Abstract
Objective
We compare the effectiveness of fluoxetine and desipramine treatment in a prospective double-blind pharmacogenetics study in Mexican-Americans and examine the role of whole-exome functional gene variations in their antidepressant response.
Method
232 Mexican-Americans who met DSM-VI diagnostic criteria for major depressive disorder (MDD) were randomly assigned to an 8-week of double-blind desipramine (50–200 mg/day) or fluoxetine (10–40 mg/day) treatment after a one-week placebo lead-in period. Outcome measures included the Hamilton Depression (HAM-D), Hamilton Anxiety, and Beck Depression Inventory. Whole exome genotyping data were obtained for 36 remitters and 29 non-responders at week 8.
Results
Our analysis showed that fluoxetine treatment produced greater HAM-D score reduction, higher response/remission rates, shorter time to response/remission, and lower incidences of anticholinogeric and cardiovascular side effect events when compared to desipramine treatment. Pharmacogenetics analysis showed that exm-rs1321744 achieved exome-wide significance for treatment remission (P=1.98×10−06; FDR=0.05). This variant is located in a brain methylated DNA immunoprecipitation sequencing site suggesting that it might be involved in epigenetic regulation of neuronal gene expression. This and two other common gene variants provided a highly accurate cross-validated predictive model for treatment remission of MDD [Receiver Operating Characteristic (ROC) = 0.95].
Conclusions
Compared with desipramine, fluoxetine treatment showed a more rapid reduction of HAM-D score and lower incidence of side effects in a population comprised primarily of first generation Spanish speaking only Mexican-American individuals with MDD. Our pharmacogenetics approach strongly implicates the role of functional variants in antidepressant treatment response. Further, independent studies are needed for replication and validation.
Keywords: depression, pharmacogenomics, drug response, antidepressants, desipramine, fluoxetine, Mexican-American population, methylation sites, epigenetic
INTRODUCTION
Efficacy and side effect data for psychotropic medications in the U.S. have been primarily investigated in non-Hispanic Caucasian populations. Currently, the Hispanic population is the largest ethnic minority group in the U.S. representing over 37 million people (1). Within this group, almost 70% are Mexican-Americans. Although this population is growing dramatically in the U.S., there is insufficient research regarding psychiatric diagnosis and treatment in this group (2).
Major depressive disorder is a serious public health issue worldwide, with a lifetime prevalence of 10% to 20% in the general population (3). In their findings, Vega et al. have reported U.S. born Mexican-Americans to have a 14.8% lifetime prevalence of major depression (4). Though Hispanics have participated in antidepressant treatment studies, it has been difficult to ascertain whether there are any major differences in antidepressant efficacy in that population. This may be explained by the inclusion of several Hispanic subgroups in an attempt to illustrate a “Hispanic response” (5), methodological differences, which makes it difficult to compare multiple studies, and very small sample sizes, which may result in increased Type II errors.
A retrospective review suggested that pharmacokinetic factors play a role in the differential sensitivity to tricyclic antidepressants (TCAs) in depressed Puerto Rican American females as compared to Anglo females, resulting in greater efficacy, higher adverse drug reactions (ADRs) and higher drop out rates in the former group (6). Open label antidepressant studies found that nefazodone had similar efficacy but higher dropouts in a predominantly Hispanic Caribbean female sample as compared to non-Hispanics from previous nefazodone trials (7), and no efficacy difference in two selective serotonin reuptake inhibitors (SSRIs), but a higher drop out rate in Mexican-American females, even though there were more severe ADRs reported in the non-Hispanic group (8). A more recent study between reported no differences in efficacy, ADRs or drop out rates between Hispanic and non-Hispanic HIV patients (9).
Three recent meta-analyses studies on pharmacogenetics of antidepressants in MDD have not being able to show genome-wide significant variations (10–12). The non-synonymous single nucleotide polymorphism (nsSNP) rs6265 in the BDNF (brain-derived neutrophic factor) gene, may have a minor impact in susceptibility to MDD (13) and antidepressant drug response (11); however, the overall conclusion of these meta-analysis studies is that the results so far do not support any major effect of any single gene variations in the pharmacogenetics of antidepressants in MDD. In the present work, we have focused on functional SNPs based on: 1) The likely significance of a functional SNP in the BDNF gene (rs6265) in genetic and pharmacogenetics of MDD; 2) Our own recent work which proposed a predictive framework for the diagnosis of MDD using interactions of multiple functional SNPs and environmental factors (Wong et al 2012); 3) A growing body of evidence supporting the involvement of epigenetic mechanisms in MDD and antidepressant action (14–18).
We present here data on the efficacy and ADR profiles of desipramine and fluoxetine, two extensively used off-patent antidepressants, and new pharmacogenetics leads that could advance our understanding of genetic variants implicated in antidepressant treatment response.
METHOD
The study protocol was approved by the University of California Los Angeles and University of Miami Institutional Review Boards, and the ANU Human Ethics Committee. This was a single site prospective double-blind 8-week trial with fluoxetine and desipramine conducted in the greater Los Angeles area. All subjects had an initial medical evaluation consisted of a detailed history, physical examination, and blood collection for routine testing and genotyping, and if enrolled, continued with two study consecutive phases: 1) A 1-week single-blind placebo lead-in phase to minimize the effect of placebo response; 2) Subsequent random assignment to two treatment groups: fluoxetine 10–40 mg/day or desipramine 50–200 mg/day with weekly follow-up visits to assess their clinical status. Written informed consent was obtained after complete description of the study to the participants. Given the proven efficacy of these antidepressant medications, a placebo lead-in period followed by active treatment for all patients was utilized in order to minimize human subjects at risk (19, 20).
Participants
All subjects met the following inclusion criteria: 1) ≥3 out of 4 grandparents born in Mexico (21); 2) DSM-IV diagnosis of current, unipolar major depressive episode (22); 3) 21-Item Hamilton Depression Rating Scale (HAM-D) (23) score of ≥18 with item #1 (depressed mood) rated ≥2; 4) age between 18–70 years.
Subjects were not able to participate in this study if they had the following exclusion criteria: 1) any axis I disorder other than major depressive disorder (e.g. dementia, psychotic illness, bipolar disorder, adjustment disorder) or primary anxiety disorders; 2) active medical illnesses that could be related to the ongoing depression (e.g. untreated hypothyroidism, recent myocardial infarction or cerebrovascular incident within the previous six months, uncontrolled hypertension or diabetes); 3) current suicidal ideation with a plan and strong intent, or recent serious suicide attempt; 4) pregnant, lactating, or women of childbearing age not using contraception; 5) history of electroconvulsive therapy in the previous six months; 6) current use of medications with central nervous system activity which interfere with EEG activity or any other antidepressant treatment within 2 weeks prior to enrollment; 7) history of poor response to therapeutic treatment with desipramine or fluoxetine; 8) illicit drug use and/or alcohol abuse in the previous three months; 9) current enrollment in counseling or psychotherapy treatment (24).
Recruitment and Outcome Measures
All subjects were recruited by advertisements in bilingual newspapers, radio, and television. Informed consent, questionnaires and assessment scales were given in their preferred language (English or Spanish). In addition, clinical staff also participated in health fairs, conferences, and cable network programs. Some subjects were also referred by regional outpatient community clinics.
The presence of a current major depressive episode was determined by the Structured Clinical Interview for Diagnosis (SCID) (25). The mean Kappa score for sensitivity and specificity among raters was between 0.84–0.85. In addition, a research psychiatrist confirmed their DSM-IV diagnoses. Symptom severity was rated by experienced, bilingual clinical personnel using Spanish or English versions of the HAM-D (23), Hamilton Anxiety Rating Scale (HAM-A) (26), Global Assessment Scale (GAS) (27), Beck Depression Inventory (BDI) (28) and Center for Epidemiological Depression Rating Scale (CESD) (29). Subjects also completed an acculturation questionnaire to determine their language preference, education level, and generation status.
Interventions and Treatment
All participants received one week of single-blind placebo to minimize the effect of placebo response, which was defined as 25% or greater decrease in HAM-D score compared to the screening visit and/or HAM-D score less than 18. The remaining subjects were randomly assigned to receive either fluoxetine or desipramine in a 1:1 ratio for an 8-week double-blind phase. Subjects initially received 10 mg fluoxetine/50 mg desipramine per day, which increased at week 2 to 20 mg fluoxetine/100 mg desipramine per day. At week 4, doses were increased in subjects who showed less than a 25% decrease in their HAM-D scores to 30 mg fluoxetine/150 mg desipramine per day. At week 6, doses were increased to 40mg fluoxetine/200mg desipramine per day if HAM-D >12. The staff and participants were aware of dose escalation, which happened only if the previous dose was well tolerated, but they were blind to the drug. At the end of the study, subjects were referred to a psychiatric clinic of their choice for follow-up treatment. Random antidepressant blood levels were collected to ascertain medication adherence but not to obtain therapeutic levels.
Statistical Analysis
Our analyses included all randomized subjects who received at least one week of drug and were performed using SAS version 9.1.3 (SAS Institute, Cary, N.C). The primary outcome measure consisted of the 21-item HAM-D. At the end of the 8-week double-blind phase, each subject was assessed according to the HAM-D score reduction from week 0 to week 8. The secondary outcomes of interest were changes in HAM-A, BDI, CESD, and GAS scores. Categorical classification was defined as: remission if HAM-D<8, response if HAM-D reduction ≥ 50%, and non-response if HAM-D reduction <50%. Remission and response were compared between treatment groups. Descriptive statistics for continuous variables were presented as means with standard deviations and summary statistics for discrete variables were presented as percentages. Student-t tests were used to compare the means of age, acculturation score and baseline clinical measurements. Chi-square or Fisher exact tests were used to compare the percentage of demographic characteristics and side effect events.
For repeated continuous outcome measures analyses, the likelihood-based mixed-effects model as the primary analysis of efficacy was used to assess differences between treatment groups in changes from baseline across 8 weeks. The model included the categorical effects of treatment, week at treatment, treatment-by-week interaction and gender, as well as continuous covariates of baseline score and age. Mixed-effects model for repeated measures (MMRM) analyses of changes from baseline were conducted using PROC MIXED in SAS. A SAS macro was written for covariance structure selection by comparing Akaike’s Information Criterion (AIC) and Schwarz’s Bayesian Criterion (BIC) using Compound Symmetry (CS), Unstructured (UN), first order Autoregressive [AR(1)] and Huynh-Feldt (HF) covariance structures based on “the smaller is better” criterion. For each outcome comparison, the covariance structure was used if the sum of AIC and BIC is smallest.
For repeated analyses of dichotomous outcomes (remission and response), the modified Poisson regression model with robust error variance estimated by generalized estimating equation (GEE) approach (30) was used to estimate the adjusted relative risk (aRR) and performed with PROC GENMOD in SAS. To compare the time to response or remission between the two treatment groups, Cox regression analysis was performed with PROC TPHREG in SAS using “DISCRETE” option to handle ties in the event time. Gender, age and baseline scores were included in all models as covariates. A two-side P≤ 0.05 was considered for statistical significance for all the analyses.
Pharmacogenetics Procedures
Whole Exome Genotyping
Genomic DNA of 65 completers, 36 remitters and 29 non-responders at week 8, were subject to whole exome genotyping performed by the Australian Genome Research Facility Ltd (AGRF, Melbourne, Australia), an Illumina® Certified Service Provider (CSPro) for the Infinum Genotyping Service. We used the Illumina® HumanExome BeadChip-12v1_A which covers putative functional exonic variants selected from over 12,000 individuals. The exonic content consists of >250,000 markers representing diverse populations—including European, African, Chinese, and Hispanic individuals—and a range of common conditions, such as type 2 diabetes, cancer, metabolic, and psychiatric disorders. Samples with calls below of the Illumina® expected 99% SNP call rate were excluded. To test genotyping reliability and quality, an individual was duplicated. The Identity by Descent (IBD) between all pairs of individuals was estimated and used for quality control.
Quality Control and Filtering
GenomeStudio data was imported to SVS 7.6.7, Golden Helix’s® (Golden Helix, Inc. Bozeman, MT, USA (http://www.goldenhelix.com), an integrated collection of analytic tools for managing, analyzing, and visualizing multifaceted genomic and phenotypic data.
Parameters for excluding markers from analyses included: (i) deviations from the Hardy-Weinberg equilibrium with P<2×10−7 (0.05/250.000 markers) in both cases and controls (this avoids the exclusion of major effect causal variants), (ii) a genotype call rate >90%, (iii) more than two alleles, and (iv) monoallelism. Genotype and allelic frequencies were estimated by maximum likelihood.
Genetic Stratification Analysis
We estimated the Inbreeding Coefficient (f) in order to detect the presence of hidden biological relatives in the sample, which might reduce the independence of the data. The Fixation Index (Fst) between pairs of subpopulations, cases and controls, was estimated to evaluate the potential presence of genotype stratification (micro differentiation); a common cause of spurious associations. Independently, an additional correction of putative population stratification was applied with 10 principal component analyses (PCA) to normalize genotypic data by its actual standard deviation.
Exome-Wide Association Analysis
The genotypic (additive model) and allelic tests of association were applied. Multiple test correction to determine exome-wide significance was performed using the false discovery rate approach (FDR). Mixed linear models where applied as a tool to include in the analyses fixed factors (sex, age, treatment) and random effects (family or population structure), and to contrast with other analytical tools, different from PCA, the effects of potential inbreeding (by inclusion of the kinship matrix as defined by IBD). We did apply the single-locus mixed model (SLMM) (assumes that all loci have a small effect on the trait) and the multi-locus mixed model (MLMM) (assumes that several loci have a large effect on the trait) as implemented in SVS 7.6.7, Golden Helix’s®. Separation of variants in common and rare was based on Myers criterion of 1% and data from the 1000 genomes was used for this filtering step. Linkage disequilibrium analysis was also implemented using SVS 7.6.7.
Advance Recursive Partitioning (tree-based) Approach (ARPA
Rao has suggested that recursive partitioning techniques should be highly recommended for genetic dissection of complex traits (31). ARPA is widely used in predictive analyses as it accounts for non-linear and interaction effects, and it offers fast solutions to reveal hidden complex substructures and provides truly non-biased statistically significant analyses of high dimension seemingly unrelated data (32).
ARPA accounts for the effect of hidden interactions better than alternative methods and is independent of the type of data, i.e., categorical, continuous, ordinal, etc., and of data distribution types, i.e., fitting or not fitting normality (31). Furthermore, results supplied by tree-based analytics are easy to interpret visually and logically (32). Therefore, to generate the most comprehensive and parsimonious classificatory model to predict MDD remission after treatment, we applied ARPA using a set of different modules implemented in the Salford Predictive Modeler® (SPM) software, namely, CART, Random Forest, and Tree-Net (http://www.salford-systems.com). SPM is a highly accurate, and ultra-fast analytics and data-mining platform for creating predictive, descriptive, and analytical models from databases of any size, complexity, or organization. One important advantage of SPM when compared to other available software is that SPM is able to use raw data with sparse or empty cells (a common problem when dealing with genetic data).
CART is a non-parametric approach whereby a series of recursive subdivisions separate the data by dichotomization (33). The aim is to identify, at each partition step, the best predictive variable, and its best corresponding splitting value, while optimizing a splitting statistical criterion, so the data set is successfully divided into increasingly homogeneous subgroups (33). We used a battery of different statistical criteria as splitting rules, e.g., Gini Index, Entropy, and Twoing, to determine the splitting rule decreasing the relative cost of the tree the most while increasing the prediction accuracy of target variable categories (33). The best split at each dichotomous node was chosen by either a measure of between-node dissimilarity or iterative hypothesis testing of all possible splits to find the most homogeneous split (lowest impurity) (33). Similarly, we used a wide range of empirical probabilities (priors) to model numerous scenarios recreating the distribution of the targeted variable categories in the population (33). Following this iterative process, each terminal node was assigned to a class outcome (remitter or non-responder).
To avoid finishing with an over fitted CART predictive model (a common problem in CART analysis), and to ensure that the final splits were well substantiated, we applied tree pruning (33). During the procedure, predictor variables that were close competitors (surrogate predictors with comparable overall classification error to the optimal predictors) are pruned to eliminate redundant commonalities among variables, so the most parsimonious tree would have the lowest misclassification rate for an individual not included in the original data (33).
Furthermore, to exactly identify the most important set of variables predicting MDD remission, we applied the Random Forest methodology using a bagging strategy (34). The Random Forest strategy differs from CART in the use of a limited number of variables to derive each node while creating hundred to thousand of trees (34). This strategy has proven to be immune to the over fitting generated by CART (34). In the Random Forest strategy, variables that appeared repeatedly as predictors in the trees were identified. The misclassification rate was recorded for each approach.
The TreeNet strategy was used to complement the CART and Random Forest analyses because it reaches a level of accuracy that is usually not attainable by single models (CART) or by ensembles such as bagging (Random Forest) (35). The TreeNet algorithm generates thousands of small decision trees built in a sequential error-correcting process converging on an accurate model (35).
To obtain honest assessments of the derived models and have a better view of their performance on future unseen data, we applied a cross–validation strategy where both training with all the data and then indirectly testing with all the data was performed. To do so, we randomly divided the data into 10 separate partitions (folds). This strategy allowed us to have a cross-validation and review the stability of results across multiple replications (33). A cross-validation is a model validation technique for assessing how the results of a statistical analysis will generalize to an independent data set and the n-fold cross-validation technique is designed to get the most out of datasets that are too small to accommodate a hold-out or test sample. For our specific problem, we use a maximization algorithm that allows us the validation of our whole genome analysis original results by considering n-fold subsamples of the original set. So we use cross-validation to be able to both train with all the data and then indirectly test with all the data as well.
We also applied a categorical approach to link the set of genotypes of the associated marker (rs1321744) by using Latent Class Analysis to identify unobservable subgroups within the subset of completers who were genotyped (remitters and non-responders).
RESULTS
Disposition and Baseline Characteristics of Patients
We phone interviewed 4,323 people to schedule 1,223 structured clinical interviews. A total of 338 participants were enrolled, of those 232 (112 on desipramine and 120 on fluoxetine) received at least one dose of drug and were included in the intent-to-treat (ITT) analysis (Fig 1S); 166 subjects completed 8 weeks of treatment (Fig 1S) and 66 did not complete the study either because they dropped out, were non-adherent, or were terminated (34% on desipramine and 23% on fluoxetine; χ2 =3.19, df=1, P=0.074). No significant differences were found in mean retention time in non-completers (t=0.33, df=64, P=0.742), and in demographic and baseline clinical characteristics between treatment groups (P≥ 0.156, Table 1S). Our participants were primarily (60%) first generation Spanish speaking only Mexican-Americans with 6–12 years of education (Table 1S).
Primary Outcome –HAM-D Score
Table 1 summarizes the mean HAM-D score changes (least-square means after adjustment for age, gender, and baseline score). MMRM analysis revealed a between-subject effect of treatment (F=4.45, df=1, 227, P=0.036; LS-means change difference=1.0, 95% confidence interval [CI]=0.1–1.9), within-subject effect of treatment time (F =76.38, df=1,1312, P<0.001), and no interaction between treatment and time (F =1.60, df=7,131, P=0.133). At the endpoint (week 8) fluoxetine showed an advantage over desipramine of 2.6 (95% CI=1.1–4.2; F=10.71, df=1,131, P=0.001). No gender-specific effect was found (F ≤ 0.54, df=1, 227, P≥0.463) on HAM-D score reductions but significant effects were found in baseline HAM-D scores (F ≥ 23.27, df=1, 227, P<0.001). MMRM revealed an age effect on HAM-D reduction (coefficient=−0.1; F=5.89, df=1, 227, P=0.016).
Table 1.
Least-Squares Mean (LS-Mean) of Changes from Baseline in Mexican-American Patients with Major depression Who Received Desipramine or Fluoxetinea
| Group | Week | HAM-D
|
HAM-A
|
BDI
|
CESD
|
GAS
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | LS-Mean | SE | P | N | LS-Mean | SE | P | N | LS-Mean | SE | P | N | LS-Mean | SE | P | N | LS-Mean | SE | P | ||
| Longitudinal analysis of observed datab | |||||||||||||||||||||
| Desipramine | 1 | 111 | −1.5 | 0.5 | 0.447 | 107 | −0.8 | 0.6 | 0.516 | 100 | −1.9 | 0.6 | 0.690 | 109 | −1.0 | 0.8 | 0.415 | ||||
| Fluoxetine | 1 | 120 | −2.0 | 0.5 | 116 | −1.3 | 0.6 | 111 | −1.6 | 0.6 | 115 | −1.8 | 0.8 | ||||||||
| Desipramine | 2 | 105 | −4.2 | 0.5 | 0.836 | 102 | 0.2 | 1.5 | 0.054 | 96 | −4.9 | 0.7 | 0.153 | 100 | −3.1 | 0.8 | 0.382 | ||||
| Fluoxetine | 2 | 113 | −4.3 | 0.5 | 111 | −3.9 | 1.5 | 104 | −3.4 | 0.7 | 106 | −4.1 | 0.8 | ||||||||
| Desipramine | 3 | 99 | −6.2 | 0.5 | 0.848 | 98 | −2.8 | 0.6 | 0.061 | 92 | −6.8 | 0.8 | 0.780 | 95 | −5.7 | 0.8 | 0.732 | ||||
| Fluoxetine | 3 | 110 | −6.0 | 0.5 | 105 | −4.4 | 0.6 | 97 | −6.5 | 0.8 | 104 | −6.1 | 0.8 | ||||||||
| Desipramine | 4 | 97 | −7.3 | 0.5 | 0.633 | 95 | −3.8 | 0.6 | 0.143 | 88 | −8.5 | 0.9 | 0.567 | 92 | −7.0 | 0.8 | 0.906 | 96 | 7.7 | 0.9 | 0.413 |
| Fluoxetine | 4 | 106 | −7.6 | 0.5 | 101 | −5.1 | 0.6 | 96 | −7.8 | 0.9 | 103 | −7.1 | 0.8 | 103 | 8.7 | 0.9 | |||||
| Desipramine | 5 | 86 | −9.3 | 0.6 | 0.531 | 88 | −5.7 | 0.7 | 0.065 | 84 | −10.1 | 0.9 | 0.916 | 82 | −8.5 | 0.9 | 0.024 | ||||
| Fluoxetine | 5 | 100 | −9.8 | 0.5 | 97 | −7.5 | 0.7 | 95 | −10.3 | 0.9 | 91 | −11.1 | 0.8 | ||||||||
| Desipramine | 6 | 82 | −10.0 | 0.6 | 0.159 | 81 | −5.8 | 0.7 | 0.016 | 77 | −9.7 | 0.9 | 0.045 | 75 | −8.6 | 0.9 | 0.006 | ||||
| Fluoxetine | 6 | 95 | −11.1 | 0.5 | 89 | −8.2 | 0.7 | 88 | −12.1 | 0.8 | 86 | −11.9 | 0.8 | ||||||||
| Desipramine | 7 | 75 | −10.5 | 0.6 | <.001 | 74 | −6.1 | 0.7 | <.001 | 75 | −11.8 | 0.8 | 0.035 | 71 | −10.2 | 0.9 | 0.005 | ||||
| Fluoxetine | 7 | 93 | −13.2 | 0.5 | 88 | −9.7 | 0.7 | 83 | −14.2 | 0.8 | 86 | −13.7 | 0.9 | ||||||||
| Desipramine | 8 | 74 | −12.0 | 0.6 | 0.001 | 72 | −7.4 | 0.7 | <.001 | 71 | −12.2 | 0.9 | 0.014 | 72 | −11.1 | 0.9 | 0.064 | 64 | 18.5 | 1.0 | 0.002 |
| Fluoxetine | 8 | 92 | −14.6 | 0.5 | 90 | −10.6 | 0.6 | 85 | −15.3 | 0.9 | 81 | −13.4 | 0.9 | 83 | 22.9 | 0.9 | |||||
Least-squares mean change from baseline was calculated after adjustment for gender, age and baseline score. SE=Standard error.
Mixed-effects model for repeated measures (MMRM) included all available observations on all subjects at all time points.
Table 2 presents the clinical response and remission rates after adjusting for age, sex, and baseline HAM-D score. At the endpoint, the fluoxetine group had 1.3–1.4-fold significantly higher response or remission rate. Survival analysis revealed that fluoxetine produced a faster response (HR=2.01, 95%CI=1.40–3.13; χ2=13.08, df=1, P<0.001) or remission (HR=1.57, 95%CI=0.98–2.51; χ2=3.54, df=1, P=0.060; Fig. 2S).
Table 2.
Clinical Remission and Response Rates in Mexican-American Patients with Major Depression Who Received Desipramine or Fluoxetinea
| Treatment | Week | N | Remission | Response | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Rate % | RR | 95% CI | P | Rate % | RR | 95% CI | P | |||
|
|
||||||||||
| Longitudinal analysis of observed datab | ||||||||||
| Desipramine | 1 | 111 | 0.9 | Reference | 1.7 | Reference | ||||
| Fluoxetine | 1 | 120 | 1.7 | 1.90 | 0.17–21.08 | 0.600 | 3.3 | 1.87 | 0.35–10.10 | 0.466 |
| Desipramine | 2 | 105 | 3.7 | Reference | 11.9 | Reference | ||||
| Fluoxetine | 2 | 113 | 3.5 | 0.95 | 0.24–3.72 | 0.936 | 9.4 | 0.79 | 0.37–1.68 | 0.534 |
| Desipramine | 3 | 99 | 3.9 | Reference | 15.2 | Reference | ||||
| Fluoxetine | 3 | 110 | 5.5 | 1.40 | 0.40–4.84 | 0.598 | 14.9 | 0.98 | 0.52–1.83 | 0.945 |
| Desipramine | 4 | 97 | 4.4 | Reference | 21.6 | Reference | ||||
| Fluoxetine | 4 | 106 | 10.2 | 2.33 | 0.81–6.69 | 0.117 | 30.7 | 1.42 | 0.90–2.26 | 0.135 |
| Desipramine | 5 | 86 | 11.9 | Reference | 36.0 | Reference | ||||
| Fluoxetine | 5 | 100 | 19.9 | 1.67 | 0.84–3.33 | 0.143 | 50.2 | 1.39 | 1.00–1.94 | 0.049 |
| Desipramine | 6 | 82 | 21.4 | Reference | 44.0 | Reference | ||||
| Fluoxetine | 6 | 95 | 25.0 | 1.17 | 0.68–2.00 | 0.575 | 59.4 | 1.35 | 1.02–1.79 | 0.036 |
| Desipramine | 7 | 75 | 31.3 | Reference | 59.0 | Reference | ||||
| Fluoxetine | 7 | 93 | 51.0 | 1.63 | 1.11–2.39 | 0.012 | 75.1 | 1.27 | 1.04–1.56 | 0.020 |
| Desipramine | 8 | 74 | 43.1 | Reference | 60.7 | Reference | ||||
| Fluoxetine | 8 | 92 | 59.1 | 1.37 | 1.01–1.87 | 0.046 | 79.7 | 1.31 | 1.09–1.59 | 0.005 |
|
|
||||||||||
| HR | 95% CI | P | HR | 95% CI | P | |||||
|
|
||||||||||
| Survival analysis of observed datac | ||||||||||
| Desipramine | time to event | 112 | Reference | Reference | ||||||
| Fluoxetine | time to event | 120 | 1.57 | 0.98–2.51 | 0.060 | 2.01 | 1.40–3.13 | <0.001 | ||
Rates were estimated after adjustment for gender, age and baseline score; RR=Relative Risk; HR=Hazard Ratio.
Generalized estimating equation (GEE) model for repeated measures analysis included all available observations.
Cox regression model for survival analysis included all subjects with event (response or remission) or right censoring time.
Secondary Outcomes – HAM-A, BDI, CES-D and GAS Scores
The results of MMRM revealed a significant between-subject effect of treatment on the difference in mean changes in HAM-A, BDI and GAS scores (Table 1). The fluoxetine group showed better improvement in HAM-A, BDI, CESD, and GAS scores than the desipramine group. Significant interactive effects of treatment and time were found BDI (F=2.09, df=7,199, P=0.005) and GAS scores (F=4.92, f=7,135, P=0.028). The fluoxetine group had smaller BDI drop in the first 4 weeks and greater reduction in the last 3 weeks (weeks 6–8). CESD scores showed treatment effect (F=5.30, df=1,211, P=0.022) at weeks 5–7, but not at week 8. Baseline scores showed significant effects on specific score changes (P<0.05). No gender effect was found, and age showed a significant effect only on GAS increase (MMRM: F=4.59, df=1,190, P=0.033 and LOCF: F=10.39, df=1, 198, P=0.002).
Adverse Drug Reactions (ADRs)
Overall, ADR data indicated that 78% (182/232) of subjects receiving treatment had one or more side effects (Table 2S). There were no differences in the overall occurrence of ADRs between treatment groups (90/112 in desipramine vs 92/120 in fluoxetine; χ2 =0.47, df =1, P=0.495). However, compared by category, desipramine treated patients showed significant higher occurrence of anticholinogeric ADRs (χ2 =4.96, df =1, P=0.026), cardiovascular ADRs (χ2 =7.22, df=1, P=0.007), perspiration, tingling/parasthesias, blurred vision, orthostatic by pulse, and palpitations (χ2, P<0.05).
Exome-Wide Association Analysis
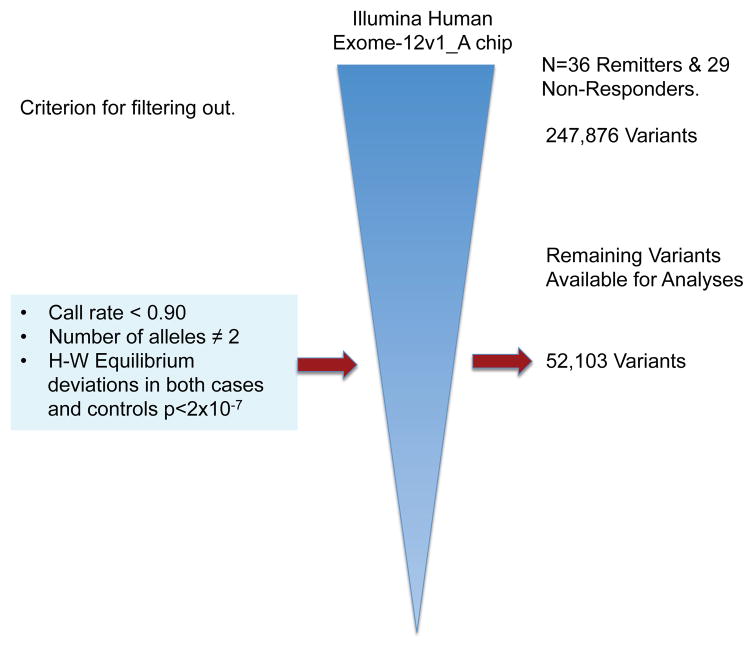
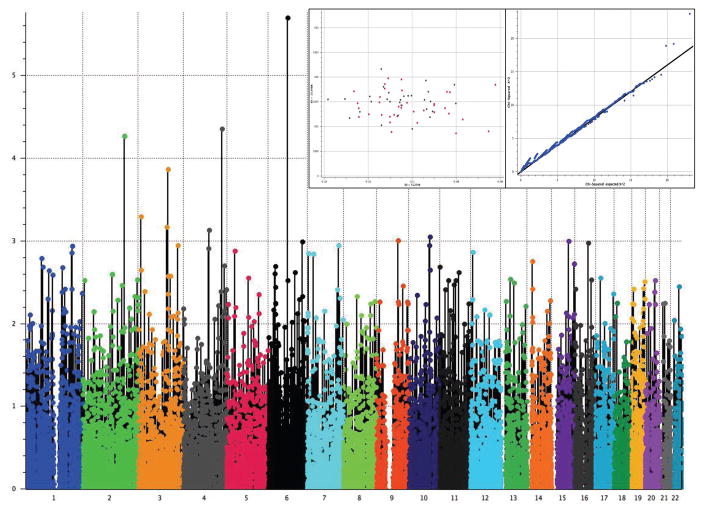
After filtering out markers not meeting either quality control criteria or variability requirements, 52,103 variants remained for further analyses (Fig. 1). The estimation of the Fst coefficient reported a value of 0.001 that according to Price et al. (36), may be corrected by PCA using 50,000 SNPs. After PCA correction, we found that a single SNP marker located in chromosome (Chr) 6 (exm-rs1321744) achieved exome-wide significance (P=1.98 X10−6 and FDR=0.05, Fig. 2 & Table 3). This marker is located in a methylated DNA immunoprecipitation sequencing site (MeDIP-seq raw data) obtained using postmortem human frontal cortex gray matter (UCSC Genome Browser, http://genome.ucsc.edu), which suggest its involvement in epigenetic regulation of neuronal gene expression. Interestingly, other top markers, which did not attain exome-wide significance, are also located in methylated DNA sites (Table 3). Linkage disequilibrium (LD) block harboring exm-rs1321744 does not harbor any gene; however, there are three genes surrounding exm-rs1321744: TBX18, NT5E, and SNX14.
Fig 1. Process used to filter out SNP Exomic Variants Genotyped with the Illumina® HumanExome BeadChip-12v1_A in a Mexican-American Cohort of Patients with Major Depressive Disorder (MDD) under Antidepressive Treatment to Evaluate Pharmacogenetic Response As Remitters (n=36) and Non-Responders (n=29).
From 247,876 SNPs 195,773 were discarded because they were either monoallelic, had more than two alleles, had a call rate lower than 90%, or their genotype proportions deviated from the expected ones as defined by the Hardy-Weinberg equilibrium theorem in both cases (Remitters) and controls (Non-responders), P<2 × 10−7. The remaining 52,103 SNPs were used for Exome-Wide Association Analysis.
Fig 2. Manhattan Plot to Evaluate Pharmacogenetic Response As Remitters (n=36) and Non-responders (n=29).
Genotype model (additive) after correcting stratification by principal component analysis (PCA) was used. A unique significant peak at chromosome 6 was found significant after correction for multiple comparisons by FDR. In the inset, the principal component analysis shows absence of stratification between remitters and non-responders (right box), and a Q-Q plot depicts the fitting of χ2 against the χ2 expected distribution (left box).
Table 3.
Exome-Wide Association Analysis results
| Marker | Chr. | Position | Ref/Alt Allele |
Major Allele |
Fisher's Exact P |
Fisher's Exact FDR |
Allele Freq. (Cases) |
Allele Freq. (Controls) |
Classification | Gene(s) | HGVS Coding |
HGVS Protein |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| exm-rs1321744a,b | 6 | 85,891,744 | T/C | C | 1.98E-06 | 0.05 | 0.19 | 0.45 | Intergenic | |||
| exm-rs16867321b | 2 | 181,362,379 | C/T | C | 5.37E-05 | 0.42 | 0.58 | 0.28 | Intergenic | |||
| exm433050 | 4 | 169,083,694 | A/C | A | 4.41E-05 | 0.51 | 0.50 | 0.38 | Coding Nonsynonymous | ANXA10 | c.211A>C | p.Met71Leu |
| exm-rs6583826b | 10 | 94,347,830 | G/A | A | 8.85E-04 | 1.00 | 0.57 | 0.24 | Intergenic | |||
| exm2259477b | 9 | 93,072,009 | C/T | C | 9.88E-04 | 1.00 | 0.57 | 0.28 | Intergenic | |||
| exm2265531b | 3 | 174,131,106 | A/G | A | 1.13E-03 | 1.00 | 0.39 | 0.43 | Intergenic | |||
| exm660951b | 7 | 138,455,988 | A/G | G | 1.13E-03 | 1.00 | 0.39 | 0.43 | Coding | ATP6V0A4 | c.5C>T | p.Ala2Val |
| exm345346b | 3 | 124,731,689 | T/A | T | 6.76E-04 | 1.00 | 0.31 | 0.33 | Coding Nonsynonymous | HEG1 | c.2734A>T | p.Thr912Ser |
| exm2270591 | 7 | 27,223,563 | G/T | T | 1.43E-03 | 1.00 | 0.49 | 0.48 | Intronic | HOXA11 | ||
| exm-rs3729931 | 3 | 12,626,516 | G/A | C | 5.04E-04 | 1.00 | 0.54 | 0.26 | Intronic | RAF1 |
Exome-wide significance.
MeDIP-seq Raw Signal indicates that this is a DNA methylation site (UCSC Genome Browser, http://genome.ucsc.edu/).
Data from postmortem human frontal cortex gray matter of a 57 year-old male. This was done to investigate the role of intragenic, tissue-specific CpG island methylation plays in controlling gene expression. Hardy-Weinberg Equilibrium exact P-values were greater than 0.05.
ARPA
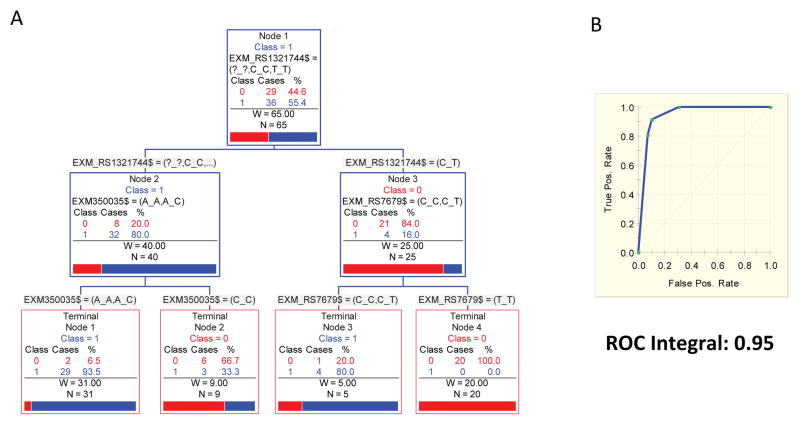
The ARPA generated a classification tree where the five main splitters variants were: exm-rs1321744, exm-rs350035, and exm-rs7679 (Fig. 3A). Both, left and right terminal nodes have a conspicuous power to discriminate between remitters (category 1) and non-responder (category 0). The ROC (Receiver Operating Characteristic) integral to evaluate the predictive accuracy of this derived model is 0.9454, which described the high sensitivity and specificity of the tree to discriminate between remitters and non-responders (Fig. 3B). As presented in Supplementary Figure 3S, the TreeNet and Random Forest analyses shows that after 150 simulated trees, the classification error is lower than 10% for both the training and the testing sample, and the ROC curve converge to values higher than 90% for both the training and the testing samples. Furthermore, the Random Forest and the TreeNet analyses, which disclosed the same set of variables as the main predictors of MDD remission and also discard the possibility of model over fitting described for the CART analyses (Fig. 3S).
Fig 3. Results of Advance Recursive Partitioning (tree-based) Approach (ARPA): A.
Reconstructed classificatory tree showing those variants involved in the process of branching. B. The ROC (Receiver Operating Characteristic) integral = 0.9454.
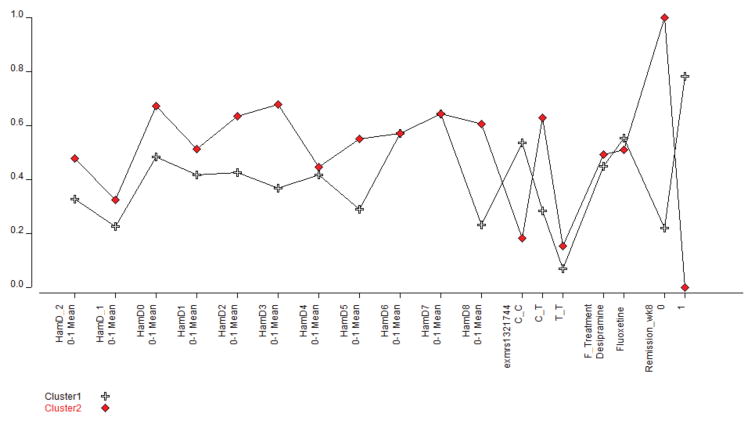
Finally we applied a categorical approach to link the set of genotypes of the associated marker (exm-rs1321744) using Latent Class Analysis that shows how the patterns of remission as depicted by the Hamilton scores, split the set of patients in two significantly different clusters associated to the three genotypes CC, CT and TT (Figure 4). Cluster 1, constituted by remitters and depicted by crosses is associated to the CC genotype, exm-rs1321744. In red, cluster 2 constituted by non-responders is associated to the CT genotype, exm-rs1321744. Treatment with either desipramine or fluoxetine did not produce any significant additional splitting of these two clusters.
Fig 4. Patterns of Hamilton Depression (HAM-D) scores split the set of patients in two significantly different clusters associated to the exm-rs1321744 genotypes.
Cluster 1 depicted by crosses is constituted by remitters and associated to the CC genotype. Cluster 2 in red is constituted by non-responders and associated with CT genotype. Treatment with either desipramine or fluoxetine did not produce any significant additional splitting of these two clusters.
DISCUSSION
In this single site double-blind antidepressant trial of predominantly first generation Mexican-American patients, we found that both fluoxetine and desipramine were effective; however, fluoxetine-treated patients had significantly better HAM-D, HAM-A, BDI, and GAS scores than desipramine-treated patients across all analytical approaches. The advantage of fluoxetine over desipramine was also evidenced by a faster response time in the survival analysis, lower occurrence of anticholinogeric and cardiovascular ADRs, and a smaller drop out rate. These clinical outcomes contrast with previous studies showing similarities in efficacy between SSRIs and TCAs (37) and no difference in the response rate to treatment in intention-to-treat analysis between SSRIs and TCAs (38).
Our results are in concordance with data showing higher dropped out rates in TCA-treated subjects due to either lack of efficacy or adverse reactions (30.0 vs. 24.7%)(38). TCAs have historically been found to have moderate to severe ADRs (39); SSRIs are associated with milder ADRs that may diminish as treatment continues and also exhibit lower toxicity and lower lethality when taken in an overdose situation (40). Our overall comparable tolerability between desipramine and fluoxetine could be related to the close monitoring of our patients, which improve TCAs outcomes in depression (41). Our data supported a higher dropout rate and occurrence of anticholinergic and cardiovascular side effects in desipramine-treated patients. Previous studies have indicated that Africans are more sensitive to TCAs (42, 43), and populations with higher African genetic admixture rates, such as Puerto Rican Americans, have increased ADRs and dropout rates with TCAs (44). This could also have contributed to an increased sensitivity to TCAs in our Hispanic subgroup with an African genetic admixture.
To our knowledge, this is the first double blind, randomized, placebo lead-in trial conducted in the U.S. comparing the clinical efficacy and tolerability of antidepressants in depressed individuals of Mexican descent. The reasons for the lack of prospective randomized clinical trials in ethnic minorities are multiple and include difficulties in recruitment and retention of appropriate subjects, and significantly less adherent to antidepressant therapy during the initial one hundred day period (45). Considerable resources were needed for our study. Recruitment was challenging despite the fact that over 8 million persons of Mexican descent live in California (1), including 3 million in Los Angeles County. We conducted more then 4300 telephone interviews and scheduled 1223 screening visits in order to obtain 166 study completers. A World Health Organization study showed that, treatment with TCAs was more cost-effective than those with SSRIs in fourteen different populations (including Mexico and the USA), particularly in lower-income sub regions (46). However, in our study fluoxetine treatment produced a better and faster response than desipramine in first generation Mexican-Americans with mild to moderate MDD, suggesting that fluoxetine may constitute a better drug of first choice for patients of Mexican descent.
Our whole exome genotyping approach identified one functional intergenic SNP in Chr 6 with exome-wide association with remission (P=1.98 X10−6, FDR=0.05), which is remarkable given the small number of remitters (n=36) and non-responders (n=29). Mexicans are an admixture of Europeans, Native Americans, and Africans but a considerable proportion of their ancestry is Caucasian (47). For European descendent populations, P≤7.2X10−8 is regarded as compellingly significant for a genome-wide effect (48) but this assumption is appropriate for hypotheses that are tested on a genome scale (49). Our results strongly support the involvement of common functional variants in antidepressant drug response, specifically of brain methylation sites. Not much is known about the function of the intergenic exm-rs1321744, but growing clinical and pre-clinical evidence has implicated brain epigenetic changes in stress, depression, and antidepressant action (14–18); however, that body of work has focused mainly on the hypothalamic-pituitary-adrenal axis (HPA) regulation.
Even though linkage disequilibrium (LD) block harboring exm-rs1321744 does not harbor any gene, there are three genes surrounding the significant associated peak namely: TBX18, NT5E, and SNX14. TBX18 encodes a member of an evolutionary conserved family of transcription factors that plays a crucial role in embryonic development. NT5E encodes a plasma protein membrane that catalyzes the conversion of extracellular nucleotides to membrane-permeable nucleosides. SNX14 encodes a member of the sorting nexin family that is involved in intracellular trafficking. The encoded protein also contains a regulator of G protein signaling domain that act as GTPase activating proteins for G alpha subunits of heterotrimeric G proteins.
Our ARPA results suggest that the phenotype for antidepressant response is polygenic, as tree analysis showed that 3 common functional SNPs could predict remitter/non-responder status with 94% accuracy in our population. The top main splitter exm-rs1321744 was discussed above but, intriguingly, the other main splitter variants (exm-rs7679 and exm-rs350035) are located in regions relevant to lipoprotein function; rs7679 (in the PCIF1 gene, Chr20) has genome-wide association with blood lipoprotein concentrations (50), and intergenic exm-rs350035 in Chr5 is 10 nucleotides away from rs351629, a genome-wide SNP associated with triglycerides phenotype (51) (source DBGap).
Limitations of our study include:
The genotyping was only done on 65 subjects - out of 166 who completed 8 weeks of treatment, out of 232 ITT subjects. We used Student-t test to evaluate differences between completers who were genotyped and completers without genotyping. We found no differences in their week 8 HAM-D scores Table 3S (online supplementary material). These results support our findings against the presence of any ascertainment bias given that they show that the sample is representative of the whole set.
Blood levels were not included as co-variates, as they were randomly collected at different times of the day. Due to logistics, some patients could only come for follow-ups in the evening.
The absence of placebo is another important limitation. However, as the efficacies of desipramine or fluoxetine were well known when we designed our study, it would be ethically problematic to justify a placebo arm. To minimize placebo response in our study, patients received placebo in the first week of their treatment. Placebo was given single-blinded; patients knew that they were going to receive one week of placebo at some point in their treatment course, but staff knew that the placebo week was in first week of treatment. In our study, we determined a priori that placebo response was a reason to remove participants from this study. Therefore, we believe that in our study SNPs are likely to predict response to antidepressant treatment and not improvement.
Our study population has following distinctive characteristics: 1) The phenotype of drug response was evaluated with great detail in a specific subgroup of Hispanics in a single site by the same bilingual clinical research team and 2) A cohort of predominantly first generation Mexican-Americans, which standardized the stress exposure level to a significant and chronic one related to immigration factors (52, 53). Further studies are required to confirm these findings and validate them in other ethnic populations.
Supplementary Material
Fig 1S. Flowchart of Study Participants, Random Assignment and Dropouts in a Clinical Trial of Desipramine and Fluoxetine Treatment for Mexican-Americans with Major Depressive Disorder.
Fig 2S. Survival Curve Showing Time to Remission or Response for Depressed Mexican-American Patients Receiving Desipramine (N=112) or Fluoxetine (N=120).
Fig 3S. A. Classification errors curves obtained by the TreeNet analyses for the training and the test samples. After 150 trees, classification error rates fell down than 10% and remain very stables in both samples. B. ROC curves obtained for 200 trees showing that ROC integrals for the training and the test samples are higher than 0.90 after the analysis of 150 trees, and they remain very stable, replicating what was found for the classification error rates and from the CART analyses. C. Importance of variables for prediction as defined by Random Forest. These variables replicate what was found by the Exome-Wide Association and CART analyses that found the variant exm-rs1321744 as the one with the highest importance in predicting remission of MDD symptoms.
Acknowledgments
We are grateful for the contributions of Fiona O’Kirwan, Israel Alvarado, Rita Jepson, Lorraine Garcia-Teague, Patricia Reyes, Isabel Rodriguez, Gabriela Marquez, and the UCLA GCRC staff.
Funding sources: NIH grants GM61394, RR017365, MH062777, RR000865, RR16996, and DK063240, German-Australian Institute of Translational Medicine, and institutional funds from the Australian National University, Flinders University and the South Australian Health and Medical Research Institute.
Footnotes
Disclosures: The authors declare no competing interest and no income from pharmaceutical companies; an intellectual property application has been prepared to include the pharmacogenetics findings of this work.
Clinical trial registration information: This study was approved by the University of California, Los Angeles, University of Miami Institutional Review Boards, and the Australian National University Human Ethics Committee, and it has been registered in the public database clinicaltrials.gov (NCT00265291).
References
- 1.Bureau US. The Hispanic Population in the United States (2001) Washington, DC: U.S. Census Bureau; Mar, 2000. [Google Scholar]
- 2.Rogler LH, Malgady RG, Rodriguez O, Rogler LH, Malgady RG, Rodriguez O. Hispanics and mental health: a framework for research. 1989. [Google Scholar]
- 3.Wong ML, Licinio J. Research and treatment approaches to depression. Nat Rev Neurosci. 2001;2:343–51. doi: 10.1038/35072566. [DOI] [PubMed] [Google Scholar]
- 4.Vega WA, Kolody B, Aguilar-Gaxiola S, Alderete E, Catalano R, Caraveo-Anduaga J. Lifetime prevalence of DSM-III-R psychiatric disorders among urban and rural Mexican Americans in California. Arch Gen Psychiatry. 1998;55:771–8. doi: 10.1001/archpsyc.55.9.771. [DOI] [PubMed] [Google Scholar]
- 5.Mendoza R, Smith MW. In: The Hispanic Response. Ruiz P, editor. Washington, D.C: American Psychiatric Press; 1982. pp. 55–89. [Google Scholar]
- 6.Marcos LR, Cancro R. Pharmacotherapy of Hispanic depressed patients: clinical observations. Am J Psychother. 1982;36:505–12. doi: 10.1176/appi.psychotherapy.1982.36.4.505. [DOI] [PubMed] [Google Scholar]
- 7.Sanchez-Lacay JA, Lewis-Fernandez R, Goetz D, Blanco C, Salman E, Davies S, et al. Open trial of nefazodone among Hispanics with major depression: efficacy, tolerability, and adherence issues. Depress Anxiety. 2001;13:118–24. doi: 10.1002/da.1027. [DOI] [PubMed] [Google Scholar]
- 8.Alonso M, Val E, Rapaport MH. An open-label study of SSRI treatment in depressed hispanic and non-Hispanic women. J Clin Psychiatry. 1997;58:31. doi: 10.4088/jcp.v58n0106c. [DOI] [PubMed] [Google Scholar]
- 9.Currier MB, Molina G, Kato M. Citalopram treatment of major depressive disorder in Hispanic HIV and AIDS patients: a prospective study. Psychosomatics. 2004;45:210–6. doi: 10.1176/appi.psy.45.3.210. [DOI] [PubMed] [Google Scholar]
- 10.Fabbri C, Di Girolamo G, Serretti A. Pharmacogenetics of antidepressant drugs: An update after almost 20 years of research. Am J Med Genet B Neuropsychiatr Genet. 2013;162:487–520. doi: 10.1002/ajmg.b.32184. [DOI] [PubMed] [Google Scholar]
- 11.Niitsu T, Fabbri C, Bentini F, Serretti A. Pharmacogenetics in major depression: A comprehensive meta-analysis. Prog Neuropsychopharmacol Biol Psychiatry. 2013;45:183–94. doi: 10.1016/j.pnpbp.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 12.GENDEP I, MARS I, STAR*D I. Common genetic variation and antidepressant efficacy in major depressive disorder: a meta-analysis of three genome-wide pharmacogenetic studies. Am J Psychiatry. 2013;170:207–17. doi: 10.1176/appi.ajp.2012.12020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Licinio J, Dong C, Wong ML. Novel sequence variations in the brain-derived neurotrophic factor gene and association with major depression and antidepressant treatment response. Arch Gen Psychiatry. 2009;66:488–97. doi: 10.1001/archgenpsychiatry.2009.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott E, Ezra-Nevo G, Regev L, Neufeld-Cohen A, Chen A. Resilience to social stress coincides with functional DNA methylation of the Crf gene in adult mice. Nat Neurosci. 2010;13:1351–3. doi: 10.1038/nn.2642. [DOI] [PubMed] [Google Scholar]
- 15.Zimmermann N, Zschocke J, Perisic T, Yu S, Holsboer F, Rein T. Antidepressants inhibit DNA methyltransferase 1 through reducing G9a levels. Biochem J. 2012;448:93–102. doi: 10.1042/BJ20120674. [DOI] [PubMed] [Google Scholar]
- 16.Menke A, Klengel T, Binder EB. Epigenetics, depression and antidepressant treatment. Curr Pharm Des. 2012;18:5879–89. doi: 10.2174/138161212803523590. [DOI] [PubMed] [Google Scholar]
- 17.Massart R, Mongeau R, Lanfumey L. Beyond the monoaminergic hypothesis: neuroplasticity and epigenetic changes in a transgenic mouse model of depression. Philos Trans R Soc Lond B Biol Sci. 2012;367:2485–94. doi: 10.1098/rstb.2012.0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vialou V, Feng J, Robison AJ, Nestler EJ. Epigenetic mechanisms of depression and antidepressant action. Annu Rev Pharmacol Toxicol. 2013;53:59–87. doi: 10.1146/annurev-pharmtox-010611-134540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baldwin D, Broich K, Fritze J, Kasper S, Westenberg H, Moller HJ. Placebo-controlled studies in depression: necessary, ethical and feasible. Eur Arch Psychiatry Clin Neurosci. 2003;253:22–8. doi: 10.1007/s00406-003-0400-2. [DOI] [PubMed] [Google Scholar]
- 20.Kim SY, Holloway RG. Burdens and benefits of placebos in antidepressant clinical trials: a decision and cost-effectiveness analysis. Am J Psychiatry. 2003;160:1272–6. doi: 10.1176/appi.ajp.160.7.1272. [DOI] [PubMed] [Google Scholar]
- 21.Hazuda HP, Comeaux PJ, Stern MP, Haffner SM, Eifler CW, Rosenthal M. A comparison of three indicators for identifying Mexican Americans in epidemiologic research. Methodological findings from the San Antonio Heart Study. Am J Epidemiol. 1986;123:96–112. doi: 10.1093/oxfordjournals.aje.a114228. [DOI] [PubMed] [Google Scholar]
- 22.Diagnostic and Statistical Manual of Mental Disorders. Washington D.C: American Psychiatric Association; 1994. [Google Scholar]
- 23.Hamilton M. A rating scale for depression. J Neurol Neurosurg Psychiatry. 1960;23:56–62. doi: 10.1136/jnnp.23.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Licinio J, O'Kirwan F, Irizarry K, Merriman B, Thakur S, Jepson R, et al. Association of a corticotropin-releasing hormone receptor 1 haplotype and antidepressant treatment response in Mexican-Americans. Mol Psychiatry. 2004;9:1075–82. doi: 10.1038/sj.mp.4001587. [DOI] [PubMed] [Google Scholar]
- 25.First M, Spitzer RL, Gibbom M, Williams JB. Structured Clinical Interview for Axis I DSM-IV Disorders-Patient Edition (SCID-I/P, version 2) New York, NY: Biometrics Research Department, NY State Psychiatric Institute; 1994. [Google Scholar]
- 26.Hamilton M. The assessment of anxiety states by rating. Br J Med Psychol. 1959;32:50–5. doi: 10.1111/j.2044-8341.1959.tb00467.x. [DOI] [PubMed] [Google Scholar]
- 27.Endicott J, Spitzer RL, Fleiss JL, Cohen J. The global assessment scale. A procedure for measuring overall severity of psychiatric disturbance. Arch Gen Psychiatry. 1976;33:766–71. doi: 10.1001/archpsyc.1976.01770060086012. [DOI] [PubMed] [Google Scholar]
- 28.Beck AT, Ward CH, Mendelson M, Mock J, Erbaugh J. An inventory for measuring depression. Arch Gen Psychiatry. 1961;4:561–71. doi: 10.1001/archpsyc.1961.01710120031004. [DOI] [PubMed] [Google Scholar]
- 29.Roberts RE. Reliability of the CES-D Scale in different ethnic contexts. Psychiatry research. 1980;2:125–34. doi: 10.1016/0165-1781(80)90069-4. [DOI] [PubMed] [Google Scholar]
- 30.Zou G. A modified poisson regression approach to prospective studies with binary data. Am J Epidemiol. 2004;159:702–6. doi: 10.1093/aje/kwh090. [DOI] [PubMed] [Google Scholar]
- 31.Rao DC. CAT scans, PET scans, and genomic scans. Genet Epidemiol. 1998;15:1–18. doi: 10.1002/(SICI)1098-2272(1998)15:1<1::AID-GEPI1>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 32.Wong ML, Dong C, Andreev V, Arcos-Burgos M, Licinio J. Prediction of susceptibility to major depression by a model of interactions of multiple functional genetic variants and environmental factors. Mol Psychiatry. 2012:624–33. doi: 10.1038/mp.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breiman L, Friedman JH, Olshen RA, Stone CH. Classification and Regression Trees. Belmont, CA: Wadsworth International Group, Inc; 1984. [Google Scholar]
- 34.Breiman L. Random Forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 35.Friedman JH. Greedy Function Approximation: a Gradient Boosting Machine. Ann Stat. 2001;29:1189–232. [Google Scholar]
- 36.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 37.MacGillivray S, Arroll B, Hatcher S, Ogston S, Reid I, Sullivan F, et al. Efficacy and tolerability of selective serotonin reuptake inhibitors compared with tricyclic antidepressants in depression treated in primary care: systematic review and meta-analysis. BMJ (Clinical research ed. 2003 May 10;326(7397):1014. doi: 10.1136/bmj.326.7397.1014. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steffens DC, Krishnan KR, Helms MJ. Are SSRIs better than TCAs? Comparison of SSRIs and TCAs: a meta-analysis. Depr Anxiety. 1997;6:10–8. doi: 10.1002/(sici)1520-6394(1997)6:1<10::aid-da2>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt LG, Grohmann R, Muller-Oerlinghausen B, Ochsenfahrt H, Schonhofer PS. Adverse drug reactions to first- and second-generation antidepressants: a critical evaluation of drug surveillance data. Br J Psychiatry. 1986;148:38–43. doi: 10.1192/bjp.148.1.38. [DOI] [PubMed] [Google Scholar]
- 40.Peretti S, Judge R, Hindmarch I. Safety and tolerability considerations: tricyclic antidepressants vs. selective serotonin reuptake inhibitors. Acta Psychiatr Scand Suppl. 2000;403:17–25. doi: 10.1111/j.1600-0447.2000.tb10944.x. [DOI] [PubMed] [Google Scholar]
- 41.Katon W, Von Korff M, Lin E, Walker E, Simon GE, Bush T, et al. Collaborative management to achieve treatment guidelines. Impact on depression in primary care. JAMA. 1995;273:1026–31. [PubMed] [Google Scholar]
- 42.Kilonzo GP, Kaaya SF, Rweikiza JK, Kassam M, Moshi G. Determination of appropriate clomipramine dosage among depressed African outpatients in Dar es Salaam, Tanzania. Cent Afric J Med. 1994;40:178–82. [PubMed] [Google Scholar]
- 43.Ziegler VE, Biggs JT. Tricyclic plasma levels. Effect of age, race, sex, and smoking. JAMA. 1977;238:2167–9. doi: 10.1001/jama.238.20.2167. [DOI] [PubMed] [Google Scholar]
- 44.Hanis CL, Hewett-Emmett D, Bertin TK, Schull WJ. Origins of U.S. Hispanics. Implications for diabetes. Diabetes Care. 1991;14:618–27. doi: 10.2337/diacare.14.7.618. [DOI] [PubMed] [Google Scholar]
- 45.Sleath B, Rubin RH, Huston SA. Hispanic ethnicity, physician-patient communication, and antidepressant adherence. Compr Psychiat. 2003;44:198–204. doi: 10.1016/S0010-440X(03)00007-5. [DOI] [PubMed] [Google Scholar]
- 46.Chisholm D, Sanderson K, Ayuso-Mateos JL, Saxena S. Reducing the global burden of depression: population-level analysis of intervention cost-effectiveness in 14 world regions. Br J Psychiatry. 2004;184:393–403. doi: 10.1192/bjp.184.5.393. [DOI] [PubMed] [Google Scholar]
- 47.Manichaikul A, Palmas W, Rodriguez CJ, Peralta CA, Divers J, Guo X, et al. Population structure of Hispanics in the United States: the multi-ethnic study of atherosclerosis. PLoS Genet. 2012;8:e1002640. doi: 10.1371/journal.pgen.1002640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 2008;32:227–34. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bush WS, Moore JH. Chapter 11: Genome-wide association studies. PLoS Comput Biol. 2012;8:e1002822. doi: 10.1371/journal.pcbi.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kathiresan S, Manning AK, Demissie S, D'Agostino RB, Surti A, Guiducci C, et al. A genome-wide association study for blood lipid phenotypes in the Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S17. doi: 10.1186/1471-2350-8-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cuellar I, Bastida E, Braccio SM. Residency in the United States, subjective well-being, and depression in an older Mexican-origin sample. J Aging Health. 2004;16:447–66. doi: 10.1177/0898264304265764. [DOI] [PubMed] [Google Scholar]
- 53.Escobar JI, Hoyos Nervi C, Gara MA. Immigration and mental health: Mexican Americans in the United States. Harv Rev Psychiatry. 2000;8:64–72. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig 1S. Flowchart of Study Participants, Random Assignment and Dropouts in a Clinical Trial of Desipramine and Fluoxetine Treatment for Mexican-Americans with Major Depressive Disorder.
Fig 2S. Survival Curve Showing Time to Remission or Response for Depressed Mexican-American Patients Receiving Desipramine (N=112) or Fluoxetine (N=120).
Fig 3S. A. Classification errors curves obtained by the TreeNet analyses for the training and the test samples. After 150 trees, classification error rates fell down than 10% and remain very stables in both samples. B. ROC curves obtained for 200 trees showing that ROC integrals for the training and the test samples are higher than 0.90 after the analysis of 150 trees, and they remain very stable, replicating what was found for the classification error rates and from the CART analyses. C. Importance of variables for prediction as defined by Random Forest. These variables replicate what was found by the Exome-Wide Association and CART analyses that found the variant exm-rs1321744 as the one with the highest importance in predicting remission of MDD symptoms.