Abstract
Human growth factor receptor-bound protein-7 (GRB7) is a pivotal mediator involved in receptor tyrosine kinase signaling and governing diverse cellular processes. Aberrant upregulation of GRB7 is frequently associated with the progression of human cancers. However, the molecular mechanisms leading to the upregulation of GRB7 remain largely unknown. Here, we propose that the epigenetic modification of GRB7 at the post-transcriptional level may be a crucial factor leading to GRB7 upregulation in ovarian cancers. Methods: The upstream miRNA regulators were predicted by in silico analysis. Expression of GRB7 was examined by qPCR, immunoblotting and immunohistochemical analyses, while miR-193a-3p levels were evaluated by qPCR and in situ hybridization in ovarian cancer cell lines and clinical tissue arrays. MS-PCR and pyrosequencing analyses were used to assess the methylation status of miR-193a-3p. Stable overexpression or gene knockdown and Tet-on inducible approaches, in combination with in vitro and in vivo tumorigenic assays, were employed to investigate the functions of GRB7 and miR-193a-3p in ovarian cancer cells. Results: Both miR-193a-3p and its isoform, miR-193b-3p, directly targeted the 3' UTR of GRB7. However, only miR-193a-3p showed a significantly inverse correlation with GRB7-upregulated ovarian cancers. Epigenetic studies revealed that methylation-mediated silencing of miR-193a-3p led to a stepwise decrease in miR-193a-3p expression from low to high-grade ovarian cancers. Intriguingly, miR-193a-3p not only modulated GRB7 but also ERBB4, SOS2 and KRAS in the MAPK/ERK signaling pathway to enhance the oncogenic properties of ovarian cancer cells in vitro and in vivo. Conclusion: These findings suggest that epigenetic silencing of miR-193a-3p by DNA hypermethylation is a dynamic process in ovarian cancer progression, and miR-193a-3p may be explored as a promising miRNA replacement therapy in this disease.
Keywords: DNA hypermethylation, miR-193a-3p, GRB7, MAPK signaling, ovarian cancer.
Introduction
Ovarian cancer is one of the most lethal gynecological malignancies. Most ovarian cancer cases have a poor prognosis and exhibit a highly lethal rate at an advanced stage. High-grade ovarian cancers have a particularly poor prognosis and are extremely aggressive. An understanding of the molecular mechanisms, such as the characterization of genetic/epigenetic alterations of oncogenes/tumor suppressor genes, could, therefore, facilitate the early diagnosis and treatment of this highly malignant disease.
Growth factor receptor bound protein 7 (GRB7) is an adaptor protein that plays an important role in mediating signal transduction from multiple cell surface receptors to various downstream signaling pathways. Emerging evidence indicates that GRB7 is overexpressed in multiple human cancers, usually accompanied by the overexpression of ErbB2 1, 2. Clinicopathological analyses have shown that overexpressed GRB7 is correlated with a metastatic phenotype 1, 3. Overexpression of GRB7 enhances the migration ability of cancer cells. Thus, GRB7 may be used as a potential tumor marker or therapeutic target for therapeutic purposes 4, 5. Conversely, inhibitors of GRB7 are currently used preclinically for the treatment of cancers 6. One promising anti-GRB7 peptide, G7-18NATE, can bind to the GRB7 SH2 domain specifically 3, 7. This inhibitor can prevent GRB7 from binding to other activated tyrosine kinases 7. Indeed, SKBR3 and ZR-7530 drugs can exert significant inhibition of cell growth and migration in ErbB2-positive breast cancer 3, 8. These reports provide strong evidence that GRB7 can be used as a tumor marker and a promising target for cancer therapy. We have previously reported that GRB7 and its isoform, GRB7v, are overexpressed and associated with high-grade ovarian cancers 9. Akin to other human cancers, GRB7 promotes cell proliferation, cell migration and cell invasion, while GRB7v only increases cell proliferation and cell anchorage-independent growth capacities 9. With the treatment of specific kinase inhibitors, we demonstrated that both GRB7 and GRB7v are associated with high-grade tumors and exert distinct tumorigenic functions by regulating different signaling pathways in ovarian cancer cells. However, the underlying mechanism leading to overexpression of GRB7 remains unclear.
MicroRNAs (miRNAs) are a class of small noncoding RNAs (19-25 nucleotides) that enable negative modulation of the expression of protein-coding genes by binding, with imperfect base pairing, to complementary sequences of target sites in messenger RNAs (3' untranslated regions, 3'-UTR) with the RNA induced silencing complex (RISC) 10, 11. This phenomenon leads to a global regulation of gene expression in cells 12. In this study, we found that the GRB7 protein levels were remarkably higher than their mRNA levels in some ovarian cancer cell lines, indicating that there is an aberrant post-transcriptional alteration in GRB7. In an in silico study using 3 bioinformatics algorithms, we discovered that miR-193a-3p directly regulates GRB7 but is usually downregulated by DNA hypermethylation during ovarian cancer development and progression. The epigenetic silencing of miR-193a-3p leads to the aberrant upregulation of GRB7 and related oncogenes in MAPK/ERK signaling. Our study provides insights into the development of miR-193a-3p as a miRNA-based oncological therapeutic approach for targeting MAPK/ERK signaling to treat human ovarian cancer.
Materials and Methods
Cell lines and Human tissues
The human ovarian cancer cell lines TOV21G, SKOV3, ES-2, and HEK293 were purchased from the American Type Culture Collection (ATCC). OVISE and OVTOKO were purchased from the National Institute of Biochemical Innovation, JCRB Cell Bank. Two ovarian cancer cell lines, A2780s and A2780cp, were kindly provided by Prof. Benjamin K Tsang, University of Ottawa. Additionally, three human ovarian cancer cell lines, OVCA420, OVCA429 and OVCA433, as well as two humans immortalized ovarian surface epithelial cell lines (HOSEs), HOSE11 and HOSE96, were kindly provided by Prof. Georg SW Tsao, The University of Hong Kong. All cell lines were cultured in DMEM or RPMI 1640 medium (Sigma-Aldrich Corp., St. Louis, MO, USA) containing 10% fetal bovine serum (Invitrogen) and incubated in 5% CO2 at 37 °C. All cell lines were authenticated by in-house STR DNA profiling analysis, and mycoplasma contamination was tested. The clinical ovarian cancer patient samples were collected from the Queen Mary Hospital (Hong Kong) and immediately snap-frozen in liquid nitrogen and stored at -80 °C. Written informed consent was obtained from the participants, and the study was approved by Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster (HKU/HA HKW IRB) (Institutional Review Board number: UW11-298).
Plasmids and cell transfection
The miR-193a-3p and miR-193b-3p-expressing constructs were established by PCR amplification of genomic DNA and ligated into the pmR-ZsGreen1 plasmid (Clontech, Mountain View, CA) to generate pmR-ZsGreen1-miR-193a-3p or pmR-ZsGreen1-miR-193b-3p plasmids. The PCR DNA fragment containing miR-193a-3p precursor was also subcloned into pmRi-mCherry vector to generate the inducible miR-193a-3p expression construct pT-193a. The GRB7-expressing plasmid pEGFP/GRB7 was reported previously 9, and pCMV6-GRB7 was generated by subcloning the GRB7 full-length cDNA fragment from a Puc57-GRB7 plasmid (purchased from BGI-Shenzhen, China) into pCMV6-Entry vector (Origene). The SOS2-expressing plasmid (pCGN-SOS2) was obtained from Addgene (plasmid# 32921). Lipofectamine 3000 (Invitrogen) was used for cell transfection following the protocol. Stable ovarian cancer cells overexpressing miR-193a-3p were harvested after 14 days of G418 selection and verified by QPCR. Ovarian cancer cells with stable GRB7 knockdown were infected with lentiviral particles carrying GRB7 shRNA plasmid (Santa Cruz). After puromycin selection for 1 week, the stable GRB7 knockdown cells in the pool were verified by Western blot analysis. The double-stable doxycycline-inducible miR-193a-3p system was established according to the manufacturer's instructions using the Mir-X Inducible miRNA System (Clontech). Surviving Tet-On-miR-193a single clones were selected, expanded and screened for doxycycline-induced (1 μg/ml, Clontech) miR-193a-3p expression by QPCR.
RNA isolation and Quantitative RT-PCR
Total RNA was extracted from cell lines and human tissue samples using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA). First-strand complementary DNA was synthesized with a universal cDNA Synthesis Kit (Exiqon, Denmark). miRNA quantification by real-time PCR was performed using miRCURY LNA™ Universal RT microRNA PCR (Exiqon). Prime sets obtained from Exiqon included hsa-miR-193a-3p (Product No. 204591), hsa-miR-193b-3p (Product No. 204226) and the internal normalization control SNORD48 (Product No. 203903). Each sample was run in triplicate on a ViiA 7 Real-Time PCR System (Thermo Scientific).
Methylation-specific PCR (MS-PCR) and Pyrosequencing analysis
Genomic DNA was extracted from cell lines or clinical samples using the Wizard Genomic DNA Purification Kit (Promega). The DNA was then modified with sodium bisulfate using the EZ DNA Methylation-Gold Kit (Zymo Research, Orange, California, USA) following the protocol described by the manufacturer. The MS-PCR primers were designed using MethPrimer software and used to amplify and determine the specific methylated region on genomic DNA by DNA MyTaq TMHS Mix Reaction Buffer (Bioline, London, UK). A pyrosequencing assay was performed to detect the methylated CpG islands within an 800-bp region in the advanced miR-193a-3p at the Centre for Genome Sciences of The University of Hong Kong. Primers for pyrosequencing were designed using PSQ Assay Design Software (version 1.0.6, Biotage).
Western Blot analysis
Cells were harvested and lysed in cell lysis buffer (Cell Signaling technology, #9803) and analyzed by Western blotting as described previously 13. Antibodies against ERBB4 (4795, Cell Signaling), GRB7 (sc-13954, Santa Cruz), KRAS (ab180772, Abcam), SOS2 (ab137199, Abcam), ERK (4795, Cell Signaling), phosphorylated ERK (4795, Cell Signaling) and β-actin (1:10000, AC-74, Sigma Chemical Co) were used.
Immunohistochemistry (IHC) and in situ hybridization (ISH)
The commercial ovarian cancer tissue array (OVC1021) was purchased from Pantomics (Pantomics Inc., San Francisco, CA). The slides were incubated with the primary polyclonal anti-GRB7 antibody (H70, Santa Cruz Biotechnology, Inc.) at a 1:50 dilution. Further details of the procedure have been described previously 13. In situ hybridization (ISH) was performed with the miRCURY LNATM microRNA ISH Optimization Kit 5 (FFPE) (Exiqon) to detect the expression of miR-193a-3p according to the manufacturer's instructions.
Proximity ligation assay (in situ PLA)
The proximity ligation assay (in situ PLA) was carried out using the Duolink in situ PLA kit (Sigma-Aldrich) according to the manufacturer's protocol. Briefly, SKOV3 cells were grown on chamber slides and transfected with pCMV6-MYC-GRB7 or pCGN-HA-SOS2 constructs. Transfected cells were then fixed, permeabilized and blocked in blocking solution in a humidified chamber for 1 hour at 37°C. Mouse anti-Myc (Sigma-Aldrich) or mouse anti-HA (Santa Cruz Biotechnology) with appropriate anti-rabbit primary antibodies (ERBB4, SOS2 or KRAS) were added at a 1:200 dilution in antibody diluent and incubated overnight at 4°C. After Duolink® PLA probe incubation, ligation and amplification, the slides were embedded in Duolink® in situ mounting medium with DAPI, and images were acquired using a Carl Zeiss LSM 800 confocal microscope.
Luciferase reporter assay
Luciferase constructs were created by ligating part of the 3'UTR sequences containing the potential miR-193a-3p binding sites of ERBB4, GRB7, KRAS and SOS2 genes into the pmirGLO plasmid (Promega, Madison, WI) (Supplementary Fig. S2). HEK293 cells were seeded into a 24-well plate and cotransfected with the pmirGLO-3'UTR plasmid of the above plasmids or vector control using LipofectamineTM 3000 (Invitrogen). Luciferase activities were detected using the Dual-Luciferase Assay Kit (Promega).
Cell viability assay, in Vitro cell migration and invasion assays
Cell viability was determined using an XTT assay. Briefly, approximately 1,000 - 2,000 cells were subcultured in a 96-well plate. XTT was measured at days 0, 1, 2 3 and 4 post-plating using the Cell Proliferation Kit II (Roche Biosciences, Indianapolis, IN, USA). Cell migration and invasion assays were performed using Transwell cell migration assay kits (Corning, New York, NY, USA) and BioCoat™ Matrigel® Invasion Chambers (Corning), respectively. For doxycycline-induced miR-193a-3p expression, cells were pretreated with or without 1 μg/ml doxycycline. After 48 h, the cells were trypsinized and placed in the upper chamber filled with serum-free culture medium with or without doxycycline (1 μg/ml) and allowed to migrate or invade.
In vivo tumorigenicity
Four-week-old BALB/C-nu/nu female mice were used for intraperitoneal injection. SKOV3 cells stably expressing the Tet-On-miR-193a in pT-193a or vector control, SKOV3 with the GRB7 stable knockdown clone or the scrambled control (2× 106) were injected into nude mice (N = 5 per group) as described previously 13. For inducible miR-193a-3p expression, nude mice in each group received 2 mg/ml doxycycline via the drinking water for 20 days. Drinking water containing doxycycline was changed every two days for the 4-week period of tumor regression. All mice were sacrificed on Day 60 after injection of the cells. All animals were housed and handled experimentally according to the guidelines approved by the Committee on the Use of Live Animals in Teaching and Research of The University of Hong Kong CULATR 3960-16.
Statistical analysis
Unless otherwise stated, all experiments were verified at least three independent times. Values are presented as the mean ± SEM, and the 2-tailed Student's t test was used for comparisons. Data were plotted using GraphPad Prism 6 software (GraphPad Software, Inc., CA, USA). Statistical analyses were carried out with SPSS software (version 16.0). A receiver operating characteristic (ROC) curve was used to determine the cut-off points for the ISH and IHC data. The Fisher's exact test (for parametric data) and the Mann-Whitney test (for non-parametric data) were used. P≤ 0.05 was considered significant.
Results
Loss of miR-193a-3p is attributed to overexpression of GRB7 in ovarian cancer cells
GRB7 is localized in the 17q12 locus, and its amplification has been reported in numerous human cancers 14, 15. Based on the Cancer Genome Atlas (TCGA) data sets in cBioPortal (www.cbioportal.org), the results also indicated a higher expression rate of GRB7 in ovarian cancer compared with other tumor sets (Fig. 1A). These data were corroborated by our previous study 9, in which we found that GRB7 and its isoform, GRB7v, were overexpressed and associated with high-grade ovarian cancers 9. Further analyses showed that both GRB7 and GRB7v promoted cell proliferation by activating the MAPK/ERK signaling pathway 9. However, we found a disparity in the expression of GRB7 protein and mRNA by real-time quantitative RT-PCR (qPCR) and Western blot analyses of a panel of ovarian cancer cell lines, suggesting an aberrant post-transcriptional alteration in GRB79.
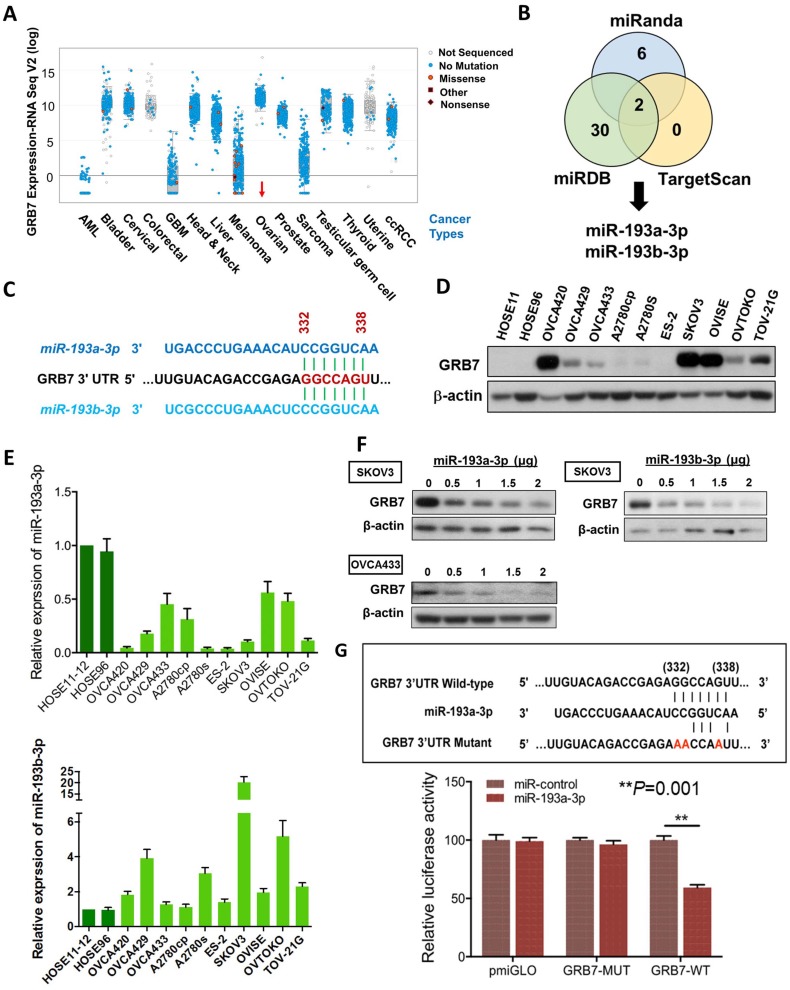
Figure 1.
The expression of miR-193a-3p is downregulated and inversely correlates with GRB7 expression in ovarian cancers. (A) Comparison of GRB7 expression in human ovarian cancer with other tumor types using the cBioPortal database. The dots represent clinical cases, and the line in the upper and lower box plot represents the upper and lower quartile of the relative mRNA levels of all samples. (B) The Venn diagram shows miR-193a-3p and miR-193b-3p commonly predicted by miRDB, TargetScan, and Miranda. (C) A schematic diagram showing the same binding site of miR-193a-3p and miR-193b-3p at position 332-338 of human GRB7 3' UTR. (D) Western blot analysis of GRB7 expression in ovarian cancer cell lines and HOSEs. (E) qPCR analysis of miR-193a-3p (upper) and miR-193b-3p (lower) expression in ovarian cancer cell lines. SNORD48 was used as an internal control. (F) Western blot analysis showing that transient transfection of pmR-ZsGreen1-miR-193a-3p or pmR-ZsGreen1-miR-193b-3p reduced the expression of GRB7 in SKOV3 or OVCA433 cells in a dose-dependent manner. (G) A schematic diagram showing the putative miR-193a-3p binding site in wild-type and mutant GRB7 3'UTR (Upper). Luciferase reporter assay showing the relative luciferase activity of wild-type or mutant GRB7 3'UTR targeted by a miR-193a-3p using pmR-ZsGreen1-miR-193a-3p plasmid in HEK293 cells (Lower).
In this in silico study using 3 bioinformatics algorithms, miRDB, miRanda, and TargetScan Human 5.2, we identified miR-193a-3p and its isoform, miR-193b-3p, by specifically targeting the same conserved site (GGCCAGT) at position 332-338 in the 3' UTR (length: 3894 of the human GRB7 gene (NM_001030002) (Fig. 1B & 1C). Using Western blot and qPCR analyses, we found that the miR-193a-3p and GRB7 levels in most ovarian cancer cells lines, as well as HOSEs, were inversely correlated, whereas the levels of miR-193b-3p were higher than HOSEs in all ovarian cancer cells and had a lower correlation with GRB7 protein levels (Fig. 1D & 1E). Of note, the expression of GRB7 was low in some ovarian cancer cell lines, such as OVCA429, A2780s and OVTOKO, which had relatively low levels of miR-193a-3p. However, we found that these cell lines had relatively high levels of miR-193b-3p (Fig. 1D & 1E), which indicated that both miR-193a-3p and miR-193b-3p exerted an equivalent translational suppression effect on GRB7 because they had the same binding site in the 3'UTR of GRB7 (Fig. 1C). Indeed, transient transfection of either miR-193a-3p or miR-193b-3p into SKOV3 or OVCA433 cells could remarkably reduce GRB7 expression in a dose-dependent manner (Fig. 1F). However, based on the expressions pattern of both miR-193a-3p and miR-193b-3p versus the protein level of GRB7 in ovarian cancer cell lines and HOSEs, the downregulated miR-193a-3p seemed to play a more important role in GRB7 post-transcriptional regulation.
To validate the post-transcriptional suppressive effect of miR-193a-3p, we employed a luciferase reporter assay with the wild-type and mutant 3'UTR of GRB7. The results showed that pmiR-193a-3p could significantly attenuate the signal from the luciferase reporter fused to the wild-type miR-193a-3p target sequence (GGCCAGT) of the GRB7 3'UTR compared with the empty vector control (Fig. 1G). Conversely, this suppressive effect was absent in the miR-193a-3p target mutant (AACCAAT) (Fig. 1G). This finding indicated that GRB7 was the target of miR-193a-3p and that the upregulation of GRB7 was attributed to the loss of miR-193a-3p in ovarian cancer cells.
MiR-193a-3p suppresses not only GRB7 but also MAPK/ERK signaling in ovarian cancer cells
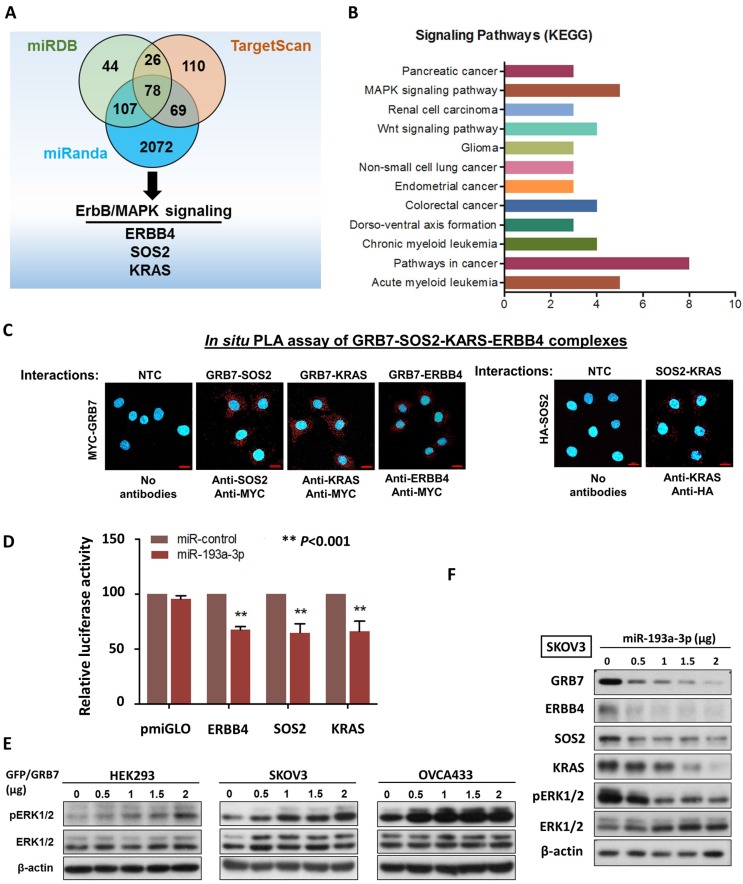
It is well known that a single miRNA can modulate a cohort of downstream gene targets. Hence, to examine other putative downstream targets of miR-193a-3p that may mediate the oncogenic capacities of ovarian cancer cells, and in silico study using three bioinformatics algorithms (miRDB, miRnada and TargetScan) was applied and identified 78 potential genes (Fig. 2A) (Supplementary Table S1). Among these potential targets, MAPK/ERK signaling was the dominant signaling pathway in cancers by KEGG (Kyoto Encyclopedia of Genes and Genomes) according to the Database for Annotation, Visualization and Integrated Discovery (DAVID) (Fig. 2B). ERBB4, SOS2 and KRAS have been defined as key components of this signaling pathway for modulating ERK activity 16 (Supplementary Fig. S3). The formation of the GRB2-SOS complex and the conversion of GDP to GTP through interactions with RAS are crucial steps in the activation of the MAPK/ERK signaling cascade 17, 18. Consistently, by employing a proximity ligation assay (in situ PLA), GRB7 was shown to directly interact with endogenous SOS2 and KRAS in the cytosol of OVCA433 though transient transfection of Myc-tagged GRB7-expressing plasmid and anti-Myc plus either anti-SOS2, anti-KRAS or anti-ERBB4 antibodies (Fig. 2C). Additionally, by transient transfection of HA-tagged SOS2-expressing plasmid into SKOV3 cells, the PLA results also provided structural evidence that SOS2 bound to KRAS and could switch RAS-GDP (off) to RAS-GTP (on) as previously reported, similarly to SOS-RAS signaling 19 (Fig. 2C). Luciferase reporter assays demonstrated that transient transfection of miR-193a-3p could suppress 30-35% of luciferase signals in ERBB4-, SOS2- and KRAS-3'UTR, compared with a 45% reduction of the GRB7-3'UTR luciferase signal (Fig. 2D & 1G). Our previous findings have shown that GRB7 is a key modulator of MAPK/ERK signaling 9, 20. Indeed, transient transfection of GFP/GRB7 could increase pERK1/2 levels in HEK293, SKOV3 and OVCA433 cells in a dose-dependent manner (Fig. 2E). In contrast, the reintroduction of miR-193a-3p could inhibit the protein levels not only of GRB7 but also of ERBB4, SOS2, KRAS and ERK (Fig. 2F). Taken together, these findings suggest that miR-193a-3p could target other factors, e.g., ERBB4, SOS2 and KRAS, in the MAPK/ERK signaling cascade, although it predominantly targeted GRB7 in ovarian cancer cells (Supplementary Fig. S3).
Figure 2.
MiR-193a-3p targets not only GRB7, but other key factors along MAPK/ERK signaling pathway in ovarian cancer cells. (A) The Venn diagram displays 77 common putative targets of miR-193a-3p predicted from the three algorithms, miRDB, miRanda and TargetScan Human 5.2, including three putative targets, ERBB4, KRAS and SOS2, involved in the MAPK/ERK signaling pathway. (B) Functional annotation analysis using the DAVID functional annotation tool (http://david.abcc.ncifcrf.gov/) showing the main signaling pathways associated with these 78 common targets of miR-193a-3p. (C) Duolink proximity ligation assay (PLA) with fluorescence and confocal microscopy showing the interactions between GRB7 and SOS2 and between GRB7 and KRAS (red dots) by transient transfection of GRB7-expressing plasmid (Upper). Anti-Myc, anti-SOS2 and anti-KRAS were used to detect Myc-tagged GRB7 and endogenous SOS2 and KRAS. Transient transfection of HA-SOS2 also showing the interaction between SOS2 and KRAS (red dots) using PLA and anti-HA as well as anti-KRAS (Lower). Scale bar, 20 μm. n = 3 independent experiments. (D) Relative luciferase activity of luciferase reporters with wild-type ERBB4, SOS2 and KRAS 3'UTRs co-transfected with miR-193a-3p. (E) Western blot analysis showing that transient transfection of GFP/GRB7-expressing plasmid profoundly elevated ERK activity in HEK293, SKOV3 and OVCA433 cells. (F) Western blot analysis indicating that transient transfection of pmR-ZsGreen1-miR-193a-3p reduced the expression of GRB7, ERBB4, SOS2, KRAS and pERK1/2 in SKOV3 cells. Cell lysates of all the above cell transfectants were harvested 24 h after cell transfection.
Identification of DNA methylation leads to the downregulation of miR-193a-3p in ovarian cancer cells
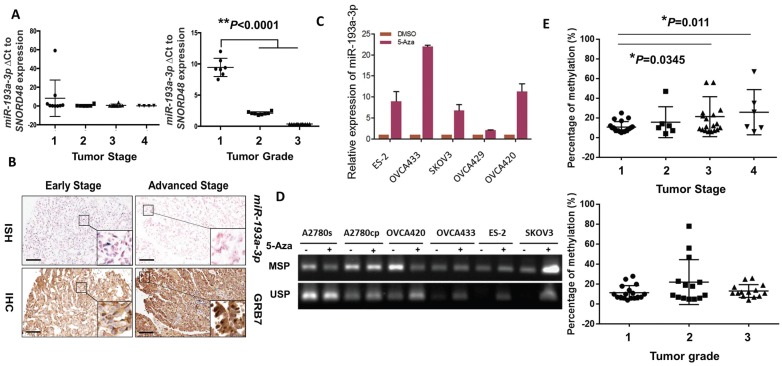
We next evaluated the expression levels of miR-193a-3p in clinical ovarian cancer samples (N = 27). Interestingly, we found that the expression pattern of miR-193a-3p showed a stepwise decrease from the early to the late tumor stage (1 to 4) and from low to high grade (1 to 3) (P<0.0001) (Fig. 3A), supporting a remarkable loss of miR-193a-3p expression during the development and progression of ovarian cancer. Given that there was an inverse relationship between miR-193a-3p and GRB7 in ovarian cancer cell lines, it was of interest to evaluate their expression patterns in clinical samples. Immunohistochemistry (IHC) and in situ hybridization (ISH) were performed using a commercial human ovarian cancer tissue array (OVC1021, Pantomics) to evaluate the expression of GRB7 and miR-193a-3p, respectively. The results demonstrated that low expression of miR-193a-3p was significantly correlated with high expression of GRB7 (N=97, P=0.003) (Fig. 3B) (Table 1). Notably, the low expression of miR-193a-3p (<4-fold) was significantly associated with advanced-stage cancers (P=0.048) (Fig. 3B) (Table 1). However, there was no significant correlation between the low level of miR-193a-3p and tumor grading or metastasis (Table 1). These findings indicated that the loss of miR-193a-3p led to a high expression level of GRB7 during ovarian cancer development.
Figure 3.
DNA methylation leads to downregulation of miR-193a-3p during the tumor development and progression of ovarian cancer cells. (A) qPCR analysis revealing a stepwise decrease in miR-193a-3p expression from Stages 1 to 4 and Grades 1 to 3 in ovarian cancers. (B) Representative photographs showing an inverse correlation between miR-193a-3p and GRB7 expression by ISH and IHC analyses for serous subtype ovarian cancer samples (early stage = Stage 1 and Grade 2, and advanced stage = Stage 2 and Grade 3) on a commercial ovarian cancer tissue array (OVC1021, Pantomics). Scale bar, 100 μm. (C) The expression of miR-193a-3p could be restored in ovarian cancer cells upon treatment with 5-Aza-dc (5 μM) for 4 days. (D) Verification of miR-193a-3p promoter methylation by MS-PCR in ovarian cancer cells upon 5-Aza-dC treatment. Treatment with 5-Aza-dC (5 μM) for 3-4 days. (E) Pyrosequencing analysis showing a progressive increase in DNA methylation with the tumor stage (Upper) and grade (Lower) of ovarian cancers.
Table 1.
Clinico-pathological correlation of the expression of miR193a-3p in an ovarian cancer tissue array (OVC1021). The 4-fold and 3-fold cut-off points of miR-193a-3p and GRB7, respectively, were determined by ROC analysis.
| Parameters n(=97) | miR-193a-3p expression | |||
|---|---|---|---|---|
| ≤ 4-fold | >4-fold | p | ||
| Grade | ||||
| Low (1+2) | 50 | 31 (62%) | 19 (38%) | |
| High (3) | 46 | 37 (80%) | 9 (20%) | 0.071 |
| Stage | ||||
| Early (1) | 48 | 29 (60%) | 19 (40%) | |
| Late (2+3) | 49 | 39 (80%) | 10 (20%) | 0.048* |
| Metastasis | ||||
| Yes | 24 | 20 (83%) | 4 (17%) | |
| No | 73 | 48 (66%) | 25 (34%) | 0.127 |
| GRB7 | ||||
| ≤ 3 folds | 38 | 20 (53%) | 18 (47%) | |
| > 3 folds | 59 | 48 (81%) | 11 (19%) | 0.003* |
We next investigated the molecular mechanisms leading to the downregulation of miR-193a-3p in ovarian cancers. As the miR-193a-3p precursor is located on chromosome 17q11.2, where loss of heterozygosity (LOH) is rare in ovarian cancer 21, 22, we therefore examined the range and content of miR193a-3p expression under the control of DNA methylation. Upon treatment with a demethylating agent, 5-Aza-dc (5 µM for 4 days), and evaluation by qPCR, the expression of miR-193a-3p was found to be upregulated in all the tested ovarian cancer cell lines (Fig. 3C). A further in-depth investigation using methylation-specific PCR (MS-PCR) analysis confirmed the silencing of miR-193a-3p in the above cell lines with hypermethylation of the miR-193a-3p promoter (Supplementary Fig. S1) (Fig. 3D). Indeed, pyrosequencing analysis showed a progressive increase in DNA methylation with a higher tumor stage (*P=0.0345 and **P=0.011) (Fig. 3E). However, there was no significant relationship between the methylation level and the tumor grade, confirming the IHC data showing that the downregulated miR-193a-3p was correlated with ovarian cancer development. Taken together, these findings suggested that the loss of miR-193a-3p was due to epigenetic alterations during the development of ovarian cancer.
MiR-193a-3p suppresses ovarian cancer cell growth and migratory/invasive capacities
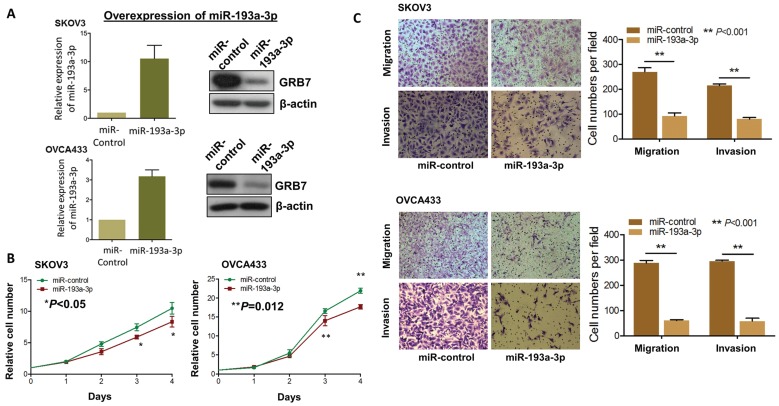
Since aberrant activation of the MAPK/ERK signaling pathway plays key roles in numerous oncogenic processes 23, 24, we postulated that re-expression of miR-193a-3p could impede the activities of this signaling and its associated oncogenic capacities. Thus, we generated stable miR-193a-3p-expressing transfectants in SKOV3 and OVCA433 cells, and the expression levels were checked by qPCR analysis, while the expression levels of GRB7 were tested by Western blot analysis (Fig. 4A). Simultaneously, their cellular functional effects were compared. As expected, re-expression of miR-193a-3p could markedly inhibit cell proliferation by 20-25% in SKOV3 (P<0.05) and OVCA433 cells (P=0.0012) (Fig. 4B). Similarly, miR-193a-3p also exhibited suppressive effects on the migratory ability by 65-80% (P<0.001) and on invasive ability by 60-80% (P<0.001) in SKOV3 and OVCA433 cells using Transwell cell migration/invasion assays (Fig. 4C). These data implied that re-expression of miR-193a-3p could suppress MAPK/ERK signaling, including GRB7, while mediating cell proliferation and migration/invasion capacities in ovarian cancer cells.
Figure 4.
Re-introduction of miR-193a-3p suppresses cell proliferation, cell migration and invasion capacities of ovarian cancer cells. (A) qPCR and Western blot analyses of miR193a-3p and GRB7 expression in stable miR-193a-3p-expressing SKOV3 and OVCA433 cells. SNORD48 was used as an internal control. (B) XTT proliferation assay, (C) Transwell cell migration and invasion assays showing that stable re-expression of miR-193a-3p inhibited cell proliferation rates, cell migration and invasion abilities of SKOV3 and OVCA433 ovarian cancer cells.
Knockdown of GRB7 imitates the effect of miR-193a-3p on the inhibition of ovarian cancer oncogenic capacities
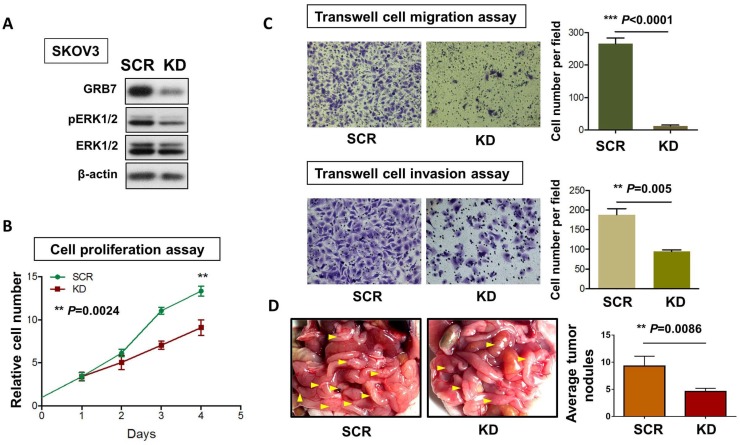
To examine the critical role of GRB7 in MAPK/ERK oncogenic signaling in ovarian cancer cells, we employed a shRNAi approach to knock down endogenous GRB7 in SKOV3 ovarian cancer cells. Western blot analysis showed that depletion of GRB7 by shRNAi caused a >60% loss of endogenous GRB7 in SKOV3, accompanied by a reduction of the pERK1/2 level (Fig. 5A). However, this reduction was lower than that of the ectopic expression of miR-193a-3p (Fig. 2F). Knockdown of GRB7 also remarkably inhibited cell proliferation by ~40% (P=0.0024) (Fig. 5B) and cell migration capacity by 95% (P<0.0001), as well as cell invasion capacity by 45% (P=0.005) (Fig. 5C). We further examined the functional effect of GRB7 in ovarian cancer cells in vivo by administering an intraperitoneal (i.p.) injection of GRB7-knockdown SKOV3 cells (Fig. 5D). Reduced numbers of tumor nodules were detected in the GRB7-knockdown SKOV3 group compared with the control (P=0.0086) (Fig. 5D). Consistent with our previous studies 9, 20, these findings confirmed that GRB7 had a tumorigenic effect on ovarian cancer cells.
Figure 5.
The functional role of GRB7 knockdown on cell proliferation, migration, invasion, and in vivo tumorigenicity. (A) Western blot analysis showing the levels of GRB7 knockdown (KD) and scrambled control (SCR) in SKOV3 cells. (B) XTT proliferation assay and (C) Transwell cell migration and invasion assays showing that knockdown of GRB7 inhibited cell proliferation, migration and invasion. (D) The 3-4-week-old female nude mice were i.p. injected with GRB7 knockdown or scrambled control SKOV3 cells and photographed on Day 60. Yellow arrows indicate the location of tumor nodules. The number of tumor nodules on the last day in each group are illustrated by bar charts (N=5).
Induced expression of miR-193a-3p suppresses ovarian cancer growth and tumor colonization
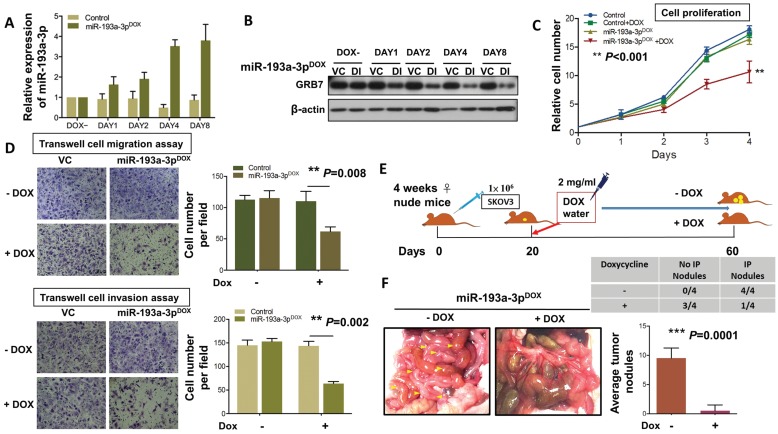
To extend the study of the precise functional role of miR-193a-3p in the tumor growth and progression of ovarian cancer, we introduced a doxycycline (Dox)-inducible system, pT-193a, that allows the induction of miR-193a-3p by the addition of Dox in a highly metastatic cell line, SKOV3 25, 26. qPCR analysis revealed increased expression of miR-193a-3p, and the maximum expression was achieved (3.7-3.9-fold) on Day 4 (Fig. 6A). Consistent with this observation, the induced expression of miR-193a-3p was able to inhibit the expression level of GRB7 in a time-dependent manner, with the highest suppression observed on Day 4 (Fig. 6B). The XTT cell proliferation assay showed that Dox-induced miR-193a-3p could suppress cell growth by >40% by compared with the controls ± Dox or miR-193a-3p-inducible SKOV3 cells without Dox induction (P<0.001) (Fig. 6C). Moreover, using Transwell cell migration/invasion assays, we demonstrated that Dox-induced miR-193a-3p expression significantly attenuated cell migration/invasion capacities by more than 50% compared with the controls (P=0.008 and P=0.002) (Fig. 6D). These data support the notion that the loss of cell growth or cell migration/invasion capacities is attributed to the re-expression of miR-193a-3p. Furthermore, we examined the functional role of miR-193a-3p in ovarian cancer cells in vivo by intraperitoneal (i.p.) injection of miR-193a-3p-inducible SKOV3 cells into nude mice. When palpable tumors were formed on Day 20, miR-193a-3p was induced by administration of Dox in water (Fig. 6E). After 40 days of Dox administration to the water, all the mice were healthy. However, we observed an almost absence or a trace number of tumor nodules disseminated in the peritoneum and other organs in the peritoneal cavity of nude mice that received the Dox-containing water (Fig. 6F). When compared with the GRB7-knockdown system, tumor size and number of tumor nodules were significantly reduced in the mice with Dox-induction of miR-193a-3p (P=0.0001) (Fig. 6F). These findings demonstrated a convergence such that the loss of miR-193a-3p could enhance oncogenic capacities via activation of MAPK/ERK signaling, facilitating tumor colonization of metastatic ovarian cancer in peritoneal metastases.
Figure 6.
DOX-induced miR-193a-3p inhibits cell migration and invasion in vitro and results in complete inhibition of tumor growth. (A) qPCR and (B) Western blot analysis showing induction of the expression of GRB7 by 1 μg/ml doxycycline (Dox) in a time-dependent manner. SNORD48 was used as an internal control. (C) XTT cell proliferation assay showing the effect of inducible miR-193a-3p on SKOV3 cells. (D) Transwell cell migration/invasion assays demonstrating the suppressive effect of inducible miR-193a-3p on the migration and invasion capacities of SKOV3 ovarian cancer cells. The induction of miR-193a-3p for cell proliferation and cell migration/invasion using 1 μg/ml Dox. (E) Schematic overview of miR-193a-3p expression in mice and its effect on the suppression of ovarian tumor growth in the intraperitoneal cavity of nude mice. (F) The 3-4-week-old female mice received an intraperitoneal injection (i.p.) of 1x106 miR-193a-3p-inducible SKOV3 cells. When palpable tumors formed on Day 20, all mice were treated with or without 2 mg/mL Dox in the drinking water until they were sacrificed on Day 60. Representative photographs of tumor nodules collected after induction of miR-193a-3p on Day 60. The table shows that only 1 out of 4 mice had tumor nodules in the Dox-inducible miR-193a-3p group, while all mice (4 out of 4) developed tumor nodules in the control group. The number of tumor nodules on the last day is illustrated for each group by bar charts (N=4).
Discussion
Aberrant upregulation of GRB7 is pervasive in many human cancers through the aberrant activation of MAPK/ERK signaling 1, 3, 9, 27. Accumulating evidence indicates that targeting GRB7 is a promising approach to impede tumor aggressiveness 4-6. However, most studies only characterized the functional roles and attempted to explore small molecules to block the activity of GRB7 and its downstream signaling 3, 7, yet the underlying molecular mechanisms leading to the upregulation of GRB7 in human cancer cells are relatively ill-defined. In this study, we provide the first evidence that miR-193a-3p plays a role as a tumor suppressor by directly targeting GRB7. Importantly, we show that the aberrant upregulation of GRB7 is attributed to epigenetically downregulating miR-193a-3p during tumor development and progression in ovarian cancer. Furthermore, we show that miR-193a-3p targets not only GRB7 but also other key components that modulate the MAPK/ERK signaling cascade in ovarian cancer cells. Interestingly, the re-introduction of miR-193a-3p exerted a stronger anti-tumor effect than just silencing GRB7 solely in ovarian cancer cells. Therefore, miR-193a-3p may be used as a miRNA-targeted therapeutic modality for ovarian cancer.
GRB7 gene amplification has been documented in numerous human cancers, such as testicular neoplasms, esophageal squamous cell carcinoma (ESCC), breast and ovarian cancers 20, 27-29. As GRB7 and HER2 are co-localized on the 17q12-21 amplicon, the upregulation of both genes is correlated with more aggressive breast carcinomas 30. GRB7 was also co-amplified with another adjacent gene, ERBB2, in aggressive Barrett's carcinoma and gastric cancer 1, 31. These findings suggest that the increased DNA copy number or amplicon is the main factor in the upregulation of GRB7 mRNA in human cancers. Two recent studies using immunohistochemical analysis (IHC) and fluorescence in situ hybridization (FISH) have shown that the protein level is positively correlated with the copy number of GRB7 30, 32. However, this is not the only conclusion based on the aberrant upregulation of GRB7 in human cancers. In this study, further in-depth investigations using Western blotting and qPCR analyses revealed a discrepancy between the protein and mRNA levels of GRB7, demonstrating the presence of aberrant post-transcriptional alterations of GRB7 in ovarian cancer cells. An in silico study revealed that miR-193a-3p and miR-193b-3p are the main targets of human GRB7. Although miR-193a-3p and miR-193b-3p are located on 17q11.2 and 16p13.12, respectively, both miRNAs share the same binding of the seed sequence in the 3' UTR (length: 387) of the GRB7 gene and, therefore, able to interfere with the translational efficiency of GRB7 in ovarian cancer cells. Intriguingly, by comparing their expression patterns, we found that miR-193b-3p expression was relatively higher in both GRB7-low or high-expressing ovarian cancer cell lines, as well as HOSEs. Conversely, miR-193a-3p was frequently downregulated and was inversely correlated with the high expression of GRB7 in ovarian cancer cell lines. Among these ovarian cancer cell lines, we noted that some cells, e.g., OVCA429, A2780s and OVTOKO, should express higher levels of GRB7. However, extremely high expression of miR-193b-3p further reduced the levels of GRB7 in these cell lines. Based on the expression levels of GRB7 and both miR-193a-3p and miR-193b-3p in ovarian cancer cells, the frequently downregulated miR-193a-3p was deemed the main player in determining the expression of GRB7, while miR-193b-3p acted solely as a subsidiary to miR-193a-3p in further modulate the expression of GRB7. This finding is consistent with recent evidence showing that miRNAs work together as a team and co-modulate the expression of target genes in cellular processes or tumor progression 33. Therefore, we focused our attention on deciphering the mechanism leading to the downregulation of miR-193a-3p and its role as a tumor suppressor-type miRNA in ovarian cancer.
It is well known that miRNAs usually act as oncogenes or tumor suppressors to control many cellular events in tumor development and progression 34. Similar to tumor suppressors, dysregulation of tumor suppressor-type miRNAs by epigenetic silencing is often observed in human cancers 35. Hypermethylation of CpG islands of promoters is the major epigenetic aberration in the inactivation of these tumor suppressors 36. In the present study, we found that miR-193a-3p was frequently downregulated in ovarian cancer cells. Accumulating evidence indicates that the miR-193a-3p precursor is located on chromosome 17q11.2, where the loss of heterozygosity (LOH) is rare in ovarian cancer 21, 22. We supposed that the downregulated miR-193a-3p was epigenetically silenced by DNA hypermethylation. As expected, hypermethylation of miR-193a-3p could be evidenced using 5-Aza-dC to mediate the restoration of miR-193a-3p, MS-PCR and pyrosequencing analyses on CpG islands of the promoter of the miR-193a-3p precursor. Correspondingly, CpG island hypermethylation of miR-193a-3p was observed in ovarian cancer clinical samples, and the progressive increase in the hypermethylation status was significantly correlated with the tumor stage of ovarian cancers. These data support not only the notion that DNA hypermethylation epigenetic silencing of miR-193a-3p but also epigenetic silencing of miR-193a-3p and upregulation of GRB7 actively occur during the tumor development and progression of ovarian cancers. Indeed, previous results showing the clinicopathological correlation of IHC and ISH analyses of miR-193a-3p and GRB7 support the above conclusion.
Aberrant activation of MAPK/ERK signaling is critical for controlling cell proliferation, cell survival, metastasis and drug resistance in more than 85% human cancers 37. Upon stimulation, this signaling pathway is initiated by upstream receptors, e.g., ErbBs that transmit the signals via GRB2, SOS and GRBs to activate the small GTPase RAS 38. The active RAS recruits RAFs to the cellular membrane, where the activated RAFs phosphorylate MEK1/2, which in turn, activates ERK1/2 38. The phosphorylated ERK1/2 further phosphorylates RSK, and both are translocated to the nucleus to transcriptionally elevate the expression of numerous oncogenes with different oncogenic properties 39. Mounting evidence has indicated that genetic/epigenetic alterations lead to the hyperactivation of MAPK/ERK signaling during tumor development and progression 40, 41. Due to its pivotal role in cancers, this phenomenon has attracted large efforts by a number of research groups or pharmaceutical companies to explore inhibitors targeting different tiers of this cascade 42. For example, the BRAF inhibitors vemurafenib (ZELBORAFTM, Roche-Genetech) and debrafenib (TAFINLARr, GSK), as well as the MEK inhibitors selumetinib (AZD6244, AstraZeneca/Array BioPharma) and trametinib (GSK1120212, GSK), are now in Phase II clinical trials for all tumors 38. GRB7 acts as a signaling adaptor that interacts with upstream receptors and modulates downstream signaling targets such as ERK 9. Therefore, it is also an important targetable factor in human cancers. In fact, several research groups have attempted to explore novel and highly specific inhibitors to block the activity of GRB7 in human cancers 43, 44. One promising inhibitor, the isanti-GRB7 peptide G7-18NATE, can suppress GRB7 binding to other activated tyrosine kinases 3, 7. However, all these inhibitors are solely focused on GRB7. In the present study, we provide another avenue for the silencing of GRB7 and its cancer-associated signaling. We show that the knockdown of GRB7 and re-introduction of miR-193a-3p can remarkably suppress tumor cell growth and cell migration/invasion almost equivalently. This finding may be explained by the equivalent inhibitory effect on MAPK/ERK signaling activity. However, when comparing the effect on tumor growth in vivo, we showed that re-expression of miR-193a-3p provides stronger inhibition of tumor dissemination and colonization of ovarian cancer cells in the murine peritoneal cavity than depletion of GRB7 alone. This finding indicates that miR-193a-3p not only targets GRB7 but also other key components, ERBB4, SOS2 and KRAS, leading to a convergence of complete suppression in the aberrantly activated MAPK/ERK signaling pathway in human cancers including ovarian cancer 38, 45. Moreover, due to the nature of miRNAs, one miRNA is able to target a network of multiple downstream signaling cascades. This signaling may be involved in promoting in vivo tumor growth, such as cell survival, angiogenesis and even cross-talk with the tumor microenvironment 46. Indeed, using the Database for Annotation, Visualization and Integrated Discovery (DAVID) functional annotation bioinformatics microarray analysis, the results indicate that miR-193a-3p influences a greater spectrum of cancer-related signaling pathways than GRB7 alone. These findings pave the way for the development of miR-193a-3p as a miRNA-based therapeutic approach for ovarian cancer treatment.
In summary, our results show that the upregulated GRB7 in some ovarian cancers is due to the epigenetic silencing of miR-193a-3p through DNA hypermethylation. Biochemical and functional analyses indicate that the re-introduction of miR-193a-3p profoundly inhibits tumorigenesis of ovarian cancer cells in vitro and in vivo by targeting GRB7 and the MAPK/ERK signaling cascade. These findings suggest that miR-193a-3p, a tumor suppressor miRNA, may potentially be employed in miRNA-based oncological therapies.
Supplementary Material
Supplementary Table S1: The list of 78 targets of miR-193a-3p commonly found by three algorithms; miRDB, miRnada and Target Scan. Supplementary Figure S1: A schematic diagram showing the distribution of CpG sites along the first 800 bp of the promoter region of pre-miR-193a-3p. The promoter regions of pre-miR-193a-3p for Methylation-specific PCR (MSP) and pyrosequencing are indicated. Supplementary Figure S2: Schema of miR-193a-3p binding sites in the corresponding 3'UTR sequence of the putative binding site in wild-type ERBB4, SOS2 and KRAS. The capital letters of each genes' 3'UTR represent the seed sequence of the miR-193a-3p binding site. Supplementary Figure S3: A schematic diagram showing miR-193a-3p targets ERBB4, GRB7, KRAS and SOS2 in modulating ERK activity for different oncogenic capacities in ovarian cancer cells.
Acknowledgments
We thank Prof. Benjamin Tsang (Department of Obstetrics and Gynaecology, The University of Ottawa) and Prof. George Tsao (Department of Anatomy, The University of Hong Kong) for providing various human ovarian cancer and HOSE cell lines. This study was supported by HKU Seed Fund for Basic Research (200811159046).
Statement of author contributions
D.C. and K.C. designed research; D.C., K.C., M.L., C.M., and R.C. performed the experiments; D.C., T.L., D.X., S.N. and K.C. contributed new reagents-analytic tools; D.C., K.C., K.K.C., H.Y., and H.N. analyzed and interpreted data; D.C. and K.C. wrote the manuscript. All authors were involved in editing the manuscript and had final approval of the submitted and published versions.
Abbreviations
- GRB7
growth factor receptor-bound protein-7
- miRNAs
microRNAs
- 3'-UTR
3' untranslated regions
- RISC
RNA induced silencing complex
- HOSEs
humans immortalized ovarian surface epithelial cell lines
- HKU/HA HKW IRB
Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster
- TCGA
the cancer genome atlas
- quantitative RT-PCR
q-PCR
- ISH
in situ hybridization
- IHC
immunohistochemistry
- ROC
receiver operating characteristic
- KEGG
kyoto encyclopedia of genes and genomes
- LOH
loss of heterozygosity
- i.p.
intraperitoneal
- Dox
doxycycline
- ESCC
esophageal squamous cell carcinoma
- FISH
fluorescence in situ hybridization
- DAVID
database for annotation, visualization and integrated discovery.
References
- 1.Walch A, Specht K, Braselmann H, Stein H, Siewert JR, Hopt U. et al. Coamplification and coexpression of GRB7 and ERBB2 is found in high grade intraepithelial neoplasia and in invasive Barrett's carcinoma. Int J Cancer. 2004;112:747–53. doi: 10.1002/ijc.20411. [DOI] [PubMed] [Google Scholar]
- 2.Tanaka S, Mori M, Akiyoshi T, Tanaka Y, Mafune K, Wands JR. et al. Coexpression of Grb7 with epidermal growth factor receptor or Her2/erbB2 in human advanced esophageal carcinoma. Cancer Res. 1997;57:28–31. [PubMed] [Google Scholar]
- 3.Tanaka S, Pero SC, Taguchi K, Shimada M, Mori M, Krag DN. et al. Specific peptide ligand for Grb7 signal transduction protein and pancreatic cancer metastasis. J Natl Cancer Inst. 2006;98:491–8. doi: 10.1093/jnci/djj105. [DOI] [PubMed] [Google Scholar]
- 4.Shen TL, Guan JL. Grb7 in intracellular signaling and its role in cell regulation. Front Biosci. 2004;9:192–200. doi: 10.2741/1229. [DOI] [PubMed] [Google Scholar]
- 5.Nadler Y, Gonzalez AM, Camp RL, Rimm DL, Kluger HM, Kluger Y. Growth factor receptor-bound protein-7 (Grb7) as a prognostic marker and therapeutic target in breast cancer. Ann Oncol. 2010;21:466–73. doi: 10.1093/annonc/mdp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pero SC, Daly RJ, Krag DN. Grb7-based molecular therapeutics in cancer. Expert Rev Mol Med. 2003;5:1–11. doi: 10.1017/S1462399403006227. [DOI] [PubMed] [Google Scholar]
- 7.Ambaye ND, Pero SC, Gunzburg MJ, Yap M, Clayton DJ, Del Borgo MP, Structural Basis of Binding by Cyclic Nonphosphorylated Peptide Antagonists of Grb7 Implicated in Breast Cancer Progression. J Mol Biol; 2011. [DOI] [PubMed] [Google Scholar]
- 8.Pero SC, Shukla GS, Cookson MM, Flemer S Jr, Krag DN. Combination treatment with Grb7 peptide and Doxorubicin or Trastuzumab (Herceptin) results in cooperative cell growth inhibition in breast cancer cells. Br J Cancer. 2007;96:1520–5. doi: 10.1038/sj.bjc.6603732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Chan DW, Liu VW, Chiu P, Ngan HY. Differential functions of growth factor receptor-bound protein 7 (GRB7) and its variant GRB7v in ovarian carcinogenesis. Clin Cancer Res. 2010;16:2529–39. doi: 10.1158/1078-0432.CCR-10-0018. [DOI] [PubMed] [Google Scholar]
- 10.Ambros V. The evolution of our thinking about microRNAs. Nat Med. 2008;14:1036–40. doi: 10.1038/nm1008-1036. [DOI] [PubMed] [Google Scholar]
- 11.Rigoutsos I. New tricks for animal microRNAS: targeting of amino acid coding regions at conserved and nonconserved sites. Cancer Res. 2009;69:3245–8. doi: 10.1158/0008-5472.CAN-09-0352. [DOI] [PubMed] [Google Scholar]
- 12.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38(Suppl):S8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 13.Mak CS, Yung MM, Hui LM, Leung LL, Liang R, Chen K. et al. MicroRNA-141 enhances anoikis resistance in metastatic progression of ovarian cancer through targeting KLF12/Sp1/survivin axis. Mol Cancer. 2017;16:11. doi: 10.1186/s12943-017-0582-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito M, Kato Y, Ito E, Fujimoto J, Ishikawa K, Doi A. et al. Expression screening of 17q12-21 amplicon reveals GRB7 as an ERBB2-dependent oncogene. FEBS letters. 2012;586:1708–14. doi: 10.1016/j.febslet.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Sparano JA, Goldstein LJ, Childs BH, Shak S, Brassard D, Badve S. et al. Relationship between quantitative GRB7 RNA expression and recurrence after adjuvant anthracycline chemotherapy in triple-negative breast cancer. Clin Cancer Res. 2011;17:7194–203. doi: 10.1158/1078-0432.CCR-10-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12:9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- 17.Lowenstein EJ, Daly RJ, Batzer AG, Li W, Margolis B, Lammers R. et al. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell. 1992;70:431–42. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- 18.Walker F, Kato A, Gonez LJ, Hibbs ML, Pouliot N, Levitzki A. et al. Activation of the Ras/mitogen-activated protein kinase pathway by kinase-defective epidermal growth factor receptors results in cell survival but not proliferation. Mol Cell Biol. 1998;18:7192–204. doi: 10.1128/mcb.18.12.7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Margarit SM, Sondermann H, Hall BE, Nagar B, Hoelz A, Pirruccello M. et al. Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell. 2003;112:685–95. doi: 10.1016/s0092-8674(03)00149-1. [DOI] [PubMed] [Google Scholar]
- 20.Chan DW, Hui WW, Cai PC, Liu MX, Yung MM, Mak CS. et al. Targeting GRB7/ERK/FOXM1 signaling pathway impairs aggressiveness of ovarian cancer cells. PLoS One. 2012;7:e52578. doi: 10.1371/journal.pone.0052578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasmant E, Masliah-Planchon J, Levy P, Laurendeau I, Ortonne N, Parfait B. et al. Identification of genes potentially involved in the increased risk of malignancy in NF1-microdeleted patients. Mol Med. 2011;17:79–87. doi: 10.2119/molmed.2010.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dimova I, Orsetti B, Negre V, Rouge C, Ursule L, Lasorsa L. et al. Genomic markers for ovarian cancer at chromosomes 1, 8 and 17 revealed by array CGH analysis. Tumori. 2009;95:357–66. doi: 10.1177/030089160909500315. [DOI] [PubMed] [Google Scholar]
- 23.Delire B, Starkel P. The Ras/MAPK pathway and hepatocarcinoma: pathogenesis and therapeutic implications. Eur J Clin Invest. 2015;45:609–23. doi: 10.1111/eci.12441. [DOI] [PubMed] [Google Scholar]
- 24.Giltnane JM, Balko JM. Rationale for targeting the Ras/MAPK pathway in triple-negative breast cancer. Discov Med. 2014;17:275–83. [PubMed] [Google Scholar]
- 25.Wu GJ, Zeng GF. METCAM/MUC18 is a novel tumor and metastasis suppressor for the human ovarian cancer SKOV3 cells. BMC Cancer. 2016;16:136. doi: 10.1186/s12885-016-2181-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ponnusamy MP, Lakshmanan I, Jain M, Das S, Chakraborty S, Dey P. et al. MUC4 mucin-induced epithelial to mesenchymal transition: a novel mechanism for metastasis of human ovarian cancer cells. Oncogene. 2010;29:5741–54. doi: 10.1038/onc.2010.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nencioni A, Cea M, Garuti A, Passalacqua M, Raffaghello L, Soncini D. et al. Grb7 upregulation is a molecular adaptation to HER2 signaling inhibition due to removal of Akt-mediated gene repression. PLoS One. 2010;5:e9024. doi: 10.1371/journal.pone.0009024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skotheim RI, Monni O, Mousses S, Fossa SD, Kallioniemi OP, Lothe RA. et al. New insights into testicular germ cell tumorigenesis from gene expression profiling. Cancer Res. 2002;62:2359–64. [PubMed] [Google Scholar]
- 29.Sawada G, Niida A, Hirata H, Komatsu H, Uchi R, Shimamura T. et al. An Integrative Analysis to Identify Driver Genes in Esophageal Squamous Cell Carcinoma. PLoS One. 2015;10:e0139808. doi: 10.1371/journal.pone.0139808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bivin WW, Yergiyev O, Bunker ML, Silverman JF, Krishnamurti U. GRB7 Expression and Correlation With HER2 Amplification in Invasive Breast Carcinoma. Appl Immunohistochem Mol Morphol; 2016. [DOI] [PubMed] [Google Scholar]
- 31.Varis A, Wolf M, Monni O, Vakkari ML, Kokkola A, Moskaluk C. et al. Targets of gene amplification and overexpression at 17q in gastric cancer. Cancer Res. 2002;62:2625–9. [PubMed] [Google Scholar]
- 32.Zeng M, Yang Z, Hu X, Liu Y, Yang X, Ran H. et al. Grb7 gene amplification and protein expression by FISH and IHC in ovarian cancer. Int J Clin Exp Pathol. 2015;8:11296–304. [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Sun Q, Zhang Z, Ge S, Han ZG, Chen WT. Loss of microRNA-143/145 disturbs cellular growth and apoptosis of human epithelial cancers by impairing the MDM2-p53 feedback loop. Oncogene. 2013;32:61–9. doi: 10.1038/onc.2012.28. [DOI] [PubMed] [Google Scholar]
- 34.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nature reviews Drug discovery. 2017;16:203–22. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 35.Cui X Chen X Wang W Chang A Yang L Liu C et al. Epigenetic silencing of miR-203 in Kazakh patients with esophageal squamous cell carcinoma by MassARRAY spectrometry Epigenetics; 2017. 0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang G, Weisenberger DJ. DNA methylation aberrancies as a guide for surveillance and treatment of human cancers. Epigenetics. 2017;12:416–32. doi: 10.1080/15592294.2017.1311434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Luca A, Maiello MR, D'Alessio A, Pergameno M, Normanno N. The RAS/RAF/MEK/ERK and the PI3K/AKT signalling pathways: role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin Ther Targets. 2012;16(Suppl 2):S17–27. doi: 10.1517/14728222.2011.639361. [DOI] [PubMed] [Google Scholar]
- 38.Burotto M, Chiou VL, Lee JM, Kohn EC. The MAPK pathway across different malignancies: a new perspective. Cancer. 2014;120:3446–56. doi: 10.1002/cncr.28864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 40.Masliah-Planchon J, Garinet S, Pasmant E. RAS-MAPK pathway epigenetic activation in cancer: miRNAs in action. Oncotarget. 2016;7:38892–907. doi: 10.18632/oncotarget.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feo F, Pascale RM, Simile MM, De Miglio MR, Muroni MR, Calvisi D. Genetic alterations in liver carcinogenesis: implications for new preventive and therapeutic strategies. Crit Rev Oncog. 2000;11:19–62. [PubMed] [Google Scholar]
- 42.Maik-Rachline G, Seger R. The ERK cascade inhibitors: Towards overcoming resistance. Drug Resist Updat. 2016;25:1–12. doi: 10.1016/j.drup.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Gunzburg MJ, Kulkarni K, Watson GM, Ambaye ND, Del Borgo MP, Brandt R. et al. Unexpected involvement of staple leads to redesign of selective bicyclic peptide inhibitor of Grb7. Sci Rep. 2016;6:27060. doi: 10.1038/srep27060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Porter CJ, Matthews JM, Mackay JP, Pursglove SE, Schmidberger JW, Leedman PJ. et al. Grb7 SH2 domain structure and interactions with a cyclic peptide inhibitor of cancer cell migration and proliferation. BMC Struct Biol. 2007;7:58. doi: 10.1186/1472-6807-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ni Y, Zhang S. Erbb4 Signaling: an overlooked backup system? Cell cycle. 2015;14:1623. doi: 10.1080/15384101.2015.1032653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Donzelli S, Cioce M, Muti P, Strano S, Yarden Y, Blandino G. MicroRNAs: Non-coding fine tuners of receptor tyrosine kinase signalling in cancer. Semin Cell Dev Biol. 2016;50:133–42. doi: 10.1016/j.semcdb.2015.12.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1: The list of 78 targets of miR-193a-3p commonly found by three algorithms; miRDB, miRnada and Target Scan. Supplementary Figure S1: A schematic diagram showing the distribution of CpG sites along the first 800 bp of the promoter region of pre-miR-193a-3p. The promoter regions of pre-miR-193a-3p for Methylation-specific PCR (MSP) and pyrosequencing are indicated. Supplementary Figure S2: Schema of miR-193a-3p binding sites in the corresponding 3'UTR sequence of the putative binding site in wild-type ERBB4, SOS2 and KRAS. The capital letters of each genes' 3'UTR represent the seed sequence of the miR-193a-3p binding site. Supplementary Figure S3: A schematic diagram showing miR-193a-3p targets ERBB4, GRB7, KRAS and SOS2 in modulating ERK activity for different oncogenic capacities in ovarian cancer cells.