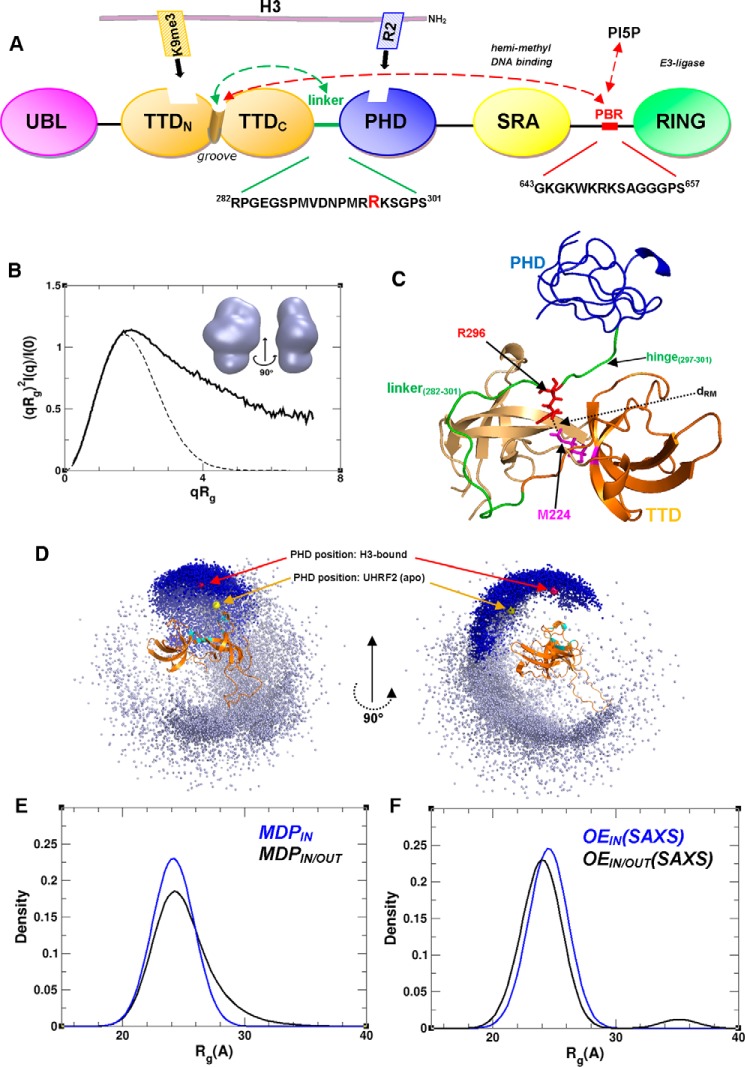
Figure 1.
A, UHRF1 domain arrangement showing the basic function of each domain. The 20-residue TTD–PHD interdomain linker sequence is displayed (with the critical Arg296 in red), as well as that of the PBR, which has been implicated in allosteric regulation of histone binding through its reversible interaction with the TTD groove or PI5P (14). B, comparison of the experimental dimensionless Kratky plot for TTD–PHD (solid line) with the theoretical plot for a globular protein (dashed line). Inset, ab initio SAXS-predicted molecular envelope of the module is shown from two points of view. C, ribbon representation of TTD–PHD based on its structure in the H3-bound state (PDB code 3ASK) (10). The Cα atoms of Arg296 and Met224 are used as reference positions for the linker and groove, respectively, to assess their relative positioning (dRM) in various TTD–PHD conformers. D, the position of PHD centers of mass (calculated as the average position of the atoms in the PHD, weighted according to their mass) in TTD–PHD structures from MDPIN (dark blue spheres) and MDPIN/OUT (dark and light blue spheres) superimposed with the TTD (as a ribbon diagram). TTD residues that bind to the H3 peptide are displayed in cyan. The red sphere shows the PHD center of mass in the H3-bound UHRF1 TTD–PHD (PDB code 3ASK) (10). The yellow sphere shows the PHD center of mass in apo TTD–PHD of UHRF2 (crystal structure, PDB code 4TVR). E, Rg distribution in MDPIN/OUT and MDPIN. F, Rg distribution in OEIN/OUT(SAXS) and OEIN(SAXS).
Histone and DNA binding by UHRF1 is regulated by long-range interdomain and linker-domain interactions within the full-length protein (14–17). Gelato et al. (14) showed that a polybasic region (PBR-UHRF1643–657) in the linker between the SRA and RING (really interesting and new gene) domains regulates the transition between PHD- and TTD–mediated histone reader states, through its reversible binding to the TTD groove or the phospholipid PI5P (see Fig. 1A). The PHD has also been reported to interact with the SRA domain in a UHRF1 state where histone binding is restricted (15, 16). These studies suggest that disruption of interdomain interactions could be a mechanism to pharmacologically target UHRF1. A detailed structural and dynamic picture of how these putative large-scale intramolecular rearrangements give rise to altered UHRF1-binding states remains elusive.
In this report we describe an integrated biophysical approach to characterize the scope of interdomain motion exhibited by the TTD–PHD histone reader module and its recognition of H3K9me3. In its apo form, we find that the unit is highly dynamic, populated with both extended and compact states. Using a fragment-based drug discovery approach, we identified a compound, 4-benzylpiperidine-1-carboximidamide (BPC) that binds to the open conformation, thereby preventing efficient recognition of H3K9me3.
Results
The UHRF1 TTD–PHD reader is highly dynamic in solution
To assess the conformational heterogeneity of the TTD–PHD histone reader module, we used small angle X-ray scattering (SAXS) in solution. The SAXS-derived ab initio molecular envelope is extended in shape with dimensions of ∼87 × 47 × 23 Å (Fig. 1B). The dimensionless Kratky plot is bell-shaped, with a maximum at a position that is shifted to higher coordinate values than expected for a globular protein and with poor convergence at high q values; this indicates the presence of flexibility/disorder. Furthermore, the average Rg value (24.5 Å) (Table 1) is more than 25% larger than the theoretical value expected for a globular protein of the same mass.
Table 1.
SAXS parameters for UHRF1 TTD–PHD with and without the presence of BPC
| SAXS parameters | TTD–PHDa | TTD–PHD/BPCb |
|---|---|---|
| I(0)c | 0.057 | 0.021 |
| Rg (Å)d | 24.5 | 27.1 |
| Rg (Å) reale | 24.9 | 27.2 |
| Dmax (Å)f | 83 | 94 |
| Vcg | 274.3 | 284.4 |
| Mwh | 25.5 (27.9) | 24.2 (28.1) |
| NSD (SAXS envelope)i | 0.75 ± 0.07 | 0.79 ± 0.02 |
a UHRF1126–366.
b UHRF1126–366 with BPC (4 mm, containing 4% DMSO).
c Intensity at q = 0.
d Based on Guinier fit.
e From GNOM (36).
f Maximum distance between atoms from GNOM.
g Volume of correlation (37).
h Mw estimated from SAXS using Vc (37). The Mw expected from the sequence is shown in parentheses.
i NSD, normalized spatial discrepancy. The values are the average and standard deviation from 15 runs of DAMMIF (38).
In the published crystal structure of H3K9me3-bound TTD–PHD, the relative orientation of the two domains is fixed and stabilized by independent TTD and PHD interactions with the histone peptide and through the binding of the linker within the TTD groove (10), for which Arg296 is a key residue (Fig. 1C). Our SAXS analysis indicates that there is no specific orientation of the two domains relative to one another in the apo state. The level of interdomain motion within the reader module depends on conformational flexibility mediated by the linker and can be principally defined by two distinct types of mobility. In the first case, the Arg296-containing linker is bound within the TTD groove, and the mobility of the two domains is mediated largely by a 5-residue flexible “hinge” region (UHRF1297–301) (Fig. 1C). In the second case, the entire linker (UHRF1282–301) is flexible and exists in both TTD-bound and unbound states. The degree to which these two types of mobility are reflected in the dynamic behavior of the TTD–PHD unit was explored.
We employed an ensemble fitting approach in which molecular dynamics combined with rigid-body modeling was used to generate an initial set of conformations that approximate the conformational space available to the TTD–PHD reader. Then using the SES method (18), SAXS and NMR relaxation data were used to identify the dominant conformational states within the structural pool. We generated two initial sets of TTD–PHD conformations. The first set (molecular dynamics pool-IN (MDPIN)) contains 6,000 TTD–PHD conformations generated with the linker bound to the TTD groove. The second set (MDPIN/OUT) contains the entire MDPIN pool and an additional 10,000 conformations with the linker displaced from the groove (Fig. 1D). SAXS data fitting was used to generate two optimal ensembles: OEIN(SAXS) and OEIN/OUT(SAXS), from their respective starting structural pools. Both ensembles fit the SAXS data equally well (χsaxs = 0.21). The Rg distribution of OEIN(SAXS) has a broad peak centered at Rg = 24.5 Å, similar to its initial starting pool (MDPIN), whereas the OEIN/OUT(SAXS) displays a bimodal Rg distribution, with major (Rg = 24.0 Å) and minor (Rg = 35 Å) peaks corresponding to compact and extended reader conformations (Fig. 1, E and F). The SAXS-fitted OEs largely reproduce the conformational space of their starting pools, but in comparison to each other, do not overlap, as would be expected if the TTD–PHD unit possessed restricted flexibility (supplemental Fig. S1). Our fitting of the SAXS data suggests that OEIN/OUT(SAXS) reflects the dynamic range of TTD–PHD in solution more accurately than OEIN(SAXS) and provides evidence for TTD–PHD states where the groove is exposed.
NMR relaxation data reflect partially coupled TTD–PHD interdomain motion
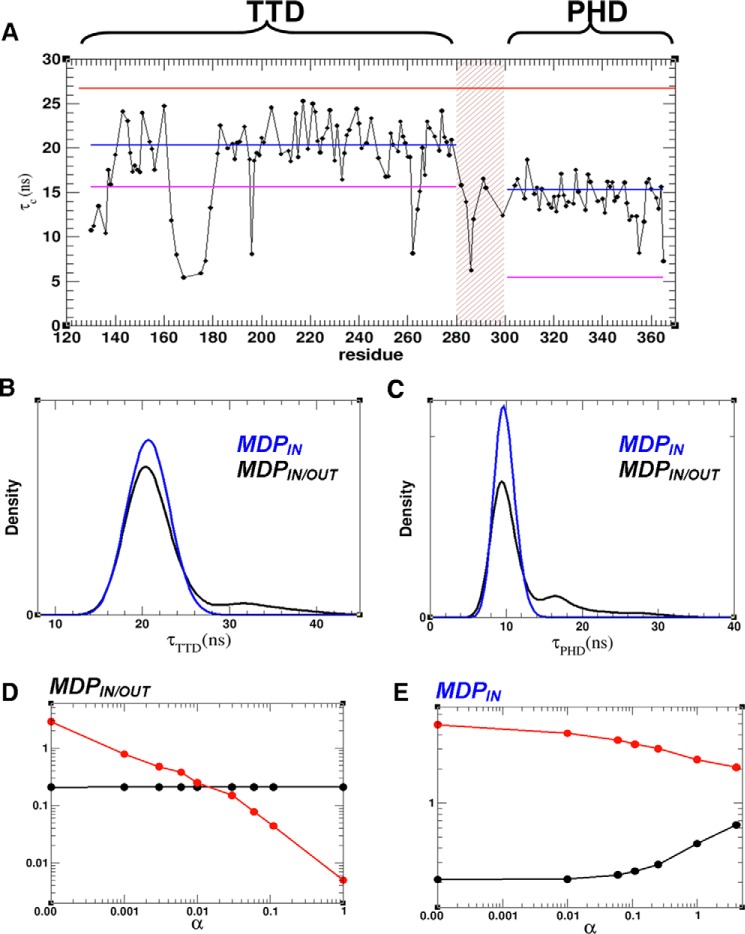
TROSY-based 1H/15N/13C triple-resonance backbone spectra were acquired to assign backbone TTD–PHD resonances (in a ∼80% deuterated sample). We could assign 203 amide resonances, of which 134 were located in the TTD, 11 in the linker and 58 in the PHD (supplemental Fig. S2). The assignments were used as the basis for residue-specific 15N relaxation and 1H-15N heteronuclear NOE measurements (supplemental Fig. S2 and supplemental Table S2). NOE values averaged across the ordered parts of the two domains are approximately the same (0.73 ± 0.06), with reduced values in loops (e.g. residues 163–180) and in the linker, indicating higher flexibility for these regions. The two domains exhibit different T1 and T2 values, and this is most evident comparing their local correlation time (τc) determined for each residue (from T1/T2 ratios) (Fig. 2A and supplemental Table S2). The results of fitting NMR relaxation data to a standard diffusion model using the program MODELFREE (19) are shown in Table 2. The data fit better when the TTD and PHD are considered individually rather than as a single unit with the two domains rigidly attached (see reduced χ2 in Table 2), which indicates the presence of interdomain flexibility. The best fit for both domains was obtained with an axial symmetric diffusion model where τm = 20.32 ± 0.65 ns for the TTD, and τm = 15.36 ± 0.81 ns for the PHD (Table 2 and Fig. 2A). The domains are tumbling with different rates; however, the rotational correlation times values predicted by HYDRONMR (20) indicate that the domain motion is coupled (supplemental Table S3 and Fig. 2A).
Figure 2.
Interdomain flexibility of the TTD–PHD module from NMR relaxation and SAXS data. A, local correlation time (τc) determined for each residue from experimental T1/T2 ratios (black spheres). The apparent global correlation times of the two domains derived by fitting to a standard diffusion model for each domain separately (blue horizontal line). The theoretical τc values predicted by HYDRONMR (20) for full-length TTD–PHD (red horizontal line) and for individual domains separately (magenta horizontal line). B and C, distribution of HYCUD-predicted (22) global τc values in MDPIN/OUT and MDPIN for the TTD (B) and PHD (C). D and E, goodness of fit of MDPIN/OUT (D) and MDPIN (E) to SAXS (χsaxs, black circles) and NMR (χrelax, red circles) data shown as a function of the weighting factor α. Ensemble fitting to both SAXS and NMR data were performed simultaneously by minimizing residual χ2 = χsaxs2 + α · χrelax2 at different values of α.
Table 2.
Rotational diffusion parameters of the TTD and PHD within UHRF1 TTD–PHD
The diffusion parameters were determined by fitting the known crystal structure (PDB code 3ASK) to 15N-relaxation data (acquired at 800 MHz) using Modelfree4.15 (19). Fitting was performed for the full-length construct and each domain individually.
| TTD | PHD | TTD–PHDa | |
|---|---|---|---|
| τm (ns)b | 20.32 ± 0.65 | 15.36 ± 0.81 | 15.65 ± 0.07 |
| Anisotropyc | 1.17 ± 0.05 | 1.41 ± 0.06 | 1.37 ± 0.04 |
| Reduced (c2)d | 4.4 | 3.9 | 23.1 |
a UHRF1126–366.
b Overall rotational correlation time, τm = 1/2(Dpar + 2Dper), where Dpar and Dper are parallel and perpendicular principal values of the axially symmetric rotational diffusion tensor, respectively.
c Degree of anisotropy of diffusion tensor, Dpar/D2per.
d Goodness of fit.
The HYCUD approach was recently developed to predict rotational correlation times of globular domains within flexible modular systems (21, 22). We used our two MD-generated pools of TTD–PHD structures (MDPIN and MDPIN/OUT) to carry out HYCUD-based predictions of the effective rotational correlation times of the TTD (τTTD) and PHD (τPHD). Using MDPIN, both τPHD and τTTD-predicted distributions are bell-shaped curves with peaks positioned at ∼ 10 and 21 ns, respectively (Fig. 2, B and C), whereas the distributions derived from predictions using MDPIN/OUT are right-skewed with the major peaks extending up to ∼35 and ∼45 ns for τPHD and τTTD, respectively. The predicted average τc values for the TTD from both MDPIN/OUT and MDPIN pools are in good agreement with the experimental value, but those for the PHD are smaller and in better agreement with the value predicted from MDPIN/OUT (Tables 2 and 3 and supplemental Table S3).
Table 3.
Global structural parameters and goodness of fit to the experimental data for different ensembles of TTD–PHD
| Data used | OEIN/OUTa |
MDPIN/OUTb | MDPINb | |
|---|---|---|---|---|
| SAXS + NMR | SAXS | |||
| χsaxsc | 0.21 | 0.21 | 0.93 | 0.52 |
| <τTTD> (ns)d | 20.9 ± 2.0 | 21.1 ± 1.7 | 22.0 ± 4.7 | 20.6 ± 1.9 |
| <τPHD> (ns)d | 15.0 ± 8.1 | 9.9 ± 1.3 | 11.9 ± 4.9 | 9.7 ± 1.0 |
| Rg (Å)e | 24.1 ± 2.6 | 24.3 ± 2.4 | 24.8 ± 2.0 | 24.0 ± 0.9 |
| Dmax (Å)e | 83.3 ± 8.4 | 85.6 ± 6.8 | 85.4 ± 7.9 | 85.8 ± 6.4 |
| Pboundf (%) | 51 | 57 | 40 | 100 |
a OEs selected from MDPIN/OUT using the SES method (18).
b MDPIN/OUT and MDPIN with uniform weights.
c Goodness of fit for fitting to SAXS data.
d Average and standard deviation values of the correlation time; τc for each member of the ensemble was predicted using the HYCUD method (21, 22).
e Average and standard deviation.
f Percentage of structures with the linker bound to the TTD groove.
Ensemble fitting using both SAXS and NMR data indicates that the TTD–PHD module adopts compact and extended conformations
Using the HYCUD method, the correlation time (τc) of a domain is calculated as a simple average from predicted τc values for all members of an initial structural ensemble, each with equal weighting. As we demonstrated above, the HYCUD-predicted τPHD and τTTD values do not agree with experimentally determined values. We therefore modified the HYCUD-based prediction algorithm by introducing non-uniform weights for conformers in the ensemble and then used the SES method to optimize and combine SAXS and NMR relaxation data to estimate appropriate weights. For this purpose, the discrepancy between the predicted and experimental data is measured χ2 = χsaxs2 + α · χrelax2, where χsaxs and χrelax measure the goodness of fit to SAXS and relaxation data, respectively (Equation 1), and the parameter α regulates the contribution of the relaxation data (see “Experimental procedures” for details). The results of SES fitting of the initial MDPIN/OUT and MDPIN pools at different values of the parameter α are shown in Fig. 2. In the case of MDPIN/OUT, increasing the value of α from 0 to 1 gradually results in progressive improvement of relaxation data fitting, whereas SAXS data fitting remains the same. At α = ∼0.01, an OE is generated from fitting MDPIN/OUT to the SAXS and NMR data equally well (Fig. 2D). In contrast, we failed to find any value of α, such that MDPIN can be fitted satisfactorily to both SAXS and NMR data (Fig. 2E).
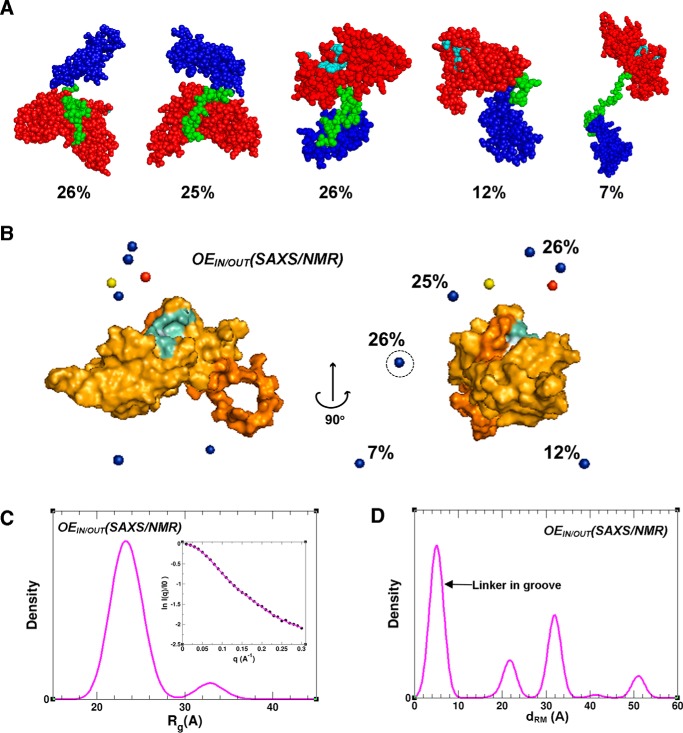
We performed recovery of solution ensembles of the TTD–PHD by fitting MDPIN/OUT to SAXS and NMR data (α = 0.06). The optimal ensemble, OEIN/OUT(SAXS/NMR), fits the SAXS data well (χsaxs = 0.21), and likewise, the predicted τPHD (∼15.0 ns) and τTTD (∼20.9 ns) are in good agreement with experimental values (Tables 2 and 3). The five most populated states comprising 96% of this ensemble are shown in Fig. 3A. The structures can be divided into two groups based on the positioning of the linker. In the first group, which we call the “bound” state, the linker occupies the TTD groove. These conformers cluster around either the observed H3K9me3-bound TTD–PHD crystal structure, or interestingly, the domain arrangement seen in apo TTD–PHD of UHRF2 (Fig. 3B and supplemental Fig. S3). All conformers in the bound state are compact (Fig. 3, C and D), with an Rg of ∼22–24 Å, which corresponds to the major Rg distribution peak. In the second group, that we call the “open” state, the TTD groove is solvent-exposed. In this state, there are both compact and extended conformers that correspond to the major Rg distribution peak (∼24 Å) and to the minor peak centered at ∼33 Å (Fig. 3C). The relative population of bound states in OEIN/OUT(SAXS/NMR) is 51% (Table 3).
Figure 3.
Dynamic ensemble of the TTD–PHD reader in solution derived from SAXS and NMR data. A, the most populated (based on weighted %) conformers from OEIN/OUT(SAXS/NMR). The TTD, PHD, and linker are colored in red, blue, and green, respectively. TTD residues that form the aromatic cage are displayed in cyan. B, the position of PHD centers of mass in the most populated conformers from OEIN/OUT(SAXS/NMR) (blue spheres) superimposed with the TTD (as a surface representation). Residues that bind to the H3 peptide are displayed in cyan. The red sphere shows the PHD center of mass in H3-bound UHRF1 TTD–PHD (PDB code 3ASK) (10), and the yellow sphere shows the PHD center of mass in apo TTD–PHD of UHRF2 (PDB code 4TVR). The average populations (%) are displayed. (See also supplemental Fig. S3 for comparison.) C, Rg distribution in OEIN/OUT(SAXS/NMR). The inset shows the experimental SAXS profile plotted with the theoretical profile averaged over the ensemble. D, the distribution of Cα-Cα distance between residues Arg296 and Met224 (i.e. dRM; refer to Fig. 1C diagram) in OEIN/OUT(SAXS/NMR).
Allosteric modulation of the TTD–PHD module with small molecules
There is strong evidence that the 15-residue PBR (UHRF1643–657) between the SRA and RING domains is involved in the allosteric regulation of TTD–PHD histone binding. This occurs through competitive displacement of the linker from the TTD groove (Fig. 1A). In full-length UHRF1, this results in the failure of UHRF1 to recognize H3K9me3 caused by a transition from TTD–mediated to PHD–mediated histone binding (14). We performed a screen of fragment-sized small molecules to identify compounds that could, in a manner analogous to the PBR, bind to the TTD groove, block linker–groove interactions, and promote open TTD–PHD conformations.
We designed a fragment library containing 2,040 compounds that was initially screened against isolated TTD (UHRF1121–286) using a fluorescence polarization (FP) assay that tracked the displacement of a N-terminally tagged H3K9me3 peptide (H3K9me3(1–25)). From this screen, eight putative TTD-binding hits were identified. Analysis of amide peak movement in the (1H-15N) HSQC spectra of the TTD in the presence of the fragments indicates that the binding of two, BPC and a tricyclic imine, occurs in the groove. Because of limited commercial availability of the tricyclic amine, we focused only on BPC. The binding of this compound occurs close to (or within) the Arg296-binding pocket; we observe significant perturbation of Trp238 and Phe278 resonances that form part of the pocket, and in Gly236, which is directly adjacent to it (Fig. 4A and supplemental Fig. S5). We also see chemical shift changes in non-surface exposed residues that are close to or are in contact with the groove. A similar amide peak perturbation pattern is observed when the TTD is titrated with a 15-residue peptide corresponding to the PBR, resulting from its binding in the groove (supplemental Fig. S5). Further characterization of BPC binding to isolated TTD was performed using ITC and DSF, with an estimated KD of 50 μm (supplemental Fig. S5) and a calculated ligand efficiency of 0.38 (which is defined as the binding energy per heavy atom).
Figure 4.
BPC and PBR peptide binding promote open TTD–PHD conformations. A, BPC (with its chemical structure displayed) binds close to the Arg296-binding pocket. Asp142, Glu153, Ala208, Met224, Trp238, and Phe278 combine to form the pocket in the TTD groove. Residues showing significant chemical shift perturbations upon BPC binding are colored orange, and those that are not assigned in this construct are in cyan. Met224 (blue), at the base of the pocket, is not significantly shifted. B, (1H-15N) TROSY overlays show that titration of TTD–PHD with either BPC or PBR peptide (at ∼250 μm protein/750 μm ligand) show substantial broadening and/or movement of linker resonances (boxed). Some TTD residues (brackets) at the interface of the groove (e.g. Gly236) are shifted or broadened as well. PHD resonances remain largely unaffected by binding (ovals). For clarity, only well resolved resonances are labeled. C, normalized pair distance distribution function P(r) calculated from SAXS data for TTD–PHD bound to BPC (4 mm, 4% DMSO) and for apo TTD–PHD. D, comparison of the Rg distribution in OEIN/OUT(SAXS) and OEBPC(SAXS) derived from SAXS data for apo and BPC-bound TTD–PHD, respectively. E, distribution of dRM (refer to Fig. 1C diagram) in the OEIN/OUT(SAXS) and OEBPC(SAXS). F, ITC curves show a cooperative profile for TTD–PHD binding to H3K9me3(1–15). G and H, the presence of BPC (at 1.5 mm, 4% DMSO, ∼ 30:1 fragment:protein) (G) or PBR peptide (at 1.5 mm, ∼30:1 peptide:protein) (H) reduces the TTD–PHD binding affinity for the histone peptide. This is consistent with a putative shift from a cooperative to a PHD–mediated binding mode.
We used NMR and SAXS analysis to assess whether BPC could promote open conformations of TTD–PHD, in a manner comparable with that of the UHRF1 PBR. In (1H-15N) TROSY spectra of the reader titrated with BPC, significant conformational broadening and/or movement of amide peaks can be observed in residues spanning the entire length of the linker. Exchange broadening is clearly observed for Arg282, Gly284, Asp291, Asn292, Met294, and Ser301 amide resonances at 1:3 protein/BPC ratios, with perturbations also seen with Gly286 and Gly299 (Fig. 4 and supplemental Fig. S6). TTD residues that form part of or are close to the Arg296-binding pocket (e.g. the 236GFW238 triad and Ala208) also exhibit exchange broadening and/or chemical shift changes, consistent with compound binding close to this site (supplemental Fig. S6). PHD resonances are, by comparison, unaffected by the presence of BPC, and in a separate titration, we confirmed there was no interaction between the fragment and this domain (supplemental Fig. S8). A strikingly similar peak broadening/perturbation pattern is observed in spectra of TTD–PHD when it is titrated with the 15-residue PBR peptide (Fig. 4B and supplemental Fig. S6).
SAXS profiles of TTD–PHD collected in the presence of BPC demonstrate an increase in the overall dimensions resulting from BPC binding (Tables 1, supplemental Table S1, and Fig. 4C). The Rg-based Kratky plot is shifted to higher coordinate values with respect to its position for apo TTD–PHD (supplemental Fig. S4). Also, the pair-distance distribution function exhibits a broad extended tail with a shallow secondary shoulder observed at ∼50 Å (Fig. 4C), indicating extended reader conformations. We performed SES fitting of SAXS data collected for TTD–PHD in the presence of BPC using MDPIN/OUT. The optimal ensemble, OEBPC(SAXS), fits the data reasonably well in the q range of 0 < q < 0.2. The Rg distribution is bimodal with the major peak at ∼25 Å and a minor peak at ∼34 Å (Fig. 4D). In comparison with the Rg distribution of the optimal ensemble generated for apo TTD–PHD (i.e. OEIN/OUT(SAXS)), the position of the major peak is shifted by 1 Å, and the minor peak has a larger height, indicating that there is a higher percentage of extended conformations. Interestingly, the distance that specifies the relative position of the linker with respect to the TTD groove (dRM) shows that the 6,000 starting structures in the pool in which the linker is bound to the groove make zero contribution to OEBPC(SAXS) (Fig. 4E). Taken together, these data indicate that BPC binds in the TTD groove and disrupts the interaction with the linker, shifting the ensemble equilibrium toward less compact structures. These less-compact, “linker-out” conformers should be unable to bind to H3K9me3 peptides in a cooperative manner (as in PDB code 3ASK (10, 23)) and, in addition, be unable to bind via the isolated TTD because groove-H3K9me3 contacts found to be essential for this binding mode, as reported by Nady et al. (9), are blocked.
To evaluate the effect of BPC on histone binding by the reader module, we compared the binding affinity of TTD–PHD with H3K9me3 peptide in the presence of both BPC and PBR peptide (Fig. 4). Apo TTD–PHD was found to exhibit cooperative H3K9me3(1–15) binding (KD = 1.2 μm, n = 1.0) resulting from dual TTD-/PHD–mediated interaction, consistent with previous studies (10) (Fig. 4F and supplemental Fig. S9). The increased KD (∼5-fold) for H3K9me3 in the presence of BPC (Fig. 4G) or PBR peptide (Fig. 4H) is fully consistent with a transition from a cooperative (i.e. PHD– and TTD–dependent) binding mode to one mediated by the PHD only.
Discussion
Large-scale intramolecular rearrangements play a critical role in UHRF1 function, consistent with a dynamic framework in which its conformational equilibria are shifted in response to the chromatin state and aggregate presence of other proteins and cellular factors with modulating influences (14–16). One noted example of this conformational modulation is that induced by the lipid PI5P, which regulates the transition between TTD- and PHD–mediated histone-binding states through its reversible interaction with the PBR element (UHRF1643–657) (14) (Fig. 1A). Structural insight into large-scale UHRF1 intramolecular rearrangements has been obscure.
Our study shows that the UHRF1 TTD–PHD histone reader is highly dynamic in its apo state, with clear evidence of open and extended conformations (Fig. 5). We have applied a novel integrated approach where both SAXS and NMR relaxation data are used to determine OEs from starting pools of conformers derived from MD simulations and rigid-body modeling. Our results indicate that TTD–PHD structures in which the linker is bound to the TTD groove are not favored over those where it is unbound. Linker positioning has been shown to be a critical determinant of histone-binding behavior, because cooperative, high-affinity interaction necessitates that it occupy the groove (10). Interestingly, even when the linker is positioned in the groove, there is evidence of additional flexibility mediated by the hinge region (UHRF1297–301). The linker-bound structures (Fig. 5A) in our OE are clustered around two configurations: in the first, the domain orientation is similar to that seen in the cooperative binding mode (PDB code 3ASK), whereas in the second, the domain orientation is similar to that seen in the crystal structure of apo UHRF2 TTD–PHD (PDB code 4TVR). Open conformers where the linker is out of the groove (Fig. 5B) reveal a vulnerability of the TTD to allosteric regulation by competitive binding of entities such as the PBR or drug-like small molecules (Fig. 5D).
Figure 5.
Apo and H3-binding states of the TTD–PHD histone reader. The apo TTD–PHD OE is consistent with conformers in which the linker is positioned both in (A) and out (B) of the TTD groove. When the linker is in the groove, the relative positions of the domains have two favorable orientations: the first is very similar to the H3-bound state described by Arita et al. (10) where histone binding is cooperatively mediated by the PHD and TTD (C); the second resembles the orientation adopted by apo TTD–PHD in its paralog UHRF2. Both extended and compact conformers are populated when the linker is out of the groove (B) and provide a clear mechanism for histone-binding states mediated independently by the PHD and/or TTD (C). Gelato et al. (14) described the formation of a PBR-bound state where the linker is forced out of the groove as a mechanism for allosteric regulation of histone binding; BPC can similarly promote open reader states (D).
UHRF1 is a potential therapeutic target, because it is essential for the maintenance of DNA methylation patterns and highly expressed in most cancers (2, 4). To date, only one UHRF1 inhibitor has been identified, a uracil derivative that interferes with the SRA domain (24). Because H3K9me3 binding is a requirement for UHRF1 function (25) and can be disrupted by the PBR through its association with the TTD (14), we hypothesized that allosteric disruption of the cooperative (23), high-affinity binding mode of the TTD–PHD module may be an attractive strategy for small molecule antagonists of UHRF1 function. An important aspect of intramolecular interactions is the high effective concentration of interacting regions within a single macromolecule. Such effects may pose a challenge for the development of drug-like small molecules that can efficiently compete with these interactions. Here, using an FP H3-peptide displacement assay with the isolated TTD, we identified the small fragment BPC as a linker-competitive binder to the TTD groove, with a KD close to 50 μm (supplemental Figs. S5 and S8). Saturating concentrations of this highly soluble compound can induce open conformers of the TTD–PHD module and reduce its affinity for H3K9me3 peptides (Fig. 4 and supplemental Figs. S5 and S6). BPC should prove useful as a tool for in vitro investigations that seek to relate open TTD–PHD conformations with specific UHRF1-binding modes. The small size and high ligand efficiency of the compound suggest that it can be further optimized for potency. An attractive approach could be to link compounds such as BPC that bind in the TTD groove, with those designed for interaction with the aromatic cage, which recognizes the trimethyl lysine of H3K9me3. Recent successes in identifying small molecule antagonists of methyl lysine reader domains bodes well for this approach (26–28).
Experimental procedures
Protein expression and purification
UHRF1126–366 corresponding to the TTD–PHD module and UHRF1121–286 corresponding to the TTD were expressed and purified as described previously (9). TTD–PHD was purified with and without an N-terminal His tag, and in cases where the tag was removed, this was by overnight incubation with TEV protease at 4 °C (9). For isotopically labeled proteins used for NMR spectroscopy (15N-labeled, 15N/13C-labeled, or 15N/13C/2H-labeled), cells were grown in M9 minimal medium supplemented with [15N]ammonium chloride (1 g/liter), [13C]glucose (2 g/liter) when required, and 80% D2O when required.
NMR spectroscopy
NMR samples were buffered in 20 mm sodium phosphate (pH 7.5), 150 mm NaCl, 5 mm DTT, 5 mm β-mercaptoethanol, 2 mm TCEP, and 10 μm ZnSO4. The protein concentration was between 200 and 300 μm for all samples. The data were collected at 25 °C on Bruker spectrometers equipped with cryoprobes and operating at 500, 600, or 800 MHz. The assignment of backbone TTD–PHD resonances was accomplished using the ABACUS method (29) for which standard backbone and 15N-edited NOESY spectra were collected using 15N/13C/2H- or 15N/13C-labeled protein. All 3D experiments were acquired using non-uniform sampling and processed using the software MDDGUI (30) or qMDD (31). 15N-Labeled TTD–PHD was used to acquire 15N T1 and T2 relaxation and 1H-15N heteronuclear NOE measurements (32) collected using standard Bruker pulse schemes in an interleaved manner. For T1 measurements, the variable delay was set to 0.1, 0.4, 0.8, 1.5, 2.0, 2.5, 3.5, and 5 s. For T2 measurements, the variable delay was set to 16, 32, 48, 64, 80, 96, 128, and 144 ms. The D1 was 3 s for all experiments. Reported values were the average from two measurements. All spectra were processed using NMRPipe (33) and analyzed with SPARKY (34).
SAXS data collection and analysis
All SAXS samples were buffered in 20 mm Tris (pH 7.5), 150 mm NaCl, 5 mm DTT, 5 mm β-mercaptoethanol, 2 mm TCEP and 10 μm ZnCl2, and data were collected at concentrations ranging from ∼1 to 5 mg/ml. Measurements were carried out at Beamline 12-ID-B of the Advanced Photon Source, Argonne National Laboratory. The energy of the X-ray beam was 14 Kev (wavelength λ = 0.8856 Å), and two setups (small- and wide-angle X-ray scattering) were used simultaneously in which the sample to 2M detector distance was adjusted to achieve scattering q values of 0.006 < q < 2.6 Å−1, where q = (4π/λ)sinθ, and 2θ is the scattering angle. To reduce radiation damage and obtain good statistics, thirty 2D images were recorded for each buffer or sample solution using a flow cell, with an accumulated exposure time of 0.4–2.0 s. No radiation damage was observed as confirmed by the absence of systematic signal changes in sequentially collected X-ray scattering images. The scattering profile of the protein was calculated by subtracting the background buffer contribution from the sample buffer profile using the program PRIMUS (ATSAS package, EMBL) (35). Concentration series measurements for a sample were carried out to remove the scattering contribution caused by interparticle interactions and to extrapolate the data to infinite dilution. Guinier analysis and the experimental radius of gyration (Rg) estimation from the data of infinite dilution were performed using PRIMUS. The pair distance distribution function, P(r), and the maximum dimension of the protein, Dmax, in real space were calculated with the indirect Fourier transform using the program GNOM (36). To avoid underestimation of the molecular dimension and consequent distortion in low resolution structural reconstruction, the parameter Dmax, the upper end of distance r, was chosen such that the resulting PDDF has a short, near zero-value tail at large r. The Rg from P(r) analysis was also reported. The molecular weights were estimated using Vc (37) in the q range of 0 < q < 0.3 Å−1. Fifteen ab initio shape reconstructions (molecular envelopes) were generated using DAMMIF (38) and averaged with DAMAVER (39). The structural models were superimposed and overlaid with the averaged envelope using SUPCOMP (40). The theoretical scattering intensity of a structural model was calculated and fitted to the experimental scattering intensity using CRYSOL (41).
Structural characterization of TTD–PHD using SAXS and NMR data
We used an ensemble approach for the structural characterization of the TTD–PHD in solution by utilizing the SES protocol (18). The strategy on which the SES method is based consists of two main steps: 1) generate the initial ensemble of conformations to approximate the conformational space available for a system in solution and 2) find the optimal population weight for each member of the initial ensemble that minimizes the discrepancy between the ensemble-predicted and the observed experimental data. The goodness of the ensemble fit is measured as χ2. We used the following expression for χ2,
| (Eq. 1) |
where
| (Eq. 2) |
| (Eq. 3) |
where Iexp(q) is the experimental SAXS scattering intensity, Nq is the number of experimental points, σsaxs(q) is the experimental error, Icalck is the scattering intensity predicted for the kth conformation, τTTDexp and τPHDexp are the experimental rotational correlation times for the TTD and PHD, respectively, σTTD and σPHD are the experimental errors, τTTDk and τPHDk are correlation times predicted for the kth conformation, Nens is the number of conformations in the initial ensemble, wk is the population weight associated with the kth conformation in the ensemble, and α is the weighting factor for the NMR relaxation data. In the case α = 0, only SAXS data is used to optimize weights wk. Equation 1 can be represented in the matrix form,
| (Eq. 4) |
where matrix C is of size (Nq + 2, Nens) and consists of predicted SAXS and NMR relaxation data for all members of the initial ensemble, matrix B consists of corresponding experimental values, ||·|| is the vector l2-norm, and w is the vector of weights. To construct the C matrix, the 30-point SAXS profiles in the range 0 < q < 0.3 Å−1 were predicted using CRYSOL, and two overall rotational correlation times, one for the TTD and one for the PHD, were predicted using HYCUD (21, 22). The ill-posed problem of finding vector of weights, w, that minimizes χ2(w, α) under the condition wk ≥ 0 is solved using the SES approach (18). We used the SES module of the ARMOR package to generate a number of sparse solutions for different ensemble sizes with multiorthogonal matching pursuit algorithm. The ARMOR output was analyzed to select the optimal ensemble size using the l-curve (18), and the vector of the optimal weights, w, was calculated by averaging over top near optimal solutions with similar value of χ2.
RBPIN/OUT and RBPIN ensemble generation
Rigid-body pools were generated using RANCH (42). In these simulations, ordered parts of the TTD–PHD were assumed to be rigid, whereas disordered parts were represented by random chains. RBPIN/OUT was generated by assuming that the His tag (in constructs containing it), the flexible TDDN loop (UHRF1163–180), and the entire linker (UHRF1282–301) are disordered. RBPIN was generated assuming that the His tag (for constructs containing it), the flexible TDDN loop (UHRF1163–180), and the five-residue hinge region of the linker (UHRF1297–301) are disordered. Both pools consist of 30,000 TTD–PHD conformations.
MDPIN/OUT and MDPIN generation
The initial pools of TTD–PHD conformations were obtained in two steps. In the first step, we performed all-atom MD simulations of TTD–PHD. Eight replica MD trajectories, each ∼150 ns long, were generated at 300 K. Four of these replicas were started from different conformations with the TTD groove occupied by the linker, whereas the other four replicas were started from conformations with the linker displaced from the groove. No bound-to-unbound or unbound-to-bound transitions of the linker were observed along the MD trajectories. Over the course of the simulations, the two domains retained their overall structure, with no ordered-to-disordered transitions, as indicated by the low root mean square deviation for TTD and PHD backbone atoms (∼3.5 and 3.3 Å, respectively) in MD-generated conformers. This is in agreement with NMR data (supplemental Fig. S7). Each 10-ps frame was saved during MD simulations, which resulted in 65,200 conformations of the TTD–PHD with the linker positioned in the groove and 55,460 conformations with the linker out of the groove. In the second step, we performed k-means clustering of the generated conformations using metrics that specify the relative position of the TTD and PHD, yielding 6,000 and 8,000 clusters of conformers with the linker in and out of the groove, respectively. To improve on relatively poor sampling of the conformers with the linker out of the groove in MD simulations, we also added 2,000 additional clusters, produced initially by rigid-body modeling, of open/extended TTD–PHD conformers. The representative structures of these clusters were used to construct MDPIN/OUT and MDPIN.
All-atom molecular dynamics simulations
A modified Generalized Born implicit solvent model (43) was exploited in the MD simulations to accelerate sampling of the conformational space for each of the systems. All simulations used an integration step of 2 fs with fixed bonds between hydrogen atoms and heavy atoms. The temperature was controlled by carrying out Langevin dynamics with the damping coefficient set to 2 or 5 ps−1. The cut-off for non-bonded Lennard-Jones and electrostatic interactions was set to 18 Å. The ionic strength was set to 0.15 m. All simulations were performed using NAMD 2.9 code (44) with the AMBER Parm99SB parameter set (45). A zinc AMBER Force Field (46) was used for PHD residues that coordinate three zinc ions.
FP, ITC, and DSF measurements
For FP assays, the TTD (UHRF1121–286) was buffered in 20 mm Tris (pH 8.5), 50 mm NaCl, 3% DMSO, and 0.01% TX100. N-terminal FITC-labeled H3K9me3(1–25) peptide was synthesized and purified by Tufts University Core Services (Boston, MA). Titrations and compound binding assays were performed in a 10-μl volume at a constant labeled peptide concentration of 0.04 μm. For compound screening and titrations, a non-saturating fixed protein concentration of 8 μm TTD was used. FP assays were performed in 384-well Axygen plates using a Synergy 4 microplate reader (BioTek). An excitation wavelength of 485 nm and an emission wavelength of 528 nm were used.
For ITC measurements of TTD–PHD (UHRF1126–366) interaction with H3K9me3, samples were dialyzed into a buffer containing 10 mm HEPES (pH 7.5), 150 mm NaCl, 1 mm TCEP, and 30 μm ZnCl2. H3K9me3(1–15) peptide, in the same buffer, was brought to a concentration of 0.50 mm. A preliminary peptide injection of 0.06 μl was followed by subsequent 2-μl injections into the sample cell containing 167 μl of 50 μm TTD–PHD. Where indicated, 1.5 mm BPC or 1.5 mm PBR peptide was included in the injection syringe and sample cell. The reported KD and n values are based on the average from two measurements. For ITC measurements of TTD (UHRF1121–286) interaction with BPC, the protein was buffered in 20 mm Tris (pH 7.4), 50 mm NaCl, and 5% DMSO. BPC was brought to a concentration of 2.5 mm and injected into the sample cell containing 40 μm protein. The data were acquired on a Nano ITC from TA Instruments at 25 °C and fitted with an independent-binding site model using NanoAnalyze software (v3.7.0).
DSF measurements were performed with a Light Cycler 480 II instrument from Roche Applied Science. TTD (UHRF1121–286) was buffered at 0.1 mg/ml in 0.1 m HEPES (pH 7.5), 0.15 m NaCl, and 5× Sypro Orange. Sypro Orange was purchased from Invitrogen as a 5,000× stock solution, and it was diluted 1:1,000 to yield a 5× working concentration. Experiments were run in the absence and presence of 3 mm BPC. DSF was carried out by increasing the temperature from 20 to 95 °C at a heating rate of 4 °C/min, and data points were collected at 1 °C intervals. The temperature scan curves were fitted to a Boltzmann sigmoid function, and the Tm values were obtained from the midpoint of the transition as described previously (47).
Author contributions
R. S .H. acquired and analyzed NMR data. A. L. interpreted NMR relaxation data, analyzed SAXS data, and performed structural modeling. L. F. acquired and processed SAXS data under the supervision of Y.-X. W. A. I. performed the initial fragment screen with UHRF1-TTD under the supervision of P. J. B. G. S. characterized the interaction of BPC with UHRF1-TTD. D. I. performed the ITC experiments. S. D., L. K., and M. S. O. designed constructs and prepared samples for SAXS, NMR analysis, and binding studies. R. S. H., A. L., and C. H. A. wrote the manuscript. C. H. A. supervised the project.
Supplementary Material
Acknowledgments
We thank Elena Dobrovetsky for help with UHRF1 protein expression, Dr. Xiaobing Zuo (Argonne National Laboratory) for expert support with SAXS measurements, and Masoud Vedadi for helpful discussions. We acknowledge use of the SAXS Core facility at the Laboratory of Cancer Research, National Cancer Institute.
The operation of the Arrowsmith NMR facility is supported by the Princess Margaret Cancer Centre of the University Health Network. The shared Beamline 12-ID-B resource was allocated under Agreement PUP-24152 between the National Cancer Institute and the Argonne National Laboratory. Use of the Advanced Photon Source, a U.S. Department of Energy Office of Science User Facility, was operated by the Argonne National Laboratory under Contract DE-AC02–06CH11357. The Structural Genomics Consortium is a registered charity (Number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada through Ontario Genomics Institute (Grant OGI-055), Innovative Medicines Initiative (EU/EFPIA) (ULTRA-DD Grant 115766), Janssen, Merck & Co., Novartis Pharma AG, Ontario Ministry of Research, Innovation and Science, Pfizer, São Paulo Research Foundation-FAPESP, Takeda, and the Wellcome Trust. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Tables S1–S3 and Figs. S1–S9.
- PHD
- plant homeodomain
- TTD
- tandem tudor domain
- PBR
- polybasic region
- BPC
- 4-benzylpiperidine-1-carboximidamide
- MDP
- molecular dynamics pool
- OE
- optimal ensemble
- PDB
- Protein Data Bank
- SAXS
- small angle X-ray scattering
- MD
- molecular dynamics
- FP
- fluorescence polarization
- TCEP
- tris-(2-carboxyethyl) phosphine hydrochloride
- TROSY
- transverse relaxation-optimized spectroscopy
- ITC
- isothermal titration calorimetry
- DSF
- differential scanning fluorimetry.
References
- 1. Tauber M., and Fischle W. (2015) Conserved linker regions and their regulation determine multiple chromatin-binding modes of UHRF1. Nucleus 6, 123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alhosin M., Omran Z., Zamzami M. A., Al-Malki A. L., Choudhry H., Mousli M., and Bronner C. (2016) Signalling pathways in UHRF1-dependent regulation of tumor suppressor genes in cancer. J. Exp. Clin. Cancer Res. 35, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Unoki M. (2011) Current and potential anticancer drugs targeting members of the UHRF1 complex including epigenetic modifiers. Recent Pat. Anticancer Drug Discov. 6, 116–130 [DOI] [PubMed] [Google Scholar]
- 4. Bronner C., Achour M., Arima Y., Chataigneau T., Saya H., and Schini-Kerth V. B. (2007) The UHRF family: oncogenes that are drugable targets for cancer therapy in the near future? Pharmacol. Ther. 115, 419–434 [DOI] [PubMed] [Google Scholar]
- 5. Wang F., Yang Y.-Z., Shi C.-Z., Zhang P., Moyer M. P., Zhang H.-Z., Zou Y., and Qin H.-L. (2012) UHRF1 promotes cell growth and metastasis through repression of p16(ink4a) in colorectal cancer. Ann. Surg. Oncol. 19, 2753–2762 [DOI] [PubMed] [Google Scholar]
- 6. Zhou L., Shang Y., Jin Z., Zhang W., Lv C., Zhao X., Liu Y., Li N., and Liang J. (2015) UHRF1 promotes proliferation of gastric cancer via mediating tumor suppressor gene hypermethylation. Cancer Biol. Ther. 16, 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rajakumara E., Wang Z., Ma H., Hu L., Chen H., Lin Y., Guo R., Wu F., Li H., Lan F., Shi Y. G., Xu Y., Patel D. J., and Shi Y. (2011) PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol. Cell 43, 275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lallous N., Legrand P., McEwen A. G., Ramón-Maiques S., Samama J.-P., and Birck C. (2011) The PHD finger of human UHRF1 reveals a new subgroup of unmethylated histone H3 tail readers. PLoS One 6, e27599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nady N., Lemak A., Walker J. R., Avvakumov G. V., Kareta M. S., Achour M., Xue S., Duan S., Allali-Hassani A., Zuo X., Wang Y.-X., Bronner C., Chédin F., Arrowsmith C. H., and Dhe-Paganon S. (2011) Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein. J. Biol. Chem. 286, 24300–24311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arita K., Isogai S., Oda T., Unoki M., Sugita K., Sekiyama N., Kuwata K., Hamamoto R., Tochio H., Sato M., Ariyoshi M., and Shirakawa M. (2012) Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1. Proc. Natl. Acad. Sci. U.S.A. 109, 12950–12955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheng J., Yang Y., Fang J., Xiao J., Zhu T., Chen F., Wang P., Li Z., Yang H., and Xu Y. (2013) Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein. J. Biol. Chem. 288, 1329–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Trotzier M.-A., Bronner C., Bathami K., Mathieu E., Abbady A.-Q., Jeanblanc M., Muller C. D., Rochette-Egly C., and Mousli M. (2004) Phosphorylation of ICBP90 by protein kinase A enhances topoisomerase IIα expression. Biochem. Biophys. Res. Commun. 319, 590–595 [DOI] [PubMed] [Google Scholar]
- 13. Yang J., Liu K., Yang J., Jin B., Chen H., Zhan X., Li Z., Wang L., Shen X., Li M., Yu W., and Mao Z. (2017) PIM1 induces cellular senescence through phosphorylation of UHRF1 at Ser311. Oncogene 36, 4828–4842 [DOI] [PubMed] [Google Scholar]
- 14. Gelato K. A., Tauber M., Ong M. S., Winter S., Hiragami-Hamada K., Sindlinger J., Lemak A., Bultsma Y., Houliston S., Schwarzer D., Divecha N., Arrowsmith C. H., and Fischle W. (2014) Accessibility of different histone H3-binding domains of UHRF1 is allosterically regulated by phosphatidylinositol 5-phosphate. Mol. Cell 54, 905–919 [DOI] [PubMed] [Google Scholar]
- 15. Fang J., Cheng J., Wang J., Zhang Q., Liu M., Gong R., Wang P., Zhang X., Feng Y., Lan W., Gong Z., Tang C., Wong J., Yang H., Cao C., et al. (2016) Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition. Nat. Commun. 7, 11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harrison J. S., Cornett E. M., Goldfarb D., DaRosa P. A., Li Z. M., Yan F., Dickson B. M., Guo A. H., Cantu D. V., Kaustov L., Brown P. J., Arrowsmith C. H., Erie D. A., Major M. B., Klevit R. E., et al. (2016) Hemi-methylated DNA regulates DNA methylation inheritance through allosteric activation of H3 ubiquitylation by UHRF1. eLife e17101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang Z.-M., Rothbart S. B., Allison D. F., Cai Q., Harrison J. S., Li L., Wang Y., Strahl B. D., Wang G. G., and Song J. (2015) An allosteric interaction links USP7 to deubiquitination and chromatin targeting of UHRF1. Cell Rep. 12, 1400–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Berlin K., Castañeda C. A., Schneidman-Duhovny D., Sali A., Nava-Tudela A., and Fushman D. (2013) Recovering a representative conformational ensemble from underdetermined macromolecular structural data. J. Am. Chem. Soc. 135, 16595–16609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mandel A. M., Akke M., and Palmer A. G. 3rd (1995) Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J. Mol. Biol. 246, 144–163 [DOI] [PubMed] [Google Scholar]
- 20. García de la Torre J., Huertas M. L., and Carrasco B. (2000) HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J. Magn. Reson. 147, 138–146 [DOI] [PubMed] [Google Scholar]
- 21. Rezaei-Ghaleh N., Klama F., Munari F., and Zweckstetter M. (2013) Predicting the rotational tumbling of dynamic multidomain proteins and supramolecular complexes. Angew. Chem. Int. Ed. Engl. 52, 11410–11414 [DOI] [PubMed] [Google Scholar]
- 22. Rezaei-Ghaleh N., Klama F., Munari F., and Zweckstetter M. (2015) HYCUD: a computational tool for prediction of effective rotational correlation time in flexible proteins. Bioinformatics 31, 1319–1321 [DOI] [PubMed] [Google Scholar]
- 23. Xie S., Jakoncic J., and Qian C. (2012) UHRF1 double tudor domain and the adjacent PHD finger act together to recognize K9me3-containing histone H3 tail. J. Mol. Biol. 415, 318–328 [DOI] [PubMed] [Google Scholar]
- 24. Myrianthopoulos V., Cartron P. F., Liutkevičiūtė Z., Klimašauskas S., Matulis D., Bronner C., Martinet N., and Mikros E. (2016) Tandem virtual screening targeting the SRA domain of UHRF1 identifies a novel chemical tool modulating DNA methylation. Eur. J. Med. Chem. 114, 390–396 [DOI] [PubMed] [Google Scholar]
- 25. Rothbart S. B., Krajewski K., Nady N., Tempel W., Xue S., Badeaux A. I., Barsyte-Lovejoy D., Martinez J. Y., Bedford M. T., Fuchs S. M., Arrowsmith C. H., and Strahl B. D. (2012) Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat. Struct. Mol. Biol. 19, 1155–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. James L. I., Barsyte-Lovejoy D., Zhong N., Krichevsky L., Korboukh V. K., Herold J. M., MacNevin C. J., Norris J. L., Sagum C. A., Tempel W., Marcon E., Guo H., Gao C., Huang X.-P., Duan S., et al. (2013) Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain. Nat. Chem. Biol. 9, 184–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. He Y., Selvaraju S., Curtin M. L., Jakob C. G., Zhu H., Comess K. M., Shaw B., The J., Lima-Fernandes E., Szewczyk M. M., Cheng D., Klinge K. L., Li H.-Q., Pliushchev M., Algire M. A., et al. (2017) The EED protein-protein interaction inhibitor A-395 inactivates the PRC2 complex. Nat. Chem. Biol. 13, 389–395 [DOI] [PubMed] [Google Scholar]
- 28. Stuckey J. I., Dickson B. M., Cheng N., Liu Y., Norris J. L., Cholensky S. H., Tempel W., Qin S., Huber K. G., Sagum C., Black K., Li F., Huang X.-P., Roth B. L., Baughman B. M., et al. (2016) A cellular chemical probe targeting the chromodomains of polycomb repressive complex 1. Nat. Chem. Biol. 12, 180–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lemak A., Steren C. A., Arrowsmith C. H., and Llinás M. (2008) Sequence specific resonance assignment via Multicanonical Monte Carlo search using an ABACUS approach. J. Biomol. NMR 41, 29–41 [DOI] [PubMed] [Google Scholar]
- 30. Orekhov V. Y., Ibraghimov I., and Billeter M. (2003) Optimizing resolution in multidimensional NMR by three-way decomposition. J. Biomol. NMR 27, 165–173 [DOI] [PubMed] [Google Scholar]
- 31. Kazimierczuk K., and Orekhov V. Y. (2011) Accelerated NMR spectroscopy by using compressed sensing. Angew. Chem. Int. Ed. Engl. 50, 5556–5559 [DOI] [PubMed] [Google Scholar]
- 32. Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., and Kay L. E. (1994) Backbone dynamics of a free and a phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 33. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 34. Goddard T., and Kneller D. G. (2004) SPARKY 3, University of California, San Francisc [Google Scholar]
- 35. Konarev P. V., Petoukhov M. V., Volkov V. V., and Svergun D. I. (2006) ATSAS 2.1, a program package for small-angle scattering data analysis. J. Appl. Crystallogr. 39, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Feigin L. A., and Svergun D. I. (1987) Structure analysis by small-angle X-ray and neutron scattering, Plenum Press, New York [Google Scholar]
- 37. Rambo R. P., and Tainer J. A. (2013) Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496, 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Franke D., and Svergun D. I. (2009) DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Volkov V. V., and Svergun D. I. (2003) Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kozin M. B., and Svergun D. I. (2001) Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 34, 33–41 [Google Scholar]
- 41. Svergun D., Barberato C., and Koch M. H. J. (1995) CRYSOL: a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 42. Tria G., Mertens H. D., Kachala M., and Svergun D. I. (2015) Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ. 2, 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Onufriev A., Bashford D., and Case D. A. (2000) Modification of the generalized born model suitable for macromolecules. J. Phys. Chem. B 104, 3712–3720 [Google Scholar]
- 44. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., and Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., and Simmerling C. (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65, 712–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Peters M. B., Yang Y., Wang B., Füsti-Molnár L., Weaver M. N., and Merz K. M. (2010) Structural survey of zinc containing proteins and the development of the zinc AMBER force field (ZAFF). J. Chem. Theory Comput. 6, 2935–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Niesen F. H., Berglund H., and Vedadi M. (2007) The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2, 2212–2221 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.