Abstract
Background and Aims: Esophageal squamous cell carcinoma (ESCC), a major histologic subtype of esophageal cancer, is increasing in incidence, but the genetic underpinnings of this disease remain unexplored. The aim of this study is to identify the recurrent genetic changes, elucidate their roles and discover new biomarkers for improving clinical management of ESCC.
Methods: Western blotting and immunohistochemistry were performed to detect the expression level of RHCG. Bisulfite genomic sequencing (BGS) and methylation-specific PCR (MSP) were used to study the methylation status in the promoter region of RHCG. The tumor-suppressive effect of RHCG was determined by both in-vitro and in-vivo assays. Affymetrix cDNA microarray was used to identify the underlying molecular mechanism.
Results: RHCG was frequently downregulated in ESCCs, which was significantly correlated with poor differentiation (P = 0.001), invasion (P = 0.003), lymph node metastasis (P = 0.038) and poorer prognosis (P < 0.001). Demethylation treatment and bisulfite genomic sequencing analyses revealed that the downregulation of RHCG in both ESCC cell lines and clinical samples was associated with its promoter hypermethylation. Functional assays demonstrated that RHCG could inhibit clonogenicity, cell motility, tumor formation and metastasis in mice. Further study revealed that RHCG could stabilize IκB by decreasing its phosphorylation, and subsequently inhibit NF-κB/p65 activation by blocking the nuclear translocation of p65, where it acted as a transcription regulator for the upregulation of MMP1 expression.
Conclusions: Our results support the notion that RHCG is a novel tumor suppressor gene that plays an important role in the development and progression of ESCC.
Keywords: ESCC, RHCG, Metastasis, NF-κB, MMP1.
Introduction
Esophageal squamous cell carcinoma (ESCC), a major histologic subtype of esophageal cancer, is one of the most common malignancies and ranks as the sixth leading cause of cancer related death in the world 1. ESCC is characterized by its remarkable geographic distribution, with more than half of ESCC cases worldwide occurring in China. Linzhou and the nearby counties in Henan province of Northern China have the highest incidence rate of ESCC in the world 2, 3. Similar to other types of cancers, ESCC is a complex disease with a multistep process of genetic alterations, including activation of oncogenes and inactivation of tumor suppressor genes (TSG) 4. Therefore, it is important to better understand the underlying mechanisms by identifying the recurrent genetic changes and characterizing genes involved in the pathogenesis of ESCC.
Downregulation is a common feature of TSG, which can be caused by allele deletion, promoter hypermethylation, histone deacetylation and posttranscriptional silencing by microRNA. Therefore, comparing gene expression profiles between tumor and non-tumor tissues by using cDNA microarray, and characterizing downregulated genes in tumor specimens are useful strategies to identify TSGs. Recently, Affymetrix cDNA microarray has been used to identify deregulated genes between 4 pairs of ESCC tumors and their adjacent non-tumor tissues, and one pair of esophageal dysplasia tissue and adjacent normal tissue. About 200 downregulated genes were identified in ESCC tumors including Rh family, C glycoprotein (RHCG). RHCG belongs to the Rhesus (Rh) family and was first identified as Rh blood group antigens in human erythroid cells. The Rh proteins are closely related to the family of ammonia transporter proteins that is characterized by the presence of 12 transmembrane-spanning segments 5, 6. Therefore, Rh family proteins fall into two functionally distinct groups: ammonia transporting Rh glycoproteins (RHAG, RHBG, and RHCG) and nontransporting Rh proteins (RHD and RHCE). Erythroid RHD is recognized for its key role in blood group incompatibility (Rh+, Rh-), where Rh- individuals can develop an immune response upon exposure to Rh+ blood 7. The nonerythroid Rh glycoproteins, RHBG and RHCG, as ammonia transporters, are widely expressed in different tissues in which ammonia metabolism is necessary for normal biological functioning, such as kidneys, liver, central nervous system, and gastrointestinal tract 8-12. While RHCG was reported to be highly expressed in human esophagus, cervix, oral cavity and skin, its physiological functions are not well documented. Accordingly, recent reports found that RHCG was frequently downregulated in primary ESCC and tongue squamous cell carcinoma compared with their corresponding normal mucosa 13, 14; however, the effects of RHCG downregulation on cancer development and progression are still not yet explored. In the present study, we evaluated the expression status of RHCG and its promoter methylation in primary ESCCs and ESCC cell lines. Functional assays with RHCG ectopic-expressing or RHCG-silenced ESCC cell lines were performed to characterize the biological effects of RHCG in ESCC tumorigenicity and metastasis, both in vitro and in vivo. The tumor-suppressive mechanism of RHCG and its potential to be a new biomarker in ESCC were also addressed.
Results
RHCG is a novel tumor suppressor gene in human ESCC
To identify deregulated genes during ESCC development, Affymetrix cDNA microarray was applied to compare differentially expressed genes between 4 pairs of ESCC tumors and corresponding adjacent non-tumor tissues (#301, #327, #351 and #363), and one pair of esophageal dysplasia tissue and adjacent normal tissue (#314) (GSE100942). About 250 deregulated genes were identified and listed in Table S1. According to previous reports and functional prediction of these genes, 30 candidates were chosen for further screening in ESCC clinical samples and cell lines, and RHCG was selected for subsequent investigations throughout this study. The expression of RHCG was dramatically reduced in 4 ESCC tumors compared with their non-tumor tissues, while its expression remained unchanged in dysplasia tissue (Fig. 1A). To determine whether downregulation of RHCG was a common event in ESCCs, qRT-PCR was performed to compare expression level of RHCG in 45 paired ESCC samples (tumor vs. non-tumor tissues). The result showed that expression of RHCG was significantly downregulated in tumor tissues compared with adjacent non-tumor tissues (paired t test, P < 0.0001; Fig. 1B). Compared with their paired non-tumor tissues, downregulation of RHCG was detected in 40/45 (88.9%) of ESCC tissues.
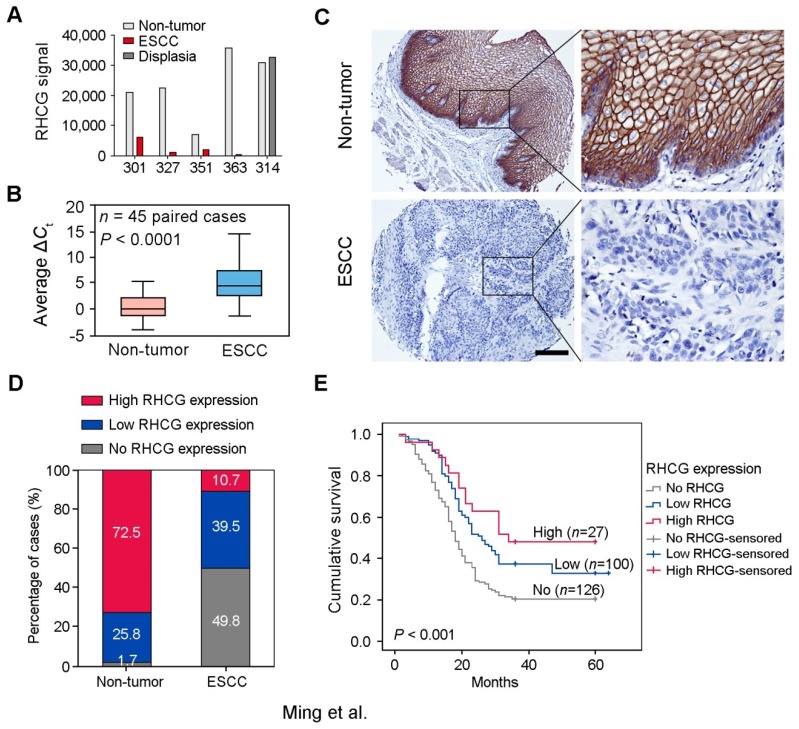
Figure 1.
Downregulation of RHCG in clinical ESCCs. (A) RHCG signal was detected by cDNA microarray in four paired ESCC/nontumor samples (#301, #327, #351 and #363), and one pair of esophageal dysplasia tissue and adjacent normal tissue (#314). (B) RHCG expression in matched nontumor and ESCC tumor samples (n = 45) as detected by qRT-PCR. GAPDH was used as an internal control. The data displayed as average ΔCt values. Boxes in the box plot contain the values between the 25th and 75th percentiles. The lines across the boxes indicate the median. (C) Representative IHC staining of RHCG in ESCC tumor and paired nontumor squamous epithelial tissue (Scale bar = 200 µm). (D) Bar chart summary of the distribution of different RHCG expression levels in nontumor versus tumor for all informative cases on the TMA. (E) Kaplan-Meier survival analysis comparing the overall survival time of ESCC patients with different RHCG expression levels. Statistics: B, paired t test; E, Kaplan-Meier analysis and log-rank test.
To investigate the clinical significance of RHCG expression in ESCC, expression of RHCG was analyzed by immunohistochemistry (IHC) on a tissue microarray (TMA) consisting of 300 paired ESCC and non-tumor specimens (Fig. 1C). Almost three quarters of all informative non-tumor samples displayed high RHCG expression level (171/236, 72.5%), whereas its expression was absent in half of the informative tumor samples (126/253, 49.8%; Fig. 1D). Non-informative samples including lost samples and unrepresentative samples were not included in data compilation. A clinicopathologic association study based on 253 informative ESCC samples showed that the absence of RHCG expression was significantly associated with presence of invasion (Pearson χ² test, P = 0.003), advanced clinical stage (Pearson χ² test, P < 0.001), lymph node metastasis (Pearson χ² test, P = 0.038) and poor differentiation (Pearson χ² test, P = 0.001; Table 1). In a Kaplan-Meier survival analysis comparing patients with different RHCG expression levels, high RHCG expression was significantly associated with long survival time (Log-rank test, P < 0.001). Among patients with different RHCG expression levels, increased survival was observed with enhanced RHCG expression (Fig. 1E). Patients with tumor expressing high RHCG expression had the longest mean survival time of 39.6 months, whereas those without RHCG expression had the worst prognosis, with a mean overall survival of 25.2 months. Multivariate Cox regression analysis further revealed that loss of RHCG was an independent prognostic marker for the overall survival of ESCC patients (hazard ratio, 0.541; 95% confidence interval, 0.395-0.743; P < 0.001; Table S2).
Table 1.
Association of RHCG expression with clinicopathologic features in 253 primary ESCCs
| Features | Total | RHCG expression | P-valueb | |
|---|---|---|---|---|
| Negative | Positive | |||
| Sex | ||||
| Female | 114 | 61 (53.5%) | 53 (46.5%) | |
| Male | 139 | 65 (46.8%) | 74 (53.2%) | 0.286 |
| Age, years | ||||
| <60 | 134 | 67 (50%) | 67 (50%) | |
| ≥60 | 119 | 59 (49.6%) | 60 (50.4%) | 0.947 |
| Differentiation | ||||
| Well/moderate | 188 | 82 (43.6%) | 106 (56.4%) | |
| Poor | 65 | 44 (67.7%) | 21 (32.3%) | 0.001 |
| Invasion | ||||
| Absent | 89 | 33 (37.1%) | 56 (62.9%) | |
| Present | 164 | 93 (56.7%) | 71 (43.3%) | 0.003 |
| Clinical Stagea | ||||
| Early stage I-II | 161 | 66 (41%) | 95 (59%) | |
| Advanced stage III-IV | 92 | 60 (65.2%) | 32 (34.8%) | <0.001 |
| Lymph node metastasis | ||||
| Absent | 135 | 59 (43.7%) | 76 (56.3%) | |
| Present | 118 | 67 (55.8%) | 51 (44.2%) | 0.038 |
aAJCC/UICC TNM staging system.
bPearson χ² test.
The RHCG promoter region is frequently hypermethylated in ESCC
In addition to clinical samples, we have also determined the expression of RHCG by Western blot analysis in 1 immortalized esophageal epithelial cell line (NE1) and 9 ESCC cell lines. The expression of RHCG was found to be absent or downregulated in all ESCC cell lines as compared with NE1 cells (Fig. 2A). Since downregulation of TSG in cancer is often associated with epigenetic regulation including promoter hypermethylation and histone deacetylation, we next studied the effects of DNA demethylation and histone acetylation in RHCG expression. EC109 and KYSE180 cells were treated with different concentrations of the DNA methyltransferase inhibitor (5-zaz-dC) or the histone acetylation agents trichostatin A (TSA). qPCR analysis showed a dose-dependent restoration of RHCG expression after demethylation treatment with 5-aza-dC (Fig. 2B). Treatment with the histone acetylation agent TSA did not significantly increase the expression level of RHCG, suggesting that DNA methylation, but not histone modification, is involved in RHCG inactivation in ESCC.
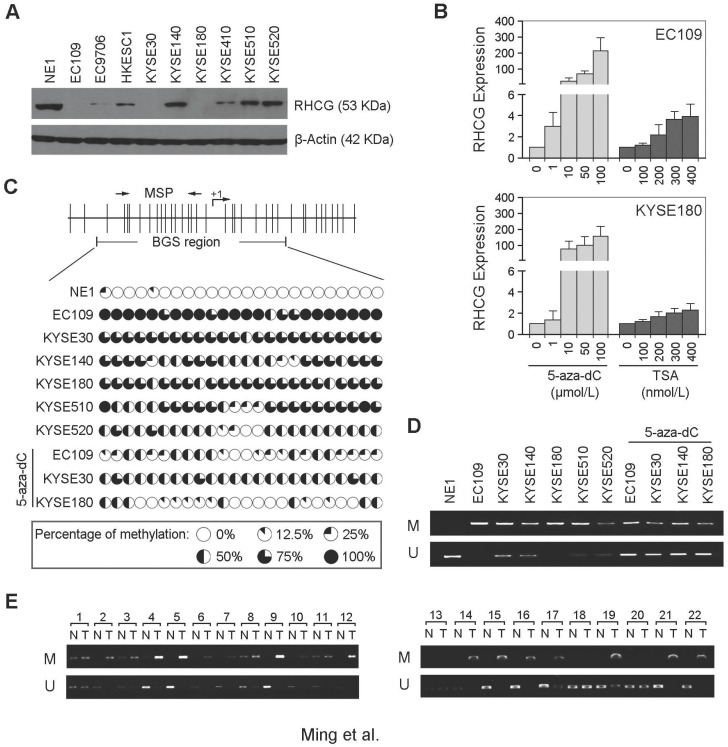
Figure 2.
RHCG promoter region is frequently hypermethylated in ESCCs. (A) Measurement of RHCG expression levels in a panel of ESCC cell lines compared with squamous epithelial cell line NE1 by Western blotting. (B) qRT-PCR analysis of RHCG expression after demethylation or histone acetylation treatment with 5-aza-dC or TSA in ESCC cell lines without RHCG expression. (C) High-resolution mapping of the methylation status of individual CpG site in the RHCG promoter by BGS in normal cell line NE1 and ESCC cell lines with or without 5-aza-dC treatment. A 270-bp region spanning the CpG island with 24 CpG sites was analyzed. Each CpG site analyzed is shown at the top row as a short vertical bar. The percentage of methylation was determined as percentage of methylated CpGs from 10 to 15 randomly sequenced clones and is displayed in the pie charts. (D-E) Representative MSP analysis of RHCG in esophageal cell lines with or without 5-aza-dC treatment, and paired primary ESCCs/nontumor tissues (T, tumor tissue; N, non-tumor tissue; M: methylation; U: unmethylation).
To investigate the role of aberrant promoter hypermethylation in RHCG silencing, BGS and MSP were performed to examine the methylation status of the RHCG promoter region. A 10 kb sequence directly upstream of the RHCG gene was analyzed by CpG Island Searcher (http://www.cpgislands.com) and MethPrimer (http://www.urogene.org/methprimer) for potential CpG islands. Only one CpG island was predicted by both programs (-193 nt to +77 nt), as shown in Fig. 2C. BGS analysis was conducted at this CpG island and found a high density of methylation in ESCC cell lines, especially in the RHCG-absent EC109 and KYSE180 cells. In contrast, methylation was rarely detected in the same CpG sites in the normal esophageal cell line NE1 (Fig. 2C). As compared with untreated cells, methylation was significantly reduced in EC109, KYSE30 and KYSE180 cells treated with 5-aza-dC. We have also performed MSP to investigate the methylation status of RHCG in the same panel of cell lines. Methylated allele of RHCG was detected in EC109 and KYSE180 cell lines, which were negative for the expression of RHCG, whereas no methylated allele was observed in NE1 (Fig. 2D). These data were highly consistent with the BGS result. Then, we investigated the methylation frequency of RHCG promoter in 22 primary ESCC tumors and their paired non-tumor tissues by MSP. Methylation of RHCG was detected in 19/22 (86.4%) of the primary ESCCs. In contrast, methylation was only found in 5/22 (22.7%) of the paired non-tumor tissues (Fig. 2E). Taken together, these observations strongly suggested that DNA promoter hypermethylation is implicated in RHCG inactivation in ESCC.
RHCG has strong tumor suppressive function
To investigate whether RHCG possesses tumor suppressive function, EC109 and KYSE180 cells without RHCG expression were stably transduced with lentivirus packaged with either a RHCG-expressing vector or an empty vector (EV) control to generate RHCG-overexpressing or control cells. Similarly, KYSE520 cells with high RHCG expression were lentivirally transduced with either RHCG-specific short hairpin RNA (shRNA) or a non-target control (NTC) to generate cells with RHCG stably repressed (KYSE520 shRHCG-1 or shRHCG-3) or control cells (KYSE520-NTC). Overexpression and knockdown of RHCG were confirmed at both mRNA and protein levels by PCR and Western blotting, respectively (Fig. 3A and 3B). Tumor suppressive function of RHCG was assessed by foci formation assay, soft agar assay and tumor xenograft experiment. Compared with control cells, RHCG-overexpressing cells displayed lower foci formation frequencies (Fig. 3C) and reduced colony forming abilities in soft agar (Fig. 3D). Conversely, silencing RHCG expression revealed an opposing effect (Fig. 3C and 3D).
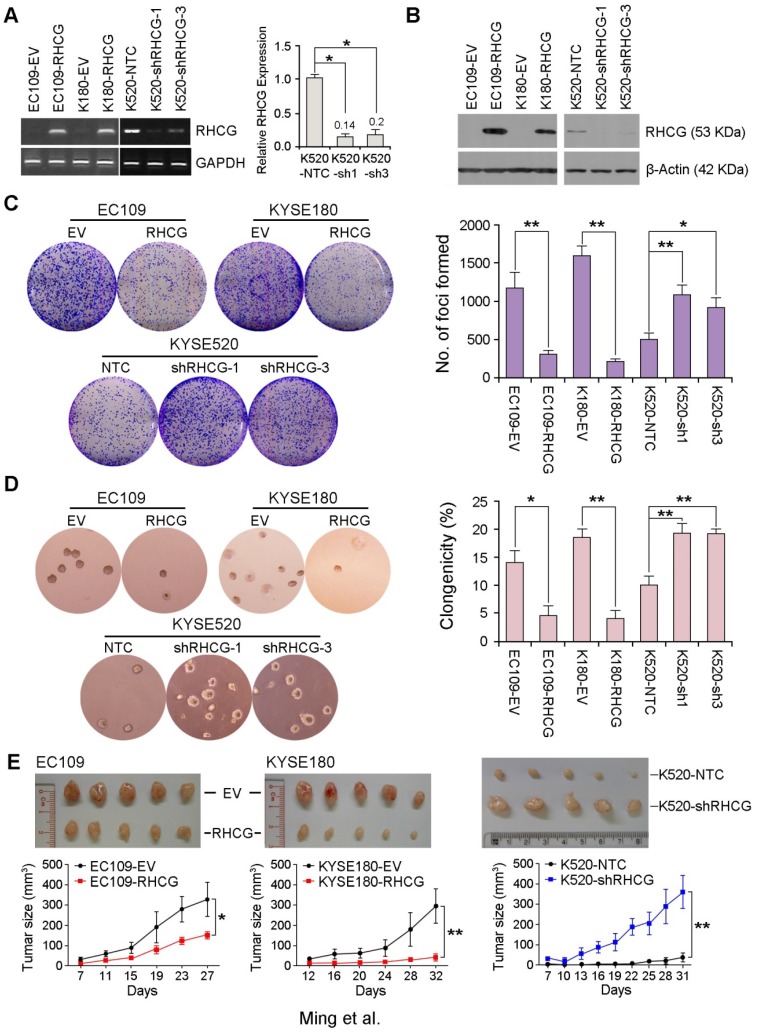
Figure 3.
RHCG from ESCC cell lines suppresses tumor formation. Stable overexpression or repression of RHCG by lentiviral transduction was confirmed in both genomic and proteomic levels by PCR (A) and Western blotting (B), respectively. (C-D) Representative images and summaries of foci formation (C) and soft agar assays (D) performed in RHCG-overexpressed or -repressed cells as compared with their controls. The values indicate the mean±SD of three independent experiments. (E) Representative images of the subcutaneous tumors formed in nude mice following injection of RHCG-overexpressed or -repressed clones and their respective controls. Tumor growth curves are summarized in the line chart. The average tumor volume is expressed as the mean±SD of five mice. Statistics: A, C, D and E, Student t test. *, P<0.05; **, P<0.001.
To further investigate the in vivo tumor suppressive ability of RHCG, RHCG-transfected cells and control cells were subcutaneously injected into the right and left dorsal flank of nude mice, respectively. Tumor induced by control cells showed significantly shorter latency and larger mean tumor volume than tumors induced by RHCG-expressing cells (Fig. 3E, left). In contrast, mice injected with RHCG-silenced KYSE520 cells formed much larger tumors than those formed by NTC cells (Fig. 3E, right). These results demonstrated that RHCG has a strong tumor suppressive ability both in vitro and in vivo.
RHCG suppresses invasion and lymph node metastasis in ESCC
As loss of RHCG was significantly associated with tumor invasion and lymph node metastasis in ESCC patients, the effect of RHCG on cell motility and tumor metastasis was studied by wound-healing, cell migration and invasion assays. The wound-healing assay showed that RHCG could dramatically inhibit tumor cell mobility (Fig. 4A). Cell migration and invasion assays also demonstrated that RHCG-transfected cells had suppressed cell migration and invasion abilities, as compared with control cells (Fig. 4B). Once endogenous RHCG was silenced by shRNA, the abilities of cell migration and invasion were significantly increased (Fig. 4A and 4B).
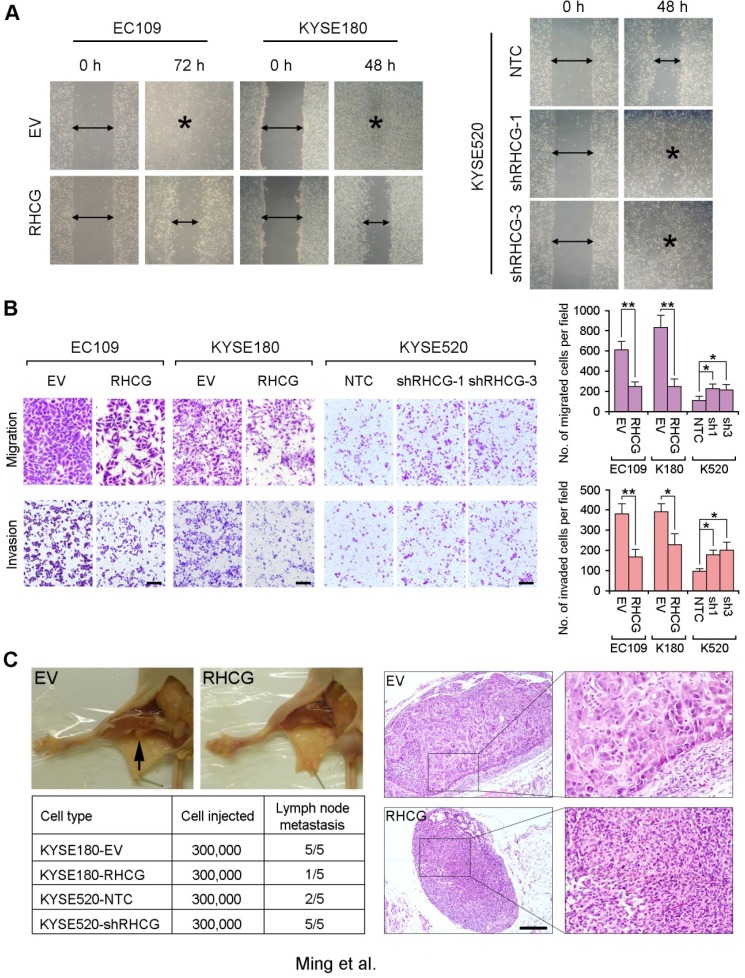
Figure 4.
RHCG suppresses in vivo lymph node metastasis of ESCC. (A) Representative images of wound healing in RHCG-overexpressed or -repressed cells as compared with their controls. (B) Representative images and summary of migration and invasion assays in RHCG-overexpressed or -repressed cells as compared with their controls. The values are expressed as the mean±SD of three independent experiments. Scale bar = 200 µm. (C) Representative images of lymph node metastasis formed in NOD/SCID mice injected with RHCG-overexpressed or -repressed cells. Black arrow indicates the swollen popliteal lymph node. Lymph nodes invaded by tumor cells were confirmed by H&E staining (Scale bar = 100 µm). The tumor metastasis rate is shown in the table section. Statistics: B, Student t test. *, P<0.05; **, P<0.001.
To further investigate the effect of RHCG on tumor metastasis, a lymph node metastasis animal model was performed by injecting RHCG-transfected KYSE180 cells into the right hind foot-pad of NOD/SCID mice. Empty vector-transfected cells were used as controls. In the lymph node metastasis model, popliteal lymph nodes, which represent the sentinel lymph node for this model, were examined 15. Swollen popliteal lymph nodes were observed in all 5 mice injected with control cells, which was confirmed by H&E staining. Only 1/5 of mice injected with RHCG-KYSE180 cells displayed tumor metastasis (Fig. 4C). Conversely, lymph node metastasis was observed in all mice injected with KYSE520-shRHCG cells, whereas 2/5 mice from the control group showed tumor metastasis. In order to minimize the number of mice to be used, two RHCG-specific shRNAs treated cells were equally mixed together as KYSE520-shRHCG group for in vivo study.
mRNA profiling identifies an altered network in RHCG-expressing KYSE180 cells
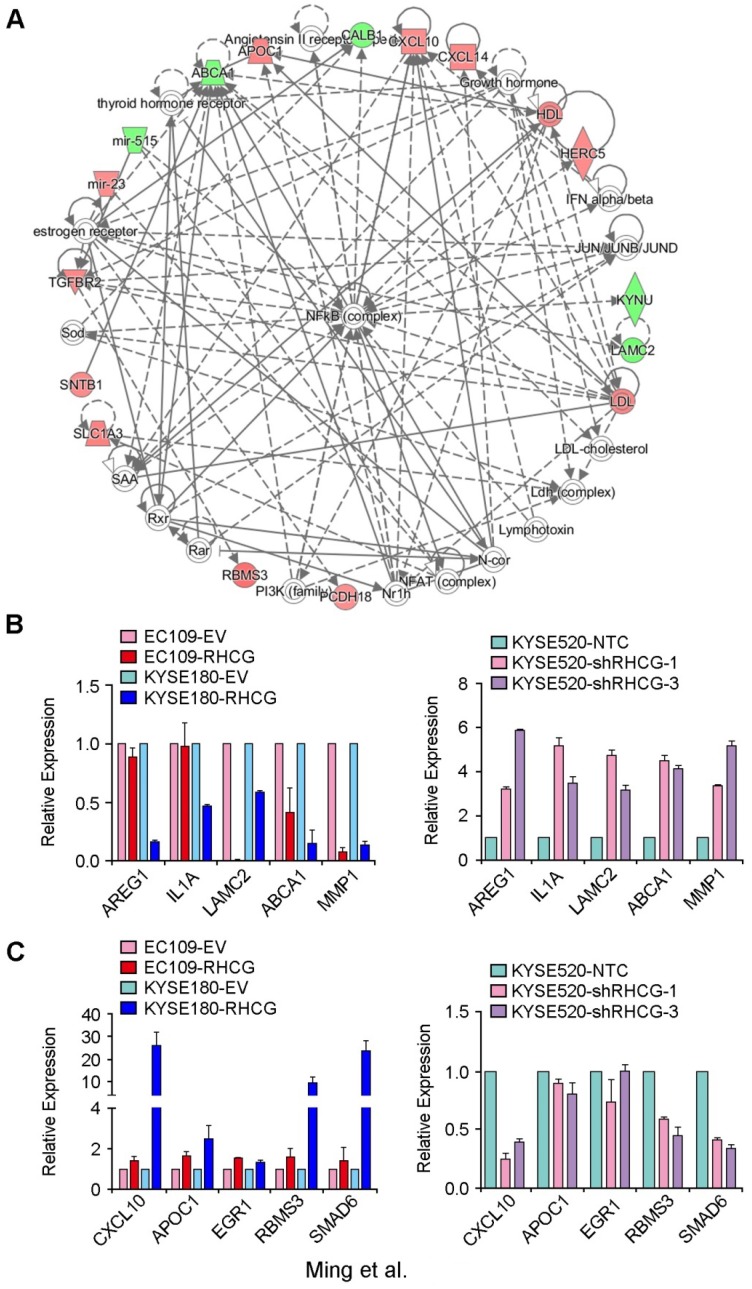
In an attempt to characterize the molecular mechanism by which RHCG inhibits tumor formation and metastasis in ESCC, a genome-wide mRNA expression profiling screen was used to compare gene expression profiles between RHCG-overexpressing KYSE180 cells and their control cells. Using a fold change of 1.5, 125 differentially expressed genes were identified, including 67 upregulated genes and 58 downregulated genes (Table S3). Further analysis by IPA software identified a significantly altered functional network on the nuclear factor κB (NF-κB) complex (Fig. 5A). To confirm whether the deregulated genes were associated with the expression of RHCG, 5 downregulated genes (AREG1, IL1A, LAMC2, ABCA1 and MMP1) and 5 upregulated genes (CXCL10, APOC1, ERG1, RBMS3 and SMAD6) were selected for validation in both RHCG-expressing cells (EC109 and KYSE180) and RHCG-silenced KYSE520 cells. The results from qRT-PCR indicated that almost all downregulated genes were negatively associated with RHCG expression, in particular, a strong inverse correlation was found in MMP1 expression (Fig. 5B). On the other hand, the expression of upregulated genes was positively correlated with RHCG expression (Fig. 5C). These results indicated that the mRNA expression profiling screening data were reliable.
Figure 5.
Deregulated genes in RHCG-overexpressed KYSE180 cells (A) A significantly deregulated NF-κB network in RHCG-KYSE180 cells identified by genome-wide expression profiling and Ingenuity Pathway Analysis (IPA). Genes in green and red indicate under- and over-expressed genes found in RHCG-KYSE180 cells compared to their negative counterparts. Five under- (B) or over-expressed (C) genes in RHCG-KYSE180 cells were selected to confirm their expression in RHCG overexpressed and suppressed cell lines by qRT-PCR. GAPDH was used as an internal control.
RHCG suppresses ESCC metastasis via inhibiting NF-κB signaling
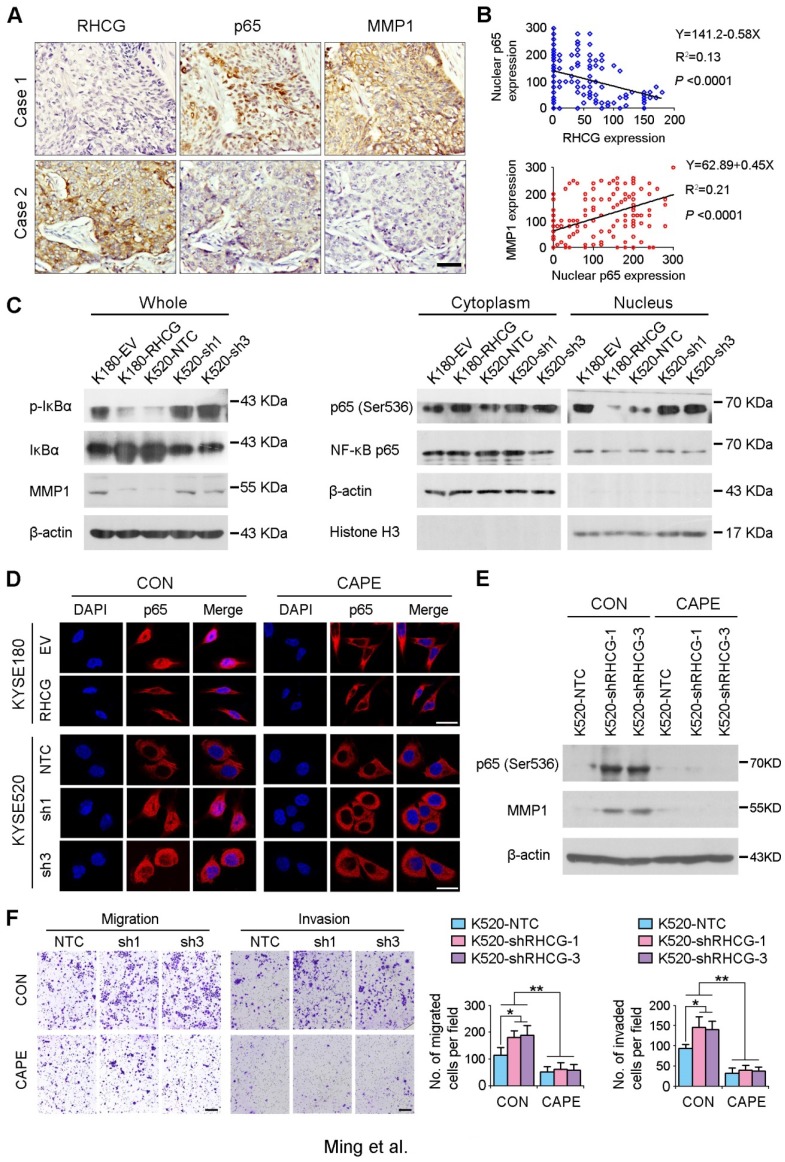
As NF-κB complex member p65 has previously been reported to be overexpressed in ESCCs 16, and NF-κB/p65 induces the production of MMPs including MMP1, which subsequently orchestrate the malignant phenotype of ESCC 17, 18, we therefore hypothesize that RHCG may downregulate MMP1 through inhibiting NF-κB signaling to suppress ESCC progression. To demonstrate this hypothesis, we checked the correlation of RHCG, p65 and MMP1 in clinical specimens of ESCC. IHC staining with antibodies of p65 and MMP1 was performed in the same ESCC TMA. A specific correlation between p65 nuclear location and MMP1 expression was observed in the ESCC samples without RHCG expression, while the nuclear translocation of p65 and the expression of MMP1 in the RHCG-expressing cases were rarely observed (Fig. 6A). Linear regression analysis was applied to evaluate the correlation between RHCG and nuclear expression of p65, as well as between nuclear p65 and MMP1. A negative correlation was observed between RHCG and nuclear p65 (R2 = 0.13; P < 0.0001), while a positive correlation was found between nuclear p65 and MMP1 (R2 = 0.21, P < 0.0001, Fig. 6B). Clinicopathologic feature studies showed that nuclear expression of p65 and upregulation of MMP1 were significantly associated with advanced clinical stage, lymph node metastasis in ESCC (Table S4-S5), and shorter overall survival rate (Fig. S1). These observations prompted us to investigate whether RHCG suppresses MMP1 expression via inhibiting NF-κB signaling.
Figure 6.
RHCG suppresses ESCC metastasis via inhibiting NF-κB signaling and MMP1 expression. (A) IHC staining for RHCG, p65 and MMP1 expression in the clinical ESCC samples. Scale bar = 50 μm. (B) The correlation between nuclear p65 and RHCG or MMP1 expression was detected by IHC staining in TMA containing 300 ESCC samples. Linear regression lines and Pearson correlation significance are shown. (C) Western blot analysis was used to detect whole cell, cytoplasm and nucleus level of NF-κB signaling and MMP1. β-actin was used as loading controls for whole protein and cytoplasm protein, while Histone H3 was used as a loading control for nuclear protein. (D) Representative images of indicated cells detected by immunofluorescent staining with p65 antibody (red color). Nuclei were counterstained with DAPI (blue color). Cells were treated with DMSO (CON) or 2 µM Caffeic acid phenethyl ester (CAPE). Scale bar = 20 μm. (E) The western blotting result showed that CAPE treatment inhibited activation of p65 and the expression of MMP1 in RHCG-suppressed KYSE520 cells. (F) Representative images and summary of migration and invasion assays in RHCG-repressed cells treated with or without CAPE. The values are expressed as the mean ± SD of three independent experiments. Scale bar = 200 μm. Statistics: B, Pearson correlation coefficients; F, ANOVA with post hoc test. *, P<0.05; **, P<0.001.
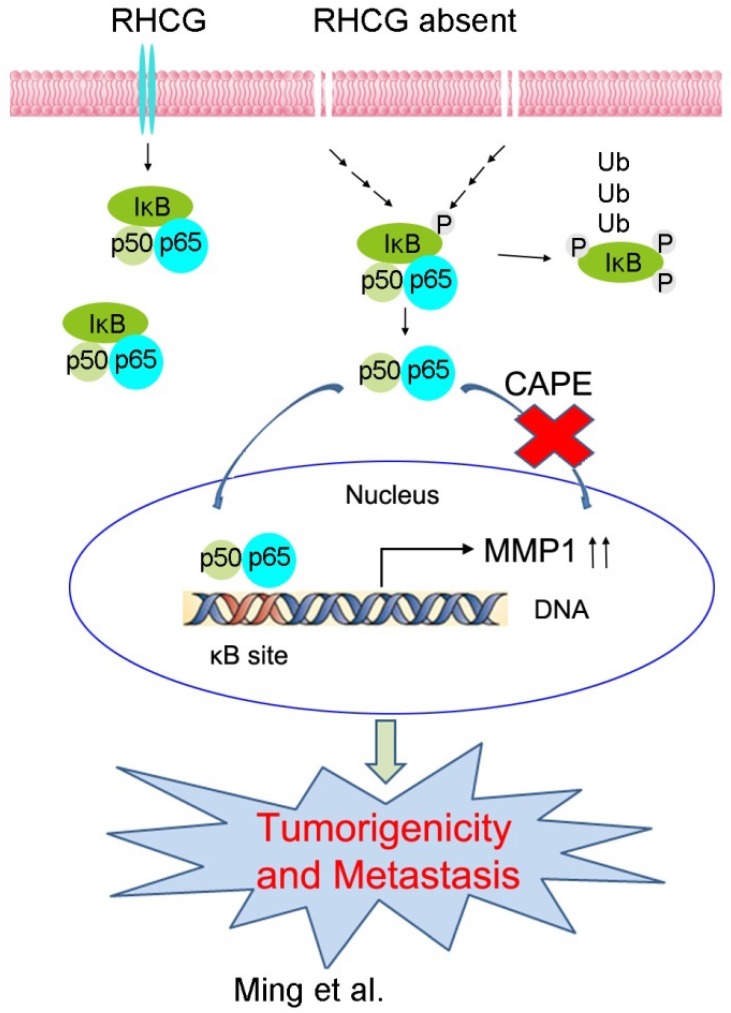
In resting state, NF-κB/p65 protein is inhibited by IκB in the cytoplasm. When IκB is phosphorylated and subsequently degraded, NF-κB/p65 protein will be activated and translocated to the nucleus for target gene regulation. To test whether RHCG suppresses MMP1 expression via inhibition of NF-κB signaling, we detected IκB/p65 complex by Western blotting in RHCG upregulated- and downregulated-cells. Consistent with qRT-PCR result, RHCG could suppress MMP1 expression, while the opposing effect was observed when RHCG was silenced by shRNA (Fig. 6C, left). From the whole protein result, we found that RHCG expression could protect IκB from phosphorylation and degradation (Fig. 6C, left). Then, we determined the phosphorylated and total IKKα/β in RHCG upregulated- and downregulated-cells, which are the classical upstream regulators of IκB/p65; however, we could not observe any difference in both cell lines (Fig. S2A). To explore whether NF-κB/p65 protein translocates to nucleus upon degradation of IκB, we first successfully isolated cytoplasm protein and nucleus protein from the RHCG upregulated- and downregulated- cells, as confirmed by the presence of β-actin and Histone H3 in cytoplasm and nucleus, respectively. The result demonstrated that the active form of NF-κB/p65 in nucleus was significantly decreased in RHCG-expressing cells, whereas it was dramatically elevated in RHCG-silenced cells, compared with their control cells (Fig. 6C, right). A similar result was observed by immunofluorescence staining, where RHCG expression inhibited the nuclear translocation of p65 (Fig. 6D, left). To further validate NF-κB/p65 is the downstream target of RHCG, Caffeic Acid Phenethyl Ester (CAPE, Selleckchem), a selective NF-κB inhibitor that suppresses the nuclear import of p65, was applied to block this signaling. The IF staining demonstrated that p65 nuclear translocation was significantly blocked in RHCG-silenced cells after treatment with CAPE (Fig. 6D, right). Accordingly, CAPE treatment could also inhibit p65 phosphorylation and MMP1 expression in RHCG-silenced cells (Fig. 6E). Consequently, CAPE treated RHCG-silenced cells showed lower foci formation frequencies (Fig. S2B) and reduced colony forming abilities in soft agar (Fig. S2C). Meanwhile, the abilities of migration and invasion of RHCG-silenced cells were also suppressed after treatment with CAPE (Fig. 6F). These results demonstrated that RHCG suppressed MMP1 expression in ESCC cells, which was dependent on NF-κB/p65 signaling (Fig. 7).
Figure 7.
A schematic diagram illustrating the proposed RHCG-regulated tumor-suppressive mechanism in ESCC tumorigenicity and metastasis.
Discussion
RHCG is an integral membrane protein that belongs to the Rh family and forms homotrimer in plasma membrane. As a member of ammonia transporters, Rh glycoproteins exert important biological and physiological functions in various tissues, e.g., kidneys, liver, brain and gastrointestinal tract 5, 8-11, 19. In addition, RHCG is also highly expressed in human squamous epithelia of esophagus, cervix, and oral cavity; however, the biological functions of RHCG in these tissues have not been explored. Recently, downregulation of RHCG was frequently observed in squamous cell carcinoma from esophagus and tongue 13, 14. To date, the relationship between inactivation of RHCG and the development of squamous cell carcinoma is still unknown. In the present study, downregulation of RHCG was frequently detected in primary ESCCs, which was significantly associated with invasion (P = 0.003), advanced clinical stage (Pearson χ² test, P < 0.001), lymph node metastasis (P = 0.038), poor differentiation (P = 0.001) and poorer prognosis (P < 0.001). Further study found that promoter hypermethylation played a crucial role in the downregulation of RHCG in ESCCs.
In the present study, we investigated the tumor suppressive function of RHCG by both in vitro and in vivo functional assays. Ectopic expression of RHCG in ESCC cell lines EC109 and KYSE180 cells could effectively suppress foci formation, colony formation in soft agar, and tumor formation in nude mice, while RHCG silencing in KYSE520 cells could promote these functions. Our clinical correlation study demonstrated that the downregulation of RHCG was significantly associated with ESCC invasion and lymph node metastasis. Since ESCC primarily exhibits metastasis through the lymphatic system 20, 21, we constructed a lymph node metastasis model to investigate whether RHCG could suppress tumor metastasis in ESCC 15. Both in vitro and lymph node metastasis animal model revealed that RHCG could suppress cell migration, invasion and tumor metastasis.
To explore the molecular mechanism(s) underlying the tumor suppressive functions of RHCG, cDNA microarray was applied to identify potential genes and signaling pathways regulated by RHCG by comparing the expression profiles between RHCG- and empty vector-transfected KYSE180 cells. Interestingly, NF-κB signaling activity was inhibited in RHCG-transfected cells. The NF-κB complex comprises a family of transcription factors involved in regulating a wide variety of biological responses. Aberrant or constitutive NF-κB activation has been detected in many human malignancies and this signaling pathway has been seen as an attractive therapeutic target for cancer treatment 22-25. Recent studies found that NF-κB/p65 promotes tumorigenicity and metastasis of ESCC by up-regulating MMP1 and MMP9 16, 18. Accordingly, high expressions of MMP1 were frequently found in ESCC and were closely associated with advanced TNM stage and lymph node metastasis. A functional study also revealed that ectopic expression of MMP1 promoted tumor growth and metastasis in ESCC 18. Our data showed that the expression of MMP1 was downregulated in RHCG-expressing cells. In addition, our clinical data showed that expression of MMP1 in ESCC tissues was positively correlated with the nuclear translocation of p65, which was significantly associated with advanced clinical stages and the presence of invasion and lymph node metastasis in primary ESCCs. These findings are consistent with a previous report that NF-κB/p65 could directly bind to the promoter of MMP1 and regulate its expression 17. Further molecular study found that downregulation of RHCG could increase the phosphorylation of IκB for its proteasomal degradation and thus, promote the subsequent activation of NF-κB/p65 and nuclear translocation. Once p65 was translocated into the nucleus, it acted as a transcription regulator and up-regulated MMP1 expression. It is still not known, however, which signals were mediated by transmembrane protein RHCG for the phosphorylation of IκB.
Although different treatment strategies have been developed, the survival rate of ESCC patients remains unsatisfactory 26-28. Therefore, novel risk markers to guide disease management of ESCC patients are required to tailor treatment to improve their prognosis. As downregulation of RHCG has been significantly associated with poorer prognosis of patients with ESCC, RHCG may be one of the novel prognostic biomarkers in ESCC patients. To further validate the potential clinical application value, univariable and multivariable Cox proportional hazard regression analysis was performed to determine whether downregulation of RHCG is an independent factor for predicting prognosis. The result showed that the downregulation of RHCG was an independent factor and the most influential factor in predicting ESCC outcome. In summary, our findings highlighted a prognostic potential of RHCG, which acts as a potent TSG and exerts a pivotal role in inhibiting tumorigenicity and metastasis in ESCC.
Materials and Methods
Cell lines
Immortalized normal esophageal epithelial cell line NE1 was obtained from Professor George Tsao's laboratory (Department of Anatomy, The University of Hong Kong) in 2006. Chinese ESCC cell lines (HKESC1, EC109 and EC9706) and six Japanese ESCC cell lines (KYSE30, KYSE140, KYSE180, KYSE410, KYSE510, and KYSE520) were kindly provided by Professor Srivastava (Department of Pathology, The University of Hong Kong) in 2004. These adherent cells were trypsinized and expanded for 2 to 4 weeks when confluency reached 70% to 80%. Mycoplasma infection was detected by DNA stain and PCR, and contamination was not found in these cell lines.
Primary ESCC specimens
Primary ESCC specimens (300 pairs) and their adjacent nontumor esophageal tissues, used for a tissue microarray (TMA), were collected from patients who underwent esophageal resection at Linzhou Cancer Hospital (Henan, China). Forty-five paired ESCC primary/nontumor tissues, used for DNA and RNA extraction, were collected from ESCC patients at Sun Yat-Sen University Cancer Center (Guangzhou, China). None of these patients received preoperative treatment. Histological examination was carried out by pathologists, and diagnosis was made based on the microscopic features of the carcinoma cells. Tumors were graded using the American Joint Committee on Cancer (AJCC)/International Union Against Cancer (UICC) tumor staging system. Informed consent was obtained from all patients before the collection of esophageal specimens, and samples used in this study were approved by the committees for ethical review of research involving human subjects at Zhengzhou University (Zhengzhou, China), and Sun Yat-Sen University (Guangzhou, China).
Animals
The study protocol was approved by and performed in accordance with the Committee of the Use of Live Animals in Teaching and Research at the University of Hong Kong. For in vivo tumorigenic experiment, the cell number used for BALB/cAnN-nu (nude) mice subcutaneous injection was 1×106 (EC109 and KYSE180), and 2×105 (KYSE520). Tumor size was measured twice a week, and tumor volumes were calculated as: V (mm3) = L×W2×0.5. For in vivo metastasis assay, each experimental group consisted of five 4-week-old NOD/SCID mice. Briefly, 3×105 cells in 20 µL PBS were injected into the right hind foot-pad of each mouse. All of the mice were euthanized 6 weeks after injection. The right side popliteal lymph nodes were excised and embedded in paraffin.
Tissue microarray construction and immunohistochemistry
Three tissue array blocks with 300 paired primary/nontumor samples were constructed. Briefly, H&E stained slides from each case were used as a guide to identify areas representing different stages of the tumor. The tissues were prepared using a tissue arraying instrument (Beecher Instruments, Silver Spring, MD). A 0.6 mm-diameter cylinder of tissue was removed and re-embedded into a predetermined position in a recipient paraffin block. Immunohistochemistry (IHC) staining was performed using the standard streptavidin-biotin-peroxidase complex method. Briefly, paraffin sections were deparaffinized and rehydrated. Slides were heated for antigen retrieval for 15 min in 10 mM citrate (pH 6.0). Sections were incubated with primary antibody at 4°C overnight. EnVision Plus System-HRP (DAB; DAKO) was used according to manufacturer's instruction, followed by Mayer's hematoxylin counterstaining. Stained slides were imaged on an AperioScanscope® CS imager (Vista, CA). The intensity of positive staining was scored as: 0, negative; 1, weak; 2, moderate; 3, strong. The total score was determined by the following formula: Staining Index (SI) = intensity × positive percentage × 100. According to ROC curve analyses, the optimum cutoff value for p65 and MMP1 was 100, so SI < 100 was considered low expression, and S ≥ 100 was considered as high expression. Since half of the ESCC samples did not express RHCG, the samples were divided into negative and positive groups; the positive group was subdivided into low expression (SI < 100) and high expression (SI ≥ 100) groups.
DNA extraction and promoter methylation analysis
Genomic DNA was extracted by phenol-chloroform method followed by bisulfite modification using the EpiTECT Bisulfite Kit (Qiagen). Primers for bisulfite genomic sequencing (BGS) and methylation-specific PCR (MSP) are listed in Table S6. For BGS, the template was amplified by PCR for 35 cycles and then cloned into the pGEM-T Easy vector (Promega) and sequenced as individual clones by Beijing Genomics Institute. The sequence data was analyzed by BiQ analyzer (Max Planck Institute). For MSP, the template was amplified by PCR for 35 cycles, and then analyzed by DNA gel electrophoresis. Cell treatment with 5-aza-2'-deoxycytidine (5-aza-dC) or trichostatin A (TSA) is described in Supplemental Methods.
In vitro tumorigenic assays
Anchorage-dependent growth was assessed by foci formation assay. Briefly, 2×103 cells were seeded in a 6-well plate. Surviving colonies (>50 cells/colony) were stained with 1% crystal violet (Sigma-Aldrich) and counted after one week. Anchorage-independent growth was assessed by colony formation in soft agar. In brief, 5×103 cells were mixed thoroughly in 0.35% low melting soft agar (BD Biosciences) and were grown in a 6-well plate precoated with 0.5% agar. After two weeks, colonies (≥ 10 cells) were counted under a microscope in 10 fields per well and photographed.
Migration and invasion assays
Migration and invasion assays were performed in 24-well millicell hanging insert (Millipore) or 24-well BioCoat Matrigel Invasion Chambers (BD Biosciences). In brief, 1×105 cells were seeded to the top chamber and 10% FBS in medium was added to the bottom chamber as a chemoattractant. After 24 h or 48 h incubation, the number of cells that invaded through the membrane (migration) or Matrigel (invasion) was counted in 10 fields and imaged using SPOT imaging software (Nikon).
cDNA microarray analysis
Expression profiling of ESCC clinical samples was performed with HG-U133-PLUS 2 (Affymetrix). Altered gene expression patterns of KYSE180-EV and KYSE180-RHCG were compared using Affymetrix GeneChip Human Gene 2.0 ST Array. The data were summarized with the Expression Console™ software (Affymetrix), and further analyzed using GeneSpring GX (Affymetrix) and Ingenuity Pathway Analysis (IPA; Ingenuity Systems) software. Differentially expressed transcripts are listed in Table S1 and Table S3. Microarray data are available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE100942.
Statistical analysis
Statistical analysis was carried out with SPSS v. 16 (Chicago, IL). Pearson χ² test was used to analyze the association of RHCG expression with clinicopathologic parameters. Kaplan-Meier plot and log-rank test were used for survival analysis. Univariable and multivariable Cox proportional hazard regression models were used to analyze independent prognostic factors. Student t test and ANOVA with post hoc test were used for most studies as indicated in the figure legends. The Pearson correlation coefficients and linear regression analysis were used to evaluate the correlation of RHCG, nuclear p65 and MMP1 in the clinical samples. The data are presented as the mean±SD of three independent experiments. The P values are denoted as *P < 0.05, and **P < 0.001 in all figures.
Acknowledgments
This work was supported by grants from the Hong Kong Research Grant Council (RGC) grants including Collaborative Research Funds (C7038-14G), and General Research Funds (17143716); grants from Health and Medical Research Fund (02131876), National Natural Science Foundation of China (81272416, 81472250 and 81472255), Guangdong Esophageal Cancer Institute (M201511), and the Science and Technology Planning Project of Guangdong Province (No.2013B021800163).
Accession code
cDNA microarray data have been submitted to Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE100942.
Abbreviations
- BGS
bisulfite genomic sequencing
- CAPE
caffeic acid phenethyl ester
- ESCC
esophageal squamous cell carcinoma
- IκB
inhibitor of kappa B
- MSP
methylation-specific PCR
- NF-κB
nuclear factor kappa B
- RHCG
Rh family, C glycoprotein
- TSG
tumor suppressor genes.
Supplementary Material
Supplementary figures and tables.
References
- 1.Ferlay J, Shin HR, Bray F. et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol. 2013;19:5598–606. doi: 10.3748/wjg.v19.i34.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lao-Sirieix P, Fitzgerald RC. Screening for oesophageal cancer. Nat Rev Clin Oncol. 2012;9:278–87. doi: 10.1038/nrclinonc.2012.35. [DOI] [PubMed] [Google Scholar]
- 4.Montesano R, Hollstein M, Hainaut P. Genetic alterations in esophageal cancer and their relevance to etiology and pathogenesis: a review. Int J Cancer. 1996;69:225–35. doi: 10.1002/(SICI)1097-0215(19960621)69:3<225::AID-IJC13>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 5.Lee HW, Verlander JW, Handlogten ME. et al. Expression of the rhesus glycoproteins, ammonia transporter family members, RHCG and RHBG in male reproductive organs. Reproduction. 2013;146:283–96. doi: 10.1530/REP-13-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang CH, Peng J. Evolutionary conservation and diversification of Rh family genes and proteins. Proc Natl Acad Sci USA. 2005;102:15512–7. doi: 10.1073/pnas.0507886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colin Y, Cherif-Zahar B, Le Van Kim C. et al. Genetic basis of the RhD-positive and RhD-negative blood group polymorphism as determined by Southern analysis. Blood. 1991;78:2747–52. [PubMed] [Google Scholar]
- 8.Weiner ID, Verlander JW. Ammonia transport in the kidney by Rhesus glycoproteins. Am J Physiol Renal Physiol. 2014;306:F1107–20. doi: 10.1152/ajprenal.00013.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gruswitz F, Chaudhary S, Ho JD. et al. Function of human Rh based on structure of RhCG at 2.1 A. Proc Natl Acad Sci USA. 2010;107:9638–43. doi: 10.1073/pnas.1003587107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiner ID, Miller RT, Verlander JW. Localization of the ammonium transporters, Rh B glycoprotein and Rh C glycoprotein, in the mouse liver. Gastroenterology. 2003;124:1432–40. doi: 10.1016/s0016-5085(03)00277-4. [DOI] [PubMed] [Google Scholar]
- 11.Handlogten ME, Hong SP, Zhang L. et al. Expression of the ammonia transporter proteins Rh B glycoprotein and Rh C glycoprotein in the intestinal tract. Am J Physiol Gastrointest Liver Physiol. 2005;288:G1036–47. doi: 10.1152/ajpgi.00418.2004. [DOI] [PubMed] [Google Scholar]
- 12.Quentin F, Eladari D, Cheval L. et al. RhBG and RhCG, the putative ammonia transporters, are expressed in the same cells in the distal nephron. J Am Soc Nephrol. 2003;14:545–54. doi: 10.1097/01.asn.0000050413.43662.55. [DOI] [PubMed] [Google Scholar]
- 13.Ye H, Yu T, Temam S. et al. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics. 2008;9:69. doi: 10.1186/1471-2164-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen BS, Xu ZX, Xu X. et al. RhCG is downregulated in oesophageal squamous cell carcinomas, but expressed in multiple squamous epithelia. Eur J Cancer. 2002;38:1927–36. doi: 10.1016/s0959-8049(02)00190-9. [DOI] [PubMed] [Google Scholar]
- 15.Qian CN, Berghuis B, Tsarfaty G. et al. Preparing the "soil": the primary tumor induces vasculature reorganization in the sentinel lymph node before the arrival of metastatic cancer cells. Cancer Res. 2006;66:10365–76. doi: 10.1158/0008-5472.CAN-06-2977. [DOI] [PubMed] [Google Scholar]
- 16.Wang F, He W, Fanghui P. et al. NF-kappaBP65 promotes invasion and metastasis of oesophageal squamous cell cancer by regulating matrix metalloproteinase-9 and epithelial-to-mesenchymal transition. Cell Biol Int. 2013;37:780–8. doi: 10.1002/cbin.10089. [DOI] [PubMed] [Google Scholar]
- 17.Wang YP, Liu IJ, Chiang CP. et al. Astrocyte elevated gene-1 is associated with metastasis in head and neck squamous cell carcinoma through p65 phosphorylation and upregulation of MMP1. Mol Cancer. 2013;12:109. doi: 10.1186/1476-4598-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu M, Hu Y, Zhang MF. et al. MMP1 promotes tumor growth and metastasis in esophageal squamous cell carcinoma. Cancer Lett. 2016;377:97–104. doi: 10.1016/j.canlet.2016.04.034. [DOI] [PubMed] [Google Scholar]
- 19.Han KH, Croker BP, Clapp WL. et al. Expression of the ammonia transporter, rh C glycoprotein, in normal and neoplastic human kidney. J Am Soc Nephrol. 2006;17:2670–9. doi: 10.1681/ASN.2006020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho JW, Choi SC, Jang JY. et al. Lymph Node Metastases in Esophageal Carcinoma: An Endoscopist's View. Clin Endosc. 2014;47:523–9. doi: 10.5946/ce.2014.47.6.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stacker SA, Baldwin ME, Achen MG. The role of tumor lymphangiogenesis in metastatic spread. FASEB J. 2002;16:922–34. doi: 10.1096/fj.01-0945rev. [DOI] [PubMed] [Google Scholar]
- 22.Palayoor ST, Youmell MY, Calderwood SK. et al. Constitutive activation of IkappaB kinase alpha and NF-kappaB in prostate cancer cells is inhibited by ibuprofen. Oncogene. 1999;18:7389–94. doi: 10.1038/sj.onc.1203160. [DOI] [PubMed] [Google Scholar]
- 23.Gilmore TD. Introduction to NF-kappaB: players, pathways, perspectives. Oncogene. 2006;25:6680–4. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- 24.Karin M, Cao Y, Greten FR. et al. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–10. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 25.Dolcet X, Llobet D, Pallares J. et al. NF-kB in development and progression of human cancer. Virchows Arch. 2005;446:475–82. doi: 10.1007/s00428-005-1264-9. [DOI] [PubMed] [Google Scholar]
- 26.Allum WH, Stenning SP, Bancewicz J. et al. Long-term results of a randomized trial of surgery with or without preoperative chemotherapy in esophageal cancer. J Clin Oncol. 2009;27:5062–7. doi: 10.1200/JCO.2009.22.2083. [DOI] [PubMed] [Google Scholar]
- 27.Nakajima M, Kato H. Treatment options for esophageal squamous cell carcinoma. Expert Opin Pharmacother. 2013;14:1345–54. doi: 10.1517/14656566.2013.801454. [DOI] [PubMed] [Google Scholar]
- 28.Kato H, Nakajima M. Treatments for esophageal cancer: a review. Gen Thorac Cardiovasc Surg. 2013;61:330–5. doi: 10.1007/s11748-013-0246-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.