Abstract
WRKY, a plant-specific transcription factor family, plays vital roles in pathogen defense, abiotic stress, and phytohormone signalling. Little is known about the roles and function of WRKY transcription factors in response to rust diseases in wheat. In the present study, three TaWRKY genes encoding complete protein sequences were cloned. They belonged to class II and III WRKY based on the number of WRKY domains and the pattern of zinc finger structures. Twenty-two DNA–protein binding docking complexes predicted stable interactions of WRKY domain with W-box. Quantitative real-time-PCR using wheat near-isogenic lines with or without Lr28 gene revealed differential up- or down-regulation in response to biotic and abiotic stress treatments which could be responsible for their functional divergence in wheat. TaWRKY62 was found to be induced upon treatment with JA, MJ, and SA and reduced after ABA treatments. Maximum induction of six out of seven genes occurred at 48 h post inoculation due to pathogen inoculation. Hence, TaWRKY (49, 50, 52, 55, 57, and 62) can be considered as potential candidate genes for further functional validation as well as for crop improvement programs for stress resistance. The results of the present study will enhance knowledge towards understanding the molecular basis of mode of action of WRKY transcription factor genes in wheat and their role during leaf rust pathogenesis in particular.
Electronic supplementary material
The online version of this article (10.1007/s13205-017-1064-3) contains supplementary material, which is available to authorized users.
Keywords: Wheat, Transcription factor, WRKY, Leaf rust, Abiotic stress
Introduction
The WRKY (single amino acid letters that code for tryptophan, arginine, lysine and tyrosine) transcription factors (TFs) belong to a large superfamily WRKY-GCM1 TFs (Babu et al. 2006; Wei et al. 2012). Members of this family contain approximately 60 amino acids comprising at least one conserved DNA-binding region, designated the WRKY domain that consists of the highly conserved WRKYGQK heptapeptide sequence and a zinc finger-like motif. Based on the number of WRKY domains and the type of zinc finger-like motif, WRKY TFs have been classified into three classes (Eulgem et al. 2000). WRKY proteins with two WRKY domains were classified as class I. Class II WRKY is marked by the presence of one WRKY domain with C2H2 type zinc-finger structure. This class is further divided into subclasses a–e based upon additional amino acid motifs present outside the WRKY domain. The class III WRKY proteins also carry a single domain but differ from class I and II in its altered C2HC zinc finger motif (C–X7–C–X23–H–X–C) (Ulker and Somssich 2004). The WRKY zinc finger motif (C–Xm–C–Xn–H–X–H/C), in class I and subclass IIc members had an m value of four while n ranged from 21 to 23. The subclass IIa, IIb, IId and IIe proteins had an m value of five and n value of either 23 or 24. The subclass IIIa had an altered motif of C–X7–C–X23–H–X–C. The motif in the subclass IIIb proteins are rather variable, with the value of m ranging from six to nine, and n ranged from 23 to 28 (Zhu et al. 2013; Pan and Jiang 2014). WRKY factors have a stereotypic binding preference to a DNA element termed the W-box that comprises of sequences 5′TTGACC/T3′. The binding of WRKY proteins to their W-boxes is a characteristic feature involving biotic and abiotic stress responses. Several studies have revealed crucial roles of WRKY TFs in several biotic as well as different types of abiotic stresses (Okay et al. 2014; Satapathy et al. 2014; Kayum et al. 2015; Ding et al. 2016; He et al. 2016).
Plants have developed diverse defense mechanisms to fight infections and to combat diseases (Glazebrook and Ton 2007). The plant innate immunity is mainly triggered against any pathogen attack and a response is generated in the form of either PTI [pathogen-associated molecular pattern (PAMP)-triggered immunity] or ETI (effector-triggered immunity), driven by plant disease resistance proteins (Chisholm et al. 2006). Both PTI and ETI can generate local as well as systemic acquired resistance (SAR) (Durrant and Dong 2004). Host resistance is governed by hypersensitive response (HR) at the site of invasion involving genetically controlled cell death, hence offering resistance by isolating the pathogen infested cells from rest of undamaged host tissue. The defense response is a complex signalling network which may be triggered by defense signalling molecules. Also, an extensive and complex transcriptional reprogramming generates responses leading towards activation and repression of numerous defense-related genes and TFs like WRKY (Eulgem and Somssich 2007; Pandey and Somssich 2009; Choura et al. 2015). WRKY TFs act as a key regulator of pathogen-triggered alterations in gene expression and are governed by MAPKs (Kuhn et al. 2016). WRKY TFs can act as both activators and repressors, before or after hormone (SA/JA/ET) signalling pathways. Many WRKY genes have been extensively identified and characterised in several plant species including the model plant Arabidopsis (Eulgem et al. 2000), rice (Wu et al. 2005), Brachypodium distachyon (Tripathi et al. 2012), physic nut (Xiong et al. 2013) and cotton (Ding et al. 2015). To date, 74 WRKY members have been identified in Arabidopsis genome (Eulgem et al. 2000), 102 in rice (Wu et al. 2005), 55 in cucumber (Ling et al. 2011) and 105 in poplar (He et al. 2012). It has been proposed that majority of WRKY gene belonging to class I and II emerged before the divergence of monocot and dicot plants, whereas group III genes appeared relatively later (Wu et al. 2005). High levels of microsynteny of WRKY class I DNA sequences among four Gramineae plants B. distachyon, Oryza sativa, Sorghum bicolor and Zea mays revealed the origin, evolution and gene duplication events (Jin et al. 2016).
Bread wheat (Triticum aestivum L.) provides one-fifth of food calories and proteins to the world population (FAO 2016) and its demand is expected to rise by 60% in the developing countries by 2050 (Rosegrant and Agcaoili 2010). The large allohexaploid (2n = 6x = 42) bread wheat genome (~ 16.94 Gb) comprising of three homoeologous A, B and D sub-genomes that originated from related progenitor species, provide significant challenges for molecular and functional genomics-based improvement of wheat (Choulet et al. 2010). The rust diseases have often threatened wheat production worldwide (McIntosh and Pretorius 2011) and mainly leaf rust, caused by Puccinia triticina Eriks., is recognized as a devastating pathogen, accounting for ~ 10% yield loss annually (Dean et al. 2012). The obligate parasitic biotrophic lifestyle of rust fungi establishes a long-term feeding relationship with living host cells by minimizing damage to host tissue. Biotrophic pathogens avoid or suppress plant defense response and take control of host cell organization and metabolism. After penetration of cell wall of living host cells, a specialized structure haustorium is formed that does not breach, but invaginates the host plasma membrane. The pathogen derives nutrients from the living host cell through haustoria and secretes ‘effectors’ and other signalling molecules that mask and manipulate the host immune response (Koeck et al. 2011).
The WRKY TFs have also been studied in wheat with respect to phylogeny, biotic and abiotic stresses (Jiang et al. 2017). Functional analysis of wheat WRKY1 gene exposed to simulated drought and ABA stress condition significantly up-regulated the gene TaWRKY1 that mediates stomata movement and leaf water retention capacity (Ding et al. 2016). Ectopic over-expression of the gene in tobacco along with knockdown of an ABA receptor gene suggested stomatal movement-mediated water regulation activity of TaWRKY1. Forty-eight putative drought-induced WRKY genes were identified from de novo transcriptome sequencing of wheat plants subjected to or without drought stress (He et al. 2016). Of these 48 genes, two genes (TaWRKY1 and TaWRKY33) were found to respond to multiple abiotic stresses. Overexpression of these genes in Arabidopsis activated several stress-related downstream genes. Okay et al. (2014) identified 160 WRKY proteins using in silico approaches; of them 10 WRKY TFs were found to be drought responsive as studied from an RNA-Seq dataset. Seven out of 10 of these TaWRKY genes showed differential relative expression in leaf and root tissues of drought-tolerant and -susceptible wheat cultivars. In one of our earlier studies, TaWRKY1B was functionally characterised (Kumar et al. 2014). Infection with a virulent race of leaf rust pathogen to wheat near-isogenic lines (NILs) carrying Lr28 (resistant) or without Lr28 (susceptible), resulted in 146 and 12-fold increase, respectively, of the TaWRKY1B gene expression indicating a protective role of the gene. In another previous study, 100 WRKY TFs were identified from wheat transcriptomes of which 45 were already known, and the rest 55 were novel (Satapathy et al. 2014). Of the novel TaWRKY genes, 22 were complete, whereas 17 were devoid of initiation/termination codons and 16 lacked the zinc-finger motif, which is essential for binding to W-box. Fifteen of these complete TaWRKY genes belonged to class II, whereas seven belonged to class III WRKY TF.
The present study was undertaken with the aim to characterize some of the WRKY genes identified in our earlier study (Satapathy et al. 2014) both at the molecular and functional level and study their response to induced biotic and abiotic stresses. Here, we describe the alterations of WRKY gene expression in wheat NILs in response to leaf rust as well as to several abiotic stress. A comprehensive network analysis of the selected WRKY genes revealed their functional partners.
Materials and methods
RNA isolation and cDNA conversion
The wheat HD2329 line containing seedling leaf rust resistant gene Lr28 was used to extract RNA for gene characterization studies. Seeds were germinated and grown in a greenhouse at BIT Mesra maintained at optimal conditions (22 °C, 16 h light at 300 lx; and 8 h of darkness). RNA was isolated from 12-day-old whole wheat plants using TRI Reagent (Molecular Research Center, Inc., USA) as recommended by the manufacturer. The integrity of the isolated RNAs was confirmed by electrophoresis on denaturing formaldehyde-1.2% agarose gel. The isolated RNA was treated with 1 unit of deoxyribonuclease I (Thermo Scientific, Lithuania) at 37 °C for 30 min. RNA quantification was performed on a BioPhotometer (Eppendorf AG, Germany). 2 µg of total RNA was used for the first-strand cDNA synthesis using Transcriptor First Strand cDNA Synthesis Kit (Roche, Germany) following the manufacturer’s instructions.
Primer designing, molecular cloning and sequencing
Primers were designed using Primer Express 2.0 software (Applied Biosystems, USA) and checked for specificity by Primer Blast using sequences of T. aestivum WRKY TFs (TaWRKY56, TaWRKY60 and TaWRKY62) whose complete coding sequences are available at NCBI (http://www.ncbi.nlm.nih.gov). DNA amplification reactions were assembled in 20 µl containing 100 ng of cDNA as template, 1 unit of Taq DNA polymerase (Fermentas GmbH, Germany), and 10 pmol of forward and reverse primers each (Online Resource 1). DNA amplifications were performed at 95 °C for 5 min followed by 30 cycles at 94 °C for 45 s, annealing for TaWRKY56 and TaWRKY60 at 60 °C and TaWRKY62 at 50 °C for 1 min, 72 °C for 2 min and a final extension at 72 °C for 30 min. The amplified products were gel purified, cloned in pTZ57R/T (Thermo Scientific, Lithuania) via T/A cloning and plasmids from five independent clones of each transformation were sequenced from both ends using M13 primers. The phylogenetic tree was constructed for TaWRKY proteins using UPGMA hierarchical clustering method and Poisson model was selected. Bootstrap values were calculated from 1000 iterations and based on the phylogenetic tree, representative WRKY domain proteins were classified.
Networking of proteins from cloned WRKY genes
A functional interacting network of proteins was performed for the in silico translated protein sequences obtained from cloned TaWRKY56, 60 and 62 genes using STRING 10 software (Franceschini et al. 2013) with a confidence value of 0.15. Since specific functions of these wheat WRKY genes are not yet assigned and are also not available in STRING database, the networking was performed with structural analogous sequences of Arabidopsis thaliana WRKY genes.
Estimation of synonymous and non-synonymous substitution rates
Synonymous (Ks) and non-synonymous (Ka) substitution rates were calculated for orthologous genes by the PAL2NAL server (http://www.bork.embl.de/pal2nal/) and period of divergence was calculated using the equation T = Ks/(2 × 9.1 × 10−9) × 10−6 million years ago (Mya) (Wang et al. 2015a).
Homology modelling of TaWRKY proteins
The 22 newly identified TaWRKY TF protein sequences in our earlier study (Satapathy et al. 2014) were used for protein modelling. The amino acid sequences were retrieved in fasta format. The total length of 22 TaWRKY proteins is listed in Table 1. The initial 3D model of WRKY domain of T. aestivum was constructed by MODWEB (https://modbase.compbio.ucsf.edu/modweb/) using the alignment between WRKY domain and the template protein. Modweb is used for homology or comparative modelling of protein three-dimensional structure that accepts one or more sequences to calculate models based on the best available template structure at protein data bank (PDB). The templates used along with PDB IDs for each of the 22 TaWRKY proteins are listed in Table 1. It was observed that in 17 TaWRKY proteins (TaWRKY46- 48, TaWRKY53-65 and 67) the template used was Arabidopsis WRKY with PDB ID-2ayd; TaWRKY49 and TaWRKY52 used templates with PDB ID-2dic; TaWRKY51 and 66 proteins matched with templates of PDB ID-iwj2, whereas TaWRKY50 matched with template of PDB ID-4ke2. The Discrete Optimized Protein Energy (DOPE) score was considered in selecting the most reliable model.
Table 1.
Detailed information on 22 modelled TaWRKY proteins
| Sl. no. | TaWRKY protein | Accession no. | Conserved WRKY domain sequence | WRKY group | Target region | Target length | Template PDB ID | Template region | E value | GA341 | z-DOPE |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. | TaWRKY46 | CJ928769 | WRKYGQK | IIc | 1–61 | 154 | 2ayd | 306–366 | 6e−10 | 1 | − 3878.1 |
| 2. | TaWRKY47 | GH726468 | WRKYGQK | IId | 80–140 | 150 | 2ayd | 304–363 | 0 | 1 | − 0.73 |
| 3. | TaWRKY48 | GH721668 | WRKYGQK | IId | 129–189 | 199 | 2ayd | 304–363 | 0 | 1 | − 0.73 |
| 4. | TaWRKY49 | DR741555 | WKKYGQK | IIc | 66–103 | 137 | 2dic | 54–93 | 0.047 | 0.12 | 0.64 |
| 5. | TaWRKY50 | DR741560 | WRKYGQK | IIIb | 83–211 | 269 | 4ke2 | 26–151 | 0 | 0.01 | − 0.13 |
| 6. | TaWRKY51 | CJ540255 | WRKYGQK | IIc | 72–159 | 156 | 1wj2 | 29–86 | 1e26 | 0 | − 3835.9 |
| 7. | TaWRKY52 | DR733802 | WKKYGQK | IIc | 65–102 | 134 | 2dic | 54–93 | 8e−12 | 0.12 | − 4187.5 |
| 8. | TaWRKY53 | DR731588 | WRKYGQK | IIc | 77–131 | 154 | 2ayd | 304–361 | 0 | 1 | − 0.23 |
| 9. | TaWRKY54 | HX150922 | WRKYGKK | IIc | 120–188 | 215 | 2ayd | 294–366 | 0 | 0.99 | − 0.52 |
| 10. | TaWRKY55 | HX153398 | WRKYGKK | IIc | 107–176 | 203 | 2ayd | 294–366 | 0 | 1 | − 0.73 |
| 11. | TaWRKY56 | HX157194 | WRKYGKK | IIc | 103–175 | 220 | 2ayd | 293–367 | 0 | 1 | − 0.01 |
| 12. | TaWRKY57 | CJ583190 | WRKYGQK | IIc | 75–140 | 171 | 2ayd | 294–363 | 0 | 1 | − 0.46 |
| 13 | TaWRKY58 | CK213748 | WRKYGQK | IIIa | 113–177 | 223 | 2ayd | 306–368 | 0.000063 | 0.77 | 0.32 |
| 14. | TaWRKY59 | HX002635 | WRKYGQK | IId | 51–111 | 121 | 2ayd | 304–363 | 0 | 1 | − 0.73 |
| 15. | TaWRKY60 | HX157222 | WRKYGKK | IIc | 76–149 | 194 | 2ayd | 293–367 | 0 | 1 | − 0.14 |
| 16. | TaWRKY61 | CJ642031 | WRKYGQK | IId | 80–140 | 148 | 2ayd | 304–363 | 0 | 1 | − 0.92 |
| 17. | TaWRKY62 | BE404156 | WRKYGEK | IIIb | 42–128 | 174 | 2ayd | 240–325 | 4e−09 | 0.29 | − 3847.7 |
| 18. | TaWRKY63 | DR741654 | WRKYGQK | IIIa | 117–175 | 270 | 2ayd | 306–361 | 0 | 0.78 | − 0.3 |
| 19. | TaWRKY64 | CV778409 | WRKYGQK | IIIa | 20–75 | 205 | 2ayd | 105–160 | 3e−14 | 0.68 | − 5105.5 |
| 20. | TaWRKY65 | DR741684 | WRKYGQK | IIIa | 117–175 | 265 | 2ayd | 306–361 | 0 | 0.78 | − 0.73 |
| 21. | TaWRKY66 | DR731931 | WRKYGQK | IIc | 49–113 | 117 | iwj2 | 399–466 | 0 | 1 | 0.64 |
| 22. | TaWRKY67 | DR741779 | WRKYGQK | IIIa | 20–61 | 270 | 2ayd | 105–146 | 7e−15 | 0.46 | − 3891.4 |
GA341 is a model score derived from statistical potentials and value > 0.7 indicates a reliable model, i.e. > 95% probability of having the correct fold. DOPE score is an atomic distance-dependent statistical score
The more negative values indicate better models
Validation of the models
Each model was validated using the Structural Analysis and Verification Server (http://nihserver.mbi.ucla.edu/SAVES/) (Cheatham et al. 1995). Stereochemical quality and accuracy of the new modelled structure (model3.pdb) were evaluated with PROCHECK (Laskowski et al. 1993) by Ramachandran plot analysis. The root mean square deviation (RMSD) between the backbone atoms of TaWRKY proteins and those of the template was calculated and stereochemical qualities of modelled protein structures were checked by inspection of the Psi/Phi Ramachandran plot using PROCHECK. The compatibility of the model with its sequence was measured by verify-3D graph (Eisenberg et al. 1997).
Modelling of DNA
DNA model for 26 nucleotide sequences with W-box (CTGACC) at the center was prepared using 3D DART web server (http://haddock.chem.uu.nl/dna) available at HADDOCK software web portal (de Vries et al. 2007) to get different conformations of DNA structure. While modelling DNA structure, HADDOCK formatting options at 3D DART were used to obtain a DNA model acceptable at HADDOCK web server for docking. The nucleic acid type and conformation was selected as DNA-duplex and canonical B-form helix, respectively. Parameters set were as follows-the number of repeats 1 and a bend angle of 60°.
Docking of TaWRKY proteins with DNA
The modelled protein and DNA was docked using HADDOCK server’s Easy Interface (de Vries et al. 2007). Intrinsic flexibility of DNA is a major problem which has hampered the development of efficient protein-DNA docking methods. But HADDOCK server has developed protocols for flexible protein-DNA docking. The protein and DNA PDB files were uploaded, active residues for both were mentioned and passive residues were automatically defined around the active residues. The protocol comprised of two sequential docking runs: in the semi-flexible refinement state of first docking run, DNA flexibility is allowed for all nucleotides and the residues of the protein at the predicted interface. The resulting solutions are analysed and subsequently used to generate a library of pre-bent and twisted DNA structures that serve as input for the second docking round. The docked structures were visualized using DISCOVERY STUDIO 4.1 (Accelrys Software Inc. 2011).
Plant material, pathogen inoculation and sample collection
A pair of wheat NILs involving seedling leaf rust resistant Lr28 gene in the background of wheat cultivar HD2329, and thus included HD2329 with Lr28 gene [resistant (Nest Immune infection type: 0–0;)] and HD2329 (susceptible, infection type + 3). The leaf rust resistance gene Lr28 is derived from Aegilops speltoides (Tauschii) and was found to be effective against all pathotypes of the leaf rust pathogen in India (Bipinraj et al. 2011). It is located on long arm of wheat chromosome 4AL. This gene has no undesirable linkage drag and is associated with improved yield and bread-making quality of wheat. Seeds were germinated on autoclaved composite soil (peat, sand, soil, 1:1:1), grown to single leaf stage (approximately 7 days after germination) in growth chamber under ideal conditions of temperature (22 °C), relative humidity (80%), and light periods (16 h day with 300 lx light; 8 h dark periods) at National Phytotron Facility, IARI, New Delhi, India. Puccinia triticina pathotype 77–5 was selected as experimental leaf rust pathogen and its inoculum was obtained from the Indian Institute of Wheat and Barley Research, Regional Station, Flowerdale, Shimla. The P. triticina pathotype 77–5 is the most prominent and devastating race in all parts of the Indian subcontinent, therefore used as a preferred pathogen in this study. The seedling avirulence/virulence formula of this race is P Lr9, Lr18, Lr19, Lr24, Lr25, Lr28, Lr29, Lr32, Lr41, Lr45/p Lr1, Lr2, Lr3, Lr10, Lr11, Lr12, Lr13, Lr14, Lr15, Lr16, Lr17, Lr18, Lr20, Lr22, Lr23, Lr26, Lr27? Lr31, Lr33, Lr34, Lr36, Lr37, Lr42, Lr43, Lr44, Lr46, Lr48, Lr49 (Kaur et al. 2008).
The pathogen inoculum was prepared by mixing urediniospores of P. triticina pathotype 77–5 and talcum powder (ratio 1:1) and applied gently on open moist leaves of HD2329 + Lr28 and HD2329 plants. Separate sets of plants were also mock inoculated with only talcum powder and used as a control. After inoculation, moisting of the growth chamber was performed and plants were placed under a high humidity of 90% for 24 h post inoculation (hpi) in the dark to facilitate infection and later transferred to the normal growth conditions as mentioned earlier. Leaf samples from five plants of each treatment were collected at 0 (just before inoculation), 12, 24, 48, 72 and 168 hpi, immediately dipped in liquid nitrogen and RNA was isolated from individual leaves using TRI-REAGENT.
Abiotic stress treatments
HD2329 + Lr28 seeds were aseptically germinated on Murashige and Skoog (1962) (MS) agar medium in a plant tissue culture room under a 16-h light/8-h dark photoperiod at 23 °C. Stress treatments were given 14 days post germination. Five ml each of 5 µM abscisic acid (ABA), 5 mM methyl jasmonate (MJ), 5 mM Salicylic acid (SA) and 1 mM Jasmonic acid (JA) was sprayed on fully opened leaves. Leaf samples from five plants of each treatment were collected at 0, 1, 2, 4, 8, 12 and 24 h post phytohormone treatment, immediately dipped in liquid nitrogen and RNA isolation was undertaken from each leaf separately using TRI-REAGENT.
Selection of target defense genes and endogenous control
Seven WRKY genes (TaWRKY49, TaWRKY50, TaWRKY52, TaWRKY53, TaWRKY55, TaWRKY57 and TaWRKY62) identified during our earlier study (Satapathy et al. 2014) were selected for expression profiling (Online Resource 2). Wheat Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was selected as an endogenous control to normalize differences in input RNAs and check efficiencies of reverse transcription among the various samples.
Designing gene-specific probes and primers for qRT-PCR studies
Full-length mRNA coding sequences for each selected wheat WRKY gene was downloaded from NCBI (http://www.ncbi.nlm.nih.gov) and entered in the Universal Probe Library (UPL) assay design centre (http://www.universalprobelibrary.com) to design 9 nucleotides long Locked Nucleic Acid (LNA) based short hydrolysis probes as well as forward and reverse primers (Online Resource 2). The probes and primers were obtained from Roche Diagnostics GmbH, Mannheim, Germany.
qRT-PCR reaction conditions for probe-based Real-time PCR
The qRT-PCR of the selected wheat WRKY genes was performed for all the samples collected after pathogen inoculation and phytohormone treatment at designated time-points. Complementary DNAs from all the samples were synthesized using Transcriptor First Strand cDNA Synthesis Kit (Roche, Germany) following the manufacturer’s instructions and were quantified on a BioPhotometer (Eppendorf AG, Germany). The qRT-PCR experiments were performed according to Singh et al. (2012) and Kumar et al. (2014). All qRT-PCR experiments were performed using the gene-specific combination of each probe and primer pairs at an optimized concentration (Online Resource 2). The samples were run with three technical and three biological replicates to minimize variations. Instrument operation, data acquisition and processing were performed employing the Sequence Detection System v1.2.2 software (Applied Biosystems, USA). Fluorescence signals were collected at each polymerization step and a threshold constant (CT) value was calculated from the amplification curve by selecting the optimal ΔRn in the exponential region of the amplification plot (Paolacci et al. 2009). Gene expression levels were computed relative to the expression of the reference gene GAPDH at same time points using the 2−ΔΔCT method. To emphasize the significant difference between resistant (HD2329 + Lr28) mock vs infected interaction and resistant (HD2329 + Lr28) infected vs susceptible (HD2329) infected interaction t test was performed for all the selected genes.
Annotation of orthologous clusters
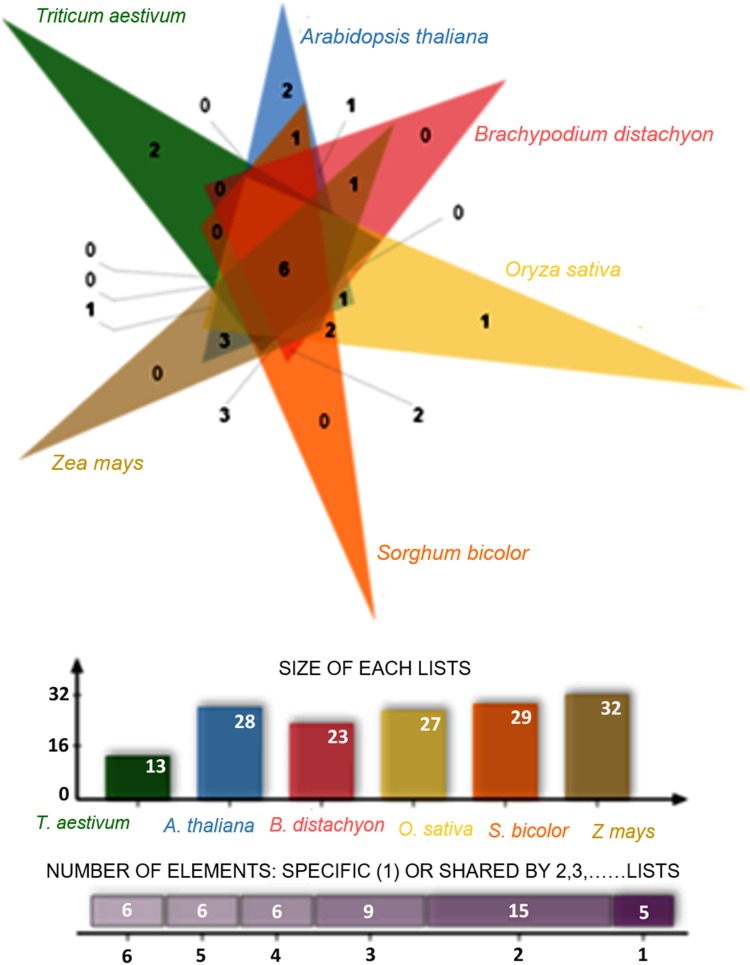
OrthoVenn clustering was performed to identify wheat WRKY gene clusters enriched in five plant species including B. distachyon, O. sativa, S. bicolor, A. thaliana and Z. mays. Pairwise sequence similarities between all input protein sequences were calculated with an E value cut-off of 1e−5. An inflation value (−I) of 1.5 was used to define orthologous cluster structure (Wang et al. 2015b).
Results
Cloning and analysis of cloned sequences
The cloned amplified products were of 655, 599 and 600 bp for TaWRKY56, TaWRKY60 and TaWRKY62, respectively (Online Resource 3 and 4). The VecScreen program at NCBI was used to remove the vector sequences. The forward and reverse-complement orientations were compared to assemble the best possible contigs and analysed using sequence analysis software Sequencher 5.1 (Gene Codes Corporation, MI, USA). The parameters used for contig assembly were Minimum Match Percentage of 99% and Minimum Overlap of 20 bases. The obtained sequences were compared with sequences available at GenBank using BLASTN. BLASTX was also performed to detect conserved domains (Online Resource 5). All three WRKY proteins showed deviations in amino acid compositions of the conserved WRKY domain. KT865877 and KT865878 contain WRKYGKK domain, whereas KT865879 contain WRKYGEK domain. The cloned cDNA sequences for TaWRKY56, 60 and 62 were submitted to NCBI with Accession IDs-KT865877, KT865878 and KT865879, respectively.
Phylogenetic tree of the cloned sequences along with the other newly identified 19 TaWRKY proteins was prepared following UPGMA using MEGA5 software, depicted that KT865877 and KT865878 belong to class IIc and KT865879 belong to class IIIb of the WRKY proteins (Fig. 1). The phylogenetic tree comprised of six clades. The major classes found were class IIc, IId, IIIa and IIIb.
Fig. 1.
Phylogenetic tree constructed from deduced amino acid sequences of cloned TaWRKY genes along with 19 newly identified WRKY proteins using MEGA5.2 software
Protein–protein networking
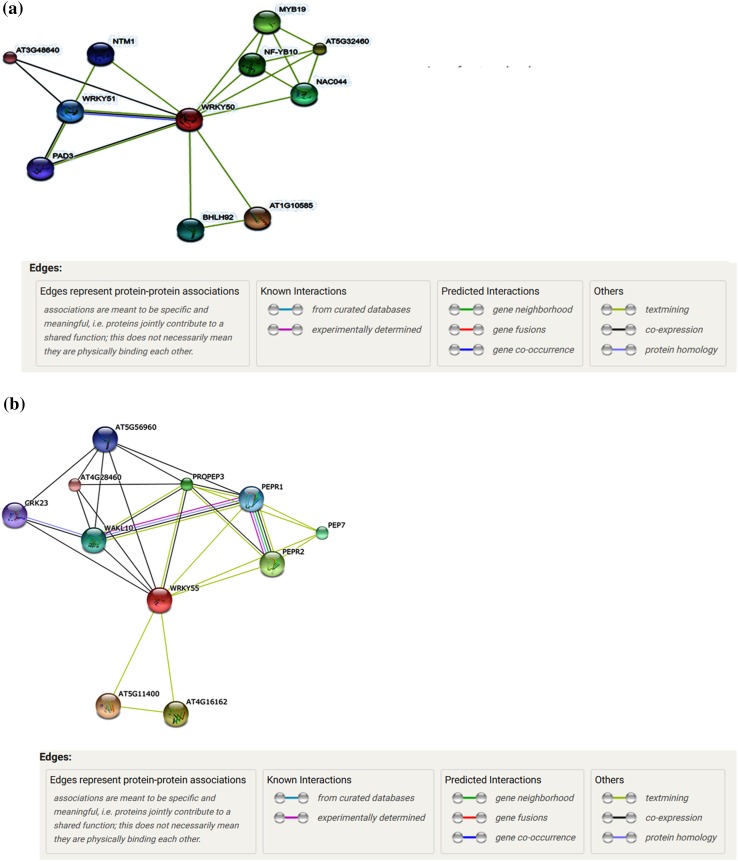
In order to predict functional units that comprised of proteins encoded by different genes, direct and indirect interactions between TaWRKY56, 60 and 62 proteins were derived using search tools in STRING functional association databases (Fig. 2a). Gene annotations for TaWRKY56 and 60 genes showed their involvement in stilbenoid, diarylheptanoid, gingerol and secondary metabolite biosynthesis pathways as well as limonene and pinene degradation pathways. Eleven nodes were obtained with 30 edges and clustering coefficient of 0.856 was obtained indicating significant interactions in the case of TaWRKY56 and 60. Both proteins showed similarity with A. thaliana WRKY50 hence, networking analysis predicted functional partners such as ATG10585 (bHLH protein); AT5G32460 (TF B3 family protein); MYB19 (MYB domain); NFYB10 (component of NF-Y/HAP TF complex); NAC044 (NAC domain protein); WRKY51 (WRKY DNA binding domain [5′-(T)TGAC(CT)-3′]; NTMI (NAC with transmembrane motif 1); AT3G48640 (uncharacterized protein); bHLH92 (TF bHLH contains 247 amino acids) and PAD3cytP45071B15 (multifunctional enzyme involved in biosynthesis of indole derived phytoalexin camalexin). Biological processes include systemic acquired resistance, SA-mediated signalling pathway, MAPK cascade, regulation of defense response and hypersensitive response, amino acid transport and cytokinin-activated signalling pathway. Molecular function includes nucleic acid binding activity and catalytic activity. Cellular components showed localizations in the nucleus, endoplasmic reticulum, intracellular membrane-bounded organelle.
Fig. 2.
STRING analysis for a TaWRKY56 and TaWRKY60 proteins b TaWRKY62 protein. Line colour indicates the type of interactions; line thickness indicates the strength of data support; line shape indicates the predicted mode of action; network nodes represent proteins. Each node represents all proteins produced by a single protein-encoding gene locus. Coloured nodes represent query proteins of first shell of interactors. White nodes represent second shell of interactors. Empty nodes represent proteins of unknown 3D structure. Filled nodes represent some 3D structure is known or predicted. Edges represent protein–protein associations
Similarly, for wheat WRKY62, networking was performed using A. thaliana WRKY55 protein. Eleven nodes with 27 edges and clustering coefficient of 0.84 were obtained indicating high significant interactions in TaWRKY62 (Fig. 2b). Functional partners predicted were ATG511400 (protein kinase family protein); ATG4G16162 (leucine rich repeat [LRR] family protein of 176 amino acids); PEPR1 and PEPR2 (PEP1 receptor 1 and 2 acts as receptor for PEP defense peptides, respectively); PROPEP3 (elicitor peptide 3 an elicitor of plant defense); PROPEP7 (elicitor peptide 7, an elicitor of plant defense); WAKL10 (WALL associated kinase Ser/Thr protein kinase that may function as signalling receptor); ATG5G56960 (bHLH TF); CRK23 [cysteine-rich (receptor like kinase) RLK]; AT4G28460 (uncharacterized protein). TaWRKY62 showed a response to JA, wounding, defense, stress, transmembrane receptor protein tyrosine kinase signalling pathway, cellular response to hormone stimulus, signal transduction. Molecular function includes cyclase activity, transmembrane receptor protein serine/threonine kinase activity, phosphotransferase activity. Cellular component displayed localization in the intracellular membrane-bounded organelle, plasma membrane.
Synonymous and non-synonymous substitution rate analysis
In order to understand the evolutionary constraints acting on wheat WRKY gene family, Ka/Ks ratio was determined for the three cloned TaWRKY gene orthologs (Table 2). In general, Ka/Ks ratio of 1 indicates that the genes are drifting neutrally, Ka/Ks > 1 indicates accelerated evolution with positive selection, whereas Ka/Ks < 1 indicates functional constraint with a negative or purifying selection of genes. All the Ka/Ks ratios from three TaWRKY orthologous pairs were less than 1. Hence, these genes have been subjected to strong purifying selection and they are slowly evolving at the protein level.
Table 2.
Determination of synonymous (Ks) substitution rates and divergence time
| Sl. no. | TaWRKY genes | K s | Ka/Ks | Time of divergence (Mya) |
|---|---|---|---|---|
| 1 | TaWRKY56 | 0.3037 | 0.188 | 16.68 |
| 2 | TaWRKY60 | 37.2730 | 0.0089 | 2047.96 |
| 3 | TaWRKY62 | 0.7280 | 0.1274 | 40.00 |
Mya million years ago
Homology modelling of TaWRKY proteins and validation of the models
The final modelled structure for all novel TaWRKY proteins considered for the study was visualized using the Discovery Studio 4.1 software (Online Resource 6). Modelled structures contained four antiparallel β sheets and three random coil structures except for TaWRKY50 protein which has only three coiled structures.
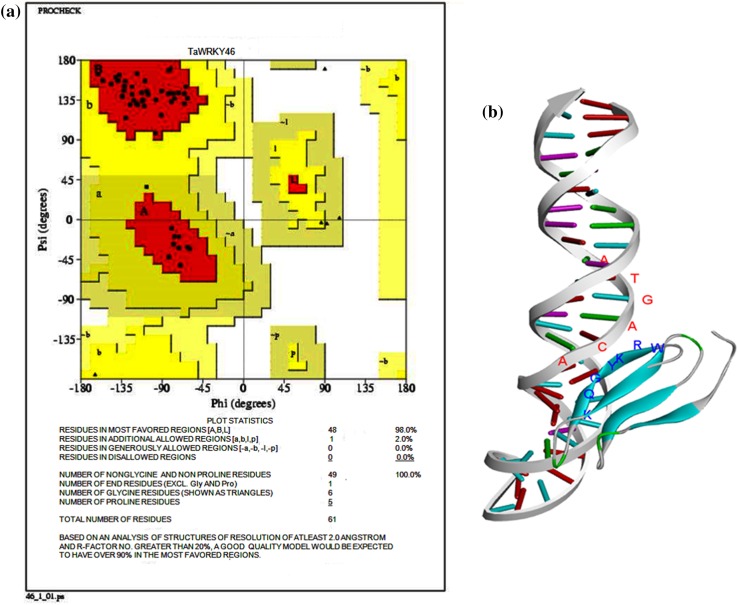
The protein structures obtained were validated by Ramachandran’s Plot in SAVES Server (Fig. 3a and Online Resource 7). The Ramachandran plots display the phi–psi torsion angles for all residues in the structures. Glycine residues are separately identified by triangles as they are not restricted to the regions. Darkest areas correspond to the “core” regions representing the most favourable combination of phi–psi values. The percentage of residues in the “core” regions is one of the best guides to stereo-chemical quality. In our study, more than 90% of residues of novel WRKY proteins were in the core region except TaWRKY51 and 62. The model selected was the best one having maximum core region and less disallowed region with minimum energy. The superimposition of TaWRKY protein with its corresponding template provided RMSD values (Table 3) along with Ramachandran plot statistics for the 22 TaWRKY proteins. The modelled structures were crosschecked and evaluated with different online servers. The stereochemical quality and accuracy of newly modelled structure (model3.pdb) were evaluated with PROCHECK. The chosen model was again analysed by VERIFY 3D, which analyzes the compatibility of an atomic model (3D) with its own amino acid sequence (1D). Percentage of the residues that had an average 3D–1D score > 0.2 in VERIFY 3D (Online Resource 8).
Fig. 3. a.
Ramachandran plot for TaWRKY46 protein as visualized by SAVES software. b Docked model of TaWRKY46 for protein–DNA interaction using HADDOCK server
Table 3.
Z-score RMS and Ramachandran plot statistics for 22 modelled TaWRKY proteins
| Sl no. | TaWRKY protein name | Z-score RMS in A° | Ramachandran plot statistics | |||
|---|---|---|---|---|---|---|
| Residues in most favoured region (%) | Residues in additional allowed region (%) | Residues in generously allowed region (%) | Residues in disallowed region (%) | |||
| 1. | TaWRKY46 | 1.207 | 96 | 2 | 0 | 0 |
| 2. | TaWRKY47 | 1.236 | 95.8 | 4.2 | 0 | 0 |
| 3. | TaWRKY48 | 1.236 | 95.8 | 4.2 | 0 | 0 |
| 4. | TaWRKY49 | 1.631 | 96.7 | 3.3 | 0 | 0 |
| 5. | TaWRKY50 | 1.643 | 99 | 1 | 0 | 0 |
| 6. | TaWRKY51 | 1.741 | 87.5 | 10.4 | 2.1 | 0 |
| 7. | TaWRKY52 | 1.631 | 96.7 | 3.3 | 0 | 0 |
| 8. | TaWRKY53 | 1.506 | 91.7 | 8.3 | 0 | 0 |
| 9. | TaWRKY54 | 1.405 | 91.7 | 8.3 | 0 | 0 |
| 10. | TaWRKY55 | 1.422 | 98.4 | 1.6 | 0 | 0 |
| 11. | TaWRKY56 | 1.524 | 96.8 | 3.2 | 0 | 0 |
| 12. | TaWRKY57 | 1.466 | 96.6 | 3.4 | 0 | 0 |
| 13. | TaWRKY58 | 1.430 | 89.7 | 8.6 | 1.7 | 0 |
| 14. | TaWRKY59 | 1.236 | 95.8 | 4.2 | 0 | 0 |
| 15. | TaWRKY60 | 1.742 | 93.8 | 6.2 | 0 | 0 |
| 16. | TaWRKY61 | 1.273 | 93.6 | 6.4 | 0 | 0 |
| 17. | TaWRKY62 | 1.466 | 87.7 | 8.2 | 1.4 | 2.7 |
| 18. | TaWRKY63 | 1.413 | 91.8 | 8.2 | 0 | 0 |
| 19. | TaWRKY64 | 1.574 | 100 | 0 | 0 | 0 |
| 20. | TaWRKY65 | 1.413 | 91.8 | 8.2 | 0 | 0 |
| 21. | TaWRKY66 | 78.267 | 89.1 | 10.9 | 0 | 0 |
| 22. | TaWRKY67 | 1.595 | 97.1 | 2.9 | 0 | 0 |
DNA modelling
Twenty-six nucleotide sequences that include various W-Box variants and expected to bind to the conserved WRKY domain were selected for DNA modelling by 3D-DART (Online Resource 9). DNA model was visualized through the Discovery Studio software (Online Resource 10). Different conformations of DNA structures were obtained of which the most stable form was used for docking. 3D-structural models of DNA are required to serve as templates for homology modelling, as starting structures for macro-molecular docking or as a scaffold for NMR structure calculations (Table 4).
Table 4.
Docking results of TaWRKY46
| Parameters | Value |
|---|---|
| HADDOCK score | − 126.8 ± 1.2 |
| Cluster size | 111 |
| RMSD from the overall lowest-energy structure | 1.3 ± 0.8 |
| Van der Waals energy | − 50.2 ± 3.6 |
| Electrostatic energy | − 499.6 ± 32.0 |
| Desolvation energy | 23.0 ± 4.5 |
| Restraints violation energy | 3.4 ± 1.36 |
| Buried surface area | 1210.6 ± 48.2 |
| Z score | − 1.5 |
DNA–protein interaction study
The optimized model for protein and DNA were docked to illustrate the interaction between W-Box and WRKY domain using HADDOCK Easy Interface (Fig. 3b and Online Resource 11). The values obtained for different parameters are within the acceptable range as described at HADDOCK web server which performed the docking process keeping in consideration the highly flexible nature of DNA molecule and suitable protocols were chosen for the process accordingly (Table 4 and Online Resource 12). The process of docking resulted in different docked clusters that were ranked in order of their stability. According to HADDOCK, the top cluster is the most reliable. Its Z-score estimates the number of times the standard deviations are from the average of the cluster is located in terms of the score (the more negative the better).
Expression analysis of TaWRKY genes in response to leaf rust pathogenesis
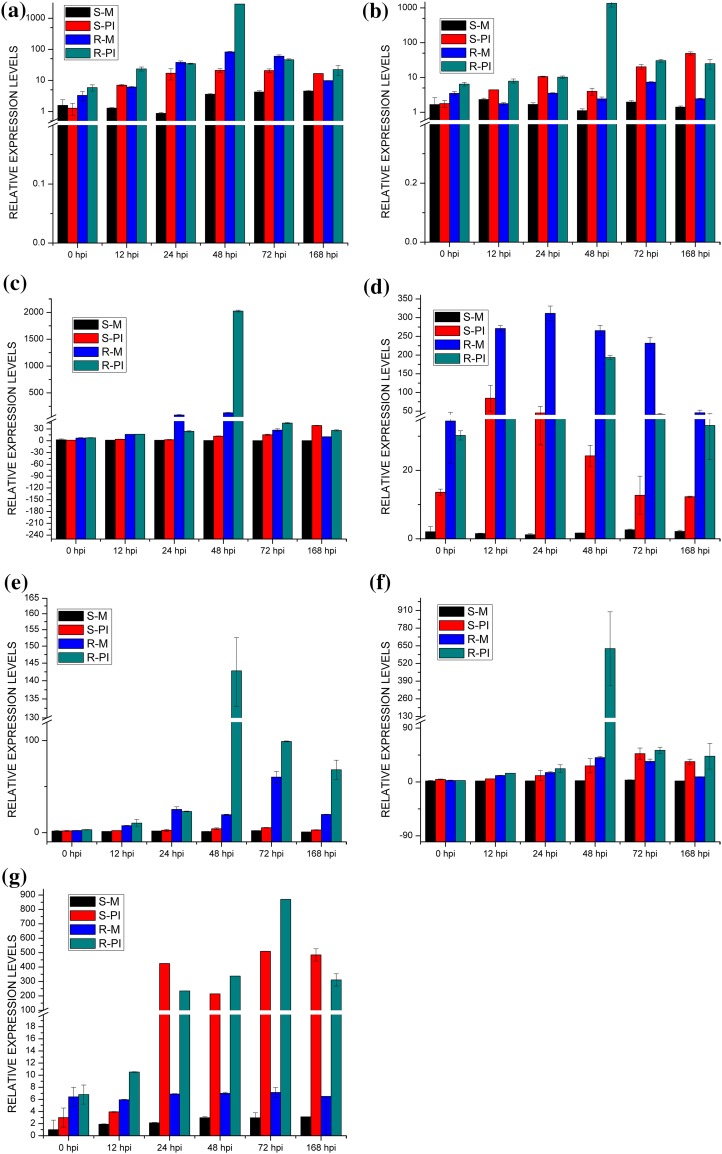
The expression of six TaWRKY genes (TaWRKY49, 50, 52, 55, 57 and 62) used for qRT-PCR study was induced at 48 hpi in the resistant NIL showing 7- to 1481-fold increase in transcript abundance; thereafter expression declined rapidly (Fig. 4a–c, e–g). In the susceptible counterpart, transcripts (TaWRKY49 and 55) and (TaWRKY52, 57 and 62) showed maximum up-regulation at 48 and 168 hpi, respectively. Another transcript (TaWRKY50) showed up-regulation of the corresponding gene after pathogen inoculation in the resistant NIL compared with the susceptible NIL, and its expression gradually increased from 48 hpi, peaked at 72 hpi and declined at 168 hpi. Similarly, transcript (TaWRKY53) showed up-regulation after leaf rust inoculation in the resistant NIL compared with the susceptible NIL, and its expression peaked at 12 hpi and declined at 24 hpi. TaWRKY62 showed up-regulation at 24 and 72 hpi in case of susceptible and resistant counterparts. Only one transcript (TaWRKY53) was not induced upon pathogen inoculation and hence exhibited more or less generalized expression, which is apparently not associated with the pathogenesis (Fig. 4d). Clearly, seven transcripts (TaWRKY49, 50, 51, 52, 55, 57 and 62) are responsive to the virulent pathogen in the resistant NIL. The temporal expression at different time points during compatible and incompatible interactions in susceptible and resistant NILs after inoculation also generated differential expression patterns. The details of maximum expression for each of the six target genes are presented in Table 5. ΔCT (mean ± SD values) of each TaWRKY genes in susceptible and resistant plants (mock and pathogen-inoculated) are presented in Online Resource 13. The t values of targeted genes at the point of maximum expression in selected plants are summarized in Online Resource 14 where critical t value is 4.604 at a confidence level of P < 0.01. The obtained t values showed a significant difference from a range of critical t value, indicating a significant difference in expression of the targeted genes as defense response at the point of maximum expression in selected plants.
Fig. 4.
Relative gene expression profiles of WRKY genes a WRKY49, b WRKY50, c WRKY52, d WRKY 53, e WRKY55, f WRKY57, g WRKY62 during pathogen-induced biotic stress. Expression profiles of susceptible (HD2329) and resistant (HD2329 + Lr28) wheat plants in response to leaf rust infection compared with mock-inoculated controls. The time points correlated with the formation and development of different infection structures as described by Hu and Rijkenberg (1998) are shown at the bottom. Relative expression is expressed as fold changes relative to mock-inoculated controls. The values on the y-axis indicate relative expression, whereas the x-axis denotes the selected time points (hour post inoculation). Data represent means of two replicate reactions from three biological repeats with error bars indicating standard deviations (SD)
Table 5.
Summary of results of expression profiling of seven target genes in the wheat NILs (HD2329, HD2329 + Lr 28) under mock and pathogen-inoculated conditions
| Target gene | HD2329 mock | HD2329 inoculated | HD2329 + Lr28 mock | HD2329 + Lr28 inoculated | Figure 4 |
|---|---|---|---|---|---|
| TaWRKY49 | Onefold, 0 hpi | Sixfold, 48 hpi | Onefold, 72 hpi | 35-fold, 48 hpi | a |
| TaWRKY50 | – | Tenfold, 72 hpi | – | 556-fold, 48 hpi | b |
| TaWRKY52 | 1.6-fold, 0 hpi | 111-fold, 168 hpi | 3.6-fold, 24 hpi | 16-fold, 48 hpi | c |
| TaWRKY53 | – | 55-fold, 12 hpi | 24 hpi, Eightfold | – | d |
| TaWRKY55 | – | 3.5-fold, 48 hpi | Onefold, 24 hpi | Sevenfold, 48 hpi | e |
| TaWRKY57 | – | 20-fold, 168 hpi | Onefold, 0 hpi | 15.5-fold, 48 hpi | f |
| TaWRKY62 | 2.6-fold, 72 hpi | 52-fold, 48 hpi | 46-fold, 48 hpi | 1481-fold, 48 hpi | g |
Maximum expression (fold) and time-point (hpi) are mentioned
Expression analysis in response to abiotic stress
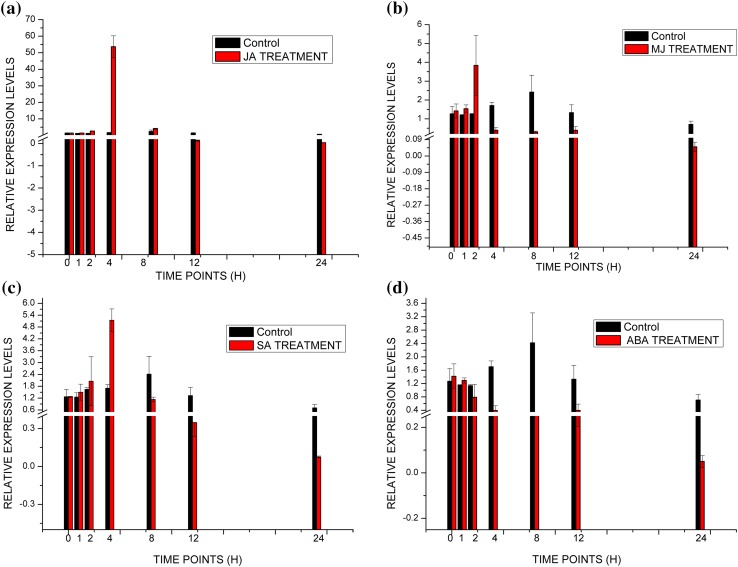
As wheat plants manifest many abiotic stress tolerance characteristics, we studied the expression pattern of TaWRKY62 gene during JA, MJ, SA and ABA treatments (Fig. 5). TaWRKY62 showed maximum expression at 4 h post treatment with JA and SA phytohormones, while 2 h post treatment with the phytohormone MJ (Fig. 5a–c). Whereas, the gene was down-regulated up on treatment with ABA (Fig. 5d). It was observed that expression gradually declined at 8, 12 and 24 h post phytohormone treatment with JA and SA treatments. In case of MJ treatment, expression declined at 4, 8, 12 and 24 h (Fig. 5a–c). ΔCT (mean ± SD values) of TaWRKY62 gene is presented in Online Resource 15. The results suggest that TaWRKY62 is induced upon treatment with JA, MJ and SA and reduced after ABA treatments. Since fewer reports were available in the study of WRKY expression patterns during abiotic stress in wheat, this comprehensive expression profile would invoke investigations on the role of WRKY in imparting phytohormone treatment based stress tolerance in wheat.
Fig. 5.
Relative expression levels of TaWRKY62 on treatment with a JA, b MJ, c SA and d ABA Relative expression is expressed as fold changes relative to mock-inoculated controls. The values on the y-axis indicate relative expression, whereas the x-axis denotes the selected time points (hour post inoculation). Data represent means of two replicate reactions from three biological repeats with error bars indicating standard deviations (SD)
Annotation of orthologous clusters
WRKY proteins of 103 O. sativa, 82 B. distachyon, 94 S. bicolor and 125 Z. mays were retrieved from GRASSIUS TF database and 72 WRKY proteins of A. thaliana were retrieved from AGRIS database for identification of orthologous wheat WRKY gene clusters. The species formed 47 clusters and 42 orthologous clusters. Clusters formed in different species were 13 in wheat, 28 in A. thaliana, 23 in Brachypodium, 27 in O. sativa, 29 in Sorghum and 32 in Z. mays (Fig. 6). The information obtained from comparisons of orthologous clusters can serve as raw material for taxonomic classification and phylogenetic studies of organisms, hence shedding light on the mechanisms underlying the molecular evolution of genes and genomes.
Fig. 6.
Identification of wheat WRKY gene clusters in orthologous species of Brachypodium distachyon, Arabidopsis thaliana, Oryza sativa, Sorghum bicolor and Zea mays
Discussion
The present study was carried out to reveal DNA–protein interactions and expression pattern of WRKY TF genes using wheat NILs in response to leaf rust infection and abiotic stresses. A network of WRKY proteins was also developed in this study. DNA–protein interactions play vital roles in key biological process and regulation of gene expression. Transcriptional regulation is one among these processes in which TFs bind to specific DNA-binding sequences to either activate or repress the expression of their regulated genes. Previous studies have shown that molecular docking can obtain accurate complex structures for protein–protein, protein–peptide, and protein–ligand interactions (Wang et al. 2015a). But, DNA–protein docking still lags behind due to limited knowledge of the interactions and remains one of the challenging problems in the field of structural bioinformatics. DNA certainly exhibits large conformational changes upon binding to a protein, which can greatly alter the shape of the interaction surface. The DNA–protein docking study predicted stable interactions of all 22 WRKY proteins with W-Box in the present study.
Gene expression patterns offer indications for determining the function of genes. The results obtained in the present study indicated that due to pathogen inoculation, maximum induction of six out of seven genes occurred at 48 hpi, whereas for TaWRKY53 expression levels were reduced when compared to mock-inoculated controls. Further, the expressions of four genes (TaWRKY49, 50, 55 and 62) were highly expressed in the presence of Lr28 than in its absence. The obtained expression pattern of the genes was assigned probable functions according to coordinated expression level and sites of action during P. triticana pathogenesis; which involved formation and development of different infection structures (Hu and Rijkenberg 1998). The sequencial events occurring during leaf rust infestation involves formation of sub-stmatal vesicles (SSVs) at 12 hpi, primary infection hyphae at 24 hpi and haustorial mother cell (HMC) at 48 hpi. The appresoria collapses at 72 hpi and intracellular hyphal development starts. The SSVs ramify to adjacent cells at 168 hpi and thereby infest wheat leaves in susceptible wheat culticars that eventually affects plant growth and grain filling. Presence of leaf rust resistance genes defer or prohibit ingression of the leaf rust pathogen during incompatible interactions, thus shielding the plant. The present study was focused on the events taking place immediately after the inoculation, with more intermediate time-points. Therefore, gene expression patterns in leaves of wheat NILs during different stages of infection progression bestowed knowledge on the protective roles of WRKY TFs.
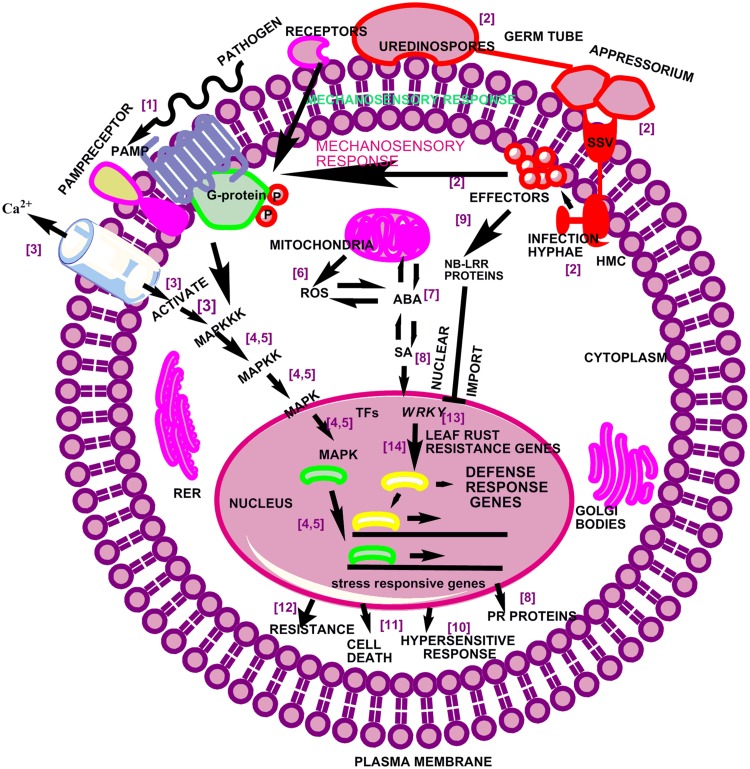
Based on the obtained results a working model has been proposed for the selected WRKY genes (Fig. 7). Cellular defense signalling is triggered by recognition of pathogen-derived PAMPs via distinct plasma membrane receptors. These plasma membrane-localized receptors present on leaf surface initiate mechano-sensory responses that activate local defense reactions to stimulate systemic responses, including activation of early responsive genes (Singh et al. 2012). These early responsive genes regulate Ca2+ ion concentration and trigger MAP kinase cascades that govern WRKY and other TF genes. These genes, in turn, actuate defense and stress-responsive genes. Subsequently, ROS in association with SA mediates the establishment of systemic defenses (SAR). The rapidity of ROS production and the potential for H2O2 to freely diffuse across membranes suggest that ROS could function as an intercellular or intracellular second messenger that establishes different defensive barriers against the pathogens. The stress-responsive genes further stimulate the downstream genes in terms of cell death, resistance, PR proteins and HR based on up- and down-regulation of respective genes. Also, the plant NBS-LRR proteins detect pathogen-associated effector molecules and subsequently activate host defense (Chandra et al. 2016) (Fig. 7). The WRKY Group III has already been considered to be associated with various defense responses in Arabidopsis (Kalde et al. 2003), rice (Shimono et al. 2007) and wheat (Bahrini et al. 2011; Kumar et al. 2014).
Fig. 7.
A model depicting the role of WRKY genes in response to leaf rust fungal pathogenesis. Cellular defense signalling is triggered by recognition of pathogen-derived PAMPs via distinct plasma membrane-localized receptors. Mechano-sensory responses transduce MAP kinase cascades that regulate WRKY genes that subsequently activate defense gene expression. SSV substomatal vesicles, HMC haustorial mother cell, G-protein guanine nucleotide-binding proteins, PAMP pathogen-associated molecular pattern, PR proteins pathogenesis-related proteins, TFs transcription factors, RER rough endoplasmic reticulum, NB-LRR proteins nucleotide binding-leucine-rich repeat proteins. [1] Jones and Dangl (2006), [2] Bolton et al. (2008), [3] Gao et al. (2014), [4] Pandey and Somssich (2009), [5] Adachi et al. (2015), [6] Yoshioka et al. (2011), [7] Cao et al. (2011), [8] Durant and Dong (2004), [9] De Young and Innes (2006), [10] Zvereva and Pooggin (2012), [11] Coll et al. (2011), [12] Gawehns et al. (2013), [13] Rushton et al. (2010), [14] Satapathy et al. (2014)
A better understanding of the molecular function of WRKY TF genes may allow coordination of the defense activity of genes during infection progression and help in the development of effective strategies for durable rust resistance. Several approaches including cDNA-amplified fragment length polymorphism (cDNA-AFLP) profiling (Zhang et al. 2003; Dhariwal et al. 2011), suppression subtractive hybridization (SSH) profiling (Thara et al. 2003), expressed sequence tag (EST) profiling (Hu et al. 2007), high-throughput microarray profiling (Fofana et al. 2007) and serial analysis of gene expression (SAGE) profiling (Poole et al. 2008) have been employed to characterize the key defense genes involved in rust resistance. Quantitative real-time PCR (qRT-PCR) has also been successfully utilized to detect and quantify specific transcripts in response to leaf rust in our earlier studies (Singh et al. 2012; Kumar et al. 2014).
TaWRKY62 was induced by SA, MJ and JA suggesting its possible role in plant development via different phytohormone signalling pathways. In response to SA treatment, TaWRKY62 expresses threefold higher than control at 2 h post-treatment. The same WRKY gene expressed 31.56-fold higher than control at 4 h post-JA treatment and threefold higher than control at 4 h post-MJ treatment. Kruskal–Wallis one-way analysis of variance statistical test presented a P value of 0.423 indicating a significant difference in expression of target genes on phytohormone-treated and control plants. WRKY genes are well recognized as significant players in plant responses to abiotic stresses; thus, suggesting TaWRKY genes might also be involved in defense-related processes or stress responses (Okay et al. 2014; He et al. 2016; Ding et al. 2016). Protein networking of TaWRKY56, 60 and 62 TF genes in this study indicated their probable roles in defense response in plants. Identifying the overlap among orthologous clusters of TaWRKY proteins enabled to elucidate the function and evolution of WRKY proteins across multiple species.
Conclusions
Differential gene expression analysis performed in this study distinctly indicated that wheat plants responded to leaf rust pathogen in a coordinated manner. During the interaction between wheat and P. triticina, maximum activity involving the highest expression of the target genes occurred at 48 hpi in pathogen-inoculated resistant plants. Hence these TaWRKY genes during incompatible interactions exhibited their positive roles against P. triticina-induced leaf-rust in wheat. Hence it can be assumed that the TaWRKY49, 50, 52, 55, 57 and 62 might also have a function in leaf-rust induced biotic stress. It might be imperative that these WRKY proteins are regulatory proteins that bind to W-Boxes of other WRKY and defense proteins to control their expression. This current investigation of WRKY genes provides a platform for further exploring the function of WRKY genes in order to suggest candidate genes for enhancing biotic and abiotic stress tolerance in wheat crops.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by BTIS Net Sub DIC (BT/BI/04/065/04) and CoE-TEQIP-II (Grant no. NPIU/TEQIP II/FIN/31/158). LS is grateful to Department of Science and Technology-INSPIRE (Fellowship/2011/318) and DK to Council of Scientific and Industrial Research [9/554 (0026) 2010-EMR-I] for Ph.D. fellowship. The authors acknowledge Dr. KV Prabhu for providing phytotron facility at IARI, New Delhi, India.
Abbreviations
- ABA
Abscisic acid
- CTAB
Cetyl trimethyl ammonium bromide
- EST
Expressed sequence tag
- ETI
Effector-triggered immunity
- HPI
Hours post inoculation
- HR
Hypersensitive response
- JA
Jasmonic acid
- MJ
Methyl jasmonate
- MS
Murashige and Skoog
- NIL
Near-isogenic line
- PTI
Pathogen-associated molecular pattern (PAMP)-triggered immunity
- SA
Salicylic acid
- TF
Transcription factor
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s13205-017-1064-3) contains supplementary material, which is available to authorized users.
References
- Adachi H, Nakano T, Miyagawa N, Ishihama N, Yoshioka M, Katou Y, Yaeno T, Shirasu K, Yoshioka H. WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell. 2015;27:2645–2663. doi: 10.1105/tpc.15.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu MM, Iyer LM, Balaji S, Aravind L. The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucl Acids Res. 2006;34:6505–6520. doi: 10.1093/nar/gkl888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrini I, Sugisawa M, Kikuchi R, Ogawa T, Kawahigashi H, Ban T, Handa H. Characterization of wheat transcription factor, TaWRKY45, and its effect on Fusarium head blight resistance in transgenic wheat plants. Breed Sci. 2011;61:121–129. doi: 10.1270/jsbbs.61.121. [DOI] [Google Scholar]
- Bipinraj A, Honrao B, Prashar M, Bhardwaj S, Rao S, Tamhankar S. Validation and identification of molecular markers linked to the leaf rust resistance gene Lr28 in wheat. J Appl Genet. 2011;52:171–175. doi: 10.1007/s13353-010-0026-9. [DOI] [PubMed] [Google Scholar]
- Bolton MD, Kolmer JA, Garvin DF. Wheat leaf rust caused by Puccinia triticina. Mol Plant Pathol. 2008;9:563–575. doi: 10.1111/j.1364-3703.2008.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao FY, Yoshioka K, Desveaux D. The role of ABA in plant-pathogen interactions. J Plant Res. 2011;489–499:2011. doi: 10.1007/s10265-011-0409-y. [DOI] [PubMed] [Google Scholar]
- Chandra S, Singh D, Pathak J, Kumari S, Kumar M, Poddar R, Balyan HS, Gupta PK, Prabhu KV, Mukhopadhyay K. De novo assembled wheat transcriptomes delineate differentially expressed host genes in response to leaf rust infection. PLoS One. 2016;11(2):e0148453. doi: 10.1371/journal.pone.0148453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheatham TI, Miller J, Fox T, Darden T, Kollman P. Molecular dynamics simulations on solvated biomolecular systems: the particle mesh Ewald method leads to stable trajectories of DNA, RNA, and proteins. J Am Chem Soc. 1995;117:4193–4194. doi: 10.1021/ja00119a045. [DOI] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124(4):803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Choulet F, Wicker T, Rustenholz C, Paux E, Salse J, Leroy P, Schlub S, Paslier MCL, Magdelenat G, Gonthier C, Couloux A, Budak H, Breen J, Pumphrey M, Liu S, Kong X, Jia J, Gut M, Brunel D, Anderson JA, Gill BS, Appels R, Keller B, Feuillet C. Megabase level sequencing reveals contrasted organization and evolution patterns of the wheat gene and transposable element spaces. Plant Cell. 2010;22:1686–1701. doi: 10.1105/tpc.110.074187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choura M, Rebai A, Masmoudi K. Unraveling the WRKY transcription factors network in Arabidopsis thaliana by an integrative approach. Netw Biol. 2015;5:55–61. [Google Scholar]
- Coll NS, Epple P, Dangl J. Programmed cell death in the plant immune system. Cell Death Differ. 2011;18:1247–1256. doi: 10.1038/cdd.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries SJ, Van D, Krzeminski ADJ, Dijk M, Van TM, Hsu A, Wassennar V, Bovin AMJJ. HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins Struct Funct Bioinform. 2007;69:726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- De Young BXJ, Innes RW. Plant NBS-LRR proteins in pathogen sensing and host defense. Nat Immunol. 2006;7:1243–1249. doi: 10.1038/ni1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R, Van-Kan JAL, Pretorius ZA, Hammond-Kosack KE, Pietro AD, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, Foster GD. The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. 2012;13:414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhariwal R, Vyas S, Bhaganagare GR, Jha SK, Khurana JP, Tyagi AK, Prabhu KV, Balyan HS, Gupta PK. Analysis of differentially expressed genes in leaf rust infected bread wheat involving seedling resistance gene Lr28. Funct Plant Biol. 2011;38:1–14. doi: 10.1071/FP10091. [DOI] [PubMed] [Google Scholar]
- Ding M, Chen J, Jiang Y, Lin L, Cao Y, Wang M, Zhang Y, Rong J, Ye W. Genome-wide investigation and transcriptome analysis of the WRKY gene family in Gossypium. Mol Genet Genom. 2015;290:151–171. doi: 10.1007/s00438-014-0904-7. [DOI] [PubMed] [Google Scholar]
- Ding W, Fang W, Shi S, Zhao Y, Li X, Xiao K. Wheat WRKY type transcription factor gene TaWRKY1 is essential in mediating drought tolerance associated with an ABA-dependent pathway. Plant Mol Biol Rep. 2016 [Google Scholar]
- Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Lüthy R, Bowie JU. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396–404. doi: 10.1016/S0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Somssich IE. Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol. 2007;10(4):366–371. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY super family of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- FAO (2016) Food and Agriculture Organization of the United Nations. http://www.fao.org. Accessed 25 Oct 2016
- Fofana B, Banks TW, McCallum B, Strelkov SE, Cloutier S. Temporal gene expression profiling of the wheat leaf rust pathosystem using cDNA microarray reveals differences in compatible and incompatible defense pathways. Int J Plant Genom. 2007;2007:17542. doi: 10.1155/2007/17542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Cox KL, Jr, He P. Functions of calcium—dependant kinases in plant innate immunity. Plants. 2014;3:160–176. doi: 10.3390/plants3010160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawehns F, Cornelissen BJ, Takken FL. The potential of effector-target genes in breeding for plant innate immunity. Microb Biotechnol. 2013;6:223–229. doi: 10.1111/1751-7915.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J, Ton J. Biotic interactions recurring themes and expanding scales. Curr Opin Plant Biol. 2007;10:331–334. doi: 10.1016/j.pbi.2007.06.005. [DOI] [Google Scholar]
- He H, Dong Q, Shao Y, Jiang H, Zhu S, Chen B, Xiang Y. Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 2012;31:1199–1217. doi: 10.1007/s00299-012-1241-0. [DOI] [PubMed] [Google Scholar]
- He GH, Xu JY, Wang YX, Liu JM, Li PS, Chen M, Ma YZ, Xu ZS. Drought-responsive WRKY transcription factor genesTaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 2016 doi: 10.1186/s12870-016-0806-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Rijkenberg FHJ. Scanning electron microscopy of early infection structure formation by Puccinia recondita f. sp. tritici on and in susceptible and resistant wheat lines. Mycol Res. 1998;102:391–399. doi: 10.1017/S0953756297005054. [DOI] [Google Scholar]
- Hu G, Linning R, McCallum B, Banks T, Cloutier S, Butterfield Y, Liu J, Kirkpatrick R, Stott J, Yang G, Smailus D, Jones S, Marra M, Schein J, Bakkeren G, Yamazaki Y. Generation of a wheat leaf rust, Puccinia triticina, EST database from stage-specific cDNA libraries. Mol Plant Pathol. 2007;8:451–467. doi: 10.1111/j.1364-3703.2007.00406.x. [DOI] [PubMed] [Google Scholar]
- Jiang J, Ma S, Ye N, Jiang M, Cao J, Zhang J. WRKY transcription factors in plant responses to stresses. JIPB. 2017;59:86–101. doi: 10.1111/jipb.12513. [DOI] [PubMed] [Google Scholar]
- Jin J, Kong J, Qiu J, Zhu H, Peng Y, Jiang H. High level of microsynteny and purifying selection affect the evolution of WRKY family in Gramineae. Dev Genes Evol. 2016;226:15–25. doi: 10.1007/s00427-015-0523-2. [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kalde M, Barth M, Somssich IE, Lippok B. Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Mol Plant-Microb Interact. 2003;16(4):295–305. doi: 10.1094/MPMI.2003.16.4.295. [DOI] [PubMed] [Google Scholar]
- Kaur S, Bansal UK, Khanna R, Saini RG. Genetics of leaf and stripe rust resistance in a bread wheat cultivar Tonichi. J Genet. 2008;87:191–194. doi: 10.1007/s12041-008-0030-6. [DOI] [PubMed] [Google Scholar]
- Kayum MA, Jung HJ, Park JI, Ahmed N, Saha G, Yang TJ, Nou IS. Identification and expression analysis of WRKY genes under biotic and abiotic stresses in B. rapa. Mol Genet Genome. 2015;290:79–95. doi: 10.1007/s00438-014-0898-1. [DOI] [PubMed] [Google Scholar]
- Koeck M, Hardham AR, Dodds PN. The role of effectors of biotrophic and hemibiotrophic fungi in infection. Cell Microbiol. 2011;13:1849–1857. doi: 10.1111/j.1462-5822.2011.01665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn H, Kwaaitaal M, Kusch S, Acevedo-Garcia J, Wu H, Panstruga R. Biotrophy at its best: novel findings and unsolved mysteries of the Arabidopsis-powdery mildew pathosystem. Arabidopsis Book. 2016;14:e0184. doi: 10.1199/tab.0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Kapoor A, Singh D, Satapathy L, Singh AK, Kumar M, Prabhu KV, Mukhopadhyay K. Functional characterization of a WRKY transcription factor of wheat and its expression analysis during leaf rust pathogenesis. Funct Plant Biol. 2014;41:1295–1309. doi: 10.1071/FP14077. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- Ling J, Jiang WJ, Zhang Y, Yu HJ, Mao ZC, Gu XF, Huang SW, Xie BY. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom. 2011;12:471. doi: 10.1186/1471-2164-12-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh RA, Pretorius ZA. Borlaug global rust initiative provides momentum for wheat rust research. Euphytica. 2011;179:1–2. doi: 10.1007/s10681-011-0389-y. [DOI] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Okay S, Derelli E, Unver T. Transcriptome-wide identification of bread wheat WRKY transcription factors in response to drought stress. Mol Genet Genom. 2014;289:769–781. doi: 10.1007/s00438-014-0849-x. [DOI] [PubMed] [Google Scholar]
- Pan LJ, Jiang L. Identification and expression of the WRKY transcription factors of Carina papaya in response to abiotic and biotic stresses. Mol Biol Rep. 2014;41:1215–1225. doi: 10.1007/s11033-013-2966-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SP, Somssich IE. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009;150(4):1648–1655. doi: 10.1104/pp.109.138990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolacci AR, Tanzarella OA, Porceddu E, Ciaffi M. Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol Biol. 2009;10:11. doi: 10.1186/1471-2199-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole RL, Barker GL, Werner K, Biggi GF, Coghill J, Gibbings JG, Berry S, Dunwell JM, Edwards KJ. Analysis of wheat SAGE tags reveals evidence for widespread antisense transcription. BMC Genom. 2008;9:475. doi: 10.1186/1471-2164-9-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosegrant MW, Agcaoili M. Global food demand, supply, and price prospects to 2010. Washington DC: International Food Policy Research Institute; 2010. [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY Transcription factors. Trends Plant Sci. 2010;15:247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Satapathy L, Singh D, Ranjan P, Kumar D, Kumar M, Prabhu KV, Mukhopadhyay K. Transcriptome-wide analysis of WRKY transcription factors in wheat and their leaf rust responsive expression profiling. Mol Genet Genom. 2014;289:1289–1306. doi: 10.1007/s00438-014-0890-9. [DOI] [PubMed] [Google Scholar]
- Shimono M, Sugano S, Nakayama A, Jiang CJ, Ono K, Toki S, Takatsuji H. Rice WRKY45 plays a crucial role in benzothiadiazole inducible blast resistance. Plant Cell. 2007;19:2064–2076. doi: 10.1105/tpc.106.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D, Bhaganagare G, Bandopadhyay R, Prabhu KV, Gupta PK, Mukhopadhyay K. Targeted spatiotemporal expression based characterization of state of infection and time-point of maximum defense in wheat NILs during leaf rust infection. Mol Biol Rep. 2012;39:9373–9382. doi: 10.1007/s11033-012-1801-y. [DOI] [PubMed] [Google Scholar]
- Thara VK, Fellers JP, Zhou JM. In planta induced genes of Puccinia triticina. Mol Plant Pathol. 2003;4:51–56. doi: 10.1046/j.1364-3703.2003.00142.x. [DOI] [PubMed] [Google Scholar]
- Tripathi P, Rabara RC, Langum TJ, Boken AK, Rushton DL, Boomsma DD, Rinerson CI, Rabara J, Reese RN, Chen X, Rohila JS, Rushton PJ. The WRKY transcription factor family in Brachypodium distachyon. BMC Genom. 2012;13:270. doi: 10.1186/1471-2164-13-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol. 2004;7(5):491–498. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Wang C, Liu J, Luo F, Deng Z, Hu QN. Predicting target-ligand interactions using the protein-ligand binding site and ligand sub structures. BMC Syst Biol. 2015 doi: 10.1186/1752-0509-9-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Feng L, Zhu Y, Li Y, Yan H, Xiang Y. Comparative genomic analysis of the WRKY III gene family in Populus, grape, Arabidopsis and rice. Biol Direct. 2015;10:48. doi: 10.1186/s13062-015-0076-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei KF, Chen J, Chen YF, Wu LJ, Xie DX. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 2012;19:153–164. doi: 10.1093/dnares/dsr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KL, Guo ZJ, Wang HH, Li J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 2005;12:9–26. doi: 10.1093/dnares/12.1.9. [DOI] [PubMed] [Google Scholar]
- Xiong W, Xu X, Zhang L, Wu P, Chen Y, Li M, Jiang H, Wu G. Genome-wide analysis of the WRKY gene family in physic nut (Jatropha curcas L.) Gene. 2013;524:124–132. doi: 10.1016/j.gene.2013.04.047. [DOI] [PubMed] [Google Scholar]
- Yoshioka H, Mase K, Yoshioka M, Kobayashi M, Asai S. Regulatory mechanisms of nitric oxide and reactive oxygen species generation and their role in plant immunity. Nitric Oxide. 2011;25:216–222. doi: 10.1016/j.niox.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Zhang L, Meakin H, Dickinson M. Isolation of genes expressed during compatible interactions between leaf rust (Puccinia triticina) and wheat using cDNA-AFLP. Mol Plant Pathol. 2003;4:469–477. doi: 10.1046/j.1364-3703.2003.00192.x. [DOI] [PubMed] [Google Scholar]
- Zhu X, Liu S, Meng C, Qin L, Kong L, Guangmin X. WRKY transcription factors in wheat and their induction by biotic and abiotic stress. Plant Mol Biol Rep. 2013;31:1053–1067. doi: 10.1007/s11105-013-0565-4. [DOI] [Google Scholar]
- Zvereva AS, Pooggin MM. Silencing and innate immunity in plant defense against viral and non-viral pathogens. Viruses. 2012;4:2578–2597. doi: 10.3390/v4112578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.