Summary
Bacterial microcompartments (MCPs) are protein organelles that typically house toxic or volatile reaction intermediates involved in metabolic pathways. Engineering bacteria to express exogenous MCPs will allow these cells to gain useful functions involving molecule compartmentalization. We cloned a 38 kb region from the Salmonella enterica serovar Typhimurium genome containing the pdu 1,2 propanediol (1,2 PD) utilization and cob/cbi genes using the FRT‐Capture strategy to clone and transfer large genomic segments. We transferred this clone to a range of Gram‐negative bacteria and found the clone to be functional for 1,2 PD metabolism in a variety of species including S. Typhimurium Δpdu, Escherichia coli, Salmonella bongori, Klebsiella pneumoniae, Cronobacter sakazakii, Serratia marcescens, and different Pseudomonas species. We successfully isolated MCPs expressed from the clone from several, but not all, of these strains, and we observed this utilizing a range of different media and in the absence of protease inhibitor. We also present a mini‐prep protocol that allows rapid, small‐scale screening of strains for MCP production. To date, this is the first analysis of cloned, exogenous microcompartment expression across several different Gram‐negative backgrounds and provides a foundation for MCP use in a variety of bacterial species using a full, intact clone.
Introduction
Bacterial microcompartments (MCPs) are protein‐based organelles that serve to compartmentalize certain metabolic reactions utilizing toxic or volatile molecules (Chowdhury et al., 2014; Bobik et al., 2015). The MCPs are generally composed of proteins that form an outer shell that is selectively permeable to house targeted enzymes and metabolic substrates and intermediates (Chowdhury et al., 2014, 2015; Bobik et al., 2015). Examples of characterized systems that utilize MCPs are carboxysomes for CO2 fixation in cyanobacteria and other species, 1,2 propanediol utilization in various enteric bacteria, and the ethanolamine utilization pathway expressed by a number of diverse bacteria (Chowdhury et al., 2014; Bobik et al., 2015). Several other MCP systems proposed to be involved with a range of other metabolic pathways have also been identified via genomics and bioinformatics (Abdul‐rahman et al., 2013; Jorda et al., 2013; Axen et al., 2014; Chowdhury et al., 2014; Bobik et al., 2015). The nature of MCPs makes them amenable for bacterial engineering in different biotechnology applications such as the ability to utilize novel metabolism, compartmentalize toxic or volatile molecules for downstream applications, express nanoscale bioreactors and potentially facilitate vaccine design strategies (Tsai and Yeates, 2011; Chen and Silver, 2012; Kim and Tullman‐Ercek, 2013; Chowdhury et al., 2014; Bobik et al., 2015).
The shell of the S. Typhimurium Pdu MCP is composed of protein subunits PduA, B, J, K, M, N, T and U (Bobik et al., 1999, 2015; Cheng et al., 2011; Chowdhury et al., 2014; Sinha et al., 2014; Jorda et al., 2015). PduA appears to play a central role in forming a selectively permeable pore that allows 1,2 PD entry into the MCP while restricting escape of the toxic compound propionaldehyde (Chowdhury et al., 2015). The enzymes for the 1, 2 PD utilization pathway are PduC, D, E, L, P, Q and W (Chowdhury et al., 2014; Bobik et al., 2015). The metabolism of 1, 2 PD requires coenzyme B12, and therefore, enzymes involved with coenzyme B12 assimilation and recycling are also associated with the Pdu MCP (Johnson et al., 2001; Sampson et al., 2005; Fan et al., 2009; Chowdhury et al., 2014; Bobik et al., 2015). The structural and regulatory genes for the Pdu MCP are found in the S. Typhimurium chromosome on a contiguous 23‐gene segment that is immediately adjacent to the cob/cbi genes used for synthesis of coenzyme B12 (on a contiguous 20‐gene segment) (Fig. 1A) (Bobik et al., 1999; McClelland et al., 2001).
Figure 1.
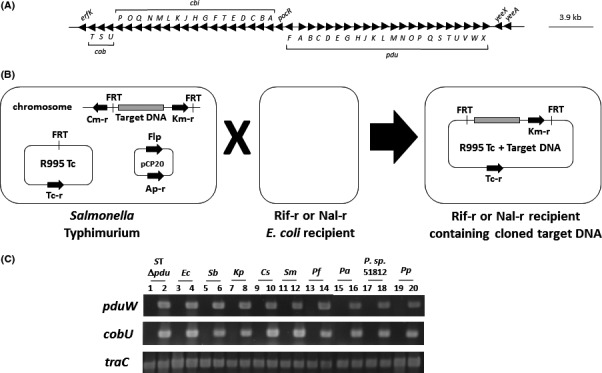
Cloning of the S. Typhimurium pdu and cob/cbi genes using FRT‐Capture. Panel A: A gene map of the targeted S. Typhimurium pdu and cob/cbi genomic segment cloned via FRT‐Capture. Panel B: A schematic diagram of the FRT‐Capture technique (not drawn to scale). FRT sites were inserted into regions flanking the target DNA (i.e. the pdu/cob/cbi region) in S. Typhimurium. Plasmids R995 (containing a single FRT site) and pCP20 (expressing FLP recombinase) were sequentially introduced into this strain. This allows FLP‐mediated excision of the target DNA and insertion into R995. The cloned target DNA construct is isolated upon conjugation to a fresh E. coli recipient strain containing appropriate counterselection (such as Rif‐R or Nal‐R). Panel C: PCR analysis of R995 + pdu ST in a range of Gram‐negative bacterial backgrounds. The pduW and cobU genes are specific for the cloned DNA segment, and the traC gene is present on the R995 vector. PCR products were analysed using 1.5% agarose gel electrophoresis followed by staining with SYBR Safe and UV light visualization. Abbreviations: ST Δpdu = Salmonella Typhimurium Δpdu; Ec = Escherichia coli; Sb = Salmonella bongori; Kp = Klebsiella pneumoniae; Cs = Cronobacter sakazakii; Sm = Serratia marcescens; Pf = Pseudomonas fluorescens; Pa = Pseudomonas aeruginosa; P. sp. 51812 = Pseudomonas species 51812; Pp = Pseudomonas putida. Odd numbered lanes are R995‐containing, and even numbered lanes are R995 + pdu ST‐containing.
Cloning and transfer of MCP‐encoding genes would allow engineering of target bacterial strains (potentially in a range of genera) to achieve novel MCP‐related functions as discussed above. In addition, such experiments allow an evolutionary analysis of pdu genes in which we can ask: Are the pdu genes from a given species expressed and functional in any other genera/species as opposed to having evolved to be restricted in these functions to the species of origin? Previous reports describing cloned Pdu MCP genes are present in the literature (Parsons et al., 2008; Sargent et al., 2013; Matsubara et al., 2016). Sargent et al. synthesized and cloned a subset of the S. Typhimurium pdu genes and observed MCP formation in an Escherichia coli background (Sargent et al., 2013). Parsons et al. cloned the pdu genes from Citrobacter freudii using a cosmid library approach, transferred the clone to an E. coli background and observed MCP formation in this species (Parsons et al., 2008). Matsubara et. al. cloned a subset of the Klebsiella pneumoniae pdu genes and coupled them with a Shimwellia blatte 1,2 PD synthetic pathway in E. coli to achieve 1‐propanol production in this background (Matsubara et al., 2016). In this study, we describe the first cloning of the entire, contiguous S. Typhimurium pdu/cob/cbi gene cluster (performed using a convenient in vivo approach termed FRT‐Capture) and the transfer of this clone to a range of different Gram‐negative genera to allow the first systematic analysis of MCP formation in a variety of bacterial species. An advantage of the present approach is the use of the entire contiguous genomic segment that contains the endogenous transcriptional and translational sequences with associated regulatory sequences. We report MCP expression from this clone across a range of bacterial genera and what appears to be robust recovery of MCPs from sample permissive strains using a range of different bacterial media.
Results
Cloning of the intact S. Typhimurium pdu/cob/cbi gene segment
To study Pdu MCP formation across bacterial genera for potential bacterial engineering purposes, we cloned a contiguous 38 kb segment of the S. Typhimurium genome containing the pdu and cob/cbi genes (please see Fig. 1 and Experimental Procedures for details). We used the FRT‐Capture method which allows in vivo cloning of large DNA segments and utilizes the broad host range plasmid vector R995 for convenient transfer of the clone to a wide variety of Gram‐negative bacteria (Santiago et al., 2011; Wilson et al., 2013). We transferred the R995 + pdu ST clone (or R995 as vector control) to a range of Gram‐negative strains including S. Typhimurium Δpdu, Escherichia coli, Salmonella bongori, Klebsiella pneumoniae, Cronobacter sakazakii, Serratia marcescens, and 4 species of Pseudomonas (P. aeruginosa, P. fluorescens, P. putida, and P. species 51812). Plasmid DNA was isolated from each of the transconjugant strains, and PCR analysis was used to confirm the transfer of the correct corresponding plasmid (Fig. 1, Panel C).
Functional expression of the pdu genes from R995 + pdu ST
To demonstrate functional expression of the pdu genes from R995 + pdu ST, we plated the indicated strains on MacConkey medium containing 1,2 PD as the carbon source and supplemented with coenzyme B12. If 1,2 PD utilization is accomplished by any R995 + pdu ST strain, the result will be a visual pink/red colony colour on this medium (as compared to the corresponding control strain). For all R995 + pdu ST strains except P. putida, clearly visible pink/red colony colour was observed compared with the R995 vector control strain (Fig. 2, Panel A). For P. putida, the R995 + pdu ST and R995 strains looked virtually identical on this medium (Fig. 2, Panel A). When comparing the K. pneumoniae strains, we observed that the R995 control strain displayed a pink colour on this medium that was, however, less intense than that observed for the R995 + pdu ST strain (which was darker pink) (Fig. 2, Panel A). This is likely due to the previously documented presence of pdu utilization genes in the K. pneumoniae genome that is causing a background level of 1,2 PD metabolism (Bobik et al., 1997, 1999; Honjo et al., 2015). To test whether the 1,2 PD metabolism displayed by R995 + pdu ST is dependent on coenzyme B12, we streaked the same strains to MacConkey medium containing 1,2 PD but without coenzyme B12. We observed no pink colony colour for any of the strains on this medium thus confirming that the 1,2 PD utilization directed by R995 + pdu ST is dependent on coenzyme B12 as would be expected (Fig. S1). These results indicate that R995 + pdu ST expresses functional MCPs and endows a range of Gram‐negative bacteria the ability to metabolize 1,2 PD.
Figure 2.
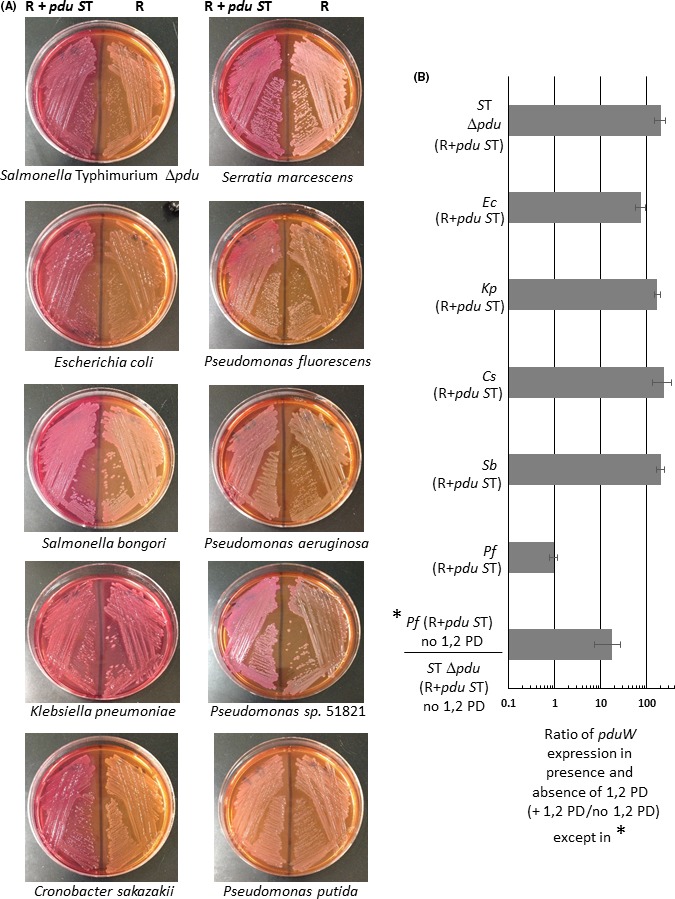
Expression analysis of pdu genes from R995 + pdu ST. Panel A: The indicated bacterial species containing either R995 + pdu ST or the R995 vector control were streaked onto MacConkey medium supplemented with 1,2 PD and coenzyme B12. Utilization of 1,2 PD will result in a pink/red colony colour on this medium. Panel B: RT‐qPCR analysis of pdu gene expression from R995 + pdu ST is shown. The indicated bacterial species containing R995 + pdu ST were grown in the presence or absence of 1,2 PD, and the total RNA was harvested for each sample. The RNA was converted to cDNA and then used as template in qPCR analysis with primers targeting the pduW or korB genes (the latter being a gene present on the R995 vector backbone). The qPCR product levels were normalized to the level of the korB gene, and a ratio of pduW levels in the presence and absence of 1,2 PD was calculated to give a fold difference in expression between the two growth conditions. Species abbreviations correspond to those in the Fig. 1 legend.
Previous studies have shown that expression of the pdu genes requires the presence of 1,2 PD (Bobik et al., 1992; Chen et al., 1995; Kim et al., 2014). To determine whether the pdu genes on R995 + pdu ST display this same pattern of expression, we performed RT‐qPCR with total RNA isolated from R995 + pdu ST strains grown with and without 1,2 PD. In five of the six strains tested, we observed the predicted induction of pdu gene transcription in the presence of 1,2 PD (Fig. 2, Panel B). In the P. fluorescens background, we did not observed a difference in pdu gene expression between the two conditions (Fig. 2, Panel B). However, when normalized signal was compared between P. fluorescens and S. Typhimurium Δpdu strains (both containing R995 + pdu ST) under conditions without 1,2 PD, we observed 10‐fold greater signal in the P. fluorescens strain (Fig. 2, Panel B). This result was also obtained if the other backgrounds in the ‘without 1,2 PD’ conditions were used to compare to the P. fluorescens background (data not shown). This indicates that the pdu genes are expressed above uninduced levels in the P. fluorescens background in both the presence and absence of 1,2 PD. This observation suggests that the regulatory circuit that results in lower pdu gene expression in the absence of 1,2 PD is not working properly in the P. fluorescens background. It also indicates that maximal expression of the pdu genes in P. fluorescens may be about 10‐fold lower than that observed in strains where the 1,2 PD induction was observed (comparing the approximately 100‐fold induction values of the other strains to the approximately 10‐fold P. fluorescens value above S. Typhimurium Δpdu in the absence of 1,2 PD). Thus, of the six strains tested, five demonstrated pdu gene regulation via the presence and absence of 1,2 PD while one demonstrated what appears to be (relatively lower) constitutive expression that is not altered by the presence of 1,2 PD.
Isolation of MCPs from strains containing R995 + pdu ST
To demonstrate that the MCPs expressed from R995 + pdu ST could be isolated from different bacteria, we performed MCP purification using the various bacterial species containing R995 + pdu ST. We used a modified strategy based on a previously established MCP purification protocol (Sinha et al., 2012) (please see Experimental Procedures for details). This protocol uses 5.7‐fold less culture volume (70 ml versus 400 ml with the 70 ml providing approximately 1 × 1011 cells) and takes approximately 2 h compared with 4 h of the previous protocol with comparable yields (approximately 1 mg in both cases) (Sinha et al., 2012). When analysed via SDS‐PAGE and Coomassie staining, we found that the R995 + pdu ST MCP samples contained several bands running at sizes consistent with known S. Typhimurium Pdu proteins that were absent from samples obtained from corresponding R995 control strains (Fig. 3). To confirm the identity of the protein bands in the R995 + pdu ST samples, we subjected them to mass spectrometry analysis which positively identified S. Typhimurium Pdu proteins running at the predicted sizes (Fig. 3). The SDS‐PAGE analysis revealed purified MCPs expressed from R995 + pdu ST in S. Typhimurium Δpdu, E. coli, S. bongori, K. pneumoniae, C. sakazakii and P. fluorescens. Intriguingly, we did not recover MCPs in preps from S. marcescens, P. aeruginosa and P. putida where preps displayed bands that ran at locations not corresponding to known Pdu proteins and were commonly found in the corresponding control background strains (data not shown). In the P. species 51812 background, we recovered MCP structures (visualized via sds‐page and electron microscopy) from both the R + pdu ST strain and control strain indicating that this species expresses an endogenous MCP‐like structure (which made interpretation of our MCP preps difficult for this strain background) (data not shown).
Figure 3.
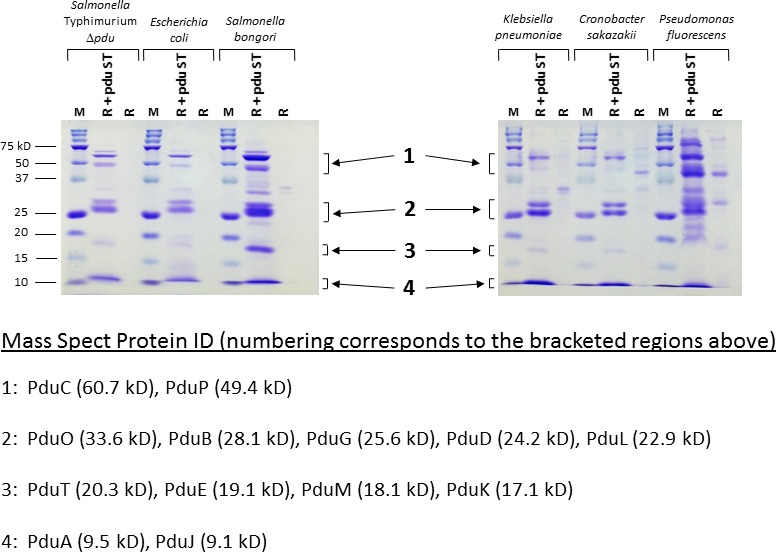
SDS‐PAGE analysis of isolated MCPs from R995 + pdu ST strains. The indicated strains containing either R995 + pdu ST or the R995 vector were grown in Pdu medium, and the MCPs were isolated from the cultures as described in Experimental Procedures. An aliquot of each MCP prep (corresponding to approximately 7–15 μg protein for the R995 + pdu ST preps) was run via SDS‐PAGE and stained with Coomassie Blue. ‘M’ stands for the protein marker (BioRad Precision Plus Kaleidoscope, 15 μg loaded). Protein band locations indicated by the numbered brackets were excised from R995 + pdu ST samples and subjected to Nano‐LC‐MS/MS mass spectrometry analysis. The protein IDs obtained from mass spectrometry analysis of samples cut from gels at the approximate indicated locations are provided as noted.
To visualize the isolated MCPs, we performed transmission electron microscopy analysis on the MCP samples (please see Experimental Procedures for details). We clearly observed MCPs in samples from S. Typhimurium Δpdu, E. coli, S. bongori, K. pneumoniae, C. sakazakii and P. fluorescens containing R995 + pdu ST (Fig. 4). We did not observe these structures in samples from the same species containing the R995 vector control (Fig. 4). We measured the diameter of the purified MCP structures in the TEM images for each species, and we found the average for these measurements to be between 88–95 nm (Fig. S2). These data are consistent with previously published numbers from similar MCP measurements (Parsons et al., 2008; Cheng et al., 2011; Sinha et al., 2012).
Figure 4.
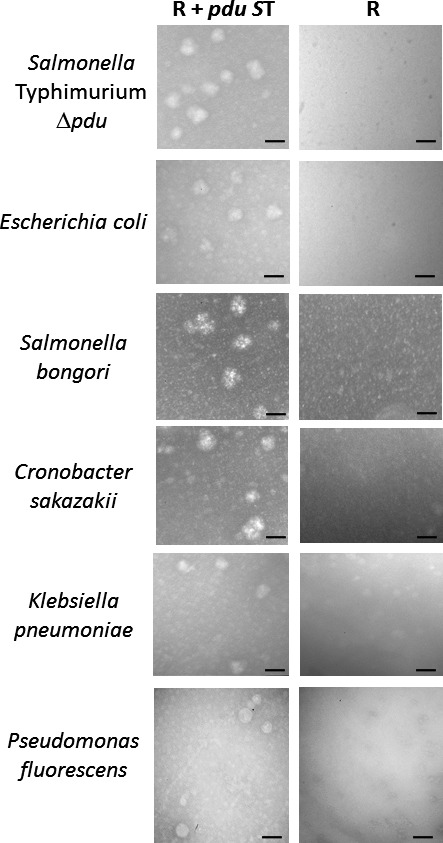
Transmission electron microscopy analysis of isolated MCPs. Samples from the indicated MCP preps were stained with 4% uranyl acetate and visualized using TEM. The size bar in each micrograph corresponds to 100 nm.
Use of different growth media reveals robust R995 + pdu ST MCP expression and isolation
Previous reports of Pdu MCP expression and isolation primarily utilize a minimal medium for bacterial growth, presumably to assure 1,2 PD‐specific expression of the pdu genes in the absence of glucose (since glucose has been shown to repress pdu gene expression) (Ailion et al., 1993; Chen et al., 1995; Kim et al., 2014). This could also be due to various researchers (understandably) following established convention in the field in which minimal media were previously shown to be effective and reproducible for Pdu MCP expression. In the experimental data described in this report above, we also used a minimal medium for our initial analysis across species (termed ‘Pdu medium’). To explore the possibility that MCP expression from R995 + pdu ST could be obtained using rich media, we grew strains containing R995 + pdu ST in various rich media (supplemented with 1,2 PD) and performed MCP isolation and SDS‐PAGE analysis. The results from species S. bongori and C. sakazakii are displayed in Fig. 5, and these data indicate a robust expression and recovery of MCPs in a range of different complex media (Fig. 5). We observed similar results in these media for the other R995 + pdu ST strains analysed in Fig. 3 as well (see Table S2 for data from LB media). This demonstrates that the glucose levels present in these media are below the level needed to repress expression of the pdu genes from R995 + pdu ST and that there appears to be a wide flexibility for functional growth media that can be used with this clone. Previously established protocols have included both 1,2 PD and protease inhibitor to be present during the entirety of the MCP preparation (Parsons et al., 2008; Sinha et al., 2012). While the purpose of protease inhibitor is self‐explanatory, we assume that the presence of 1,2 PD during cell lysis and MCP purification is meant to stabilize the MCP in the case that any of the Pdu proteins need the initial substrate to maintain proper conformation for an intact MCP. However, we hypothesized that the MCP protein parts would be tightly interlocked such that the MCP structure (and its contents) would be inherently stable during purification such that 1,2 PD and protease inhibitor would not be necessary for the isolation protocol. Therefore, we performed the MCP isolation protocol described in Experimental Procedures in the absence of 1,2 PD and protease inhibitor for cell lysis and purification. We observed essentially identical results comparing preps performed in the presence and absence of 1,2 PD and protease inhibitor during purification further demonstrating the robust nature of MCP isolation from R995 + pdu ST strains (Fig. 5).
Figure 5.
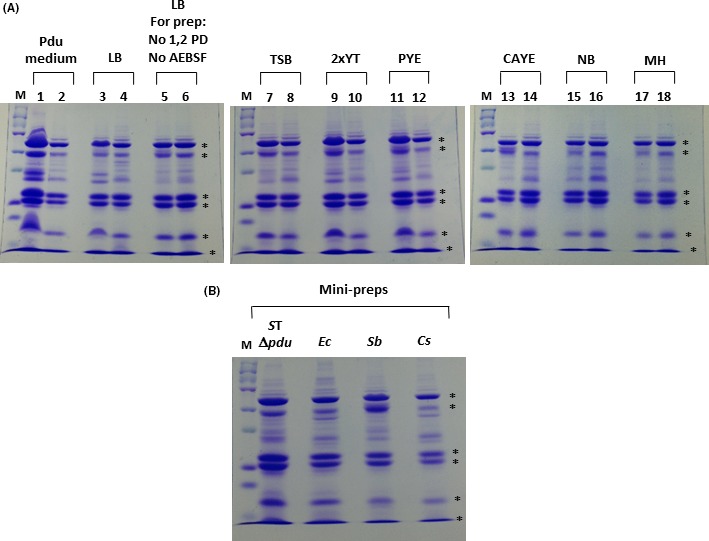
Analysis of R995 + pdu ST MCP isolation using different growth media and a mini‐prep protocol. Panel A: S. bongori and C. sakazakii strains containing R995 + pdu ST were grown in the indicated media supplemented with 1,2 PD, and the MCPs were isolated from each culture. An aliquot of each MCP prep (corresponding to approximately 10–20 μg protein) was run via SDS‐PAGE and stained with Coomassie Blue. The ‘M’ lane is protein marker (15 μg), odd numbered lanes contain samples from S. bongori R995 + pdu ST, and even numbered lanes contain samples from C. sakazakii R995 + pdu ST. Abbreviations correspond to those indicated in the Experimental Procedures for each medium. Control strains containing the R995 vector did not display MCP bands when grown and analysed in these media (data not shown). The asterisks on the right side of each gel indicate bands corresponding to known S. Typhimurium Pdu proteins. The sample ‘LB, For prep: no 1,2 PD, no AEBSF’ indicates MCP preps obtained from cells grown in LB medium (containing 1,2 PD) but with no 1,2 PD or protease inhibitor present during the purification procedure. Panel B: The indicated strains containing R995 + pdu ST were grown in LB medium supplemented with 1,2 PD, and the MCPs from each culture were isolated using a mini‐prep protocol as described in Experimental Procedures. An aliquot of each MCP mini‐prep was run via SDS‐PAGE and stained with Coomassie Blue. Species abbreviations correspond to those indicated in the legend for Fig. 1.
Mini‐prep isolation of MCPs from R995 + pdu ST strains
We reasoned that MCP isolation from R995 + pdu ST strains would be amenable to a smaller scale protocol using fewer cells, smaller buffer volumes and a microfuge (so as to effectively be an MCP ‘mini‐prep’). We designed such a protocol (described in Experimental Procedures) using approximately sevenfold less cells for starting material and processed using 1.5 ml microfuge tubes in a microfuge. The mini‐prep protocol yielded comparable MCPs as compared to previous preps (though total MCP yield was lower as would be expected and discussed in Experimental Procedures) (Fig. 5). The MCP mini‐prep allows convenient MCP isolation from several cultures processed at the same time, and the mini‐prep will be applicable to other MCP systems as well (such as carboxysomes and the ethanolamine MCP).
Discussion
We have cloned the entire, contiguous pdu/cob/cbi gene segment from the S. Typhimurium genome onto the plasmid vector R995 which can conveniently transfer to a range of Gram‐negative species. This allows relatively straightforward use of the Pdu MCP for cellular engineering in different bacterial backgrounds. It also allows the analysis of MCP expression, function, and isolation across species as an evolutionary study (i.e. has the S. Typhimurium pdu gene system evolved to be restricted to function in only S. Typhimurium or can it be functionally expressed and isolated from a range of species?). We observed functional MCP expression and isolation from S. Typhimurium Δpdu, E. coli, S. bongori, K. pneumoniae, C. sakazakii and P. fluorescens. Interestingly, we observed MCP function in S. marcescens and P. aeruginosa, but we did not observed successful MCP isolation from these species. For some reason, it could be that the MCP is not stable enough for isolation or requires additional specific protocol treatment in these bacteria. We did not observe evidence for Pdu MCP functional expression in P. putida. Further study is required to determine the reason for this such as testing the transcription and translation of specific pdu genes on R995 + pdu ST in P. putida. In the P. species 51812 background, we observed the presence of copious endogenous MCP‐like structures expressed in the control strain. We do not know the nature of these structures, and further characterization is required to determine their composition and function. We found that pdu gene transcription from R995 + pdu ST in P. fluorescens was not regulated by the presence/absence of 1,2 PD (i.e. it appeared to be constitutively expressed regardless of 1,2 PD status). The PocR transcriptional activator (encoded in the cloned pdu/cob/cbi segment) plays a central role in the induction of pdu genes in the presence of 1,2 PD (Bobik et al., 1992; Ailion et al., 1993; Chen et al., 1995). In addition, the Crp protein is required for full induction of the pdu genes under both aerobic and anaerobic conditions (Bobik et al., 1992; Chen et al., 1995). It is possible that PocR and/or Crp have altered activity at the pdu promoter(s) in the P. fluorescens background such that the result is constitutive pdu gene expression from R995 + pdu ST. Alternatively, a separate, heterologous gene regulator in P. fluorescens could be acting to induce pdu gene expression in the absence of 1,2 PD (or acting in combination with altered PocR and Crp activity) in this species. Such observations regarding altered expression and activity across different species are an intriguing source of further study with R995 + pdu ST.
We observed robust expression and isolation of Pdu MCPs from R995 + pdu ST strains in a range growth media, and we found that MCP isolation can be achieved in a mini‐prep format using a tabletop microfuge. In addition, we found that the presence of 1,2 PD and protease inhibitor during the purification protocol is not necessary to achieve successful MCP isolation. These results indicate the potential for convenient flexibility and utility of the R995 + pdu ST clone (or similar MCP gene clones) for future applications across different bacteria. Such applications could possibly include providing useful metabolic options for desired species or enabling MCP‐based nanobiotechnology across bacteria. Strategies and approaches towards these ends will be developed for future studies involving this clone.
Experimental procedures
Bacterial strains and plasmids
Please refer to Table 1 for a list of strains and plasmids used in this study. For routine growth of strains for cloning and maintenance, either Lennox broth (LB) or M9 minimal media were used (Lennox, 1955; Green and Sambrook, 2012). Antibiotics were used at the following concentrations (μg ml−1): kanamycin 50, tetracycline 5, rifampicin 75, nalidixic acid 5, ampicillin 200, chloramphenicol 10.
Table 1.
Strains and plasmids used in this study
| Strain | Species | Comments |
|---|---|---|
| χ3477 | Salmonella Typhimurium | Gift from Roy Curtiss III |
| χ3477 Δpdu | Salmonella Typhimurium | This study |
| TOP10 Rif | Escherichia coli | Rif‐R isolate of Invitrogen strain |
| TOP10 Nal | Escherichia coli | Nal‐R isolate of Invitrogen strain |
| ATCC 43975 | Salmonella bongori | American type culture collection |
| ATCC 13883 | Klebsiella pneumoniae | American type culture collection |
| ATCC 29544 | Cronobacter sakazakii | American type culture collection |
| ATCC 14041 | Serratia marcescens | American type culture collection |
| ATCC 13525 | Pseudomonas fluorescens | American type culture collection |
| PAK pilA | Pseudomonas aeruginosa | Wilson et al. (2007) |
| ATCC 51812 | Pseudomonas sp. 51812 | American type culture collection |
| ATCC 49128 | Pseudomonas putida | American type culture collection |
Cloning of the S. Typhimurium pdu and cob/cbi genes using FRT‐Capture
The region targeted for cloning via FRT‐Capture is diagrammed in Fig. 1A (Santiago et al., 2011; Wilson et al., 2013). Using primers listed in Table S1, we designed PCR products containing single FRT sites to insert at regions flanking the S. Typhimurium pdu/cob/cbi region (in the erfK and yeeA genes) (Fig. 1A,B). This was achieved using standard recombineering reagents and techniques (Datsenko and Wanner, 2000). Plasmid R995 Tc (containing a single FRT site) was introduced into this strain followed by introduction of plasmid pCP20 which expresses the FLP recombinase (Datsenko and Wanner, 2000; Santiago et al., 2011). This resulted in excision of the pdu/cob/cbi gene segment and insertion of this segment into R995 Tc, and this construct could be isolated using conjugation to a fresh E. coli background containing a counterselection (such as TOP10 Rif‐R or TOP10 Nal‐R) (Fig. 1B). PCR was used to confirm successful cloning and conjugation of the targeted genomic segment (Fig. 1C). The strain S. Typhimurium Δpdu was obtained as a by‐product of this procedure as this strain was easily isolated after FLP treatment (by screening pCP20‐cured isolates for Km‐S and Cm‐R).
Transfer of R995 + pdu ST to Gram‐negative recipients
An E. coli strain containing R995 + pdu ST was used as a donor in conjugative transfer to various Gram‐negative recipients as described previously (Wilson et al., 2004; Wilson and Nickerson, 2006). All transconjugants were selected on M9 medium (since the donor E. coli strain is auxotrophic) containing Tc and Km, except the conjugation to S. Typhimurium Δpdu which was selected on LB Cm Tc Km. Plasmid DNA was isolated from each strain to verify transfer via PCR (Fig. 1C).
Testing of R995 + pdu ST gene expression
MacConkey media containing 1,2 PD as a carbon source were formulated as follows (per litre): 20 g Bacto peptone, 5 g NaCl, 10 ml 1,2 PD (Sigma #398039), 75 mg neutral red, 15 g Bacto agar. Coenzyme B12 (Sigma #CO884) was added as indicated to a concentration of 500 nM. Stock solutions and agar plates containing coenzyme B12 were stored protected from light. RT‐qPCR was performed as described previously (Wilson et al., 2008; Soni et al., 2014; Herman et al., 2016) using total RNA harvested from the indicated strains (grown either with or without 1,2 PD) which was then converted to cDNA and analysed using pduW and korB primers indicated in Table S1.
Isolation and analysis of MCPs expressed from R995 + pdu ST
We modified a commonly referenced Pdu MCP isolation protocol (Sinha et al., 2012) as follows: strains containing R995 + pdu ST or R995 vector were routinely grown overnight in 100 ml of Pdu medium (1× M9 salts, 1 mM MgSO4, 0.5% succinate, 0.5% 1,2 PD, 2 mg ml−1 casamino acids) containing Km and Tc selection. Seventy millilitres (35 ml in each of two Oak Ridge tubes) of the cultures was centrifuged at 12 000 g to pellet the cells, the supernatant was entirely removed, and the pellets in each tube were resuspended in 7.5 ml buffer A (50 mM Tris‐HCl pH = 7.5, 500 mM KCl, 12.5 mM MgCl2, 0.5% 1,2 PD, 5 mM beta‐mercaptoethanol, 0.4 mM protease inhibitor AEBSF [Sigma #A8456], 100 μg ml−1 lysozyme [Sigma #L3790], 2 units ml−1 DNase I [New England Biolabs #M0303S]) in each tube. The starting amount of cells for each prep was typically between 6.3 × 1010 and 1.4 × 1011 CFU total, determined by serial dilution and plating of samples for CFU counts (or using OD600 measurement correlated to known CFU amounts for each strain). Three millilitres of B‐PER lysis reagent (Thermo Fisher #90084) was added to each tube and mixed gently, and the tubes were incubated with rotation at room temperature for 1 h. The samples were centrifuged at 12 000 g for 5 min to pellet cell debris, the supernatant transferred to fresh tubes, and the samples then centrifuged at 20 000 g for 30 min to pellet MCPs. After careful removal of the entire supernatant, the pellets were resuspended and combined in a total volume of 500 μl buffer B (50 mM Tris‐HCl pH = 7.5, 50 mM KCl, 5 mM MgCl2, 1% 1,2 PD, 0.4 mM AEBSF) and transferred to a single microfuge tube. The sample was then centrifuged at highest speed in a microfuge (16 000 g) for one minute to pellet any additional insoluble material, and the supernatant was transferred to a fresh microfuge tube (which represented the final sample). For analysis, we would typically load 1%–3% of total yield on a 12% SDS‐PAGE gel followed by Coomassie Blue staining, with total yield being typically between 500 and 2000 micrograms of protein. To identify proteins from SDS‐PAGE gels, bands were excised from the gels and subjected to Nano‐LC‐MS/MS mass spectrometry analysis (Alphalyse, Inc., Palo Alto, CA). Briefly, the proteins obtained from excised gel bands were reduced, carbamidomethylated, and subsequently digested with trypsin. The resulting peptides were concentrated via lyophilization and redissolved for injection into a Dionex nano‐LC system followed by MS/MS analysis on a Bruker Maxis Impact QTOF instrument. The MS/MS spectra were used for Mascot‐based searching of UniProt and NCBI protein databases containing more than 80 million known non‐redundant protein sequences. For the MCP mini‐preps, strains were grown overnight with plasmid selection in 25 ml of media, and then 10 ml of culture was evenly distributed across eight microfuge tubes and pelleted in a microfuge at 16 000 g for 5 min at 4°C. After complete removal of the supernatant via aspiration, 500 μl of buffer A containing B‐PER was added to each tube, and the cells were fully resuspended. After rotation at room temperature for 45 min, the samples were microfuged at 12 000 g for 3 min at 4°C to pellet cell debris. After this spin, the supernatants for each sample were pooled into four microfuge tubes (now containing approximately 1 ml each), and the samples were microfuged at 16 000 g for 1 h at 4°C. After this spin, the supernatants were removed, and the pellets were resuspended and pooled in a total of 150 μl of buffer B. Typical total protein yield from the mini‐prep was 100–200 μg. All MCP preps were stored at either 4°C or at −80°C.
Electron microscopy
Transmission electron microscopy (TEM) was performed using standard protocols as described previously (Jennings et al., 2012). MCP samples were spotted onto carbon‐coated copper grids (300 mesh), negatively stained using 4% uranyl acetate and visualized using an Hitachi H‐7600 TEM microscope at 80 kV. To quantify MCP size, the diameters of at least 50 individual MCPs were measured in random fields of view in TEM images for the indicated species. The widest aspect of a given MCP diameter was measured for this analysis.
Different media used for MCP growth and isolation
The different growth media tested for support of MCP expression and isolation from R995 + pdu ST strains are as follows (ingredients given per litre): Pdu medium (recipe provided above), Lennox LB broth (10 g tryptone, 5 g yeast extract, 5 g NaCl), tryptic soy broth (TSB) (15 g tryptone, 5 g soytone, 5 g NaCl), 2xYT broth (16 g tryptone, 10 g yeast extract, 5 g NaCl), PYE broth (10 g peptone, 10 g yeast extract, 5 g NaCl), CAYE broth (20 g casamino acids, 10 g yeast extract, 5 g NaCl), nutrient broth (NB) (4.5 g beef extract, 7.5 g peptone, 5 g NaCl), Mueller Hinton broth (MH) (17.5 g casein acid hydrolysate, 3 g beef extract, 1.5 g starch). All media were supplemented with 1,2 PD to 0.5%. Control strains containing the R995 vector grown in these media did not display the MCP bands via SDS‐PAGE and Coomassie staining (data not shown).
Conflict of interest
None declared.
Supporting information
Table S1. DNA primers used in this study.
Table S2. MCP yields from R995 + pdu ST‐containing bacteria.
Fig. S1. The utilization of 1,2 PD directed by R995 + pdu ST depends on coenzyme B12.
Fig. S2. Diameters of MCPs isolated from different bacterial species containing R995 + pdu ST.
Acknowledgements
We gratefully acknowledge the Biology Department at Villanova University for their generous support of this work. We thank members of the Biology Department for discussions, reagents and use of equipment during this work, particularly Nancy Peltier for assistance in the Villanova Imaging Center at Mendel Hall.
Microbial Biotechnology (2018) 11(1), 199–210
Funding Information
Villanova University.
References
- Abdul‐rahman, F. , Petit, E. , and Blanchard, J. (2013) The distribution of polyhedral bacterial microcompartments suggests frequent horizontal transfer and operon reassembly. J Phylogen Evol Biol 1: 1–7. [Google Scholar]
- Ailion, M. , Bobik, T. A. , and Roth, J. R. (1993) Two global regulatory systems (Crp and Arc) control the cobalamin/propanediol regulon of Salmonella typhimurium . J Bacteriol 175: 7200–7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axen, S. D. , Erbilgin, O. , and Kerfeld, C. A. (2014) A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS Comput Biol 10: e1003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobik, T. A. , Ailion, M. , and Roth, J. R. (1992) A single regulatory gene integrates control of vitamin B12 synthesis and propanediol degradation. J Bacteriol 174: 2253–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobik, T. A. , Xu, Y. , Jeter, R. M. , Otto, K. E. , and Roth, J. R. (1997) Propanediol utilization genes (pdu) of Salmonella typhimurium: three genes for the propanediol dehydratase. J Bacteriol 179: 6633–6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobik, T. A. , Havemann, G. D. , Busch, R. J. , Williams, D. S. , and Aldrich, H. C. (1999) The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B(12)‐dependent 1, 2‐propanediol degradation. J Bacteriol 181: 5967–5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobik, T. A. , Lehman, B. P. , and Yeates, T. O. (2015) Bacterial microcompartments: widespread prokaryotic organelles for isolation and optimization of metabolic pathways. Mol Microbiol 98: 193–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, A. H. , and Silver, P. A. (2012) Designing biological compartmentalization. Trends Cell Biol 22: 662–670. [DOI] [PubMed] [Google Scholar]
- Chen, P. , Ailion, M. , Bobik, T. , Stormo, G. , and Roth, J. (1995) Five promoters integrate control of the cob/pdu regulon in Salmonella typhimurium . J Bacteriol 177: 5401–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, S. , Sinha, S. , Fan, C. , Liu, Y. , and Bobik, T. A. (2011) Genetic analysis of the protein shell of the microcompartments involved in coenzyme B12‐dependent 1,2‐propanediol degradation by Salmonella . J Bacteriol 193: 1385–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury, C. , Sinha, S. , Chun, S. , Yeates, T. O. , and Bobik, T. A. (2014) Diverse bacterial microcompartment organelles. Microbiol Mol Biol Rev 78: 438–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury, C. , Chun, S. , Pang, A. , Sawaya, M. R. , Sinha, S. , Yeates, T. O. , and Bobik, T. A. (2015) Selective molecular transport through the protein shell of a bacterial microcompartment organelle. Proc Natl Acad Sci USA 112: 2990–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko, K. A. , and Wanner, B. L. (2000) One‐step inactivation of chromosomal genes in Escherichia coli K‐12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, C. , Fromm, H. J. , and Bobik, T. A. (2009) Kinetic and functional analysis of L‐threonine kinase, the PduX enzyme of Salmonella enterica . J Biol Chem 284: 20240–20248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, M. , and Sambrook, J. (2012) Molecular Cloning. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Herman, A. , Serfecz, J. , Kinnally, A. , Crosby, K. , Youngman, M. , Wykoff, D. , and Wilson, J. W. (2016) The bacterial iprA gene is conserved across Enterobacteriaceae, is involved in oxidative stress resistance, and influences gene expression in Salmonella enterica Serovar Typhimurium. J Bacteriol 198: 2166–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo, H. , Tsuruno, K. , Tatsuke, T. , Sato, M. , and Hanai, T. (2015) Dual synthetic pathway for 3‐hydroxypropionic acid production in engineered Escherichia coli . J Biosci Bioeng 120: 199–204. [DOI] [PubMed] [Google Scholar]
- Jennings, M. E. , Quick, L. N. , Ubol, N. , Shrom, S. , Dollahon, N. , and Wilson, J. W. (2012) Characterization of Salmonella type III secretion hyper‐activity which results in biofilm‐like cell aggregation. PLoS ONE 7: e33080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C. L. , Pechonick, E. , Park, S. D. , Havemann, G. D. , Leal, N. A. , and Bobik, T. A. (2001) Functional genomic, biochemical, and genetic characterization of the Salmonella pduO gene, an ATP:cob(I)alamin adenosyltransferase gene. J Bacteriol 183: 1577–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorda, J. , Lopez, D. , Wheatley, N. M. , and Yeates, T. O. (2013) Using comparative genomics to uncover new kinds of protein‐based metabolic organelles in bacteria. Protein Sci 22: 179–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorda, J. , Liu, Y. , Bobik, T. A. , and Yeates, T. O. (2015) Exploring bacterial organelle interactomes: a model of the protein‐protein interaction network in the Pdu microcompartment. PLoS Comput Biol 11: e1004067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, E. Y. , and Tullman‐Ercek, D. (2013) Engineering nanoscale protein compartments for synthetic organelles. Curr Opin Biotechnol 24: 627–632. [DOI] [PubMed] [Google Scholar]
- Kim, E. Y. , Jakobson, C. M. , and Tullman‐Ercek, D. (2014) Engineering transcriptional regulation to control Pdu microcompartment formation. PLoS ONE 9: e113814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennox, E. S. (1955) Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1: 190–206. [DOI] [PubMed] [Google Scholar]
- Matsubara, M. , Urano, N. , Yamada, S. , Narutaki, A. , Fujii, M. , and Kataoka, M. (2016) Fermentative production of 1‐propanol from d‐glucose, l‐rhamnose and glycerol using recombinant Escherichia coli. J Biosci Bioeng 122: 421–426. [DOI] [PubMed] [Google Scholar]
- McClelland, M. , Sanderson, K. E. , Spieth, J. , Clifton, S. W. , Latreille, P. , Courtney, L. , et al (2001) Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413: 852–856. [DOI] [PubMed] [Google Scholar]
- Pansegrau, W. , Lanka, E. , Barth, P. T. , Figurski, D. H. , Guiney, D. G. , Haas, D. , et al (1994) Complete nucleotide sequence of Birmingham IncP alpha plasmids. Compilation and comparative analysis. J Mol Biol 239: 623–663. [DOI] [PubMed] [Google Scholar]
- Parsons, J. B. , Dinesh, S. D. , Deery, E. , Leech, H. K. , Brindley, A. A. , Heldt, D. , et al (2008) Biochemical and structural insights into bacterial organelle form and biogenesis. J Biol Chem 283: 14366–14375. [DOI] [PubMed] [Google Scholar]
- Sampson, E. M. , Johnson, C. L. , and Bobik, T. A. (2005) Biochemical evidence that the pduS gene encodes a bifunctional cobalamin reductase. Microbiology 151: 1169–1177. [DOI] [PubMed] [Google Scholar]
- Santiago, C. P. , Quick, L. N. , and Wilson, J. W. (2011) Self‐transmissible IncP R995 plasmids with alternative markers and utility for Flp/FRT cloning strategies. J Microbiol Biotechnol 21: 1123–1126. [DOI] [PubMed] [Google Scholar]
- Sargent, F. , Davidson, F. A. , Kelly, C. L. , Binny, R. , Christodoulides, N. , Gibson, D. , et al (2013) A synthetic system for expression of components of a bacterial microcompartment. Microbiology 159: 2427–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, S. , Cheng, S. , Fan, C. , and Bobik, T. A. (2012) The PduM protein is a structural component of the microcompartments involved in coenzyme B(12)‐dependent 1,2‐propanediol degradation by Salmonella enterica . J Bacteriol 194: 1912–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, S. , Cheng, S. , Sung, Y. W. , McNamara, D. E. , Sawaya, M. R. , Yeates, T. O. , and Bobik, T. A. (2014) Alanine scanning mutagenesis identifies an asparagine‐arginine‐lysine triad essential to assembly of the shell of the Pdu microcompartment. J Mol Biol 426: 2328–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni, A. , O'Sullivan, L. , Quick, L. N. , Ott, C. M. , Nickerson, C. A. , and Wilson, J. W. (2014) Conservation of the low‐shear modeled microgravity response in Enterobacteriaceae and analysis of the trp genes in this response. Open Microbiol J 8: 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, S. J. , and Yeates, T. O. (2011) Bacterial microcompartments insights into the structure, mechanism, and engineering applications. Prog Mol Biol Transl Sci 103: 1–20. [DOI] [PubMed] [Google Scholar]
- Wilson, J. W. , and Nickerson, C. A. (2006) A new experimental approach for studying bacterial genomic island evolution identifies island genes with bacterial host‐specific expression patterns. BMC Evol Biol 6: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J. W. , Figurski, D. H. , and Nickerson, C. A. (2004) VEX‐capture: a new technique that allows in vivo excision, cloning, and broad‐host‐range transfer of large bacterial genomic DNA segments. J Microbiol Methods 57: 297–308. [DOI] [PubMed] [Google Scholar]
- Wilson, J. W. , Coleman, C. , and Nickerson, C. A. (2007) Cloning and transfer of the Salmonella pathogenicity island 2 type III secretion system for studies of a range of Gram‐negative genera. Appl Environ Microbiol 73: 5911–5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J. W. , Ott, C. M. , Quick, L. , Davis, R. , Honer zu Bentrup, K. , Crabbe, A. , et al (2008) Media ion composition controls regulatory and virulence response of Salmonella in spaceflight. PLoS ONE 3: e3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J. W. , Santiago, C. P. , Serfecz, J. , and Quick, L. N. (2013) Recombineering and conjugation as tools for targeted genomic cloning In Genetic Manipulation of DNA and Protein – Examples from Current Research. Figurski D. D. (ed). Rijeka, Croatia: In Tech, pp. 437–450. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. DNA primers used in this study.
Table S2. MCP yields from R995 + pdu ST‐containing bacteria.
Fig. S1. The utilization of 1,2 PD directed by R995 + pdu ST depends on coenzyme B12.
Fig. S2. Diameters of MCPs isolated from different bacterial species containing R995 + pdu ST.


