ABSTRACT
The detection of prions is difficult due to the peculiarity of the pathogen, which is a misfolded form of a normal protein. The specificity and sensitivity of detection methods are imperfect in complex samples, including in excreta. Here, we combined optimized prion amplification procedures with a statistical method that accounts for false-positive and false-negative errors to test deer saliva for chronic wasting disease (CWD) prions. This approach enabled us to discriminate the shedding of prions in saliva and the detection of prions in saliva—a distinction crucial to understanding the role of prion shedding in disease transmission and for diagnosis. We found that assay sensitivity and specificity were indeed imperfect, and we were able to draw several conclusions pertinent to CWD biology from our analyses: (i) the shedding of prions in saliva increases with time postinoculation, but is common throughout the preclinical phase of disease; (ii) the shedding propensity is influenced neither by sex nor by prion protein genotype at codon 96; and (iii) the source of prion-containing inoculum used to infect deer affects the likelihood of prion shedding in saliva; oral inoculation of deer with CWD-positive saliva resulted in 2.77 times the likelihood of prion shedding in saliva compared to that from inoculation with CWD-positive brain. These results are pertinent to horizontal CWD transmission in wild cervids. Moreover, the approach described is applicable to other diagnostic assays with imperfect detection.
KEYWORDS: CWD, RT-QuIC, false negative, false positive, occupancy modeling, prion
INTRODUCTION
Prions, the causative agents of transmissible spongiform encephalopathies, are unusual pathogens that require innovative approaches for diagnosis. A prion (PrPSc) is an abnormal protease-resistant conformation of a normal protein (PrPC). The repeated conversion of PrPC to PrPSc results in the spread of prions throughout the body (1). Common detection methods for other infectious agents are not applicable to prions. DNA-based detection methods (e.g., quantitative PCR) are not effective, because both diseased and normal individuals have the gene for PrPC. RNA-based detection methods (e.g., reverse transcriptase quantitative PCR) are not useful because the transcription of the gene is not consistently modified during disease (2, 3). Protein-based detection methods (e.g., immunoblot, enzyme-linked immunosorbent assays) require substantial modifications to differentiate between PrPC and PrPSc (4). The early diagnosis of prion diseases would facilitate more informative clinical trials in humans and more effective disease management strategies for wildlife and farmed cervids.
The diagnosis of human prion disease traditionally relies on autopsy, and then immunoblot or immunohistochemistry (IHC) that is made specific for PrPSc by treating the sample with proteinase K (thereby destroying PrPC in the sample) (5). The diagnosis of chronic wasting disease (CWD) in deer and elk followed a similar approach for many years—PrPSc was detected in postmortem tissue by immunoblot, enzyme-linked immunosorbent assay, or IHC (1, 6). A recent movement toward antemortem diagnoses for Creutzfeldt-Jakob disease (CJD) and CWD has led to the adoption of real-time quaking-induced conversion (RT-QuIC) as a sensitive diagnostic method for a variety of sample types. RT-QuIC has a very low limit of detection because it amplifies tiny amounts of PrPSc by facilitating the misfolding of recombinant PrPC. PrPSc is detected by the intercalation of the amyloid-binding dye thioflavin T (ThT), which results in a detectable shift in the fluorescence emission spectrum. Since its inception, RT-QuIC has been used for the detection of CJD prions in human cerebral spinal fluid and nasal brushings and CWD prions in urine, feces, saliva, and other tissues (7–12).
CWD-infected cervids likely shed prions in excreta throughout the course of disease (9–11), and prion-positive saliva, urine, and feces are infectious to other deer (13–15). Though diagnoses are made with brain or lymphoid tissue, we hypothesize that prions in excreta are an essential component of the horizontal transmission of CWD within cervid populations (16). Horizontal transmission is the most likely explanation for high CWD prevalence estimates (6 to 70%) among free-ranging cervids in North America (17, 18). Excreta not only play important biological roles in CWD transmission but are noninvasively accessible antemortem. We are particularly interested in the detection of prions in saliva, because saliva appears to have the highest prion concentration of any excreta in deer and because salivary glands contain prions fairly early in disease (10, 19, 20). Saliva is an easily obtained diagnostic specimen used in a variety of conditions, including oral diseases, human immunodeficiency virus infection, hepatitis A virus, Epstein-Barr virus, rubella virus, and breast cancer (21).
Previously, we observed variability in the detection of prions in the saliva of preclinical and symptomatic deer infected with CWD (10). However, the variation was not explained by biological factors of interest (e.g., time since inoculation, deer characteristics, etc.) when the data were examined without accounting for imperfect detection. Failing to account for false-positive and -negative errors can bias estimates of interest (shedding, prevalence, or sensitivity) and can mask biological patterns. We hypothesize that the variation in prion detection is due either to the sensitivity limits of RT-QuIC and the presence of assay inhibitors in saliva or to biological variability in shedding; thus, we used a novel modeling approach to distinguish between shedding and detection (B. M. Brost, B. A. Mosher, and K. A. Davenport, in review).
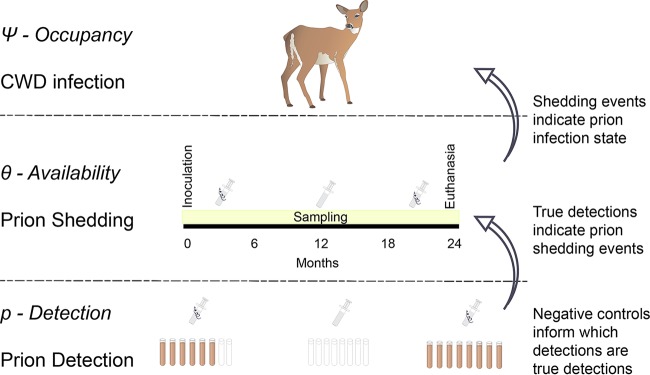
We use an occupancy modeling framework (22), a statistical method that uses repeat surveys (RT-QuIC replicates in this case) of a single sample at one point in time to account for imperfect detection (22, 23). Occupancy modeling was developed to estimate the probability that a site was occupied by a given wildlife species when the detection of that species was imperfect, but it has also been applied to the detection of pathogens to obtain unbiased estimates of prevalence and test sensitivity (24). We extended the traditional multiscale occupancy model to account for false-positive detections in addition to false-negative detections (see Materials and Methods). We assumed that all deer in our study (Table 1) were infected with prions, which was confirmed by necropsy at the end of the study. Our research question was whether deer were consistently shedding prions throughout the course of disease or there was variation in shedding (Fig. 1) that could be attributed to various hypothesized factors (Table 2). To understand variation in shedding, it was critical to account for both false-positive and false-negative detection errors. We used negative-control samples to account for the probability of false-positive reactions and multiple replicates to understand the probability of false negatives. We used 198 saliva samples collected longitudinally from 45 experimentally inoculated white-tailed deer (Odocoileus virginianus), deer we knew were “occupied by” prions. However, in the case of unknown disease status, the model can predict the likelihood of occupancy (prion infection). We used this model to estimate the sensitivity and specificity of RT-QuIC detection and to identify important drivers of prion shedding.
TABLE 1.
Summary of the characteristics of the deer included in the study
| Characteristic or groupa | No. (%) of deer (n = 45) |
|---|---|
| 96GG | 35 (78) |
| 96GS | 10 (22) |
| Male | 23 (51) |
| Female | 22 (49) |
| PTA concn | 17 (38) |
| IOME concn | 28 (62) |
| Blood-IP | 4 (9) |
| Blood-IV | 13 (29) |
| Brain-aerosol | 6 (13) |
| Brain-IC | 1 (2) |
| Brain-PO | 11 (24) |
| Contact | 4 (9) |
| Saliva-PO | 6 (13) |
96GG, deer homozygous for glycine at codon 96, the most susceptible genotype; PTA, saliva concentration by phosphotungstic acid; IOME, saliva concentration by iron oxide magnetic extraction; IP, intraperitoneal; IV, intravenous; IC, intracerebral; PO, per os (oral).
FIG 1.
Schematic framework for multiscale occupancy model. We adopted a multiscale occupancy modeling framework, where our top tier (deer level, ψ) represents the CWD infection status of an individual deer, the second tier (sample level, θ) represents individual saliva samples over time, when the deer may or may not be shedding. The third tier (detection, p) indicates RT-QuIC detection in each replicate from an individual saliva sample. Because we have false-positive and false-negative errors, it is possible that a saliva sample with no prions (no shedding, middle sample) can produce positive replicates in RT-QuIC and that a sample that does contain prions (shedding, left sample) can produce negative replicates in RT-QuIC. Deer and syringe images courtesy of the Integration and Application Network, University of Maryland Center for Environmental Science (http://ian.umces.edu/symbols/).
TABLE 2.
Covariates for occupancy model
| Covariatea | Scale parameterb | Prediction | Reference(s) |
|---|---|---|---|
| Sex | θ | Males will have higher probability of shedding (θ) because males have a higher CWD incidence in the wild. The difference is likely due to behaviors exhibited by male deer, which may or may not be replicated in exptl conditions in castrated deer. | 33, 34 |
| Genotype (codon 96) | θ | 96GG deer will have a higher probability of shedding because they are more susceptible to disease and because prion replication occurs earlier than in those with 96GS/96SS. | 5, 19 |
| Mos postinoculation | θ | Deer are more likely to shed prions later in the disease course because the prion load in the animal is higher. | 19 |
| Tonsil biopsy specimen positive | θ | Tonsil biopsy can be used to monitor disease progression. We predict that a deer with a positive tonsil biopsy specimen would be more likely to shed prions in its saliva because the deer has a broader prion distribution than a tonsil biopsy specimen-negative individual. | 45 |
| Inoculum/route of inoculation | θ | Deer inoculated with excreta (saliva, etc.) will have a higher probability of shedding than deer inoculated with brain material. We hypothesize that excreted prions are structurally different from CNS prions and these prions may preferentially be shed. | |
| Concn technique | p | The assay type will affect the probability of detection (p). Of the two methods of sample concn, we predict that IOME will result in a higher probability of detection. | 45 |
| Storage duration | p | Increased storage duration will decrease the probability of detection due to sample degradation. |
We selected covariates and predicted their effects a priori. We described each covariate, the levels of the model impacted by the covariate, and the direction of the effect.
θ indicates shedding/availability (Fig. 1) and p indicates detection.
MATERIALS AND METHODS
Deer inclusion criteria.
We only used deer that were diagnosed as CWD positive (CWD+) by immunohistochemistry or Western blotting of the obex region of the brain stem collected during necropsy. Immunohistochemistry (20) and Western blotting (25) were performed as previously described. Deer included in the study are summarized in Table 1. All statistical conclusions in the paper are derived from a sample size of 45 deer.
White-tailed deer studies.
White-tailed deer were from the University of Georgia Warnell School of Forestry and Natural Resources (a region where CWD is not endemic) and were housed in an indoor CWD research facility at Colorado State University. Spanning 15 years, deer were exposed to CWD as follows (Table 1). (i) Saliva (50 ml total, per os [PO]) was administered over 6 to 12 doses to 6 deer (13, 26). (ii) Whole blood (150 to 315 ml total, intraperitoneal [IP]) was given as a single bolus dose to 4 deer (26). (iii) Whole blood and blood fractions (B cells/platelets/peripheral blood mononuclear cell [PBMC], intravenous [IV]) were given as single bolus doses to 13 deer (27). (iv) Brain (1g, intracranial [IC] or PO) was administered as a single dose to one deer (13, 28). (v) Brain (2 ml, 5% CWD+ brain homogenate, aerosolization) was given as two doses, 1 week apart, to 6 deer (29). (vi) One contact deer was continually exposed to bedding and water buckets transferred daily from CWD+ deer rooms (26). (vi) Additional contact deer (n = 3) were exposed through direct cohousing with CWD-inoculated deer. The CWD-inoculated deer (n = 3) were administered 315 ml CWD+ whole blood via the IP route and held for 1 month before introducing the naive (contact) cohort. Whole blood for inoculation was obtained and pooled from three CWD+ deer from a previous study at the facility. Deer were cohoused for 19 months, during which excreta were collected at 3- to 6-month intervals to monitor disease progression. The contact deer were euthanized after 19 months of cohousing.
All deer were maintained in accordance with protocols approved by the Colorado State University Institutional Animal Care and Use Committee. Longitudinal saliva samples were collected approximately every 3 months until euthanasia due to clinical signs or study duration. Saliva was obtained from anesthetized deer by inserting a 3-ml syringe into the cheek pouch closest to the ground and aspirating. Samples were aliquoted and frozen at −80°C until testing. Baseline saliva samples (obtained immediately before inoculation) were used as negative controls.
Tonsil biopsy.
Tonsil biopsy specimens were collected using a modified version of the protocol described in reference 30. The oral cavities of anesthetized deer were held open using a mouth gag, and the tonsils were visualized by depressing the tongue with a laryngoscope. Endoscopic forceps were used to collect two biopsy specimens from a single tonsil at each time point. One biopsy specimen was frozen at −80°C for Western blotting and one biopsy specimen was fixed in either 10% formalin or paraformaldehyde-lysine-periodate (PLP) for immunohistochemistry.
Western blotting.
Biopsy tissues and brains from euthanized animals were homogenized, and prions were detected according to the following protocols.
(i) Saliva-oral, blood-IV, blood-IP, contact, and brain-IC groups.
Tissues were homogenized in NP-40 buffer, protease treated, and separated on a polyacrylamide gel. Proteins were transferred to a polyvinylidene fluoride (PVDF) membrane, probed with BAR224 primary antibodies and a horseradish peroxidase (HRP)-conjugated goat anti-mouse secondary, detected using an Amersham ECL detection system, and visualized on a digital GelDoc (13, 26, 27).
(ii) Brain-PO group.
Tissues were processed according to the previously described protocol (31). Briefly, tissues were homogenized in 1× phosphate-buffered saline (PBS)-Triton X buffer, protease treated, separated on a polyacrylamide gel, and transferred to a PVDF membrane. HRP-conjugated BAR224 was used to probe the membrane, which was developed with an ECL-plus detection kit (GE) and visualized on a digital GelDoc (31).
(iii) Brain-aerosol group.
Tissues were examined with a slightly modified version of the previous protocol, wherein a SNAP i.d. system (Millipore) was used for rapid Western blotting (25).
Immunohistochemistry.
We used adaptations of a previously described protocol to perform IHC on fixed, embedded, and sectioned biopsy specimens or brain from euthanized deer (32). Briefly, for deer in the saliva-PO, blood-IV, blood-IP, contact, and brain-IC cohorts, slides were processed through the Ventana autostainer (Tuscon, AZ) using BAR224 primary antibodies, biotinylated or HRP goat anti-mouse secondary, and the Ventana Red Map stain kit (13, 26, 27). For deer in the brain-PO cohort, sections were stained using a previously described tyramide signal amplification (TSA) protocol (28, 33). For the brain-aerosol cohort, tissue staining was performed on a Dako autostainer using HRP-conjugated BAR224 with an AEC (3-amino-9-ethylcarbazole) chromogen (31).
Production of recombinant PrP.
We expressed truncated Syrian hamster (SH) PrPC for RT-QuIC as previously reported (11). We added BL21 cells containing the sequence for the expression of amino acids 90 to 231 of SH PrPC to 5 ml LB medium, grew the cultures overnight, and then added the bacteria to 1L LB medium with autoinduction reagents [final concentrations, 0.5 M (NH4)2SO4, 1 M KH2PO4, 1 M Na2HPO4, 0.5% glycerol, 0.05% glucose, 0.2% α-lactose, and 0.001 M MgSO4]. When the optical density at 600 nm (OD600) reached approximately 1.7, we lysed the cells and purified the inclusion bodies according to the manufacturer's protocol with BugBuster and Lysonase (EMD-Millipore).
To purify recombinant PrP (rPrP), we solubilized the inclusion bodies in 8 M guanidine hydrochloride (GdnHCl) and 100 mM Na2HPO4 at room temperature overnight in an end-over-end rotator. We mixed the denatured rPrP slurry with Superflow nickel resin (Qiagen) and refolded the rPrP on the column and eluted it as previously reported (34).
RT-QuIC with iron oxide bead magnetic extraction.
We included four replicates each of four negative controls (baseline/preinoculation saliva samples), one positive control, and 19 unknown samples on each 96-well plate. We thawed single-use aliquots of saliva at room temperature, and then diluted each sample 1:100 in 1× PBS. We added 10 μl iron oxide beads (∼9 μm; Bangs Laboratory, IN) to 500 μl diluted saliva and incubated them for 1 h on an end-over-end rocker. We magnetically separated the beads, removed the supernatant, and resuspended the beads in 10 μl 0.1% SDS in 1× PBS. We added 2 μl of beads to each well of a 96-well plate containing 10 μg recombinant PrP, 320 mM NaCl, 1.0 mM EDTA, 20 mM NaH2PO4, and 10 μM thioflavin T. Each RT-QuIC experiment lasted 40 h, with cycles comprising 1 min of shaking (double orbital shaking at 700rpm) and 1 min of rest on a BMG Fluostar microplate reader. Fluorescence was recorded every 15 min (excitation, 450 nm; emission, 480 nm) using a gain of 1,700. Each sample was tested on two separate plates for a total of eight replicates. A replicate was considered positive if the fluorescence in the well crossed the threshold (5 standard deviations above the mean baseline fluorescence) within the course of the 40-h experiment.
The negative-control samples were used to monitor for the spontaneous conversion of rPrPC to amyloid. RT-QuIC is sensitive and specific with common sample types, including brain and lymph node biopsy specimens, but can produce false-positive (or -negative) reactions, especially when the sample is complex (e.g., saliva, urine, and feces) (9–11, 35).
RT-QuIC with PTA concentration.
Saliva samples were tested after sodium phosphotungstic acid (PTA) concentration as previously described (10). Briefly, we added 100 μl saliva to freshly prepared 4% PTA for a final concentration of 0.3% PTA. We incubated the samples for 1 h at 37°C with shaking at 1,700 rpm and then pelleted the protein precipitate by centrifugation at 17,000 × g for 30 min. We resuspended the PTA pellets in 10 μl 1× PBS with 0.1% SDS and added 2 μl of each sample to an RT-QuIC plate well.
Occupancy model.
We used a multiscale occupancy model (36, 37) to accommodate multiple longitudinally collected saliva samples per inoculated deer and multiple RT-QuIC replicates per saliva sample. The multiscale model is useful for estimating the probability of prion occurrence at two levels, namely, within the study unit (i.e., the deer) and within a longitudinal sample (i.e., saliva sample) (Fig. 1). We interpret the probability of prion occurrence at the deer level as the probability of CWD infection (ψ) and the probability of prion occurrence at the saliva sample level as the probability of shedding, given the animal was infected (θ) (Fig. 1). Although occupancy models were originally developed only to account for false-negative errors, we used a recent extension that also accommodates false-positive errors (B. M. Brost, B. A. Mosher, and K. A. Davenport, in review). This approach enables us to estimate the probability of a true detection (that is, of detecting prions when the saliva sample contains them [p]) and the probability of a false positive (that is, of obtaining a false detection in an RT-QuIC replicate, whether or not the sample contains prions [ϕ]). See Appendix SA in the supplemental material for additional details and a full model specification.
We fixed parameter ψ (the probability of CWD infection) to 1 for this analysis because our inclusion criteria required deer to have evidence of prion disease at euthanasia (specifically, positive Western blot or IHC of the obex region of the brainstem); however, this parameter can be estimated and may be of primary interest (interpreted as prevalence; sensu that in references 38 and 39) when the infection status is unknown. We focused on identifying factors that influence the probability that an infected animal is shedding prions. We made a priori predictions about the factors we thought would influence shedding probability (Table 2), which included the animal's sex and genotype, the inocula and route used to infect the animal, the time postinoculation, and deer tonsil biopsy specimen status associated with each saliva sample. Months postinoculation and tonsil biopsy specimen status were highly correlated and interesting for different reasons. Months postinoculation is a relatively precise measurement of disease progression, whereas tonsil biopsy specimen status may be useful in cases of natural infection. To consider the effect of each covariate representing disease progression, we ran two models: one that contained months postinoculation (and all other shedding covariates) and one that replaced months postinoculation with the sample's tonsil biopsy specimen status. In addition, we modeled the detection probability (p) as a function of the concentration technique used (PTA or iron oxide bead magnetic extraction [IOME]), the sample storage duration, and the interaction of these covariates in order to accommodate the variation in sensitivity associated with these two methods (Table 2). As we show in Fig. 3, the storage time and concentration technique are confounded; samples tested by PTA were all stored for less time than samples tested by IOME. Because the concentration technique and storage time are confounded, it is impossible to isolate the effects of time and technique on detection, and comparisons of the efficacy of PTA and IOME cannot be made.
FIG 3.
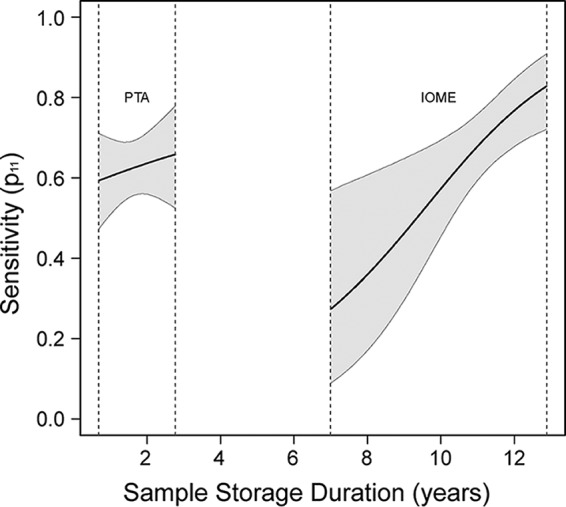
Sensitivity is imperfect. We compared two methods for the concentration of saliva before testing by RT-QuIC and assessed the effect of sample storage duration (i.e., amount of time stored at −80°C). Because older samples were all tested with IOME and newer samples were all tested with PTA, it is impossible to model the effects of method and storage time separately. However, we can assess their combined effects on sensitivity. The black lines indicate the average probability of detection (or sensitivity) and the gray shaded regions represent the 95% credible intervals. Vertical dotted lines indicate the ranges of sample storage duration for our samples.
Hypotheses.
We hypothesized that male deer would be more likely to shed prions in their saliva (Table 2). Population studies indicate that male deer have a higher CWD prevalence than female deer, a phenomenon attributed to behavioral differences between the sexes (40, 41). Because the exposure to the pathogen was ensured by our experimental inoculation, we could differentiate the effect of sex on shedding from its effect on CWD exposure or infection.
We hypothesized that shedding would increase as the disease progressed, because prions spread through the body early in the course of disease and likely replicate in local tissue environments (such as oropharyngeal lymphoid tissues) (19, 20, 42). Therefore, the prion burden in oropharyngeal tissues is likely to increase with time, creating a larger pool of prions to be shed in saliva (Table 2).
We collected and tested tonsil biopsy specimens throughout the CWD incubation period. We classified saliva samples as having been collected on or after the sampling period when the tonsil biopsy specimen was first confirmed CWD+ or before that time. We hypothesized that deer with a positive tonsil biopsy specimen were more likely to shed prions than deer with a negative biopsy specimen result (Table 2).
The most common polymorphism associated with CWD susceptibility is a glycine-serine substitution at codon 96. 96GG homozygotes are the most susceptible and have the most common genotype, while 96GS heterozygotes have intermediate susceptibility and 96SS homozygotes are least susceptible. Specifically, GS deer can be infected with CWD but have extended incubation periods and disease courses compared to those of 96GG deer (1, 20). Therefore, we hypothesized that 96GS deer would be less likely to shed prions than 96GG deer (Table 2).
We inoculated deer with a variety of inoculum sources and via several routes over 15 years of experiments in the natural host. We hypothesized that prions in excreta (or in the periphery in general) may be adapted to the periphery in some way and would thereby result in more frequent and consistent prion shedding (Table 2).
We used two approaches to concentrate saliva samples before testing by RT-QuIC. Phosphotungstic acid (PTA) precipitates protein from a complex sample, enabling the concentration of large volumes (100 μl) of saliva into an assayable volume (10 μl). Iron oxide metal extraction (IOME) involves the binding of prions (and other components of saliva) to iron oxide beads, which can be resuspended in much smaller volumes. We hypothesized that IOME would result in a higher probability of detection than concentration by PTA (Table 2) (35).
Our saliva samples were collected over a period of 15 years and the samples had variable storage durations at −80°C. We hypothesized that the probability of detection would decrease with increased storage duration, a consequence of sample degradation over long storage periods (Table 2).
Model implementation.
We fit the models described using a Markov chain Monte Carlo (MCMC) algorithm in R (code provided in Text S1; R Development Core Team 2012).
RESULTS
Observations from raw data suggested variable shedding and/or detection of prions in saliva.
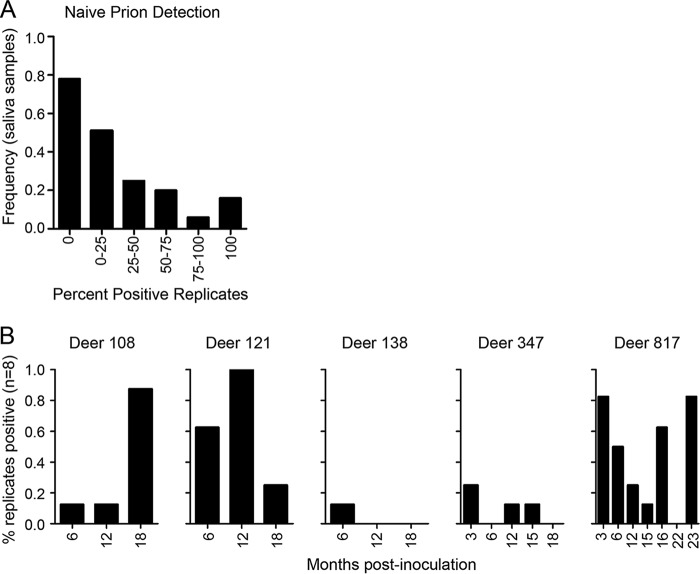
In 52% of our saliva samples, we detected prions in at least one but not all replicates, which suggested imperfect sensitivity (Fig. 2A; see also Tables S1 and S2 in the supplemental material). It was also common to detect prions in saliva during one sampling period but not in a subsequent sampling period (Fig. 2B and Tables S1 and S2). Inconsistent detection within replicates of a single sample and over time for a single deer prompted us to investigate the sensitivity of RT-QuIC for the detection of prions in saliva. We have observed false-positive errors in RT-QuIC (in our negative controls), and so we analyzed the data using a method that accommodates false-positive results (9–11).
FIG 2.
Representative RT-QuIC data for saliva from CWD-infected deer over the course of observation. Each sample was tested in quadruplicates in two experiments (n = 8 replicates total). (A) The numbers of samples with 0%, 0 to ≤25%, 25% to ≤50%, 50 to ≤75%, 75 to <100%, or 100% positive replicates (raw, or naive, results) are indicated by the y axis. (B) The percentages of replicates that were positive for individual samples are indicated by bars. The x axis specifies the months postinoculation; a label on the x axis with no bar indicates that a sample was tested, and no replicates were positive. The observations of imperfect detection (A) and apparent randomness in shedding (B) in the raw data, as depicted here, motivated us to pursue a modeling approach that accounts for detection error.
Prion detection in saliva with RT-QuIC is imperfect.
The probability of a positive RT-QuIC reaction given that the sample is positive (p) and the probability of a false-positive RT-QuIC reaction, whether or not a sample contains prions (ϕ), influence the likelihood of a positive RT-QuIC result. Sensitivity (p) was less than 100% (Fig. 3). Contrary to our prediction, the sensitivity of both methods increased with increasing sample storage time. We suspect this relationship is due to freeze denaturation and the degradation of the RT-QuIC assay inhibitor that is present in deer saliva. Specificity (1 − ϕ)for PTA was 96.2% (95% confidence interval [CI], 91.9% to 98.9%) and the specificity for IOME was 90.8% (95% CI, 85.9% to 94.9%).
Shedding of prions in saliva increases with disease progression.
Occupancy modeling enabled us to account for imperfect detection and obtain unbiased estimates of shedding. Consistent with our a priori predictions (see Materials and Methods), the probability of shedding (θ) increased with time postinoculation, regardless of sex, genotype, route of inoculation, or inoculum type (slope, 0.580 [95% CI, 0.11 to 1.07]) (Fig. 4A). For example, deer inoculated with saliva via the oral route had a probability of shedding of 0.79 (95% CI, 0.49 to 0.97) at 3 months postinoculation (the first sampling period) and a probability of shedding of 0.96 (95% CI, 0.86 to 1.00) at 26 months postinoculation (the last sampling period we tested). In one other example, deer inoculated with blood via the intravenous route had a probability of shedding of 0.07 (95% CI, 0.01 to 0.19) at 3 months postinoculation and a probability of shedding of 0.37 (95% CI, 0.10 to 0.71) at 26 months postinoculation (Fig. 4A).
FIG 4.
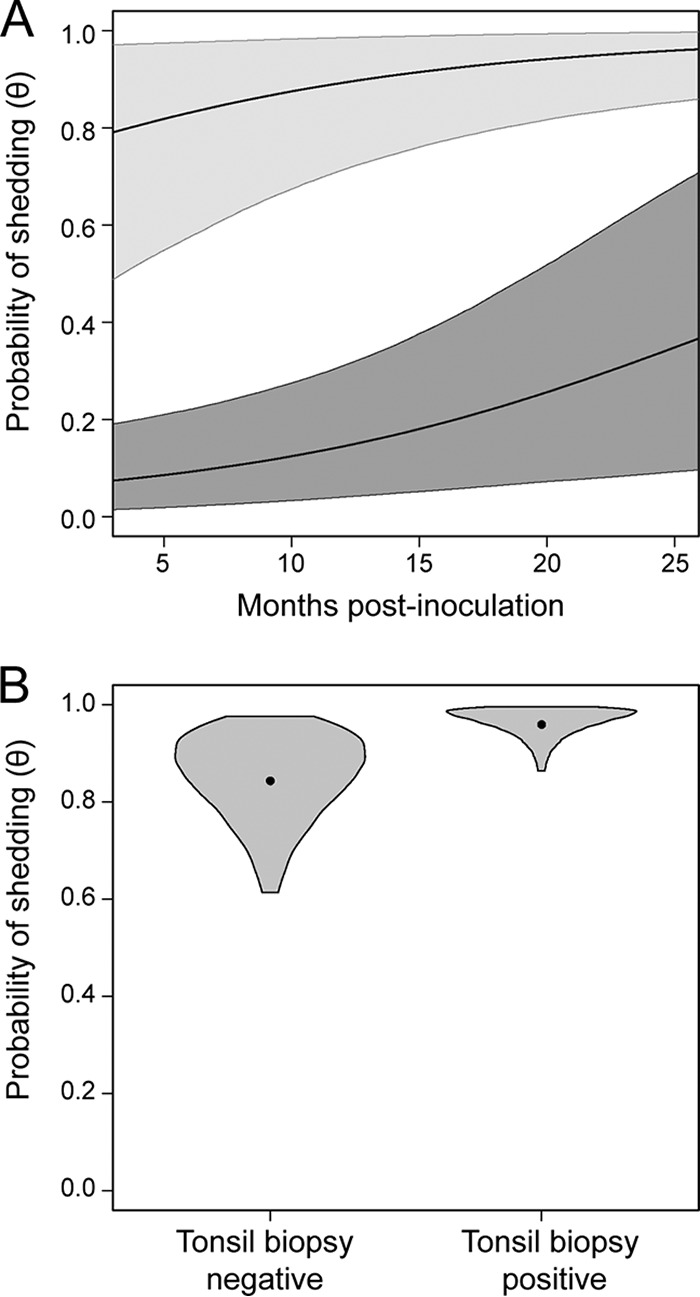
Probability of prion shedding in saliva increases with time. (A) The mean probability of prion shedding in saliva for a male GG deer inoculated with saliva via the oral route is indicated by the higher black line, and the light gray shaded region represents the 95% credible interval. The mean probability of prion shedding in saliva for a male GG deer inoculated with blood via the IV route is indicated by the lower black line, and the dark gray shaded region represents the 95% credible interval. The x axis indicates months postinoculation, from the date of the last inoculation or the date the contact deer were initially exposed to prion-infected deer. (B) We compared the probabilities of shedding in tonsil biopsy specimen-positive and tonsil biopsy specimen-negative samples. The values are derived from male GG deer inoculated with saliva via the oral route. Violin plots portray our knowledge of the effect of tonsil biopsy specimen status on prion shedding. The widths of the violins are proportional to the probabilities of a particular value of shedding probability being the true value, and the heights of the violins span the 95% credible intervals for the probability of shedding (i.e., there is a 95% probability that the true values of prion shedding are encompassed by the violins). The dots represents the “posterior” means and are a point estimate for shedding probability in each case.
We also found a positive relationship between prion shedding and tonsil biopsy specimen status. The probability of prion shedding in saliva for a tonsil biopsy specimen-positive (male, GG genotype, saliva-inoculated) deer was 0.96 (95% CI, 0.86 to 1.00) and the probability of prion shedding for a tonsil biopsy specimen-negative (male, GG, saliva-inoculated) deer was 0.84 (95% CI, 0.61 to 0.98) (Fig. 4B). These data imply that a positive tonsil biopsy specimen result is very suggestive that the deer is shedding prions but that a negative biopsy specimen result does not preclude shedding.
Neither genotype at codon 96 nor sex is associated with differences in prion shedding in saliva.
Contrary to our predictions, we found no difference in the probabilities of shedding in saliva for 96GG and 96GS deer (Fig. 5A). Similarly, we observed no difference in shedding between male and female deer (Fig. 5B).
FIG 5.
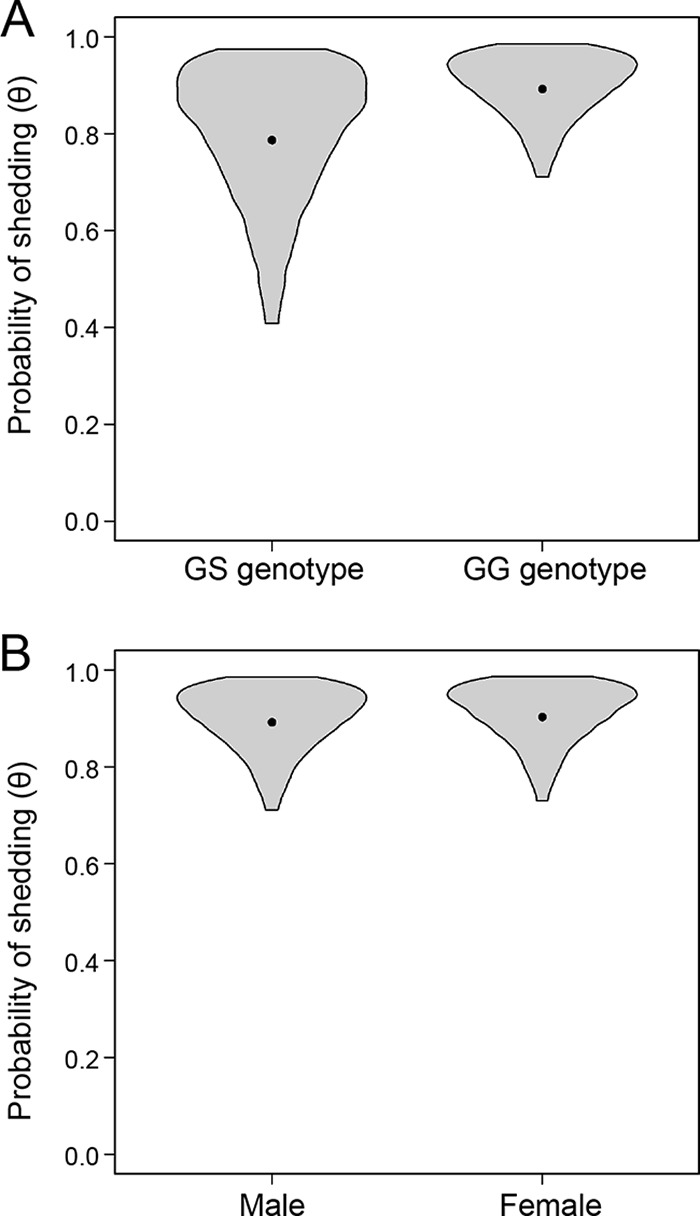
Probability of shedding is not associated with the genotype at codon 96 or with sex. (A) We compared the probabilities of prion shedding in saliva from deer homozygous for glycine at codon 96 (GG, the more susceptible genotype) and deer heterozygous at codon 96 (GS, the more resistant genotype). The values are derived from male deer inoculated with saliva via the oral route. (B) We compared the probabilities of prion shedding in saliva between male and female deer. The values are derived from GG deer inoculated with saliva via the oral route. Violin plots portray our knowledge of the effect of sex or genotype on prion shedding. The widths of the violins are proportional to the probabilities of a particular value of shedding probability being the true value, and the heights of the violins span the 95% credible intervals for the probability of shedding (i.e., there is a 95% probability that the true values of prion shedding are encompassed by the violins). The dots represents the posterior means and are a point estimate for shedding probability in each case.
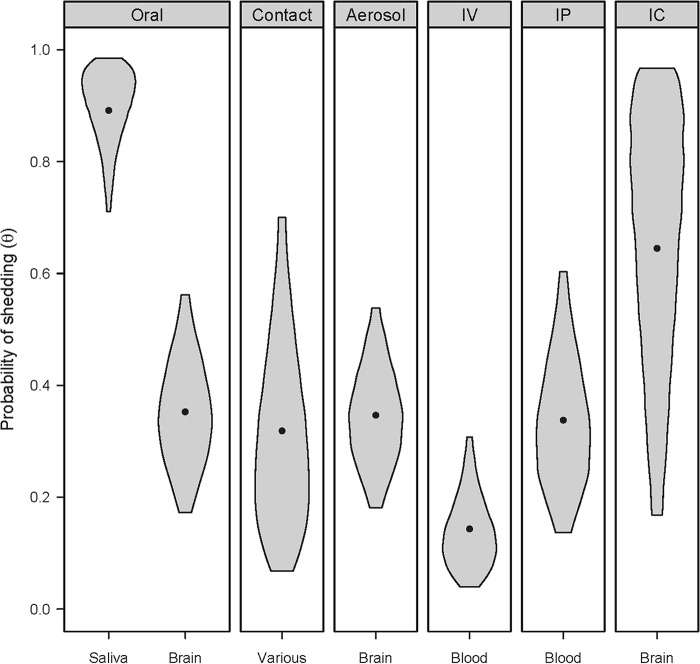
Deer inoculated with saliva (via the oral route) were more likely to shed prions in their saliva than deer inoculated via other route/inocula combinations.
Inoculation with saliva resulted in a much higher probability of shedding (θ = 0.89 [95% CI, 0.71 to 0.99]) than inoculation with brain via the same (oral) route (θ = 0.35 [95% CI, 0.17 to 0.56]) (Fig. 6). A variety of doses were used in these studies, and some of the doses of brain material were very high (up to 10 g). Despite the very high doses of CWD+ brain, brain-inoculated deer were still less likely to shed prions in their saliva than deer exposed to modest prion titers in saliva (see Materials and Methods) (13).
FIG 6.
Deer inoculated with CWD+ saliva via the oral route were more likely to shed prions in their saliva than those with other inoculum/route combinations. The probabilities of shedding for GG male deer in each source/route group were compared. Violin plots portray our knowledge of the effect of inoculum source/route on prion shedding. The widths of the violins are proportional to the probabilities of a particular value of shedding probability being the true value, and the heights of the violins span the 95% credible intervals for the probability of shedding (i.e., there is a 95% probability that the true values of prion shedding are encompassed by the violins). The dots represents the posterior means and are a point estimate for shedding probability in each case.
DISCUSSION
The occupancy modeling approach we describe here separates detection and shedding, processes that are otherwise confounded in diagnostic approaches with imperfect sensitivity and specificity. Moreover, this approach enabled us to test the importance of a number of covariates that may impact shedding or detection, including the sample concentration method, disease progression, genetic susceptibility, sex, inoculum type, and route of inoculation. Despite the use of one of the most sensitive assays in the prion field (RT-QuIC), such a modeling approach is necessary to separate variation in detection from variation in shedding and to obtain accurate estimates of prion shedding. It is also an important step toward understanding chronic wasting disease transmission and improving antemortem diagnosis.
Saliva likely plays an important role in the horizontal transmission of CWD, and its accessibility makes it an appealing diagnostic specimen. However, the assay sensitivity, even with the current optimizations of RT-QuIC, is too low to permit naive interpretation. We confirmed that the sensitivity of RT-QuIC for the detection of prions in saliva was low and increased with storage time regardless of the concentration technique used. Saliva contains an inhibitor for RT-QuIC that requires some samples to be diluted substantially before detection is possible (K. A. Davenport, unpublished data), and we hypothesize that the inhibitor is freeze-denatured during long periods of storage at −80°C (Fig. 3). Until the inhibitor can be identified and removed, the testing of saliva with RT-QuIC will have imperfect sensitivity. On the other hand, PTA improves specificity, but it is likely that some components remain in saliva that induce spontaneous misfolding of the recombinant PrPC (43). Like other diagnostic techniques, RT-QuIC would benefit from improvements in sensitivity that do not sacrifice specificity. Regardless of assay optimization and replicate size, all sampling methods are imperfect, and analysis with this occupancy modeling approach can account for imperfect sensitivity and specificity to generate unbiased estimates of shedding (and prevalence in other studies).
We predicted that the probability of prion shedding would increase with time, as measured either by months postinoculation or by tonsil biopsy. Indeed, there was a positive relationship between both metrics and prion shedding in saliva. Deer shed prions throughout much of the course of the disease, particularly in the preclinical phase. For example, the mean value for the onset of clinical signs was 19.5 months postinoculation and the probability of shedding was high well before that time.
Homozygotes for glycine at amino acid 96 (96GG) and male deer have increased likelihoods of CWD infection in natural populations (1, 40); however, the effects of genotype and sex on prion shedding were not evaluated before our study. We observed no difference in salivary prion shedding between female and castrated male deer, which were equally represented within experiments. This result can be interpreted in two ways. First, it is possible that the increased CWD prevalence in male deer is due to behaviors that increase the individual's susceptibility to infection, which are irrelevant in our experimental design. Second, it is possible (though seemingly unlikely) that intact male deer have a biological difference in disease progression compared to that in castrated male deer.
Similarly, 96GG deer have a higher CWD prevalence and faster disease progression but have the same probability of shedding prions in their saliva as do 96GS deer. The 96GG polymorphism likely increases the propensity of PrPC to misfold to PrPSc (N. J. Haley, R. Rielinger, K. A. Davenport, K. O'Rourke, G. Mitchell, and J. A. Richt, in press), and prions spread through the bodies of 96GG deer faster than through 96GS deer (20), but our data suggest that prions are not shed in saliva more frequently by 96GG deer. Although 96GS deer are more resistant to CWD (44), our data suggest that a CWD+ 96GS deer is just as infectious to its herd mates as is a CWD+ 96GG deer. Since 96GS deer typically have a longer incubation period, we hypothesize that 96GS deer shed more prions in saliva in a lifetime than 96GG deer, which contributes to the contamination of the environment.
CWD spreads readily among natural populations, but experimental inoculations have not been able to recapitulate horizontal transmission with appropriate inoculum or with realistic doses. Natural exposure probably involves exposure to prions from excreta (saliva, feces, and urine in the environment) via the oral route, while most experimental infections have been via a bolus of brain material as the inoculum (28, 29, 44–46). The data presented here compare the effects of the inoculum type and the route of inoculation on shedding. We hypothesized that prions in excreta may be adapted in some way to horizontal transmission. The dramatic difference in the probabilities of shedding we observed between deer inoculated with saliva versus brain (both via the oral route) supports our hypothesis. Deer inoculated with saliva are 2.77 times more likely (95% CI, 1.55 to 5.15) to shed prions in their saliva than deer inoculated with nonexcreta. Perhaps CWD spreads so easily among natural populations because the shed pathogen is adapted for transmission.
Besides the source of prions, the other difference between natural exposure and experimental inoculations is the dose. We exposed our deer to substantial doses of saliva (though the total dose was still far below that contained in the brain inoculum) and were able to compare the effects of large doses of saliva versus large doses of brain. We were unable to directly compare the difference between brain and saliva at very low doses. The deer exposed to CWD by contact were likely exposed to very low doses of excreta, but we have no group exposed to very low doses of brain to compare. Therefore, it is difficult to assess the low probability of shedding among the contact-exposed deer. Similarly, we were unable to test the effect of multiple inoculations versus a single inoculation in this model; however, the saliva group received multiple doses, whereas brain homogenate was delivered as a single dose. We are currently examining whether multiple doses may increase the propensity for infection (47, 48).
Though our study included saliva samples collected from a large number of deer, we faced several challenges. First, the deer in our study were inoculated, tested, and euthanized over a 15-year period. The studies were designed to answer a number of research questions but were not designed to compare the shedding or detection of prions in saliva or the covariates that affect either. Therefore, we do not have deer that represent every combination of route of inoculation and inoculum type, nor are the numbers of deer in each group equal. Our detection methods are also not evenly distributed over the range of sample storage durations (Fig. 3). Nonetheless, our sample sizes were sufficient to obtain reasonable estimates of shedding probability, and we were able to draw interesting conclusions from the combinations of route and inoculum available. We collected and stored all saliva samples consistently, but there seems to be an effect of storage time on detection probability (Fig. 3). False-negative (from inhibition or degraded sample quality) and false-positive errors increase the uncertainty in the estimates and inferences we make. However, the modeling framework presented here accounts for both types of observation errors to provide unbiased conclusions.
We developed an innovative approach to a stubborn diagnostic challenge, i.e., the differentiation of truly negative samples (due to a lack of shedding, in this case) from test-negative true positives, even in the face of imperfect specificity. The use of a negative-control data set to inform the probability of false positives is broadly applicable to diagnosticians, who rely on negative controls to monitor assay performance. This approach can be applied in any system with imperfect pathogen detection. In the case of samples from wild cervids (or for any other naturally occurring disease), our model can be applied as long as multiple replicates of a diagnostic test are performed and a negative control is present. We assessed the effects of the inoculum and inoculation route (because we had that information and are interested in CWD pathogenesis), although diagnosticians could ask questions about detection, shedding, or prevalence as a function of other covariates of interest (e.g., detection methods, tissue types, study areas, etc.). An understanding of the occurrence of false-negative and false-positive detection errors is critical for understanding pathogen prevalence and shedding and underlies sound scientific understanding of the transmission process and disease course. We outline a framework that explicitly accounts for these detection errors and generate unbiased inferences on shedding and prevalence that will be relevant to laboratory or field samples alike. Finally, our results are pertinent to understanding CWD pathogenesis and for controlling the spread of CWD in the environment.
Supplementary Material
ACKNOWLEDGMENTS
We thank Byron Caughey for the Syrian Hamster PRNP plasmid and guidance in the establishment of RT-QuIC in our laboratory. We thank Nikki Buhrdorf for compiling many of the samples and assisting in the performance of many of the RT-QuIC assays. We further thank Larissa Bailey and Thierry Chambert for their helpful discussions about model design and implementation and four anonymous reviewers for comments that strengthened the manuscript.
This work was supported by NIH F30OD021442-02, NIH N01AI25491, NIH R01NS047433, and NIH R01NS061902.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01243-17.
REFERENCES
- 1.Haley NJ, Hoover EA. 2015. Chronic wasting disease of cervids: current knowledge and future perspectives. Annu Rev Anim Biosci 3:305–325. doi: 10.1146/annurev-animal-022114-111001. [DOI] [PubMed] [Google Scholar]
- 2.Larska M, Polak MP, Zmudzinski JF, Torres JM. 2010. Comparison of mRNA expression levels of selected genes in the brain stem of cattle naturally infected with classical and atypical BSE. Brain Res 1351:13–22. doi: 10.1016/j.brainres.2010.07.035. [DOI] [PubMed] [Google Scholar]
- 3.Llorens F, Ansoleaga B, Garcia-Esparcia P, Zafar S, Grau-Rivera O, Lopez-Gonzalez I, Blanco R, Carmona M, Yague J, Nos C, Del Rio JA, Gelpi E, Zerr I, Ferrer I. 2013. PrP mRNA and protein expression in brain and PrP(c) in CSF in Creutzfeldt-Jakob disease MM1 and VV2. Prion 7:383–393. doi: 10.4161/pri.26416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang S-C, Li R, Wang C, Pan T, Liu T, Rubenstein R, Barnard G, Wong B-S, Sy M-S. 2003. Guanidine hydrochloride extraction and detection of prion proteins in mouse and hamster prion diseases by ELISA. J Pathol 199:534–541. doi: 10.1002/path.1294. [DOI] [PubMed] [Google Scholar]
- 5.Puoti G, Bizzi A, Forloni G, Safar JG, Tagliavini F, Gambetti P. 2012. Sporadic human prion diseases: molecular insights and diagnosis. Lancet Neurol 11:618–628. doi: 10.1016/S1474-4422(12)70063-7. [DOI] [PubMed] [Google Scholar]
- 6.Haley NJ, Carver S, Hoon-Hanks L, Henderson DM, Davenport KA, Bunting E, Gray S, Trindle B, Galeota J, LeVan I, Dubovos T, Shelton P, Hoover EA. 2014. Chronic wasting disease detection in the lymph nodes of free-ranging cervids by real time quaking-induced conversion. J Clin Microbiol 52:3237–3243. doi: 10.1128/jcm.01258-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orrú CD, Bongianni M, Tonoli G, Ferrari S, Hughson AG, Groveman BR, Fiorini M, Pocchiari M, Monaco S, Caughey B, Zanusso G. 2014. A test for Creutzfeldt–Jakob disease using nasal brushings. N Engl J Med 371:519–529. doi: 10.1056/NEJMoa1315200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orrú CD, Groveman BR, Hughson AG, Zanusso G, Coulthart MB, Caughey B. 2015. Rapid and sensitive RT-QuIC detection of human Creutzfeldt-Jakob disease using cerebrospinal fluid. mBio 6:e02451-14. doi: 10.1128/mBio.02451-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henderson DM, Davenport KA, Haley NJ, Denkers ND, Mathiason CK, Hoover EA. 2014. Quantitative assessment of prion infectivity in tissues and body fluids by RT-QuIC. J Gen Virol 96(Pt 1):210–219. doi: 10.1099/vir.0.069906-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson DM, Denkers ND, Hoover CE, Garbino N, Mathiason CK, Hoover EA. 2015. Longitudinal detection of prion shedding in saliva and urine by chronic wasting disease-infected deer by real-time quaking-induced conversion. J Virol 89:9338–9347. doi: 10.1128/jvi.01118-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henderson DM, Manca M, Haley NJ, Denkers ND, Nalls AV, Mathiason CK, Caughey B, Hoover EA. 2013. Rapid antemortem detection of CWD prions in deer saliva. PLoS One 8:e74377. doi: 10.1371/journal.pone.0074377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng YC, Hannaoui S, John TR, Dudas S, Czub S, Gilch S. 2016. Early and non-invasive detection of chronic wasting disease prions in elk feces by real-time quaking induced conversion. PLoS One 11:e0166187. doi: 10.1371/journal.pone.0166187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathiason CK, Powers JG, Dahmes SJ, Osborn DA, Miller KV, Warren RJ, Mason GL, Hays SA, Hayes-Klug J, Seelig DM, Wild MA, Wolfe LL, Spraker TR, Miller MW, Sigurdson CJ, Telling GC, Hoover EA. 2006. Infectious prions in the saliva and blood of deer with chronic wasting disease. Science 314:133–136. doi: 10.1126/science.1132661. [DOI] [PubMed] [Google Scholar]
- 14.Haley NJ, Mathiason CK, Zabel MD, Telling GC, Hoover EA. 2009. Detection of sub-clinical CWD infection in conventional test-negative deer long after oral exposure to urine and feces from CWD+ deer. PLoS One 4:e7990. doi: 10.1371/journal.pone.0007990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haley NJ, Seelig DM, Zabel MD, Telling GC, Hoover EA. 2009. Detection of CWD prions in urine and saliva of deer by transgenic mouse bioassay. PLoS One 4:e4848. doi: 10.1371/journal.pone.0004848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller MW, Williams ES. 2003. Prion disease: horizontal prion transmission in mule deer. Nature 425:35–36. doi: 10.1038/425035a. [DOI] [PubMed] [Google Scholar]
- 17.Miller MW, Williams ES, McCarty CW, Spraker TR, Kreeger TJ, Larsen CT, Thorne ET. 2000. Epizootiology of chronic wasting disease in free-ranging cervids in Colorado and Wyoming. J Wildl Dis 36:676–690. doi: 10.7589/0090-3558-36.4.676. [DOI] [PubMed] [Google Scholar]
- 18.Selariu A, Powers JG, Nalls A, Brandhuber M, Mayfield A, Fullaway S, Wyckoff CA, Goldmann W, Zabel MM, Wild MA, Hoover EA, Mathiason CK. 2015. In utero transmission and tissue distribution of chronic wasting disease-associated prions in free-ranging Rocky Mountain elk. J Gen Virol 96:3444–3455. doi: 10.1099/jgv.0.000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haley NJ, Mathiason CK, Carver S, Zabel M, Telling GC, Hoover EA. 2011. Detection of chronic wasting disease prions in salivary, urinary, and intestinal tissues of deer: potential mechanisms of prion shedding and transmission. J Virol 85:6309–6318. doi: 10.1128/JVI.00425-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoover CE, Davenport KA, Henderson DM, Denkers ND, Mathiason CK, Soto C, Zabel MD, Hoover EA. 2017. Pathways of prion spread during early chronic wasting disease in deer. J Virol 91:e00077-17. doi: 10.1128/JVI.00077-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiappin S, Antonelli G, Gatti R, De Palo EF. 2007. Saliva specimen: a new laboratory tool for diagnostic and basic investigation. Clin Chim Acta 383:30–40. doi: 10.1016/j.cca.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 22.MacKenzie DI, Nichols JD, Lachman GB, Droege S, Royle JA, Langtimm CA. 2002. Estimating site occupancy rates when detection probabilities are less than one. Ecology 83:2248–2255. doi: 10.1890/0012-9658(2002)083[2248:ESORWD]2.0.CO;2. [DOI] [Google Scholar]
- 23.MacKenzie DI, Nichols JD, Royle JA, Pollock KH, Bailey LL, Hines JE. 2006. Occupancy estimation and modeling. Inferring patterns and dynamics of species occurrence. Academic Press, Cambridge, MA. [Google Scholar]
- 24.Lachish S, Gopalaswamy AM, Knowles SCL, Sheldon BC. 2012. Site-occupancy modelling as a novel framework for assessing test sensitivity and estimating wildlife disease prevalence from imperfect diagnostic tests. Methods Ecol Evol 3:339–348. doi: 10.1111/j.2041-210X.2011.00156.x. [DOI] [Google Scholar]
- 25.Denkers ND, Telling GC, Hoover EA. 2011. Minor oral lesions facilitate transmission of chronic wasting disease. J Virol 85:1396–1399. doi: 10.1128/JVI.01655-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathiason CK, Hays SA, Powers J, Hayes-Klug J, Langenberg J, Dahmes SJ, Osborn DA, Miller KV, Warren RJ, Mason GL, Hoover EA. 2009. Infectious prions in pre-clinical deer and transmission of chronic wasting disease solely by environmental exposure. PLoS One 4:e5916. doi: 10.1371/journal.pone.0005916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathiason CK, Hayes-Klug J, Hays SA, Powers J, Osborn DA, Dahmes SJ, Miller KV, Warren RJ, Mason GL, Telling GC, Young AJ, Hoover EA. 2010. B cells and platelets harbor prion infectivity in the blood of deer infected with chronic wasting disease. J Virol 84:5097–5107. doi: 10.1128/JVI.02169-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goñi F, Mathiason CK, Yim L, Wong K, Hayes-Klug J, Nalls A, Peyser D, Estevez V, Denkers N, Xu J, Osborn DA, Miller KV, Warren RJ, Brown DR, Chabalgoity JA, Hoover EA, Wisniewski T. 2015. Mucosal immunization with an attenuated Salmonella vaccine partially protects white-tailed deer from chronic wasting disease. Vaccine 33:726–733. doi: 10.1016/j.vaccine.2014.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denkers ND, Hayes-Klug J, Anderson KR, Seelig DM, Haley NJ, Dahmes SJ, Osborn DA, Miller KV, Warren RJ, Mathiason CK, Hoover EA. 2013. Aerosol transmission of chronic wasting disease in white-tailed deer. J Virol 87:1890–1892. doi: 10.1128/JVI.02852-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wild MA, Spraker TR, Sigurdson CJ, O'Rourke KI, Miller MW. 2002. Preclinical diagnosis of chronic wasting disease in captive mule deer (Odocoileus hemionus) and white-tailed deer (Odocoileus virginianus) using tonsillar biopsy. J Gen Virol 83:2629–2634. doi: 10.1099/0022-1317-83-10-2629. [DOI] [PubMed] [Google Scholar]
- 31.Denkers ND, Seelig DM, Telling GC, Hoover EA. 2010. Aerosol and nasal transmission of chronic wasting disease in cervidized mice. J Gen Virol 91:1651–1658. doi: 10.1099/vir.0.017335-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spraker TR, Miller MW, Williams ES, Getzy DM, Adrian WJ, Schoonveld GG, Spowart RA, O'Rourke KI, Miller JM, Merz PA. 1997. Spongiform encephalopathy in free-ranging mule deer (Odocoileus hemionus), white-tailed deer (Odocoileus virginianus) and Rocky Mountain elk (Cervus elaphus nelsoni) in northcentral Colorado. J Wildl Dis 33:1–6. doi: 10.7589/0090-3558-33.1.1. [DOI] [PubMed] [Google Scholar]
- 33.Seelig DM, Mason GL, Telling GC, Hoover EA. 2011. Chronic wasting disease prion trafficking via the autonomic nervous system. Am J Pathol 179:1319–1328. doi: 10.1016/j.ajpath.2011.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davenport KA, Henderson DM, Bian J, Telling GC, Mathiason CK, Hoover EA. 2015. Insights into chronic wasting disease and bovine spongiform encephalopathy species barriers by use of real-time conversion. J Virol 89:9524–9531. doi: 10.1128/jvi.01439-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denkers ND, Henderson DM, Mathiason CK, Hoover EA. 2016. Enhanced prion detection in biological samples by magnetic particle extraction and real-time quaking-induced conversion. J Gen Virol 97:2023–2029. doi: 10.1099/jgv.0.000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nichols JD, Bailey LL, O'Connell AF Jr, Talancy NW, Campbell Grant EH, Gilbert AT, Annand EM, Husband TP, Hines JE. 2008. Multi-scale occupancy estimation and modelling using multiple detection methods. J Appl Ecol 45:1321–1329. doi: 10.1111/j.1365-2664.2008.01509.x. [DOI] [Google Scholar]
- 37.Schmidt BR, Kéry M, Ursenbacher S, Hyman OJ, Collins JP. 2013. Site occupancy models in the analysis of environmental DNA presence/absence surveys: a case study of an emerging amphibian pathogen. Methods Ecol Evol 4:646–653. doi: 10.1111/2041-210X.12052. [DOI] [Google Scholar]
- 38.Eads DA, Biggins DE, Doherty PF Jr, Gage KL, Huyvaert KP, Long DH, Antolin MF. 2013. Using occupancy models to investigate the prevalence of ectoparasitic vectors on hosts: an example with fleas on prairie dogs. Int J Parasitol Parasites Wildl 2:246–256. doi: 10.1016/j.ijppaw.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elmore SA, Huyvaert KP, Bailey LL, Iqbal A, Su C, Dixon BR, Alisauskas RT, Gajadhar AA, Jenkins EJ. 2016. Multi-scale occupancy approach to estimate Toxoplasma gondii prevalence and detection probability in tissues: an application and guide for field sampling. Int J Parasitol 46:563–570. doi: 10.1016/j.ijpara.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 40.Farnsworth M, Wolfe LLL, Hobbs NT, Burnham KP, Williams ES, Theobald DM, Conner MM, Miller MW. 2005. Human land use influences chronic wasting disease prevalence in mule deer. Ecol Appl 15:119–126. doi: 10.1890/04-0194. [DOI] [Google Scholar]
- 41.Garlick MJ, Powell JA, Hooten MB, MacFarlane LR. 2014. Homogenization, sex, and differential motility predict spread of chronic wasting disease in mule deer in southern Utah. J Math Biol 69:369–399. doi: 10.1007/s00285-013-0709-z. [DOI] [PubMed] [Google Scholar]
- 42.Davenport KA, Hoover C, Bian J, Telling GC, Mathiason CK, Hoover EA. 2017. PrPC expression and prion seeding activity in the alimentary tract and lymphoid tissue of deer. PLoS One 12:e0183927. doi: 10.1371/journal.pone.0183927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henderson DM, Tennant JM, Haley NJ, Denkers ND, Mathiason CK, Hoover EA. 2017. Detection of chronic wasting disease prion seeding activity in deer and elk feces by real-time quaking-induced conversion. J Gen Virol 98:1953–1962. doi: 10.1099/jgv.0.000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller MW, Wolfe LL, Sirochman TM, Sirochman MA, Jewell JE, Williams ES. 2012. Survival patterns in white-tailed and mule deer after oral inoculation with a standardized, conspecific prion dose. J Wildl Dis 48:526–529. doi: 10.7589/0090-3558-48.2.526. [DOI] [PubMed] [Google Scholar]
- 45.Sigurdson CJ, Spraker TR, Miller MW, Oesch B, Hoover EA. 2001. PrPCWD in the myenteric plexus, vagosympathetic trunk and endocrine glands of deer with chronic wasting disease. J Gen Virol 82:2327–2334. doi: 10.1099/0022-1317-82-10-2327. [DOI] [PubMed] [Google Scholar]
- 46.Sigurdson CJ, Williams ES, Miller MW, Spraker TR, O'Rourke KI, Hoover EA. 1999. Oral transmission and early lymphoid tropism of chronic wasting disease PrPres in mule deer fawns (Odocoileus hemionus). J Gen Virol 80:2757–2764. doi: 10.1099/0022-1317-80-10-2757. [DOI] [PubMed] [Google Scholar]
- 47.Diringer H, Roehmel J, Beekes M. 1998. Effect of repeated oral infection of hamsters with scrapie. J Gen Virol 79:609–612. [DOI] [PubMed] [Google Scholar]
- 48.Gravenor MB, Stallard N, Curnow R, McLean AR. 2003. Repeated challenge with prion disease: the risk of infection and impact on incubation period. Proc Natl Acad Sci U S A 100:10960–10965. doi: 10.1073/pnas.1833677100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.