SUMMARY
Many neurodegenerative proteinopathies share a common pathogenic mechanism: the abnormal accumulation of disease-related proteins. As growing evidence indicates that reducing the steady-state levels of disease-causing proteins mitigates neurodegeneration in animal models, we developed a strategy to screen for genes that decrease the levels of tau, whose accumulation contributes to the pathology of both Alzheimer’s disease (AD) and progressive supranuclear palsy (PSP). Integrating parallel cell-based and Drosophila genetic screens, we discovered that tau levels are regulated by Nuak1, an AMPK-related kinase. Nuak1 stabilizes tau by phosphorylation specifically at Ser356. Inhibition of Nuak1 in fruit flies suppressed neurodegeneration in tau-expressing Drosophila, and Nuak1 haploinsufficiency rescued the phenotypes of a tauopathy mouse model. These results demonstrate that decreasing total tau levels is a valid strategy for mitigating tau-related neurodegeneration and reveal Nuak1 to be a novel therapeutic entry point for tauopathies.
INTRODUCTION
In many neurodegenerative conditions, normally soluble proteins accumulate and form filamentous insoluble aggregates over time (Chiti and Dobson, 2006). Alzheimer disease (AD), the most prevalent neurodegenerative disease, is characterized by extracellular deposits of amyloid beta (Aβ) plaques and the intracellular formation of neurofibrillary tangles (NFTs) made from hyperphosphorylated tau (Hardy and Selkoe, 2002). Tau undergoes many posttranslational modifications, predisposing it to self-assembly and accumulation in NFTs in AD and also in several other neurodegenerative diseases, including progressive supranuclear palsy (PSP) and frontotemporal dementia. Mutations in the gene which encodes for tau, microtubule-associated protein tau (MAPT), cause familial frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17), directly implicating tau dysfunction in neurodegenerative processes (Clark et al., 1998; Hutton et al., 1998; Pittman et al., 2006). Furthermore, a copy number variation (CNV) consisting of a complete duplication of the MAPT locus has been described in a patient with frontotemporal dementia (Rovelet-Lecrux and Campion, 2012). These observations suggest that abnormal forms of tau or even elevated levels of wild-type tau are sufficient to cause neuropathology.
Although the molecular mechanism by which tau accumulation causes neuronal dysfunction and death is not fully understood, useful clues can be gleaned from studies of other neurodegenerative proteinopathies such as amyotrophic lateral sclerosis, Huntington’s disease and the spinocerebellar ataxias, all of which share the formation of globular insoluble aggregates of mutant or wild-type protein (SOD1, HTT, and ataxins, respectively). In each case, the data clearly indicate that decreasing the accumulation of these proteins, usually by genetic manipulation, can reverse pathology in animal models for the disease (Harper et al., 2005; Miller et al., 2005; Williams and Paulson, 2008; Xia et al., 2004; Zu et al., 2004). For instance, studies of conditional tauopathy mouse models exhibit accumulation of NFTs in neurons associated with neurodegeneration, and have shown that suppressing expression of the conditional mutant tau gene improves memory and halts neuronal loss (Santacruz et al., 2005). Moreover, partial reduction of tau during early development in mice is well tolerated, increases resistance to chemically induced seizures, and markedly diminishes Aβ-driven neuronal and cognitive impairment in vivo (Morris et al., 2011; Roberson et al., 2007; Small and Duff, 2008). Therefore, we hypothesized that decreasing tau levels might be a feasible therapeutic approach for tauopathies, as we have pursued for other proteinopathies (Park et al., 2013). Consequently, we set out to identify druggable targets that modulate tau levels.
To take advantage of the unbiased, functional nature of genetic screens (Westbrook et al., 2008) and compensate for the weaknesses inherent in any given model system, we utilized a genetic approach that we previously developed to successfully modulate the levels of glutamine-expanded ATXN1, which causes spinocerebellar ataxia type 1 (SCA1) (Park et al., 2013). We used a cross-species strategy: complementary forward genetic screens that targeted the levels of human tau in a tauopathy Drosophila model and in a human cell line that stably expresses tau. Because phosphorylation is important for tau accumulation and aggregation (Wang and Mandelkow, 2016), and because kinases are readily targeted with pharmacological agents (Noble et al., 2004), we chose to screen the human kinome. Our screen identified a previously unknown kinase that regulates tau levels by phosphorylating a site not previously known to affect protein levels. Additional studies in a mouse model demonstrate that reducing tau levels safely reverses tau-induced phenotypes and pathology.
RESULTS
Nuak1 influences steady-state levels of tau protein
To identify regulators of tau levels in human cells, we engineered a human-medulloblastoma-derived cell line with a transgene encoding human tau fused with monomeric green fluorescent protein (tau-GFP). To distinguish modifiers that regulate tau protein levels from those that regulate transgene transcription, we placed the gene encoding DsRed upstream of an internal ribosomal entry site (IRES) driving independent translation of tau-GFP fusion protein (DsRed-IRES-tau-GFP). The DsRed to GFP fluorescence ratio by flow cytometry analysis serves as a proxy for tau-GFP protein levels while controlling for fluctuations in transcription of the transgene (Figure S1A).
We used our cell system to test the effects of individual short interfering RNAs (siRNAs) targeting every known human kinase and kinase-like gene (1,908 siRNAs, 636 genes; test performed in triplicates) on tau levels (Figure S1B). In parallel, we performed a genetic screen in a tauopathy Drosophila model expressing four-repeat wild-type human tau, in which fruit flies develop an external eye (ommatidial) phenotype in response to tau toxicity (Figure S1C). We screened a total of 704 inducible RNAi alleles targeting the Drosophila kinome (337 genes) for those that would modulate degeneration induced by human wild-type tau, using the readily observable ommatidial phenotype as a reliable read-out.
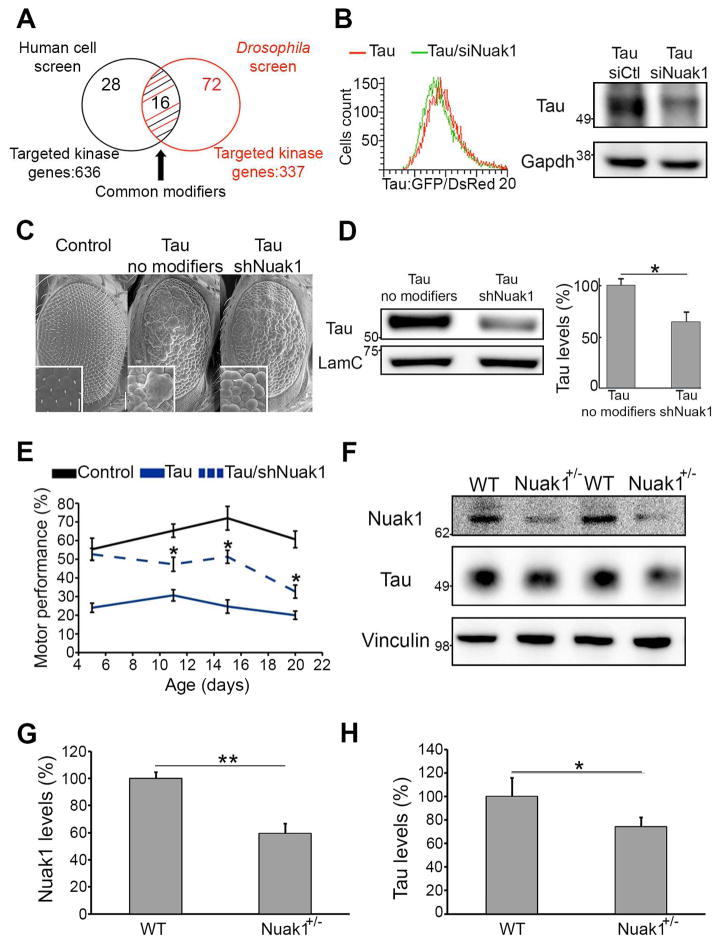
The cell-based screen revealed 44 human modifier genes that reduced tau levels and the Drosophila screen uncovered 88 genes that ameliorated tau-induced degeneration. We identified 16 modifiers common to both screens (Figure 1A and Figure S1D) and subjected them to a number of additional criteria before pursuing further validation (Figure S1E). One hit that satisfied all our conditions was Nuak1: Nuak1 down-regulation decreases tau levels in the human cell line not only in the GFP/DsRed ratio by flow cytometry but also by western blot analysis (Figure 1B), and knocking down the Drosophila Nuak1 homolog also resulted in a decrease of tau levels consistent with the observed suppression of eye degeneration (Figure 1C and D, Figure S2A). These results indicate that partial inhibition of Nuak1 is sufficient to reduce aberrant tau levels.
Figure 1. Integrated genetic screens identified Nuak1 as a regulator of tau levels.
A) Venn diagram of the identified gene candidates. B) Histogram showing the distribution of Tau-GFP/DsRed ratio abundance in cells treated with siRNA for Nuak1 compared to control. Western blot showing a tau level decrease in Tau-GFP/DsRed cells upon treatment with siRNA for Nuak1. C) Scanning electron microscopy images of Drosophila eyes showing suppression of wild-type human tau-induced degeneration by reduced levels of Nuak1. D) Western blot showing a decrease in tau protein upon lowering the levels of Drosophila Nuak1. E) Decreased levels of Drosophila Nuak1 homolog suppressed motor impairment in tau animals. Error bars, s.d.m. *p<0.05. F–H) Western blot quantifications showing a tau level decrease in Nuak1+/− mouse brains in comparison with three-month-old WT littermates (n=5). Data are represented as mean ± s.d.m. *p<0.05, **p<0.01. Student’s T-test. See also Figures S1 and S2.
We therefore sought to validate the Nuak1 effect using a functional behavioral read-out in the nervous system. Neuronal-specific expression of human wild-type tau in the Drosophila impairs motor performance in a way that can be easily quantified using a climbing assay. We found that knockdown of the Drosophila homolog of Nuak1 suppressed these human tau-induced motor deficits (Figure 1E and Video S1). We next tested the effect of an increase in Nuak1 levels, and found Nuak1-overexpressing tau flies showed a substantial worsening of the motor deficit. No motor abnormalities were observed in wild-type flies when Nuak1 was over expressed, demonstrating that under these conditions Nuak1 does not induce toxicity by itself but only augments tau toxicity (Figure S2B).
We next tested the effect of Nuak1 deletion in mice. Since Nuak1+/− mice are viable and fertile (Figure S2C–S2D), we were able to analyze the brains of 3-month-old animals. Western blot analysis revealed that a 50% reduction in Nuak1 was sufficient to decrease tau levels in the brain (Figure 1F–H). In parallel we compared the levels of tau-encoding mRNA in the Nuak1+/− and wild-type mice. No differences between genotypes were observed, confirming that Nuak1 affects tau levels post-translationally (Figure S2E).
Nuak1 is associated with tau accumulation in human tauopathy post-mortem brain tissue
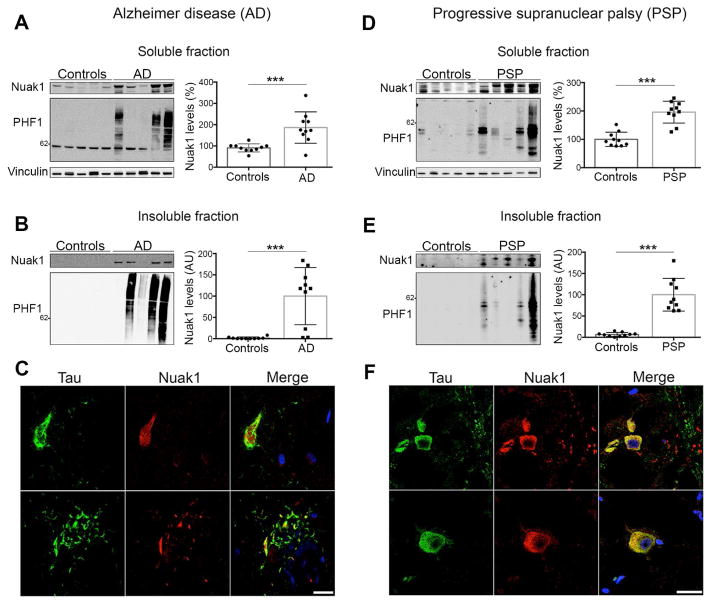
To determine if Nuak1 is associated with tau in the context of human tauopathies, we biochemically and pathologically characterized Nuak1 in post-mortem brains from Alzheimer cases and age-matched controls (Table S1). We analyzed the RIPA soluble fraction and observed an increase in the levels of Nuak1 in comparison with controls (Figure 2A). When we analyzed the insoluble fraction, which was enriched with tau fibrillar material, we detected the presence of Nuak1 only in the AD cases (Figure 2B). This result strongly suggests Nuak1 is associated with highly insoluble tau aggregates. To confirm this, we performed double immunofluorescence (IF) in AD brain sections and age-matched controls using an anti-tau antibody and an anti-Nuak1 antibody. Fluorescence images of AD brain sections show that Nuak1 co-localizes with tau NFTs (Figure 2C, top panel) as well as with tau neuropil threads (Figure 2C, bottom panel) while age-matched controls show that Nuak1 is normally located in the cytoplasm and within the nucleus (Figure S3A), as previously reported (Sun et al., 2013). This result supports the notion that Nuak1 is associated with pathological tau accumulation in Alzheimer disease.
Figure 2. Nuak1 levels are increased and associated with tau pathology in human AD and PSP.
A) Nuak1 levels are elevated in soluble fractions of AD cases in comparison with age-matched controls. B) Nuak1 is also elevated in AD insoluble fractions in relation with age-matched controls. C) Double staining in AD brain sections showed co-localization of tau (green) and Nuak1 (red) in NFTs (top panel) and Neuropil threads. D) Nuak1 levels are elevated in soluble fractions of PSP cases in comparison with age-matched controls. E) Nuak1 is also elevated in PSP insoluble fractions in comparison with age-matched controls. F) Double staining in PSP brain sections showed co-localization of tau (green) and Nuak1 (red) in globose NFTs. Scale bar, 20 μm. For all quantifications, n=10, mean ± s.d.m. ***p<0.001, **p<0.01. Student’s T-test. See also Figure S3.
To establish whether this association between Nuak1 and tau occurs only in the context of AD or also exists in other types of tauopathies, we performed similar evaluation in post-mortem brain tissue from PSP patients. Biochemical analysis of brain soluble fractions in PSP cases (Table S1) showed an increase in the levels of Nuak1 in comparison with age-matched controls (Figure 2D). When the insoluble fractions were analyzed, Nuak1 was detected only in PSP cases but not in controls (Figure 2E), suggesting that Nuak1 is also associated with tau aggregates in the context of PSP. Double IF on PSP brain sections demonstrated that Nuak1 co-localized with globose type-NFTs, which is a pathological hallmark of PSP (Figure 2F). Double IF on age-matched controls for this set of cases once again reveal that Nuak1 is located in the nucleus and the cytoplasm in normal physiological conditions (Figure S3B). Nuak1 is thus strongly associated with tau in a human disease context.
Finally, to determine if the increase of Nuak1 levels is due to an increase in the expression of Nuak1 versus the stability or activity of the protein, we measured the levels of Nuak1-encoding mRNA in AD cases, PSP cases and the respective age-matched controls. The qRT-PCR analysis shows a decrease in the levels of Nuak1-encoding mRNA in AD as opposed to age-matched controls and no differences between PSP cases and its age-matched controls (Figure S3C–S3D). These results indicate that the increase in Nuak1 protein levels is due to an increase in protein stability or activity rather than transcription. In addition, the decrease in Nuak1-encoding mRNA in AD cases suggests a compensatory mechanism at late stages of the disease. In parallel, we analyzed available RNA seq data from AD and PSP patients. RNA seq data generated by the Mayo Brain Gene Expression (MayoEGWAS and Mayo Pilot studies) (Zou et al., 2012) revealed that Nuak1-encoding mRNA does not statistically change in AD and PSP cases in comparison with control cases (Figure S3E), which supports the conclusion that Nuak1 levels are likely increased due to protein stability.
Nuak1 regulates tau levels by phosphorylating Ser356
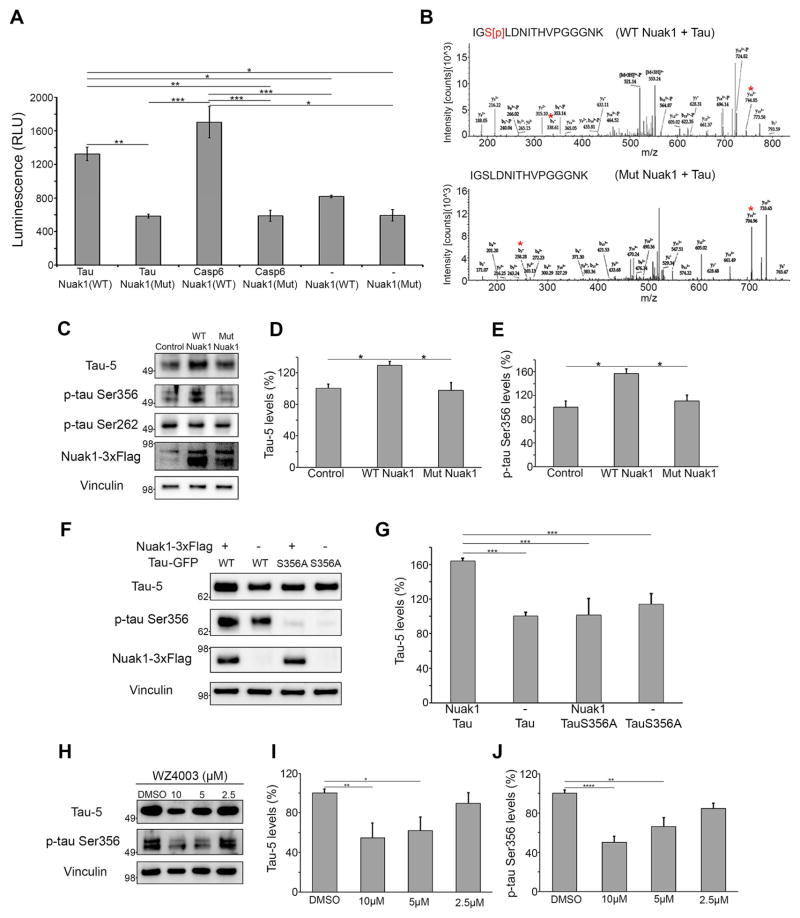
To determine whether Nuak1 directly regulates tau via phosphorylation, recombinant wild-type Nuak1 and tau were evaluated in an in vitro kinase assay. Notably, Nuak1 directly phosphorylates tau (Figure 3A), while mutant Nuak1 lacking kinase activity failed to phosphorylate tau (Figure 3A). As a positive control we performed the kinase assay with Nuak1 and Caspase-6, a known Nuak1 substrate (Suzuki et al., 2004). To identify the sites phosphorylated by Nuak1, we repeated the in vitro kinase assay followed by mass spectrometry. The LC-MS/MS data revealed Nuak1 directly phosphorylates tau at Serine356 (the amino acid number is based on the longest isoform (isoform 2, amino acids 1–441) (Figure 3B). Nuak1 did not phosphorylate tau at Thr212, Thr231 Ser214, Ser262 or Ser396, which are known sites for phosphorylation by other AMPK-related kinases (Mairet-Coello et al., 2013; Vingtdeux et al., 2011; Yoshida and Goedert, 2012) (data not shown).
Figure 3. Nuak1 phosphorylates tau at Serine356 and regulates tau levels.
A) In vitro kinase assay with purified recombinant tau, wild type active Nuak1, mutant inactive Nuak1 and Caspase-6, confirmed Nuak1 directly phosphorylated tau (n=6). B) Mass Spec analysis from in vitro kinase assays showed Nuak1 phosphorylated tau at Ser356. *Indicated the b and y ions for the modified as well as non-modified peptide, which confirms phosphorylation on Serine 356. C) Western blot analysis of lysates from neuroblastoma cells transfected with active Nuak1-3xflag (WT Nuak1) or mutant K84M kinase dead Nuak1-3xflag (Mut Nuak1) (n=9). D) Graph showing western blot quantification of total tau levels measured with tau-5 antibody. E) Graph showing western blot quantification of p-tau Ser356 levels. F) Western blot analysis of lysates from Neuroblastoma cells transfected with wild-type tau-GFP (WT) or mutant S356A tau (S356A) and with or without active Nuak1-3xflag. G) Graph showing western blot quantification of total tau levels measured with tau-5 antibody. H) Western blot analysis of lysates from neuroblastoma cells treated with 10, 5 and 2.5 μM of WZ4003 for 24 hours (n=6). I) Graph showing western blot quantification of total tau levels measured with tau-5 antibody. J) Graph showing western blot quantification of p-tau Ser356 levels. For all quantifications, *p<0.05, **p <0.01, ***p<0.001, ANOVA followed by Bonferroni’s post hoc test. See also Figures S4 and S5.
To determine whether Nuak1 phosphorylates tau in living cells, we transfected a neuroblastoma cell line with either wild-type Nuak1 or the kinase-dead Nuak1 mutant K84M (Suzuki et al., 2006). Nuak1 overexpression led to a 30% increase in tau levels (Figure 3C and D), and a >50% increase in the levels of tau phosphorylated at Ser356 (Figure 3C and E), but no changes in the levels of Ser262 phosphorylated tau (Figure 3C). Overexpression of the K84M mutant had no effect on tau levels (Figure 3C and D). Decreasing endogenous levels of Nuak1 from neuroblastoma cells induced a significant decrease in the levels of phospho-Ser356 tau (Figure S4A–S4D), confirming the physiological effect of Nuak1 on tau phosphorylation. Because tau phosphorylation at Ser262/Ser356 contributes to tau hyperphosphorylation at AD-related SP/ST sites (Ando et al., 2016; Nishimura et al., 2004), we also tested whether tau phosphorylation at Ser356 by Nuak1 initiates a cascade of hyperphosphorylation. Indeed, we found Nuak1 overexpression increases the levels of tau phosphorylated at Thr231 and Ser396/Ser404 (Figure S4E–S4G)
To confirm that the effect of Nuak1 on tau levels was dependent on Ser356 phosphorylation, we co-expressed Nuak1 with wild-type tau or mutant S356A tau. Western blot analysis showed Nuak1 has a strong effect on wild-type tau levels but no effect over the levels of mutant S356A tau (Figure 3F and G). To determine if Nuak1 alters tau levels through stabilization of tau by phosphorylation at Ser356, we generated doxycycline-inducible cell lines expressing wild type tau or mutant S356A tau. Our protein stability measurements demonstrated that wild type tau has a higher half-life than the mutant S356A tau. Furthermore, over-expression of Nuak1 increases wild type tau half-life but does not have any effect over mutant S356A tau stability (Figure S5A–S5B). It has been previously reported that the carboxy terminus of the Hsp70-interacting protein CHIP binds and ubiquitinates tau within the microtubule-binding domain, which promotes tau proteasomal degradation (Dickey et al., 2007; Petrucelli et al., 2004). Whenever tau is phosphorylated at Ser262/Ser356, however, CHIP is unable to bind or ubiquitinate tau, and tau is thereby hindered from undergoing proteasomal degradation (Dickey et al., 2007; Dickey et al., 2008). Therefore, we performed an immunoprecipitation assay to test if a similar mechanism occurs when tau is phosphorylated at Ser356 by Nuak1. Indeed, CHIP binding and ubiquitination of tau are interrupted by the effects of Nuak1 on tau (Figure S5C).
Having established the role of Nuak1 in phosphorylating tau at Ser356 using molecular and genetic approaches, we evaluated the effect of pharmacologic inhibition of Nuak1. Cells treated with the Nuak1 inhibitor WZ4003 (Banerjee et al., 2014) showed decreased levels of total and phospho Ser356 tau (Figure 3H–J), confirming that Nuak1’s effect on tau levels is conferred by its kinase activity. Together with the genetic data, these results strongly support the hypothesis that Nuak1 regulates tau levels. These findings also validate the genetic screen strategy and provide evidence that tau levels can be modulated pharmacologically.
Reducing Nuak1 rescues the phenotypes in a tauopathy mouse model
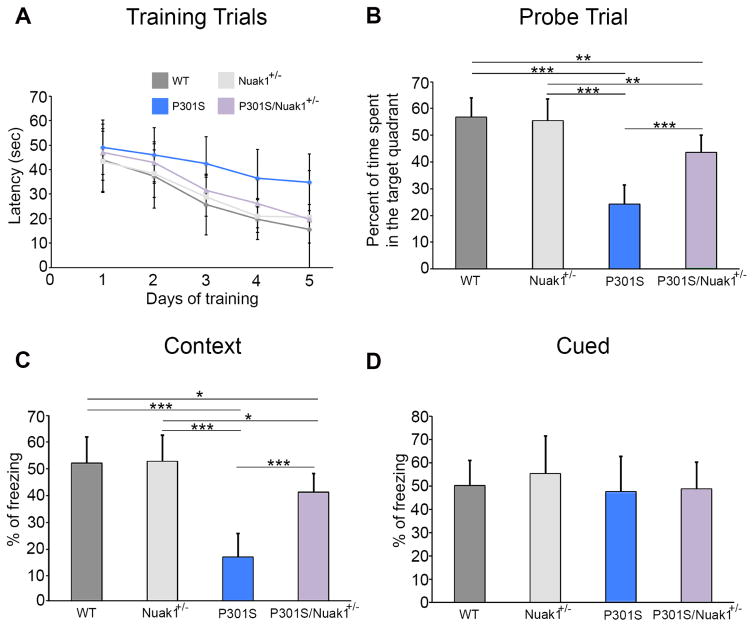
To determine if the decrease in tau levels observed in Nuak1+/− mice was functionally penetrant in a disease model, we bred Nuak1+/− mice to tau P301S transgenic mice, which bear human tau with the P301S mutation and recapitulate many features of human tauopathies (Yoshiyama et al., 2007). The offspring were aged and evaluated using comprehensive behavioral and pathological evaluation.
To test the effect of deleting one Nuak1 allele on memory, 7.5-month-old progeny were assayed in the Morris water maze. During the training phase, the latency to find the platform was somewhat increased in the P301S mice in comparison with wild-type, Nuak1+/−, and P301S/Nuak1+/− mice, although this difference was not statistically significant (Figure 4A). In the probe trial, a 50% reduction of Nuak1 in P301S mice improved memory retention, as shown by the higher percentage of time that the P301S/Nuak1+/− spent in the target quadrant in comparison with P301S mice (Figure 4B). Because all the mice performed equally well during the visible platform trial (Figure S6A), it was clear that all four genotypes have the ability to see and to swim efficiently. In addition, all four groups displayed comparable swim speeds (Figure S6B), so the decrease in time that the P301S mice spent in the target quadrant during the probe trial was due to their poorer ability to use and remember spatial cues.
Figure 4. Reduction of Nuak1 by 50% reverses memory deficits in tau P301 transgenic mice.
A) Morris water maze analysis of invisible platform training sessions expressed as the latency to find the platform. B) Probe trial for water morris maze. P301S mice showed less time in the target quadrant. C) Contextual fear conditioning results. Freezing was statistically reduced in the P301S in context testing. D) Cued fear conditioning. No statistical difference was observed between groups. For all experiments, n=20, mean ± s.d.m. *p<0.05, **p<0.01 and ***p<0.001. ANOVA followed by Bonferroni’s post hoc test. See also Figures S6A–S6B
Next, we performed contextual and cued fear conditioning tests, which are largely dependent on the hippocampus and amygdala, respectively (Takeuchi et al., 2011). Interestingly, the P301S mice exhibited a significantly lower percentage of freezing compared to wild-type and Nuak1+/− mice during the contextual fear assay (Figure 4C). The P301S/Nuak1+/− mice showed better memory in the contextual fear test than the P301S mice (Figure 4C). No differences were observed between any groups on the cued fear test (Figure 4D). Therefore, decreasing the levels of Nuak1 by 50% reverses the spatial learning deficit in P301S mice.
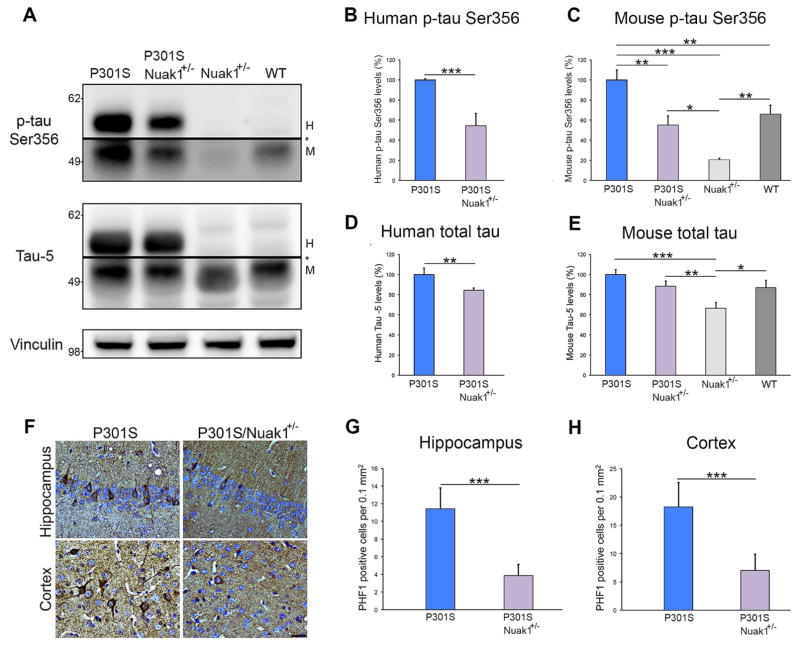
After the behavioral studies, mice were sacrificed and brains were collected for biochemical and immunohistochemical analyses. Western blot was used to investigate whether reducing the levels of Nuak1 also decreased total and phosphorylated human mutant tau in P301S mice. Indeed, P301S/Nuak1+/− mice presented lower levels of human phosphorylated tau at Ser356 than P301S mice (Figure 5A and B). The P301S/Nuak1+/− mice also showed lower levels of endogenous murine phospho Ser356 tau than the P301S mice (Figure 5A and C). Remarkably, this phospho form of tau was almost absent in the Nuak1+/− mice (Figure 5A and C). When we measured total levels of tau using the tau-5 antibody, P301S/Nuak1+/− mice presented lower levels of total human tau than P301S mice (Figure 5A and D). When we analyzed the levels of total endogenous mouse tau, we observed that only the Nuak1+/− mice showed a decrease in total mouse tau levels in comparison with the other three groups (Figure 5A and E). Considering that tau phosphorylation at Ser262/Ser356 is required to initiate the pathogenic cascade of tau hyperphosphorylation (Ando et al., 2016; Nishimura et al., 2004), we measured the levels of tau phosphorylated at the disease-associated residue Thr231 and Ser396/Ser404. Western blot analysis revealed that P301S/Nuak1+/− mice presented lower levels of phosphorylated tau at Thr231 and Ser396/Ser404 than P301S mice (Figures S6C–S6D). To assess the degree of tau phosphorylation due to changes in total tau levels, we measured the levels of phospho Ser356, phospho Thr231 and phospho Ser396/Ser404 relative to those of total tau. The relative ratio of phospho Ser356 to total tau was significantly decreased in P301S/Nuak1+/− mice in comparison with P301S mice (Figure S6E). No differences were observed between groups when the relative ratios of phospho Thr231 and phospho Ser396/Ser404 to total tau were measured (Figures S6F–S6G). These results confirm that the effect of Nuak1 over tau levels is mainly due to tau phosphorylation at Ser356 by Nuak1.
Figure 5. Nuak1 down-regulation prevents tau accumulation and NFT formation in P301S mice.
A) Western blot analysis of brain homogenates from four different genotypes. H=human tau, M=mouse tau. * indicated change in exposure of the membrane. B) Graph showing western blot quantification of human p-tau Ser356 levels. C) Graph showing western blot quantification of endogenous mouse p-tau Ser356 levels. D) Graph showing western blot quantification of total human tau measured with tau-5 antibody. E) Graph showing western blot quantification of endogenous mouse total tau levels. F) PHF1 immunostaining of hippocampal and cortex neurons in P301S and P301S/Nuak1+/− sections. Scale bar 25 μm. G) Graph showing immunostaining quantification of PHF1 positive neurons in the hippocampus. H) Graph showing immunostaining quantification of PHF1 positive neurons in the cortex. For all experiments, n=8, mean ± s.d.m. *p<0.05, **p<0.01 and ***p<0.001. For western blot quantification we utilized ANOVA followed by Bonferroni’s post hoc test. For staining quantification Student’s T-test was utilized. See also Figures S6C–S6G.
For immunohistochemical analysis we stained brain sections from P301S and P301S/Nuak1+/− mice with the PHF1 antibody, which recognizes pathological-associated tau phosphorylated at epitopes Serine 396 and Serine 404 (Otvos et al., 1994). We found that eliminating one copy of mouse Nuak1 reduced the number of PHF1-positive neurons in the hippocampus and cortex of P301S mice (Figure 5F–H). This result demonstrated that a minor decrease in tau levels at early stages has a pervasive effect on the amount of pathological PHF1-positive NFTs.
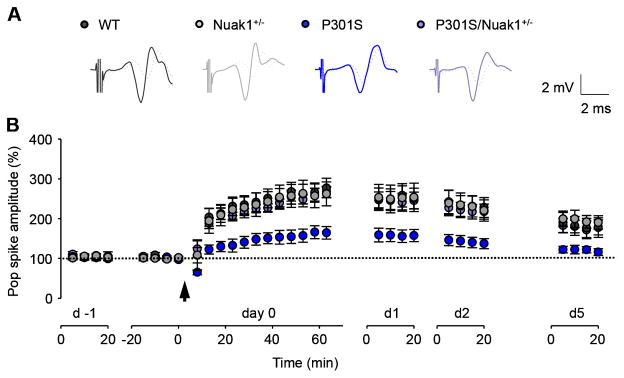
Finally, in a second cohort, we measured long-term synaptic plasticity of freely moving mice. Long-term synaptic plasticity (e.g., long-term potentiation, LTP) serves as a neural substrate for learning and memory (Malenka and Bear, 2004; Whitlock et al., 2006). We investigated LTP over several days in freely moving WT, P301S, Nuak1+/− and P301S/Nuak1+/− mice (Figure 6A and B). Before and after LTP induction, evoked responses were monitored in the perforant path recorded in the dentate gyrus. Following two days of baseline recording, tetanic stimulation of the perforant path induced significant potentiation of the population spike for multiple days in all groups (day 0, P < 0.001 versus baselines, respectively). Human P301S tau expression severely impaired hippocampal LTP over time in P301S mice 1 h after induction in comparison with the WT and Nuak1+/−, but a 50% decrease in Nuak1 expression completely rescued the impairment of synaptic plasticity in P301S/Nuak1+/− mice. We observed no difference in LTP between P301S/Nuak1+/− mice and WT or Nuak1+/− controls.
Figure 6. Nuak1 down-regulation restores hippocampal synaptic plasticity in the dentate gyrus of freely moving mice.
A), Superimposed traces of the perforant path recorded in the dentate gyrus 5 min before (dotted lines) and 55 min after (dark gray, gray, blue, or purple) LTP induction. B), Summary of in vivo LTP in WT (n = 7), P301S (n = 8), Nuak1+/− (n = 9) and P301S/Nuak1+/− mice (n = 7). LTP induction potentiated the population spikes in all the groups (one-way ANOVA on day 0: p < 0.001 for all groups). Two-way repeated measures ANOVA revealed significant main effects of population spike amplitudes among the four groups on day 0 (p = 0.01), day 1 (p < 0.05), day 2 (p < 0.05), and day 5 (p < 0.05). Tukey posthoc indicate LTP was impaired in P301S mice compared to WT or Nuak1+/− controls (day 0, P = 0.001; day 1, P = 0.015; day 2, P = 0.009; a trend on day 5, P = 0.058). Nuak1 rescued the LTP impairment in P301S/Nuak1+/− mice compared to P301S mice (day 0, P = 0.004; day 1, P = 0.024; day 2, P = 0.024). There was no difference of LTP between P301S/Nuak1+/− mice and WT or Nuak1+/− controls (P > 0.05 for all the tested days). Arrow, LTP induction. Data are mean ± s.e.m.
DISCUSSION
The data reported here show that Nuak1, an AMPK-related kinase, modulates tau levels in human cells, Drosophila, and mouse model systems. This kinase was highly associated with tau accumulation in two different tauopathies, AD and PSP, which strongly suggests that Nuak1 has a role in tau accumulation in a disease context, making this kinase a valuable target for further validation. We also demonstrated that hippocampal LTP is severely impaired in freely moving P301S mice and that decreasing Nuak1 levels or activity reverses several deficits in a tauopathy mouse model.
Previous studies reported that MARK2 phosphorylated tau at Ser262/Ser356 and AMPK directly phosphorylated tau at Ser262 but not at Ser356, demonstrating the important role this family of kinases plays in tau physiological functions (Dickey et al., 2007; Mairet-Coello et al., 2013). To our knowledge Nuak1 is the first kinase to be identified that exclusively phosphorylates tau at Ser356 but not at Ser262. More importantly, we show that such phosphorylation directly affects tau levels.
Nuak1 directly phosphorylates tau at Ser356, which is located in the microtubule-binding domain (Buee et al., 2000). Tau phosphorylation at Ser262 and Ser356, both within the microtubule-binding domain, causes tau to be released from the microtubules, which in turn leads to microtubule destabilization (Biernat and Mandelkow, 1999). These are physiological phosphorylations that are able to regulate microtubule dynamics, which is important for axonal growth and other processes that require neuronal plasticity (Biernat and Mandelkow, 1999). Although phosphorylation at Ser356 plays a necessary role in the regulation of normal tau function, previous studies have demonstrated that phosphorylation at this site is required to initiate the pathogenic cascade of hyperphosphorylation on additional sites by other kinases associated with tau aggregation and formation of NFTs (Nishimura et al., 2004). This last observation holds true in the context of Nuak1, where we showed how tau phosphorylation at Ser356 by Nuak1 primes tau for the subsequent phosphorylation at other pathological sites. Taking into consideration that Nuak1 decreases tau ubiquitination and its binding to CHIP, which are essential for tau proteosomal degradation (Dickey et al., 2007; Dickey et al., 2008), and that Nuak1 is highly associated with tau pathology in human tauopathies; it is possible that direct phosphorylation of tau at Ser356 by Nuak1 triggers a cascade of events that promote or inhibit several post-translational modifications key to increasing tau stability, promoting its accumulation, aggregation and formation of NFTs. This common mechanism for tau pathogenesis may have certain differences according to the nature of each tauopathy. For instance in AD, Aβ amyloid plays an important role in the activation of different kinases which subsequently phosphorylate tau (Ma et al., 2009; Takashima et al., 1998; Zempel et al., 2010). Alternatively in PSP, which is solely characterized by presence of tau pathology, the accumulation or activation of Nuak1 and other kinases could be triggered by a different mechanism. This point could explain the variability of Nuak1 levels between AD and PSP cases and even within cases of the same disease.
Our in vivo genetic interaction study demonstrated that reduction of Nuak1 by 50% was sufficient to decrease total tau levels and reverse the deficits in a tauopathy mouse model. This result is rather remarkable, considering that human tau is expressed in the P301S mouse model at five times the levels of endogenous mouse tau (Yoshiyama et al., 2007). It is thus possible that even a modest (less than 50%) reduction of Nuak1 could exert a strong effect on endogenous tau levels
Previous efforts in the tauopathy field have aimed to develop screening strategies for the discovery of therapeutic targets focused on tau aggregation or pathological hyper-phosphorylation (Cavallini et al., 2013; Pickhardt et al., 2005). Our study focused on a much earlier stage in disease pathogenesis, the point at which tau levels begin to increase beyond the neuron’s ability to degrade the protein. This strategy allowed us to identify Nuak1, a kinase that phosphorylates tau at a physiological site such as Ser356. This site represents only a minor fraction of total phospho-sites, but still acts as a key trigger for tau accumulation and aggregation (Biernat and Mandelkow, 1999; Biernat et al., 2002; Nishimura et al., 2004). The fact that a small reduction in tau levels was sufficient to have prominent effects on multiple disease-like phenotypes substantiates screening for modifiers of tau levels as a strategy to subdue disease or even prevent its development.
EXPERIMENTAL PROCEDURES
Generation of stable cell lines
DsRed-IRES-tau:EGFP cell line was generated as previously described (Park et al., 2013). Briefly, the construct was cloned into a pHAGE vector. Lentiviral packaged clones were infected into Daoy cells and then selected with puromycin and ran through Aria II (BD Biosciences) for selection of cells that show expression of DsRed and GFP.
Cell-based kinase siRNA screen
Daoy DsRed-IRES-tau:EGFP cells were split into 96-well plates. On the next day, each siRNA (kinase siRNA library from Invitrogen) was transfected at 20 nM with 0.08 μl of transfection reagent (Dharmacon) into corresponding wells and incubated for 72 h. Before running FACS analysis (LSR II, BD Biosciences), the cells were trypsinized and suspended in PBS with 5% FBS.
Drosophila kinase screen
For the screen we used a transgenic line that expresses a four-repeat wild type isoform of human tau under the control of a GMR-Gal4 driver. We obtained homologs for all human kinases in Drosophila from the Vienna Drosophila RNAi Center (VDRC). All crosses for the screen were done at 28°C for external eye phenotype and at 26.5°C for analysis of the retina. Flies were processed for scanning electron microscopy and paraffin sections of the retina as previously described (Park et al., 2013). Additional lines were obtained from Bloomington Stock center (Indiana).
Cell culture, over-expression, siRNA transfections and inhibitor treatment
Daoy stable cell lines and 2C neuroblastoma cell lines were cultured in DMEM or F12/MEM (Invitrogen) with 10% FBS (Invitrogen) respectively. siRNAs (Invitrogen) were transfected with DharmaFECT (Dharmacon) and incubated for 3 days before analysis. Human wild-type Nuak1 and kinase dead Nuak1 (K84M) were cloned into a 3xflag-CMV plasmid (Sigma-aldrich). GFP-fused human tau (gift of K. Ashe, Addgene plasmid no. 46904) was utilized to generated mutant tau S356A (QuikChange XL Site-Directed Mutagenesis Kit). All plasmids were transfected with Lipofectamine 2000 (Invitrogen) and incubated for 48 hours. 2C cells were treated with Nuak1 inhibitor (WZ4003, abcam) for 24 hours.
Cell lysate preparation and immunoblot analysis
Before collection, cells were washed with PBS and lysed on ice for 20 min in RIPA buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40 and 5mM EDTA) supplemented with protease inhibitors (Roche). The cell lysates were then centrifuged at 13,200 rpm for 20 min at 4°C, and the supernatants were analyzed by western blot. Primary antibodies used: anti-tau (tau-5, 1:1000, abcam, RRID:AB_304171), anti-pSer356 tau (1:750, abcam, RRID:AB_10586459),), anti-pSer262 tau (1:750, abcam, RRID:AB_2139717), Anti-Nuak1 (1:800, abcam, RRID:AB_1267723) and Anti vinculin (1:10000, Sigma-aldrich, RRID:AB_477629).
Human and mouse brain samples preparation and immunoblot analysis
Postmortem brain tissues from subjects with AD, PSP and control subjects were provided in the form of frozen blocks by the Brain Resource Center at Johns Hopkins. AD cases consisted of pathologically severe AD, stage V–VI. Each brain was homogenized in RIPA buffer with a protease inhibitor cocktail (Roche) and a dilution of brain:RIPA of 1:10 (w/v). Samples were then centrifuged at 13,200 rpm for 15 min at 4°C. The supernatants were portioned into aliquots, snap-frozen, and stored at −80°C until analyzed. The RIPA insoluble pellet was treated with formic acid by mixing samples with 88% formic acid for 1 hour at room temperature (The volume of 88% FA was ¼ of the volume used for RIPA). Samples were then diluted with distilled water to obtain the same volume used in RIPA. Samples were then lyophilized for 24 hours. Freeze-dried samples were reconstituted in PBS using the same volume that was originally used for RIPA. Samples were then sonicated for 30 seconds. Finally, samples were mixed with running buffer, run on a gel, and analyzed by Western blot. Primary antibodies used: anti-Nuak1 (1:800), PHF1 (1:500, gift from Peter Davies, RRID:AB_2315150) and anti-vinculin (1:10000), tau-5 (1:1000) and anti-pSer356 tau (1:750).
Brain sections immunofluorescence (IF)
Paraffin sections were deparaffinized, rehydrated, and washed in 0.01 M PBS 1X 3 times for 5 min each time. After blocking in normal goat serum for 1 hr, sections were incubated overnight with tau-5 antibody (1:100) and anti-Nuak1 antibody (1:100). The next day, the sections were washed in PBS 1X 3 times for 10 min each time and then incubated with goat anti-mouse Alexa Fluor 488 (1: 700; Invitrogen, RRID:AB_2534069) and goat anti-rabbit Alexa Fluor 568 (1:700; Invitrogen, RRID:AB_2534094) for 1 hr. Sections were washed and mounted in Vectashield mounting medium with DAPI (Vector Laboratories, RRID:AB_2336790). The sections were examined using a Zeiss LSM 710 confocal microscope.
Mouse models
All procedures for mouse animal use were approved by the Institutional Animal Care and Use Committee for Baylor College of Medicine and Affiliates. The PS19 mouse model, which over expresses human tau with the P301S mutation, was directly purchased from Jackson laboratories (Stock No:008169, RRID:IMSR_JAX:024841). Nuak1 heterozygous embryos were provided by the RIKEN Center for Developmental Biology in Japan (RRID:MGI:3771062). The Transgenic Mouse Core Rederivation at Baylor College of Medicine performed embryo pre-implantation.
Morris water maze (MWM)
Morris water maze was performed as previously described (Takeuchi et al., 2011). A circular pool (120 cm in diameter) was filled with water (21±1 °C), in which non-toxic white tempera paint was mixed to make the surface opaque. For the invisible platform test, a white colored platform was placed at the center in one of four quadrants of the pool (southwest area) and submerged 1 cm below the water surface so that it was invisible at water level. The location of the platform was fixed at the same quadrant while the start position of swimming was varied. Mice were given 4 trials per day for 5 consecutive days, during which they were allowed to find the platform within 60 seconds. Each trial was separated by an inter-trial interval of 1–2 min, which was adopted through all the tests. Once the mouse located the platform, it was permitted to stay on it for 10 seconds. If the mouse did not find the platform within 60 seconds, it was guided to the platform and placed on it for 20 seconds. To evaluate the spatial reference memory, all mice were given a probe trial 24 hours after the last trial of the invisible training test which consisted of removing the platform from the pool and allowing the mice to swim for 60 sec in its search. A record was kept of the swimming time (sec) in the pool quadrant where the platform had previously been placed. During the visible platform test, a colored platform was placed in the quadrant 1 cm above the surface of the water, and its location was always varied randomly in each trial. All mice were subjected to 8 blocks with 4 trials per block during 1 day. Swim speed (cm/sec), latency time to find the platform (sec), and the time that each mouse swam in the target quadrant were recorded by video camera and analyzed by a computer-controlled video tracking system (Ethovision XT, by Noldus Information Technology, Leesburg, VA).
Fear conditioning
Fear conditioning was performed as previously described (Takeuchi et al., 2011). Each mouse was placed in a sound-attenuated chamber and allowed to explore freely for 2 min. An 80-dB white noise, the conditioned stimulus (CS), was presented for 30 sec followed by a mild (2 sec, 0.72 mA) foot shock, the unconditioned stimulus (US). Two more CS–US pairings were presented with 2-min interstimulus intervals. Context testing was conducted 1 day after conditioning in the same chamber. Cued testing with altered context was conducted on the same day, following the context testing, using a triangular box made of white opaque Plexiglas, which was located in a different room. Data acquisition, control of stimuli (i.e., tones and shocks), and data analysis were performed automatically using Coulbourn/Actimetrics FreezeFrame3 System.
Surgery and electrophysiology
Induction and recording of hippocampal synaptic plasticity in vivo were conducted as previously published with a few modifications (Davis et al., 1997; Hao et al., 2015; Tang and Dani, 2009). Mice were secured on a stereotaxic frame (David Kopf) with 1–2% isoflurane as anesthesia. Under aseptic conditions, the recording electrode (Teflon-coated tungsten wire, bare diameter 50 μm, A-M Systems) was surgically aimed at the dentate gyrus (1.8 – 2.0 mm posterior, 1.4 – 1.6 mm lateral of bregma, 2.2 – 2.3 mm below the skull) while a concentric stimulating electrode (same sort of tungsten wire as for recordings) was implanted ipsilaterally in the medial perforant path (0.2 mm posterior and 2.8–3.0 mm lateral of lambda, 1.0–1.3 mm below the dura) (Paxinos and Franklin, 2001). Evoked potentials of the performant path recorded in the dentate were used to guide the final positions of both electrodes. Dental cement was used to anchor the electrode assembly that was connected to a unity gain preamplifier, and the connecting device for chronic recordings. After at least two weeks of recovery from surgical implantation, mice were transported and habituated to the recording system during each of the 4 days prior to starting the LTP test. Signals were amplified (100x), filtered (bandpass, 0.1–5 kHz), digitized at 10 kHz, and stored on disk for off-line analysis (pClamp10 and 1440A; Molecular Devices). The time course of LTP tests is across 7 days (day -1 to day 5). Test responses elicited by monophasic pulses (0.1 ms duration) were recorded for 20-min periods on consecutive days at an intensity that evoked 40% of the maximal population spike. Following two days of stable baseline, a tetanus was delivered to the perforant path for LTP induction. Pulse width was doubled during tetani, which consisted of 6 series of 6 trains of 6 stimuli at 400 Hz, 200 ms between trains, 20 s between series. Responses were measured for 60 min after tetanus and again, for 20 min at 24 h, 48 h, and 120 h after tetanus. Since the latency of the population spike usually decreases following LTP induction, it is impractical to compare the initial slope of the fEPSP (field excitatory postsynaptic potential) before and after LTP induction in awake animals (Jones et al., 2001; Malleret et al., 2001). Accordingly, we quantified the amplitude of the population spikes (Hao et al., 2015; Tang and Dani, 2009). Data were averaged every 5 min and normalized to the baseline measured over the 10 min before tetanic stimulation and presented as mean ± standard error of mean. Two-way repeated measures ANOVA (between groups) or one-way ANOVA (within group) followed by Tukey posthoc were used for data analysis.
Statistical analyses
Experimental analysis and data collection were performed in a blinded fashion. P values were determined using the appropriate statistical method via GraphPad Prism, as described throughout the manuscript. For simple comparisons, Student’s T-Test was used. For multiple comparisons, ANOVA followed by the appropriate post hoc analysis were utilized. Data is presented as mean ± s.d.m. *, ** and *** denote P < 0.05, P < 0.01 and P < 0.001, respectively. To identify the primary screen hits, we calculated the whole-screen mean and selected the siRNAs that decreased the DsRed-IRES-tau:EGFP ratio below 2 standard deviations. For the confirmation screen, we performed three independent experimental sets and compared the effect of siRNAs within each set to the internal siRNA controls by analysis of variance followed by Dunnet’s and Tukey’s post-hoc tests to select for the siRNAs that were significantly decreased.
Supplementary Material
Acknowledgments
We thank the members of the Zoghbi and Botas laboratories for suggestions and discussions, and V. Brandt for critical reading of the manuscript. This work was supported by the Howard Hughes Medical Institute, the Robert A and Renee E Belfer Family Foundation, The Hamill Foundation, The Chapman Foundation and grant NIH/NINDS R01 NS027699-17. The NIH/NINDS 3R01 NS027699-25S1 and 1K22NS092688-01 to C.A.L.R. Texas Alzheimer’s Research and Care Consortium-Investigator Grant Program to H.Y.Z and J.B. The Darrel K Royal foundation grant to I.A-R. M.W.C.R. wants to thank The Canadian Institutes of Health Research Fellowship (201210MFE-290072-173743). We also appreciate the assistance of Drs. Jun Qin and Sung Yun Jung at the Mass Spectrometry-Proteomics Core Laboratory (MS-PCL), the confocal microscopy, neuroconnectivity and mouse behavioral cores of Baylor College of Medicine (BCM) Intellectual and Developmental Disabilities Research Center (1U54 HD083092). AD and PSP tissues for this research were provided by the Johns Hopkins University Morris Udall Parkinson’s Disease Center of Excellence (NINDS P50 NS38377) and Alzheimer Disease Research Center (NIA P50 AG05146).
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, 6 figures, one table and one video.
AUTHOR CONTRIBUTIONS
C.A.L.R. and H.Y.Z. conceived the study, designed experiments, analyzed and interpreted the data and wrote the manuscript. M.D.H, I.A-R and J.B. conceived, designed, performed and interpreted Drosophila screen and validation experiments. C.A.L.R., J.P., L.N., and A.D.M. performed molecular and biochemical experiments. M.W.C.R. and P.J-N. performed the cell-based screens. C.A.L.R., L.V.V and L.S. performed mouse genotyping, histology, mouse behavior and microscopy. J.T., S.H. and Z.W. designed, performed and interpreted in vivo electrophysiology. J.C.T. provided reagents and aided in data interpretation. T.F.W. provided reagents and input for the cell based screen.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ando K, Maruko-Otake A, Ohtake Y, Hayashishita M, Sekiya M, Iijima KM. Stabilization of Microtubule-Unbound Tau via Tau Phosphorylation at Ser262/356 by Par-1/MARK Contributes to Augmentation of AD-Related Phosphorylation and Abeta42-Induced Tau Toxicity. PLoS genetics. 2016;12:e1005917. doi: 10.1371/journal.pgen.1005917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Buhrlage SJ, Huang HT, Deng X, Zhou W, Wang J, Traynor R, Prescott AR, Alessi DR, Gray NS. Characterization of WZ4003 and HTH-01-015 as selective inhibitors of the LKB1-tumour-suppressor-activated NUAK kinases. The Biochemical journal. 2014;457:215–225. doi: 10.1042/BJ20131152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernat J, Mandelkow EM. The development of cell processes induced by tau protein requires phosphorylation of serine 262 and 356 in the repeat domain and is inhibited by phosphorylation in the proline-rich domains. Molecular biology of the cell. 1999;10:727–740. doi: 10.1091/mbc.10.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernat J, Wu YZ, Timm T, Zheng-Fischhofer Q, Mandelkow E, Meijer L, Mandelkow EM. Protein kinase MARK/PAR-1 is required for neurite outgrowth and establishment of neuronal polarity. Molecular biology of the cell. 2002;13:4013–4028. doi: 10.1091/mbc.02-03-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buee L, Bussiere T, Buee-Scherrer V, Delacourte A, Hof PR. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain research Brain research reviews. 2000;33:95–130. doi: 10.1016/s0165-0173(00)00019-9. [DOI] [PubMed] [Google Scholar]
- Cavallini A, Brewerton S, Bell A, Sargent S, Glover S, Hardy C, Moore R, Calley J, Ramachandran D, Poidinger M, et al. An unbiased approach to identifying tau kinases that phosphorylate tau at sites associated with Alzheimer disease. The Journal of biological chemistry. 2013;288:23331–23347. doi: 10.1074/jbc.M113.463984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annual review of biochemistry. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Clark LN, Poorkaj P, Wszolek Z, Geschwind DH, Nasreddine ZS, Miller B, Li D, Payami H, Awert F, Markopoulou K, et al. Pathogenic implications of mutations in the tau gene in pallido-ponto-nigral degeneration and related neurodegenerative disorders linked to chromosome 17. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:13103–13107. doi: 10.1073/pnas.95.22.13103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S, Bliss TV, Dutrieux G, Laroche S, Errington ML. Induction and duration of long-term potentiation in the hippocampus of the freely moving mouse. Journal of neuroscience methods. 1997;75:75–80. doi: 10.1016/s0165-0270(97)00053-8. [DOI] [PubMed] [Google Scholar]
- Dickey CA, Kamal A, Lundgren K, Klosak N, Bailey RM, Dunmore J, Ash P, Shoraka S, Zlatkovic J, Eckman CB, et al. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. The Journal of clinical investigation. 2007;117:648–658. doi: 10.1172/JCI29715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey CA, Koren J, Zhang YJ, Xu YF, Jinwal UK, Birnbaum MJ, Monks B, Sun M, Cheng JQ, Patterson C, et al. Akt and CHIP coregulate tau degradation through coordinated interactions. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3622–3627. doi: 10.1073/pnas.0709180105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao S, Tang B, Wu Z, Ure K, Sun Y, Tao H, Gao Y, Patel AJ, Curry DJ, Samaco RC, et al. Forniceal deep brain stimulation rescues hippocampal memory in Rett syndrome mice. Nature. 2015;526:430–434. doi: 10.1038/nature15694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, Yang L, Kotin RM, Paulson HL, Davidson BL. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- Jones MW, Errington ML, French PJ, Fine A, Bliss TV, Garel S, Charnay P, Bozon B, Laroche S, Davis S. A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nature neuroscience. 2001;4:289–296. doi: 10.1038/85138. [DOI] [PubMed] [Google Scholar]
- Ma QL, Yang F, Rosario ER, Ubeda OJ, Beech W, Gant DJ, Chen PP, Hudspeth B, Chen C, Zhao Y, et al. Beta-amyloid oligomers induce phosphorylation of tau and inactivation of insulin receptor substrate via c-Jun N-terminal kinase signaling: suppression by omega-3 fatty acids and curcumin. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2009;29:9078–9089. doi: 10.1523/JNEUROSCI.1071-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairet-Coello G, Courchet J, Pieraut S, Courchet V, Maximov A, Polleux F. The CAMKK2-AMPK kinase pathway mediates the synaptotoxic effects of Abeta oligomers through Tau phosphorylation. Neuron. 2013;78:94–108. doi: 10.1016/j.neuron.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Malleret G, Haditsch U, Genoux D, Jones MW, Bliss TV, Vanhoose AM, Weitlauf C, Kandel ER, Winder DG, Mansuy IM. Inducible and reversible enhancement of learning, memory, and long-term potentiation by genetic inhibition of calcineurin. Cell. 2001;104:675–686. doi: 10.1016/s0092-8674(01)00264-1. [DOI] [PubMed] [Google Scholar]
- Miller TM, Kaspar BK, Kops GJ, Yamanaka K, Christian LJ, Gage FH, Cleveland DW. Virus-delivered small RNA silencing sustains strength in amyotrophic lateral sclerosis. Annals of neurology. 2005;57:773–776. doi: 10.1002/ana.20453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris M, Maeda S, Vossel K, Mucke L. The many faces of tau. Neuron. 2011;70:410–426. doi: 10.1016/j.neuron.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura I, Yang Y, Lu B. PAR-1 kinase plays an initiator role in a temporally ordered phosphorylation process that confers tau toxicity in Drosophila. Cell. 2004;116:671–682. doi: 10.1016/s0092-8674(04)00170-9. [DOI] [PubMed] [Google Scholar]
- Noble ME, Endicott JA, Johnson LN. Protein kinase inhibitors: insights into drug design from structure. Science. 2004;303:1800–1805. doi: 10.1126/science.1095920. [DOI] [PubMed] [Google Scholar]
- Otvos L, Jr, Feiner L, Lang E, Szendrei GI, Goedert M, Lee VM. Monoclonal antibody PHF-1 recognizes tau protein phosphorylated at serine residues 396 and 404. Journal of neuroscience research. 1994;39:669–673. doi: 10.1002/jnr.490390607. [DOI] [PubMed] [Google Scholar]
- Park J, Al-Ramahi I, Tan Q, Mollema N, Diaz-Garcia JR, Gallego-Flores T, Lu HC, Lagalwar S, Duvick L, Kang H, et al. RAS-MAPK-MSK1 pathway modulates ataxin 1 protein levels and toxicity in SCA1. Nature. 2013;498:325–331. doi: 10.1038/nature12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. The Mouse Brain in Stereotaxic Coordinates. San Diego, CA: Academic Press; 2001. [Google Scholar]
- Petrucelli L, Dickson D, Kehoe K, Taylor J, Snyder H, Grover A, De Lucia M, McGowan E, Lewis J, Prihar G, et al. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Human molecular genetics. 2004;13:703–714. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- Pickhardt M, von Bergen M, Gazova Z, Hascher A, Biernat J, Mandelkow EM, Mandelkow E. Screening for inhibitors of tau polymerization. Current Alzheimer research. 2005;2:219–226. doi: 10.2174/1567205053585891. [DOI] [PubMed] [Google Scholar]
- Pittman AM, Fung HC, de Silva R. Untangling the tau gene association with neurodegenerative disorders. Human molecular genetics. 2006;15(Spec No 2):R188–195. doi: 10.1093/hmg/ddl190. [DOI] [PubMed] [Google Scholar]
- Roberson ED, Scearce-Levie K, Palop JJ, Yan F, Cheng IH, Wu T, Gerstein H, Yu GQ, Mucke L. Reducing endogenous tau ameliorates amyloid beta-induced deficits in an Alzheimer’s disease mouse model. Science. 2007;316:750–754. doi: 10.1126/science.1141736. [DOI] [PubMed] [Google Scholar]
- Rovelet-Lecrux A, Campion D. Copy number variations involving the microtubule-associated protein tau in human diseases. Biochemical Society transactions. 2012;40:672–676. doi: 10.1042/BST20120045. [DOI] [PubMed] [Google Scholar]
- Santacruz K, Lewis J, Spires T, Paulson J, Kotilinek L, Ingelsson M, Guimaraes A, DeTure M, Ramsden M, McGowan E, et al. Tau suppression in a neurodegenerative mouse model improves memory function. Science. 2005;309:476–481. doi: 10.1126/science.1113694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small SA, Duff K. Linking Abeta and tau in late-onset Alzheimer’s disease: a dual pathway hypothesis. Neuron. 2008;60:534–542. doi: 10.1016/j.neuron.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Gao L, Chien HY, Li WC, Zhao J. The regulation and function of the NUAK family. Journal of molecular endocrinology. 2013;51:R15–22. doi: 10.1530/JME-13-0063. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Kusakai G, Kishimoto A, Shimojo Y, Miyamoto S, Ogura T, Ochiai A, Esumi H. Regulation of caspase-6 and FLIP by the AMPK family member ARK5. Oncogene. 2004;23:7067–7075. doi: 10.1038/sj.onc.1207963. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Ogura T, Esumi H. NDR2 acts as the upstream kinase of ARK5 during insulin-like growth factor-1 signaling. The Journal of biological chemistry. 2006;281:13915–13921. doi: 10.1074/jbc.M511354200. [DOI] [PubMed] [Google Scholar]
- Takashima A, Honda T, Yasutake K, Michel G, Murayama O, Murayama M, Ishiguro K, Yamaguchi H. Activation of tau protein kinase I/glycogen synthase kinase-3beta by amyloid beta peptide (25–35) enhances phosphorylation of tau in hippocampal neurons. Neuroscience research. 1998;31:317–323. doi: 10.1016/s0168-0102(98)00061-3. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Iba M, Inoue H, Higuchi M, Takao K, Tsukita K, Karatsu Y, Iwamoto Y, Miyakawa T, Suhara T, et al. P301S mutant human tau transgenic mice manifest early symptoms of human tauopathies with dementia and altered sensorimotor gating. PloS one. 2011;6:e21050. doi: 10.1371/journal.pone.0021050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Dani JA. Dopamine enables in vivo synaptic plasticity associated with the addictive drug nicotine. Neuron. 2009;63:673–682. doi: 10.1016/j.neuron.2009.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vingtdeux V, Davies P, Dickson DW, Marambaud P. AMPK is abnormally activated in tangle- and pre-tangle-bearing neurons in Alzheimer’s disease and other tauopathies. Acta neuropathologica. 2011;121:337–349. doi: 10.1007/s00401-010-0759-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Mandelkow E. Tau in physiology and pathology. Nature reviews Neuroscience. 2016;17:22–35. doi: 10.1038/nrn.2015.1. [DOI] [PubMed] [Google Scholar]
- Westbrook TF, Hu G, Ang XL, Mulligan P, Pavlova NN, Liang A, Leng Y, Maehr R, Shi Y, Harper JW, et al. SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature. 2008;452:370–374. doi: 10.1038/nature06780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock JR, Heynen AJ, Shuler MG, Bear MF. Learning induces long-term potentiation in the hippocampus. Science. 2006;313:1093–1097. doi: 10.1126/science.1128134. [DOI] [PubMed] [Google Scholar]
- Williams AJ, Paulson HL. Polyglutamine neurodegeneration: protein misfolding revisited. Trends in neurosciences. 2008;31:521–528. doi: 10.1016/j.tins.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H, Mao Q, Eliason SL, Harper SQ, Martins IH, Orr HT, Paulson HL, Yang L, Kotin RM, Davidson BL. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nature medicine. 2004;10:816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Goedert M. Phosphorylation of microtubule-associated protein tau by AMPK-related kinases. Journal of neurochemistry. 2012;120:165–176. doi: 10.1111/j.1471-4159.2011.07523.x. [DOI] [PubMed] [Google Scholar]
- Yoshiyama Y, Higuchi M, Zhang B, Huang SM, Iwata N, Saido TC, Maeda J, Suhara T, Trojanowski JQ, Lee VM. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron. 2007;53:337–351. doi: 10.1016/j.neuron.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Zempel H, Thies E, Mandelkow E, Mandelkow EM. Abeta oligomers cause localized Ca(2+) elevation, missorting of endogenous Tau into dendrites, Tau phosphorylation, and destruction of microtubules and spines. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2010;30:11938–11950. doi: 10.1523/JNEUROSCI.2357-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou F, Chai HS, Younkin CS, Allen M, Crook J, Pankratz VS, Carrasquillo MM, Rowley CN, Nair AA, Middha S, et al. Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS genetics. 2012;8:e1002707. doi: 10.1371/journal.pgen.1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Duvick LA, Kaytor MD, Berlinger MS, Zoghbi HY, Clark HB, Orr HT. Recovery from polyglutamine-induced neurodegeneration in conditional SCA1 transgenic mice. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2004;24:8853–8861. doi: 10.1523/JNEUROSCI.2978-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.