Abstract
Lipidic cubic phase (LCP) has been widely recognized as a promising membrane-mimicking matrix for biophysical studies of membrane proteins and their crystallization in a lipidic environment. Application of this material to a wide variety of membrane proteins, however, is hindered due to a limited number of available host lipids, mostly monoacylglycerols (MAGs). Here, we designed, synthesized and characterized a series of chemically stable lipids resistant to hydrolysis, with properties complementary to the widely used MAGs. In order to assess their potential to serve as host lipids for crystallization, we characterized the phase properties and lattice parameters of mesophases made of two most promising lipids at a variety of different conditions by polarized light microscopy and small-angle X-ray scattering. Both lipids showed remarkable chemical stability and an extended LCP region in the phase diagram covering a wide range of temperatures down to 4 °C. One of these lipids has been used for crystallization and structure determination of a prototypical membrane protein bacteriorhodopsin at 4 °C and 20 °C.
Graphical abstract
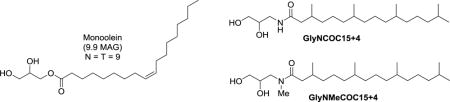
Chemical structures of monoolein (9.9 MAG) and two new LCP lipids characterized in this study: GlyNCOC15+4 and GlyNMeCOC15+4.
We have designed, synthesized and characterized a series of chemically stable lipids resistant to hydrolysis capable of forming a lipidic cubic phase. The phase properties and lattice parameters of mesophases made of two most promising lipids were characterized. One of these lipids has been used for crystallization and structure determination of a prototypical membrane protein bacteriorhodopsin at 4 °C and 20 °C.
INTRODUCTION
Membrane proteins reside in lipid bilayers of biological membranes, where they play critical roles in mediating cellular processes. The knowledge of their three-dimensional structures is essential for deciphering the underlying mechanisms of intercellular interactions, signal transduction, transport of ions and molecules, and other functions. X-ray crystallography represents the most successful technique for structure determination, however, it requires the availability of highly ordered protein crystals, growing which in case of membrane proteins is challenging. Structural biology of membrane proteins received a significant boost after the introduction of unique crystallization methods using lipidic mesophases, such as a lipidic cubic phase (LCP)1. LCP is an artificial membrane-mimicking gel-like material that forms spontaneously upon mixing approximately equal volumes of specific lipids and an aqueous buffer2. Solubilized membrane proteins can be reconstituted into the lipid bilayer of LCP after purification relieving them from their detergent micelle coating. Adding a precipitant to the lipid/protein mixture triggers crystal nucleation and growth3. LCP crystallization offers several advantages over the conventional vapor-diffusion methods, such as an increased stability of proteins, a tighter crystal packing, and a better tolerance to impurities4. As a result, LCP crystallization method often yields well-ordered crystals diffracting to higher resolution compared to crystals grown in detergent micelle solutions5.
After the first demonstration of LCP crystallization with bacteriorhodopsin in 19961, the method has undergone continuous development. Special tools, assays and instrumentation for handling of LCP have been introduced2. Automation and miniaturization of the crystallization process enabled high-throughput screening of a vast range of precipitant conditions6. Advances in the LCP crystallization method have contributed to the structure determinations of about a hundred different membrane proteins from diverse families, including enzymes, transporters, ion channels and G protein-coupled receptors7. It was shown that LCP can be used for direct crystallization of membrane proteins from membranes8, amphipols9 and nanodiscs10,11. Future progress in the application of this method to crystallization of a wider variety of membrane proteins, however, may be hindered due to a limited number of available lipids that that support formation of a stable LCP at or below room temperature. The identity of the host lipid affects such LCP parameters as the bilayer thickness, solvent channel diameter, curvature and fluidity of the bilayer. Consequently, stability, diffusion, nucleation and crystallization of the embedded membrane protein targets critically depend on the selection of the LCP host lipid.
Currently, monoacylglycerols (MAGs) represent the most widely used lipid class for crystallization of membrane proteins by the LCP method12. MAGs are derivatives of the tri-hydric alcohol glycerol, in which the primary hydroxyl group is esterified with a long-chain fatty acid, typically monounsaturated. Different monounsaturated MAGs are often described by an N.T MAG shorthand notation13, where N (Neck) is the number of carbon atoms between the lipid’s polar head and the double bond, and T (Tail) is the number of carbon atoms from the double bond to the terminal methyl. While serving successfully as LCP host lipids, MAGs are chemically unstable and susceptible to hydrolysis of the ester linkage between the glycerol head group and the fatty acid under alkaline and acidic conditions leading to production of fatty acid14,15. The same process occurs slowly even at mid-range pH values, and can be accelerated by trace amounts of lipases, potentially modifying the properties of LCP or even converting it into a non-LCP phase16, thereby affecting crystal nucleation and growth. Additionally, alkyl chain migration from the primary 1-OH to the secondary 2-OH group also occurs in MAGs during incubation in aqueous solution, resulting in the accumulation of 2-MAGs that modulate LCP properties17,18.
The most commonly used MAG is monoolein (9.9 MAG) (Scheme 1), which supports crystallization of many different membrane proteins19. Several other MAGs with varying N and T numbers have been developed for low temperature crystallization and other specific crystallization projects7,20,21. Recently, Salvati et al.22 substituted the double bond of monoolein with a cyclopropyl group, which extended the cubic phase formation region to a lower temperature range and supported the crystallization of bacteriorhodopsin (bR) at 4 °C.
Scheme 1.
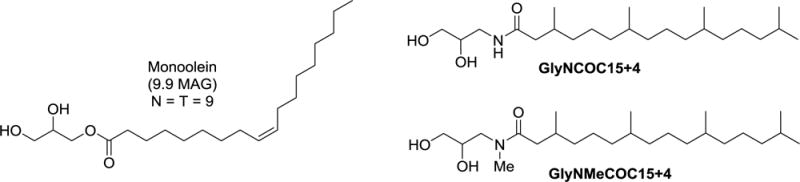
Chemical structures of monoolein (9.9 MAG) and two new lipids (GlyNCOC15+4 and GlyNMeCOC15+4) characterized in this study.
In this study, we have designed, synthesized and tested a panel of 33 non-hydrolysable amide-linked lipids with cis-olefinic or isoprenoid tails for their ability to form LCP and support crystallization. Isoprenoid lipids have previously been successfully used in LCP crystallization studies23 and have displayed distinct structural properties24,25 making them a useful compliment to the standard MAG lipids set. Our goal was to develop chemically stable lipids that form a stable cubic phase under a broad range of conditions including extreme pH and low temperature. We streamlined lipid screening to quickly identify most promising LCP lipids. With two promising hits found (GlyNCOC15+4 and GlyNMeCOC15+4, where Gly refers to the glycerol headgroup, NCO or NMeCO – to the amide- or N-methyl amide- linkers and C15+4 – to the isoprenoid chain with 15 carbon groups and 4 branched methyls), and one of them (GlyNCOC15+4) selected for further crystallization studies, we crystallized a prototypical membrane protein bacteriorhodopsin26 at 20 °C and 4 °C and solved the corresponding structures. Initial attempts to crystallize the A2A adenosine (A2AAR) G protein-coupled receptor (GPCR) using the same host lipid produced small crystals. This work, thus, contributes to the ongoing efforts of expanding the lipid toolbox for LCP crystallization of membrane proteins.
EXPERIMENTAL SECTION
Synthesis of GlyNCOC15+4 and GlyNMeCOC15+4
Phytol (5.0 g, 16.9 mmol) and Raney-Ni (50% slurry in H2O, 1.0 g, 8.4 mmol) were mixed in 25 mL ethanol (Scheme S1a). The reaction atmosphere was then exchanged to H2 and stirred for 3 days at room temperature. The suspension was filtered through celite and washed with methanol. After the removal of solvent, dihydro-phytol (3,7,11,15-tetramethylhexadecan-1-ol) was left as a colorless oil (5.0 g, 99%). Dihydro-phytol (5.0 g, 16.8 mmol) was dissolved in 120 mL acetone and the solution was cooled down to 0 °C. Then Jones reagent (2.7 M, 7.5 mL, 20.2 mmol) was added dropwise over half an hour with stirring at the same temperature. The reaction was quenched in half an hour by adding isopropanol. The mixture was then filtered through celite, washed with acetone, and concentrated in vacuo. The residue was purified by silica-gel column chromatography to give phytanic acid (3,7,11,15-tetramethylhexadecanoic acid) as a colorless oil (4.7 g, 91%).
Phytanic acid (160 mg, 0.51 mmol), 3-aminopropane-1,2-diol (60 mg, 0.66 mmol), N-Ethyl-N′-(3-dimethylaminopropyl)carbodiimide (EDC, 196 mg, 1.02 mmol) and 1-hydroxybenzotriazole(HOBt, 156 mg, 1.02 mmol) were mixed in 3 mL DMF (Scheme S1b). N,N-diisopropylethyl amine (DIEA, 0.35 mL, 2.04 mmol) was then added at 0 °C. The reaction mixture was stirred at room temperature for 1 day. Then H2O and ethyl acetate were added. The reaction mixture was washed sequentially with H2O and a saturated solution of sodium chloride. The organic layer was separated, dried over Na2SO4, filtered, and concentrated in vacuo. The residue was purified by silica-gel column chromatography to give GlyNCOC15+4 as a colorless oil (175 mg, 88%). GlyNMeCOC15+4 was prepared similarly as GlyNCOC15+4 except for the using of 3-(methylamino)propane-1,2-diol as a reactant (Scheme S1a,c).
The 1H and 13C NMR spectra for GlyNCOC15+4 and GlyNMeCOC15+4 are shown in Figure S1. Both GlyNCOC15+4 and GlyNMeCOC15+4 could be efficiently scaled up for synthesis at multigrams scale.
GlyNCOC15+4: 1H NMR (600 MHz, CDCl3) δ 6.95 (s, br, 1H), 4.40 (s, br, 2H), 3.74-3.70 (m, 1H), 3.54-3.47 (m, 2H), 3.41-3.24 (m, 2H), 2.24-2.20 (m, 1H), 1.96-1.90 (m, 2H), 1.52-1.46 (m, 1H), 1.37-0.99 (m, 20H), 0.88 (d, J = 6.1 Hz, 3H), 0.84 (d, J = 6.6 Hz, 6H), 0.81 (d, J = 6.6 Hz, 6H). 13C NMR (125 MHz, CDCl3) δ 175.1, 71.2, 63.9, 44.5, 44.4, 42.3, 39.6, 37.7, 37.6, 37.5, 33.0, 31.1, 28.2, 25.0, 24.7, 22.9, 22.8, 19.9, 19.8, 19.7, 19.6.
GlyNMeCOC15+4: 1H NMR (600 MHz, CDCl3) δ 3.96 (br, 2H), 3.81-3.77 (m, 1H), 3.52-3.41 (m, 4H), 3.06 (s, 2.7H), 2.94 (s, 0.3H), 2.33-2.27 (m, 1H), 2.12-2.08 (m, 1H), 1.96-1.92 (m, 1H), 1.51-1.44 (m, 1H), 1.34-0.98 (m, 20H), 0.90-0.88 (m, 3H), 0.82 (d, J = 6.7 Hz, 6H), 0.80 (d, J = 6.7 Hz, 6H). 13C NMR (125 MHz, CDCl3) δ 175.1, 70.5, 63.5, 51.0, 40.8, 40.7, 39.4, 37.8, 37.5, 37.4, 37.3, 32.8, 30.4, 28.0, 24.8, 24.5, 22.8, 22.7, 19.9, 19.9, 19.7, 19.6.
The amide bond in GlyNMeCOC15+4 exists in a mixture of cis- and trans- configurations at an approximate ratio of 1:9, based on the 1H NMR assignment.
Small-angle X-ray scattering
Samples in capillaries
Initial SAXS samples were prepared by mixing corresponding lipids with an excess buffer (25 mM HEPES pH 7.0; 2:3 v/v ratio, lipid:buffer) at room temperature using a syringe mixer27. The resulting lipidic mesophases were then transferred from the Hamilton syringe into X-ray-transparent capillaries (d = 1.5 mm) that were flame-sealed afterwards. The capillaries were placed in a temperature-controlled holder and SAXS data were collected at 4 °C, 10 °C, 20 °C, 30 °C and 40 °C. The samples were incubated at 4 °C for 1 h before collecting data at this temperature and for 30 min at subsequent temperatures in the heating direction. SAXS experiments were conducted on a Rigaku SAXS instrument equipped with a pinhole camera attached to a rotating Cu anode X-ray high-flux beam generator (MicroMax 007-HF) which operated at 40 kV and 30 mA (1200 W). A multiwire gas-filled area detector Rigaku ASM DTR Triton 200 (diameter 200 mm, pixel size 200 μm) was placed at a distance of 203 cm from the sample. The entire optical path of the X-ray beam was kept in vacuum during data collection. The detector covered Q-range was 0.006 – 0.2 Å−1 (Q = 4π sinθ/λ, where λ is the wavelength and 2θ is the scattering angle). The beam size was 400 μm and the exposure time ranged from 1800 s to 2500 s.
Samples of GlyNCOC15+4 at different hydration levels were prepared the same way except for the different lipid:water ratios. SAXS data for these samples were collected at room temperature.
Samples in 96-well sandwich plates
Samples were prepared in special X-ray transparent 96-well sandwich plates made of two 140 μm-thick COC films (TOPAS 8007×4) with a 130 μm-thick perforated double-sticky spacer 468MP (3M) between them and mounted on a rigid PMMA frame (Figure S2). Lipids were first mixed with water at a ratio of 3:2 v/v using a lipid syringe mixer to form LCP. Then LCP was distributed by 50 nL per well in 96-well sandwich plates and covered with 800 nL of precipitant solutions using an NT8-LCP crystallization robot (Formulatrix). Analysis was made with a home-made “FRAP pH 7” screen28, and commercial “MemStart & MemSys”, “MemGold” (Molecular Dimensions) and “Wizard Cubic LCP” (Rigaku) screens. For each combination of lipid and screen two crystallization plates were prepared that differed in the direction of sample dispensing (from A1 to H12 and from H12 to A1) to reduce potential effects of sample dehydration29. Coordinates of LCP drops in each plate were manually identified after taking their images with an automatic FRAP imager (Formulatrix) and then translated to the SAXS holder coordinates. SAXS data for each well were collected by the Rigaku instrument. 2D scattering profiles were collected using automatic sample holder. The beam size of 400 μm and 100 s exposure time per well were used for this data collection. All experiments were performed at 20 °C. The scattering data were analyzed with the SAXSGUI package (Rigaku) to convert 2D scattering images into 1D radially-averaged scattered curves. Phase identification and lattice parameters determination were performed as previously described29.
Polarized Light Microscopy (PLM)
Lipids were first mixed with water at a ratio of 1:2 (v/v) using a lipid syringe mixer to form a homogenous phase. The resultant material was deposited on a microscope glass slide and covered with a cover slip to form a thin layer. Samples were tested for birefringence using Nikon SMZ1500 microscope equipped with two rotating linear polarizing filters placed before and after the sample. The samples that didn’t change light polarization were likely isotropic and formed either LCP or sponge phase.
Expression, purification and crystallization of bR and A2AAR
bR
Wild type bR was expressed in Halobacterium salinarum, isolated and purified using the standard protocol30. In brief, bR was solubilized from purple membranes of Halobacterium salinarum with 50 mM sodium phosphate buffer (pH 6.9) containing 1.2% (w/v) β-octylglucoside (OG) and purified by SEC using a Superdex 200 16/60 column (GE Healthcare). The protein was concentrated using a 50 kDa MWCO Amicon Centrifugal Device (Millipore). bR concentration was determined by absorbance at 550 nm on a NanoDrop spectrophotometer using the extinction coefficient ε550 = 5.8 × 104 M−1cm−1. For crystallization, 30 mg/ml of bR in OG was mixed with GlyNCOC15+4 in 2:3 ratio (v/v) using coupled syringes device27 to create LCP. LCP drops of 200 nl volume were dispensed in each well of 96-well glass sandwich plates (Marienfeld) using a manual LCP dispenser (Hamilton) and covered with 0.8 μl of precipitant. The precipitant screen was based on literature conditions1 using a concentration gradient around the condition: 1.9-3.0 M sodium/potassium phosphate buffer (pH5.6), 3.5% v/v methylpentanediol and 0.5% w/v OG. No further screening and optimization of crystallization conditions were performed.
A2AAR
A2AAR was expressed and purified according to previously published protocol31. In short, the third intracellular loop of A2AAR between residues L209 and G218 was replaced with a thermostabilized apocytochrome b562RIL (BRIL) and the C-terminal residues 317-412 were truncated. HA and Flag tags were added to the N-terminus of the protein and 10xHis tag was placed on the C-terminus. The protein was expressed in insect sf9 cells using Bac-to-Bac Baculovirus Expression System (Invitrogen). Insect cell membranes were disrupted by thawing frozen cell pellets in a hypotonic buffer containing an EDTA-free complete protease inhibitor cocktail (Roche). Extensive washing of the isolated raw membranes was performed by repeated dounce homogenization and centrifugation in the same hypotonic buffer (~2-3 times), and then in a high osmotic buffer (~3-4 times) to remove soluble and membrane associated proteins. Prior to solubilization, purified membranes were thawed on ice in the presence of 4 mM theophylline (Sigma), 2.0 mg/ml iodoacetamide (Sigma), and an EDTA-free complete protease inhibitor cocktail (Roche). After incubation for 30 min at 4 °C, membranes were solubilized by incubation in the presence of 0.5% (w/v) n-dodecyl-β-D-maltopyranoside (DDM) (Anatrace) and 0.1% (w/v) cholesteryl hemisuccinate (CHS) (Sigma) for 2.5 h at 4 °C. The unsolubilized material was removed by centrifugation at 150,000 ×g for 45 min. The supernatant was incubated with TALON IMAC resin (Clontech) in the buffer containing 50 mM HEPES (pH 7.5), 800 mM NaCl, 0.5% (w/v) DDM, 0.1% (w/v) CHS, and 20 mM imidazole. The protein was purified on TALON resin using 10 column volumes (CV) of wash buffer 1 (50 mM HEPES (pH 7.5), 800 mM NaCl, 10% (v/v) glycerol, 25 mM imidazole, 0.1% (w/v) DDM, 0.02% (w/v) CHS, 10 mM MgCl2, 8 mM ATP (Sigma) and 25 μM ZM241385) followed by 5 CV of wash buffer 2 (50 mM HEPES (pH 7.5), 800 mM NaCl, 10% (v/v) glycerol, 50 mM imidazole, 0.05% (w/v) DDM, 0.01% (w/v) CHS and 25 μM ZM241385). The receptor was eluted with 25 mM HEPES (pH 7.5), 800 mM NaCl, 10% (v/v) glycerol, 220 mM imidazole, 0.025% (w/v) DDM, 0.005% (w/v) CHS and 25 μM ZM241385.
To make 9:1 (w/w) GlyNCOC15+4/cholesterol mixture, the lipids were added together and dissolved in a minimal volume of chloroform. The excess of chloroform was removed by evaporation under a stream of nitrogen gas, and the lipid mixture was fully dried under vacuum (<0.1 Torr) for over 12 hours.
Protein samples of A2AAR in complex with ZM241385 were reconstituted into lipidic cubic phase (LCP) by mixing it with molten GlyNCOC15+4/cholesterol at a 2:3 (v/v, protein/lipid) ratio using a syringe mixer. Due to a relatively high viscosity of the lipid mixture, it was transferred into the syringe mixer using a spatula. Crystallization trials were performed in 96-well glass sandwich plates (Marienfeld) by an NT8-LCP crystallization robot (Formulatrix) using 50 nl protein-laden LCP overlaid with 0.8 μl precipitant solution in each well. Crystals were obtained overnight in 25-28% (v/v) PEG 400, 0.04 to 0.06 M sodium thiocyanate, 2% (v/v) 2,5-hexanediol, 100 mM sodium citrate pH 5.0.
Crystallographic data collection and structure determination
Synchrotron diffraction data was collected on the 23 ID-D beamline at the Advanced Photon Source equipped with a Pilatus3 6M detector. Datasets were collected using 20×20 μm2 beam, 0.2 sec exposure time and oscillation angle 0.2° without attenuation of the beam intensity. bR_20C dataset was obtained by merging the data collected on 4 crystals and bR_4C dataset was obtained by merging the data from 5 crystals. The data was indexed, merged and integrated with XDS32, molecular replacement solutions were found with Phaser33 using PDB ID 1XJI as a model. The models were refined using multiple cycles of refinement with phenix.refine34 using NCS restraints followed by manual rebuilding in Coot35.
Initially, bR crystals grown at 4 °C were indexed in C2221 space group, however the initial Rfree after molecular replacement was above 40% and didn’t improve after rebuilding and refinement. Subsequently, the data was re-indexed in P21, where the L-test has shown a perfect pseudo-merohedral twinning with a twin law h,-k,-h-l. After re-indexing the correct MR solution could be found and the structure was refined to Rfree = 26.5% using phenix.refine with the twin law h,-k,-h-l. The data obtained from the crystals grown at 20 °C was indexed in R3 space group with no twinning detected by the L and NZ tests performed in phenix.Xtriage. Data processing and refinement statistics is shown in Table 1.
Table 1.
Crystallographic data collection and structure refinement statistics. Values in parentheses correspond to the highest resolution shell.
| BR_20C (5VN7) | BR_4C (5VN9) | |||
|---|---|---|---|---|
| Data collection | ||||
| Wavelength (Å) | 1.033 | 1.033 | ||
| Number of crystals | 4 | 5 | ||
| Resolution range (Å) | 28.52 - 2.70 (2.80 - 2.70) | 28.34 - 2.59 (2.69 - 2.59) | ||
| Space group | R 3 | P 21 | ||
| Unit cell, a b c (Å) α β γ (°) | 107.05 107.05 149.86 90 90 120 | 50.07 109.63 56.56 90 116.3 90 | ||
| Total reflections | 51,454 (4,126) | 40,533 (1,396) | ||
| Unique reflections | 17,181 (1,716) | 15,034 (833) | ||
| Multiplicity | 3.0 (2.4) | 2.7 (1.7) | ||
| Completeness (%) | 97 (96) | 90 (49) | ||
| Mean I/sigma(I) | 7.4 (1.8) | 5.3 (1.7) | ||
| Wilson B-factor (Å2) | 52.8 | 46.5 | ||
| R-merge | 0.151 (0.645) | 0.143 (0.317) | ||
| CC1/2 | 0.988 (0.476) | 0.987 (0.76) | ||
| Refinement | ||||
| R-work/ R-free | 0.196/0.225 | 0.223/0.265 | ||
| Clashscore | 4.4 | 5.5 | ||
| A | B | A | B | |
| Number of non-hydrogen atoms | 1,748 | 1,740 | 1,766 | 1,746 |
| protein | 1,728 | 1,720 | 1,735 | 1,720 |
| retinal | 20 | 20 | 20 | 20 |
| water | 0 | 0 | 11 | 6 |
| Average B-factor | 51.8 | 67.5 | 39.3 | 36.4 |
| protein | 51.9 | 67.5 | 39.4 | 36.5 |
| retinal | 42.5 | 49.5 | 35.4 | 33.2 |
| water | – | – | 38.1 | 33.7 |
| RMS bonds (Å) | 0.004 | 0.003 | ||
| RMS angles (°) | 0.64 | 0.58 | ||
| A | B | A | B | |
| Ramachandran favored (%) | 99.1 | 97.3 | 98.6 | 98.6 |
| Ramachandran allowed (%) | 0.9 | 2.7 | 1.4 | 1.4 |
| Ramachandran outliers (%) | 0 | 0 | 0 | 0 |
RESULTS AND DISCUSSION
Design and synthesis of novel lipids
Currently, only several lipids have been extensively characterized for their ability to form LCP under conditions amenable to membrane protein crystallization14,20,21,24,25,36,37. The complexity of lipid-solvent interactions and a poor understanding of the relationship between the lipid structure and its phase behavior makes it challenging to rationally design a lipid with specific physical-chemical and phase properties suitable for LCP crystallization. Given the labile nature of the ester bond in MAGs, we set our goals to discover more chemically stable LCP lipids as part of our efforts to advance the utility of the LCP technology. Based on our prior experience, simple substitution of the ester group of monoolein with a non-hydrolysable amide or ether group results in formation of non-LCP liquid crystalline phases at room temperature and excess hydration conditions. We speculated that the change of the ester linkage to the amide or ether group may lead to altered lipid hydration and hydrogen-bond patterns. A subsequent search for new lipids was therefore mainly focused on the alteration of the polar groups while using traditional cis-olefinic and isoprenoid chains. Following this approach, we have synthetized and evaluated 33 new amide-linked lipids (Table S1) with two of them found to form stable LCP suitable for membrane protein crystallization studies.
Polarized Light Microscopy and SAXS analysis of the designed lipids
To quickly identify lipids suitable for membrane protein crystallization, all synthesized lipids were first pre-screened for their ability to form LCP at excess hydration and 20 °C by mixing them with water (lipid:water = 1:2 v/v) using a syringe mixer27 and analyzing the samples under cross-polarized light (Table S1). Two lipids containing a branched isoprenoid chain, GlyNCOC15+4 (lipid 16) and GlyNMeCOC15+4 (lipid 18) (Scheme 1), formed a gel-like phase and showed no birefringence under cross-polarized light microscopy, indicative of a cubic phase formation. Both of these lipids contain glycerol as their polar head group, similar to monoolein, connected to an isoprenoid acyl chain through an amide bond. To determine the phase state, these two lipids were further subjected to SAXS analysis at five different temperatures between 4 °С and 40 °С (Figure S3). SAXS measurements also allow to obtain lattice parameters for each identified phase. We observed that samples made of GlyNCOC15+4 formed cubic-Pn3m phase in the entire temperature range from 4 to 40 °С, while samples made of GlyNMeCOC15+4 formed mixtures of lamellar and cubic phases at low temperatures, which turned into a single cubic-Pn3m phase at 30 °С and 40 °С. The full hydration boundary for the cubic-Pn3m phase made of GlyNCOC15+4 at 20 °С was found to be 44±2% (v/v water) and the lattice parameter of the fully hydrated cubic-Pn3m phase 106.2±1.0 Å (Table S2).
Both lipids that formed LCP in excess hydration were further subjected to an extensive screening for their compatibility with typical conditions encountered during crystallization trials using a high-throughput LCP-SAXS method29. For these tests we used three commercial screens: “MemStart&MemSys” and “MemGold” screens from Molecular Dimensions and “Emerald Cubic LCP” screen from Rigaku, as well as an in-house made “FRAP pH 7” screen (see Methods section). Phase identities and unit cell parameters were determined from SAXS patterns for each screened condition. Lipids GlyNCOC15+4 and GlyNMeCOC15+4 formed LCP in a wide range of precipitant conditions with predominantly Pn3m and Ia3d symmetries and cell parameters ranging from 68 Å to 128 Å for cubic-Pn3m and from 106 Å to 156 Å for cubic-Ia3d phases (Figure S4). The LCP composed of GlyNCOC15+4 was found to be compatible with most conditions from the commercial screens, which were not designed specifically for LCP crystallization (MemStart&MemSys and MemGold), and thus contain many conditions not compatible with a monoolein-based LCP29,38. The LCP composed of GlyNMeCOC15+4 was generally much less stable, being compatible with around 50% conditions of the MemStart&MemSys and 25% conditions of the MemGold screens (solutions containing PEG with molecular weight 4,000 Da and higher turned the phase into lamellar). While both monoolein and GlyNCOC15+4 are fully compatible with the Wizard Cubic LCP screen, GlyNMeCOC15+4 displayed low compatibility mostly due to the presence of high molecular weight PEGs in this screen. Overall, GlyNCOC15+4 exhibited superior LCP compatibility with a broader variety of crystallization screening conditions compared to both GlyNMeCOC15+4 and monoolein38.
In general, we observed an increased stability of GlyNCOC15+4 and GlyNMeCOC15+4 towards highly alkaline and acidic buffers. Monoolein, the most widely used lipid for LCP crystallization, can be typically used only in the pH range between 2-3 and 8-9 (depending on the nature of the precipitant salt used), limiting its application for the proteins that favor crystallization conditions outside of this range. We have tested GlyNCOC15+4 with different buffers in the range from pH=2.2 to pH=11, where it demonstrated a remarkable stability and LCP remained intact for at least four weeks (Figure 1). Therefore, substitution of the ester linkage with an amide greatly improved the stability of lipids both at harsh chemical conditions and during long incubations at normal conditions.
Figure 1.
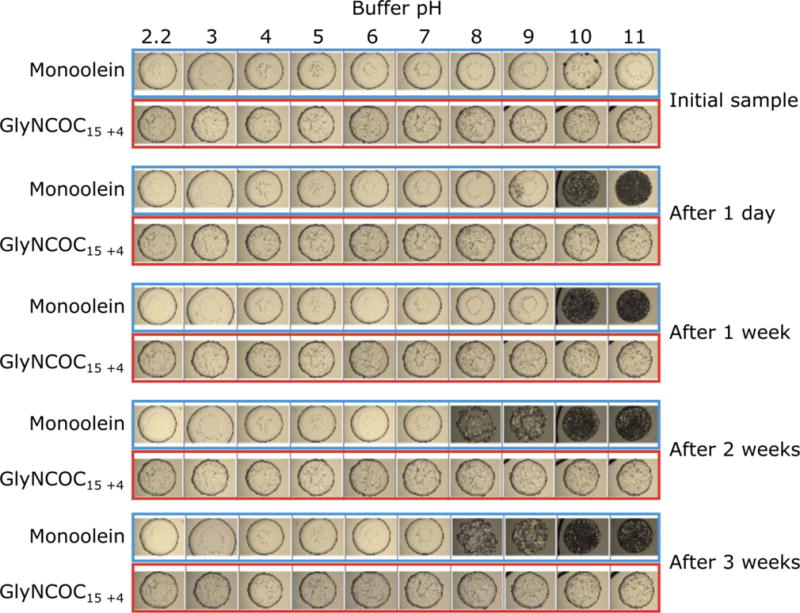
Stability of LCP made of GlyNCOC15+4 and monoolein under a range of pH values (2.2-11) over time. Both lipids were mixed with water to produce LCP, and the LCP boluses were covered with solutions containing 150 mM NaCl, 30% PEG400 and 100 mM of the respective buffer. The buffer solutions were taken from the StockOptions pH screen (Hampton).
LCP lattice parameters of GlyNCOC15+4
We used the SAXS data for GlyNCOC15+4 to estimate the lipid membrane thickness and the diameter of the water channels for the cubic-Pn3m phase at 20 °C and maximum hydration level.
The lipid length, l, is related to the LCP lattice parameter, a, and the volume fraction of lipids in LCP, Φl, through the following equation39:
where A0 is the area of the membrane surface in the unitary unit cell, and χ is the Euler-Poincare parameter. In case of the cubic-Pn3m phase A0=1.919, χ=-2. Using the estimated Φl of 0.56±0.01 at the full hydration boundary and the measured lattice parameter of 106.2±1.0 Å, we derived the lipid length l=16.3±0.3 Å and the bilayer thickness d=2·l=32.6±0.6 Å. The radius of the aqueous channel in LCP can then be further estimated from these parameters using the following formula36:
resulting in rw = 25.2±0.5 Å.
Comparing these numbers with the bilayer thickness, d=34.6 Å, and the radius of the aqueous channel, rw=25.3 Å, reported previously for the monoolein-based cubic Pn3m phase36,40 we conclude that the LCP formed by GlyNCOC15+4 has a slightly smaller bilayer thickness and a similar water channel diameter.
Crystallization of membrane proteins using designed lipid
The choice of a host lipid is undoubtedly an important parameter in LCP crystallization trials41. We tested the newly synthesized lipids for their ability to support crystallization of membrane proteins. GlyNCOC15+4 was chosen for the crystallization trials, as it formed stable LCP in the widest temperature and precipitant composition space. bR was used for initial crystallization tests, as it is a well-characterized membrane protein, for which high-resolution structures are available in the Protein Data Bank (PDB). Crystals of bR were obtained after incubating LCP crystallization plates at two different temperatures: 4 °C and at 20 °C (Figure 2), simply by precipitant screening around conditions that are known to support bR crystallization in monoolein1 (see Methods). With this observation, we did not attempt to further extend and optimize the screens. Crystals appeared in 3-5 days and continued to grow for several weeks at both crystallization temperatures. At 20 °C crystals grew to a maximal size of 40 μm and had a hexagonal shape, similar to bR crystals grown in monoolein42,43. The crystals grown at 4 °C were rhombic and, generally, smaller in size (~20-30 μm).
Figure 2.
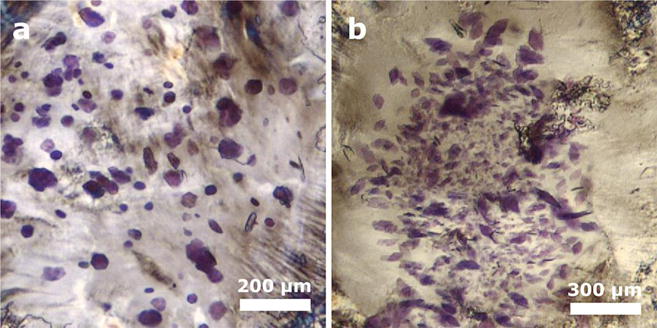
Crystals of bacteriorhodopsin obtained by LCP crystallization using GlyNCOC15+4 as the host lipid. (a) Crystals grown at 20 °C. (b) Crystals grown at 4 °C.
X-ray crystallographic data for both types of crystals were collected at the GM/CA facility at the Advanced Photon Source, Argonne, IL (Table 1). Data obtained from four hexagonal crystals grown at 20 °C were indexed in the R3 space group and merged together at the resolution of 2.7 Å. A PDB search indicated that bR has never been previously crystallized in such a space group. Typically, hexagonal crystals of bR belong to the space group P63, where bR molecules are packed as trimers in the plane of the lipidic membrane, with one molecule per asymmetric unit (ASU) (Figure 3a, d). In our case, however, there are two bR molecules per ASU, one of which forming a trimer together with its symmetry mates around the three-fold symmetry axis similar to the trimers in the P63 space group bR structures. Meanwhile, the second bR molecule is attached peripherally to the trimer at all three sides (Figure 3b, e), thus forming a hexamer of bR molecules. There are no crystal contacts between hexamers in the membrane plane resulting in a loose crystal packing with the Matthews coefficient Vm of 3.08 Å3/Da and an estimated solvent content of 60% compared to 2.21 Å3/Da and 45% correspondingly for bR crystallized in the P63 space group (PDB ID 1MD2)43. Consistent with the absence of crystal contacts in the membrane plane, the outer parts of the hexamer-forming proteins have increased B-factors compared to the hexamer core (Figure 3h). In the stacking direction, on the contrary, the crystal packing is very tight with a minimal spacing between protein layers.
Figure 3.
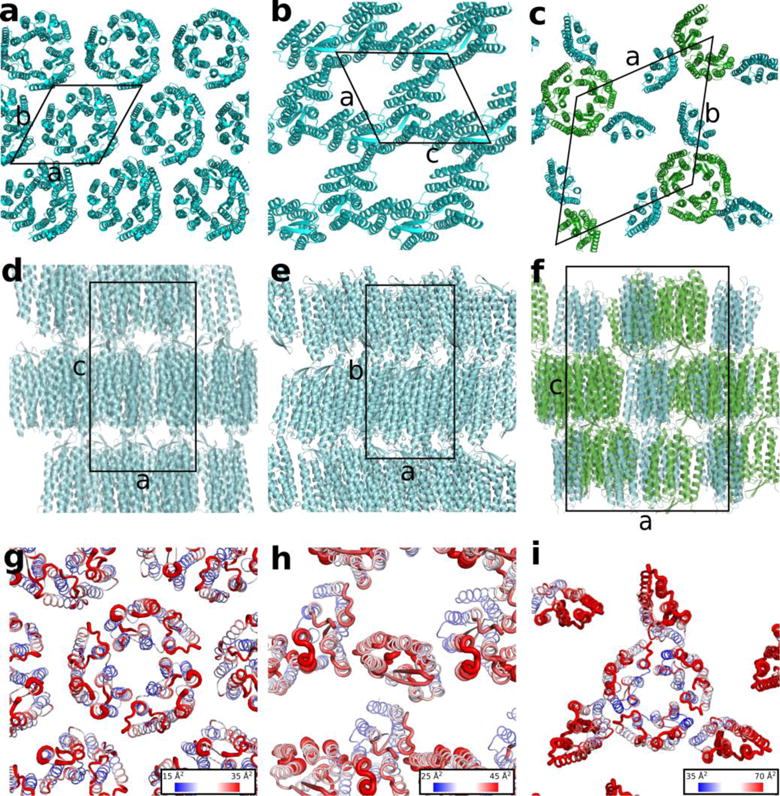
Crystal packing of bR crystallized using monoolein, PDB ID 1M0L (a, d, g) and GlyNCOC15+4 as a host lipid for LCP at 4 °C (b, e, h) and 20 °C (c, f, i) as viewed
(a, b, c) in the membrane plane,
(d, e, f) along the membrane,
(g, h, i) in the membrane plane with B factors mapped as both thickness of the lines and as a gradient coloring with the blue color corresponding to the lowest values and the red color corresponding to the highest values.
The core bR trimer for the 20 °C structure is shown in green and the peripheral protomers are shown in cyan (panels c and f). Unit cells are outlined with solid black lines.
The overall bR structure within the trimer core of the hexamer is very similar to the previous bR structures (Cα atoms RMSD = 0.40 Å with PDB ID 1M0L). However, the tight packing between crystal layers has a big impact on the structure of the extracellular loop between helices B and C (BC loop) of the distal protomers in the hexamer. This BC loop interacts with the same protomers of the neighboring layer in a crystal and adopts a distinct conformation compared to the core protomers and all the previously published structures (Figure 4). Phe71 plays a crucial role in this interaction protruding out into the adjacent membrane layer and anchoring the loop to the cytoplasmic side of the neighboring protein.
Figure 4.
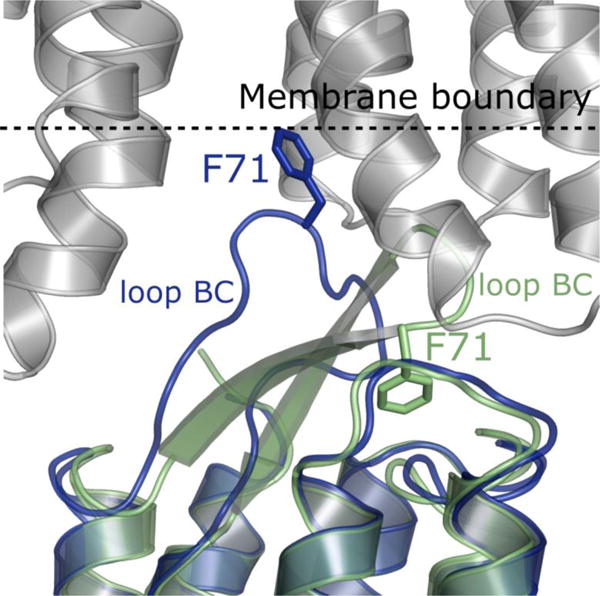
Crystal contacts between bR molecules crystallized at 20 °C. A bR protomer from the trimeric core (shown in green) was superimposed with a peripheral protomer (shown in dark blue). The structure of the BC loop is clearly different in the respective protomers. Molecules of the core bR trimer show a typical configuration of the BC loop containing a β-hairpin motif. On the other hand, the peripheral bR protomers exhibit an extended loop conformation without the β-hairpin, which would clash with its crystal neighbor, and instead with F71 positioned to potentially stabilize the packing by a hydrophobic interaction with the next membrane layer. The symmetry mate molecules are shown in light grey and the membrane boundary is shown as a dashed line. The membrane boundary was calculated using the Orientations of Proteins in Membranes Server (http://opm.phar.umich.edu/).
Data collected from bR crystals grown at 4 °C were indexed in P21 space group. A full X-ray diffraction dataset in this case was obtained by combining data from five crystals grown in similar conditions at 2.6 Å resolution. Interestingly, the crystal packing is very similar to that observed for bR crystallized from bicelles at 37 °C44. The ASU contains two molecules of bR assembled in an antiparallel manner. The overall structure of both molecules is similar to that of published before (Cα atoms RMSD = 0.49 Å with PDB ID 1KME).
After successful bR crystallization, we tested GlyNCOC15+4 for its ability to support crystallization of a GPCR: A2A adenosine receptor (A2AAR). Several structures of this receptor containing a fusion protein, either a cysteine-less lysozyme from T4 phage (T4L) or a thermostabilized apo-cytochrome b562 (BRIL), have been previously obtained using LCP crystallization method with monoolein (9.9 MAG) doped with cholesterol as the host lipid45–47. Since GPCRs typically require cholesterol binding for their stability, we mixed GlyNCOC15+4 with 10% cholesterol by weight to ensure typical crystallization conditions for this family of receptors. We used the same engineered A2AAR-BRIL construct as in our previous work31 and obtained hits (Figure 5) with very small (~1 μm) crystals forming with a high nucleation rate and in similar precipitant conditions as previously published31. Despite the efforts to optimize crystallization conditions by varying PEG400 and salt concentrations, the crystal size could not be further improved. Although such crystals are too small for data collection at a synchrotron source, they could potentially be suitable for serial femtosecond crystallography (SFX) with an X-ray free electron laser (XFEL)48,49.
Figure 5.
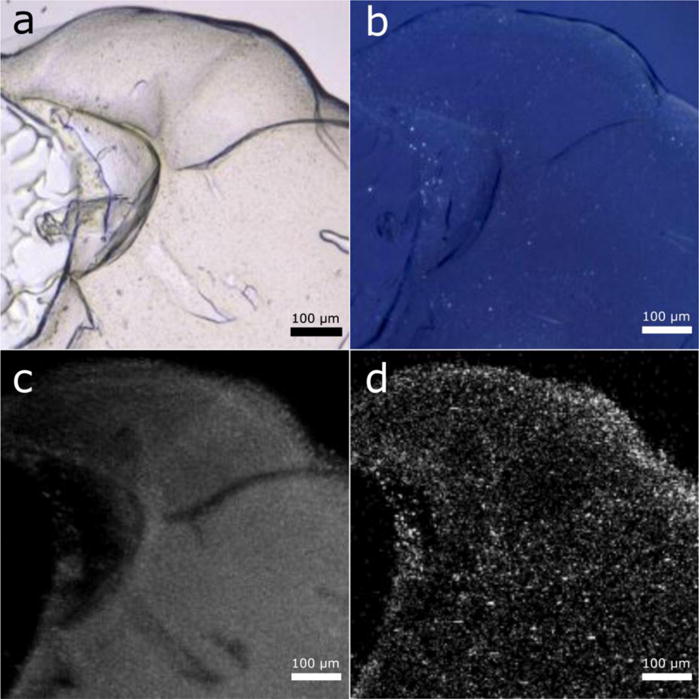
Crystal hits obtained from crystallization of A2AAR using GlyNCOC15+4 as the LCP host lipid.
Brightfield mode
Cross-polarized light
Two-photon UV fluorescence mode
SHG (second harmonic generation) mode
In this work, we synthesized dozens of non-hydrolysable lipids and characterized them for their potential to be used as host lipids for LCP crystallization of membrane proteins. By examining subgroups of structures containing an identical aliphatic chain, we showed that minor structural variation in the polar region have a dramatic impact on the lipid hydration and corresponding phase properties. Two of these lipids, GlyNCOC15+4 and GlyNMeCOC15+4, formed stable LCP at room temperature and excess hydration. Substitution of the ester bond between the lipid head and the tail with an amide bond as expected greatly improved the overall resistance towards hydrolysis during prolonged incubation. While different by only a single methyl, GlyNCOC15+4 demonstrated the ability to sustain LCP in a wider range of conditions, and was further used as a host lipid for crystallization of two membrane proteins.
Analysis of the phase behavior with various screens typically employed for membrane protein crystallization confirmed that both GlyNCOC15+4 and GlyNMeCOC15+4 are suitable to be used as host lipids for LCP crystallization, although the latter lipid forms LCP under substantially fewer conditions than the former one. In contrast to monoolein-based LCP, most conditions induced shrinkage of the cubic-Pn3m phase for both tested lipids. For example, in case of the FRAP pH 7 screen the lattice parameter of the cubic-Pn3m phase was on averages smaller by 15 Å for GlyNCOC15+4 and by 20 Å for GlyNMeCOC15+4 (Figure S4). Few conditions, especially those with high concentrations of alcohols swelled the cubic-Pn3m phase and occasionally transformed it into a sponge phase. We expect that new and a broader range of screens could be developed for further evaluation of these lipids, especially for GlyNCOC15+4.
Since LCP made of GlyNCOC15+4 was found to be compatible with a wider array of conditions compared to GlyNMeCOC15+4, we tested whether this lipid can support crystallization of a prototypical membrane protein bR and A2AAR. The crystals of bR were grown both at 20 °C and 4 °C and the respective structures were solved at 2.7 Å and 2.6 Å resolution. The structure of bR at 20 °C revealed novel crystal packing in the R3 space group, while at 4 °C crystal packing was very similar to that previously obtained for bR crystallized in bicelles at 37 °C. These observations emphasized the importance of the host lipid structure and properties on the mechanism and outcome of membrane protein crystallization in LCP.
Therefore, based on the SAXS results and crystallization experiments we conclude that the new lipids can be a useful alternative to MAGs as the host lipids for LCP crystallization, in particularly, at harsh conditions and at low temperatures.
Supplementary Material
Acknowledgments
This work was supported by the Ministry of Education and Science of the Russian Federation (RFMEFI58716X0026) and the National Institutes of Health (P50 GM073197 and R01 GM098538).
Footnotes
Supporting information
The Supporting information is available free of charge on the ACS Publications website at DOI: Chemical structures of synthesized lipids, physical properties of lipidic mesophases, synthesis schemes for GlyNMeCOC15+4 and GlyNCOC15+4, NMR spectra, assembly of SAXS sandwich plates, SAXS patterns, phase identities and lattice parameters of lipidic mesophases.
Accession codes
The coordinates and structure factors were deposited in the Protein Data Bank with the accession codes 5VN7 (bR at 20 °C) and 5VN9 (bR at 4 °C).
References
- 1.Landau EM, Rosenbusch JP. Lipidic cubic phases: a novel concept for the crystallization of membrane proteins. Proc Natl Acad Sci USA. 1996;93:14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cherezov V. Lipidic cubic phase technologies for membrane protein structural studies. Curr Opin Struct Biol. 2011;21:559–566. doi: 10.1016/j.sbi.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caffrey M. On the mechanism of membrane protein crystallization in lipidic mesophases. Cryst Growth Des. 2008;8:4244–4254. doi: 10.1021/cg101384p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caffrey M. A lipid’s eye view of membrane protein crystallization in mesophases. Curr Opin Struct Biol. 2000;10:486–497. doi: 10.1016/s0959-440x(00)00119-6. [DOI] [PubMed] [Google Scholar]
- 5.Kang HJ, Lee C, Drew D. Breaking the barriers in membrane protein crystallography. Int J Biochem Cell Biol. 2013;45:636–644. doi: 10.1016/j.biocel.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 6.Ishchenko A, Abola E, Cherezov V. Membrane Proteins Production for Structural Analysis. Springer; New York: 2014. Lipidic cubic phase technologies for structural studies of membrane proteins; pp. 289–314. [Google Scholar]
- 7.Caffrey M. A comprehensive review of the lipid cubic phase or in meso method for crystallizing membrane and soluble proteins and complexes. Acta Crystallogr Sect F, Struct Biol Commun. 2015;71:3–18. doi: 10.1107/S2053230X14026843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nollert P, Royant A, Pebay-Peyroula E, Landau EM. Detergent-free membrane protein crystallization. FEBS Lett. 1999;457:205–208. doi: 10.1016/s0014-5793(99)01014-5. [DOI] [PubMed] [Google Scholar]
- 9.Polovinkin V, Gushchin I, Sintsov M, Round E, Balandin T, Chervakov P, Schevchenko V, Utrobin P, Popov A, Borshchevskiy V, Mishin A, Kuklin A, Willbold D, Chupin V, Popot J-L, Gordeliy V. High-resolution structure of a membrane protein transferred from amphipol to a lipidic mesophase. J Membr Biol. 2014;247:997–1004. doi: 10.1007/s00232-014-9700-x. [DOI] [PubMed] [Google Scholar]
- 10.Nikolaev M, Round E, Gushchin I, Polovinkin V, Balandin T, Kuzmichev P, Shevchenko V, Borshchevskiy V, Kuklin A, Round A, Bernhard F, Willbold D, Büldt G, Gordeliy V. Integral membrane proteins can be crystallized directly from nanodiscs. Cryst Growth Des. 2017;17:945–948. [Google Scholar]
- 11.Broecker J, Eger BT, Ernst OP. Crystallogenesis of membrane proteins mediated by polymer-bounded lipid nanodiscs. Structure. 2017;25:384–392. doi: 10.1016/j.str.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Caffrey M. Membrane protein crystallization. J Struct Biol. 2003;142:108–132. doi: 10.1016/s1047-8477(03)00043-1. [DOI] [PubMed] [Google Scholar]
- 13.Misquitta Y, Caffrey M. Rational design of lipid molecular structure: a case study involving the C19:1c10 monoacylglycerol. Biophys J. 2001;81:1047–1058. doi: 10.1016/S0006-3495(01)75762-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clogston J, Rathman J, Tomasko D, Walker H, Caffrey M. Phase behavior of a monoacylglycerol: (myverol 18-99K)/water system. Chem Phys Lipids. 2000;107:191–220. doi: 10.1016/s0009-3084(00)00182-1. [DOI] [PubMed] [Google Scholar]
- 15.Coleman BE, Cwynar V, Hart DJ, Havas F, Mohan JM, Patterson S, Ridenour S, Schmidt M, Smith E, Wells AJ. Modular approach to the synthesis of unsaturated 1-monoacyl glycerols. Synlett. 2004;2004:1339–1342. [Google Scholar]
- 16.Clogston J, Craciun G, Hart DJ, Caffrey M. Controlling release from the lipidic cubic phase by selective alkylation. J Control Release. 2005;102:441–461. doi: 10.1016/j.jconrel.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Cherezov V, Clogston J, Misquitta Y, Abdel-Gawad W, Caffrey M. Membrane protein crystallization in meso: lipid type-tailoring of the cubic phase. Biophys J. 2002;83:3393–3407. doi: 10.1016/S0006-3495(02)75339-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murgia S, Caboi F, Monduzzi M, Ljusberg-Wahren H, Nylander T. Lipid and Polymer-Lipid Systems. Springer Berlin Heidelberg; Berlin, Heidelberg: 2002. Acyl migration and hydrolysis in monoolein based systems; pp. 41–46. [Google Scholar]
- 19.Kulkarni CV, Wachter W, Iglesias-Salto G, Engelskirchen S, Ahualli S. Monoolein: a magic lipid? Phys Chem Chem Phys. 2011;13:3004–3021. doi: 10.1039/c0cp01539c. [DOI] [PubMed] [Google Scholar]
- 20.Misquitta Y, Cherezov V, Havas F, Patterson S, Mohan JM, Wells AJ, Hart DJ, Caffrey M. Rational design of lipid for membrane protein crystallization. J Struct Biol. 2004;148:169–175. doi: 10.1016/j.jsb.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Misquitta LV, Misquitta Y, Cherezov V, Slattery O, Mohan JM, Hart D, Zhalnina M, Cramer WA, Caffrey M. Membrane protein crystallization in lipidic mesophases with tailored bilayers. Structure. 2004;12:2113–2124. doi: 10.1016/j.str.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 22.Salvati Manni L, Zabara A, Osornio YM, Schöppe J, Batyuk A, Plückthun A, Siegel JS, Mezzenga R, Landau EM. Phase behavior of a designed yclopropyl analogue of monoolein: implications for low-temperature membrane protein crystallization. Angew Chem Int Ed Engl. 2015;54:1027–1031. doi: 10.1002/anie.201409791. [DOI] [PubMed] [Google Scholar]
- 23.Borshchevskiy V, Moiseeva E, Kuklin A, Büldt G, Hato M, Gordeliy V. Isoprenoid-chained lipid β-XylOC 16+4 - A novel molecule for in meso membrane protein crystallization. J Cryst Growth. 2010;312:3326–3330. [Google Scholar]
- 24.Yamashita J, Shiono M, Hato M. New lipid family that forms inverted cubic phases in equilibrium with excess water: molecular structure-aqueous phase structure relationship for lipids with 5,9,13,17-tetramethyloctadecyl and 5,9,13,17-tetramethyloctadecanoyl chains. J Phys Chem B. 2008;112:12286–12296. doi: 10.1021/jp8029874. [DOI] [PubMed] [Google Scholar]
- 25.Hato M, Yamashita J, Shiono M. Aqueous phase behavior of lipids with isoprenoid type hydrophobic chains. J Phys Chem B. 2009;113:10196–10209. doi: 10.1021/jp902883q. [DOI] [PubMed] [Google Scholar]
- 26.Lanyi JK. Bacteriorhodopsin. Annu Rev Physiol. 2004;66:665–688. doi: 10.1146/annurev.physiol.66.032102.150049. [DOI] [PubMed] [Google Scholar]
- 27.Cheng A, Hummel B, Qiu H, Caffrey M. A simple mechanical mixer for small viscous lipid-containing samples. Chem Phys Lipids. 1998;95:11–21. doi: 10.1016/s0009-3084(98)00060-7. [DOI] [PubMed] [Google Scholar]
- 28.Xu F, Liu W, Hanson MA, Stevens RC, Cherezov V. Development of an automated high throughput LCP-FRAP assay to guide membrane protein crystallization in lipid mesophases. Cryst Growth Des. 2011;11:1193–1201. doi: 10.1021/cg101385e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joseph JS, Liu W, Kunken J, Weiss TM, Tsuruta H, Cherezov V. Characterization of lipid matrices for membrane protein crystallization by high-throughput small angle X-ray scattering. Methods. 2011;55:342–349. doi: 10.1016/j.ymeth.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dencher NA, Heyn MP. Methods in Enzymology. Vol. 88 Elsevier: 1982. Biomembranes Part I: Visual Pigments and Purple Membranes II. [Google Scholar]
- 31.Liu W, Chun E, Thompson AA, Chubukov P, Xu F, Katritch V, Han GW, Roth CB, Heitman LH, IJzerman AP, Cherezov V, Stevens RC. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337:232–236. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Briggs J, Chung H, Caffrey M. The temperature-composition phase diagram and mesophase structure characterization of the monoolein/water system. J Phys II. 1996;6:723–751. [Google Scholar]
- 37.Imura T, Hikosaka Y, Worakitkanchanakul W, Sakai H, Abe M, Konishi M, Minamikawa H, Kitamoto D. Aqueous-phase behavior of natural glycolipid biosurfactant mannosylerythritol lipid A: sponge, cubic, and lamellar phases. Langmuir. 2007;23:1659–1663. doi: 10.1021/la0620814. [DOI] [PubMed] [Google Scholar]
- 38.Cherezov V, Fersi H, Caffrey M. Crystallization screens: compatibility with the lipidic cubic phase for in meso crystallization of membrane proteins. Biophys J. 2001;81:225–242. doi: 10.1016/S0006-3495(01)75694-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qiu H, Caffrey M. Lyotropic and thermotropic phase behavior of hydrated monoacylglycerols: structure characterization of monovaccenin. J Phys Chem B. 1998;102:4819–4829. [Google Scholar]
- 40.Caffrey M, Li D, Dukkipati A. Membrane protein structure determination using crystallography and lipidic mesophases: recent advances and successes. Biochemistry. 2012;51:6266–6288. doi: 10.1021/bi300010w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li D, Shah STA, Caffrey M. Host lipid and temperature as important screening variables for crystallizing integral membrane proteins in lipidic mesophases. Trials with diacylglycerol kinase. Cryst Growth Des. 2013;13:2846–2857. doi: 10.1021/cg400254v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schobert B, Cupp-Vickery J, Hornak V, Smith S, Lanyi J. Crystallographic structure of the K intermediate of bacteriorhodopsin: conservation of free energy after photoisomerization of the retinal. J Mol Biol. 2002;321:715–726. doi: 10.1016/s0022-2836(02)00681-2. [DOI] [PubMed] [Google Scholar]
- 43.Borshchevskiy V, Round E, Erofeev I, Weik M, Ishchenko A, Gushchin I, Mishin A, Willbold D, Büldt G, Gordeliy V. Low-dose X-ray radiation induces structural alterations in proteins. Acta Crystallogr D Biol Crystallogr. 2014;70:2675–2685. doi: 10.1107/S1399004714017295. [DOI] [PubMed] [Google Scholar]
- 44.Faham S, Bowie JU. Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J Mol Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 45.Jaakola V-P, Griffith MT, Hanson MA, Cherezov V, Chien EYT, Lane JR, Ijzerman AP, Stevens RC. The 2.6 Angstrom Crystal Structure of a Human A2A Adenosine Receptor Bound to an Antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi H-J, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang C, Jiang Y, Ma J, Wu H, Wacker D, Katritch V, Han GW, Liu W, Huang X-P, Vardy E, McCorvy JD, Gao X, Zhou XE, Melcher K, Zhang C, Bai F, Yang H, Yang L, Jiang H, Roth BL, Cherezov V, Stevens RC, Xu HE. Structural basis for molecular recognition at serotonin receptors. Science. 2013;340:610–614. doi: 10.1126/science.1232807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu W, Wacker D, Gati C, Han GW, James D, Wang D, Nelson G, Weierstall U, Katritch V, Barty A, Zatsepin NA, Li D, Messerschmidt M, Boutet S, Williams GJ, Koglin JE, Seibert MM, Wang C, Shah STA, Basu S, Fromme R, Kupitz C, Rendek KN, Grotjohann I, Fromme P, Kirian RA, Beyerlein KR, White TA, Chapman HN, Caffrey M, Spence JCH, Stevens RC, Cherezov V. Serial femtosecond crystallography of G protein-coupled receptors. Science. 2013;342:1521–1524. doi: 10.1126/science.1244142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu W, Ishchenko A, Cherezov V. Preparation of microcrystals in lipidic cubic phase for serial femtosecond crystallography. Nat Protoc. 2014;9:2123–2134. doi: 10.1038/nprot.2014.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


